- 1Key Laboratory of Vegetation Ecology of the Ministry of Education, Jilin Songnen Grassland Ecosystem National Observation and Research Station, Institute of Grassland Science, Northeast Normal University, Changchun, China
- 2Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin, China
Introduction: Grazing ruminant production has the risk of degrading the environment beyond natural recovery due to their production of enteric methane (CH4) which is the main contributor to the increase in global CH4 emissions. In particular, grasslands are currently experiencing severe saline-alkaline degradation that is prevalent in arid and semi-arid grassland areas globally. Yet, the impact of grassland saline-alkaline degradation-induced alterations in plant resources on herbivore, and subsequent CH4 emissions, remain underexplored.
Methods: Here we examined these effects by feeding domestic ruminant-sheep with plants from undegraded (UG), moderately degraded (MG), and severely degraded grasslands (SG), focusing on rumen key microbes and nutrition process.
Results: Our results showed that moderately and severely saline-alkaline degradation of grasslands differently influences rumen key microbes associated with CH4 synthesis, thereby affecting CH4 emissions of ruminants. Specifically, the relative abundance of Treproema that can competitively inhibit the CH4 production was significantly increased in MG-fed sheep, which resulted in reduced CH4 emissions. Conversely, the relative abundance of Methanosphaera that positively related to CH4 production was significantly increased in SG-fed sheep, which resulted in increased CH4 emissions. Forage resources in severely degraded grasslands exhibited extremely high sodium (Na) content, while high forage diversity was found in moderately degraded grassland. Further, we found that increased Na intake has a significant influence on the abundance of Methanosphaera.
Discussion: Taken together, our study provides novel insights into the underlying mechanism of the CH4 emissions induced by saline-alkaline degradation in ruminant herbivores; the increase in Na intake induced by grassland saline-alkaline degradation could be an important factor affecting rumen Methanosphaera thereby CH4 emissions by livestock. Our findings suggest that increasing grassland saline-alkaline degradation worldwide will greatly change the risk of CH4 emissions from grazing ruminants depending on the degree of degradation, which should be incorporated into future consideration of grassland carbon budgets.
1 Introduction
Livestock production, particularly ruminant production, carries the risk of deteriorating the environment beyond natural recovery (1). This is because the enteric methane (CH4) from ruminants is a major contributor to the increase in global CH4 emissions, which greatly increases the risk of global warming (2). Notably, ruminants from grazing systems, usually have higher CH4 emissions relative to stable-fed ruminants with high intake of nutrient-dense rations (3–5). More importantly, estimates suggest that approximately 50 percent of the grasslands in the world have already shown degradation to some extent as a result of climate change and human activities (6). Particularly saline-alkaline degradation, which is prevalent in arid and semi-arid grassland areas globally (7), is likely to further greatly increase CH4 emissions from grazing ruminants. However, the impact of saline-alkaline grassland degradation induced alterations in plant resources on the CH4 of grazing ruminants, which are an important component of managed grassland ecosystems (8), still remains poorly understood.
Methanogenesis is a normal process that mostly occurs during the anaerobic fermentation of feed by a microbial consortia in the rumen (9). In the fermentation process, the carbohydrates in the feed are first fermented by rumen fungi and bacterial communities to produce energetic substrates utilized by the host animal, as well as carbon dioxide (CO2) and hydrogen (H2). Subsequently, the CO2 and H2 generated are mainly converted to CH4 by rumen methanogens, whereas can also be converted to useful metabolites by specific bacteria (10). For the methanogenic pathway, rumen archaea including Methanobacterium, Methanobrevibacter, Methanomicrobium, Methanoculleus, Methanosarcina and Methanosphaera have been demonstrated to exhibit strong CH4-producing capacities by converting H2 and CO2 into CH4 (11, 12). Conversely, certain rumen bacterial genera, such as Ruminococcus, Butyribacterium, Clostridium and Treponema, have been shown to compete with methanogens for H2 and CO2, redirecting these substrates toward the synthesis of alternative metabolic products, such as acetate (13). In addition, protozoa populations, as the engineers of rumen microbial ecosystem, are also able to provide H2 for methanogens to produce CH4 (14). Therefore, the production of CH4 is driven by the interaction of methanogens with other rumen microbes, including protozoa, bacteria, and fungi. Evidence from experimental study has demonstrated the interaction process of various rumen microbes was mainly modulated by diet nutrient profiles (15), and thus, saline-alkaline grassland degradation may affect the CH4 emissions of grazing ruminants by altering their nutrition intake and further affecting the rumen microbial community. Indeed, a previous study found that the fiber content in grassland plant resources showed a linear decrease with increasing grassland degradation level due to the decrease in perennial plants and increase in annual plants (16, 17). The decrease in fiber intake could reduce the CH4 emissions of ruminants by affecting the relative abundance of fungi and bacteria in the rumen to reduce the supply of CO2 and H2 available to methanogens (18). Further study also found that the fat content in grassland plant resources showed a linear increase with increasing grassland degradation level (19). The increase in fat intake could reduce the CH4 emissions of ruminants through competition of unsaturated fatty acids for H2 (20, 21). Meanwhile, the decreased calcium content in grassland plant resources induced by saline-alkaline degradation also can result in more CH4 emissions, on average, of ruminants via increasing the relative abundance of methanogens in the rumen (22–24). Furthermore, different levels of grassland saline-alkaline degradation induced alterations in plant diversity, salt content and secondary metabolites may also affect the CH4 emissions of ruminants. Consequently, the process underlying the effects of grassland saline-alkaline degradation on the CH4 emissions from grazing ruminants is extremely complex and unpredictable.
Here, we examined the effects of grassland saline-alkaline degradation on ruminant CH4 emissions together with the underlying impact mechanisms by feeding small-tailed sheep diets that simulated forage availability in grasslands with different levels of degradation.
2 Materials and methods
2.1 Animals, experimental design, and diets
This experiment was conducted at the experimental base of the Institute of Animal Husbandry, Heilongjiang Academy of Agricultural Sciences (45°42′N, 126°38′E). All animal-based experiments were conducted in accordance with the principles and responsibilities outlined in the Northeast Normal University’s (Changchun, China) guidelines for animal research. Twelve male small-tailed Han sheep of similar weight (30.49 ± 4.31 kg, mean ± SD) were selected for this study. They were each assigned to 1 of 3 dietary treatments which represent different degrees of degraded grasslands classified by the biomass of grassland plants (Areas with 40–60% vegetation cover were deemed as moderately degraded, and those with less than 40% vegetation cover were regarded as heavily degraded) (25): (1) undegraded grassland plants (UG); (2) moderately degraded grassland plants (MG); or (3) severely degraded grassland plants (SG). Plant samples from grasslands degraded to different levels were obtained from the Songnen grassland in Jilin, China (44°45′N, 123°45′E) and plants were cut and baled for feeding to sheep. The grasslands in the area have already shown saline-alkaline degradation to different levels as a result of climate change and overgrazing. The climate of the area is continental with average annual temperatures ranging from 2.4°C to 2.7°C and precipitation ranging from 300 mm to 500 mm, most of which occurs between June and August (data from Changling County Climate Station, Jilin Province).
