- 1Institute of Animal Husbandry and Veterinary Science, Jiangxi Academy of Agricultural Sciences, Nanchang, China
- 2Department of Animal Science, Jiangxi Biological Vocational College, Nanchang, China
Introduction: Jiangxi Province possesses abundant genetic resources of local pig breeds, whose effective conservation is essential for maintaining biodiversity and sustainable utilization. Streptococcus suis is an important zoonotic pathogen that continuously threatens swine production systems and public health globally. This study aimed to investigate the epidemiological characteristics of S. suis among local pig breeds in Jiangxi Province.
Methods: An investigation was conducted on the prevalence and antimicrobial resistance profiles of S. suis in six local pig breeds from Jiangxi Province, including Gandong black pig, Hang pig, Ganxi two-end-black pig, Dongxiang spotted pig, Yushan black pig, and Binhu black pig.
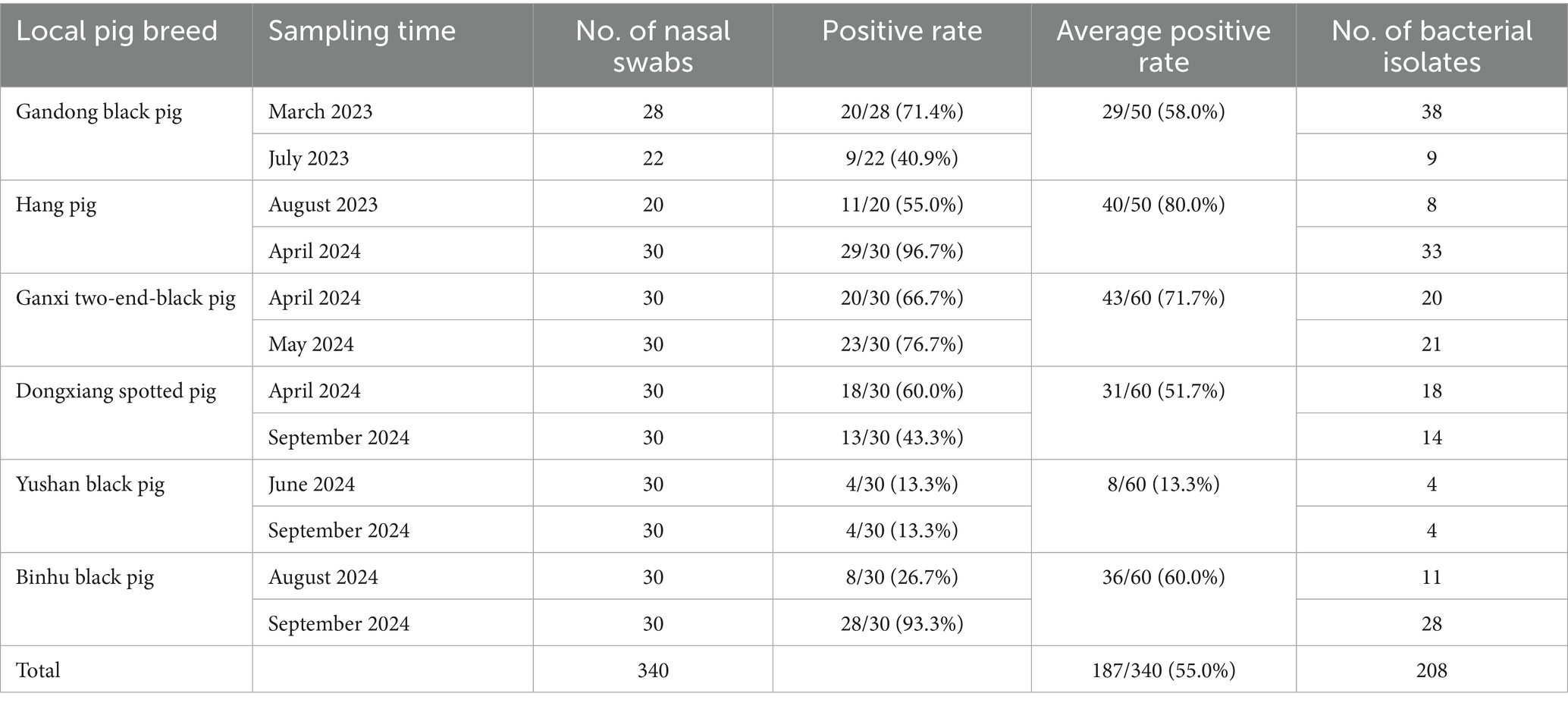
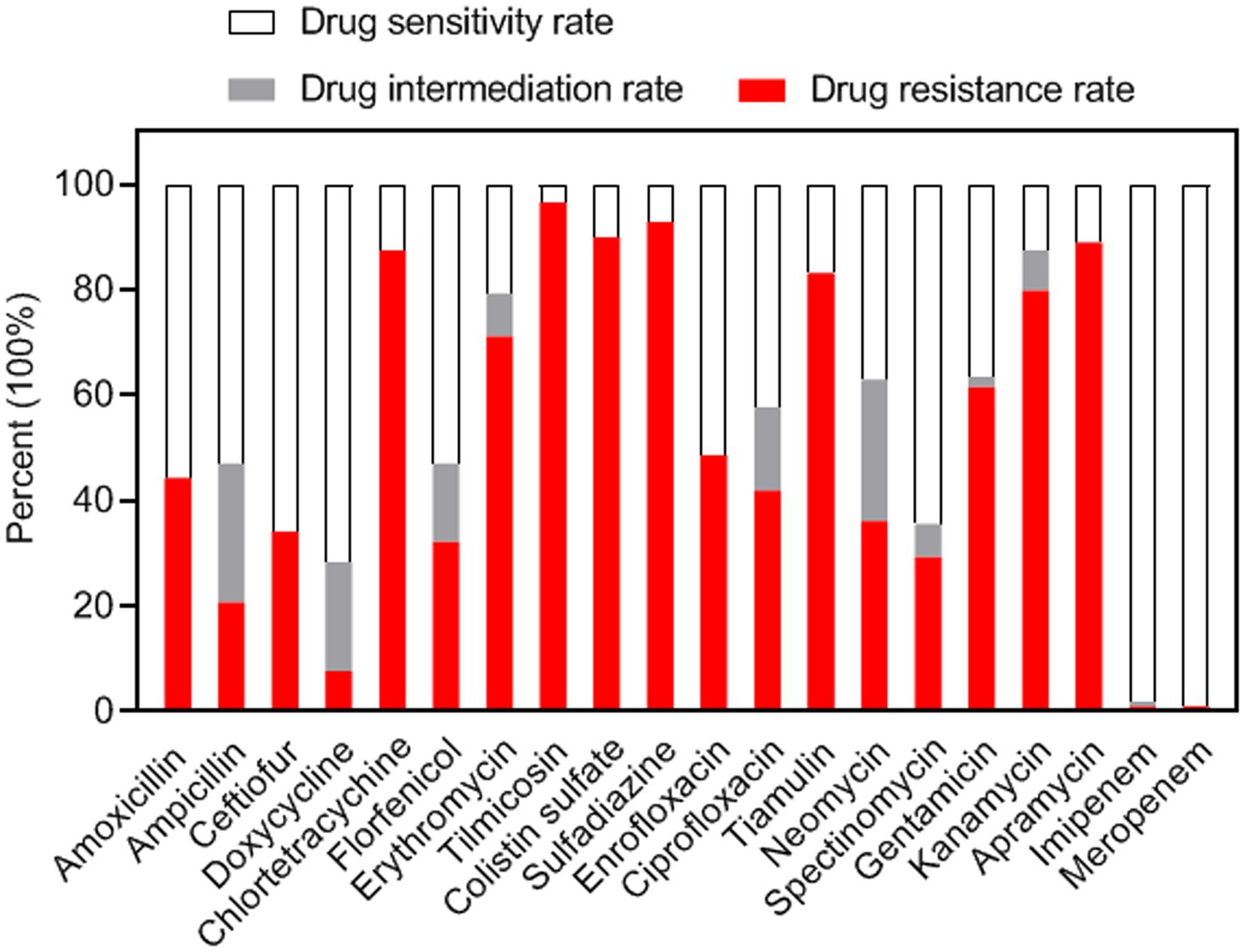
Results: A total of 340 porcine nasal swabs were collected from six local pig breeds. 208 S. suis strains were isolated from 187 samples, with an overall isolation rate of 55.0%. The positive isolation rates of the six local breeds were 58.0, 80.0, 71.7, 51.7, 13.3, and 60.0%, respectively. Antimicrobial susceptibility testing revealed that >98% of isolates were susceptible to carbapenems (meropenem and imipenem), followed by doxycycline (71.6%), ceftiofur (65.9%), spectinomycin (64.4%), and amoxicillin (55.8%). High resistance rates were observed for tilmicosin (96.6%), sulfadiazine (92.8%), colistin (89.9%), apramycin (88.9%), chlortetracycline (87.5%), tiamulin (83.2%), and kanamycin (79.8%). 100% of isolates exhibited multidrug resistance, with significant variations in resistance patterns among breeds. Genotypic analysis identified ermB (macrolides), tetL (tetracyclines), and Sul2 (sulfonamides) as the predominant resistance determinants.
Discussion: Multidrug-resistant S. suis strains have become widespread among local pig breeds. This study could provide evidence-based guidance for developing effective prevention and control strategies against S. suis infections and protecting valuable genetic resources of indigenous pig breeds.
Introduction
Through thousands of years of domestication and natural selection, Chinese local pig breeds have developed distinct genetic characteristics, contributing to global genetic diversity and the sustainable development of the pig industry (1). Compared with commercial Western pig breeds, Chinese local pigs typically exhibit superior resistance to coarse feed and environmental stresses (2). Jiangxi Province is home to one of the largest populations of native pig breeds in China, accounting for approximately one-tenth of the country’s local pig genetic resources (3). However, over the past decade, many of these local breeds have faced near extinction due to multiple factors, including competition from commercial Western pig breeds, small population sizes, and threats from epidemic diseases (2). Effective conservation of these breeds is essential for their preservation and future utilization.
