- 1State Key Laboratory of Biocontrol, School of Life Science, Sun Yat-sen University, Guangzhou, China
- 2Higher Education Mega Center, School of Life Science, Sun Yat-sen University, Guangzhou, China
Apoptosis is a tightly regulated mechanism of cell death that plays important roles in various biological processes including biological evolution, multiple system development, anticancer, and viral infections. Swine enteropathogenic coronaviruses invade and damage villous epithelial cells of the small intestine causing severe diarrhea with high mortality rate in suckling piglets. Transmissible gastroenteritis virus (TGEV), Porcine epidemic diarrhea virus (PEDV), Porcine deltacoronavirus (PDCoV), and Swine acute diarrhea syndrome coronavirus (SADS-CoV) are on the top list of commonly-seen swine coronaviruses with a feature of diarrhea, resulting in significant economic losses to the swine industry worldwide. Apoptosis has been shown to be involved in the pathogenesis process of animal virus infectious diseases. Understanding the roles of apoptosis in host responses against swine enteropathogenic coronaviruses infection contribute to disease prevention and control. Here we summarize the recent findings that focus on the apoptosis during swine coronaviruses infection, in particular, TGEV, PEDV, PDCoV, and SADS-CoV.
Introduction
Apoptosis, also known as programmed cell death, is a ubiquitous mode of cell death known to be responsible for clearance of unwanted, injured, or virus-infected cells (1, 2). Cells undergoing apoptosis are accompanied by characteristic morphological changes, including cell shrinkage and deformation, chromatin condensation, nuclear fragmentation, and plasma membrane blebbing, which forms the apoptotic body containing the fragments of nucleus or cytoplasm (3). Cell apoptosis is an active process, which involves a series of genes activation and expression, various proteins regulation. To date, it was reported that there are two main apoptotic pathways: the extrinsic/death receptor pathway (4) and the intrinsic/mitochondrial pathway (5). Death receptors belong to the tumor necrosis factor receptor (TNFR) superfamily (6). Members of the TNFR family are type I membrane surface receptors, which include the Fatty acid synthetase receptor (FasR), TNFR, Death receptor (DR)3, DR4, DR5, etc. (7–11). Their ligands belong to type II membrane proteins, which include Fatty acid synthetase ligand (FasL), TNF-α Apo3 ligand (Apo3L), Apo2 ligand (Apo2L), etc. (7–11). Upon death receptor-ligand binding, the adapter protein Fas-associated death domain (FADD) is recruited by death domain, then associates with pro-cysteinyl aspartic acid protease (caspase)-8 via dimerization of the death effector domain and a death-inducing signaling complex (DISC) is formed to activate caspase-8, then caspase-8 results in the caspase-3 activation (12–14). Once caspase-3 is activated, the execution phase of apoptosis is triggered (12–14). The mitochondrial pathways involve some chemical or physical stimulus factors leading to change of the permeability of mitochondrial membrane, resulting in release of the cytochrome c (cyt c) or other apoptotic molecules into the cytoplasm cavity and activation of downstream caspases to initiate apoptosis (15, 16). In addition, T-cell mediated cytotoxicity involved with perforin-granzyme to kill target cells is known as another apoptotic pathway (17).
It is known that many viruses can evolve various sophisticated strategies to modulate apoptosis as a critical armament to complete their replication cycle (18), reflected in the relationship between viral infection and cell apoptosis is bidirectional. Viruses could hijack host's apoptotic pathway to delay apoptotic response, providing sufficient time for maximizing progeny virus production (19). On the other hand, viruses could induce apoptosis to enable the release and dissemination of viral progeny for further invasion to the neighboring cells at the late stages of viral infection (18).
Porcine coronaviruses (CoVs) are significant enteric and respiratory pathogens of swine. Six porcine CoVs have so far been identified: transmissible gastroenteritis virus (TGEV) (20), porcine respiratory coronavirus (PRCV) (21), porcine epidemic diarrhea virus (PEDV) (22), and swine acute diarrhea syndrome coronavirus (SADS-CoV) (23) in the Alphacoronavirus genus; porcine hemagglutinating encephalomyelitis virus (PHEV) (24) in the Betacoronavirus genus; porcine deltacoronavirus (PDCoV) (25) in the Deltacoronavirus genus. The clinical signs of swine enteropathogenic CoVs including TGEV, PEDV, PDCoV, and SADS-CoV are characterized by severe watery diarrhea with subsequent dehydration in pigs of all ages, and a high mortality rate in suckling piglets (26–29). The molecular surveillance studies indicated that swine enteropathogenic CoVs were common viral pathogen of pigs around the world (30–44) (Figure 1). In addition, co-infections of these diarrhea-associated viruses were commonly found in pig farms (45). For swine enteropathogenic CoVs infection prevention, the administration of vaccines and antiviral drugs are important tool. Currently a number of vaccines and antiviral drugs, such as killed, live-attenuated vaccine, shRNA expression vector are widely used to prevent swine enteropathogenic CoVs infection (46–49). However, it still can not stop the swine enteropathogenic CoVs outbreak, due to they are not optimal in terms of safety and efficacy, and large-scale infections still occur, resulting in the death of large numbers of piglets, causing huge economic losses to the pig industry (23, 50–52), indicating that a deeper understanding of the pathogenesis of swine enteropathogenic CoVs is needed to develop more effective prevention and control measures.
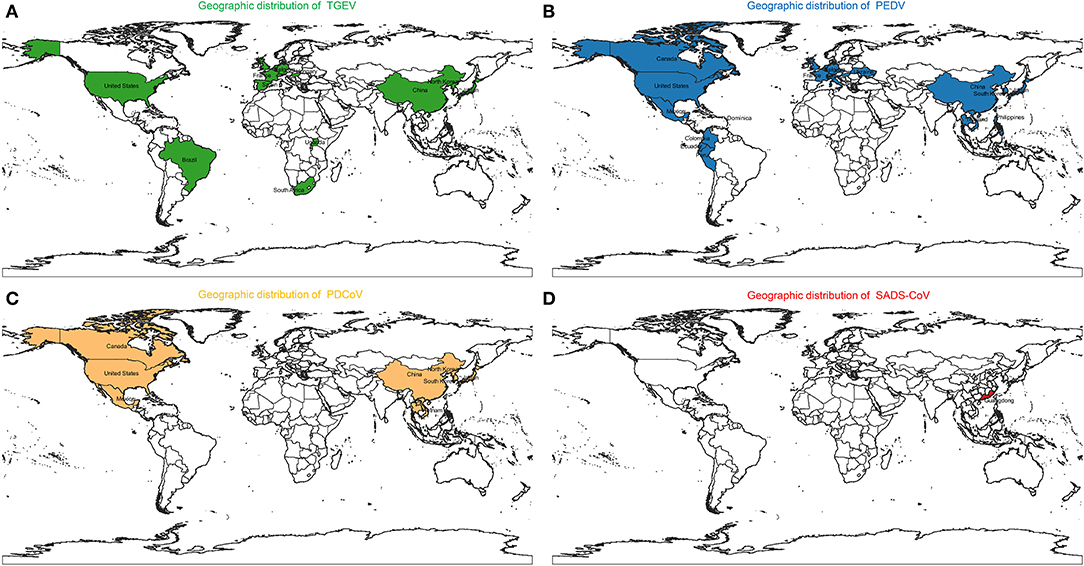
Figure 1. Geographic distribution of (A) Transmissible gastroenteritis virus (TGEV), (B) Porcine epidemic diarrhea virus (PEDV), (C) Porcine deltacoronavirus (PDCoV), and (D) Swine acute diarrhea syndrome coronavirus (SADS-CoV).
