- 1Guangxi Key Laboratory of Veterinary Biotechnology, Key Laboratory of China (Guangxi)-ASEAN Cross-Border Animal Disease Prevention and Control, Guangxi Veterinary Research Institute, Ministry of Agriculture and Rural Affairs of China, Nanning, China
- 2College of Animal Science and Technology, Guangxi University, Nanning, China
- 3Guangxi National Farm Yongxin Animal Husbandry Group Co., Ltd., Nanning, China
- 4Yulin Animal Disease Prevention and Control Center, Yulin, China
Introduction: Mycoplasma hyopneumoniae (Mhp) infection significantly challenges Guangxi’s pig farms, yet its prevalence and molecular characteristics remain poorly understood. This study aimed to define circulating Mhp genotypes and their distribution in the region.
Methods: From 2022–2023, 1,362 pig lung samples were randomly collected from 14 Guangxi regions. Mhp was detected using TaqMan Real-time PCR. Strong positive samples underwent multilocus sequence typing (MLST) of adk, rpoB, and tpiA genes to assess genetic relationships.
Results: Of 1,362 samples, 655 (48.1%) were Mhp-positive. MLST amplification succeeded for 61 samples, revealing 27 sequence types (24 novel) across all 14 regions. Phylogenetic analysis indicated predominant circulation of Mhp types I and V. High Mhp incidence and substantial genetic diversity were observed.
Discussion: This study provides comprehensive analysis of Mhp in Guangxi, revealing high prevalence and genetic diversity dominated by types I and V. These findings expand understanding of Mhp epidemiology in China and offer a theoretical foundation for developing prevention and control strategies in Guangxi.
Introduction
Mycoplasma hyopneumoniae (Mhp) is the etiological agent of Mycoplasma pneumonia (MPS), also called enzootic pneumonia (EP) in swine, commonly referred to as porcine “wheezing disease” (1). This disease exhibits high morbidity but low mortality, and can appear year-round. Clinical signs primarily include dyspnea, abdominal breathing, lethargy, and coughing, along with decreased growth performance and increased feed conversion ratio (2, 3). Mhp can also act synergistically with other pathogens (4, 5), such as porcine circovirus type 2 and porcine reproductive and respiratory syndrome virus, to produce porcine respiratory disease complex (PRDC) (6, 7), thereby complicating prevention and control efforts, and presenting a challenge to the worldwide swine industry. Guangxi Province, a critical region for swine production in China, faces challenges due to Mycoplasma hyopneumoniae (Mhp) infections across its diverse pig farms. Zhang et al. identified Guangxi as a high-prevalence area for Mhp in China during a 2019 outbreak, reporting substantial genetic diversity, including eight novel ST types. Li (8) reported widespread Mhp infection across Chinese pig farms, with a national antigen-positive rate of 19.19%. Guangxi contributed the largest sample size, exhibiting an infection rate comparable to the national average. However, a comprehensive understanding of the prevalence and molecular characteristics of these infections throughout the region remains elusive. Multilocus sequence typing (MLST) has emerged as the gold standard for high-resolution molecular epidemiology of Mhp, enabling robust strain differentiation, phylogenetic analysis, and inter-study comparisons via global databases (9, 10). This method defines stable sequence types (STs) based on nucleotide variations in core housekeeping genes. The analysis of the adk, rpoB, and tpiA genes specifically provides discriminatory power equivalent to the original 7-gene scheme for Mhp (11), making it highly practical for large-scale surveillance and genotyping directly from clinical samples.
This study randomly sampled 1,362 pig lung tissue from 14 cities in Guangxi for pathogen detection. The targeted gene sequences obtained by sequencing were connected in series according to the order of adk-rpoB-tpiA. Genetic evolution tree was drawn for genetic polymorphism analysis to explain the genotype and distribution of Mhp in Guangxi pig herds.
Materials and methods
Samples
Between 2022 and 2023, 1,362 lung samples (one per pig) were collected opportunistically from swine at various abattoirs across 14 municipalities in Guangxi Province, negating the need for ethical approval. In order to ensure the scientificity and fairness of sampling and minimize selection bias, we carried out meticulous and rigorous sampling work in 14 cities in Guangxi Province according to multi-dimensional factors such as geographical distribution characteristics, breeding scale and production capacity. In the process of pig farm selection, we comprehensively referred to the breeding record information provided by the agricultural departments of various cities and divided all farms into three categories: large, medium and small according to the breeding scale. Then, a stratified sampling method is used to randomly select a certain number of farms from each level according to an equal proportion, so as to ensure that farms of different sizes have the opportunity to be included in the sample category.
At the same time, the geographical span of each city is fully considered to ensure that the selected farms cover all major breeding clusters to avoid bias caused by geographical concentration. Specifically, samples originated from the following locations: Nanning (n = 282), Liuzhou (n = 42), Guilin (n = 81), Wuzhou (n = 110), Beihai (n = 76), Fangchenggang (n = 80), Qinzhou (n = 49), Guigang (n = 99), Yulin (n = 97), Baise (n = 98), Hezhou (n = 15), Hechi (n = 100), Laibin (n = 168), and Chongzuo (n = 65).
DNA extraction, RT-qPCR detection
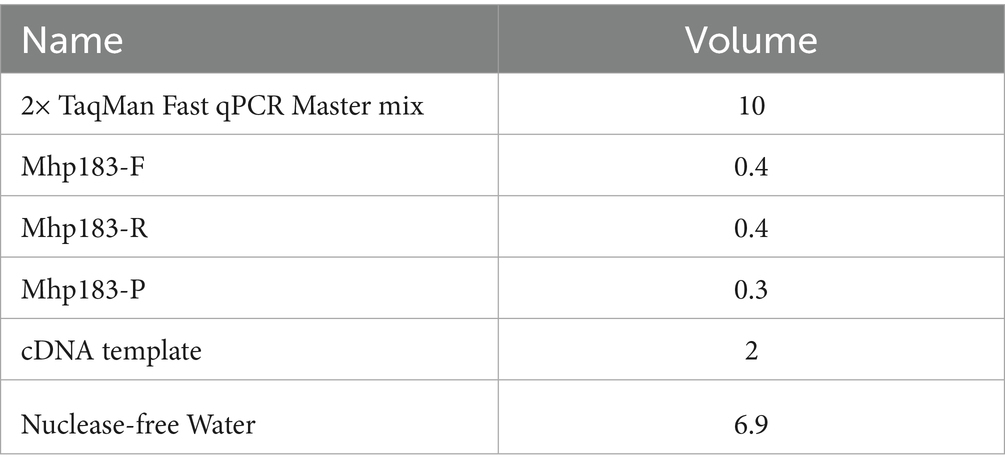
Lung tissue (0.5 g) was collected in a 2 mL enzyme-free Eppendorf tube and minced. Four grinding beads and approximately 1 mL of physiological saline were added to the tube. Each sample was homogenized twice at 70 Hz for 70 s utilizing a tissue homogenizer. DNA from Mhp was extracted from the homogenates utilizing an Automatic Nucleic Acid Extraction Machine (Zybio Technology Company, Chongqing Province, China). Mhp was detected utilizing a highly specific and sensitive qPCR assay (12). Primers and probe sequences were as follows: Mhp 183-F: CAAAGCGAGTATGAAGAACAAGAAA; Mhp 183-R: GTCATCATTGGGTGGCTAAGT; Mhp183-ROX-TCCAGGAAGTCAAGGTAACTAGTGACCA-BHQ. qPCR reactions were carried out in a 20 μL reaction volume consisting of 10 μL 2× Taqman Fast qPCR Mastermix (Sangong, Shanghai, China), 0.4 μL each of primers Mhp 183-F and Mhp 183-R, 0.3 μL of probe Mhp 183-P, 4 μL of cDNA, and 4.8 μL of nuclease-free water. Samples exhibiting strong positive results (Ct-value <30) were eligible for direct genotyping (13).
