- 1Food Science and Engineering College of Inner Mongolia Agricultural University, Hohhot, China
- 2Department of Pediatrics, Inner Mongolia Maternal and Child Health Care Hospital, Hohhot, China
- 3Inner Mongolia Autonomous Region Hospital of Traditional Chinese Medicine, Hohhot, China
Cryptosporidium spp. are apicomplexan parasites that can cause diarrhea in humans and animals, posing a health risk to both animals and humans. Molecular epidemiological analysis provides essential data for understanding Cryptosporidium transmission, treatment, and control. In this study, SSU rRNA was used to determine the prevalence of Cryptosporidium in cattle. A total of 847 fecal samples were collected from 16 farms in central and western Inner Mongolia (Hohhot, Ordos, Bayan Nur and Baotou), and 15.94% (135/847) of samples were positive. Overall, Cryptosporidium was detected in all seasons. Calves up to 2 months of age were found with the highest rate of infection (33.33%). In older animals, there was a significant decline in infection rates with increasing age. The species, C. parvum (n = 105), C. andersoni (n = 21) and C. bovis (n = 9) were detected, individually, or in mixed infections involving two or three Cryptosporidium spp., and five subtypes of C. parvum (IIdA17G1, IIdA17G2, IIdA18G1, IIdA19G1, IIdA20G1) were identified. Our findings provide data to support the epidemiological control of Cryptosporidium infection in cattle.
1 Introduction
Cryptosporidium spp. are apicomplexan parasites that can infect a variety of hosts including humans and animals, and mainly exist in the intestines of the host (1). Cryptosporidiosis has been the second leading cause of diarrheal disease and death in infants after rotavirus (2). Being infected can cause severe diarrhea and a compromised immune system (3, 4). Cryptosporidiosis can be transmitted either directly between hosts via the fecal-oral route or indirectly through ingestion of water or food contaminated with Cryptosporidium (5). Cattle is a common species infected with Cryptosporidium. There are more than 50 species and 120 genotypes for the diagnosis of Cryptosporidium infection through existing molecular techniques, among which C. parvum, C. bovis, C. andersoni, and C. ryanae are the main species that infect cattle (6, 7).
Cryptosporidium infection can reduce the productivity of cattle, cause severe diarrhea and even death, which seriously hindering the development of husbandry (8, 9). Until November 29, 2023, the first Cryptosporidium vaccine for calves has only been approved by the European Union. The development of more safe and effective new drugs and vaccines against Cryptosporidium is ongoing (10), so it is necessary to acquire the prevalence of Cryptosporidium in cattle. Cryptosporidium prevalence surveys in cattle have been conducted worldwide. In a rural settlement in the Northwest region of São Paulo, Brazil, the infection rate in calves from January to June was 7.36% (17/231) (11). In Buffaloes in Sylhet, Bangladesh, the positive rate of Cryptosporidium was found to be 9.18%, and the calves under 6 month was as high as 22.61% (12). Of the 1,283 samples of bovine manure collected from 97 farms in New Zealand, the positive rate for Cryptosporidium was 13.7% (13). In Jiangxi Province in China, the prevalence is 12.8% (71/556) (14), in Beijing (2.55%, 21/822) (15), in Hebei Province (9.2%, 66/718) (16).
Infection with Cryptosporidium in cattle may be related to age (9, 17, 18) and season (4, 19). Calves under 6 months of age are more susceptible to Cryptosporidium infection than adult cattle (14, 20). Depending on the characteristics of the region, the summer and winter are generally the peak seasons for Cryptosporidium infection (20, 21). Explore and identify the prevalence of Cryptosporidium species at different ages and seasons in specific areas and take appropriate measures against them, on the one hand, it is conducive to reducing economic losses, and on the other hand, it is of great significance to local public health. However, the Inner Mongolia Autonomous Region, a major animal husbandry province in China, has been less investigated for cattle infection with Cryptosporidium. Therefore, we chose to investigate the prevalence of Cryptosporidium in the central and western regions of Inner Mongolia, where animal husbandry is more concentrated. The data obtained provides an important basis for the prevention and control of Cryptosporidium infection.
2 Materials and methods
2.1 Ethics statements
The experimental procedure is in accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China (approval number 2020–075-1). The samples were collected with the prior consent of the farm owner. No cattles or other animals were injured during the collection of fecal samples.
2.2 Study area and sample collection
Between July 2020 and July 2021, a total of 847 stool samples of dairy cattle were collected from 16 farms in central and western Inner Mongolia (Hohhot-265, Ordos-181, Bayan Nur-196 and Baotou-205) randomly (Supplementary Figure S1), which include female sample (n = 410) and male sample (n = 437). The age of sampled animals ranged from 8 days to 8.2 years old. Fecal samples were collected from the rectum of cattle using disposable gloves, placed in a 15 mL centrifuge tube, and immediately transferred to the laboratory and stored at 4°C before DNA extraction.