Streptococcus suis is a Gram-positive bacterial pathogen that causes porcine respiratory tract infections and severe invasive diseases, including meningitis, septicemia, and arthritis (4). As an important swine pathogen, S. suis is responsible for substantial economic losses in the global swine industry (5). Moreover, it has emerged as an important zoonotic agent, with frequent cases of human transmission, particularly in Asia (5). The major outbreaks of S. suis in China in 1998 and 2005 brought global attention to this pathogen (6). Continued reports of human cases across 22 Chinese provinces further emphasize the persistent threat of this zoonotic infection (7). Consequently, S. suis infections in pig populations threaten not only agricultural sustainability but also public health (8). Although antimicrobial prophylaxis and therapy have been successful in controlling these infections, the emergence and spread of antibiotic-resistant S. suis strains elevate the risks of treatment failure in both pigs and humans (9). Therefore, monitoring antimicrobial resistance (AMR) profiles of S. suis in specific regions is essential for optimizing effective antimicrobial therapies and tracking the development of bacterial drug resistance (10).
In recent years, antimicrobial resistance (AMR) phenotypes and genotypes of S. suis isolates from clinically healthy commercial three-way crossbred pigs (2017–2019) in Jiangxi Province, China, have been characterized (11). However, data on S. suis in local pig breeds within this Province remain unknown. In the present study, S. suis strains were isolated from nasal cavities of local breed piglets and evaluated their antimicrobial susceptibility profiles. These findings may provide evidence-based guidance for S. suis infection prevention and control, while contributing to the conservation of porcine germplasm resources in China.
Materials and methods
Clinical sample collection
From March 2023 to September 2024, 340 nasal swab samples were collected from clinically healthy weaned piglets (aged 30–76 days) representing six local breeds in Jiangxi Province, China: Gandong black pig, Hang pig, Ganxi two-end-black pig, Dongxiang spotted pig, Yushan black pig, and Binhu black pig (Figure 1). Samples were obtained from two distinct batches per breed with sterile cotton swabs (Table 1). Swabs were then stored in 1.5 mL of transport buffer within bacterial sampling tubes (Youkang Biotechnology, Beijing, China) and transported to the laboratory at a low temperature.
Bacterial isolation and identification
The liquid samples were cultured aerobically on Columbia agar (Hopebio, Qingdao, China) plates supplemented with 5% sterile defibrinated sheep blood (Hopebio) at 37°C for 24 h. Alpha-hemolytic colonies exhibiting typical morphology were subjected to Gram staining. Suspected S. suis colonies were then analyzed by PCR targeting the 16S rRNA gene using primer pair 27F/1492R (Table S1). The amplification used the following cycling parameters: one cycle of 98°C for 8 min, followed by 30 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min. Amplification products were sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing. The resulting sequences were analyzed using NCBI’s Nucleotide BLAST with default parameters.
