- 1Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Fisheries College of Guangdong Ocean University, Zhanjiang, China
- 2Guangdong Provincial Engineering Research Center for Aquatic Animal Health Assessment, Shenzhen Public Service Platform for Evaluation of Marine Economic Animal Seedings, Shenzhen Institute of Guangdong Ocean University, Shenzhen, China
Fish nocardiosis is a chronic, systemic, granulomatous disease in aquaculture. Nocardia seriolae has been reported to be one of the main pathogenic bacteria of fish nocardiosis. There are few studies on the associated virulence factors and pathogenesis of N. seriolae. Alanine dehydrogenase (ALD), which may be a secreted protein, was discovered by analysis using bioinformatics methods throughout the whole genomic sequence of N. seriolae. Nevertheless, the roles of ALD and its homologs in the pathogenesis of N. seriolae are not demonstrated. In this study, the function of N. seriolae ALD (NsALD) was preliminarily investigated by gene cloning, host cell subcellular localization, secreted protein identification, and cell apoptosis detection. Identification of the extracellular products of N. seriolae via mass spectrometry (MS) analysis revealed that NsALD is a secreted protein. In addition, subcellular localization of NsALD-GFP recombinant protein in fathead minnow (FHM) cells showed that the strong green fluorescence co-localized with the mitochondria. Moreover, apoptosis assays demonstrated that the overexpression of NsALD induces apoptosis in FHM cells. This study may lay the foundation for further exploration of the function of NsALD and facilitate further understanding of the pathogenic mechanism and the associated virulence factors of N. seriolae.
Introduction
Fish nocardiosis, a chronic systemic granulomatous disease, has great influence on both marine and freshwater aquaculture industry (1, 2). Nocardia salmonicida, Nocardia asteroids, and Nocardia seriolae have been isolated from diseased fish and confirmed as the pathogenic bacteria of fish nocardiosis (3). Remarkably, N. seriolae has been most frequently reported as the main pathogen in the last 30 years. Nocardia seriolae can infect the immunodeficient fish via wounds, the gills, and feeds (4). The symptoms of diseased fish include skin ulceration and serious sarcoidosis caused by a mass of white nodules in the gills, spleen, liver, head kidney, and trunk kidney (5). According to reports, N. seriolae was able to infect about 42 kinds of marine and freshwater fish, such as blotched snakehead (Channa maculata), amberjack (Seriola dumerili), yellowtail (Seriola quinqueradiata), snubnose pompano (Trachinotus blochii), golden pompano (Trachinotus ovatus), largemouth bass (Micropterus salmoides), large yellow croaker (Larimichthys crocea), and red drum (Sciaenops ocellatus) (3, 6–8).
The virulence factors and pathogenic mechanisms of N. seriolae–host interaction are not fully studied. It was reported that the virulence of Nocardia species is related to their resistivity to oxidative damage of macrophages, inhibition of a combination of phagosomes and lysosomes, alteration of lysosomal enzymes in phagocytes, and neutralization of the acidification of the phagosome, and some extracellular products seemed to participate in the above processes in Nocardia asteroides (9–12). Our previous studies also indicated that the MTSP3141, GluNS, NsHLP, PLC, SOD, robl/LC7, PTP, and NlpC/P60 of N. seriolae are able to lead to apoptosis in fish cells and are the potential virulence factors of N. seriolae. MTSP3141 and GluNS were also confirmed to be secreted proteins and mitochondrial targeting secretory proteins (MTSPs) (7, 13–15). The disease mechanisms of Nocardia sp. have been demonstrated to be varied and complicated and needed further clarification.
In this study, an alanine dehydrogenase (ALD) homolog of N. seriolae (NsALD) was amplified by gene cloning. Then, the secreted proteins were identified and the subcellular localization of NsALD and its involvement in disease mechanisms were evaluated. This study will put forward further clarification of the role of NsALD during infection and facilitate new insights into the molecular pathogenicity of N. seriolae.
Materials and Methods
Bacterial Strains, Cell Line, and Plasmids
Nocardia seriolae ZJ0503 strain (16), fathead minnow (FHM) cells (17), plasmid PEGFP-N1, plasmid PCDNA3.1-His A, and Escherichia coli DH5α were conserved in our laboratory. Nocardia seriolae ZJ0503 isolated from diseased fish was cultured in optimal medium to clone the ALD gene. Escherichia coli DH5α transformed by plasmid DNA was cultured to extract large numbers of endotoxin-free plasmids. FHM cells (17) transformed by endotoxin-free plasmids were cultured in Leibovitz's L15 medium with 10% fetal bovine serum for apoptosis assays. The endotoxin-free plasmids pEGFP-N1 and pCDNA3.1-His A were used for subcellular localization and overexpression, respectively.
Gene Cloning and Sequence Bioinformatics Analysis of NsALD
Nocardia seriolae ZJ0503 was collected after 5 days of cultivation in optimal medium to extract genomic DNA using TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). NsALD was cloned via PCR with two pairs of primers designed using Primer Premier 5.0 software, pEGFP-ALD-F/R and pcDNA-ALD-F/R, shown in Table 1. Based on the whole-genome sequence data of the N. seriolae strain ZJ0503 (accession no. NZ_JNCT01000022), BLAST sequence analysis was carried out through NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). The amino acid sequence for NsALD was predicted and the physicochemical property predicted using ExPASy software (http://www.expasy.org/). Multiple sequence alignment was done with DNAMAN software. A biological evolutionary tree was constructed using the evolutionary analysis software MEGA 6.0. LocTree 3 (https://rostlab.org/services/loctree3/) and PSORT II Prediction (https://psort.hgc.jp/form2.html) were utilized to predict the subcellular localization and signal peptides.
Plasmid Construction
Using the two pairs of primers listed in Table 1, the gene NsALD was cloned into pcDNA3.1/His A and pEGFP-N1 vectors to explore the molecular function and subcellular localization of NsALD in vitro. The constructed recombinant plasmids were subsequently confirmed by sequencing.