2.3 DNA extraction
DNA was extracted using a TIAN amp DNA Stool Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. DNA was eluted in 50 μL of double distilled water (ddH2O) and stored at −20°C for subsequent Polymerase Chain Reaction (PCR) analysis.
2.4 Cryptosporidium detection, genotyping and subtyping
Individual DNAs were subjected to nested PCR-based amplification and sequencing of regions of SSU rRNA. The first primer set was CryF1(5’-TTCTAGAGCTAATACATGCG-3′) and CryR1(5’-CCCTAATCCTTCGAAACAGGA-3′). The second primer set was CryF2 (5′-GGAAGGGTTGTATTTATTAGATAAAG-3′) and CryR2 (5′-AAGGAGTAAGGA ACAACCTCCA-3′). The amplifying fragments were 1325 bp and 840 bp, respectively. And the PCR cycle conditions for the primary and the secondary as described previously (22). Test-positive, test-negative and no-template controls were included in every round of every PCR run. 1% agarose gel electrophoresis was used to analyze the secondary PCR products and GoodViewTM (SBS, Beijing, China) staining with DL2000 (Tiangen, Beijing, China) as a size marker. The products were visualized under ultraviolet light using Gel imaging system Gel DocTM XR + (BIO-RAD, USA). The positive secondary PCR products were purified using TIANgel Midi Purification Kit (Tiangen, Beijing, China) following the manufacture’s protocol, then samples were sent to Sangon Biotech (Shanghai) Co., Ltd. be sequenced. The C. parvum positive samples were further subtyped according to the 60-kDa glycoprotein (gp60) gene (23).
2.5 Sequence analysis
