- 1Museum of Vertebrate Zoology, University of California, Berkeley, Berkeley, CA, United States
- 2Department of Biology, San Francisco State University, San Francisco, CA, United States
- 3Berkeley Natural History Museums, University of California, Berkeley, Berkeley, CA, United States
- 4US Department of Agriculture, Forest Service, Pacific Northwest Research Station, Corvallis, OR, United States
Emerging infectious diseases have been especially devastating to amphibians, the most endangered class of vertebrates. For amphibians, the greatest disease threat is chytridiomycosis, caused by one of two chytridiomycete fungal pathogens Batrachochytrium dendrobatidis (Bd) and Batrachochytrium salamandrivorans (Bsal). Research over the last two decades has shown that susceptibility to this disease varies greatly with respect to a suite of host and pathogen factors such as phylogeny, geography (including abiotic factors), host community composition, and historical exposure to pathogens; yet, despite a growing body of research, a comprehensive understanding of global chytridiomycosis incidence remains elusive. In a large collaborative effort, Bd-Maps was launched in 2007 to increase multidisciplinary investigations and understanding using compiled global Bd occurrence data (Bsal was not discovered until 2013). As its database functions aged and became unsustainable, we sought to address critical needs utilizing new technologies to meet the challenges of aggregating data to facilitate research on both Bd and Bsal. Here, we introduce an advanced central online repository to archive, aggregate, and share Bd and Bsal data collected from around the world. The Amphibian Disease Portal (https://amphibiandisease.org) addresses several critical community needs while also helping to build basic biological knowledge of chytridiomycosis. This portal could be useful for other amphibian diseases and could also be replicated for uses with other wildlife diseases. We show how the Amphibian Disease Portal provides: (1) a new repository for the legacy Bd-Maps data; (2) a repository for sample-level data to archive datasets and host published data with permanent DOIs; (3) a flexible framework to adapt to advances in field, laboratory, and informatics technologies; and (4) a global aggregation of Bd and Bsal infection data to enable and accelerate research and conservation. The new framework for this project is built using biodiversity informatics best practices and metadata standards to ensure scientific reproducibility and linkages across other biological and biodiversity repositories.
Introduction
The amphibian vertebrate lineage evolved over 360 million years ago, and has survived multiple mass extinction events, yet today amphibians are the most endangered class of vertebrates and may be harbingers of a new sixth mass-extinction event (1). Emerging infectious diseases have been especially devastating to amphibians (1–3). Chytridiomycosis is a potentially lethal amphibian skin disease caused by one of two chytridiomycete fungal pathogens, Batrachochytrium dendrobatidis (Bd) and Batrachochytrium salamandrivorans (Bsal). Bd chytridiomycosis, was first discovered over two decades ago (4–6), and later in 2013, Bsal chytridiomycosis was detected (7). A growing body of research has shown that amphibian susceptibility to these diseases varies phylogenetically, geographically, and is influenced by synergisms with abiotic and biotic factors (8, 9). However, a comprehensive understanding of the lethal, sublethal, and benign effects of these fungal pathogens and their long-term effects on vertebrates in class Amphibia is still incomplete. Chytridiomycosis has raised alarms and the World Organization of Animal Health (OIE) has listed Bd and Bsal as reportable pathogens (10) on the global stage. Bd, which includes multiple genetic lineages (11, 12), has spread across continents likely by human actions, and in some regions it has invaded naive host populations causing epizootics (epidemics in wildlife) that affect hundreds of species (13–15). The discovery of chytridiomycosis and the documentation of its impacts on amphibians have fundamentally altered the way scientists view emerging infectious diseases, their contributions to global biodiversity losses, and biodiversity conservation approaches to emerging disease threats (3, 8, 16).
We are facing a rapidly changing scientific knowledge landscape for amphibian emerging infectious diseases in the 21st Century (7), which has increased the challenges for reporting and tracking advances (6). Indeed, the relatively recent discovery of the Bsal pathogen (7) has shown that we must be nimble in our approach to data management and analysis and adapt to new diseases and new technologies. Can we predict the data management needs of the next emerging disease that will infect amphibians or other wildlife species? What we do know is that sharing data and responding rapidly is essential to disease mitigation. As a scientific community we can heed lessons learned from our collective experience in Bd research for the last two decades. For example, we know that Bd can devastate entire amphibian populations (13) and entire amphibian communities quickly (14, 17). In outbreaks in Panama, 50% of local amphibian species were extirpated (17), and in Peru, 40% of species were extirpated (14). The advent of the discovery of Bsal motivated the formation of the North American Bsal Task Force (18), which included representatives from governmental, academic, and advocacy organizations in a broad coalition across the USA, Canada, and Mexico. Two of us (MK, DO) lead the Bsal Task Force Data Management Working Group. Our Bsal Task Force discussions clearly showed consensus for coordinated efforts in planning for when, not if, Bsal would eventually be detected in North America (19, 20). All parties agreed it is essential to track, archive, and quickly share sampling efforts for Bsal.
The first effort to compile Batrachochytrium occurrences for online access and mapping, beginning in 2007, resulted from a collaboration between the USDA Forest Service and Imperial College London known as Bd-Maps (21). The chytridiomycosis research community soon turned to Bd-Maps as the main source for compiled global data on Bd (21). This has been a labor-intensive aggregation of global Bd sampling efforts including >33,000 data records at >14,000 unique site coordinates to date (22). Based on both the Bd-Maps data and unique records recently summarized by Castro Monzon et al. (23), Bd has been detected in 1,375 of 2,525 (55%) species (composed of 88% of frog families, 100% of salamander families, and 70% of caecilian families) and 93 of 134 (69%) countries sampled to date [see (22) for details]. Over time, the labor-intensive methodology and unfunded infrastructure of the Bd-Maps became unsustainable. A new urgency arose when Bsal was discovered (7), and we embraced the challenge of how best to share data as quickly as it was produced and verified. Aggregating disease data can immediately address fundamental questions about where sampling effort has been applied, where disease occurrences are documented, and which species are affected, in addition to identifying active researchers to facilitate collaboration. In particular, it has become increasingly important to not only document known occurrences of Bd and Bsal, but also known instances of negative data (i.e., samples tested for Bd and/or Bsal that did not find evidence of infection), which by themselves may not be suitable for publication in many peer-reviewed journals. These data are critically important, however, in predictive distribution modeling [e.g., (22, 24)], and in examining host species traits such as phylogeny, habitat use, or behavior that may explain host infection or disease susceptibility [e.g., (25, 26)], and help understand the synergisms of co-occurring chytrid fungi [e.g., (27)]. If samples have linked genetic and genomic data, pathogenic fungal migration and evolution can be examined, revealing new insights on virulence, novel introductions, and origins of pathogens using phylodynamics [e.g., (11, 28–30)]. In addition, negative data can help describe the timing of pathogen invasion, which can also help in our understanding of present-day dynamics (31).
We introduce here an open-access repository and archive for Batrachochytrium data called the Amphibian Disease Portal (https://amphibiandisease.org) that addresses two urgent needs: (1) to create a sustainable, modernized repository to aggregate and rapidly share data on the fungal pathogens of amphibians Bd and Bsal; and (2) to upgrade and migrate the Bd-Maps datasets to a new repository that can continue to grow.
