- 1Department of Statistics, School of Agriculture and Science, College of Agriculture, Engineering and Science, University of KwaZulu-Natal, Durban, South Africa
- 2Department of Statistics, School of Agriculture and Science, College of Agriculture, Engineering and Science, University of KwaZulu-Natal, Pietermaritzburg, South Africa
- 3Centre for the AIDS Programme of Research in South Africa (CAPRISA), School of Laboratory Medicine and Medical Sciences, University of KwaZulu-Natal, Durban, South Africa
- 4Nelson R Mandela School of Medicine, University of KwaZulu-Natal, Durban, South Africa
HIV and tuberculosis (TB) remain closely linked public health threats in sub-Saharan Africa, with South Africa bearing the highest burden of both diseases. In KwaZulu-Natal, where HIV prevalence peaks among individuals aged 15–49, HIV-induced immunosuppression significantly increases TB risk. Despite their biological interplay, HIV and TB are often analysed separately. This study jointly modelled CD4 cell count and TB diagnosis using a Bayesian latent variable approach to examine their interdependence among HIV-positive individuals. Data were drawn from 7,776 HIV-positive individuals aged 15–49 participating in two population-based cross-sectional surveys (2014–2016) under the HIPSS project. A Bayesian multivariate latent variable model jointly estimated CD4 cell count (continuous) and TB diagnosis (binary) using a probit link. Model fitting was conducted in R using the brms package with Hamiltonian Monte Carlo sampling. The analysis revealed a moderate negative correlation (−0.38) between predicted CD4 cell counts and TB probabilities, supporting the inverse biological relationship between immune suppression and TB risk. Antiretroviral therapy (ARV) use was significantly associated with improved immune status and reduced TB risk. Other key factors, such as male sex, lower educational attainment, and high viral load, were linked to both increased TB susceptibility and lower CD4 cell counts. These findings demonstrate the utility of joint Bayesian modelling in capturing the interdependence of comorbid outcomes and highlight the clinical and policy relevance of integrated HIV–TB programming. They support targeted screening, early treatment initiation, and resource prioritisation for at-risk populations in high-burden settings like KwaZulu-Natal.
1 Introduction
The dual burden of human immunodeficiency virus (HIV) and tuberculosis (TB) remains a major public health challenge globally, particularly in sub-Saharan Africa, where the epidemics intersect with devastating impact. South Africa is among the most severely affected countries, with KwaZulu-Natal province at the epicentre of the HIV–TB syndemic. Recent estimates indicate that over 20% of adults in the province are living with HIV, and TB remains the leading cause of death among people living with HIV (PLHIV) (1, 2).
CD4 cell count is a critical biomarker of immune function, and its decline during HIV infection significantly increases susceptibility to TB. Studies show that PLHIV with CD4 cell counts below 200 cells/mm3 face several-fold higher odds of developing TB compared to those with higher counts (3). Conversely, TB infection can accelerate HIV progression through sustained immune activation (4, 5). This bidirectional interplay underscores the need for analytical approaches that can jointly model these interdependent outcomes.
However, most epidemiological studies analyse TB status (binary) and CD4 cell count (continuous) separately, typically using standard regression or mixed-effects models (6, 7). These approaches often overlook latent correlations and shared unobserved risk factors, potentially biasing estimates and weakening inference.
Joint modelling strategies address this gap by simultaneously estimating multiple correlated outcomes (8). While such methods have been applied in HIV progression and survival analysis (9) and for binary outcomes (10), they are seldom used in HIV–TB research. A few recent studies have jointly modelled TB diagnosis and CD4 cell count outcomes using multivariate approaches, particularly within a Bayesian framework. For instance Gebre and Hussen (11), examined co-infection risks using joint modelling and concluded that ignoring the correlation between CD4 cell count and TB outcomes may underestimate joint disease burden. Another study by Mchunu et al. (12) also utilised joint modelling to examine the association between CD4 cell count and the risk of death in TB/HIV data. However, both studies did not account for shared latent factors that may influence both conditions simultaneously.
Beyond regression-based joint models, a variety of techniques, such as multivariate latent class models, shared frailty models, and joint survival models, have enhanced infectious disease research. For instance Mugwanyaet al. (13), identified adherence profiles using latent class analysis in PrEP users in Kenya. Work by Kazibwe et al. (14) applied a frailty model to explore predictors of TB incidence among PLHIV in Nigeria, while Mollel et al. (15) used frailty-adjusted Poisson models to assess the effect of TB co-infection and spatial clustering on HIV mortality in Tanzania. Similarly Gumede-Moyo et al. (16), employed multivariate survival models to study ART retention and mortality in Zambia.
Parallel advances in mechanistic modelling have explored syndemic dynamics and behavioural feedback mechanisms. These include models of media-driven psychological fear (17), prevalence-dependent awareness (18), and COVID-19, related treatment disruption (19, 20). Demographic-focused models, such as fractional gender-structured frameworks (21), have also revealed heterogeneity in co-infection risk. However, these models typically do not incorporate individual-level clinical biomarkers or support joint statistical inference.
To address persistent gaps in modelling the interplay between HIV progression and TB risk, we apply a Bayesian multivariate latent variable model to jointly analyse binary TB diagnosis and continuous log-transformed CD4 cell count among PLHIV in KwaZulu-Natal. This flexible framework accommodates mixed outcome types, accounts for latent correlations, and enables full posterior inference. Unlike mechanistic epidemic models, our approach provides statistically grounded, data-driven insights into the co-dynamics of immune suppression and TB risk, tailored to clinical and public health applications.
Bayesian latent variable models are particularly suited to complex epidemiological data, offering advantages such as the incorporation of prior knowledge, robustness to sparse or hierarchical structures, and the ability to model unobserved confounders (e.g., stigma, nutritional status, or healthcare access) that influence both HIV progression and TB susceptibility (22). These properties make them powerful tools for integrated analysis in syndemic contexts like HIV–TB.
This study makes a novel contribution by being among the first to apply a Bayesian multivariate probit latent variable framework to jointly model CD4 cell count and TB diagnosis in a high-burden, real-world setting. Previous work has typically modelled these outcomes separately, used less flexible methods, or lacked explicit handling of latent dependence and unobserved heterogeneity. By addressing these limitations, our approach enhances both inferential accuracy and epidemiological relevance, offering a new analytical lens to support targeted interventions and future research in syndemic disease management.
2 Materials and methods
2.1 Sources of data and study population
This study is based on secondary data from two consecutive, population-based cross-sectional surveys conducted under the HIV Incidence Provincial Surveillance System (HIPSS). HIPSS is a large-scale surveillance initiative aimed at monitoring HIV incidence and prevalence in KwaZulu-Natal, South Africa. The first survey was conducted between 11 June, 2014, and 18 June, 2015, and the second between 8 July, 2015, and 7 June, 2016. Both surveys were carried out in the Vulindlela (a rural area) and Greater Edendale (a peri-urban area) within the uMgungundlovu District.
To ensure representativeness, HIPSS employed a multi-stage probability sampling method. From 600 enumeration areas (EAs), 591 EAs with at least 50 households were eligible. Of these, 221 EAs were randomly selected for the 2014 survey and 203 for the 2015 survey. Within each selected EA, households were systematically sampled, and one eligible individual from each household was randomly chosen after providing written informed consent. The geographic coordinates of all sampled households were captured using GPS technology to support spatial analysis and minimise selection bias.
Data quality was rigorously maintained through a series of checks. During the first month of fieldwork, data were monitored daily, followed by monthly checks for 6 months, and then quarterly assessments. The Mobenzi Researcher system (Durban, South Africa) enabled real-time tracking of field activities, protocol compliance, and data integrity. Automated systems flagged inconsistencies immediately for correction. The dataset also includes laboratory-verified HIV test results from peripheral blood samples, enhancing its epidemiological value. All data underwent centralised management, rigorous quality control, and completeness verification.
The 2014 HIPSS survey included 9,812 participants (6,265 females and 3,547 males), while the 2015 survey included 10,236 participants (6,431 females and 3,805 males), for a combined total of 20,048 individuals aged 15–49. Among these, 7,839 tested HIV positive, and 7,776 had valid CD4 cell count measurements. Participants with missing CD4 cell count data (n = 63) were excluded from this analysis, resulting in a final sample of 7,776. To address missingness in other variables, we employed multiple imputation by chained equations (MICE) using the mice package in R. The imputation model included all variables used in the analysis to preserve multivariable relationships and ensure congeniality. While some risk of bias remains due to unmeasured confounding or potential violations of the missing at random (MAR) assumption, the robust sampling design, inclusion of auxiliary variables, and principled handling of missing data enhance the validity and generalisability of our findings.
The choice to focus on individuals aged 15 to 49 years is grounded in both epidemiological and policy considerations. This age group encompasses the sexually active and economically productive population, who are at the highest risk for HIV acquisition and TB co-infection. It also aligns with national surveillance efforts and UNAIDS reporting standards, allowing for comparability with broader public health data and making the results directly relevant for intervention planning in high-burden regions like KwaZulu-Natal.
2.2 Study variables
Two dependent variables were included in this study. The first one was CD4 count (continuous), which was log-transformed prior to modelling. Log transformation helps stabilise variance, improve normality assumptions, and enhance the interpretability of regression coefficients in the context of continuous outcomes. By transforming CD4 cell counts, the model achieves a better fit and more reliable inference under the assumption of normally distributed residuals.
The second response variable was TB, which was categorised as a binary outcome as shown in Equation 1:
The selection of explanatory variables was guided by prior literature, theoretical relevance, and data availability. Key demographic (age and gender), socioeconomic (education and income), and behavioural (ARV use, alcohol use) factors were included to control for confounding and to investigate their role in shaping health outcomes among people living with HIV. Variables such as viral load suppression and STI diagnosis were also included, given their biological association with immune response and TB susceptibility. The year of the survey was added to adjust for temporal effects.