PCR was performed with TaKaRa Ex Taq® polymerase using the following PCR program: pre-denaturation at 95°C for 5 min, 34 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 3 min. The PCR products of NsALD were electrophoresed on 1% agarose gel and purified using EasyPure PCR Purification Kit (TransGen, Beijing, China). The purified PCR products were digested by the corresponding restriction enzymes, ligated into the eukaryotic vectors pEGFP-N1 and pcDNA3.1/His A, and then transformed into competent E. coli DH5α cells. The different constructs were confirmed by corresponding restriction enzyme digestion (Table 1) and DNA sequencing by Guangzhou Sangon Biological Engineering & Technology and Service Co. Ltd. Finally, the recombinant plasmids named as pEGFP-ALD and pcDNA-ALD.
Preparation and Identification of Extracellular Products
After culturing in optimal medium for 3–5 days, single colonies were selected to prepare bacterial suspension. Sterilized cellophane was laid flat on the modified medium plate as close to the medium as possible. Of the bacterial suspension, 100 μl was spread evenly on the medium covered with cellophane and incubated at 28°C for 3–5 days. The cellophane with colonies from the medium was removed with tweezers and the colonies on the cellophane rinsed into a sterile beaker with sterile phosphate-buffered saline (PBS). The collected liquid was transferred into a 50-ml centrifuge tube at 8,000 × g at 4°C for 20 min. After centrifugation, the supernatant was sterilized using a 0.22-μm microporous membrane and then transferred into a dialysis bag. Dialysis was performed at 4°C for 10–16 h in ultrapure water, during which the dialysate was changed 3 times. After dialysis, the samples were transferred into a 50-ml centrifuge tube and the liquid was frozen into solid at −80°C before freeze drying in a vacuum freeze dryer. The freeze-dried extracellular product samples were sent to a related biological company for protein shotgun LC-MS identification.
Cell Transfection and Subcellular Localization
Plasmids pEGFP-ALD and pEGFP-N1 were extracted in large quantities via Endo-Free Plasmid Mini Kit I D6948 (Omega Bio-Tek, Norcross, GA, USA). Cell transfection was carried out to explore the subcellular localization of NsALD in host cells using the Lipofectamine 3000 reagent. At 48 h post-transfection (hpt), pEGFP-ALD-transfected FHM cells and pEGFP-N1-transfected cells were both stained with 4′,6-diamidino-2-phenylindole (DAPI) and Mito Tracker Red according to the protocols. After staining and washing with sterilized PBS, the cells were examined under a fluorescence microscope.
Detection of Cell Apoptosis Induced by Overexpression of NsALD Protein
To determine whether the overexpression of NsALD induces apoptosis in fish cells, transient transfection of FHM cells was carried out using pcDNA-ALD as an experimental group or plasmid pcDNA3.1/His A as a control group. The overexpression of NsALD in FHM cells was verified by Western blot. Then, FHM cells were dyed with DAPI at 48 hpt and observed using a fluorescence microscope. Furthermore, the caspase-3 activity and mitochondrial membrane potential (ΔΨm) of the transfected cells were detected, respectively (18) with a caspase-3 colorimetric assay kit K106-25 (BioVision, Milpitas, CA, USA) and a mitochondrial membrane potential assay kit with JC-1 (Beyotime, Shanghai, China).
Quantitative Analysis of the mRNA Expression of Apoptosis-Related Genes
Quantitative PCR primers (Table 2) for the genes Bcl-2, Bax, Bid, and Bad of FHM cells were designed using Primer Premier 5.0 software. FHM cells transfected with pcDNA-ALD and pcDNA3.1/His A were collected at 0, 24, 48, and 72 hpt. Total RNA was extracted using the TransZol Up Plus RNA Kit (TransGen) and cDNA was synthesized with the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen) for real-time quantitative PCR (RT-qPCR) with PerfectStart® Green qPCR SuperMix (TransGen). The effect of transfection on the expressions of apoptosis-related genes was calculated by LightCycler 96 software for RT-qPCR. Each test was performed with the β-actin gene as the internal control, and there were 3 replicates. The PCR reaction volume was 10 μl, 1 μl for each primer (10 μM), 1 μl for cDNA, 2 μl for PCR grade water, and 5 μl for Green qPCR SuperMix. The PCR procedures for the four apoptosis-related genes and β-actin were as follows: 95°C for 2 min, 40 cycles of 95°C for 75 s, and 55°C for 1 min.
Data Analysis
The comparative 2(−ΔΔCt) method was used to calculate the relative expression levels of the four apoptosis-related genes. The data obtained were analyzed with one-way ANOVA using GraphPad Prism 8.0 software. The data were edited and plotted into histograms, and the expression of each apoptosis-related gene at 0 hpt was calculated as 1. Asterisks show statistical significance, as follows: p > 0.05, not significant; *p < 0.05, significant; and **p < 0.01, extremely significant.
Results
Sequence Characterization and Bioinformatics Analysis of NsALD
NsALD was cloned via PCR using pEGFP-ALD-F/R and pcDNA-ALD-F/R (Figure 1A). Sequencing analysis showed that the total length of the open reading frame (ORF) of NsALD was successfully acquired, which encoded an alanine dehydrogenase homolog. The results of sequencing analysis revealed that the ORF of the NsALD gene (GenBank: 61148960) was 1,188 bp encoding 395 amino acid sequences, with alanine dehydrogenase/PNT, N-terminal domain, and alanine dehydrogenase NAD-binding and catalytic domains, which belong to the NADB-Rossmann superfamily (Figure 1B). For subcellular localization, NsALD was predicted to locate in the mitochondria with 81% expected accuracy using LocTree 3, while it was predicted to locate in the cytoplasm with 94.1% expected accuracy using PSORT II Prediction. No signal peptide or transit peptide was found. Schematic representation of the prosites demonstrated the protein NsALD to comprise two complexity domains (30–163 and 168–377) and nine kinds of functional sites (Figure 1C).