Although swine enteropathogenic CoVs might infect multiple organs in pigs, the intestinal tract is the major target organ, where virus replication is limited to intestinal villus epitheial cells (29, 53). Diarrhea caused by pathogen infection associates with viral damage to intestinal epithelial cells, which plays an important role in the nutrition absorption (54), causes a breach of mucosal physical barriers and reduction of enzyme activities, leading to electrolyte imbalances, nutrient decomposition and absorption anomalies (53, 55). It has been reported that apoptosis, which occurs in the infection course of many CoVs (56, 57), is involved in viral pathogenesis and disease processes that promote cell death and tissue injury (58, 59). In this review, the roles of apoptosis in the pathogenesis and control of TGEV, PEDV, PDCoV, and SADS-CoV will be discussed, which may provide some clues to further understandings of pathogenesis of swine enteropathogenic CoVs.
Apoptosis Associated With Transmissible Gastroenteritis Virus (TGEV)
Virus Characteristics of TGEV
TGEV is an enveloped, single-stranded, positive-sense RNA virus with a genome of appropriately 28 kb in length (20, 60). The full-length genome of TGEV is arranged in the order of: 5′ UTR-ORF1a/1b-S-3a-3b-E-M-N-7-3′ UTR, containing nine open reading frames (ORFs) encoding four structure proteins (S, M, N, E) and five non-structure proteins (61). The nsp1 protein of TGEV can efficiently suppress protein synthesis in mammalian (62). The nsp3 protein of TGEV can cleave a peptide mimicking the cognate nsp2|nsp3 cleavage site based on its papain-like protease 1 (PL1(pro)) domain (63). The N protein of TGEV belongs to a multifunctional phosphoprotein, which can package the RNA genome into a helical ribonucleoprotein, regulate viral RNA synthesis, and modulate of host cell metabolism (64). E protein promotes TGEV maturation in the secretory pathway (65). S1 and M proteins play a role in viral replication (66, 67). In addition, M protein can affect TGEV-induced IFN-α production (68).
The Role of Apoptosis in TGEV Infection
It was reported that TGEV invades and replicates in villous epithelial cells to provoke villous atrophy, causing severe diarrhea, and dehydration in piglets is the central event in the pathogenesis of TGEV infection (69, 70). Apoptosis plays a important role in the pathogenesis process of animal virus infectious diseases (71–73). Many studies shown that TGEV infection could induce apoptosis in PK-15 cells, swine testicular (ST) cells, swine kidney cells, MDCK-APN cells (canine kidney cell line expressing porcine APN) or human rectal tumor cells (HRT18, expressing porcine APN) (Table 1) (74–82, 91), which associates with intracellular molecules, such as p53, reactive oxygen species (ROS), mitochondrial apoptosis-inducing factor (AIF), poly (ADP-ribose) polymerase (PARP), and caspases (74, 82). Interestingly, TGEV may not induce apoptotic death of intestinal villous enterocytes in vivo (77). TGEV infection could decrease p300/CBP, down-regulate MDM2, and promote p53 phosphorylation at serine 15, 20, and 46, resulting in accumulation and activation of p53, then p53 induced ROS accumulation which leads to mitochondrial oxidative damage to release cyt c to the cytosol and thereby activate apoptosis in PK-15 cells (79, 91). In addition, TGEV infection up-regulated FasL to activate caspase-8 and cleaved Bid to tBid which was transferred to the mitochondria, resulting in release of cyt c into the cytoplasm to activate caspase-9 (78). Moreover, TGEV could down-regulate Bcl-2, increase the expression of Bax, and promote the transfer of Bax from cytoplasm to mitochondria (78). Then, mitochondria released cyt c to activate caspase-9, and finally caspase-9 activates caspase-3 to induce cell apoptosis (78). microRNAs (miRNAs) play a key role in the regulation of virus-induced apoptosis (80). For instance, miR-27b can directly target the 3′ UTR of runt-related transcription factor 1 (RUNX1) mRNA to regulate the expression of RUNX1 in PK-15 cells (80). It has been found that TGEV infection can down-regulate the expression of miR-27b in host cells, thereby down-regulating the expression of RUNX1 and activating the Bax regulated expression of caspase-9/3 to induce cell apoptosis (80). These results suggest that TGEV can induce apoptosis through both extrinsic and intrinsic pathways (Figure 2) in PK-15 cells. Further analysis of the key viral proteins induced by TGEV showed that N protein could activate p53, p21 to eliminate cyclin B/cdc2, promote the transfer of Bax to mitochondria, lead to the release of cyt c from mitochondria, and activate caspase-9/3 to induce cell apoptosis (81) (Figure 2). Interestingly, N protein was cleaved at the position of VVPD359 by activated caspase-6/7 during TGEV-induced apoptosis (76) in human rectal tumor cell line, indicating that N protein plays a very important role in the interaction between virus and host during apoptotic process. It is well-known that viruses could manipulate apoptosis to complete their life cycle. It was reported that mitophagy induced by DJ-1 to counteract apoptosis could promote viral infection during TGEV infection (54) (Figure 2). The above studies confirmed that there may be a certain correlation between pathogenicity and apoptosis after TGEV infection. More research is needed on how apoptosis affects the proliferation and spread of TGEV during viral infection.
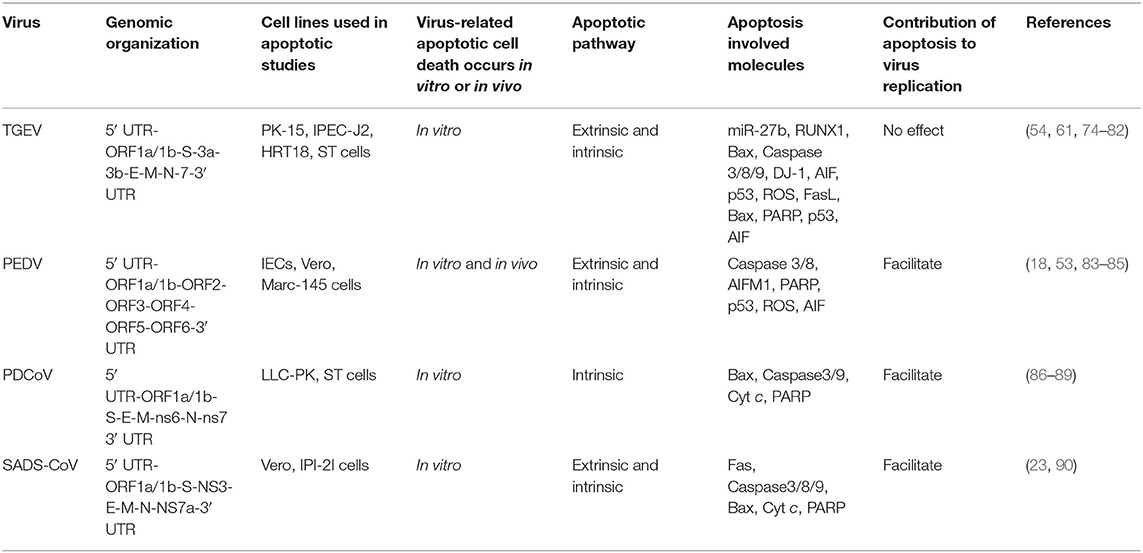
Table 1. Compare apoptotic cell death caused by the four swine enteropathogenic CoVs and the related mechanisms.
