- 1Institute of Veterinary Medicine and Immunology, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2Research Center of Avian Diseases, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, Sichuan, China
- 4Engineering Research Center of Southwest Animal Disease Prevention and Control Technology, Ministry of Education of the People's Republic of China, Chengdu, Sichuan, China
- 5Hulunbuir Agricultural and Livestock Product Quality and Safety Center, Hulunbuir, Inner Mongolia, China
Effector protein functions of Type III secretion system (T3SS) encoded by Salmonella pathogenicity islands 2 (SPI-2) have not been fully characterized in Salmonella enterica serovar Choleraesuis. This study characterized 21 effectors of SPI-2 T3SS of S. Choleraesuis in terms of macrophage survival and virulence in mice via construction of various gene mutant strains. Eight effector genes including sseF, sseJ, sifB, sseK, sifA, sopD2, steC, and steD contributed to bacterial survival in macrophage cell line RAW264.7; whereas only sopD2 also promoted bacterial virulence in mice like other three effector genes sseL, steA, and spiC. The mutant strain, ΔsopD2, ΔsseL, ΔsteA, or ΔspiC, led to higher mouse survival compared to the wild-type strain post-oral infection, while their bacterial loads in spleen and liver were not reduced except the ΔspiC that was undetectable in mouse tissues. Then, the triple-gene mutant strain ΔsseLΔsopD2ΔsteA was constructed and found to be virulence attenuated with a compromised colonization ability. Finally, immunization of this mutant orally induced robust serum IgG responses and provided 40% protection against lethal S. Choleraesuis challenge. Our study highlights the critical role of four SPI-2 T3SS effectors in S. Choleraesuis pathogenesis.
1 Introduction
Salmonella enterica serovar Choleraesuis (S. Choleraesuis) is a zoonotic pathogen causing swine paratyphoid, characterized by enterocolitis and septicemia, which imposes substantial economic burdens on global swine husbandry (1, 2). Although S. Choleraesuis is adapted to pigs, it is also a major cause of life-threatening septicemia, particularly in children and immunocompromised individuals in East Asia and Europe (3, 4). Human infections frequently arise from direct contact with infected swine or ingestion of contaminated pork-derived products (1, 5). Due to the excessive use of antibiotics and environmental diversity, the emergence of multidrug-resistant S. Choleraesuis strains has become increasingly prevalent (6–8).
Vaccination represents the most cost-effective prophylactic strategy for disease control, effectively reducing antibiotic use and retarding the emergence of antibiotic resistance (9). The live vaccine strain C500 obtained by chemical mutagenesis has been used in China for more than 40 years to prevent paratyphoid fever in piglets (10). However, it still has non-negligible side effects related to residual toxicity, leading to adverse reactions in animals after vaccination and the genetic background of the vaccine is still unclear (11). Notably, there is no available vaccine for human use to date. Therefore, it is urgent and necessary to devise an innovative and efficacious vaccine against this important pathogen.
Understanding the mechanisms underlying bacterial pathogenesis is essential for the development of live attenuated vaccines. The Salmonella pathogenicity islands 2 (SPI-2) type III secretion system (T3SS) has been found to be essential for bacterial virulence of S. Typhimurium (12). This system promotes bacterial replication within membrane-bound Salmonella-containing vesicles (SCV) in host macrophages via production of various effector proteins (13). Loss of some effector genes sseF, sseG, and sseM that maintain bacterial nutrient acquisition within vesicles significantly reduces the replication ability of S. Typhimurium in host cells (14, 15). Effectors SpvB and SteC manipulate the actin cytoskeleton, affecting bacterial replication and subsequently impacting bacterial virulence (16, 17). Mutations in SPI-2 T3SS effectors can attenuate virulence, positioning them as promising candidates for live attenuated vaccines (18).
Despite their recognized importance, the specific roles of individual SPI-2 T3SS effectors in S. Choleraesuis virulence remain poorly characterized. This study aims to characterize SPI-2 T3SS effectors of S. Choleraesuis via construction of various single effector gene deletion strains. The mutant strains were systematically evaluated for their intracellular survival in macrophages, growth curves and swimming, and virulence in mice. Then, a triple mutant strain (ΔsseLΔsopD2ΔsteA) was constructed based on virulence effectors and its protection efficacy was finally evaluated.
2 Materials and methods
2.1 Bacterial strains and growth conditions
A complete list of all bacterial strains and plasmids utilized in the experiments is provided in Supplementary Tables S1, S2. Salmonella Choleraesuis CVCC2139 was referred to as the wild-type (WT) strain for genetic manipulation to construct the indicated mutants. The Escherichia coli (E. coli) SM10 λ pir strain (19) served as the host for transferring suicide plasmids. All bacterial strains were cultured in Luria Bertani (LB) broth or agar at 37°C containing the appropriate antibiotics: 25 μg/mL chloramphenicol, 100 μg/mL ampicillin, and 50 μg/mL 2,6-diaminopimelic acid. For counterselecting mutant constructs via the sacB gene system, NaCl-free LB agar supplemented with 12.5% (w/v) sucrose was used.
2.2 Construction of the S. Choleraesuis mutant and complemented strains
Twenty-two S. Choleraesuis mutants were generated via allelic exchange, employing the suicide plasmid pRE112 as previously detailed (20). The primer sequences designed for gene deletion or complementation in S. Choleraesuis strains are detailed in Supplementary Table S3. To generate the ΔsseJ mutant, upstream and downstream homologous arms were PCR-amplified using primer pairs DsseJ-1F/1R and DsseJ-2F/2R, respectively. These PCR products were joined by overlap PCR and subsequently cloned and inserted into the suicide vector pRE112 (21) through seamless cloning generating plasmid pRE112-ΔsseJ, which carries a deletion of the entire sseJ gene. The pRE112-ΔsseJ plasmid was subsequently introduced into the WT strain via conjugation. This process involved chloramphenicol-mediated positive selection and sacB-mediated sucrose sensitivity screening for the generation of the markerless mutant strain ΔsseJ. Furthermore, to complement sseJ gene in the ΔsseJ, the coding sequence of sseJ were amplified with the primer sseJ-F/R. Then, the PCR product was inserted into the plasmid of pCZb1 (20) via a Seamless Cloning Kit (Sangon Biotech, Shanghai, China), generating plasmid pCZb1-sseJ. Following, the recombinant plasmid was transformed into the mutant strain ΔsseJ to construct the complemented strain named C-ΔsseJ. The same method was applied to constructions of other gene mutants and complemented strains.
2.3 Detection of growth curves of S. Choleraesuis strains
The S. Choleraesuis WT and mutant strains were inoculated in 5 mL of LB broth at 37°C with shaking at 180 rpm/min overnight. The following day, cultures of each strain were normalized to an OD600 of 0.05 and then cultured in LB broth at 37°C with shaking at 180 rpm/min. The OD600 of each culture was measured every 2 h for 12 h.
