- 1College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
- 2Seoul Metropolitan Government Research Institute of Public Health and Environment, Seoul, Republic of Korea
We investigated the antimicrobial resistance (AMR) rates and profiles of Staphylococcus spp. and Escherichia coli (E.coli) isolated from dogs and cats in Seoul, South Korea during 2021–2023. We analyzed AMR profiles of Staphylococcus spp. and E. coli isolated from 2,180 samples (1,859 canine and 321 feline) collected from 36 veterinary clinics in Seoul, South Korea, as part of the Korean Veterinary Antimicrobial Resistance Monitoring System (2021–2023). A total of 484 Staphylococcus spp. isolates and 158 E. coli isolates were identified and used for AMR test. Staphylococcus spp. isolates exhibited the highest resistance to penicillin in both dogs (85%) and cats (29.81%). Multidrug-resistant (MDR) Staphylococcus spp. was more prevalent in dogs (65%) than in cats (14.42%), with three S. pseudintermedius isolates from dogs and a S. pseudintermedius isolate from a cat showing resistance to eight antibiotic classes. Methicillin-resistant S. pseudintermedius (MRSP) constituted 105 out of 284 S. pseudintermedius isolates (36.97%) in dogs and seven strains out of 14 (50%) in cats. E. coli isolates demonstrated the highest resistance to cefalexin in both dogs (61.72%) and cats (56.67%). The prevalence of MDR E. coli was higher in dogs (37.5%) than in cats (26.67%). This study highlights the concerning prevalence of AMR in commensal or potentially opportunistic pathogens from companion animals, particularly in dogs. It is crucial to promote the prudent use of antimicrobials in companion animals and ensure the ongoing monitoring of trends in antimicrobial-resistant bacteria to mitigate the selection and spread of antibiotic-resistant bacteria between humans and companion animals.
1 Introduction
Antimicrobial resistance (AMR) is recognized by the World Health Organization (WHO) as one of the top global public health and development threats (1, 2). AMR in animals has increased due to the overuse and misuse of antibiotics not only in food-producing animals (3–5) but also in companion animals (6). In particular, antimicrobial agents used in human medicine have been frequently used in small animal veterinary practice, with a significant reliance on broad-spectrum agents (5). This perspective has drawn increased attention to companion animals, mainly dogs and cats, as potential reservoirs of AMR due to their close contact with humans and frequent exposure to broad-spectrum antimicrobials (3, 5). In recent years, the emergence of multidrug-resistant (MDR) bacteria in dog and cat populations has posed a significant threat, contributing to therapeutic failures and prolonged hospitalization periods (7–9). In response, the World Organization for Animal Health (WOAH) revised its international standards on AMR, expanding its scope to include companion animals. These revised standards provide specific recommendations for the prudent use of critically important antimicrobial agents in veterinary medicine to protect both animal and human health1. Therefore, the investigation of AMR in companion animals is crucial from both veterinary and human medicine perspectives.
To monitor AMR, indicator bacteria such as Staphylococcus spp. and Escherichia coli (E. coli) have been used across various sources, including companion animals (1), pig farms (10), wildlife (11), raw milk (12), foodstuffs (13), patients with bacteremia (14), and hospital environments (15). E. coli is a commensal or potentially opportunistic pathogen commonly found in the intestinal tract of animals and humans (10). Similarly, different Staphylococcus spp. are part of the commensal microbiota but can also cause diseases ranging from abscesses and mastitis to septicemia (10). In recent decades, the prevalence of AMR in Staphylococcus spp., particularly methicillin resistance, has significantly increased in both human and veterinary patients (16).
According to a report from the Seoul Digital Foundation, 612,000 of South Korea’s 3,500,000 registered pet dogs (17.5%) reside in Seoul, and the ratio of pet dogs to households in Seoul is 14.9%, indicating that more than one in 10 households owns a pet dog.2 Densely populated cities like Seoul can serve as a reservoir for antibiotic-resistant bacteria, as dogs and cats live in close proximity to humans, increasing the risk of microorganism transmission (17). This study aimed to investigate the AMR rates and resistance profiles of Staphylococcus spp. and E. coli isolated from dogs and cats in Seoul, South Korea, between 2021 and 2023, and as part of the Korean Veterinary Antimicrobial Resistance Monitoring System.
2 Materials and methods
2.1 Sample collection
Clinical samples were collected from 36 veterinary clinics in Seoul participating in the Korean Veterinary Antimicrobial Resistance Monitoring System between 2021 and 2023 (15 clinics in 2021, 15 in 2022, and 20 in 2023). Samples included diarrhea, skin swabs, nasal swabs, and urine collected from dogs and cats exhibiting symptoms relevant to these sample types. In total, 1,859 samples were collected from dogs and 321 from cats (Table 1). However, cat urine samples were excluded from the monitoring system due to challenges associated with sample collection. All samples were placed on ice immediately after collection and transported to the Seoul Research Institute of Public Health and Environment within 6 h.
