- 1Department of Pathobiology (Microbiology Section), University of Veterinary and Animal Sciences, Lahore, Pakistan
- 2Department of Pathobiology (Pathology Section), University of Veterinary and Animal Sciences, Lahore, Pakistan
- 3Department of Pathobiology (Parasitology Section), University of Veterinary and Animal Sciences, Lahore, Pakistan
- 4Institute of Bacterial Infections and Zoonoses, Friedrich-Loeffler-Institut, Jena, Germany
Colibacillosis associated with colistin-resistant avian pathogenic Escherichia coli (E. coli) poses a threat to both food security and public health. The potential horizontal transmission of mobilized colistin-resistant (mcr) genes facilitates the co-emergence of Klebsiella pneumoniae. This study aimed to determine the prevalence, molecular detection, analyze the antibiogram and identify associated risk factors for colistin-resistant E. coli and Klebsiella pneumoniae isolated from broiler chicken in three districts of Punjab province, Pakistan. In total, 230 visceral organ samples were collected from 13 different chicken farms located in Sargodha, Jhang and Toba Tek Singh in Pakistan. Following isolation, the broth microdilution test was used to confirm phenotypic colistin resistance. Polymerase chain reaction was used to detect mcr-1 and mcr-2 genes associated with colistin resistance. Antimicrobial susceptibility test against 11 antibiotics was performed using the Kirby-Bauer disk diffusion method. Risk factors associated with colistin-resistant bacteria, including host attributes, farm management practices, environmental and agent characteristics, were analyzed. The prevalence of colistin-resistant E. coli and K. pneumoniae was 24.78% (95% CI, 19.6–30.7%) and 3.04% (95% CI, 1.5–6.1%), respectively. The prevalence of colistin-resistant E. coli varied between cities at 42, 23.61 and 5.55% for Jhang, Sargodha and Toba Tek Singh, respectively. The detection frequency of mcr-1 gene, 42.1% (24/57), was significantly (p < 0.01) higher than that of the mcr-2 gene, 14.03% (8/57). Phylogenetic analysis of lipid A phosphoethanolamine transferase sequences revealed greater similarity with mcr-1.5 variant. Isolates were found resistant to amoxicillin-clavulanic acid (84.21%), cefotaxime (70.17%), and trimethoprim-sulfamethoxazole (73.68%). The multivariate logistic regression predicted preceding viral infection of the respiratory tract as a significant association (OR = 4.808, p < 0.01), whereas daily removal/culling of dead/diseased chicken (OR = 0.308, p = 0.01) was a protective factor against the emergence of colistin-resistant strains. These findings indicate that the emergence of colistin-resistant strains deteriorate colibacillosis control efforts in poultry and serves as a possible reservoir for zoonotic infections.
1 Introduction
Avian colibacillosis is an infectious disease in chickens caused by avian pathogenic Escherichia coli (E. coli) (APEC). Colibacillosis is characterized by multisystemic expression of lesions including airsacculitis, perihepatitis, pericarditis, salpingitis, peritonitis, cellulitis, omphalitis and osteoarthritis (1). Colibacillosis poses significant economic losses to the poultry industry worldwide, in terms of mortality, weight loss, decreased egg production, lower hatchability, carcass contamination and costs of prophylaxis and treatment (2). Control of colibacillosis is reliant on strict biosecurity practices, vaccination and antibiotic treatment. Despite standard biosecurity practices adopted on poultry farms, E. coli continues to maintain and evolve into diverse strains within the hen house environment via fecal contamination, as the bacterium originates from the avian gut microbiota (3). Due to the diverse plethora of strains and vaccine efficacy only against homologous strains, no single vaccine is effective against all strains; thus, flock-specific autogenous bacterins are often developed for effective prophylaxis (4). Therefore, in most low- and middle-income countries, colibacillosis is treated with the use of antibiotics (2, 5). However, the injudicious use of antibiotics in poultry production and natural evolutionary mechanisms in pathogenic bacteria have resulted in the emergence of multidrug-resistant (MDR) strains of E. coli (6, 7). This situation intensifies the challenges of controlling avian colibacillosis.
Antimicrobial resistance (AMR) is a phenomenon of global concern. The use of antimicrobials in food-animal production accounts for 73% of the antimicrobials sold globally (8). Antimicrobials are consumed mainly in terms of achieving productivity goals, maintaining good farm hygiene, and disease control and prevention purposes (8). The importance of antimicrobial resistance genes (ARGs) can never be overemphasized when it comes to the horizontal transfer of genetic determinants of resistance within the populations of pathogenic bacteria, which is critical to the health of both humans and animals (9). Apart from infecting chickens, the resistant clones of Klebsiella pneumoniae (K. pneumoniae) and E. coli have public health significance, as these bacteria can be transmitted from poultry to humans through poultry-origin food products and environmental contamination (10).
Colistin (Polymyxin E) is a cationic polypeptide, broad-spectrum antibiotic, mainly active against Gram-negative bacteria. Despite its nephrotoxicity potential, colistin is considered a last resort antibiotic for treating multidrug-resistant (MDR) Gram-negative bacterial infections due to the unavailability of new antibiotics (11). However, members of the Enterobacteriaceae such as E. coli, Salmonella spp., and K. pneumoniae are becoming increasingly resistant to colistin. One of the earlier known mechanisms of colistin resistance involves chromosomal mutations that activate the two-component regulatory systems PhoP-PhoQ and PmrA-PmrB, causing changes in lipopolysaccharide (LPS) structure, leading to loss of affinity for colistin attachment (12, 13). Further, in 2015, the mechanism of horizontal transmission of colistin resistance was first reported in E. coli strains of chicken and porcine origin and K. pneumoniae strains of human origin (14). Reportedly, a plasmid harboring the mobilized colistin resistance (mcr-1) gene, which encodes the phosphoethanolamine transferase enzyme and adds phosphoethanolamine to lipid A of LPS, causes a reduction in net-negative charge of the outer membrane of Gram-negative bacteria, resulting in loss of colistin affinity (14, 15). More recently, mcr gene variants (mcr-1 to mcr-10) have been reported in multiple bacterial species, originating from different sources such as animals, humans, food and the environment (16). These findings highlight the diversity and potential of mcr gene to rapidly disseminate colistin resistance.
The medical importance of colistin-resistant bacteria has been well understood. However, little is known about the potential of colistin-resistant avian pathogenic E. coli, causing colibacillosis in chickens in low-and middle-income countries like Pakistan. Considering the economic impact and dangers to food security, the present study was conducted to understand the gravity of colibacillosis caused by colistin-resistant E. coli in broiler chicken. Therefore, this study aimed to determine the prevalence, molecular characterization, antibiogram, and associated risk factors for colistin-resistant E. coli and K. pneumoniae in colibacillosis-infected chickens in three districts of Punjab province of Pakistan.
2 Materials and methods
2.1 Collection of specimens and survey data
In this study, 13 broiler chicken farms located in three cities, Sargodha, Jhang and Toba Tek Singh in Punjab province of Pakistan, were investigated from February 2023 to November 2023. Broiler farms in these specific areas were selected through purposive sampling due to their diagnostic records indicating sporadic colibacillosis outbreaks. A total of 230 multi-organ samples (liver, cecum, heart and lungs) were collected from necropsied broiler chickens with a history of clinical signs and gross lesions (perihepatitis, pericarditis, peritonitis, airsacculitis and omphalitis) associated with colibacillosis (Table 1). All tissue samples were collected in sterile vials, properly labeled and shipped with ice packaging to the Microbiology Research Laboratory, Department of Pathobiology, College of Veterinary and Animal Sciences, Jhang campus, University of Veterinary and Animal Sciences, Lahore, Pakistan. In order to study associated risk factors, information related to farming practices, birds’ health, medication history and farm biosecurity practices was collected from farm managers via a semi-structured questionnaire-based method during interviews, followed by direct observation where possible. All information was gathered and processed in a pre-consented manner and as per the ethical guidelines of the intradepartmental ethical review committee of the University of Veterinary and Animal Sciences, Lahore.
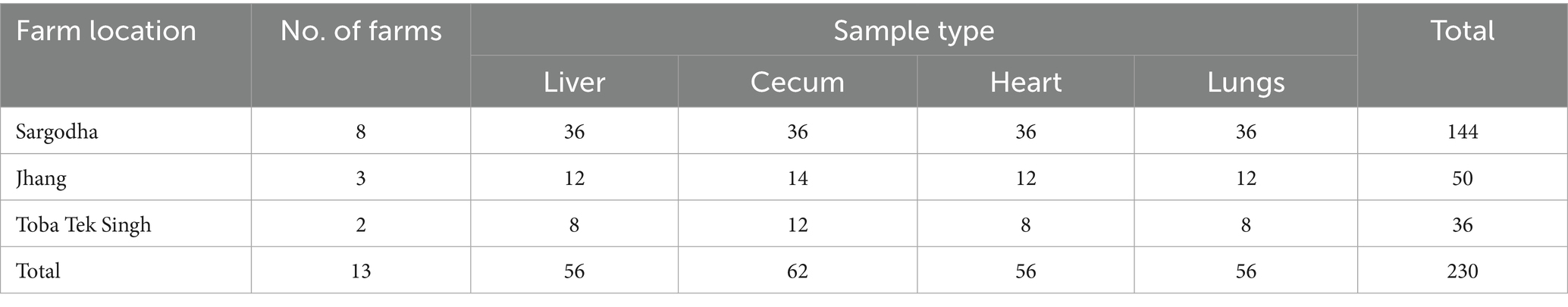
Table 1. Distribution of the visceral organ samples collected from necropsied chickens in the study area.
2.2 Isolation and identification of E. coli and K. pneumoniae
For isolation of colistin-resistant bacteria, a previously described method with a few modifications was used (17). Briefly, the collected samples were pre-enriched by inoculation into 10 mL of tryptone soy broth (CM0129, Oxoid, UK) supplemented with 4 μg/mL colistin (Sigma-Aldrich, USA) for selective isolation of colistin-resistant strains and incubated aerobically at 37°C for 24 h. One hundred μL of pre-enriched tryptone soy broth was streaked onto MacConkey agar (Oxoid, Hampshire, UK) supplemented with 4 μg/mL colistin and incubated at 37°C for 24 h. One representative colony from each MacConkey agar plate was subjected to biochemical tests to identify E. coli and K. pneumoniae species via the analytical profile index (API)-20E kit (bioMérieux, Craponne, France). Aliquots of identified cultures were preserved and stored as 50% glycerol stocks at −21°C until further use.