2.4 Swimming assay
The swimming motility phenotypes of wild-type (WT) and mutant bacterial strains were evaluated using a previously described protocol (22). Cultures of each strain were grown in LB broth to an optical density (OD600) of 0.6–0.8. Bacteria were harvested by centrifugation, washed twice with PBS, and resuspended in the same buffer. Subsequently, 3 μL of the bacterial suspension was applied as droplets to LB agar plates containing 0.25% agar. After incubation at 37°C for 6 h, the diameter of the bacterial migration zone was measured to assess swimming motility.
2.5 Adhesion, invasion and intracellular survival of S. Choleraesuis in macrophages
RAW264.7 macrophages were plated at a density of 5 × 105 cells per well in 24-well plates containing DMEM (Gibco, NY, USA) supplemented with 10% FBS (Tian Hang, Hangzhou, China) and 1% penicillin-streptomycin. S. Choleraesuis WT or mutant strains were added at a multiplicity of infection (MOI) of 100. Following a 30-min incubation in a 5% CO2 at 37°C incubator to facilitate bacterial adhesion, cell monolayers were washed thrice with PBS to remove non-adherent bacterial cells. The adherent bacteria were then released by lysing the cells with 0.2% Triton X-100, and their numbers were enumerated via serial dilution and colony counting.
For the invasion assay, after the 30-min adhesion step, fresh DMEM was added and cells were incubated for an additional 90 min at 37°C under 5% CO2. After incubation, the supernatant was discarded, cells were washed twice with PBS and lysed using 0.2% Triton X-100 to enumerate intracellular bacteria.
For intracellular survival analysis, following the invasion step, DMEM supplemented with 10 ng/mL gentamicin was used to eliminate extracellular bacteria. T = 0 h was defined as the initial time point following invasion. At designated time points (T = 0 h and 24 h), serial dilutions of the resulting cell lysates were then plated onto MacConkey agar (Coolaber, Beijing, China) plates for bacterial enumeration and incubated at 37°C for 24 h to count colony-forming units (CFUs).
2.6 Colonization and virulence of S. Choleraesuis mutant strains in mice
Female Kunming mice (6 weeks old) were procured from Dashuo Experimental Animal Ltd. (Chengdu, China) and underwent a 1-week acclimation period before experimental procedures. Bacterial colonization and virulence phenotypes were evaluated using methodologies reported in prior studies (20, 22). Ten mice were orally inoculated with PBS or approximately 1 × 108 CFU of S. Choleraesuis WT strain or each mutant strain. Then, spleens and livers were collected from 5 mice at 6 days post-infection. The samples were weighed, ground in PBS, and the bacterial suspensions were serially diluted and spread onto MacConkey agar (Coolaber, Beijing, China) to enumerate viable CFUs, which were expressed as log10 CFU/g. The remaining 5 mice in each group were observed for survival for 1 month after infection.
2.7 Measurement of 50% Lethal dose (LD50) of the ΔsseLΔsopD2ΔsteA strain
The LD50 of the ΔsseLΔsopD2ΔsteA was determined as previously described (23). 10-fold serial dilutions of the CFU of the ΔsseLΔsopD2ΔsteA strain were orally inoculated into groups of Kunming mice (n = 5/per dose). Animals were monitored daily for 30 days post-infection to assess survival rates. The median lethal dose (LD50) was determined using the Reed-Muench method. To ensure humane endpoints, mice exhibiting severe distress—characterized by labored breathing, tremors, unresponsiveness to tactile stimuli, or inability to access food/water—were humanely euthanized via CO2 inhalation. Deceased animals were immediately subjected to sterilization, sealed in biohazard bags, and transferred to the Sichuan Agricultural University Laboratory Animal Center for compliant biosafety disposal.
2.8 Immunization and challenge
Female Kunming mice (6–8 weeks old) were randomly divided into three groups (n = 20/group). The experimental group received an oral gavage of 1 × 109 CFU ΔsseLΔsopD2ΔsteA in 200 μL PBS, while the control group received an equal volume of PBS alone. A booster immunization was administered on day 14 using the same protocol. Serum samples were collected via retro-orbital bleeding on days 7 and 21 from six randomly selected mice per group. On day 42, all mice were orally challenged with 10-fold LD50 of S. Choleraesuis CVCC2139. Five mice per group were euthanized on day 6 post-challenge, and samples from the spleen and liver were collected for measurement of bacterial loads. Survival of the remaining mice was monitored and recorded daily for 30 days.
2.9 Enzyme-linked immunosorbent assay (ELISA)
Antibody titers against inactivated S. Choleraesuis antigens were quantified using an indirect ELISA protocol as previously described (24). In brief, 100 μL of 109 CFU/ml of the heat-killed S. Choleraesuis antigens was added to wells of a 96-well ELISA plate coated with antigen with overnight incubation at 4°C. The next day, the plates were washed three times with PBST followed by blocking with 5% BSA (BD, San Diego, CA) in PBS at 37°C for 2 h. Following antigen coating and blocking, the plate was washed three times with PBST. Serum samples, diluted 1:200 in blocking buffer (5% BSA in PBS), were added to each well (100 μL/well) and incubated at 37°C for 1 h in a humidified chamber. The plate was then washed five times with PBST to remove unbound antibodies. Subsequently, 100 μL of HRP-conjugated goat anti-mouse IgG (Abclonal, Wuhan, China), diluted 1:5,000 in antibody diluent, was added to each well and incubated at 37°C for 1 h. After five additional PBST washes, the plate was ready for substrate development. 100 μL of TMB substrate solution (Macgene, Shanghai, China) was added, and the plates were incubated in the dark at 25°C for 10 min. After adding 50 μL of 2 M H2SO4 to stop the reaction, absorbance was measured at 450 nm using a Bio-Rad microplate reader (Bio-Rad, California, USA).
2.10 Ethics statement
All animal procedures were conducted in strict adherence to the Guide for the Care and Use of Laboratory Animals published by China's Ministry of Science and Technology. The study protocol was approved by the Animal Ethics Committee of Sichuan Agricultural University and the Sichuan Laboratory Animal Management Committee (permit number: SYXK2019-187), ensuring compliance with national and institutional welfare guidelines.
2.11 Statistical analysis
Data are presented as the mean ± standard deviation (SD) and analyzed using one-way analysis of variance (ANOVA) followed by Tukey's post-hoc multiple-comparison test with GraphPad Prism software (GraphPad Software, California, USA). Statistical significance was defined as P < 0.05. All in vitro experiments were independently repeated three times to ensure reproducibility.