The spliced gene sequences were compared with available DNA sequencing of Cryptosporidium in GenBank database using online basic local alignment search tool (http://www.ncbi.nlm.nih.gov/ BLAST) to identify the infected Cryptosporidium species. The phylogenetic tree was constructed using the Neighbor-Join method (NJ) of MEGA11 (24), and the reliability of the phylogenetic tree was estimated by the Bootstrap test, with 1,000 replicates.
2.6 Statistical analysis
Statistical analysis was carried out by one-way ANVOA with Graphpad Prism 9.3.1. When p < 0.05, the difference was considered significant.
3 Results
3.1 Prevalence of Cryptosporidium spp.
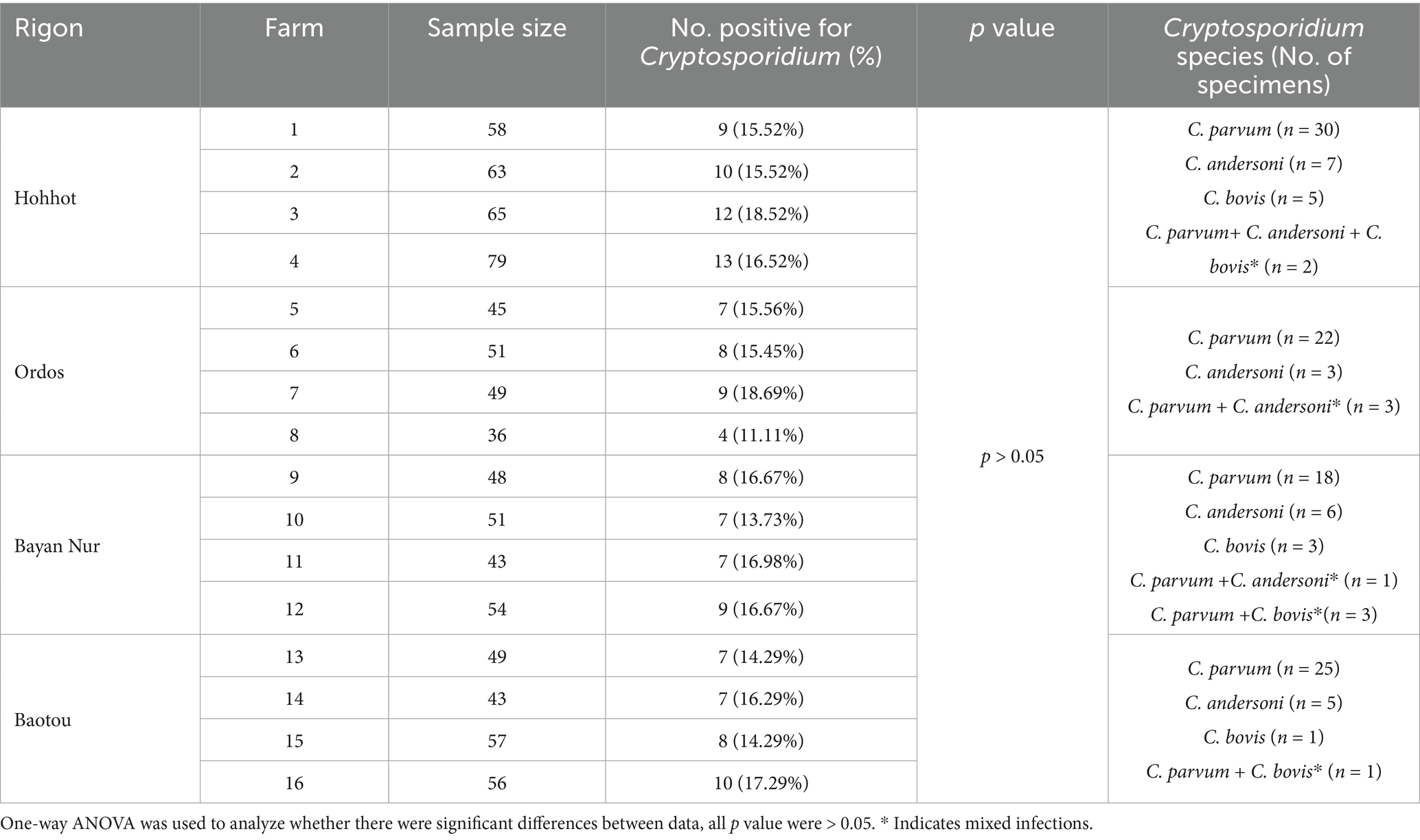
847 fecal samples from cattle were collected from 16 farms in midwestern Inner Mongolia, and 135 (15.94%) samples were positive for Cryptosporidium DNA, which Hohbot 13.95% (44/265), Ordos 21.67% (28/181), Bayan Nur 11.18% (31/196), Baotou 18.54% (32/205) (Table 1). The positive samples were C. parvum (n = 105), C. andersoni (n = 21) and C. bovis (n = 9), along with a mixed infection involving two or three Cryptosporidium species.
3.2 Cryptosporidium prevalence in different seasons in cattle
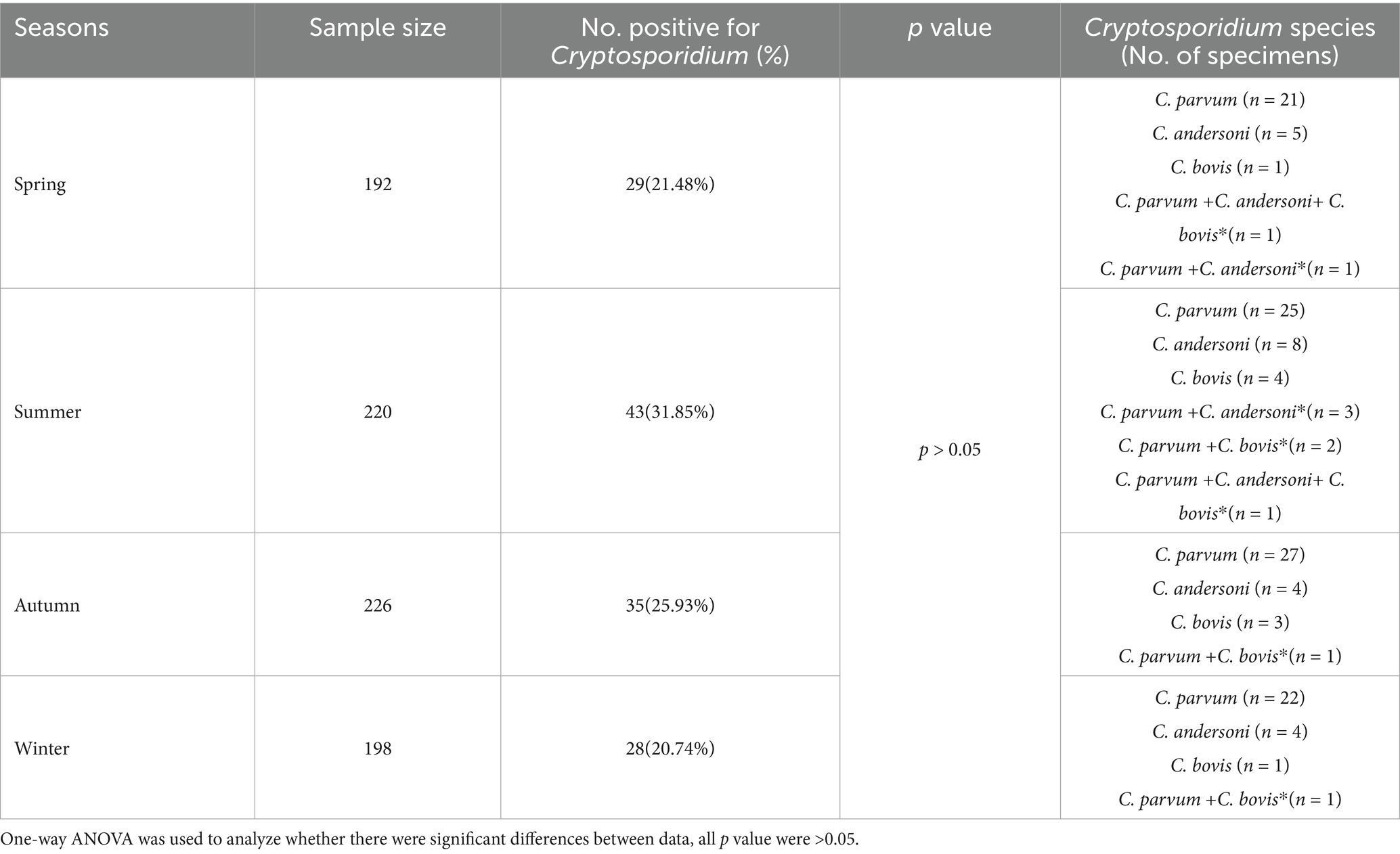
Overall, Cryptosporidium was detected in all seasons. Specifically, Cryptosporidiosis occurred mainly in summer (31.85%), followed by 25.93% in autumn, 21.48% in spring and 20.74% in winter (Table 2). Infections between seasons are still dominated by C. parvum, there are more co-infections in summer.
