- 1Department of Veterinary Parasitology, College of Veterinary and Animal Sciences, Wayanad, India
- 2Department of Veterinary Biochemistry, College of Veterinary and Animal Sciences, Wayanad, India
In the present study, 111 blood samples were collected from apparently healthy cats belonging to four districts of Kerala, southern India, and they were investigated for the presence of hemoparasites and hemoplasmas by light microscopic examination and polymerase chain reaction (PCR). The microscopic examination of the Giemsa-stained blood smears did not reveal any parasites/organisms. However, PCR followed by nucleotide sequencing could detect 10 (9.01%) out of 111 samples infected with Hepatozoon felis, 3 (2.70%) with Cytauxzoon spp., and 10 (9.01%) with Mycoplasma spp. None of the samples revealed amplicons specific for the Babesia spp. and Trypanosoma evansi. The phylogenetic analysis of 18S ribosomal RNA (rRNA) gene sequences of H. felis revealed the existence of two different populations of H. felis circulating in the blood of infected cats. The phylogenetic tree was constructed based on 18S rRNA gene sequences of Cytauxzoon spp. and revealed that these isolates formed a distinct clade and do not cluster with any of the isolates from other countries. Among the 10 samples positive for Mycoplasma spp. infections, 7 were detected positive for Candidatus Mycoplasma haemominutum, two for Mycoplasma haemofelis, and one for Candidatus Mycoplasma turicensis. Phylogenetic analysis of 16S rRNA gene sequences of Mycoplasma spp. showed no distinct geographical grouping of the sequences. The sequences of M. haemofelis, Candidatus M. haemominutum, and Candidatus M. turicensis identified in the study clustered along with their respective isolates from around the world. To the best of our knowledge, this study forms the first report of molecular detection of Cytauxzoon spp. and Candidatus M. turicensis in cats from India.
Introduction
Diseases caused by hemoprotozoan and hemoplasmal organisms are emerging problems in cats in many parts of the world (1, 2). These diseases are mostly fatal vector-borne diseases and are quickly disseminated (1, 3). The increased occurrence of feline vector-borne diseases in Europe was speculated for the reasons like climatic change, increased vector population, drug resistance in vector/pathogen population, and increased international transport of man and animals (3, 4).
Feline hemoprotozoans include hemogregarines like Hepatozoon felis, Hepatozoon canis; piroplasm-causing organisms such as Cytauxzoon felis, Cytauxzoon manul, Babesia felis, Babesia cati, Babesia herpailuri, and Babesia vogeli; and hemoflagellates such as Trypanosoma evansi, Trypanosoma cruzi, Trypanosoma brucei, Trypanosoma congolense, and Trypanosoma rangeli (5, 6). Mycoplasma haemofelis, Candidatus Mycoplasma haemominutum, Candidatus Mycoplasma turicensis, and Candidatus Mycoplasma haematoparvum-like (1) are the hemotropic mycoplasmas. Among them, M. haemofelis and Candidatus M. haemominutum were originally classified under Anaplasmataceae, order Rickettsiales, and were previously known as Haemobartonella felis large and small forms, respectively (7, 8).