3 Results
3.1 Roles of SPI-2 T3SS effectors of S. Choleraesuis in bacterial adhesion to, invasion into and survival within macrophages
Twenty-one effector genes (sseJ, sseG, slrP, sseF, gtgE, gogB, sspH, gtgA, sifA, sifB, sseK, steA, steC, sseL, sopD2, spiC, sseI, pipB, pipB2, sopD, steD) were screened and each of them was deleted from the WT S. Choleraesuis strain CVCC2139. The ssaV mutant strain (ΔssaV) was also constructed as a positive control as the SsaV is a structural component forming the inner ring of the SPI-2 T3SS injectosome that is involved in effector protein translocation (25). Then, the mutant strains were compared to the WT strain in terms of the ability of bacteria to adhere to, invade and survive within RAW264.7 macrophages. Deletion of the SPI-2 T3SS effector genes neither affected bacterial adhesion to nor changed bacterial invasion into macrophages (Figures 1A, B). Nevertheless, bacterial replication in the mutant strain ΔssaV, ΔsseF, ΔsseJ, ΔsifB, ΔsseK, ΔsifA, ΔsopD2, ΔsteC, or ΔsteD was significantly decreased compared to that in the WT strain, while loss of either of the other 13 genes had on adverse effects (Figure 1C). Gene complementation in trans in the mutant strains fully or partially restored the WT phenotypes (Figure 1C). This finding indicated that effector genes including sseF, sseJ, sifB, sseK, sifA, sopD2, steC, and steD promoted bacterial survival in macrophages.
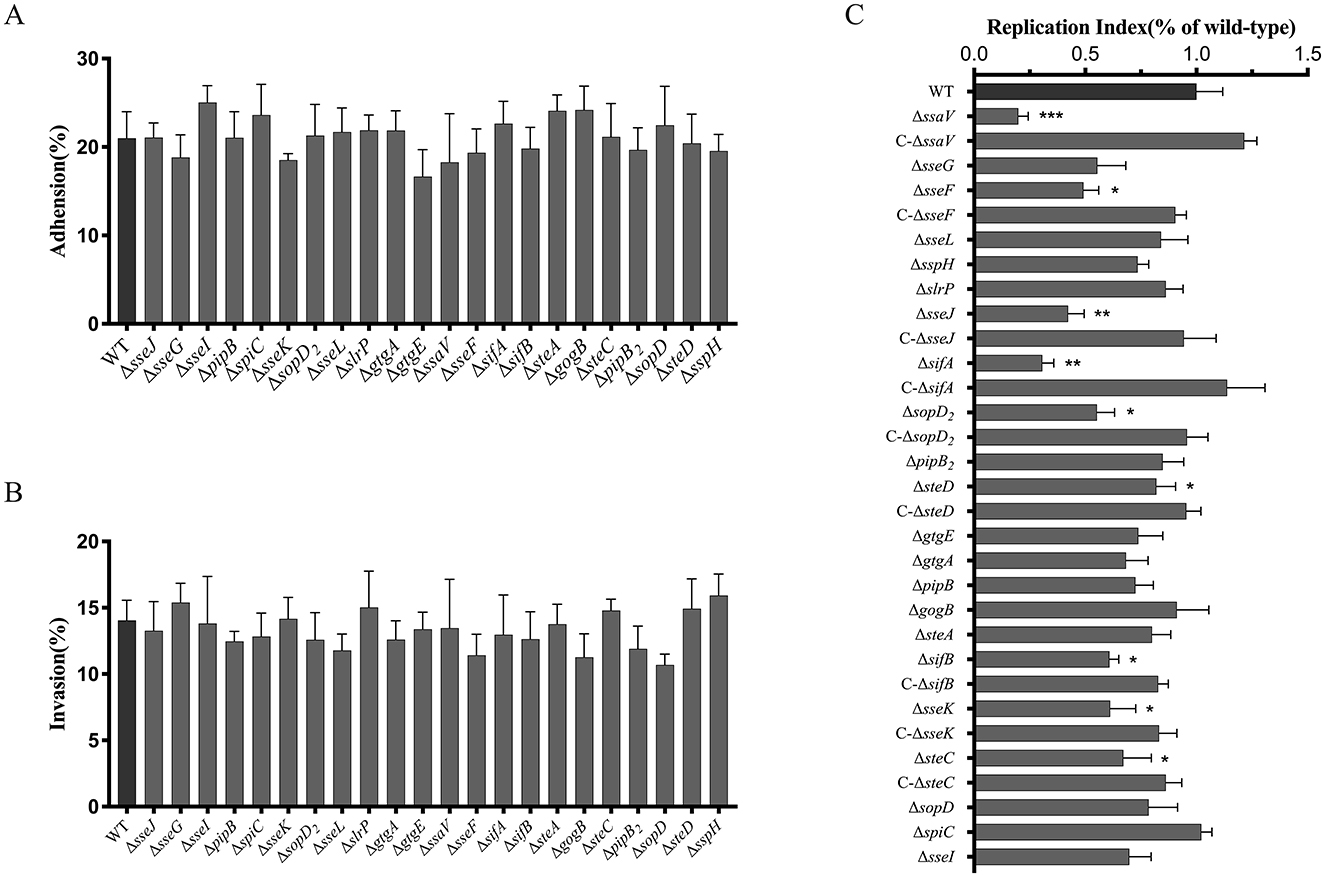
Figure 1. Roles of SPI-2 T3SS effectors of S. Choleraesuis in bacterial adhesion to (A), invasion into (B) and survival (C) within macrophages. (A and B) RAW264.7 cells were infected with WT or mutant strains (MOI = 100) for 30 min at 37°C. After PBS washing, adherent bacteria were quantified by direct lysis and CFU counting. The adhesion rate = (Nadherent/Ninitial × 100%). For invasion, cells were further incubated for 1 h to invade, then lysed by adding 0.2% triton X-100 then lysed for CFU counts. The invasion rate = (Ninvasive/Nadherent × 100%). C. For intracellular survival, invaded cells by the S. Choleraesuis were further cultured for 24 h. Then cells were lysed and bacterial counts were measured. Intracellular bacteria were quantified as replication index (Nsurvival/Nadherent × 100%). Asterisks above the error bars indicate significant differences compared with the WT group. *P < 0.05; **P < 0.01; ***P < 0.001.
3.2 Roles of SPI-2 T3SS effectors of S. Choleraesuis in bacterial growth and swimming
The 21 S. Choleraesuis mutant strains were subjected to detection of growth curves and swimming. Compared to the WT strain, the growth rates of the ΔgtgA, ΔsteA, ΔspiC, ΔsopD2, ΔslrP, ΔpipB2, ΔsopD, and ΔsteD strains were significantly reduced in LB broth at 37°C (Figure 2A). In contrast, the remaining 14 mutants exhibited similar growth rates to the WT strain (Figures 2B, C). This finding suggested that the effector genes gtgA, steA, spiC, sopD2, slrP, pipB2, sopD, and steD promotes the in vitro growth of S. Choleraesuis. Also, swimming of the ΔsseJ, ΔslrP, ΔsifB, and ΔsopD2 mutants were significantly enhanced compared to that of the WT strain, whereas the ΔpipB2 and ΔsspH strains exhibited reduced motility (Figure 2D). Complementation of sseJ, slrP, sifB, sopD2, pipB2and sspH in corresponding mutants restored WT swimming phenotype. Therefore, the effector genes sseJ, slrP, sifB, and sopD2 restrain the swimming ability of S. Choleraesuis, whereas sspH and pipB2 positively influence motility.
