- 1Institute of Animal Science and Veterinary Medicine, Wuhan Academy of Agricultural Sciences, Wuhan, China
- 2Laboratory of Genetic Breeding, Reproduction and Precision Livestock Farming and Hubei Provincial Center of Technology Innovation for Domestic Animal Breeding, School of Animal Science and Nutritional Engineering, Wuhan Polytechnic University, Wuhan, China
The embryonic loss during early stage of gestation is one of the major causes of infertility for domestic ruminants, causing huge economic losses to pasture. Maternal recognition of pregnancy and implantation are the crucial process for determining the successful establishment and development of pregnancy in cattle. The research on molecular mechanisms of pregnancy recognition will facilitate illustrating the complex process of pregnancy establishment and help to improve pregnancy outcomes. In this study, we performed transcriptomic analysis of primary bovine endometrial epithelial cells (BEND) with or without IFNT and hormones intervention through RNA sequencing. We eventually identified 608 differentially expressed genes (DEGs) including 409 up-regulated genes and 199 down-regulated genes in IFNT and hormones-treated group compared with control group. Gene Ontology (GO) enrichment analysis demonstrated that the majority of DEGs were implicated in immune system process, response to external stimulus, response to cytokine, regulation of response to stress. Results from KEGG analysis showed a significant enrichment of NOD-like receptor signaling pathway, antigen processing and presentation, necroptosis, oxidative phosphorylation, RIG-I-like receptor signaling pathway. Additionally, a set of promising candidate genes, including (USP18, STAT1, PSMB8, IFIH1, MX2, IFI44, DHX58, CASP8, DRAM1, CXCR4), were characterized by constructing an integrated interaction network. Specifically, the mRNA expression of HOXA11, PTGS1 and PTGS2 were remarkably suppressed by silencing DRAM1 under IFNT and hormone administration, thus speculating that DRAM1 might play a crucial role in early pregnancy by regulating endometrial function. The results of this study depicted a relatively comprehensive transcriptional profiles of BEND in response to IFNT and hormones, which contributes to a better understanding of gene interaction network and underlying regulatory mechanisms in endometrium of ruminants during early pregnancy.
1 Introduction
Reproductive performance is a key determinant affecting the economic benefits of pasture. Pregnancy failure will retard the reproductive cycles, increase feeding costs and cause enormous economic losses in the livestock sector. In domestic ruminants, the establishment of pregnancy involves multiple physiological and biochemical processes, including pregnancy recognition, embryo implantation and placentation. It is well known that the fertilization rate in ruminants is considerably higher than pregnancy rate due to the vast majority of pregnancy loss occurring during the peri-implantation period, which encompasses the maternal recognition of pregnancy process ranging from days 16 to 25, contributing to approximately 30–35% embryo loss in cattle (1–4). Maternal pregnancy recognition (MRP) is a physiological process whereby the embryo signals its presence to maternal system, thus preventing the regression of corpus luteum (CL), prolonging the lifespan of CL and secreting progesterone (P4) continuously (5, 6). Therefore, the moderate communication between conceptus and maternal uterus is crucial for the establishment and maintenance of pregnancy.
With regard to the establishment of pregnancy in ruminants, endometrial epithelium requires the coordinated orchestration of conceptus-derived interferon-tau (IFNT), and maternally derived estradiol (E2) and P4 (7–9). IFNT is the primary signal of pregnancy recognition, exclusively secreted by mononuclear cells of conceptus trophectoderm during the early gestation period in ruminant species (10). Accompanied by the progression of pregnancy, IFNT is expressed from days 10 to days 21–25 with maximal peaks on days 17–20 in cattle, which acts on the endometrium through a paracrine manner and causes changes of prostaglandin secretion patterns during pregnancy recognition (11). The expression of estrogen receptor (ESR1) and oxytocin receptor (OXTR) are suppressed by IFNT in endometrial epithelial cells, thereby preventing the luteolytic pulses of prostaglandin F2α (PGF2α) from uterus (12). Moreover, P4 can inhibit the expression of ESR1 by binding to progesterone receptor (PR), subsequently downregulating OXTR in endometrial luminal and superficial glandular epithelial cells (13). The combinatorial and sequential action of P4 and IFNT ultimately restrains the expression of ESR1 and OXTR in endometrium, which inhibits the release of PGF2α and prevents luteolysis. IFNT can also significantly increase the secretion of PGE2 in endometrial stromal cells to prohibit luteolysis and maintain pregnancy (14). In addition, various available evidences indicated that IFNT regulate the expression of numerous P4-induced genes implicated in embryonic development, implantation, uterine receptivity and immune tolerance (15–18). The expression of several classical interferon stimulated genes (ISGs) in endometrium such as ISG15, RSAD2 and OAS1 can be upregulated by IFNT through the JAK–STAT-IRF signaling pathway, that play an essential role in modulating conceptus elongation, implantation and immunity during pregnancy (19). IFNT also signal via MAPK, PI3K signaling pathway independent of STAT1 to regulate the expression of multiple non-canonical ISGs including WNT7A, CST3, HIF2A associated with endometrium-secreted cytokines that promote the proliferation and migration of trophectoderm cells (20). During the peri-implantation period of pregnancy, the nutrients in uterine histotroph are required for embryonic survival, development and adhesion. The expression of glucose transporters SLC2A1, SLC5A1 and SLC5A11, as well as amino acid transporters SLC7A1 and SLC7A2 are remarkably enhanced in endometrial luminal and superficial glandular epithelium of ewes from days 10 to 20 of pregnancy under the stimulation of P4 and IFNT, which functions prominently for conceptus growth and elongation (21, 22). A recent study revealed that the expression of CASP7, CASP8, CASP4 and NLRP3 in uterus from 18-day gestation cattle are dramatically higher than that of non-pregnant uterus, and IFNT also stimulates the expression of CASP8 and CASP11 in endometrial epithelial cells (23), the apoptosis and pyroptosis-related genes are speculated to involve in IFNT-induced pregnancy recognition and embryo implantation. While plenty of genes have been identified during peri-implantation period in ruminants, the gene-interaction network and underlying molecular mechanisms regulating MRP remain intricate, which need to be further elucidated. Herein, RNA-seq was applied to dissect the changes of gene expression profile in bovine endometrial epithelial cells (BEND) under the combinatory treatment of IFNT, P4 and E2, the regulatory network during MRP was uncovered by metabolic pathway analysis, that provides a reference for clarifying the molecular mechanisms of MRP in cattle.
2 Materials and methods
2.1 Bovine endometrial epithelial cell isolation and primary culture
Fresh and healthy uteri without endometritis from Holstein cows were acquired from a surrounding abattoir (Wuhan, China). The health and estimated estrous cycle of bovine were objectively confirmed by individual records of well-being and examination of veterinary surgeon from Wuhan Academy of Agricultural Sciences. This experimental processes were approved by the Institutional Animal Use Committee of Institute of Animal Science and Veterinary Medicine, Wuhan Academy of Agricultural Sciences.
Primary bovine endometrial epithelial cells were isolated and cultured in accordance with protocols previously described with some minor changes (24, 25). The uteri were eviscerated within 30 min after exsanguination, and immediately rinsed 3 times with ice-cold PBS to remove residual blood and dirt on the surface. After be immersed in sterile PBS, the uteri were kept on ice and transported to the laboratory within 1 h. The endometrium tissue was dissected into strips, which were placed in serum-free DMEM/F12 (Gibco, United States) supplemented with 50 U/L penicillin, 50 μg/mL streptomycin (Gibco, United States), 50 μg/mL gentamycin (Sangon Biotech, Shanghai, China) under sterile conditions. Then, the strips were cut into about 1mm3 pieces and washed 3 times with sterile PBS. Tissue pieces were digested with PBS containing 0.1% collagenase I (Sigma, United Kingdom) and incubated in a constant-temperature shaker at 80 r/min for 1 h at 37°C. The suspension was filtered with a 40 μm mesh, the filtrate was centrifuged to remove the supernatant at 1000 r/min for 10 min at room temperature, and resuspended in DMEM/F12 supplemented with 10% FBS (Gibco, United States). After washing five times, the cell pellets were resuspended with complete culture medium composed of 90% DMEM/F12, 10% FBS, 50 U/L penicillin and 50 μg/mL streptomycin, and dispersed into single cell suspension. Cells were seeded into a 10 cm culture dish which was kept in a water-jacked incubator at 37°C with 5% CO2. The medium was changed every 48 h. Subculturing was conducted when the cells reached approximately 80–90% confluence. Cell morphology was recorded and photographed under an inverted microscope.