Cats having access to the outdoors are more vulnerable to these infections as a result of exposure to a variety of ectoparasites that may transmit these diseases (1, 9, 10). Ticks play an important role in the transmission of babesiosis, hepatozoonosis, and cytauxzoonosis (11–13). Bloodsucking insects such as Tabanus, Stomoxys, Atylotus, and Lyperosia act as vectors for trypanosomosis (14). Feline hemoplasmas are transmitted by Ctenocephalides felis, blood transfusion, and also by fighting, scratching, and biting with other cats. Other routes of infection include transuterine and transmammary transmission (1).
The population of pet cats in India was estimated to be around 2 million in 2018 (https://www.statista.com/statistics/1061172/india-population-of-pet-cats). The majority of them are feral or semidomesticated, which wander in the area around the house from where they may get food and shelter. Rearing domestic cats with a good pedigree has increased recently.
There were only a few documented reports on the molecular detection of these infectious agents in cats from India (15, 16). Hence, the present communication focuses on the generation of baseline information regarding the presence and distribution of hemoparasites and hemoplasmas in cats of southern India.
Materials and Methods
Study Area and Samples
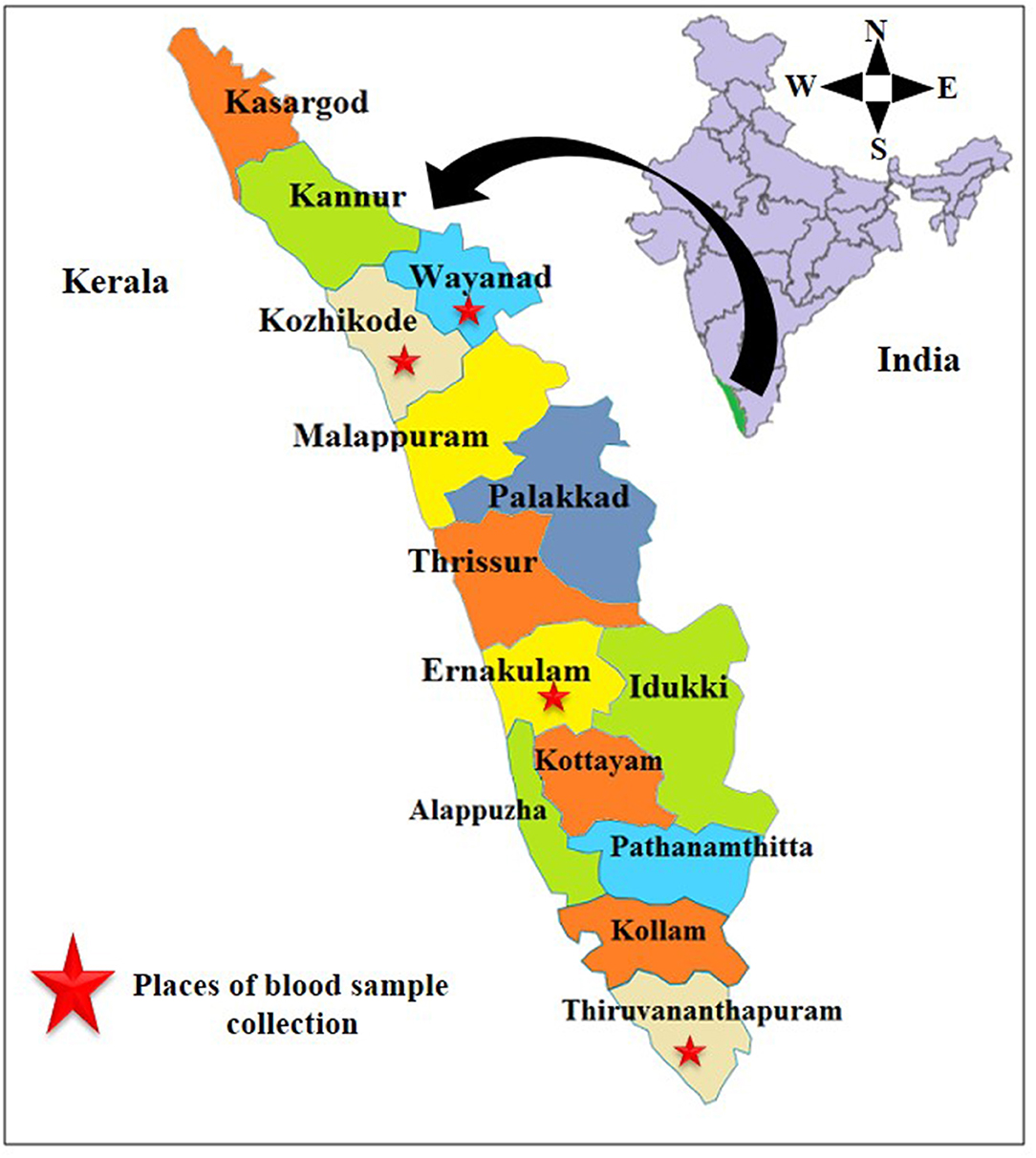
For this study, 111 blood samples were collected from cats belonging to four districts of Kerala, India, viz., Wayanad, Kozhikode, Ernakulam, and Thiruvananthapuram (Figure 1) in the period between March 2018 and June 2019. Whole blood samples were collected in ethylenediaminetetraacetic acid (EDTA) vials from femoral or medial saphenous veins. Thin blood smears were prepared using a drop of this blood.
Staining
Thin peripheral blood smears were fixed in methanol, then stained with diluted (1:10) Giemsa's stain (Merck Life Science, Mumbai) for 45 min. The blood smears were washed with water and air dried. The stained blood smears were examined under the oil immersion objective (100× ) of the light microscope (Leica DM1000 LED, Germany) for the presence of parasites. A minimum of 150 fields were examined thoroughly before declaring a sample as negative.
Genomic DNA Extraction and Quantification
Genomic DNA was isolated from blood samples collected in EDTA vials using DNeasy® blood and tissue kit (Qiagen, Germany), according to the manufacturer's protocol. Extracted DNA was eluted in 100 μL of DNA elution buffer. The DNA concentration was determined using a NanoDrop® 2000C spectrophotometer (Thermo Scientific, USA) and stored at −20°C for further analysis.
Polymerase Chain Reaction
The genomic DNAs isolated from these samples were used for PCR, in an automated thermal cycler with a heated lid (Eppendorf, Germany). All PCRs were carried out in a final reaction volume of 25 μL containing 0.2 mM deoxyribonucleotide triphosphates (dNTPs) (Thermoscientific, Lithuania), 1 U DyNAzyme II DNA polymerase (Thermo Scientific, USA), 10×PCR buffer (containing MgCl2 at a final concentration of 1.5 mM), 20 ng of template DNA, and 10 pmol each of forward and reverse primers. The details of the primers used, the amplification conditions, and the amplicon size are shown in Table 1.
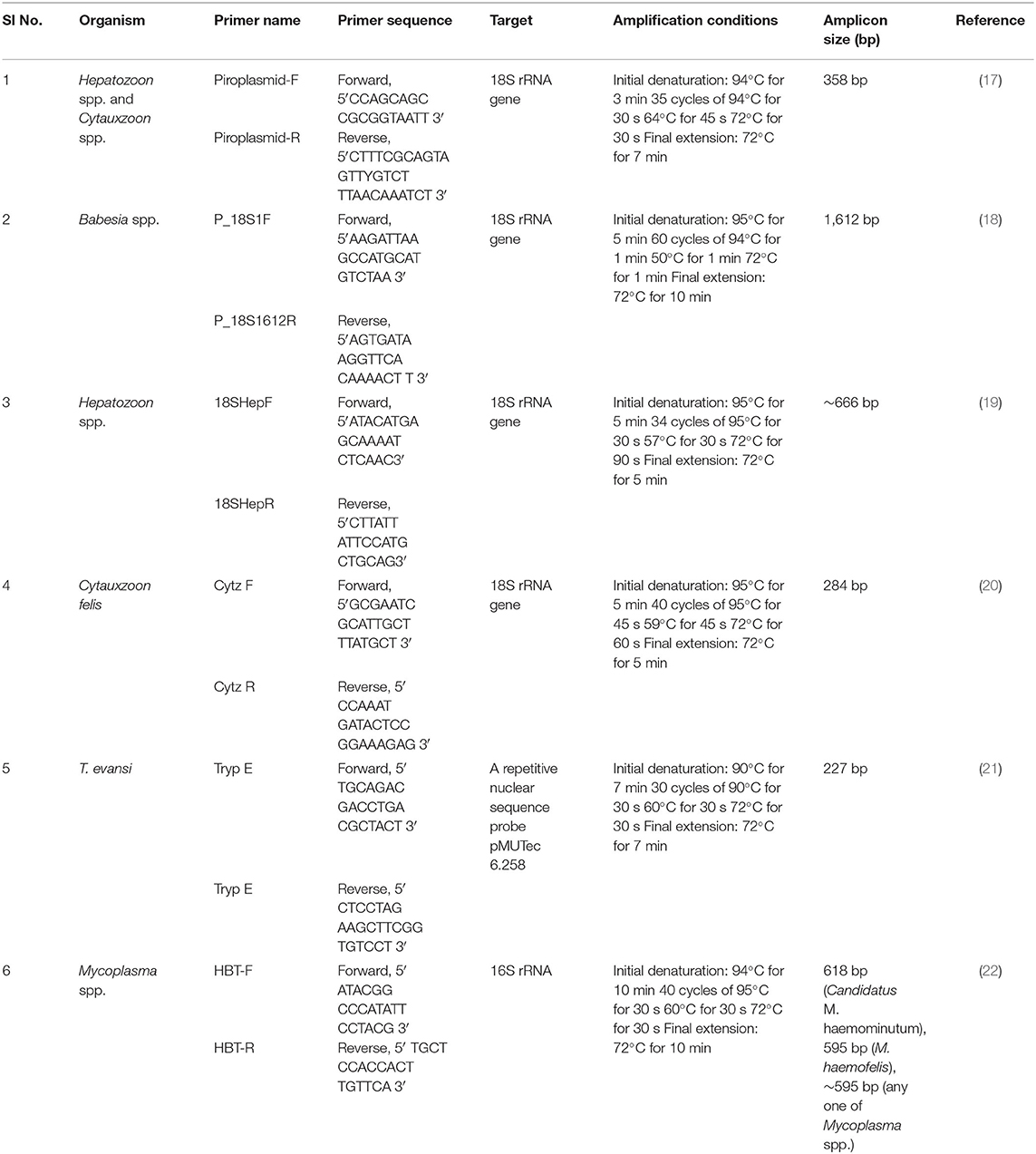
Table 1. The details of primers used for the PCR, the amplification conditions, and the amplicon size.