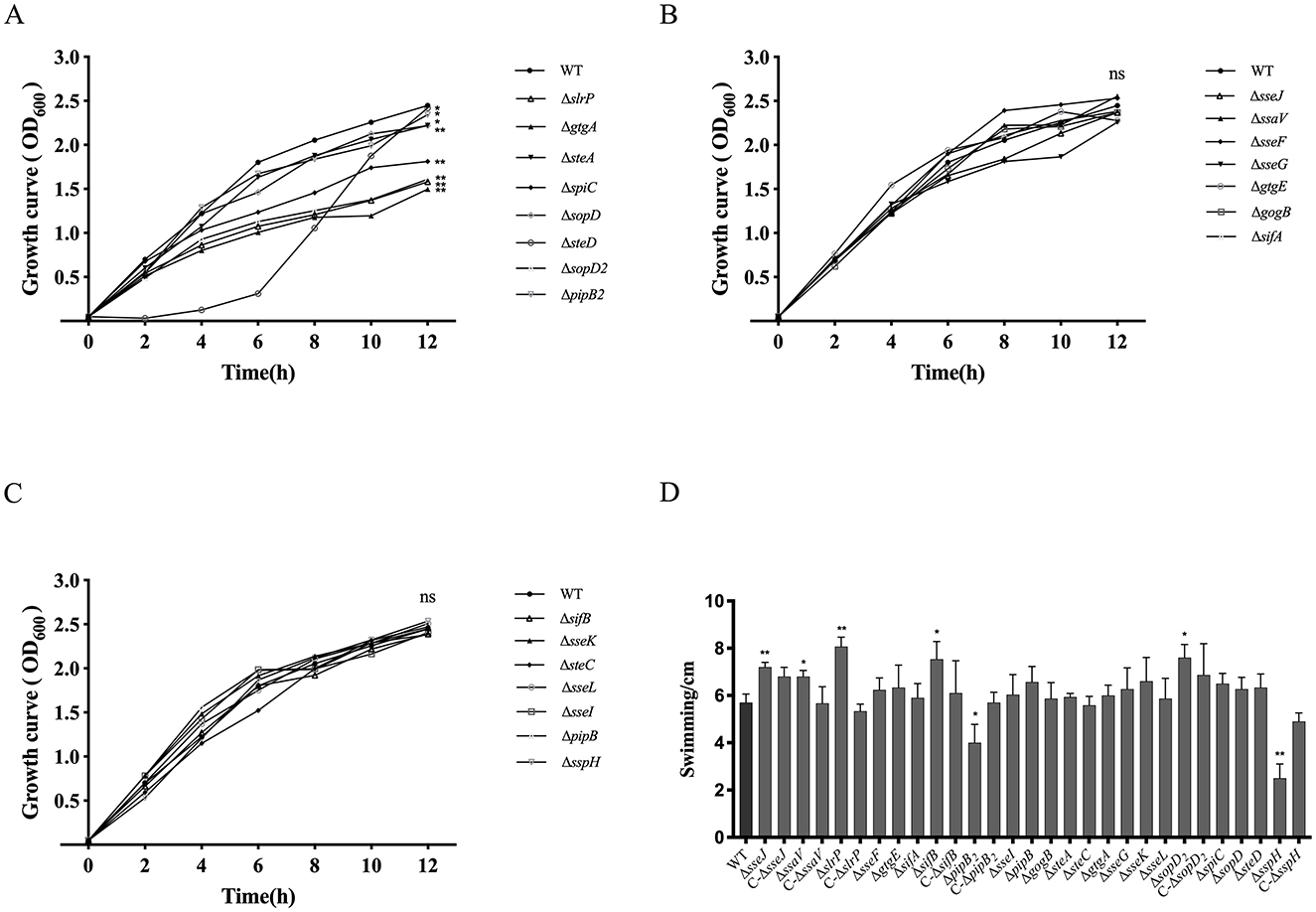
Figure 2. Detection of growth curves and swimming of S. Choleraesuis strains. (A–C) Growth curves of S. Choleraesuis WT and mutant strains were analyzed by measuring OD600 every 2 h for 12 h. (D) Swimming assay. The WT strain, 22 mutant strains and complemented strains were cultured in LB to OD600 = 0.6–0.8, harvested and suspended in PBS. Three microliters of bacterial suspension were spotted onto the center of a soft agar plate. Then, colony diameters were measured after 6 h of incubation at 37°C. An asterisk above the error bar shows a significant difference from the WT group. *P < 0.05; **P < 0.01.
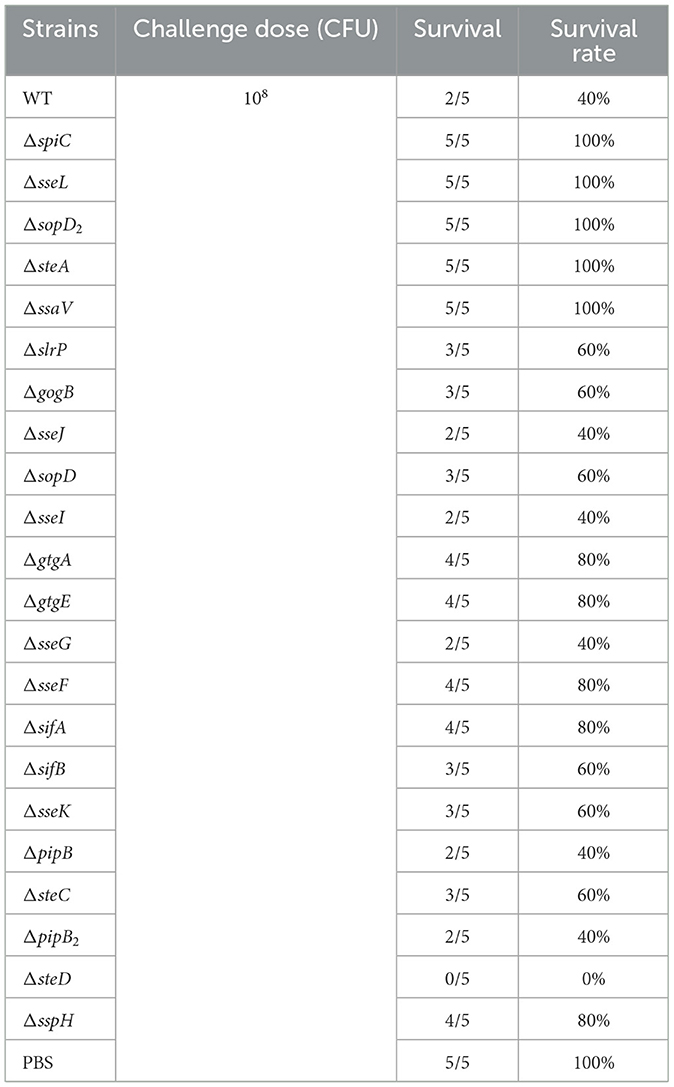
3.3 Roles of SPI-2 T3SS effectors of S. Choleraesuis in bacterial virulence in mice
To determine roles of the SPI-2 T3SS effector in virulence of S. Choleraesuis, mice were orally administered with 1 × 108 CFU of the WT strain or each of the mutant strains. The survival of mice was monitored for 1 month. The WT strain led to 40% of survival post-infection, while the five strains including ΔspiC, ΔssaV, ΔsseL, ΔsopD2, and ΔsteA caused no death (Table 1). Also, ΔgtgA, ΔgtgE, ΔsseF, ΔsifA, and ΔsspH resulted in increased survival (80%), by contrast, all mice succumbed to the ΔsteD infection. The remaining strains caused a 40% or 60% survival (Table 1). Thus, SPI-2 T3SS effector genes sseL, sopD2, and spiC, and the gene ssaV contributed remarkably to S. Choleraesuis virulence in mice, functioning as virulence genes. To further detect the roles of the five virulence genes in bacterial colonization, mice were inoculated with 108 CFU of the WT or mutant strains, then bacterial loads in liver and spleen were measured 6 days post-infection. No bacteria were recovered from the ΔspiC and ΔssaV groups. However, the bacterial loads of ΔsseL, ΔsopD2, and ΔsteA in liver and spleen tissues were comparable to those of the wild-type strain (Figures 3A, B). This finding suggests that the spiC and ssaV contribute to the colonization of S. Choleraesuis in the liver and spleen of mice, while sseL, sopD2, and steA are not involved in bacterial colonization.
