- 1Key Laboratory of Livestock Disease Prevention of Guangdong Province, Key Laboratory for Prevention and Control of Avian Influenza and Other Major Poultry Diseases, Ministry of Agriculture and Rural Affairs, Guangdong Provincial Observation and Research Station for Animal Disease, Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
Porcine circoviruses disease (PCVD), caused by porcine circovirus (PCVs), is an important swine disease characterized by porcine dermatitis, nephrotic syndrome and reproductive disorders in sows. However, diseases caused by PCV2, PCV3, or PCV4 are difficult to distinguish, so a simple, rapid, accurate and high-throughput diagnostic and identification method is urgently needed to differentiate these three types. In this study, specific primers and probes were designed based on the conserved region sequences of the Rep gene of PCV2, and the Cap gene of PCV3 and PCV4. A multiplex qPCR assay was developed and optimized that the limit of detection concentration could reach as low as 3.8 copies/μL, with all correlation coefficients (R2) exceeding 0.999. Furthermore, the method showed no cross-reaction with other crucial porcine viral pathogens, and both intra-repeatability and inter-reproducibility coefficients of variation were below 2%. The assay was applied to the detection of 738 pig samples collected from 2020 to 2021 in Guangdong Province, China. This revealed positive infection rates of 65.18% for PCV2, 29.27% for PCV3, and 0% for PCV4, with a PCV2/PCV3 co-infection rate of 23.17%. Subsequently, complete genome sequences of 17 PCV2 and 4 PCV3 strains were obtained from the above positive samples and pre-preserved positive circovirus samples. Nucleotide sequence analysis revealed that the 17 PCV2 strains shared 96.7–100% complete nucleotide identity, with 6 strains being PCV2b and 11 strains being PCV2d; the 4 PCV3 strains shared 98.9–99.4% complete nucleotide identity, with 2 strains being PCV3a-1 and 2 strains being PCV3b. This research provides a reliable tool for rapid PCVs identification and detection. Molecular epidemiological investigation of PCVs in pigs in Guangdong Province will help us to understand PCV2 and PCV3 epidemiological characteristics and evolutionary trends.
1 Introduction
Porcine circoviruses (PCVs) are circular, non-enveloped, single-stranded DNA viruses that are prevalent worldwide and pose a serious challenge to pig industry development (1). Circoviruses have the smallest genome length of all known animal viruses, ranging from 1758 to 2000 nt and containing two major open reading frames (ORFs). The ORF1 gene is located on the genome sense strand and encodes the replication-associated protein (Rep), while the ORF2 gene is located in the opposite orientation and encodes the capsid protein (Cap), which determines the antigenic characteristics of circoviruses (2). Currently, four species of PCVs have been identified, including PCV1, PCV2, PCV3, and PCV4 (3–5). PCV1 is generally considered non-pathogenic and is widespread in pigs; it also plays as a source of contamination of cell lines (6). PCV2 is considered a causative agent of porcine circovirus diseases (7). Based on phylogenetic analysis of the genome and ORF2 sequences, PCV2 can be divided into 8 subtypes (PCV2a-PCV2h) (8). Among them, PCV2a, PCV2b, and PCV2d are transmitted in pig populations worldwide, whereas PCV2c is currently isolated only from samples from Brazil and Denmark, and PCV2e is mainly distributed in the United States and Mexico (9, 10). PCV3 was initially detected in pigs with porcine dermatitis and nephropathy syndrome (PDNS) in the United States using high-throughput sequencing in 2016 (2, 11). It is not yet apparent how PCV3 contributes to illness pathogenesis or whether disease-related damage can occur. In addition, PCV3 has been found in a variety of animals, including wild boar and farmed pigs (12, 13). Currently, there are several PCV3 genotyping methods, for example, based on the consistent differences in the amino acid positions 24 and 27 of the capsid protein, PCV3 is classified into three genotypes (PCV3a, PCV3b, and PCV3c). There is no apparent correlation between PCV3 subtypes and their source strains (8, 14, 15). Finally, PCV4 was initially detected in Hunan Provence, China, in 2019, and has been reported in recent years in Guangxi Province, Henan Province and Sichuan Province in China (5, 16–18). It has been suspected to be associated with severe clinical conditions, such as PDNS, respiratory signs, and enteric manifestations (16, 19).
PCVs infection in pigs can be either a single genotype or a co-infection with multiple genotypes. Due to their similar clinical symptoms, molecular biology methods are usually required for differential diagnosis in clinical settings (20). Currently, commonly used PCVs detection methods include enzyme-linked immunosorbent assay (2, 21), and conventional PCR. Although these methods can detect PCV, they are time-consuming, cumbersome, and unable to simultaneously identify multiple pathogens, so cannot fully meet practical production needs. Therefore, in this study, a TaqMan triple real-time PCR assay for rapid differential detection of PCV2, PCV3 and PCV4 was established and also applied in an epidemiological PCVs investigation in Guangdong Province from 2020 to 2021. Complete genome sequence amplification and phylogenetic analysis of PCV2 and PCV3 were performed on the 738 samples collected from pig farms in Guangdong Province from 2020 to 2021 and PCV-positive samples kept in our laboratory from 2015 to 2021. Our aim was to provide detection tools and relevant references for clinical diagnosis and epidemiologic investigation of PCVs.
2 Materials and methods
2.1 Primers and probes
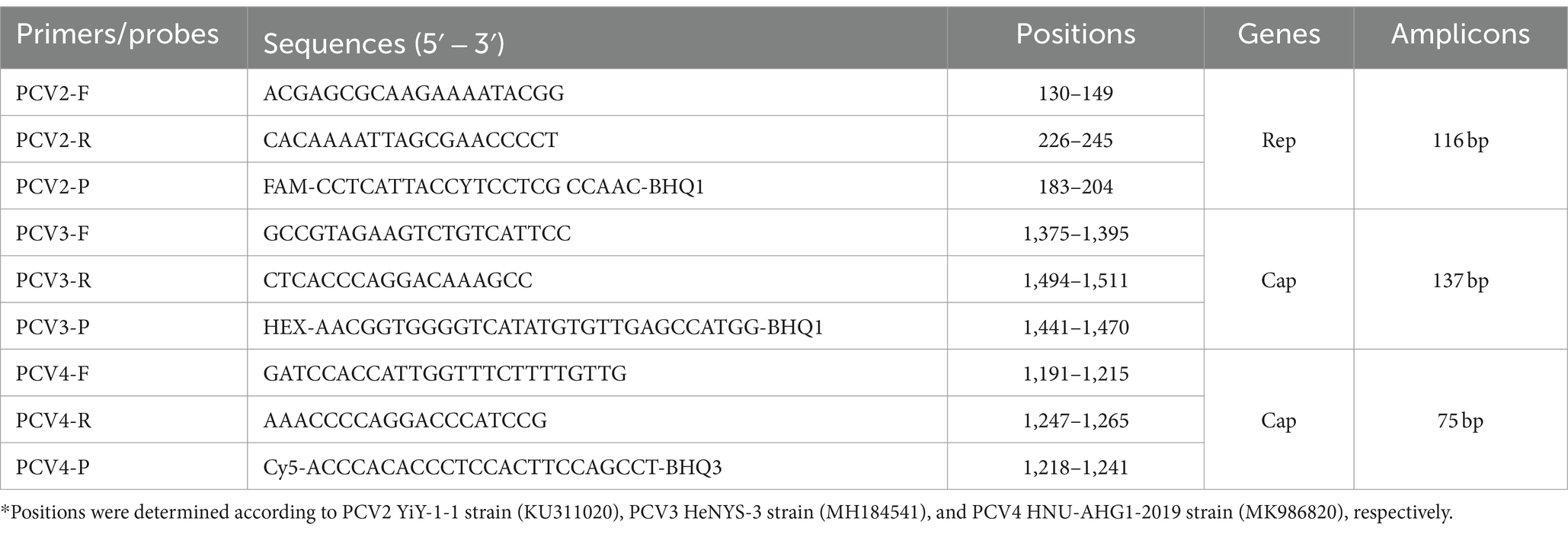
A total of 282 PCV complete genomes (175 PCV2, 82 PCV3, and 25 PCV4) were downloaded from the GenBank database. Using MEGA-X software, a multiple sequence alignment was performed to select specific conserved regions. The conserved region of Rep was chosen for designing the primer and probe for PCV2, and the Cap conserved region was chosen for PCV3 and PCV4. Specific primers and probes for each type were designed at the conserved sequences using primer 6.0 software, and qPCR amplification fragment sizes were as follows: PCV2 (116 bp), PCV3 (137 bp) and PCV4 (75 bp). All primer and probe sequences synthesized by Beijing Tsingke Biotech Co., Ltd. (Beijing, China) are shown in Table 1.