Positive Controls
The DNA isolated from the blood samples (Qiagen DNeasy® blood and tissue kit, Germany) of infected dogs (diagnosed based on microscopical examination of Giemsa's stained blood smears) presented to the Teaching Veterinary Clinical Complex (TVCC), College of Veterinary and Animal Sciences, Pookode, Wayanad were used as positive controls for genus-specific PCRs for Hepatozoon spp., Babesia spp., and species-specific PCR for T. evansi. Polymerase chain reactions specific for Mycoplasma spp. and C. felis were standardized without any positive controls, as they were rarely reported previously from the state.
Sequencing and Sequence Analysis
Products of polymerase chain reactions (18S rRNA and 16S rRNA) were purified using NucleoSpin® Gel and PCR Clean-Up Kit (Macherey-Nagel, Germany) as per the manufacturer's protocol. They were sent to the AgriGenome Labs Private Ltd., Cochin, Kerala, for automated nucleotide sequencing by Sanger dideoxy method with both the forward and reverse primers. The resulting sequences were examined for the overlapping peaks suggestive of coinfection using Bioedit software (23) before the comparison of the new sequence of each isolate to other published sequences available in the GenBank using NCBI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Unique sequences were deposited in the GenBank database.
Phylogenetic Analysis
For the phylogenetic analysis, the nucleotide sequences were aligned using ClustalW (24) with the previously published sequences in the GenBank. Aligned sequences were trimmed to the same length (with gaps) from which phylogenetic trees were constructed based on the neighbor-joining (NJ) tree method using the program MEGA X.0 (25) with the suitable models [18S rRNA for H. felis: Tamura three-parameter model; 18S rRNA for Cytauxzoon spp.: Tamura three-parameter model + gamma distribution (T92 + G = 0.18); 16S rRNA gene of Mycoplasma spp.: Kimura two-parameter model]. The reliability of the topologies was tested by bootstrapping with 1,000 replications.

Table 2. Sex, breed, and age-wise distribution of hemoparasitic disease and hemotropic mycoplasmosis in domestic cats of Kerala.
Results
Microscopical Examination
The light microscopy examination of Giemsa's stained peripheral blood smears under oil immersion (100 × ) could not detect any hemoparasites and hemoplasmas in the blood smears of 111 cats examined.
PCR and Sequence Analysis
None of the samples revealed amplicons specific for the Babesia spp. and T. evansi. A 358-bp fragment of the 18S rRNA gene of Hepatozoon/Cytauxzoon species was amplified by PCR using the piroplasm-specific primers from the blood of 13 cats out of the 111 samples examined. Sequencing followed by NCBI-BLAST analysis revealed an identity of 99.4–100% to H. felis (JN584475, MK724001) for the 10 sequences and identity of 92.3–92.6% to C. felis (GU903911) for the three sequences. These 13 samples were used for confirmation using primers specific for the amplification of the 18S rRNA of Hepatozoon spp. and C. felis. Ten samples identified as H. felis with piroplasmid primers were further confirmed for monoinfection using primers specific for Hepatozoon spp. Three samples detected positive, as Cytauxzoon spp. did not amplify the desired amplicon when using primers specific for Hepatozoon spp., revealing monoinfection in these samples, too. None of the samples produced amplicons specific for the 18S rRNA of C. felis.
Amplicons (595, 618, and ~595 bp) specific for the 16S rRNA gene of Mycoplasma spp. were amplified by the PCR from the blood samples of 10 cats out of the 111 samples examined. Sequencing revealed that two sequences showed an identity of 98.5–100% to M. haemofelis (MK632346, KU645929), seven sequences with an identity of 98.8–99.8% to Candidatus M. haemominutum (KU645934, KR905451, MK632386, MK632392) and one sequence with an identity of 99.8% to Candidatus M. turicensis (KR905459).
Mixed infection due to the presence of both H. felis and Candidatus M. haemominutum was identified in 4 sample (3.6%) out of 111 DNA samples by PCR. These cats were from Wayanad (one), Ernakulam (one), and Thiruvananthapuram (two) districts of Kerala. Mixed infection due to different protozoans was not detected in any samples tested in the present study. The occurrence of infection due to hemoparasites and hemotropic mycoplasmas (Table 2) was slightly higher in male compared to female cats. The non-descript cats harbored more infectious organisms than Persian cats. Moreover, cats belonging to the age group of 1–2 years showed a higher prevalence.
