- 1College of Animal Science, Xinjiang Agricultural University, Urumqi, China
- 2College of Food Science and Pharmacy, Xinjiang Agricultural University, Urumqi, China
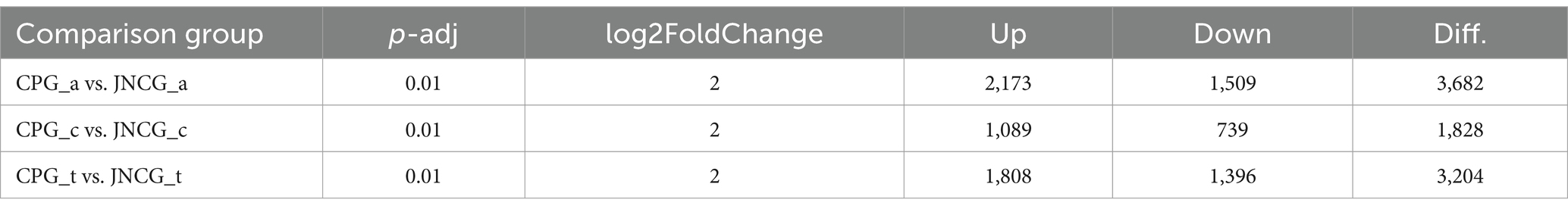
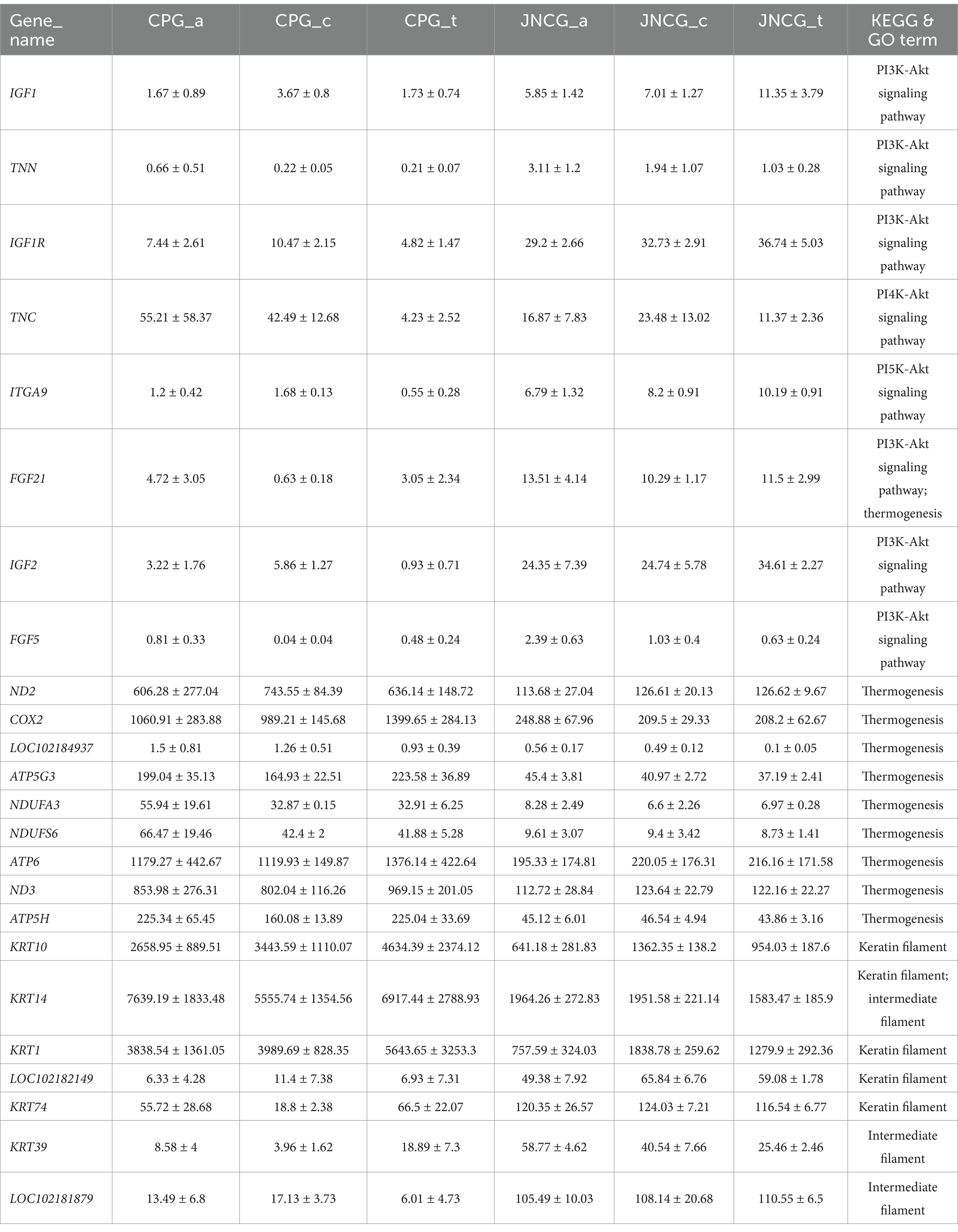
Cashmere goats are excellent livestock breeds known for producing high-quality cashmere fibers from secondary hair follicles. In this study, we aimed to explore the key RNA molecules responsible for the differences in cashmere quality between Jiangnan cashmere goats (JNCG) and Changthangi pashmina goats (CPG). Skin transcriptomic data from the anagen, catagen, and telogen stages of hair follicle growth were retrieved from the SRA database for both JNCG and CPG. Bioinformatics analyses were conducted to identify key molecular differences underlying the variation in cashmere fiber quality. The results showed that there were 4,942 differentially expressed genes (DEGs) between JNCG and CPG through differential analysis, and the DEGs were mainly enriched in PI3K-Akt signaling pathway, Thermogenesis, ECM-receptor interaction in KEGG through functional enrichment analysis, and GO entries were mainly enriched in keratin filament, intermediate filament, keratinization. Twenty-four key candidate genes including IFG1, IGF1R, FGF5, FGF21, ND2, COX2, KRT10, KRT39, and KRT74 were further mined through pathways and entries. These genes play an important role in the development of secondary hair follicles and the formation of cashmere quality in cashmere goats, providing a theoretical basis for the genetic improvement of cashmere goats in the future.
1 Introduction
The cashmere goat is a breed primarily bred for cashmere production, while also being used for both meat and cashmere, making it an important component of the livestock industry. Cashmere goats are found in countries such as China, India, Iran, Pakistan, and Mongolia. The hair of cashmere goats is unique, characterized by a typical heterogeneous coat. Primary hair follicles (PHFs) produce coarse, medullated hair, while secondary hair follicles (SHFs) produce cashmere, which is fine and non-medullated. Cashmere is produced by the SHFs in the skin of cashmere goats, and the fibers are soft, fine, lustrous, warm, comfortable, lightweight, and elastic, making it the highest quality natural fiber material in the textile industry, often referred to as the “fiber gem” or “soft gold” (1–3).
Jiangnan cashmere goat (JNCG) is a newly developed breed, with the male parent being the Liaoning cashmere goat and the female parent being the Xinjiang goat, primarily aimed at increasing cashmere production while maintaining fiber fineness and ensuring post-harvest body weight. It has white-colored cashmere, high yield, stable genetic traits, and is adapted to the arid, semi-arid, and shrubland environments of extreme ecological regions in China, with a cashmere yield of 350–500 g and an average fiber diameter of approximately 15.2 μm (4, 5). The Changthangi pashmina goat (CPG) is a genetically significant breed, adapted to the harsh, cold, dry, and high-altitude climatic conditions of the Ladakh region in India, producing the world’s finest and most expensive animal fiber (pashmina), with an average fiber diameter ranging from 11–14 μm and a cashmere yield of 70–500 g (6, 7).