We applied the variance inflation factor (VIF) to check for multicollinearity before fitting the model. All the VIF values were quite low, all less than 1.5, showing that multicollinearity was not a significant concern in the fitted model.
2.3 Statistical analysis
To jointly model the continuous CD4 cell count and the binary TB diagnosis, we adopted a Bayesian multivariate latent variable framework. This approach simultaneously accounts for the distinct nature of each outcome while capturing shared latent influences, such as unobserved immune vulnerability, socioeconomic conditions, or healthcare access disparities.
2.3.1 Bayesian joint modelling framework
We jointly model CD4 cell count and TB diagnosis outcomes using a Bayesian multivariate framework to account for their potential latent correlation. Let denote the log-transformed CD4 cell count (a continuous outcome), and the TB diagnosis status (a binary outcome) for individual , with covariate vectors and , respectively.
2.3.1.1 Likelihood specification
For the continuous CD4 cell count outcome, we assume a Gaussian distribution, specified in Equation 2:
For the binary TB outcome, modelled using a probit link, the specification is given in Equation 3:
where is the inverse standard normal CDF (probit link), and are the respective regression coefficients associated with the covariates and respectively, and is an independent and identically distributed random error term that captures residual variability not explained by the model, assumed to follow a normal distribution with mean 0 and variance .
2.3.1.2 Priors
We assign weakly informative priors to all model parameters:
The choice of priors in the model offers several key advantages. The normal priors for fixed effects provide weak regularisation, allowing the regression coefficients to vary while still centring them around zero, which is useful when prior information about the predictors is limited or uncertain. This approach strikes a balance between flexibility and constraining the model to avoid overfitting (23). Similarly, the Student-t priors on the intercepts and residual standard deviations are particularly advantageous in handling potential outliers and providing robust estimates, as the heavy tails of the Student-t distribution allow the model to accommodate extreme values without being overly influenced by them (24). This robustness is crucial when modelling real-world data that may contain outliers or unusual patterns. The normal prior on random effects helps to model individual-level deviations in a hierarchical structure, ensuring that the random effects are not overfitted, while also promoting stability and better generalisation of the model (22). Finally, the Student-t prior for the variance parameter adds flexibility to the modelling of the random effects’ variance, providing the model with the ability to adapt to varying levels of uncertainty in individual deviations. Overall, these priors allow for a well-regularised model that balances flexibility with robustness, ensuring stable estimates and reliable inference even in the presence of outliers or limited prior information.
To assess the robustness of our model to prior assumptions, we conducted a sensitivity analysis by re-fitting the joint model using alternative prior distributions, Normal, Half-Normal, Cauchy, and Half-Cauchy, for key parameters. Model performance under each specification was evaluated using the expected log predictive density (ELPD), estimated via leave-one-out cross-validation (LOOCV). ELPD values are computed relative to the base model with Student-t priors. Higher values indicate better out-of-sample predictive performance. The results are summarised in Table 1.
As summarised in Table 1, all alternative prior specifications yielded comparable fits to the default Student-t priors, with only minor differences in expected log predictive density (ELPD). The largest deviation (ΔELPD = −0.6) occurred under the Cauchy prior, which also exhibited slower convergence and signs of potential multimodality, consistent with the behaviour of heavy-tailed priors. Overall, the similar predictive performance across priors supports the robustness of the model to reasonable prior choices. These findings suggest that the default prior did not unduly influence posterior inference. Furthermore, model diagnostics indicated no evidence of convergence issues or multimodality for any specification, reinforcing the stability of the MCMC estimation process.
2.3.1.3 Joint model formulation
The joint likelihood for individual
is given by Equation 4:
Across all n individuals the full likelihood is expressed in Equation 5:
With .
2.3.1.4 Posterior distribution
The posterior distribution of the parameters is proportional to the product of the likelihood and the prior distributions:
With P (.) indicating prior densities for parameters.
2.3.2 Bayesian latent variable model
The Bayesian latent variable framework is well-suited for analysing multivariate outcomes where at least one of them may be binary, unobserved, or subject to measurement error. In this study, we jointly modelled a continuous outcome (CD4 cell count) and a binary outcome (TB diagnosis) using a shared latent variable structure. This framework allows for the estimation of the correlation between the latent propensity for TB and the observed CD4 cell count, capturing shared variance potentially driven by underlying biological mechanisms or sociodemographic factors.
To represent the binary TB outcome within a latent structure, we use a probit formulation in which the observed TB diagnosis arises from an unobserved continuous latent variable . Specifically, we define:
Continuous outcome (CD4 cell count) given in Equation 6:
Latent TB propensity, defined in Equation 7:
The observed TB outcome is then determined as Equation 8:
Where is the latent variable capturing shared unobserved heterogeneity between outcomes.
2.3.3 Joint latent variable formulation
To explicitly model the residual dependence between the continuous and binary outcomes, we assume the error terms and follow a bivariate normal distribution:
This formulation introduces a latent correlation parameter , which captures the residual association between CD4 count and the latent TB propensity after accounting for observed covariates. This enhances the model’s flexibility in representing complex biological interdependencies.
The posterior distribution of the parameters is proportional to the product of the likelihood and the prior distributions:
2.4 Parameter estimation
Building on the latent variable specification from Section 2.3.2, we now describe the estimation procedure used to fit the joint model.
Let denote the observed data for individuals, where is the continuous CD4 count and is the binary TB status.
The likelihood for the observation is shown in Equation 9:
The full likelihood over all individuals is then given in Equation 10:
Where is the cumulative distribution function (CDF) of the standard normal distribution.
Taking logs, the full log-likelihood is expressed in Equation 11:
Expanding the expression we get Equation 12:
With parameter vector defined in Equation 13:
The posterior distribution is then defined as:
Where represents the prior distributions assigned to the model parameters.
Due to the non-conjugacy introduced by the probit link and latent terms, this posterior is not analytically tractable. Bayesian inference was therefore conducted using Markov Chain Monte Carlo (MCMC) methods, specifically Hamiltonian Monte Carlo (HMC) as implemented in Stan, accessed through the brms package in R. This approach efficiently samples from high-dimensional posterior distributions while maintaining convergence and computational stability. Importantly, MCMC allowed us to explore the full joint posterior distribution of model parameters, enabling robust inference despite the lack of closed-form solutions.
2.5 Model diagnostics
Posterior inference was carried out using Markov Chain Monte Carlo (MCMC) sampling, with diagnostics and validation techniques to ensure reliable estimation. Convergence was assessed using the Gelman–Rubin statistic (R-hat) and effective sample size metrics, supplemented by trace and posterior density plots to inspect mixing and distributional behaviour. Posterior predictive checks were used to evaluate how well the model replicated observed data for both outcomes, CD4 cell count and TB diagnosis. Additionally, residual plots were generated to identify model misfit. To assess the joint structure, the correlation between predicted TB probabilities and CD4 cell counts was examined, validating the contribution of shared random effects in capturing latent dependencies.
2.6 Software and implementation
All modelling was conducted in R version 4.4.0, using the brms package, which provides a high-level interface to Stan for efficient Hamiltonian Monte Carlo (HMC) sampling. The model specified separate likelihoods for the two outcomes: a Gaussian likelihood for continuous CD4 cell count, and a Bernoulli likelihood with a probit link for binary TB diagnosis. A shared random effect structure was used to jointly model the two outcomes, while controlling for a consistent set of covariates across both components.
3 Empirical results
3.1 Descriptive statistics and joint model results
In this study of 7,776 HIV-positive individuals residing in Vulindlela and Greater Edendale areas, the distribution of CD4 cell counts (log-transformed) indicated generally preserved immune function. The mean CD4 count was 6.13 with a standard deviation of 0.64, while the median was 6.23. CD4 cell count values ranged from 2.40 to 7.58, with an interquartile range of 5.82 to 6.57, suggesting moderate variability around the central values.
Regarding tuberculosis (TB), a total of 2,155 individuals were diagnosed with TB, yielding an overall TB prevalence of 27.7% (95% CI: 26.7–28.7%). This substantial co-infection rate highlights the ongoing dual burden of TB and HIV in this high-prevalence setting. Table 2 presents a detailed stratification of TB prevalence and CD4 cell count distribution across key demographic, socioeconomic, and clinical covariates.