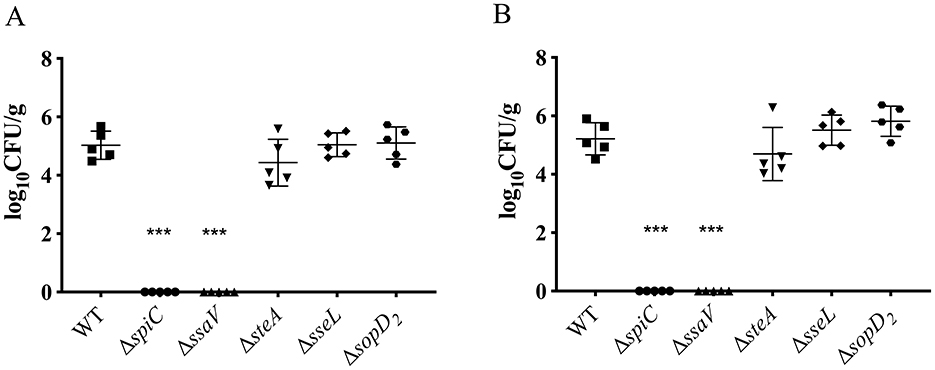
Figure 3. Detection of bacterial loads in mice after infection with S. Choleraesuis strains. Kunming mice (n = 5/group) were inoculated with S. Choleraesuis WT strain and mutant strains orally, respectively. Then, the bacterial loads in spleens (A) and livers (B) were measured and calculated as log10 CFU/g. An asterisk above the error bar indicates a significant difference from the WT group. /ast/ast/astP < 0.0001.
3.4 Virulence and colonization of the triple mutant strain ΔsseLΔsopD2ΔsteA of S. Choleraesuis mutants in mice
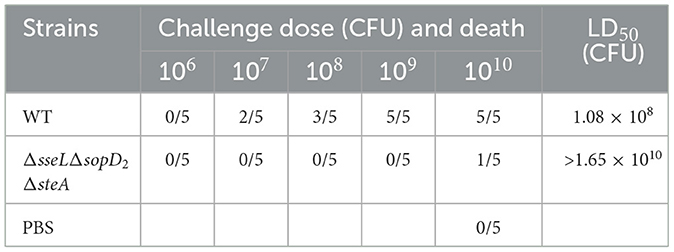
Although the two genes spiC and ssaV play a significant role in bacterial virulence, their mutant strains lost colonization ability in mice, hinting their poor immunogenicity. To develop a suitable live attenuated strain, the other three virulence genes sseL, sopD2, and steA were deleted from the WT strain simultaneously, generating a triple mutant ΔsseLΔsopD2ΔsteA. This strain colonized of the spleen and liver at a significantly lower level than the WT strain post oral infection (Figure 4). Also, the LD50 of the triple mutant was >1.65 × 1010 CFU, demonstrating at least a 150-fold reduction in virulence compared with that of the wild-type strain with the LD50 of 1.08 × 108 CFU (Table 2).
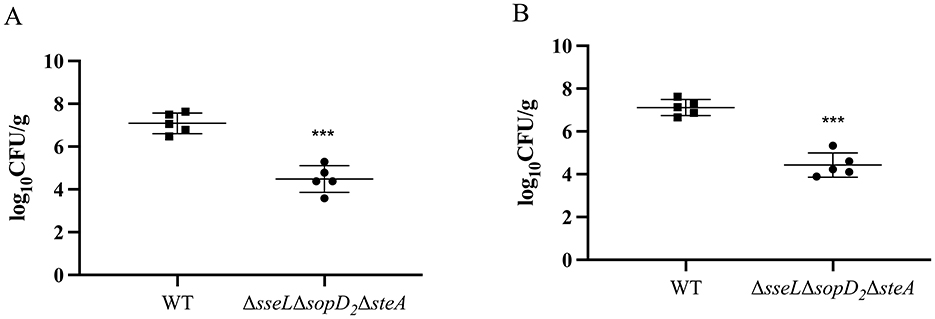
Figure 4. Colonization of the triple mutant strain ΔsseL ΔsopD2 ΔsteA of S. Choleraesuis in mice. Mice were inoculated with S. Choleraesuis WT strain or the ΔsseL ΔsopD2 ΔsteA orally. Bacterial loads in spleen (A) and liver (B) tissues were quantified as log10 CFU/g. The asterisk above the error bar indicates a significant difference compared with the WT group. ***P < 0.001.
3.5 Protection efficacy of the triple mutant strain ΔsseLΔsopD2ΔsteA
To evaluate the vaccine potential of the attenuated strain ΔsseLΔsopD2ΔsteA, mice were orally administered with 109 CFU of the vaccine strain twice with an interval of 14 days and then were challenged orally with a lethal dose of the S. Choleraesuis WT strain 28 days post-second immunization. Immunization with the ΔsseLΔsopD2ΔsteA induced significantly higher serum IgG responses to whole bacterial antigens than with the PBS group on Day 7 and 21 post-immunization (Figure 5A). Following the challenge, the bacterial loads in the spleen and liver of the ΔsseLΔsopD2ΔsteA group were significantly lower than those in the PBS group (Figures 5B, C). Furthermore, all the mice in the PBS control group died, whereas 40% of the mice in the immunized group survived (Figure 5D). Thus, immunization with the ΔsseLΔsopD2ΔsteA strain induced a robust antibody response, significantly reduced the tissue loads of the challenge strain, and provided 40% protection efficacy against lethal S. Choleraesuis infection.