SHFs are unique, renewable organs. After birth, the SHFs of cashmere goats undergo cyclical growth phases of anagen, catagen, and telogen annually. The growth and development of these follicles are closely related to the length, fineness, and yield of cashmere fibers (8, 9). Numerous studies have shown that the cyclical growth and quality formation of SHFs in cashmere goats are regulated by various molecular factors, including hormonal regulation, such as melatonin (MT) (10), prolactin (PRL) (11), and growth factors, such as insulin-like growth factors (IGFs) (12) and fibroblast growth factors (FGFs) (13). In addition, keratin genes (KRTs) (14), keratin-associated proteins (KRTAPs) (15), and bone morphogenetic proteins (BMPs) (16) have been shown to regulate hair follicle cycles. The PI3K-Akt signaling pathway, MAPK signaling pathway, and cell cycle pathways also play important roles in hair follicle development (17, 18).
This study aims to select JNCG from China and the CPG from India as research subjects. Through the use of public databases, we analyzed the transcriptome data of skin tissues from both JNCG and CPG at different growth stages (anagen, catagen, and telogen). This bioinformatics analysis was used to identify transcriptomic differences between the two breeds and explore the key RNA molecules that contribute to the differences in cashmere quality. The findings will provide a theoretical foundation for improving cashmere fiber quality and breeding strategies for high-quality cashmere goats.
2 Materials and methods
2.1 RNA-Seq datasets
The RNA-Seq datasets was retrieved from the NCBI SRA database, specifically targeting skin RNA-Seq data for Chanthangi pashmina goat (CPG) and Jiangnan cashmere goat (JNCG). A total of three BioProjects were included: PRJNA688899, PRJNA480975, and PRJNA778726, comprising 44 samples in total. Among these, 30 samples were from CPG, including 16 from anagen, 2 from catagen, and 12 from telogen stages; while 14 samples were from JNCG, including 8 from anagen, 3 from catagen, and 3 from telogen stages (Table 1). All RNA-Seq data were generated using the Illumina platform with paired-end sequencing. Detailed data can be found in Supplementary Table 1.
2.2 Sequencing data quality assessment and quality control
To ensure the quality and reliability of the data analysis, raw data were evaluated and subjected to quality control to obtain clean reads. FastQC (v0.12.1) and Trimmomatic (v0.39) were used for quality control, with Trimmomatic’s key parameters set as: PE-phred33 ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.
2.3 Reference sequence alignment analysis
The reference genome and gene annotation files used in this study were from the goat reference genome (version: GCF_001704415.1_ARS1). Data analysis was performed using a high-performance computing node (Inspur, NF5280M6). The clean data for each sample were aligned to the reference genome using HISAT2 (v2.1.0) (19), with the following parameters: -p 5 --dta --phred33 --no-unal-t --un-conc-gz. BAM files were generated using Samtools (v1.6) (20), with the main parameter: samtools sort-O bam -@ 2-o. The BAM files were processed using StringTie (v2.2.1) (21), generating GTF files with the parameters: stringtie-e -B-p 5-G -o-A. A Python script was then used to process the StringTie output, calculate sample counts, and FPKM. Linux scripts and Python code are detailed in Supplementary Material 1.
2.4 Differential RNA expression analysis
The expression matrix of the samples was analyzed for differential expression significance. The analysis was conducted to identify differential genes between CPG and JNCG skin tissue samples at different stages: CPG_a vs. JNCG_a, CPG_c vs. JNCG_c, and CPG_t vs. JNCG_t. Differential expression analysis was performed using DESeq2 (v1.44.0) (22, 23) in R (v4.4.1), with the threshold for significant differential expression set at log2|(fold change)| >2 and adjusted p-value (p-adj) <0.01. Volcano plots of differential genes were generated using the ggplot2 (v3.5.1). Venn analysis was performed to obtain the union of the differential genes across all three groups, and a clustering heatmap of the differential genes was created using the pheatmap (v1.0.12).
2.5 Functional enrichment analysis
Functional enrichment analysis of all differential genes was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The DAVID (24) database was used for both GO and KEGG enrichment analysis, with a p-value <0.05 considered significant. A gene clustering heatmap of key pathways and functions was generated, and protein–protein interaction (PPI) network analysis was performed for these gene sets using the online STRING protein interaction database1 (25), with the species selected as Capra hircus.
3 Results
3.1 RNA-Seq data set analysis
In this study, RNA-Seq data from 44 samples were analyzed after quality control and filtering, a total of 1,928,029,516 reads were obtained. The clean data were mapped to the goat reference genome, with 1,881,690,918 reads successfully aligned, resulting in an overall alignment rate of 97.59%. The mapping rate for individual samples ranged from 93.46 to 98.22%, with an average mapping rate of 97.51% for CPG and 97.76% for JNCG (Table 2). The distribution of gene expression in each sample was analyzed using the FPKM values, and a violin plot of FPKM distribution was generated for all samples (Figure 1). The distribution of log10 (FPKM + 1) values was found to be consistent in size and distribution patterns across all groups.
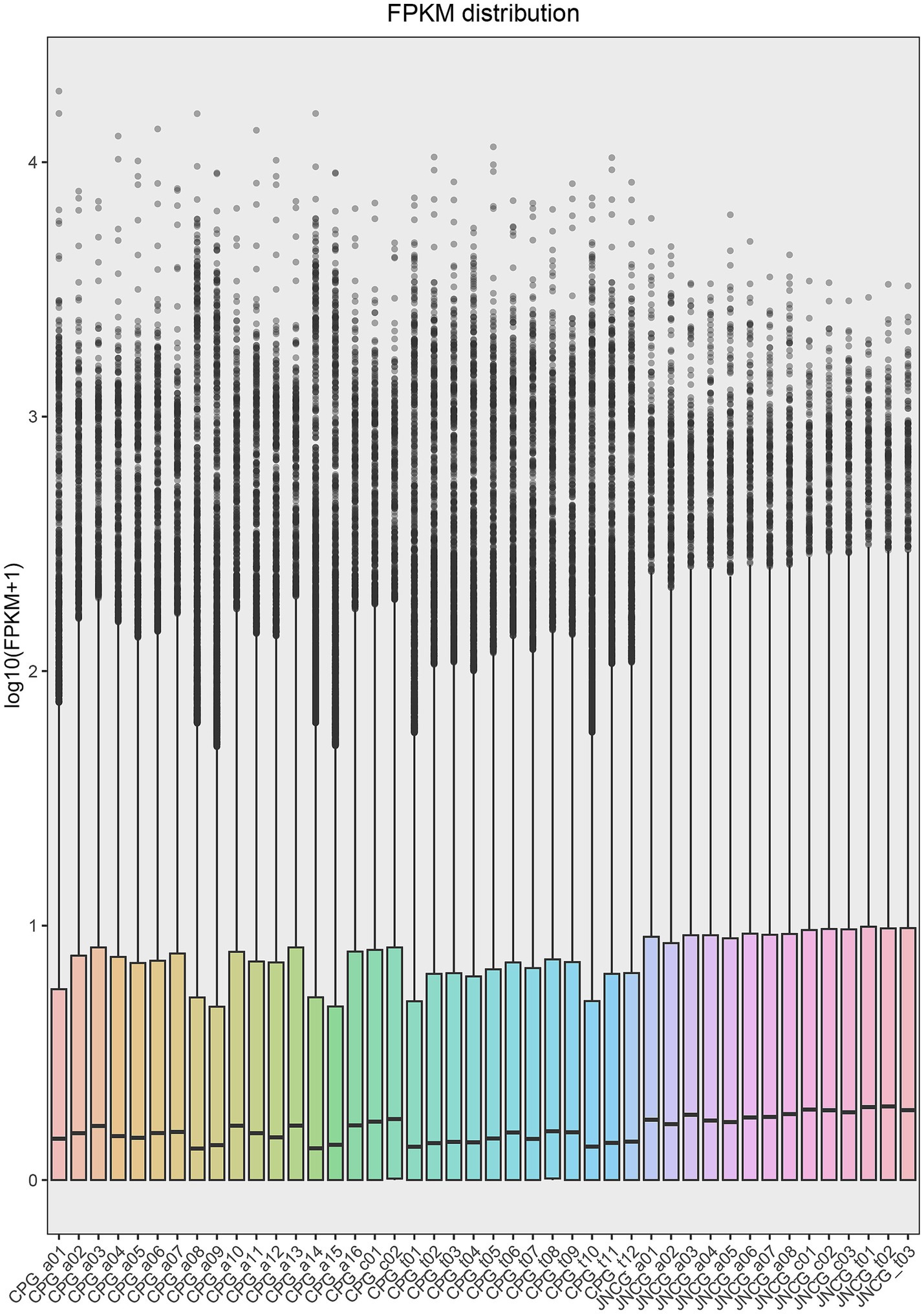
Figure 1. FPKM distribution map. The x-axis represents the samples, and the y-axis represents log10 (FPKM + 1).
