- 1College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Chengdu Center for Animal Disease Prevention and Control, Chengdu, China
- 3Key Laboratory for Animal Disease Resistant Nutrition of the Ministry of Education, Animal Nutrition Institute, Sichuan Agricultural University, Chengdu, China
- 4Key Laboratory of Agricultural Bioinformatics, Ministry of Education, Sichuan Agricultural University, Chengdu, China
Porcine circovirus (PCV) typically causes severe immune suppression in pigs, leading to mixed clinical infections with various pathogens that can cause significant harm to the pig industry. PCV has four subgenotypes, with PCV4 being an emerging virus that requires investigation due to its potential for epidemic outbreaks. Therefore, there is a need to develop a method that can detect all four PCV strains simultaneously. In this study, four pairs of specific primers and TaqMan probes were designed based on the conserved sequence of the PCV1–4 ORF2 gene to establish a PCV1–4 TaqMan multiplex real-time quantitative PCR method. The novel method was compared to six commercial testing kits for its efficacy. Then, a total of 595 mixed samples of spleen and lymph node collected from 12 districts in Chengdu from July to December 2021 were tested using the novel method. The results showed that the novel PCV1–4 TaqMan multiplex real-time quantitative PCR detection method has satisfied specificity, sensitivity, and repeatability. The positive rates of PCV1, PCV2, and PCV3 in Chengdu were 2.18%, 31.60%, and 15.29%, respectively, while no positive PCV4 was detected. The mixed infection rate of PCV2 and PCV3 was 5.21%. Our novel method may be as a potential method for PCV1–4 detection. Currently, PCV2 is the main epidemic PCV subtype in Chengdu, while the potential threat of PCV4 should also be considered.
1 Introduction
The pig industry is one of the pillar industries in the world, playing a crucial role in ensuring people's livelihoods and promoting economic growth (1, 2). However, the industry has been facing challenges due to the prevalence of porcine circovirus (PCV), which is often referred to as the “invisible killer,” that cause recessive and mixed infections of multiple pathogens without typical clinical symptoms (3). PCV can be subdivided into four genotypes, namely porcine circovirus type 1–4 (PCV1–4), based on differences in nucleotide homology. Among these, PCV4 is a novel circovirus that was first discovered in 2019 and is currently endemic only in Asia, mainly in China (4–7).
The complete genome size of PCV is ~1.7–2.0 kb (8). The ORF2 is located on the negative strand of the virus and encodes the capsid protein (Cap), which is one of the most crucial antigenic sites among all four subtypes of PCV (9, 10). As the outermost structural component of the virus, the Cap protein directly interacts with the host immune system, resulting in substantial immunological pressure (11–13). Consequently, it undergoes a high rate of amino acid variation in order to evade immune responses (14). When comparing the amino acid sequences of Cap proteins from PCV subtypes 1–4, the homology ranges from 24.5 to 64% (7, 9). Since the significant antigenic response and the lower genetic relatedness of ORF2, it can be used for detecting and distinguishing different subtypes of PCV (15–17).
Currently, commercial diagnostic kits for PCV primarily focus on detecting PCV2 (18), which may overlook the detection of other subtypes (19). Therefore, there is a need to develop a method that can detect all four PCV strains simultaneously. Additionally, it is unclear about the specific infection situation in Chengdu, which is one of the most renowned pig farming regions in Southwest China (20). This study established a TaqMan multiplex real-time quantitative PCR (qPCR) detection method for PCV1–4, evaluated its amplification efficiency and accuracy, and compared it with a commercial detection kit. Furthermore, the method was preliminarily applied in Chengdu.
2 Materials and methods
2.1 Primer probe design
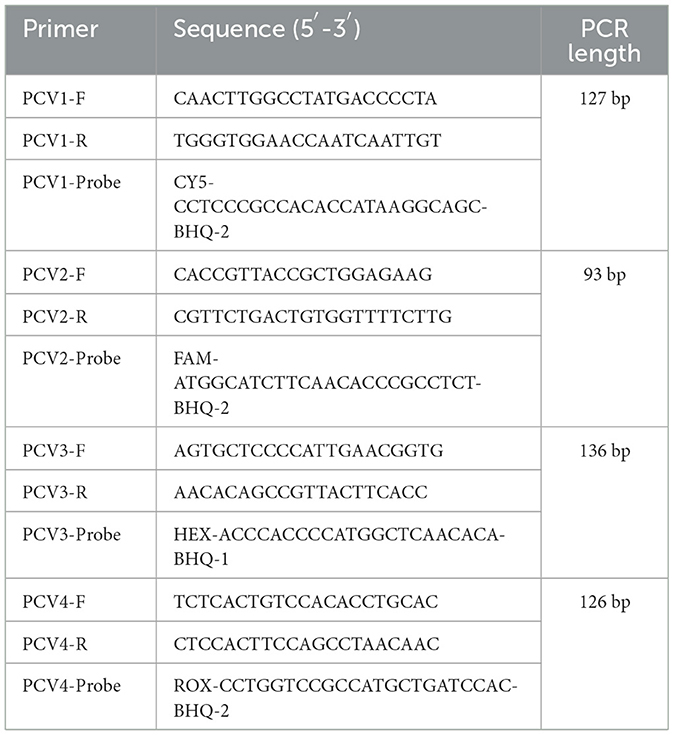
ORF2 gene sequences of PCV1–4 from different countries published in NCBI GeneBank were selected for sequence alignment using SnapGene software (GSL Biotech; available at snapgene.com). The highly conserved region in ORF2 gene was selected to design primer probes using OLIGO 7 primer analysis software (21). The designed primers were compared with NCBI for Blast to verify the specificity. Primers and probes were synthesized by Qink Bio (Chongqing) Co., Ltd., listed in Table 1.