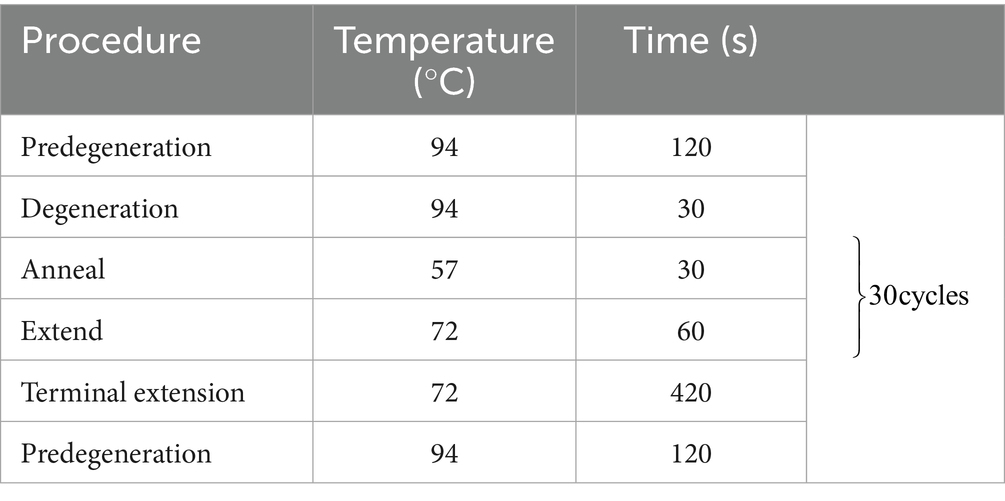
The reaction system is as follows:
The reaction system is as follows:
MLST
Nucleotide sequence data for the adk, ropB, and tpiA genes were generated from Mhp-positive samples through Sanger sequencing. These genes were amplified utilizing primers and conditions previously described by Mayor et al. Amplicons were resolved by electrophoresis on a 1.5% agarose gel and purified utilizing the SanPrep Column DNA Gel Extraction Kit (Sangon, Shanghai, China).
Sequencing was performed by Sangon Biotech (Shanghai, China). The resulting sequence data were deposited in the PubMLST database for sequence type (ST) assignment. Each housekeeping gene receives a unique allele number, and the combination of the three alleles determines a specific ST.
Guangxi, China
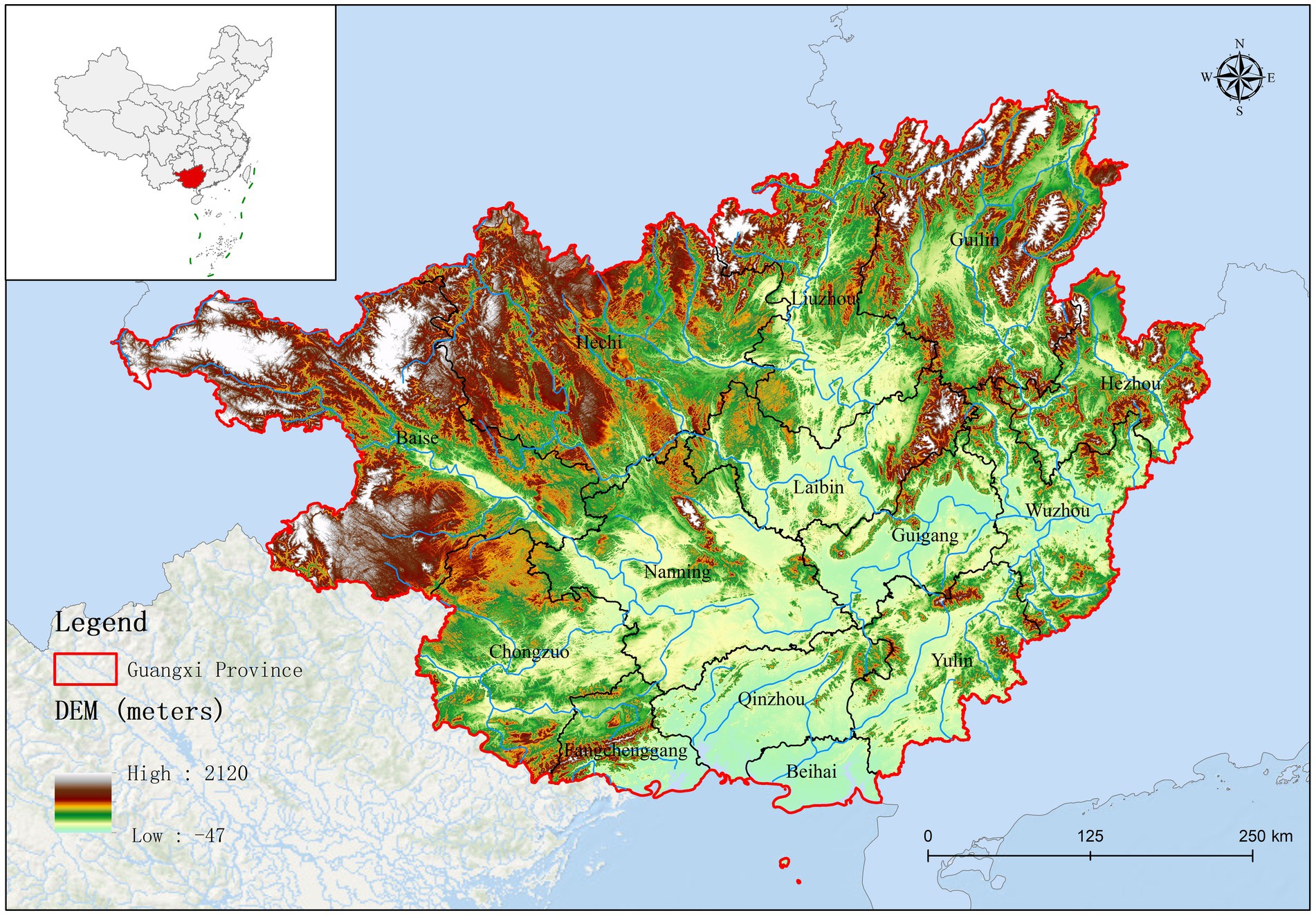
Guangxi Province is located in southern China, bordered by Yunnan to the west, Guizhou to the north, Hunan to the northeast, and Guangdong to the southeast. Its topography comprises a diverse range of landscapes, from mountains and karst formations to rivers and fertile plains. As presented in Supplementary Figure 1, the northern region of Guangxi (Baise, Hechi, Liuzhou, and Guilin) is primarily mountainous. Guangxi plays a crucial role in China’s pork production; the region has a large agricultural sector, with pig farming as a major component. In effect, Guangxi ranks as a leading province for pig farming and is a significant contributor to the national pork supply.
Data analysis
We obtained the allele data for adk, rpoB, tpiA and ST from the PubMLST database.1 We calculated Simpson’s index of diversity values utilizing the website.2 We carried out sequence alignment and cluster analysis through Molecular Evolutionary Genetics Analysis (MEGA version 6.0) utilizing the neighbour-joining method (Kimura 2-parameter model).