3.2 Differential RNA expression analysis
Differential expression analysis was performed on the count data of CPG and JNCG for three different growth phases, and volcano plots were generated (Figures 2a–c). The results of differential analysis are presented in Table 3. In the anagen, 2,173 genes were upregulated, and 1,509 genes were downregulated between CPG and JNCG. In the catagen, 1,089 genes were upregulated, and 739 genes were downregulated. In the telogen, 1,808 genes were upregulated, and 1,396 genes were downregulated. Venn analysis (Figure 2d) revealed a total of 4,942 differentially expressed genes, with 1,277 genes differentially expressed in all three phases, 1,218 genes in two phases, and 2,447 genes in one phase only. A clustering heatmap of the FPKM data for all differentially expressed genes (Supplementary Table 2) is shown in Figure 2e, where clear expression differences between the CPG and JNCG groups were observed.
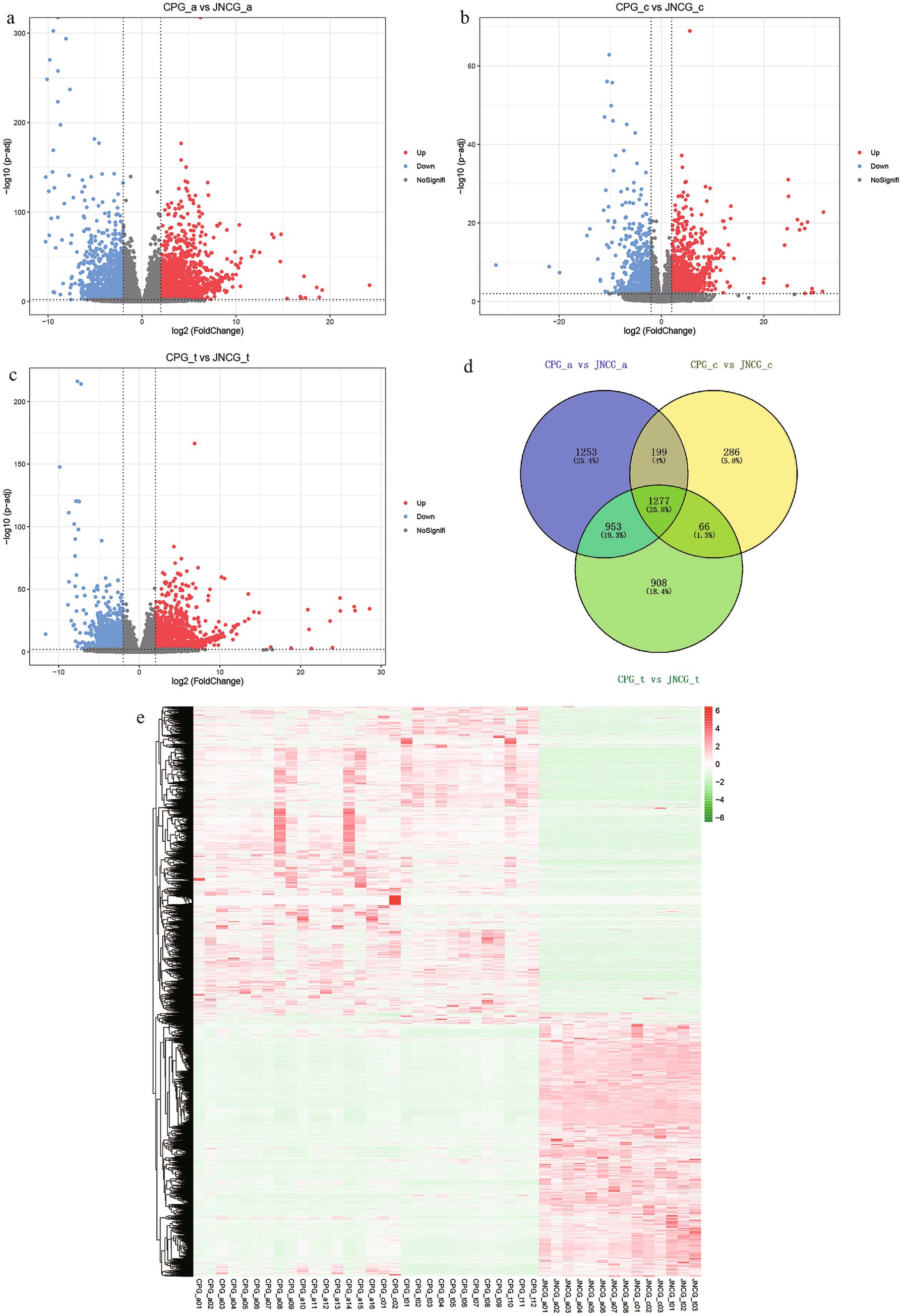
Figure 2. Differential expression results. (a) Volcano plot for CPG_a vs. JNCG_a. (b) Volcano plot for CPG_c vs. JNCG_c. (c) Volcano plot for CPG_t vs. JNCG_t. (d) Venn diagram for three phases of differential expression. (e) Clustering heatmap of all differentially expressed genes.
3.3 Differential RNA expression analysis
Functional enrichment analysis was performed for the 4,942 differentially expressed genes. GO enrichment analysis (Figure 3a and Supplementary Table 3) showed that 2,418 genes were enriched in molecular function (MF), with 70 MF categories identified, of which 55 were significantly enriched, mainly related to structural constituent of skin epidermis, calcium ion binding, dynein intermediate chain binding, and DNA binding. A total of 2,301 genes were enriched in biological process (BP), with 149 BP categories, 97 of which were significantly enriched, primarily in processes like keratinization, intermediate filament organization, natural killer cell activation in immune response, and inflammatory response. In cell component (CC), 2,584 genes were enriched across 45 CC categories, with 36 significantly enriched, mainly related to keratin filament, intermediate filament, and extracellular space.