The endometrial epithelial cells were purified by differential attachment method following three to five passages. Then, the purity of epithelial cell culture was confirmed by immunofluorescence detection of cytokeratin 18. Briefly, the primary endometrial epithelial cells were plated on glass coverslips in six-well culture dishes. The cells were fixed in 4% paraformaldehyde for 15 min after reaching 50–60% confluence, and permeabilized with 0.5% Triton X-100 (prepared in PBS) for 20 min at room temperature. Non-specific binding was blocked by pre-treating the cells with 3% BSA (Sigma, United States) for 30 min at room temperature. Subsequently, the cells were incubated with primary antibody CK-18 (Servicebio, Wuhan, China) at 4°C overnight, followed by incubation with the secondary fluorescently-labeled Cy3 antibody (Servicebio, Wuhan, China) for 1 h at room temperature, after which the cells were washed three times in PBS. The nuclei were counterstained with DAPI (Thermo, United States). Fluorescent images were obtained using a laser scanning confocal microscope (NIKON Eclipse Ti, Tokyo, Japan).
2.2 Cell treatment
As shown in Figure 1A, the BEND at logarithmic growth stage were inoculated in 6 cm dishes and cultured with DMEM/F12 containing 10% FBS and 1% penicillin–streptomycin in humidified atmosphere with 5% CO2 at 37°C. Once reaching 50–60% confluence, the cells were divided into three groups of three replicates and rinsed with PBS. For the groups specified for E2, P4 and IFNT treatment, the medium were replaced by fresh complete culture medium supplemented with E2 (10−9 M, Sigma) and P4 (10−7 M, Sigma, United States), and continued culturing for 12 h. Subsequently, the cells were treated with IFNT (100 ng/mL, Cell sicences, United States) for an additional 12 h.
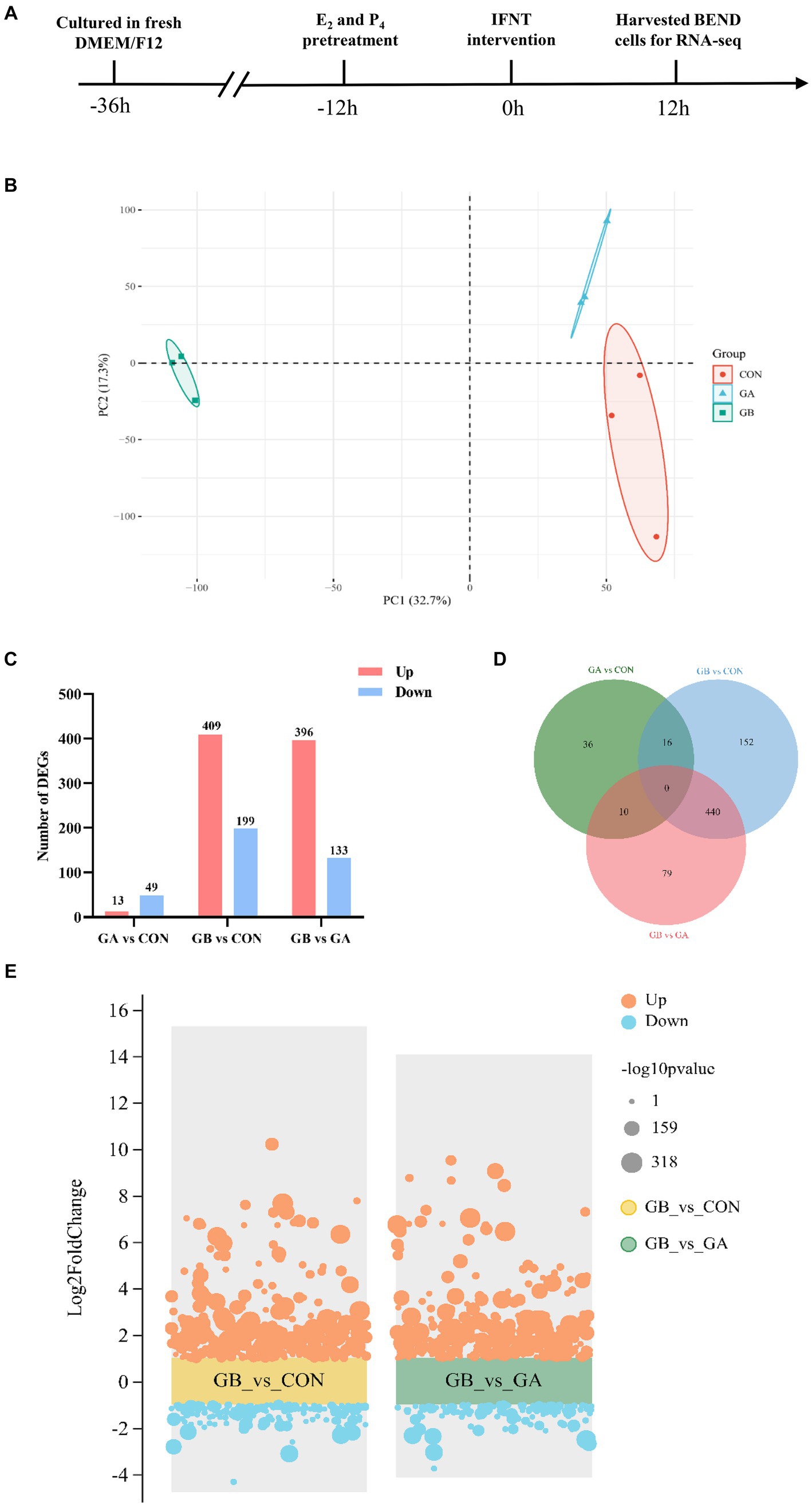
Figure 1. Identification of differentially expressed genes. (A) The workflow of sample preparation for sequencing; (B) PCA analysis of the expressed transcripts. The X-axis is the first principal component, and the Y-axis is the second principal component. Different shapes and colors represent different groups; (C) Statistical analysis of DEGs in GA vs. CON, GB vs. CON, GB vs. GA groups; (D) Venn diagram of the number of common DEGs in GA vs. CON, GB vs. CON, GB vs. GA groups; (E) Volcano plot of DEGs identified in GB vs. CON, GB vs. GA groups. The orange and blue dots represent up- and down-regulated genes, respectively. The dots size indicates the corresponding -log10p value, a larger dot represents a smaller P value.
2.3 RNA extraction and quality assessment
Total RNA extraction from BEND were performed with TRIzol reagent (Invitrogen, United States), and genomic DNA was removed using DNase I (Invitrogen, USA) according to the manufacturer’s instructions (HiPure Universal RNA Mini Kit, Magen). RNA degradation and contamination were monitored on 1% agarose gels. The quantity and purity of RNA was measured using Qubit 3.0 (Thermo Fisher Scientific, MA, United States) and Nanodrop One (Thermo Fisher Scientific, MA, USA). RNA integrity was accurately assessed using the Agilent 2,100 system (Agilent Technologies, Waldbron, Germany).
2.4 Library preparation and sequencing
The mRNAseq library was generated using the ALFA-SEQ Directional RNaLib Prep Kit following the manufacturer’s recommendations. In brief, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using RT Buffer interruption within the ALFA-SEQ Directional RNaLib Prep Kit. First-strand cDNA was synthesized using random hexamer primers and M-MuLV reverse transcriptase (RNase H), followed by second-strand cDNA synthesis using DNA polymerase I and RNase H. The remaining overhangs were converted into blunt ends by exonuclease/polymerase activity. After adenylation of the 3′ end of the DNA fragments, X-Fold diluted VAHTS Adapter-S with a hairpin loop structure were ligated to prepare for hybridization. To preferentially select cDNA fragments of 150–200 bp in length, these library fragments were selected by AMPure XP beads (GE Sera-Mag Magnetic Speed-beads carboxyl-modified). Then, PCR was performed with 2× PfuMax HiFi PCR ProMix (EnzyValley), primers with index VAHTS Multiplex Oligos Set 4 for Illumina. Finally, AMPure XP beads (GE Sera-Mag Magnetic Speed-beads carboxyl-modified) was used to purify the PCR products, and the library insert sizes were evaluated on the Qsep400 high-throughput nucleic acid protein analysis system (Houze Biotechnology Co., Ltd., Hangzhou, China). The clustering of the index-coded samples was performed on a cBot Cluster Generation System. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2,500 platform and 150 bp paired-end reads were generated. The sequencing data generated in this study were deposited in the NCBI GEO database (accession number GSE253481).
2.5 Data quality control and reference genome alignment
Use fastp (v0.21.0) (https://github.com/OpenGene/fastp) to process the raw data in fastq format to obtain clean data (clean reads). Clean reads were mapped to the NCBI Rfam database, and rRNA sequences were removed by Bowtie2 (v2.33) (https://github.com/BenLangmead/bowtie2). The reference genome (ARS-UCD1.2) and annotation files were downloaded directly from the genome website. The sequences after ribosome removal were aligned to the reference genome by Hisat2 (v2.2.1) software (https://github.com/ infphilo/ hisat2).