Methods and Materials
In the creation of this online resource, we considered a broad range of users, facilitated by discussions with the Bsal Task Force, the AmphibiaWeb steering committee, and members of natural history museums and other institutions that provide biodiversity informatics data to users online. When planning how to store, structure, and share the data we considered the needs of conservation biologists, disease ecologists, evolutionary biologists, resource managers, and many others. We aimed to follow several principles to achieve these goals. The Amphibian Disease Portal was developed to: (1) prioritize structured data, data quality, and online accessibility, maximizing its usefulness and accessibility with modern web technologies and standards; (2) offer tangible benefits to users who are contributing their data (for the purposes of this paper, we refer to researchers who submit data as “contributors,” and those who access data for summaries or downloading for analysis as “portal users”); (3) be sustainable and cost-efficient for both maintenance and users (contributors and portal users will not be charged); and (4) support reproducible and replicable (i.e., repeatable methods with the same or comparable data, respectively, to produce the same results) data-driven science.
We initiated development in 2015, and the final architecture for the portal is described here. The overall architecture can be divided as a database “backend” and a user interface “frontend.” Many of our core goals align with an established metadata repository, the Genomic Observatories Metadatabase or GEOME (32), especially with respect to biological data management and structured, internet-accessible data. The GEOME repository offers a suite of user-friendly tools to manage and aggregate standardized biological sample data with its derivative genetic data, including associated geospatial, diagnostic, and publication context data [see [32] for core architecture]. We adopted the GEOME platform as the database and validation service for the ADP's “backend.” To meet the needs of research access and data access, we developed a dedicated “frontend,” a user-friendly website where all users can interact, visualize, and discover data. In addition, we created a middleware application programming interface (API) that communicates with the GEOME “backend.” Together, they comprise of the frontend, API, and backend components forming the Amphibian Disease Portal (Figure 1); all code is open-source and available on Github in two code repositories licensed as GNU General Public License (33), one for its frontend website (https://github.com/BNHM/AmphibiaWebDiseasePortal) and one for the API (https://github.com/BNHM/AmphibiaWebDiseasePortalAPI). We describe how we address and meet the principles and goals of the project with respect to its infrastructure.
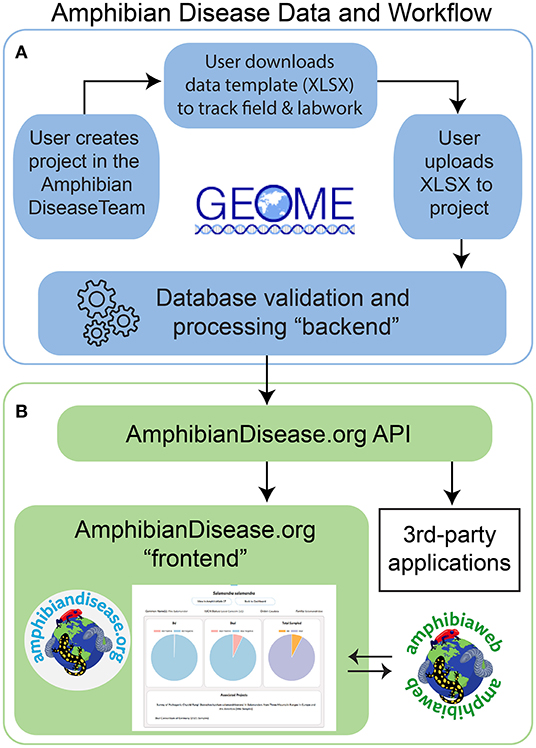
Figure 1. Amphibian Disease Portal data and work flow. (A) Users begin the process of contributing data by creating a project in Geome under the Amphibian Disease Team; user can download a data template for field and laboratory work. Once data gathering is complete, user uploads to the project for processing at Geome, and data are now available via the AmphibianDisease API. (B) The “frontend” includes the website AmphibianDisease.org where aggregated data can be graphed, mapped, queried, and downloaded. The AmphibianDisease API allows data to be accessible by any third-party application that can integrate web services like R Statistics. AmphibiaWeb site reciprocally links to Amphibian Disease species charts when data are available for a given species.
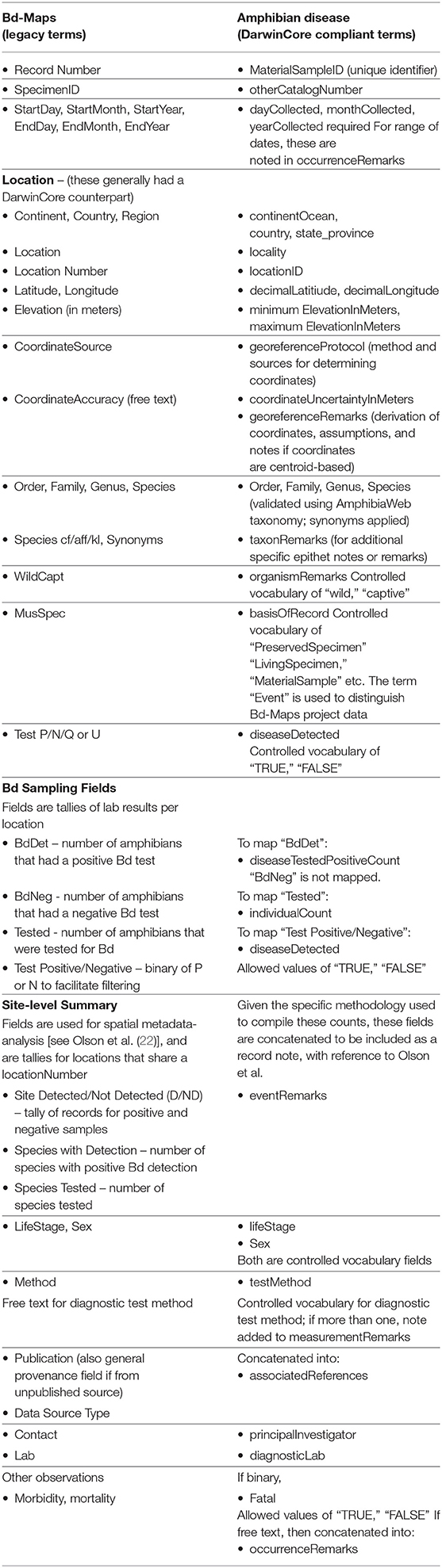
The Amphibian Disease Portal has four interwoven Goals. (1) A key aim of the repository is to provide structured data, data quality, and online accessibility. GEOME is founded on a principle of providing interlinked and machine-readable data over the Internet, while drawing on standardized vocabularies from the scientific community. We built the repository with a focus on biological samples, and recognized the need to integrate data that may be housed in separate databases, especially gene-sequence data, which is critical in furthering our understanding of Bd and Bsal disease dynamics utilizing the field of phylodynamics (30). To achieve this goal, the portal uses DarwinCore (34) and MIxS (35) metadata standards, now common among biodiversity data repositories. The GEOME validation process checks data requirements such as appropriate diagnostic fields for Bd and Bsal, compliance with the AmphibiaWeb taxonomy (36), and controlled vocabularies for country, disease type, sample type, and basis of record. (Examples of controlled vocabularies are included in Table 1 and are lists of terms from which users can choose). Datasets with the DarwinCore fields of institutionID, collectionID, and catalogNumber constitute a global unique identifier for a cataloged specimen in a natural history museum collection, and may be linked to a biodiversity aggregator (e.g., Global Biodiversity Information Facility, GBIF); likewise, contributors may provide a Uniform Resource Identifier (or URI, e.g., a unique web address) for the field “associatedSequences” to link to external genetic repositories (i.e., National Center for Biotechnology Information, NCBI), for genomic Sequence Read Archive (SRA) or other molecular data. Contributors can choose to submit data directly to SRA through GEOME using its “FASTQ” module, in which case the sample is automatically linked between GEOME and SRA using the material Sample Identifier.