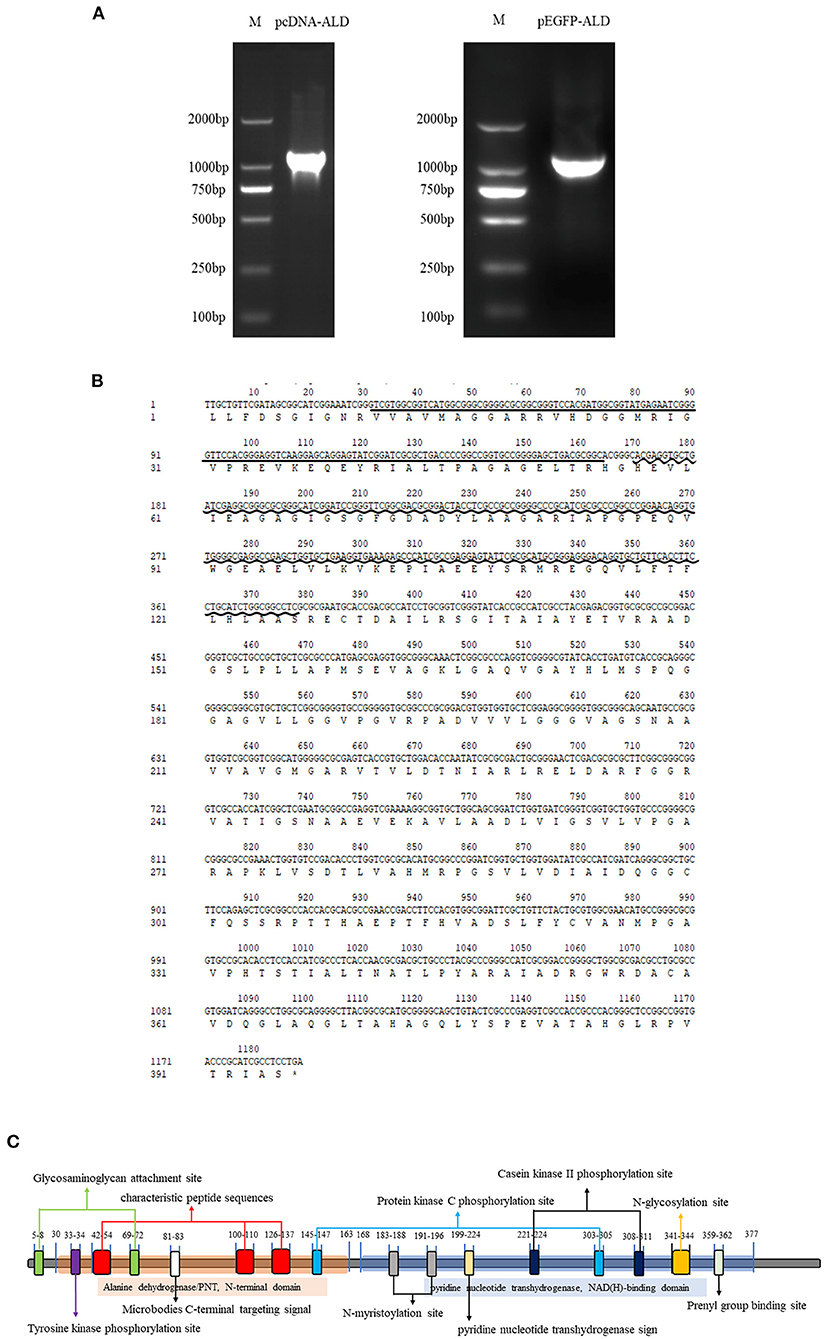
Figure 1. Sequence and structure analysis of the alanine dehydrogenase of Nocardia seriolae (NsALD). (A) NsALD was amplified via PCR using the primers pEGFP-ALD-F/R and pcDNA-ALD-F/R. (B) NsALD nucleic acid sequence and its derived amino acid sequence. The straight line indicates the ALD/PNT, N-terminal domain and the wavy line shows the ALD NAD-binding and catalytic domains. (C) Schematic representation of the prosites of protein NsALD. The NsALD protein comprised two complexity domains (30–163 and 168–377) and nine kinds of functional sites.
BLAST protein analysis showed that the NsALD amino acid sequence had high homology with other ALD sequences in actinomycetes, with 92.41, 79.05, 76.15, 68.29, 66.67, 62.60, 52.30, 50.68, and 53.66% identity to the ALDs of Nocardia concava, N. asteroides, N. salmonicida, Rhodococcus kunmingensis, Mycobacterium tuberculosis H37Rv, Streptomyces coelicolor A3(2), Shewanella oneidensis MR-1, Microcystis aeruginosa, and Deinococcus radiodurans R1, respectively (Figure 2A). A biological evolutionary tree was constructed with the amino acid sequences of 12 bacterial ALDs, which shows that NsALD had a rather high homology among the Nocardia species (Figure 2B). Structure analysis showed a high similarity between the ALDs from N. seriolae and Nocardia concave (Figure 2C).