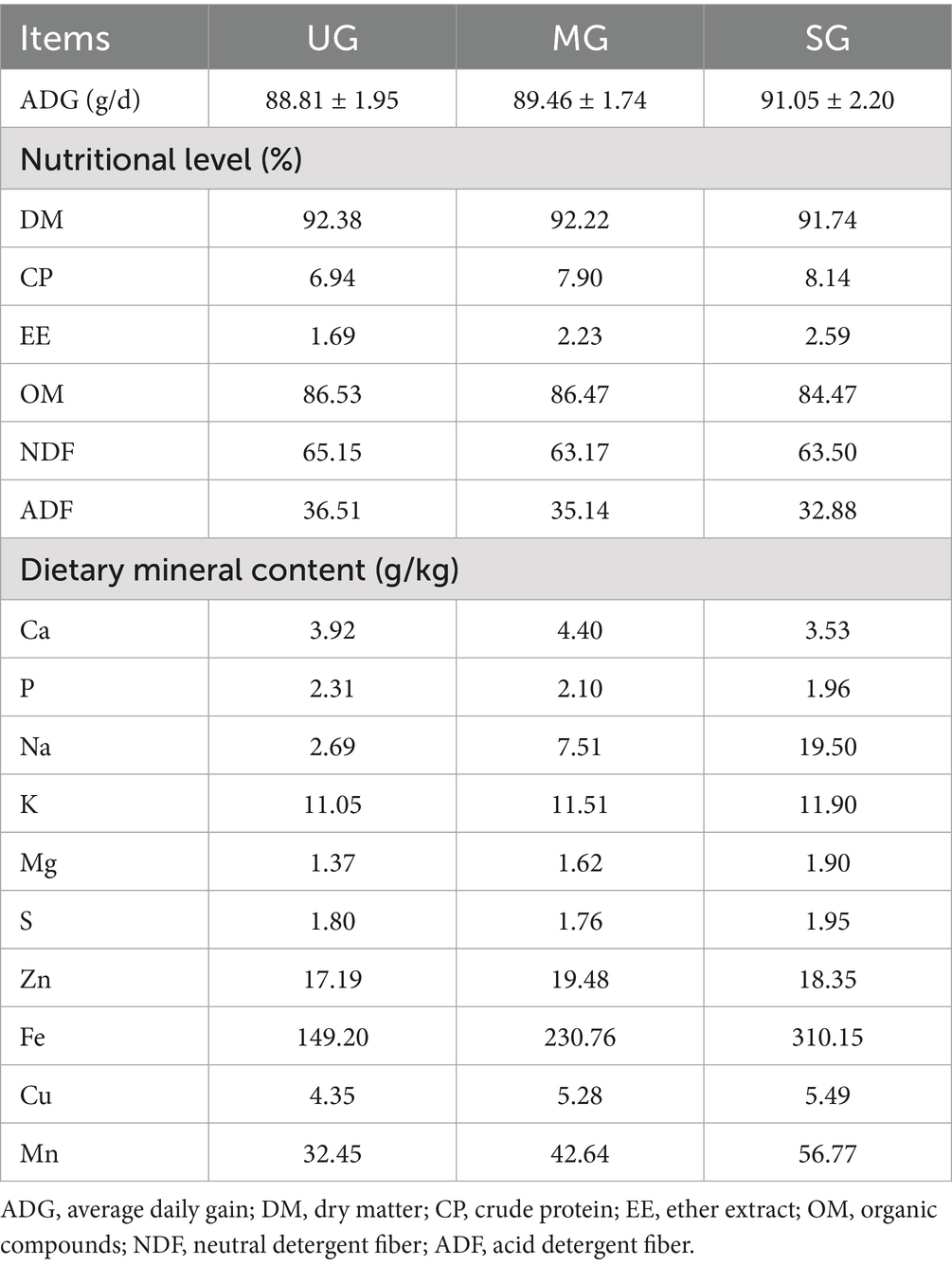
We established three 400-m parallel transects at 50-m intervals in grasslands with different levels of degradation (UG: undegraded grassland plants, MG: moderately degraded grassland plants, and SG: severely degraded grassland plants). Next, we determined the locations of 100 cm × 100 cm quadrants every 40-m along each transect. We used these quadrants to measure the composition and proportion of plants, and then plant samples were harvested at a cutting height of approximately 10 cm. The diets of the sheep were formulated according to the types and proportions of plants in actual grasslands degraded to different levels, as presented in Figure 1. Throughout the 8-week feeding trial, the sheep were fed individually in cages, which were spacious enough for them to stand and lie down in. The sheep were weighed on the final day of the trial period to determine average daily weight gain (ADG) which calculated by dividing the difference in body weight by the number of days. The sheep were fed twice per day, and allowed to feed and drink ad libitum throughout the experiment period.
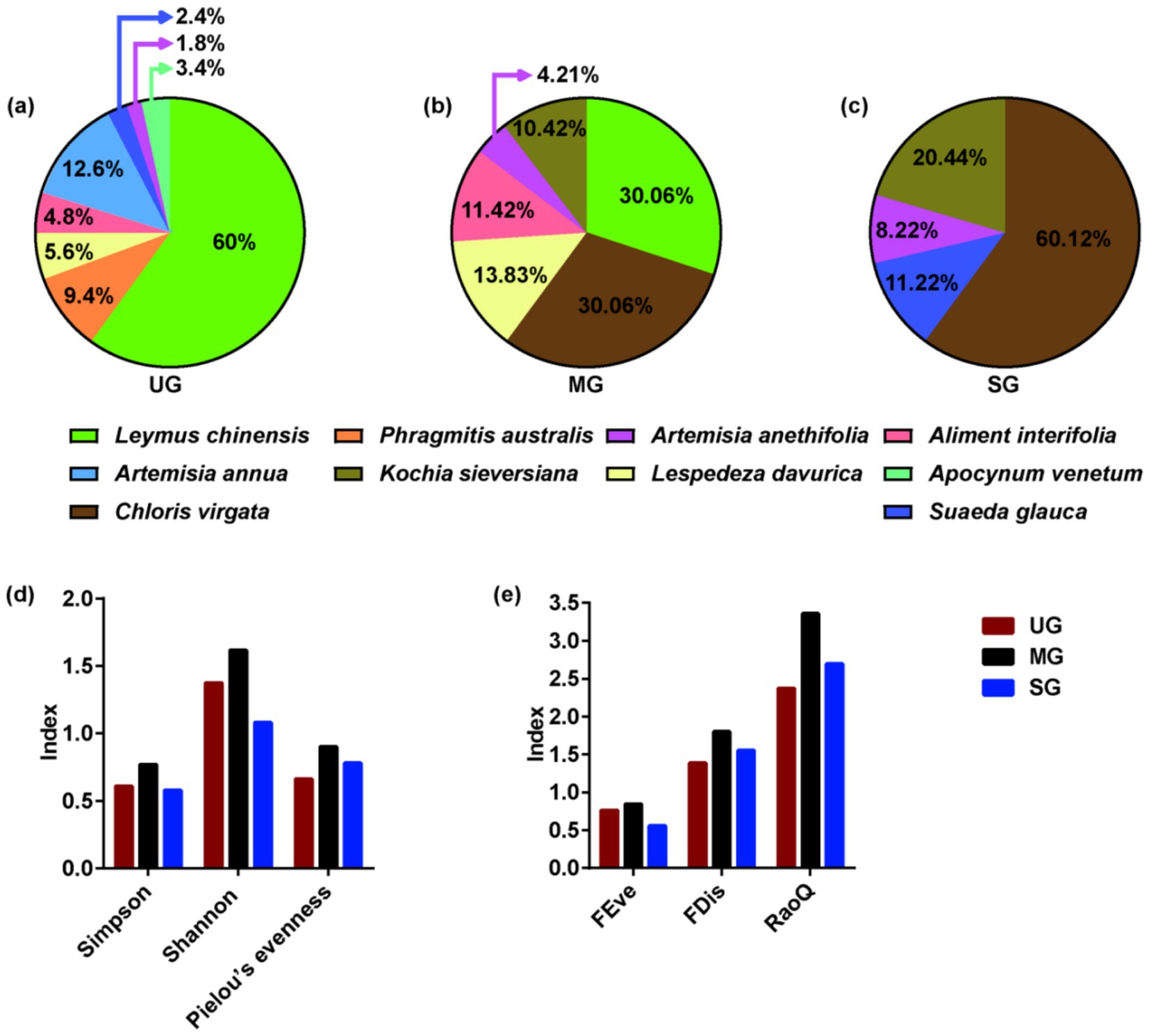
Figure 1. Plant composition (a–c), species diversity (d), and functional diversity (e) of grasslands with different levels of saline-alkaline degradation. UG undegraded grassland, MG moderately degraded grassland, SG severely degraded grassland, FEve functional evenness, FDis functional dispersion, RaoQ Rao’s quadratic entropy index.