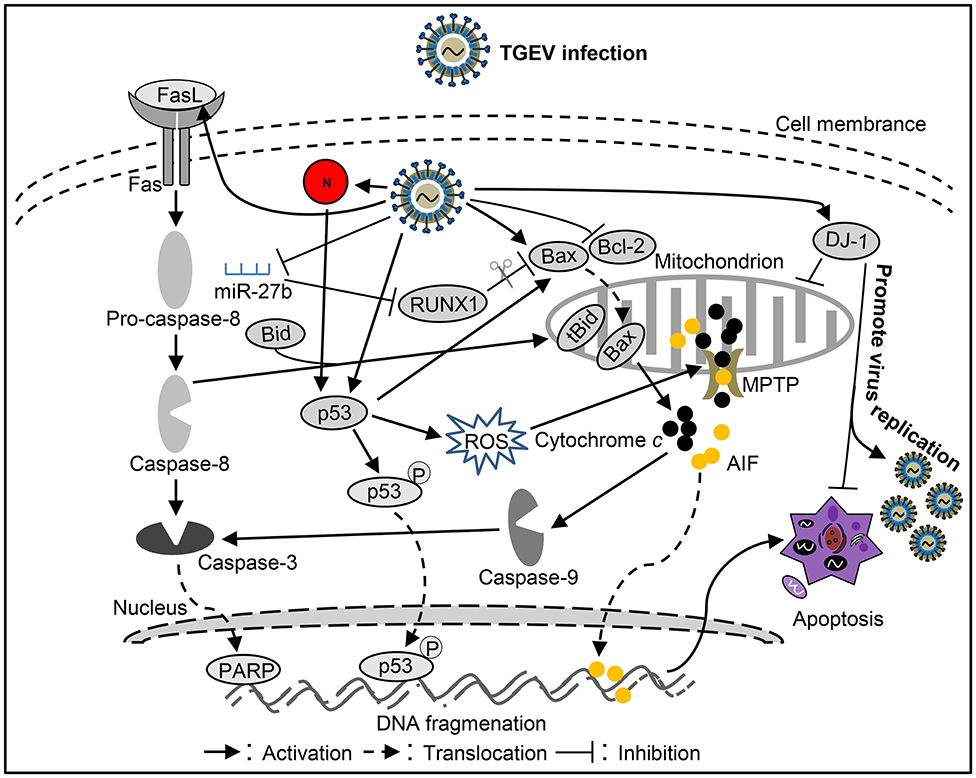
Figure 2. Diagram of the roles of apoptosis in the pathogenesis of Transmissible gastroenteritis virus (TGEV) infection.
Apoptosis Associated With Porcine Epidemic Diarrhea Virus (PEDV)
Virus Characteristics of PEDV
PEDV is an enveloped, single-stranded, positive-sense RNA virus with a genome of appropriately 28 kb in length (22). The viral genome is sequentially composed of 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), ORF2-6, and 3′ UTR (84). The ORF 1a/1b cover the 5′-proximal two-thirds of the genome coding for replicase polyprotein (pp) la and pp1ab, respectively (84, 92). These pp1a and pp1ab polyproteins can be cleaved by internal proteases generating sixteen non-structural proteins, namely nsp1-16 (85). Moreover, the genome of PEDV encodes four structural proteins including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, while ORF3 encodes an accessory protein (84). The functional form of the S protein is a trimer, which protrude from the viral membrane thus providing typical crown appearance of the CoVs (93). It functions during cell entry by binding to cellular receptors and causing fusion of the viral and host cell membranes (93). During maturation, the S protein is cleaved into a receptor-binding subunit S1 and a membrane-fusion subunit S2 (84). The E protein has ion channel activity and plays an important role in virion morphogenesis (93, 94). The M protein is the main component of the viral envelope and interacts with all structural components during viral particle assembly (93). The N protein packages the genomic RNA to form the helical nucleocapsid (RNP) (93). ORF3 protein was known to be related to PEDV pathogenicity (93). In addition, N and ORF3 are involved in viral replication (95, 96). Furthermore, the encoded N, M, E, ORF3, PLP2, nsp 1, nsp 3, nsp 5, nsp 7, nsp 14, nsp 15, nsp 16 proteins can antagonize Interferon-β production (97–100).
The Role of Apoptosis in PEDV Infection
PEDV infection can damages pig intestinal epithelial tissue and interfere with epithelial mucosal cell function, resulting in abnormal nutrient absorption and diarrhea (53). This phenomenon may be related to apoptosis caused by PEDV infection. Apoptosis of cells in the lamina propria or submucosa of PEDV-infected jejunum or ileum was increased (18, 101). However, PEDV may not induce apoptosis death of intestinal villous enterocytes in vivo (50), like TGEV and PDCoV. AIF could translocate to the nucleus to cause apoptosis after PEDV infection in Vero cells (18). Moreover, PEDV could promote p53 phosphorylation at serine 20 and subsequent translocation to the nucleus, leading to p53 activation and thereby apoptosis in Vero cells (83). During this process, ROS also accumulates to promote apoptosis (83). In addition, apoptosis was mediated by activation of caspase-8 and caspase-3 in the late stage of PEDV-infected Vero cells (102). Treatment with the inhibitor of pro-apoptotic molecule could significantly inhibit PEDV infection (18), indicating that apoptosis plays an important role in the PEDV pathogenicity. However, the host cells showed an anti-apoptotic effect through LTBR during PEDV infection (103). By analysis cell apoptosis induced by PEDV-related viral proteins, it was found that M, nsp1, nsp2, nsp5, nsp6, nsp7, nsp9, nsp13, and S1 proteins can induce apoptosis, among which S1 is the critical apoptotic-inducing protein in Vero cells, but the detailed molecular mechanism is still unclear (85). On the contrary, PEDV encoded ORF3 could inhibit cell apoptosis to promote virus proliferation (93), indicating that PEDV applies different strategies to regulate cell apoptosis in different stages of infection to complete viral proliferation. Although some studies have been conducted on the influence of PEDV on apoptosis (Figure 3), many questions remain unclear, such as how does S1 induce apoptosis? And what role does S1 portein induced-apoptosis play in virus-caused diarrhea? Clarifying these issues will help explain how PEDV causes diarrhea in pigs.
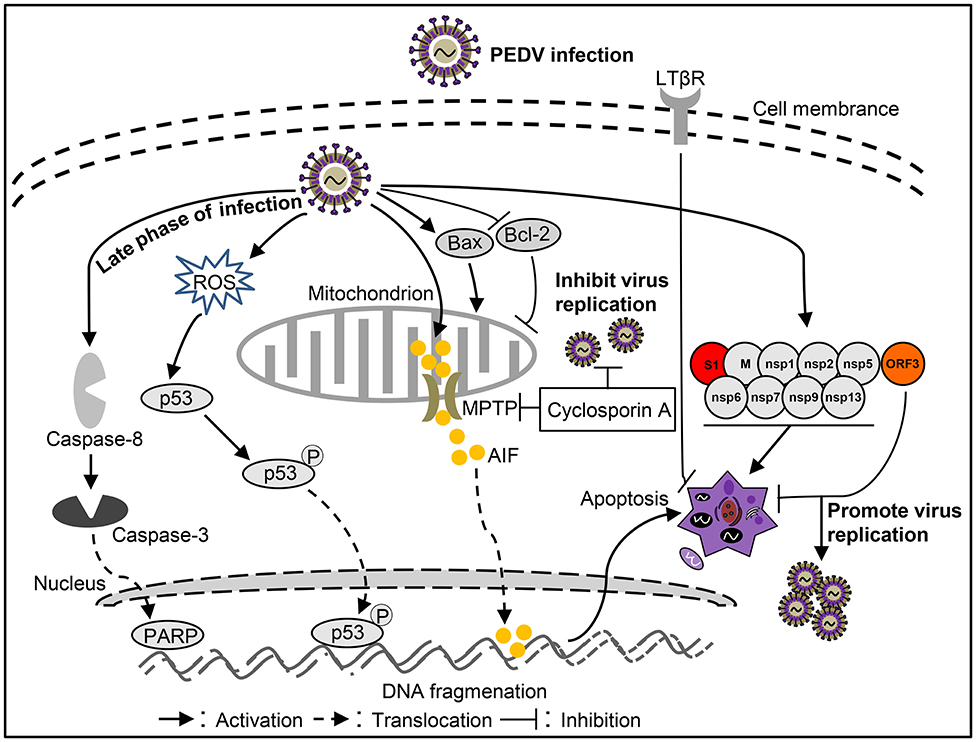
Figure 3. Diagram of the roles of apoptosis in the pathogenesis of Porcine epidemic diarrhea virus (PEDV) infection.