3.3 Cryptosporidium epidemics of different ages and sex in cattle
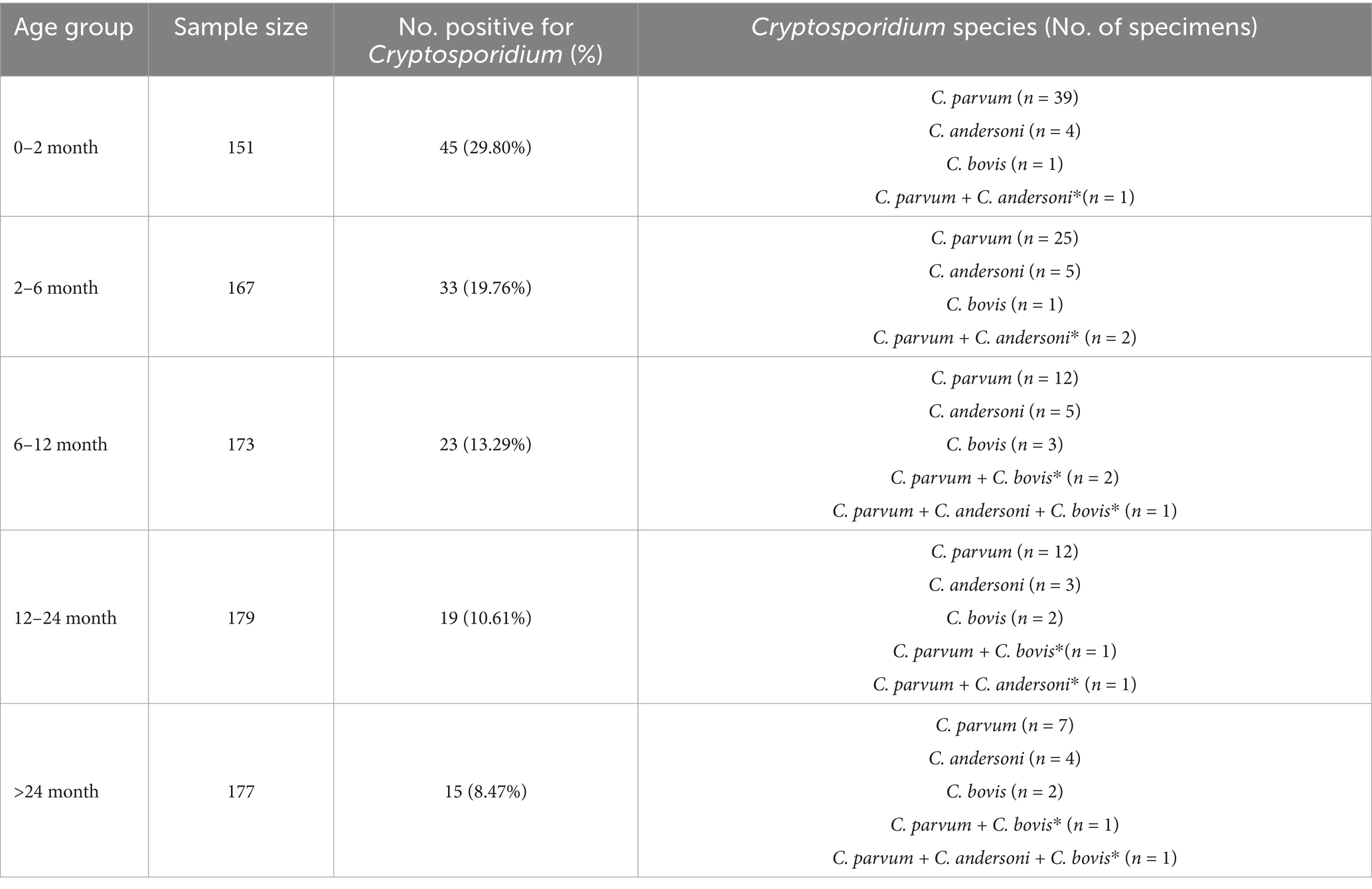
There exists a variation in the infection rate among cattle at varying stages of age, with the increase of age, the number of mixed infections gradually rises (Table 3). Calves within the age range of 0 to 2 months exhibit the highest infection rate (29.80%), with a significant subsequent decline as age progresses, in addition, there were significant differences in infection rates between some age groups (Supplementary Figure S2a). The prevalence of Cryptosporidium in male cattle was 16.25% (71/437) and female cattles 15.61% (64/410). There were no statistical differences between the sexes (Supplementary Figure S2b).
