- 1Department of Veterinary Clinical Medicine, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
- 2Department of Veterinary Pharmacy, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
- 3Department of Microbiology, College of Medicine and Health Science, University of Gondar, Gondar, Ethiopia
Schmallenberg virus (SBV) is an arthropod-borne virus that emerged recently in northwestern Europe in 2011 that affects domestic and wild ruminants and induces abortion, stillbirth, and newborns with congenital anomalies. Since its discovery, SBV has spread very rapidly to too many countries in the world. The overall serological investigation of SBV is needed to improve modeling predictions and assess the overall impact on ruminant animals, which helps to design interventions for control and prevention strategies. Thus, this study aimed to estimate the overall serological assay of SBV in both domestic and wild ruminants around the world. This systematic review was conducted as per the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. International databases were employed To search for relevant articles. The pooled prevalence with a 95% confidence interval was calculated with a random effects model. The Cochran’s Q test, τ2, and I2 were used to assess the sources of heterogeneity. In the current meta-analysis, a total of 41 articles were included. The overall pooled proportion of SBV in domestic and wild ruminants was 49 and 26%, respectively. Substantial heterogeneity was observed in studies on domestic ruminants (I2 = 99.7%; p < 0.01) and studies on wild ruminants (I2 = 97.9%; p < 0.01). The pooled prevalence of SBV was significantly associated with publication time, detection techniques, and species of animals. According to the subgroup analysis, the highest pooled prevalence of SBV was reported in cattle (59%), followed by sheep (37%) and goat (18%). In addition to the subgroup analysis based on publication year, the pooled prevalence of SBV infection has become endemic since 2013 (49%) among domestic animals in the world. Of the diagnostic tests used, the highest anti-SBV antibodies (66%) were detected by a virus neutralization test. In this meta-analysis, the major wild animals that were infected by SBV were red deer, roe deer, fallow deer, mouflon, and wild boar. The highest sub-pooled prevalence of SBV was found in roe deer (46%), followed by fallow deer (30%), red deer (27%), mouflon (22%), and wild boar (11%). In general, the prevalence of SBV was high in cattle among domestic ruminants and in roe deer among wild animals. According to the current information provided by this meta-analysis, evidence-based risk management measures should be established to restrict SBV spread in both domestic and wild ruminants.
Introduction
Schmallenberg virus was detected for the first time in November 2011 in plasma samples collected from cows displaying fever and diarrhea and farmed near the town of Schmallenberg, Germany (1); hence, it is named Schmallenberg virus. The first acute infections associated with SBV were reported in August 2011, while the first malformations in stillborn animals caused by this virus were detected in The Netherlands in December 2011 (2). Since then, nine countries have reported congenital malformations and stillbirths associated with the presence of SBV as of May 2012. The appearance of SBV was first noted in Belgium toward the end of December 2011, followed by the United Kingdom on 22 January 2012. France detected its first SBV case on 25 January 2012, after the virus genome was discovered by RT-qPCR in brain samples from misshapen lambs born on farms situated in the territorial divisions of “Moselle” and “Meurthe-et-Moselle” in northeastern France (3). It infects several domestic and wild animal species, including cattle, sheep, and goats (4) in alpaca (5) red deer, roe deer (6), fallow deer, moose, bison (7), wild boar (8), and several zoo animals (9). Spain reported the first outbreaks of Schmallenberg disease (SBD) in sheep in 2012 (10).
The diagnosis of SBV infection in living adult animals is difficult for veterinarians.
The infection caused by SBV in adult ruminants is primarily asymptomatic or displays symptoms such as febrile syndrome, especially in cattle. These observable clinical manifestations may also be observed in other viral infections, such as bovine herpes virus type 1, bluetongue virus, and foot-and-mouth disease virus. The direct diagnosis of SBV infection can be realized by performing real-time quantitative polymerase chain reaction (RT-qPCR) developed by Friedrich-Loeffler institute in 2011 on the L segment of the SBV genome (11). A protocol targeting the S segment was later developed and showed higher sensitivity. Besides RT-qPCR, the detection of anti-SBV antibodies present in the serum of infected animals can be performed by the indirect method of SBV infection diagnosis. The virus neutralization test (VNT) and enzyme-linked immunosorbent assay (ELISA) have been developed as tools for serological diagnosis.
Schmallenberg virus infection is also classified as one of the emerging infectious diseases of ruminants, with the transmission mediated by Culicoides midges (12). The virus is part of the Orthobunyavirus genus, which belongs to the Bunyaviridae family. The virus is closely related to the Akabane, Aino, and Shamonda viruses. A variety of Culicoides groups, including Culicoides obsoletus complex, Culicoides chiopterus, and Culicoides dewulfi, were linked to the transmission of SBV in Europe during the virus outbreak in 2011/12 (13). Apparent clinical signs of SBV infection in adult cattle are reported to be short-lived. These include loss of appetite, hyperthermia, diarrhea, and reduction in milk production. Infection during the certain critical period of pregnancy between days 47 and 162 of gestation (14) causes neonatal malformation affecting the neuro-musculoskeletal systems (1, 15). The syndrome is known as arthrogryposis-hydranencephaly syndrome (AHS) and is characterized by arthrogryposis, severe torticollis, ankylosis, kyphosis, lordosis, scoliosis, brachygnathia inferior, and neurological disorders. Most of the anomalies were observed in cases of abortions and stillbirths, while some calves may be born alive with various pathologies and behavioral abnormalities (16).
In a study by Veldhuis et al. (17), seropositivity to SBV was significantly associated with decreased reproductive performances and repeat breeder. Since the report of Hoffmann et al. (1), serological and molecular evidence of the virus has been available from several European countries, including Belgium, France, Greece, the UK, Italy, Spain, Luxembourg, Denmark, Poland, Sweden, and Switzerland (15, 18), Turkey (19), and China (20). According to the European Food Safety Authority (EFSA), SBV infection had also been confirmed in approximately 9,000 ruminant herds across Europe, and nearly half of them were reported in France.
In a Belgian sheep farm where SBV emerged and spread between mid-September and mid-October 2011, ewes lambing in January 2012 gave birth to 17% (28/163) of stillborn or newborn lambs presenting typical deformities, while ewes lambing in March 2012 gave birth to only 5% (8/150) of aborted fetuses, and in May 2012, no impact on lambing was observed. In Spain, SBV circulation has been found regionally in livestock, with seroprevalence values ranging between 54.4 and 75.6% (10). The high seroprevalence detected in some species (up to 80%) raises the question of whether wild ruminants play a role in the maintenance of SBV in Europe (21).
In Spain, there are seven wild ruminant species, with red deer (Cervus elaphus) and roe deer (Capreolus capreolus) being the most widely distributed. Fallow deer (Dama dama), mouflon (Ovis aries musimon), and Barbary sheep (Ammotragus lervia) are less abundant species, with locally significant populations, while Southern chamois (Rupicapra pyrenaica) and Iberian wild goat (Capra pyrenaica) are more frequent in the mountain ecosystems of northern and Mediterranean Spain, respectively (22). Serological investigations of wildlife animals represent a cornerstone of disease surveillance (23), especially when virus shedding is only transient and the time frame is unknown since antibodies remain detectable for longer periods.
Using recently developed ELISA tools, several serological surveys revealed a large exposure of wild-living ruminant species in Europe, particularly in roe deer (Capreolus capreolus) and red deer (Cervus elaphus), with seroprevalences ranging from 20 to 90% (2, 6, 24). In Africa, the available research evidence on SBV in cattle is few; however, seroprevalence as high as 61% in Tanzania (25), 56.6% in Ethiopia (26), and 100% in Mozambique (27) were reported. In sub-Saharan Africa, research studies have suggested a possible occurrence of the virus in both wild and domesticated livestock (25, 28).
The distribution of the virus has an ecological association, with the occurrence being higher (2.5 times) in midland than highland. The prevalence of the virus was found to be 72.3% in aborted animals, 53.5% in non-aborted animals, and 71.4 in the birth of weak calves in Ethiopia (29). Populations of some wild species have expanded in recent decades, mainly because of ongoing changes in land use and more intensive game management practices (30). These epidemiological scenarios have been shown to increase the risk of disease transmission among sympatric species (31). In recent times, meta-analyses are a powerful statistical tool used to summarize existing evidence, systematize, and inform specific decisions as well as that could be extended to incorporate economic considerations in a decision analysis framework. It is a more precise estimation of the effect size of an event compared with only one study result (32). This investigation was important for scientific evidence for the researchers and baseline survey collectors regarding the dairy herd reproductive infectious diseases. Furthermore, it supports intervention regarding the prevention and control of the virus. Therefore, this systematic review and meta-analysis aimed to provide an overall estimate of the seroprevalence of SBV in both domestic and wild animals around the globe.
Methods
The literature search was conducted from 12 August to 20 September 2023. A systematic assessment of published articles reporting the overall proportion of SBV was performed based on the PRISMA checklist (33) (Supplementary material 1). The major working protocol was performed in seven key steps: study eligibility criteria, information sources, search strategy, outcome variable, data extraction, study quality assessment, data synthesis, and statistical analysis. A comprehensive search strategy was made to identify all relevant studies. Databases such as PubMed, Google Scholar, Web of Science, snowball searching from retrieved articles, and other manual methods were used for literature searches to select included studies by two authors (MD and AS) independently. The research question was “What is the pooled prevalence of SBV in domestic and wild ruminants in the world?”
The following Mesh terms were used in electronic database search: “Schmallenbergvirus,” “SBV,” “PrevalenceofSchmallenberg virus” “epidemiology of Schmallenberg virus,” “Schmallenberg virus in cattle,” “Schmallenberg virus in sheep,” “Schmallenberg virus in goat,” “Schmallenberg virus in domestic ruminants,” Schmallenberg virus in wild ruminants, “Schmallenberg virus in wild animals,” “Schmallenberg virus in red deer,” “Schmallenberg virus in roe deer,” “Schmallenberg virus in wild goat,” Schmallenberg virus in wild boar,” Schmallenberg virus in bison, “Schmallenberg virus in buffalo,” “Schmallenberg virus in fallow deer,” and “Schmallenberg virus in mouflon.” Even we have searched articles based on the combination of these words with specific continents and countries as the context. Articles were written in English. All identified studies were imported to Medley software to remove duplicates and scientific citations from the references.
Study eligibility criteria
Inclusion criteria
The search was performed by three field experts (Microbiology, Veterinary Pharmacy, and Veterinary Clinical Medicine) to avoid authors’ bias. This meta-analysis includes all of the primary descriptive studies that have been published in the English language that document the occurrence of SBV in domestic and wild ruminants. The inclusion criteria included articles with a clear estimation of the prevalence of SBV. Study animals became domestic and wild ruminants around the globe at any year. Samples had to be collected from animals that had not been experimentally infected.
Exclusion criteria
Studies that do not have clear and detailed estimates of the proportion of SBV in the affected host were also excluded. Review articles, duplicates, abstract only, qualitative studies, or only KAP (knowledge, attitude, and practice) questionnaire-based studies, book chapters, case reports, editorials, newsletters, and forum discussions, among others were excluded. Intervention studies that lacked baseline data on the association between animal exposure and disease were excluded from the meta-analysis.
Information sources
The literature search was conducted from August to September 2023. Database sources such as PubMed/PubMed, HINARI, Web of Science, Google, and Google Scholar were used. The included studies were reported from continents in any study year.
Data extraction
The relevant data were extracted independently by two investigators (MD and AS). Quantitative and qualitative data extraction from the included studies was performed and presented in the form of a table in an Excel spreadsheet. The extracted components encompassed the name of the primary author along with the year of publication, year of study, country, and all domestic and wild species of ruminants, the total number of animals (N), the number of seropositive animals (primary outcome interest), diagnostic methods (ELISA, PCR, VNT), data collection techniques, and ethical considerations. Disagreements were resolved by discussion and consultation with a third author.