2.3 Phenotypic confirmation of colistin-resistant E. coli and K. pneumoniae
For phenotypic screening of colistin-resistant isolates, isolated E. coli and K. pneumoniae were thoroughly swabbed on Mueller-Hinton agar (Oxoid, Hampshire, UK) plates, a colistin disc (10 μg) was applied, and plates were incubated at 37°C for 20 h. All isolates with a diameter of zone of inhibition ≤10 mm were tested further for minimum inhibitory concentration (MIC). MIC was determined via the broth microdilution method by following the guidelines provided by the Clinical Laboratory Standards Institute (CLSI) (18). Briefly, cation-adjusted Mueller-Hinton broth with colistin concentrations (0.25–64 μg/mL) was used, and test cultures with adjusted turbidity equivalent to a 0.5 McFarland standard (1:100 dilution) were used as inoculum. E. coli (ATCC 8739) was used as a control organism. Test cultures with MIC value ≥4 μg/mL were considered as colistin-resistant isolates (18).
2.4 Antimicrobial susceptibility test
Antimicrobial susceptibility test was performed by using the Kirby-Bauer disk diffusion method. A panel of 11 antibiotic discs including Streptomycin (S-10 μg), Gentamicin (CN-10 μg), Amoxicillin-clavulanic acid (AMC-20/10 μg), Cefotaxime (CTX-30 μg), Ciprofloxacin (CIP-5 μg), Enrofloxacin (ENR-5 μg), Chloramphenicol (C-30 μg), Tetracycline (TE-30 μg), Imipenem (IPM-10 μg), Meropenem (MEM-10 μg) and Trimethoprim-sulfamethoxazole (SXT-1.25/23.75 μg) (Oxoid, Hampshire, UK) was applied. Isolated E. coli and K. pneumoniae with adjusted turbidity equivalent to 0.5 McFarland standard were swabbed on Mueller-Hinton agar (MHA) plates (Oxoid, Hampshire, UK)., The selected antibiotic discs were applied using sterile forceps and plates were left at room temperature for 30 min, followed by incubation aerobically at 37°C for 24 h. The diameter of zone of inhibition was measured in millimeters and interpreted as resistant, intermediate, or susceptible as per the CLSI criteria (18).
2.5 Genomic DNA extraction and molecular detection of mobilized colistin-resistant (mcr) genes
Genomic DNA was extracted from 1 mL of an overnight incubated tryptone soy broth culture of colistin-resistant bacteria. DNA was extracted by using the GeneJET genomic DNA Purification kit (K0721, ThermoFisher Scientific, USA) following the manufacturer’s instructions. Polymerase chain reaction (PCR) was conducted using previously reported primer sets to detect mobilized colistin-resistant (mcr) genes including mcr-1 (Forward: 5′-AGTCCGTTTGTTCTTGTGGC-3′, reverse: 5′-AGATCCTT GGTCTCGGCTTG-3′) and mcr-2 (Forward: 5′ AGCCGAGTCT AAGGACTTGATGAATTTG-3′, reverse: 5′ GCGGTATCGACAT CATAGTCATCTTG-3′) with generation of PCR product of 320 bp and 576 bp size, respectively (19, 20). Briefly, a total of 50 μL mono-plex PCR reaction mixture was prepared by mixing 25 μL master mix (WizPure™ PCR 2X, W1401-2, Korea), 2 μL each primer (10 μM), 4 μL template DNA and 17 μL of nuclease-free water. Running conditions for amplification in thermal cycler (Biorad, T100, USA) were as follows: Initial denaturation for 15 min at 94°C, 25x (denaturation for 30 s at 94°C, annealing for 90 s at 58°C and extension for 60 s at 72°C) with a final cycle of extension step for 10 min at 72°C. Nuclease-free water was substituted for template DNA in PCR negative controls. DNA from multidrug-resistant (MDR) strains K. pneumoniae strain MASJG8 (GenBank: OP744534.1) and E. coli strain MASMS_A3 (GenBank: ON736876.1) was used as mcr-1 and mcr-2 positive controls, respectively. These strains were isolated from our previous studies and maintained at the Microbiology laboratory of the College of Veterinary and Animal Sciences, Jhang. PCR products were electrophoresed in a 1.2% agarose gel at 100 volts for 45 min using the Mupid-One electrophoresis system (Nippon Genetics, Tokyo, Japan). Agarose gel was stained with ethidium bromide (0.5 μg/mL). Gel images were captured and processed using a gel documentation system (Syngene, Cambridge, UK).
2.6 Lipid a phosphoethanolamine transferase (mcr gene product) phylogenetic analysis
Selected PCR amplicons of mcr-1 gene were processed for Sanger sequencing by a commercial service provider, Beijing Genomics Institute (BGI), Shenzhen (518083), China. Sequencing results were submitted to the GenBank database of the National Center for Biotechnology Information (NCBI). A phylogenetic analysis was performed by using corresponding partial protein sequences of lipid A phosphoethanolamine transferase (mcr-1 product) obtained in the present study, including GenBank accession numbers WPF45700.1 and WPF45701.1. The comparator and reference sequences included in phylogenetic analysis were obtained from the NCBI public database. This selection was made using the BLAST-p program. The sequences included a reference sequence of mcr-1 (HBY7764053.1). The comparator sequences included mcr-1.3 (WP077064885.1), mcr-1.5 (APM84488.1), mcr-1.8 (WP085562407.1), mcr-2.1 (WHI19688.1, WHF75690.1), mcr-2.8 (QXM27672.1), mcr-3.1 (BBA91300.1, WBW54110.1), mcr-4 (QDF67528.1) and mcr-4.3 (AYJ09357.1). Class A beta-lactamase CTX-M-1 (WHD27734.1) sequence was added as an outgroup. Whelan and Goldman (WAG) was computed by MEGA software (v 12.0.11) as the best-fit amino acids substitution model based on the lowest BIC (Bayesian information criterion) score (21). A phylogenetic tree was constructed using the maximum likelihood method with bootstraps (500) and the WAG substitution model using MEGA 12 software (22).
2.7 Descriptive statistical and associated risk factors analysis
Numerical data was put in Microsoft Excel 365 to calculate percentages and mean values. Regional and comparative prevalence and confidence intervals (CI) of colistin-resistant bacteria were determined using EpiTools (V 2.0) epidemiological calculators (23). Prevalence and mean difference were tested for significance by using one-way analysis of variance (ANOVA) and an independent t-test by considering p-values less than 0.05 (p < 0.05) as statistically significant. The association of individual risk factors with colistin-resistant bacteria was determined by univariate logistic regression analysis using JASP (Version 0.17.2) (24) to calculate odds ratios (ORs), confidence intervals and p-values. Only risk factors with p ≤ 0.15 in the univariate logistic regression were selected for inclusion in the final multivariate logistic regression analysis. Selected risk factors were further analyzed via multivariate logistic regression in JASP, wherein both “enter” and automated “backward selection” models were built (24) only factors with p < 0.05 at 95% confidence intervals were considered significantly associated with the detection of colistin-resistant bacteria.
3 Results
3.1 Prevalence of colistin-resistant bacteria
Typical colibacillosis gross lesions, such as severe pericarditis, perihepatitis, airsacculitis and peritonitis, were consistently observed in chickens that died of colistin-resistant strains of avian pathogenic E. coli (APEC) (Figure 1). The overall prevalence of colistin-resistant bacteria in collected samples was 27.83% (95% CI, 22.4–33.9%). In a total of 230 collected samples, 57 (24.78%) and 7 (3.04%) isolates were identified as colistin-resistant E. coli and K. pneumonia, respectively (Table 2). The MIC of colistin for E. coli and K. pneumonia ranged between 4–16 μg/mL and 4–8 μg/mL, respectively (Supplementary data). The prevalence of colistin-resistant E. coli was 24.78% (95% CI, 19.6–30.7%), which was significantly (p = 0.01) higher than colistin-resistant K. pneumoniae, 3.04% (95% CI, 1.5–6.1%). The prevalence of colistin-resistant E. coli varied significantly (p = 0.02) between the three studied cities, with the highest in Jhang, 42% (21/50), followed by Sargodha, 23.61% (34/144) and Toba Tek Singh, 5.55% (2/36). Isolation frequency of colistin-resistant E. coli was highest in liver samples, 41.07% (23/56), followed by cecum at 33.87% (21/62), heart at 14.28% (8/56) and lungs at 8.93% (5/56).