2.6 Differential expression analysis
RSEM (v1.3.3) (https://github.com/deweylab/RSEM) was applied to obtain the read counts for each gene. Fragments per kilobase of exon per million mapped reads (FPKM) of genes in each sample were calculated to estimate gene expression levels. Differentially expressed genes between two groups (three biological replicates per group) were screened with DESeq2 package (v1.34.0) (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html). The resulting p-values were adjusted using the Benjamini-Hochberg method for controlling the false discover rate (FDR). |log2 (Fold Change)| > 1 and FDR < 0.05 were set as thresholds for significantly differential expressed genes (DEGs). These genes were used for subsequent bioinformatic analysis.
2.7 GO and KEGG enrichment analyses of differentially expressed genes
The function annotation of DEGs, including cellular component (CC), molecular function (MF) and biological process (BP), were analyzed using Gene Ontology (GO) database (http://www.geneontology.org). Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) was engaged to systematically analyze the biochemical metabolic and signal transduction pathways in which the DEGs are involved. GO and KEGG enrichment analysis of DEGs were performed using the clusterProfiler (v4.2.2) (http://www.Bioconductor.org/packages/release/bioc/html/clusterProfiler.html) for function and signaling pathway annotation. GO terms and KEGG pathways with FDR < 0.05 were considered significantly enriched for DEGs.
2.8 PPI network analysis
A Protein–protein interaction (PPI) network was constructed for DEGs, based on the STRING database (http://string-db.org). The constructed networks were based on known interactions in the selected reference species (cow) with a combined interaction score above 0.4 as selection threshold. Cytoscape software (version 3.10.1) was applied to visualize the PPI networks.
2.9 qRT-PCR
The expression profiles of selected DEGs were validated to confirm the repeatability and reproducibility of gene expression data obtained by RNA sequencing, which were accomplished by SYBR Green-based qRT-PCR using specific primers designed by an online tool (https://primer3.ut.ee/) and a subset of samples from those utilized for RNA sequencing. 1 μg of RNA was reverse transcribed into cDNA, using a reverse transcription kit (with gDNA wiper, Vazyme, China) following the manufacturer’s instructions. The relative expression level of genes were determined by qRT-PCR using the AceQ qPCR SYBR Green Master Mix (Vazyme, China) with the following program: pre-degeneration at 95°C for 5 min, 40 cycles of degeneration at 95°C for 10s and annealing at 60°C for 30s. Data acquisition and analysis were performed on a CFX96 Connect real-time PCR detection system using CFX manager software (Bio-Rad, United States). The sequences of primers synthesized by Tsingke Biotechnology Co., Ltd. (China) are listed in Supplementary Table S1. Relative mRNA expression levels of target genes were normalized against the relative quantity of ACTB mRNA, which was based on threshold cycle (Ct) values using a comparative 2-△△Ct method. qRT-PCR was carried out in three biological replicates.
2.10 RNA interference
RNA interference of DRAM1 was executed using small interfering RNA (siRNA). BEND was transiently transfected with control siRNA or siRNA-DRAM1 by Lipofectamine 3,000 (Invitrogen, United States) according to the manufacturer’s manual. The sequence of siRNA- DRAM1 was sub5’-UCCUGCAAUCCGUCAUCUCUUdTdT-3′. qPCR was employed to detect the knockdown efficiency of DRAM1 in BEND.
2.11 Statistical analysis
Experimental data are presented as mean ± standard deviation (SD). All data were representative for one of three independently repeated experiments. The statistical analyses were carried out by GraphPad Prism software 8.0 (GraphPad Prism Software Inc., CA, United States). Unpaired two-tailed t test (Student’s t-test) was devoted to make comparisons between two independent groups. For comparison of statistical differences among multiple groups (>2), one-way analysis of variance (one-way ANOVA) with multiple comparisons tested by Tukey’s post hoc test was performed. p < 0.05 was considered statistically significant.
3 Results
3.1 Identification of differentially expressed genes
The identification of primary BEND was performed by immunofluorescence, which showed that the epithelial-specific marker cytokeratin is positive in primary BEND (Supplementary Figure S1). The flow schema of sample preparation for sequencing is shown in Figure 1A. Triplicate mRNAseq libraries from three groups of BEND, which includes cells of control group (CON), cells treated with E2 and P4 (GA), cells treated with E2, P4 and IFNT (GB), were generated and sequenced to yield 65.15Gb of data. After filtering the sequencing adapter sequence and low-quality reads, a total of 62,289,909, 67,196,670, 63,332,621 clean reads with >93.67% of Q30 and exceeding 51.74% of the GC content were acquired from CON, GA and GB group, respectively, that were used for the succedent analyses. The mean value of clean reads ratio was 88.66, 88.25 and 89.55% for the CON, GA and GB group, respectively (Supplementary Table S2). Approximately 98.18–98.38% of clean reads were mapped to the reference genome after removal of reads aligned with rRNA (Supplementary Table S3). Unmapped reads (1.62–1.82%) were excluded from further analyses. Accordingly, 17,162 mRNA transcripts were detected in the 9 samples. Principal component analysis (PCA) which clusters similar samples together and accounts for the origin of variance in data was performed with two elements: PC1 (32.7%) and PC2 (17.3%), indicating high similarity between samples in the group (Figure 1B). All these results demonstrated the reliability of RNA sequencing data, that could be used in further analyses.
The differential gene expression profile between groups was determined with DESeq. A total of 608 DEGs were identified in GB group compared to the CON group, among which 409 genes were upregulated and 199 genes were downregulated; 529 DEGs composed of 396 upregulated genes and 133 downregulated genes were identified in GB group compared to the GA group; 62 DEGs containing 13 upregulated genes and 49 downregulated genes were detected in GA group compared to the CON group (Figure 1C). In addition, a venn diagram of DEGs in three comparison groups was plotted, which showed that 440 genes are co-differentially expressed in both comparison groups (GB vs. CON, GB vs. GA) (Figure 1D). The volcano plot was constructed to visualize the distribution and fold change of these DEGs identified in GB vs. CON and GB vs. GA (Figure 1E) groups.
3.2 Cluster analysis of differentially expressed genes
The hierarchical cluster analysis of DEGs was applicated to identify the expression level of genes with high correlation among samples. Heatmaps depicting the DEGs of GB vs. CON, GB vs. GA groups are shown in Figures 2A,B, respectively. Meanwhile, we plotted a heatmap of 440 DEGs that were commonly present in both comparison groups, which revealed that most of the DEGs exhibit similar differential expression trends and patterns among samples (Figure 2C).
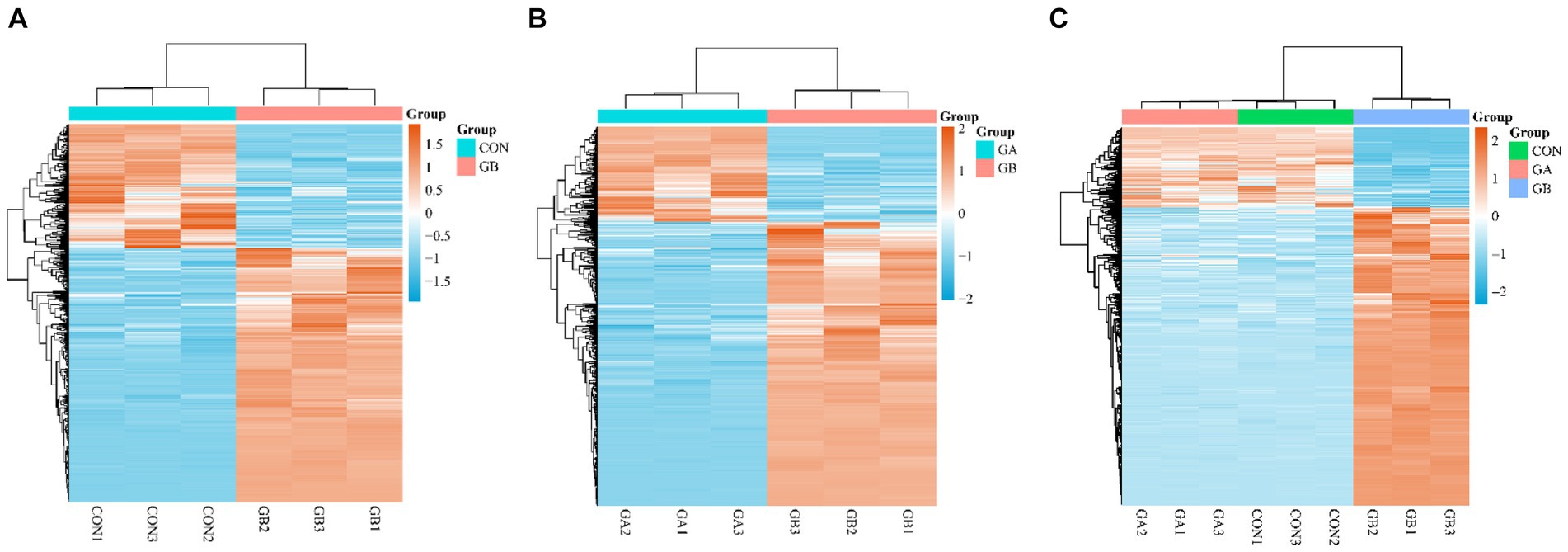
Figure 2. Cluster analysis of DEGs. Heatmap of DEGs in the pair of GB vs. CON (A), GB vs. GA (B), common DEGs in GB vs. CON and GB vs. GA (C). The expression level was calculated by log2 (FPKM) presented as gradient color barcode at the right top. The orange and blue color indicate up- and down-regulated genes, respectively.
