1
Laboratoire Psychologie de la Perception, CNRS and Université Paris Descartes, Paris, France
2
Département d’Etudes Cognitives, Ecole Normale Supérieure, Paris, France
“Brian” is a simulator for spiking neural networks (http://www.briansimulator.org
). The focus is on making the writing of simulation code as quick and easy as possible for the user, and on flexibility: new and non-standard models are no more difficult to define than standard ones. This allows scientists to spend more time on the details of their models, and less on their implementation. Neuron models are defined by writing differential equations in standard mathematical notation, facilitating scientific communication. Brian is written in the Python programming language, and uses vector-based computation to allow for efficient simulations. It is particularly useful for neuroscientific modelling at the systems level, and for teaching computational neuroscience.
Computational modelling of neural networks plays an increasing role in neuroscience research. Writing simulation code from scratch (typically in Matlab or C) can be very time-consuming and in all but the simplest cases requires expertise in programming and knowledge of neural simulation algorithms. This can discourage researchers from investigating non-standard models or more complex network structures.
Several successful neural simulators already exist (Brette et al., 2007
), such as Neuron (Carnevale and Hines, 2006
) and Genesis (Bower and Beeman, 1998
) for compartmental modelling, and NEST (Gewaltig and Diesmann, 2007
) for large scale network modelling. These simulators implement computationally efficient algorithms and are widely used for large-scale modelling and complex biophysical models. However, computational efficiency is not always the main limiting factor when simulating neural models. Efforts have been made in several simulators to make it as easy as possible, such as Neuron’s NMODL language described in Hines and Carnevale (2000) and graphical user interface. In many practical cases; however, it still takes considerably more time to write the code than to run the simulations. Minimising learning and development time rather than simulation time implies different design choices, putting more emphasis on flexibility, readability, and simplicity of the syntax.
There are various projects underway to address these issues. All of the major simulation packages now include Python interfaces (Eppler et al., 2008
; Hines et al., 2009
) and the PyNN project (Davison et al., 2008
) is working towards providing a unified interface to them. Because it is both easy and powerful, Python is rapidly becoming the standard high-level language for the field of computational neuroscience, and for scientific computing more generally (Bassi, 2007
).
We took those approaches one step further by developing a new neural simulator, Brian, which is an extension package for the Python programming language (Goodman and Brette, 2008
). A simulation using Brian is a Python program either executed from a script or interactively from a Python shell. The primary focus is on making the development of a model by the user as rapid and flexible as possible. For that purpose, the models are defined directly in the main script in their mathematical form (differential equations and discrete events). This design choice addresses the three issues mentioned earlier: flexibility, as the user can change the model by changing the equations; readability, as equations are unambiguous and do not require any specific knowledge of the Brian simulator to understand them; and simplicity of the syntax, as models are expressed in their original mathematical form, with little syntax to learn that is specific to Brian. Computational efficiency is achieved using vector-based computation techniques.
We expect that the availability of this new tool will have the strongest impact in two areas: firstly in systems neuroscience, by making it easy to explore spiking models with custom architectures and properties, and secondly in teaching computational neuroscience, by making it possible to write the code for a functional neural model within a single tutorial, and without strong programming skills required. In this review, we start by describing the core principles of Brian, then we discuss a few concrete examples for which Brian is particularly useful. Finally, we outline directions for future research.
Brian can be downloaded from http://briansimulator.org
. It is open source and freely available under the CeCILL license (compatible with the GPL license).
The main goal of the Brian project is to minimise the development time for a neural model, and in particular the time spent writing code, so that scientists can spend their time on the details of their model rather than the details of its implementation. The development time also includes the time spent learning how to use the simulator. Ease of learning is important because it opens the possibility for people who would not previously have considered computational modelling to try it out. It is also helpful for teaching (see Teaching). More generally, it is desirable to minimise the time it takes to read and understand someone else’s code. It helps peers to verify and evaluate research based on computational modelling, and it makes it easier for researchers to share their models. Finally, in order to minimise the time spent on writing code, the translation of the model definition into code should be as direct as possible. Execution speed and memory usage of the code is also an issue, but only inasmuch as it puts a constraint on what can be done with it. It is therefore most important in the case of large neural networks.