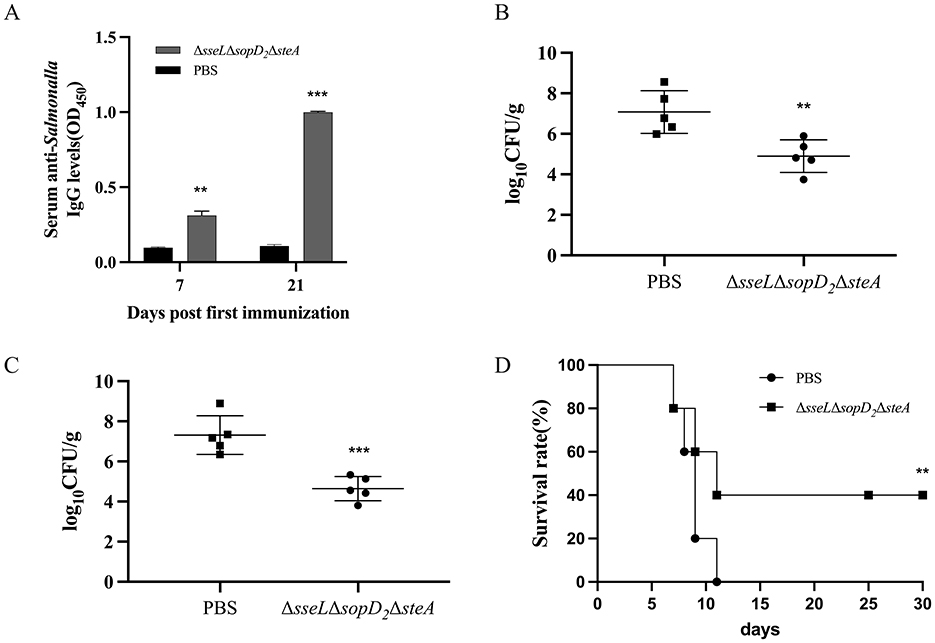
Figure 5. Antibody responses and protection efficacy induced by the triple mutant strain ΔsseL ΔsopD2 ΔsteA. Mice were immunized orally with the attenuated strain ΔsseL ΔsopD2 ΔsteA twice and then challenged with the S. Choleraesuis WT strain 28 days post-second immunization. Indirect ELISA was used to detect serum IgG levels against whole bacterial antigens 7 days and 21 days post-immunization (A). Bacterial loads in the spleen (B) and liver (C) were measured 6 days post-challenge. (D) Survival rates of mice were monitored over a 30-day period after challenge. Asterisks above the error bars indicate significant differences compared with the PBS group. **P < 0.01; ***P < 0.001.
4 Discussion
Our comprehensive analysis of SPI-2 T3SS effectors in S. Choleraesuis reveals multifaceted roles of individual effectors in intracellular survival, systemic virulence, motility, and growth, offering mechanistic insights into how this pathogen adapts to host defenses and establishes infection.
None of detected SPI-2 T3SS effectors was involved in the adhesion and invasion process of S. Choleraesuis to the macrophage RAW264.7, which is in line with previous studies on S. Typhimurium (26–29). We also observed notable effects of effectors on bacterial motility and growth. Several mutants exhibited reduced growth rates or altered swimming motility, indicating that SPI-2 effectors also influence global physiological fitness. Such effects may be mediated via metabolic reprogramming or indirect transcriptional regulation. For instance, pipB2 has been shown to alter host cytoskeletal tension and organelle dynamics, which may feed back to bacterial stress signaling (30). Reduced motility may compromise their ability to penetrate mucus layers or disseminate systemically, further contributing to attenuation (31). The spiC mutant strain of S. Enteritidis exhibits stronger swimming ability (32); whereas deletion of the spiC of S. Choleraesuis did not affect swimming. These contradictions suggested that some effector molecules exhibit functional heterogeneity across different Salmonella serovars.
Eight effector genes sseF, sseJ, sifB, sseK, sifA, sopD2, steC, and steD significantly enhanced the survival of S. Choleraesuis in macrophages, which highlights the core function of SPI-2 T3SS effector in maintaining intracellular survival and is largely consistent with previous studies. Most of these effectors are known to modulate SCV maturation, membrane dynamics, or host trafficking pathways, helping bacteria to evade lysosomal degradation and acquire nutrients. For instance, sifA stabilizes the SCV membrane and recruits kinesin-1 via SKIP (33), while sseF and sseG facilitate microenvironment construction (34). The involvement of sopD2 and steD suggests a multi-effector strategy to subvert host immunity: sopD2 interferes with Rab GTPase-mediated trafficking (35), while steD downregulates MHC II surface expression through host ubiquitination machinery (36). However, SteC of S. Typhimurium barely affects bacterial proliferation in macrophages (37). Interestingly, steA and pipB2, although previously implicated in vacuole positioning and actin remodeling (30, 38), had minimal impact on intracellular survival in S. Choleraesuis, hinting at possible functional redundancy or compensation by other effectors in this serovar.
All the effectors associated with intracellular survival except for sseJ and steD contribute to the virulence of S. Choleraesuis in mice, while three virulence determinants (spiC, sseL, and steA) were not related to bacterial intracellular survival. A previous study also found that protein SseL was shown to enhance the virulence of S. Pullorum by suppressing host NF-κB signaling but not affect the intracellular bacterial survival (39). These findings indicated a lack of strong correlation between intracellular replicative capacity and systemic virulence.
