- 1Avian Diseases Laboratory, College of Veterinary Medicine, Konkuk University, Seoul, South Korea
- 2KHAV Co., Ltd., Seoul, South Korea
- 3Wildlife Health Laboratory, College of Veterinary Medicine, Konkuk University, Seoul, South Korea
Infectious bronchitis virus (IBV) has evolved through various mutation mechanisms, including antigenic drift and recombination. Four genotypic lineages of IBVs including GI-15, GI-16, GI-19, and GVI-1 have been reported in Korea. In this study, we isolated two IBVs from chicken farms, designated IBV/Korea/289/2019 (K289/19) and IBV/Korea/163/2021 (K163/21), which are two distinct natural recombinant viruses most likely produced by genetic reassortment between the S1 gene of K40/09 strain (GI-19 lineage) and IBV/Korea/48/2020 (GI-15 lineage) in co-infected commercial chickens. Comparative sequence analysis of hypervariable regions (HVRs) revealed that the K289/19 virus had similar HVR I and II with the K40/09 virus (100% and 99.2% nucleotide sequence identity, respectively), and HVR III with the IBV/Korea/48/2020 virus (100% nucleotide sequence identity). In contrast, the K163/21 virus had HVR I and II similar to the IBV/Korea/48/2020 virus (99.1% and 99.3% nucleotide sequence identity, respectively), and HVR III to the K40/09 virus (96.6% nucleotide sequence identity). The K289/19 virus exhibited similar histopathologic lesions, tissue tropism in trachea and kidney, and antigenicity with the parental K40/09 virus. The K163/21 exhibited similar pathogenicity and tissue tropism with the K40/09 virus, which were similar results with the isolate K289/19. However, it showed a lower antigenic relatedness with both parental strains, exhibiting R-value of 25 and 42, respectively. The continued emergence of the novel reassortant IBVs suggests that multiple recombination events have occurred between different genotypes within Korea. These results suggest that antigenic profiles could be altered through natural recombination in the field, complicating the antigenic match of vaccine strains to field strains. Enhanced surveillance and research into the characteristics of newly emerging IBVs should be carried out to establish effective countermeasures.
1. Introduction
Infectious bronchitis virus (IBV) is a pathogen associated with acute respiratory tract diseases in chickens. The IBV is highly contagious and can also affect multiple organs according to their pathotypes, such as the respiratory tract, kidneys, and reproductive tract. Chickens infected with IBV show respiratory signs, reduced egg production, weight loss, and decreased weight gain. Mortality may vary depending on the IBV strain, secondary bacterial infection, or coinfection with other viruses. It is economically important to control this infection because of its detrimental effects on poultry production (1, 2).
High antigenic diversity of IBVs and the extensive emergence of variants are problematic, resulting in poor cross-protection by the available vaccine strains (3–5). The spike (S) protein of IBV, comprising ~3.4 kb, is a major inducer of virus-neutralizing antibodies and an important factor in determining tissue tropism (6), which is post-translationally cleaved to S1 and S2 subunits. The S2 subunit is associated with membrane fusion (6, 7). The S1 subunit, responsible for host cell attachment, has a receptor binding domain and hypervariable regions (HVR). HVRs (HVR I, II, and III) are associated with neutralizing antibodies, virulence of the virus, and tissue tropism (8–12). The HVR I is a major epitope inducing neutralizing antibodies and associated with tissue tropism for respiratory tract (12, 13). In addition, a previous study reported that the HVR II is related to kidney affinity (14). Because of these characteristics of the S1 subunit, IBV genotypes have been classified based on the S1 gene (15–17).
According to the classification system of IBV lineages defined by Valestro et al. (15), IBVs isolated in Korea are classified into four genotypic lineages: GI-15, GI-16, GI-19, and GVI-1 (16). The GI-15, previously designated as Korean group I (K-I), is associated with respiratory diseases in chickens (18). The GI-19 includes nephropathogenic viruses that can be divided into three subgroups: KM91-like, QX-like, and K40/09-like (17). The KM91-like subgroup, also known as Korean group II a (K-IIa), have been detected from 1990's with nephropathogenicity in chickens (19, 20). The QX-like subgroup, also known as Korean group II b (K-IIb), causes outbreaks in chickens globally, including South Korea (15, 17). The K40/09-like subgroup was reported as the Korean new cluster I in our previous study, which is produced by recombination between the KM91-like virus and QX-like virus (21). The GI-16 strains were isolated in Korea during 2003–2006, designated as Korean group III (K-III) (20), which have caused respiratory syndrome and nephropathogenic diseases in chicken farms (22, 23). The GVI-1, reported as the Korean new cluster II causes clinical signs mainly in the respiratory tract of chickens (24, 25). Diverse recombinant genotypes of IBVs in Korea suggests that multiple recombination have occurred between different genotypes in Korea.
In this study, we isolated two novel IBVs which are natural recombinant between the GI-15 and GI-19 lineages. We investigated their pathobiological characteristics, such as pathogenicity, antigenicity, and tissue tropism in chickens.
2. Materials and methods
2.1. Virus isolation and propagation
The IBVs used in this study were K40/09, IBV/Korea/48/2020, K289/19, and K163/21. The K40/09 (K40/09-like subgroup of GI-19) and IBV/Korea/48/2020 (B4-like subgroup of GI-15) viruses were isolated from chickens in Korea (17, 21). The isolate K289/19 was isolated from 46-day-old broiler breeders showing respiratory signs and nephritis. The isolate K163/21 was isolated from a 77-day-old layer that showed respiratory signs. All IBVs used in this study were propagated in 10-day-old specific-pathogen free (SPF) embryonated chicken eggs for 48 h (21). The allantoic fluid was harvested from the inoculated eggs and stored at −70°C until use. Before using the allantoic fluid, quantification of viruses was conducted by titration to calculate 50% embryo infectious dose (EID50) (26).
2.2. PCR, sequencing, and phylogenetic analysis
Viral RNA was extracted from harvested allantoic fluid using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The S1 gene of each IBV isolate was amplified using a OneStep RT-PCR Kit (Qiagen) according to the manufacturer's instructions and using two pairs of primers (S1 forward F: CGGAACAAAAGACMGACTTAGT and S1 forward R: CWGTACCATTAACAAARTAAGCMAG; S1 rear F: TGTGTATTTTAAAGCAGGTGGACC and S1 rear R: GTTTGTATGTACTCATCTGTAAC). The reaction was conducted in a ProFlex PCR System (Applied Biosystems, Forest City, CA, USA) at 50°C for 30 min; 95°C for 15 min, and 35 cycles of 94°C for 60 s, 53°C for 60 s, 72°C for 120 s, and a final extension step at 72°C for 7 min. The PCR products were purified using a GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced using Sanger sequencing (Macrogen Co., Ltd., Seoul, South Korea). The obtained nucleotide sequences of the S1 gene were assembled and aligned with the prototype of different lineages of IBV and reference strains (15, 17) shown in Supplementary Table S1 using Geneious Prime® 2022.1.1 software (https://www.geneious.com/). A phylogenetic tree was constructed in MEGA version 10.2.5, using the neighbor-joining method with 1000 bootstrap replicates.