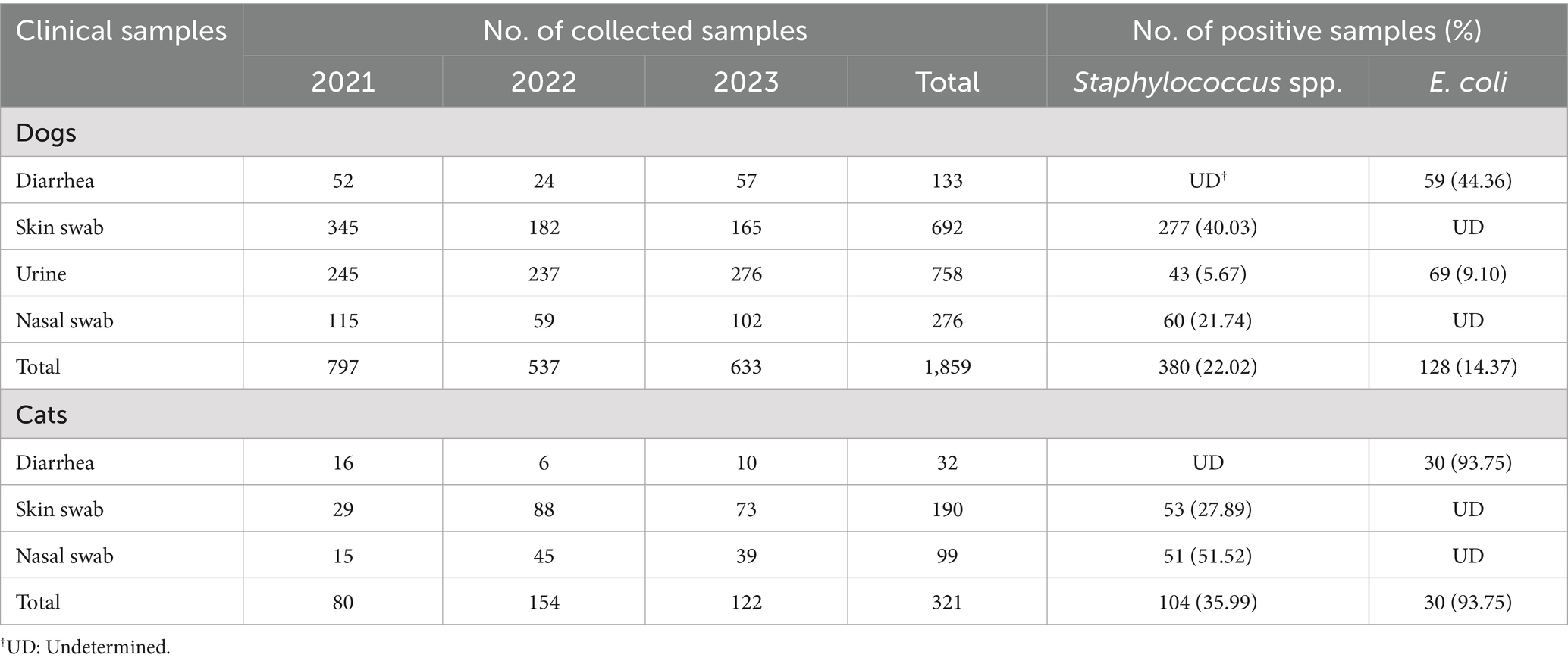
Table 1. The number of clinical samples from dogs and cats collected between 2021 and 2023 and prevalence of Staphylococcus spp. and E. coli in the samples.
2.2 Bacteria isolation and identification
Skin and nasal swabs, urine samples from dogs, as well as skin and nasal swabs from cats, were collected for the isolation of Staphylococcus spp. Diarrheic fecal samples from both dogs and cats, along with urine from dogs, were used to isolate E. coli. Diarrhea samples were collected using transported fecal swabs (470 CE, Copan Italy Spa, Brescia, Italy) and plated onto eosin methylene blue agar (Thermo Fisher Scientific, Milan, Italy) for E. coli isolation. Other clinical samples were inoculated onto MacConkey agar and blood agar (Thermo Fisher Scientific) using sterilized cotton swabs. All culture plates were incubated aerobically at 37°C overnight. After incubation, presumptive E. coli and Staphylococcus spp. colonies were identified using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). For MALDI-TOF analysis, a single colony was directly applied onto the target plate, overlaid with 1 μL of matrix solution. The sample was allowed to air-dry at room temperature prior to analysis. Spectra acquisition was performed using the Bruker MALDI-TOF system (Bruker Corporation, Billerica, MA, USA) following the manufacturer’s protocol. Identification of isolates was carried out with the MALDI Biotyper MBT Compass 4.1 software. Isolates yielding a score ≥2.0 were considered reliably identified at the species level, while those with scores ranging from 1.7 to 1.99 were classified at the genus level and subjected to further confirmation using the VITEK 2 Compact system (bioMérieux, Marcy-l’Étoile, France).
2.3 Antibiotic susceptibility test
Isolates collected in 2021 and 2022 were tested using the Sensititre™ MIC panel system (Thermo Fisher Scientific), with COMPGP1F for Staphylococcus spp. and COMPGN1F for E. coli to determine the minimum inhibition concentrations (MICs) of the isolates. Isolates collected in 2023 were analyzed using the VITEK 2 Compact system, employing GP80 AST cards for Staphylococcus spp. and GN97 AST cards for E. coli. In this study, we report the results of antimicrobial susceptibility testing for 13 agents common to both systems: for Staphylococcus spp.—gentamicin, oxacillin + 2% NaCl, penicillin, cefovecin, chloramphenicol, clindamycin, enrofloxacin, marbofloxacin, pradofloxacin, nitrofurantoin, erythromycin, trimethoprim/sulfamethoxazole, and amoxicillin/clavulanic acid; and for E. coli—amikacin, gentamicin, ampicillin, imipenem, cefovecin, chloramphenicol, doxycycline, tetracycline, enrofloxacin, marbofloxacin, pradofloxacin, trimethoprim/sulfamethoxazole, and amoxicillin/clavulanic acid.