2.2 Virus and clinical samples
All viruses used for specificity validation were obtained from commercial vaccines purchased from Jilin Zhengye Biological Products Co., Ltd. (Jilin, China), including Porcine Reproductive and Respiratory Syndrome virus (PRRSV, HuN4-F112), Pseudorabies virus (PRV, Bartha-K61), Porcine Transmissible Gastroenteritis virus (PTGV), Porcine Epidemic Diarrhea virus (PEDV), Porcine Rotavirus (PoRV), and Classical Swine Fever virus (CSFV).
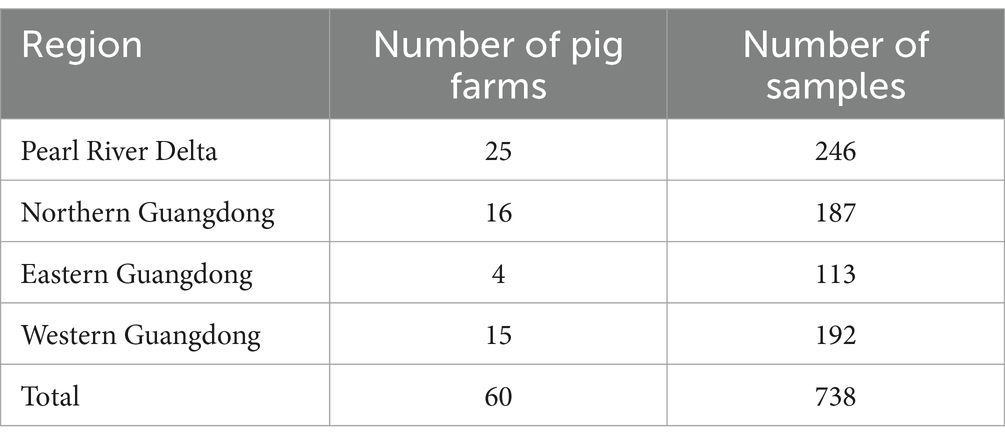
During 2020–2021, a total of 738 samples were collected from swine farms in Guangdong province and preserved in our laboratory, including 326 nasopharyngeal swab samples and 412 blood samples (Table 2). Molecular epidemiological investigation of the above samples was conducted using the established multiplex qPCR assay established in this study. Furthermore, complete genome amplification and genetic evolutionary analysis were also performed on the positive samples detected in 2020–2021 as described above and PCV-positive samples kept in our laboratory from 2015 to 2021.
2.3 DNA/RNA extraction and reverse transcription
RNA/DNA was extracted from samples or vaccines using a RaPure Viral RNA/DNA Kit (Magen, Guangzhou, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized using extracted viral RNA and a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). DNA and cDNA products were stored at −80°C.
2.4 Recombinant plasmid construction
To construct the recombinant standard plasmid, the multiplex qPCR assay amplification products (PCV2-PCV3-PCV4) were connected together as a standard sequence for synthesis (Tsingke, Beijing, China). The synthesized sequence was cloned into the pUC57 vector and transformed into DH5α competent cells. Then, the bacterial cultures were shaken and grown for 16 h at 37°C. The recombinant plasmid was purified using a GeneJET Plasmid Miniprep Kit (Thermo Scientific, USA) and the concentration was determined using Qubit 4.0 (Thermo Scientific, USA). The final plasmid obtained was named PUC57-PCVs. The plasmid concentration was converted into a copy number using the following formula:
2.5 Multiplex qPCR assay optimization
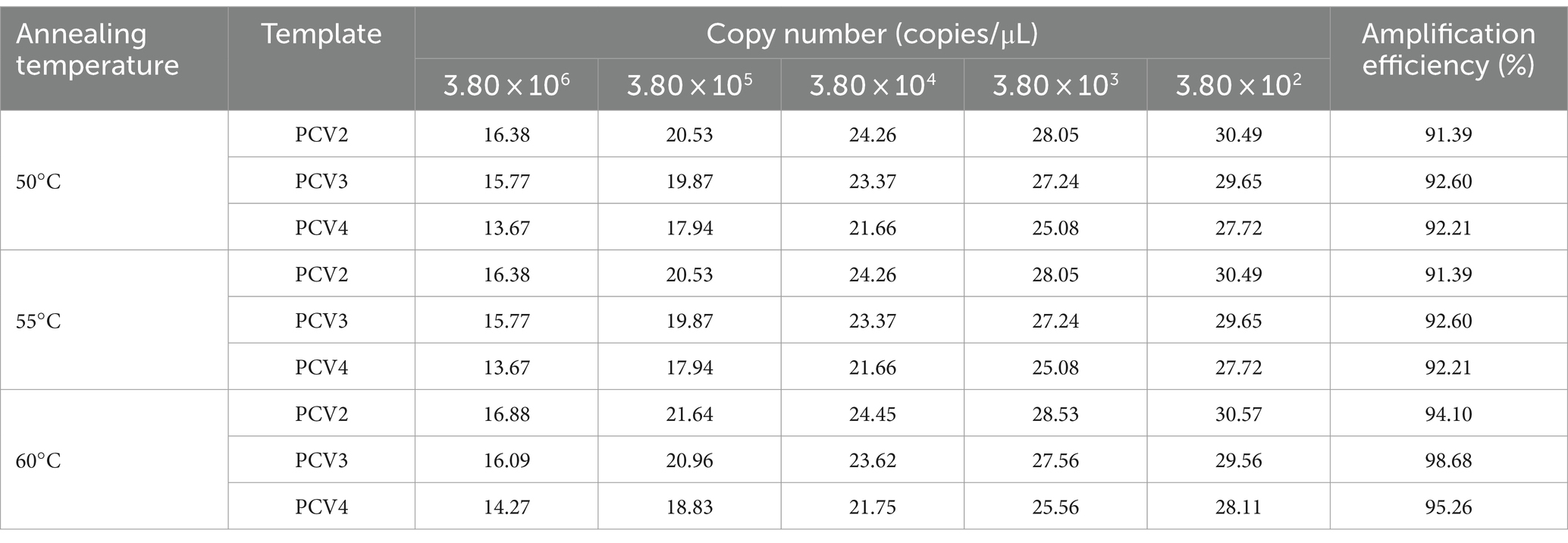
The multiplex qPCR conditions were optimized, including annealing temperature, primer and probe concentrations. The reaction system was 20 μL, including 10 μL of 2 × AceQ Universal U+ Probe Master Mix V2 (Vazyme, Nanjing, China), 2 μL of templates, different final concentrations of primers, probes, and nuclease-free water. The Optimization of the reaction conditions was performed utilizing a matrix approach with annealing temperatures ranging from 50°C to 60°C; final primer concentrations spanning from 100 nM to 500 nM in nine incremental dilutions; and final probe concentrations set at 50, 100, 150, and 200 nM, respectively. Amplification conditions were 37°C for 2 min, pre-denaturation at 95°C for 300 s, followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s. The qPCR instrument fluorescence channels were configured as follows: channel 1, FAM; channel 2, HEX; and channel 3, Cy5. Fluorescence signals were collected by a LightCycler® 96 Instrument (Roche Applied Science, Penzberg, Germany). Standard plasmids were diluted from 3.8 × 106 copies/μL to 3.8 × 102 copies/μL as amplification templates, and the system was optimized by generating the lowest threshold cycle (Ct) and the highest cycle increase (ΔRn) for each specific fluorescent signal.