2.2 Sample collection
The feed offered and refused for individual sheep was weighed each day to calculate the daily dry matter intake (DMI). Samples of various plants and formulated diets were collected once a week and later pooled by week. Subsequently, all feed samples were dried at 65°C for 72 h, milled to pass through a 1-mm mesh screen, then stored in sealed bags (150 mm × 220 mm) at 4°C until chemical composition was determined.
Ruminal fluid samples were collected on the final day of the experiment before the morning feeding via an oral stomach tube equipped with a vacuum pump. Approximately 10 mL of ruminal fluid initially collected was discarded to reduce the chance of contamination of the fluid sample in the stomach tube with saliva. After that, 4 layers of cheesecloth were used to strain the ruminal fluid, and the pH was measured immediately. Then, 10 mL of the filtrate is acidified by mixing with 2 mL of metaphosphoric acid (25%, wt/vol) and the mixture was centrifuged at 3,000 × g for 15 min. The supernatant was separated and stored at −20°C until volatile fatty acids (VFA) was determined. Another 4 mL of filtrate was immediately stored in liquid nitrogen until the microbial community was analyzed.
2.3 Analysis of the nutritional composition of feeds
Samples of each plant and diets were sent to the Animal Nutrition Laboratory of Northeast Agricultural University (Harbin, China) for nutrient analysis using wet chemistry methods. Contents of the dry matter (DM, method 934.01), ash (method 942.05), ether-extract (EE, method 920.39) and crude protein (CP, method 988.05) in feeds were assayed in accordance with the procedures of AOAC International (26). The neutral detergent fiber (NDF) and acid detergent fiber (ADF) contents were measured based on the method described by previous study (27), which the heat-stable α-amylase was used to treat the feeds. The mineral element content was measured using an inductively coupled plasma-optical emission spectrometer (ICP-6800S, Shanghai Meixi Instrument Co., Ltd., China).
2.4 Analysis of ruminal fermentation parameters and microbial community
Concentrations of acetate and propionate in the rumen fluid samples were determined using gas chromatography (Shimadzu GC-2010, Japan) (28).
Total DNA in the rumen fluid was extracted using a DNA extraction kit (Shanghai Shengye Biotech, China). The V3 and V4 region of bacteria and archaea was used for 16S rRNA gene sequencing while the fungus was used for 18S rRNA gene sequencing, with all amplicon libraries preparation and sequencing performed on a MiSeq platform (Illumina, San Diego, CA, United States). The fluorescent quantitative polymerase chain reaction (PCR) amplification reaction conditions were pre-denaturation 98°C (30 s); denaturation 98°C (15 s), annealing temperature 50°C (30 s); extension temperature 72°C (30 s), 30 cycles, and final extension 72°C (5 min). After being recycled from a 1.8% agarose gel, the PCR products were purified using an OMEGA DNA purification column (Gene Company Limited). The purified products were quantified using a Quant-iT PicoGreen dsDNA Assay Kit (Gene Company Limited) in accordance with the kit’s instructions. The DNA fragments were paired-end sequenced by an Illumina miseq/novaseq. The DADA2 method was used to perform the steps of de-priming, mass filtering, denoising, splicing, and de-chimerization. Sequencing data were processed using MacQIIME version 1.9.1. Primers and homopolymer runs (maximum length, 8) of the joined sequences were trimmed after paired-end forward and reverse reads were joined. Only sequences ≥400 bp in length, with both an average quality score ≥ 25 and ambiguous bases ≤6 remained for downstream analysis. UCHIME (29) was used for de novo chimera checking, and USEARCH (30) was used for operational taxonomic unit (OTU) identification in order to identify similar sequences that had >97% similarity. BLAST (31) was used to assign representative sequences for bacterial and archaeal OTUs to the Greengenes16S rRNA gene database [version gg_13_8; (32)] and RIM-DB database (33), respectively.
2.5 In vitro rumen fermentation
The in vitro fermentation device adopted in this study was the artificial rumen simulating system MC-ABSF-II (Beijing Mancang Technology Co., Ltd., China). The protocol used for the in vitro incubation was described by Li et al. (34). Sheep rumen fluid was collected prior to morning feeding, filtered through four layers of cheesecloth, and combined with artificial saliva (39°C) at a 2:1 ratio (buffer: ruminal fluid, v: v) (35). Dispensing the 150 mL of buffered ruminal fluid into 200 mL incubation flasks that had been preheated. In each incubation flask, 2 g of each substrate were mixed with the buffered ruminal fluid. The mixture was then incubated for 24 h at 39°C in a hot water bath shaker. Using a real-time in vitro fermentation system (made by Jilin Academy of Agricultural Sciences, model Qtfxy-6), the milliliter (mL) of CH4 from each flask was monitored.
2.6 Statistical analysis
R Statistical Software (v4.1.2; (36)) was used to analyze all of the experiment’s data. One-way ANOVA (LSD) were used to analyze the effects of grassland degradation on nutrient intake, rumen fermentation parameters and methane emission of sheep. To analyze the effects of nutrient intake to rumen microbial relative abundance, we used linear mixed effects model (LMMs). In this model, nutrient intakes were taken as fixed factors and grassland types (undegraded grassland plants, moderately degraded grassland plants and severely degraded grassland plants) were taken as random factors. Specifically, the equation of model is Yᵢj = Xᵢjβ + Zᵢjμi + ϵᵢj, where Yᵢj is the observed response for the j-th observation within group i, Xᵢj is a vector of covariates associated with the fixed effects for the j-th observation in group i, β is the population mean which represents fixed-effect factors, Zᵢj is a vector of variables associated with the random effects for the j-th observation in group i, μi is intercepts relative to population mean across different groups which means variability between groups, ϵᵢj is the error term for the j-th observation in group i representing the unexplained variability within the group. Statistically significant differences among treatments were analyzed using Tukey’s test. Significant differences were declared at p ≤ 0.05, and marginally significant differences were defined at 0.05 < p ≤ 0.10.
3 Results
3.1 Growth performance and rumen fermentation
The results (Figure 2a) for in vitro fermentation showed that the CH4 production in the SG group was higher than that of UG group (p < 0.05). Conversely, the sheep from MG group produced significantly less CH4 than that of UG group (p < 0.05; Figure 2a). However, no difference for H2 production was observed among the three groups (p > 0.10), indicating variation in carbon was driving differences. In addition, we further observed that ratio of acetate concentrations to propionate concentrations, ruminal total volatile fatty acids and acetate concentrations in the SG group was significantly decreased, but their levels significantly increased in MG group compared with the UG group (p < 0.01; Figure 2c). However, the pH (Figure 2b) and propionate concentration (Figure 2c) in the rumen were not altered among the three groups (p > 0.10). Similarly, we did not observe any differences in the average daily gain of sheep among the groups (Table 1).