For the Sensititre MIC panel, bacterial suspensions were adjusted to a 0.5 McFarland standard. The inoculum (10 μL) was added to a tube containing 11 mL of Sensititre™ Cation-Adjusted Mueller-Hinton Broth with TES and thoroughly mixed, and 50 μL was dispensed into each well of a 96-well plate. The plates were sealed and incubated aerobically at 35°C for 18–24 h. The results were interpreted using the Sensititre Vizion™ Digital MIC Viewing System. All results were interpreted according to the guidelines of the Korean Animal and Plant Quarantine Agency, which are based on Clinical and Laboratory Standards Institute (CLSI) documents VET01S (18) and M100 breakpoints (19) (Supplementary Table 1).
For the VITEK 2 system, bacterial suspensions were adjusted to a 0.6 McFarland standard. For gram-negative isolates, 145 μL of the suspension was added to 3 mL of sterile saline; for gram-positive isolates, 280 μL was used. The inoculum of Staphylococcus spp. was loaded onto a GP80 AST card, and that of E. coli was loaded onto a GN97 AST card. AMR results were interpreted based on the “Result – Expertised” data provided by the system’s Advanced Expert System.
Intermediate susceptibility results were not classified as resistant. In prevalence analyses, only isolates explicitly categorized as “resistant” were considered resistant. Intermediate results were grouped with susceptible isolates unless otherwise stated. MDR was defined as resistance to at least one antimicrobial agent in three or more different antimicrobial classes, excluding intrinsic resistance, per the criteria proposed by Magiorakos et al. (20).
Quality control was performed for the Sensititre™ MIC panel system using the following ATCC reference strains following the manufacturer’s instructions: S. aureus ATCC 29213, Enterococcus faecalis (E. faecalis) ATCC 29212, E. coli ATCC 25922, E. coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, and Klebsiella pneumoniae ATCC 700603. For the VITEK 2 Compact system, E. faecalis ATCC 29212 and E. coli ATCC 25922 were used. All quality control results were within CLSI-defined acceptable ranges throughout the study period.
2.4 mecA polymerase chain reaction (PCR)
To distinguish methicillin-resistant Staphylococcus pseudintermedius (S. pseudintermedius, MRSP), the presence of the mecA gene was investigated in all oxacillin-resistant S. pseudintermedius isolates using PCR, following a modified protocol by Lee et al. (21).
For direct colony PCR, individual colonies grown on blood agar plates were used directly as templates, and sterilized water was included as a negative control. Each PCR reaction was performed in a final volume of 50 μL, containing 0.2 μM of mecA-specific primers (mecA-MR1: 5′-ATGAGATTAGGCATCGTTCC-3′ and mecA-MR2: 5′-TGGATGACAGTACCTGAGCC-3′) (21) and 25 μL of 2 × EmeraldAmp® GT PCR Master Mix (Takara, Otsu, Japan). The PCR conditions were as follows: initial denaturation at 94°C for 5 min; followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and a final extension at 72°C for 7 min. PCR products were analyzed by electrophoresis on 2% agarose gels containing SafeView™ Classic stain (ABM, Richmond, BC, Canada). Samples showing a positive band at 554 bp were subjected to gel extraction using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific), and the identity of the mecA gene was confirmed by sequencing.
2.5 Statistical methods
Statistical analysis was performed using MedCalc® Statistical Software version 23.0.6. Differences in antibiotic resistance rates between clinical sample types from dogs and cats were evaluated using a two-way chi-square test, with p-values < 0.05 considered significant.
3 Results
3.1 Prevalence of Staphylococcus spp. in the samples
The prevalence and species distribution of Staphylococcus spp. isolated from clinical samples collected from dogs and cats are summarized in Tables 1, 2. Staphylococcus spp. were identified in 380 out of 1,726 samples (22.02%) obtained from dogs, which included 692 skin swabs, 758 urine samples, and 276 nasal swabs. In cats, Staphylococcus spp. were detected in 104 out of 289 samples (35.99%), consisting of 190 skin swabs and 99 urine samples. Among canine samples, the highest isolation rates were observed in skin swabs (40.03%), followed by nasal swabs (21.74%) and urine samples (5.67%). In cats, nasal swabs exhibited the higher isolation rate (51.52%) than skin swabs (27.89%) (Table 1).