3.3 GO and KEGG enrichment analyses
We performed gene ontology (GO) annotation of DEGs to characterize the functional roles of these genes in biological pathways. For GO enrichment analysis, GO terms with adjusted p-value <0.05 were designated as significantly enriched. A large number of DEGs were enriched in the GO biological process category. 17 GO terms from Process Ontology were significantly enriched in GB vs. CON group, such as immune system process, response to external biotic stimulus, defense response, response to cytokine, antigen processing and presentation (Supplementary Table S5). 43 GO terms from Process Ontology were significantly enriched in GB vs. GA group, including immune system process, multi-organism process, regulation of immune system process, regulation of response to stress, negative regulation of cellular protein metabolic process (Supplementary Table S6). The GO terms with most significant enrichment in GB vs. CON (Figure 3A), GB vs. GA (Figure 3B) are displayed as circle diagrams.
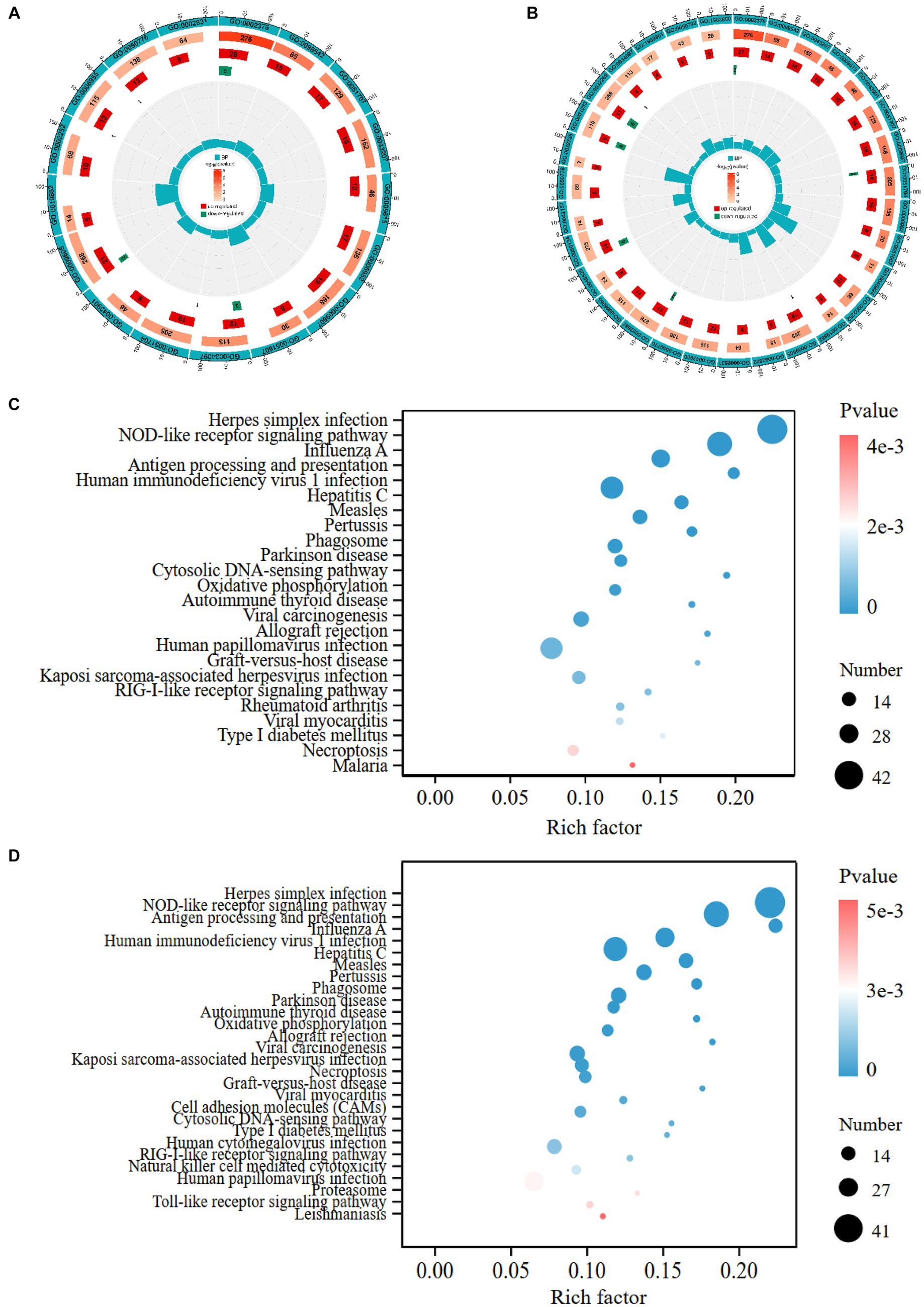
Figure 3. GO and KEGG enrichment analyses of DEGs. The circle diagrams of GO enrichment analysis of DEGs in the pair of GB vs. CON (A), GB vs. GA (B). The first circle shows significantly enriched GO term from biological process ontology; the second circle indicates the number and value of p of the GO term in each classification gene background; the third and fourth circle indicate the number of up- and down-regulated genes in each category of GO term, respectively; the fifth circle indicates enrichment factor value of each GO term. The bubble plots display the significantly enriched KEGG pathways in the pair of GB vs. CON (C), GB vs. GA (D). The size of dot represents the number of DEGs annotated in each pathway term, the color of dot denotes p-value.
KEGG enrichment analysis of DEGs was conducted to calculate the abundant metabolic pathways and signal transduction pathways. KEGG pathways were considered significantly enriched with adjusted p-value <0.05. Overall, 348 DEGs were assigned to 24 significantly enriched KEGG pathways in GB vs. CON group, which contains herpes simplex infection, NOD-like receptor signaling pathway, influenza A, antigen processing and presentation, measles, parkinson disease, cytosolic DNA-sensing pathway, oxidative phosphorylation, RIG-I-like receptor signaling pathway, necroptosis (Supplementary Table S7). Moreover, 305 DEGs were allotted to 28 significantly enriched KEGG pathways in GB vs. GA group, such as herpes simplex infection, NOD-like receptor signaling pathway, human immunodeficiency virus 1 infection, human papillomavirus infection, influenza A, phagosome, cell adhesion molecules (CAMs), Toll-like receptor signaling pathway (Supplementary Table S8). Advanced bubble charts reveal the significantly enriched KEGG pathways of DEGs in GB vs. CON (Figure 3C), GB vs. GA (Figure 3D).
3.4 PPI network analysis of differentially expressed genes
A PPI network composed of 260 nodes connected via 2018 edges was generated by Cytoscape software based on STRING database to illustrate the interactions between DEGs. The results showed that a few node genes interact closely with other genes, including STAT1, IFIH1, MX2, IFI44, USP18, DHX58, PSMB8, CASP8, DRAM1, CXCR4 (Figure 4). Some of these highly connected node genes are rarely reported to correlate with early pregnancy, that need to be concerned and further investigated.