3.4 Subtypes of Cryptosporidium parvum
Three species, C. parvum, C. andersoni and C. bovis, were detected. The subtypes of C. parvum were analyzed by gp60 gene sequences. Nucleotide sequences of the 18S rRNA gene fragment from Cryptosporidium were deposited into the GenBank database under accession numbers OR506389-OR506409, PP800188, PP784678 and PP784681. Five subtypes of C. parvum were identified, including, IIdA17G1, IIdA17G2, IIdA18G1, IIdA19G1, IIdA20G1. The phylogenetic tree was mapped using 18S rRNA gene sequences obtained from all currently known Cryptosporidium spp. (Figure 1).
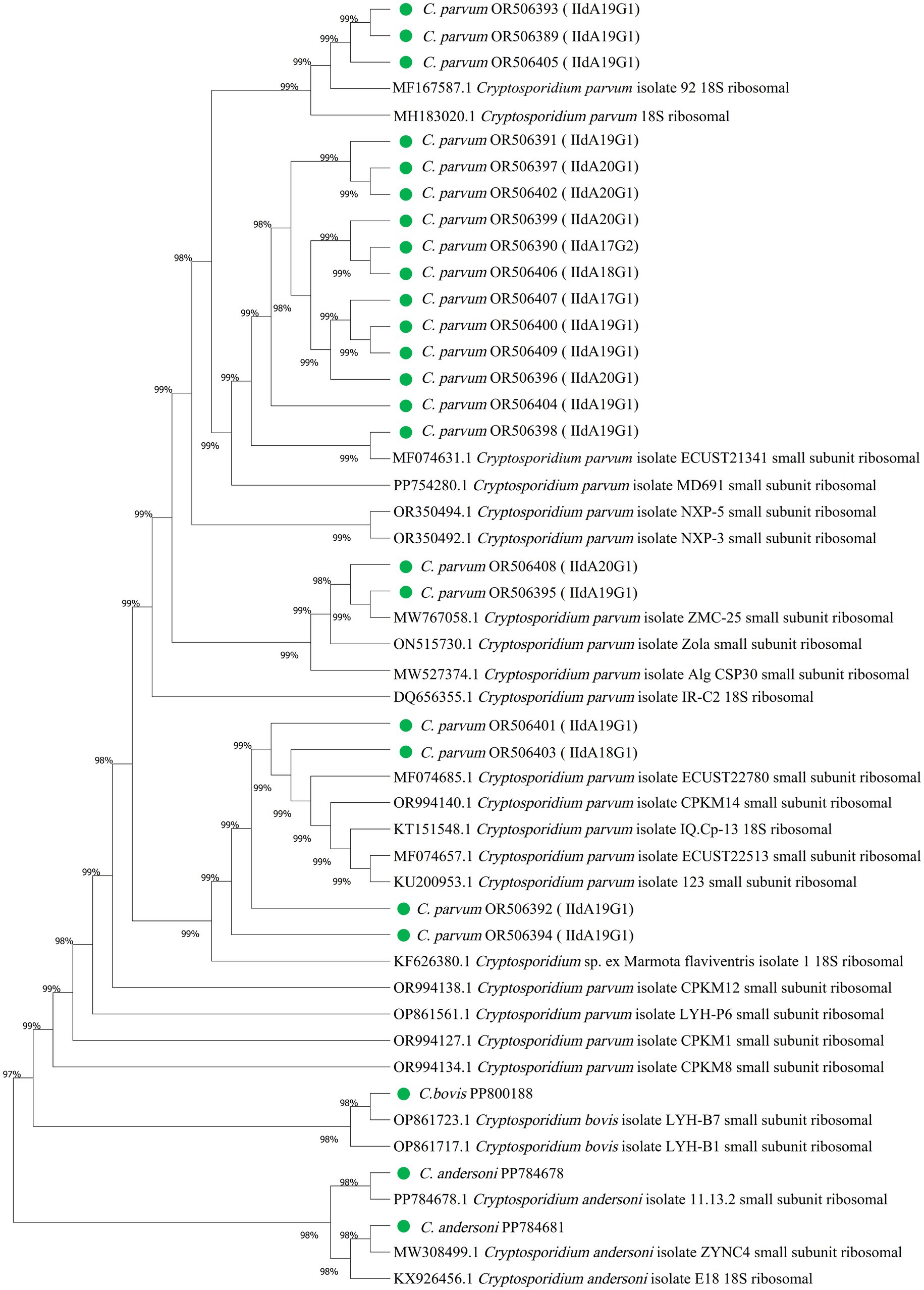
Figure 1. Phylogenetic relationships among Cryptosporidium spp. and genotypes according to the neighbor-joining analysis of a fragment from the partial SSU rRNA sequence. OR506389-OR506409, PP800188, PP784678 and PP784681 were Cryptosporidium isolates from cattle in Inner Mongolia. Sequences of other Cryptosporidium spp. and genotypes were obtained from GenBank.