Study quality assessment
Quality assessment was performed to verify the methodological quality of this systematic review (MD and BD). The quality assessment of the included studies was assessed by the Appraisal tool for Cross-Sectional Studies (AXIS) quality tool (34). This quality assessment tool includes different items including study design, sample size justification, sample representativeness, target population, the use of validated measures, diagnosis of statistical methods, sample selection, sample frame, discussion of non-response bias, reporting of funding, and conflicts of interest.
Data synthesis and statistical analysis
A meta-analysis was carried out using the R software using the “meta prop” function of the ‘meta’ package (35) and “metaphor” (36) in R software. The pooled prevalence and 95% confidence intervals were estimated using the random effects model based on the restricted maximum likelihood approach (REML), which computes within- and between-study variabilities. It was used to perform the overall meta-analysis (overall effect size), pooled odds ratio, heterogeneity, and weight of each study. In addition, graphs and tables were used to demonstrate the prevalence. Considering that the outcome variable is binary (i.e., SBV positive or negative and given only for single groups, the only possible parameter to measure effect size was the raw proportion with 95% confidence intervals) (32). In a logistic-normal random-effect regression model, the logit transformation was utilized to estimate the pooled proportions, and a mixed-effect logistic regression model was employed for the subgroup analysis.
Investigation of heterogeneity
The Cochran’s Q test (reported as the p-value), τ2 (between-study variance), and inverse variance index (I2) were used to assess the sources of heterogeneity, which describes the percentage of observed total variation between studies that is due to heterogeneity rather than chance. As explained by Higgins and Thompson (37), the I2 index was estimated to represent low, moderate, and high heterogeneity, if this corresponds to I2 values of 25, 50, and 75%, respectively. Heterogeneity was deemed to be statistically significant if the I2 value exceeded 50% and the Q test revealed a p-value of less than 0.10. The degree of study heterogeneity has been evaluated using a forest plot diagram. The forest plot diagram displayed the weights, magnitude of effects, and 95% confidence intervals for each study.
Subgroup sets
To determine specific between-study variability, a subgroup analysis of the proportion of the SBV in ruminants was performed based on study year, methods of diagnosis study location, and species of the animals.
Publication bias assessment
Publication bias is usually evaluated through a funnel plot in which asymmetry can be assessed visually, beggar rank, and the Egger test. Thus, in our case, publication bias was assessed using funnel plot diagrams and Egger’s regression test.
Sensitivity and influential analysis
Sensitivity analysis of studies was performed to evaluate the effect of each study on the pooled result. The results showed that the studies were the prime determinants of the pooled result.
Results
Article search results
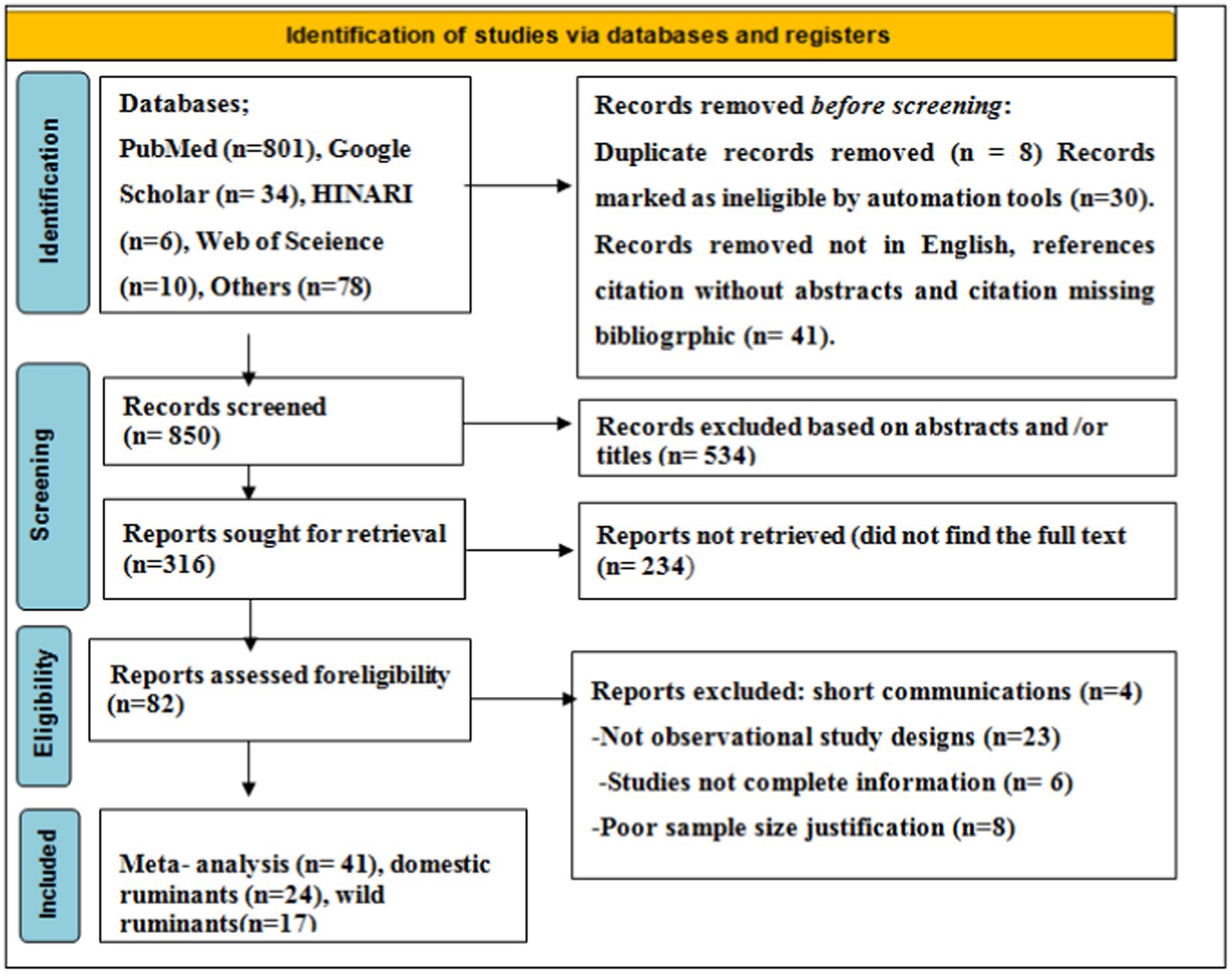
As shown in the PRISMA 2020 flowchart (Figure 1), a total of 929 articles in various electronic databases and other methods were searched, from which 8 were excluded after article duplication assessment (n = 8), 30 records were marked as ineligible by automation tools (n = 30), and 41 records were removed for other reasons (n = 41). Among 850 articles, 534 articles were excluded by article title and abstract screening, 316 studies were reports searched for retrieval, and 233 articles were reports not retrieved. A total of 82 (n = 82) articles were reports being evaluated for eligibility, and 41 (n = 41) of them were excluded for various reasons. Finally, 41 (n = 41) studies were included for meta-analysis.
Overview of included studies for both domestic and wild ruminants (n = 41)
The characteristics of the studies about SBV are intricately described in a step-by-step manner. The study animals comprised of cattle which are of all ages, both genders, wild and domestic ruminants in the world. A total of 41 independent articles (domestic, n = 24, wild, n = 17) were considered for the analysis of all pooled prevalence SBV in wild and domestic ruminants. A meta-analysis was carried out separately for the prevalence of SBV in domestic and wild ruminants.
In domestic ruminants, the included studies for this meta-analysis were conducted and published in different parts of the world between 2011 and 2022. We noticed that the same article was used multiple times due to different species of animals. The study designs were cross-sectional, and the types of samples were serum (blood) and brain tissue. Therefore, it helps to reduce the variability between studies.
The included domestic ruminants were cattle, goats, and sheep with a total number of 909,938 animals and 42,306 SBV positive. In this systematic review, 9,497 cattle (38) served as the maximum sample size, and 19 cattle were used as the minimum sample size (39). Methods of diagnosis tests were ELISA, VNT, and rRT-PCR to examine the presence of the SBV. The majority of these studies were conducted in Europe (Spain, Belgium, France, Greece, etc.) and Africa; four studies were included from Tanzania, Ethiopia, Nigeria, and Mozambique. Moreover, the characteristics of the included studies are depicted in Table 1.
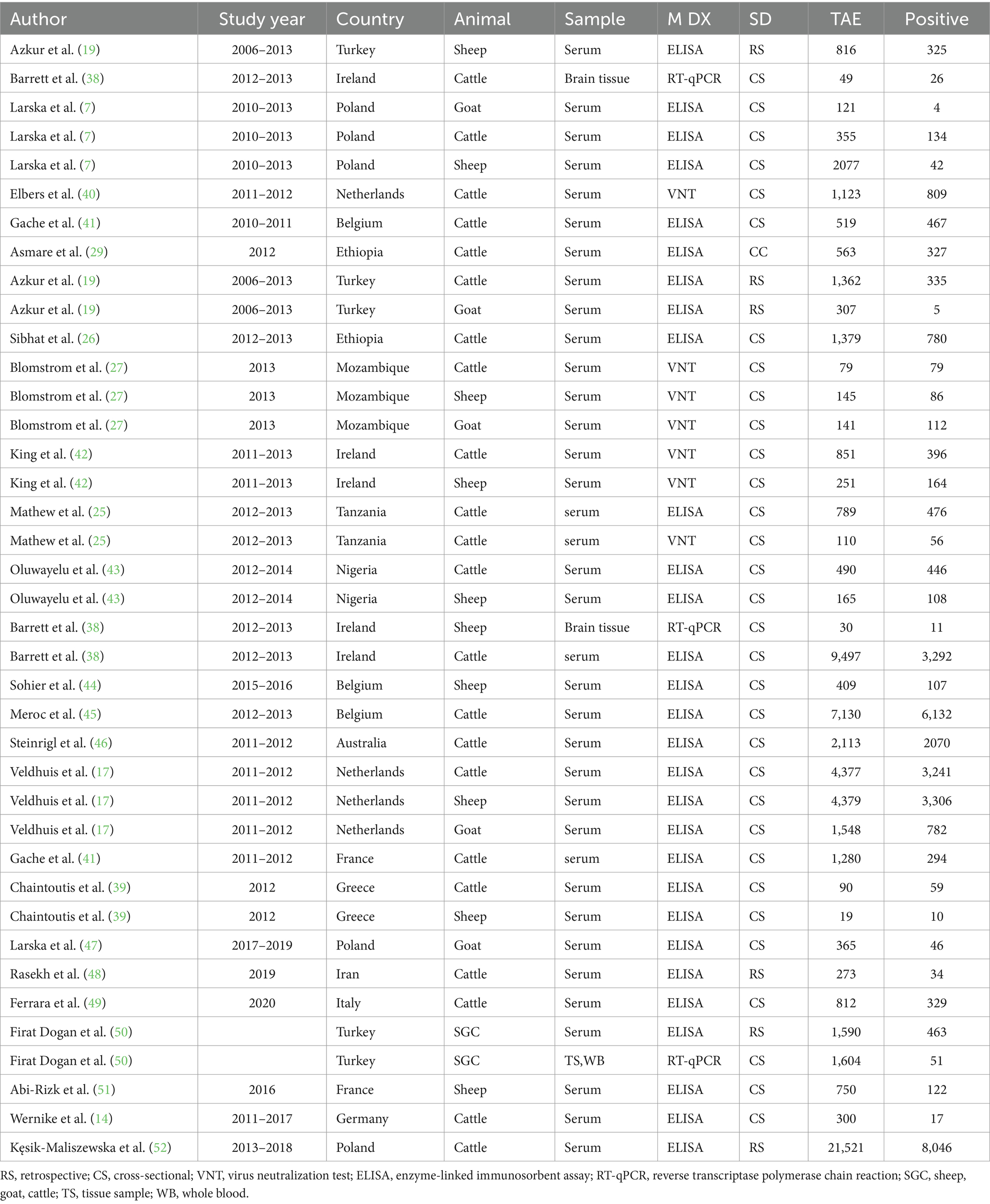
Table 1. The characteristics of studies included in the meta-analysis (n = 40) for domestic ruminants.