2.3. Recombination analysis
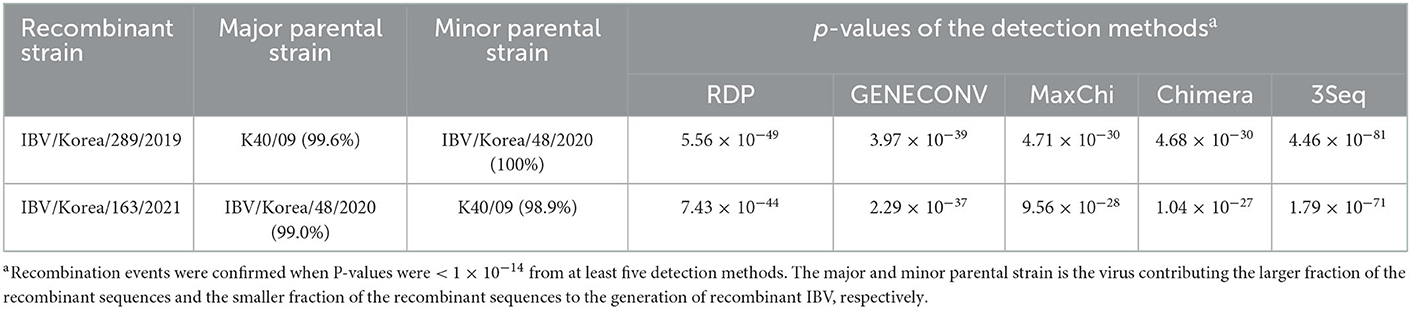
The Recombination Detection Program (RDP) 4 software (v. 4.39) was used to identify putative recombination events using several detection methods: RDP, GENECONV, BootScan, MaxChi, Chimera, SiScan, Phylpro, LARD, and 3Seq (27). Recombination events where at least five detection methods showed a p-value < 1 × 10−14 were accepted (28). After identifying their putative parental strains from 59 reference sequences, the S1 gene sequences of the IBV isolates were compared with the sequences of their putative parental strains. Nucleotide sequences from the S1 gene were used to generate similarity plot using the Simplot software (v. 3.5.1), with a window size of 200 bp and a step size of 20 bp (29).
2.4. Pathogenicity and tissue tropism investigation
One-day-old SPF chickens (n = 150) were randomly divided into five groups (Group 1: K40/09; Group 2: IBV/Korea/48/2020; Group 3: K289/19; Group 4: K163/21; Group 5: Negative control; n=30 in each group) and maintained separately in isolation cabinets (Three Shine, Daejeon, Korea). All chickens used in this study were obtained from Namduk SPF (Incheon, Republic of Korea). Chickens in the treated group were inoculated with a virus with a 105 EID50 per chick via the ocular route. Chickens from the negative control group were inoculated with sterile phosphate-buffered saline (PBS) for the same volume as the virus inoculated groups. At 5 days post-inoculation (5 dpi), twenty chicks were sacrificed from each group. The upper, middle, and lower regions of tracheal and kidney tissues were collected from ten necropsied chickens from each group for histopathological examination. Ciliary loss, inflammatory response of the trachea, and severity of kidney inflammation were scored as previously described (30). Tracheal and kidney tissues were collected from remaining 10 sacrificed chickens from each group to isolate virus. Tracheal and kidney tissues were homogenized and diluted to 10% (w/v) in PBS containing 400 mg/mL gentamicin. The supernatants of the homogenized tissue samples were clarified by centrifugation at 3,000 rpm (107 × g) for 10 min, filtered using a 0.45 μm Minisart syringe filter (Sartorius, Göttingen, Germany), followed by propagationin 10-day-old SPF embryonated chicken eggs at 37°C for 72 h. After incubation, the allantoic fluid was harvested and viral RNA was extracted. Viral replication in trachea and kidney samples was confirmed using real-time reverse transcription-PCR (rRT-PCR) as previously described (31).
Ten of thirty chicks from each group were observed for clinical signs, morbidity, and mortality for 2 weeks after experimental infection. Clinical signs were monitored twice daily for clinical signs, including rales, nasal discharge, coughing, eye irritation, depression, and watery diarrhea. The morbidity and mortality rates were monitored daily. If mortality occurred, the dead chickens were necropsied to observe gross lesions.
2.5. Antigenicity study using cross-neutralization test
Allantoic fluids of the K40/09, IBV/Korea/48/2020, K289/19, and K163/21 viruses were inactivated with 0.1% formaldehyde for 24 h at room temperature (20–22°C). Inactivated allantoic fluids were emulsified with Montanid ISA70 (Seppic, Paris, France) at ratio of 3:7 (v/v) to be injected into SPF chickens to produce antisera (32).
Two-week-old SPF chickens (n = 20) were randomly separated into four groups and each oil-emulsified inactivated virus (0.5 mL) was intramuscularly inoculated into each group. Two weeks after inoculation, all groups were given a booster of the same inactivated virus. Two weeks after boosting, hyperimmune antisera were collected from each group and inactivated at 56°C for 30 min. Virus cross-neutralization tests were performed using SPF embryonated eggs with the Alpha method according to the OIE Terrestrial Manual (33). Briefly antisera and the ten-fold dilutions of the allantoic fluids, which began from the titer of 107 EID50, were mixed and incubated at 37°C for 30 min. After incubation, the mixtures were inoculated into five SPF embryonated chicken eggs at each dilution. Eggs were inoculated with the corresponding titers of the virus with PBS in parallel. Endpoints were calculated using the Reed and Muench method (34). The neutralization index (NI) and antigenic relatedness (R-value) were calculated as previously described (19).
2.6. Statistical analyses
The ciliary loss and inflammatory response scores of test groups were compared with the score of negative group using Dunnett's multiple comparisons test. Re-isolation rate of the inoculated virus among the groups was analyzed using one-tailed Fisher's exact test. Statistical significance was set at p < 0.05. All statistical analyses were performed using GraphPad Prism version 9.2.0 software (San Diego, CA, USA).
2.7. Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Konkuk University of South Korea (permission number KU22113, July 18, 2022).
3. Results
3.1. Phylogenetic, recombination and genomic analysis
Phylogenetic analysis of the S1 gene revealed that the K289/19 and K163/21 viruses were genetically distinct from previously identified IBVs in South Korea. These viruses did not belong to the QX-like, KM91-like, or K40/09-like and GI-15 subgroups in the phylogeny (Figure 1).