2.2 Primer probe design plasmids synthesis
A set of plasmids containing the complete genomes of PCV1–4 were designed based on the reference sequences for PCV1–4 types available in GenBank (accession numbers: EF533941, AF201310, MG778698, MT311852, respectively). These plasmids were synthesized (Qink Bio, Chongqing Co., Ltd.) at a concentration of 100 ng/μl, named pUC57-PCV1-Complete, pUC57-PCV2-Complete, pUC57-PCV3-Complete, and pUC57-PCV4-Complete, respectively.
The pClone007 Simple Vector Kit was used to perform T-A ligation. Trelief TM 5α competent cells were used for transformation immediately following the instructions. The transformed cells were incubated in LB medium containing 50 μg/ml Ampicillin at 37°C for 12 h. Monoclonal colonies were then inoculated in LB liquid medium and shaken overnight.
2.3 Optimization of TaqMan multiplex real-time fluorescence quantitative PCR amplification reaction conditions
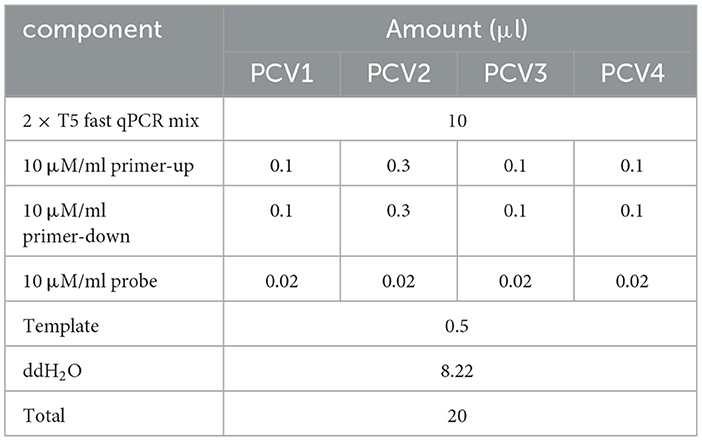
The TaqMan multiplex real-time fluorescence quantitative PCR amplification was performed using the specific primers and probes designed in this experiment, with the recombinant plasmid DNA as the template. The amplification was performed with the reaction conditions: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 10 s and annealing/extension at 60°C for 50 s. The original reaction system shown in Table S1.
A matrix optimization was performed for the parameters, including the concentrations of primers (0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, and 0.5 μM), probe (0.01, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35 μM), annealing temperatures (65, 64, 63, 62, 61, 60, 59, and 58°C) and cycle number (30, 35, and 40). The smallest Ct value was used for determining the excellent reaction conditions. When the Ct values were the same, the higher annealing temperature was selected.
2.4 Verification of the novel method
The positive standard plasmids were diluted to 1 × 103-1 × 107 copies/μl. 0.5 μl of each plasmid were selected as the templates for reaction system. The negative control template was 0.5 μl of dd H2O. Multiple quantitative PCR amplification was performed according to the optimized reaction conditions. The amplification results were analyzed using Bio-Rad CFX Maestro, a quantitative PCR data analysis software, to establish standard curves. The standard plasmids were further diluted to 1 × 104-1 × 100 copies/μl, that were selected as the template for performing the sensitivity test.
In order to better simulate clinical examination and study its specificity and repeatability, we randomly diluted different concentrations of the four viral plasmids, detected their plasmid concentrations by A260/A280 (Qubit 4 Fluorometer, Thermo Fisher Scientific). DNA or reversed cDNA of PRV, ASFV, PPV, and PRRSV, PoRV, PEDV and PK-15 cell line and blank DH5α competent cells were selected as templates for verify the specificity of TaqMan multiplex real-time PCR method. The TaqMan multiple real-time PCR amplification was performed for three repetitions.