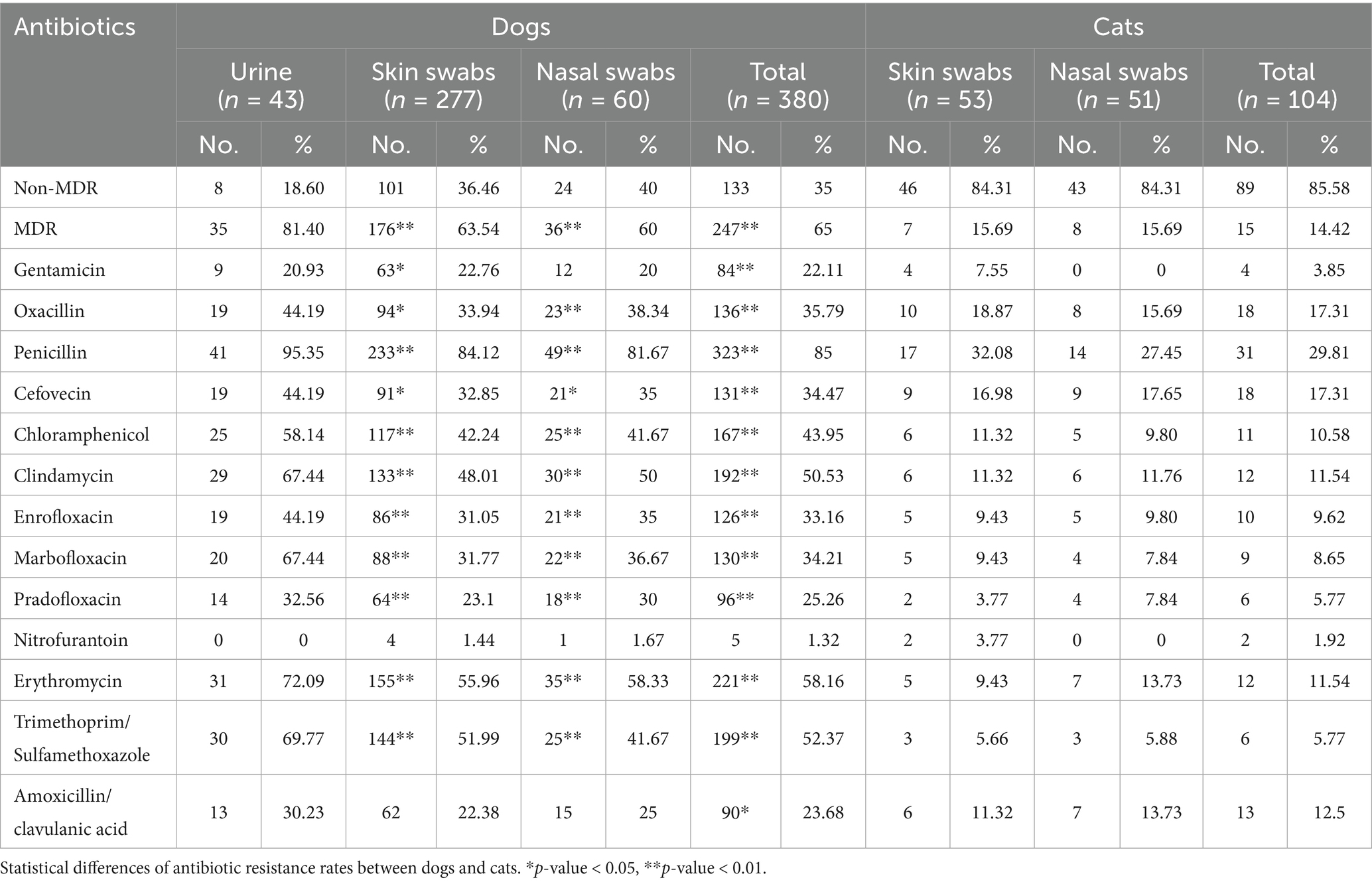
Table 2. Antibiotic resistance of Staphylococcus spp. isolated from dogs and cats in this study (n = 484).
S. pseudintermedius was the most prevalent Staphylococcus species isolated from dogs, accounting for 284 out of 380 isolates (74.74%) across all sample types (Supplementary Table 2). S. schleiferi was the second most frequently isolated species in urine and skin swab samples, while S. felis was the second most common in nasal swabs. In cats, S. felis was predominant, comprising 61 out of 104 isolates (58.65%) across all sample types. S. pseudintermedius was the second most prevalent species in skin swabs and S. aureus was the second most frequently detected species in nasal swabs (Supplementary Table 2). The distribution of Staphylococcus species varied by sample type, with nasal swabs showing the highest species diversity in both dogs and cats (Supplementary Table 2).
3.2 Antibiotic resistance of Staphylococcus spp.
The antibiotic resistance rates and the MIC distribution of Staphylococcus spp. in dogs and cats are summarized in Table 2 and Supplementary Table 3. Overall, 247 out of 380 isolates (65%) from dogs and 15 out of 104 isolates (14.42%) from cats were resistant to at least one agent in three or more antimicrobial categories, classifying them as MDR (Table 2). Statistical analysis revealed significantly higher overall MDR rates as well as MDR rates by sample type, in dogs than in cats (p < 0.00001). Penicillin exhibited the highest resistance rate across all sample types in isolates from both dogs and cats, while nitrofurantoin showed the lowest resistance rate (Table 2). In dogs, isolates exhibited the highest resistance to penicillin (85%), followed by erythromycin (58.16%), trimethoprim/sulfamethoxazole (52.37%), and clindamycin (50.53%). In cats, resistance was most frequently observed to penicillin (29.81%), followed by oxacillin and cefovecin (17.31% each). Comparative analysis of Staphylococcus spp. isolates from the same clinical sample types in dogs and cats revealed that resistance rates to most tested antibiotics were significantly higher in isolates from dogs than from cats, with the exception of nitrofurantoin (p < 0.05) (Table 2).