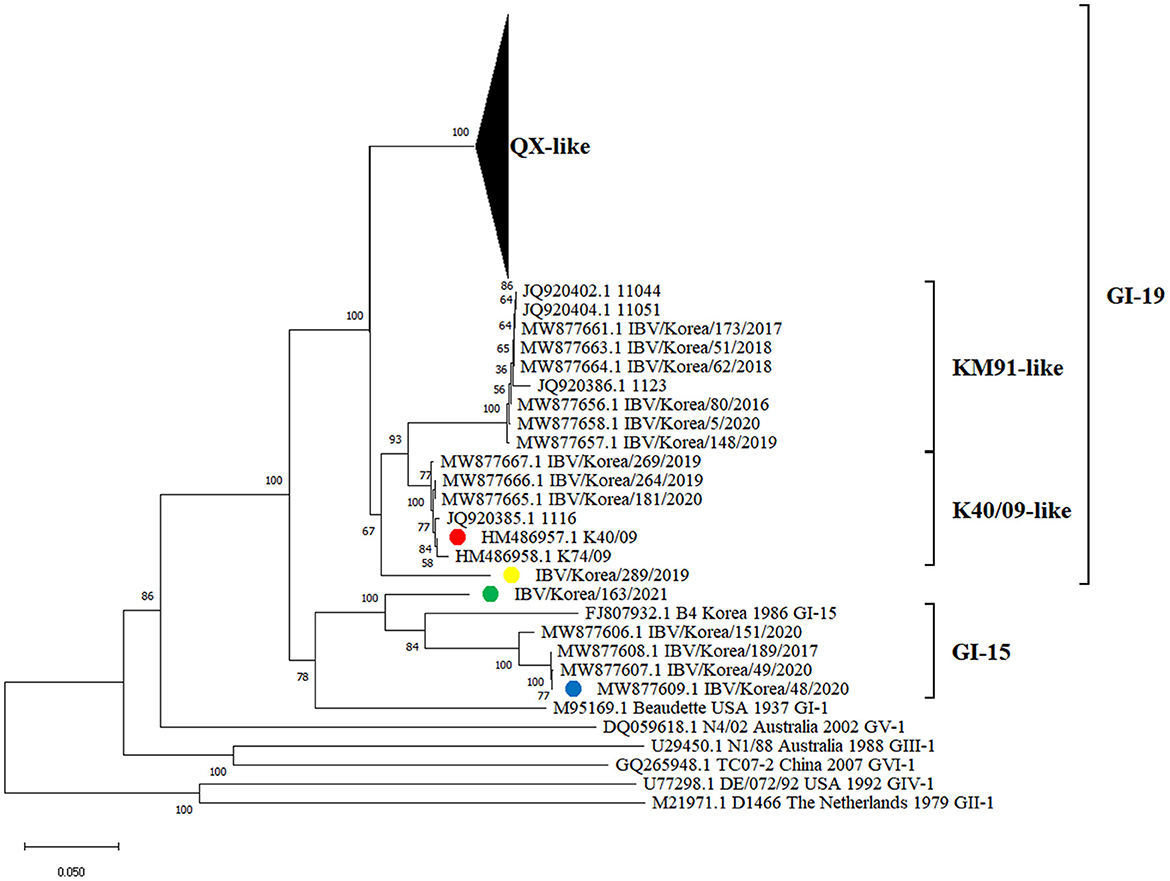
Figure 1. Phylogenetic tree based on the S1 gene nucleotide sequences of IBVs. The IBVs newly isolated in this study are highlighted in yellow and green circles (plural). Strains highlighted in red and blue circles (plural) are putative parental strains of isolates, which are identified as progenitors of recombination events using RDP4 software in this study.
Recombination events in K289/19 and K163/21 were identified using RDP4 software (Table 1). Five different methods in RDP4 and Simplot analysis showed that their putative parental strains are most likely the K40/09 strain of GI-19 and the IBV/Korea/48/2020 strain of GI-15 (p value < 1 × 10−14) (Figure 2). Other putative recombination events were excluded, because they did not satisfy the criteria for recombination event described above (p < 1 × 10−14 at least five detection methods).
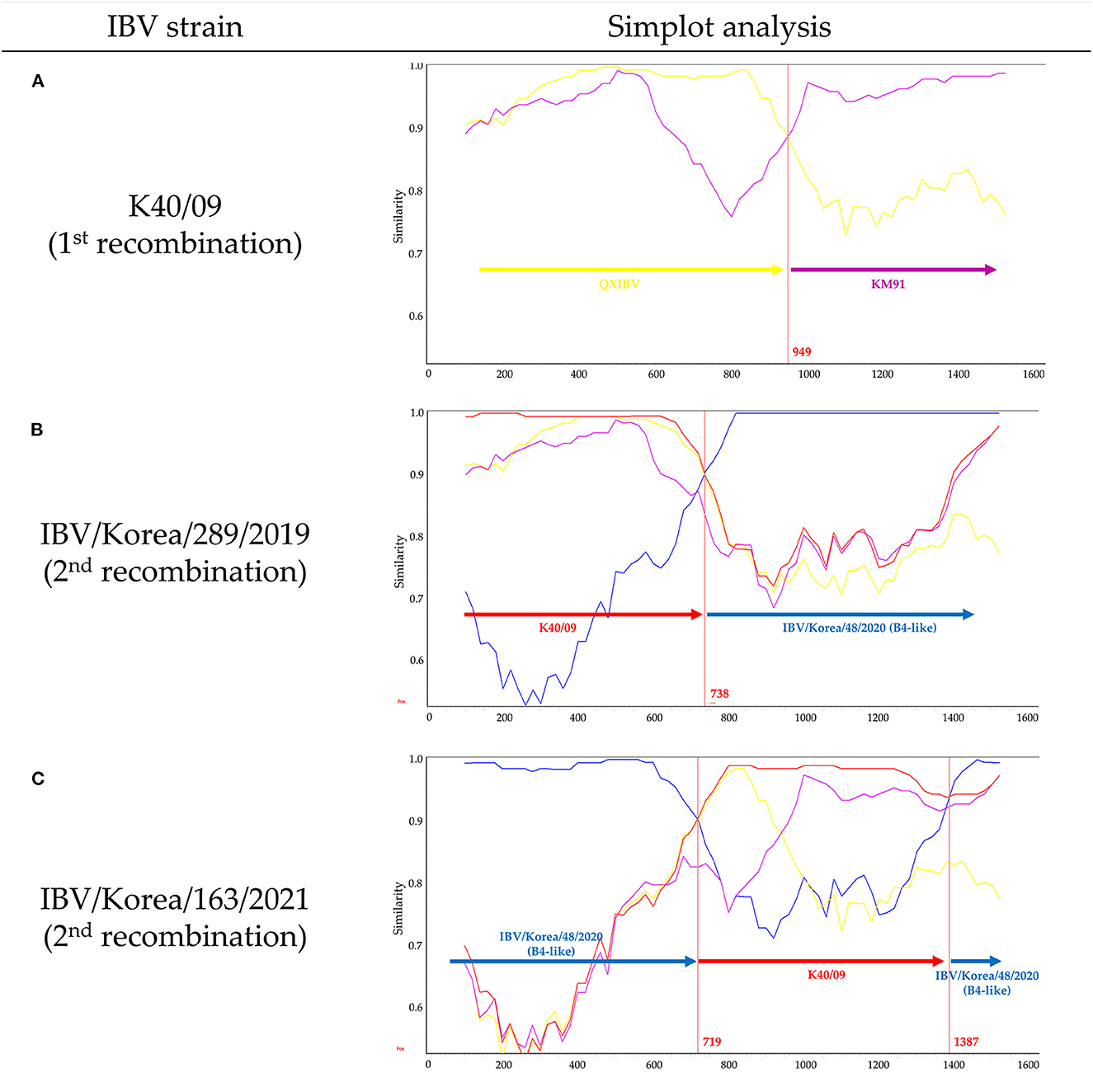
Figure 2. Evolutionary history of novel IBVs produced by multiple recombination events in Korea. Simplot analysis was performed with a window size of 200 bp and a step size of 20 bp. (A) Similarity plot of K40/09 strain, which was reported as recombinant between QXIBV (yellow) and KM91 (purple). (B) Similarity plot of K289/19. (C) Similarity plot of K163/21. Putative parental strains are highlighted in red (K40/09 strain) and blue (IBV/Korea/48/2020 strain). Red vertical lines and numbers indicate the crossover breakpoints and their nucleotide positions.