(2) We prioritized tangible benefits to the user to incentivize the contributions of researchers to the repository. Therefore, we considered the needs of the contributor foremost, including how and when to make a dataset publicly accessible. Many datasets are created for publication, and may have pre-publication restrictions (e.g., a graduate student's dissertation); hence, contributors can make newly uploaded datasets private and thus not searchable or accessible until a later date (e.g., when the study is published).
Even before uploading data, contributors may benefit from project planning and data-management tools provided by the portal. In GEOME, the Amphibian Disease Portal is identified as a “Team,” and all Bd and Bsal projects that are part of the team adhered to the database rules of the portal. These projects share not only database rules for Bd and Bsal samples but customized and customizable data templates. Contributors are encouraged to download and use these templates in Microsoft Excel format (XLSX) for data collection in the field and lab (See Supplementary Material for example templates). Instructions are included in each template file where fields and controlled vocabulary are defined and are clearly labeled. Once completed, the same file can be used to upload directly to the database without complicated formatting requirements. The portal has a minimum set of required fields and many optional fields from which users can customize their own template. Currently the Amphibian Disease Portal provides two different template configurations to any participating project: one designed for samples from museum specimens, and another addressing catch-and-release specimens from the field. We will continue to add new templates as new use cases arise, such as one for environmental samples and for laboratory or captive specimens. Along with an online guideline and instructions, we aimed to make data management easier during the data-collection phase.
Closing the cycle of a project for contributors is usually the publication step; journals are increasingly requiring authors to provide their data in an accessible archive. Data uploaded to the portal are provided with archival resource keys (ARK), a form of digital object identifier (DOI), generated by GEOME using California Digital Library's EZID service (37), and have satisfied data access requirements for journals to ensure that the data are citable, accessible, and credited.
(3) We designed the portal for flexibility, accessibility, and sustainability. By using representational state transfer (REST) web services (or the application programming interface, API), we display database-supplied data in easy-to-use formats, which appear on the Amphibian Disease Portal website via open access JavaScript libraries. Having data accessible through web services also allows third-party applications (e.g., R Statistics software) and other programming tools (e.g., Python) access to data without requiring secured database access. The portal's programming interface scripts are written in Python and allow for specialized processing such as matching taxonomy and synonyms from the AmphibiaWeb database. Using web services or APIs for the website has other development advantages, as well. We can deploy web developers as they are available from other projects for maintenance or let multiple developers work on new features for the site without relying solely on a database administrator, who has different skills and more strict access requirements. Thus, the architecture of the Amphibian Disease Portal allows for nimble management and enhancements as funding and needs change, providing a more sustainable project.
(4) Lastly, we aimed to support reproducible and replicable data-driven science. Reproducibility in science, under which independent researchers can repeat study results, requires access to the original sample data, yet published papers rarely contain those data and instead provide summarized data. Often the burden of providing the original data falls on scientists who have neither the time nor capacity to adequately store and retrieve data on request [e.g., (38)]. Replication in science, when applying comparable datasets to methods to test outcomes, requires access to datasets collated in a comparable manner, which also requires access to the original data. Submitting data to discipline-specific repositories compliant with well-known metadata standards, such as this portal, will reduce the future burden on the scientists who produced the data, and will provide aggregated data for re-use in potentially novel studies.
Results
The outcome of this needs-based effort is the Amphibian Disease Portal, an online site with a user-friendly interface created in partnership with the GEOME repository. We established a user workflow that is easy to follow, cyber-secure, and makes data discoverable and accessible in a single site (Figure 1). Data and workflow include these steps: (1) registered contributors initiate projects based on their study and can use customized data templates in MS Excel format for project management (the same template, when completed after field and lab work is done, can be uploaded to their project); (2) uploaded data are validated against the portal's database rules (“backend”), which are defined in the template (feedback during this process is designed to help contributors correct and successfully load their data); (3) when the data are made public by the contributor, either immediately on upload or subsequent to publication, they will then be harvested and processed by the portal's web service; and 4) the data from public projects become output to the Amphibian Disease Portal (“frontend”) and other third-party applications (e.g., R program analyses).
The main website (https://amphibiandisease.org) is comprised of: (1) a basic map-based query interface allowing for filtered or spatial searches and mapping; (2) a dashboard of summary statistics and dynamic charts by country and by species; (3) search interfaces for projects, species, and datasets; (4) various other information pages such as how to contribute data and a blog (Figure 2). Each species in the portal has a dedicated dashboard page to display aggregated Bd and Bsal samples with links to contributing projects. Graphs of species data (Figure 2B) are dynamic data-driven and reciprocally linked by URL to the respective species account page in AmphibiaWeb (https://amphibiaweb.org). The scripts enabling these reciprocal links are adaptable to other external websites as well.
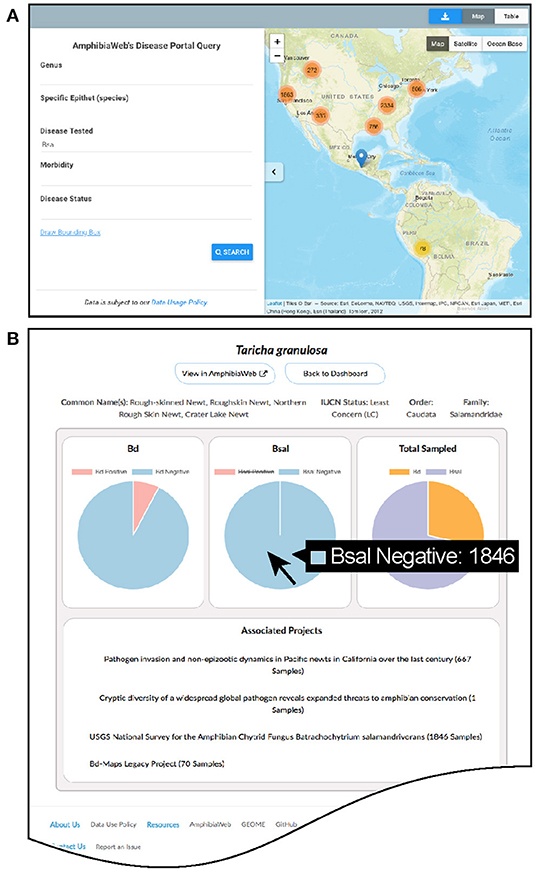
Figure 2. Examples of the Amphibian Disease portal's user-friendly pages. (A) Basic query and interactive mapping interface showing zoom-dependent clustering of data points with counts. (B) Dynamic graphing page for species showing proportion and counts of Batrachochytrium dendrobatidis (Bd) and B. salamandrivorans (Bsal) data with relevant links to projects (example species Taricha granulosa). Hovering over pie chart will display the specific counts of samples. All graphs in the Data Dashboard section of the website are enabled by the Amphibian Disease web services.
With respect to the integration of the legacy Bd-maps dataset into the portal, our challenge in modernizing and migrating the Bd-Maps data is to overcome the differences in their original conception and data structure. Instead of a biological samples-based approach, Bd-Maps database compiles locality-based summaries by species as reported in diverse ways in the literature. Entries generally were not submitted by authors but compiled from literature queries or sent to the data manager as unpublished observations (21, 22). We elucidate the transition with an important migration step: matching all Bd-Maps fields to the DarwinCore standard equivalent used in the Amphibian Disease Portal. Table 1 compares the two respective metadata schemas.