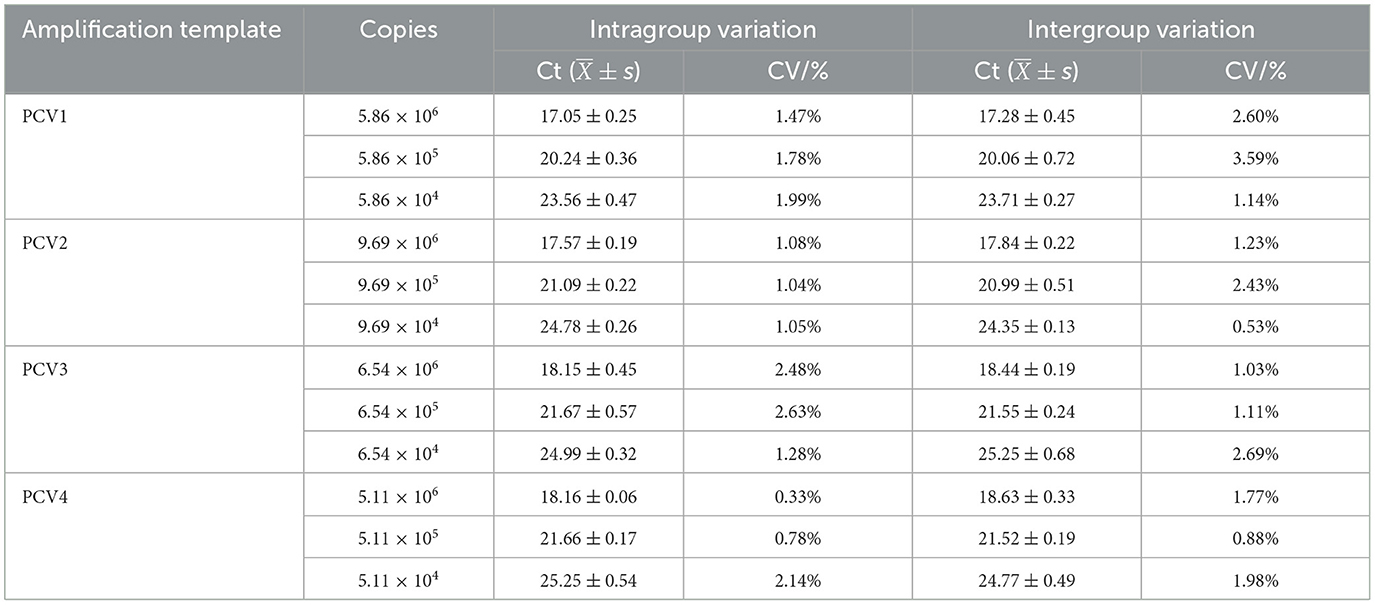
The above positive standards were collected and tested three times at every week interval under the same reaction conditions, which was used as the repeated test between groups. The intragroup and intergroup coefficients of variation were calculated to verify the reproducibility of the established TaqMan multiplex real-time PCR method.
2.5 Comparison of our novel method to six commercial kits
For further confirming the detection accuracy of our method, six commercial TaqMan real-time fluorescence quantitative PCR detection kits for PCV were purchased from three different manufacturers, named A1, A2, B1, B2, C1, and C2 (specific parameters for each kit listed in Table S2). The plasmids containing complete genomes of PCV1–4 were random diluted (Table S3) and then detected by our method and all the commercial kits. Please note the specific parameters may be various according to the manufacturer manual. Thirty mixed samples of spleen and lymph node collected from Chengdu humane slaughterhouse were also detected as blind samples.
2.6 Preliminary application of our method for Chengdu
From July to December 2021, a total of 595 spleen and lymph node mixed samples from humane slaughterhouse were collected from 15 districts in Chengdu. The positive control used the plasmids containing the complete genomes of PCV1–4 described previously. The preliminary application of our method for Chengdu was performed for the detection of PCV1–4.
3 Results
3.1 Preliminary TaqMan multiplex real-time fluorescence quantitative PCR
The amplification results showed typical amplification curves for all four positive plasmids, while not for negative control, indicating that the preliminary designed primers and probes have good potential for further optimization (Figure 1).