These goals led us to make the following two choices: firstly, to use a standard programming language that is both intuitive and well established, and secondly, to let users define models in a form that is as close as possible to their mathematical definitions. As in other simulation projects, we identified Python as an ideal choice for the programming language. Python is a well established language that is intuitive, easy to learn and benefits from a large user community and many extension packages (in particular for scientific computing and visualisation). While other simulators use Python as an interface to lower level simulation code, the Brian simulator itself is written in Python. The most original aspect of Brian is the emphasis on defining models as directly as possible by providing their mathematical definition, consisting of differential equations and discrete events (the effect of spikes) written in standard mathematical notation. One of the simplest examples is a leaky integrate-and-fire neuron, which has a single variable V which decays to a resting potential V0 over time according to the equation τdV/dt = −(V − V0). If this variable reaches a threshold V > VT the neuron fires a spike and then resets to a value VR. If neuron i is connected to neuron j with a synaptic weight Wij then neuron i firing causes Vj to jump to Vj + Wij. This is represented in Brian with the following code for a group of N neurons with all-to-all connectivity:

As can be seen in this code, the thresholding condition and the reset operation are also given in standard mathematical form. In fact it is possible to give arbitrary Python expressions for these, including calling user-defined functions. This gives greater generality, but using only mathematical expressions improves clarity. In addition to the mathematical definitions, the physical units are specified explicitly for all variables, enforcing dimensional consistency (here variable V has units of volts). In order to avoid ambiguities and make the code more readable, there is no standard scale for physical quantities (e.g. mV for voltages) and the user provides the units explicitly. For example, the potential of the neurons is set to −70 mV by writing
G.V=−70*mV or equivalently G.V=−.07*volt.Making differential equations the basis of models in Brian may seem like a fundamental restriction. For example, we may want to define an α post-synaptic current by g(t) = (t/τ)e1 − t/τ rather than a differential equation. However, this is just the integrated formulation of the associated kinetic model: 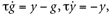 where presynaptic spikes act on y (that is, cause an instantaneous increase in y). More generally, biophysical neural models can almost always be expressed as differential equations, whose solutions are the post-synaptic potentials or currents.
where presynaptic spikes act on y (that is, cause an instantaneous increase in y). More generally, biophysical neural models can almost always be expressed as differential equations, whose solutions are the post-synaptic potentials or currents.
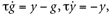 where presynaptic spikes act on y (that is, cause an instantaneous increase in y). More generally, biophysical neural models can almost always be expressed as differential equations, whose solutions are the post-synaptic potentials or currents.
where presynaptic spikes act on y (that is, cause an instantaneous increase in y). More generally, biophysical neural models can almost always be expressed as differential equations, whose solutions are the post-synaptic potentials or currents.In the same way, short-term plasticity (Tsodyks and Markram, 1997
) and spike-timing-dependent plasticity (STDP, Song et al., 2000
) can also be defined by differential equations with discrete events. In fact, although STDP is often described by the modification of the synaptic weight for a given pair of presynaptic and postsynaptic spikes, this phenomenological description is ambiguous because it does not specify how pairs interact (e.g. all-to-all or nearest neighbours). These considerations motivated our choice to define all models unambiguously with differential equations and discrete events.
Brian emphasises this approach more strongly than any other simulator package, and it has some crucial benefits. Foremost, it separates the definition of the model from its implementation, which makes the code highly reproducible and readable. The equations define the models unambiguously, independently of the simulator itself, which makes it easy to share the code with other researchers, even though they might use a different simulator or no simulator at all. Second, it minimises the amount of simulator specific syntax that needs to be learnt. For example, users do not need to look up the names of the model parameters since they define them themselves. Third, simulating custom models is as simple as simulating standard models. In the following, we illustrate these features in a few concrete cases.
In this section, we discuss several examples that illustrate the key features of Brian.
Investigating a New Neuron Model
Several recent experimental (Badel et al., 2008
) and theoretical studies (Deneve, 2008
) have suggested that the threshold for spike initiation should increase after each spike and decay to a stationary value. A simple model is given by a differential equation for the threshold VT: τtdVT/dt = VT0 − VT, and an instantaneous increase at spike time: VT ← VT + δ. This model can be simulated directly with Brian, even though it is not a predefined model:

Two things can be noted in this code: the threshold condition can be any boolean condition on the variables of the model (not only a fixed threshold), and the reset can be any sequence of Python instructions.