The K40/09 strain is a recombinant strain derived from QXIBV and KM91 (21) (Figure 2A). Single crossover breakpoint in the S1 gene of K289/19 virus was identified at the nucleotide position 738 bp; 1–738 bp was similar to the K40/09 strain, followed by IBV/Korea/48/2020 strain (Figure 2B). However, two crossover breakpoints were identified in the S1 gene of K163/21 virus, at nucleotide positions 719 bp and 1387 bp; the 719–1387 bp region in the S1 gene of IBV/Korea/48/2020 strain was substituted by the partial S1 gene of K40/09 strain (Figure 2C). Briefly, the S1 genes of two isolates were produced by recombination between same parental strains, the K40/09 strain and IBV/Korea/48/2020, but in different patterns.
The nucleotide sequence identity of the S1 gene of each recombinant was compared with the GI-15 and GI-19 IBVs identified previously in Korea. Nucleotide sequence identities of K289/19 were 81.9–87.8% with the GI-15 IBVs, and 85.9–91.6% with the GI-19 IBVs. Nucleotide sequence identities of K163/21 were 84.6–91.6% with the GI-15 IBVs, and the 78.7–86.8% with GI-19 IBVs (data not shown). Sequence analysis of the HVR I, II, and III in the S1 gene showed that the HVR I and II of K289/19 were similar to the K40/09 strain, except one substitution at 120th amino acid (S120R), but HVR III was similar to the IBV/Korea/48/2020 strain (Figure 3). In contrast, the HVR I and II of the isolate K163/21 was similar to the IBV/Korea/48/2020 strain, except two substitutions at 65th and 122th amino acids (E65D and N122T). The HVR III of the isolate K163/21 was similar to the K40/09 strain except two substitutions at 291th and 294th amino acids (N291D and H294N).
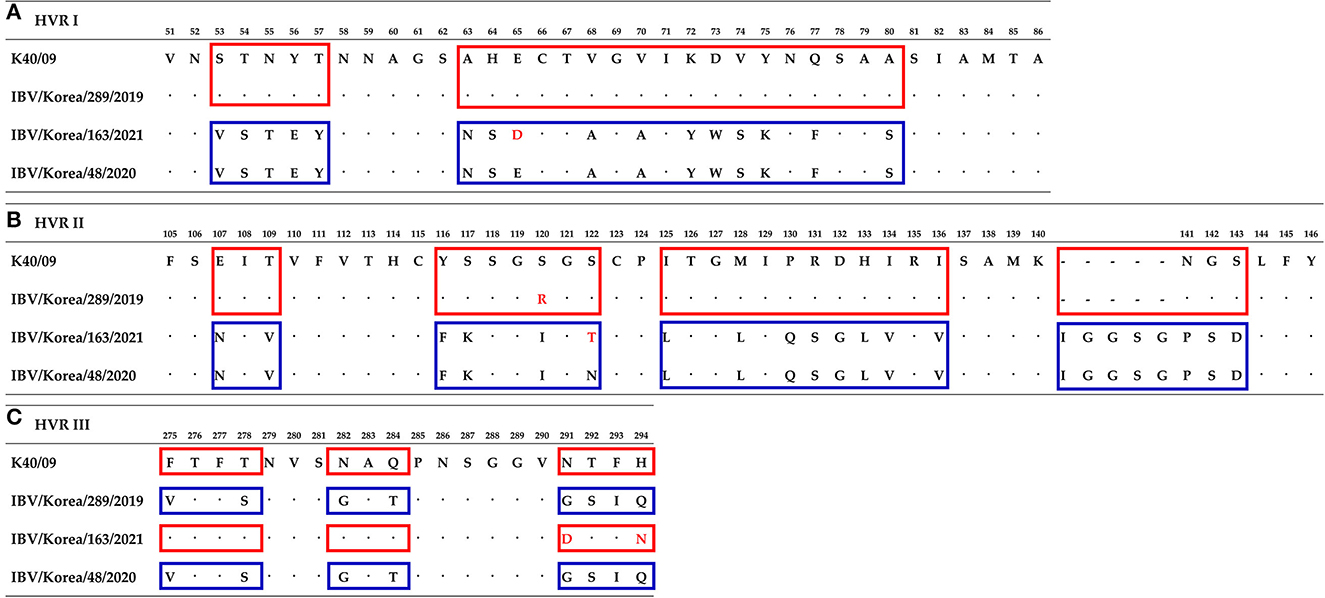
Figure 3. Comparison of amino acids sequences of hypervariable region at S1 gene between isolates and their parental strains. Numbering is based on isolate K289/19. (A) Hypervariable region I. (B) Hypervariable region II. (C) Hypervariable region III. Similar amino acids sequences with the K40/09 strain are highlighted in red boxes. Blue boxes indicate amino acids sequences similar to the IBV/Korea/48/2020 strain. Substitutions of amino acids are highlighted with red font.