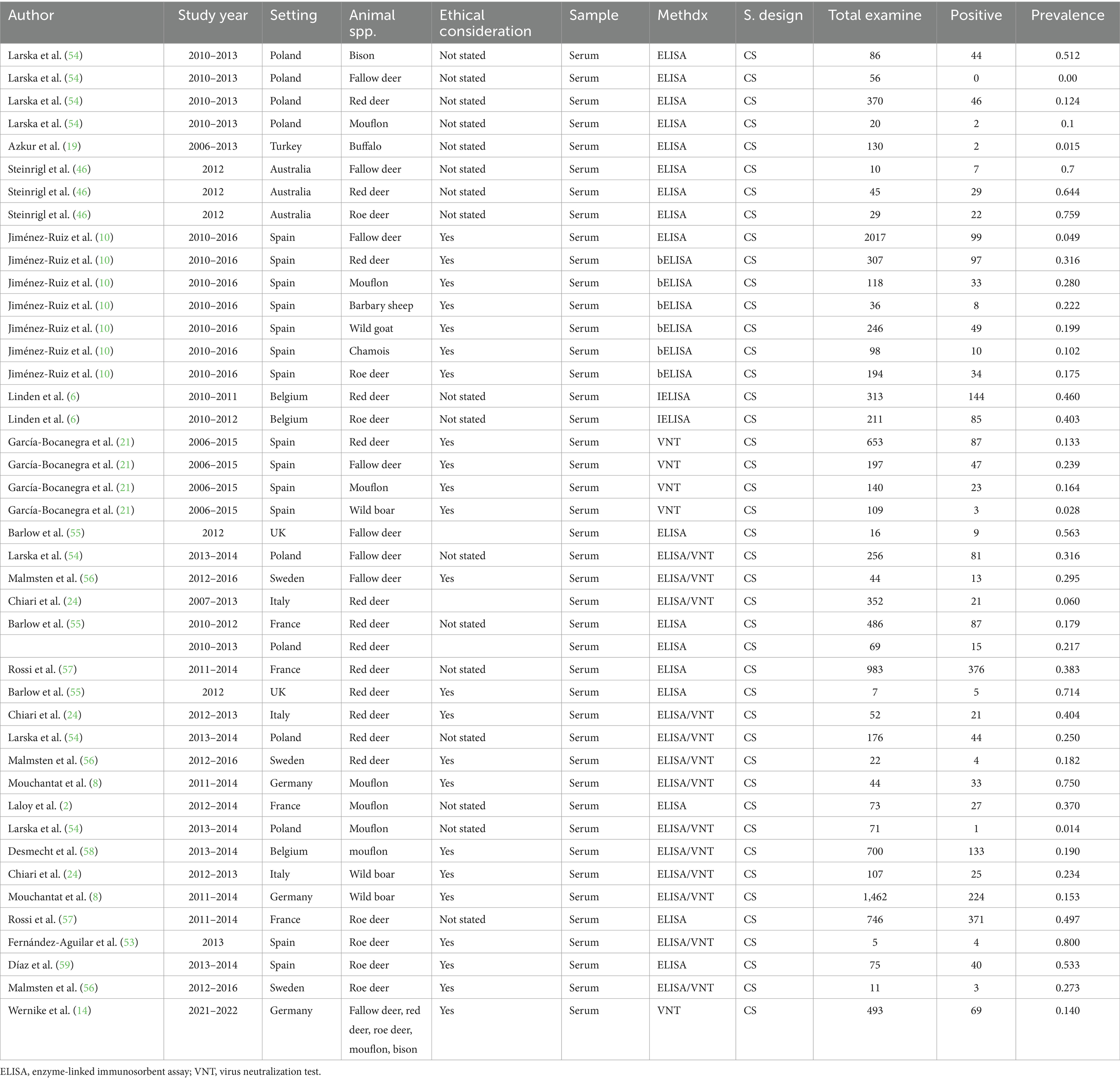
In wild ruminants, the major wild ruminants in this study comprise fallow deer, red deer, roe deer, and mouflon. However, some studies included wild boar, wild goat, bison, buffalo, chamois, and Barbary sheep. Of the included studies (n = 24), one study (14) in Germany put only the overall prevalence (fallow deer, red deer, roe deer, mouflon, bison) of the SBV infection in wild animals and did not estimate the prevalence for each wild animal species. There were a total of 11,579 wild animals with 2,477 SBV positive. In this review, 2017 (10) cattle served as the maximum sample size, and five cattle were used as the minimum sample size (53). Methods of diagnosis tests were ELSA, VNT, and rRT-PCR to examine the presence of the SBV. The majority of these studies were conducted in Europe (Spain, Poland, France, Italy, and Germany). Furthermore, the details of the included studies are presented in Table 2.
Meta-analysis, testing heterogeneity, and bias assessment
In the meta-analysis of studies regarding SBV, a total of 41 independent articles were incorporated in both domestic and wild ruminants (domestic, n = 24, wild, n = 17). However, it should be noted that certain articles were utilized multiple times due to their relevance in similar years but in different animal species. Therefore, a total of 40 dependent and independent articles were included in the case of domestic ruminants, while 42 articles were included in wild ruminants.
Result of meta-analysis for domestic ruminants
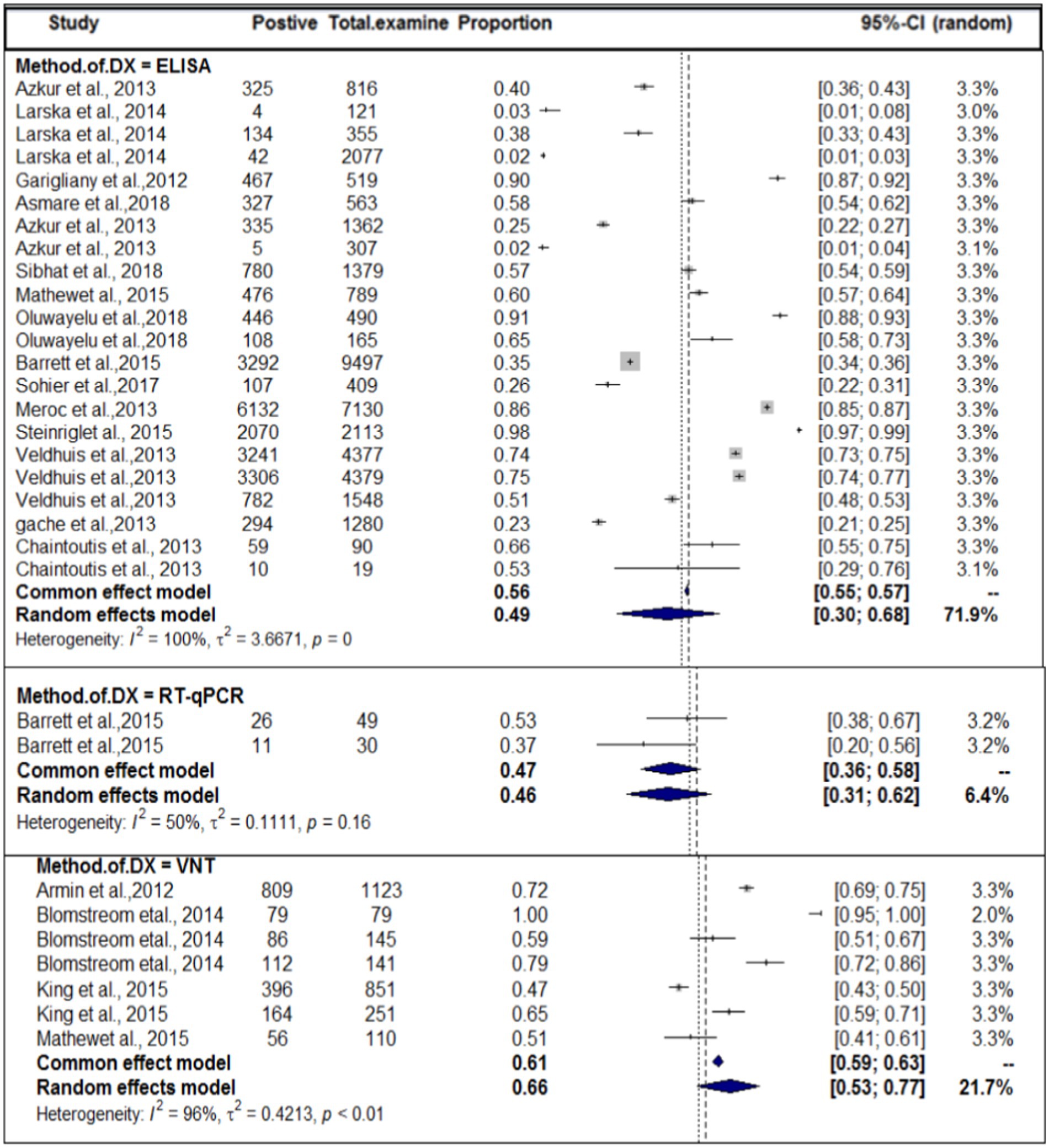
In the context of domestic ruminants, the included studies exhibited a high level of heterogeneity (I2 = 99.7%: τ2 = 3.0276; p < 0.01), and the estimated pooled proportion of SBV among overall domestic ruminants was calculated to be 49% (95% CI: 41–657%; Figure 2). The between-study variability was statistically significant (Q = 12262.29, DF = 39, p < 0.0001). Similarly, the heterogeneity and outliers of studies were plotted, and the expectation of all studies is not falling within 95% of the range bounded by the two confidence interval lines (Figure 3).