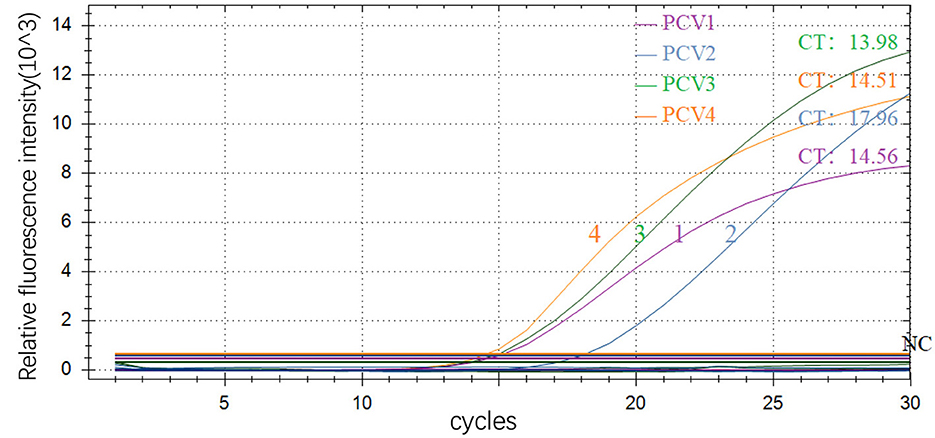
Figure 1. Preliminary amplification results of multiple real-time fluorescence quantitative PCR 1–4, PCV1, PCV2, PCV3 and PCV4; NC: negative control.
For optimization of reaction conditions, at 59–61°C under the same template concentration, the amplification efficiency and fluorescence intensity were consistent, with little change of Ct value. Since higher annealing temperature may reduce non-specific amplification, the annealing temperature was finally chosen to be 61°C. For the final primer concentration optimization, PCV1, PCV3, and PCV4 required a primer concentration of 0.05 μmol/ml, while PCV2 required a concentration of 0.15 μmol/ml. The probe concentration was set to 0.01 μmol/ml for all targets (Table 2).
After optimization, the amplification curve (Figure 2A) and standard curve (Figure 2B) of TaqMan multiplex real-time PCR showed a good linear relationship between Ct value (y) and positive standard copy number (x) in the gradient concentration range of 107-103 copies/μl. The standard curves corresponding to the species target genes were as follows: PCV1: y = −3.293x + 36.838, R2 = 1.000, amplification efficiency (E) = 101.2%; PCV2: Y = −3.515x + 38.358, R2 = 0.998, amplification efficiency (E) = 92.5%; PCV3: y = −3.025x + 36.828, R2 = 0.999, amplification efficiency (E) = 114.1%; PCV4: y = −3.342x + 38.392, R2 = 0.999, amplification efficiency (E) = 99.2%.
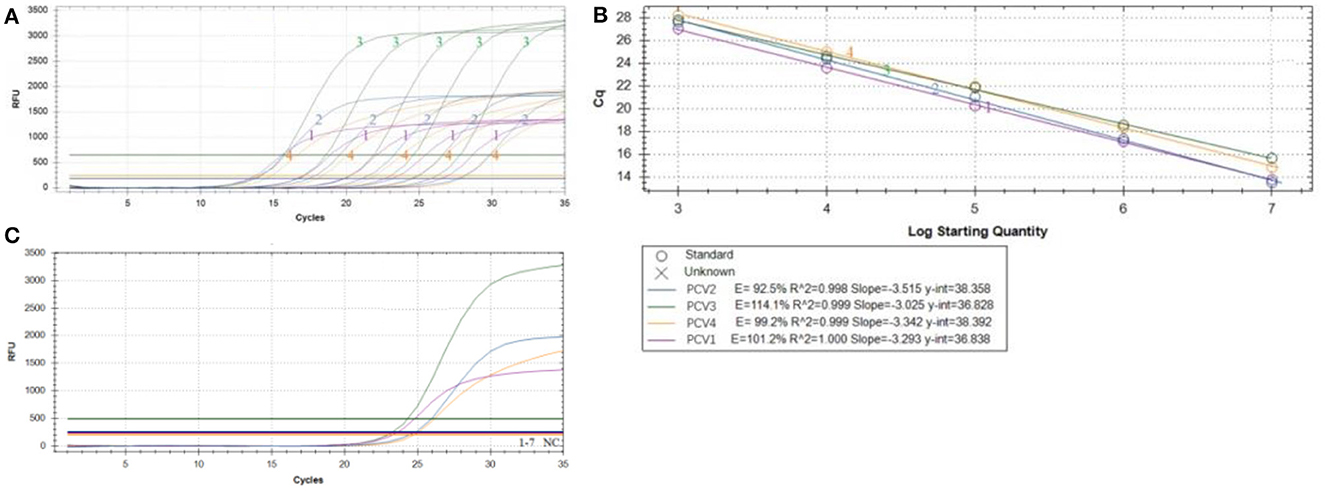
Figure 2. Multiplex qPCR amplification curve. The amplification curve (A) and standard curve (B) of TaqMan multiplex real-time PCR using positive standard plasmid with concentration gradient of 1 × 103-1 × 107copies/μl; (C) Specificity of multiplex qPCR amplification of PCV1–4 PRV, ASFV, PPV, PRRSV, PoRV, PEDV and PK-15 cell, please note the original Plasmid concentration of PCV1–4 were: PCV1 5.86 × 101 copies/μl, PCV2 9.69 × 102 copies/μl, PCV3 6.54 × 101 copies/μl, PCV4 5.11 × 101 copies/μl, respectively.