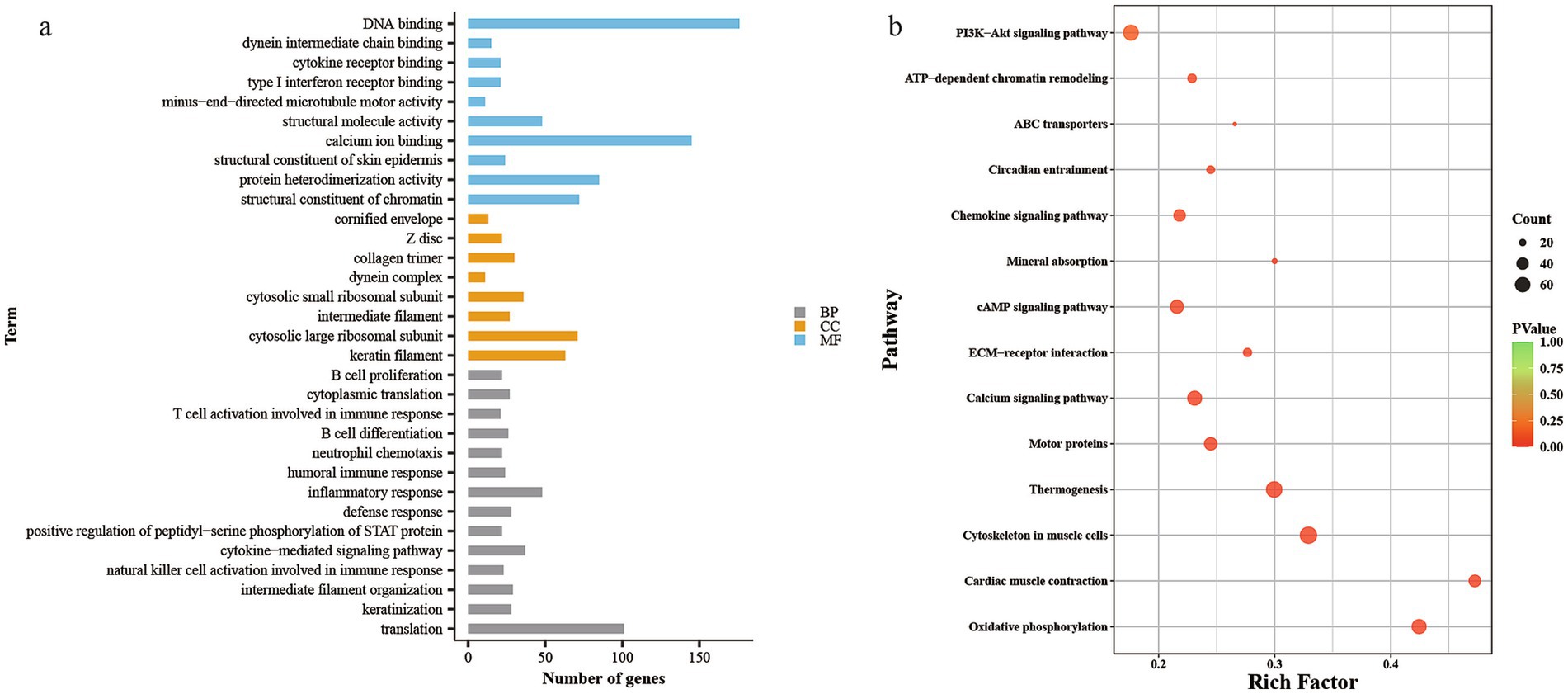
Figure 3. Differential mRNAs GO analysis histogram and KEGG analysis of bubble map. (a) GO analysis histogram, (b) KEGG analysis of bubble map.
KEGG pathway enrichment analysis (Figure 3b and Supplementary Table 4) revealed that 1,396 genes were enriched in 60 KEGG pathways, with 51 pathways significantly enriched. The differentially expressed genes were primarily enriched in pathways related to thermogenesis, PI3K-Akt signaling, ECM-receptor interaction, cAMP signaling, and ABC transporters, all of which have been previously reported in studies of secondary hair follicles.
3.4 Keratin and intermediate filament-related GO category analysis
In GO enrichment analysis, several keratin-related categories were identified in BP and CC, including keratinization (GO:0031424), intermediate filament organization (GO:0045109), keratin filament (GO:0045095), and intermediate filament (GO:0005882), all of which are related to fiber formation in cashmere and hair. Analysis of 84 genes in these categories (Figure 4a) revealed that these genes could be classified into three groups. The first group, with higher expression in CPG than JNCG, included genes like KRT14, BFSP2, KRT79, and KRT5. The second group had lower expression in CPG than JNCG, including KRT74, KRT39, and LOC102182149 (epiplakin). The third group included genes like KRT1, KRT10, and KRT4. Expression trends of key genes (Figures 4b–g) were analyzed, showing that KRT1, KRT10, and KRT14 had higher expression in CPG than in JNCG. KRT1 and KRT10 showed lower expression in the anagen and catagen phases in CPG, while in JNCG, their expression was higher in anagen and telogen compared to catagen. KRT39, KRT74, and LOC102182149 exhibited higher expression in JNCG compared to CPG, with a trend of decreasing expression in JNCG from anagen to telogen, whereas the trend in CPG was reversed, with rapid upregulation in catagen.
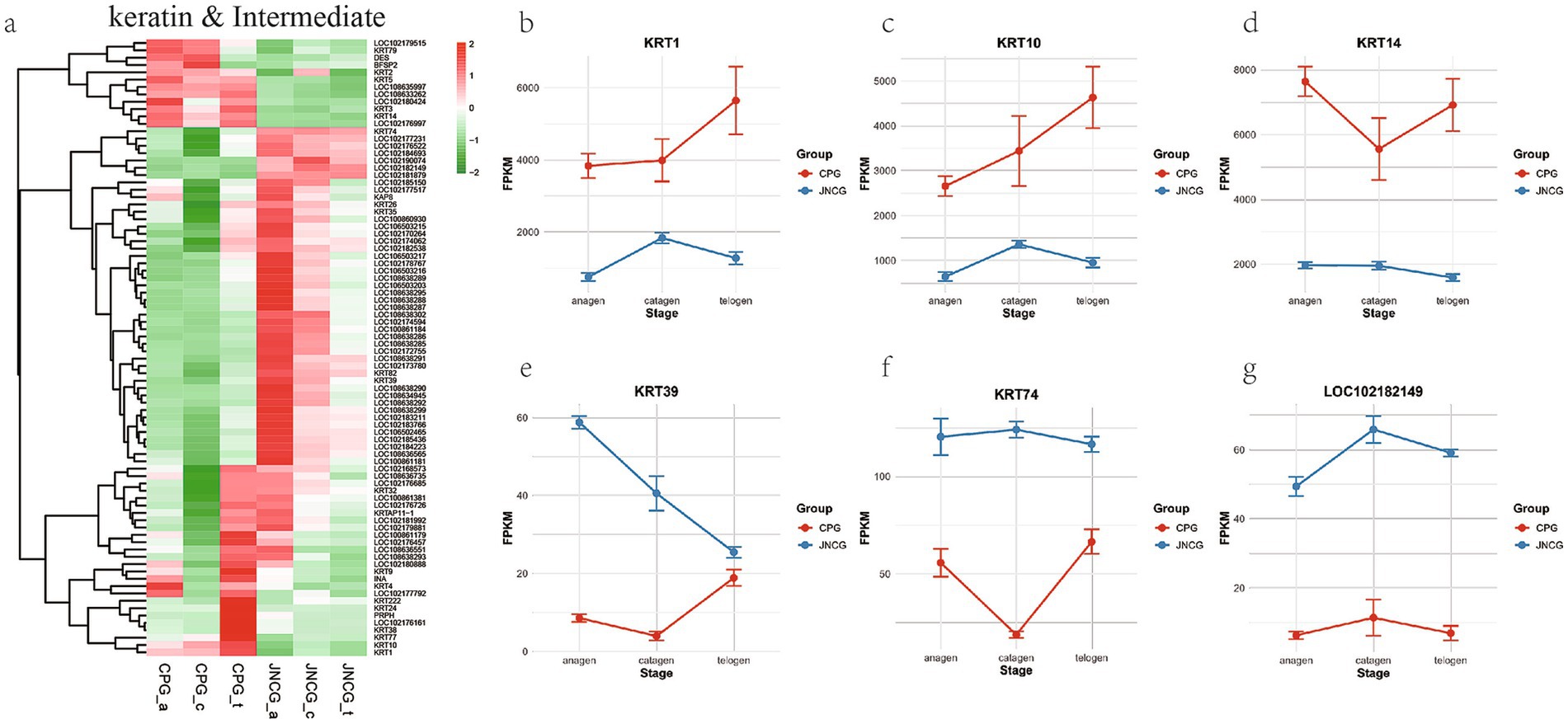
Figure 4. Gene expression analysis for keratin and intermediate filament GO categories. (a) Gene clustering heatmap. (b–g) Expression trend maps of some key mRNAs.