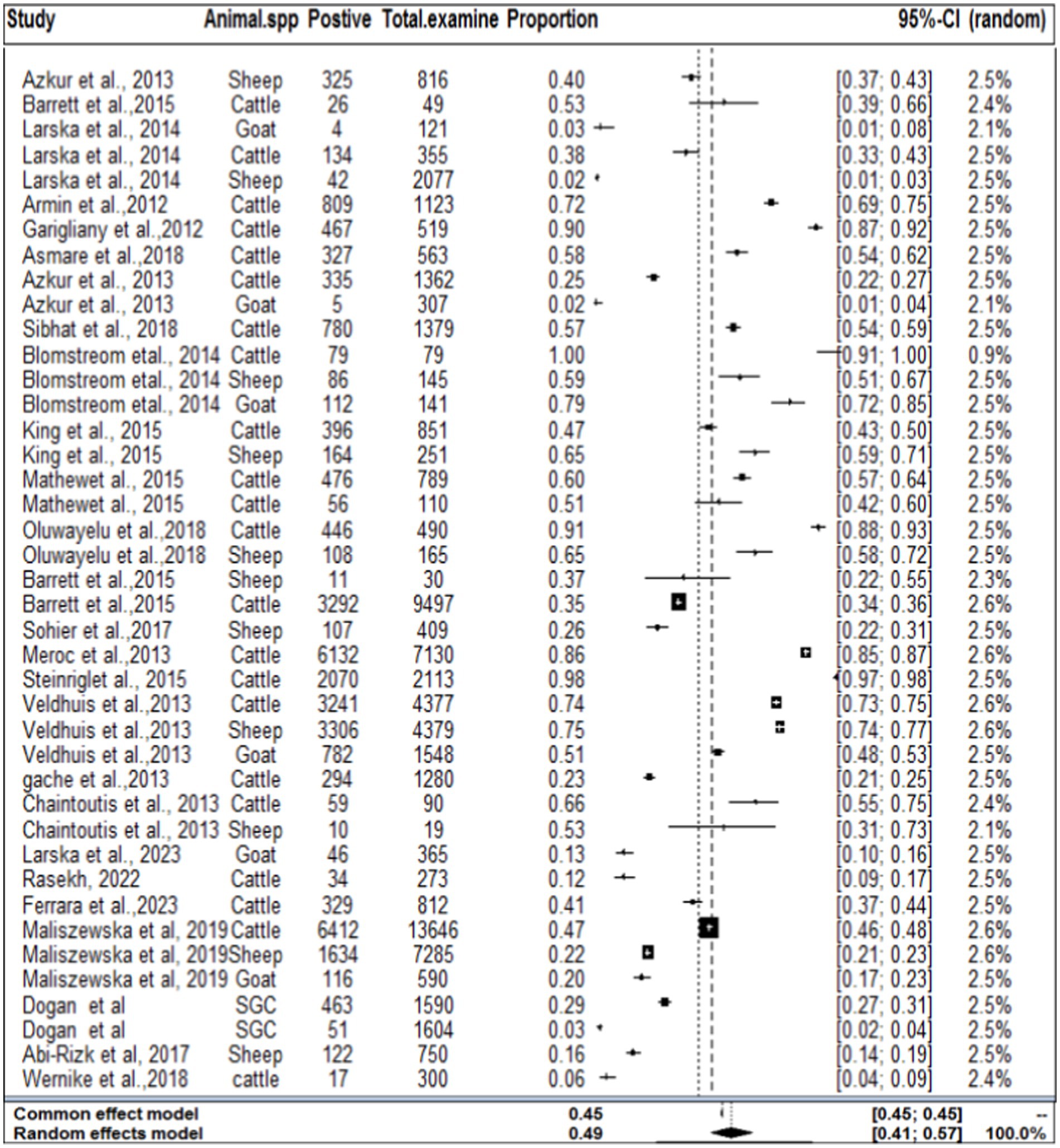
Figure 2. Forest plot plots with random-effects meta-analysis of SBV infection in domestic ruminants.
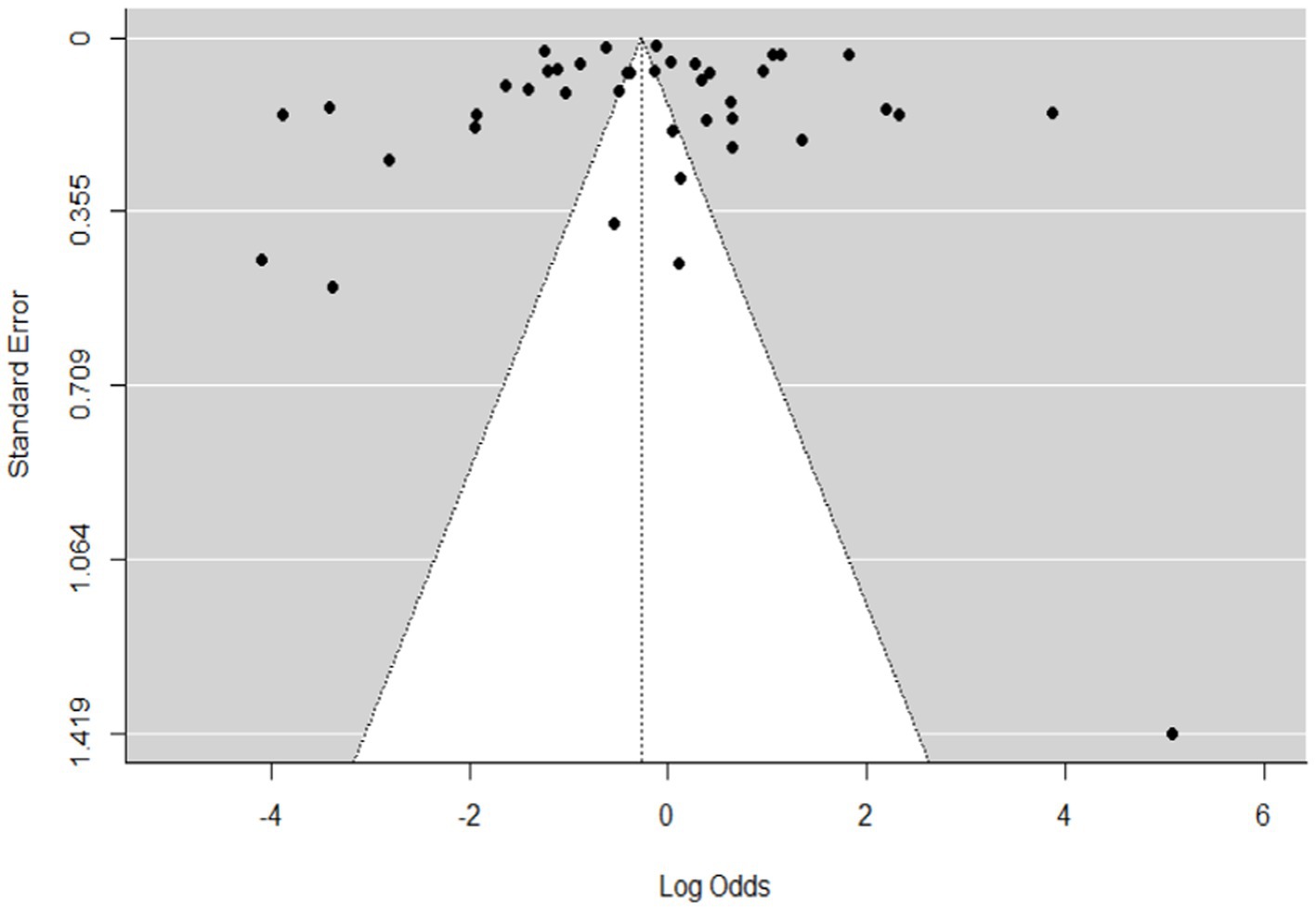
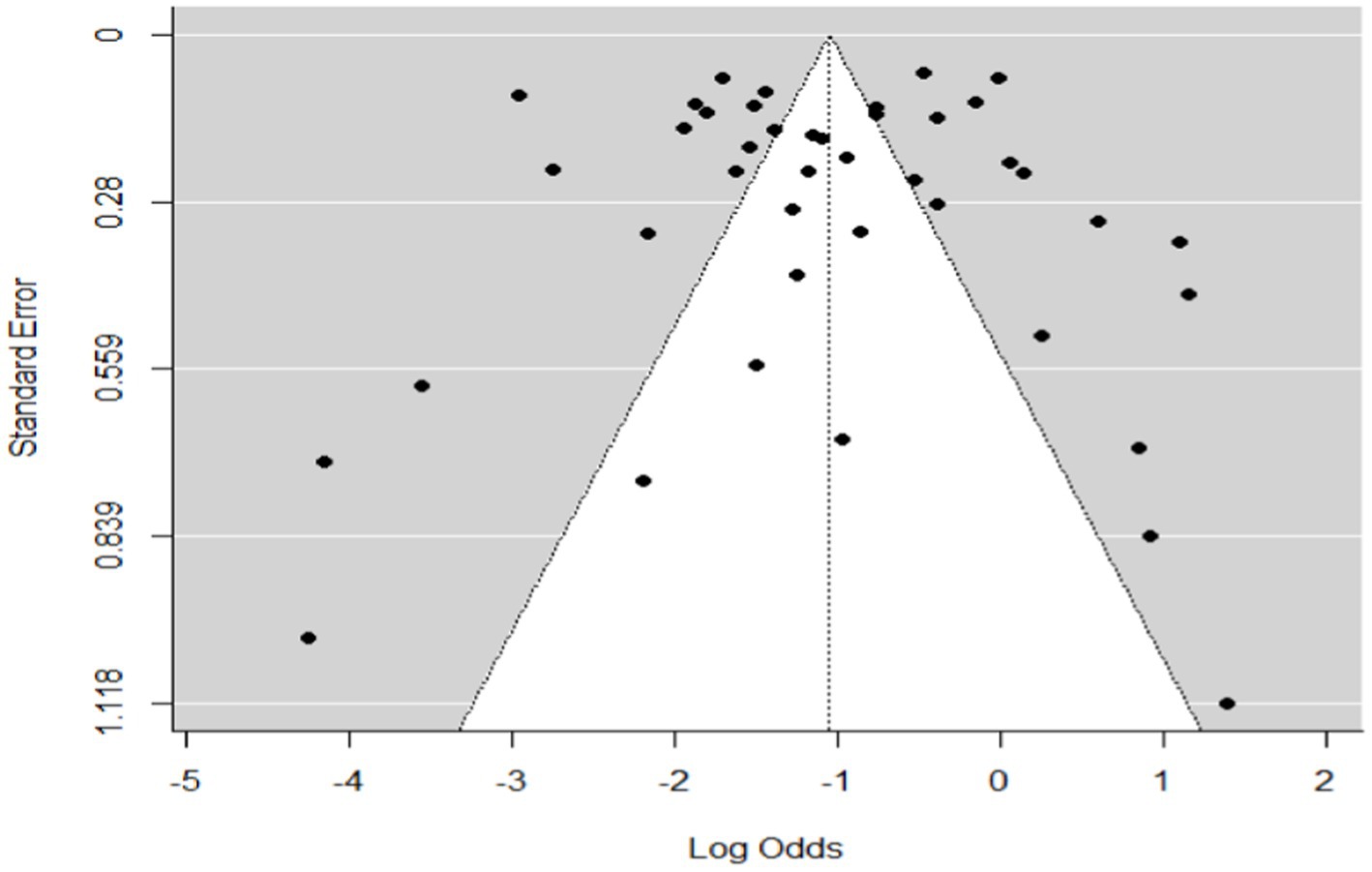
Figure 3. Funnel plot plots the standard error by log odds of the proportion of SBV domestic ruminants.
The regression test for funnel plot asymmetry was done using a mixed-effects meta-regression model and standard error as a predictor (Eger’s test, b = −0.2035 (CI: −0.6588, 0.2518), z = 0.1008, p = 0.9197). The test for funnel plot asymmetry was also performed via rank correlation test for funnel plot asymmetry (Kendall’s tau = −0.0073, p = 0.9555).
Subgroup analysis results
In the current meta-analysis, heterogeneity was explored through a subgroup analysis. Subgroup analyses were conducted based on study year, species of the host or study animals, and methods of diagnosis. All of the subgroup analyses of exploratory outcomes showed considerable heterogeneity (I2 > 98). Significant statistical heterogeneity in the subgroup analysis reveals a likely interaction among exploratory variables.