Antibiotic drugs
A total of 20 commercially available antimicrobial agents for veterinary and human use were prepared for antimicrobial susceptibility testing, including: penicillin [amoxicillin (AMX) and ampicillin (AMP)], cephalosporins [ceftiofur (FUR)], tetracyclines [doxycycline (DX) and chlortetracycline (CT)], phenicols [florfenicol (FFC)], macrolides [erythromycin (E) and tilmicosin (TIL)], polypeptides [colistin sulfate (CS)], sulfonamides [sulfadiazine (SUZ)], quinolones [enrofloxacin (ENR) and ciprofloxacin (CIP)], diterpenes [tiamulin (T)], aminoglycosides [neomycin (N), spectinomycin (SPT), gentamicin (GM), kanamycin (K), and apramycin (AP)], and carbapenems [imipenem (IMI) and meropenem (MRP)]. Antibiotic discs containing CT were obtained from HiMedia Laboratories (Mumbai, India), while all other discs were sourced from Liofilchem S.r.l. (Roseto degli Abruzzi, Italy). The drug concentrations in each disc are detailed in Table S2.
Antimicrobial susceptibility assay
The antimicrobial susceptibility of all isolates was determined using the Kirby-Bauer disk diffusion method on Mueller-Hinton (MH) agar (Hopebio) supplemented with 5% newborn bovine serum (Sijiqing, Hangzhou, China), following the standardized protocol recommended by the Clinical and Laboratory Standards Institute (CLSI) (12). Briefly, S. suis strains were cultured on MH agar plates at 37°C under 5% CO2 for 12 h. Selected colonies were suspended in normal saline and adjusted to a 0.5 McFarland standard. Bacterial suspensions were uniformly spread on MH plates using sterile cotton swabs. Antibiotic discs were aseptically placed using a disc dispenser, and plates were incubated at 37°C in 5% CO2 for 18 h. Streptococcus pneumoniae ATCC 49619 served as the quality control strain. Isolates were categorized as susceptible, intermediate, or resistant based on established criteria (Supplementary Table S2). For three carbapenem-resistant S. suis strains, minimum inhibitory concentrations (MICs) of imipenem and meropenem were determined using MIC Test Strips (Liofilchem).
Notably, since species-specific CLSI breakpoints for S. suis are currently unavailable, the standards established for S. pneumoniae (non-meningitis isolates) were adopted in this study.
Identification of antibiotic resistance genes
All S. suis isolates were cultured overnight at 37°C in tryptone soy broth (Hopebio) supplemented with 5% newborn bovine serum. Genomic DNA was extracted using bacterial DNA kits (Cwbiotech, Beijing, China) following the manufacturer’s instructions. PCR amplification was performed to detect resistance genes for macrolides (ermA, ermB, and mefA) (13), tetracyclines (tetL, tetO, tetK, and tetM) (14), aminoglycosides [aph(3′)], and sulfonamides (Sul1, Sul2, and Sul3) (15). The 11 primer sets, along with their expected amplicon sizes and annealing temperatures, are listed in Supplementary Table S1. The PCR cycling conditions were as follows: 1 cycle at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, Tm for 30 s, and 72°C for 1 min. PCR products were analyzed using electrophoresis on 1.0% (wt/vol) agarose gels.
Statistical analysis
Data visualization was conducted with GraphPad Prism 8.0 software (San Diego, USA). Statistical analysis was performed using IBM SPSS Statistics 25.0 (Amonk, USA). Intergroup comparisons were analyzed by Chi-square tests with false discovery rate (FDR) correction for multiple comparisons in SPSS. A two-tailed FDR-adjusted p-value <0.05 was considered statistically significant.
Ethics statement
All sampling procedures were approved by the Animal Ethics Committee of the Institute of Animal Husbandry and Veterinary Medicine, Jiangxi Academy of Agricultural Science (2020-JXAAS-XM-01).
Results
Prevalence of S. suis in local pig breeds
According to current Chinese agricultural industry standards, the Gandong black pig, Hang pig, Dongxiang spotted pig, and Binhu black pig are classified as endangered breeds; the Yushan black pig as a vulnerable protected breed; and the Ganxi two-end-black pig as a low-risk breed (Figure 1) (3). From March 2023 to September 2024, 340 porcine nasal swabs from six local pig breeds were obtained for S. suis isolation. From each sample, five suspected bacterial colonies were subjected to Gram staining and PCR identification. Results revealed an overall S. suis carrier rate of 55.0% (187/340; Table 1), with breed-specific rates of 58.0% (29/50), 80.0% (40/50), 71.7% (43/60), 51.7% (31/60), 13.3% (8/60), and 60.0% (36/60) (χ2 = 62.05, p < 0.001), demonstrating statistically significant differences among breeds. A total of 208 S. suis strains were stored and used for subsequent antimicrobial susceptibility assays.
Antimicrobial susceptibility profiles
The antimicrobial susceptibility profiles of 208 S. suis strains were assessed using the disc diffusion method with 20 antimicrobial agents. Inhibition zone diameters for all tested antibiotics are presented in Supplementary Table S3. Results demonstrated high susceptibility rates (>98%) to MRP (99.0%) and IMI (98.1%), followed by DX (71.6%), FUR (65.9%), SPT (64.4%), and AMX (55.8%) (Figure 2). Conversely, high resistance frequencies were observed to TIL (96.6%), SUZ (92.8%), CS (89.9%), AP (88.9%), CT (87.5%), T (83.2%), and K (79.8%) (Figure 2). The highest intermediate resistance level was observed for N (26.9%).
Notably, three carbapenem-resistant strains demonstrating resistance to imipenem and/or meropenem were identified. Results of additional MIC testing confirmed these strains exhibited resistance to imipenem and/or meropenem (Supplementary Table S4). The emergence of carbapenem-resistant strains warrants particular concern given the limited therapeutic options available for possible infections.