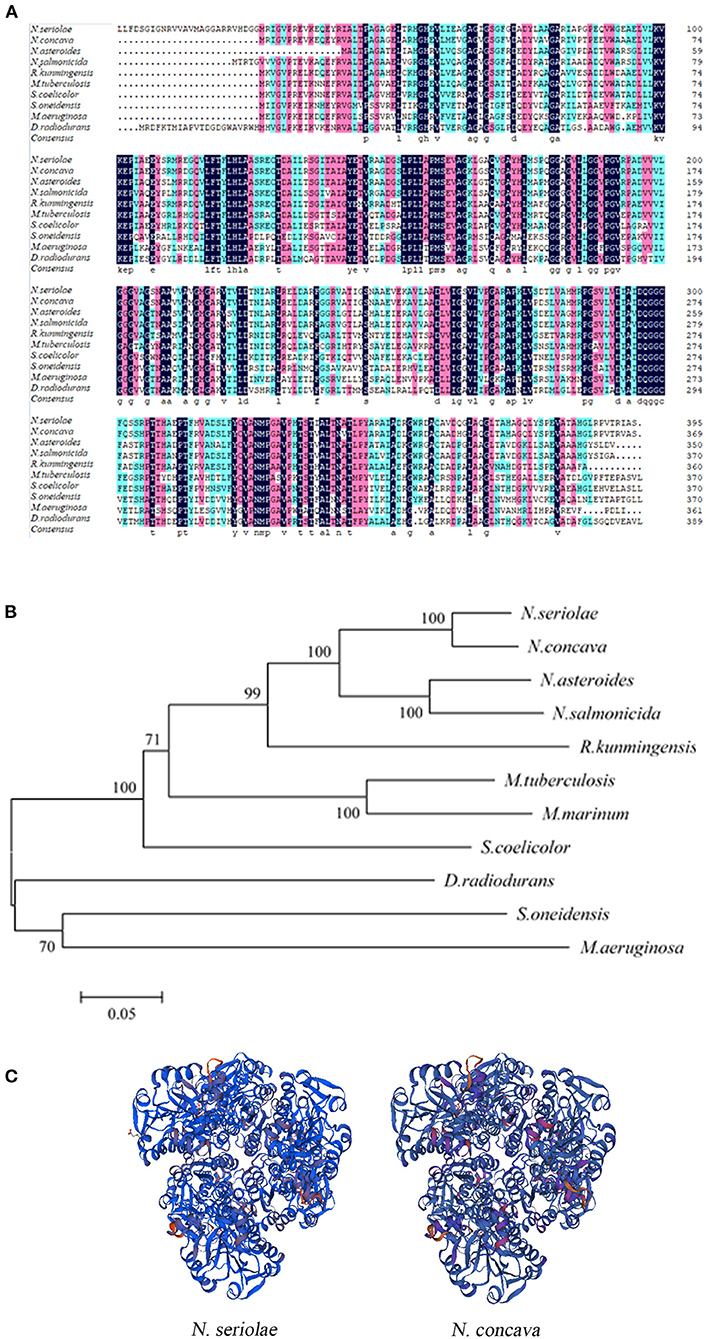
Figure 2. Multiple sequence alignment, construction of a phylogenetic tree, and the three-dimensional structure of Nocardia seriolae alanine dehydrogenase (NsALD) protein. (A) Multiple alignment of the deduced amino acid sequences of NsALD protein among different species. Black shaded backgrounds indicate 100% homology, pink shaded backgrounds indicate >75% homology, and blue shaded backgrounds indicate >50% homology. (B) Construction of a phylogenetic tree among N. seriolae and other species with protein NsALD homologous sequences. The protein sequences were aligned using DNAMAN software, and the non-rooted neighbor joining tree was generated by the MEGA 5.0 program. Number at branch points indicates bootstrap support. (C) Structure analysis of NsALD from N. seriolae. The predicted three-dimensional structure of NsALD protein from N. seriolae (left) and Nocardia concava (right). The diagrams were generated using SWISS-MODEL online. GenBank accession numbers are shown as follows: N. seriolae ZJ0503, WP_033090406.1; N. concava, WP_040806553.1; Nocardia asteroides, SFM85941.1; Nocardia salmonicida, WP_062992005.1; Rhodococcus kunmingensis, WP_068269631.1; Mycobacterium tuberculosis H37Rv, NP_217296.1; Mycobacterium marinum, WP_020728350.1; Streptomyces coelicolor A3(2), NP_626044.1; Shewanella oneidensis MR-1, YP_007001355.1; Microcystis aeruginosa, WP_012268249.1; and Deinococcus radiodurans R1, NP_295618.1.
Detection of Subcellular Localization of NsALD in Transfected Cells
To explore the subcellular localization of the NsALD protein, the recombinant plasmid pEGFP-ALD was transfected into FHM cells, which was tested by green fluorescence signals. The nucleus was shown as blue fluorescence and the mitochondria was displayed as red fluorescence. In pEGFP-N1-transfected cells, the green fluorescence was dispersed in the whole cell of FHM cells, the mitochondria were distributed in the perinuclear cytoplasm, and the nucleus margin was smooth and had no apoptosis feature (Figure 3, right). Differently, in pEGFP-ALD-transfected cells, nuclear shrinkage appeared, the mitochondria aggregated near the nucleus, and a strong green fluorescence of ALD-GFP exhibited an aggregated distribution near the nucleus and overlapped with the location of the mitochondria, which indicated that the protein NsALD was co-localized with the mitochondria (Figure 3, left).

Figure 3. Subcellular localization of the Nocardia seriolae alanine dehydrogenase (NsALD) protein in fathead minnow (FHM) cells. Green fluorescence shows ALD-GFP or GFP, red fluorescence shows the mitochondria, and blue fluorescence shows the nucleus. Left panels are pEGFP-ALD and right panels are pEGFP-N1. In pEGFP-N1-transfected cells, the green fluorescence was dispersed in the whole cell of FHM cells, the mitochondria were distributed in the perinuclear cytoplasm, and the edge of the nucleus was smooth and had no apoptosis characteristics. In pEGFP-ALD-transfected cells, nuclear shrinkage appeared, the mitochondria aggregated near the nucleus, and a strong green fluorescenc of ALD-GFP exhibited an aggregated distribution near the nucleus and overlapped with the distribution of the mitochondria.
Secreted Protein Identification to NsALD
The secreted proteins of N. seriolae were acquired and identified with shotgun mass spectrometry. The results revealed that three characteristic peptide sequences of NsALD (IALTPAGAGELTR, SGITAIAYETVR, and VKEPIAEEYSR) were tested with a confidence ≥99%, which demonstrated that NsALD is a secreted protein of Nocardia seriolae.
Apoptosis Detection Induced by the Overexpression of NsALD
To explore whether the NsALD protein has an influence on the apoptosis of fish cells, the control plasmid and the recombinant plasmid pcDNA-ALD were transfected into FHM cells. At 48 hpt, several apoptotic bodies can be observed in the transfected FHM cells (Figure 4A), and Western blot showed that the NsALD protein was expressed (Figure 4B). Apoptotic bodies were counted to calculate the apoptosis rate. The result revealed that there were highly significant differences between the experimental group and the control group (Figure 4C). As exhibited in Figure 4D, detection of caspase-3 activity revealed that the maximum value appeared at 48 hpt, which was a 1.63-fold increase compared to that of the control group. Furthermore, the ΔΨm values of NsALD-overexpressing cells compared with those of control cells were reduced remarkably. The ΔΨm values of NsALD-overexpressing cells decreased to minimum values at 48 hpt, a decrease of 0.43-fold compared to the control group (Figure 4E).