2.6 Standard curve construction
Based on the optimal reaction system, the standard plasmid was diluted 10-fold from 3.8 × 106 copies/μL to 3.8 × 102 copies/μL, and multiplex qPCR was performed with five concentrations of the standard plasmid as templates. The standard curve was plotted using Prism software.
2.7 Specificity, sensitivity, repeatability assay
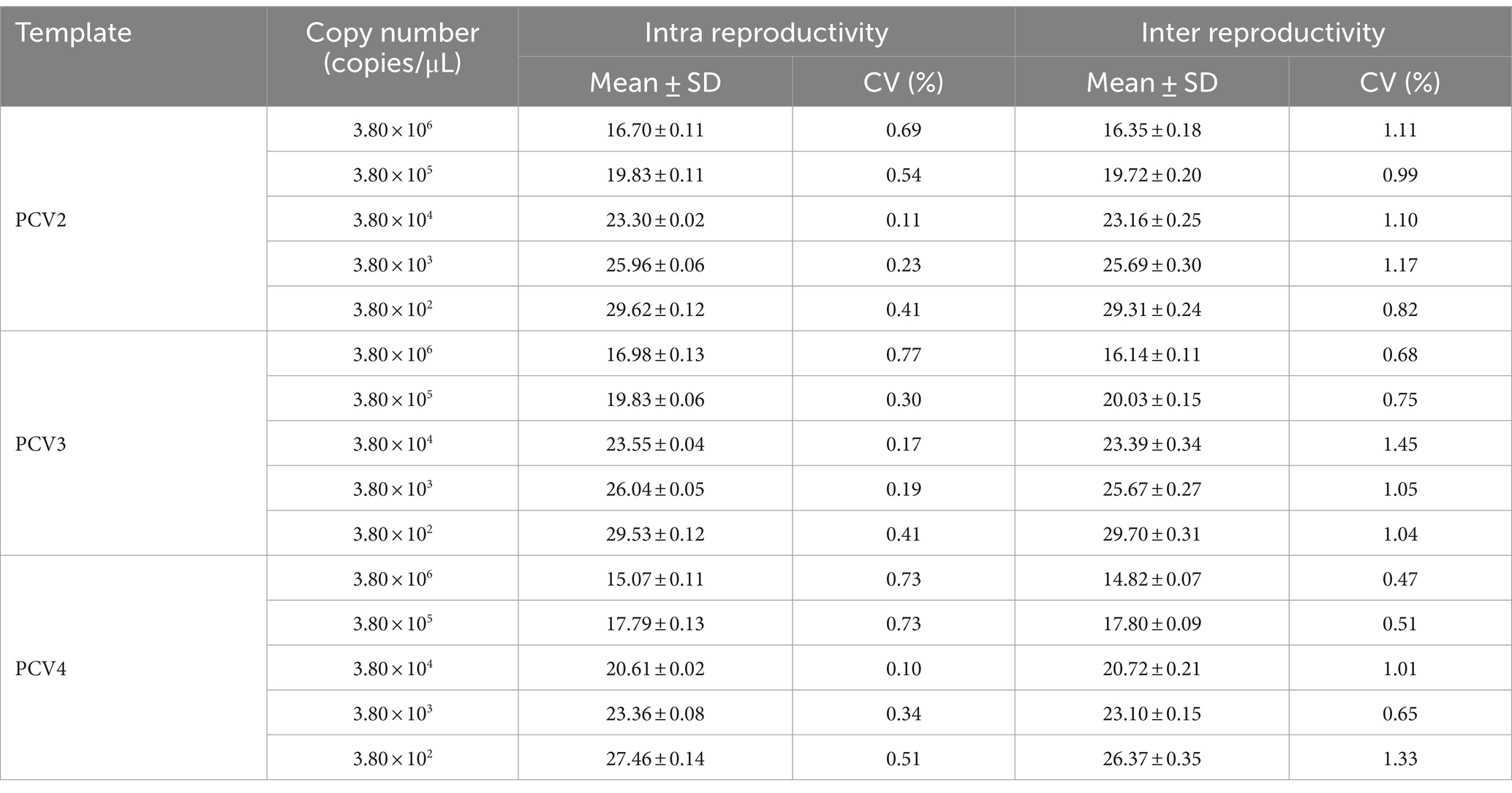
To assess the specificity of the multiplex qPCR assay, the PUC57-PCVs standard plasmid was used as the positive control and ddH2O as the negative control, DNA/cDNA of PRRSV, PEDV, CSFV, PTGV, PoRV and PRV extracted from the vaccine strains were used as templates for the qPCR assay detection. To determine the sensitivity, the standard plasmid was diluted 10-fold from 3.8 × 106 copies/μL to 3.8 × 100 copies/μL, qPCR amplification was performed in an optimal reaction system, and three replicates of each concentration were performed to confirm the detection limit of this multiplex qPCR assay. Intra-assay and inter-assay reproducibility was assessed using standard plasmids (3.8 × 106 copies/μL to 3.8 × 102 copies/μL) as templates. For intra-assay reproducibility, three parallel replicates of each dilution were performed daily under the same conditions, and for inter-assay reproducibility, each dilution was tested by six independent experiments performed by two operators on different days according to the MIQE guidelines (22). Coefficients of variation of the Ct values were calculated based on the intra-assay or inter-assay results.
2.8 Clinical sample detection
From 2020 to 2021, 738 clinical samples, including 326 nasopharyngeal swabs and 412 blood samples, were collected from swine farms in Guangdong Province, China. These samples were subsequently analyzed using an established multiplex qPCR assay to determine infection rates.
2.9 Complete genome amplification and sequencing
PCV2 (23) and PCV3 (20) complete genomes were amplified using the primers described previously. The reaction system was 50 μL, containing 25 μL of 2 × Platinum™ II Taq Hot-Start DNA Polymerase (Thermo Scientific, USA), 2 μL of template DNA, 200 nM primers, and the remaining was added to ddH2O. The PCR reaction for PCV2 was executed by pre-denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 56°C for 15 s, and 68°C for 20 s. PCV3 amplification conditions were similar to those of PCV2, except that the PCV3 primer set was annealed at 57°C. The purified PCR products were cloned using a pMD18-T vector cloning kit (Takara, Dalian, China), and propagated in DH5α competent cells (Takara, Dalian, China) according to the manufacturer’s instructions. Positive clones were sequenced by Beijing Tsingke Biotech Co., Ltd. The obtained complete genome sequences were edited and assembled using DNAstar V7.1 software.
2.10 Sequence analysis
A total of 17 PCV2 and 4 PCV3 complete genomes were obtained in this study (Supplementary Table 1) and uploaded to NCBI GenBank with accession numbers MZ667312 (PCV2), MZ667314-MZ667329 (PCV2) and MZ667331-MZ667334 (PCV3). The complete PCV3 coding sequence (ORF1 + ORF2) was analyzed. Non-coding regions were deleted and the coding sequence was split into two parts, ORF1 (Rep) and ORF2 (Cap). Because ORF2 was in the opposite orientation, respective ORFs were downloaded and spliced separately. PCV3 subtypes were proposed by Li et al. (24). The PCV2 strain ORF2 sequence and the complete PCV3 coding sequence were aligned with the relevant reference sequences in GenBank using the ClustalW function of the Molecular Evolutionary Genetics Analysis software (MEGA, version 11.0), respectively. Phylogenetic trees were generated by the neighbor-joining (NJ) method in MEGA 11.0 with a p-distance model, and a bootstrap of 1,000 replicates.