Apoptosis Associated With Porcine Deltacoronavirus (PDCoV)
Virus Characteristics of PDCoV
PDCoV is an enveloped, single-stranded, positive-sense RNA virus with a genome of appropriately 25 kb in length (26). The genome organization of PDCoV are in the order of: 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), spike (S), envelope (E), membrane (M), non-structural protein 6 (ns6), nucleocapsid (N), non-structural protein 7 (ns7), and 3′ UTR (86, 87). The diverse dimerization forms of nsp9 protein could enhance their nucleic acid binding affinity (104). ns6 protein is an important virulence factor of PDCoV (105). It is known that N, nsp5, nsp15, ns6 contribute to inhibit interferon-β production (106–109).
The Role of Apoptosis in PDCoV Infection
PDCoV is a newly discovered virus that causes severe clinical diarrhea and intestinal pathological damage in piglets (88), but the pathogenesis of PDCoV infection is still largely unknown. Current studies showed that PDCoV infection could promote Bax translocation and mediate mitochondrial outer membrane permeabilization (MOMP), resulting in specific relocation of the mitochondrial cyt c into the cytoplasm, thus activating caspase-9/3 to initiate apoptosis in ST cells (88). These results indicate that PDCoV mediates cell apoptosis through a caspase-dependent endogenous apoptotic pathway (Figure 4). Moreover, apoptosis caused by PDCoV contributes to viral protein translation and the caspase-dependent intrinsic apoptosis pathway in PDCoV-infected ST cells is also used for facilitation of viral replication (88). Interestingly, PDCoV induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Another form of cell death, necrosis, has been found in PDCoV-infected swine intestinal enterocytes as well as in the porcine small intestinal epithelial cell line, IPEC-J2 in vitro (89). The above results indicate that a better model of cellular infection is needed to reflect the infection in vivo. In-depth study on the molecular mechanism of cell death caused by PDCoV infection will help to analyze the pathogenesis of PDCoV. In addition, the key proteins responsible for cell death caused by the virus still need to be further studied, which will help to identify virulence factors and provide guidance for prevention and control PDCoV infection.
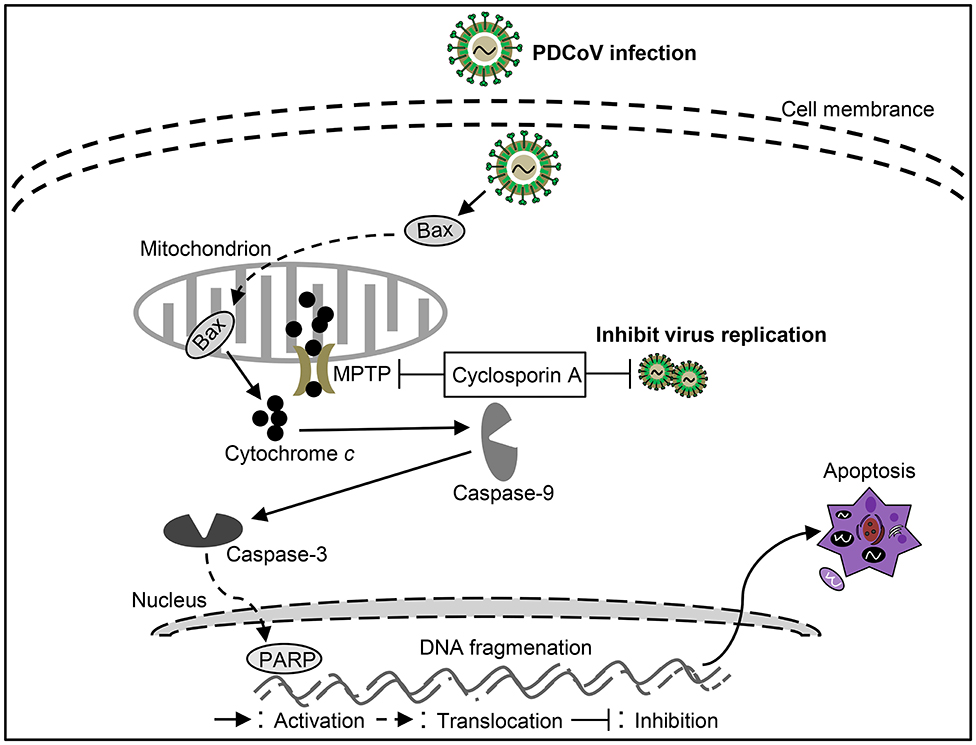
Figure 4. Diagram of the roles of apoptosis in the pathogenesis of Porcine deltacoronavirus (PDCoV) infection.
Apoptosis Associated With Swine Acute Diarrhea Syndrome Coronavirus (SADS-CoV)
Virus Characteristics of SADS-CoV
SADS-CoV, also named PEAV (110) and SeACoV (111), is an enveloped, single-stranded positive-sence RNA virus (110). The full-length genome of SADS-CoV is about 27 kb (110), arranged in the order of: 5′ UTR-ORF1a/1b-S-NS3-E-M-N-NS7a-3′ UTR (112). It is known that the S protein has many important characteristics in CoVs, such as virus attachment and entry, and induction of neutralizing antibodies in vivo (113). Of note, compare to other reported CoVs, SADS-CoV has the smallest S protein including 1,129 amino acids (110). To date, the papain-like protease 2 (PLP2) domain of nsp3 was shown to be able to cleave nsp1 proteins and also peptides mimicking the nsp2/nsp3 cleavage site and also have deubiquitinating and deISGynating activity (114). The function of other viral proteins of SADS-CoV remains to be further explored.
The Role of Apoptosis in SADS-CoV Infection
As another newly identified swine intestine CoV, detail information of the pathogenic mechanism of SADS-CoV remains unclear. It was reported that SADS-CoV infection could increased apoptosis in the small intestinal epithelial cell line IEC in vitro (90). SADS-CoV infection could up-regulate FasL, subsequentially activates caspase-8/3 to lead to apoptosis in Vero and IPI-2I cells (90). Moreover, activated caspase-8 could cleave Bid, then the cleaved Bid translocated to mitochondria participating in the destruction of mitochondria integrity and cyt c release to cytosol, which in turn facilitates caspase-9/3 activation thus result in apoptosis (90). In addition, SADS-CoV infection triggers Bax recruitment into the mitochondria, leading to cyt c but not AIF release into cytoplasm to induce apoptosis through mitochondrial permeability transition pore (MPTP), which involve with cyclophilin D (CypD) in these processes (90). These results suggest SADS-CoV-induced apoptosis were mediated by both extrinsic and intrinsic pathways (Figure 5). The viral replication was affected with the inhibitors of caspase-8 or capases-9, indicating that SADS-CoV-induced apoptosis contributes to viral replication (90). Although it has been demonstrated well in SADS-CoV induced apoptosis, the function of viral protein in SADS-CoV-induced apoptosis and the exact mechanism underlying remains unclear. More efforts to elucidate the molecular mechanisms of SADS-CoV-induced apoptosis will help to explore the pathogenesis of SADS-CoV infection.