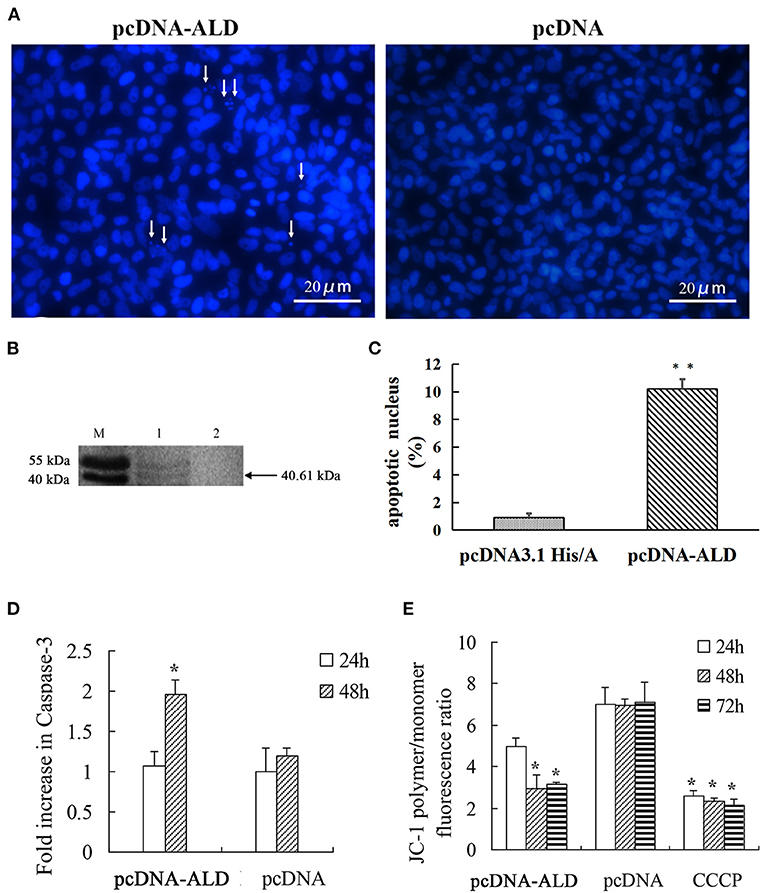
Figure 4. Apoptosis characteristics in pcDNA-ALD-transfected fathead minnow (FHM) cells. (A) Overexpression of protein NsALD (Nocardia seriolae alanine dehydrogenase) in FHM cells. White arrows indicate apoptotic bodies. (B) Confirmation of NsALD expression in FHM cells by Western blot. M, marker. 1, NsALD; 2, pcDNA. (C) Percentage of apoptotic body in plasmid pcDNA-ALD- or pcDNA-transfected cells. Error bars indicate SD (**p < 0.01). (D) Fold increase in caspase-3 after plasmid pcDNA-ALD or pcDNA transfection of FHM cells. FHM cells transfected with plasmids were collected at indicated points after transfection and the levels of cleaved caspase-3 measured. Error bars indicate SD (*p < 0.05). (E) Detection of ΔΨm values. FHM cells transfected with plasmid pcDNA-ALD or pcDNA were collected at indicated time points after transfection and the values accessed using JC-1. Untransfected cells treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP) were positive controls. Data are expressed as the JC-1 polymer/monomer fluorescence ratio. Error bars indicate SD (*p < 0.05).
Quantitative Detection of Apoptosis-Related Genes in FHM Cells
The expression level of each apoptosis-related gene at 0 hpt was considered as the control group. The expressions of Bad and Bax genes increased significantly at 24–72 hpt, with peak values of 10.5-fold at 72 hpt and 2.9-fold at 48 hpt, respectively. The Bid gene increased significantly at 48–72 hpt, reaching a peak value of 2.5-fold at 72 hpt. Interestingly, the expression of Bcl-2, which is an anti-apoptotic gene, showed no change during 0–48 hpt, while it increased significantly at 72 hpt. Since a high Bax/Bcl-2 ratio is related to cell apoptosis, the Bax/Bcl-2 ratio was calculated according to their expressions. The Bax/Bcl-2 ratio increased significantly at 24–48 hpt, with a peak value of 10.5-fold at 48 hpt, and then it decreased to 0.5 times that of the control group at 72 hpt (Figure 5). This meant that, although the expression of Bcl-2 increased significantly, the Bax/Bcl-2 ratio is still low at 72 hpt.