3.2 Sensitivity, specificity and repeatability of our novel detect method
The TaqMan multiplex real-time PCR method showed a good minimum copy number detection limit for each of the PCV targets: PCV1 (5.86 × 101 copies/μl), PCV2 (9.69 × 102 copies/μl), PCV3 (6.54 × 101 copies/μl), and PCV4 (5.11 × 101 copies/μl). Specificity testing demonstrated that only the PCV plasmid yielded typical amplification curves, while other pathogens and cells (including PRV, ASFV, PPV, PRRSV, PoRV, PEDV, and PK-15 cells) did not show amplification curves, indicating the method's good specificity (Figure 2C). For repeatability, the intragroup coefficient of variation ranged from 0.33 to 2.48%, and the intergroup coefficient of variation ranged from 0.53 to 3.59% (Table 3), all of which were < 5%, indicating that the established method exhibited good repeatability.
3.3 Comparison of our novel method to six commercial kits
The comparison between our novel method and six different commercial test kits revealed that the lowest detectable copy number ranged from 5 × 101 copies/μl to 5 × 103 copies/μl in different genotypes. In most cases, our novel method detected similar or fewer copies than the commercial test kits (Table S4). The coefficient of variation for all seven methods ranged from 0.52 to 2.86%, indicating good reproducibility and suggesting that both the novel method and the commercial test kits are reliable (Table S5). Using 30 clinical mixed spleen and lymph node samples, we found that the novel method and test kits provided similar results to the standard plasmids. Of the 30 tissue samples, the agreement rates between our novel method and the test kits ranged from 73.33 to 96.67%, with significant correlation coefficients of 0.877–0.977 (Table S6). However, no samples were detected with PCV4. Overall, our novel method performed comparably or even better than the commercial test kits, indicating that it could be a viable alternative for PCV detection in clinical samples.
3.4 Clinical detection using the TaqMan multiplex real-time fluorescence quantitative PCR
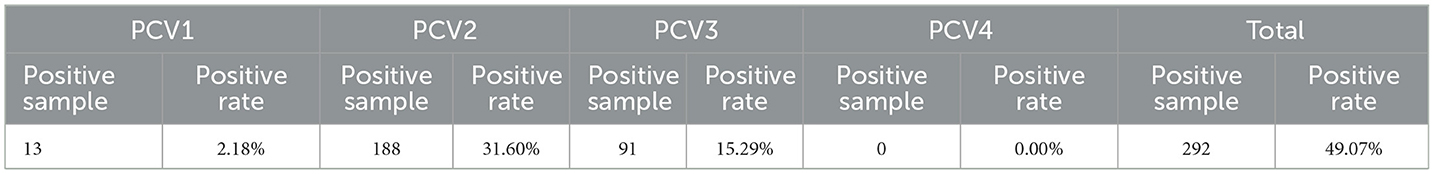
Using the TaqMan multiplex real-time fluorescence quantitative PCR method established in this study, we tested a total of 595 spleen and lymph node mixed samples from dead pigs sent to harmless treatment plants from slaughterhouse in 15 districts in Chengdu city from July to December 2021 for PCV. The total number of PCV-positive samples was 292, resulting in a positive rate of 49.07%, with a mixed infection positive rate of 6.39% (Table 4). The infection was still dominated by PCV2 and PCV3 mixed infection, accounting for approximately 81% of the total mixed infections.
4 Discussion
PCV1 was first identified as a PK15 porcine kidney cell line contaminant in 1974. PCV2 and PCV3 were subsequently isolated and identified in 1991 and 2016, respectively (9, 22). Both types of viruses are highly infectious to pigs and often cause PMWS, PDNS, PRDC, and PNP in clinical practice. PCV4 was first discovered in Hunan Province in 2019 (9, 23, 24) and has been detected in several other provinces in China as well as in South Korea (25, 26).
In this study, a convenient PCV1–4 TaqMan multiplex real-time PCR detection method was established based on the same principle as ordinary PCR. However, this multiplex PCR method adds multiple primer pairs to the reaction system to obtain multiple target genes through specific amplification of different templates or different regions of the same template. Real-time quantitative PCR methods are mainly divided into SYBR dye method and TaqMan probe method (27). Compared with ordinary PCR and SYBR dye method, TaqMan probe method avoids false positives caused by primer dimer, does not require a dissolution curve, and has high specificity, making it possible to detect multiple pathogens at the same time (28, 29).