3.5 PI3K-Akt signaling pathway analysis
The PI3K-Akt signaling pathway is a crucial pathway in secondary hair follicle development, regulating hair growth and quality. Analysis of 65 genes enriched in this pathway (Figure 5a) revealed two distinct gene groups. One group showed higher expression in CPG, including genes like TNC and GNGT1, while the other group showed higher expression in JNCG, including IGF1, IGF2, IFG1R, ITGA9, FGF5, FGF21, and TNN. A PPI network analysis (Figure 5b) showed direct interactions between several proteins, with ITGB3 at the center linking two protein networks. IGF1, IGF2, IFG1R, FGF5, and FGF21 were closely interconnected, while TNC, TNN, and ITGA9 formed another tight network. The expression trends for these genes (Figures 5c–j) revealed consistent patterns for IGF1, IGF2, IFG1R, and ITGA9, with higher expression levels in JNCG than CPG. In JNCG, expression was higher in telogen compared to anagen and catagen, while in CPG, the highest expression was observed in catagen. The expression trends for FGF5, FGF21, and TNN were similar, with higher levels in JNCG than in CPG, with the highest expression of FGF5 in anagen in both groups.
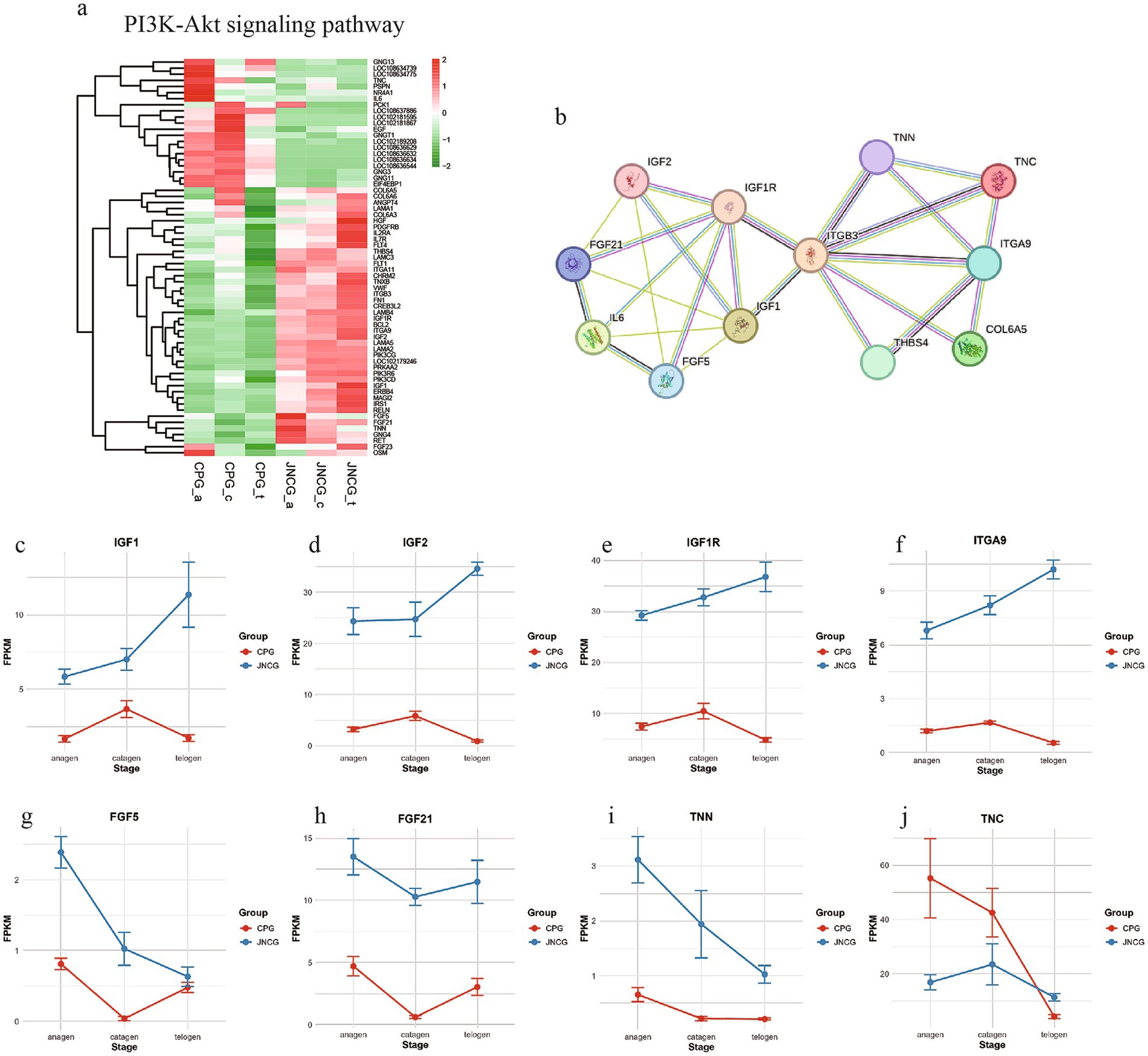
Figure 5. Gene expression analysis in the PI3K-Akt signaling pathway. (a) Gene clustering heatmap. (b) PPI analysis. (c-j) Expression trend maps of some key mRNAs.
3.6 Thermogenesis pathway analysis
Thermogenesis is associated with heat production, and the production of cashmere in goats is also aimed at protecting against cold. Different breeds of goats exhibit distinct cashmere production mechanisms due to environmental and coat differences. Analysis of 71 genes enriched in this pathway (Figure 6a) revealed two main groups. One group showed higher expression in CPG, including genes such as ATP6, ND2, ND3, and COX2, which are related to energy metabolism, indicating that CPG, living in a colder environment, may have upregulated thermogenesis-related genes. The other group showed higher expression in JNCG, including FGF21 and BMP8B. PPI network analysis (Figure 6b) revealed strong interactions between proteins in this pathway. Expression trends of selected genes (Figures 6c–j) indicated higher expression of ATP6, ATP5G3, ND3, and COX2 in CPG, with the highest expression observed in telogen. JNCG showed lower expression for these genes. NDUFA3 and NDUFS6 exhibited the highest expression in CPG during anagen. LOC102184937 (cytochrome c oxidase subunit 6B2) showed higher expression in CPG, with both CPG and JNCG showing the highest expression in anagen.