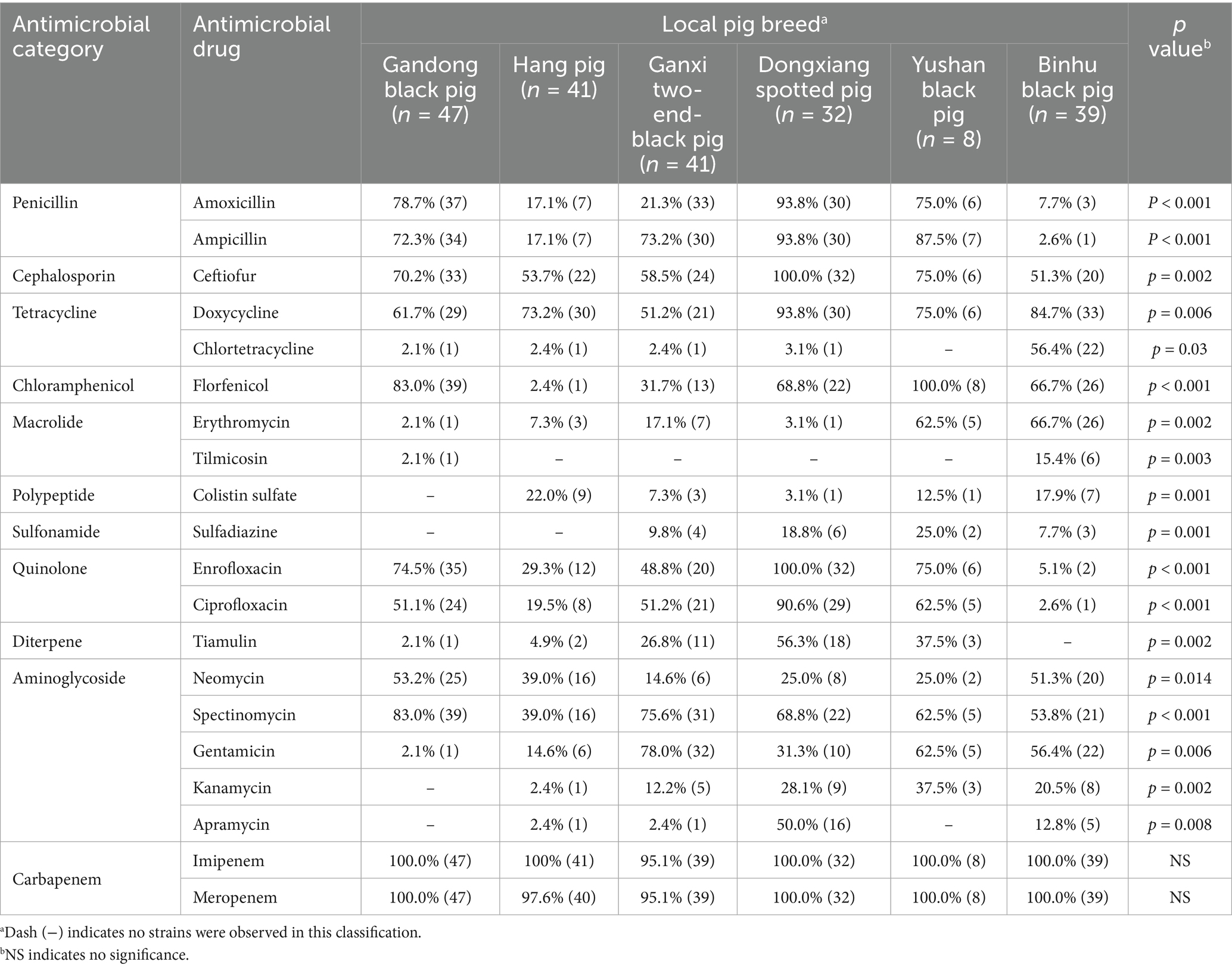
Drug sensitivity of S. suis from different local pig breeds
S. suis strains exhibited similarly high resistance rates to imipenem (IMI) and meropenem (MRP), while susceptibility to other antibiotics varied significantly across isolates from the six breeds (Table 2). For instance, AMX susceptibility rates were 78.7% (37/47) in Gandong black pigs, 17.1% (7/41) in Hang pigs, 21.3% (33/41) in Ganxi two-end-black pigs, 93.8% (30/32) in Dongxiang spotted pigs, 75.0% (6/8) in Yushan black pigs, and 7.7% (3/39) in Binhu black pigs (p < 0.001). Similarly, enrofloxacin (ENR) resistance rates were 74.5% (35/47), 29.3% (12/41), 48.8% (20/41), 100.0% (32/32), 75.0% (6/8), and 5.1% (2/39), respectively (p < 0.001).
We further analyzed inhibition zone diameters for six key antibiotics (AMX, AML, FUR, FFC, ENR, and CIP), which are commonly used to control S. suis infections in swine production (16). While Hang pig isolates showed high resistance to all six drugs, Dongxiang spotted pig isolates displayed markedly lower resistance (Figure 3). This disparity indicates a significant correlation between bacterial isolation source (i.e., pig breed) and antimicrobial susceptibility profiles.