Specificity and sensitivity are two basic characteristics of PCR diagnostic reagents and also two important indexes that directly determine the performance of the reagents (30). Factors affecting these characteristics in PCR reactions include primer design, primer concentration, annealing temperature, substrate concentration, elongation time (31–33). Generally, excessively high primer concentration leads to increased non-specific amplification products, while excessively low primer concentration does not meet the needs of the exponential amplification period, both of which affect amplification efficiency. Similarly, excessively high or low probe concentration affects the accuracy of experimental results (34). Therefore, this experiment optimized primer and probe concentrations using the matrix method to excessive primers and probes. In this study, amplification efficiency, fluorescence signal intensity, and non-specific amplification of primer probes with different concentrations were analyzed to determine the two groups of primer probe concentrations with the optimal ratio. Increasing the annealing temperature can reduce non-specific binding between the primer and template, make the DNA double strands unchain more thoroughly, and improve the amplification efficiency and specificity of PCR reaction, but reducing the annealing temperature can improve the amplification yield. The temperature gradient of 58–65°C was set in this experiment, and the final annealing temperature was chosen as 61°C. The number of PCR cycles mainly depends on the concentration of template DNA, which is generally 25–35 times. In this experiment, the number of cycles was optimized under the above optimized reaction conditions, and 35 cycles were chosen.
Current studies on PCV real-time PCR methods include several studies that have established minimum detection limits for PCV1, PCV2, PCV3, and PCV4. For instance, Li et al. (35) established minimum detection limits of 1 × 101 copies/μl and 1 × 102 copies/μl for PCV1 and PCV2, respectively, using a dual real-time PCR method. Moreover, Li et al. (36) established the lowest detection limits of 2.9 × 100 copies/μl and 2.25 × 101 copies/μl for PCV2 and PCV3, respectively, using a dual real-time fluorescence quantification method. Chen et al. (4) established a quadruplex real-time PCR method for PCV1–4, with a lowest copy number detection limit of 2.8 × 101 copies/μl for each genotype. In this study, the lowest copy number detection limit of PCV2 was 9.69 × 102 copies/μl, which was slightly higher than that of previous studies. Nonetheless, this method still meets the needs of clinical detection.
By utilizing the TaqMan multiplex real-time fluorescence quantitative PCR method developed in our study, a total of 292 samples tested positive for PCV, resulting in a positivity rate of 49.07%. Among these samples, there were 38 cases of mixed PCV infections, with a mixed infection positivity rate of 6.39%. Notably, PCV2 and PCV3 co-infections were prevalent among the mixed infections, accounting for 31 out of the 38 cases. When compared to findings from other researchers, our study showed consistent positivity rates for both PCV single infections and mixed infections. However, the regional distribution of PCV yielded intriguing results, revealing that PCV2 and PCV3 were detected in all districts and cities, except for the five urban areas (Wuhou, Qingyang, Jinjiang, Chenghua, and Jinniu) where pig farming activities were absent. Furthermore, these areas exhibited comparatively high positivity rates. Although, the potential impact of varying viral loads at different tissue locations may result the variation of detection rates, our detect in Chengdu is relatively similar with other regions of China (37–39). This suggests that PCV remains a hidden threat to pig populations in the Chengdu region. Although pigs in Chengdu have not been infected with PCV4, protective measures should still be taken. Farmers should continue to strengthen the management of feeding and the feeding environment to reduce the cross-infection of viruses.
In conclusion, our novel method may provide for PCV1–4 detection. Currently, PCV2 is the main popular in Chengdu, while the potential threat of PCV4 should also be considered.
Data availability statement
Publicly available datasets were analyzed in this study, with the NCBI GenBank accession numbers: EF533941, AF201310, MG778698, MT311852.
Author contributions
YM: Writing – original draft, Investigation, Writing – review & editing. DH: Writing – review & editing, Conceptualization, Investigation, Methodology, Writing – original draft. YZ: Writing – original draft, Investigation, Validation, Writing – review & editing. ML: Writing – review & editing, Methodology, Resources. JY: Resources, Writing – review & editing. HZ: Writing – review & editing, Investigation. HLi: Writing – original draft. ZZhon: Writing – review & editing, Methodology. HLiu: Writing – original draft, Data curation. GP: Writing – review & editing, Resources. LZ: Resources, Writing – review & editing. XZ: Writing – review & editing, Resources, Supervision. ZZhou: Writing – review & editing, Conceptualization, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Doctoral program of Chengdu Agricultural College (2023001) and the Central Government Funds of Guiding Local Scientific and Technological Development (grant 2022ZYDFO36).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1337461/full#supplementary-material