Results
Survey on Mhp positivity rate in Guangxi
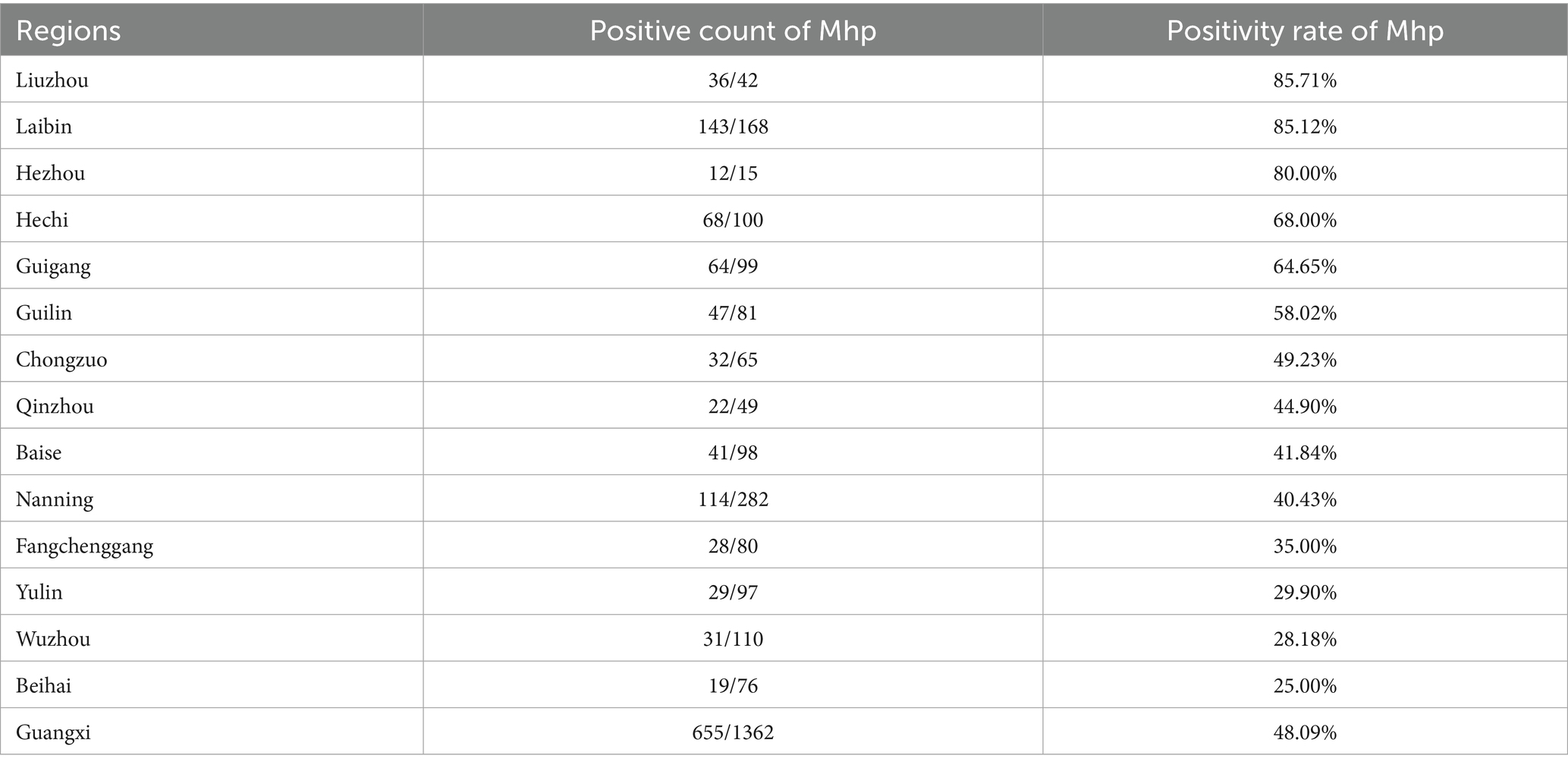
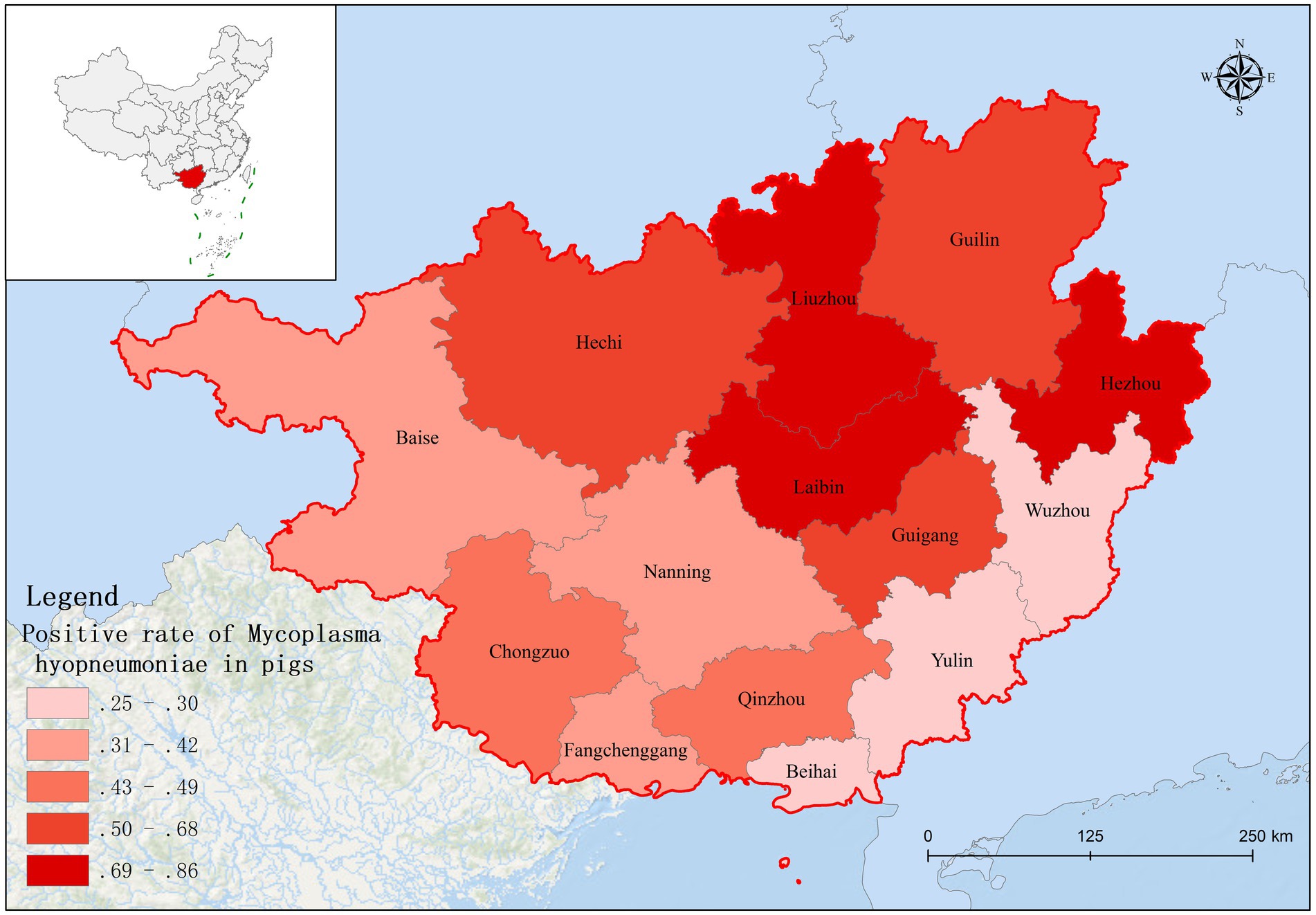
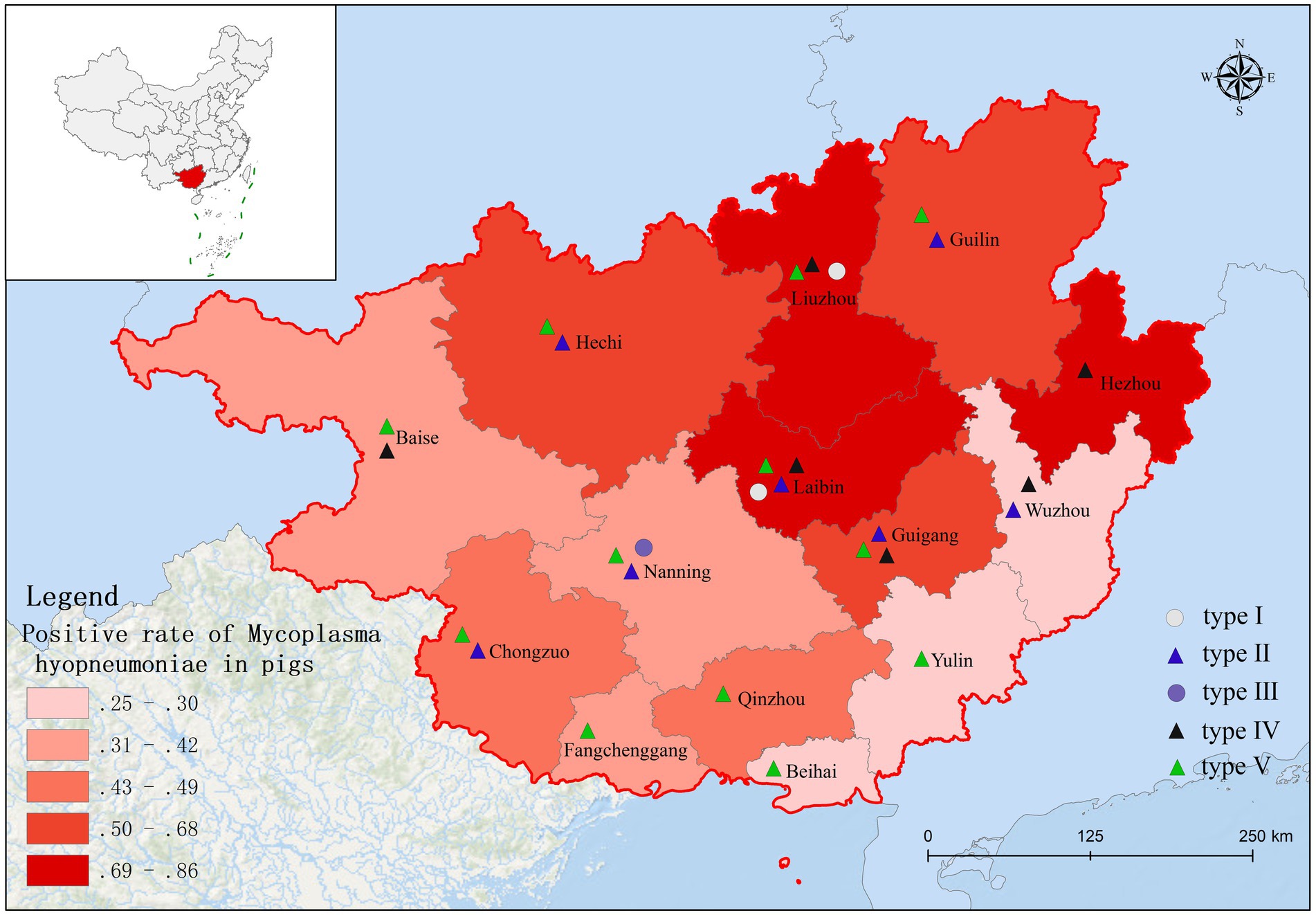
Based on fluorescence quantitative PCR results, we identified 655 Mhp-positive samples out of 1,362 lung tissue samples collected from 14 municipalities in Guangxi. Six regions demonstrated notably high positive rates: Liuzhou 85.71% (36/42), Hezhou 80.00% (12/15), Laibin 85.12% (143/168), Hechi 68.00% (68/100), Guigang 64.65% (64/99), and Guilin 58.02% (47/81). These six regions all maintained positivity rates above 50%. Across the entire Guangxi region, the overall Mhp positivity rate reached 48.09% (655/1,362). We discovered three regions with low Mhp positivity rates: Yulin 29.90% (29/97), Wuzhou 28.18% (31/110), and Beihai 25.00% (19/76), with all these regions indicating rates below 30% (Table 1). Our spatial analysis indicated that in the northern part of the Guangxi region, Laibin, Liuzhou, Hezhou, Guilin, and Hechi displayed higher Mhp positivity rates. In contrast, the southeastern areas of Wuzhou, Yulin, and Beihai, which lie next to Guangzhou, demonstrated relatively lower Mhp positivity rates (Figure 1). Our terrain analysis demonstrated that the northern regions of Guangxi, which had the previously mentioned higher Mhp-positive rates, featured higher terrain and primarily mountainous landscapes (Figure 2).
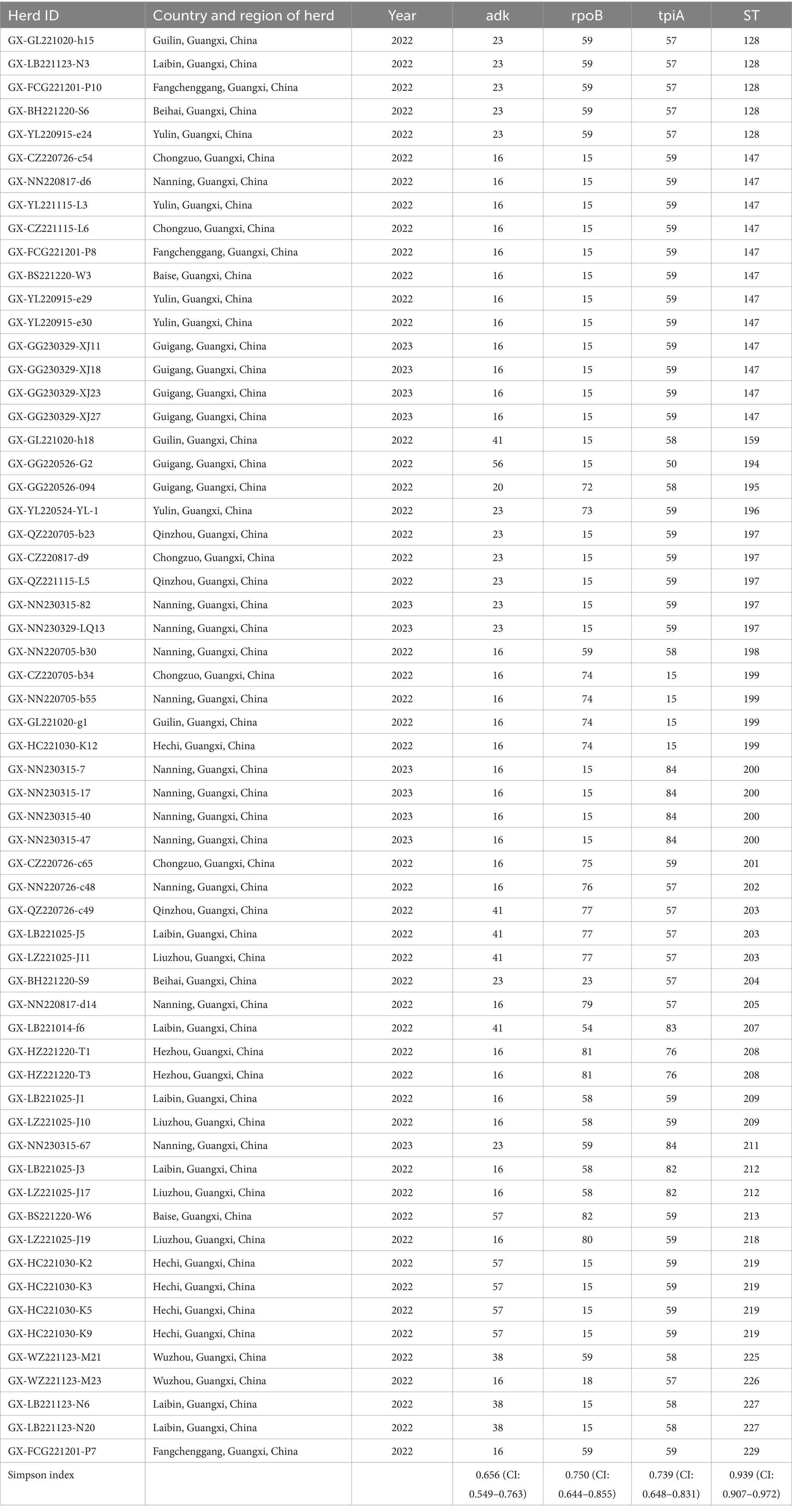