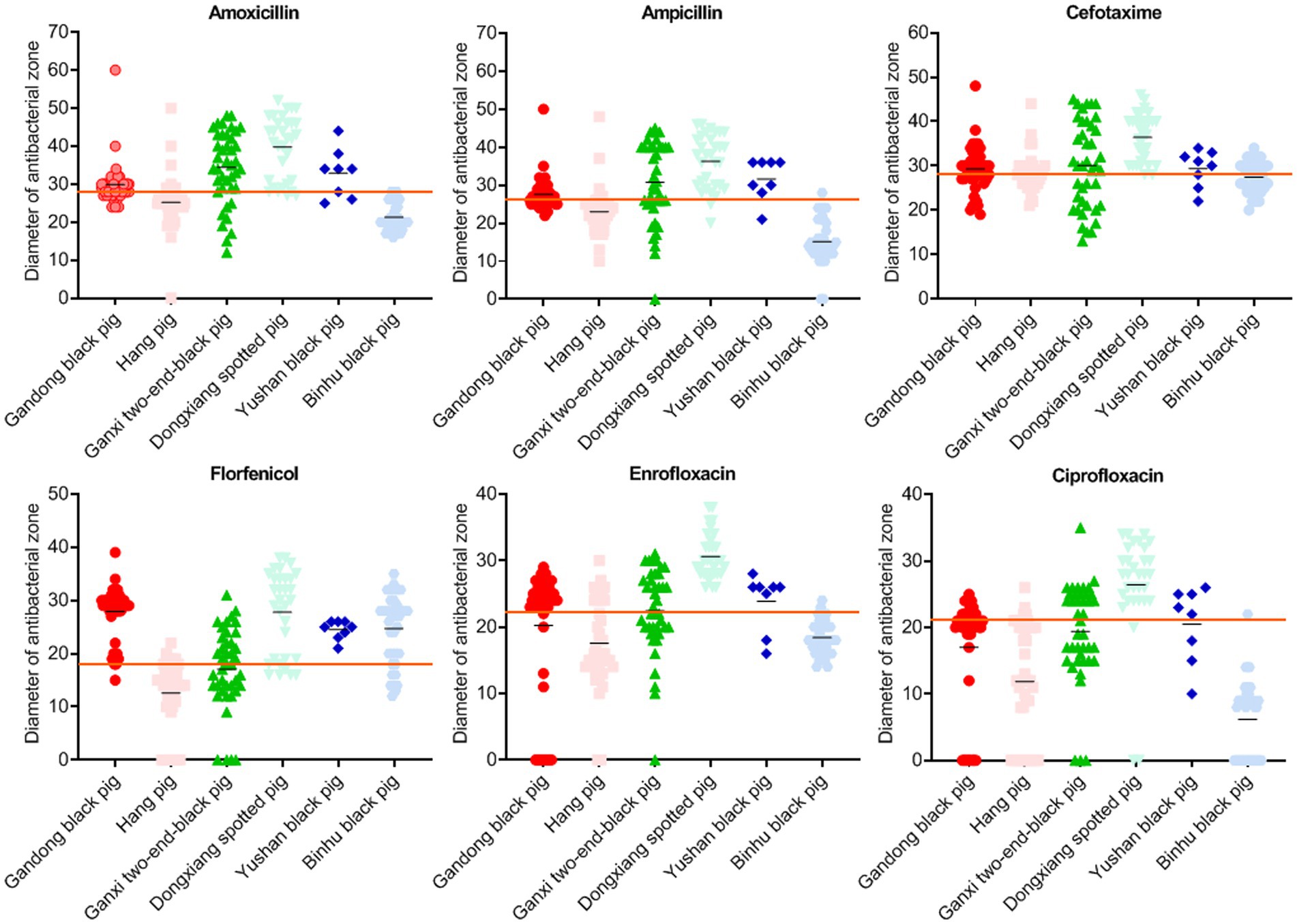
Figure 3. Inhibition zone diameters of bacteria isolated from six local pig breeds against specific drugs. X-axes of scatter diagrams indicate the six local pig breeds. Short black lines represent average values. Long red lines represent sensitivity thresholds of specific antimicrobials.
Except for IMI and MRP, the most sensitive drugs for S. suis in Gandong black pigs were FFC (83.0%) and SPT (83.0%), in Hang pigs was DX (73.2%), in Ganxi two-end-black pigs was GM (78.0%), in Dongxiang spotted pigs were FUR (100.0%) and ENR (100.0%), in Yushan black pigs was FFC (100.0%), and in Binhu black pig was DX (84.7%) (Table 2). These findings provide evidence-based guidance for clinical prevention and treatment strategies.
Multidrug resistance
The standardized definition of multidrug resistance refers to bacterial isolates exhibiting non-susceptibility to at least three antimicrobial categories (17). All S. suis isolates in this study demonstrated resistance to a minimum of four antimicrobial categories (Table S3), with 100.0% classified as multidrug-resistant (Figure 4). In addition, 38.5% (80/208) of the isolates displayed the most prevalent antimicrobial resistance pattern (CT-E-TIL-CS-SUZ-T-GM-AP). Furthermore, 21.2% (44/208) of isolates exhibited resistance to nine antimicrobial drugs, while 20.7% (43/208) showed resistance to eight antimicrobial drugs, representing the predominant resistance profiles observed.
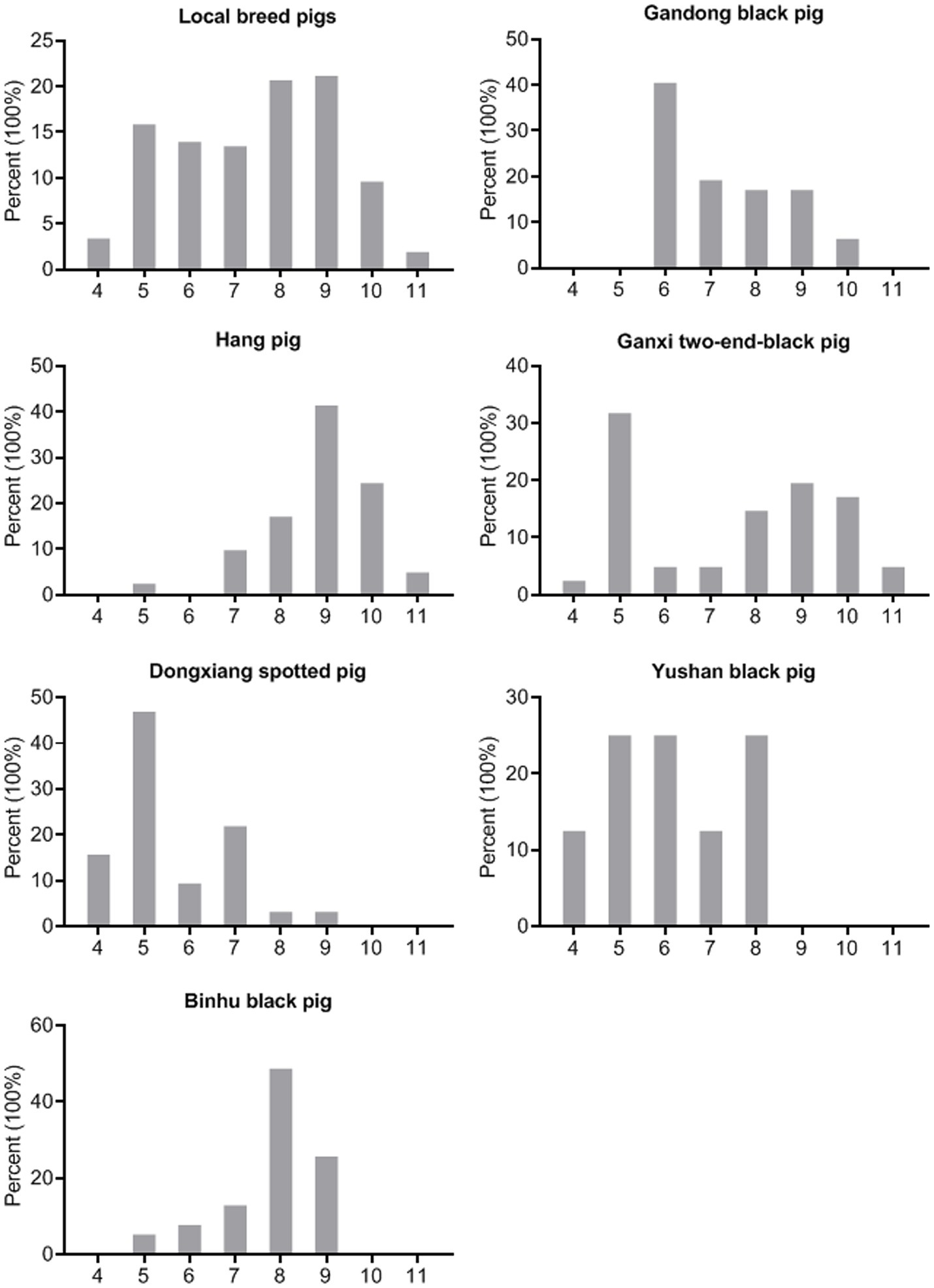
Figure 4. Multidrug resistance of the S. suis isolates (n = 208). X-axes of seven histograms indicate resistance of the S. suis isolates to 4–11 categories of antimicrobials.