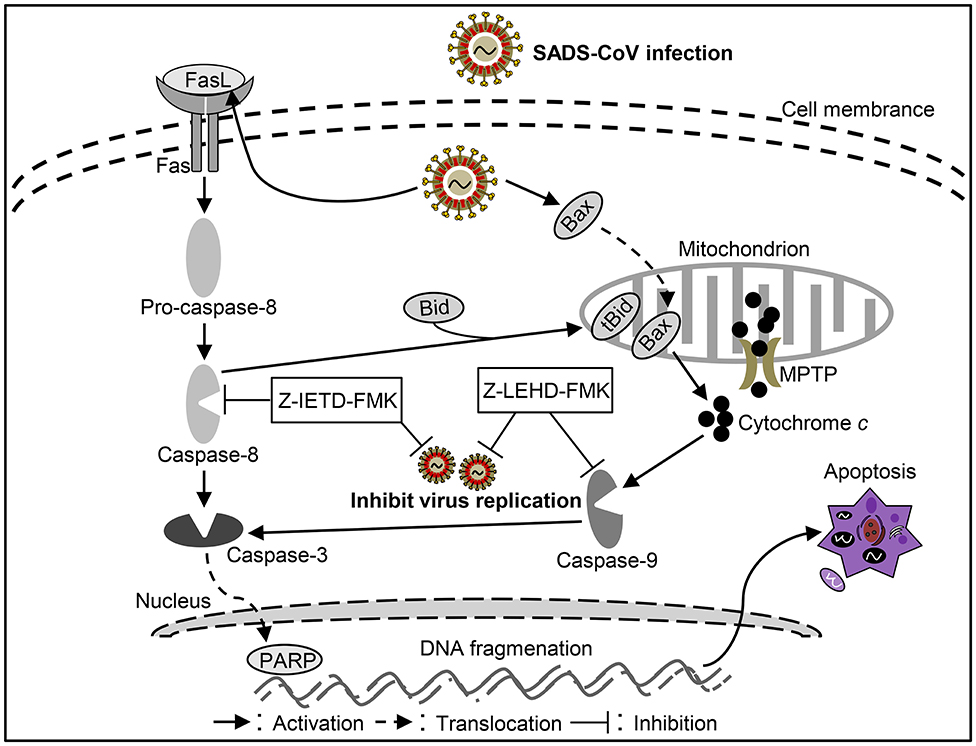
Figure 5. Diagram of the roles of apoptosis in the pathogenesis of Swine acute diarrhea syndrome coronavirus (SADS-CoV) infection.
Other Mechanisms Are Related to the Pathogenesis of Swine Enteropathogenic CoVs
Innate immunity is thought to be the first line of host defense against a wide variety of pathogenic infections (115). Of note, type I interferon (IFN-α/β), as important cytokines of innate immunity induced by virus invasion, could establish an anti-viral state in infected sites (116). In order to infect the organism and cause pathogenicity, the virus must break through the anti-viral state of the organism. It was reported that PEDV, PDCoV and SADS-CoV can inhibit the up-regulated expression of type I interferon through a variety of different mechanisms (100, 115, 117–120), thus leading to virus infection, indicating that the inhibition of type I interferon might relate to the pathogenesis of these viruses. In addition, inhibition of anti-viral status to promote viral infection might contribute to the occurrence of apoptosis (121). Interestingly, unlike PEDV, PDCoV, and SADS-CoV, TGEV infects the body can promote the up-regulated expression of IFN-β (122). IFN-β has been reported to induce apoptosis (123, 124). Whether the apoptosis induced by TGEV is related to the upregulation of IFN-β needs further study.
Conclusions
Lines of evidence indicate that apoptosis play critical roles in the pathogenesis of swine enteropathogenic CoVs. Most of the information is gleaned from the studies on the apoptosis of TGEV, PEDV, PDCoV, or SADS-CoV infections in vitro and in vivo. Viral proteins, such as N from TGEV, S1 from PEDV, are involved in regulation of virus-mediated apoptosis production, which might provide some clues to determine the virulence factors and serve as the targets of antiviral drugs. However, the roles of apoptosis in host response to swine enteropathogenic CoVs infection alone or co-infections are still far from elucidation and need to be further investigated. In addition, the apoptotic forms and mechanisms caused by these four swine enteropathogenic CoVs are different (Table 1), which whether involved with the differences of pathogenicity also needs further study. In brief, further investigation into the role of apoptosis in these swine enteropathogenic CoVs is conducive to elucidate of the pathogenesis of viral infections and develop an appropriate strategy for the prevention and control of swine diarrhea diseases.
Author Contributions
ZX collected the data and wrote the paper. YZ revised the paper. YC checked and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (#31902248), Doctoral Initiative Project of Natural Science Foundation of Guangdong Province (#2018A030310158) and Guangdong Basic and Applied Basic Research Foundation (#2019B1515210026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bortner CD, Cidlowski JA. Cellular mechanism for the repression of apoptosis. Annual Rev Pharmacol Toxicol. (2002) 42:259–81. doi: 10.1146/annurev.pharmtox.42.083101.143836
2. Pandya NM, Jain SM, Santani DD. Apoptosis: a friend or foe? Transplant Proc. (2006) 33:2414–6. doi: 10.1016/S0041-1345(01)02057-7
3. Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, et al. Classification of cell death: recommendations of the nomenclature committee on cell death. Cell Death Differ. (2005) 12:1463–7. doi: 10.1038/sj.cdd.4401724
4. Schmitz I, Kirchhoff S, Krammer PH. Regulation of death receptor-mediated apoptosis pathways. Int J Biochem Cell Biol. (2000) 32:1123–36. doi: 10.1016/S1357-2725(00)00048-0
5. Gibson CJ, Davids MS. BCL-2 antagonism to target the intrinsic mitochondrial pathway of apoptosis. Clin Cancer Res. (2015) 21:5021–9. doi: 10.1158/1078-0432.CCR-15-0364
6. Jeong M, Lee EW, Seong D, Seo J, Kim JH, Grootjans S. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene. (2017) 36:458–70. doi: 10.1038/onc.2016.215
7. Chicheportiche Y, Bourdon PR, Xu H, Hsu YM, Scott H, Hession C, et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem. (1997) 272:32401–10. doi: 10.1074/jbc.272.51.32401
8. Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. (1998) 281:1305–8. doi: 10.1126/science.281.5381.1305
9. Peter ME, Krammer PH. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr Opin Immunol. (1998) 10:545–51. doi: 10.1016/S0952-7915(98)80222-7
10. Suliman ALA, Datta R, Srivastava RK. Intracellular mechanisms of TRAIL: apoptosis through mitochondrial-dependent and -independent pathways. Oncogene. (2001) 20:2122–33. doi: 10.1038/sj.onc.1204282
11. Rubio-Moscardo F, Blesa D, Mestre C, Siebert R, Balasas T, Benito A, et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. (2005) 106:3214–22. doi: 10.1182/blood-2005-05-2013
12. Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. Embo J. (1995) 14:5579–88. doi: 10.1002/j.1460-2075.1995.tb00245.x
13. Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. (2000) 12:599–609. doi: 10.1016/S1074-7613(00)80211-3
14. Kischkel FC, Lawrence DA, Tinel A, Leblanc H, Virmani A, Schow P, et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem. (2001) 276:46639–46. doi: 10.1074/jbc.M105102200
15. Jiang S, Cai J, Wallace DC, Jones DP. Cytochrome c-mediated apoptosis in cells lacking mitochondrial DNA: signaling pathway involving release and caspase 3 activation is conserved. J Biol Chem. (1999) 274:29905–11. doi: 10.1074/jbc.274.42.29905
16. Li CY, Lee JS, Ko YG, Kim JI, Seo JS. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. J Biol Chem. (2000) 275:25665–71. doi: 10.1074/jbc.M906383199
17. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. (2007) 35:495–516. doi: 10.1080/01926230701320337
18. Kim Y, Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. (2014) 460–1:180–93. doi: 10.1016/j.virol.2014.04.040
19. Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. J Gen Virol. (1999) 53:577–628. doi: 10.1146/annurev.micro.53.1.577