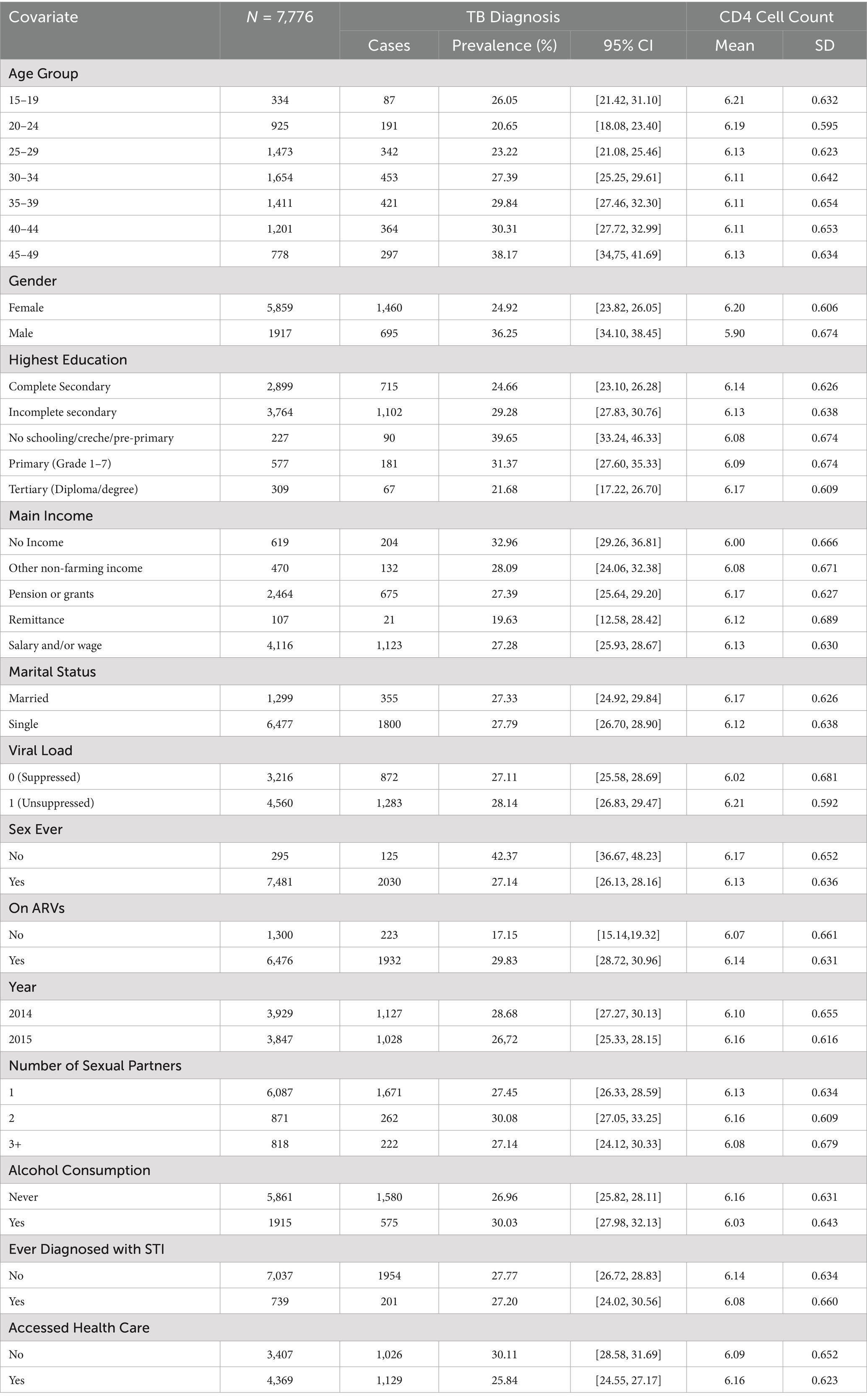
Table 2. Descriptive summary statistics for both TB and CD4 Cell count by each covariate among HIV-positive individuals in Vulindlela and greater Edendale areas in Umgungundlovu municipality.
Results from Table 2 reveal significant variations in TB prevalence among HIV-positive individuals across demographic, socioeconomic, and clinical characteristics. TB prevalence tended to be higher among older age groups, males, individuals with lower education levels, and those without a source of income. Elevated prevalence was also observed among individuals not accessing healthcare services, those who consumed alcohol, and those who had never had sex. These patterns suggest that social vulnerability, limited healthcare access, and certain behavioural or clinical factors may contribute to an increased risk of TB in this population.
Table 2 also highlights differences in log-transformed CD4 cell counts across similar subgroups. Higher CD4 cell counts were generally observed among younger individuals, females, those with higher education levels, and individuals on ART. Socioeconomic factors such as having a source of income and accessing healthcare were also positively associated with CD4 cell count. These findings underscore the complex interplay between social determinants, clinical care, and immune function in the study population.
Overall, Table 2 provides summary statistics for both TB prevalence and CD4 cell count distribution across all covariates. While these descriptive findings offer useful preliminary insights into potential associations, the results from the joint multivariate Bayesian model are emphasised for their capacity to account for both shared and outcome-specific predictors, as well as the potential directional relationship between TB prevalence and immune status. This modelling approach enables a more robust and integrated interpretation of the data.
To jointly model TB diagnosis and CD4 cell count among HIV-positive individuals aged 15–49 years in KwaZulu-Natal, a Bayesian multivariate model was fitted using the brms package in R. Table 3 presents the results, including estimated average marginal effects (AMEs) for the binary outcome of TB diagnosis and posterior means for the continuous outcome of CD4 cell count, each accompanied by their respective 95% credible intervals. This joint modelling approach quantifies the independent effects of sociodemographic and clinical covariates on both outcomes, while accounting for potential confounding and shared influences. Covariates were deemed statistically significant if their 95% credible intervals did not include zero.
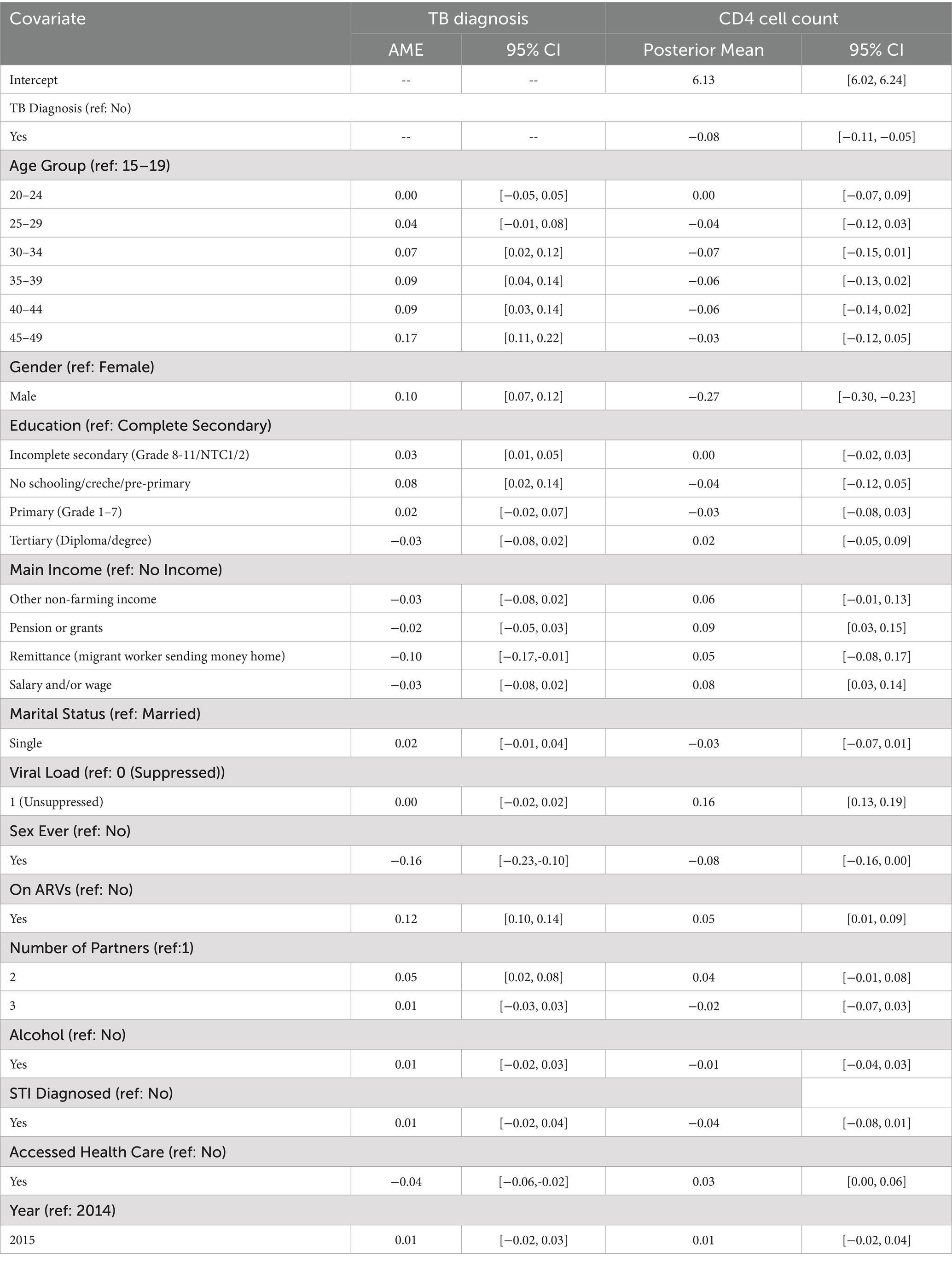
Table 3. Estimated average marginal effects and posterior means with 95% credible intervals for covariates associated with TB diagnosis and CD4 cell count among HIV-positive individuals.
Focusing first on TB diagnosis, several sociodemographic and clinical covariates showed significant associations with the likelihood of being diagnosed with TB. Age emerged as a strong predictor of TB diagnosis. Compared to individuals aged 15–19 years, those aged 30–34 had a 7-percentage point higher probability of being diagnosed with TB (AME: 0.07, 95% CrI: 0.02 to 0.12). This probability increased among those aged 35–39 and 40–44, with both groups showing a 9-percentage point increase (AME: 0.09, 95% CrI: 0.04 to 0.14 and 0.09, 95% CrI: 0.03 to 0.14, respectively). The highest marginal effect was observed in the 45–49 age group, with a 17-percentage point higher likelihood of TB diagnosis compared to the reference group (AME: 0.17, 95% CrI: 0.11 to 0.22).