Fields for date, taxonomy, and geography such as latitude and longitude, for example, were relatively straightforward to map. Three Bd-Maps fields that tally counts of total positive Bd samples (“BdDet”), total samples (“Tested”) and total number of samples with low zoospore levels which made them questionable (“BdQues” or “BdUnc”) required different accommodations, and are important to Bd-Maps based analyses [e.g., (21, 22)]. These fields were dependent on the “Location Number” field, which is matched to the portal field “locationID.” To make these data more usable in future studies, “BdDet” is mapped to a new field “diseaseTestedPositiveCount” and “Tested” is mapped to “individualCount.” Together they can be used to estimate prevalence (“diseaseTestedPositiveCount”/“individualCount”) for any given spatial aggregation or help recreate Bd-Maps by summarizing results for all records sharing a given “locationID.” Many of the other Bd-Maps data, especially those specialized for previous analysis are not abandoned but shifted to remarks fields. The Amphibian Disease Portal field “occurrenceRemarks” contains observations on morbidity and mortality; if mortality was noted, the portal field “fatal” was marked as “TRUE.” Details on the source and derivation for a Bd-Maps locality are captured in “georeferenceProtocol” and “georeferenceRemarks.” Other Bd-Maps notes on data sources (e.g., museum vs. field) or record type (species records as opposed to full records; 22) are concatenated into “occurrenceRemarks.” Finally, meta-analysis observations on number of Bd positive and negative species at a location are compiled into “eventRemarks” such that each record indicates whether a site has the disease Detected (D) or Not Detected (ND), number of species with positive Bd detection and the total number of species tested, separated by pipes for subsequent parsing by users; for example, “D | 2 | 4” indicates “Bd Detected, 2 positive out of 4 tested”. (For details of the Bd-Maps legacy database fields, refer to Appendix S1 of 22.)
Although usage, interest, and records in the portal are in their early growth stages, we observe trends from early user submissions, which may reflect broader research priorities and reveal useful patterns and gaps in effort. A total of 62,045 samples from 2,760 taxa and 128 countries are shown in 5-degree bins in Figure 3, including 34,267 records from the legacy Bd-Maps database (22) and excluding private datasets. Of these initial samples, about a quarter (26%) of the samples are derived from voucher specimens deposited in natural history museum collections, whereas the majority of records come from field surveillance and sampling of live amphibians globally. The repository includes the results from the first US-wide survey for Bsal (from May 2014 through August 2017) conducted by the US Geological Survey (39); more than 11,000 amphibians from reserves across the US were tested for Bsal and all were Bsal-negative, providing a valuable baseline for future Bsal monitoring in North America. Lastly, one contributor has submitted pathogen sampling data from an ongoing monitoring program that uses environmental DNA (eDNA) analyses, in which organismal DNA either from shed skin cells or directly from microbes is found in terrestrial or aquatic habitats [e.g., (40, 41)]. The portal is flexible in its structure to accommodate these types of samples while adhering to metadata standards. The dynamic displays will be adapted to differentiate these records, which undoubtedly will grow in usage especially as a means of early detection.
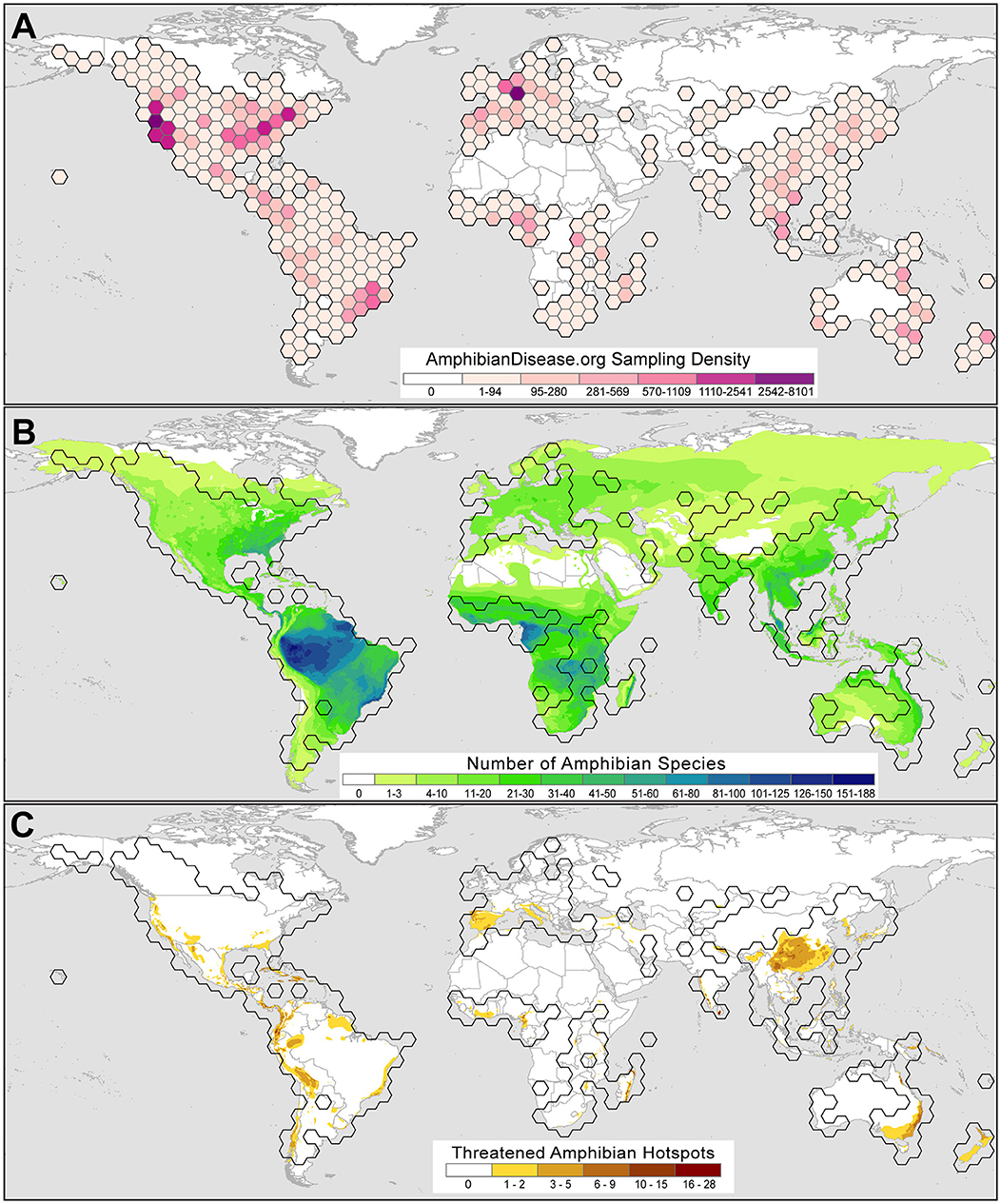
Figure 3. Amphibian Disease sampling point data for the chytrid fungal pathogens Batrachochytrium dendrobatidis (Bd) and B. salamandrivorans (Bsal) summarized by 5-degree latitude and longitude bins as of June 2021. (A) Bd and Bsal sampling is worldwide and reflects prominent sampling efforts for Bsal in the USA by the US Geological Survey (39) and in Germany by the Bsal Consortium Germany (42). Sampling strength is classified using natural breaks with Jenks optimization where darker colors denote greater numbers of samples per bin. (B) Bd and Bsal global sampling with respect to amphibian species alpha-diversity, where darker gradient denotes greater number of amphibian species. (C) Bd and Bsal global sampling with respect to threatened amphibian species, where darker gradient denotes greater number of threatened amphibian species. Range map sources: AmphibiaWeb (2021; https://amphibiaweb.org) and IUCN Red List (2021; https://www.iucnredlist.org). Threatened species status: IUCN Red List (IUCN 2021, accessed June 10, 2021).