As shown in Figure 4, the predominant multidrug resistance profiles varied among breeds: resistance to six antimicrobial categories in Gandong black pigs (40.4%, 19/47), nine categories in Hang pigs (41.5%, 17/41), five categories in Ganxi two-end-black pigs (31.7%, 13/41), five categories in Dongxiang spotted pigs (46.9%, 15/32), five/six/eight categories in Yushan black pigs (25.0%, 2/8), and eight categories in Binhu black pigs (48.7%, 19/39). These findings demonstrate substantial variation in multidrug resistance patterns of S. suis across different indigenous pig breeds.
Moreover, the three carbapenem-resistant strains exhibited extensive drug resistance to all 11 antimicrobial categories tested, with one strain being resistant to all 20 drugs (Table S3).
Detection of drug resistance genes
Due to the high frequency of resistance to macrolides, tetracyclines, aminoglycosides, and sulfonamides, all strains were screened for corresponding resistance genes using PCR assays (Figure 5). The most prevalent macrolide resistance gene was ermB (87.0%, 181/208), followed by mefA (7.69%, 16/208). Among the tetracycline resistance genes, tetL (73.6%), tetO (54.8%), and tetM (37.0%) were detected, whereas tetK was absent (0%). The aminoglycoside resistance gene aph(3′) was identified in 26% (54/208) of the isolates. Additionally, Sul2 was the predominant sulfonamide resistance determinant in S. suis, present in 94.2% (196/208) of the strains.
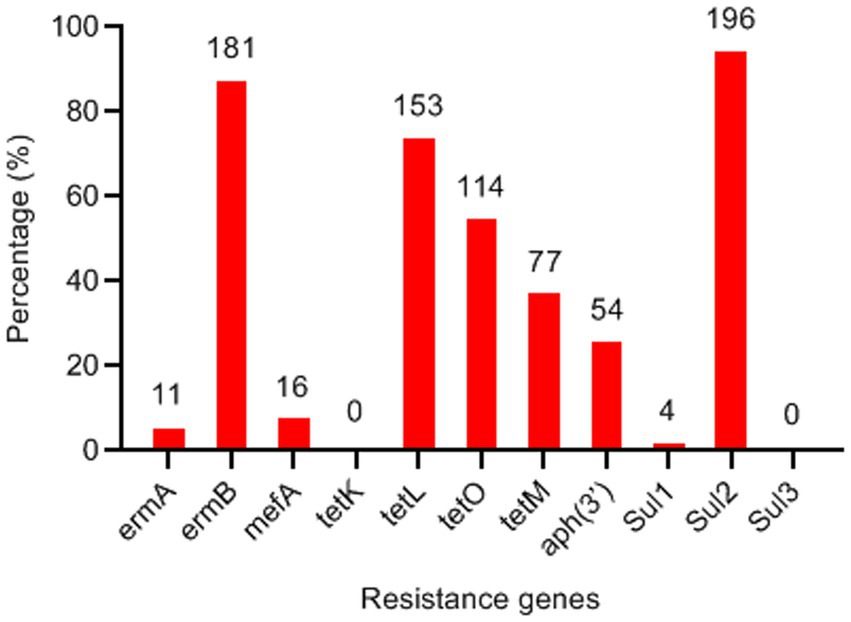
Figure 5. Detection frequency of antimicrobial resistance genes in Streptococcus suis isolates (n = 208). The numbers on the columns indicate the number of S. suis isolates possessing the corresponding resistance genes.
Discussion
Disease prevention and control are critical for protecting the germplasm resources of Chinese local pig breeds. S. suis naturally colonizes the upper respiratory tract of healthy pigs and is commonly detected in commercial pig populations across most provinces of China (7). Previous studies indicate that S. suis prevalence is highest in smallholder farms, with pigs under 6 months of age being more susceptible to infection than older pigs (18). Additionally, the zoonotic risk posed by S. suis remains underappreciated, even after the two largest outbreaks in China in 1998 and 2005, and its prevalence is particularly concerning in regions with intensive pig farming (19). Given this background, we investigated the prevalence and antimicrobial resistance of S. suis in six local pig breeds in Jiangxi Province. This study aims to reduce disease risks in the local pig breed production system and provide baseline data to enhance biosecurity measures.
The positive carrier rate of S. suis in the six pig breeds ranged from 13.3 to 80.0%, with an average of 58.0%. This rate is higher than both the 34.08% prevalence previously reported in Jiangxi Province’s commercial pigs (11) and the 15.8% prevalence observed in smallholder farms in the Philippines (20). These findings suggest that local pig populations in Jiangxi Province are at high risk of S. suis infection.
Antimicrobial drugs have long been proven effective in promoting growth and preventing bacterial infections. However, S. suis has rapidly developed antibiotic resistance through horizontal gene transfer (21). Multidrug-resistant S. suis strains have been identified across all serotypes, time periods, and geographical regions worldwide (22). The results of antimicrobial susceptibility assays revealed that 100% of the isolates in this study were multidrug-resistant. The human-use-only antimicrobials IMI and MRP remained the most effective agents. However, high resistance rates were observed to commonly used veterinary antimicrobial drugs against S. suis, including AMX (44.2%), AMP (47.1%), SUZ (92.8%), E (79.3%), ENR (48.6%), CIP (57.7%), and T (83.2%). These findings raise serious concerns about potential treatment failures in the future.