Investigating Plasticity Mechanisms
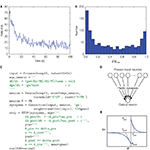
The mechanisms of spike-timing-dependent plasticity are an active line of research, so that many different models coexist and we might expect new models to emerge in the near future. Figure 1
shows a Brian script replicating some of the results of Song et al. (2000)
, where a neuron receives random inputs through plastic synapses. The STDP rule is exponential with all-to-all interactions between spike pairs and results in a bimodal stationary distribution of synaptic weights. Although the exponential STDP rule can be most simply used in Brian with the
ExponentialSTDP object, the most general way to define such a mechanism in Brian is to give its equations. As is shown in Figure 1
, a synaptic plasticity model is defined by three components: differential equations for synaptic variables (here Apre and Apost); events at presynaptic spike times; and events at postsynaptic spike times. This formalism encompasses a very broad class of plasticity rules. For example, triplet-based STDP rules (Pfister and Gerstner, 2006
) can be implemented by inserting a third differential equation; and nearest-neighbour rules can be obtained by replacing the presynaptic code A_pre+=dA_pre by A_pre=dA_pre and the postsynaptic code A_post+=dA_post by A_post=dA_post.Figure 1. Implementing spike-timing-dependent plasticity with Brian. The source code in panel (C) shows the complete implementation of the network shown in panel (D), adapted from Song et al. (2000)
. A set of N = 1000 neurons (the object named
input in the code) firing spikes at random times with Poisson statistics are connected via excitatory synapses (the object synapses) to a leaky integrate-and-fire neuron (the object neuron evolving according to the differential equationsin the string eqs_neuron). The initially random synaptic weights are modified according to the STDP rule (the object stdp) in panel (E): for all pairs of pre- and post-synaptic firing times tpre, tpost, the weight of the synapse is modified by an amount Δw depending on the difference in spike times Δt = tpost − tpre. The equation for Δw is Δpree−Δt/τpre when Δt > 0 or ΔposteΔt/τpost when Δt < 0. This is equivalent to the following system of equations. Each synapse has two variables Apre, Apost evolving according to the equations τpredApre/dt = −Apre and τpostdApost/dt = −Apost (the eqs string in the STDP declaration). A presynaptic spike causes Apre to increase by Δpre and the weight w to jump by Apost (the pre string), and a postsynaptic spikes causes Apost to increase by Δpost and w by Apre (the post string). The STDP rule causes the population firing rate shown in panel (A) to adapt, and the final distribution of synaptic weights shown in panel (B) to tend towards a bimodal distribution.Modelling a Functional Neural System
Figure 2
shows a Brian script adapted from Stürzl et al. (2000)
, which models the neural mechanisms of prey localisation by a sand scorpion. This model is fairly complex and includes in particular noise and delays, which would make equivalent code in Matlab or C very long, whereas the full script takes only about 20 lines with Brian (plus the definition of parameter values). The script illustrates the fact that the code is close to the mathematical definition of the model, which makes it relatively easy to understand. Many (if not most) neurocomputational studies at the systems level are not published along with the code, which might be because the authors find their code less informative and usable for readers than the article describing it. Code readability has several benefits: firstly, the code can be provided to the reviewers, who can run it or at least understand precisely how the authors produced the figures; secondly, the code can be published along with the article and provide information to readers even though they might not be familiar with the simulator; and thirdly, readers can work from the authors’ code without rewriting everything from scratch.