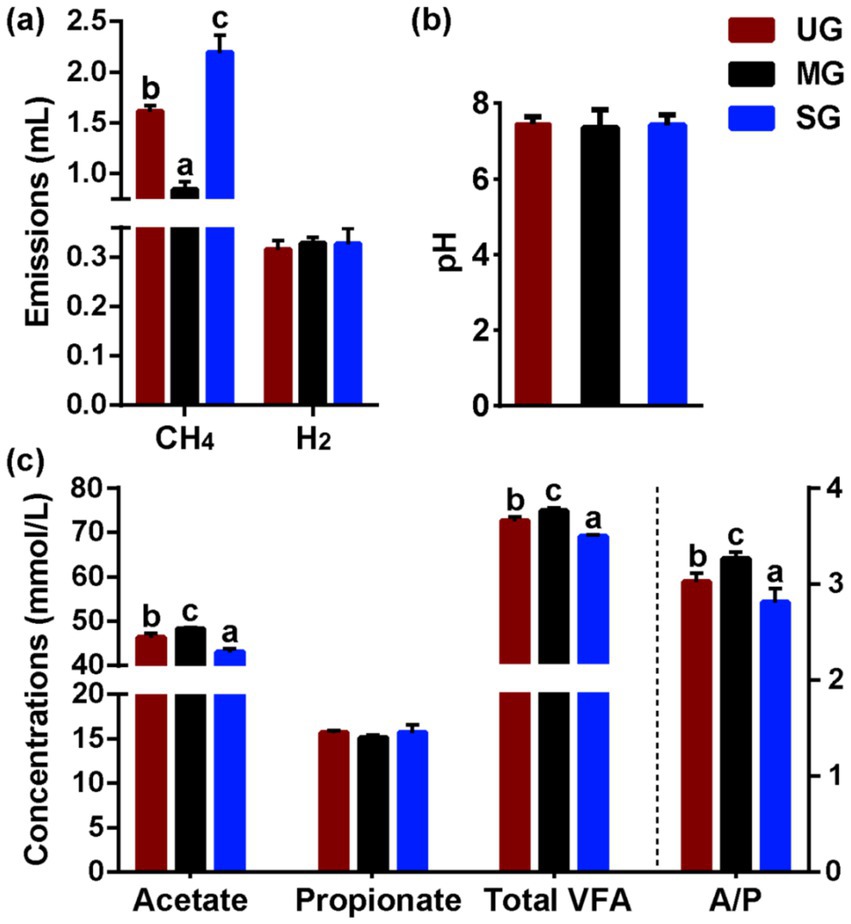
Figure 2. Rumen fermentation parameters in sheep fed diets simulating different levels of grassland saline-alkaline degradation. (a) methane (CH4) and hydrogen (H2) emissions in vitro rumen experiment; (b,c) pH and volatile fatty acids (VFA) profile in vivo feeding experiment; UG undegraded grassland, MG moderately degraded grassland, SG severely degraded grassland, A/P ratio of acetate to propionate; values with different letters indicate significant differences (p < 0.05).
3.2 Variation in rumen microbial community
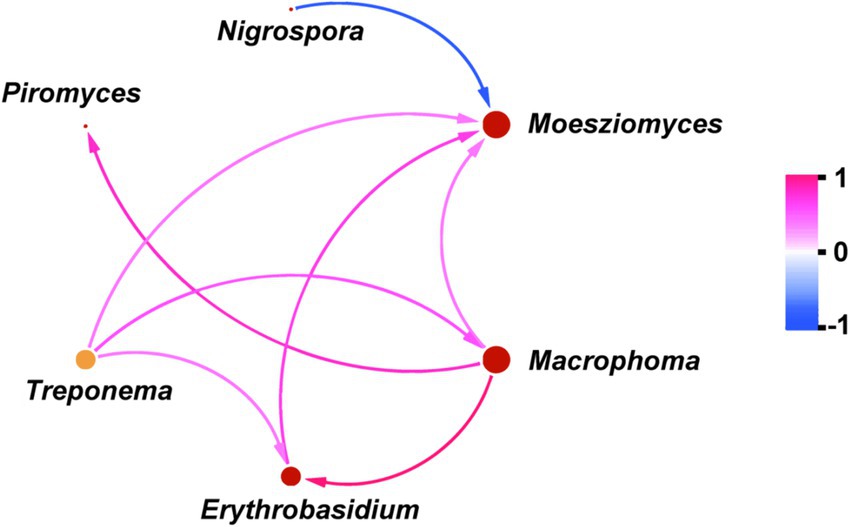
We next focused particularly on the variation in microbes involved in the competition for carbon nutrition. Notably, we observed the relative abundance of Treproema that capable of converting carbon nutrition into acetic acid in the rumen of sheep in the MG group was significantly higher than that of sheep in the UG and SG groups (p < 0.05; Figure 3a). In addition, sheep in the SG group had higher ruminal Methanosphaera, which is a crucial role in the methane production by using carbon nutrition in relative abundance compared with the UG and MG groups (p < 0.05; Figure 3b). Among the different fungal genera, the relative abundance of Macrophoma, Moesziomyces and Erythrobasidium in the MG group increased significantly compared with the UG and SG groups, whereas the relative abundance of Nigrospora in sheep fed MG and SG diets decreased significantly compared with those sheep fed UG diet (p < 0.05; Figure 3c). Meanwhile, the Piromyces in relative abundance increased linearly with increasing levels of grassland saline-alkaline degradation (UG vs. MG vs. SG, all p < 0.05).
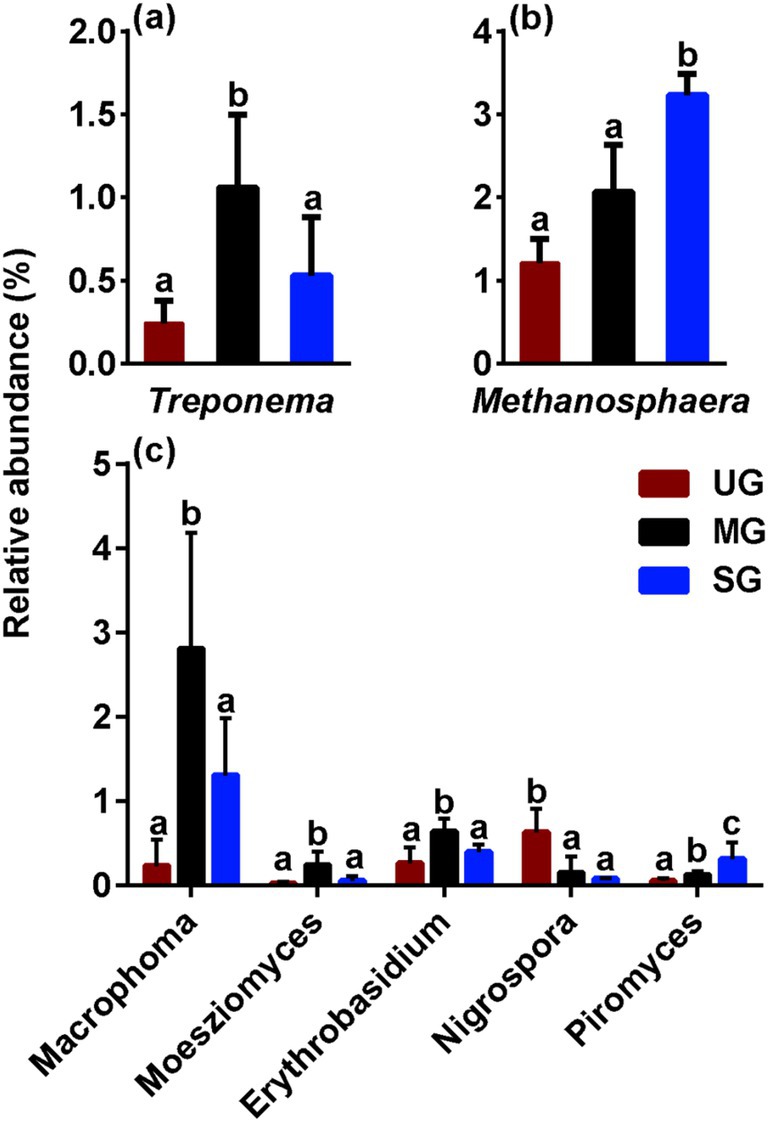
Figure 3. The relative abundance of key rumen bacterial (a), archaeal (b), and fungal (c) genus in sheep fed diets simulating different levels of grassland saline-alkaline degradation. UG undegraded grassland, MG moderately degraded grassland, SG severely degraded grassland; values with different letters indicate significant differences (p < 0.05).
3.3 Correlation among various key microbes
To investigate the underlying causes of the observed variation in carbon-nutrient competing microbes, we firstly conducted an analysis of the microbial network structure. Figure 4 showed that the Moesziomyces play the most important role among these key microbes, as it not only had a positive effect on Macrophoma, Erythrobasidium and Treproema, but also had a negative effect on Nigrospora (p < 0.05). Meanwhile, the Macrophoma and Erythrobasidium also had a positive effect on Treproema (p < 0.05). However, there was no significant effect of Nigrospora and Erythrobasidium on the Treproema (p > 0.10). Furthermore, our observations revealed that Methanosphaera was not integrated into the microbial network diagram, suggesting a potential insusceptibility of this genus to microbial interaction effects.