Discussion
The great amphibian epizootic, caused by Bd and Bsal, is on track to becoming a significant factor in an unfolding mass extinction event as extensive as the previous five recorded in Earth's geologic history (1, 43). Unlike previous extinction events, however, this one is unusual in that it is occurring over a significantly shorter time period (43), and at least for amphibians, disease is a major factor (1, 9, 23). Globally, amphibian chytridiomycosis has changed the way that we think about and understand wildlife disease (8, 44). For example, when amphibians were first reported to be mysteriously declining and disappearing in the late 1970s and early 1980s in Australia, Central America, and western North America (45), few imagined so much of the decline could be driven by epizootics caused by a single pathogen [(46, 47) but see (48)]. At the time, our relatively poor understanding of the biology of Chytridiomycete fungi and a general perception that a fungal pathogen could not drive populations or species to extinction, likely contributed to the fact that it took nearly 20 years to identify Bd, discovered in 1998 (4) and described in 1999 (5), as a proximal cause of amphibian declines and extinctions.
The study of chytridiomycosis has led to significant breakthroughs in our understanding of the factors that can lead to outbreaks (epizootics) and population collapse, or non-fatal pathogen infections of hosts (9, 49). Yet, our ability to predict infection outcomes has been hampered because of the great variation in the timing of when this pathogen invaded different continents, the array of species traits in the numerous amphibian hosts, the biotic conditions under which it can infect amphibians, and the multiple genetic lineages differing significantly in virulence (8, 11, 12). Studies have described widely divergent host-pathogen dynamics in this system. Some characterize Bd host dynamics as stable enzootics where hosts do not succumb (50), whereas in other settings, even though they may consist of the same host species, living in the same nearby environment, Bd-host dynamics are characterized as epizootic, and hosts suffer mass die-offs (13). It has become evident that our ability to understand this disease and predict the outcome of infection relies on many complex variables including but not limited to the timing of pathogen invasion (31). Thus, understanding present-day disease dynamics may require describing the past. The Amphibian Disease Portal contains critical archival Bd data that provide unique historic insights. Cheng et al. (51) demonstrated that amphibian museum specimens can be successfully tested for the presence of Bd, and thus opened the door to studies of Bd temporal dynamics over long timescales (over a century). Other studies followed using museum specimens to help describe Bd distributions in the past (31, 52–65), revealing the unique value of museum specimen collections to disease ecology, and the importance of negative data. This realization has set next priorities to develop a voucher specimen lookup service, which will facilitate incorporation of collection records in data templates and increase data quality.
Currently, the data from the growing base of portal users show that global amphibian disease sampling efforts are uneven (Figure 3A); baseline Bd and Bsal sample data cover some amphibian hotspots but only lightly sample others (Figure 3B). For example, the Amazon basin is the most species-rich region for amphibians and yet sampling is sparse. Likewise, there are few samples from the range of the Western Ghats (India), which has high beta-diversity. However, a baseline of Bd and Bsal data for the Appalachian region in southeastern North America is forming, which is a global hotspot for salamander diversity (24, 66). The portal can visualize Bd and Bsal sampling in areas where threatened amphibian species occur [species with IUCN Red List status of Critically Endangered, Endangered or Vulnerable, (67)] and may help prioritize monitoring and surveillance efforts for the Bd and Bsal pathogens (Figure 3C). Regions where an accumulation of threatened species are known and baseline Bd and Bsal data have been collected include Central America south through the Andean cordillera (S. America), regions in which studies have increased our understanding of chytrid disease dynamics [e.g., (13, 51, 68, 69)].
Outcomes of chytridiomycosis also are influenced by host species (70, 71), pathogen lineage (72, 73), host community (74, 75), host microbiome (49, 76–79), abiotic conditions (75, 80), and host and pathogen population genetics (11, 12, 72). This complex reality requires collaboration between research groups allowing for sharing and visualization of original, raw data (not summarized data), in ways that are not currently possible with the peer-review-based, publication-based science that is currently the norm. The Amphibian Disease Portal provides a platform to share data on pathogen lineage, host traits, and a suite of metadata associated with where the samples were collected (various abiotic and biotic factors) as well as other potential cofactors that may yet be identified.
Disease ecology, in particular the study of emerging infectious diseases, is decidedly hampered by the standard peer-reviewed science approach because negative data are not typically published. For emerging infectious diseases, knowing that a host population was negative for a pathogen before pathogen invasion and emergence is critically important. Researchers need to be able to share pre-publication data with trusted collaborators (password protected). In conservation emergencies, they should also be able to share data in a completely open format (when appropriate) in ways that many current peer-review-based, scientific publications do not allow. Knowing whether a host population is naïve to an invading pathogen completely alters the prediction of disease outcome (50). If hosts are naïve to a pathogen, they are predicted to be much more vulnerable to epizootic dynamics and host mortality (50) than if they have previous experience with the pathogen (50, 81). The Amphibian Disease Portal provides that missing scientific platform to archive host/pathogen data by providing unique and citable digital object identifiers (DOI), making them available for the benefit of science and for the conservation of species. The uploaded data in the portal must follow strict, yet simple, rules that ensure compatibility of data across studies. Researchers must upload specific details that do not vary across projects but are given the opportunity to provide additional data (e.g., host size and weight) that might prove to be important after further analysis. The portal also gives researchers the option of storing their data in a private, password-protected environment when necessary; however, if the situation is deemed an emergency, researchers can quickly make the data available to the public. With the creation of the Amphibian Disease Portal, we show that we can harness new technologies to increase collaboration and communication among scientists globally using a workflow that is simple, sustainable, and low-cost. We provide a place to rapidly access data not only from published papers, but also from researchers who are willing to share data not yet published. The Amphibian Disease Portal enables digital object identifier assignments for these data regardless of whether they are published in a journal and allows scientists to cite their data on a stable platform that is permanently accessible. Because the portal stores original data and not summarized data, researchers are able to access the raw sample data and potentially analyze them in novel ways. This is significant because it means that data will be more easily compiled and compared across different studies. For research that is global in scope, such as studies of emerging infectious diseases (e.g., the great amphibian panzootic), being able to access and share verified data across studies is essential. In addition, the portal provides a flexible framework that allows for new research results to be added quickly and efficiently. For example, new studies are showing that eDNA methods (82) can detect the presence of Bd in aquatic systems, and this could greatly increase sampling across geographic space and could help direct limited resources toward organismal surveys in key areas. Additionally, the portal can link to existing biodiversity datasets to integrate access to genomic/genetic raw sequence data that could facilitate research on lineages of Bd pathogens (11, 12, 83–85). Thus, the portal utilizes critical functions of modern biodiversity data repositories and promotes open science practices. The lessons of the past decades of emerging infectious disease studies in amphibians have focused our attention on a few focal pathogens with the recognition that we need to expand our temporal and genomic investigations. The Amphibian Disease Portal is designed to maximize compatibility, access, and value of data to understand the current disease dynamics and to predict and adapt to future disease threats.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at: https://amphibiandisease.org and https://geome-db.org/workbench/project-overview?projectId=291.