Phylogeny
Hepatozoon felis
The phylogenetic tree for Hepatozoon spp. (Figure 2) based on 18S rRNA sequences revealed five clades (clades 1, 2, 3, 4, and 5). The first clade consisted of H. felis sequences from cats of Spain and Israel. Among the 10 isolates of Hepatozoon spp., nine (Wayanad isolate 2; Kozhikode isolates 1, 2, and 3; Ernakulam isolate 1; Thiruvananthapuram isolate 1, 2, 3, and 4 with accession numbers MN227268, MN227269, MN227270, MN227271, MN227272, MN227273, MN227274, MN227275, MN227276) clustered with H. felis isolates reported from cats of Hyderabad, India in clade 2. The sequences of Hepatozoon ursi from the black bear of Japan and the sloth bear of India occupied the third clade. All the H. canis sequences from dogs of India, Brazil, Taiwan, and Spain and wild dogs from India were clustered in clade 4. Wayanad isolate 1 of H. felis identified in the present study (MN227267) formed a separate clade (clade 5) along with other H. felis isolates of domestic cats from Japan and Austria as well as isolates from an Indian lion, a wild cat from Bosnia and Herzegovina, the flat-headed cat from Thailand, and Korean leopard cat.
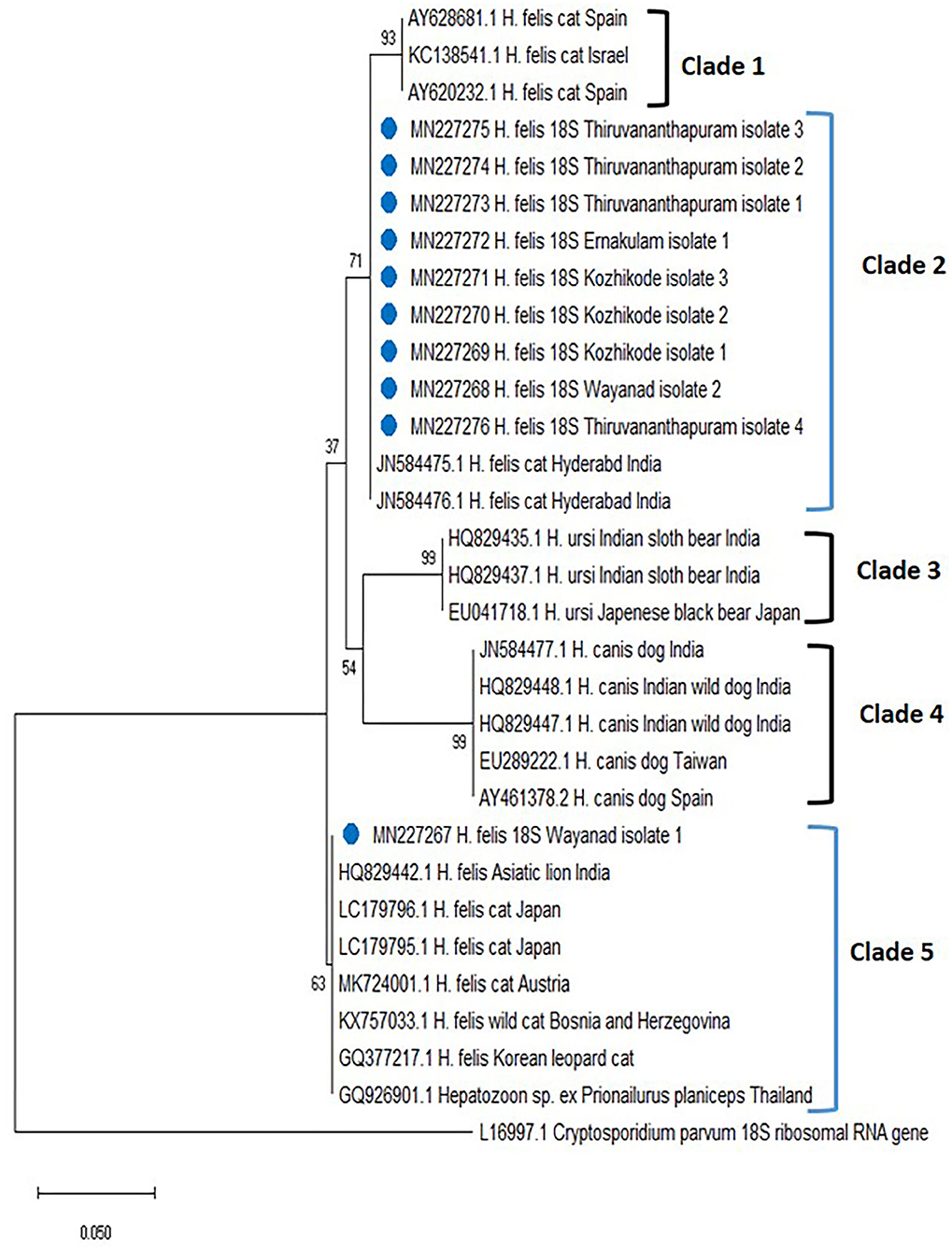
Figure 2. Phylogenetic tree constructed using partial 18S ribosomal RNA (rRNA) gene sequence of Hepatozoon spp. amplified using piroplasmid primers. The evolutionary history was inferred by using the neighbor-joining (NJ) tree based on the Tamura three-parameter model (T92). The analysis involved 31 nucleotide sequences comprising of the field isolates from the current study (indicated by the blue circle) and previously published sequences in the GenBank. Evolutionary analyses were conducted in Mega X.
Cytauxzoon spp.
The phylogenetic tree was constructed based on 18S rRNA gene sequences (MN252095, MN252096, MN252097) of Cytauxzoon spp. (Figure 3) and revealed five clades. Clade 1 comprised of C. felis isolates from domestic cats of the USA and South Africa. Sequences of C. felis from Ocelot (Leopardus pardalis) and Northern Tiger Cat (Leopardus tigrinus) of Brazil, clustered in clade 2. The three isolates of Cytauxzoon species from Kerala (Kozhikode isolate 1, 2, and 3) formed a separate clade 3. Cytauxzoon manul from Pallas cats of Mongolia was grouped in clade 4. Cytauxzoon species of undetermined status (Cytauxzoon sp. European strain) from Spain, Italy, and France were clustered into clade 5.