3.4 Nutrient intake
We further analyzed the significance of differences in nutrient intake among different groups of sheep. We found no significant differences in DM intake (Figure 5a)among the groups of sheep (p > 0.05), but the CP (Figure 5b) and EE (Figure 5c) intake of sheep in the SG group was higher (p < 0.05). Additionally, there were no significant differences in NDF intake (Figure 5d) among the groups of sheep (p > 0.05), while the ADF (Figure 5e) intake of sheep in the SG group was lower (p < 0.05). Notably, the Na intake (Figure 5f) of sheep increased linearly with the increasing levels of grassland saline-alkaline degradation (UG vs. MG vs. SG, all p < 0.05).
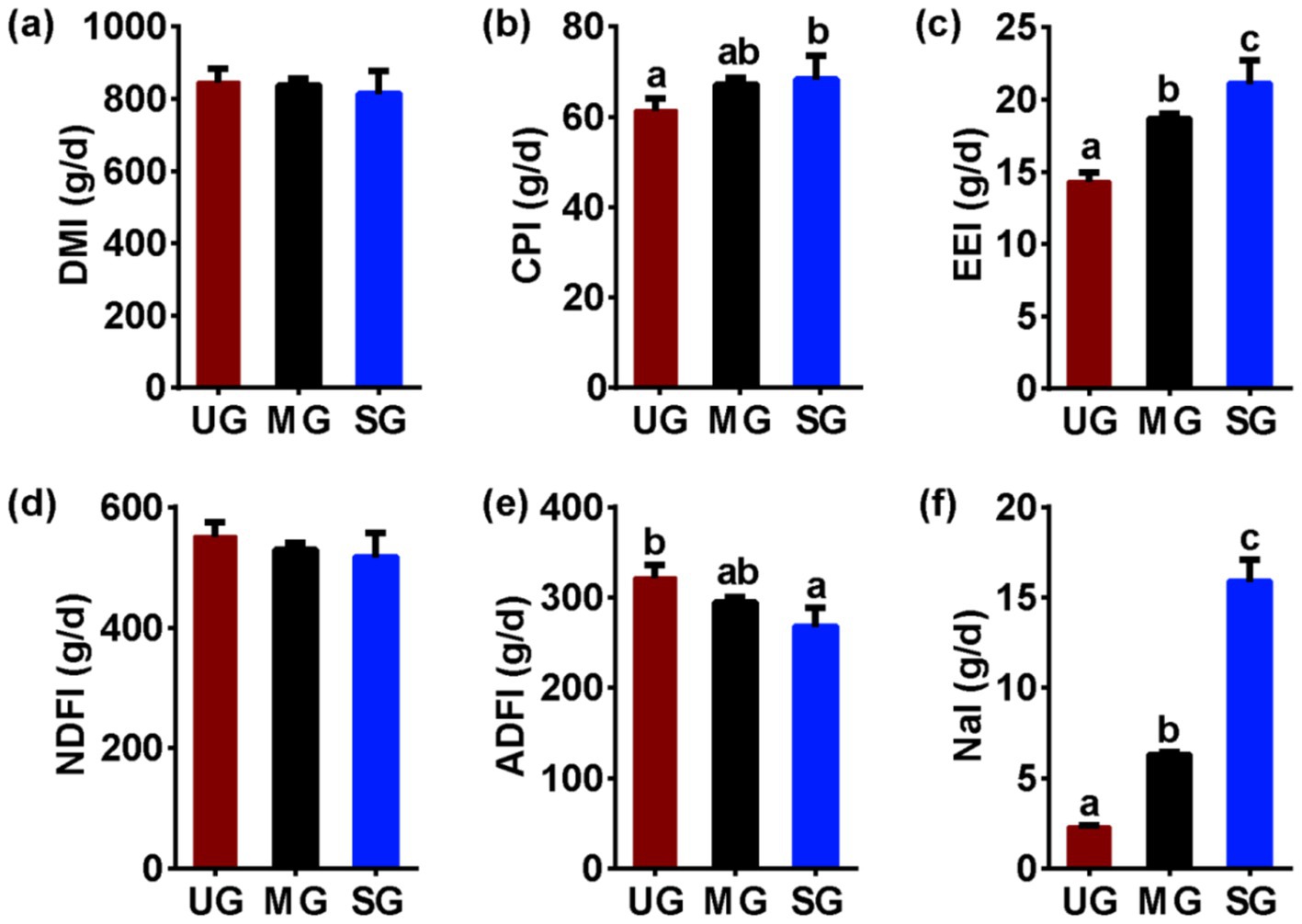
Figure 5. The nutrient intake in sheep fed diets simulating different levels of grassland saline-alkaline degradation. UG undegraded grassland, MG moderately degraded grassland, SG severely degraded grassland, DMI dry matter intake (Figure 5a), CPI, crude protein intake (Figure 5b), EEI ethyl ether extract intake (Figure 5c), NDFI neutral detergent fiber intake (Figure 5d), ADFI acid detergent fiber intake (Figure 5e), NaI Na intake (Figure 5f). values with different letters indicate significant differences (p < 0.05).
3.5 The effects of nutrient intake on Methanosphaera in the rumen of sheep
To identify the nutritional factors most likely to influence the variation in Methanosphaera, we next focused on analyzing the correlation between the nutritional resources consumed by those sheep with significant differences and Methanosphaera (Table 2). We found a significant effect of Na intake on Methanosphaera relative abundance (p < 0.01), but no significant effect of other nutrient intake on the Methanosphaera relative abundance was observed.
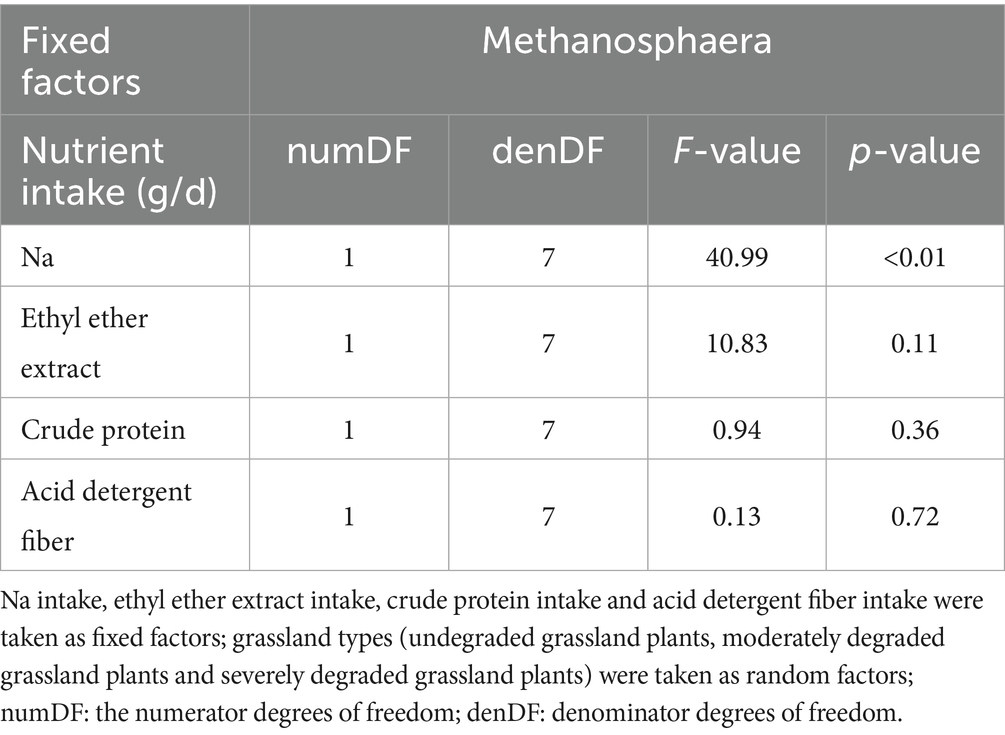
Table 2. Summary of linear mixed effects models analyzing the effects of nutrient intake to Methanosphaera relative abundance in the rumen.