The positive carrier rate, antimicrobial susceptibility, and multidrug resistance patterns of S. suis varied among local pig breeds. The widespread emergence of resistant bacterial strains typically results from antimicrobial selective pressure (23). Consequently, the observed interbreed differences in bacterial characteristics may be attributed to not only porcine genetic factors but also variations in farm management levels and medication strategies. The highest isolation rates of S. suis occurred in Hang pigs (80.0%) and Ganxi two-end-black pigs (71.7%). Furthermore, these two populations contained carbapenem-resistant strains that demonstrated multidrug resistance across all tested antimicrobial categories. The emergence of carbapenem-resistant S. suis represents a significant public health concern due to its implications for both veterinary and human clinical treatment (24, 25). Carbapenem resistance in Streptococcus spp. has been increasingly reported in human clinical cases (26, 27) and diseased pigs (22) in recent years. These findings underscore the need for current S. suis control strategies to emphasize both appropriate antimicrobial selection and prudent usage (10). In this study, we have identified the most effective antibiotics against S. suis infection for each local pig breed, findings that may help mitigate the impact and transmission of resistant S. suis strains.
Resistance to macrolides is primarily mediated by ribosomal methylases encoded by erm genes, or less commonly, by macrolide efflux pumps encoded by mef genes (13). The ermB gene was the most prevalent resistance gene in S. suis isolates, observed in both locally bred pigs in this study and commercial pigs (11). Previous studies have demonstrated that tetO represents the predominant tetracycline resistance gene in S. suis isolates from pigs and humans in Italy, Japan, Korea, and Jiangxi Province (11). In contrast, our results revealed tetL as the most prevalent tetracycline resistance gene among S. suis isolates from locally bred pigs. Sulfonamide resistance is known to be conferred by sul genes (15). The high detection rate of Sul2 (94.2%) is consistent with the resistance rate of the isolates to SUZ (92.8%).
Notably, the complete information of the other resistance genes and mobile genetic elements in these strains remains uncharacterized. As previously described, mobile genetic elements harboring multiple AMR genes and virulence factors facilitate the horizontal dissemination of AMR determinants across the intra- and inter-species (11). Multidrug-resistant S. suis not only poses a direct threat to swine production and public health but also serves as a reservoir for AMR gene transmission to clinically relevant streptococcal pathogens, including Streptococcus agalactiae, Streptococcus pneumoniae, and Streptococcus pyogenes (28). Therefore, further genome sequencing and bioinformatics analysis would provide more detailed information about these S. suis strains and would be more helpful for developing targeted prevention strategies and control interventions.
Conclusion
This study provides the first investigation of S. suis carrier rates and detailed antimicrobial resistance profiles among six representative local pig breeds in Jiangxi Province, demonstrating a high prevalence in clinically healthy piglets. Antimicrobial susceptibility assays revealed widespread multidrug resistance among S. suis isolates, with significant breed-specific variations in therapeutic efficacy of antimicrobials used for S. suis prevention and treatment. These findings highlight the urgent need for coordinated government-farmer collaborations to implement antimicrobial stewardship programs, establish continuous AMR surveillance systems, and advance research into resistance mechanisms to develop novel prevention strategies against S. suis infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by Institute of Animal Husbandry and Veterinary Medicine, Jiangxi Academy of Agricultural Science (2020-JXAAS-XM-01). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
M-FT: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – original draft. C-LL: Data curation, Methodology, Resources, Writing – original draft. Q-YZ: Data curation, Project administration, Resources, Writing – original draft. J-ZW: Formal analysis, Software, Writing – original draft. J-WH: Methodology, Writing – original draft. M-CW: Resources, Writing – original draft. F-LZ: Visualization, Writing – original draft. H-YJ: Conceptualization, Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by Jiangxi Provincial Natural Science Foundation (20242BAB20313), the National Natural Science Foundation of China (32460884), the Basic Research and Talent Training Program of Jiangxi Academy of Agricultural Sciences (JXSNKYJCRC202307, JXSNKYJCRC202217), and the Jiangxi Provincial Pig Industry Technology System (JXARS-10).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1582461/full#supplementary-material
References
1. Zhao, QB, Oyelami, FO, Qadri, QR, Sun, H, Xu, Z, Wang, QS, et al. Identifying the unique characteristics of the Chinese indigenous pig breeds in the Yangtze River Delta region for precise conservation. BMC Genomics. (2021) 22:151. doi: 10.1186/s12864-021-07476-7
2. Wu, J, Liu, R, Li, H, Yu, H, and Yang, Y. Genetic diversity analysis in Chinese miniature pigs using swine leukocyte antigen complex microsatellites. Anim Biosci. (2021) 34:1757–65. doi: 10.5713/ab.20.0637
3. Wan, C, Liu, C, Zhou, C, Xiong, LG, and Ji, H. The current status and development strategies of local pig germplasm resource protection in Jiangxi Province. Chin Livest Poult Breed. (2024) 20:43–9.
4. Tram, G, Jennings, MP, Blackall, PJ, and Atack, JM. Streptococcus suis pathogenesis-a diverse array of virulence factors for a zoonotic lifestyle. Adv Microb Physiol. (2021) 78:217–57. doi: 10.1016/bs.ampbs.2020.12.002
5. Votsch, D, Willenborg, M, Weldearegay, YB, and Valentin-Weigand, P. Streptococcus suis—the “two faces” of a pathobiont in the porcine respiratory tract. Front Microbiol. (2018) 9:480. doi: 10.3389/fmicb.2018.00480
6. Lv, R, Zhang, W, Sun, Z, Si, X, Dong, H, and Liu, X. Current prevalence and therapeutic strategies for porcine Streptococcus suis in China. Appl Environ Microbiol. (2025) 91:e0216024. doi: 10.1128/aem.02160-24
7. Liu, P, Zhang, Y, Tang, H, Wang, Y, and Sun, X. Prevalence of Streptococcus suis in pigs in China during 2000-2021: a systematic review and meta-analysis. One Health. (2023) 16:100513. doi: 10.1016/j.onehlt.2023.100513
8. Osei, EK, Mahony, J, and Kenny, JG. From farm to fork: Streptococcus suis as a model for the development of novel phage-based biocontrol agents. Viruses. (2022) 14:1996. doi: 10.3390/v14091996
9. Uruen, C, Garcia, C, Fraile, L, Tommassen, J, and Arenas, J. How Streptococcus suis escapes antibiotic treatments. Vet Res. (2022) 53:91. doi: 10.1186/s13567-022-01111-3
10. Varela, NP, Gadbois, P, Thibault, C, Gottschalk, M, Dick, P, and Wilson, J. Antimicrobial resistance and prudent drug use for Streptococcus suis. Anim Health Res Rev. (2013) 14:68–77. doi: 10.1017/S1466252313000029
11. Tan, MF, Tan, J, Zeng, YB, Li, HQ, Yang, Q, and Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J Appl Microbiol. (2021) 130:797–806. doi: 10.1111/jam.14831
12. CLSI. M100: performance standards for antimicrobial susceptibility testing. 32nd ed. Wayne, PA: Clinical and Laboratory Standards Institute (2022).