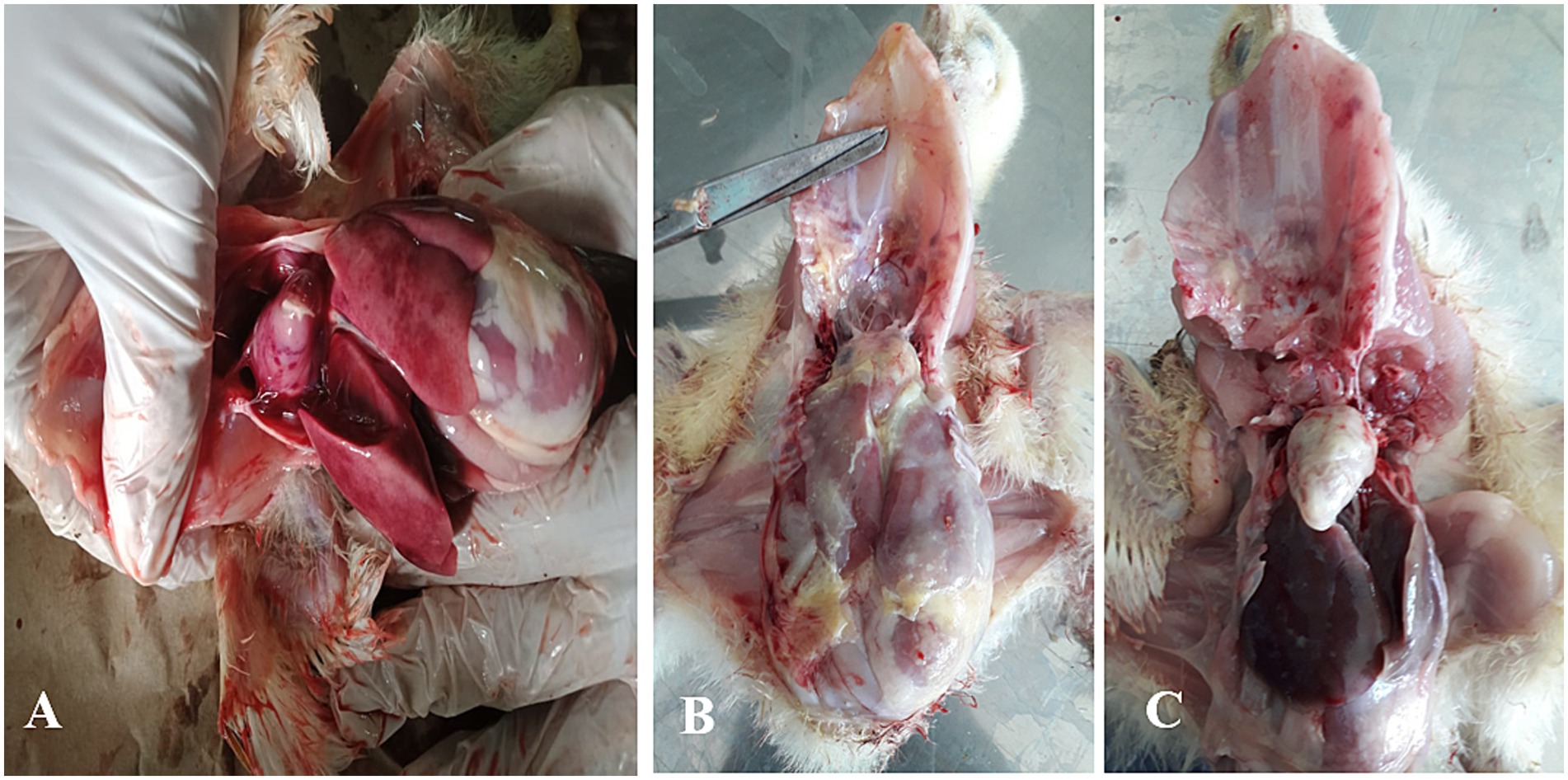
Figure 1. Description of various gross lesions found in necropsy examination of chicken infected with colibacillosis caused by colistin-resistant E. coli. (A) Multiple necrotic foci are shown on the surface of the heart and liver associated with pericarditis and perihepatitis. (B) Fibrous exudate diffusely deposited on the surface of the heart and liver. (C) Severe pericarditis marked by creamy fibrous discharge covering the heart surface.
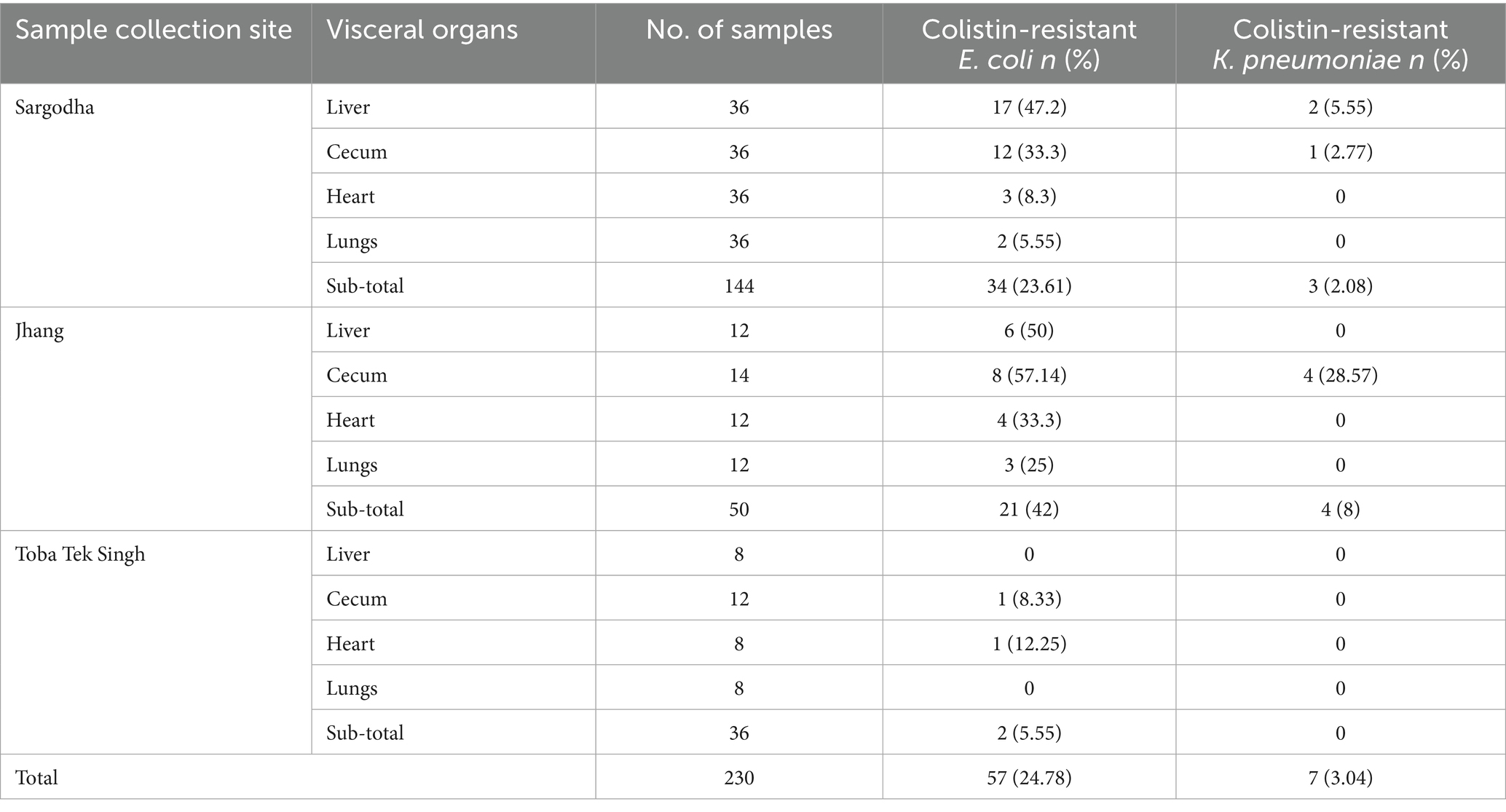
Table 2. Prevalence of colistin-resistant strains of E. coli and K. pneumoniae recovered from chicken visceral organs in different cities.
3.2 Detection and frequency of mcr-1 and mcr-2 genes in colistin-resistant bacteria
The mcr-1 and mcr-2 genes were detected using PCR by amplification of 320 bp and 576 bp products, respectively (Figure 2). In E. coli isolates, the detection frequency of mcr-1 gene was 42.1% (24/57), which was significantly (p < 0.01) higher than that of mcr-2 at 14.03% (8/57). However, in 7% (4/57) of E. coli isolates, the co-presence of both genes was observed in four isolates (7.01%) (Table 3). In 40.62% (26/64) colistin-resistant isolates, neither mcr-1 nor mcr-2 genes were detected. The mcr-1 gene was identified in two isolates of K. pneumoniae, while the mcr-2 gene was not present in any of the seven K. pneumoniae isolates.
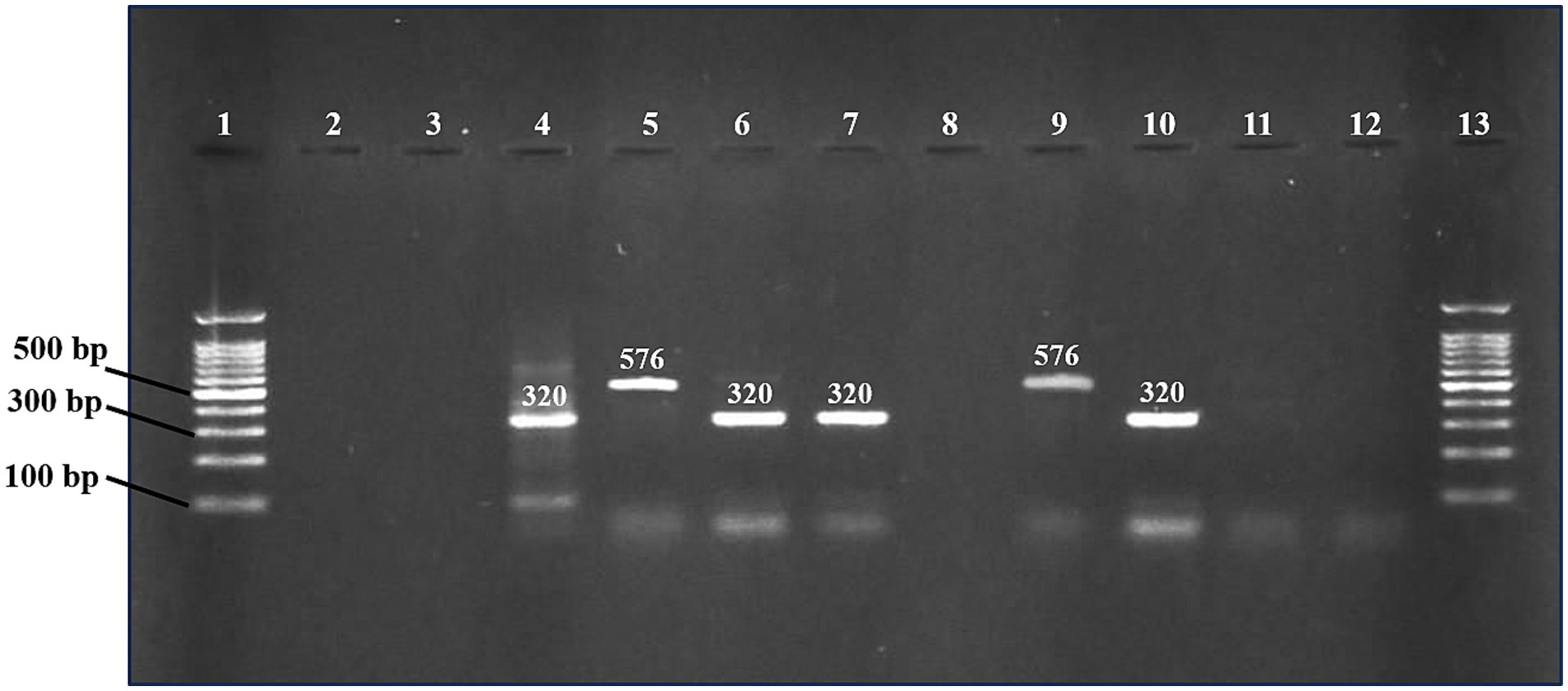
Figure 2. PCR products were detected on a 1.2% agarose gel for mcr-1 and mcr-2 genes. Lane 1 and 13 (DNA ladder 100 bp, Solis BioDyne, Tartu, Estonia). Lanes 2 and 3 contain negative controls for mcr-1 and mcr-2, respectively. Lanes 4 and 5 contain positive controls for mcr-1 and mcr-2, respectively. Lanes 6, 7, and 10 contain positive samples for mcr-1 gene (320 bp). Lane 9 contains a positive sample for mcr-2 gene (576 bp). Negative samples (lanes 8, 11, and 12).