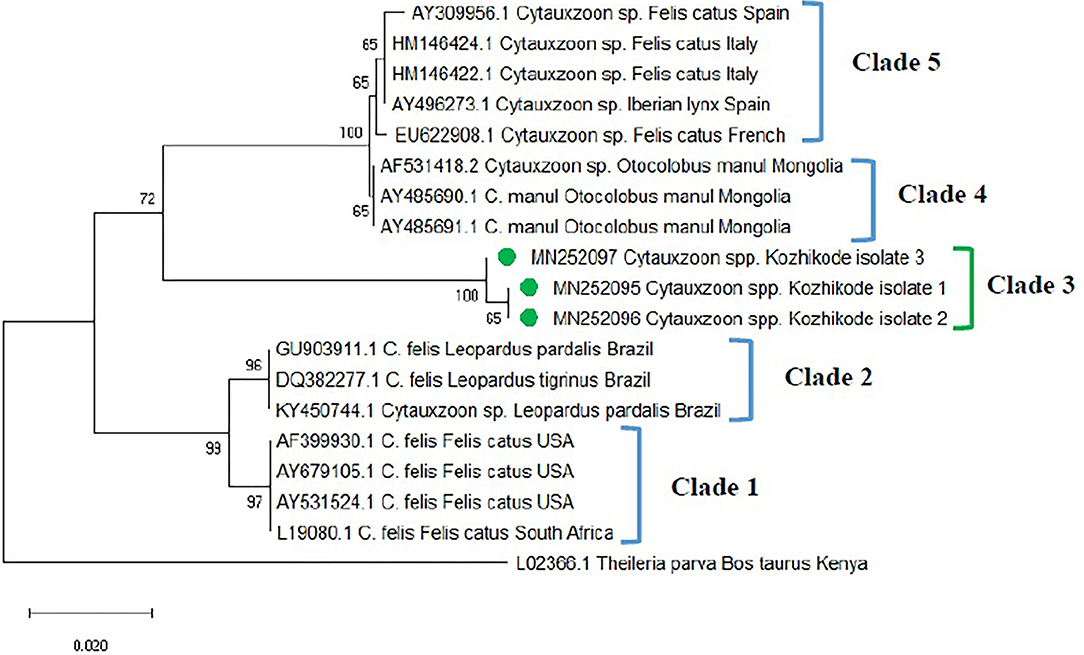
Figure 3. Phylogenetic tree constructed using partial 18S ribosomal RNA (rRNA) gene sequence of Cytauxzoon spp. amplified using piroplasmid primers. The evolutionary history was inferred by using the neighbor-joining (NJ) tree based on the Tamura three-parameter model + gamma distribution (T92 + G). A discrete Gamma distribution parameter (+G = 0.18) was used to model evolutionary rate differences among the sites. The analysis involved 19 nucleotide sequences comprising of the field isolates from the current study (indicated by the green circle) and previously published sequences in the GenBank. Evolutionary analyses were conducted in Mega X.
Mycoplasma spp.
The phylogenetic tree was constructed based on the 16S rRNA for Mycoplasma spp. [revealed three clades, in which the Mycoplasma spp. isolates of Kerala fit into three different clades (clades 1, 2, and 3)] (Figure 4). One sequence of Candidatus M. turicensis isolate of Kerala (Thiruvananthapuram isolate 1 with accession number MN240801) clustered in clade 1 along with Candidatus M. turicensis isolates from other countries [Australia (DQ464423; DQ464425; DQ464417), South Africa (DQ464424; DQ464419; DQ464422), UK (DQ464420; DQ464421), and Switzerland (DQ157150). Two M. haemofelis isolates (MN240855, MN240856) from Kerala (Wayanad isolates 1 and 2) were grouped in clade 2 comprising M. haemofelis isolates from other countries (South Africa (AF548631), USA (AY069948; AF178677), UK (AY150984), and Australia (AY150977). Seven isolates of Candidatus M. haemominutum from Kerala, viz., Wayanad isolate 1 (MN240862), Wayanad isolate 2 (MN240863), Kozhikode isolate 1 (MN240864), Ernakulam isolate 1 (MN240865) and Ernakulam isolate 2 (MN240866), Thiruvananthapuram isolate 1 (MN240867), and Thiruvananthapuram isolate 2 (MN240868) were clustered in clade 3 along with Candidatus M. haemominutum isolates from other countries (Israel, UK, USA, and South Africa).