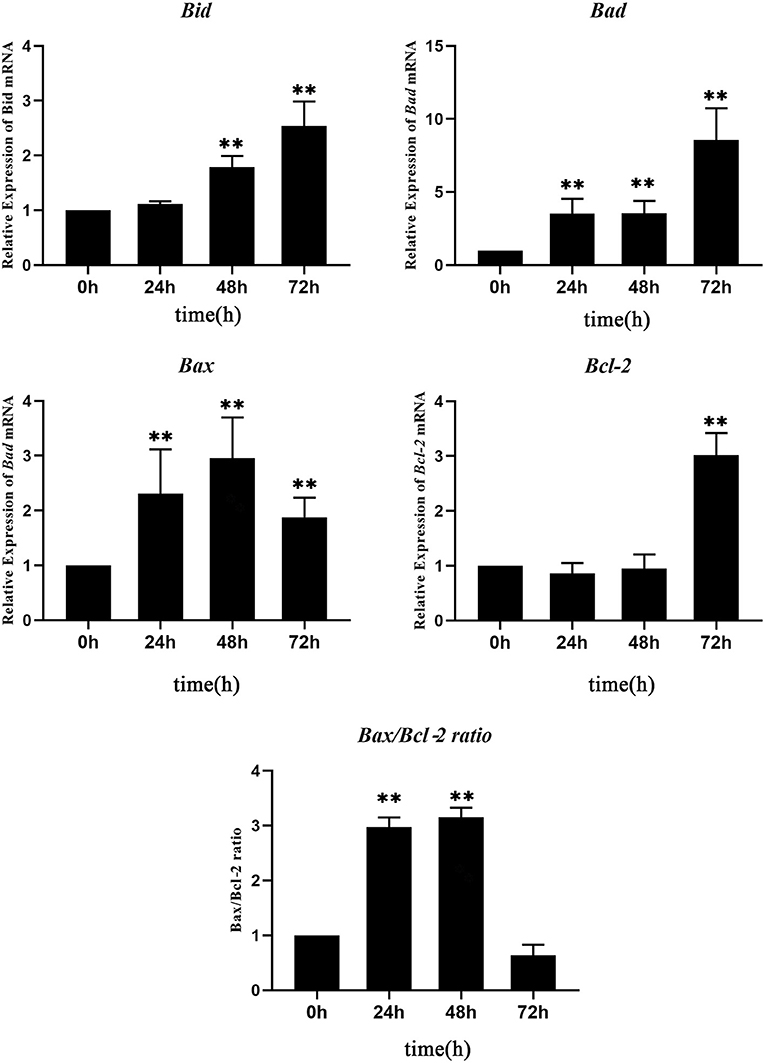
Figure 5. Real-time quantitative PCR (RT-qPCR) analysis of the expressions of apoptosis-related genes in Nocardia seriolae alanine dehydrogenase (NsALD)-overexpressing fathead minnow (FHM) cells. Significant differences are indicated by **p < 0.01.
Discussion
Alanine dehydrogenase (ALD) is a microbial enzyme that catalyzes the reversible oxidation of the deamination of alanine to pyruvate. ALD interconversion between alanine and pyruvate is the core of microbial metabolism (19). ALD was firstly found in Bacillus subtilis by Wiame and Pierard (20). The pathogenicity of bacteria is closely associated with its pathogenic factors. However, only a few studies have been conducted on the virulence factor of N. seriolae. Interestingly, in some bacteria, ALD was found to be an antigen-secreting protein in vitro and was regarded as a virulence factor of pathogenic bacteria. For instance, ALD was found in M. tuberculosis culture filtrate and identified as one of its main antigens (21). Transcriptional induction of the ALD gene has been observed upon infection of leopard frogs with Mycobacterium marinum, which indicated that ALD may be crucial during M. marinum infection (22). So far, information about the regulation of ALD and its possible role in pathogenesis is limited (18), and no study related to the ALD of N. seriolae (NsALD) has been reported.
In this study, NsALD was obtained and identified as a secreted protein without a signal peptide. It has been reported that the ALD of M. tuberculosis is a predominant antigen in its culture filtrate, which means that ALD is a secreted protein of M. tuberculosis; the ALD of M. tuberculosis also lacks a signal peptide (23). NsALD was found to co-localize with the mitochondria in this research. Some proteins targeting the mitochondria contain a transit peptide, while no transit peptide was found in the NsALD protein by bioinformatic prediction. Mitochondrial targeting of bacterial proteins has been reported. Some proteins from parasitic microorganisms such as rickettsial postulated peptidase (RPP) have been discovered to be similar to the protein subunit of mitochondrial processing peptidase (MPP) (24). Additionally, some bacterial virulence factors contain N-terminal mitochondrial targeting signals (25, 26). The host cells are invaded by certain proteins of Gram-negative bacteria through the type 3 secretion system (TTSS) and are co-positioned with the mitochondria of host cells (25–27). Helicobacter pylori VacA-targeted mitochondria was associated with the formation of anion-selective channels in mitochondrial membranes (28). Further research on the mitochondria-targeting mechanism of NsALD is needed.
The mitochondria is an important organelle with various important functions, such as participating in innate and adaptive immunity and acting as the signal center in cell apoptosis (29), and is also the key target of bacterial invasion (30). Most of the identified bacterial proteins targeting the mitochondria are involved in cell apoptosis, which is consistent with the core effect of the mitochondria in apoptosis regulation (31). For example, the hlyA gene of enteropathogenic E. coli (EPEC) encodes hemolysin, which is targeted to the mitochondria and triggers the apoptosis of host cells via mitochondrial pathways (25). In the process of infection with macrophages, the secreted SopA protein of Salmonella enterica locates in the mitochondria to activate the caspase-1 independent pathway via the TTSS and leads to macrophage death (32). Cytotoxin VacA related to gastric epithelial lesions is a virulence factor of pathogenic H. pylori (33). It was found that p34, a subunit of VacA, specifically locates in host mitochondria and induces the release of host cytochrome C, which activates caspase-3 and leads to cell death (34). In this study, typical apoptosis characteristics, such as apoptotic bodies, ΔΨm values dropping, and caspase-3 activation, were examined in NsALD-expressed cells, which indicated that NsALD induces apoptosis in host cells.
In our previous work, the secreted virulence factors of N. seriolae have also been identified to induce cell apoptosis. The phospholipase C of N. seriolae was a secreted protein and induced apoptosis in FHM cells (15). The MTSP3141 of N. seriolae was proven to be a secreted protein, co-located with the mitochondria, and had a 30-amino acid transit peptide at the N-terminal (35). Glutamyl endopeptidase was a secreted protein without a transit peptide, localized in the mitochondria, and was also a virulence factor of N. seriolae that induced apoptosis (36). In addition, the superoxide dismutase (SOD) and histone-like DNA-binding protein (HLP) of N. seriolae was also identified as secreted proteins that lead to apoptosis in host cells (7, 13). Taken together, secreted protein-induced apoptosis is associated with N. seriolae infection.