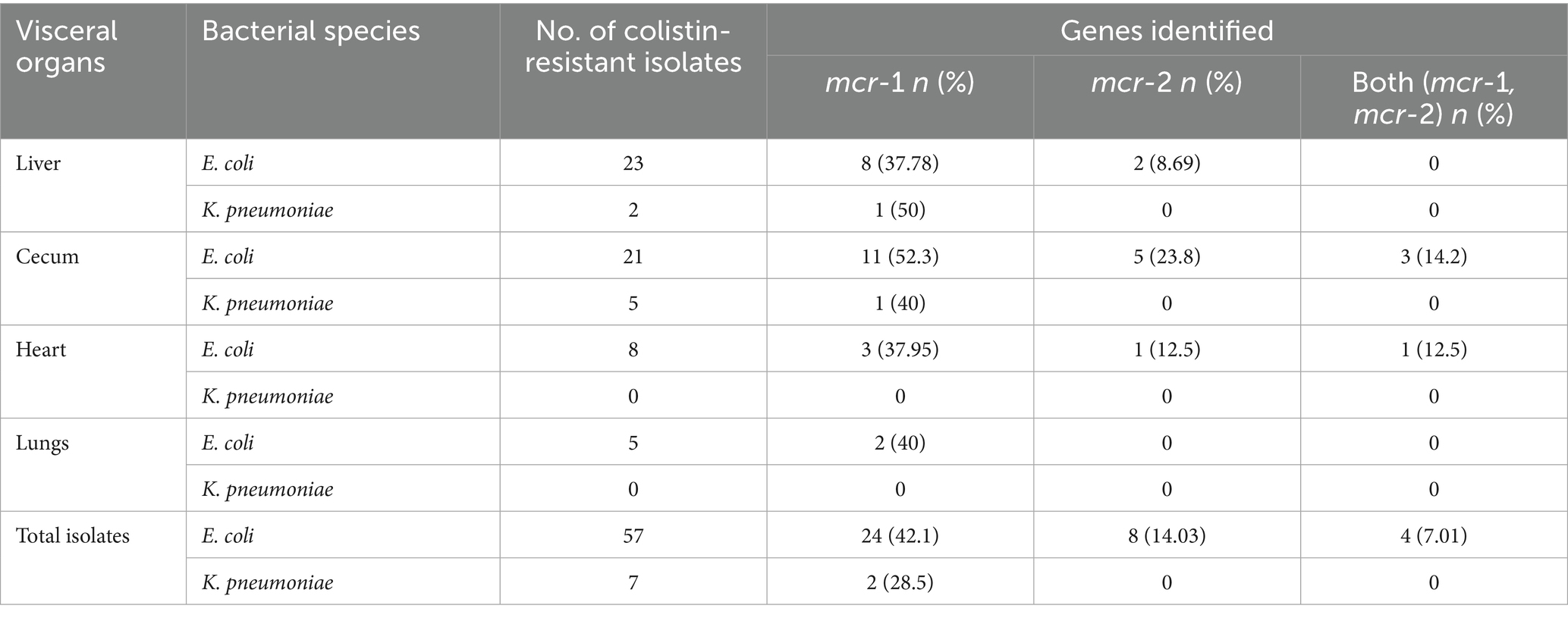
Table 3. Frequency distribution of mcr-1 and mcr-2 genes detected in phenotypically confirmed colistin-resistant E. coli and K. pneumoniae.
3.3 Phylogenetic analysis of lipid a phosphoethanolamine transferase sequences
The nucleotide sequences of mcr gene products were sequenced and submitted to the National Center for Biotechnology Information (NCBI) with accession numbers OR680710 and OR680711. The corresponding amino acid sequences of mcr gene product, lipid A phosphoethanolamine transferase, obtained have accession numbers of WPF45700.1 and WPF45701.1. The maximum likelihood-based phylogenetic tree presented four clades, where mcr-1 and mcr-2 were found to be the most closely related variants, as compared to mcr-3 and mcr-4 variants. The sequences of the present study were grouped in mcr-1 clade and showed genetic relatedness to mcr-1.5 variant (Figure 3).
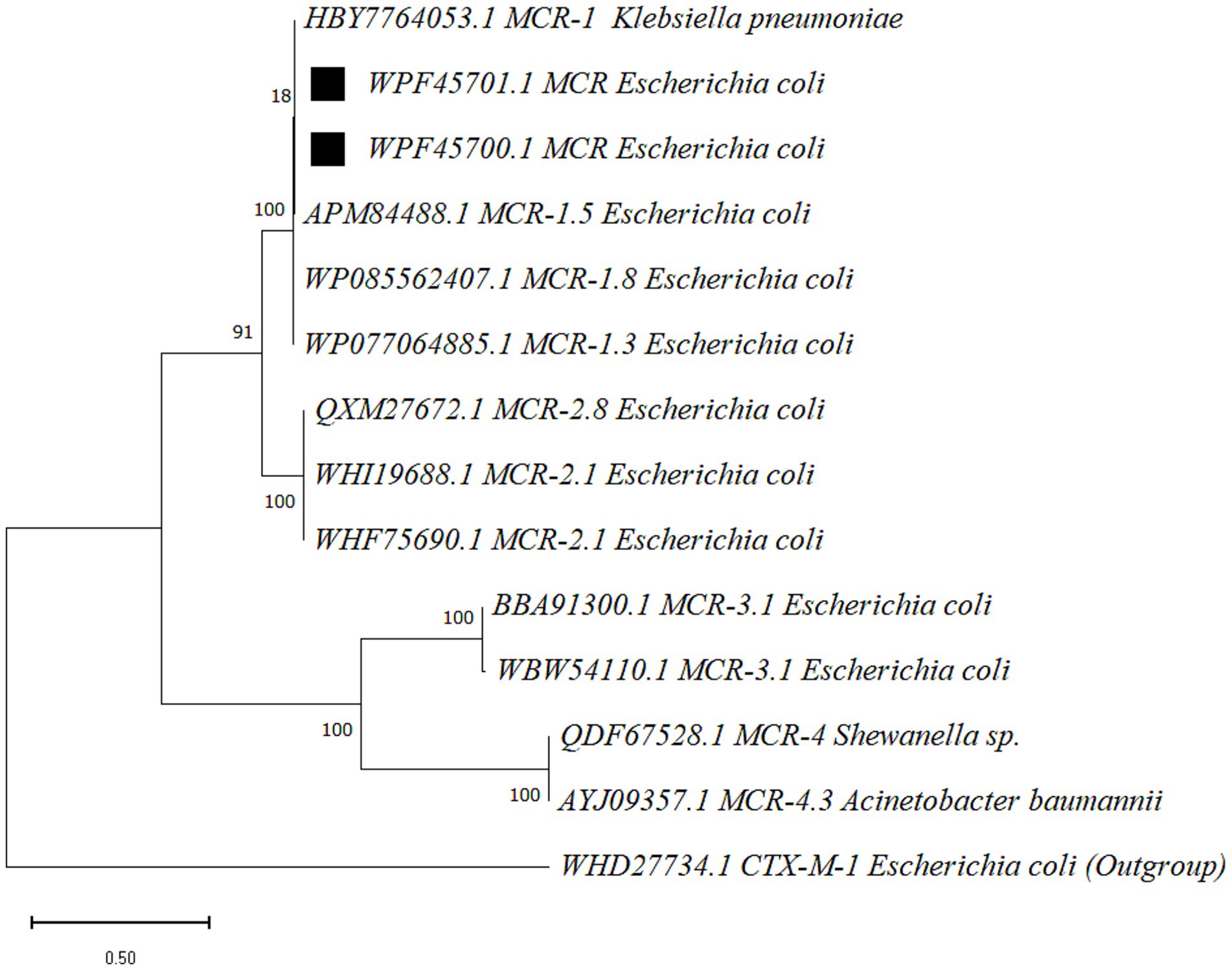
Figure 3. The phylogenetic tree was constructed for mcr gene product, lipid A phosphoethanolamine transferase, via MEGA (v 12.0.11) with the maximum likelihood method (bootstraps 500) and the WAG substitution model. Sequences obtained in the present study are marked with black squares. Node labels represent the bootstrap support values (percentage), and the scale bar indicates 0.50 amino acid substitutions per site.