3.2. Pathogenicity investigation and tissue tropism
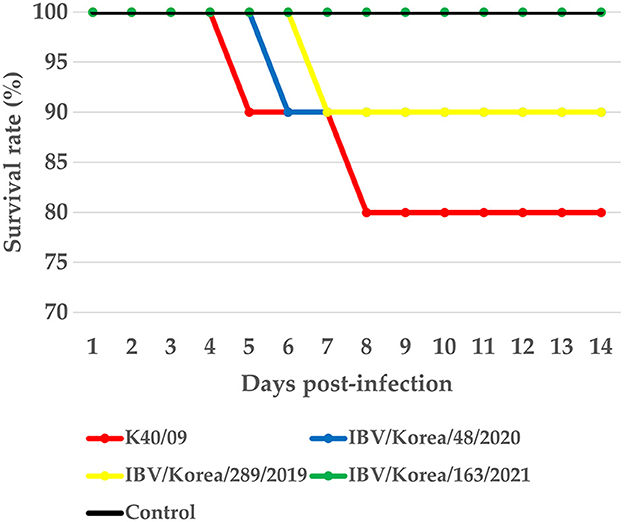
Mortality was not observed in the K163/21 challenged group or the negative control group (Figure 4). The chickens challenged with the IBV/Korea/48/2020 or K289/19 viruses exhibited 10% mortality (1/10) at 6 dpi and 7 dpi, respectively. The chickens inoculated with K40/09 strain showed 20% mortality (2/10). Mild exudates and petechial hemorrhage in the trachea were commonly observed in the dead chickens. Nephritis and urate deposition were only observed in the dead chickens challenged with the K289/19 and the K40/09 viruses, indicating nephropathogenicity of these viruses.
Histopathological examination (Figure 5) and virus re-isolation test (Table 2) were performed at 5 dpi to evaluate the pathogenicity and tissue tropism of each virus. For the ciliary loss score of upper trachea, the score of the K40/09, IBV/Korea/48/2020, K289/19 group were higher than the negative control group, followed by the score of the K163/21 group. However, there was no statistical difference compared to the negative control group (p > 0.05). Likewise, for the ciliary loss score of middle trachea, the scores of inoculated groups were higher than that of the negative control group, but there was no significant difference compared with negative control group. However, for the ciliary loss score of lower trachea, the score of K289/19 challenged group was the highest (p<0.05), followed by the K40/09 challenged group and the IBV/Korea/48/2020 challenged group. The K163/21 had a slightly higher score compared to the negative control group. Based on the results of the scoring of ciliary loss, it seems that the K289/19 was more pathogenic than its parental strains, but the K163/21 was less pathogenic than its parental strains.
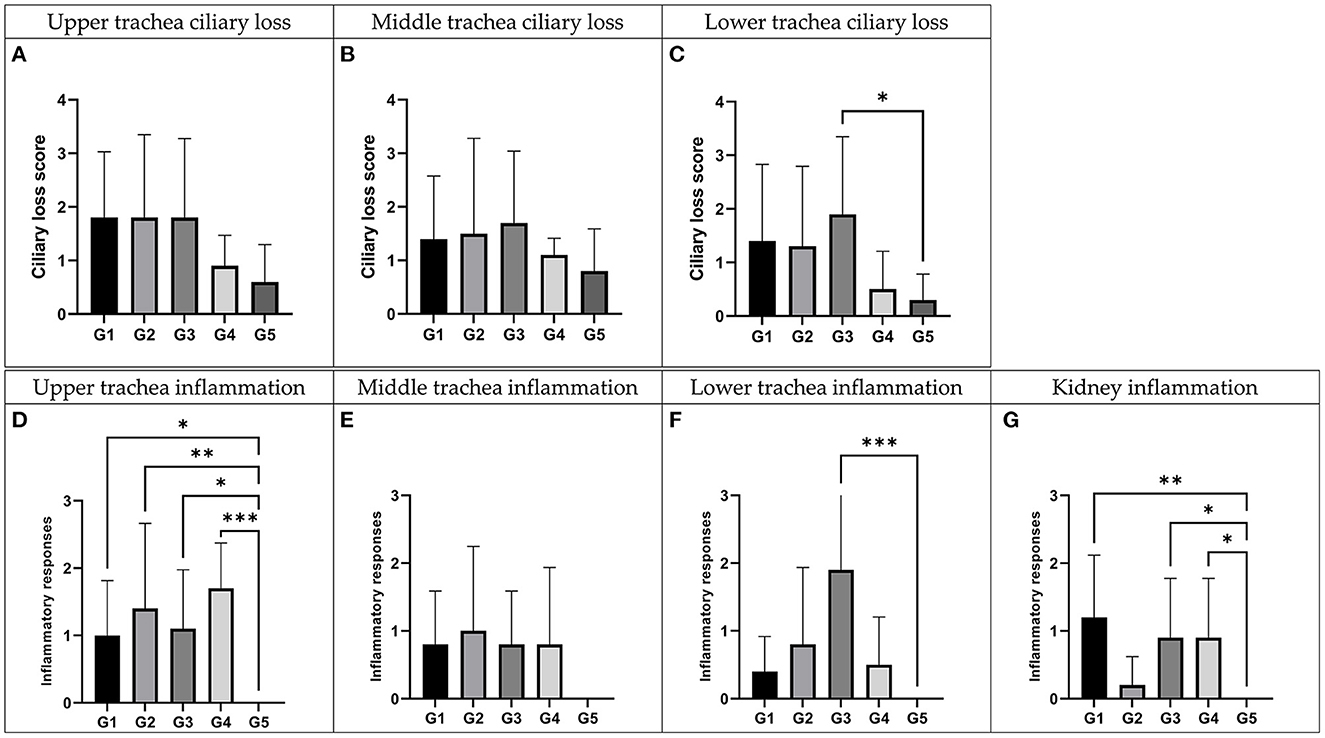
Figure 5. Histopathologic examination of recombinant strains and their parent strains. Trachea and kidney samples were collected at 5 days post-inoculation. G1: K40/09 challenged group, G2: IBV/Korea/48/2020 challenged group, G3: IBV/Korea/289/2019 challenged group, G4: IBV/Korea/163/2021 challenged and G5: negative control group. (A–C) Ciliary damages were compared with negative control. (D–G) Inflammation responses were estimated comparing with negative control. *P < 0.05, **P < 0.01, and ***P < 0.001, compared to the negative control group as determined using Dunnett's multiple comparisons test.
All the groups inoculated with each virus showed significantly higher inflammation score than the negative control group in the upper trachea (P < 0.05). Chickens inoculated with each virus showed higher inflammation score than the negative control group in the middle trachea, but there was no statistically significant difference (P > 0.05). For the inflammation score in the lower trachea, the score of K289/19 challenged group was only significantly higher than the negative control group (P < 0.001). Collectively, all inoculated viruses were identified to be pathogenic in trachea, although virulence of each virus was slightly different. The inflammation score in kidney tissues of IBV/Korea/48/2020 challenged group was similar to the negative control group. However, the K40/09 group and two novel recombinant viruses showed statistically higher inflammation scores in kidney compared to the negative control group (P < 0.05).