Of 655 PCR-positive samples (Ct <30) subjected to amplification of key MLST genes (adk, rpoB, tpiA), only 61 (9.3%) were successful, yielding a 90.7% failure rate. Figure 3 presents the gel electropherograms depicting the simultaneous amplification of these three housekeeping genes. Analysis of the alleles from the successfully amplified samples indicated the identification of two novel adk alleles (56, 57), 10 novel rpoB alleles (72, 73, 74, 75, 76, 77, 79, 80, 81, 82), and three novel tpiA alleles (82, 83, 84). These 61 samples were grouped into 27 ST types based on the allelic profiles of adk, rpoB, and tpiA (128, 147, 159, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 207, 208, 209, 211, 212, 213, 218, 219, 225, 226, 227, 229). Of these STs, 24 are newly described. Specifically, ST147 and ST128 were prevalent, observed in six geographically proximate cities (Baise, Chongzuo, Nanning, Fangchenggang, Yulin, and Guigang) and five more geographically dispersed cities (Nanning, Guilin, Laibin, Fangchenggang, Beihai, and Yulin), respectively. In addition, ST197, ST199, and ST203 were found in multiple regions. The Simpson index of diversity for STs is 0.939 (Table 2). Phylogenetic analysis of ST sequences resolved the 61 strains into five genotypes (types I, II, III, IV, and V). Type I comprised strains from Switzerland, Hungary, and Thailand, as well as two strainsfrom this study. Type II comprised 10 strains from this study, representing seven regions of Guangxi: Nanning, Hechi, Chongzuo, Guilin, Wuzhou, Laibin, and Guigang. This distribution suggests that type Mhp is prevalent in half of Guangxi. In addition, a phylogenetic