20. Doyle LP, Hutchings LM. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. (1946) 108:257–9.
21. Wesley RD, Woods RD, Hill HT, Biwer JD. Evidence for a porcine respiratory coronavirus, antigenically similar to transmissible gastroenteritis virus, in the United States. J Vet Diagn Invest. (1990) 2:312–7. doi: 10.1177/104063879000200411
22. Pensaert MB, Bouck PD. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. (1978) 58:243–7. doi: 10.1007/BF01317606
23. Peng Zhou HF, Tian L, Xing-Lou Y, Wei-Feng S, Wei Z, Yan Z, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. (2018) 556:255–8. doi: 10.1038/s41586-018-0010-9
24. Sasseville MJ, Boutin M, Gélinas AM, Dea S. Sequence of the 3'-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J Gen Virol. (2002) 83:2411. doi: 10.1099/0022-1317-83-10-2411
25. Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11
26. Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. (2016) 226:71–84. doi: 10.1016/j.virusres.2016.05.028
27. Lu X, Yang Y, Wang J, Jing Y, Qian Y. Impact of TGEV infection on the pig small intestine. Virology J. (2018) 15:102. doi: 10.1186/s12985-018-1012-9
28. Wen Z, Xu Z, Zhou Q, Li W, Wu Y, Du Y, et al. Oral administration of coated PEDV-loaded microspheres elicited PEDV-specific immunity in weaned piglets. Vaccine. (2018) 36:6803–9. doi: 10.1016/j.vaccine.2018.09.014
29. Xu Z, Zhang Y, Gong L, Huang L, Lin Y, Qin J, et al. Isolation and characterization of a highly pathogenic strain of Porcine enteric alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound Emerg Dis. (2019) 66:119–30. doi: 10.1111/tbed.12992
30. Bereiter M, Hasler J, Keller H. Transmissible gastroenteritis (TGE) in Switzerland: antibody persistence after infection and seroepidemiologic studies of the significance of the TGE virus as the cause of diarrhea. Schwzer Archiv Für Tierhlkunde. (1988) 130:237–48.
31. Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. (2012) 44:167–75. doi: 10.1007/s11262-012-0713-1
32. Carvajal A, Arguello H, Martinez-Lobo FJ, Costillas S, Miranda R, Pj GDN, et al. Porcine epidemic diarrhoea: new insights into an old disease. Porcine Health Manag. (2015) 1:12. doi: 10.1186/s40813-015-0007-9
33. Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. (2016) 226:20–39. doi: 10.1016/j.virusres.2016.05.023
34. Sun D, Wang X, Wei S, Chen J, Feng L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci. (2016) 78:355–63. doi: 10.1292/jvms.15-0446
35. Diep NV, Sueyoshi M, Izzati U, Fuke N, Teh APP, Lan NT, et al. Appearance of US-like porcine epidemic diarrhoea virus (PEDV) strains before US outbreaks and genetic heterogeneity of PEDVs collected in Northern Vietnam during 2012-2015. Transbound Emerg Dis. (2018) 65:e83–93. doi: 10.1111/tbed.12681
36. Hsu TH, Liu HP, Chin CY, Wang C, Zhu WZ, Wu BL, et al. Detection, sequence analysis, and antibody prevalence of porcine deltacoronavirus in Taiwan. Arch Virol. (2018) 163:3113–7. doi: 10.1007/s00705-018-3964-x
37. Jang G, Kim SH, Lee YJ, Kim S, Lee DS, Lee KK, et al. Isolation and characterization of Korean porcine deltacoronavirus strain KNU16-07. J Vet Sci. (2018) 19:577–81. doi: 10.4142/jvs.2018.19.4.577
38. Lara-Romero R, Gomez-Nunez L, Cerriteno-Sanchez JL, Marquez-Valdelamar L, Mendoza-Elvira S, Ramirez-Mendoza H, et al. Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013-2016. Virus Genes. (2018) 54:215–24. doi: 10.1007/s11262-017-1528-x
39. Suzuki T, Shibahara T, Imai N, Yamamoto T, Ohashi S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect Genet Evol. (2018) 61:176–82. doi: 10.1016/j.meegid.2018.03.030
40. Xu Z, Zhong H, Zhou Q, Du Y, Chen L, Zhang Y, et al. A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol Sin. (2018) 33:131–41. doi: 10.1007/s12250-018-0003-8
41. Chen F, Knutson TP, Rossow S, Saif LJ, Marthaler DG. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci Rep. (2019) 9:3953. doi: 10.1038/s41598-019-40564-z
42. Perez-Rivera C, Ramirez-Mendoza H, Mendoza-Elvira S, Segura-Velazquez R, Sanchez-Betancourt JI. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg Dis. (2019) 66:1436–41. doi: 10.1111/tbed.13193
43. Yang YL, Yu JQ, Huang YW. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. (2020) 285:198024. doi: 10.1016/j.virusres.2020.198024
44. Lorincz M, Biksi I, Andersson S, Cságola A, Tuboly T. Sporadic re-emergence of enzootic porcine transmissible gastroenteritis in Hungary. Acta Vet Hung. (2014) 62:125–33. doi: 10.1556/avet.2013.043
45. Zhang F, Luo S, Gu J, Li Z, Li K, Yuan W, et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet Res. (2019) 15:470. doi: 10.1186/s12917-019-2212-2
46. Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol. (2017) 206:45–51. doi: 10.1016/j.vetmic.2016.11.029
47. Li K, Li H, Bi Z, Song D, Zhang F, Lei D, et al. Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector. Antiviral Res. (2019) 166:11–8. doi: 10.1016/j.antiviral.2019.03.010
48. Gao X, Zhao D, Zhou P, Zhang L, Li M, Li W, et al. Characterization, pathogenicity and protective efficacy of a cell culture-derived porcine deltacoronavirus. Virus Res. (2020) 282:197955. doi: 10.1016/j.virusres.2020.197955
49. Xu Z, Liu Y, Peng P, Liu Y, Huang M, Ma Y, et al. Aloe extract inhibits porcine epidemic diarrhea virus in vitro and in vivo. Vet Microbiol. (2020) 249:108849. doi: 10.1016/j.vetmic.2020.108849
50. Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. (2015) 204:134–43. doi: 10.1016/j.tvjl.2015.02.017
51. Jung K, Hu H, Saif LJ. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. (2016) 226:50–9. doi: 10.1016/j.virusres.2016.04.009
52. Guo R, Fan B, Chang X, Zhou J, Zhao Y, Shi D, et al. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology. (2020) 545:24–32. doi: 10.1016/j.virol.2020.03.001
53. Wang XY, Zhao TQ, Xu DP, Zhang X, Ji CJ, Zhang DL. The influence of porcine epidemic diarrhea virus on pig small intestine mucosal epithelial cell function. Arch Virol. (2018) 164:83–90. doi: 10.1007/s00705-018-4061-x