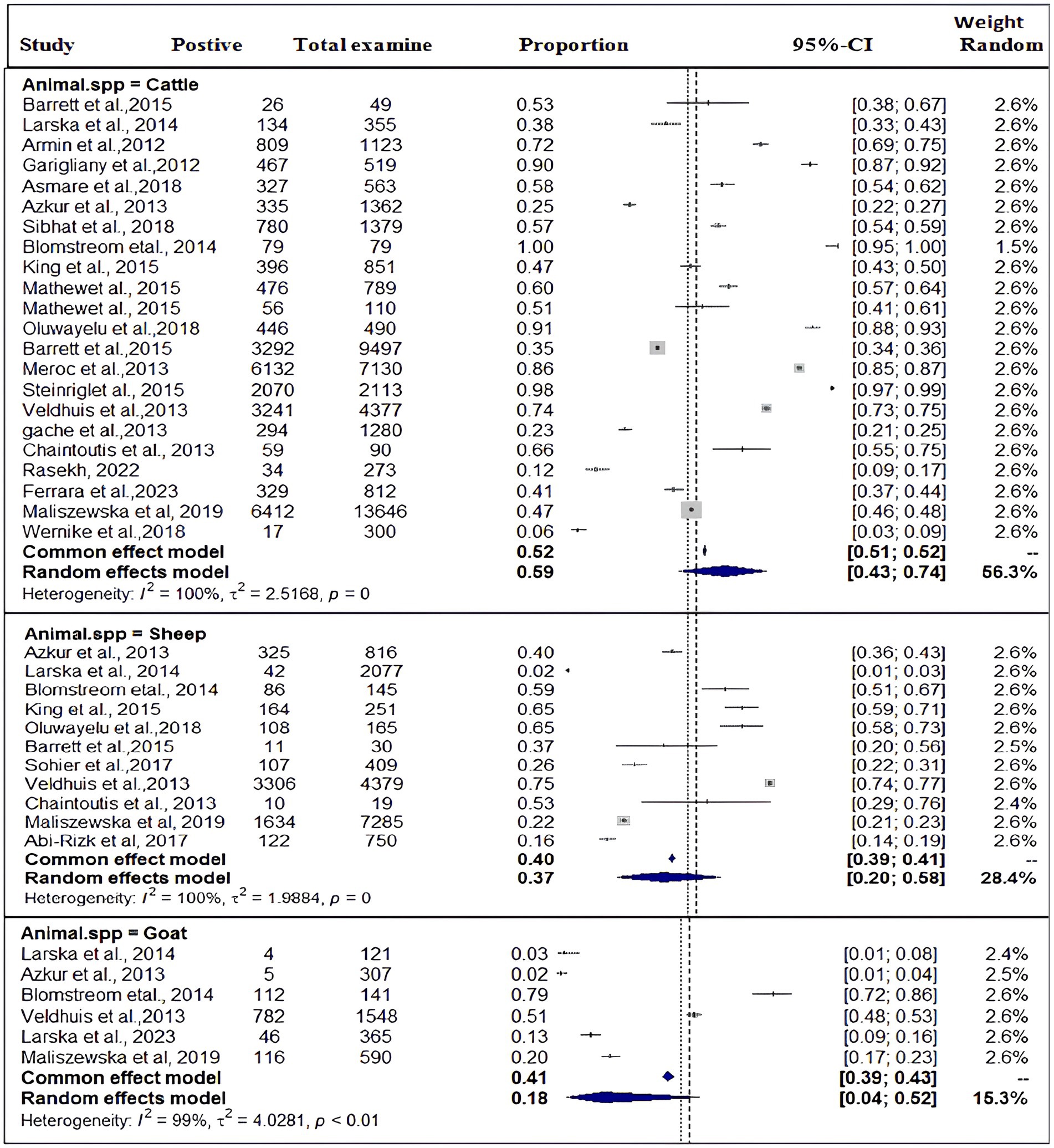
Based on the species of ruminant animals, the included studies were categorized into three groups: cattle (n = 22), sheep (n = 11), and goat (n = 6). Significant discrepancies were found in the subgroup analysis of the most SBV prevalence by animal type. As shown in Figure 4, the subgroup analysis revealed that the pooled prevalence of SBV in cattle was 59% (95% CI: 43–74%) and (I2 = 100%: τ2 = 2.5168=; p = 0), followed by sheep at 37% (95% CI: 20–58%) and (I2 = 100%: τ2 = 1.9884; p = 0.0) and goat at 18% (95% CI: 4–52%) and (I2 = 99%: τ2 = 4.0281; p < 0.01).
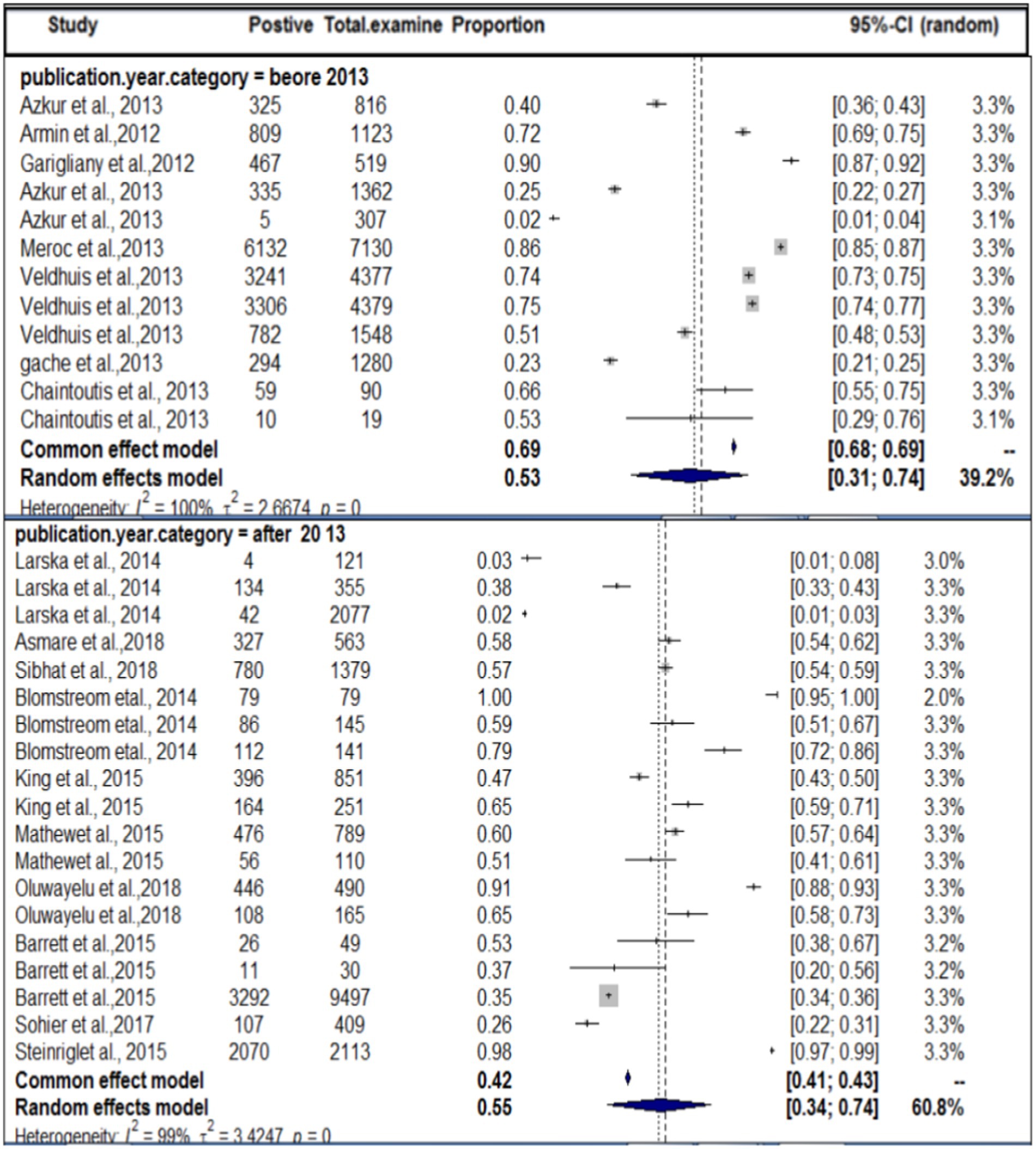
In terms of the year of publication, as shown in Figure 5, the subgroup analysis found that the pooled prevalence of SBV virus in domestic ruminants was 27% (95% CI: 25–56%) with 99% degree of heterogeneity (I2) and (τ2 = 3.1833: p < 0.01) in the up to or before 2013 group, 49% (95% CI: 25–56%) in the 2013 or later group with 98% degree of heterogeneity (I2).
In terms of the method of diagnosis, subgroup analysis of the method of diagnosis was carried out. The method of diagnosis was categorized into VNT, ELISA, and RT-qPCR. In this case, we also encountered considerable heterogeneity (I2 > 50) in each group. The highest study heterogeneity (I2 = 100) was revealed in the diagnosis of SBV by ELISA methods. The sub-pooled prevalence (66%, Figure 6) of SBV was highest in the diagnosis method of VNT, followed by ELISA (49%) and RT-qPCR (46%). The subgroup difference test suggested that there was a statistically significant group effect (Q = 23.49; DF = 2; p < 0.0001).
Result of meta-analysis and subgroup for wild ruminants
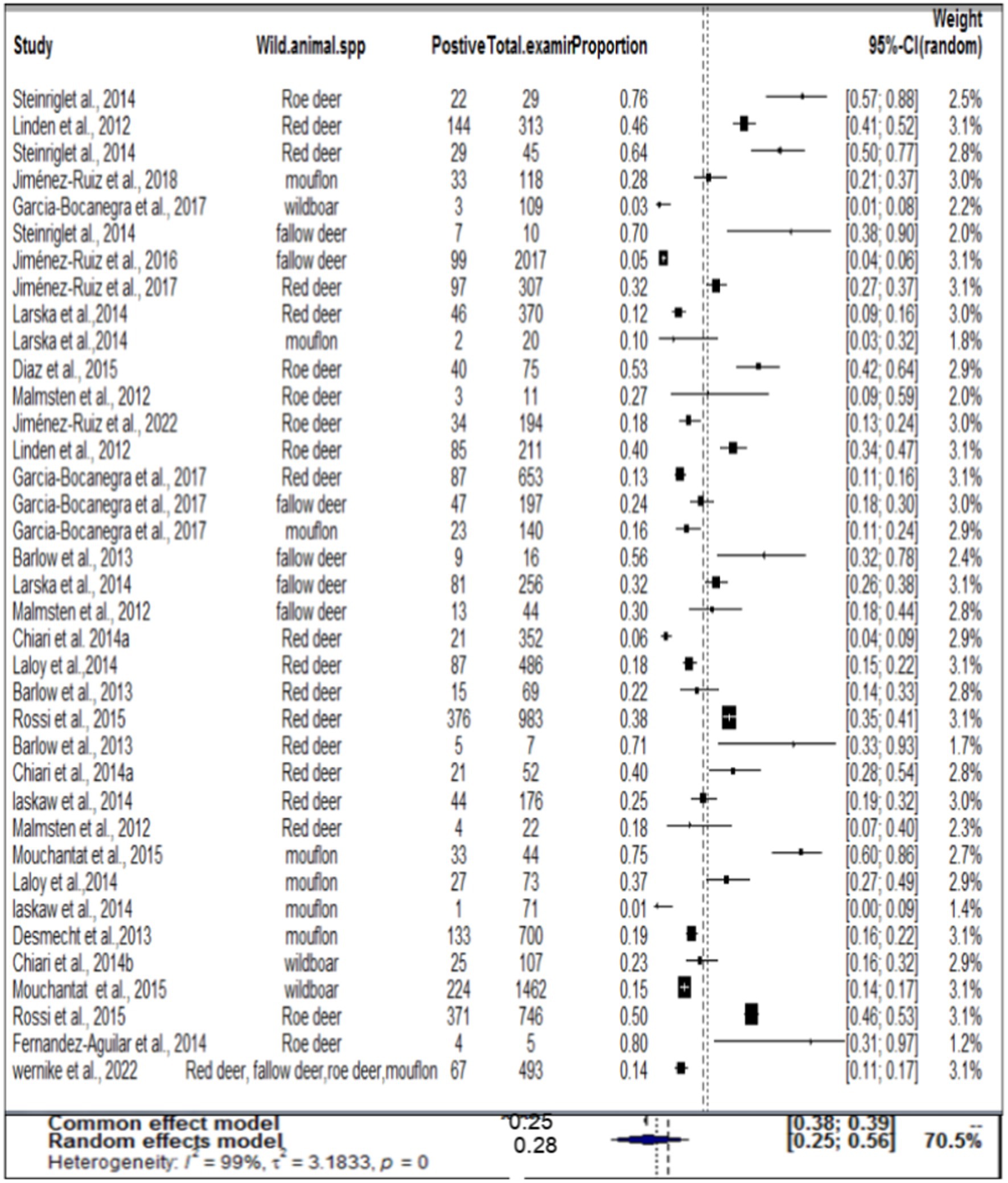
In the case of wild ruminants, the combined prevalence of SBV in wild ruminants was found to be 26%, with a degree of heterogeneity of 97.9% (Figure 7). Funnel plot asymmetry was also conducted for the studies included in wild ruminants, using Egger’s regression test (b = −1.0658, CI: −1.5391 to −0.5924, z = 0.0768, p = 0.9388) and rank correlation test for funnel plot asymmetry (Kendall’s tau = 0.0685, p = 0.5319). Therefore, both the funnel plot (Figure 8), Egger’s regression test, and rank correlation test revealed that there was no asymmetrical distribution of studies, indicating that smaller studies were not likely to be overlooked.