relationship was observed between the UK strain J, the US strain 232, and the following strains from this study: ST194, ST227, ST225, ST198, ST199, and ST159. Type III was found only in the Nanning area, along with strains from Thailand, Canada, Australia, and South Korea. Type IV consisted of eight strains from this study, detected in Liuzhou, Laibin, Guigang, Wuzhou, Hezhou, and Baise. These strains demonstrated a closer evolutionary relationship with a 2016 French strain, a 2017 Thai strain, and a 2020 Jiangsu strain than with the Swiss strains belonging to a different genotype. Type V, the largest genotype in this study, was identified in nearly every region of Guangxi. This genotype also included a 2018 Jiangxi strain (JX486), 2019 Guangxi strains (GX23, GX8-2, and GXF10), 2020 Guangdong strains (GD-22, GD-18), and a 2020 Jiangsu strain (JS-10) (Figure 4). Geospatial analysis demonstrated a limited distribution of type I, exclusively in Laibin and Liuzhou, whereas type II exhibited a broader presence across Guangxi, represented by an “x” shaped distribution across the northeastern and southwestern regions. The four type III strains originated from a single farm located in Nanning. Type IV was primarily located in the northeastern quadrant of Guangxi, including Liuzhou, Hezhou, Wuzhou, Laibin, and Guigang. Type V was observed in all areas with the exception of Wuzhou and Hezhou (Figure 5).
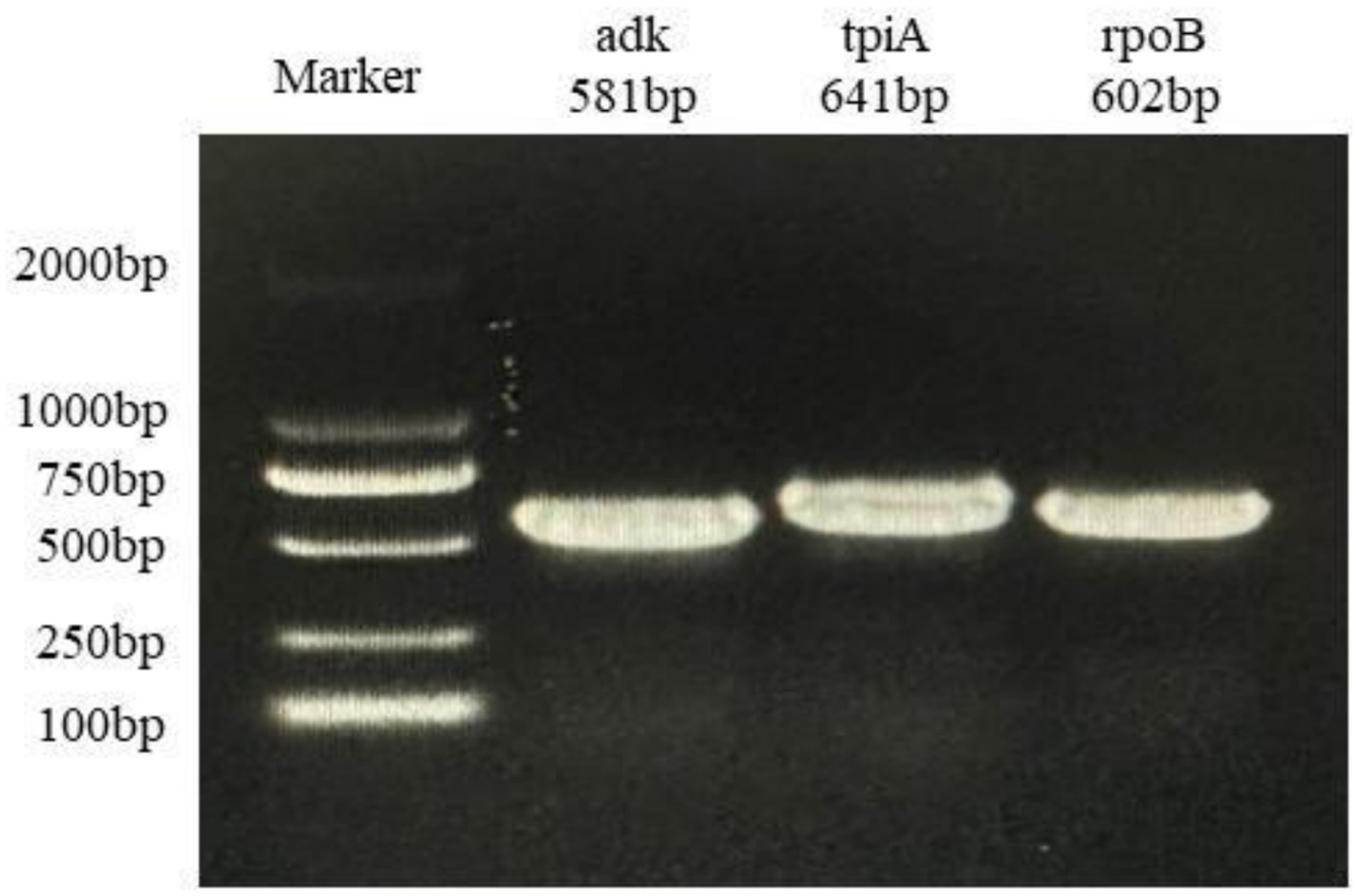
Figure 3. Agarosegel electrophoresis of PCR amplification of Mhp housekeeping gene adk, tpiA, and rpoB.