3 Results
3.1 Optimization of qPCR reaction conditions
Annealing temperatures of 50, 55, and 60°C, primer final concentrations of 100–500 nM and probe final concentrations of 100–250 nM were selected for multiplex qPCR system optimization. Based on the optimization results, the annealing temperature was determined to be 60°C (Table 3), final primer concentrations for PCV2, PCV3, and PCV4 were 300, 350 and 200 nM, respectively (Supplementary Tables 2–4), and final concentrations of all three probes were 100 nM (Supplementary Table 5). Therefore, the final amplification system was 10 μL of 2× AceQ Universal U+ Probe Master Mix V2, 300 nM PCV2 primer, 350 nM PCV3 primer, and 200 nM PCV4 primer, each with 100 nM for each probe, 2 μL of template, and ddH2O was added to a final volume of 20 μL. Reaction conditions were 37°C for 2 min, pre-denaturation at 95°C for 300 s, followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s.
3.2 Multiplex qPCR assay standard curve
The recombinant standard plasmid was diluted to 3.8 × 106 to 3.8 × 102 copies/μL in a 10-fold gradient, and multiplex qPCR assay was performed according to the optimal reaction system and procedure. The standard curve was established with the obtained Ct values as the y axis coordinates and the logarithm of the plasmid concentration as the x axis coordinates. All standard curves had good correlation coefficients and amplification efficiency. The slopes of PCV2, PCV3 and PCV4 were −3.383, −3.331 and −3.345, respectively; the correlation coefficient (R2) and amplification efficiency (Eff%) of the equation were as follows: PCV2 (R2 = 0.9991, Eff% = 97.51), PCV3 (R2 = 0.9992, Eff% = 99.62), PCV4 (R2 = 0.9991, Eff% = 99.05) (Figure 1).
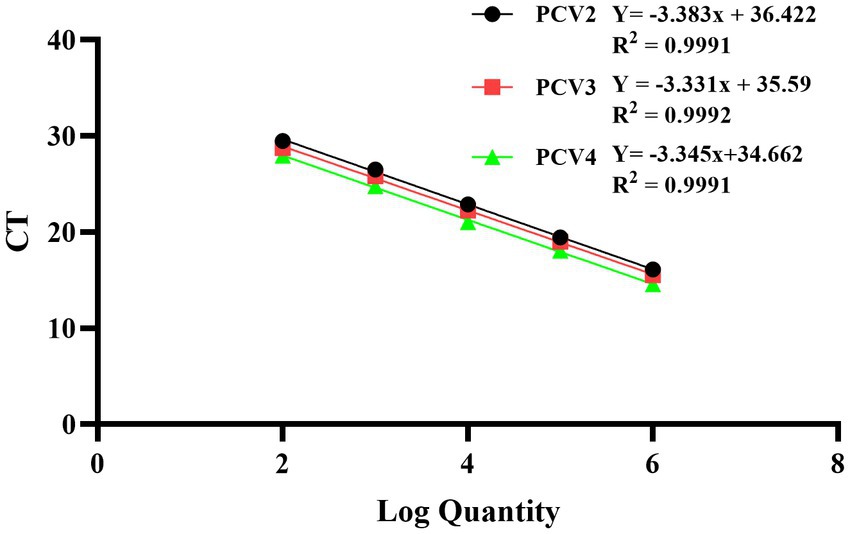
Figure 1. Standard curves of multiplex qPCR assay. Serial 10-fold dilutions of the standard plasmid were generated at final concentrations of 3.8 × 106–3.8 × 102 copies/μL. Data are from three independent studies. The threshold cycles (Ct) from the multiplex qPCR assay are plotted against the log numbers of the standards. Mean ± standard deviations are shown for each individual study.
3.3 Multiplex qPCR assay specificity
To assess the specificity of the multiplex qPCR, the recombinant standard plasmid PUC57-PCVs was used as a positive control. DNA/cDNA extracted from PRRSV, PEDV, CSFV, PTGV, PoRV, and PRV vaccines were used as templates, and ddH2O was used as a negative control. qPCR was performed using an optimal reaction system. Specific fluorescent signals for FAM, HEX and Cy5 were detected only when the PUC57-PCVs standard plasmid was used as a template. However, DNA and cDNA of other pathogens such as PRRSV, PEDV CSFV, and PRV were not amplified (Figure 2). These results support high specificity of the multiplex qPCR assay.
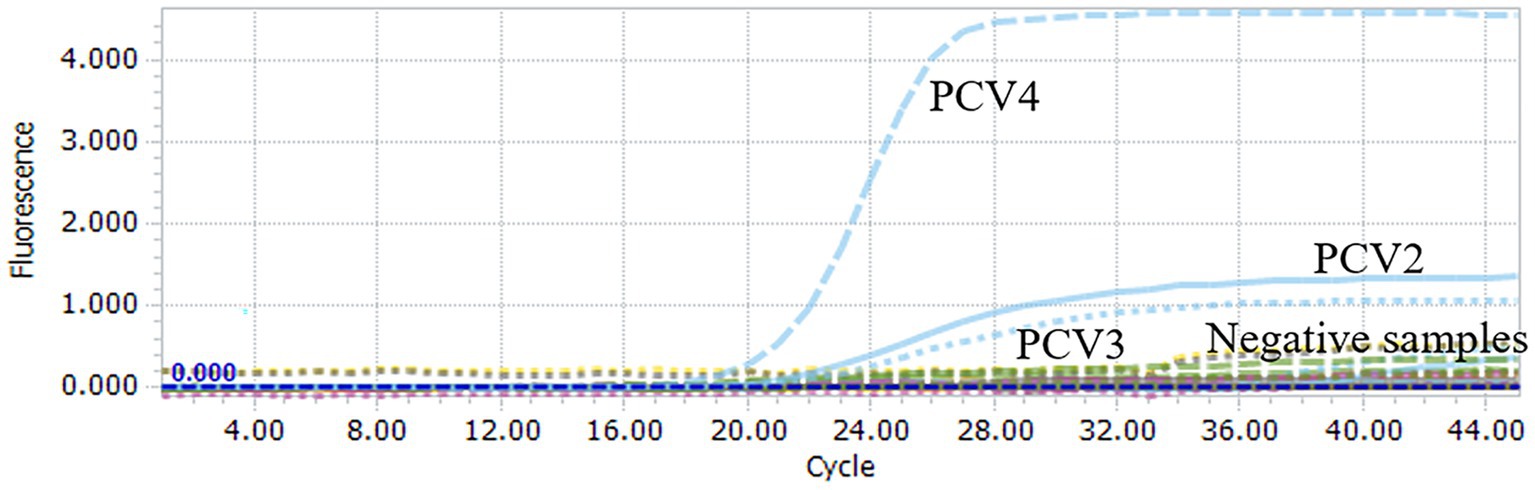
Figure 2. Specificity analysis of multiplex qPCR assay. When PUC57-PCVs plasmid was used, FAM, HEX and Cy5 fluorescent signals were specifically detected, respectively. No specific fluorescence signal was obtained when testing other viruses (PRRSV, PRV, PTGV, PEDV, PoRV, CSFV) and negative controls.
3.4 Multiplex qPCR assay sensitivity
To further assess the sensitivity of the multiplex qPCR assay, multiplex qPCR was performed using standard plasmids with concentrations ranging from 3.8 × 106 to 3.8 × 100 copies/μL. The results show that the detection limits of the assay were 3.8 × 100 copies/μL for PCV2, PCV3 and PCV4 (Figure 3), suggesting that the multiplex qPCR assay has a good sensitivity.