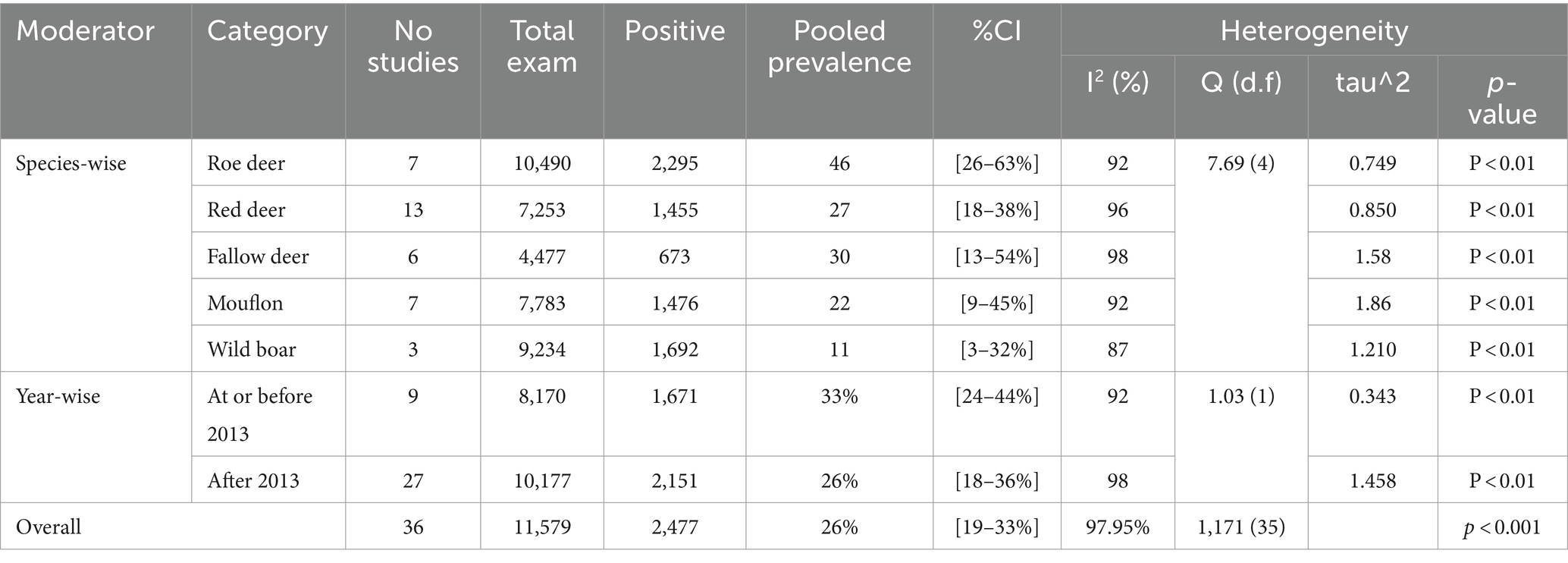
Regarding wild ruminants, a subgroup analysis was conducted based on the publication year and type or species of wild animal. The pooled prevalence of SBV infection in roe deer was found to be 33% (95% CI: 24–44%) in the group up to or before 2013, and 26% (95% CI 18–36%) in the group after the publication year of 2013 (Table 3). The estimated pooled prevalence of SBV infection was 46% (95% CI 26–63%) in tested roe deer, 30% (95% CI 13–54%) in fallow deer, 27% (95% CI 18–38%) in red deer, 22% (95% CI 9–45%) in mouflon, and 11% (95% CI 3–32%) in wild boar (Table 3). Among the included studies, a high level of variability was observed in studies focusing on fallow deer, with an I2 value of 98 and a p-value of <0.01.
Discussion
To the best of our knowledge, this is the first meta-analysis of the global prevalence of SBV infection in wild and domestic ruminants, although many investigations regarding the prevalence of SBV infection in ruminants in different countries have been conducted. The overall serological investigations of SBV are needed to improve modeling predictions and assess the overall impact of SBV in ruminant animals. Wildlife animals may be susceptible to multiple infectious agents of public health or veterinary relevance, thereby potentially forming a reservoir that bears the constant risk of re-introduction into the human or livestock population. A meta-analysis was performed to estimate the pooled prevalence of SBV infection in domestic and wild ruminants in the world. The current meta-analysis includes 34 articles, and it found that the overall pooled prevalence of SBV was 54 and 26% in domestic and wild ruminants, respectively.
In terms of domestic ruminants, according to this finding, the pooled prevalence of SBV (49%) was very high. This finding was comparable to the results (39) in sheep in Greece (53%) (38), in cattle in Ireland (53%) (25), in cattle in Tanzania (51%), and (17) in goat in the Netherlands (50%). The consistency in the results between the three serological surveys, in itself, suggests that there was limited bias in sample selection, equal vaccine cover, and equal exposure to the vector. The result was lower than the results reported by Refs. (27, 45, 46, 60) in cattle in the Netherlands (72%), Mozambique (79.4%), Australia (98%), and Belgium (86%), respectively. In contrast, the estimated pooled result was much higher compared with Refs. (7, 19, 44). in Poland (2%), Turkey (24%), and Belgium (26%), respectively.
However, some of the researchers have found different findings in the same area, and the same time, and even in the same species, for instance, the percentages of seropositive cattle reached 90.8% (8) and 99.76% (19) in Belgium and France up to 79% (7). The seroprevalence reported at the same time in sheep and goats differed significantly. For example, SBV seroprevalence in small ruminants did not exceed 58.7 and 43.8% in Germany. In Greece, the difference between the percentages of seropositive samples from cattle (64.45%) and sheep (5.3%) also was significant. A British study has also shown rather low seropositivity in sheep flocks investigated. No seropositive ewes were found in some flocks despite confirmation of SBV infection in the lambs originating from the same flocks and grazing close to seropositive cattle farms. In the Netherlands, the seroprevalence varied depending on the production type with 98.5, 63.4, 89.5, and 50.8% of seropositive animals among beef cattle, dairy cattle, sheep, and goats, respectively (23). These differences may be related to the availability of vaccination, the season of sample collection, the geographical location farm, the clinical stage of the virus during sampling, farm management systems, and the density of the vector (culicoides). Probably, environmental characteristics such as humidity and variation in temperature during the night and day affect the density of the vector, influencing the possible transmission and consequently the prevalence of SBV in both domestic and wild ruminants.
A Scottish SBV model indicates that the introduction of SBV must occur relatively early in the vector season to bring about extensive spread (61). Furthermore, stocking densities, manure in farms, land use, and meteorological conditions are considered to account for the differences in infection pressure (62). Moist soils rich in organic matter such as those created by dumping manure and other farm wastes may well support the breeding of Culicoides spp. Agroecological differences between the locations of the farms had a significant effect on the seroprevalence of SBV. Farms located in the midland areas had higher seroprevalence than those located in the highlands. This finding could be associated with favorable environmental conditions created by factors, including warmer average temperatures and moisture that support insect breeding and multiplications in the low-lying areas (63). Virus transmission and spread are possible at temperatures around 15°C with an optimum temperature range between 18°C and 19°C due to vector limitations (64).
In the subgroup analysis, the overall prevalence of SBV in cattle (59%) was highest compared with small ruminants, which is consistent with the result reported by Chaintoutis et al. (39) in cattle in Greece and (25) in cattle in Tanzania. Several studies have shown that cattle are more attractive to biting midges than other ruminant species, which makes them the ideal sentinel species for a midge-borne disease. However, a search of the literature indicates that, to date, there is no direct evidence that such resistance or susceptibility mechanisms of cattle for SBV. The current meta-analysis has revealed that the pooled prevalence of SBV was higher in sheep (37%) compared with goats (18%). It was proposed that this discrepancy in seropositivity between sheep and goats in Belgium and the Netherlands is a result of the animals’ husbandry practices, with sheep being more commonly kept outdoors (17). Furthermore, the timing of the reproductive season, particularly in sheep flocks with early breeding seasons (which coincided with the active season of the Culicoides vector), has also been identified as a risk factor for congenital Schmallenberg disease (65).
By study year subgroup analysis, a considerable increase in the seropositivity rate was observed in animals that were tested after 2013 compared with animals tested in or before 2013, indicating a re-circulation of SBV and seroconversion of naive young stock. May also related to the probability of the population of the vector density, if the vector density is increased in a specific area the disease may be transmitted and increase the probability of exposure. The virus entered a virgin population within 13 years, became endemic. This high seroprevalence is also the result of prolonged immunity (demonstrated in several studies) and continuous exposure (in many countries, the vector season is now very prolonged). Climatic conditions, local densities of vectors, the competence of vector species, and the topography have been recognized as factors that can affect the infection rate among species of farm animals. The occurrence of very cold weather, combined with wet and windy climatic conditions, is likely to result in a delay in the resumption of midge activity (66). The interactivity of Culicoides (where SBV replicates to transmissible levels within the midge) is dependent on the extrinsic incubation period (EIP), which is believed to vary between 9 and 41 days depending on the microclimatic temperatures found on farms (67). In Germany, it has been reported that midge activity is minimal during cold weather (68), whereas in warmer conditions, SBV has the potential to spread rapidly. This observation may be attributed to temperature-dependent factors, such as the midge biting rate, extrinsic incubation period, and vector mortality rates, all of which are known to influence the transmission of SBV between animals (68). The adult midges from the Culicoides obsoletus group possess a striking ability to endure long periods of time without a blood meal. Specifically, they can survive for up to 10 days at 4°C and up to 92 days at temperatures ranging between 17°C and 35°C (69). These findings suggest that infected midges have the potential to persist during the colder months of the year and subsequently infect hosts once temperatures rise to levels that are more conducive for virus transmission. This proposition is further substantiated by the presence of evidence suggesting the transmission of SBV (Schmallenberg virus) in Germany during the winter of 2013 (14).
The second objective was related to the distribution of SBV in wild animals. In this analysis, the pooled prevalence of SBV virus in wild animals was 24%. In terms of wild animal species, the peak seroprevalences observed in roe deer (46%) are similar to those reported in roe deer in Belgium and France (6, 57). High seroprevalence, ranging between 27.3 and 80.0%, was recently also detected in roe deer in different regions of Spain during the 2013–2014 period. In the last few years, serosurveys have revealed widespread exposure to SBV among wild artiodactyl species in different countries. The prevalence of SBV has been observed in roe deer (Capreolus capreolus) (ranging from 17.3 to 75.9%) (10, 46), red deer (Cervus elaphus) (ranging from 6.0 to 71.4%) (24, 55) in Australia and UK, fallow deer (Dama dama) (ranging from 5 to 70%) (10, 46) in Australia and Spain, bison (Bison bonasus) (51%) (7) in Poland, chamois (R. rupicapra) (10%) (10) and mouflon (Ovis aries musimon) (ranging from 10 to 75%) (8, 54) in Poland and Germany, and wild boar (Sus scrofa) (ranging from 3 to 23.4%) (21, 24) in Spain and Italy. SBV seropositivity was determined only from samples positive by both bELISA and VNT. Several sera positive by bELISA could not be tested by VNT due to serum cytotoxicity, so seroprevalence may have been slightly underestimated. Similar peak seroprevalences were also reported in domestic species from the Netherlands, Belgium, Germany, Sweden, and Spain (24, 53, 70).