Figure 2. Brian implementation of a model of prey localisation in the sand scorpion, adapted from Stürzl et al. (2000)
. The movement of the prey causes a surface wave [function
wave in the code in panel (C)] which is detected by mechanoreceptors [the red points in panel (A)] at the ends of each of the scorpion's legs. The mechanoreceptors are modelled by noisy, leaky integrate-and-fire neurons with an input current defined by the surface wave displacement at the ends of the legs (object legs in the code, defined by the equations eqs_legs. These neurons send an excitatory signal (the object synapses_ex) to corresponding command neurons (the blue points) modelled by leaky integrate-and-fire neurons (object neurons with equations eqs_neuron), which also receive delayed inhibitory signals (the object synapses_inh) from the three legs on the other side (the for loop). A wave arriving first at one side of the scorpion will cause an excitatory signal to be sent to the command neurons on that side causing them to fire, and an inhibitory signal to the command neurons on the other side, stopping them from firing when the wave reaches the other side. The result is that command neurons are associated to the directions of the corresponding legs, firing at a high rate if the prey is in that direction. Panel (B) shows the firing rates for the eight command neurons in a polar plot for a prey at an angle of 60 degrees relative to the scorpion.Teaching
We end this section with the experience of one of the authors (Romain Brette) on teaching a computational neuroscience course which included a weekly two hour tutorial on computers (http://www.di.ens.fr/∼brette/coursneurones/coursneurones.html
). A single tutorial was dedicated to learning Python and Brian, while every subsequent one illustrated the lecture which preceded it. For example, in one tutorial the students implemented a spiking version of the sound source localisation model of Jeffress (1948) and another sound localisation model that was recently proposed for small mammals (McAlpine et al., 2001
). In another tutorial, the students implemented the synfire chain model described in Diesmann et al. (1999)
. The year before, when students used Scilab for simulations (a free equivalent of Matlab), the synfire chain model was a 2-month student project. This experience illustrates the fact that choosing to minimise the time spent on learning and writing code has qualitative implications: what was before a full project can now be done by students in a single tutorial, allowing for a much richer course content.
With the Brian project, we chose a different emphasis from previous simulation projects: flexibility, readability, and simplicity of the syntax. Two choices followed from those goals: firstly, Brian is an extension package of Python, a simple and popular programming language, and secondly Brian is equation-oriented, that is models are written in a form that is as close as possible to their mathematical expression, with differential equations and discrete events. This makes Brian a convenient simulation tool for exploring new spiking neural models, modelling complex neural models at the systems level, and teaching computational neuroscience.
Python is an interpreted language, and although it is fast there is an overhead for every Python operation. Brian can achieve good performance by using the technique of vectorisation, similar to the same technique familiar to Matlab users, which makes the interpretation overhead small relative to overall simulation times for large networks. Brian is typically much faster than simulations coded in Matlab, and a little slower than simulations coded in C (Goodman and Brette, 2008
). Brian is proportionally slower for small networks, which seems like a reasonable trade off given that smaller networks tend to take less time to run in absolute terms than larger networks.
Currently, the main limitation of Brian is that it cannot run distributed simulations (although independent simulations can be run with job scheduling software such as Condor (Frey et al., 2002
). Our plan for the future is to include some support for parallel processing with the same goals of simplicity and accessibility. For that purpose, we are focusing on using graphics cards with general purpose graphics processing units (GPUs), an inexpensive piece of hardware (around several hundred dollars) consisting of a large number of parallel processing cores (in the hundreds for the latest models (Luebke et al., 2004
)). Using these cards gives the equivalent of a small parallel cluster in a single machine at much lower cost. Early work in the case of auditory filtering and the solution of nonlinear differential equations gives an indication of the speed improvements to be gained from using GPUs. Applying a bank of 6000 auditory filters (approximately the number of cochlear filters in the human auditory system) was around 7 times faster with a GPU than with a CPU, and for an even larger bank of filters approached 10–12 times faster (GPUs generally perform better as the data set gets larger). For solving nonlinear differential equations, a key step in the simulation of many biophysically realistic neuron models, GPU performance was between 2 and 60 times better than the CPU depending on the equations and the number of neurons.
We initiated the Brian project with the goal of providing a simple simulation tool to explore spiking neural models without spending too much time on their implementation. Our hope is that our efforts in developing Brian will make spiking neuron models more widely accessible for systems neuroscience research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was partially supported by the French ANR (ANR HIT), the CNRS and the Ecole Normale Supérieure. The authors would like to thank all those who have been involved in testing Brian and making suggestions for improving it.
Python: A high-level programming language. Programs can be run from a script or interactively from a shell (as in Matlab). It is often used for providing a high-level interface to low-level code. The Python community has developed a large number of third party packages, including NumPy and SciPy, which are commonly used for efficient numerical and scientific computation.
Brette, R., Rudolph, M., Carnevale, T., Hines, M., Beeman, D., Bower, J. M., Diesmann, M., Morrison, A., Goodman, P. H., Harris, F. C., Zirpe, M., Natschläger, T., Pecevski, D., Ermentrout, B., Djurfeldt, M., Lansner, A., Rochel, O., Vieville, T., Muller, E., Davison, A. P., Boustani, S. E., and Destexhe, A. (2007). Simulation of networks of spiking neurons: a review of tools and strategies. J. Comput. Neurosci. 23, 349–398.