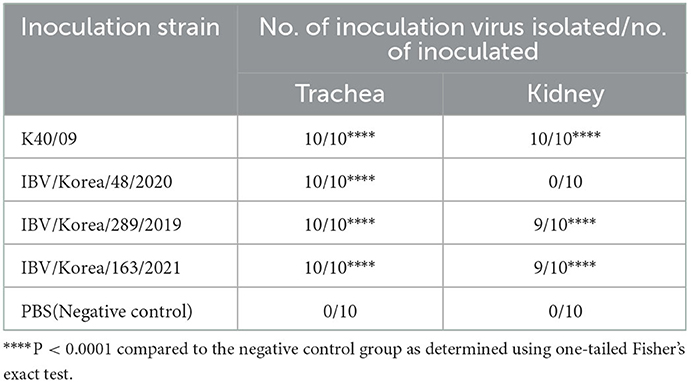
Similar results were also identified from the virus re-isolation test (Table 2). The positive rate of IBV was 100% in trachea of all groups, suggesting efficient infection and robust replication of all IBVs in the upper respiratory tract of chickens. However, the positive rates for the kidney tissues varied among the groups. Chickens inoculated with K40/09 showed a 100% re-isolation rate from kidney tissue samples. Chickens infected with K289/19 or K163/21 exhibited 90% positive rate from kidney tissues. However, the IBV/Korea/48/2020 group and the negative control group showed 0% positive rate from kidney tissues.
3.3. Antigenicity study using cross-neutralization test
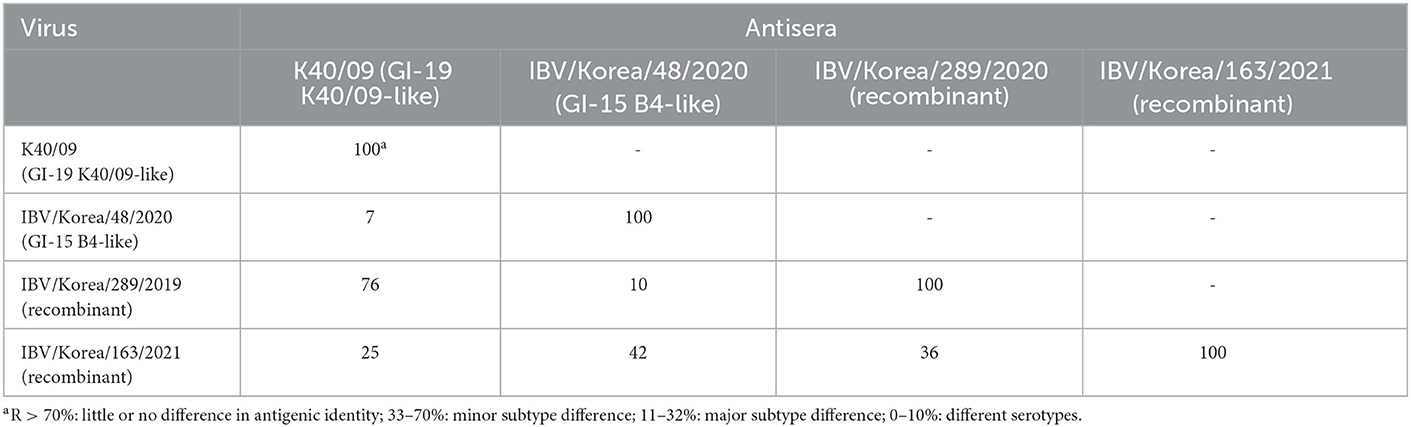
Cross-neutralization tests were performed using the recombinant viruses and their putative parental strains. NI values retrieved from the cross-neutralization tests were converted to R-values using the calculation method described by Archetti and Horsfall (35). R-values implicate antigenic relatedness between two viruses. R-values >70% indicate the same serotype, R-values between 33 and 70% indicate subtype with a minor difference, between 11 and 32% indicate a subtype with a major difference, and R-values <11% indicate a different serotype (19). Recombinant viruses showed different antigenic properties as shown in Table 3. The two parental strains, K40/09 and IBV/Korea/48/2020, were antigenically different (7%). The isolate K289/19 was antigenically close with the K40/09 strain (76%), but different with the IBV/Korea/48/2020 strain (10%). The antigenic identity of K163/21 was identified to be major subtype difference from K40/09 strain (25%) and minor subtype difference from K289/19 (36%) and IBV/Korea/48/2020 (42%).
4. Discussion
The IBV evolves through a variety of mutation mechanisms, not only accumulations of point mutations but also recombination events, which makes it difficult to perfectly prevent the infection of IBVs (3–5). In particular, the recombination between field strains and vaccine strains contributes to the sudden emergence of various strains (36). In this study, we isolated two novel recombinant IBVs (K289/19 and K163/21) and investigated the genetic characteristics based on the S1 gene and pathobiological features in chickens. Our data showed that these viruses were produced by recombination between same progenitors, the GI-19-like virus and GI-15-like virus in different patterns. Since two recombinants were identified as progenies of identical parental strains based on the S1 gene, we had assumed that these viruses may have similar pathobiological characteristics to one of their parental strains. However, the pathogenicity, tropism, and antigenicity were not identical to those of parental strains. Although the S1 subunit is a major determinant of pathogenicity, tissue tropism, and antigenicity of IBV (8–12), genetic changes in S1 gene could not explain molecular mechanism of these phenotypical changes since other genes such as S2 gene and replicase gene can also alter their pathobiological characteristics (37, 38). Whole genome sequencing would be required to better understand the molecular mechanism for these findings.
The HVRs of S1 gene are highly diverse and often associated with antigenicity, pathogenicity, and tissue tropism (13, 39). In particular, it has been reported that the HVR II is associated with nephropathogenicity (14). The K289/19, harboring HVR II derived from the nephropathogenic IBV, exhibited kidney affinity. Unexpectedly, the K163/21, harboring HVR II derived from the non-nephropathogenic IBV, also showed nephropathgenicity in chickens. It implies that the HVR II of S1 gene is not the only factor that determine the tissue tropism of IBV. Furthermore, other studies have suggested that genetic changes in other structural or non-structural protein genes could contribute to the pathogenicity and tissue tropism of coronaviruses (14, 37, 40–42). Therefore, further studies are needed to clarify the determinants of pathogenicity and tissue tropism.