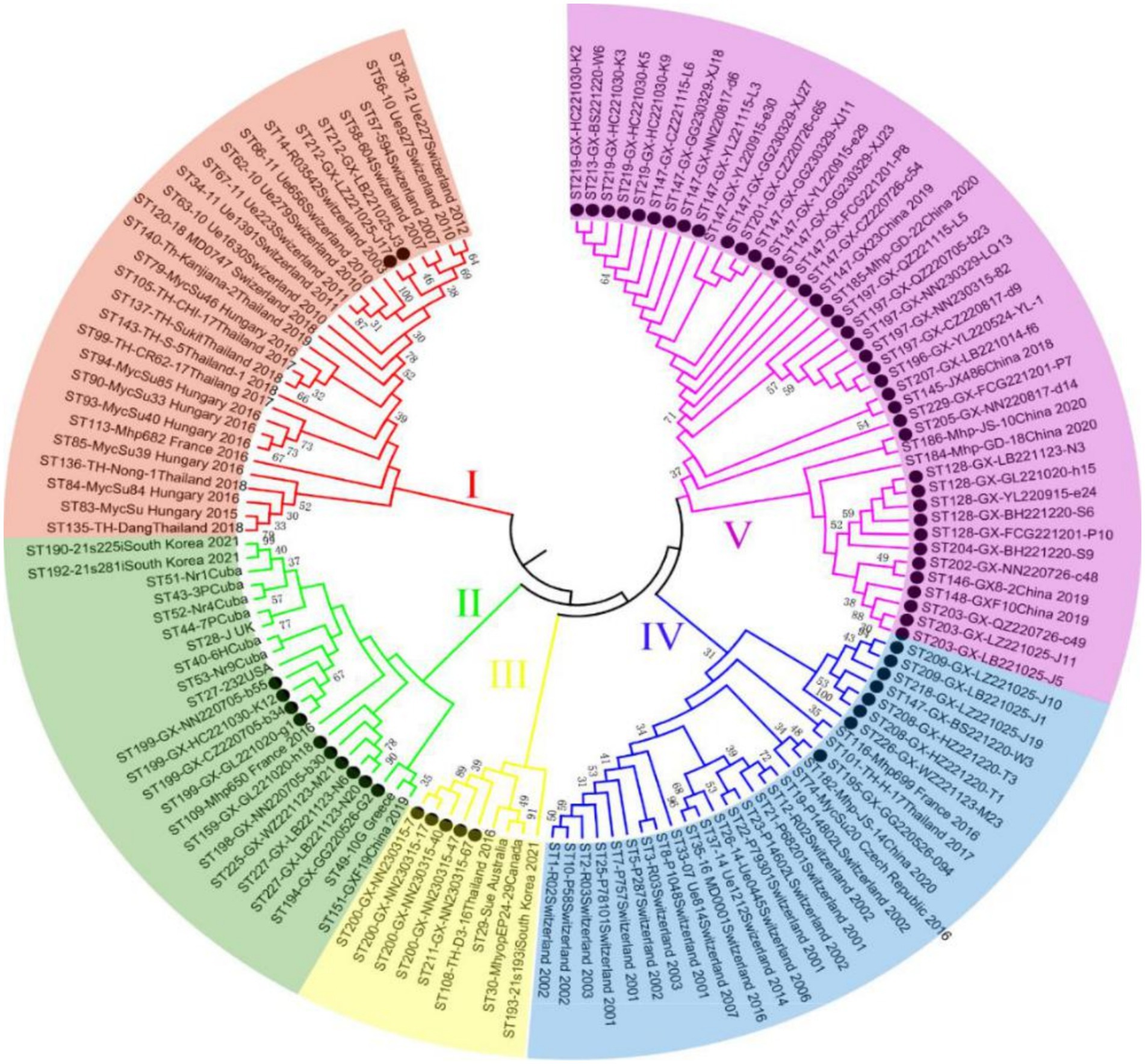
Figure 4. The neighbor-joining tree of selected sequence types (STs) based on the adk, rpoB, and tpiA sequences of Mhp in Guangxi. ● Represents the isolates in this experiment.
Discussion
A total of 1,362 samples were randomly collected from 14 different regions throughout Guangxi Province. Analysis by real-time fluorescence quantitative PCR identified 655 samples as positive for Mycoplasma hyopneumoniae (Mhp). This finding clearly demonstrates a serious Mhp infection problem in Guangxi. In particular, the positive rates in Liuzhou, Laibin, Hezhou, Hechi, Guigang, and Guilin exceeded 50%, indicating that these areas are the most severely affected by Mhp infection in Guangxi. The relatively high positive rates in Liuzhou, Laibin, Hezhou, Hechi, and Guilin may be closely related to their geographical characteristics. Compared to southern Guangxi, the northern part of the province has a higher latitude, lower temperatures, more mountainous terrain, and greater humidity. Previous work by Goodwin (14) and Browne et al. (15) has presented that Mhp is more likely topersist in environments with low temperatures and high humidity (16). Studies have demonstrated a strong association between the prevalence of this pathogen and seasonal variations, as well as a significant effect of climate factors (17, 18). For instance, research conducted in Belgium and the Netherlands found that the infection rate of Mycoplasma hyopneumoniae in swine differed seasonally, with a significantly higher infection rate in pigs born in autumn (19). Moreover, environmental factors, including temperature and humidity, greatly affect disease transmission. The spread of the pathogen may be facilitated by the creation of bioaerosols (20), notably during cold and wet seasons (18, 21). While vaccination has significantly offered some protection to pigs against Mhp, these protective effects are known to differ significantly across pig herds. This variability may arise from a range of factors, including the Mhp infection level in the herd, the age of infection onset in pigs, and even potential variations among Mhp isolates (22, 23). Notwithstanding such variability, vaccination is still broadly considered the most effective method for managing Mhp infection (24). These vaccines primarily reduce clinical signs by stimulating an immune response; however, they cannot entirely prevent infection or viral shedding (25, 26). It is important to understand that, while vaccination can successfully reduce clinical signs and lung lesions in infected pigs, its ability to limit the transmission of Mhp is quite restricted (22, 27). This limited capacity to control Mhp spread may be a key factor contributing to the challenges in effectively managing and controlling Mhp. In addition, a positive Mhp test result currently does not restrict the commercial trade of live pigs. Guangxi is a major pig-producing province in China, and live pig trade extends to adjacent provinces such as Guangdong and Jiangxi. This expansive trade network may contribute to the spread of Mhp across different regions. Pig trade is also an important route for the spread of pathogens between countries, which is common on many continents (28). Currently, Mhp is widespread in Guangxi Province. Specifically, Mhp positive rates in Guigang, Chongzuo, and Nanning are comparatively high. This might be due to that these areas are major pig production hubs in Guangxi. These combined factors likely contribute to the high Mhp infection rate and genetic diversity in the province. In this research, we performed a detailed genotyping analysis analyzing Mhp strains located in Guangxi, China. In our collection of 655 positive samples, we observed that 61 samples demonstrated concurrent amplification patterns of the adk, rpoB, and tpiA genes. When conducting multilocus sequence typing (MLST) assays of Mhp, the standard protocol traditionally requires that seven housekeeping genes must be amplified in a parallel fashion. Nevertheless, extensive research has consistently demonstrated that the discriminatory capabilities of the three genes mentioned above match those achieved with all seven housekeeping genes (29–31). For an overwhelming number of samples in this study, we regularly encountered situations where, likely due to insufficient nucleic acid content of Mhp present in the tissue samples or because of various PCR inhibitors in the samples (9, 32), we could not achieve successful simultaneous amplification and sequencing of all seven housekeeping genes. Our analysis indicated new sequence type (ST) distribution patterns throughout the 14 regions that constitute Guangxi Province. We discovered that particular ST classifications, specifically ST197, ST199, and ST203, were located across multiple separate regions. Across Guangxi Province, a considerable distance of approximately 520 kilometers separates Guilin (in the northeast) and Chongzuo (in the southwest). This geographic distribution suggests that distance alone does not significantly impede Mhp transmission.