Gender also had a significant effect, with males showing a 10-percentage point higher probability of TB diagnosis than females (AME: 0.10, 95% CrI: 0.07 to 0.12). Education was another important factor. Individuals with incomplete secondary education were slightly more likely to be diagnosed with TB (AME: 0.03, 95% CrI: 0.01 to 0.05), while those with no formal schooling or only pre-primary education had a more substantial increase in risk (AME: 0.08, 95% CrI: 0.02 to 0.14), compared to those who completed secondary education.
Economic status, as measured by the main source of income, was also associated with TB diagnosis. Participants who reported receiving remittances from migrant workers had a significantly lower probability of TB diagnosis (AME: –0.10, 95% CrI: −0.17 to −0.01) compared to those with no income, suggesting a potential protective effect of financial support.
Interestingly, individuals who had ever had sexual intercourse were significantly less likely to be diagnosed with TB than those who had not (AME: –0.16, 95% CrI: −0.23 to −0.10). This may reflect confounding by age, marital status, or health-seeking behaviour. Additionally, those who were on antiretroviral therapy (ARVs) had a 12-percentage point higher probability of being diagnosed with TB (AME: 0.12, 95% CrI: 0.10 to 0.14), potentially indicating better case detection among those already engaged in HIV care services.
Furthermore, individuals who had two sexual partners had a significantly increased probability of TB diagnosis compared to those with only one partner (AME: 0.05, 95% CrI: 0.02 to 0.08), suggesting a possible link between behavioural risk factors and TB exposure.
Lastly, access to health care services was associated with a reduced probability of TB diagnosis. Participants who had accessed health care had a 4-percentage point less likely to be diagnosed with TB (AME: –0.04, 95% CrI: −0.06 to −0.02), which may reflect the protective role of preventive care, early treatment, or improved health literacy among health service users.
Turning to CD4 cell count, several covariates were significantly associated with immune status as measured by CD4 levels. Individuals diagnosed with TB had significantly lower CD4 cell counts (posterior mean = −0.08; 95% CrI: −0.11 to −0.05), underscoring the immunosuppressive impact of TB co-infection. Males had notably lower CD4 cell counts compared to females (posterior mean = −0.27; 95% CrI: −0.30 to −0.23), reflecting potential biological or behavioural disparities in immune status or healthcare utilisation.
Socioeconomic variables showed a consistent pattern. Participants receiving a pension or grants (posterior mean = 0.09; 95% CrI: 0.03 to 0.15) or earning a salary or wage (posterior mean = 0.08; 95% CrI: 0.03 to 0.14) had significantly higher CD4 cell counts relative to those with no income, suggesting a positive link between financial stability and immune health.
Interestingly, individuals with unsuppressed viral load had higher CD4 cell counts (posterior mean = 0.16; 95% CrI: 0.13 to 0.19) than those with suppressed viral load. This counterintuitive finding may reflect the timing of CD4 cell count and viral load measurements (e.g., early ART initiation before viral suppression is achieved), or confounding factors such as recent seroconversion or treatment adherence patterns.
Being on antiretroviral therapy (ARVs) was associated with modestly improved CD4 cell counts (posterior mean = 0.05; 95% CrI: 0.01 to 0.09), affirming the benefit of treatment, although the modest effect size may reflect late initiation or suboptimal adherence in some individuals.
Access to healthcare services was marginally associated with higher CD4 cell counts (posterior mean = 0.03; 95% CrI: 0.00 to 0.06). While the lower bound of the credible interval touches zero, the positive direction and biological plausibility suggest a potential, albeit modest, benefit of healthcare access on immune function.
Finally, individuals who had ever had sex showed slightly lower CD4 cell counts (posterior mean = −0.08; 95% CrI: −0.16 to 0.00). Although the upper bound of the CrI includes zero, the direction of association may reflect behavioural or demographic factors such as age or sexual health risk profiles. Given the borderline nature of these results, they should be interpreted with caution and viewed as suggestive rather than conclusive.
Having examined the associations between participant characteristics and both TB diagnosis and CD4 cell count using a joint Bayesian multivariate framework (as shown in Table 3), it is essential to assess the adequacy and reliability of the fitted model. Model diagnostics provide critical insights into convergence, goodness-of-fit, and the robustness of posterior estimates. This step ensures that the inferences drawn from the joint model are statistically sound and not influenced by poor model performance, inadequate mixing, or convergence issues.
3.2 Model fit and diagnostics
This section presents a comprehensive evaluation of model fit and convergence to validate the credibility of the Bayesian estimates from the joint model. Diagnostics include trace and density plots to assess MCMC chain mixing, R-hat and effective sample sizes for sampling efficiency, residual plots to evaluate fit, posterior predictive checks (PPCs) to examine predictive performance, and an analysis of the correlation between predicted TB probabilities and CD4 cell counts to investigate joint outcome behaviour.
3.2.1 Convergence diagnostics
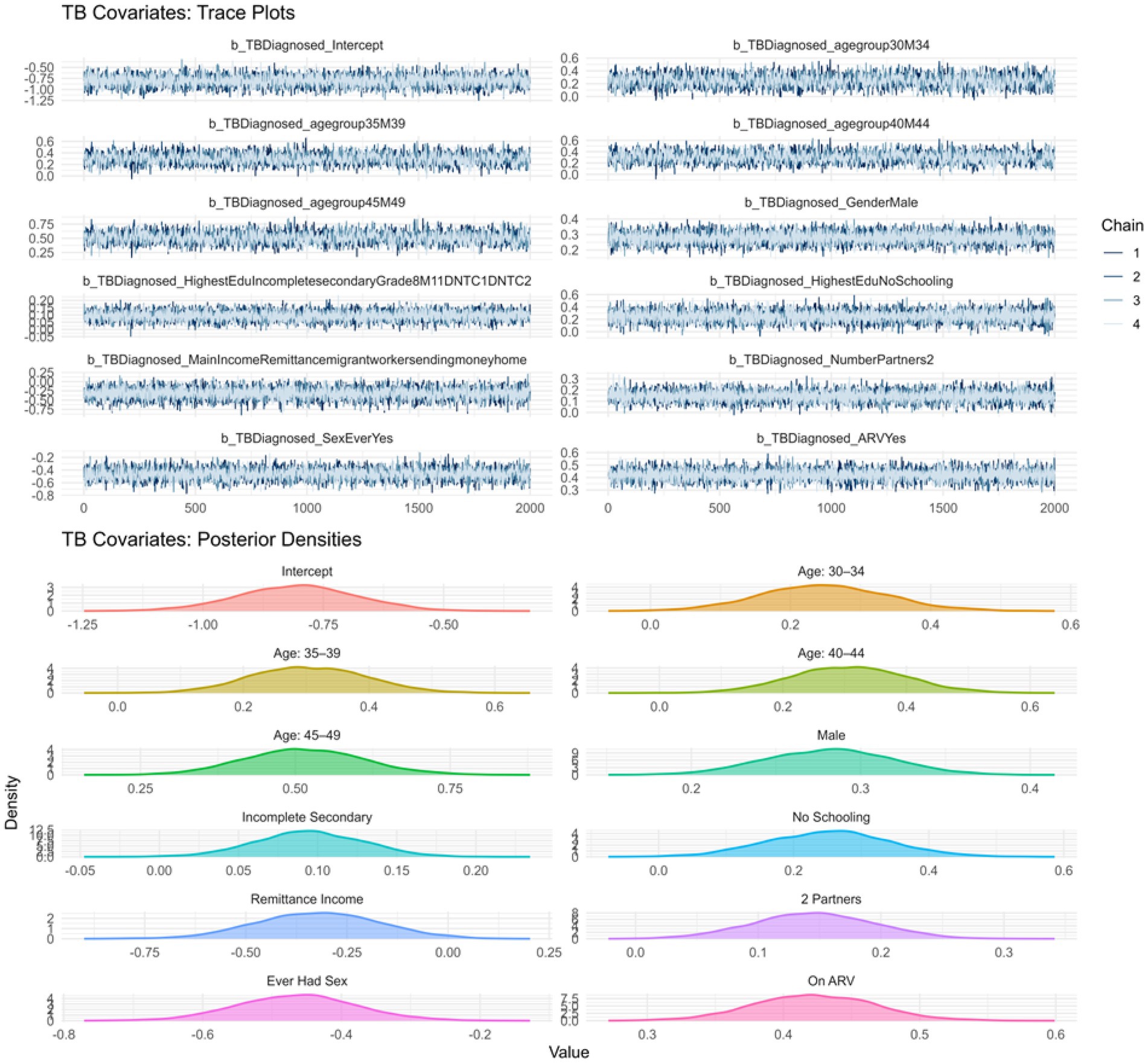
Trace and density plots were used to assess convergence and posterior distribution behaviour for both sub models. Figure 1 displays the trace and density plots for the TB sub model. The chains exhibit consistent mixing without divergences or trends, and the density plots show smooth, unimodal distributions, suggesting good convergence and stable posterior estimation.