References
1. Lassaletta L, Estellés F, Beusen AH, Bouwman L, Calvet S, Van Grinsven HJ, et al. Future global pig production systems according to the shared socioeconomic pathways. Sci Total Environ. (2019) 665:739–51. doi: 10.1016/j.scitotenv.2019.02.079
2. Zhong S, Li J, Chen X, Wen HJ. A multi-hierarchy meta-frontier approach for measuring green total factor productivity: an application of pig breeding in China. Socioecon Plann Sci. (2022) 81:101152. doi: 10.1016/j.seps.2021.101152
3. Segalés J, Allan GM, Domingo M. Porcine circovirus diseases. Anim Health Res Rev. (2005) 6:119–42. doi: 10.1079/AHR2005106
4. Chen N, Xiao Y, Li X, Li S, Xie N, Yan X, et al. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu Province of China from 2016 to 2020. Transbound Emerg Dis. (2021) 68:1615–24. doi: 10.1111/tbed.13833
5. Wang F, Guo X, Ge X, Wang Z, Chen Y, Cha Z, et al. Genetic variation analysis of chinese strains of porcine circovirus type 2. Virus Res. (2009) 145:151–6. doi: 10.1016/j.virusres.2009.05.015
6. Franzo G. Segalés J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE. (2018) 13:e0208585. doi: 10.1371/journal.pone.0208585
7. Zhang D, Bai C, Ge K, Li Y, Gao W, Jiang S, et al. Establishment of an Sybr green-based real-time PCR assay for porcine circovirus type 4 detection. J Virol Methods. (2020) 285:113963. doi: 10.1016/j.jviromet.2020.113963
8. Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, et al. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg Dis. (2018) 65:264–6. doi: 10.1111/tbed.12639
9. Zhang HH, Hu WQ Li JY, Liu TN, Zhou JY, Opriessnig T, Xiao CT. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, Hunan Province, China. Transbound Emerg Dis. (2020) 67:1057–61. doi: 10.1111/tbed.13446
10. Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2017) 91. doi: 10.1128/JVI.01879-16
11. Ji W, Zhang X, Niu G, Chen S, Li X, Yang L, et al. Expression and immunogenicity analysis of the capsid proteins of porcine circovirus types 2 to 4. Int J Biol Macromol. (2022) 218:828–38. doi: 10.1016/j.ijbiomac.2022.07.204
12. Zhou J, Li H, Yu T, Li J, Dong W, Ojha NK, et al. Protein interactions network of porcine circovirus type 2 capsid with host proteins. Front Microbiol. (2020) 11:1129. doi: 10.3389/fmicb.2020.01129
13. Meng X-JJARAB. Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. (2013) 1:43–64. doi: 10.1146/annurev-animal-031412-103720
14. Finsterbusch T, Mankertz A. Porcine circoviruses—small but powerful. Virus Res. (2009) 143:177–83. doi: 10.1016/j.virusres.2009.02.009
15. Butler J, Lager K, Golde W, Faaberg KS, Sinkora M, Loving C, et al. Porcine reproductive and respiratory syndrome (PRRS): an immune dysregulatory pandemic. Immunol Res. (2014) 59:81–108. doi: 10.1007/s12026-014-8549-5
16. Sahoo M, Pathak M, Patel SK, Saikumar G, Upmanyu V, Thakor JC, et al. Pathomorphology, immunohistochemical, and molecular detection of an atypical porcine dermatitis and nephropathy syndrome (PDNS) due to PCV-2d-2 in naturally affected grower pigs of India. Microb Pathog. (2022) 171:105738. doi: 10.1016/j.micpath.2022.105738
17. Maity HK, Samanta K, Deb R, Gupta VK. Revisiting porcine circovirus infection: recent insights and its significance in the piggery sector. Vaccines. (2023) 11:1308. doi: 10.3390/vaccines11081308
18. Liu C, Liu Y, Feng H, Zhao B, Chen Y, Huang H, et al. PCV cap proteins fused with calreticulin expressed into polymers in Escherichia coli with high immunogenicity in mice. BMC Vet Res. (2020) 16:1–10. doi: 10.1186/s12917-020-02527-9
19. Helke KL, Ezell PC, Duran-Struuck R, Swindle MM. Biology and diseases of swine. In:Anderson LC, Otto G, Pritchett-Corning KR, , editors. Laboratory Animal Medicine. Amsterdam: Elsevier (2015), p. 695–769. doi: 10.1016/B978-0-12-409527-4.00016-X
20. De Barcellos MD, Grunert KG, Zhou Y, Verbeke W, Perez-Cueto F, Krystallis AJA, et al. Consumer attitudes to different pig production systems: a study from Mainland China. Agric Human Values. (2013) 30:443–55. doi: 10.1007/s10460-012-9416-4
21. Rychlik W. Oligo 7 Primer Analysis Software. In: PCR Primer Design (2007). p. 35−59. doi: 10.1007/978-1-59745-528-2_2
22. Zhang L, Luo Y, Liang L, Li J, Cui S. Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in China detected by duplex nanoparticle-assisted PCR. Infect Genet Evol. (2018) 60:1–6. doi: 10.1016/j.meegid.2018.02.006
23. Tian RB, Zhao Y, Cui JT, Zheng HH, Xu T, Hou CY, et al. Molecular detection and phylogenetic analysis of porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound Emerg Dis. (2021) 68:276–82. doi: 10.1111/tbed.13714
24. Sun W, Du Q, Han Z, Bi J, Lan T, Wang W, et al. Detection and genetic characterization of porcine circovirus 4 (PCV4) in Guangxi, China. Gene. (2021) 773:145384. doi: 10.1016/j.gene.2020.145384
25. Nguyen VG, Do HQ, Huynh TML, Park YH, Park BK, Chung HC. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound Emerg Dis. (2022) 69:538–48. doi: 10.1111/tbed.14017
26. Grau Roma L. New Insights on the Epidemiology of Postweaning Multisystemic Wasting Syndrome (PMWS). Barcelona: Universitat Autònoma de Barcelona. (2009).