Infected and asymptomatically infected pigs are the primary infection reservoirs, with sows and latently infected pigs representing the key vectors in farms. Mhp spreads horizontally across long distances through direct pig contact, or through contaminated excretions, oral secretions, and even aerosols (33, 34). Therefore, the inter-regional movement of pigs likely contributes to Mhp dissemination. Zhang et al. (35) first identified ST147 in Guangxi in 2019. However, another study indicated that ST128 was the major ST circulating in Guangxi during that period. By early 2022, Yiming (36) confirmed ST128 as the epidemic strain across seven infected pig farms in Guangxi. This study also identified ST197, ST199, and ST203 in various locations throughout Guangxi. Considering the limited sample size, we could only establish a preliminary prevalence trend for these STs. Future research with a larger sample cohort is necessitated to confirm and further investigate the concurrent dominance of multiple STs in Guangxi Province (37). Analysis of the genetic evolutionary tree confirmed the classification of the 61 tested strains into five genotypes (I, II, III, IV, and V). The results indicate that genotype V first appeared in other regions of China several years ago (35) and is now widely distributed throughout both Guangxi and Guangdong Provinces. In addition, the strong genetic similarity observed between the strains isolated in Guangxi and those in other provinces is likely connected to Guangxi’s prominent role as a major pig-producing region in China. The transport and sale of pigs from Guangxi to other provinces, including Guangdong and Jiangxi, may offer a mechanism for the spread of Mhp. This interprovincial movement of pigs offers a plausible explanation for the observation that the majority of genotype V strains from Guangxi analyzed in this study demonstrate a close genetic relationship with strains identified in Guangdong. In the preceding study, we identified ST128 and ST147 in 5 and 6 different regions across Guangxi, respectively, with both indicating significant prevalence throughout the entire province. Our analysis also indicated that ST199, ST197, and ST203 were detected across multiple regions, indicating clear patterns of epidemic spread in Guangxi. A critical point to emphasize is that these five ST types all originated from genotypes II and V. However, our research demonstrated no significant correlation between the geographical origins of the samples and the ST types of the isolated strains, as we found various ST types present in individual regions, while the same ST types appeared across different geographical locations. These observations align closely with findings documented in the majority of international research papers (29), indicating widespread and extensive dissemination of Mhp across Guangxi Province. Our data also demonstrated that Mhp has maintained its presence in Guangxi over an extended time period, leading to the formation of unique independent clusters. Based on these comprehensive findings, we can draw the logical conclusion that the genetic diversity of Mhp observed in Guangxi acts as one of the key factors contributing to the high infection rates of Mhp observed throughout this geographical area.
Conclusion
Based on the analysis of 1,362 randomly collected samples from Guangxi, China, our results indicated a high incidence rate of Mhp in the region. The study identified ST147 and ST128 as the primary genotypes, which were detected across six and five regions of Guangxi, respectively. Among the newly identified STs, ST197, ST199, and ST203 demonstrated up as the current trending genotypes in Guangxi. All these ST types fell in cluster II and cluster V, which represented the main genotypes currently spreading throughout Guangxi. This research stands as one of the most thorough analyses into the genotypes and geographical distribution of Mhp in Guangxi’s swine populations. Our findings have significantly advanced our knowledge regarding the incidence rates and molecular typing of Mhp in this region, contributing valuable data to China’s local Mhp epidemiological database while offering a scientific foundation for implementing Mhp control and prevention strategies in Guangxi, China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP123456–OP123527.
Ethics statement
The animal study protocol was approved by the Institutional Ethics Committee. All samples were collected post-mortem from commercial slaughterhouses, in accordance with national regulations that exempt material obtained from routine food production processes from requiring specific animal welfare approval. The study was conducted in full compliance with local legislation and institutional requirements.
Author contributions
YZ: Writing – review & editing. SZ: Data curation, Writing – review & editing. LL: Writing – review & editing, Investigation. YQ: Investigation, Writing – review & editing. KC: Writing – review & editing, Methodology. ZC: Supervision, Writing – review & editing. BL: Writing – original draft, Data curation. QD: Data curation, Writing – original draft. YP: Writing – review & editing. YHu: Investigation, Writing – original draft. ZD: Investigation, Writing – review & editing. KJ: Writing – review & editing. XY: Writing – review & editing. YHe: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the district science and technology bureau scientific research and technology development programme project of Guangxi Key Technologies R&D Program (AB2506938), Guangxi Key Technologies R&D Program (AB25069246), Basic scientific research business expense special topic of Guxnagxi (Guike Specialized 23-4), Agricultural Science and Technology Self-financed Project Mission Statement of Guangxi (Z2023033), Scientific Research and Technology Development Program of Nanning (20232039), Innovation Drive Development Special Funds Program of Guangxi (Guike AA18118051, Guike AA17204057) and Special Fund for Guangxi Innovation Team Building of National Modern Agricultural Industry Technology System (nycytxgxcxtd-2023-15-02).
Conflict of interest
LL and KC were employed by Guangxi National Farm Yongxin Animal Husbandry Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1619301/full#supplementary-material
Abbreviations
EP, Enzootic pneumonia; MLST, Multilocus sequence typing; ST, Sequence type; Mhp, Mycoplasmahyopneumoniae; PRDC, Porcine respiratory disease syndrome.
Footnotes
References
1. Maes, D, Segales, J, Meyns, T, Sibila, M, Pieters, M, and Haesebrouck, F. Control of Mycoplasma hyopneumoniae infections in pigs. Vet Microbiol. (2008) 126:297–309. doi: 10.1016/j.vetmic.2007.09.008
2. Deeney, AS, Maglennon, GA, Chapat, L, Crussard, S, Jolivet, E, and Rycroft, AN. Mycoplasma hyopneumoniae evades phagocytic uptake by porcine alveolar macrophages in vitro. Vet Res. (2019) 50:51. doi: 10.1186/s13567-019-0667-6
3. Thacker, EL. Diagnosis of Mycoplasma hyopneumoniae. Anim Health Res Rev. (2004) 5:317–20. doi: 10.1079/ahr200491
4. Hansen, MS, Pors, SE, Jensen, HE, Bille-Hansen, V, Bisgaard, M, Flachs, EM, et al. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J Comp Pathol. (2010) 143:120–31. doi: 10.1016/j.jcpa.2010.01.012
5. Zhang, H, Lunney, JK, Baker, RB, and Opriessnig, T. Cytokine and chemokine mRNA expression profiles in tracheobronchial lymph nodes from pigs singularly infected or coinfected with porcine circovirus type 2 (PCV2) and Mycoplasma hyopneumoniae (MHYO). Vet Immunol Immunopathol. (2011) 140:152–8. doi: 10.1016/j.vetimm.2010.11.019
6. Bourry, O, Fablet, C, Simon, G, and Marois-Créhan, C. Efficacy of combined vaccination against Mycoplasma hyopneumoniae and porcine reproductive and respiratory syndrome virus in dually infected pigs. Vet Microbiol. (2015) 180:230–6. doi: 10.1016/j.vetmic.2015.09.015