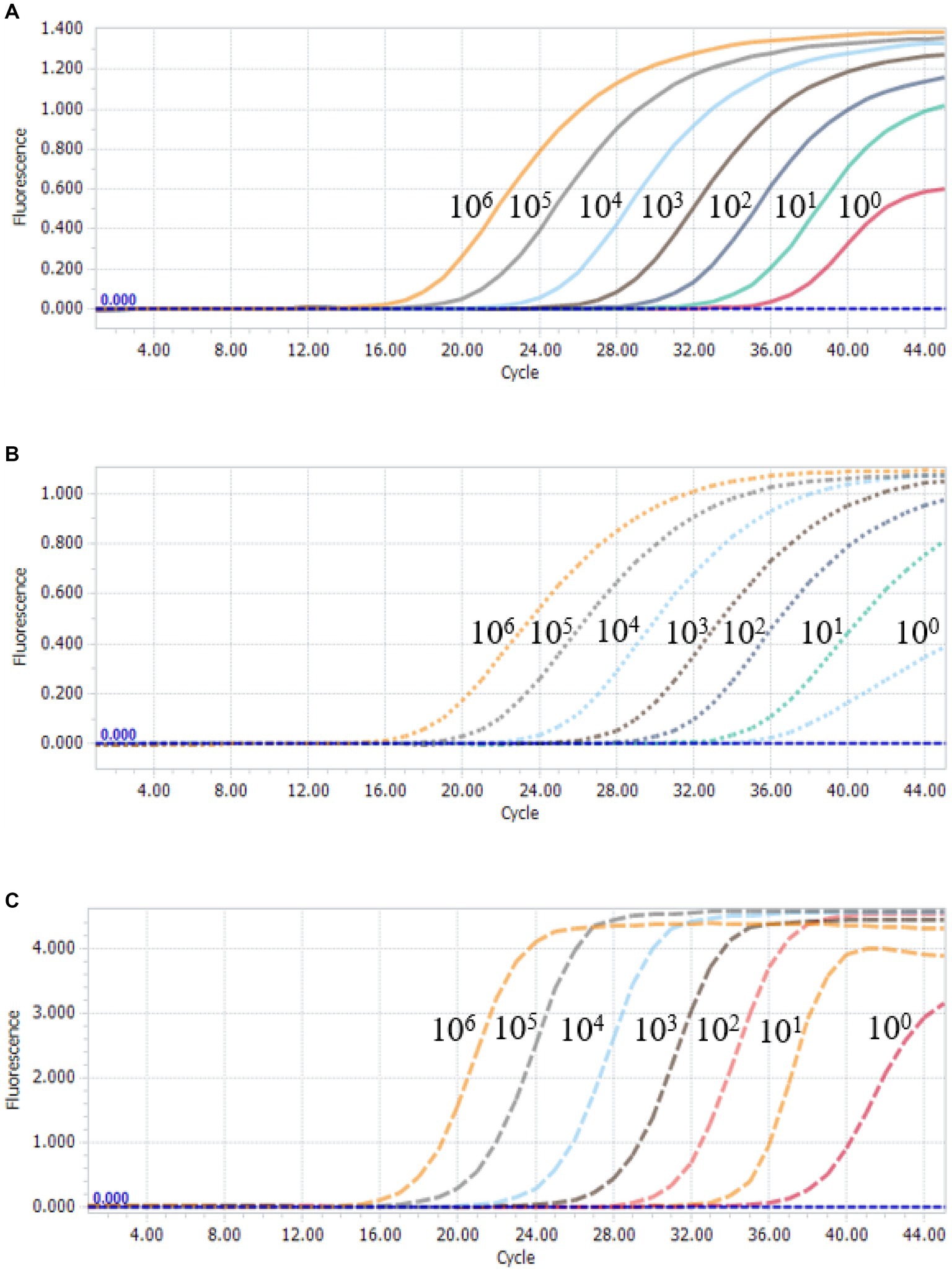
Figure 3. Sensitivity analysis of multiplex qPCR assay. Serial 10-fold serial dilutions of the standard plasmid were generated at final concentrations of 3.8 × 106–3.8 × 100 copies/μL. The multiplex qPCR assay has the detection limit at 3.8 × 100 copies/μL for each species of PCVs. (A) Sensitivity analysis of PCV2. (B) Sensitivity analysis of PCV3. (C) Sensitivity analysis of PCV4.
3.5 Multiplex qPCR assay repeatability
To assess the reproducibility of the multiplex qPCR assay, standard plasmids with concentrations ranging from 3.8 × 106 to 3.8 × 102 copies/μL were tested, respectively. Intra- and inter-assay coefficients of variation (CV) of Ct values for the multiplex qPCR assay ranged from 0.10 to 0.77% and 0.47 to 1.45%, respectively (Table 4). These results indicated that the multiplex qPCR assay established in this study has good reproducibility and reliability.
3.6 Clinical sample detection
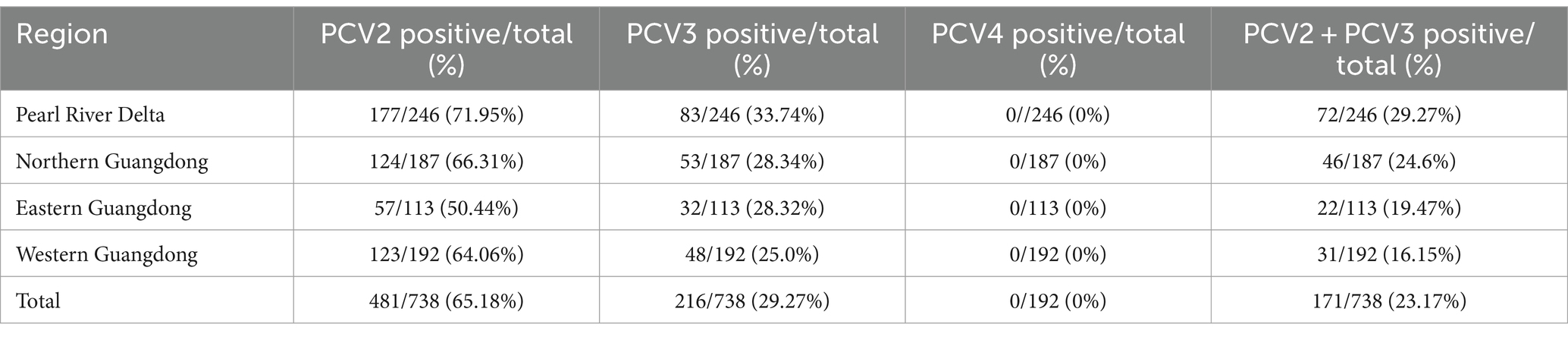
738 samples collected from different swine farms in Guangdong Province during 2020–2021 were tested using the method established in this study. Among the 738 samples, PCV2 infection rate was 65.18% (481/738), PCV3 infection rate was 29.27% (216/738), and PCV2/PCV3 co-infection rate was 23.17% (171/738) (Table 5). In the Pearl River Delta, western Guangdong, eastern Guangdong and northern Guangdong, PCV2 infection rate ranged from 50.44 to 71.95%, PCV3 infection rate ranged from 25.0 to 33.74%, and PCV2 and PCV3 co-infection rate ranged from 16.15 to 29.27%. For nasopharyngeal swabs, PCV2 infection rate was 70.25% (229/326), PCV3 infection rate was 25.77% (84/326), and PCV2/PCV3 co-infection rate was 21.17% (69/326). For blood samples, infection rate for PCV2 was 61.17% (252/412), 32.04% for PCV3 (132/412), and for PCV2/PCV3 co-infection rate was 24.76% (102/412) (Table 6). PCV4 was not detected in the above samples.