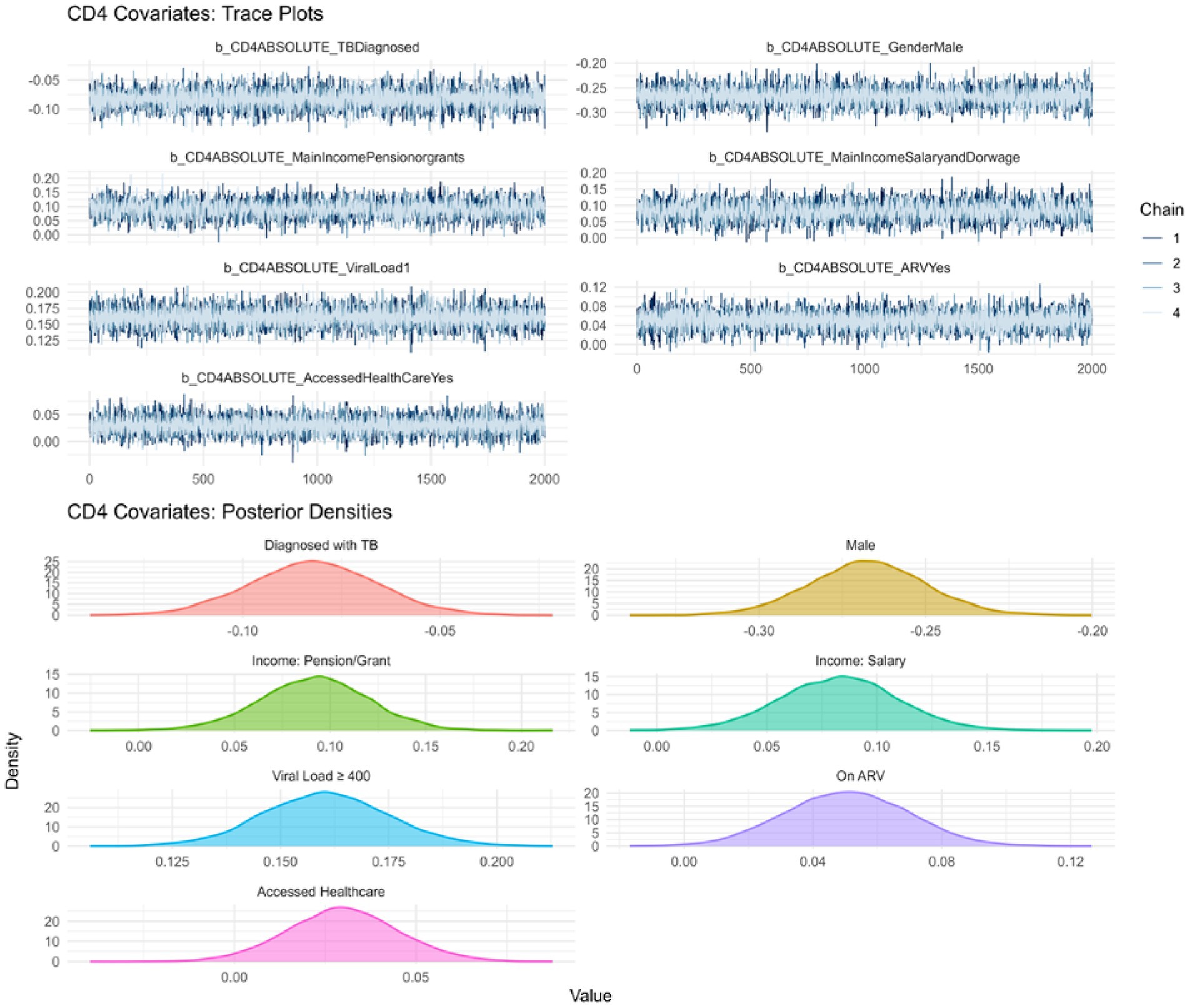
Similarly, Figure 2 presents the trace and density plots for the CD4 cell count sub model, showing well-mixed chains and smooth posterior distributions, confirming that both sub models yielded robust and interpretable estimates.
To further confirm sampling stability, we examined R-hat values and effective sample sizes (ESS). All R-hat values for both outcomes were exactly 1.00, indicating excellent chain convergence. Bulk and Tail ESS values exceeded the recommended threshold of 1,000, supporting the reliability and efficiency of the sampling process.
3.2.2 Residual analysis
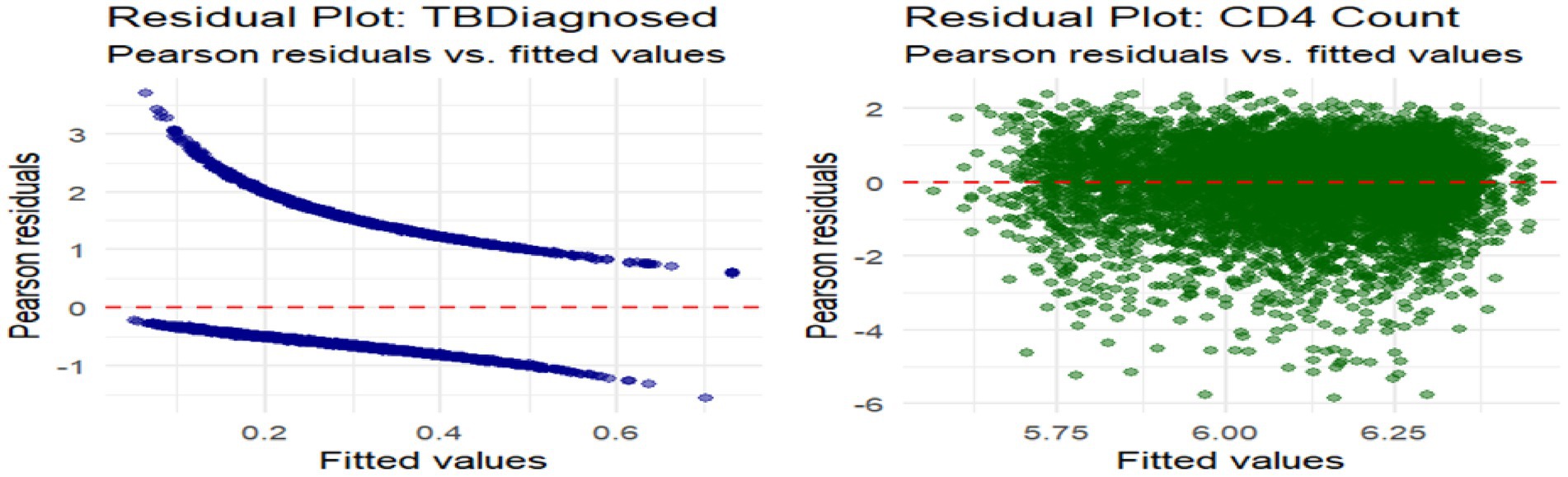
Model residuals were analysed to inspect fit for each outcome. Figure 3 shows Pearson residuals plotted against fitted values. The TB diagnosis model exhibits a mild curvature in residuals, hinting at minor model misspecification or unaccounted non-linearity. However, residuals largely remain within acceptable bounds. In contrast, the CD4 cell count model displays a symmetric residual pattern centred around zero, with no systematic deviation, suggesting a well-fitting model.
3.2.3 Posterior predictive checks and model calibration
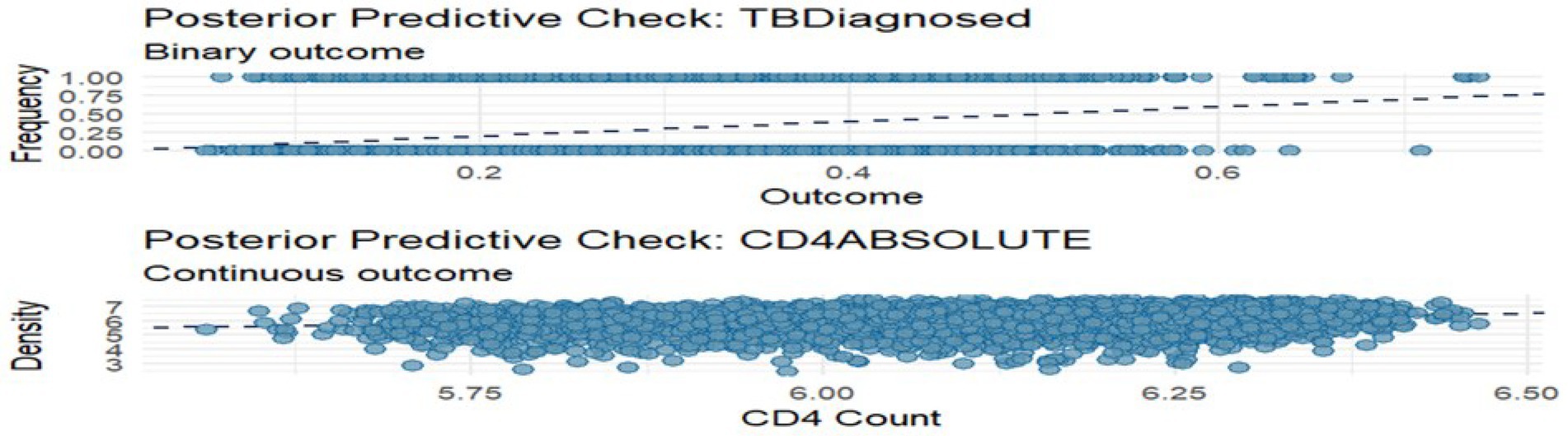
Posterior predictive checks (PPCs) were conducted to assess the model’s ability to replicate key features of the observed data. Figure 4 shows a density overlay and scatter average comparison for both outcomes. For TB diagnosis, predicted probabilities aligned well with observed outcomes, particularly showing higher predicted risk for TB-positive individuals. For CD4 cell counts, the overlay of observed and predicted densities revealed a good fit, though the upper tail was slightly underestimated.