54. Zhu L, Mou C, Xing Y, Jian L, Qian Y. Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget. (2015) 7:27122–41. doi: 10.18632/oncotarget.8345
55. Rogers DG, Andersen AA. Intestinal lesions caused by two swine chlamydial isolates in gnotobiotic pigs. J Vet Diagn Invest. (1996) 8:433–40. doi: 10.1177/104063879600800405
56. Li FQ, Tam JP, Liu DX. Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology. (2007) 365:435–45. doi: 10.1016/j.virol.2007.04.015
57. Krähling V, Stein DA, Spiegel M, Weber F, Mühlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol. (2009) 83:2298–309. doi: 10.1128/JVI.01245-08
58. Collins M. Potential roles of apoptosis in viral pathogenesis. Am J Respir Crit Care Med. (1995) 152:S20–24. doi: 10.1164/ajrccm/152.4_Pt_2.S20
59. Zhai Y, Ding N. MicroRNA-194 participates in endotoxemia induced myocardial injury via promoting apoptosis. Eur Rev Med Pharmacol. (2018) 22:2077–83. doi: 10.26355/eurrev_201804_14739
60. Curtis KM, Yount B, Baric RS. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J Virol. (2002) 76:1422–34. doi: 10.1128/JVI.76.3.1422-1434.2002
61. Enjuanes L, Dediego ML, lvarez E, Deming D, Sheahan T, Baric R. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. (2008) 133:45–62. doi: 10.1016/j.virusres.2007.01.021
62. Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol. (2011) 85:638–43. doi: 10.1128/JVI.01806-10
63. Wojdyla JA, Manolaridis I, Van Kasteren PB, Kikkert M, Snijder EJ, Gorbalenya AE, et al. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J Virol. (2010) 84:10063–73. doi: 10.1128/JVI.00898-10
64. Yount B, Curtis KM, Baric RS. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol. (2000) 74:10600–11. doi: 10.1128/JVI.74.22.10600-10611.2000
65. Ortego J, Ceriani JE, Patino C, Plana J, Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. (2007) 368:296–308. doi: 10.1016/j.virol.2007.05.032
66. Peng Y, Shilei H, Zhou Y, Luyi X, Wang K, Yang Y, et al. UBXN1 interacts with the S1 protein of transmissible gastroenteritis coronavirus and plays a role in viral replication. Vet Res. (2019) 50:28. doi: 10.1186/s13567-019-0648-9
67. Song Z, Yang Y, Wang L, Wang K, Ran L, Xie Y, et al. EIF4A2 interacts with the membrane protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Res Vet Sci. (2019) 123:39–46. doi: 10.1016/j.rvsc.2018.12.005
68. Laude H, Gelfi J, Lavenant L, Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J Virol. (1992) 66:743–9. doi: 10.1128/JVI.66.2.743-749.1992
69. Enjuanes L, van der Zeijst BAM. Molecular Basis of Transmissible Gastroenteritis Virus Epidemiology. Springer US (1995). doi: 10.1007/978-1-4899-1531-3_16
70. Gu JP, Yue XW, Xing R, Li CY, Yang ZB. Progress in genetically engineered vaccines for porcine transmissible gastroenteritis virus. Rev Méd Vét. (2012) 163:107–11.
71. Helen E, Grant M. Apoptosis: an innate immune response to virus infection. Trends Microbiol. (1999) 7:160–5. doi: 10.1016/S0966-842X(99)01487-0
72. Wen LS, Hsu HW, Liao MH, Long HL, Liu HJ. Avian reovirus σC protein induces apoptosis in cultured cells. Virology. (2004) 321:65–74. doi: 10.1016/j.virol.2003.12.004
73. Chen S, Cheng AC, Wang MS, Xi P. Detection of apoptosis induced by new type gosling viral enteritis virus in vitro through fluorescein annexin V-FITC/PI double labeling. World J Gastroenterol. (2008) 014:2174–8. doi: 10.3748/wjg.14.2174
74. Eleouet JF, Chilmonczyk S, Besnardeau L, Laude H. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J Virol. (1998) 72:4918–24. doi: 10.1128/JVI.72.6.4918-4924.1998
75. Sirinarumitr T, Kluge JP, Paul PS. Transmissible gastroenteritis virus induced apoptosis in swine testes cell cultures. Arch Virol. (1998) 143:2471–85. doi: 10.1007/s007050050477
76. Eléouët JF, Slee EA, Saurini F, Castagné N, Poncet D, Garrido C, et al. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and−7 during TGEV-induced apoptosis. J Virol. (2000) 74:3975–83. doi: 10.1128/JVI.74.9.3975-3983.2000
77. Kim B, Kim O, Tai JH, Chae C. Transmissible gastroenteritis virus induces apoptosis in swine testicular cell lines but not in intestinal enterocytes. J Comp Pathol. (2000) 123:64–6. doi: 10.1053/jcpa.2000.0386
78. Ding L, Xu X, Huang Y, Li Z, Zhang K, Chen G, et al. Transmissible gastroenteritis virus infection induces apoptosis through FasL- and mitochondria-mediated pathways. Vet Microbiol. (2012) 158:12–22. doi: 10.1016/j.vetmic.2012.01.017
79. Ding L, Zhao X, Huang Y, Du Q, Dong F, Zhang H, et al. Regulation of ROS in transmissible gastroenteritis virus-activated apoptotic signaling. Biochem Biophys Res Commun. (2013) 442:33–7. doi: 10.1016/j.bbrc.2013.10.164
80. Zhao X, Song X, Bai X, Fei N, Huang Y, Zhao Z, et al. miR-27b attenuates apoptosis induced by transmissible gastroenteritis virus (TGEV) infection via targeting runt-related transcription factor 1 (RUNX1). Peer J. (2016) 4:e1635. doi: 10.7717/peerj.1635
81. Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, et al. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun. (2014) 445:497–503. doi: 10.1016/j.bbrc.2014.02.039
82. Hong M. p53- and ROS-mediated AIF pathway involved in TGEV-induced apoptosis. J Vet Med Sci. (2018) 80:1775–81. doi: 10.1292/jvms.18-0104
83. Xu X, Xu Y, Zhang Q, Yang F, Yin Z, Wang L, et al. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet Microbiol. (2019) 232:1–12. doi: 10.1016/j.vetmic.2019.03.028
84. Kocherhans R, Bridgen A, Ackermann M, Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. (2001) 23:137–44. doi: 10.1023/A:1011831902219
85. Chen Y, Zhang Z, Li J, Gao Y, Zhou L, Ge X, et al. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol J. (2018) 15. doi: 10.1186/s12985-018-1078-4
86. Fangzhou C, Yinxing Z, Meizhou W, Xugang K, Yao L, He Q. Full-Length Genome Characterization of Chinese Porcine Deltacoronavirus Strain CH/SXD1/2015. Shanxi: Genome Announcements (2015).
87. Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. (2015) 482:51–9. doi: 10.1016/j.virol.2015.03.024
88. Lee Y, Lee C. Porcine deltacoronavirus induces caspase-dependent apoptosis through activation of the cytochrome c -mediated intrinsic mitochondrial pathway. Virus Res. (2018) 253:112–23. doi: 10.1016/j.virusres.2018.06.008
89. Jung K, Hu H, Saif LJ. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet Microbiol. (2016) 182:57–63. doi: 10.1016/j.vetmic.2015.10.022
90. Zhang J, Han Y, Shi H, Chen J, Zhang X, Wang X, et al. Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D- dependent. Emerg Microbes Infect. (2020) 9:439–56. doi: 10.1080/22221751.2020.1722758
91. Huang Y, Ding L, Li Z, Dai M, Zhao X, Li W, et al. Transmissible gastroenteritis virus infection induces cell apoptosis via activation of p53 signalling. J Gen Virol. (2013) 94:1807–17. doi: 10.1099/vir.0.051557-0
92. Lee C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virology J. (2016) 13:19. doi: 10.1186/s12985-016-0465-y
93. Si F, Hu X, Wang C, Chen B, Wang R, Dong S, et al. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. (2020) 12:214. doi: 10.3390/v12020214
94. Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta. (2011) 1808:510–5. doi: 10.1016/j.bbamem.2010.05.006
95. Shiyi Ye ZL, Fangzhou C, Wentao L, Xiaozhen G, Han H, Qigai H. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. (2015) 51:385–92. doi: 10.1007/s11262-015-1257-y
96. Shi D, Shi H, Sun D, Chen J, Zhang X, Wang X, et al. Nucleocapsid interacts with NPM1 and protects it from proteolytic cleavage, enhancing cell survival, and is involved in PEDV growth. Sci Rep. (2017) 7:39700. doi: 10.1038/srep39700
97. Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. (2013) 94:1554. doi: 10.1099/vir.0.051169-0
98. Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, et al. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. (2014) 88:8936–45. doi: 10.1128/JVI.00700-14
99. Dang W, Liurong F, Yanling S, Huan Z, Gao L, Peng G, et al. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J Virol. (2015) 90:2090–101. doi: 10.1128/JVI.02514-15
100. Zhang Q, Shi K, Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. (2016) 489:252–68. doi: 10.1016/j.virol.2015.12.010
101. Curry SM, Schwartz KJ, Yoon KJ, Gabler NK, Burrough ER. Effects of porcine epidemic diarrhea virus infection on nursery pig intestinal function and barrier integrity. Vet Microbiol. (2017) 211:58–66. doi: 10.1016/j.vetmic.2017.09.021
102. Zhang X, Li P, Zheng Q, Hou J. Lactobacillus acidophilus S-layer protein-mediated inhibition of PEDV-induced apoptosis of Vero cells. Vet Microbiol. (2019) 229:159–67. doi: 10.1016/j.vetmic.2019.01.003
103. Altawaty T, Liu L, Zhang H, Tao C, Hou S, Li K, et al. Lack of LTβR increases susceptibility of IPEC-J2 cells to porcine epidemic diarrhea virus. Cells. (2018) 7:222. doi: 10.3390/cells7110222
104. Zeng Z, Feng D, Shi K, Ye G, Wang G, Fang L, et al. Dimerization of coronavirus Nsp9 with diverse modes enhances its nucleic acid binding affinity. J Virol. (2018) 92:e00692–18. doi: 10.1128/JVI.00692-18
105. Mengjia Z, Wan L, Peng Z, Dejian L, Rui L, Anan J, et al. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: a novel strategy for vaccine design. Emerg Microbes Infect. (2019) 20:20–31. doi: 10.1080/22221751.2019.1701391
106. Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, et al. Porcine deltacoronavirus Nsp5 antagonizes type i interferon signaling by cleaving STAT2. J Virol. (2017) 91:e00003–17. doi: 10.1128/JVI.00003-17
107. Fang P, Fang L, Ren J, Hong Y, Liu X, Zhao Y, et al. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J Virol. (2018) 92:e00712–18. doi: 10.1128/JVI.00712-18
108. Ji L, Li S, Zhu W, Ma J, Yan Y. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-β production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front Immunol. (2019) 10:24. doi: 10.3389/fimmu.2019.01024
109. Liu X, Fang P, Fang L, Hong Y, Zhu X, Wang D, et al. Porcine deltacoronavirus nsp15 antagonizes interferon-β production independently of its endoribonuclease activity. Mol Immunol. (2019) 114:100. doi: 10.1016/j.molimm.2019.07.003
110. Lang G, Jie L, Zhou Q, Xu Z, Li C, Yun Z, et al. A new bat-HKU2–like coronavirus in swine, China, 2017. Emerg Infect Dis. (2017) 23:1607–9. doi: 10.3201/eid2309.170915
111. Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. (2017) 211:15–21. doi: 10.1016/j.vetmic.2017.09.020
112. Lau SKP, Woo PCY, Li KSM, Huang Y, Wang M, Lam CSF, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. (2007) 367:428–39. doi: 10.1016/j.virol.2007.06.009
113. Cruz DJM, Kim CJ, Shin HJ. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. (2008) 132:0–196. doi: 10.1016/j.virusres.2007.10.015
114. Wang L, Hu W, Fan C. Structural and biochemical characterization of SADS-CoV papain-like protease 2. Protein Sci. (2020) 29:1228–41. doi: 10.1002/pro.3857
115. Xu Z, Gong L, Peng P, Liu Y, Xue C, Cao Y. Porcine enteric alphacoronavirus Inhibits IFN-α, IFN-β, OAS, Mx1, and PKR mRNA expression in infected peyer's patches in vivo. Front Vet Sci. (2020) 7:449. doi: 10.3389/fvets.2020.00449
116. Xie S, Chen XX, Qiao S, Li R, Sun Y, Xia S, et al. Identification of the RNA Pseudoknot within the 3' end of the porcine reproductive and respiratory syndrome virus genome as a pathogen-associated molecular pattern to activate antiviral signaling via RIG-I and toll-like receptor 3. J Virol. (2018) 92:18. doi: 10.1128/JVI.00097-18
117. Luo J, Fang L, Dong N, Fang P, Ding Z, Wang D, et al. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-β production. Virology. (2016) 495:10–7. doi: 10.1016/j.virol.2016.04.025
118. Shi P, Su Y, Li R, Liang Z, Dong S, Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. (2019) 265:57–66. doi: 10.1016/j.virusres.2019.03.005
119. Ji L, Wang N, Ma J, Cheng Y, Wang H, Sun J, et al. Porcine deltacoronavirus nucleocapsid protein species-specifically suppressed IRF7-induced type I interferon production via ubiquitin-proteasomal degradation pathway. Vet Microbiol. (2020) 250:108853. doi: 10.1016/j.vetmic.2020.108853
120. Zhou Z, Sun Y, Yan X, Tang X, Li Q, Tan Y, et al. Swine acute diarrhea syndrome coronavirus (SADS-CoV) antagonizes interferon-beta production via blocking IPS-1 and RIG-I. Virus Res. (2020) 278:197843. doi: 10.1016/j.virusres.2019.197843
121. Fletcher NF, Wilson GK, Murray J, Hu K, Lewis A, Reynolds GM, et al. Hepatitis C virus infects the endothelial cells of the blood-brain barrier. Gastroenterology. (2012) 142:634–43.e6. doi: 10.1053/j.gastro.2011.11.028
122. Zhou Y, Wu W, Xie L, Wang D, Ke Q, Hou Z, et al. Cellular RNA helicase DDX1 is involved in transmissible gastroenteritis virus Nsp14-induced interferon-beta production. Front Immunol. (2017) 8:940. doi: 10.3389/fimmu.2017.00940
123. Yen JH, Ganea D. Interferon beta induces mature dendritic cell apoptosis through caspase-11/caspase-3 activation. Blood. (2009) 114:1344–54. doi: 10.1182/blood-2008-12-196592
Keywords: swine enteropathogenic coronaviruses, transmissible gastroenteritis virus, porcine epidemic diarrhea virus, porcine deltacoronavirus, swine acute diarrhea syndrome coronavirus, swine, apoptosis
Citation: Xu Z, Zhang Y and Cao Y (2020) The Roles of Apoptosis in Swine Response to Viral Infection and Pathogenesis of Swine Enteropathogenic Coronaviruses. Front. Vet. Sci. 7:572425. doi: 10.3389/fvets.2020.572425
Received: 14 June 2020; Accepted: 28 October 2020;
Published: 26 November 2020.
Edited by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Kwonil Jung, The Ohio State University, United StatesFrancisco Rivera-Benítez, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), Mexico
Copyright © 2020 Xu, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchang Cao, Y2FveWNoQG1haWwuc3lzdS5lZHUuY24=
 Zhichao Xu
Zhichao Xu Yun Zhang
Yun Zhang Yongchang Cao
Yongchang Cao