Commercially available ELISA assays are sensitive, specific, and robust, but cross-reactivity with other members of the Simbu serogroup has been reported for the assay used in this study previously Consequently, when interpreting the results of this and similar ELISAs, care must be exercised because a positive result may not indicate infection with SBV but could be due to infection with another Simbu serogroup virus. While VNTs are considered to be the ‘gold standard’ for the assessment of other assays, it is well recognized that even they are prone to cross-reactivity for viruses belonging to the Simbu serogroup. As the ELISA is relatively easy to perform, requires minimal laboratory equipment, and laboratories do not need to have all reference viruses, it will be preferred in many of the regions where multiple Simbu serogroup viruses may be present. There is a need to validate ELISA kits for use in these endemic areas but this will be challenging due to the complex cross-reactivity (71).
This investigation presents compelling evidence of SBV infection in both domestic and wild ruminants worldwide. The findings of the present study appear to suggest an increase in the spread of SBV after the year 2013. Additionally, our research also indicated a high distribution of SBV in both domestic and wild ruminants. Nevertheless, certain aspects lack documented studies, while other countries have recently discovered the presence of the virus, such as Ethiopia, where the SBV was detected around the year 2018. Specifically, the current study revealed a lack of comprehensive information regarding the propagation of SBV in wild animals. In certain regions of the world, the seroprevalence of SBV is significantly high, yet it receives minimal attention, particularly about dairy cattle in African countries. This research also aids in enhancing our comprehension of the range of species susceptible to the virus and evaluating the effectiveness of current SBV serological assays in both wild and domestic ruminants. Therefore, further investigation is required concerning the spread of the virus in wild animals, as well as the implementation of preventive and control measures, such as vaccine production, in countries with high SBV prevalence.
There are several limitations in our meta-analysis and included articles. First, our systematic review only includes published articles, resulting in the potential reported bias. Second, the subgroup analysis was limited to only publication year, geographical location, or detection method as moderators for investigation of the source of heterogeneity between studies, and the included studies did not cover all wild ruminants. Third, the articles we found did not cover all regions of the world, as some countries had no published articles, leading to potential publication bias. The final limitation was that our review was not registered in the PROSPERO database.
Conclusion
The current meta-analysis demonstrates the significant rate at which the overall prevalence of SBV was higher in domestic ruminants (49%) compared with wild ruminants (26%). Within domestic ruminants, the present study reveals that the overall serological assay was greater in cattle in comparison with small ruminants. Among wild ruminants, the roe deer exhibited the highest seropositivity. This investigation includes a substantial number of European studies, indicating a better comprehension or possibly a high distribution of the intermediate host (vectors). Based on the available evidence regarding its emergence in 2011, we presume that the global coverage of the virus is extensive. Particularly, this virus will exert a significant impact on dairy industries. Therefore, it is imperative to pay close attention to this disease to counteract its rapid dissemination worldwide. Additionally, certain countries should prioritize early diagnosis for both domestic and wild ruminants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MD: Conceptualization, Formal analysis, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. AS: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. BD: Conceptualization, Formal analysis, Software, Writing – review & editing, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We gratefully acknowledge the University of Gondar for providing some of the materials and important pieces of training.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1371495/full#supplementary-material
References
1. Hoffmann, B, Schulz, C, and Beer, M. First detection of Schmallenberg virus RNA in bovine semen, Germany, 2012. Vet Microbiol. (2013) 167:289–95. doi: 10.1016/j.vetmic.2013.09.002
2. Laloy, E, Bréard, E, Sailleau, C, Viarouge, C, Desprat, A, Zientara, S, et al. Schmallenberg virus infection among Red Deer, France, 2010–2012. Int Conf Emerg Infect Dis. (2014) 20:131–4. doi: 10.3201/eid2001.130411
3. OIE (2013). Technical Fact Sheet. Schmallenberg Virus. Available at: http://www.oie.int/fr/notre-expertise-scientifique/informations-specifiques-etrecommandations/virus-schmallenberg/. (Accessed April 2, 2013).
4. van den Brom, R, Lievaart-Peterson, K, Luttikholt, S, Peperkamp, K, Wouda, W, and Vellema, P. Abortion in small ruminants in the Netherlands between 2006 and 2011. Tijdschr Diergeneeskd. (2012) 137:450–7.
5. Jack, C, Anstaett, O, Adams, J, Noad, R, Brownlie, J, and Mertens, P. Evidence of seroconversion to SBV in camelids. Vet Rec. (2012) 170:603. doi: 10.1136/vr.e3939
6. Linden, A, Desmecht, D, Volpe, R, Wirtgen, M, Gregoire, F, Pirson, J, et al. Epizootic spread of Schmallenberg virus among wild Cervids, Belgium, fall 2011. Int Conf Emerg Infect Dis. (2012) 18:2006–8. doi: 10.3201/eid1812.121067
7. Larska, M, Kęsik-Maliszewska, J, and Kuta, A. Spread of Schmallenberg virus infections in the ruminants in Poland between 2012 and 2013. Bull Vet Inst Pulawy. (2014) 58:169–76. doi: 10.2478/bvip-2014-0026
8. Mouchantat, S, Wernike, K, Lutz, W, Hoffmann, B, Ulrich, RG, Börner, K, et al. A broad spectrum screening of Schmallenberg virus antibodies in wildlife animals in Germany. Vet Res. (2015) 46:99:1–5. doi: 10.1186/s13567-015-0232-x
9. Molenaar, FM, la Rocca, SA, Khatri, M, Lopez, J, Steinbach, F, and Dastjerdi, A. Exposure of Asian elephants and other exotic ungulates to Schmallenberg virus. PLoS One. (2015) 10:1–11. doi: 10.1371/journal.pone.0135532
10. Jiménez-Ruiz, S, Arenas-Montes, A, Cano-Terriza, D, Paniagua, J, Pujols, J, Miró, F, et al. Blood extraction method by endocranial venous sinuses puncture in hunted wild ruminants. Eur J Wildl Res. (2016) 62:775–80. doi: 10.1007/s10344-016-1056-3
11. Hoffmann, B, Scheuch, M, Höper, D, Jungblut, R, Holsteg, M, Schirrmeier, H, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. (2012) 18:469–72. doi: 10.3201/eid1803.111905
12. Pawaiya, RVS, and Gupta, VK. A review on Schmallenberg virus infection: a newly emerging disease of cattle, sheep and goats. Vet Med (Praha). (2013) 58:516–26. doi: 10.17221/7083-VETMED
13. Doceul, V, Lara, E, Sailleau, C, Belbis, G, Richardson, J, Bréard, E, et al. Epidemiology, molecular virology and diagnostics of Schmallenberg virus, an emerging orthobunyavirus in Europe. Ann Rech Vet. (2013) 44:31–13. doi: 10.1186/1297-9716-44-31
14. Wernike, K, Holsteg, M, Szillat, KP, and Beer, M. Development of within-herd immunity and long-term persistence of antibodies against Schmallenberg virus in naturally infected cattle. Vet Res. (2018) 14:1–6. doi: 10.1186/s12917-018-1702-y
15. Beer, M, Conraths, FJ, and van der Poel, WHM. ‘Schmallenberg virus’ – a novel orthobunyavirus emerging in Europe. J Hyg. (2013) 141:1–8. doi: 10.1017/S0950268812002245
16. Marie, GM, Bayrou, C, Kleijnen, D, Cassart, D, and Desmecht, D. Schmallenberg Virus in Domestic. Med Vet Entomol. (2012) 18:7–9. doi: 10.3201/eid1809.120716
17. Veldhuis, AMB, van Schaik, G, Vellema, P, Elbers, ARW, Bouwstra, R, van der Heijden, HMJF, et al. Schmallenberg virus epidemic in the Netherlands: spatiotemporal introduction in 2011 and seroprevalence in ruminants. Prev Vet Med. (2013) 112:35–47. doi: 10.1016/j.prevetmed.2013.06.010
18. EFSA. EFSA assesses current knowledge of Schmallenberg virus. Vet Rec. (2014) 174:566. doi: 10.1136/vr.g3572
19. Azkur, AK, Albayrak, H, Risvanli, A, Pestil, Z, Ozan, E, Yılmaz, O, et al. Antibodies to Schmallenberg virus in domestic livestock in Turkey. Trop Anim Health Prod. (2013) 45:1825–8. doi: 10.1007/s11250-013-0415-2
20. Zhai, S-L, Lv, DH, Wen, X-H, Zhu, X-L, Yang, Y-Q, Chen, Q-L, et al. Preliminary serological evidence for Schmallenberg virus infection in China. Trop Anim Health Prod. (2018) 50:449–53. doi: 10.1007/s11250-017-1433-2
21. García-Bocanegra, I, Cano-Terriza, D, Vidal, G, Rosell, R, Paniagua, J, Jiménez-Ruiz, S, et al. Monitoring of Schmallenberg virus in Spanish wild artiodactyls, 2006–2015. PLoS One. (2017) 12:2006–15. doi: 10.1371/journal.pone.0182212
22. Palomo, L.J., Gisbert, J., and Blanco, J Y. Atlas y Libro Rojo de los mamíferos terrestres de España. Madrid: Organismo Autónomo de Parques Nacionales. (2007).
23. Michel, AL, van Heerden, H, Crossley, BM, Al Dahouk, S, Prasse, D, and Rutten, V. Pathogen detection and disease diagnosis in wildlife: challenges and opportunities. Rev Sci Tech. (2021) 40:105–18. doi: 10.20506/rst.40.1.3211
24. Chiari, M, Sozzi, E, Zanoni, M, Alborali, LG, Lavazza, A, and Cordioli, P. Serosurvey for Schmallenberg Virus in Alpine Wild Ungulates. Transbound Emerg Dis. (2012) 2014:1–3. doi: 10.1111/tbed.12158
25. Mathew, C, Klevar, S, Elbers, ARW, van der Poel, WHM, Kirkland, PD, Godfroid, J, et al. Detection of serum neutralizing antibodies to Simbu sero-group viruses in cattle in Tanzania. BMC Vet Res. (2015) 11. doi: 10.1186/s12917-015-0526-2
26. Sibhat, B, Ayelet, G, Gebremedhin, EZ, and Skjerve, E. Acta Tropica Seroprevalence of Schmallenberg virus in dairy cattle in Ethiopia. Acta Trop. (2018) 178:61–7. doi: 10.1016/j.actatropica.2017.10.024
27. Blomstrom, AL, Stenberg, H, Scharin, I, Figueiredo, J, Nhambirre, O, Abilio, AP, et al. Serological screening suggests presence of Schmallenberg virus in cattle. Sheep Goat. (2014) 61:289–92. doi: 10.1111/tbed.12234
28. Leask, R, Botha, AM, and Bath, GF. Schmallenberg virus – is it present in South Africa? J S Afr Vet Assoc. (2013) 84:1–5. doi: 10.4102/jsava.v84i1.535
29. Asmare, K, Sibhat, B, Ayelet, G, Zewdu, E, Aragaw, K, and Skjerve, E. Acta Tropica serological evidence of bovine herpesvirus-1, bovine viral diarrhea virus and Schmallenberg v irus infections in relation to reproductive disorders in dairy cattle in Ethiopia. Acta Trop. (2018) 178:236–41. doi: 10.1016/j.actatropica.2017.12.005
30. Acevedo, P, Farfán, MÁ, Márquez, AL, Delibes-Mateos, M, Real, R, and Vargas, JM. Past, present and future of wild ungulates in relation to changes in land use. Landsc Ecol. (2011) 26:19–31. doi: 10.1007/s10980-010-9538-2
31. Gortázar, C, Acevedo, P, Ruiz-Fons, F, and Vicente, J. Disease risks and overabundance of game species. Z Jagdwiss. (2006) 52:81–7. doi: 10.1007/s10344-005-0022-2
32. Borenstein, M, and Higgins, JPT. Meta-analysis and subgroups. Prevent Sci. (2013) 14:134–43. doi: 10.1007/s11121-013-0377-7
33. Page, MJ, Mckenzie, JE, Bossuyt, PM, Boutron, I, Hoffmann, TC, Mulrow, CD, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. (2021) 88. doi: 10.1186/s13643-021-01626-4
34. Downes, MJ, Brennan, ML, Williams, HC, and Dean, RS. Development of a critical appraisal tool to assess the quality of cross-sectional studies (AXIS). BMJ Open. (2016) 6:1–7. doi: 10.1136/bmjopen-2016-011458
35. Balduzzi, S, Rücker, G, and Schwarzer, G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. (2019) 22:153–60. doi: 10.1136/ebmental-2019-300117
36. Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J Stat Softw. (2010) 36:1–48.