Construction of live attenuated bacterial vaccines should balance the virulence and immunogenicity (40). However, we found that the ΔspiC of S. Choleraesuis as well as the ΔssaV was avirulent and colonized mouse liver or spleen at levels below the threshold of detection. Too-low bacterial loads in tissues imply low immunogenicity (41). Therefore, the two strains were not considered as vaccine candidates in our study. Compared to the currently licensed live attenuated vaccine strain C500 in China, which was derived through chemical mutagenesis and has been used for over four decades, the ΔsseLΔsopD2ΔsteA strain developed in this study presents both advantages and limitations. C500 has demonstrated high protection efficacy in piglets but suffers from residual virulence and has an undefined genetic background, which raise biosafety concerns and hinder its broader application, particularly in immunocompromised hosts (10). In contrast, the triple mutant strain constructed here is genetically defined and rationally attenuated by deletion of three characterized virulence genes, thereby reducing the risk of reversion and enhancing safety. However, immunization with ΔsseLΔsopD2ΔsteA conferred only moderate protection (40%) against lethal challenge in mice, which is lower than the reported protection level of C500 or other strains such as the ΔspiC mutant of S. Pullorum that offered more than 90% protection (18). This limited efficacy may be attributed to its moderate tissue colonization and immunogenicity. Therefore, further optimization is required, such as incorporating additional immunostimulatory mutations or adjuvant delivery strategies, to enhance both antigen persistence and immune protection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The animal studies were approved by the study protocol was approved by the Animal Ethics Committee of Sichuan Agricultural University and the Sichuan Laboratory Animal Management Committee (permit number: SYXK2019-187). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
RX: Writing – original draft, Investigation. XJ: Writing – original draft, Investigation. JL: Writing – review & editing, Investigation. DZ: Investigation, Writing – review & editing. ML: Writing – review & editing, Investigation. MW: Writing – review & editing, Investigation. RJ: Writing – review & editing, Investigation. SC: Investigation, Writing – review & editing. QY: Investigation, Writing – review & editing. YW: Validation, Writing – review & editing. SZ: Writing – review & editing, Validation. JH: Validation, Writing – review & editing. XO: Writing – review & editing, Validation. DS: Validation, Writing – review & editing. BT: Validation, Writing – review & editing. YH: Formal analysis, Writing – review & editing. ZW: Writing – review & editing, Formal analysis. AC: Project administration, Writing – review & editing. XZ: Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Key Research and Development Program of China (2023YFD1800200), the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System (SCCXTD-2021-18) and the Earmarked Fund for China Agriculture Research System (CARS-42-17).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1637327/full#supplementary-material
References
1. Li H, Wu Y, Feng D, Jiang Q, Li S, Rong J, et al. Centralized industrialization of pork in Europe and America contributes to the global spread of salmonella enterica. Nat Food. (2024) 5:413–22. doi: 10.1038/s43016-024-00968-1
2. Eddicks M, Hausleitner R, Eddicks L, Blutke A, Straubinger RK, Wolf G, et al. [detection of salmonella choleraesuis var. Kunzendorf in a fattening pig with septicaemic salmonellosis A case report]. Tierarztl Prax Ausg G Grosstiere Nutztiere. (2016) 44:381–7. doi: 10.15653/TPG-160300
3. Longo A, Losasso C, Vitulano F, Mastrorilli E, Turchetto S, Petrin S, et al. Insight into an outbreak of salmonella choleraesuis var. Kunzendorf in wild boars Vet Microbiol. (2019) 238:108423. doi: 10.1016/j.vetmic.2019.108423
4. Hajra D, Nair AV, Chakravortty D. Decoding the invasive nature of a tropical pathogen of concern: The invasive non-typhoidal salmonella strains causing host-restricted extraintestinal infections worldwide. Microbiol Res. (2023) 277:127488. doi: 10.1016/j.micres.2023.127488
5. Chiu C-H, Su L-H, Chu C. Salmonella enterica serotype choleraesuis: Epidemiology, pathogenesis, clinical disease, and treatment. Clin Microbiol Rev. (2004) 17:311–22. doi: 10.1128/CMR.17.2.311-322.2004
6. Luk-In S, Chatsuwan T, Pulsrikarn C, Bangtrakulnonth A, Rirerm U, Kulwichit W. High prevalence of ceftriaxone resistance among invasive salmonella enterica serotype choleraesuis isolates in Thailand: The emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. Choleraesuis isolates Int J Med Microbiol. (2018) 308:447–53. doi: 10.1016/j.ijmm.2018.03.003
7. Jacqueline C, Samper-Cativiela C, Monzon Fernandez S, Ugarte-Ruiz M, Cuesta de la Plaza I, Alvarez J, et al. Phenotypic and genetic characterization of antimicrobial resistance in salmonella enterica serovar choleraesuis isolates from humans and animals in Spain from 2006 to 2021. J Antimicrob Chemother. (2024) 79:790–800. doi: 10.1093/jac/dkae022
8. Alvarez DM, Barrón-Montenegro R, Conejeros J, Rivera D, Undurraga EA, Moreno-Switt AI, et al. review of the global emergence of multidrug-resistant salmonella enterica subsp. enterica serovar infantis. Int J Food Microbiol. (2023) 403:110297. doi: 10.1016/j.ijfoodmicro.2023.110297
9. Plotkin SA, Plotkin SL. The development of vaccines: How the past led to the future. Nat Rev Microbiol. (2011) 9:889–93. doi: 10.1038/nrmicro2668
10. Zhu L, Zhao X, Yin Q, Liu X, Chen X, Huang C, et al. Mucosal IgA and IFN-γ+ CD8 T cell immunity are important in the efficacy of live salmonella enteria serovar choleraesuis vaccines. Sci Rep. (2017) 7:46408. doi: 10.1038/srep46408
11. Ji Z, Shang J, Li Y, Wang S, Shi H. Live attenuated salmonella enterica serovar choleraesuis vaccine vector displaying regulated delayed attenuation and regulated delayed antigen synthesis to confer protection against streptococcus suis in mice. Vaccine. (2015) 33:4858–67. doi: 10.1016/j.vaccine.2015.07.045
12. Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. (2017) 15:323–37. doi: 10.1038/nrmicro.2017.20
13. Miao EA, Warren SE. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J Clin Immunol. (2010) 30:502–6. doi: 10.1007/s10875-010-9395-6
14. Yu X-J, Xie H, Li Y, Liu M, Hou R, Predeus AV, et al. Modulation of salmonella virulence by a novel SPI-2 injectisome effector that interacts with the dystrophin-associated protein complex. MBio. (2024) 15:e0112824. doi: 10.1128/mbio.01128-24