Author Contributions
MK conceived, designed, led development, and writing of the study. VV conceived, consulted, and wrote the study. JD designed and wrote the study. DO, KR, and DW consulted and wrote the study. All authors approved the final version of this manuscript.
Funding
Funding for initial development was provided by a USDA Forest Service Cooperative Agreement #15-CR-11261953-098 (University of California Berkeley) and a Belmont Forum Project NSF #1633948 to Vredenburg. The use of trade or firm names is for reader information and does not imply endorsement by the U.S. Department of Agriculture of any product or service.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank developer Philip Kahn and web developer Diana Lovette, who brought creative enthusiasm. We are indebted to early adopters of the Amphibian Disease portal: David Blackburn, Allison Byrne, Alessandro Catenazzi, and Greg Jongsma. We dedicate this paper to David Wake who passed away while we were finalizing the manuscript; his commitment to open-access data to better inform decisions and facilitate research to save amphibians has inspired generations of students and projects like this.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.728232/full#supplementary-material
References
1. Wake DB, Vredenburg VT. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. (2008) 105 (Supp 1):11466–73. doi: 10.1073/pnas.0801921105
2. Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, et al. Status and trends of amphibian declines and extinctions worldwide. Science. (2004) 306:1783–6. doi: 10.1126/science.1103538
3. IPBES. Workshop Report on Biodiversity and Pandemics of the Intergovernmental Platform on Biodiversity and Ecosystem Services. Daszak P, das Neves C, Amuasi J, Hayman D, Kuiken T, Roche B, et al. Bonn, Germany (2020).
4. Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, et al. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of australia and central america. Proc Natl Acad Sci USA. (1998) 95:9031–6. doi: 10.1073/pnas.95.15.9031
5. Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen. et sp. nov. a Chytrid Pathogenic to Amphibians. Mycologia. (1999) 91:219–27. doi: 10.1080/00275514.1999.12061011
6. Olson DH. A decade of herpetological disease papers: puzzle pieces of a bigger picture. Herpetol Rev. (2019) 50:37–40.
7. Martel A, Spitzen-van der Sluijs A, Blooi M, Bert W, Ducatelle R, Fisher MC, et al. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc Natl Acad Sci USA. (2013) 110:15325–9. doi: 10.1073/pnas.1307356110
8. Blaustein AR, Urbina J, Snyder PW, Reynolds E, Dang T, Hoverman JT, et al. Effects of emerging infectious diseases on amphibians: a review of experimental studies. Diversity. (2018) 10:1–49. doi: 10.3390/d10030081
9. Fisher MC, Garner TWJ. Chytrid fungi and global amphibian declines. Nat Rev Microbiol. (2020) 18:332–43. doi: 10.1038/s41579-020-0335-x
10. OIE [World Organization of Animal Health]. Aquatic Animal Health Code, Disease Listed by the OIE, Chapter 1.. (2019). Available online at: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/aquatic-code-online-access (accessed June 15, 2021).
11. Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, Clare F, et al. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci USA. (2011) 108:18732–6. doi: 10.1073/pnas.1111915108
12. Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, Eastman JM, et al. Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci USA. (2013) 110:9385–90. doi: 10.1073/pnas.1300130110
13. Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci USA. (2010) 107:9689–94. doi: 10.1073/pnas.0914111107
14. Catenazzi A, Lehr E, Rodriguez LO, Vredenburg VT. Batrachochytrium dendrobatidis and the collapse of anuran species richness and abundance in the upper manu national park, southeastern Peru. Conserv Biol. (2011) 25:382–91. doi: 10.1111/j.1523-1739.2010.01604.x
15. Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, Phillott AD, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth. (2007) 4:125–34. doi: 10.1007/s10393-007-0093-5
16. Garner TWJ, Schmidt BR, Martel A, Pasmans F, Muths E, Cunningham AA, et al. Mitigating amphibian chytridiomycoses in nature. Phil Trans R Soc B. (2021) 371:20160207. doi: 10.1098/rstb.2016.0207
17. Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, et al. Emerging infectious disease and the loss of biodiversity in a neotropical amphibian community. Proc Natl Acad Sci USA. (2006) 103:3165–70. doi: 10.1073/pnas.0506889103
18. North American Bsal Task Force [NABTF]. Annual Report. (2016). Available online at: https://www.salamanderfungus.org/task-force/ (accessed November 1, 2020).
19. Gray MJ, Lewis JP, Nanjappa P, Klocke B, Pasmans F, Martel A, et al. Batrachochytrium salamandrivorans: the North American Response and a Call for Action. PLoS Pathogens. (2015) 11:e1005251. doi: 10.1371/journal.ppat.1005251
20. Yap TA, Nguyen NT, Serr M, Shepack A, Vredenburg VT. Batrachochytrium salamandrivorans and the risk of a second amphibian pandemic. EcoHealth. (2017) 14:851–64. doi: 10.1007/s10393-017-1278-1
21. Olson DH, Aanensen DM, Ronnenberg KL, Powell CI, Walker SF, Bielby J, et al. Mapping the global emergence of Batrachochytrium dendrobatidis, the amphibian chytrid fungus. PLoS ONE. (2013) 8:e56802. doi: 10.1371/journal.pone.0056802
22. Olson DH, Ronnenberg KL, Glidden CK, Christiansen KR, Blaustein AR. Global patterns of the fungal pathogen Batrachochytrium dendrobatidis support conservation urgency. Duffus ALJ, Marschang RE, editors. Emerging Infections and Diseases of Herpetofauna. Front Vet Sci Zool Med. (2021) 8:685877. doi: 10.3389/fvets.2021.685877
23. Castro Monzon F, Rödel MO, Jeschke JM. Tracking Batrachochytrium dendrobatidis infection across the globe. EcoHealth. (2020) 17:270–9. doi: 10.1007/s10393-020-01504-w
24. Yap TA, MS Koo, RF Ambrose, DB Wake, Vredenburg VT. Averting a North American biodiversity crisis. Science. (2015) 349:481–2. doi: 10.1126/science.aab1052
25. Gervasi SS, Stephens PR, Hua J, Searle CL, Urbina J, Olson DH, et al. Linking ecology and epidemiology to understand predictors of multi-host responses to an emerging pathogen, the amphibian chytrid fungus. PLoS ONE. (2017) 12:e0167882. doi: 10.1371/journal.pone.0167882
26. Greenberg DA, Palen WJ, Mooers AØ. Amphibian species traits, evolutionary history and environment predict Batrachochytrium dendrobatidis infection patterns, but not extinction risk. Evol Appl. (2017) 10:1130–45. doi: 10.1111/eva.12520
27. Longo AV, Fleischer RC, Lips KR. Double trouble: co-infections of chytrid fungi will severely impact widely distributed newts. Biol Invasions. (2019) 21:2233–45. doi: 10.1007/s10530-019-01973-3
28. Basanta DM, Byrne AQ, Rosenblum EB, Piovia-Scott J, Parra-Olea G. Early presence of Batrachochytrium dendrobatidis in Mexico with a contemporary dominance of the Global Panzootic Lineage. Mol Ecol. (2020) 30:424–37. doi: 10.1111/mec.15733
29. Becker CG, Greenspan SE, Tracy KE, Dash JA, Lambertini C, Jenkinson TS, et al. Variation in Phenotype and Virulence Among Enzootic and Panzootic Amphibian Chytrid Lineages. Fungal Ecol. (2017) 26:45–50. doi: 10.1016/j.funeco.2016.11.007
30. Rife BD, Mavian C, Chen X, et al. Phylodynamic Applications in 21st Century Global Infectious Disease Research. Glob Health Res PolGHeRP. (2017) 2:13. doi: 10.1186/s41256-017-0034-y
31. Vredenburg VT, McNally SVG, Sulaeman H, Butler HM, Yap T, Koo MS, et al. Pathogen Invasion History Elucidates Contemporary Host Pathogen Dynamics. PLoS ONE. (2019) 14:e0219981. doi: 10.1371/journal.pone.0219981
32. Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C, et al. The genomic observatories metadatabase (GEOME): a new repository for field and sampling event metadata associated with genetic samples. PLoS Biol. (2017) 15:e2002925. doi: 10.1371/journal.pbio.2002925
33. Free Software Foundation. The GNU General Public License v3.0 - GNU Project - Free Software Foundation. (n.d.) Available online at: https://www.gnu.org/licenses/gpl-3.0.html (accessed January 10, 2021).
34. Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, et al. Darwin core: an evolving community-developed biodiversity data standard. PLoS ONE. (2012) 7:e29715. doi: 10.1371/journal.pone.0029715
35. Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. (2011) 29:415–20. doi: 10.1038/nbt.1823
36. AmphibiaWeb. (2020) Taxonomy and Synonym Daily List. Available online at: https://amphibiaweb.org/taxonomy/AWtaxonomy.html (accessed November 1, 2020).
37. EZID California Digital Library. (2020). Available online at: https://ezid.cdlib.org (accessed November 1, 2020).
38. Vision TJ. Open data and the social contract of scientific publishing. BioScience. (2010) 60:330–1. doi: 10.1525/bio.2010.60.5.2
39. Waddle JH, Grear DA, Mosher BA, Campbell Grant EH, Adams MJ, et al. Batrachochytrium salamandrivorans (Bsal) not detected in an intensive survey of wild North American amphibians. Sci Rep. (2020) 10:13012. doi: 10.1038/s41598-020-69486-x
40. Goldberg CS, Pilliod DS, Arkle RS, Waits LP. Molecular detection of vertebrates in stream water: a demonstration using rocky mountain tailed frogs and Idaho giant salamanders. PLoS ONE. (2011) 6:e22746. doi: 10.1371/journal.pone.0022746
41. Chestnut T, Anderson C, Popa R, Blaustein AR, Voytek M, Olson DH, et al. Heterogeneous occupancy and density estimates of the pathogenic fungus Batrachochytrium dendrobatidis in waters of North America. PLoS ONE. (2014) 9:e106790. doi: 10.1371/journal.pone.0106790
42. Vences M, Lötters S. The salamander plague in Europe—a German perspective. Salamandra. (2020) 56:169–71.
43. Barnosky AD, Matzke N, Tomiya S, Wogan G, Swartz B, Quental T, et al. Has the earth's sixth mass extinction already arrived? Nature. (2011) 471:51–7. doi: 10.1038/nature09678
44. Yong E. The Worst Disease Ever Recorded. The Atlantic (2019). Available online at: https://www.theatlantic.com/science/archive/2019/03/bd-frogs-apocalypse-disease/585862/ (accessed March 28, 2019).
45. Wake DB. Declining amphibian populations. Science. (1991) 253:860. doi: 10.1126/science.253.5022.860
46. Blaustein AR, Wake DB. The puzzle of declining amphibian populations. Sci Am. (1995) 272:52–7. doi: 10.1038/scientificamerican0495-52
47. Hero J-M, Gillespie GR. Epidemic disease and amphibian declines in Australia. Conserv Biol. (1997) 11:1023–5. doi: 10.1046/j.1523-1739.1997.96291.x
48. Laurance WF, McDonald KR, Speare R. Epidemic disease and the catastrophic decline of australian rain forest frogs. Conserv Biol. (1996) 10:406–13. doi: 10.1046/j.1523-1739.1996.10020406.x
49. Bernardo-Cravo AP, Schmeller DS, Chatzinotas A, Vredenburg VT, Loyau A. Environmental factors and host microbiomes shape host–pathogen dynamics. Trends Parasitol. (2020) 36:616–33 doi: 10.1016/j.pt.2020.04.010
50. Briggs CJ, Knapp RA, Vredenburg VT. Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. Proc Natl Acad Sci USA. (2010) 107:9695–700. doi: 10.1073/pnas.0912886107
51. Cheng TL, Rovito SM, Wake DB, Vredenburg VT. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. (2011) 108:9502–7. doi: 10.1073/pnas.1105538108
52. Vredenburg VT, Felt SA, Morgan EA, McNally SVG, Wilson S, Green SL. Prevalence of Batrachochytrium dendrobatidis in Xenopus Collected in Africa (1871–2000) and in California (2001–2010). PLoS ONE. (2013) 8:e63791. doi: 10.1371/journal.pone.0063791
53. Huss M, Huntley L, Vredenburg V, Johns J, Green S. Presence of Batrachochytrium dendrobatidis in 120 archived specimens of Lithobates catesbeianus (American Bullfrog) collected in California, 1924–2007. EcoHealth. (2013) 10:339–43. doi: 10.1007/s10393-013-0895-6
54. Zhu W, Bai C, Wang S, Soto-Azat C, Li X, Liu X, et al. Retrospective survey of museum specimens reveals historically widespread presence of Batrachochytrium dendrobatidis in China. EcoHealth. (2014) 11:241–50. doi: 10.1007/s10393-013-0894-7
55. Talley BL, Muletz CR, Vredenburg VT, Fleischer RC, Lips KR. A century of Batrachochytrium dendrobatidis in Illinois Amphibians (1888–1989). Biol Conserv. (2015) 182:254–61. doi: 10.1016/j.biocon.2014.12.007
56. Fong JJ, Cheng TJ, Battaile A, Pessier AP, Waldman B, Vredenburg VT. Early 1900s detection of Batrachochytrium dendrobatidis in Korean Amphibians. PLoS ONE. (2015) 10:e0115656. doi: 10.1371/journal.pone.0115656
57. Sette CM, Vredenburg VT, Zink AG. Reconstructing historical and contemporary disease dynamics: a case study using the california slender salamander. Biol Conserv. (2015) 192:20–9. doi: 10.1016/j.biocon.2015.08.039
58. Yap TA, Gillespie L, Ellison S, Flechas SV, Koo MS, Martinez Vredenburg VT. Invasion of the fungal pathogen Batrachochytrium dendrobatidis on California Islands. EcoHealth. (2016) 13:145–50. doi: 10.1007/s10393-015-1071-y
59. Adams AJ, Pessier AP, Briggs CJ. Rapid extirpation of a north american frog coincides with an increase in fungal pathogen prevalence: historical analysis and implications for reintroduction. Ecol Evol. (2017) 7:10216–32. doi: 10.1002/ece3.3468
60. De Léon ME, Vredenburg VT, Piovia-Scott J. Recent emergence of a chytrid fungal pathogen in California cascades frogs (Rana cascadae). EcoHealth. (2017) 14:155–61. doi: 10.1007/s10393-016-1201-1
61. Yap TA, Koo MS, Ambrose RF, Vredenburg VT. Introduced bullfrog facilitates pathogen invasion in the Western United States. PLoS ONE. (2018) 13:e0188384. doi: 10.1371/journal.pone.0188384
62. Chaukulkar S, Sulaeman H, Zink AG, Vredenburg VT. Pathogen invasion and non-epizootic dynamics in Pacific newts in California over the last century. PLoS ONE. (2018) 13:e0197710. doi: 10.1371/journal.pone.0197710
63. Rios-Sotelo G, Figueroa-Valenzuela R, Vredenburg VT. Retrospective survey reveals extreme rarity of amphibian fungal pathogen Batrachochytrium dendrobatidis in Japanese amphibians from 1890–1990s. Herpetol Rev. (2018) 49:247–52.