37. Higgins, JPT, and Thompson, SG. Quantifying heterogeneity in a meta‐analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
38. Barrett, D, More, SJ, O’Neill, R, Bradshaw, B, Casey, M, Keane, M, et al. Prevalence and distribution of exposure to Schmallenberg virus in Irish cattle during October 2012 to November 2013. Vet Res. (2015) 11:1–10. doi: 10.1186/s12917-015-0564-9
39. Chaintoutis, SC, Kiossis, E, Giadinis, ND, Brozos, CN, Sailleau, C, Viarouge, C, et al. Evidence of Schmallenberg virus circulation in ruminants in Greece. Trop Anim Health Prod. (2014) 46:251–5. doi: 10.1007/s11250-013-0449-5
40. Elbers, ARW, Meiswinkel, R, van Weezep, E, Kooi, EA, and van der Poel, WHM. Schmallenberg virus in CulicoidesBiting midges in the Netherlands in 2012. J Vet Med A Physiol Pathol Clin Med. (2015) 62:339–42. doi: 10.1111/tbed.12128
41. Gache, K, Dominguez, M, Pelletier, C, Petit, E, Calavas, D, Hendrikx, P, et al. Paper Schmallenberg virus: A seroprevalence survey in cattle and sheep, France, winter 2011-2012. Vet Rec. (2013) 173:141. doi: 10.1136/vr.101377
42. King, B, Brown, TOS, Tarlinton, R, and Daly, JM. Seroprevalence of Schmallenberg virus in the United Kingdom and the Republic of Ireland: 2011 – 2013. Vet Microbiol. (2015) 180:36–40. doi: 10.1016/j.vetmic.2015.07.025
43. Oluwayelu, D, Wernike, K, Adebiyi, A, Cadmus, S, and Beer, M. Neutralizing antibodies against Simbu serogroup viruses in cattle and sheep, Nigeria, 2012–2014. Vet Res. (2018) 14:1–7. doi: 10.1186/s12917-018-1605-y
44. Sohier, C, Michiels, R, Kapps, E, Van, E, Quinet, C, Cay, AB, et al. PT both authors contributed equally to this work SC. Res Vet Sci. (2017)
45. Meroc, A, Van, Loo H, Van, Driessche E, Czaplicki, G, Quinet, C, and Riocreux, F. Follow-up of the Schmallenberg virus Seroprevalence in Belgian cattle. (2015). 116:350–359.
46. Steinrigl, A, Schiefer, P, Schleicher, C, Peinhopf, W, Wodak, E, Bagó, Z, et al. Ruminant abortions and foetal death in Austria in 2012/2013. Prev Vet Med. (2014). doi: 10.1016/j.prevetmed.2014.03.006
47. Larska, M, Socha, W, and Rola, J. Occurrence of emerging ruminant viruses in goats in Poland. Pol J Vet Sci. (2023) 26:137–42. doi: 10.24425/pjvs.2023.145015
48. Rasekh, M, Sarani, A, and Jafari, A. First detection of Schmallenberg virus antibody in cattle population of eastern Iran. Vet Res Forum. (2022) 13:443–6. doi: 10.30466/vrf.2021.135144.3032
49. Ferrara, G, Wernike, K, Iovane, G, Pagnini, U, and Montagnaro, S. First evidence of Schmallenberg virus infection in southern Italy. BMC Vet Res [Internet]. (2023) 19:1–9. doi: 10.1186/s12917-023-03666-5
50. Firat Dogan, A, Bilal Dik, B, Bilge-Dagalp, S, Farzani, TA, Ataseven, VS, Acar, G, et al. Prevalance of Schmallenberg orthobunyavirus (SBV) infection in sampled ruminants in Turkey’s eastern Mediterranean region between 2015 and 2017. Res Vet J. (2020). doi: 10.1016/j.rvsc.2022.02.013
51. Abi-Rizk, A, Kanaan, T, and El, HJ. Seroprevalence of Schmallenberg virus and other Simbu group viruses among the Lebanese sheep. Open Vet J. (2017) 7:290–3.
52. Kęsik-Maliszewska, J, Collins, ÁB, Rola, J, Blanco-Penedo, I, and Larska, M. Schmallenberg virus in Poland endemic or re-emerging? A six-year serosurvey. Transbound Emerg Dis. (2021) 68:2188–98. doi: 10.1111/tbed.13870
53. Fernández-Aguilar, X, Pujols, J, Velarde, R, Rosell, R, López-Olvera, JR, Marco, I, et al. Schmallenberg virus circulation in High Mountain ecosystem, Spain. Emerg Infect Dis. (2014) 20:1062–4. doi: 10.3201/eid2006.130961
54. Larska, M, Krzysiak, MK, Kęsik-Maliszewska, J, and Rola, J. Cross-sectional study of Schmallenberg virus seroprevalence in wild ruminants in Poland at the end of the vector season of 2013. Vet Res. (2014) 10:1–7. doi: 10.1186/s12917-014-0307-3
55. Barlow, A, Peter Green, TB, and Healy, N. Serological confirmation of SBV infection in wild British deer. Vet Rec. (2013) 172:429. doi: 10.1136/vr.f2438
56. Malmsten, A, Malmsten, J, Blomqvist, G, Näslund, K, Vernersson, C, Hägglund, S, et al. Serological testing of Schmallenberg virus in Swedish wild cervids from 2012 to 2016. Vet Res. (2017) 13:84:1–5. doi: 10.1186/s12917-017-1005-8
57. Rossi, S, Viarouge, C, Faure, E, Gilot-Fromont, E, Gache, K, Gibert, P, et al. Exposure of wildlife to the Schmallenberg virus in France (2011-2014): higher, faster, stronger (than bluetongue)! J Vet Med A Physiol Pathol Clin Med. (2017) 64:354–63. doi: 10.1111/tbed.12371
58. Desmecht, D., Garigliany, M.M., Beer, M., Schirrmeier, H., Paternostre, J., Volpe, R., et al. (2013). Detection of antibodies against Schmallenberg virus in wild boars, Belgium, 2010–2012. In Proceedings of the 31th Congress of the International Union of Game Biologists, Brussels, Belgium, 27–29 August 2013.
59. Díaz, P, Remesar, S, Venzal, JM, Vázquez-López, ME, Fernández, G, and López, C. Occurrence of Borrelia and Borreliella species in Ixodes ricinus collected from roe deer in northwestern Spain. Med Vet Entomol. (2019) 33:427–30. doi: 10.1111/mve.12364
60. Elbers, ARW, Loeffen, WLA, Quak, S, de Boer-Luijtze, E, van der Spek, AN, Bouwstra, R, et al. Seroprevalence of Schmallenberg virus antibodies among dairy cattle, the Netherlands, winter 2011–2012. Int Conf Emerg Infect Dis. (2012) 18:1065–71. doi: 10.3201/eid1807.120323
61. Bessell, PR, Searle, KR, Auty, HK, Handel, IG, Purse, BV, and Bronsvoort, BM. Epidemic potential of an emerging vector borne disease in a marginal environment: Schmallenberg in Scotland. Sci Rep. (2013) 3:1178. doi: 10.1038/srep01178
62. Gubbins, S, Carpenter, S, Baylis, M, Wood, JL, and Mellor, PS. Assessing the risk of bluetongue to UK livestock: uncertainty and sensitivity analyses of a temperature-dependent model for the basic reproduction number. J. R. Soc. Interface. (2008) 5:363–71. doi: 10.1098/rsif.2007.1110
63. Constable, PD, Hinchcliff, KW, and Done, SH. African horse sickness. Veterinary medicine: A textbook of the diseases of cattle, horses, sheep, pigs, and goats. 11th ed. Missouri: Elsevier (2017).
65. Luttikholt, S, Veldhuis, A, van den Brom, R, Moll, L, Lievaart-Peterson, K, Peperkamp, K, et al. Risk factors for malformations and impact on reproductive performance and mortality rates of Schmallenberg virus in sheep flocks in the Netherlands. PLoS One. (2014) 9:1–8. doi: 10.1371/journal.pone.0100135
66. McCarthy, TK, Bateman, A, and Nowak, AL. National BTV vector surveillance Programme 2007–2009. Annual Report 2009/2010. (2016).
67. Haider, N, Cuellar, AC, Kjær, LJ, Sørensen, JH, and Bødker, R. Microclimatic temperatures at Danish cattle farms, 2000-2016: quantifying the temporal and spatial variation in the transmission potential of Schmallenberg virus. Parasites Vectors. (2018) 11:1–14. doi: 10.1186/s13071-018-2709-8
68. Mehlhorn, H, Walldorf, V, Klimpel, S, Schaub, G, Kiel, E, Focke, R, et al. Bluetongue disease in Germany (2007-2008): monitoring of entomological aspects. Parasitol Res. (2009) 105:313–9. doi: 10.1007/s00436-009-1416-y
69. Collins, B, Grant, J, Barrett, D, Doherty, ML, Hallinan, A, and Mee, JF. Schmallenberg virus: predicting within-herd seroprevalence using bulk-tank milk antibody titres and exploring individual animal antibody titres using empirical distribution functions (EDF). Prev Vet Med [Internet]. (2017) 143:68–78. doi: 10.1016/j.prevetmed.2017.04.013
70. Helmer, C, Eibach, R, and Tegtmeyer, PC. Survey of Schmallenberg virus (SBV) infection in German goat flocks. Epidemiology & Infection (2013). 2335–2345.
Keywords: domestic ruminants, meta-analysis, pooled prevalence, Schmallenberg virus, wild ruminants
Citation: Dagnaw M, Solomon A and Dagnew B (2024) Serological prevalence of the Schmallenberg virus in domestic and wild hosts worldwide: a systematic review and meta-analysis. Front. Vet. Sci. 11:1371495. doi: 10.3389/fvets.2024.1371495
Edited by:
Carlos Tejeda, Austral University of Chile, ChileReviewed by:
Gianmarco Ferrara, University of Naples Federico II, ItalyGiulia Franzoni, Experimental Zooprophylactic Institute of Sardinia (IZS), Italy
Copyright © 2024 Dagnaw, Solomon and Dagnew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melkie Dagnaw, bWVsa2llZGFnbmF3MzUyOEBnbWFpbC5jb20=
 Melkie Dagnaw
Melkie Dagnaw Atsede Solomon2
Atsede Solomon2