15. Yu X-J, Liu M, Holden DW. Salmonella effectors SseF and SseG interact with mammalian protein ACBD3 (GCP60) to anchor salmonella-containing vacuoles at the Golgi network. MBio. (2016) 7:e00474–16. doi: 10.1128/mBio.00474-16
16. Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol. (2003) 48:401–15. doi: 10.1046/j.1365-2958.2003.03438.x
17. Méresse S, Unsworth KE, Habermann A, Griffiths G, Fang F, Martínez-Lorenzo MJ, et al. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell Microbiol. (2001) 3:567–77. doi: 10.1046/j.1462-5822.2001.00141.x
18. Wang Y, Huang C, Tang J, Liu G, Hu M, Kang X, et al. Salmonella pullorum spiC mutant is a desirable LASV candidate with proper virulence, high immune protection and easy-to-use oral administration. Vaccine. (2021) 39:1383–91. doi: 10.1016/j.vaccine.2021.01.059
19. Rubirés X, Saigi F, Piqué N, Climent N, Merino S, Albertí S, et al. gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J Bacteriol. (1997) 179:7581–6. doi: 10.1128/jb.179.23.7581-7586.1997
20. Zhao X, Dai Q, Jia R, Zhu D, Liu M, Wang M, et al. Two novel salmonella bivalent vaccines confer dual protection against two salmonella serovars in mice. Front Cell Infect Microbiol. (2017) 7:391. doi: 10.3389/fcimb.2017.00391
21. Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. (1998) 207:149–57. doi: 10.1016/S0378-1119(97)00674-5
22. Zhao X, Dai Q, Zhu D, Liu M, Chen S, Sun K, et al. Recombinant attenuated salmonella typhimurium with heterologous expression of the salmonella choleraesuis O-polysaccharide: High immunogenicity and protection. Sci Rep. (2017) 7:7127. doi: 10.1038/s41598-017-07554-3
23. Zhao X, Yang F, Shen H, Liao Y, Zhu D, Wang M, et al. Immunogenicity and protection of a pasteurella multocida strain with a truncated lipopolysaccharide outer core in ducks. Vet Res. (2022) 53:17. doi: 10.1186/s13567-022-01008-w
24. Zhao X, Zeng X, Dai Q, Hou Y, Zhu D, Wang M, et al. Immunogenicity and protection efficacy of a Salmonella enterica serovar Typhimurium fnr, arcA and fliC mutant. Vaccine. (2021) 39:588–95. doi: 10.1016/j.vaccine.2020.11.070
25. Yu X-J, Grabe GJ, Liu M, Mota LJ, Holden DW. SsaV interacts with SsaL to control the translocon-to-effector switch in the salmonella SPI-2 type three secretion system. MBio. (2018) 9:e01149–18. doi: 10.1128/mBio.01149-18
26. Fulde M, van Vorst K, Zhang K, Westermann AJ, Busche T, Huei YC, et al. SPI2 T3SS effectors facilitate enterocyte apical to basolateral transmigration of salmonella-containing vacuoles in vivo. Gut Microbes. (2021) 13:1973836. doi: 10.1080/19490976.2021.1973836
27. Naseer N, Egan MS, Reyes Ruiz VM, Scott WP, Hunter EN, Demissie T, et al. Human NAIP/NLRC4 and NLRP3 inflammasomes detect salmonella type III secretion system activities to restrict intracellular bacterial replication. PLoS Pathog. (2022) 18:e1009718. doi: 10.1371/journal.ppat.1009718
28. Zhang K, Riba A, Nietschke M, Torow N, Repnik U, Pütz A, et al. Minimal SPI1-T3SS effector requirement for salmonella enterocyte invasion and intracellular proliferation in vivo. PLoS Pathog. (2018) 14:e1006925. doi: 10.1371/journal.ppat.1006925
29. Gulati A, Shukla R, Mukhopadhaya A. Salmonella effector SteA suppresses proinflammatory responses of the host by interfering with IκB degradation. Front Immunol. (2019) 10:2822. doi: 10.3389/fimmu.2019.02822
30. Mandal K. Review of PIP2 in Cellular Signaling, Functions and Diseases. Int J Mol Sci. (2020) 21:8342. doi: 10.3390/ijms21218342
31. Ottemann KM, Miller JF. Roles for motility in bacterial-host interactions. Mol Microbiol. (1997) 24:1109–17. doi: 10.1046/j.1365-2958.1997.4281787.x
32. Wang Y, Liu G, Zhang J, Gu D, Hu M, Zhang Y, et al. WbaP is required for swarm motility and intramacrophage multiplication of salmonella enteritidis spiC mutant by glucose use ability. Microbiol Res. (2021) 245:126686. doi: 10.1016/j.micres.2021.126686
33. Dumont A, Boucrot E, Drevensek S, Daire V, Gorvel J-P, Poüs C, et al. the host target of the salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic. (2010) 11:899–911. doi: 10.1111/j.1600-0854.2010.01062.x
34. Feng Z-Z, Jiang A-J, Mao A-W, Feng Y, Wang W, Li J, et al. The salmonella effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J Biol Chem. (2018) 293:9662–73. doi: 10.1074/jbc.RA118.002708
35. D'Costa VM, Braun V, Landekic M, Shi R, Proteau A, McDonald L, et al. Salmonella disrupts host endocytic trafficking by SopD2-mediated inhibition of Rab7. Cell reports. (2015) 12:1508–18. doi: 10.1016/j.celrep.2015.07.063
36. Bayer-Santos E, Durkin CH, Rigano LA, Kupz A, Alix E, Cerny O, et al. The salmonella effector SteD mediates MARCH8-dependent ubiquitination of MHC II molecules and inhibits T cell activation. Cell Host Microbe. (2016) 20:584–95. doi: 10.1016/j.chom.2016.09.006
37. Dai Y, Zhang M, Liu X, Sun T, Qi W, Ding W, et al. Salmonella manipulates macrophage migration via SteC-mediated myosin light chain activation to penetrate the gut-vascular barrier. EMBO J. (2024) 43:1499–518. doi: 10.15252/embj.2023100502
38. Domingues L, Holden DW, Mota LJ. The salmonella effector SteA contributes to the control of membrane dynamics of salmonella-containing vacuoles. Infect Immun. (2014) 82:2923–34. doi: 10.1128/IAI.02081-14
39. Geng S, Wang Y, Xue Y, Wang H, Cai Y, Zhang J, et al. The SseL protein inhibits the intracellular NF-κB pathway to enhance the virulence of salmonella pullorum in a chicken model. Microb Pathog. (2019) 129:1–6. doi: 10.1016/j.micpath.2019.03.024
40. Galen JE, Curtiss R. The delicate balance in genetically engineering live vaccines. Vaccine. (2014) 32:4376–85. doi: 10.1016/j.vaccine.2014.06.011
Keywords: S. choleraesuis, SPI-2, type III secretion system, virulence, vaccines
Citation: Xu R, Ji X, Lian J, Zhu D, Liu M, Wang M, Jia R, Chen S, Yang Q, Wu Y, Zhang S, Huang J, Ou X, Sun D, Tian B, He Y, Wu Z, Cheng A and Zhao X (2025) Roles of SPI-2 T3SS effectors in virulence of Salmonella Choleraesuis and Construction of a triple-gene mutant vaccine strain. Front. Vet. Sci. 12:1637327. doi: 10.3389/fvets.2025.1637327
Received: 29 May 2025; Accepted: 04 July 2025;
Published: 12 August 2025.
Edited by:
Mengmeng Zhao, Foshan University, ChinaReviewed by:
Songbiao Chen, Henan University of Science and Technology, ChinaConsuelo Pia Badilla Pino, Universidad de Las Américas, Chile
Copyright © 2025 Xu, Ji, Lian, Zhu, Liu, Wang, Jia, Chen, Yang, Wu, Zhang, Huang, Ou, Sun, Tian, He, Wu, Cheng and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=; Xinxin Zhao, eHhpbnpoYW9AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Rui Xu1,2,3,4†
Rui Xu1,2,3,4† Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Mingshu Wang
Mingshu Wang Shun Chen
Shun Chen Shaqiu Zhang
Shaqiu Zhang Juan Huang
Juan Huang Di Sun
Di Sun Bin Tian
Bin Tian Anchun Cheng
Anchun Cheng Xinxin Zhao
Xinxin Zhao