4 Discussion
Our results revealed that cattle are generally susceptible to Cryptosporidium spp. Overall, the prevalence of Cryptosporidium infection in cattle in Inner Mongolia was 15.94% (135/847), which was close to Xinjiang in Northwestern China (16.0%,82/514) (25), higher than Hebei (9.2%, 66/718) (16) in northern China, Shanghai (13.65%, 67/491), Anhui (2.4%, 23/955) in Eastern China (17). In addition, a survey of the prevalence of Cryptosporidium in various regions in China from 2008 to 2018 showed that the overall prevalence of Cryptosporidium was 17.0% (3,901/33313) (26). The Cryptosporidium was detected in four regions, there was no statistically significant. We speculated that the climate, temperature, humidity, geography, and other conditions of the sampling position were close to each other, so there was no significant difference.
Calves aged 0 to 2 months exhibited the highest infection rate (29.80%), consistent with findings in most surveys of Cryptosporidium in cattle (14, 19). There are less mixed infections in calves, which may be related to their small activity range and relatively monotonous diet. Studies have shown that the immune system of calves is not perfect, and the resistance to external pathogens is weak (27), and with age, the immune system gradually matures, and the resistance to pathogens gradually increases (28). In fact, surveys of adult cattle infected with Cryptosporidium in several countries show that the infection rate is around 10%, much lower than in calves (9), our survey also demonstrates that the Cryptosporidium infection rate decreases progressively with age, reaching 8.47% in adult cattle.
A survey of Cryptosporidium infection in cattle in Hebei Province showed a significant difference between seasons (16). However, our results showed that there was no significant difference between seasons. Our findings showed that Cryptosporidium has the most severe epidemic in summer (19.45%), followed by autumn and spring, and the lowest infection rate in winter, which is similar to the previous report in Xinjiang (4). Similar but slightly different in Ireland, the results showed higher prevalence in summer (9.7%, 7/72) and winter (9.7%, 7/72) than in spring (5.6%, 4/72) and autumn (4.2%, 3/72) (29). Interestingly, dairy cattle were significantly more likely to be infected by Cryptosporidium in winter than in summer in New York State in USA (30). There were more samples of co-infection in summer, which may be related to the temperature and humidity suitable for the reproduction of Cryptosporidium and the wide range of cattle activities. These results indicate that the seasonal prevalence of Cryptosporidium can be influenced by multiple factors, including geographical location, temperature, climate, and farm management practices.
Three species, C. parvum, C. andersoni and C. bovis were detected. Mixed infections involving two or three species also occurred, which we speculate may be related to the scattered nature of farms and the breeding environment. According to the available literature, the subtypes of C. parvum in China are mainly IId family, and then IIa family (31). IIdA14G1 and IIdA15G1 were detected in Xinjiang (4), IIdA19G1, IIdA17G1 and IIdA15G1 were detected in Beijing (15), IIdA20G1 was detected in Hebei (16). A survey of Cryptosporidium infection in cattle in central Inner Mongolia showed that the infection rate was 29.90% (151/505), involving C. parvum, C. andersoni, C. bovis and C. ryanae, the subtype of C. parvum was IIdA19G1 (32). In contrast, three species, C. parvum, C. andersoni and C. bovis were detected in our investigation. Our study covered a larger geographical range within Inner Mongolia and analyzed a greater diversity of C. parvum subtypes. By analyzing our results, five subtypes of C. parvum were detected, IIdA17G1, IIdA17G2, IIdA18G1, IIdA19G1, IIdA20G1. Taken together, we provided more comprehensive data support for Cryptosporidium infection in cattle. C. parvum belongs to zoonotic Cryptosporidium and the subtype of IId is more common in Asia, which can seriously affect public health safety. In Inner Mongolia, the livestock industry is developed, the number of cattle is huge. However, the sanitary conditions in some pastoral areas are relatively poor, so controlling the infection of domestic animals can not only increase the income of herders, but also is of great significance for the prevention and control of human Cryptosporidium infection. In the future investigations, we will expand the scope of the investigation to explore the infection of Cryptosporidium in food, water and other domestic animals in more area, and comprehensively analyze the association between the infections, so as to provide prevention and control strategies for Cryptosporidium infection.
5 Conclusion
Cryptosporidium was detected in the central and western regions of Inner Mongolia throughout the year. The infection rate was the highest in calves aged 0–2 months, and gradually decreased with age growing. In addition, there was no significant difference in infection rate between different sexes. The infected species are diverse, but C. parvum was the predominant, of which C. parvum contains five subtypes. The above results enriched the data of the epidemiological survey of Cryptosporidium in Inner Mongolia and provided data support and theoretical guidance for animal husbandry and public health safety.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number OR506389-OR506409, PP800188, PP784678 and PP784681.