4 Discussion
Our study is the first to evaluate the impact of grassland saline-alkaline degradation on herbivore CH4 emissions through feeding small-tailed sheep diets that simulated forage availability in grasslands with different levels of degradation. Our results found that the intake of moderately degraded grassland plants decreased CH4 emission in the rumen of sheep, however, the CH4 emission was higher in sheep that consumed severely degraded grassland plants (Figure 2a; UG: 1.61 mL vs. MG: 0.84 mL vs. SG: 2.19 mL). Moreover, we also further analyzed the rumen microbes, rumen fermentation parameters, and nutrient intake in sheep fed with different degraded grassland plants. These results provide a profound insight into how grassland saline-alkaline degradation affects CH4 emissions from sheep.
To our knowledge, the primary mechanism of the ruminal CH4 synthesis is the hydrogenotrophic pathway, which accounts for approximately 78% of total CH4 production (37). The abundance of methanogens and acetogenic bacteria are two important factors affecting this pathway in the rumen (10). In this pathway, methanogens can utilize H2 and CO2 to generate CH4, while acetogenic bacteria can also use the H2 to synthesize acetate (38). Thus, when the abundance of one of the microbe changes, the metabolic pathway of H2 shifts due to competition for substrates, which in turn affects CH4 production in the rumen. Our results showed that sheep fed diets simulating severely degraded grassland exhibited a notable increase in the abundance of Methanosphaera (Figure 3b), which has been proven to be a key methanogen in the rumen (13). Hence, the increased Methanosphaera in relative abundance may contribute to enhanced CH4 production via shifting the H2 toward methanogenesis pathway. Furthermore, our results also found that the relative abundance of Treproema in the rumen of sheep in the MG group was significantly higher than that of sheep in the UG and SG groups (Figure 3a). Further correlation analyses among various key microbes indicated that the increased relative abundance of Moesziomyces in the rumen of sheep fed moderately degraded grassland plants mainly driven the increasing in the relative abundance of Treproema (Figures 3c, 4). This phenomenon may be explained by the microbial food web theory proposed by Mizrahi et al. (39), who found that microbial communities undergo cascading metabolism in the rumen in a complex and coordinated manner, with continuous, cross-feeding relationships between different rumen microbes across the food web. Indeed, previous study demonstrated that Moesziomyces has the ability to degrade various plant polysaccharides by secreting plant cell wall-degrading enzymes (40). Although research on Moesziomyces in the ruminal environments remain limited to date, our findings suggest that this genus may contribute to provide more available nutrients for the Treproema to metabolize at the next trophic-like level. More importantly, previous research has recognized the Treproema is an acetogenic bacteria in the rumen, which is able to compete with methanogens for the utilization of H2 to produce acetate (41). Therefore, the increased Treproema in relative abundance may contribute to the decreased CH4 (Figure 2a) production and increased acetate concentration (Figure 3c) in the rumen of sheep fed moderately degraded grassland plants.
Evidence from experimental study has demonstrated that the rumen microbes were mainly affected by diet nutrient profiles (15), and therefore, grassland saline-alkaline degradation-induced alterations in plant resources in this study may be a key factor in causing an altered rumen microflora in sheep as described above, thereby further influencing CH4 emission. Our results found that as the level of grassland saline-alkaline degradation increased, the intake of ADF in sheep significantly decreased (Figure 5e), whereas the intake of EE significantly increased (Figure 5c). The decrease in ADF intake is often considered to reduce CH4 production in the rumen by reducing the abundance of fibrinolytic bacteria, which are major contributor for producing H2 for methanogens to synthesize CH4 (10, 42, 43). However, we found that the relative abundance of fibrinolytic bacteria (Figures 3a,c) and H2 production (Figure 2a) in the rumen of the SG group did not decrease. This observation may be attributed to the smaller changes in fiber intake between groups as compared to other studies (44, 45), suggesting that the alterations might not be substantial enough to induce variations in H2 content in the rumen. Moreover, although some fatty acids can reduce CH4 production by suppressing the number of methanogens and reducing H2 concentration through unsaturated bonds (21), the H2 content (Figure 2a) and abundance of methanogens (Figure 3b) also did not decrease with increasing EE intake in sheep. This phenomenon may be attributed to saline-alkaline grassland plants that lack specific fatty acids that reduce CH4 release from grazing animals. Consequently, we believe that grassland saline-alkaline degradation-induced alterations in ADF and EE intake was not the key factor affecting CH4 emissions in sheep. This opinion was further confirmed by the results of the linear mixed effects modeling analysis in this study, which found that there was no significant effect of ADF and EE intake on the relative abundance of Methanosphaera (Table 2).
Interestingly, we are the first to find that excessive intake of Na might be the reason for increases in abundance of Methanosphaera (Table 2), thereby increasing the CH4 emissions of sheep fed severely degraded grassland plants. Despite previous studies have not focused on the association between Na intake and rumen methanogens, published research has found that methanogenic archaea primarily inhabited areas with higher concentrations of Na (46). Similarly, adenosine triphosphate synthase inhibitors can only decrease methanogenesis under low Na concentrations (47). These studies suggested that Methanosphaera may prefer saline environments. Therefore, an increase in Na intake may be a new pathway to promote the proliferation of methanogens for CH4 production. Our other important finding is that the plant species diversity and functional diversity in moderately degraded grassland was higher than that in undegraded and severely degraded grasslands (Figure 1). An interesting study demonstrated that the provision of a more diverse nutrient source (nine polysaccharides: starch, mucin, galactan, pectin, arabinogalactan, hemicellulose, cellulose, hyaluronan, and chondroitin sulfate) to microbes enables these species to coexist in high abundance by improving the mutual complementarity of their nutritional preferences (48). Hence, more diverse nutrient availability in moderately degraded grasslands, especially fiber, may contribute to increased relative abundance of Moesziomyces, which in turn may further increase the relative abundance of Treproema, thereby reducing the CH4 production via shifting the H2 toward acetate synthesis pathway. In general, our results suggested that changes in Na content and plant diversity induced by grassland saline-alkaline degradation may be the major contributors to influencing CH4 emissions of sheep by altering their rumen microbial community (Figure 6), which is worth further exploration.