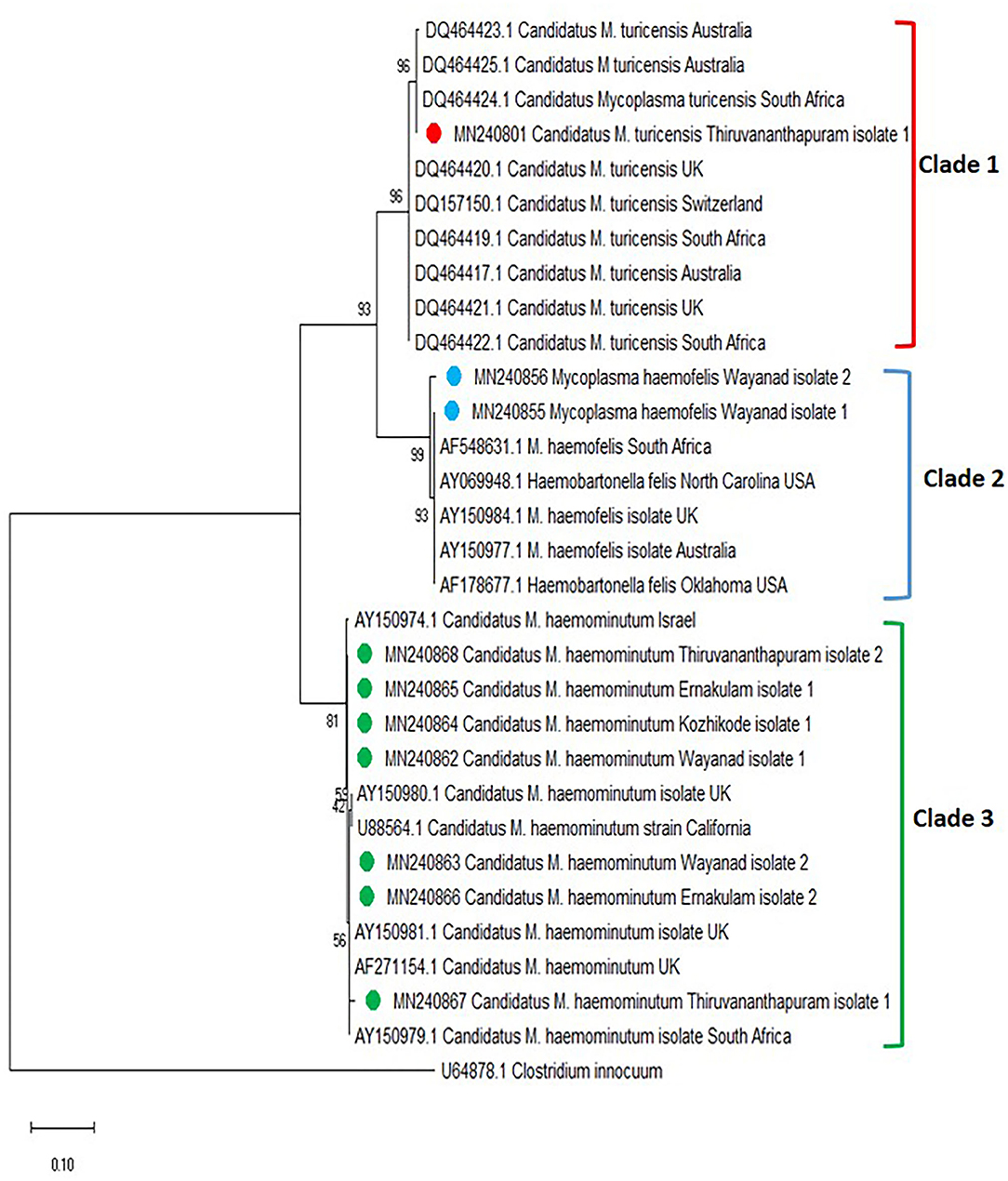
Figure 4. Phylogenetic tree constructed using partial 16S ribosomal RNA (rRNA) gene sequence of Mycoplasma spp. amplified using HBT primers. The evolutionary history was inferred by using the neighbor-joining (NJ) tree based on the Kimura two-parameter model. The analysis involved 31 nucleotide sequences comprising of the field isolates from the current study (indicated by the red, blue, and green circle) and previously published sequences in the GenBank. Evolutionary analyses were conducted in Mega X.
Discussion
No hemoparasites and hemotropic mycoplasmas could be detected based on microscopy of blood smears stained with Giemsa's stain. H. felis gamonts were difficult to be observed under a microscope, as they were less conspicuous, smaller, or low in number (11, 26, 27). The intraerythrocytic piroplasms of Cytauxzoon spp. were not detected in healthy cats that were PCR positive (6, 28). It is also believed that the examination of stained smears was not a sensitive diagnostic tool and cannot identify the three different hemoplasma species (29, 30).
In the present study, PCR and subsequent nucleotide sequencing detected 9% prevalence for H. felis in the blood samples collected from cats of Kerala. Previously, the presence of H. canis (32.3%) in stray cats of Bangkok (31) and H. felis (34.8%) and H. canis (1.3%) among domestic cats of Israel (17) were reported. Based on 18S rRNA sequences, all the H. felis field isolates detected in the present study were grouped into two different clades, clades 2 and 5. The present study also disproved the concept of genetic relatedness of Indian isolates of Hyderabad with isolates from Spain and Israel (15). Thus, there might be two different populations of H. felis in Kerala that could infect domestic cats.
The detection of Cytauxzoon spp. in three blood samples of cats in the present study forms the first report from India. C. felis was endemic solely to North America for many years, where bobcats (Lynx rufus) are believed to serve as the main hosts even though reports are available from the USA, Brazil, Spain, France, Italy, and Iraq (32, 33) for its presence in domestic cats. The “parasite” has also been described in felids originating from several Asian countries, including India. However, there are no comprehensive molecular data available, which could confirm the specific identity of these Asian isolates. The nucleotide sequences generated in the current study showed only 92% identity to the closest match in the GenBank database. Furthermore, the Indian sequences formed a separate clade (herein designated as clade 3) that is very distant from that of pathogenic C. felis isolates from the USA, Netherlands, and Brazil (6). Further, the primer sets targeting 18 S rRNA gene specific for C. felis did not reveal any amplification. The Cytauxzoon spp. detected in Kerala were from apparently healthy cats. However, Cytauxzoon felis infections (34) are highly fatal except for a few reports from asymptomatic cats (28, 33, 34). In other words, the 18S rDNA sequences confirmed in the cats tested in this study most likely belong to another Cytauxzoon species, not C. felis.
Hemoplasmosis was previously reported from different parts of the world (7, 29, 30, 35–40). In the present study, Mycoplasma spp. was detected in 10 blood samples of cats collected from all four districts of Kerala. Seven cats (6.3%) were infected with Candidatus M. haemominutum, two (1.2%) with M. haemofelis, and one (0.9%) with Candidatus M. turicensis. A previous study conducted in Thrissur, Kerala (16) detected a prevalence of 23% of Candidatus M. haemominutum and 1% of M. haemofelis by PCR. Candidatus M. haematoparvum-like organisms (0.7%) reported previously from the USA (41) were not detected in the present study. In addition, the present study reports for the first time the presence of “Candidatus M. turicensis” among the cat population in India.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Ethics Committee, College of Veterinary and Animal Sciences, Pookode. Written informed consent for participation was not obtained from the owners because the samples were collected from cats brought to the veterinary clinics are managed by registered veterinary practitioners. The blood samples were collected by the veterinarians after getting the oral consent from the pet owners for the detection of pathogenic organisms in their pets.
Author Contributions
LM, AN, CB, BA, PK, RP, and MN collected the samples, conducted the experiments, participated in the data acquisition, and drafted the manuscript. KA conceived the study and supervised the protocols. AV, CD, LJ, and RR helped in the collection of samples, data acquisition, supervision of the experiments, and review of the manuscript. All authors read and approved the manuscript.