3.4 Antimicrobial susceptibility test
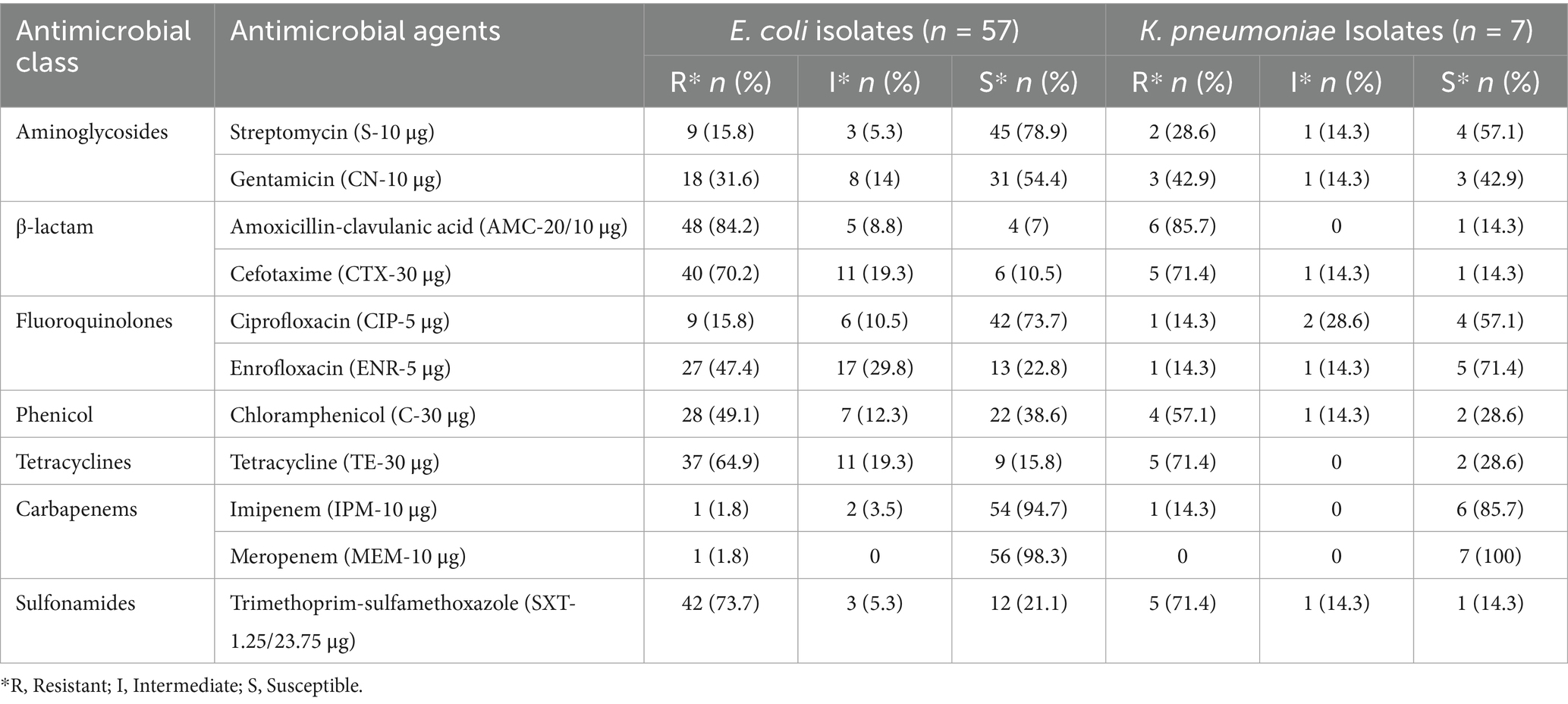
Colistin-resistant isolates of E. coli and K. pneumoniae were tested for susceptibility against 11 antimicrobial agents belonging to seven different antibiotic classes. Isolates with relatively higher resistance (≥70%) levels were found resistant to amoxicillin-clavulanic acid (84.21%), cefotaxime (70.17%) and trimethoprim-sulfamethoxazole (73.68%). Colistin-resistant E. coli showed higher susceptibility to meropenem (98.25%), imipenem (94.74%), streptomycin (78.94%) and ciprofloxacin (73.7%). The susceptibility of K. pneumoniae isolates varied from 14.29 to 100% (Table 4).
3.5 Risk factors analysis
Risk-associated factors with colistin-resistant bacteria, representing host attributes, farm management practices, environmental factors and agent characteristics, were studied. Initially, univariate logistic regression analysis was performed (Table 5). Univariate analysis found four risk factors including the use of colistin without prescription and/or bacterial sensitivity test (p = 0.132), daily removal/culling of dead/diseases chicken (p = 0.107), flock history of any preceding viral disease (p = 0.138) and bird age (p = 0.106) as statistically significant factors with p ≤ 0.15. These four risk factors were tested with multivariate logistic regression analysis. The final multivariate logistic regression model was found statistically significant (χ2 15.64, p < 0.01) with 74% accuracy. Only two risk factors were found statistically significantly (p < 0.05) associated with colistin-resistant bacteria (Table 6). These included daily removal/culling of dead/diseased chicken (OR = 0.308, 95% CI = 0.115–0.821, p = 0.019) and history of laboratory confirmed viral infection of the respiratory tract (OR = 4.808, 95% CI = 1.961–11.789, p < 0.01). Daily culling was found to be protective against the emergence of colistin-resistant bacteria. The risk of the emergence of colistin-resistant bacteria was found to be 4.8 times higher in chickens infected with viral respiratory disease than in those without a history of respiratory viral infection.
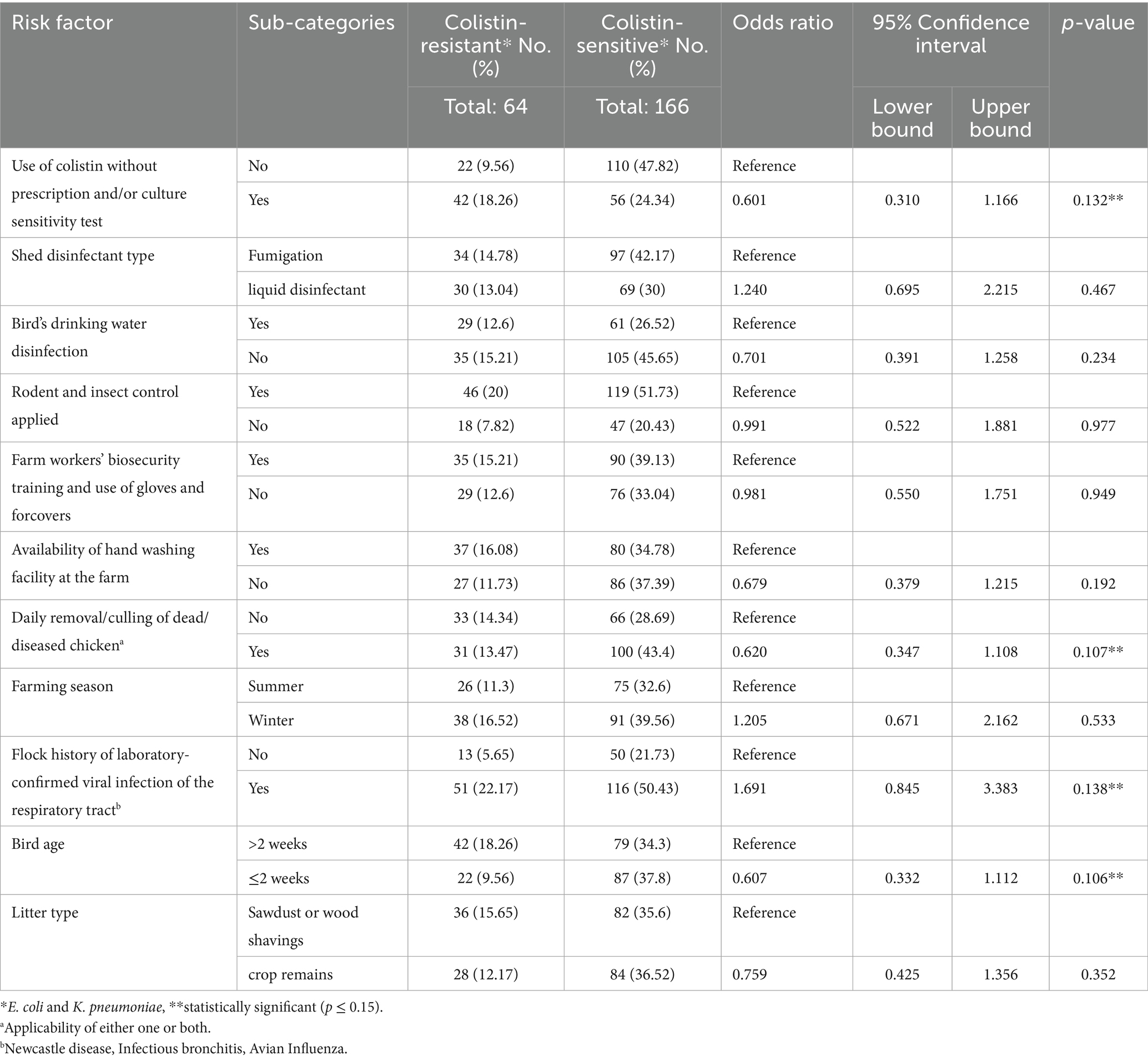
Table 5. Univariate analysis of the association of potential risk factors with the emergence of colistin-resistant bacteria in broiler chicken farms.