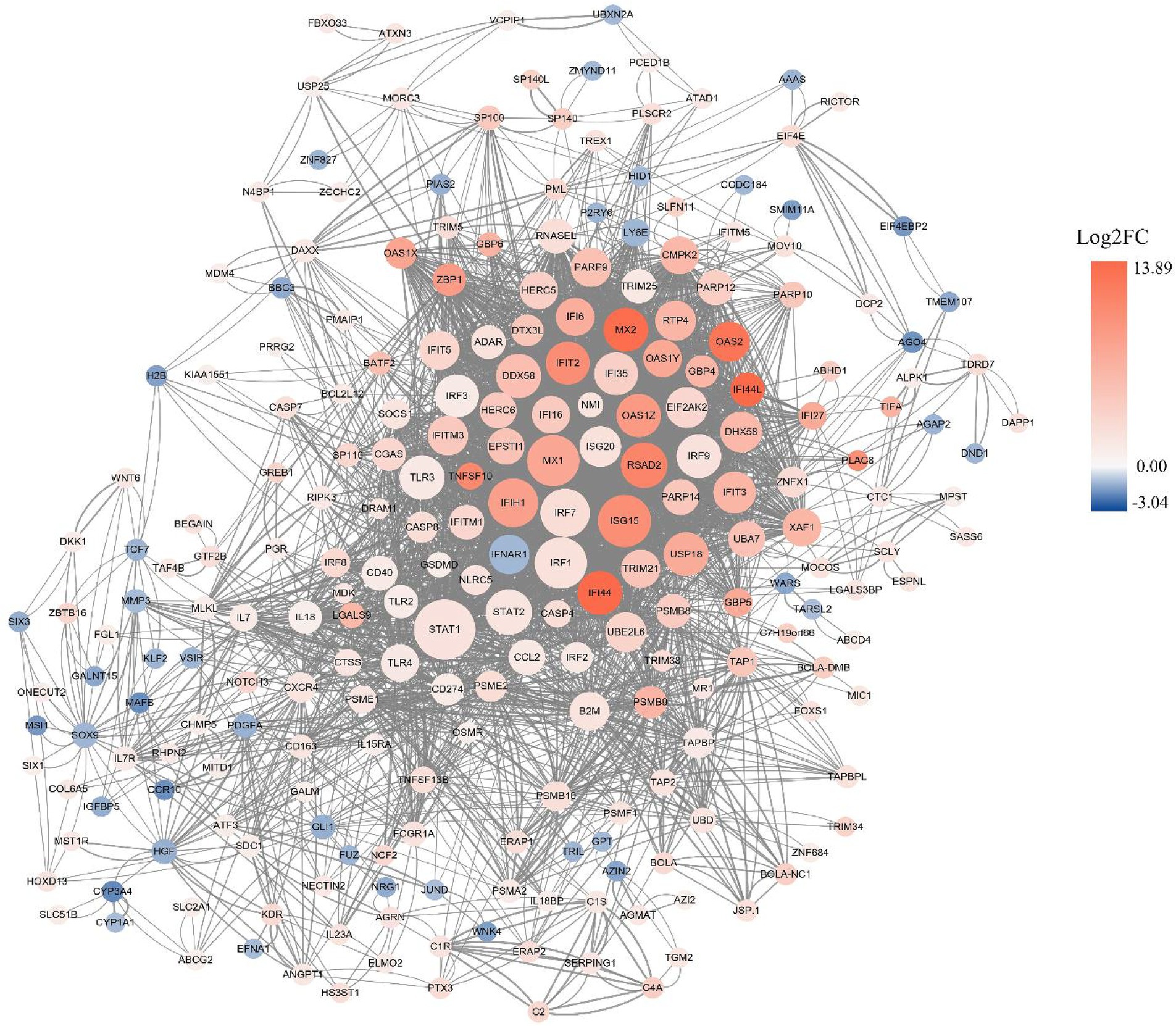
Figure 4. PPI network analysis of DEGs. The red and blue dots indicate up- and down-regulated genes, respectively. The intensity of color denotes fold-change of DEGs, and the size of dots represent node degree.
3.5 qRT-PCR verification
Considering the results of GO and KEGG enrichment analysis, 8 DEGs were randomly selected for exploring the relative expression pattern to validate the accuracy and reliability of RNA-Seq results by performing qRT-PCR. The expression trend of all these selected genes in the results of qRT-PCR were proved to be identical with RNA-seq data (Figure 5A). Meanwhile, the results of Spearman’s correlation analysis of all selected genes showed a strong positive correlation between RNA-seq data and qRT-PCR experiments (r2 > 0.65, p < 0.05), which confirmed the credibility of DEGs (Figure 5B).
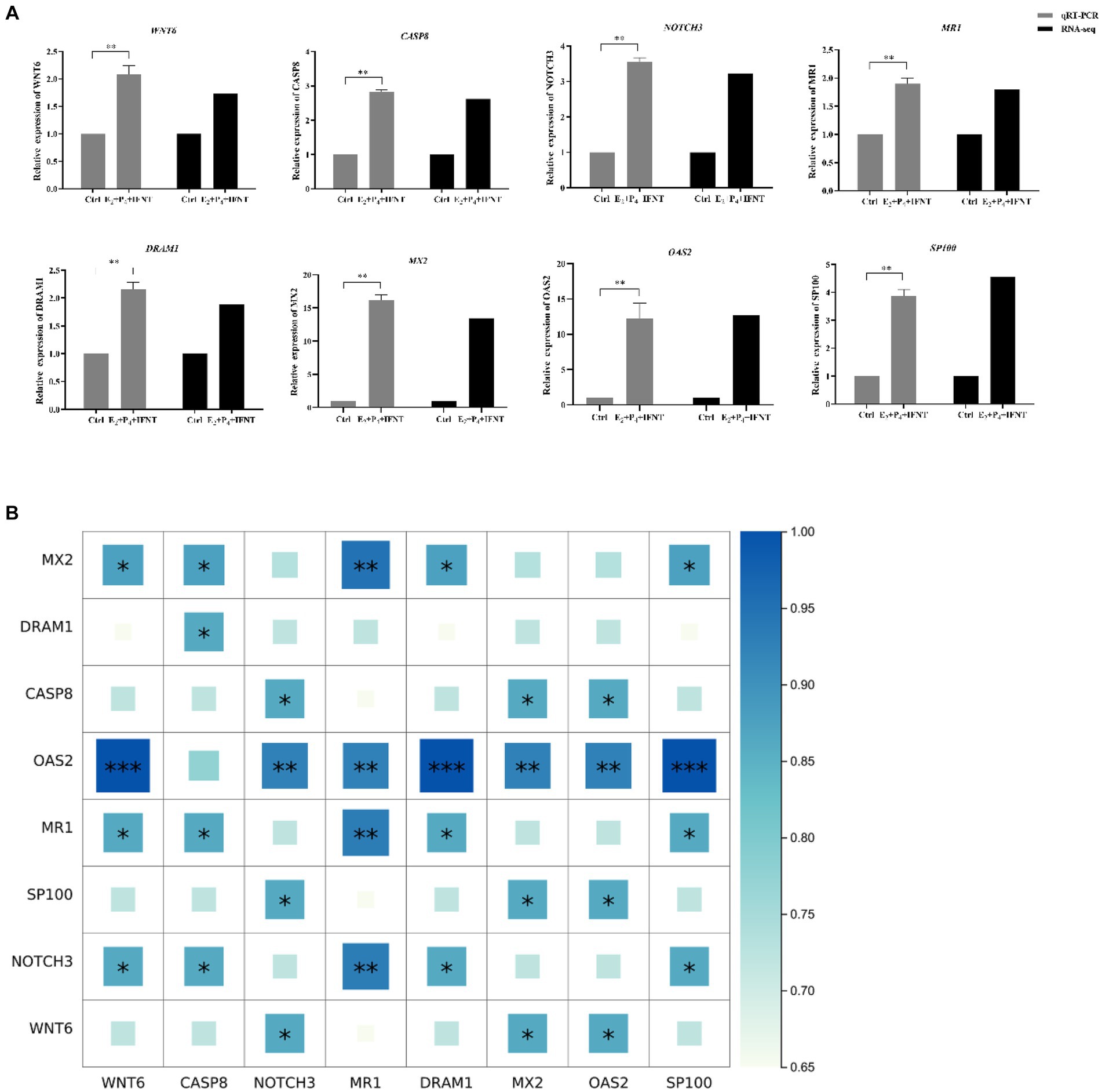
Figure 5. Expression pattern validation of selected DEGs by real-time PCR. (A) The relative expression level of target mRNA was calculated using the 2−△△Ct method and presented as the ratio of Log2 fold change between groups; (B) Spearman’s correlation analysis heatmap of RNA-seq and qRT-PCR data. “*” p < 0.05, “**” p < 0.01, “***” p < 0.001.
3.6 Effects of DRAM1 interference on expression of genes related to receptivity and prostaglandin synthesis in bovine endometrial epithelial cells
To investigate the functional role of DRAM1 during early pregnancy in ruminants, we conducted RNA interference assay to downregulate the transcript expression of DRAM1 in BEND. The results indicated that the mRNA expression of DRAM1 is prominently reduced (Supplementary Figure S2). Additionally, the mRNA expression of HOXA11, PTGS1 and PTGS2 were dramatically downregulated by silencing DRAM1 upon IFNT stimulation (Figures 6B–D), but no significant change was observed on mRNA expression of HOXA10 by suppressing DRAM1 under IFNT treatment (Figure 6A). Furthermore, even in the absence of external intervention with IFNT, the mRNA expression of PTGS1 and PTGS2 were also significantly inhibited in siDRAM1 group compared with siNC group (Figures 6C,D). Nevertheless, interfering with DRAM1 exerted no distinct influence on the mRNA expression of HOXA10 and HOXA11 in BEND without IFNT treatment (Figures 6A,B).