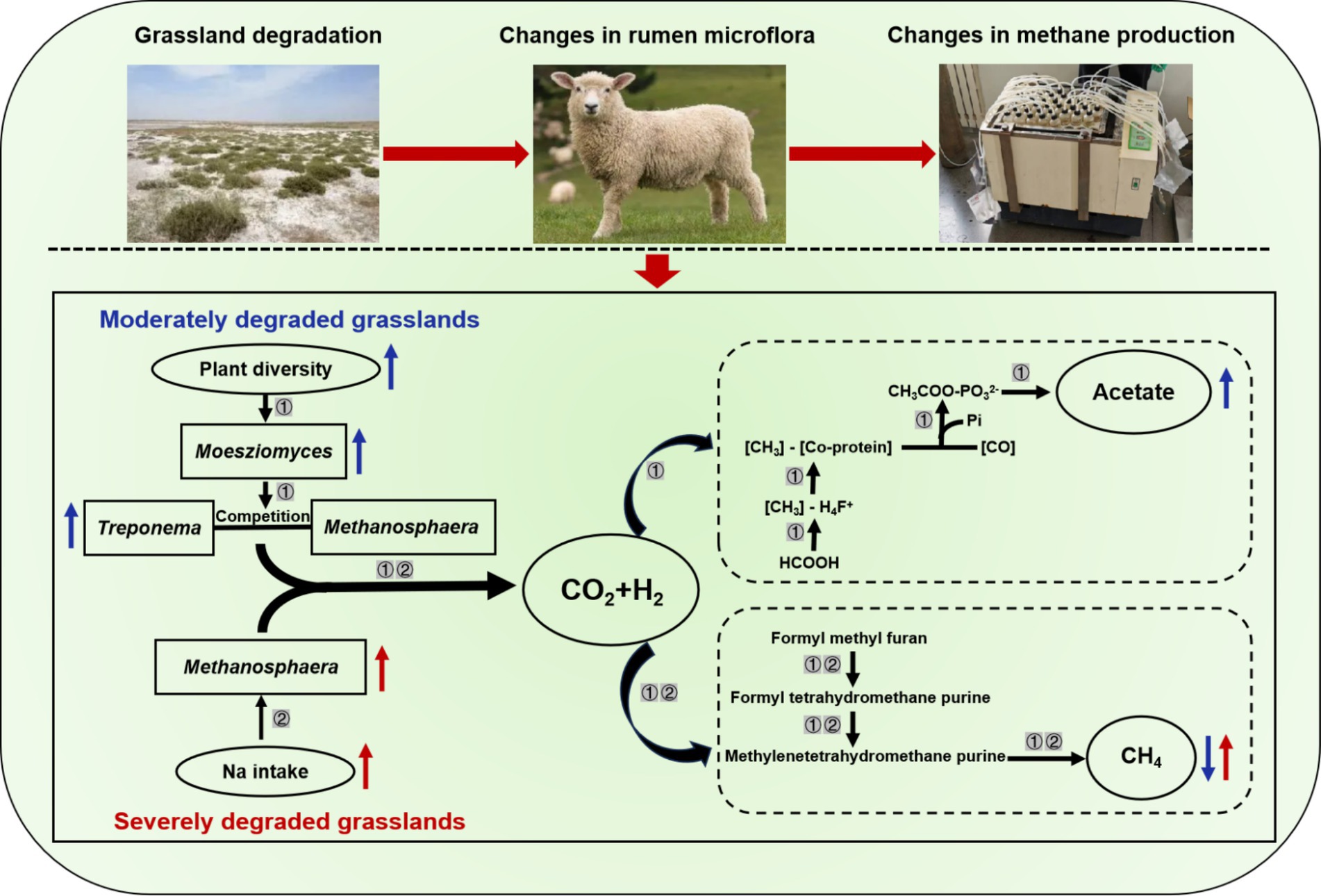
Figure 6. The underlying mechanisms of grassland saline-alkaline degradation affecting methane emissions of sheep. ① represents the pathway of influence of moderately degraded grasslands on methane emissions from sheep; ② represents the pathway of influence of severely degraded grasslands on methane emissions from sheep; and
represents the related parameter of sheep fed diets simulating moderately degraded grasslands was higher and lower (p < 0.05) than that sheep fed diets simulating undegraded grasslands, respectively;
represents the related parameter of sheep fed diets simulating severely degraded grasslands was higher (p < 0.05) than that sheep fed diets simulating undegraded grasslands.
5 Conclusion
Sheep that consume severely degraded grassland plants with excessive Na content experience an increase in the relative abundance of ruminal Methanosphaera. This shift leads to a redirection of H2 utilization toward CH4 production pathways. However, moderately degraded grassland plants increase the relative abundance of ruminal Treproema in sheep, which could inhibit the formation of CH4 via shifting the H2 toward acetate synthesis pathway. Overall, our results highlight that Na to be an important factor influencing ruminant CH4 emissions and also implicate that plant diversity as a possible another factor influencing CH4 emissions, but need to be further explored. Our study provides experimental evidence that livestock grazing on severely degraded saline-alkaline grasslands may exacerbate the adverse environmental effects of grassland degradation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Experimental Animal Welfare and Ethics Committee of Northeast Normal University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YW: Formal analysis, Investigation, Writing – original draft. XJ: Writing – review & editing, Formal analysis. GM: Writing – review & editing, Investigation. YS: Writing – review & editing, Investigation. XW: Investigation, Writing – review & editing. HS: Resources, Writing – review & editing. YL: Methodology, Writing – review & editing. LW: Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Nos. 32271642 and 32301497), the National Key Research and Development Program of China (2023YFD1301700 and 2022YFF1300604), the Program for Introducing Talents to Universities (B16011), and the Ministry of Education Innovation Team Development Plan, Grant/Award Number: 2013–373, the Fundamental Research Funds for the Central Universities (2412022QD025).
Acknowledgments
We thank the Hongjian Xu and Guangning Zhang of Northeast Agricultural University for their help with animal care. We also would like to thank Joseph Elliot at the University of Kansas for her assistance with the English language and grammatical editing of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1598973/full#supplementary-material
References
1. Roques, S, Martinez-Fernandez, G, Ramayo-Caldas, Y, Popova, M, Denman, S, Meale, SJ, et al. Recent advances in enteric methane mitigation and the Long road to sustainable ruminant production. Annu Rev Anim Biosci. (2024) 12:321–43. doi: 10.1146/annurev-animal-021022-024931
2. Mizrahi, I, and Jami, E. Review: the compositional variation of the rumen microbiome and its effect on host performance and methane emission. Animal. (2018) 12:s220–32. doi: 10.1017/S1751731118001957
3. Herrero, M, Havlík, P, Valin, H, Notenbaert, A, Rufino, MC, Thornton, PK, et al. Biomass use, production, feed efficiencies, and greenhouse gas emissions from global livestock systems. Proc Natl Acad Sci USA. (2013) 110:20888–93. doi: 10.1073/pnas.1308149110
4. Herrero, M, Henderson, B, Havlík, P, Thornton, PK, Conant, RT, Smith, P, et al. Greenhouse gas mitigation potentials in the livestock sector. Nat Clim Chang. (2016) 6:452–61. doi: 10.1038/nclimate2925
5. Hristov, AN, Oh, J, Firkins, JL, Dijkstra, J, Kebreab, E, Waghorn, G, et al. Special topics—mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J Anim Sci. (2013) 91:5045–69. doi: 10.2527/jas.2013-6583
6. Bardgett, RD, Bullock, JM, Lavorel, S, Manning, P, and Shi, H. Combatting global grassland degradation. Nat Rev Earth Environ. (2021) 2:720–35. doi: 10.1038/s43017-021-00207-2
7. Dierickx, WR. The salinity and alkalinity status of arid and semi-arid lands In: Encyclopedia of land use, land cover and soil sciences In: HV Willy, Editor. Encyclopedia of land use, land cover and soil sciences. EOLSS Publishers Co (2009). 5:163–89.