Funding
The research work was supported financially by Kerala Veterinary and Animal Sciences University, Pookode.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the veterinary doctors and pet owners for their cooperation in the collection of the samples.
References
1. Tasker S. Haemotropic mycoplasmas: what's their real significance in cats? J Feline Med Surg. (2010) 12:369–81. doi: 10.1016/j.jfms.2010.03.011
2. Díaz-Regañón D, Villaescusa A, Ayllón T, Rodríguez-Franco F, Baneth G, Calleja-Bueno L, et al. Molecular detection of Hepatozoon spp. and Cytauxzoon sp. in domestic and stray cats from Madrid, Spain. Parasit Vectors. (2017) 10:112. doi: 10.1186/s13071-017-2056-1
3. Otranto D, Dantas-Torres F, Breitschwerdt EB. Managing canine vector-borne diseases of zoonotic concern: part one. Trends Parasitol. (2009) 25:157–63. doi: 10.1016/j.pt.2009.01.003
4. Vilhena H, Martinez-Díaz VL, Cardoso L, Vieira L, Altet L, Francino O, et al. Feline vector-borne pathogens in the north and centre of Portugal. Parasit Vectors. (2013) 6:99. doi: 10.1186/1756-3305-6-99
5. Bowman DD, Hendrix CM, Lindsay DS, Barr SC. Feline Clinical Parasitology. 1st ed. Ames, IA: Iowa State University Press (2002). 469 p.
6. Alho AM, Silva J, Fonseca MJ, Santos F, Nunes C, de Carvalho LM, et al. First report of Cytauxzoon sp. infection in a domestic cat from Portugal. Parasit Vectors. (2016) 9:220. doi: 10.1186/s13071-016-1506-5
7. Foley JE, Pedersen NC. “Candidatus Mycoplasma haemominutum”, a low-virulence epierythrocytic parasite of cats. Int J Syst Evolutionary Microbiol. (2001) 51:815–7. doi: 10.1099/00207713-51-3-815
8. Neimark H, Johansson KE, Rikihisa Y, Tully JG. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of “Candidatus Mycoplasma haemofelis”, “Candidatus Mycoplasma haemomuris”, “Candidatus Mycoplasma haemosuis” and “Candidatus Mycoplasma wenyonii”. Int J Sys Evol Microbiol. (2001) 51:891–9. doi: 10.1099/00207713-51-3-891
9. Beugnet F, Marié JL. Emerging arthropod-borne diseases of companion animals in Europe. Vet Parasitol. (2009) 163:298–305. doi: 10.1016/j.vetpar.2009.03.028
10. Otranto D, Dantas-Torres F. Canine and feline vector-borne diseases in Italy: current situation and perspectives. Parasit Vectors. (2010) 3:2. doi: 10.1186/1756-3305-3-2
11. Baneth G. Perspectives on canine and feline hepatozoonosis. Vet Parasitol. (2011) 181:3–11. doi: 10.1016/j.vetpar.2011.04.015
12. Hartmann K, Addie D, Belák S, Boucraut-Baralon C, Egberink H, Frymus T, et al. Babesiosis in cats: ABCD guidelines on prevention and management. J Feline Med Surg. (2013) 15:643–646. doi: 10.1177/1098612X13489230
13. Sherrill MK, Cohn LA. Cytauxzoonosis: diagnosis and treatment of an emerging disease. J Feline Med Surg. (2015) 17:940–8. doi: 10.1177/1098612X15610681
14. Brun R, Hecker H, Lun ZR. Trypanosoma evansi and T. equiperdum: distribution, biology, treatment, and phylogenetic relationship (a review). Vet Parasitol. (1998) 79:95–107. doi: 10.1016/S0304-4017(98)00146-0
15. Pawar RM, Poornachandar A, Srinivas P, Rao KR, Lakshmikantan U, Shivaji S. Molecular characterization of Hepatozoon spp. infection in endangered Indian wild felids and canids. Vet Parasitol. (2012) 186:475–9. doi: 10.1016/j.vetpar.2011.11.036
16. Ameldev P, Tresamol PV. Molecular detection and therapeutic management of feline mycoplasmosis. IOSR J Agric Vet Sci. (IOSR-JAVS) (2017) 10:83–6. doi: 10.9790/2380-1002018386
17. Baneth G, Sheiner A, Eyal O, Hahn S, Beaufils JP, Anug Y, et al. Redescription of Hepatozoon felis (Apicomplexa: Hepatozoidae) based on phylogenetic analysis, tissue and blood form morphology, and possible transplacental transmission. Parasit Vectors. (2013) 6:102. doi: 10.1186/1756-3305-6-102
18. Wong SS, Poon RW, Hui JJ, Yuen KY. Detection of Babesia hongkongensis sp. nov. in a free-roaming Felis catus cat in Hong Kong. J Clin Microbiol. (2012) 50:2799–2803. doi: 10.1128/jcm.01300-12
19. Inokuma H, Okuda M, Ohno K, Shimoda K, Onishi T. Analysis of 18S rRNA gene sequence of a Hepatozoon detected in two Japanese dogs. Vet Parasitol. (2002) 106:265–271. doi: 10.1016/s0304-4017(02)00065-1
20. Birkenheuer AJ, Le JA, Valenzisi AM, Tucker MD, Levy MG, Breitschwerdt EB. Cytauxzoon felis infection in cats in the mid-Atlantic states: 34 cases (1998–2004). J Am Vet Med Assoc. (2006) 228:568–571. doi: 10.2460/javma.228.4.568
21. Chokesajjawattee N. DNA sequence analysis of a specific Trypanosoma evansi. DNA probe (M.Sc. thesis). Mahidol University, Bangkok (1993).
22. Criado-Fornelio A, Martinez-Marcos A, Buling-Sarana A, Barba-Carretero JC. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum, and piroplasmids in cats from southern Europe: a molecular study. Vet Microbiol. (2003) 93:307–317. doi: 10.1016/s0378-1135(03)00044-0
23. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. (1999) 41:95–8.
24. Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
25. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
26. Lloret A, Addie DD, Boucraut-Baralon C, Egberink H, Frymus T, Gruffydd-Jones T, et al. Hepatozoonosis in cats: ABCD guidelines on prevention and management. J Feline Med Surg. (2015) 17:642–4. doi: 10.1177/1098612x15589879
27. Salakij C, Sirinarumitr T, Tongthainun D. Molecular characterization of Hepatozoon species in a leopard cat (Prionailurus bengalensis) from Thailand. Vet Clin Path. (2010) 39:199–202. doi: 10.1111/j.1939-165X.2010.00216.x
28. Brown HM, Latimer KS, Erikson LE, Cashwell ME, Britt JO, Peterson DS. Detection of persistent Cytauxzoon felis infection by polymerase chain reaction in three asymptomatic domestic cats. J Vet Diagn Invest. (2008) 20:485–8. doi: 10.1177/104063870802000411
29. Fujihara M, Watanabe M, Yamada T, Harasawa R. Occurrence of “Candidatus Mycoplasma turicensis” infection in domestic cats in Japan. J Vet Med Sci. (2007) 69:1061–3. doi: 10.1292/jvms.69.1061
30. Bauer N, Balzer HJ, Thüre S, Moritz A. Prevalence of feline haemotropic mycoplasmas in convenience samples of cats in Germany. J Feline Med Surg. (2008) 10:252–8. doi: 10.1016/j.jfms.2007.12.004
31. Jittapalapong S, Rungphisutthipongse O, Maruyama S, Schaefer JJ, Stich RW. Detection of Hepatozoon canis in stray dogs and cats in Bangkok, Thailand. Ann N Y Acad Sci. (2006) 1081:479–88. doi: 10.1196/annals.1373.071
32. Suliman EG. Detection of the infection with Babesia spp. Cytauxzoon felis and Haemobaronella felis in stray cats in Mosul. Iraqi J Vet Sci. (2009) 23(Suppl. 1):49–55. Available online at: https://www.semanticscholar.org/paper/Detection-the-infection-with-Babesia-spp.-felis-and-Suliman/32af3c2714eeeab704fdd09ab4213aec0926abda
33. Wang JL, Li TT, Liu GH, Zhu XQ, Yao C, Two tales of Cytauxzoon felis infections in domestic cats. Clin Microbiol Rev. (2017) 30:861–85. doi: 10.1128/CMR.00010-17
34. Nentwig A, Meli ML, Schrack J, Reichler IM, Riond B, Gloor C, et al. First report of Cytauxzoon sp. infection in domestic cats in Switzerland: natural and transfusion-transmitted infections. Parasit Vectors. (2018) 11:292. doi: 10.1186/s13071-018-2728-5
35. Grindem CB, Corbett WT, Tomkins MT. Risk factors for Haemobartonella felis infection in cats. J Am Vet Med Ass. (1990) 196:96–9.
36. Tasker S, Helps CR, Day MJ, Harbour DA, Shaw SE, Harrus S, et al. Phylogenetic analysis of hemoplasma species: an international study. J Clin Microbiol. (2003) 41:3877–80. doi: 10.1128/JCM.41.8.3877-3880.2003
37. Inokuma H, Taroura S, Okuda M, Hisasue M, Itamoto K, Une S, et al. Molecular survey of Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum” infection in cats in Yamaguchi and surrounding areas. J Vet Med Sci. (2004) 66:1017–20. doi: 10.1292/jvms.66.1017
38. Ghazisaeedi F, Atyabi N, Salehi TZ, Gentilini F, Tamai IA, Akbarein H, et al. A molecular study of hemotropic mycoplasmas (hemoplasmas) in cats in Iran. Vet Clin Path. (2014) 43:381–6. doi: 10.1111/vcp.12166
39. Alho AM, Lima C, Latrofa MS, Colella V, Ravagnan S, Capelli G, et al. Molecular detection of vector-borne pathogens in dogs and cats from Qatar. Parasit Vectors. (2017) 10:1–5. doi: 10.1186/s13071-017-2237-y
40. Cetinkaya H, Haktanir D, Arun S, Vurusaner C. Molecular detection and prevalence of feline hemotropic mycoplasmas in Istanbul, Turkey. Acta Parasitol. (2016) 61:165–71. doi: 10.1515/ap-2016-0022
Keywords: cats, Cytauxzoon spp., Hepatozoon felis, Candidatus M. haemominutum, Mycoplasma haemofelis, Candidatus M. turicensis, phylogeny
Citation: Malangmei L, Ajith Kumar KG, Nandini A, Bora CAF, Varghese A, Amrutha BM, Kurbet PS, Pradeep RK, Nimisha M, Deepa CK, John L and Ravindran R (2021) Molecular Characterization of Hemoparasites and Hemoplasmas Infecting Domestic Cats of Southern India. Front. Vet. Sci. 7:597598. doi: 10.3389/fvets.2020.597598
Received: 21 August 2020; Accepted: 11 December 2020;
Published: 25 January 2021.
Edited by:
Donato Traversa, University of Teramo, ItalyReviewed by:
Adnan Hodžic, University of Veterinary Medicine Vienna, AustriaMian A. Hafeez, University of Guelph, Canada
Copyright © 2021 Malangmei, Ajith Kumar, Nandini, Bora, Varghese, Amrutha, Kurbet, Pradeep, Nimisha, Deepa, John and Ravindran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reghu Ravindran, ZHJyZWdodXJhdmlAeWFob28uY29t
 Lanchalung Malangmei
Lanchalung Malangmei Karapparambu Gopalan Ajith Kumar
Karapparambu Gopalan Ajith Kumar Ashwathappa Nandini
Ashwathappa Nandini Christophe Angeline Felicia Bora
Christophe Angeline Felicia Bora Anju Varghese
Anju Varghese Birur Mallappa Amrutha
Birur Mallappa Amrutha Prashant Somalingappa Kurbet
Prashant Somalingappa Kurbet Rangapura Kariyappa Pradeep
Rangapura Kariyappa Pradeep Murikoli Nimisha
Murikoli Nimisha Chundiyil Kalarickal Deepa
Chundiyil Kalarickal Deepa Lijo John
Lijo John Reghu Ravindran
Reghu Ravindran