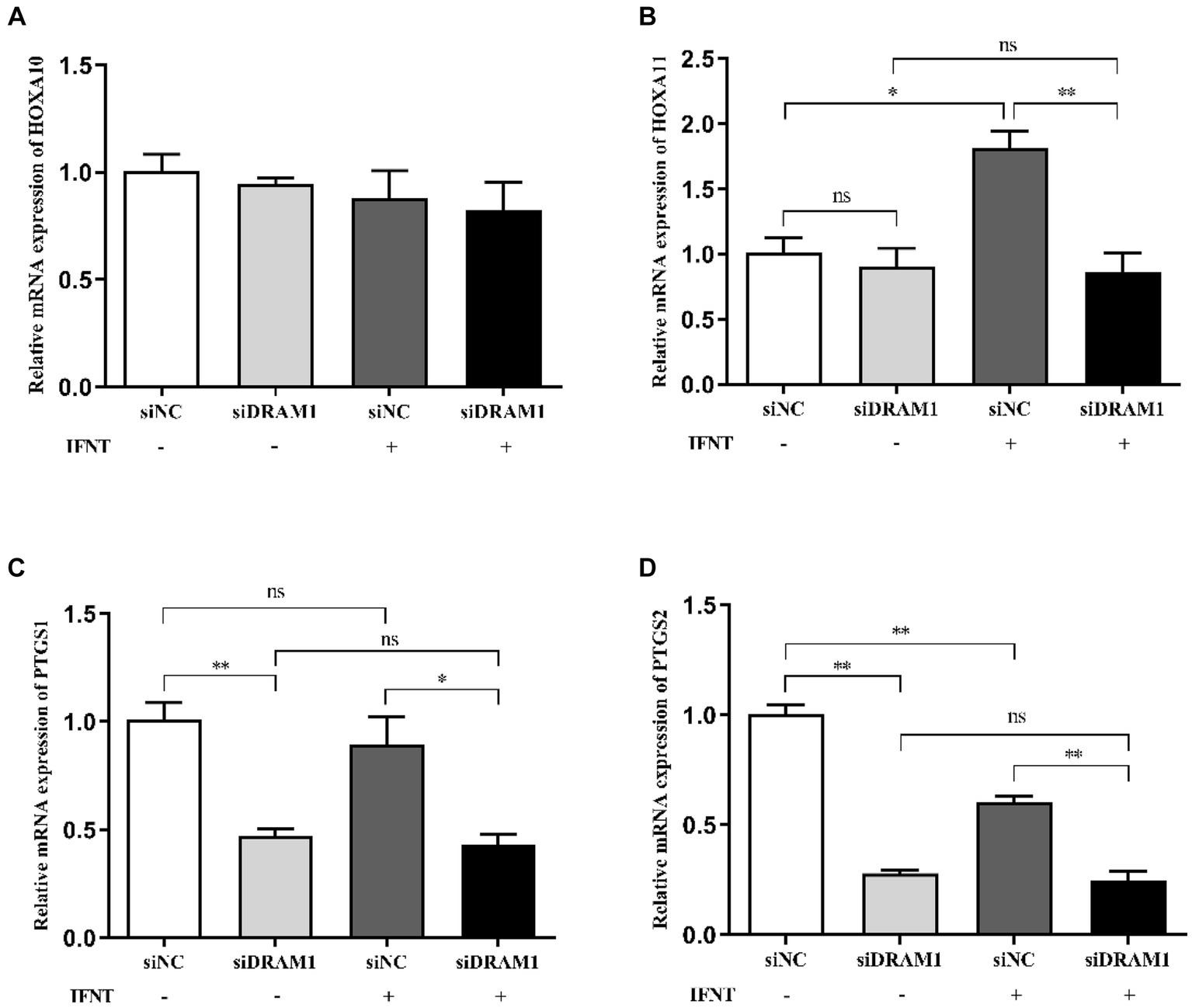
Figure 6. Effects of DRAM1 interference on expression of genes related to receptivity and prostaglandin synthesis in BEND. BEND was transiently transfected with control siRNA or siRNA-DRAM1. Subsequently, the cells were incubated with E2 and P4 followed by IFNT stimulation for 12 h. Total RNA isolated form BEND were applied to Real-time PCR. Real-time PCR analysis of the mRNA expression of endometrial receptivity marker gene HOXA10 (A) and HOXA11 (B). Real-time PCR analysis of the mRNA expression of rate-limiting enzymes of PGs synthesis PTGS1 (C) and PTGS2 (D). “*” p < 0.05, “**” p < 0.01.
4 Discussion
Pregnancy, an important driver for economic efficiency of pasture, is a complicated physiological process that requires the precise coordination of multiple molecular mechanisms involving numerous genes. However, the gene-interaction network and underlying molecular mechanisms implicated in early pregnancy remain elusive. Although there have been a few reports on the regulation of endometrium by IFNT in domestic ruminants, most studies which were performed in an immortalized bovine endometrial cell line only analyzed the role of IFNT, irrespective of the impact of hormones during early pregnancy (26, 27). In this study, we isolated and cultured primary endometrial epithelial cells of bovine (BEND), and performed transcriptomic analysis of BEND in response to IFNT, P4 and E2, which depicted a relatively comprehensive DEGs profile based on the RNA-seq dataset. Comparing our results with previous studies, we noticed some similarities and inconsistencies. Zhao et al. (27) identified 839 upregulated DEGs and 148 downregulated DEGs in bovine endometrial epithelial cells under the stimulation of IFNT for 12 h compared with untreated groups. Chaney et al. (28) analyzed the bovine endometrial epithelial transcriptomic response to recombinant IFNT and identified 622 increased DEGs, 41 decreased DEGs, among which 247 of 622 upregulated DEGs and 9 of 41 downregulated DEGs appeared in our DEGs list, and were consistent with the gene expression trend in our study. Adhikari et al. (29) compared the transcriptomic profiles of endometrial tissue of pregnant heifers (day 15–17 of gestation) with cyclic (non-bred) heifers and identified 107 DEGs, 7 of top 40 DEGs in Adhikari’s study were matched to the DEGs list of GB vs. CON group in our study; simultaneously, 6 of 7 DEGs showed consistent expression trend with our data, 1 DEG exhibited completely opposite expression trend.
GO enrichment analysis indicated that the higher number of DEGs in our data were mainly implicated in immune system process, response to external stimulus, response to cytokine, which are in agreement with the previous results reported by Zhao et al. (27). In addition, immune response and Type-1 interferon signaling were also the important enriched function in the endometrium tissue of pregnant bovine versus cyclic and nonpregnant (29). Similar results were also acquired by KEGG pathway analysis in this study, which revealed that immunity related pathways, such as herpes simplex infection, NOD-like receptor signaling pathway were the most notably enriched pathways. Meanwhile, some DEGs related with immunity and apoptotic process, such as MR1, CASP8, OAS2, MX2, SP100, were remarkably enriched in these pathways. All these experimental evidences suggest that immunity or apoptosis process induced by IFNT may play a significant role during early pregnancy in ruminants, which contributes to protect the embryo from rejection by maternal immune system.
It is well known that successful pregnancy in ruminants requires the participation of numerous genes regulated by IFNT. The results of predicted protein–protein interaction network demonstrated that a large number of DEGs of this study regulated directly or indirectly by IFNT and hormones constitute a complicated network, which might affect early pregnancy. As expected, the expression of many classical ISGs, such as ISG15, RSAD2, MX2, OAS2 and STAT1, were upregulated by IFNT and hormones in this study. However, some DEGs have rarely been reported previously to be associated with pregnancy. Ubiquitin-specific protease 18 (USP18), a critical negative regulator of type I interferon signaling pathway by competing with JAK1 to bind interferon receptors, exert deubiquitination through the ISGylation modification system, and its expression is significantly enhanced in the endometria of pregnant cow on day 16 (30, 31). Similarly, a recent study proved that the expression of USP18 is significantly increased in goat uterine tissue from days 5–18 of pregnancy, which might promote the establishment of endometrial receptivity to affect early pregnancy by regulating the JAK/STAT1 and ISGylation pathway (32). Few studies have previously reported the relationship between proteasome subunit beta type-8 (PSMB8) and pregnancy. PSMB8 was found to be an important regulator in the process of glioma cell migration, proliferation and apoptosis mediated by ERK1/2 and PI3K-AKT pathways, which shares common pathways of invasion and angiogenesis with embryo implantation (33, 34). Xie et al. (35) proved that 3’-UTR of PSMB8 targeted directly by chi-miR-451-5p of extracellular vesicles in goat uterine fluids might play an essential role in endometrial remodeling during peri-implantation. Apoptotic cell death has been shown to occur in the endometrium during mammalian preimplantation period, such as bovine (23, 36), rats (37), porcine (38). The expression of proapoptotic genes, CASP8, XAF1 and TNFSF10 were confirmed to be remarkably enhanced in the bovine endometrium during early pregnancy (36). These findings were also supported by a research conducted by Suzuki et al. (23), they demonstrated that apoptosis related genes (CASP7, CASP8, FADD) and pyroptosis related genes (CASP4, CASP11, NLRP3) are expressed at significantly higher levels in the endometrium of pregnant bovine at day 18 in comparison to non-pregnant. Meanwhile, the expression of CASP7, FADD, CASP4, CASP11 and GSDMD were considerably increased in cultured uterine epithelial cells with IFNT administration (23).The similar results were observed in our study. However, the increased activity of caspases and elevation of apoptosis were barely detected in the endometrium of early pregnant bovine (23, 36), it is presumed that the proapoptotic signaling arose by IFNT are not transduced. Thus, the mechanisms protecting the endometrium from apoptosis to allow embryonic implantation might exist, that need to be further clarified.