3.7 PCV2 genetic characteristics
We attempted Complete genome amplification from the above samples and PCV-positive samples kept in our laboratory from 2015 to 2021. A total of 17 PCV2 complete genomes were obtained, each of which was 1767 nt in size. A phylogenetic tree was constructed based on the ORF2 sequences of the 17 PCV2 strains and 35 PCV2 reference sequences (PCV2a-h) (Figure 4A). 17 PCV2 sequences we obtained could be classified into PCV2b (6 strains) and PCV2d (11 strains) subtypes. Nucleotide sequence analysis revealed that the 17 PCV2 strains shared 96.7–100% complete nucleotide identity and 94.0–100% ORF2 identity. Compared with the 35 PCV2 reference strains, the nucleotide identity of complete nucleotide and ORF2 varied 92.0 to 99.9% and 82.9 to 100%.
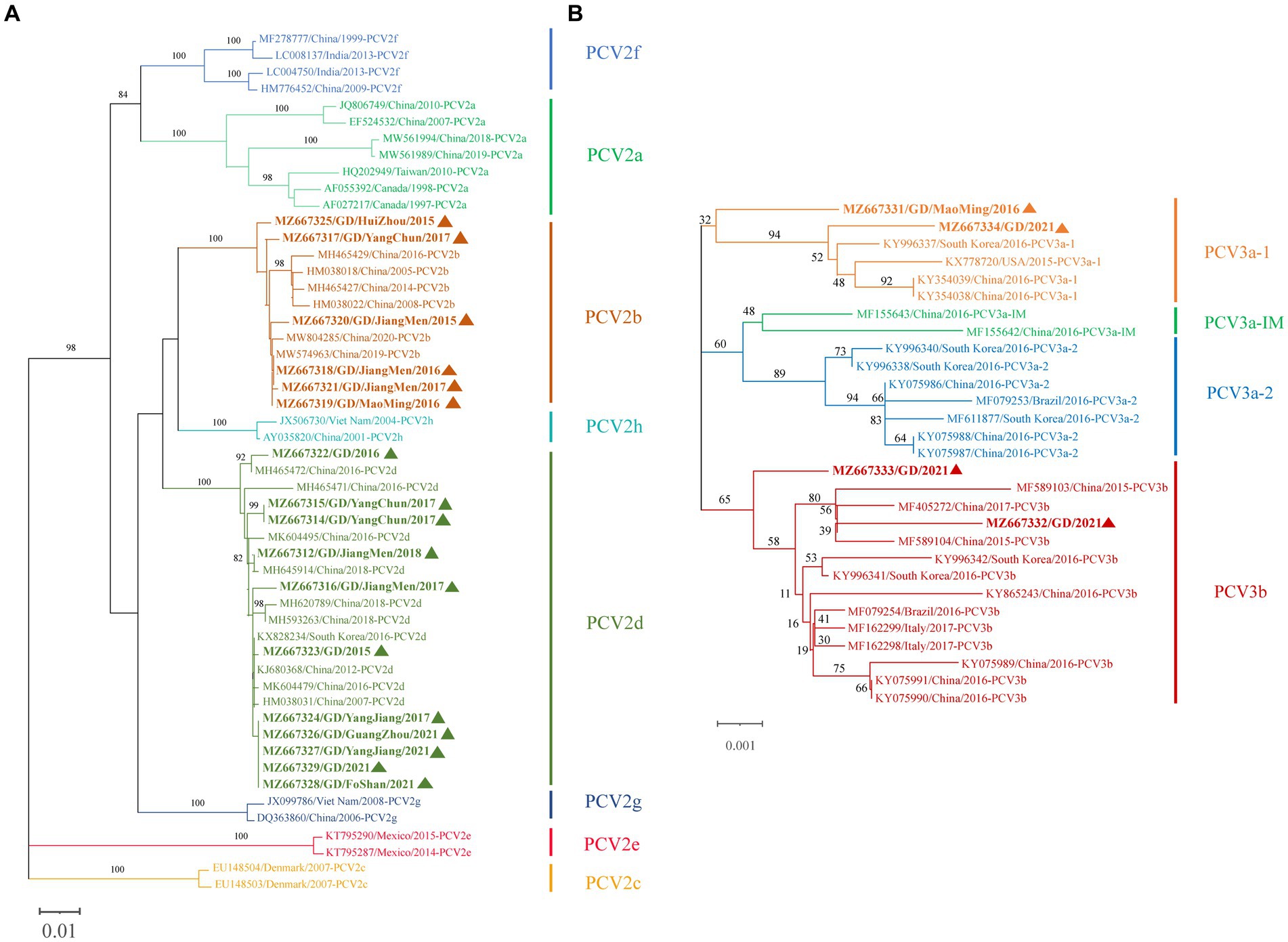
Figure 4. Phylogenetic tree based on ORF2 gene of PCV2 and the complete coding sequence (ORF1 + ORF2) of PCV3. The NJ tree was constructed using the p-distance model and bootstrapped at 1000 replicates. (A) PCV2. (B) PCV3. PCV3 isoforms were proposed by Li et al. (24). The sequences of PCV2 and PCV3 obtained in this study are marked with triangles. The different genotypes are represented by different colors as indicated in the figures.
3.8 PCV3 genetic characteristics
Similarly, we successfully obtained 4 PCV3 complete genomes, all of which was 2000 nt in size, which were submitted to NCBI GenBank. A phylogenetic tree was constructed based on the coding gene sequences (ORF1 + ORF2) of the 4 PCV3 strains and the reference sequences of 25 PCV3 strains (Figure 4B). Among the 4 PCV3 strains we obtained, two belonged to PCV3b and two belonged to PCV3a-1, which had different clustering levels. Nucleotide sequence analysis showed that the 4 PCV3 strains shared 98.9–99.4% complete nucleotide identity and 98.3–99.2% ORF2 identity. Compared with the 25 PCV3 reference strains, the nucleotide identity of complete nucleotide and ORF2 varied 98.8 to 99.7% and 97.8 to 99.7%.
4 Discussion
PCV2 is an important pathogen of porcine circovirus disease (PCVD), which is highly prevalent in swine herds and severely affects the global pig industry (7, 25). Since the discovery of PCV3 in 2016, increasing evidence suggests that this strain poses a severe threat to both sows and piglets (26–28), and the emergence of PCV4 in 2019 has further complicated porcine circovirus disease diagnosis (17, 23). The prevalence of PCV3 and PCV4 infections in pigs is increasing, although the pathogenicity of PCV3 and PCV4 in this area remains controversial (29). Several diagnosis methods have been developed for PCV, including immunohistochemistry, ELISA, RT-PCR, loop-mediated isothermal amplification (LAMP), and real-time RT-PCR. However, ELISA and immunohistochemistry are time-consuming and cannot detect viral DNA/cDNA copy numbers. Conversely, qPCR has been widely used for the diagnosis of related diseases because of its advantages over conventional PCR, such as high sensitivity and specificity for high-throughput screening and viral load quantification using small volumes. Based on the above objectives, we developed a multiplex qPCR assay for the simultaneous differential diagnosis of PCV2, PCV3, and PCV4. Through sequence alignment of the complete circovirus genome, primers and probes were finally designed for the rep gene of PCV2, the cap gene of PCV3 and PCV4. After system optimization, the primers and probes have high specificity and are not affected by other binding sites. The assay can detect PCVs plasmid at a concentration of 3.8 copies/μL, which is more sensitive than the assay established by Chen and Zou et al. (30, 31). Additionally, the coefficients of variation (CV) of the intra- and inter-group for this method ranged from 0.10 to 0.77% and 0.47 to 1.45%, providing good sensitivity and reproducibility. Thus, the newly established assay provides a relevant tool for more rapid and accurate diagnosis of different species of PCVs.