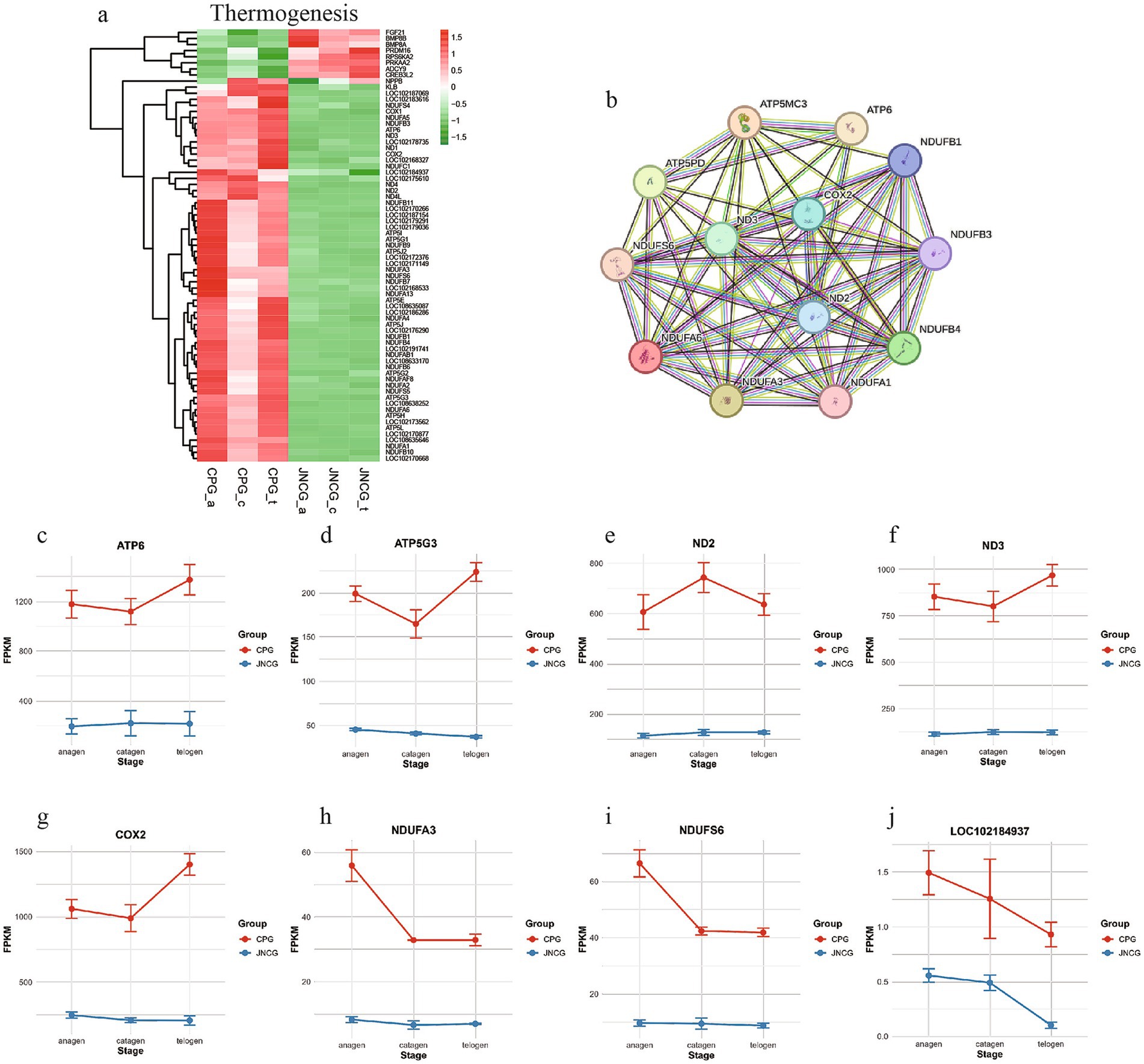
Figure 6. Gene expression analysis in thermogenesis. (a) Gene clustering heatmap. (b) PPI analysis. (c–j) Expression trend maps of some key mRNAs.
3.7 Pathway network and key gene analysis
Based on the analysis of key pathways and entries, and combining research on secondary hair follicle development, a selection of key candidate genes was identified (Table 4). These genes exhibited differential expression in the skin tissues of both cashmere goat breeds and at different stages of secondary hair follicle growth. Two key pathways, the PI3K-Akt signaling pathway and thermogenesis, were selected for further analysis, as these pathways are interrelated. Using the KEGG network, we constructed a KEGG molecular network diagram for the key genes (Figure 7). The diagram shows that several important molecules, upstream of these pathways (such as BMP8B, IFG1R, FGF5, FGF21, TNN, etc.), regulate corresponding receptor genes (such as ADCY9, IGF1R, GNG4, and ITGA9). Through indirect or direct interactions, these genes influence the PI3K and UCP1 genes, which in turn regulate ND2, COX2, ATP6, and other genes to maintain the TCA cycle. The PI3K gene further regulates the AKT and BCL2 genes.
4 Discussion
This study, through a comparative analysis of the skin transcriptomes of JNCG and CPG, reveals differences in gene expression across different follicle growth stages. These differences provide a molecular basis for understanding the variations in cashmere quality among different breeds of cashmere goats.
The secondary hair follicles of cashmere goats produce cashmere primarily to withstand cold environments. The cashmere layer in the skin efficiently helps cashmere goats maintain body temperature. Numerous studies have shown that cashmere growth is influenced by multiple factors, including breed, genetics, nutrition, age, shearing, and environment (26–29). Several studies have demonstrated that signaling pathways such as Wnt, TGF-β, PI3K-Akt, and cAMP regulate the development of secondary follicles and cashmere growth in cashmere goats (6, 30). In this study, differentially expressed genes were also enriched in the PI3K-Akt and cAMP signaling pathways, indicating that the variations in cashmere between these two breeds are influenced by these pathways, which play important roles in follicle development and cashmere fiber growth.
The PI3K-Akt signaling pathway is critical in regulating cell growth, differentiation, and survival. Among its key regulators, IGF1 is upregulated, promoting cell proliferation and facilitating the transition of SHFs dermal cells in Inner Mongolia cashmere goats from the anagen to telogen (31). Additionally, FGF5 overexpression can upregulate IGF1, and FGF5 itself acts as an inhibitor of hair growth (32, 33). Downregulation of FGF5 can influence the PI3K-Akt signaling pathway, and studies by Hu et al. (34) demonstrated that knocking in the VEGF gene at the FGF5 locus through gene editing led to increased cashmere yield and fiber length in cashmere goats. In our study, FGF5 expression in CPG was lower than in JNCG, further confirming that FGF5 regulates cashmere quality. Similarly, FGF21 influences the hair length of cashmere goats, showing a significant positive correlation with hair length (35). FGF21 knockout mice exhibit reduced body weight, slower hair growth, smaller follicle diameters, decreased follicle numbers, and lower hair density (36, 37). The differential expression patterns of IGF1, IGF1R, FGF5, and FGF21 in the skin tissues of the two cashmere goat breeds suggest that these growth factors play important roles in cashmere fiber growth and quality formation.
Additionally, we found that differentially expressed genes were significantly enriched in the thermogenesis pathway. Research by Nocelli et al. (9) showed that thermogenesis influences the transition from the growth phase to the regression phase in secondary follicles of Italian cashmere goats. Thermogenesis reflects the differences in environmental adaptation between these two cashmere goat breeds. JNCG is primarily distributed in the Aksu region of Xinjiang, China, where the environment is characterized by arid, semi-arid conditions and shrub grasslands. In contrast, CPG inhabit the Ladakh region of India, a high-altitude area exceeding 3,000 meters above sea level. This region is cold, has thin air with lower oxygen levels, and experiences extremely low temperatures in winter. The higher expression levels of thermogenesis-related genes in CPG may be associated with its adaptation to cold, high-altitude environments, where more efficient energy metabolism is required to maintain body temperature and physiological functions. In studies involving humans and mice, COX2 inhibitors have been shown to restore hair growth, indicating that hair loss is attributed to elevated COX2 enzyme activity (38). Transgenic mice overexpressing COX2 exhibit hair loss, and feeding them COX2 inhibitors suppresses COX2 activity and halts hair loss progression (39). Zhang et al. (40) investigated hedgehog spine follicles and mouse hair follicle development, revealing that COX2 acts upstream of ATP synthase, influencing energy metabolism and cell proliferation to regulate spine follicle size. In this study, COX2 and numerous ATP-related differentially expressed genes were also identified in both cashmere goat breeds. These genes exhibited higher expression levels in CPG, which may correlate with the unstable cashmere yield observed in this breed.