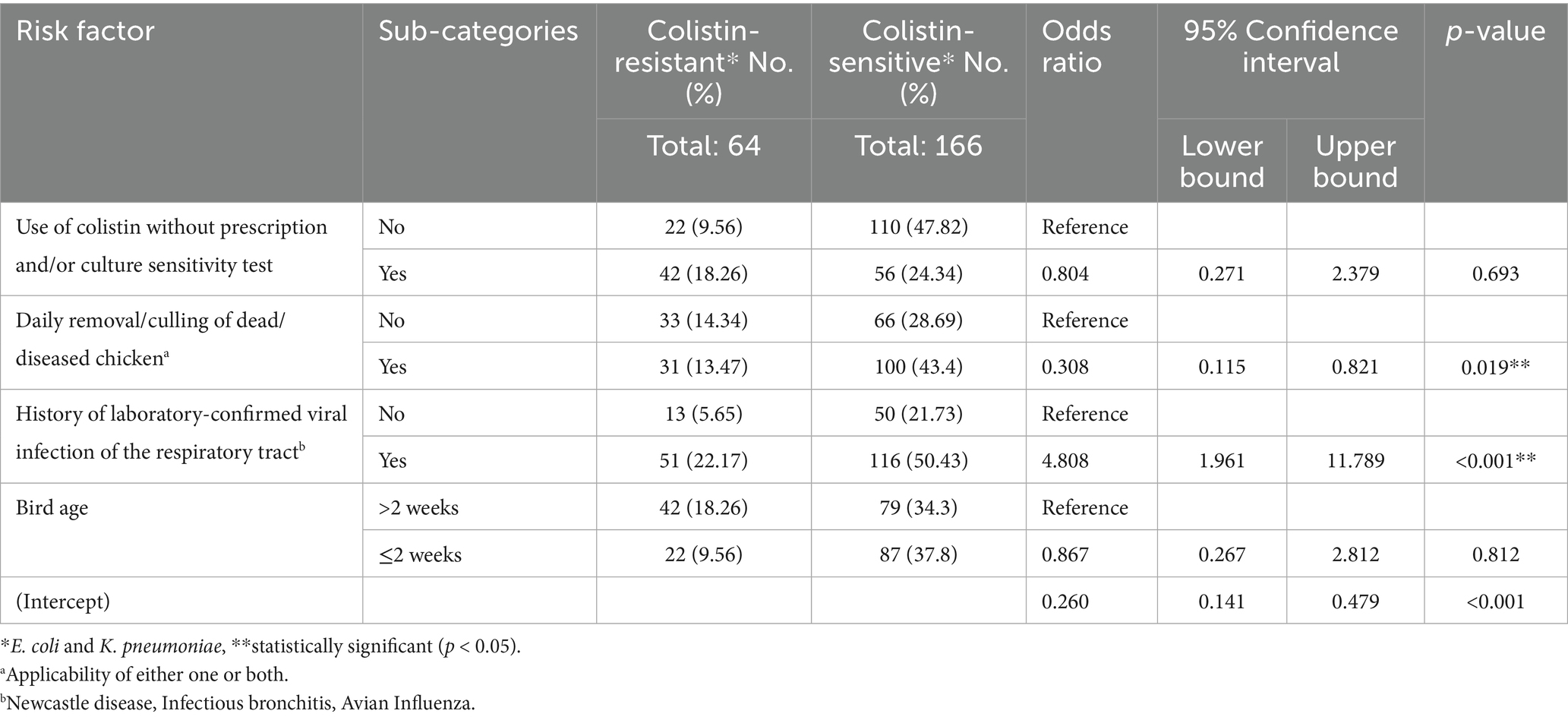
Table 6. Multivariate analysis of the association of potential risk factors with the emergence of colistin-resistant bacteria in broiler chicken farms.
4 Discussion
In this study, samples were collected from broiler birds primarily infected naturally with colibacillosis. The postmortem findings revealed organ lesions indicative of pericarditis, perihepatitis, airsacculitis and peritonitis, typically associated with avian colibacillosis. The higher prevalence of E. coli in this study is attributed to the fact that avian colibacillosis is caused by avian pathogenic E. coli (APEC). However, the detection of colistin-resistant K. pneumoniae in this study highlights the possibility of the emergence of secondary pathogens that become active following a primary respiratory tract infection caused by APEC (2). It also indicates the possible transmission of acquired antimicrobial resistance via mobile genetic elements between different pathogenic species of bacteria (25). In the present study, the prevalence of colistin-resistant E. coli, 24.78% (95% CI, 19.6–30.7%), was significantly (p = 0.01) higher than colistin-resistant K. pneumoniae, 3.04% (95% CI, 1.5–6.1%). Isolation frequency of colistin-resistant E. coli was highest in liver samples, 41.07% (23/56), followed by cecum at 33.87% (21/62), heart at 14.28% (8/56) and lungs at 8.93% (5/56). During systemic manifestations of colibacillosis infection, APEC colonizes the upper respiratory tract, trachea, and air sacs, followed by colonization of the liver and pericardium via bacteremia (2;19). Bacteremia leads to the multiplication of APEC in organs such as the liver and spleen, where reticuloendothelial tissues are found abundantly (26). Therefore, chicken liver offers a relatively higher isolation sensitivity rate for APEC. The geographical location-based differences in the prevalence of APEC were evident from the present study, interestingly, between the neighboring cities of Toba Tek Singh (5.55%) and Jhang (42%). Factors such as local farming practices, host characteristics, biosecurity, antibiotic usage patterns, vector control and water source have been identified to contribute to the prevalence of APEC at the farm level (27, 28). Similarly, the present study also identified the association of respiratory viral infections and culling management with the prevalence of colistin-resistant E. coli. The presence of multidrug-resistant and virulent strains of APEC at farms enhances the risk of transmission to the human population, either through direct contact or the consumption of contaminated chicken products (29, 30).
In this study, the mobilized colistin resistance gene mcr-1 was detected in 42.1% of colistin-resistant E. coli. The phylogenetic analysis based on two partial mcr gene product sequences showed that the amino acid sequences of lipid A phosphoethanolamine transferase were grouped into mcr-1 clade next to mcr-1.5 variant. These findings are consistent with the findings of previous studies in Pakistan, which reported mcr-1 found in bacteria isolated from a wide variety of sources, including colibacillosis-infected chicken (31), poultry farm flies and chicken meat (32). Relatively lower prevalence of colistin-resistant E. coli, 18.95 and 7% was reported in Pakistan from healthy poultry birds and livestock in two different studies and only mcr-1 gene was detected (33, 34). However, similar to our findings, a recent study targeting samples taken from colibacillosis-infected chicken reported 37% prevalence of avian pathogenic E. coli (APEC), wherein 38% isolates were positive for mcr-1 gene (31). The mcr-1 gene has been reported to be associated with the IncI2 plasmid, which harbors multiple virulence factors and is capable of horizontal gene transfer among carrier E. coli (35).
Sampling from internal organs of chicken infected with colibacillosis, as compared to cloacal swabs or fecal samples, allows for isolation of genetically diverse extraintestinal pathogenic strains (36). Hence, depending on the sampling origin, more diverse resistant strains with high sensitivity of isolation can be obtained from diseased chickens as compared to healthy chickens (37).
The mcr-2 gene was detected in 14.03% (8/57) of colistin-resistant E. coli in the present study; however, to the best of the authors’ knowledge, there is no report of mcr-2 gene detected in colibacillosis-infected broilers in Pakistan previously. The mcr-2 gene was first reported in E. coli isolates associated with diarrhea in calves and piglets in Belgium (38). The mcr-2 gene harboring plasmid IncX4 is capable of as high as 102–105-fold transfer frequencies as compared to the epidemic IncFII plasmid (38). These findings explain the rapid horizontal transmission potential of the mcr-2 genes in E. coli. In China, the mcr-2 gene was detected in colistin-resistant E. coli isolated from pigs (46.82%), chicken (14.90%) and cattle (19.05%) (39). Similarly, the mcr-2 gene has been detected in 3.4% of isolates originating from the chicken gut (3). Furthermore, the co-occurrence of both mcr-1 and mcr-2 genes has also been reported in colistin-resistant isolates from the same source (40). A study from Egypt found the prevalence of mcr-2 in bacteria isolated from resident birds, migratory birds, water sources, and humans as 1.4, 3.6, 11.1 and 9.6%, respectively (41). These studies suggest that the mcr-2 gene has evolved in bacteria of multiple sources, including farm animals, chicken, wild birds, humans and the environment. We also found the co-presence of mcr-1 and mcr-2 in 7% of colistin-resistant E. coli. In general, the mcr-2 allele has been found globally to be low in prevalence, which varies geographically. In Europe, its prevalence varied from 0.15 to 11.4% in E. coli; moreover, the co-existence of mcr-1 and mcr-2 of swine origin E. coli was reported in Germany (42). In Bangladesh, the mcr-2 prevalence in chicken has been reported as 3.4%, and co-detection with mcr-1 as 1.34% (2/149) (40). In this study, the prevalence of colistin-resistant K. pneumoniae was determined as 3.04% (7/230), whereas mcr-1 gene was detected in 28.5% (2/7) of colistin-resistant isolates. However, all K. pneumoniae isolates were negative for mcr-2 gene. Similarly, a study from Egypt found the prevalence of K. pneumoniae as 9% (9/100) in commercial chicken, and mcr-1 was detected in 18.9% samples (43).
In this study, 40.62% (26/64) colistin-resistant isolates possessed neither mcr-1 nor mcr-2 genes. The diversity of colistin resistance mechanisms explains the absence of mcr-1 and mcr-2 genes in colistin-resistant bacteria. There are 10 major variants of mcr gene (mcr-1 to mcr-10) along with sub-variants (16). In addition, chromosomal mutations lead to structural modifications of lipid A, including the addition of 4-amino-L-arabinose or cationic phosphoethanolamine (pEtN) to lipid A, which results in a reduction of negative charge on lipid A and halting colistin coupling with lipid A (25). Such changes are also attributed to the mutations in lipid A biosynthesis genes or via overexpression of chromosomally mediated two-component system genes (PmrAB and PhoPQ) (25). Therefore, different colistin-resistant strains may possess one or multiple different underlying mechanisms for such a phenotype.