7. Garcia-Morante, B, Segalés, J, Fraile, L, Pérez De Rozas, A, Maiti, H, Coll, T, et al. Assessment of Mycoplasma hyopneumoniae-induced pneumonia using different lung lesion scoring systems: a comparative review. J Comp Pathol. (2016) 154:125–34. doi: 10.1016/j.jcpa.2015.11.003
9. Kuhnert, P, and Overesch, G. Molecular epidemiology of Mycoplasma hyopneumoniae from outbreaks of enzootic pneumonia in domestic pig and the role of wild boar. Vet Microbiol. (2014) 174:261–6. doi: 10.1016/j.vetmic.2014.08.022
10. Longyu, Z, Yao, Z, Longhua, L, Jiyun, C, Caiping, M, Jie, H, et al. Isolation and identification of Mycoplasma hyopneumoniae Mhp-1 strain and its whole genome sequencing analysis. Chin J Prev Vet Med. (2024) 46:630–5. doi: 10.3969/j.issn.1008-0589.202311030
11. Mayor, D, Jores, J, Korczak, BM, and Kuhnert, P. Multilocus sequence typing (MLST) of Mycoplasma hyopneumoniae: a diverse pathogen with limited clonality. Vet Microbiol. (2008) 127:63–72. doi: 10.1016/j.vetmic.2007.08.010
12. Ying, Z, Shuo, Z, Guo-xi, Q, Long-hua, L, Ting, X, Zhong-wei, C, et al. Establishment and application of TaqMan duplex real-time PCR detection method for Mhp and Mhr. Prog Vet Med. (2024) 45:11–6. doi: 10.3969/j.issn.1007-5038.2024.02.004
13. Mayor, D, Zeeh, F, Frey, J, and Kuhnert, P. Diversity of Mycoplasma hyopneumoniae in pig farms revealed by direct molecular typing of clinical material. Vet Res. (2007) 38:391–8. doi: 10.1051/vetres:2007006
14. Goodwin, RF. Apparent reinfection of enzootic-pneumonia-free pig herds: search for possible causes. Vet Rec. (1985) 116:690–4. doi: 10.1136/vr.116.26.690
15. Browne, C, Loeffler, A, Holt, HR, Chang, YM, Lloyd, DH, and Nevel, A. Low temperature and dust favour in vitro survival of Mycoplasma hyopneumoniae: time to revisit indirect transmission in pig housing. Lett Appl Microbiol. (2017) 64:2–7. doi: 10.1111/lam.12689
16. Li, R, Hu, Y, Ge, M, Zhao, D, Yang, T, Qing, R, et al. Analysis of correlation between the detection rate of Mycoplasma hyopneumoniae in slaughter pigs and season, climate change, and presence of lung lesions. Med Weter. (2019) 75:6196–2019. doi: 10.21521/mw.6196
17. Vangroenweghe, FACJ, Labarque, GG, Piepers, S, Strutzberg-Minder, K, and Maes, D. Mycoplasma hyopneumoniae infections in peri-weaned and post-weaned pigs in Belgium and the Netherlands: prevalence and associations with climatic conditions. Vet J. (2015) 205:93–7. doi: 10.1016/j.tvjl.2015.03.028
18. Yue, W, Liu, Y, Meng, Y, Ma, H, and He, J. Prevalence of porcine respiratory pathogens in slaughterhouses in Shanxi Province, China. Vet Med Sci. (2021) 7:1339–46. doi: 10.1002/vms3.532
19. Lisgara, M, Poulaki, K, Kalogeropoulos, L, Skampardonis, V, and Katsafadou, AI. Frequency and severity of enzootic pneumonia-like lesions in Greek swine herds and their association with different vaccination protocols against Mycoplasma hyopneumoniae. J Appl Anim Res. (2022) 50:540–7. doi: 10.1080/09712119.2022.2110499
20. Shi, W, Wei, M, Wang, Q, Wang, H, Ma, C, and Shi, C. Rapid diagnosis of Mycoplasma pneumonia infection by denaturation bubble-mediated strand exchange amplification: comparison with LAMP and real-time PCR. Sci Rep. (2019) 9:896. doi: 10.1038/s41598-018-36751-z
21. Cai, HY, van Dreumel, T, McEwen, B, Hornby, G, Bell-Rogers, P, McRaild, P, et al. Application and field validation of a PCR assay for the detection of Mycoplasma hyopneumoniae from swine lung tissue samples. J Vet Diagn Invest. (2007) 19:91–5. doi: 10.1177/104063870701900115
22. Mateusen, B, Maes, D, Van Goubergen, M, Verdonck, M, and de Kruif, A. Effectiveness of treatment with lincomycin hydrochloride and/or vaccination against Mycoplasma hyopneumoniae for controlling chronic respiratory disease in a herd of pigs. Vet Rec. (2002) 151:135–40. doi: 10.1136/vr.151.5.135
23. Villarreal, I, Maes, D, Vranckx, K, Calus, D, Pasmans, F, and Haesebrouck, F. Effect of vaccination of pigs against experimental infection with high and low virulence Mycoplasma hyopneumoniae strains. Vaccine. (2011) 29:1731–5. doi: 10.1016/j.vaccine.2011.01.002
24. Meyns, T, Dewulf, J, de Kruif, A, Calus, D, Haesebrouck, F, and Maes, D. Comparison of transmission of Mycoplasma hyopneumoniae in vaccinated and non-vaccinated populations. Vaccine. (2006) 24:7081–6. doi: 10.1016/j.vaccine.2006.07.004
25. Stingelin, GM, Mechler-Dreibi, ML, Storino, GY, Sonalio, K, de Souza Almeida, HM, Petri, FAM, et al. Chemotherapeutic strategies with valnemulin, tilmicosin, and tulathromycin to control Mycoplasma hyopneumoniae infection in pigs. Antibiotics. (2022) 11:893. doi: 10.3390/antibiotics11070893
26. Storino, GY, Petri, FAM, Mechler-Dreibi, ML, Aguiar, GA, Toledo, LT, Arruda, LP, et al. Use of nanostructured silica SBA-15 as an Oral vaccine adjuvant to control Mycoplasma hyopneumoniae in swine production. Int J Mol Sci. (2023) 24:6591. doi: 10.3390/ijms24076591
27. Maes, D, Sibila, M, Kuhnert, P, Segales, J, Haesebrouck, F, and Pieters, M. Update on Mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound Emerg Dis. (2018) 65:110–24. doi: 10.1111/tbed.12677
28. Cha, S, Choi, E, Park, J, Yoon, S, Song, J, Kwon, J, et al. Molecular characterization of recent Korean porcine reproductive and respiratory syndrome (PRRS) viruses and comparison to other Asian PRRS viruses. Vet Microbiol. (2006) 117:248–57. doi: 10.1016/j.vetmic.2006.05.007
29. Balestrin, E, Kuhnert, P, Wolf, JM, Wolf, LM, Fonseca, A, Ikuta, N, et al. Clonality of Mycoplasma hyopneumoniae in swine farms from Brazil. Vet Microbiol. (2019) 238:108434. doi: 10.1016/j.vetmic.2019.108434
30. Dos, SL, Sreevatsan, S, Torremorell, M, Moreira, MA, Sibila, M, and Pieters, M. Genotype distribution of Mycoplasma hyopneumoniae in swine herds from different geographical regions. Vet Microbiol. (2015) 175:374–81. doi: 10.1016/j.vetmic.2014.11.018
31. Panneitz, AK, Braga, ER, Petri, FAM, Menegatt, JCO, Driemeier, D, Maes, D, et al. Exploring the genetic diversity of Mycoplasma hyopneumoniae in pigs with pneumonia and pleurisy at slaughter. Microorganisms. (2024) 12:1988. doi: 10.3390/microorganisms12101988
32. Kuhnert, P, Overesch, G, and Belloy, L. Genotyping of Mycoplasma hyopneumoniae in wild boar lung samples. Vet Microbiol. (2011) 152:191–5. doi: 10.1016/j.vetmic.2011.04.026
33. Calsamiglia, M, and Pijoan, C. Colonisation state and colostral immunity to Mycoplasma hyopneumoniae of different parity sows. Vet Rec. (2000) 146:530–2. doi: 10.1136/vr.146.18.530
34. Fano, E, Pijoan, C, and Dee, S. Evaluation of the aerosol transmission of a mixed infection of Mycoplasma hyopneumoniae and porcine reproductive and respiratory syndrome virus. Vet Rec. (2005) 157:105–8. doi: 10.1136/vr.157.4.105
35. Zhang, H, Wang, Y, Gao, L, Wang, Y, and Wei, R. Genotype diversity of Mycoplasma Hyopneumoniae in Chinese swine herds based on multilocus sequence typing. BMC Vet Res. (2021) 17:347. doi: 10.1186/s12917-021-03059-6
36. Yiming, G. Epidemiological investigation and immune effect evaluation of Mycoplasma pneumoniae in a pig farm of a company in Guangxi In: Master thesis. Wuhan: Huazhong Agricultural University (2023)
Keywords: Mycoplasma hyopneumoniae, genotyping, MLST, swine epidemiology, Guangxi
Citation: Zhou Y, Zhao S, Liang L, Qin Y, Cheng K, Chen Z, Lu B, Duan Q, Peng Y, Huang Y, Duan Z, Jiang K, Yang X and He Y (2025) Epidemiological investigation of porcine Mycoplasma hyopneumoniae in pig herds in Guangxi, China (2022–2023) and genetic diversity analysis based on multilocus sequence typing. Front. Vet. Sci. 12:1619301. doi: 10.3389/fvets.2025.1619301
Edited by:
Mengmeng Zhao, Foshan University, ChinaReviewed by:
Li Chaosi, Boehringer Ingelheim Animal Health (Shanghai) Co., Ltd., ChinaMengkai Cai, Guangdong Meizhou Vocational and Technical College, China
Copyright © 2025 Zhou, Zhao, Liang, Qin, Cheng, Chen, Lu, Duan, Peng, Huang, Duan, Jiang, Yang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying He, aGV5aW5nOTIxQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Ying Zhou
Ying Zhou Shuo Zhao
Shuo Zhao Longhua Liang3†
Longhua Liang3† Ke Cheng
Ke Cheng