Furthermore, a large number of KRTs, such as KRT10, KRT14, KRT39, and KRT74, were identified among the differentially expressed genes. KRT10 protein content in cashmere fibers influences fiber diameter, with higher expression levels observed in coarser cashmere compared to finer cashmere (41), and KRT10 gene expression is significantly higher in fine wool Angora rabbits (42). In our study, KRT10 expression in CPG was significantly higher than in JNCG, potentially due to the undefined cashmere fineness of the sampled JNCG individuals, which exhibited finer fibers. Additionally, KRT39 and KRT74 are highly expressed in long-haired cashmere goats and play a role in regulating hair growth (43). KRT74 expression in adult fine-wool sheep is more than twice as high as in adult coarse-wool sheep (44). In our study, KRT74 expression in JNCG was over twice that of CPG, further supporting its influence on fiber fineness and aligning with previous findings that JNCG produces finer cashmere fibers. These results highlight the critical role of keratin genes in fiber formation and hair structure development.
5 Conclusion
This study systematically analyzed the transcriptome data of JNCG and CPG skin, revealing gene expression differences in the skin tissues of the two cashmere goat breeds. The findings indicate that the PI3K-Akt signaling pathway, thermogenesis pathway, and keratin-related genes play important roles in the development of secondary hair follicles and the formation of cashmere fiber quality. A total of 24 key candidate genes, including IGF1, IGF1R, FGF5, FGF21, ND2, COX2, KRT10, KRT39, and KRT74, were identified, providing a theoretical basis for the future genetic improvement of cashmere goats.
Data availability statement
RNA-Seq data are from PRJNA688899, PRJNA480975, and PRJNA778726. The FPKM data of differentially expressed genes are shown in Supplementary Table 2. Please contact the corresponding author for other data (Gao Gong: Z2dhbzE5OTVAMTYzLmNvbQ==).
Ethics statement
The animal study was approved by Xinjiang Agricultural University Experimental Animal Welfare Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
GG: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. SB: Writing – original draft, Data curation, Writing – review & editing. XL: Data curation, Methodology, Writing – original draft. YA: Data curation, Methodology, Writing – original draft. FX: Data curation, Methodology, Writing – original draft. YS: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by Natural Science Foundation of Xinjiang Uygur Autonomous Region (2024D01B42); Talent Development Fund “Tianchi Talent” Introduction Plan Project of Xinjiang Uygur Autonomous Region (2224ZZQRCXM); China Agriculture Research System of MOF and MARA (CARS-39); College Students’ Innovation Training Program Project of Xinjiang Agricultural University (dxscx2024228).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1571803/full#supplementary-material
SUPPLEMENTARY TABLE 1 | SRA data download statistics table.
SUPPLEMENTARY TABLE 2 | FPKM data for all differentially expressed genes.
SUPPLEMENTARY TABLE 3 | GO analysis table of differentially expressed genes.
SUPPLEMENTARY TABLE 4 | KEGG enrichment analysis table of differentially expressed genes.
SUPPLEMENTARY MATERIAL 1 | Linux scripts and Python code.
Footnotes
References
1. Zhou, H, Bai, L, Li, S, Li, W, Wang, J, Tao, J, et al. Genetics of wool and cashmere fibre: progress, challenges, and future research. Animals. (2024) 14:3228. doi: 10.3390/ani14223228
2. Xu, S, Akhatayeva, Z, Liu, J, Feng, X, Yu, Y, Badaoui, B, et al. Genetic advancements and future directions in ruminant livestock breeding: from reference genomes to multiomics innovations. Sci China Life Sci. (2024) 68:934–60. doi: 10.1007/s11427-024-2744-4
3. Wang, Z, Lv, Q, Li, W, Huang, W, Gong, G, Yan, X, et al. Chromosome-level genome assembly of the cashmere goat. Sci Data. (2024) 11:1107. doi: 10.1038/s41597-024-03932-7
4. Wu, C, Qin, C, Fu, X, Huang, X, and Tian, K. Integrated analysis of lncRNAs and mRNAs by RNA-Seq in secondary hair follicle development and cycling (anagen, catagen and telogen) of Jiangnan cashmere goat (Capra Hircus). BMC Vet Res. (2022) 18:167. doi: 10.1186/s12917-022-03253-0
5. Wu, C, Yuan, L, Cao, W, Ye, X, Ma, X, Qin, C, et al. Regulation of secondary hair follicle cycle in cashmere goats by miR-877-3p targeting IGFBP5 gene. J Anim Sci. (2023) 101:101. doi: 10.1093/jas/skad314
6. Bhat, B, Yaseen, M, Singh, A, Ahmad, SM, and Ganai, NA. Identification of potential key genes and pathways associated with the pashmina fiber initiation using RNA-Seq and integrated bioinformatics analysis. Sci Rep. (2021) 11:1766. doi: 10.1038/s41598-021-81471-6
7. Bhat, B, Ganai, NA, Singh, A, Mir, R, Ahmad, SM, Majeed Zargar, S, et al. Changthangi pashmina goat genome: sequencing, assembly, and annotation. Front Genet. (2021) 12:695178. doi: 10.3389/fgene.2021.695178
8. Liu, Y, Wang, M, Liu, Z, Liang, X, Sheng, S, Dai, H, et al. Transcriptional signatures of secondary hair follicles during annual cashmere growth. Sci Data. (2024) 11:1427. doi: 10.1038/s41597-024-04316-7
9. Nocelli, C, Cappelli, K, Capomaccio, S, Pascucci, L, Mercati, F, Pazzaglia, I, et al. Shedding light on cashmere goat hair follicle biology: from morphology analyses to transcriptomic landascape. BMC Genomics. (2020) 21:458. doi: 10.1186/s12864-020-06870-x
10. Rong, Y, Ma, R, Zhang, Y, and Guo, Z. Melatonin’s effect on hair follicles in a goat (Capra Hircus) animal model. Front Endocrinol. (2024) 15:1361100. doi: 10.3389/fendo.2024.1361100
11. Zhang, L, Duan, C, Guo, Y, Zhang, Y, and Liu, Y. Inhibition of prolactin promotes secondary skin follicle activation in cashmere goats. J Anim Sci. (2021) 99:skab079. doi: 10.1093/jas/skab079
12. Liu, Y, Kong, L, Li, S, Nie, L, Gao, J, Li, S, et al. Correlation and regression analysis of GH and IGF-1 genes in Liaoning cashmere goats with body size and other production performance. J Genet Eng Biotechnol. (2024) 22:100440. doi: 10.1016/j.jgeb.2024.100440
13. Li, Y, Song, S, Zhang, Z, Liu, X, Zhang, Y, Guangxin, E, et al. A deletion variant within the FGF5 gene in goats is associated with gene expression levels and cashmere growth. Anim Genet. (2022) 53:657–64. doi: 10.1111/age.13239
14. Zhang, C, Qin, Q, Liu, Z, Xu, X, Lan, M, Xie, Y, et al. Identification of the key proteins associated with different hair types in sheep and goats. Front Genet. (2022) 13:993192. doi: 10.3389/fgene.2022.993192