Live attenuated vaccines derived from GI-15 and GI-19-like viruses have been used in Korea (30, 43). The antigenicity of the recombinant viruses was identified to be different, despite of being derived from the same parental strains. Since IBVs generally show poor cross-protective efficacy between different serotypes (44), vaccine strains should be carefully selected based on the epidemiology and antigenic properties of IBVs. It should be also noted that the pathogenicity and antigenicity of the recombinant virus in field condition could be different from laboratory animal study and insufficient efficacy of vaccines could accelerate viral evolution (45). Recently, an IBV that has highly similar recombinant pattern with the K163/21 virus was reported in Korea, which has the partial S1 gene of QX-like IBV(GI-19) between 724 and 1,102 bp in the S1 gene of GI-15 IBV (29). The emergence of such a diverse range of natural recombinant IBVs raises a concern that novel recombinants could possibly escape vaccine induced immunity and extensively spread in Korea possibly due to insufficient cross-neutralization between commercial vaccines and novel variants. Therefore, enhanced surveillance for newly emerged recombinants should be carried out, and IBV characteristics such as whole genome sequences, pathogenicity, and antigenicity should be quickly analyzed to prepare control measures.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Konkuk University IACUC.
Author contributions
C-SS: conceptualization, validation, and project administration. D-HL, H-JK, H-CL, and AC: methodology. H-JK, Y-JC, and HL: formal analysis. D-HL, H-JK, Y-JC, and HL: investigation. H-JK and H-CL: resources. H-JK and AC: data curation. H-JK: writing—original draft preparation and visualization. D-HL, H-CL, AC, and C-SS: writing—review and editing. D-HL and C-SS: supervision and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Animal Disease Management Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (grant number: 122057-2).
Acknowledgments
We thank Ye-Ram Seo from Konkuk University for technical support.
Conflict of interest
H-CL and C-SS were employed by KHAV Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1107059/full#supplementary-material
References
1. Jackwood MW, de Wit S. Infectious bronchitis. In:Swayne DE, Boulianne M, Logue CM, McDougald LR, Nair V, Suarez DL, Wit S, Grimes T, Johnson D, Kromm M, Prajitno TY, Rubinoff I, Zavala G, , editors. Diseases of Poultry. (2020). doi: 10.1002/9781119371199.ch4
2. Abdel-Moneim AS. Coronaviridae: Infectious Bronchitis Virus. In:Bayry J, , editor. Emerging and Re-emerging Infectious Diseases of Livestock. Cham: Springer International Publishing. (2017). p. 133–66.
3. Cavanagh D, Casais R, Armesto M, Hodgson T, Izadkhasti S, Davies M, et al. Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine. (2007) 25:5558–62. doi: 10.1016/j.vaccine.2007.02.046
4. Liu XL, Su JL, Zhao JX, Zhang GZ. Complete genome sequence analysis of a predominant infectious bronchitis virus (IBV) strain in China. Virus Genes. (2009) 38:56–65. doi: 10.1007/s11262-008-0282-5
5. Legnardi M, Tucciarone CM, Franzo G, Cecchinato M. Infectious bronchitis virus evolution, diagnosis and control. Vet Sci. (2020) 7:79. doi: 10.3390/vetsci7020079
6. Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. (2007) 38:281–97. doi: 10.1051/vetres:2006055
7. Bande F, Arshad SS, Bejo MH, Moeini H, Omar AR. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J Immunol Res. (2015) 2015:424860. doi: 10.1155/2015/424860
8. Moore KM, Jackwood MW, Hilt DA. Identification of amino acids involved in a serotype and neutralization specific epitope with in the s1 subunit of avian infectious bronchitis virus. Arch Virol. (1997) 142:2249–56. doi: 10.1007/s007050050239
9. Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol. (2000) 145:291–300. doi: 10.1007/s007050050024
10. Alvarado IR, Villegas P, Mossos N, Jackwood MW. Molecular characterization of avian infectious bronchitis virus strains isolated in Colombia during 2003. Avian Dis. (2005) 49:494–9. doi: 10.1637/7202-050304R.1
11. Zanaty A, Arafa AS, Hagag N, El-Kady M. Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J Virol. (2016) 5:125–34. doi: 10.5501/wjv.v5.i3.125
12. Cavanagh D, Davis PJ, Mockett APA. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. (1988) 11:141–50. doi: 10.1016/0168-1702(88)90039-1
13. Promkuntod N, van Eijndhoven RE, de Vrieze G, Grone A, Verheije MH. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. (2014) 448:26–32. doi: 10.1016/j.virol.2013.09.018
14. Bouwman KM, Parsons LM, Berends AJ, Vries RPd, Cipollo JF, Verheije MH. Three amino acid changes in avian coronavirus spike protein allow binding to kidney tissue. J Virol. (2020) 94:e01363–19. doi: 10.1128/JVI.01363-19
15. Valastro V, Holmes EC, Britton P, Fusaro A, Jackwood MW, Cattoli G, et al. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet Evol. (2016) 39:349–64. doi: 10.1016/j.meegid.2016.02.015
16. Feng K, Wang F, Xue Y, Zhou Q, Chen F, Bi Y, et al. Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013-2015. Sci Rep. (2017) 7:6576. doi: 10.1038/s41598-017-06987-2
17. Lee HC, Jeong S, Cho AY, Kim KJ, Kim JY, Park DH, et al. Genomic analysis of avian infectious bronchitis viruses recently isolated in South Korea reveals multiple introductions of GI-19 Lineage (QX Genotype). Viruses. (2021) 13:1045. doi: 10.3390/v13061045
18. Lee HJ, Youn HN, Kwon JS, Lee YJ, Kim JH, Lee JB, et al. Characterization of a novel live attenuated infectious bronchitis virus vaccine candidate derived from a Korean nephropathogenic strain. Vaccine. (2010) 28:2887–94. doi: 10.1016/j.vaccine.2010.01.062
19. Choi KS, Lee EK, Jeon WJ, Park MJ, Kim JW, Kwon JH. Pathogenicity and antigenicity of a new variant of Korean nephropathogenic infectious bronchitis virus. J Vet Sci. (2009) 10:357–9. doi: 10.4142/jvs.2009.10.4.357
20. Lee EK, Jeon WJ, Lee YJ, Jeong OM, Choi JG, Kwon JH, et al. Genetic diversity of avian infectious bronchitis virus isolates in Korea between 2003 and 2006. Avian Dis. (2008) 52:332–7. doi: 10.1637/8117-092707-ResNote.1
21. Lim TH, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, et al. An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect Genet Evol. (2011) 11:678–85. doi: 10.1016/j.meegid.2011.01.007