PCVs distribution and spread are quite prevalent among pig populations in China. According to Li et al., 15,130 samples from eight provinces were tested for PCV2 from 2018 to 2021, and overall PCV2 positivity rate was 24.04% (3,637/15,130) in some regions, with Guangdong province testing positive at a rate of 17.18% (315/1834) (32). Additionally, Nan et al. tested 573 samples for PCV2 from 132 pig farms in northern Guangdong Province from 2016 to 2021 and found a positive rate of 51.38% (297/573) (33). Recently, Zou et al. established a multiplex qPCR assay and tested 535 samples from Shandong, Zhejiang, Jiangsu and Anhui provinces in eastern China from 2020 to 2022. PCV2, PCV3 and PCV4 infection rates were 31.03, 30.09 and 30.84%, respectively, and the mixed infection rate was 28.22% (31). The above data suggests that PCV2 and PCV3 are more widely distributed and transmitted in swine herds in some regions of China. In this study, we used a newly established qPCR assay on porcine blood and nasopharyngeal swab samples collected in Guangdong Province in 2020–2021, and found that the infection rates of PCV2 and PCV3 were relatively high (65.18 and 29.27%, respectively), and PCV2/PCV3 co-infection rate of 23.17%, which was higher than that recently reported for circoviruses in Guangdong Province (34). The differences in these data can be attributed to various factors, including sample types, sample sizes, geographic location, health status, immune response, PCR detection methods, and other variables. Overall, the results of this study indicated that PCV2 and PCV3 are more widely distributed and spread in pig farms in Guangdong Province, but PCV4 was not detected in any of the clinical samples.
Since the first reported case of PCV2 infection in China in 2000, PCV2a was the predominant epidemic subtype in Chinese pig herds (35), but around 2003, PCV2b replaced PCV2a as the new major epidemic subtype (36). In recent years, PCV2d has become an emerging major prevalent genotype in China (37). In our study, 17 PCV2 strains were isolated with 96.7 to 100% complete genome sequence identity and 94.0 to 100% ORF2 identity, 6 of which belonged to PCV2b and 11 to PCV2d. Four PCV3 strains had 98.9 to 99.4% complete genome sequence identity and 98.3 to 99.2% ORF2 identity, of which 2 strains belonged to PCV3b and 2 belonged to PCV3a-1. Unfortunately, due to limitations of certain conditions, the complete genome of all the samples was not obtained so this data may not fully represent all Guangdong Province PCVs genotypes. Curiously, we did not obtain PCV2a sequence, which seems to be inconsistent with previous reports (33, 34). However, our findings indicating that PCV2d is still the main prevalent genotype, which is consistent with previous reports (38). The first case of PCV4 transmission was detected in Hunan Province in 2019, and recent studies have detected PCV4 epidemics in Henan, Jiangsu, and Shanxi provinces (5, 23). However, we did not detect PCV4 in swine samples from Guangdong Province, which may be due to the short time for sample collection, insufficient sample number and sample type. To determine the prevalence of PCV4, continuous monitoring of pigs from Guangdong Province is needed. Moreover, due to changing PCV2 subtypes and the presence of PCV3 and PCV4, the current commercial PCV2 vaccine has a relatively singular scope of action, and co-infections of PCVs with other porcine reproductive disorders have become common (39, 40). Therefore, it is necessary to conduct regular testing on swine farms to grasp the status of circovirus infection in a timely manner, in order to provide a basis for the prevention and control PCV.
5 Conclusion
We successfully established a triple TaqMan real-time PCR assay for simultaneous PCV2, PCV3 and PCV4 detection. The method exhibits excellent specificity, sensitivity and reproducibility. Additionally, molecular epidemiologic testing of PCVs conducted in Guangdong Province in 2020 to 2021 showed that the PCV2 infection rate was 65.18% (481/738), PCV3 infection rate was 29.27% (216/738), and PCV2/PCV3 co-infection rate was 23.17% (171/738). Finally, 17 PCV2 and 4 PCV3 complete genome sequences were obtained in this study. These results indicate that PCV2 and PCV3 are prevalent in swine herds in Guangdong Province, China, and that co-infections of these two viruses are common. The above data provide an important reference for PCVs epidemiological investigation and genetic variation in Guangdong Province, China.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/nuccore/; MZ667312, MZ667314-MZ667329, MZ667331-MZ667334.
Ethics statement
The animal studies were approved by Institute of Animal Health, Guangdong Academy of Agricultural Sciences. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
PZ: Formal analysis, Writing – original draft, Data curation. ZR: Methodology, Writing – original draft. XG: Formal analysis, Methodology, Writing – original draft. MZ: Data curation, Writing – original draft. YW: Methodology, Writing – original draft, Software. JC: Data curation, Writing – original draft. GW: Data curation, Writing – review & editing. HX: Data curation, Writing – original draft. RC: Data curation, Writing – original draft. SL: Methodology, Writing – original draft. XW: Conceptualization, Methodology, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following grants: the Project of Collaborative Innovation Center of GDAAS (XT202207), the Scientific and Technological Plan Projects of Guangzhou (grant numbers 2023E04J1256 and 2023B04J0137), the Planning Funds for Science and Technology of Guangdong Province (grant numbers 2021B1212050021 and 2023A1111110001), Key Laboratory of Livestock Disease Prevention of Guangdong Project (2023B1212060040), Guangdong Provincial Forestry Department’s Provincial Financial Special Fund for Ecological Forestry Construction-Wildlife Conservation, the Guangdong Province Modern Agriculture Industry Technology System Pig Innovation Team Project.
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1353439/full#supplementary-material
References
1. Mankertz, A, Caliskan, R, Hattermann, K, Hillenbrand, B, Kurzendoerfer, P, Mueller, B, et al. Molecular biology of porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol. (2004) 98:81–8. doi: 10.1016/j.vetmic.2003.10.014
2. Palinski, R, Piñeyro, P, Shang, P, Yuan, F, Guo, R, Fang, Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2017) 91:e01879-16. doi: 10.1128/JVI.01879-16
3. Lagan Tregaskis, P, Staines, A, Gordon, A, Sheridan, P, McMenamy, M, Duffy, C, et al. Co-infection status of novel parvovirus’s (PPV2 to 4) with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound Emerg Dis. (2021) 68:1979–94. doi: 10.1111/tbed.13846
4. Zhai, SL, Lu, SS, Wei, WK, Lv, DH, Wen, XH, Zhai, Q, et al. Reservoirs of porcine circoviruses: a mini review. Front Vet Sci. (2019) 6:319. doi: 10.3389/fvets.2019.00319
5. Zhang, HH, Hu, WQ, Li, JY, Liu, TN, Zhou, JY, Opriessnig, T, et al. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, Hunan province, China. Transbound Emerg Dis. (2020) 67:1057–61. doi: 10.1111/tbed.13446
6. Tischer, I, Gelderblom, H, Vettermann, W, and Koch, MA. A very small porcine virus with circular single-stranded DNA. Nature. (1982) 295:64–6. doi: 10.1038/295064a0
7. Segalés, J . Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. (2012) 164:10–9. doi: 10.1016/j.virusres.2011.10.007