15. Wang, J, Zhou, H, Hickford, JGH, Zhao, M, Gong, H, Hao, Z, et al. Identification of caprine Krtap28-1 and its effect on cashmere fiber diameter. Genes. (2020) 11:121. doi: 10.3390/genes11020121
16. Zhang, Y, Wu, K, Wang, L, Wang, Z, Han, W, Chen, D, et al. Comparative study on seasonal hair follicle cycling by analysis of the transcriptomes from cashmere and milk goats. Genomics. (2020) 112:332–45. doi: 10.1016/j.ygeno.2019.02.013
17. Diao, X, Yao, L, Wang, X, Li, S, Qin, J, Yang, L, et al. Hair follicle development and cashmere traits in albas goat kids. Animals. (2023) 13:617. doi: 10.3390/ani13040617
18. Vasu, M, Ahlawat, S, Arora, R, and Sharma, R. Deciphering the molecular drivers for cashmere/pashmina fiber production in goats: a comprehensive review. Mamm Genome. (2025) 36:162–82. doi: 10.1007/s00335-025-10109-z
19. Kim, D, Paggi, JM, Park, C, Bennett, C, and Salzberg, SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
20. Li, H, Handsaker, B, Wysoker, A, Fennell, T, Ruan, J, Homer, N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
21. Pertea, M, Pertea, GM, Antonescu, CM, Chang, TC, Mendell, JT, and Salzberg, SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. (2015) 33:290–5. doi: 10.1038/nbt.3122
22. Anders, S, and Huber, W. Differential expression analysis for sequence count data. Genome Biol. (2010) 11:R106. doi: 10.1186/gb-2010-11-10-r106
23. Love, MI, Huber, W, and Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
24. Sherman, BT, Hao, M, Qiu, J, Jiao, X, Baseler, MW, Lane, HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. (2022) 50:W216–21. doi: 10.1093/nar/gkac194
25. Szklarczyk, D, Gable, AL, Nastou, KC, Lyon, D, Kirsch, R, Pyysalo, S, et al. The string database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
26. Zhang, W, Wang, RL, Zhu, XP, Kleernann, DO, Yue, CW, and Jia, ZH. Effects of dietary copper on ruminal fermentation, nutrient digestibility and fibre characteristics in cashmere goats. Asian Australas J Anim Sci. (2007) 20:1843–8. doi: 10.5713/ajas.2007.1843
27. Yuan, C, Wang, X, Geng, R, He, X, Qu, L, and Chen, Y. Discovery of cashmere goat (Capra Hircus) microRNAs in skin and hair follicles by Solexa sequencing. BMC Genomics. (2013) 14:511. doi: 10.1186/1471-2164-14-511
28. Song, S, Yao, N, Yang, M, Liu, XX, Dong, KZ, Zhao, QJ, et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra Hircus). BMC Genomics. (2016) 17:17. doi: 10.1186/s12864-016-2449-0
29. Todini, L. Thyroid hormones in small ruminants: effects of endogenous environmental and nutritional factors. Animal. (2007) 1:997–1008. doi: 10.1017/s1751731107000262
30. Liu, F, Shi, J, Zhang, Y, Lian, A, Han, X, Zuo, K, et al. NANOG attenuates hair follicle-derived mesenchymal stem cell senescence by upregulating PBX1 and activating AKT signaling. Oxid Med Cell Longev. (2019) 2019:4286213. doi: 10.1155/2019/4286213
31. Sha, RN, Dai, B, Ren, LQ, Han, XY, Yuan, JL, and Liu, DJ. Cx43 promotes SHF-DPCs proliferation in the hair follicle of Albas cashmere goats from anagen to telogen. Res Vet Sci. (2020) 133:92–7. doi: 10.1016/j.rvsc.2020.09.002
32. He, X, Chao, Y, Zhou, G, and Chen, Y. Fibroblast growth factor 5-short (FGF5s) inhibits the activity of FGF5 in primary and secondary hair follicle dermal papilla cells of cashmere goats. Gene. (2016) 575:393–8. doi: 10.1016/j.gene.2015.09.034
33. Hébert, JM, Rosenquist, T, Götz, J, and Martin, GR. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. (1994) 78:1017–25. doi: 10.1016/0092-8674(94)90276-3
34. Hu, X, Hao, F, Li, X, Xun, Z, Gao, Y, Ren, B, et al. Generation of VEGF knock-in cashmere goat via the CRISPR/Cas9 system. Int J Biol Sci. (2021) 17:1026–40. doi: 10.7150/ijbs.55559
35. Gong, G, Fan, Y, Zhang, Y, Yan, X, Li, W, Yan, X, et al. The regulation mechanism of different hair types in inner Mongolia cashmere goat based on PI3K-AKT pathway and FGF21. J Anim Sci. (2022) 100:skac292. doi: 10.1093/jas/skac292
36. Liu, X, Zhang, P, Zhang, X, Li, X, Bai, Y, Ao, Y, et al. Fgf21 knockout mice generated using CRISPR/Cas9 reveal genetic alterations that may affect hair growth. Gene. (2020) 733:144242. doi: 10.1016/j.gene.2019.144242
37. Liu, X, Zhang, P, Zhang, XF, Li, X, Bai, Y, Jia, KR, et al. Construction of FGF21 knockout mouse models by the CRISPR/Cas9 system. Yi Chuan. (2018) 40:66–74. doi: 10.16288/j.yczz.17-011
38. Bol, DK, Rowley, RB, Ho, CP, Pilz, B, Dell, J, Swerdel, M, et al. Cyclooxygenase-2 overexpression in the skin of transgenic mice results in suppression of tumor development. Cancer Res. (2002) 62:2516–21.
39. Müller-Decker, K, Leder, C, Neumann, M, Neufang, G, Bayerl, C, Schweizer, J, et al. Expression of cyclooxygenase isozymes during morphogenesis and cycling of pelage hair follicles in mouse skin: precocious onset of the first catagen phase and alopecia upon cyclooxygenase-2 overexpression. J Invest Dermatol. (2003) 121:661–8. doi: 10.1046/j.1523-1747.2003.12473.x
40. Zhang, M, Wang, M, Jiang, J, Liu, W, Zhou, S, Wang, D, et al. COX2-ATP synthase regulates spine follicle size in hedgehogs. Int J Biol Sci. (2023) 19:4763–77. doi: 10.7150/ijbs.83387
41. Zhang, C, Qin, Q, Wang, Y, Wang, Z, and Liu, Z. Identification of key proteins related to cashmere fiber diameter by integrated proteomics and bioinformatic analyses in the Alpas and Alxa goat breeds. Genes. (2024) 15:1154. doi: 10.3390/genes15091154
42. Huang, D, Ding, H, Wang, Y, Wang, X, and Zhao, H. Integration analysis of hair follicle transcriptome and proteome reveals the mechanisms regulating wool fiber diameter in angora rabbits. Int J Mol Sci. (2024) 25:3260. doi: 10.3390/ijms25063260
43. Gong, G, Fan, Y, Li, W, Yan, X, Yan, X, Zhang, L, et al. Identification of the key genes associated with different hair types in the Inner Mongolia cashmere goat. Animals. (2022) 12:1456. doi: 10.3390/ani12111456
Keywords: cashmere goats, skin transcriptome, PI3K-Akt signaling pathway, thermogenesis, KRTs
Citation: Gong G, Bi S, Liang X, Ao Y, Xu F and Sulaiman Y (2025) Molecular mechanisms underlying cashmere quality differences between Jiangnan cashmere goats and Changthangi pashmina goats. Front. Vet. Sci. 12:1571803. doi: 10.3389/fvets.2025.1571803
Edited by:
Cuiling Wu, Xinjiang Normal University, ChinaReviewed by:
Feng Su, Shandong Agricultural University, ChinaXuefeng Fu, Xinjiang Academy of Animal Sciences, China
Copyright © 2025 Gong, Bi, Liang, Ao, Xu and Sulaiman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gao Gong, Z2dhbzE5OTVAMTYzLmNvbQ==
 Gao Gong
Gao Gong Shijie Bi2
Shijie Bi2 Yiming Sulaiman
Yiming Sulaiman