Oxacillin-resistant Staphylococcus spp. isolated from various clinical samples in dogs and cats are summarized in Table 3. For S. pseudintermedius, the most commonly isolated species from dog, oxacillin resistance was observed in 73 out of 211 isolates (34.60%) from skin swabs, 15 out of 34 isolates (44.12%) from nasal swabs, and 17 out of 39 isolates (43.59%) from urine samples. In cats, oxacillin resistance among S. pseudintermedius was recorded in 5 out of 9 isolates (55.56%) from skin swabs and 2 out of 5 isolates (40%) from nasal swabs. Oxacillin-resistant S. pseudintermedius harboring mecA gene was 101 out of 284 S. pseudintermedius isolates (35.56%) in dogs and 7 out of 14 (50%) in cats (data not shown).
The antibiotic resistance patterns of Staphylococcus spp. are summarized in Supplementary Table 4. Among the isolates, 37 out of 380 isolates (9.74%) from dogs and 65 out of 104 isolates (62.5%) from cats were susceptible to all tested antibiotics. The highest proportions of resistant strains were observed for β-lactams in dogs (48 out of 380, 12.11%) and in cats (12 out of 104, 11.54%) (Supplementary Table 4). The most prevalent MDR pattern in dogs was β-lactams-amphenicols-lincosamides-macrolides-trimethoprim/sulfamethoxazole, identified in 32 out of 247 MDR isolates (12.96%). Meanwhile in cats, the most common pattern was β-lactams-quinolones-amoxicillin/clavulanic acid, observed in 3 out of 15 MDR isolates (20%) (Supplementary Table 4). Notably, three S. pseudintermedius isolates from dogs and a S. pseudintermedius isolate from a cat exhibited resistance to 8 antibiotic classes (Supplementary Table 4).
3.3 Prevalence and antibiotic resistance of Escherichia coli
The prevalence of E. coli in dogs and cats is summarized in Table 1. In dogs, E. coli was isolated from 128 out of 891 total samples (14.37%), which included 59 out of 133 diarrhea samples (44.36%) and 69 out of 758 urine samples (9.10%). In cats, E. coli was isolated from 30 out of 32 diarrhea samples (93.75%).
The antibiotic resistance rates and the MIC distribution of E. coli from dogs and cats are presented in Table 4 and Supplementary Table 5, respectively. Overall, 48 out of 128 isolates (37.5%) from dogs and 8 out of 30 isolates (26.67%) from cats were classified as MDR. The MDR rate in urine isolates (30 out of 69, 43.48%) was higher than that in diarrhea isolates (18 out of 59, 30.51%). E. coli isolates from diarrhea samples exhibited the highest resistance rates to cefalexin, with 50 out of 59 isolates (84.75%) from dogs and 17 out of 30 isolates (56.67%) from cats, showing a significant difference between the isolates from the two host species (p <0.01) (Table 4). No resistance to amikacin or imipenem was detected across all host species and sample types (Table 4). In dogs, E. coli isolates from diarrhea samples showed no resistance to ampicillin or amoxicillin/clavulanic acid. However, urine isolates exhibited the highest resistance to ampicillin, with 43 out of 69 isolates (62.32%) displaying resistance (Table 4). In cats, high resistance rates were observed against cefovecin and cefpodoxime, with 11 out of 30 isolates (36.67%) being resistant to each. These results suggest a high antibiotic resistance to cephalosporins in isolates from cats (Table 4).
The antibiotic resistance pattern of E. coli isolates revealed that 25 out of 128 isolates (19.531%) from dogs and 10 out of 30 isolates (33.33%) from cats were susceptible to all tested antibiotics (Supplementary Table 6). The most common resistance pattern was exclusively against β-lactams, observed in 27 out of 128 isolates (21.10%) from dogs and 6 out of 30 isolates (20.0%) from cats (Supplementary Table 6). Six isolates from dogs and two isolates from cats exhibited resistance to 6 antibiotic classes (Supplementary Table 6).
4 Discussion
Seoul is the capital city of South Korea, a densely populated metropolis with a population of 9.619 million people (in 2024) living in a land area of 605.21 km2.3 According to the Korea Rural Economic Institute, the proportion of the total population that owns pets has increased from 17.4% in 2010 to 27.7% in 2020. As both human and pet populations continue to grow in Seoul, monitoring trends in antimicrobial-resistant bacteria in companion animals is increasingly important for public and animal health. In this study, we investigated the prevalence and AMR patterns of Staphylococcus spp. and E. coli isolated from clinical samples of dogs and cats collected from veterinary clinics in Seoul, South Korea, between 2021 and 2023 as part of the Korean Veterinary Antimicrobial Resistance Monitoring System. Notably, due to the transition in antimicrobial susceptibility testing methodology within the Korean Veterinary Antimicrobial Resistance Monitoring System in 2023, different antimicrobial susceptibility testing methods were used for isolates collected in different years: the Sensititre™ MIC system for isolates from 2021 to 2022 and VITEK® 2 Compact system for those from 2023. Each system employs its own panel of antimicrobial concentrations and interpretive criteria. Results from the Sensititre™ system were interpreted using CLSI guidelines, while those from the VITEK® 2 system were interpreted using the Advanced Expert System. These methodological differences may affect the comparability of MIC values and resistance patterns across years and should be considered when interpreting the findings.