Colistin-resistant isolates of E. coli and K. pneumoniae were tested for susceptibility against 11 antibiotics belonging to seven different classes of antibiotics. E. coli with relatively higher resistance (≥70%) levels were found resistant to amoxicillin-clavulanic acid (84.21%), cefotaxime (70.17%), and trimethoprim-sulfamethoxazole (73.68%). While most of the isolates remained susceptible to meropenem and Imipenem. Colistin-resistant E. coli were found sensitive to meropenem (98.25%), imipenem (94.74%), streptomycin (78.94%) and ciprofloxacin (73.7%). The susceptibility of K. pneumoniae isolates varied from 14.29 to 100%. Similar findings of the highest antibiotic resistance for ampicillin (β-lactam group) were found from a previous study, which reported carbapenem-resistant mcr-positive E. coli associated with avian colibacillosis (31). However, the present study reports a higher resistance rate to amoxicillin-clavulanic acid, thus indicating a wide spectrum of resistance via possible production of inhibitor-resistant β-lactamases and extended-spectrum beta-lactamase (ESBL) type enzymes by resistant bacteria (44). Detection of colistin-resistant and β-lactamase-producing E. coli has been isolated from chickens infected with colibacillosis in Tunisia (45). These findings explain the possible resistance mechanisms against third-generation cephalosporins such as cefotaxime. The colistin-resistant E. coli in poultry may harbor various genetic determinants that allow for the multidrug resistance phenomenon. In a previous study in Bangladesh, various genetic determinants, including tetA (for tetracycline), sul1 (for sulfonamide), aadA1 (for streptomycin), aac-3-IV (for gentamicin) and the two genes cmlA and catA1 (for chloramphenicol), were detected in chicken meat-associated multidrug-resistant E. coli (6).
The risk factors associated with the emergence of colistin-resistant E. coli and K. pneumoniae in broiler chicken flocks were studied. The final multivariate logistic regression analysis identified two risk factors that were statistically associated with colistin-resistant bacteria. The history of laboratory-confirmed preceding viral infection of the respiratory tract (Newcastle disease, Infectious bronchitis, and Avian Influenza) (OR = 4.808, 95% CI = 1.961–11.789, p < 0.01) was positively associated with colistin-resistant bacteria. While farm hygienic measures, daily removal/culling of dead/diseased chicken (OR = 0.308, 95% CI = 0.115–0.821, p = 0.019) turned out to be protective, they were found to be associated with reduced risk of emergence of the colistin-resistant bacteria. Depending on the farm management practices, geographical location, virulence of E. coli strains and immune status of birds, the risk factors associated with avian colibacillosis can vary (26, 33). The use of groundwater as the source of drinking water, failure to disinfect water channels, farms located in close proximity to other farms, distances greater than 20 meters from car parking to shed and presence of wild birds within 50 meters of the shed surrounding area were found associated with carriage of multidrug-resistant avian pathogenic E. coli strains (46, 47). However, in our findings, the use of drinking water disinfectant or groundwater as drinking water has not been identified as a statistically significant factor. These findings may indicate the partial contribution of sewerage contamination of groundwater, with variable levels in different geographical locations. However, a recent study from Jordan identified poor farm sanitary conditions and improper use of antibiotics, especially doxycycline, with the emergence of mcr-1 colistin-resistant E. coli in broilers (48). The timely and effective culling management, which contributes to overall farm hygiene, has been found to have a significant association. Similar to our findings, a previous study demonstrated that the association of preceding respiratory viral diseases caused by infectious bronchitis virus (IBV), Newcastle disease virus (NDV) and avian metapneumovirus (aMPV) aggravated the avian colibacillosis condition caused by E. coli by damaging the respiratory mucosa, tracheitis and airsacculitis (49). Preceding viral diseases are thought to enhance host susceptibility to secondary bacterial infection via multiple factors, including immune deficiency and damage to mucosal barriers.
The present study was limited to the detection of mcr-1 and mcr-2 genes only. To achieve a more comprehensive understanding of the emergence of colistin resistance in the region, future studies should aim to investigate both mcr-mediated and non-mcr-mediated resistance mechanisms.
5 Conclusion
This study contributed to the understanding of the dissemination of colistin-resistant E. coli and K. pneumoniae in chickens infected with avian colibacillosis in Pakistan. Mobilized colistin resistance gene (mcr-1) was identified as a dominant genetic determinant. However, mcr-2 was also detected in E. coli. Colistin-resistant bacteria were found resistant to 11 other types of antibiotics, predominantly amoxicillin-clavulanic acid, cefotaxime and trimethoprim-sulfamethoxazole. This study also identified the association of viral respiratory diseases and non-frequent disposal of dead birds and poor culling management as risk factors for avian colibacillosis. The detection of colistin-resistant E. coli and K. pneumoniae in chickens not only poses a significant threat to food security but also contributes to zoonotic transmission of antibiotic-resistant bacteria. Therefore, efforts must be put in place to conduct genomic epidemiological studies on colistin-resistant bacteria, legislative controls on antibiotic use in animal production and better farm management practices considering associated risk factors in the chicken farming industry in Pakistan.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Institutional Review Board and standard recommendations of the intradepartmental Ethical Review Committee and directorate of advanced studies of the University of Veterinary and Animal Sciences (UVAS), Lahore, Approval Code: MICR/LEC/23, Approval Date: 13 January 2023 and DAS/1991 Approval Date: 06 November 2023. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MS: Methodology, Conceptualization, Data curation, Writing – original draft, Writing – review & editing, Resources, Investigation. HA: Methodology, Writing – review & editing, Investigation, Data curation. SE-u-H: Writing – review & editing, Methodology, Investigation, Data curation, Resources. AK: Investigation, Methodology, Supervision, Writing – review & editing, Resources, Data curation, Writing – original draft, Conceptualization. AzR: Writing – review & editing, Methodology, Investigation. AiR: Writing – review & editing, Methodology, Investigation. MR: Methodology, Investigation, Conceptualization, Writing – review & editing. IA: Writing – review & editing, Methodology, Investigation. MQ: Writing – review & editing, Supervision, Validation. HT: Supervision, Validation, Writing – review & editing. HE-A: Conceptualization, Validation, Writing – review & editing, Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to appreciate the participation of local poultry farmers and field veterinarians, especially Dr. Muhammad Sibtain, Punjab Feeds (Ltd.), for technical consultation regarding necropsy examination and sampling from the poultry farms.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1612542/full#supplementary-material
References
1. Dziva, F, and Stevens, MP. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic in their natural hosts. Avian Pathol. (2008) 37:355–66. doi: 10.1080/03079450802216652
2. Guabiraba, R, and Schouler, C. Avian colibacillosis: still many black holes. FEMS Microbiol Lett. (2015) 362:fnv118. doi: 10.1093/femsle/fnv118
3. Ewers, C, Antao, EM, Diehl, I, Philipp, HC, and Wieler, LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic strains with zoonotic potential. Appl Environ Microbiol. (2009) 75:184–92. doi: 10.1128/Aem.01324-08
4. Sadeyen, JR, Wu, Z, Davies, H, van Diemen, PM, Milicic, A, La Ragione, RM, et al. Immune responses associated with homologous protection conferred by commercial vaccines for control of avian pathogenic Escherichia coli in turkeys. Vet Res. (2015) 46:5. doi: 10.1186/s13567-014-0132-5
5. Anyanwu, MU, Jaja, IF, Okpala, COR, Njoga, EO, Okafor, NA, and Oguttu, JW. Mobile Colistin resistance (mcr) gene-containing organisms in poultry sector in low- and middle-income countries: epidemiology, characteristics, and one health control strategies. Antibiotics (Basel). (2023) 12:1117. doi: 10.3390/antibiotics12071117
6. Rahman, MM, Husna, A, Elshabrawy, HA, Alam, J, Runa, NY, Badruzzaman, ATM, et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci Rep. (2020) 10:21999. doi: 10.1038/s41598-020-78367-2
7. Gomez, JE, Kaufmann-Malaga, BB, Wivagg, CN, Kim, PB, Silvis, MR, Renedo, N, et al. Ribosomal mutations promote the evolution of antibiotic resistance in a multidrug environment. eLife. (2017) 6:e20420. doi: 10.7554/eLife.20420
8. Van Boeckel, TP, Glennon, EE, Chen, D, Gilbert, M, Robinson, TP, Grenfell, BT, et al. Reducing antimicrobial use in food animals. Science. (2017) 357:1350–2. doi: 10.1126/science.aao1495
9. Marshall, BM, and Levy, SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. (2011) 24:718–33. doi: 10.1128/CMR.00002-11
10. Savin, M, Bierbaum, G, Hammerl, JA, Heinemann, C, Parcina, M, Sib, E, et al. ESKAPE bacteria and extended-spectrum-beta-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl Environ Microbiol. (2020) 86:e02748–19. doi: 10.1128/AEM.02748-19
11. El-Sayed Ahmed, MAE, Zhong, LL, Shen, C, Yang, Y, Doi, Y, and Tian, GB. Colistin and its role in the era of antibiotic resistance: an extended review (2000-2019). Emerg Microbes Infect. (2020) 9:868-85. doi: 10.1080/22221751.2020.1754133
12. Cannatelli, A, D'Andrea, MM, Giani, T, Di Pilato, V, Arena, F, Ambretti, S, et al. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother. (2013) 57:5521–6. doi: 10.1128/AAC.01480-13
13. Gunn, JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. (2008) 16:284–90. doi: 10.1016/j.tim.2008.03.007
14. Liu, YY, Wang, Y, Walsh, TR, Yi, LX, Zhang, R, Spencer, J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
15. Samantha, A, and Vrielink, A. Lipid a Phosphoethanolamine transferase: regulation, structure and immune response. J Mol Biol. (2020) 432:5184–96. doi: 10.1016/j.jmb.2020.04.022
16. Hussein, NH, Al-Kadmy, IMS, Taha, BM, and Hussein, JD. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol Biol Rep. (2021) 48:2897–907. doi: 10.1007/s11033-021-06307-y
17. Hassan, J, Mann, D, Li, S, Deng, X, and Kassem, II. Emergence of the mobile colistin resistance gene, mcr-1, in multidrug-resistant E. coli isolated from the fecal matter of toddlers in a community. Antimicrob Agents Chemother. (2021) 65:e00243-21. doi: 10.1128/aac.00243-21
18. CLSI. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100s. 31st ed. USA: Clinical and Laboratory Standards Institute (2021).