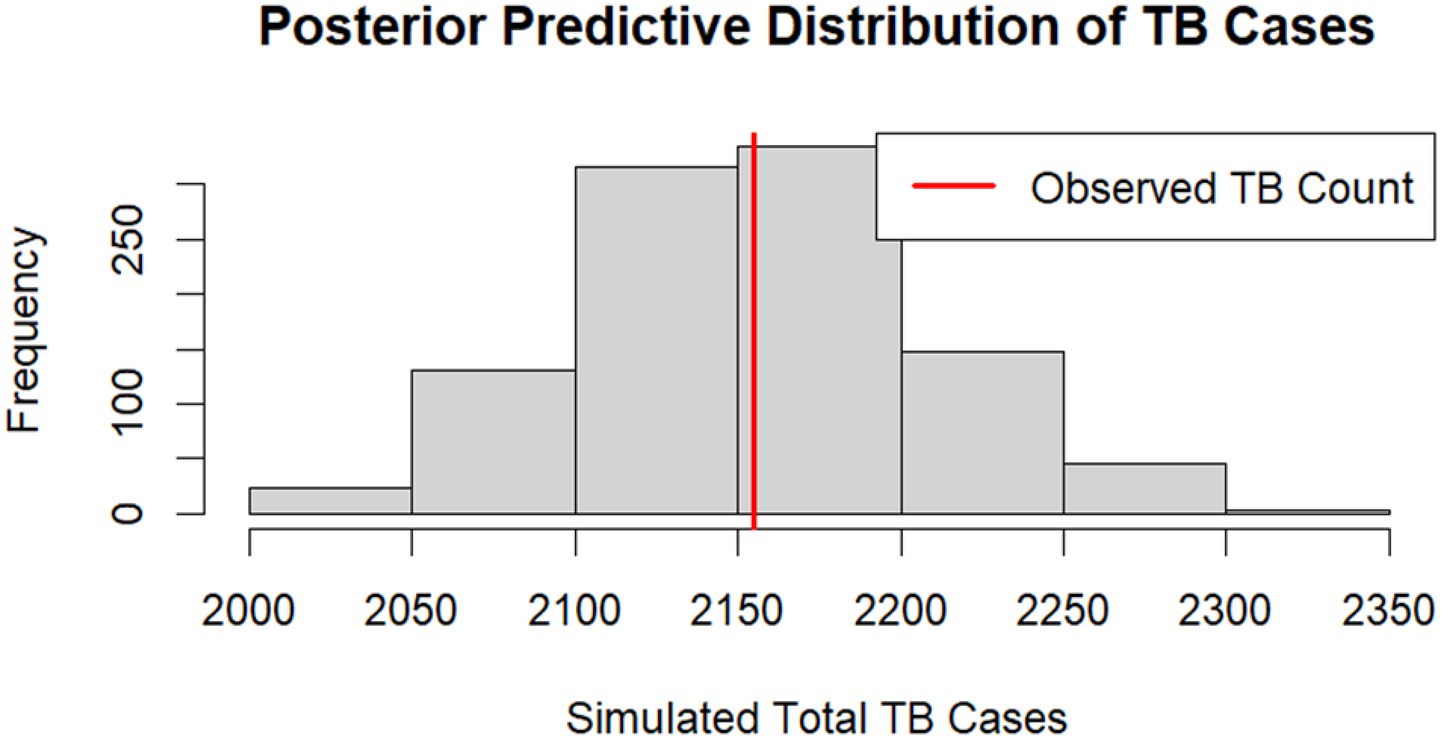
To further evaluate model calibration, we examined the full posterior predictive distributions of summary statistics. For the TB binary outcome, we compared the observed total number of TB cases to simulated totals from the posterior predictive distribution (Figure 5). The observed count fell near the centre of the simulated distribution, and the Bayesian posterior predictive p-value (PPP) was 0.52. This value suggests the model can replicate the observed TB prevalence without significant bias.
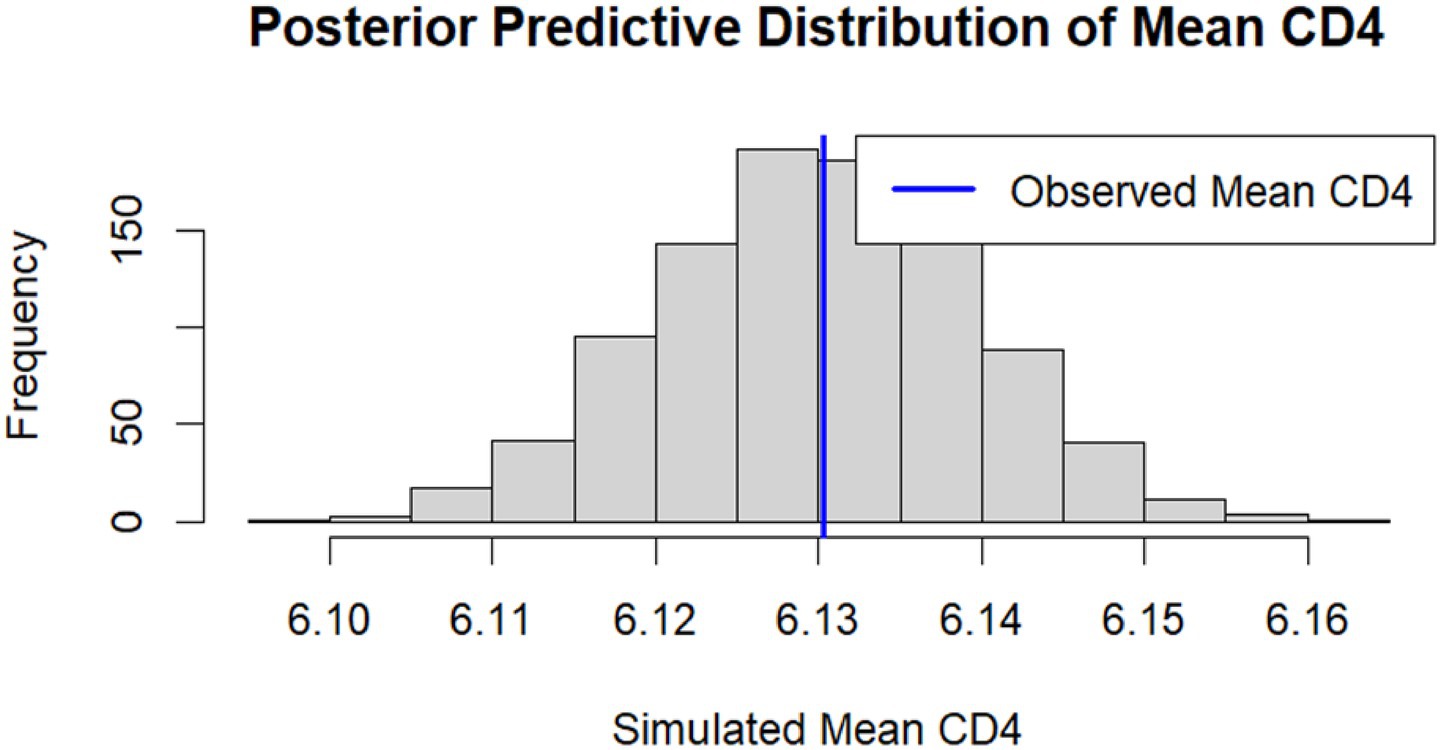
Similarly, for the CD4 cell count outcome, the distribution of simulated mean CD4 values is shown in Figure 6. The observed mean CD4 cell count (blue line) was centrally located within the posterior predictive distribution, indicating that the model accurately reproduced both the central tendency and variability of CD4 cell count levels.
3.2.4 Joint outcome behaviour and latent correlation
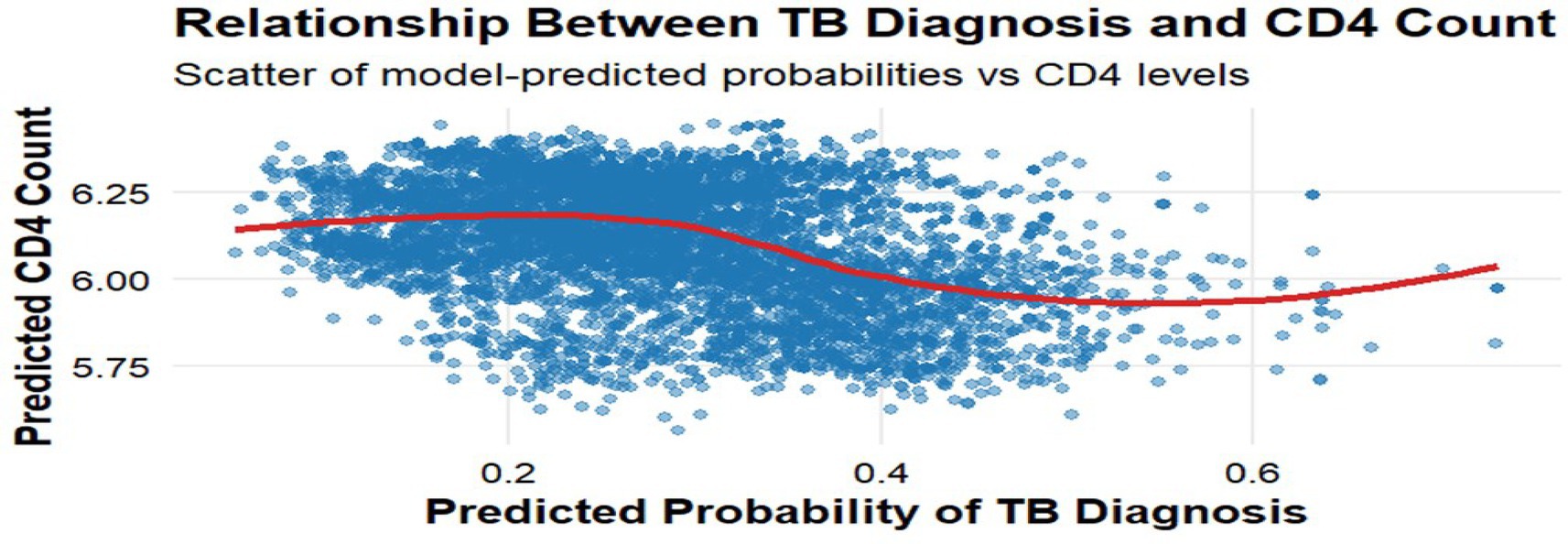
Figure 7 plots predicted TB probabilities against predicted CD4 cell counts, showing a clear inverse relationship consistent with biological expectations. A locally estimated scatterplot smoothing (LOESS) curve illustrates a steady downward trend, indicating that individuals with lower predicted CD4 cell counts had higher predicted TB probabilities. The estimated correlation between individual-level predicted means was −0.38, and the posterior distribution of this latent correlation yielded a mean of −0.36 with a 95% credible interval of (−0.47, −0.25), reinforcing the negative association between immune suppression and TB susceptibility.