Author contributions
XY: Data curation, Writing – original draft, Writing – review & editing. WG: Data curation, Writing – original draft, Writing – review & editing. RuL: Resources, Writing – original draft. RuiL: Funding acquisition, Writing – original draft. WK: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Inner Mongolia Natural Science Foundation Program of China, grant number was No. 2021MS08099.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1587302/full#supplementary-material
References
1. Buchanan, R, Wieckowski, P, Matechou, E, Katzer, F, Tsaousis, AD, and Farré, M. Global prevalence of Cryptosporidium infections in cattle: a meta-analysis. Curr Res Parasitol Vector Borne Dis. (2025) 7:100264. doi: 10.1016/j.crpvbd.2025.100264
3. Li, F, Su, J, Chahan, B, Guo, Q, Wang, T, Yu, Z, et al. Different distribution of Cryptosporidium species between horses and donkeys. Infect Genet Evol. (2019) 75:103954. doi: 10.1016/j.meegid.2019.103954
4. Zhang, K, Wu, Y, Jing, B, Xu, C, Chen, Y, Yu, F, et al. Seasonal monitoring of Cryptosporidium species and their genetic diversity in neonatal calves on two large-scale farms in Xinjiang, China. J Eukaryot Microbiol. (2021) 69:e12878. doi: 10.1111/jeu.12878
5. Xiao, LH, Fayer, R, Ryan, U, and Upton, SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. (2004) 17:72–97. doi: 10.1128/CMR.17.1.72-97.2004
6. Mirhashemi, ME, Zintl, A, Grant, T, Lucy, F, Mulcahy, G, and De Waal, T. Molecular epidemiology of Cryptosporidium species in livestock in Ireland. Vet Parasitol. (2016) 216:18–22. doi: 10.1016/j.vetpar.2015.12.002
7. O'Leary, JK, Blake, L, Corcoran, GD, Sleator, RD, and Lucey, B. Increased diversity and novel subtypes among clinical Cryptosporidium parvum and Cryptosporidium hominis isolates in southern Ireland. Exp Parasitol. (2020) 218:107967. doi: 10.1016/j.exppara.2020.107967
8. Huang, JY, Yue, DY, Qi, M, Wang, RJ, Zhao, JF, Li, JQ, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet Res. (2014) 10:292. doi: 10.1186/s12917-014-0292-6
9. Díaz, P, Navarro, E, Remesar, S, García-Dios, D, Martínez-Calabuig, N, Prieto, A, et al. The age-related Cryptosporidium species distribution in asymptomatic cattle from North-Western Spain. Animals. (2021) 11:256. doi: 10.3390/ani11020256
10. Rahman, SU, Mi, R, Zhou, S, Gong, H, Ullah, M, Huang, Y, et al. Advances in therapeutic and vaccine targets for Cryptosporidium: challenges and possible mitigation strategies. Acta Trop. (2022) 226:106273. doi: 10.1016/j.actatropica.2021.106273
11. de Matos, LVS, Neto, LD, Oliveira, BCM, Makatu, MY, Pierucci, JC, Viol, MA, et al. Molecular characterization of Cryptosporidium in calves from rural settlements in the northwest region of the state of Sao Paulo, Brazil. Semin Cienc Agrar. (2019) 40:491–6. doi: 10.5433/1679-0359.2019v40n1p491
12. Mahen, MSK, Chowdhury, MSR, Hossain, H, Hossain, MM, Islam, MR, and Rahman, MM. Investigating the infection dynamics and molecular detection of Cryptosporidium in buffaloes in Sylhet, Bangladesh. Vet Parasitol Reg Stud Rep. (2024) 52:101043. doi: 10.1016/j.vprsr.2024.101043
13. Al Mawly, J, Grinberg, A, Velathanthiri, N, and French, N. Cross sectional study of prevalence, genetic diversity and zoonotic potential of Cryptosporidium parvum cycling in New Zealand dairy farms. Parasit Vectors. (2015) 8:240. doi: 10.1186/s13071-015-0855-9
14. Li, S, Zou, Y, Wang, P, Qu, MR, Zheng, WB, Wang, P, et al. Prevalence and multilocus genotyping of Cryptosporidium spp. in cattle in Jiangxi Province, southeastern China. Parasitol Res. (2021) 120:1281–9. doi: 10.1007/s00436-021-07047-5
15. Li, FH, Wang, HY, Zhang, ZJ, Li, JQ, Wang, CR, Zhao, JF, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet Parasitol. (2016) 228:69–9. doi: 10.1016/j.vetpar.2016.03.006
16. Sun, TB, Lin, XY, Pan, SM, Chen, LF, Shi, QM, Li, WC, et al. Prevalence of Cryptosporidium spp. and Enterocytozoon bieneusi in beef cattle in the Hebei Province of China*. Med Weter. (2023) 79:1–6. doi: 10.21521/mw.6748