64. De León ME, Zumbado-Ulate H, García-Rodríguez A, Alvarado G, Sulaeman H, Bolaños F, et al. Batrachochytrium dendrobatidis infection in amphibians predates first known epizootic in costa rica. PLoS ONE. (2019) 14:e0208969. doi: 10.1371/journal.pone.0208969
65. Sette CM, Vredenburg VT, Zink AG. Differences in fungal disease dynamics in co-occurring terrestrial and aquatic amphibians. EcoHealth. (2020) 17:302–14. doi: 10.1007/s10393-020-01501-z
66. AmphibiaWeb. (2021). Database Application: species range maps. Available online at: https://amphibiaweb.org (accessed May 1, 2021).
67. Mace GM, Collar NJ, Gaston KJ, Hilton-Taylor C, Akçakaya HR, Leader-Williams N, et al. Quantification of extinction risk: IUCN's system for classifying threatened species. Conserv Biol. (2008) 22:1424–42. doi: 10.1111/j.1523-1739.2008.01044.x
68. Catenazzi A, von May R, Vredenburg VT. High prevalence of infection in tadpoles increases vulnerability to fungal pathogen in high-andean amphibians. Biol Conserv. (2013) 159:413–21. doi: 10.1016/j.biocon.2012.11.023
69. Catenazzi A, Lehr E, Vredenburg VT. Thermal physiology, disease, and amphibian declines on the eastern slopes of the andes. Conserv Biol. (2014) 28:509–17. doi: 10.1111/cobi.12194
70. Gahl MK, Longcore JE, Houlahan JE. Varying responses of northeastern North American amphibians to the chytrid pathogen Batrachochytrium dendrobatidis. Conserv Biol. (2012) 26:135–41. doi: 10.1111/j.1523-1739.2011.01801.x
71. Reeder NMM, Pessier AP, Vredenburg VT. A Reservoir species for the emerging amphibian pathogen Batrachochytrium dendrobatidis thrives in a landscape decimated by disease. PLoS ONE. (2012) 7:e33567. doi: 10.1371/journal.pone.0033567
72. Morgan JAT, Vredenburg VT, Rachowicz LJ, Knapp RA, Stice MJ, Tunstall T, et al. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. (2007) 104:13845–50. doi: 10.1073/pnas.0701838104
73. Retallick RW, Miera V. Strain differences in the amphibian chytrid Batrachochytrium dendrobatidis and non-permanent, sub-lethal effects of infection. Dis Aquat Org. (2007) 75:201–7. doi: 10.3354/dao075201
74. Searle CL, Gervasi SS, Hua J, Hammond JI, Relyea RA, Olson DH, et al. Differential host susceptibility to batrachochytrium dendrobatidis, an emerging amphibian pathogen. Conserv Biol. (2011) 25:965–74. doi: 10.1111/j.1523-1739.2011.01708.x
75. Lambertini C, Becker CG, Belasen AM, Valencia-Aguilar A, Nunes-de-Almeida CHL, Betancourt-Román CM, et al. Biotic and abiotic determinants of Batrachochytrium dendrobatidis infections in amphibians of the Brazilian Atlantic forest. Fungal Ecol. (2021) 49:100995. doi: 10.1016/j.funeco.2020.100995
76. Woodhams DC, Vredenburg VT, Simon M, Billheimer D, Shakhtour B, Shyr Y, et al. Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol Conserv. (2007) 138:390–8. doi: 10.1016/j.biocon.2007.05.004
77. Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. (2009) 3:818–24 doi: 10.1038/ismej.2009.27
78. Lam BA, Walke JB, Vredenburg VT, Harris RN. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv. (2010) 143:529–31. doi: 10.1016/j.biocon.2009.11.015
79. Vredenburg VT, Briggs CJ, Harris RN. Host-pathogen dynamics of amphibian chytridiomycosis: the role of the skin microbiome in health and disease. In: Olsen L, Choffnes ER, Relman DA, Pray L, editors. Fungal Diseases: An Emerging Threat to Human, Animal and Plant Health. Washington, DC: The National Academies Press IOM (Institute of Medicine) (2011). p. 342–55.
80. Murrieta-Galindo R, Parra-Olea G, González-Romero A, López-Barrera F, Vredenburg VT. Detection of Batrachochytrium dendrobatidis in amphibians inhabiting cloud forests and coffee agroecosystems in central Veracruz, Mexico. Eur J Wildlife Res. (2014) 60:431–9. doi: 10.1007/s10344-014-0800-9
81. Rachowicz LJ, Hero J-M, Alford RA, Taylor JW, Morgan JA, Vredenburg VT, et al. The novel and endemic pathogen hypotheses: competing explanations for the origin of emerging infectious diseases of wildlife. Conserv Biol. (2005) 19:1441–8. doi: 10.1111/j.1523-1739.2005.00255.x
82. Barnes MA, Brown AD, Daum MN, de la Garza KA, Driskill J, Garrett K, et al. Detection of the amphibian pathogens chytrid fungus (Batrachochytrium dendrobatidis) and Ranavirus in West Texas, USA, using environmental DNA. J Wildlife Dis. (2020) 56:702–6 doi: 10.7589/2019-08-212
83. Byrne AQ, Rothstein AP, Poorten TJ, Erens J, Settles ML, Rosenblum EB. Unlocking the story in the swab: a new genotyping assay for the amphibian Chytrid fungus Batrachochytrium dendrobatidis. Mol Ecol Resour. (2017) 17:1283–92. doi: 10.1111/1755-0998.12675
84. O'Hanlon SJ, Rieux A, Farrer RA, Rosa GM, Waldman B, Bataille A, et al. Recent Asian origin of Chytrid fungi causing global amphibian declines. Science. (2018) 360:621–7. doi: 10.1126/science.aar1965
Keywords: DarwinCore standards, amphibia, chytridiomycosis, MIxS standards, Bd, biodiversity informatics infrastructure, Bsal
Citation: Koo MS, Vredenburg VT, Deck JB, Olson DH, Ronnenberg KL and Wake DB (2021) Tracking, Synthesizing, and Sharing Global Batrachochytrium Data at AmphibianDisease.org. Front. Vet. Sci. 8:728232. doi: 10.3389/fvets.2021.728232
Received: 21 June 2021; Accepted: 02 September 2021;
Published: 04 October 2021.
Edited by:
Rachel E. Marschang, Laboklin GmbH Co. KG, GermanyReviewed by:
Louise A. Rollins-Smith, Vanderbilt University, United StatesDavid Lesbarreres, Laurentian University, Canada
Copyright © 2021 Koo, Vredenburg, Deck, Olson, Ronnenberg and Wake. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle S. Koo, bWtvb0BiZXJrZWxleS5lZHU=
 Michelle S. Koo
Michelle S. Koo Vance T. Vredenburg
Vance T. Vredenburg John B. Deck
John B. Deck Deanna H. Olson
Deanna H. Olson Kathryn L. Ronnenberg
Kathryn L. Ronnenberg David B. Wake1
David B. Wake1