Together, these diagnostics affirm that the joint Bayesian model is well-specified, convergent, and capable of accurately reproducing observed data for both outcomes. The consistency of the posterior predictive distributions with the observed data, along with strong convergence and biologically coherent latent structure, supports the validity and interpretability of the inferences drawn from this model.
4 Discussion
This study applied a Bayesian joint multivariate probit model to simultaneously assess predictors of tuberculosis (TB) diagnosis (binary outcome) and CD4 cell count (continuous outcome) among people living with HIV (PLHIV) aged 15–49 in KwaZulu-Natal, South Africa. By explicitly modelling the latent correlation between these outcomes, the approach captures their underlying biological and epidemiological interdependence. This represents a methodological advance over separate univariate models, which often ignore shared unobserved risk factors and may lead to biased or incomplete inferences.
Several socio-demographic and behavioural factors were significantly associated with TB diagnosis. Individuals aged 30–49 had elevated TB odds, reflecting patterns observed nationally and globally, where this economically active group faces higher HIV prevalence and cumulative TB exposure (2, 25). Male gender was also associated with increased TB risk (AME = 0.10), consistent with studies highlighting gender disparities in TB due to behavioural risks, occupational exposure, and reduced healthcare utilisation among men (26, 27).
Educational attainment emerged as a protective factor. Individuals with lower education had higher odds of TB, likely reflecting delayed care-seeking, lower health literacy, and limited awareness of TB symptoms and prevention (28). Notably, those receiving remittances were less likely to be diagnosed with TB (AME = −0.10). While literature on remittances and TB is sparse, this may suggest that financial support from external sources improves living conditions and facilitates healthcare access, mitigating TB vulnerability, echoing broader findings on socioeconomic buffers in infectious disease epidemiology (29).
Behavioural factors showed nuanced effects. Reporting two or more sexual partners was associated with increased TB risk, consistent with literature linking high-risk sexual behaviour to co-infection vulnerability and network-driven transmission (30). Conversely, having ever had sex was associated with reduced TB odds, an unexpected finding that may reflect unmeasured confounders, such as relationship stability, community engagement, or health-seeking patterns.
Antiretroviral therapy (ART) use was positively associated with TB diagnosis (AME = 0.12), likely reflecting TB immune reconstitution inflammatory syndrome (IRIS). This condition occurs when ART-induced immune recovery unmasks previously latent TB. Studies in sub-Saharan Africa and India report IRIS incidence rates ranging from 7 to 54%, particularly among individuals with low baseline CD4 cell counts or high mycobacterial burden (31). In contrast, access to healthcare was negatively associated with TB (AME = −0.04), underscoring the importance of early and sustained engagement with HIV care services (32).
CD4 count was inversely associated with TB diagnosis (posterior mean = −0.08), reflecting the well-established relationship between immune suppression and TB risk. Declining CD4 cell count levels compromise host immune responses, increasing susceptibility to both reactivation of latent TB and new infections. Work by Buziashvili et al. (33) reported similar findings in a multinational cohort, while Lu et al. (34) found TB risk declines steeply as CD4 cell counts rise above 300 cells/mm3. These results reinforce the importance of early ART initiation to preserve immune function and reduce TB risk.
In our CD4 sub-model, individuals diagnosed with TB also had lower CD4 cell counts, confirming the reciprocal clinical pattern observed in HIV–TB co-infection. This finding strengthens evidence that TB often presents in more immunocompromised individuals, and that TB itself may further depress immune function. These dynamics support public health strategies focused on early HIV diagnosis and timely ART uptake to prevent co-infection.
Gender disparities were also evident in immune status. Male participants exhibited significantly lower CD4 counts than females, potentially due to delayed ART initiation, lower healthcare utilisation, or broader structural vulnerabilities. This aligns with findings from fractional gender-structured models (21), which underscore the influence of gender in shaping HIV–TB trajectories. Our model, which found both elevated TB risk and lower CD4 cell counts among men, further highlights the need to incorporate gender-sensitive strategies in both research and intervention design.
Socioeconomic factors influenced immune outcomes as well. Participants earning wages or receiving pensions had higher CD4 cell counts, consistent with studies linking income stability to better nutritional status, ART adherence, and healthcare access (35). These findings underscore the role of economic stability in improving immunological outcomes among PLHIV.
As expected, ART use was positively associated with CD4 cell count (posterior mean = 0.05), reaffirming its central role in immune restoration (32). Interestingly, viral load showed a positive association with CD4 count (posterior mean = 0.16), a counterintuitive result that may reflect measurement timing. For instance, some participants may have had improving CD4 cell count levels but had not yet achieved full viral suppression. Alternatively, the data may include individuals in early ART stages where CD4 cell count recovery precedes virologic control. This finding warrants further exploration.
This study contributes to syndemic epidemiology by applying a joint Bayesian latent variable model to simultaneously estimate TB diagnosis and CD4 cell count, while accounting for their residual correlation. The model revealed a biologically plausible negative latent correlation (ρ = −0.38), likely capturing the influence of unmeasured shared factors, such as nutritional status, healthcare access, stigma, and co-infections, that simultaneously affect immune suppression and TB risk. Traditional models that analyse these outcomes separately often fail to account for such underlying dependencies, potentially leading to biased estimates or underestimation of uncertainty (36). In contrast, our latent variable framework improves statistical efficiency and enhances robustness by explicitly modelling unmeasured heterogeneity and residual dependencies (37), providing a clearer reflection of the interconnected nature of TB and HIV-related immunodeficiency.
While joint models have been applied in studies of HIV progression and survival (12, 38), their use in high-burden HIV–TB contexts, particularly involving mixed outcome types, remains limited. Compared to copula-based methods, such as Clayton or Frank copulas, that impose strong and often restrictive parametric assumptions about the dependency structure (39), our approach avoids the need to specify a particular copula form and is therefore more robust to model misspecification. Similarly, fully semiparametric models, although flexible, may suffer from instability and convergence challenges in high-dimensional or large-scale datasets due to weak regularisation and computational complexity (40). By combining interpretability, flexibility, and computational tractability, our Bayesian framework offers a practical and scalable alternative for modelling co-occurring health outcomes in syndemic settings.
Implemented using the brms package in R with Hamiltonian Monte Carlo (via Stan), our model supports both continuous and binary outcomes, incorporates prior information, and yields full posterior distributions for all parameters. This enhances transparency in uncertainty estimation and model interpretation, and offers improved convergence behaviour relative to traditional MCMC approaches. Such features make this framework particularly well-suited to complex, noisy datasets arising from routine surveillance systems in resource-limited settings, including many countries in East and Central Africa, where data quality and missingness pose ongoing challenges.
Finally, while recent advances in mechanistic modelling, including feedback systems, behavioural responses, and syndemic interactions, offer important system-level insights, they are often simulation-based and rely on strong structural assumptions (17, 19, 21). Our empirical Bayesian framework complements these approaches by offering a data-driven method for identifying individual-level dependencies between outcomes. This combination of empirical grounding and flexibility positions the model as a valuable tool for both predictive analytics and the design of targeted interventions in regions facing intertwined HIV and TB epidemics.
The associations observed in this study align with earlier research on the social and immunological determinants of HIV–TB co-infection. For example, our finding that lower CD4 cell count predicts higher TB risk mirrors the results of (33, 34), who reported similarly strong inverse associations in varied geographic settings. The gender disparities we observe, higher TB odds and lower CD4 cell counts among men, are consistent with gender-structured models (21), reinforcing the need for male-targeted interventions. In contrast to previous studies that analysed TB and CD4 cell count separately (35, 36), our joint latent variable approach accounts for shared unobserved risk factors, thereby enhancing both statistical efficiency and interpretability. This methodologically advances the literature by enabling joint modelling of mixed outcome types in a high-burden, real-world context. Furthermore, compared to copula-based or semiparametric models (39, 40), our framework offers a computationally stable, interpretable alternative suitable for public health application.
In summary, this study provides novel empirical insights into the immuno-epidemiological dynamics of HIV–TB co-infection and introduces a flexible, interpretable, and statistically rigorous modelling approach that can be extended to other syndemic settings.
5 Contribution of the study
This study advances HIV-TB research by applying a Bayesian joint modelling approach that simultaneously estimates TB diagnosis and CD4 cell count, capturing their latent correlation and improving estimation efficiency. It provides context-specific insights into how demographic, socioeconomic, and clinical factors, such as gender, education, income, ARV use, and healthcare access, shape TB risk and immune status. The use of rigorous diagnostics reinforces the robustness of the findings, while the inverse association between TB risk and CD4 cell count supports integrated modelling in co-infection research. These findings offer actionable evidence for targeted interventions in high-burden settings.