17. Liu, XC, Tang, L, Li, WC, Li, C, and Gu, YF. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J Vet Med Sci. (2022) 84:40–7. doi: 10.1292/jvms.21-0425
18. Meng, YW, Shu, FF, Pu, LH, Zou, Y, Yang, JF, Zou, FC, et al. Occurrence and molecular characterization of Cryptosporidium spp. in dairy cattle and dairy buffalo in Yunnan Province, Southwest China. Animals. (2022) 12:1031. doi: 10.3390/ani12081031
19. Chen, F, and Huang, KH. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J Vet Sci. (2012) 13:15–22. doi: 10.4142/jvs.2012.13.1.15
20. Das, P, Deka, D, Borthakur, SK, Roychoudhury, P, and Das, M. Studies on occurrence, molecular detection and genotyping of Cryptosporidium parvum along with associated risk factors in cattle and human from Aizawl district, Mizoram, India. Biol Rhythm Res. (2020) 51:238–53. doi: 10.1080/09291016.2018.1526501
21. Kashyap, G, Sindhoora, K, Singh, S, Banerjii, PS, Gupta, D, Kumar, P, et al. Occurrence and diagnosis of cryptosporidiosis in cattle calves with clinical diarrhoea. Biol Rhythm Res. (2021) 52:717–25. doi: 10.1080/09291016.2019.1603687
22. Xiao, LH. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. (2010) 124:80–9. doi: 10.1016/j.exppara.2009.03.018
23. Xiao, LH, Escalante, L, Yang, CF, Sulaiman, I, Escalante, AA, Montali, RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. (1999) 65:1578–83. doi: 10.1128/AEM.65.4.1578-1583.1999
24. Liang, XX, Zou, Y, Li, TS, Chen, H, Wang, SS, Cao, FQ, et al. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in Yunling cattle in Yunnan Province, southwestern China. Microb Pathog. (2021) 158:105025. doi: 10.1016/j.micpath.2021.105025
25. Qi, M, Wang, H, Jing, B, Wang, D, Wang, R, and Zhang, L. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, northwestern China. Vet Parasitol. (2015) 212:404–7. doi: 10.1016/j.vetpar.2015.07.002
26. Cai, Y, Zhang, N-Z, Gong, Q-L, Zhao, Q, and Zhang, X-X. Prevalence of Cryptosporidium in dairy cattle in China during 2008-2018: a systematic review and meta-analysis. Microb Pathog. (2019) 132:193–200. doi: 10.1016/j.micpath.2019.05.006
27. Fayer, R, Santin, M, and Trout, JM. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet Parasitol. (2007) 145:260–6. doi: 10.1016/j.vetpar.2006.12.009
28. Ares-Mazas, ME, Fernandez-da Ponte, B, Vergara-Castiblanco, CA, Freire-Santos, F, Quilez-Cinca, J, Causape-Valenzuela, AC, et al. Oocysts, IgG levels and immunoblot patterns determined for Cryptosporidium parvum in bovine examined during a visit to a farm (northeastern Spain). Vet Parasitol. (1999) 81:185–93. doi: 10.1016/S0304-4017(98)00245-3
29. Moriarty, EM, McEvoy, JM, Lowery, CJ, Thompson, HP, Finn, M, Sheridan, JJ, et al. Prevalence and characterisation of Cryptosporidium species in cattle faeces and on beef carcases at slaughter. Vet Rec. (2005) 156:165–8. doi: 10.1136/vr.156.6.165
30. Mohammed, HO, Wade, SE, and Schaaf, S. Risk factors associated with Cryptosporidium parvum infection in dairy cattle in southeastern New York state. Vet Parasitol. (1999) 83:1–13. doi: 10.1016/S0304-4017(99)00032-1
31. Gong, C, Cao, XF, Deng, L, Li, W, Huang, XM, Lan, JC, et al. Epidemiology of Cryptosporidium infection in cattle in China: a review. Parasite. (2017) 24:1. doi: 10.1051/parasite/2017001
Keywords: Cryptosporidium , cattle, subtype, prevalence, molecular epidemiological
Citation: Yan X, Guo W, Liang R, Li R and Kang W (2025) Prevalence and molecular characterization of Cryptosporidium spp. in cattle in central and Western Inner Mongolia, China. Front. Vet. Sci. 12:1587302. doi: 10.3389/fvets.2025.1587302
Edited by:
Kálmán Imre, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaReviewed by:
Rongsheng Mi, Chinese Academy of Agricultural Sciences, ChinaMd. Mahfujur Rahman, Sylhet Agricultural University, Bangladesh
Copyright © 2025 Yan, Guo, Liang, Li and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruifeng Li, bm1fZnlsaXJ1aWZlbmdAMTYzLmNvbQ==; Wenbin Kang, NDk3MjI1MDE0QHFxLmNvbQ==
†These authors have contributed equally to this work
 Xinlei Yan
Xinlei Yan Wenhui Guo1†
Wenhui Guo1† Ruifeng Li
Ruifeng Li