Damage-regulated autophagy modulator 1 (DRAM1), an evolutionarily conserved protein located on the lysosome membrane, was identified as a direct target of p53 to modulate stress-induced autophagy and critical for p53-mediated apoptosis (39). Specifically, DRAM1 is involved in the process of autophagy under the conditions of various cellular stress, such as endoplasmic reticulum stress, mitochondrial damage and nucleic acid damage (40, 41), which was considered to be a key regulator related to the balance between cell survival and apoptotic cell death. Yang et al. (42) indicated that endoplasmic reticulum stress triggered by hormone and IFNT may contribute to early pregnancy success and the regulation of endometrial function by modulating mTOR-autophagy pathway in goat endometrial epithelial cells. Previous studies have suggested that DRAM1 might play an essential regulatory role during embryo implantation and early embryonic development. Chen et al. (43) confirmed the role of DRAM1 in mitophagy, which contributes to the regulation of preeclampsia caused by high levels of oxidative stress, mitochondrial dysfunction and apoptosis in the placental tissues of mice. DRAM1 may be implicated in the regulation of proliferation, differentiation and apoptosis of placental cells, affecting the oxygen and nutrients supply of placenta. Nevertheless, the exact relationship between DRAM1 and early pregnancy, and the underlying mechanisms remain not well-understood. In this study, DRAM1 was identified to be a key node gene interacting closely with other genes by constructing a PPI network based on STRING database, and confirmed to be a DEG by performing qRT-PCR verification. Therefore, we preliminarily explored the impact of DRAM1 on expression of genes related with endometrial function. Prostaglandin-endoperoxide synthase 1 (PTGS1) and prostaglandin-endoperoxide synthase 2 (PTGS2) are rate-limiting enzymes in the biosynthesis pathway of PGs secreted by endometrium, which have crucial effects on the regulation of conceptus development and endometrial function (14, 44). Homeobox A10 (HOXA10) and Homeobox A11 (HOXA11) were proved to be involved in the regulation of endometrial receptivity, thereby increasing the embryo adhesion and implantation (45, 46). The results showed that mRNA expression of HOXA11, PTGS1 and PTGS2 were remarkably suppressed by silencing DRAM1 under IFNT and hormone administration. Therefore, we presume that DRAM1 may play a crucial role in early pregnancy by regulating endometrial function. Whereas, we did not further anatomize the specific molecular mechanisms by which DRAM1 regulates endometrial function, that will be addressed in our future work.
5 Conclusion
Taken together, our results revealed multiple promising genes and signaling pathways in BEND, which might participate in the regulation of endometrial function during the early stage of pregnancy, especially early embryo implantation. The present study provides a more comprehensive overview of transcriptional changes of BEND in response to IFNT and hormones, which may pave the way for illustrating the molecular mechanisms of pregnancy recognition and endometrial function regulation, contributing to a better understanding of early pregnancy loss in ruminants.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/; GSE253481.
Ethics statement
The animal study was approved by the Institutional Animal Use Committee of Institute of Animal Science and Veterinary Medicine, Wuhan Academy of Agricultural Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JY: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. CL: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. HC: Formal analysis, Investigation, Methodology, Writing – review & editing. MX: Investigation, Methodology, Writing – review & editing. XH: Investigation, Methodology, Writing – review & editing. ZZ: Investigation, Methodology, Writing – review & editing. QL: Investigation, Methodology, Writing – review & editing. DW: Investigation, Methodology, Writing – review & editing. LC: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Key Research and Development Program of Hubei Province (2022BBA007 and 2023BBB058), Innovation Project of Wuhan Academy of Agricultural Sciences (QNCX202305).
Acknowledgments
We would like to thank all the members of Institute of Animal Husbandry and Veterinary, Wuhan Academy of Agricultural Science for support, assistance and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1344259/full#supplementary-material
References
1. Silva-Del-Rio, N , Colloton, JD , and Fricke, PM . Factors affecting pregnancy loss for single and twin pregnancies in a high-producing dairy herd. Theriogenology. (2009) 71:1462–71. doi: 10.1016/j.theriogenology.2009.01.013
2. Wiltbank, MC , Baez, GM , Garcia-Guerra, A , Toledo, MZ , Monteiro, PL , Melo, LF, et al. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology. (2016) 86:239–53. doi: 10.1016/j.theriogenology.2016.04.037
3. Reese, ST , Franco, GA , Poole, RK , Hood, R , Fernadez, ML , Oliveira, FR, et al. Pregnancy loss in beef cattle: a meta-analysis. Anim Reprod Sci. (2020) 212:106251. doi: 10.1016/j.anireprosci.2019.106251
4. Sigdel, A , Bisinotto, RS , and Penagaricano, F . Genes and pathways associated with pregnancy loss in dairy cattle. Sci Rep. (2021) 11:13329. doi: 10.1038/s41598-021-92525-0
5. Roberts, RM , Xie, S , and Mathialagan, N . Maternal recognition of pregnancy. Biol Reprod. (1996) 54:294–302. doi: 10.1095/biolreprod54.2.294
6. Bazer, FW , Burghardt, RC , Johnson, GA , Spencer, TE , and Wu, G . Interferons and progesterone for establishment and maintenance of pregnancy: interactions among novel cell signaling pathways. Reprod Biol. (2008) 8:179–211. doi: 10.1016/s1642-431x(12)60012-6
7. Simmons, RM , Satterfield, MC , Welsh, TJ , Bazer, FW , and Spencer, TE . Hsd11b1, hsd11b2, ptgs2, and nr3c1 expression in the peri-implantation ovine uterus: effects of pregnancy, progesterone, and interferon tau. Biol Reprod. (2010) 82:35–43. doi: 10.1095/biolreprod.109.079608
8. Brooks, K , Burns, G , and Spencer, TE . Conceptus elongation in ruminants: roles of progesterone, prostaglandin, interferon tau and cortisol. J Anim Sci Biotechnol. (2014) 5:53. doi: 10.1186/2049-1891-5-53
9. Moraes, J , Behura, SK , Geary, TW , Hansen, PJ , Neibergs, HL , and Spencer, TE . Uterine influences on conceptus development in fertility-classified animals. Proc Natl Acad Sci USA. (2018) 115:E1749–58. doi: 10.1073/pnas.1721191115
10. Forde, N , and Lonergan, P . Interferon-tau and fertility in ruminants. Reproduction. (2017) 154:F33–43. doi: 10.1530/REP-17-0432
11. Spencer, TE , and Hansen, TR . Implantation and establishment of pregnancy in ruminants. Adv Anat Embryol Cell Biol. (2015) 216:105–35. doi: 10.1007/978-3-319-15856-3_7
12. Telgmann, R , Bathgate, RAD , Jaeger, S , Tillmann, G , and Ivell, R . Transcriptional regulation of the bovine oxytocin receptor gene1. Biol Reprod. (2003) 68:1015–26. doi: 10.1095/biolreprod.102.008961
13. Spencer, TE , Johnson, GA , Burghardt, RC , and Bazer, FW . Progesterone and placental hormone actions on the uterus: insights from domestic animals1. Biol Reprod. (2004) 71:2–10. doi: 10.1095/biolreprod.103.024133
14. Arosh, JA , Banu, SK , and McCracken, JA . Novel concepts on the role of prostaglandins on luteal maintenance and maternal recognition and establishment of pregnancy in ruminants. J Dairy Sci. (2016) 99:5926–40. doi: 10.3168/jds.2015-10335
15. Satterfield, MC , Bazer, FW , and Spencer, TE . Progesterone regulation of preimplantation conceptus growth and galectin 15 (lgals15) in the ovine uterus. Biol Reprod. (2006) 75:289–96. doi: 10.1095/biolreprod.106.052944
16. Song, G , Kim, J , Bazer, FW , and Spencer, TE . Progesterone and interferon tau regulate hypoxia-inducible factors in the endometrium of the ovine uterus. Endocrinology. (2008) 149:1926–34. doi: 10.1210/en.2007-1530