13. Jacobs, MR, Bajaksouzian, S, and Appelbaum, PC. Telithromycin post-antibiotic and post-antibiotic sub-MIC effects for 10 gram-positive cocci. J Antimicrob Chemother. (2003) 52:809–12. doi: 10.1093/jac/dkg437
14. Zhang, C, Zhang, Z, Song, L, Fan, X, Wen, F, Xu, S, et al. Antimicrobial resistance profile and genotypic characteristics of Streptococcus suis capsular type 2 isolated from clinical carrier sows and diseased pigs in China. Biomed Res Int. (2015) 2015:284303. doi: 10.1155/2015/284303
15. Tian, XY, Zheng, N, Han, RW, Ho, H, Wang, J, Wang, YT, et al. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb Pathog. (2019) 131:33–9. doi: 10.1016/j.micpath.2019.03.035
16. Seitz, M, Valentin-Weigand, P, and Willenborg, J. Use of antibiotics and antimicrobial resistance in veterinary medicine as exemplified by the swine pathogen Streptococcus suis. Curr Top Microbiol Immunol. (2016) 398:103–21. doi: 10.1007/82_2016_506
17. Sweeney, MT, Lubbers, BV, Schwarz, S, and Watts, JL. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother. (2018) 73:1460–3. doi: 10.1093/jac/dky470
18. Dione, M, Masembe, C, Akol, J, Amia, W, Kungu, J, Lee, HS, et al. The importance of on-farm biosecurity: Sero-prevalence and risk factors of bacterial and viral pathogens in smallholder pig systems in Uganda. Acta Trop. (2018) 187:214–21. doi: 10.1016/j.actatropica.2018.06.025
19. Zhu, S, Li, S, Wu, B, Yang, Z, Zhang, Y, Chen, J, et al. Uncovering a cryptic Streptococcus suis endemic post-outbreak: evidence of host switching to humans. Sci Total Environ. (2025) 959:178307. doi: 10.1016/j.scitotenv.2024.178307
20. Sedano, SA, Cantalejo, M, Lapitan, C, de, A, Consignado, J, Tandang, N, et al. Epidemiology and genetic diversity of Streptococcus suis in smallhold swine farms in the Philippines. Sci Rep. (2023) 13:21178. doi: 10.1038/s41598-023-48406-9
21. Kouki, A, Pieters, RJ, Nilsson, UJ, Loimaranta, V, Finne, J, and Haataja, S. Bacterial adhesion of Streptococcus suis to host cells and its inhibition by carbohydrate ligands. Biology (Basel). (2013) 2:918–35. doi: 10.3390/biology2030918
22. Lunha, K, Chumpol, W, Samngamnim, S, Jiemsup, S, Assavacheep, P, and Yongkiettrakul, S. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018-2020. Antibiotics (Basel). (2022) 11:410. doi: 10.3390/antibiotics11030410
23. Kwong, SM, Ramsay, JP, Jensen, SO, and Firth, N. Replication of staphylococcal resistance plasmids. Front Microbiol. (2017) 8:2279. doi: 10.3389/fmicb.2017.02279
24. Shen, B, Tong, L, Qiu, J, Zhong, J, Tong, Z, and Li, X. Suppurative meningitis complicated with arthritis caused by Streptococcus suis infection: a case report. Infect Drug Resist. (2024) 17:561–9. doi: 10.2147/IDR.S416341
25. Liu, T, Liu, H, and Jia, Y. Streptococcus suis meningitis in China: a case report. Front Public Health. (2024) 12:1369703. doi: 10.3389/fpubh.2024.1369703
26. Ronning, TG, Olaisen, C, As, CG, Afset, JE, and Haugan, MS. Identification and characterisation of carbapenem-resistant Streptococcus nidrosiense sp. nov. isolated from blood culture. New Microbes New Infect. (2024) 62:101473. doi: 10.1016/j.nmni.2024.101473
27. Sarangi, J, Ido, A, Ito, M, Iinuma, C, Doyama, Y, Jin, W, et al. Clinical isolates of Streptococcus mitis/oralis-related species with reduced carbapenem susceptibility, harboring amino acid substitutions in penicillin-binding proteins in Japan. Antimicrob Agents Chemother. (2024) 68:e0117923. doi: 10.1128/aac.01179-23
28. Huang, J, Ma, J, Shang, K, Hu, X, Liang, Y, Li, D, et al. Evolution and diversity of the antimicrobial resistance associated Mobilome in Streptococcus suis: a probable Mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol. (2016) 6:118. doi: 10.3389/fcimb.2016.00118
Keywords: Streptococcus suis, prevalence, antimicrobial resistance, local pig breed, Jiangxi Province
Citation: Tan M-F, Liu C-L, Zhou Q-Y, Wei J-Z, Hong J-W, Wan M-C, Zhang F-L and Ji H-Y (2025) Prevalence and antimicrobial resistance of Streptococcus suis isolated from local pig breeds in Jiangxi Province, China. Front. Vet. Sci. 12:1582461. doi: 10.3389/fvets.2025.1582461
Edited by:
Satoshi Ito, Kagoshima University, JapanReviewed by:
Ben Pascoe, University of Oxford, United KingdomFaham Khamesipour, Ministry of Health and Medical Education, Iran
Copyright © 2025 Tan, Liu, Zhou, Wei, Hong, Wan, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-Yuan Ji, ZmVpeWFuZ2poeUAxMjYuY29t
 Mei-Fang Tan
Mei-Fang Tan Chen-Long Liu1
Chen-Long Liu1 Hua-Yuan Ji
Hua-Yuan Ji