According to relevant articles, the main apoptosis pathways in fish are the extrinsic/death receptor pathway (37) and the intrinsic/Bcl-2-regulated/mitochondrial pathway (38). In addition, like mammals, fish have similar functions of conserved members of the Bcl-2 family and inherent apoptosis pathways. Among them, the Bax/Bak-like proteins (Bax, Bak, Bok, Bcl-xs, and Mtd) and the BH3-only proteins (Bad, Bid, Bik, Bim, Bmf, and Noxa) are pro-apoptotic molecules (39). According to the results of the RT-qPCR of the four apoptosis-related genes, three pro-apoptotic genes (Bax, Bad, and Bid) and the Bax/Bcl-2 ratio were significantly activated at 24–48 hpt, suggesting that NsALD could induce the apoptosis of host cells through the intrinsic/Bcl-2-regulated/mitochondrial pathway.
In this study, NsALD was verified to be a secreted protein of N. seriolae by shotgun mass spectrometry and target the mitochondria in fish cells. The overexpression of the NsALD protein caused obvious apoptotic characteristics, such as the increase of the activity of caspase-3, the decrease of the ΔΨm values in FHM cells, and the significantly induced expressions of pro-apoptotic genes, indicating that NsALD can induce cell apoptosis. This study has laid the foundation for further clarification of the function of NsALD and provided new insights into the understanding of the molecular pathogenicity of N. seriolae.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This research did not include live animals. All animal experimental procedures involved were carried out in accordance with the Regulations for Animal Experimentation of GuangDong Ocean University, and the animal facility was based on the National Institutes of Health guide for the care and use of the Laboratory (NIH Publications No. 8023).
Author Contributions
LX and YLiu designed the experiments. GC, LX, ZT, YLu, and TW performed the experiments. GC, LX, and ZT contributed to analysis. GC and LX wrote the paper. YLiu polished the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Project (2020YFD0900201), the Natural Science Foundation of Guangdong Province (2021A1515011222), the Key Research and Development Projects in Guangdong Province (2020B0202010009), and the Shenzhen Dapeng New District special fund for industry development (KJYF202001-08).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to all the laboratory members for their constructive suggestions to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.801990/full#supplementary-material
References
1. Valdez IE, Conroy DA. The study of a tuberculosis-like condition in neon tetras (hyphessobrycon innesi). II. Characteristics of the bacterium isolated. Microbiol Espanola. (1963) 16:249–53.
2. Ho PY, Byadgi O, Wang PC, Tsai MA, Liaw LL, Chen SC. Identification, molecular cloning of il-1 beta and its expression profile during Nocardia seriolae infection in largemouth bass, Micropterus salmoides. Int J Mol Sci. (2016) 17:1670. doi: 10.3390/ijms17101670
3. Xia L, Cai J, Wang B, Huang Y, Jian J, Lu Y. Draft genome sequence of Nocardia seriolae ZJ0503, a fish pathogen isolated from Trachinotus ovatus in China. Genome Announcements. (2015) 3:e01223-14. doi: 10.1128/genomeA.01223-14
4. Manrique WG, Da SCG, de Castro MP, Petrillo TR, Figueiredo MA, Belo MA, et al. Expression of cellular compo nents in granulomatous inflammatory response in piaractus mesopotamicus model. PLoS ONE. (2015) 10:e0121625. doi: 10.1371/journal.pone.0121625
5. Maekawa S, Yoshida T, Wang PC, Chen SC. Current knowledge of nocardiosis in teleost fish. J Fish Dis. (2018) 41:413–9. doi: 10.1111/jfd.12782
6. Shimahara Y, Nakamura A, Nomoto R, Itami T, Chen SC, Yoshida T. Genetic and phenotypic comparison of Nocardia seriolae isolated from fish in Japan. J Fish Dis. (2008) 31:481–8. doi: 10.1111/j.1365-2761.2008.00920.x
7. Wang W, Chen J, Liao B, Xia L, Hou S, Wang Z, et al. Identification and functional characterization of Histone-like DNA-binding protein in Nocardia seriolae (NsHLP) involved in cell apoptosis. J Fish Dis. (2019) 42:657–66. doi: 10.1111/jfd.12962
8. Rio-Odriguez RE, Ramirez-Paredes JG, Soto-Rodriguez SA, Shapira Y, Haydon DJ. First evidence of fish nocardiosis in mexico caused by Nocardia seriolae in farmed red drum (Sciaenops ocellatus, Linnaeus). J Fish Dis. (2021) 44:1117–30. doi: 10.1111/jfd.13373
9. Barry DP, Beaman BL. Nocardia asteroides strain GUH-2 induces proteasome inhibition and apoptotic death of cultured cells. Res Microbiol. (2007) 158:86–96. doi: 10.1016/j.resmic.2006.11.001
10. Camp DM, Loeffler DA, Razoky BA, Tam S, Beaman BL, LeWitt PA. Nocardia asteroides culture filtrates cause dopamine depletion and cytotoxicity in PC12 cells. Neurochem Res. (2003) 28:1359–67. doi: 10.1023/A:1024944431725
11. Beaman BL, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev. (1994) 7:213–64. doi: 10.1128/cmr.7.2.213
12. Loeffler DA, Camp DM, Qu S, Beaman BL, LeWitt PA. Characterization of dopamine-depleting activity of Nocardia asteroides strain GUH-2 culture filtrate on PC12 cells. Microb Pathog. (2004) 37:73–85. doi: 10.1016/j.micpath.2004.05.001
13. Hou S, Chen G, Wang W, Xia L, Wang Z, Lu Y. Identification of a cell-wall peptidase (NlpC/P60) from Nocardia seriolae which induces apoptosis in fathead minnow cells. J Fish Dis. (2020) 43:571–81. doi: 10.1111/jfd.13154