17. Ahn, HW , Farmer, JL , Bazer, FW , and Spencer, TE . Progesterone and interferon tau-regulated genes in the ovine uterine endometrium: identification of periostin as a potential mediator of conceptus elongation. Reproduction. (2009) 138:813–25. doi: 10.1530/REP-09-0208
18. Brooks, KE , Burns, GW , and Spencer, TE . Peroxisome proliferator activator receptor gamma (pparg) regulates conceptus elongation in sheep. Biol Reprod. (2015) 92:42. doi: 10.1095/biolreprod.114.123877
19. Spencer, TE , Sandra, O , and Wolf, E . Genes involved in conceptus-endometrial interactions in ruminants: insights from reductionism and thoughts on holistic approaches. Reproduction. (2008) 135:165–79. doi: 10.1530/REP-07-0327
20. Bazer, FW , Ying, W , Wang, X , Dunlap, KA , Zhou, B , Johnson, GA, et al. The many faces of interferon tau. Amino Acids. (2015) 47:449–60. doi: 10.1007/s00726-014-1905-x
21. Gao, H , Wu, G , Spencer, TE , Johnson, GA , and Bazer, FW . Select nutrients in the ovine uterine lumen. Ii. Glucose transporters in the uterus and peri-implantation conceptuses1. Biol Reprod. (2009) 80:94–104. doi: 10.1095/biolreprod.108.071654
22. Gao, H , Wu, G , Spencer, TE , Johnson, GA , and Bazer, FW . Select nutrients in the ovine uterine lumen. Iii. Cationic amino acid transporters in the ovine uterus and peri-implantation conceptuses1. Biol Reprod. (2009) 80:602–9. doi: 10.1095/biolreprod.108.073890
23. Suzuki, T , Sakumoto, R , Hayashi, KG , Ogiso, T , Kunii, H , Shirozu, T, et al. Involvement of interferon-tau in the induction of apoptotic, pyroptotic, and autophagic cell death-related signaling pathways in the bovine uterine endometrium during early pregnancy. J Reprod Dev. (2018) 64:495–502. doi: 10.1262/jrd.2018-063
24. Skarzynski, DJ , Miyamoto, Y , and Okuda, K . Production of prostaglandin f2α by cultured bovine endometrial cells in response to tumor necrosis factor α: cell type specificity and intracellular mechanisms1. Biol Reprod. (2000) 62:1116–20. doi: 10.1095/biolreprod62.5.1116
25. Chanrot, M , Guo, Y , Dalin, AM , Persson, E , Båge, R , Svensson, A, et al. Dose related effects of lps on endometrial epithelial cell populations from dioestrus cows. Anim Reprod Sci. (2017) 177:12–24. doi: 10.1016/j.anireprosci.2016.12.002
26. Palma-Vera, SE , and Einspanier, R . Experimental and bioinformatic analysis of cultured bovine endometrial cells (bend) responding to interferon tau (ifnt). Reprod Biol Endocrinol. (2016) 14:22. doi: 10.1186/s12958-016-0156-y
27. Zhao, G , Jiang, K , Zhang, T , Wu, H , Qiu, C , and Deng, G . Specific interferon tau gene-regulation networks in bovine endometrial luminal epithelial cells. Theriogenology. (2018) 105:51–60. doi: 10.1016/j.theriogenology.2017.09.004
28. Chaney, HL , Grose, LF , Charpigny, G , Behura, SK , Sheldon, IM , Cronin, JG, et al. Conceptus-induced, interferon tau-dependent gene expression in bovine endometrial epithelial and stromal cells†. Biol Reprod. (2021) 104:669–83. doi: 10.1093/biolre/ioaa226
29. Adhikari, B , Lee, CN , Khadka, VS , Deng, Y , Fukumoto, G , Thorne, M, et al. Rna-sequencing based analysis of bovine endometrium during the maternal recognition of pregnancy. BMC Genomics. (2022) 23:494. doi: 10.1186/s12864-022-08720-4
30. Malakhova, OA , Kim, K II, Luo, J , Zou, W , Kumar, KS , Fuchs, SY, et al. Ubp43 is a novel regulator of interferon signaling independent of its isg15 isopeptidase activity. EMBO J. (2006) 25:2358–67. doi: 10.1038/sj.emboj.7601149
31. Forde, N , Carter, F , Spencer, TE , Bazer, FW , Sandra, O , Mansouri-Attia, N, et al. Conceptus-induced changes in the endometrial transcriptome: how soon does the cow know she is pregnant?1. Biol Reprod. (2011) 85:144–56. doi: 10.1095/biolreprod.110.090019
32. Zhang, X , Gong, S , Li, H , Jiang, J , Jia, Y , Zhang, R, et al. Usp18 promotes endometrial receptivity via the jak/stat1 and the isgylation pathway. Theriogenology. (2023) 202:110–8. doi: 10.1016/j.theriogenology.2023.03.011
33. Murray, MJ , and Lessey, BA . Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Semin Reprod Endocrinol. (1999) 17:275–90. doi: 10.1055/s-2007-1016235
34. Yang, B , Song, J , Sun, H , Xing, J , Yang, Z , Wei, C, et al. Psmb8 regulates glioma cell migration, proliferation, and apoptosis through modulating erk1/2 and pi3k/akt signaling pathways. Biomed Pharmacother. (2018) 100:205–12. doi: 10.1016/j.biopha.2018.01.170
35. Xie, Y , Liu, G , Zang, X , Hu, Q , Zhou, C , Li, Y, et al. Differential expression pattern of goat uterine fluids extracellular vesicles mirnas during peri-implantation. Cell. (2021) 10:2308. doi: 10.3390/cells10092308
36. Groebner, AE , Schulke, K , Unterseer, S , Reichenbach, HD , Reichenbach, M , Büttner, M, et al. Enhanced proapoptotic gene expression of xaf1, casp8 and tnfsf10 in the bovine endometrium during early pregnancy is not correlated with augmented apoptosis. Placenta. (2010) 31:168–77. doi: 10.1016/j.placenta.2009.12.017
37. Tassell, W , Slater, M , Barden, JA , and Murphy, CR . Endometrial cell death during early pregnancy in the rat. Histochem J. (2000) 32:373–9. doi: 10.1023/A:1004069731540
38. Okano, ANIO , Ogawa, H , Takahashi, H , and Geshi, M . Apoptosis in the porcine uterine endometrium during the estrous cycle, early pregnancy and post partum. J Reprod Dev. (2007) 53:923–30. doi: 10.1262/jrd.18139
39. Crighton, D , Wilkinson, S , O'Prey, J , Syed, N , Smith, P , Harrison, PR, et al. Dram, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. (2006) 126:121–34. doi: 10.1016/j.cell.2006.05.034
40. Xie, X , Le, L , Fan, Y , Lv, L , and Zhang, J . Autophagy is induced through the ros-tp53-dram1 pathway in response to mitochondrial protein synthesis inhibition. Autophagy. (2014) 8:1071–84. doi: 10.4161/auto.20250
41. Lu, T , Zhu, Z , Wu, J , She, H , Han, R , Xu, H, et al. Dram1 regulates autophagy and cell proliferation via inhibition of the phosphoinositide 3-kinase-akt-mtor-ribosomal protein s6 pathway. Cell Commun Signal. (2019) 17:17. doi: 10.1186/s12964-019-0341-7
42. Yang, D , Jiang, T , Liu, J , Hong, J , Lin, P , Chen, H, et al. Hormone regulates endometrial function via cooperation of endoplasmic reticulum stress and mtor-autophagy. J Cell Physiol. (2018) 233:6644–59. doi: 10.1002/jcp.26315
43. Chen, G , Lin, Y , Chen, L , Zeng, F , Zhang, L , Huang, Y, et al. Role of dram1 in mitophagy contributes to preeclampsia regulation in mice. Mol Med Rep. (2020) 22:1847–58. doi: 10.3892/mmr.2020.11269
44. Nuttinck, F , Jouneau, A , Charpigny, G , Hue, I , Richard, C , Adenot, P, et al. Prosurvival effect of cumulus prostaglandin g/h synthase 2/prostaglandin2 signaling on bovine blastocyst: impact on in vivo posthatching development†. Biol Reprod. (2017) 96:531–41. doi: 10.1095/biolreprod.116.145367
45. Makker, A , Goel, MM , Nigam, D , Bhatia, V , Mahdi, AA , Das, V, et al. Endometrial expression of homeobox genes and cell adhesion molecules in infertile women with intramural fibroids during window of implantation. Reprod Sci. (2017) 24:435–44. doi: 10.1177/1933719116657196
Keywords: bovine, early pregnancy, endometrial epithelial cells, interferon tau, transcriptomic profiling
Citation: Yu J, Liu C, Chen H, Xiang M, Hu X, Zhong Z, Liu Q, Wang D and Cheng L (2024) Transcriptomic analysis of bovine endometrial epithelial cells in response to interferon tau and hormone stimulation. Front. Vet. Sci. 11:1344259. doi: 10.3389/fvets.2024.1344259
Edited by:
Kirsten E. Scoggin, University of Kentucky, United StatesReviewed by:
Hossam El-Sheikh Ali, Mansoura University, EgyptLinjun Hong, South China Agricultural University, China
Copyright © 2024 Yu, Liu, Zhong, Hu, Xiang, Chen, Liu, Wang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Cheng, Y2hlbmdsZWlfMDExQDEyNi5jb20=
 Jie Yu
Jie Yu Chenhui Liu1
Chenhui Liu1 Zhuxia Zhong
Zhuxia Zhong