Staphylococcus spp. are commensal bacteria and opportunistic pathogens capable of zoonotic transmission (22, 23). Several studies have examined the prevalence of Staphylococcus spp. in both healthy and sick humans and animals (1, 24–26). Prevalence rates vary depending on the sample type and host species: ranging from 67.3 to 73.8% in healthy dogs and cats (26), to 69% in sick dogs and cats (1), and to 82.81, 76.4, and 91% in healthy domestic cats, feral cats, and sick cats, respectively (25). In the present study, the prevalence of Staphylococcus spp. ranged from 5.67% in canine urine samples to 51.52% in feline nasal swab samples. The predominant species identified were S. pseudintermedius in dogs and S. felis in cats, which is consistent with previous studies (14, 25). In this study, Staphylococcus spp. and E. coli isolates from dogs generally exhibited higher resistance rates than those from cats. This finding aligns with the results of a previous study by Yang et al. (27), which reported a greater abundance of antibiotic-resistance genes in canine feces than in feline feces. Notably, this study identified a high prevalence of MDR in Staphylococcus spp. isolated from dogs. S. pseudintermedius poses a potential public health concern due to its ability to transfer antibiotic resistance genes to human staphylococcal species (28, 29).
According to the “2023 Use of Antimicrobials in Domestic Veterinary Clinics” survey conducted by the Animal and Plant Quarantine Agency,4 the top four antibiotics with the highest sales volumes for companion animal treatment were enrofloxacin, cefalexin, ampicillin, and penicillin G. In our study, Staphylococcus spp. and E. coli isolates from dogs and cats exhibited the highest resistance rates to penicillin and cefalexin, respectively. This finding is consistent with previous studies on E. coli strains from fecal samples of healthy dogs and cats (30) and Staphylococcus spp. strains from dogs (31, 32) in South Korea. We suggest that the frequent use of these antibiotics in companion animals has significantly contributed to the selection and maintenance of resistant bacteria in South Korea. However, comprehensive clinical metadata, including prior antimicrobial treatment history of the sampled animals, were unavailable, and uniform inclusion and exclusion criteria were not applied due to the retrospective nature of sample collection from multiple clinics. As prior antibiotic exposure is known to significantly influence the prevalence of antimicrobial resistance, the absence of this information may have limited the interpretation of resistance patterns across host species or sample types.
The use of veterinary antibiotics can promote the selection and transmission of MDR to other organisms, underscoring the needs for strategies that reduce resistance selection and preserve the clinical efficacy of antibiotics. Empirical first-line treatments for urinary or skin infections in companion animals commonly include amoxicillin/clavulanate, trimethoprim/sulfadiazine, and clindamycin. Although antimicrobials are generally not indicated in most cases of diarrhea in dogs, as they are often self-limiting and resolve with supportive fluid therapy alone, broad-spectrum antibiotics such as amoxicillin/clavulanate, cefalexin, and ampicillin are still commonly prescribed in primary veterinary practice for gastrointestinal infections presenting with diarrhea (33).
In this study, Staphylococcus spp. exhibited high resistance to trimethoprim/sulfadiazine (52.37%) and clindamycin (50.53%), suggesting limited effectiveness of these agents for empirical treatment of opportunistic Staphylococcus infections in companion animals. Additionally, amoxicillin/clavulanate and ampicillin may exert a greater selective pressure on intestinal E. coli populations than cefalexin, potentially influencing the gut microbiota and resistance dynamics. The antimicrobial resistance data generated in this study offer valuable insights to inform more judicious selection of antibiotics for the treatment of infectious diseases in companion animals. In addition, the current study did not assess inducible clindamycin resistance in Staphylococcus spp. Therefore, without confirmatory testing such as the D-zone test, isolates with inducible resistance may be incorrectly classified as clindamycin-susceptible, potentially resulting in inappropriate treatment. To improve the accuracy of antimicrobial susceptibility testing, future studies should incorporate methods to detect inducible clindamycin resistance.
Although this study reports phenotypic resistance profiles for E. coli and Staphylococcus spp., molecular mechanisms of resistance in E. coli, such as extended-spectrum β-lactamases (ESBLs), carbapenemases, and other resistance genes, were not investigated. In Staphylococcus spp., resistance gene detection was limited to mecA. Comprehensive molecular analyses using whole-genome sequencing or targeted PCR would enhance understanding of resistance mechanisms, facilitate epidemiological tracking, and support more effective antimicrobial resistance control.
Guided by the One Health concept, which connects human, animal, and environmental health, the findings of this study offer valuable insights into the epidemiology of AMR in companion animals. The results of this study emphasize the need for integrated approaches to address this shared public health challenge. The antibiotic resistance findings in bacteria isolated from companion animals are particularly significant, as Staphylococcus and E. coli are zoonotic pathogens capable of exchanging antibiotic-resistance genes with the human microbiome (24, 34). Addressing this issue requires the implementation of robust antibiotic resistance monitoring and gene analysis in both human and companion animal populations. Our study highlights the concerning prevalence of AMR in commensal or potentially opportunistic pathogens from companion animals, particularly in dogs, where high rates of MDR were observed in Seoul, South Korea. These findings underscore the need for long-term surveillance of resistance patterns in veterinary settings to prevent the spread and evolution of antibiotic-resistant bacteria among both humans and companion animals.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.