19. Rebelo, AR, Bortolaia, V, Kjeldgaard, JS, Pedersen, SK, Leekitcharoenphon, P, Hansen, IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. (2018) 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
20. Zhang, JL, Wang, JW, Chen, L, Yassin, AK, Kelly, P, Butaye, P, et al. Housefly (Musca domestica) and blow Fly (Protophormia terraenovae) as vectors of bacteria carrying colistin resistance genes. Appl Environ Microb. (2018) 84:e01736–17. doi: 10.1128/AEM.01736-17
21. Kumar, S, Stecher, G, Suleski, M, Sanderford, M, Sharma, S, and Tamura, K. MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol Biol Evol. (2024) 41:msae263. doi: 10.1093/molbev/msae263
22. Tamura, K, Stecher, G, and Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
23. Sergeant ESG. EpiTools epidemiological calculators: Ausvet (2018). Available online at: http://epitools.ausvet.com.au (Accessed June 26, 2024).
24. Team, J. JASP (version 0.17.2.1) [computer software]. (2023). Available online at: https://jasp-stats.org/ (Accessed March 10, 2024).
25. Gogry, FA, Siddiqui, MT, Sultan, I, and Haq, QMR. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front Med (Lausanne). (2021) 8:677720. doi: 10.3389/fmed.2021.677720
26. Lutful Kabir, SM. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int J Environ Res Public Health. (2010) 7:89–114. doi: 10.3390/ijerph7010089
27. Elmi, SA, Simons, D, Elton, L, Haider, N, Abdel Hamid, MM, Shuaib, YA, et al. Identification of risk factors associated with resistant Escherichia coli isolates from poultry farms in the east coast of peninsular Malaysia: a cross sectional study. Antibiotics (Basel). (2021) 10:117. doi: 10.3390/antibiotics10020117
28. Rahman, A, Rahman, CMS, Hossain, H, Elsaid, FG, Almutairi, LA, Begum, R, et al. Identification of virulence genes and multidrug resistance in Shiga-toxin producing Escherichia coli (STEC) from migratory and captive wild birds. Pak Vet J. (2024) 44:1120–1130. doi: 10.29261/pakvetj/2024.264
29. Hassan, MA, Batiha, GE, Saad, SA, and Mahrous, E. Study on enterotoxigenic Escherichia coli producing extended spectrum beta lactamase (ESBL) from chicken meat and its products. Int J Vet Sci. (2023) 12:652–8. doi: 10.47278/journal.ijvs/2022.217
30. Xue, M, Li, Z, Zhang, P, and Lei, W. Genomic characteristics of ETT2 gene clusters in avian pathogenic Escherichia coli identified by whole-genome sequencing. Pak Vet J. (2024) 44:833–9. doi: 10.29261/pakvetj/2024.240
31. Usman, M, Rasool, MH, Khurshid, M, Aslam, B, and Baloch, Z. Co-occurrence of mcr-1 and carbapenem resistance in avian pathogenic E. coli serogroup O78 ST95 from colibacillosis-infected broiler chickens. Antibiotics (Basel). (2023) 12:812. doi: 10.3390/antibiotics12050812
32. Umair, M, Hassan, B, Farzana, R, Ali, Q, Sands, K, Mathias, J, et al. International manufacturing and trade in colistin, its implications in colistin resistance and one health global policies: a microbiological, economic, and anthropological study. Lancet Microbe. (2023) 4:e264–76. doi: 10.1016/S2666-5247(22)00387-1
33. Shafiq, M, Rahman, SU, Bilal, H, Ullah, A, Noman, SM, Zeng, M, et al. Incidence and molecular characterization of ESBL-producing and colistin-resistant Escherichia coli isolates recovered from healthy food-producing animals in Pakistan. J Appl Microbiol. (2022) 133:1169–82. doi: 10.1111/jam.15469
34. Jamil, A, Zahoor, MA, Nawaz, Z, Siddique, AB, and Khurshid, M. Genetic diversity of Escherichia coli coharboring mcr-1 and extended spectrum beta-lactamases from poultry. Biomed Res Int. (2022) 2022:8224883. doi: 10.1155/2022/8224883
35. Wu, Y, Ch, W, Li, X, Li, F, Jiang, ML, Liu, ZK, et al. Characteristics of the plasmid-mediate colistin-resistance gene mcr-1 in Escherichia coli isolated from pig farm in Jiangxi. Pak Vet J. (2024) 44:1303–7. doi: 10.29261/pakvetj/2024.269
36. Antao, EM, Glodde, S, Li, G, Sharifi, R, Homeier, T, Laturnus, C, et al. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb Pathog. (2008) 45:361–9. doi: 10.1016/j.micpath.2008.08.005
37. Delannoy, S, Schouler, C, Souillard, R, Yousfi, L, Le Devendec, L, Lucas, C, et al. Diversity of strains isolated from day-old broiler chicks, their environment and colibacillosis lesions in 80 flocks in France. Vet Microbiol. (2021) 252:108923. doi: 10.1016/j.vetmic.2020.108923
38. Xavier, BB, Lammens, C, Ruhal, R, Kumar-Singh, S, Butaye, P, Goossens, H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. (2016) 21:pii=30280. doi: 10.2807/1560-7917.Es.2016.21.27.30280
39. Zhang, X, Zhang, B, Guo, Y, Wang, J, Zhao, P, Liu, J, et al. Colistin resistance prevalence in Escherichia coli from domestic animals in intensive breeding farms of Jiangsu Province. Int J Food Microbiol. (2019) 291:87–90. doi: 10.1016/j.ijfoodmicro.2018.11.013
40. Islam, S, Urmi, UL, Rana, M, Sultana, F, Jahan, N, Hossain, B, et al. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci Rep. (2020) 10:17292. doi: 10.1038/s41598-020-74402-4
41. Ahmed, ZS, Elshafiee, EA, Khalefa, HS, Kadry, M, and Hamza, DA. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob Resist Infect Control. (2019) 8:197. doi: 10.1186/s13756-019-0657-5
42. Ewers, C, Gopel, L, Prenger-Berninghoff, E, Semmler, T, Kerner, K, and Bauerfeind, R. Occurrence of mcr-1 and mcr-2 colistin resistance genes in porcine Escherichia coli isolates (2010-2020) and genomic characterization of mcr-2-positive E. coli. Front Microbiol. (2022) 13:1076315. doi: 10.3389/fmicb.2022.1076315
43. Elmonir, W, Abd El-Aziz, NK, Tartor, YH, Moustafa, SM, Abo Remela, EM, Eissa, R, et al. Emergence of colistin and carbapenem resistance in extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolated from chickens and humans in Egypt. Biology (Basel). (2021) 10:373. doi: 10.3390/biology10050373
44. Haines, RR, Putsathit, P, Hammer, KA, and Tai, AS. Activity of newest generation beta-lactam/beta-lactamase inhibitor combination therapies against multidrug resistant Pseudomonas aeruginosa. Sci Rep. (2022) 12:16814. doi: 10.1038/s41598-022-21101-x
45. Dhaouadi, S, Soufi, L, Hamza, A, Fedida, D, Zied, C, Awadhi, E, et al. Co-occurrence of mcr-1 mediated colistin resistance and beta-lactamase-encoding genes in multidrug-resistant Escherichia coli from broiler chickens with colibacillosis in Tunisia. J Glob Antimicrob Resist. (2020) 22:538–45. doi: 10.1016/j.jgar.2020.03.017
46. Awawdeh, L, Forrest, R, Turni, C, Cobbold, R, Henning, J, and Gibson, J. Risk factors associated with the carriage of pathogenic Escherichia coli in healthy commercial meat chickens in Queensland, Australia. Poultry. (2022) 1:94–110. doi: 10.3390/poultry1020009
47. Ibrahim, RA, Cryer, TL, Lafi, SQ, Basha, E-A, Good, L, and Tarazi, YH. Identification of Escherichia from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC Vet Res. (2019) 15:1–16. doi: 10.1186/s12917-019-1901-1
48. Gharaibeh, MH, Sheyab, SYA, Lafi, SQ, and Etoom, EM. Risk factors associated with mcr-1 colistin-resistance gene in Escherichia coli broiler samples in northern Jordan. J Glob Antimicrob Resist. (2024) 36:284–92. doi: 10.1016/j.jgar.2024.01.003
49. Weerts, EAWS, Matthijs, MGR, Bonhof, J, van Haarlem, DA, Dwars, RM, Gröne, A, et al. The contribution of the immune response to enhanced colibacillosis upon preceding viral respiratory infection in broiler chicken in a dual infection model. Vet Immunol Immunopathol. (2021) 238:110276. doi: 10.1016/j.vetimm.2021.110276
Keywords: Escherichia coli , Klebsiella pneumoniae , mcr , risk factors, avian colibacillosis
Citation: Saeed MA, Asif H, Ehtisham-ul-Haque S, Khan AU, Rehman Au, Rehman A, Rafique MK, Ahmed I, Qamar MF, Tomaso H and El-Adawy H (2025) Detection and risk factor analysis of avian colibacillosis associated with colistin-resistant Escherichia coli and Klebsiella pneumoniae. Front. Vet. Sci. 12:1612542. doi: 10.3389/fvets.2025.1612542
Edited by:
Vangelis Economou, Aristotle University, GreeceReviewed by:
Marja-Liisa Hänninen, University of Helsinki, FinlandSultan Ali, University of Agriculture, Faisalabad, Pakistan
Copyright © 2025 Saeed, Asif, Ehtisham-ul-Haque, Khan, Rehman, Rehman, Rafique, Ahmed, Qamar, Tomaso and El-Adawy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Adnan Saeed, YWRuYW4uc2FlZWRAdXZhcy5lZHUucGs=; Hosny El-Adawy, aG9zbnkuZWxhZGF3eUBmbGkuZGU=
 Muhammad Adnan Saeed
Muhammad Adnan Saeed Haseeb Asif
Haseeb Asif Syed Ehtisham-ul-Haque
Syed Ehtisham-ul-Haque Aman Ullah Khan
Aman Ullah Khan Aziz ur Rehman
Aziz ur Rehman Aiman Rehman1
Aiman Rehman1 Ishtiaq Ahmed
Ishtiaq Ahmed Muhammad Fiaz Qamar
Muhammad Fiaz Qamar Herbert Tomaso
Herbert Tomaso Hosny El-Adawy
Hosny El-Adawy