22. Yu L, Jiang Y, Low S, Wang Z, Nam SJ, Liu W, et al. Characterization of three infectious bronchitis virus isolates from China Associated with Proventriculus in Vaccinated Chickens. Avian Dis. (2001) 45:416–24. doi: 10.2307/1592981
23. Toffan A, Bonci M, Bano L, Bano L, Valastro V, Vascellari M, et al. Diagnostic and clinical observation on the infectious bronchitis virus strain Q1 in Italy. Vet Ital. (2013) 49:347–55. doi: 10.12834/VetIt.1303.01
24. Lim TH, Kim MS, Jang JH, Lee DH, Park JK, Youn HN, et al. Live attenuated nephropathogenic infectious bronchitis virus vaccine provides broad cross protection against new variant strains. Poult Sci. (2012) 91:89–94. doi: 10.3382/ps.2011-01739
25. Li L, Xue C, Chen F, Qin J, Xie Q, Bi Y, et al. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet Microbiol. (2010) 143:145–54. doi: 10.1016/j.vetmic.2009.11.022
26. Kint J, Maier HJ, Jagt E. Quantification of infectious bronchitis coronavirus by titration in vitro and in ovo. Methods Mol Biol. (2015) 1282:89–98. doi: 10.1007/978-1-4939-2438-7_9
27. Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. (2015) 1:vev003. doi: 10.1093/ve/vev003
28. Yan W, Qiu R, Wang F, Fu X, Li H, Cui P, et al. Genetic and pathogenic characterization of a novel recombinant avian infectious bronchitis virus derived from GI-1, GI-13, GI-28, and GI-19 strains in Southwestern China. Poult Sci. (2021) 100:101210. doi: 10.1016/j.psj.2021.101210
29. Youn SY, Lee JY, Bae YC, Kwon YK, Kim HR. Genetic and Pathogenic characterization of QX(GI-19)-recombinant infectious bronchitis viruses in South Korea. Viruses. (2021) 13:1163. doi: 10.3390/v13061163
30. Lim TH, Youn HN, Yuk SS, Kwon JH, Hong WT, Gwon GB, et al. Successful cross-protective efficacy induced by heat-adapted live attenuated nephropathogenic infectious bronchitis virus derived from a natural recombinant strain. Vaccine. (2015) 33:7370–4. doi: 10.1016/j.vaccine.2015.07.043
31. Callison SA, Hilt DA, Boynton TO, Sample BF, Robison R, Swayne DE, et al. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J Virol Methods. (2006) 138:60–5. doi: 10.1016/j.jviromet.2006.07.018
32. Radmehri M, Talebi A, Ameghi Roudsari A, Mousaviyan SM, Gholipour MAJ, Taghizadeh M. Comparative study on the efficacy of MF 59, ISA70 VG, and nano-aluminum hydroxide adjuvants, alone and with nano-selenium on humoral immunity induced by a bivalent Newcastle+Avian influenza vaccine in chickens. Arch Razi Inst. (2021) 76:1213–20. doi: 10.22092/ari.2021.356666.1887
33. OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals chapter 3. 3. 2. Avian infectious bronchitis virus. 2018. World Organisation for Animal Health fouded as OIEAvian Infectious Bronchitis Virus. [801]. Available online at: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ (accessed May, 2018).
34. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints12. Am J Epidemiol. (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
35. Archetti I, Horsfall FL Jr. Persistent antigenic variation of influenza a viruses after incomplete neutralization in ovo with heterologous immune serum. J Exp Med. (1950) 92:441–62. doi: 10.1084/jem.92.5.441
36. Bande F, Arshad SS, Omar AR, Hair-Bejo M, Mahmuda A, Nair V. Global distributions and strain diversity of avian infectious bronchitis virus: a review. Anim Health Res Rev. (2017) 18:70–83. doi: 10.1017/S1466252317000044
37. Armesto M, Cavanagh D, Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS ONE. (2009) 4:e7384. doi: 10.1371/journal.pone.0007384
38. Cheng J, Zhao Y, Xu G, Zhang K, Jia W, Sun Y, et al. The S2 Subunit of QX-type infectious bronchitis coronavirus spike protein is an essential determinant of neurotropism. Viruses. (2019) 11:972. doi: 10.3390/v11100972
39. Shan D, Fang S, Han Z, Ai H, Zhao W, Chen Y, et al. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. (2018) 250:104–13. doi: 10.1016/j.virusres.2018.04.013
40. Liu S, Zhang X, Gong L, Yan B, Li C, Han Z, et al. Altered pathogenicity, immunogenicity, tissue tropism and 3'-7kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. (2009) 27:4630–40. doi: 10.1016/j.vaccine.2009.05.072
41. Phillips JE, Jackwood MW, McKinley ET, Thor SW, Hilt DA, Acevedol ND, et al. Changes in nonstructural protein 3 are associated with attenuation in avian coronavirus infectious bronchitis virus. Virus Genes. (2012) 44:63–74. doi: 10.1007/s11262-011-0668-7
42. Lin SY, Li YT, Chen YT, Chen TC, Hu CJ, Chen HW. Identification of an infectious bronchitis coronavirus strain exhibiting a classical genotype but altered antigenicity, pathogenicity, and innate immunity profile. Sci Rep. (2016) 6:37725. doi: 10.1038/srep37725
43. Choi KS, Jeon WJ, Lee EK, Kye SJ, Park MJ, Kwon JH. Development of an attenuated vaccine strain from a korean respiratory type infectious bronchitis virus. Korean J Vet Res. (2011) 51:193–201. doi: 10.14405/kjvr.2011.51.3.193
44. Kamble NM, Pillai AS, Gaikwad SS, Shukla SK, Khulape SA, Dey S, et al. Evolutionary and bioinformatic analysis of the spike glycoprotein gene of H120 vaccine strain protectotype of infectious bronchitis virus from India. Biotechnol Appl Biochem. (2016) 63:106–12. doi: 10.1002/bab.1298
Keywords: infectious bronchitis virus, recombination, pathogenicity, antigenicity, tissue tropism
Citation: Kim H-J, Lee H-C, Cho AY, Choi Y-J, Lee H, Lee D-H and Song C-S (2023) Novel recombinant avian infectious bronchitis viruses from chickens in Korea, 2019–2021. Front. Vet. Sci. 10:1107059. doi: 10.3389/fvets.2023.1107059
Received: 24 November 2022; Accepted: 11 January 2023;
Published: 01 February 2023.
Edited by:
Christina Leyson, Agricultural Research Service (USDA), United StatesReviewed by:
Henry Muriuki Kariithi, Agricultural Research Service (USDA), United StatesANa Da Silva, University of California, Davis, United States
Copyright © 2023 Kim, Lee, Cho, Choi, Lee, Lee and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Hun Lee,  ZG9uZ2h1bmxlZUBrb25rdWsuYWMua3I=; Chang-Seon Song,
ZG9uZ2h1bmxlZUBrb25rdWsuYWMua3I=; Chang-Seon Song,  c29uZ2NzQGtvbmt1ay5hYy5rcg==
c29uZ2NzQGtvbmt1ay5hYy5rcg==
 Hyun-Jin Kim
Hyun-Jin Kim Hyuk-Chae Lee2
Hyuk-Chae Lee2 Andrew Y. Cho
Andrew Y. Cho Yun-Jeong Choi
Yun-Jeong Choi Heesu Lee
Heesu Lee Dong-Hun Lee
Dong-Hun Lee Chang-Seon Song
Chang-Seon Song