8. Franzo, G, and Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One. (2018) 13:e0208585. doi: 10.1371/journal.pone.0208585
9. He, W, Zhao, J, Xing, G, Li, G, Wang, R, Wang, Z, et al. Genetic analysis and evolutionary changes of porcine circovirus 2. Mol Phylogenet Evol. (2019) 139:106520. doi: 10.1016/j.ympev.2019.106520
10. Xia, D, Huang, L, Xie, Y, Zhang, X, Wei, Y, Liu, D, et al. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch Virol. (2019) 164:2435–49. doi: 10.1007/s00705-019-04336-4
11. Phan, TG, Giannitti, F, Rossow, S, Marthaler, D, Knutson, TP, Li, L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. (2016) 13:184. doi: 10.1186/s12985-016-0642-z
12. Sirisereewan, C, Thanawongnuwech, R, and Kedkovid, R. Current understanding of the pathogenesis of porcine circovirus 3. Pathogens. (2022) 11:64. doi: 10.3390/pathogens11010064
13. Dei Giudici, S, Franzoni, G, Bonelli, P, Angioi, PP, Zinellu, S, Deriu, V, et al. Genetic characterization of porcine circovirus 3 strains circulating in Sardinian pigs and wild boars. Pathogens. (2020) 9:344. doi: 10.3390/pathogens9050344
14. Fux, R, Söckler, C, Link, EK, Renken, C, Krejci, R, Sutter, G, et al. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol J. (2018) 15:25. doi: 10.1186/s12985-018-0929-3
15. Ouyang, T, Zhang, X, Liu, X, and Ren, L. Co-infection of swine with porcine circovirus type 2 and other swine viruses. Viruses. (2019) 11:185. doi: 10.3390/v11020185
16. Sun, W, Du, Q, Han, Z, Bi, J, Lan, T, Wang, W, et al. Detection and genetic characterization of porcine circovirus 4 (PCV4) in Guangxi, China. Gene. (2021) 773:145384. doi: 10.1016/j.gene.2020.145384
17. Tian, RB, Zhao, Y, Cui, JT, Zheng, HH, Xu, T, Hou, CY, et al. Molecular detection and phylogenetic analysis of porcine circovirus 4 in Henan and Shanxi provinces of China. Transbound Emerg Dis. (2021) 68:276–82. doi: 10.1111/tbed.13714
18. Xu, T, You, D, Wu, F, Zhu, L, Sun, XG, Lai, SY, et al. First molecular detection and genetic analysis of porcine circovirus 4 in the southwest of China during 2021-2022. Front Microbiol. (2022) 13:1052533. doi: 10.3389/fmicb.2022.1052533
19. Niu, G, Zhang, X, Ji, W, Chen, S, Li, X, Yang, L, et al. Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo. Transbound Emerg Dis. (2022) 69:e1632–41. doi: 10.1111/tbed.14498
20. Ku, X, Chen, F, Li, P, Wang, Y, Yu, X, Fan, S, et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. (2017) 64:703–8. doi: 10.1111/tbed.12638
21. Yu, S, Vincent, A, Opriessnig, T, Carpenter, S, Kitikoon, P, Halbur, PG, et al. Quantification of PCV2 capsid transcript in peripheral blood mononuclear cells (PBMCs) in vitro. Vet Microbiol. (2007) 123:34–42. doi: 10.1016/j.vetmic.2007.02.021
22. Bustin, SA, Benes, V, Garson, JA, Hellemans, J, Huggett, J, Kubista, M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
23. Xu, T, Hou, CY, Zhang, YH, Li, HX, Chen, XM, Pan, JJ, et al. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene. (2022) 808:145991. doi: 10.1016/j.gene.2021.145991
24. Li, G, He, W, Zhu, H, Bi, Y, Wang, R, Xing, G, et al. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. Adv Sci Weinh Baden. (2018) 5:1800275. doi: 10.1002/advs.201800275
25. Tischer, I, Mields, W, Wolff, D, Vagt, M, and Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. (1986) 91:271–6. doi: 10.1007/BF01314286
26. Arruda, B, Piñeyro, P, Derscheid, R, Hause, B, Byers, E, Dion, K, et al. PCV3-associated disease in the United States swine herd. Emerg Microbes Infect. (2019) 8:684–98. doi: 10.1080/22221751.2019.1613176
27. Fu, X, Fang, B, Ma, J, Liu, Y, Bu, D, Zhou, P, et al. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg Dis. (2018) 65:e296–303. doi: 10.1111/tbed.12752
28. Tan, CY, Lin, CN, and Ooi, PT. What do we know about porcine circovirus 3 (PCV3) diagnosis so far?: a review. Transbound Emerg Dis. (2021) 68:2915–35. doi: 10.1111/tbed.14185
29. Niu, G, Chen, S, Li, X, Zhang, L, and Ren, L. Advances in crosstalk between porcine circoviruses and host. Viruses. (2022) 14:1419. doi: 10.3390/v14071419
30. Chen, N, Xiao, Y, Ye, M, Li, X, Li, S, Xie, N, et al. High genetic diversity of Chinese porcine reproductive and respiratory syndrome viruses from 2016 to 2019. Res Vet Sci. (2020) 131:38–42. doi: 10.1016/j.rvsc.2020.04.004
31. Zou, J, Liu, H, Chen, J, Zhang, J, Li, X, Long, Y, et al. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of porcine circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet Sci. (2022) 10:29. doi: 10.3390/vetsci10010029
32. Li, C, Liu, G, Tong, K, Wang, Y, Li, T, Tan, X, et al. Pathogenic ecological characteristics of PCV2 in large-scale pig farms in China affected by African swine fever in the surroundings from 2018 to 2021. Front Microbiol. (2022) 13:1013617. doi: 10.3389/fmicb.2022.1013617
33. Nan, W, Wu, J, Hu, H, Peng, G, Tan, S, and Deng, Z. Prevalence and genetic diversity of porcine circovirus type 2 in northern Guangdong Province during 2016-2021. Front Vet Sci. (2022) 9:932612. doi: 10.3389/fvets.2022.932612
34. Lv, W, Cao, L, Yang, L, Wang, N, Li, Z, Huang, S, et al. The prevalence and genetic diversity of porcine circoviruses (PCVs) during 2017-2023 in Guangdong Province, China. Anim Open Access J MDPI. (2023) 13:3640. doi: 10.3390/ani13233640
35. Allan, GM, and Ellis, JA. Porcine circoviruses: a review. J Vet Diagn Investig Off Publ Am Assoc Vet Lab Diagn Inc. (2000) 12:3–14. doi: 10.1177/104063870001200102
36. Wang, Y, Noll, L, Porter, E, Stoy, C, Dong, J, Anderson, J, et al. Development of a differential multiplex real-time PCR assay for porcine circovirus type 2 (PCV2) genotypes PCV2a, PCV2b and PCV2d. J Virol Methods. (2020) 286:113971. doi: 10.1016/j.jviromet.2020.113971
37. Qu, T, Li, R, Yan, M, Luo, B, Yang, T, and Yu, X. High prevalence of PCV2d in Hunan province, China: a retrospective analysis of samples collected from 2006 to 2016. Arch Virol. (2018) 163:1897–906. doi: 10.1007/s00705-018-3823-9
38. Xu, T, Zhang, YH, Tian, RB, Hou, CY, Li, XS, Zheng, LL, et al. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in Central China. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. (2021) 94:105016. doi: 10.1016/j.meegid.2021.105016
39. Du, S, Xu, F, Lin, Y, Wang, Y, Zhang, Y, Su, K, et al. Detection of porcine circovirus type 2a and Pasteurella multocida capsular serotype D in growing pigs suffering from respiratory disease. Vet Sci. (2022) 9:528. doi: 10.3390/vetsci9100528
Keywords: porcine circoviruses, multiplex real-time PCR, prevalence, genotype, phylogenetic analysis
Citation: Zhang P, Ren Z, Gao X, Zhao M, Wang Y, Chen J, Wang G, Xiang H, Cai R, Luo S and Wang X (2024) Development and application of a TaqMan-probe-based multiplex real-time PCR assay for simultaneous detection of porcine circovirus 2, 3, and 4 in Guangdong province of China. Front. Vet. Sci. 11:1353439. doi: 10.3389/fvets.2024.1353439
Edited by:
Marta Canuti, University of Copenhagen, DenmarkReviewed by:
Chanhee Chae, Seoul National University, Republic of KoreaYang Zhan, Hunan Agricultural University, China
Copyright © 2024 Zhang, Ren, Gao, Zhao, Wang, Chen, Wang, Xiang, Cai, Luo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohu Wang, d2FuZ3hpYW9odTIwMjBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Pian Zhang
Pian Zhang Zhaowen Ren1,2†
Zhaowen Ren1,2† Mengpo Zhao
Mengpo Zhao Rujian Cai
Rujian Cai Shengjun Luo
Shengjun Luo Xiaohu Wang
Xiaohu Wang