14. Hou S, Wang W, Chen G, Xia L, Wang Z, Lu Y. Identification of a secreted superoxide dismutase (SOD) from Nocardia seriolae which induces apoptosis in fathead minnow (FHM) cells. J Fish Dis. (2021) 44:63–72. doi: 10.1111/jfd.13268
15. Xia L, Liang H, Xu L, Chen J, Bekaert M, Zhang H, et al. Subcellular localization and function study of a secreted phospholipase C from Nocardia seriolae. Fems Microbiol Lett. (2017) 364:fnx143. doi: 10.1093/femsle/fnx143
16. Huang YC, Jian JC, Wu ZH, Lu YS, Yu TJ. Isolation and identification of the pathogen causing sarcoidosis of Trachinotus ovatus. J Guangdong Ocean Univ. (2008) 4:49–53. doi: 10.3969/j.issn.1673-9159.2008.04.011
17. Gravell M, Malsberger RG. A permanent cell line from the fathead minnow (Pimephales promelas). Ann N Y Acad Sci. (1965) 126:555–65. doi: 10.1111/j.1749-6632.1965.tb14302.x
18. Feng Z, Cáceres NE, Sarath G, Barletta RG. Mycobacterium smegmatis L-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth. J Bacteriol. (2002) 184:5001–10. doi: 10.1128/jb.184.18.5001-5010.2002
19. Dave UC, Kadeppagari RK. Alanine dehydrogenase and its applications-A review. Crit Rev Biotechnol. (2019) 39:648–64. doi: 10.1080/07388551.2019.1594153
20. Wiame JM, Pierard A. Occurrence of an L(+)-alanine-dehydrogenase in Bacillus subtilis. Nature. (1955) 176:1073–5. doi: 10.1038/1761073b0
21. Hutter B, Singh M. Properties of the 40 kDa antigen of Mycobacterium tuberculosis, a functional L-alanine dehydrogenase. Biochem J. (1999) 343:669–72. doi: 10.1042/0264-6021:3430669
22. Chan K, Knaak T, Satkamp L, Humbert O, Falkow S, Ramakrishnan L. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc Nat Acad Sci. (2002) 99:3920–5. doi: 10.1073/pnas.002024599
23. Andersen AB, Andersen P, Ljungqvist L. Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect Immun. (1992) 60:2317–23. doi: 10.1128/IAI.60.6.2317-2323.1992
24. Kitada S, Uchiyama T, Funatsu T, Kitada Y, Ogishima T, Ito A, et al. protein from a parasitic microorganism, Rickettsia prowazekii, can cleave the signal sequences of proteins targeting mitochondria. J Bacteriol. (2007) 189:844–50. doi: 10.1128/JB.01261-06
25. Kenny B, Jepson M. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. (2000) 2:579–90. doi: 10.1046/j.1462-5822.2000.00082.x
26. Nougayrède JP, Donnenberg MS. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell Microbiol. (2004) 6:1097–111. doi: 10.1111/j.1462-5822.2004.00421.x
27. Papatheodorou P, Domańska G, Öxle M, Mathieu J, Selchow O, Kenny B, et al. The enteropathogenic Escherichia coli (EPEC) Map effector is imported into the mitochondrial matrix by the TOM/Hsp70 system and alters organelle morphology. Cell Microbiol. (2006) 8:677–89. doi: 10.1111/j.1462-5822.2005.00660.x
28. Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. (2012) 2:92. doi: 10.3389/fcimb.2012.00092
29. Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. (2012) 13:780–8. doi: 10.1038/nrm3479
30. Lobet E, Letesson JJ, Arnould T. Mitochondria: a target for bacteria. Biochem Pharmacol. (2015) 94:173–85. doi: 10.1016/j.bcp.2015.02.007
31. Kozjak-Pavlovic V, Ross K, Rudel T. Import of bacterial pathogenicity factors into mitochondria. Curr Opin Microbiol. (2008) 11:9–14. doi: 10.1016/j.mib.2007.12.004
32. Layton AN, Brown PJ, Galyov EE. The Salmonella translocated effector SopA is targeted to the mitochondria of infected cells. J Bacteriol. (2005) 187:3565–71. doi: 10.1128/JB.187.10.3565-3571.2005
33. Galmiche A, Rassow J. Targeting of Helicobacter pylori VacA to mitochondria. Gut Microbes. (2010) 1:392–5. doi: 10.4161/gmic.1.6.13894
34. Galmiche A, Rassow J, Doye A, Cagnol S, Chambard JC, Contamin S, et al. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. (2000) 19:6361–70. doi: 10.1093/emboj/19.23.6361
35. Chen J, Xia L, Wang W, Wang Z, Hou S, Xie C, et al. Identification of a mitochondrial-targeting secretory protein from Nocardia seriolae which induces apoptosis in fathead minnow cells. J Fish Dis. (2019) 42:1493–507. doi: 10.1111/jfd.13062
36. Wang W, Hou S, Chen G, Xia L, Chen J, Wang Z, et al. Characterization and function study of a glutamyl endopeptidase homolog from Nocardia seriolae. J Fish Dis. (2021) 44:813–21. doi: 10.1111/jfd.13311
37. Mehlen P, Bredesen DE. The dependence receptor hypothesis. Apoptosis Int J Programmed Cell Death. (2004) 9:37–49. doi: 10.1023/B:APPT.0000012120.66221.b2
38. Reed JC. Cytochrome c: Can't live with it - can't live without it. Cell. (1997) 91:559–62. doi: 10.1016/S0092-8674(00)80442-0
Keywords: Nocardia seriolae, alanine dehydrogenase, secreted protein, mitochondrial localization, apoptosis
Citation: Chen G, Tan Z, Liu Y, Weng T, Xia L and Lu Y (2022) Function and Characterization of an Alanine Dehydrogenase Homolog From Nocardia seriolae. Front. Vet. Sci. 8:801990. doi: 10.3389/fvets.2021.801990
Received: 26 October 2021; Accepted: 22 November 2021;
Published: 12 January 2022.
Edited by:
Lixing Huang, Jimei University, ChinaReviewed by:
Jie Li, Chinese Academy of Fishery Sciences (CAFS), ChinaWeiwei Zhang, Ningbo University, China
Copyright © 2022 Chen, Tan, Liu, Weng, Xia and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqun Xia, eGlhbHFAZ2RvdS5lZHUuY24=; Yishan Lu, ZmlzaGRpc0AxNjMuY29t
†ORCID: Guoquan Chen orcid.org/0000-0002-0847-0408
Liqun Xia orcid.org/0000-0001-6244-8060
 Guoquan Chen1,2†
Guoquan Chen1,2† Liqun Xia
Liqun Xia Yishan Lu
Yishan Lu