8. Jiang, X, and Wang, L. Grassland-based ruminant farming systems in China: potential, challenges and a way forward. Anim Nutr. (2022) 10:243–8. doi: 10.1016/j.aninu.2022.04.007
9. Shabat, SKB, Sasson, G, Doron-Faigenboim, A, Durman, T, Yaacoby, S, Berg Miller, ME, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. (2016) 10:2958–72. doi: 10.1038/ismej.2016.62
10. Li, QS, Wang, R, Ma, ZY, Zhang, XM, Jiao, JZ, Zhang, ZG, et al. Dietary selection of metabolically distinct microorganisms drives hydrogen metabolism in ruminants. ISME J. (2022) 16:2535–46. doi: 10.1038/s41396-022-01294-9
11. Janssen, PH, and Kirs, M. Structure of the archaeal community of the rumen. Appl Environ Microbiol. (2008) 74:3619–25. doi: 10.1128/AEM.02812-07
12. Nagaraja, TG. Microbiology of the rumen. Rumenology. (2016) 12:39–61. doi: 10.1007/978-3-319-30533-2_2
13. Nollet, L, Vande, VI, and Verstraete, W. Effect of the addition of Peptostreptococcus productus ATCC35244 on the gastro-intestinal microbiota and its activity, as simulated in an in vitro simulator of the human gastro-intestinal tract. Appl Microbiol Biotechnol. (1997) 48:99–104. doi: 10.1007/s002530051022
14. Solomon, T, Gupta, V, and Ncho, CM. Balancing livestock environmental footprints with forestry-based solutions: a review. Ecol. (2023) 4:714–30. doi: 10.3390/ecologies4040047
15. Morgavi, DP, Forano, E, Martin, C, and Newbold, CJ. Microbial ecosystem and methanogenesis in ruminants. Animal. (2010) 4:1024–36. doi: 10.1017/S1751731110000546
16. Chang, J, Peng, S, Ciais, P, Saunois, M, Dangal, SRS, Herrero, M, et al. Revisiting enteric methane emissions from domestic ruminants and their δ13cCH4 source signature. Nat Commun. (2019) 10:3420. doi: 10.1038/s41467-019-11066-3
17. Folks, DJ, Gann, K, Fulbright, TE, Hewitt, DG, DeYoung, CA, Wester, DB, et al. Drought but not population density influences dietary niche breadth in white–tailed deer in a semiarid environment. Ecosphere. (2014) 5:1–15. doi: 10.1890/ES14-00196.1
18. Martin, C, Morgavi, DP, and Doreau, M. Methane mitigation in ruminants: from microbe to the farm scale. Animal. (2010) 4:351–65. doi: 10.1017/S1751731109990620
19. Sun, H. X., Liu, C. L., and Ch, S. L. (2008). Distribution of crude fat for a halophyte (Suaeda glauca) growing in the Songnen grassland. Proceedings of the World Grassland and Grassland Conference
20. Niu, M, Kebreab, E, Hristov, AN, Oh, J, Arndt, C, Bannink, A, et al. Prediction of enteric methane production, yield, and intensity in dairy cattle using an intercontinental database. Glob Chang Biol. (2018) 24:3368–89. doi: 10.1111/gcb.14094
21. Rasmussen, J, and Harrison, A. The benefits of supplementary fat in feed rations for ruminants with particular focus on reducing levels of methane production. Int Sch Res Notices. (2011) 2011:613172. doi: 10.5402/2011/613172
22. Cherdthong, A, Wanapat, M, and Wachirapakorn, C. Effects of urea-calcium mixture in concentrate containing high cassava chip on feed intake, rumen fermentation and performance of lactating dairy cows fed on rice straw. Livest Sci. (2011) 136:76–84. doi: 10.1016/j.livsci.2010.08.002
23. Mayberry Ayberry, D, Masters, DG, and Vercoe, P. Mineral metabolism of sheep fed saltbush or a formulated high-salt diet. Small Rumin Res. (2010) 91:81–6. doi: 10.1016/j.smallrumres.2009.10.020
24. Sahoo, B, Saraswat, ML, Haque, N, and Khan, MY. Energy balance and methane production in sheep fed chemically treated wheat straw. Small Ruminant Res. (1999) 35:13–19.
25. Zhou, C, Xia, H, Yang, T, Zhang, Z, and Zheng, G. Grassland degradation affected vegetation carbon density but not soil carbon density. BMC Plant Biol. (2024) 24:719. doi: 10.1186/s12870-024-05409-6
26. AOAC International. Official methods of analysis. 17th ed. Arlington, VA: Off. Anal. Chem (2000).
27. Soest, PJV, Robertson, JB, and Lewis, BA. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J Dairy Sci. (1991) 74:3583–97. doi: 10.3168/jds.S0022-0302(91)78551-2
28. Erwin, ES, Marco, GJ, and Emery, EM. Volatile fatty acid analyses of blood and rumen fluid by gas chromatography. J Dairy Sci. (1961) 44:1768–71. doi: 10.3168/jds.S0022-0302(61)89956-6
29. Edgar, RC, Haas, BJ, Clemente, JC, Quince, C, and Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
30. Edgar, RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
31. Altschul, SF, Gish, W, Miller, W, Myers, E, and Lipman, D. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
32. McDonald, D, Price, MN, Goodrich, J, Nawrocki, EP, DeSantis, TZ, Probst, A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. (2012) 6:610–8. doi: 10.1038/ismej.2011.139
33. Seedorf, H, Kittelmann, S, Henderson, G, and Janssen, PH. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J. (2014) 2:e494. doi: 10.7717/peerj.494
34. Li, Y, Lv, JY, Wang, JH, Zhou, S, Zhang, G, Wei, B, et al. Changes in carbohydrate composition in fermented total mixed ration and its effects on in vitro methane production and microbiome. Front Microbiol. (2021) 12:738334. doi: 10.3389/fmicb.2021.738334
35. Menke, HH, and Steingass, H. Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim Res Dev. (1988) 28:7–55.
36. R Core Team, 2021. A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria. (2001). Available from: http://www.R-project.org
37. Seshadri, R, Leahy, SC, Attwood, GT, Teh, KH, Lambie, SC, Cookson, AL, et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat Biotechnol. (2018) 36:359–67. doi: 10.1038/nbt.4110
38. Greening, C, Geier, R, Wang, C, Woods, LC, Morales, SE, McDonald, MJ, et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. (2019) 13:2617–32. doi: 10.1038/s41396-019-0464-2
39. Mizrahi, I, Wallace, RJ, and Moraïs, S. The rumen microbiome: balancing food security and environmental impacts. Nat Rev Microbiol. (2021) 19:553–66. doi: 10.1038/s41579-021-00543-6
40. Soria, NW, Figueroa, AC, Díaz, MS, Alasino, RV, Yang, P, and Beltramo, DM. Identification and expression of some plant cell wall-degrading enzymes present in three ontogenetics stages of Thecaphora frezii, a peanut (Arachis hypogaea L.) pathogenic fungus. Am J Plant Sci. (2022) 13:1–22. doi: 10.4236/ajps.2022.131001
41. Leadbetter, JR, Schmidt, TM, Graber, JR, and Breznak, JA. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science. (1999) 283:686–9. doi: 10.1126/science.283.5402.686
42. Liu, Y, and Whitman, WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci. (2008) 1125:171–89. doi: 10.1196/annals.1419.019
43. Moraes, LE, Strathe, AB, Fadel, JG, Casper, DP, and Kebreab, E. Prediction of enteric methane emissions from cattle. Glob Change Biol. (2014) 20:2140–8. doi: 10.1111/gcb.12471
44. Wang, K, Nan, XM, Chu, KK, Tong, J, Yang, L, Zheng, S, et al. Shifts of hydrogen metabolism from methanogenesis to propionate production in response to replacement of forage fiber with non-forage fiber sources in diets in vitro. Front Microbiol. (2018) 9:2764–76. doi: 10.3389/fmicb.2018.02764
45. Zhang, J, Shi, H, Wang, Y, and Ji, S. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front Microbiol. (2017) 8:293131. doi: 10.3389/fmicb.2017.02206
46. Friedrich, MW, Schmitt-Wagner, D, Lueders, T, and Brune, A. Axial differences in community structure of Crenarchaeota and Curyarchaeota in the highly compartmentalized gut of the soil-feeding termite cubitermes orthognathus. Appl Environ Microbiol. (2001) 67:4880–90. doi: 10.1128/AEM.67.10.4880-4890.2001
47. De Vrieze, J, Hennebel, T, Boon, N, and Verstraete, W. Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol. (2012) 112:1–9. doi: 10.1016/j.biortech.2012.02.079
Keywords: grassland degradation, global warming, herbivore grazing, methane emissions, rumen microbes
Citation: Wang Y, Jiang X, Ma G, Sun Y, Wang X, Sun H, Li Y and Wang L (2025) Impact of grassland saline-alkaline degradation on domestic herbivore rumen microbiota and methane emissions. Front. Vet. Sci. 12:1598973. doi: 10.3389/fvets.2025.1598973
Edited by:
Yafeng Huang, Anhui Agricultural University, ChinaReviewed by:
Edward James Raynor, Colorado State University, United StatesQingbiao Xu, Huazhong Agricultural University, China
Copyright © 2025 Wang, Jiang, Ma, Sun, Wang, Sun, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Jiang, amlhbmd4NzEwQG5lbnUuZWR1LmNu; Ling Wang, d2FuZ2w4OTBAbmVudS5lZHUuY24=
 Yizhen Wang
Yizhen Wang Xin Jiang1*
Xin Jiang1*