Ethics statement
Sample collection for bacterial isolation involves procedures or treatments that fall under standard veterinary practices for diagnosing and treating animals, therefore, ethical approval was considered unnecessary. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
Y-RS: Data curation, Writing – original draft. S-yC: Methodology, Writing – original draft. SK: Data curation, Writing – original draft. K-sK: Supervision, Writing – review & editing. C-sR: Supervision, Writing – review & editing. J-YH: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the KU Research Professor Program of the Konkuk University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1563780/full#supplementary-material
Footnotes
1. ^https://www.woah.org/en/revised-standards-strengthened-actions-to-contain-amr/
2. ^https://smart.seoul.go.kr/board/41/22821/board_view.do
3. ^https://english.seoul.go.kr/seoul-views/meaning-of-seoul/4-population/
References
1. Cocca, G, Piva, S, Magno, SD, Scarpellini, R, Giacometti, F, Serraino, A, et al. Prevalence and patterns of antimicrobial resistance among Escherichia coli and Staphylococcus Spp. in a veterinary university hospital. Vet Sci. (2021) 8:308. doi: 10.3390/vetsci8120308
2. WHO. Antimicrobial resistance: global report on surveillance. Geneva: World Health Organization (2014).
3. Li, Y, Fernandez, R, Duran, I, Molina-Lopez, RA, and Darwich, L. Antimicrobial resistance in bacteria isolated from cats and dogs from the Iberian Peninsula. Front Microbiol. (2020) 11:621597. doi: 10.3389/fmicb.2020.621597
4. Xiong, W, Sun, Y, and Zeng, Z. Antimicrobial use and antimicrobial resistance in food animals. Environ Sci Pollut Res Int. (2018) 25:18377–84. doi: 10.1007/s11356-018-1852-2
5. Guardabassi, L, Schwarz, S, and Lloyd, DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother. (2004) 54:321–32. doi: 10.1093/jac/dkh332
6. Barber, DA, Miller, GY, and McNamara, PE. Models of antimicrobial resistance and foodborne illness: examining assumptions and practical applications. J Food Prot. (2003) 66:700–9. doi: 10.4315/0362-028x-66.4.700
7. Kaspar, U, von Lutzau, A, Schlattmann, A, Roesler, U, Kock, R, and Becker, K. Zoonotic multidrug-resistant microorganisms among small companion animals in Germany. PLoS One. (2018) 13:e0208364. doi: 10.1371/journal.pone.0208364
8. Marco-Fuertes, A, Marin, C, Gimeno-Cardona, C, Artal-Munoz, V, Vega, S, and Montoro-Dasi, L. Multidrug-resistant commensal and infection-causing Staphylococcus spp. isolated from companion animals in the Valencia region. Vet Sci. (2024) 11:54. doi: 10.3390/vetsci11020054
9. Teng, L, Feng, M, Liao, S, Zheng, Z, Jia, C, Zhou, X, et al. A cross-sectional study of companion animal-derived multidrug-resistant Escherichia coli in Hangzhou, China. Microbiol Spectr. (2023) 11:e0211322. doi: 10.1128/spectrum.02113-22
10. Ikwap, K, Gertzell, E, Hansson, I, Dahlin, L, Selling, K, Magnusson, U, et al. The presence of antibiotic-resistant Staphylococcus spp. and Escherichia coli in smallholder pig farms in Uganda. BMC Vet Res. (2021) 17:31. doi: 10.1186/s12917-020-02727-3
11. van den Honert, MS, Gouws, PA, and Hoffman, LC. A preliminary study: antibiotic resistance of Escherichia coli and Staphylococcus aureus from the meat and feces of various south African wildlife species. Food Sci Anim Resour. (2021) 41:135–44. doi: 10.5851/kosfa.2020.e62
12. Tyasningsih, W, Ramandinianto, SC, Ansharieta, R, Witaningrum, AM, Permatasari, DA, Wardhana, DK, et al. Prevalence and antibiotic resistance of Staphylococcus aureus and Escherichia coli isolated from raw Milk in East Java, Indonesia. Vet World. (2022) 15:2021–8. doi: 10.14202/vetworld.2022.2021-2028
13. Khan, MKR, and Malik, A. Antibiotic resistance and detection of Β-lactamase in bacterial strains of Staphylococci and Escherichia coli isolated from foodstuffs. World J Microbiol Biotechnol. (2001) 17:863–8. doi: 10.1023/A:1013857101177
14. Haindongo, EH, Ndakolo, D, Hedimbi, M, Vainio, O, Hakanen, A, and Vuopio, J. Antimicrobial resistance prevalence of Escherichia coli and Staphylococcus aureus amongst bacteremic patients in Africa: a systematic review. J Glob Antimicrob Resist. (2023) 32:35–43. doi: 10.1016/j.jgar.2022.11.016
15. Lestari, ES, Severin, JA, Filius, PM, Kuntaman, K, Duerink, DO, Hadi, U, et al. Antimicrobial resistance among commensal isolates of Escherichia coli and Staphylococcus aureus in the Indonesian population inside and outside hospitals. Eur J Clin Microbiol Infect Dis. (2008) 27:45–51. doi: 10.1007/s10096-007-0396-z
16. Authority EFS, Prevention ECfD, Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. Efsa Journal. (2021) 19:e06490.
17. Vassallo, A, Kett, S, Purchase, D, and Marvasi, M. The bacterial urban resistome: recent advances. Antibiotics (Basel). (2022) 11:512. doi: 10.3390/antibiotics11040512
18. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI supplement VET01S. 7th ed. Wayne, PA: CLSI (2024).
19. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. 35th ed. Wayne, PA: CLSI (2025).
20. Magiorakos, A-P, Srinivasan, A, Carey, RB, Carmeli, Y, Falagas, M, Giske, C, et al. Multidrug-resistant, extensively drug-resistant and Pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
21. Lee, H-J, Kim, Y-S, Kim, J-S, Cho, Y-H, Lee, K-G, Suh, J-T, et al. A study of mecA and femA of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens. Korean J Clin Pathol. (2001) 21:45–8.
22. Lozano, C, Rezusta, A, Ferrer, I, Pérez-Laguna, V, Zarazaga, M, Ruiz-Ripa, L, et al. Staphylococcus Pseudintermedius human infection cases in Spain: dog-to-human transmission. Vector-Borne Zoonotic Dis. (2017) 17:268–70. doi: 10.1089/vbz.2016.2048
23. Sips, GJ, van Dijk, MA, van Westreenen, M, van der Graaf-van, BL, Duim, B, and Broens, EM. Evidence of cat-to-human transmission of Staphylococcus Felis. J Med Microbiol. (2023) 72:001661. doi: 10.1099/jmm.0.001661
24. Thomson, P, Garcia, P, Miles, J, Isla, D, Yanez, C, Santibanez, R, et al. Isolation and identification of staphylococcus species obtained from healthy companion animals and humans. Vet Sci. (2022) 9:79. doi: 10.3390/vetsci9020079
25. Bierowiec, K, Korzeniowska-Kowal, A, Wzorek, A, Rypula, K, and Gamian, A. Prevalence of staphylococcus species colonization in healthy and sick cats. Biomed Res Int. (2019) 2019:1–10. doi: 10.1155/2019/4360525
26. Ma, GC, Worthing, KA, Ward, MP, and Norris, JM. Commensal staphylococci including methicillin-resistant Staphylococcus aureus from dogs and cats in remote New South Wales, Australia. Microb Ecol. (2020) 79:164–74. doi: 10.1007/s00248-019-01382-y
27. Yang, Y, Hu, X, Li, W, Li, L, Liao, X, and Xing, S. Abundance, diversity and diffusion of antibiotic resistance genes in cat feces and dog feces. Environ Pollut. (2022) 292:118364. doi: 10.1016/j.envpol.2021.118364
28. Ventrella, G, Moodley, A, Grandolfo, E, Parisi, A, Corrente, M, Buonavoglia, D, et al. Frequency, antimicrobial susceptibility and clonal distribution of methicillin-resistant Staphylococcus Pseudintermedius in canine clinical samples submitted to a veterinary diagnostic laboratory in Italy: a 3-year retrospective investigation. Vet Microbiol. (2017) 211:103–6. doi: 10.1016/j.vetmic.2017.09.015
29. Soedarmanto, I, Kanbar, T, Ülbegi-Mohyla, H, Hijazin, M, Alber, J, Lämmler, C, et al. Genetic relatedness of methicillin-resistant Staphylococcus Pseudintermedius (Mrsp) isolated from a dog and the dog owner. Res Vet Sci. (2011) 91:e25–7. doi: 10.1016/j.rvsc.2011.01.027
30. Moon, BY, Ali, MS, Kwon, DH, Heo, YE, Hwang, YJ, Kim, JI, et al. Antimicrobial resistance in Escherichia coli isolated from healthy dogs and cats in South Korea, 2020-2022. Antibiotics (Basel). (2023) 13:27. doi: 10.3390/antibiotics13010027
31. Yoon, JW, Lee, KJ, Lee, SY, Chae, MJ, Park, JK, Yoo, JH, et al. Antibiotic resistance profiles of Staphylococcus Pseudintermedius isolates from canine patients in Korea. J Microbiol Biotechnol. (2010) 20:1764–8.
32. Jung, WK, Shin, S, Park, YK, Noh, SM, Shin, SR, Yoo, HS, et al. Distribution and antimicrobial resistance profiles of bacterial species in stray dogs, hospital-admitted dogs, and veterinary staff in South Korea. Prev Vet Med. (2020) 184:105151. doi: 10.1016/j.prevetmed.2020.105151
33. Hall, E. Dealing with haemorrhagic diarrhoea in dogs. In Pract. (2023) 45:516–31. doi: 10.1002/inpr.369
Keywords: antibiotic resistance, companion animal, E. coli , Staphylococcus spp., multi-drug resistance
Citation: Seo Y-R, Choi S-y, Kim S, Kang K-s, Ro C-s and Hyeon J-Y (2025) Antimicrobial resistance profiles of Staphylococcus spp. and Escherichia coli isolated from dogs and cats in Seoul, South Korea during 2021–2023. Front. Vet. Sci. 12:1563780. doi: 10.3389/fvets.2025.1563780
Edited by:
Linda Antionette Bester, University of KwaZulu-Natal, South AfricaReviewed by:
Filip Boyen, Ghent University, BelgiumAnna Luiza F. V. Assumpcao, University of Arkansas System, United States
Copyright © 2025 Seo, Choi, Kim, Kang, Ro and Hyeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Yeon Hyeon, U2g3MDJAa29ua3VrLmFjLmty
†These authors share first authorship
 Ye-Ram Seo
Ye-Ram Seo Sue-young Choi2†
Sue-young Choi2† Sori Kim
Sori Kim Ji-Yeon Hyeon
Ji-Yeon Hyeon