27. Navarro E, Serrano-Heras G, Castaño M, Solera J. Real-time PCR detection chemistry. Clin Chim Acta. (2015) 439:231–50. doi: 10.1016/j.cca.2014.10.017
28. Tajadini M, Panjehpour M, Javanmard SH. Comparison of sybr green and taqman methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res. (2014) 3:85. doi: 10.4103/2277-9175.127998
29. Cikoš Š, Koppel J. Transformation of real-time PCR fluorescence data to target gene quantity. Anal Biochem. (2009) 384:1–10. doi: 10.1016/j.ab.2008.08.031
30. Bustin S, Huggett J, QPCR. primer design revisited. Biomol Detect Quantif. (2017) 14:19–28. doi: 10.1016/j.bdq.2017.11.001
31. Chen S, Sun Y, Fan F, Chen S, Zhang Y, Zhang Y, et al. Present status of microfluidic pcr chip in nucleic acid detection and future perspective. Trends Anal Chem. (2022) 157:116737. doi: 10.1016/j.trac.2022.116737
32. Trinh TND, Lee NY. Advances in nucleic acid amplification-based microfluidic devices for clinical microbial detection. Chemosensors. (2022) 10:123. doi: 10.3390/chemosensors10040123
33. Maia R, Carvalho V, Faria B, Miranda I, Catarino S, Teixeira S, et al. Diagnosis methods for COVID-19: a systematic review. Micromachines. (2022) 13:1349. doi: 10.3390/mi13081349
34. Becker K. Expanding the Scope of the T7 RNA Polymerase-A Novel Plasmid Requires T7 RNA Polymerase for Replication. Zurich: ETH Zurich (2017).
35. Li J, Shi J, Wu X, Cong X, Xu S, Yuan X, et al. Differentiation of PCV1 and PCV2 by a multiplex real-time PCR assay. Vet Rec. (2013) 173:346. doi: 10.1136/vr.101686
36. Li X, Qiao M, Sun M, Tian K. A duplex real-time PCR assay for the simultaneous detection of porcine circovirus 2 and circovirus 3. Virol Sin. (2018) 33:181–6. doi: 10.1007/s12250-018-0025-2
37. Jiang C-G, Wang G, Tu Y-B, Liu Y-G, Wang S-J, Cai X-H, et al. Genetic analysis of porcine circovirus type 2 in China. Arch Virol. (2017) 162:2715–26. doi: 10.1007/s00705-017-3414-1
38. Fu X, Fang B, Ma J, Liu Y, Bu D, Zhou P, et al. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in Southern China. Transbound Emerg Dis. (2018) 65:e296–303. doi: 10.1111/tbed.12752
Keywords: porcine circovirus, TaqMan multiplex real-time PCR, kit comparison, analysis of epidemic, genetic evolutionary analysis
Citation: Mi Y, Huang D, Zhuo Y, Li M, Yue J, Zhong H, Li H, Zhong Z, Liu H, Peng G, Zhu L, Zhou X and Zhou Z (2024) Development and preliminary application of a quadruplex real-time PCR assay for differential detection of porcine circovirus 1–4 in Chengdu, China. Front. Vet. Sci. 11:1337461. doi: 10.3389/fvets.2024.1337461
Received: 13 November 2023; Accepted: 15 April 2024;
Published: 30 April 2024.
Edited by:
Hong-Ying Chen, Henan Agricultural University, ChinaReviewed by:
Jinyang Zhang, Kunming University of Science and Technology, ChinaPeter Oba, International Livestock Research Institute, Uganda
Copyright © 2024 Mi, Huang, Zhuo, Li, Yue, Zhong, Li, Zhong, Liu, Peng, Zhu, Zhou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxiao Zhou, c3BvbmdlYm9iMTk4OTExMDRAZ21haWwuY29t; Ziyao Zhou, enpob3VAc2ljYXUuZWR1LmNu
†These authors have contributed equally to this work
 Yong Mi1†
Yong Mi1† Yong Zhuo
Yong Zhuo Zhijun Zhong
Zhijun Zhong Haifeng Liu
Haifeng Liu Guangneng Peng
Guangneng Peng Ling Zhu
Ling Zhu Ziyao Zhou
Ziyao Zhou