6 Implications of the study findings
This study highlights key clinical and public health priorities. Identifying risk factors such as male gender, low education, unstable income, and limited healthcare access calls for targeted TB and HIV interventions. The joint modelling approach supports integrated care strategies, reflecting the biological and statistical link between immunosuppression and TB risk. The observed negative correlation between TB probability and CD4 cell count reinforces the need for combined monitoring tools. These findings can guide early HIV diagnosis, timely ART initiation, and the incorporation of socioeconomic support in TB/HIV programmes, while also showcasing the utility of Bayesian methods in complex health analyses.
7 Strengths and limitations
This study’s primary strength lies in its use of a Bayesian joint modelling framework, which enables simultaneous estimation of TB diagnosis (binary) and CD4 cell count (continuous), while accounting for their latent correlation. This approach improves statistical efficiency and captures the underlying biological linkage between immunosuppression and TB risk. The analysis is further strengthened by the use of nationally representative, population-based data from a high HIV/TB burden setting, and comprehensive model diagnostics confirming good fit and convergence.
However, several limitations should be acknowledged. Firstly, the cross-sectional design precludes any inference about temporal relationships or causality. CD4 cell count is a dynamic, time-varying biomarker, and its association with TB may change over the course of disease progression or with the initiation and continuation of antiretroviral therapy (ART). Our static model cannot capture these longitudinal dynamics, limiting the interpretation of the timing and directionality of effects. Secondly, the routine health records used in the analysis may be subject to data quality issues, including under-reporting or misclassification of TB diagnoses, inconsistent CD4 cell count measurements, and inaccuracies in behavioural or self-reported variables. These sources of measurement error may affect the precision and robustness of the model estimates.
8 Future research
Future research should build on this foundation to incorporate longitudinal data, additional biomarkers (e.g., viral load trajectories), and causal structures to inform precision public health responses in high-burden regions. Spatial and predictive modelling approaches could also be used to identify geographic and individual-level TB/HIV co-infection risks, enabling more targeted interventions and personalised care.
9 Conclusion
This study applied a Bayesian joint modelling framework to simultaneously analyse TB diagnosis (binary) and CD4 cell count (continuous) outcomes among HIV-positive individuals in a high-burden setting. By accounting for the latent correlation between these interdependent health indicators, the approach provided more robust and biologically coherent inferences than would be possible through separate models.
Key findings confirmed well-established associations, including the inverse relationship between CD4 cell count and TB risk, the beneficial effects of ARV use, and the impact of socioeconomic factors on immune function. Male gender, lower education levels, and higher viral load were associated with greater TB vulnerability and poorer immune status, reflecting the complex interplay of structural, behavioural, and biological determinants.
Posterior predictive checks and diagnostic evaluations supported model adequacy and convergence, while the observed correlation between predicted TB probability and CD4 cell count (−0.38) reinforced the appropriateness of a joint multivariate approach. Unexpected findings, such as the paradoxical association between sexual activity and TB risk or the positive link between viral load and CD4 cell count, highlight areas for further longitudinal or qualitative investigation.
These results emphasise the importance of integrated care models that address both TB and HIV simultaneously and underscore the need for gender-sensitive, socioeconomically informed public health strategies. The use of joint modelling techniques provides a powerful tool for advancing our understanding of co-epidemic dynamics and guiding more targeted interventions in high-burden settings.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Biomedical Research Ethics Committee at the University of KwaZulu-Natal (Reference BF269/13). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
EC: Conceptualization, Formal analysis, Visualization, Methodology, Validation, Writing – original draft, Data curation. RC: Conceptualization, Supervision, Methodology, Writing – review & editing. JB: Writing – review & editing, Methodology, Supervision, Validation. KC: Writing – review & editing, Methodology, Supervision. AK: Writing – review & editing, Investigation.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We gratefully acknowledge CAPRISA for providing access to the dataset used in this study. We also thank the journal editors and reviewers for their valuable comments and suggestions that helped improve the quality of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HIV, Human Immunodeficiency Virus; KZN, KwaZulu Natal; HIPSS, HIV Incidence provincial surveillance system; STIs, Sexually transmitted infections; ART, Anti-retroviral therapy; GIS, Geographical information system; MCMC, Markov Chain Monte Carlo; CI, Credible/confidence interval; TB, Tuberculosis; LOESS, Locally estimated scatterplot smoothing; IRIS, Immune reconstitution inflammatory syndrome; AME, Average marginal effect; MAR, Missing at random; PPP, Posterior predictive p-value; ELPD, Expected log predictive density; LOOCV, Leave-one-out cross validation; ESS, Effective sample sizes.
References
1. UNAIDS. (2023). Global AIDS update 2023: The path that ends AIDS. Geneva: Joint United Nations Programme on HIV/AIDS. Available online at: https://www.unaids.org/en/resources/documents/2023/2023-global-aids-update [Accessed April 12, 2025].
2. World Health Organization (WHO) (2023). Global Tuberculosis Report 2023. Geneva: World Health Organization. Available online at: https://www.who.int/publications/i/item/9789240079925 [Accessed April 13, 2025].
3. Geremew, D, Gezie, LD, Tessema, ZT, Teshale, AB, and Adane, F. The spatial distribution and factors associated with HIV prevalence in Ethiopia: evidence from 2016 Ethiopian demographic and health survey. BMC Infect Dis. (2020) 20:741. doi: 10.1186/s12879-020-05439-7
4. Shen, Y. Mycobacterium tuberculosis and HIV co-infection: a public health problem that requires ongoing attention. Viruses. (2024) 16:1375. doi: 10.3390/v16091375
5. Scully, EP, and Bryson, BD. Unlocking the complexity of HIV and Mycobacterium tuberculosis coinfection. J Clin Invest. (2021) 131:e154407. doi: 10.1172/JCI154407
6. Tadesse, A, Seid, O, and Mekonnen, A. Modeling CD4 count and associated factors of HIV infected patients using linear mixed model: a case study at Debre Berhan referral hospital, Ethiopia. HIV AIDS (Auckl). (2021) 13:869–78. doi: 10.2147/HIV.S331682
7. Temesgen, Z, Gebre, M, Tadesse, A, Berhe, H, and Gebrehiwot, H. Predictors of tuberculosis and HIV co-infection among adult patients in Mekelle, Ethiopia: a case-control study. J Trop Med. (2018) 2018:7063896. doi: 10.1155/2018/7063896
8. Huang, Y, Li, C, Xu, L, and Zhang, X. Joint modeling of longitudinal and survival data: recent developments and applications. Stat Med. (2022) 41:3561–78. doi: 10.1002/sim.9434
9. Long, EF, and Mills, CW. Joint modeling of HIV disease progression and the effect of treatment: a multivariate approach. Stat Med. (2018) 37:1191–221. doi: 10.1002/sim.7577
10. Mehrotra, A, Kim, DD, Samorani, M, and Liao, JM. A multivariate probit model for analyzing binary health outcomes: an application to delays in cancer screening. Health Serv Res. (2021) 55:122–33. doi: 10.1111/1475-6773.13505
11. Gebre, T, and Hussen, A. Joint modeling of CD4 count and tuberculosis co-infection among HIV-positive patients in Ethiopia: a Bayesian approach. BMC Public Health. (2023) 23:456. doi: 10.1186/s12889-023-15287-0
12. Mchunu, N, Dlamini, T, and Manda, SOM. A Bayesian joint model to assess the association between CD4 count and risk of death among TB/HIV co-infected patients in South Africa. Int J Environ Res Public Health. (2022) 19:7895. doi: 10.3390/ijerph19137895
13. Mugwanya, KK, Palayew, A, Schaafsma, T, Irungu, EM, Bukusi, E, Mugo, N, et al. For the partners scale-up project. Patterns of PrEP continuation and coverage in the first year of use: a latent class analysis of a programmatic PrEP trial in Kenya. J Int AIDS Soc. (2023) 26:e26137. doi: 10.1002/jia2.26137
14. Kazibwe, A, Oryokot, B, Mugenyi, L, Kagimu, D, Oluka, AI, Kato, D, et al. Incidence of tuberculosis among PLHIV on antiretroviral therapy who initiated isoniazid preventive therapy: a multi-center retrospective cohort study. PLoS One. (2022) 17:e0266285. doi: 10.1371/journal.pone.0266285
15. Mollel, EW, Todd, J, Mahande, MJ, and Msuya, SE. The effect of tuberculosis infection on mortality of HIV-infected patients in northern Tanzania. Trop Med Health. (2020) 48:26. doi: 10.1186/s41182-020-00212-z
16. Gumede-Moyo, S, Filteau, S, Munthali, T, Todd, J, and Musonda, P. Implementation effectiveness of revised (post-2010) World Health Organization guidelines on prevention of mother-to-child transmission of HIV using routinely collected data in sub-Saharan Africa: a systematic literature review. Medicine (Baltimore). (2017) 96:e8055. doi: 10.1097/MD.0000000000008055
17. Abta, FA, Terefe, YA, and Teshome, DT. A simple SI-type model for HIV/AIDS with media and self-imposed psychological fear. Math Comput Simul. (2023) 208:351–65. doi: 10.1016/j.matcom.2023.01.020
18. Oladejo, NK, Fatunmbi, OO, and Okuonghae, D. Impact of nonlinear infection rate on HIV/AIDS considering prevalence dependent awareness. Heliyon. (2023) 9:e14172. doi: 10.1016/j.heliyon.2023.e14172
19. Oyelami, OA, and Bonyah, E. Vaccination impact on impending HIV-COVID-19 dual epidemic with autogenous behaviour modification: Hill-type functional response and premeditated optimization technique. Math Biosci Eng. (2024) 21:272–99. doi: 10.3934/mbe.2024013
20. Akinyemi OFBonyah, E. Impact of saturated treatments on HIV-TB dual epidemic as a consequence of COVID-19: optimal control with awareness and treatment. Math Comput Simul. (2024) 223:1–19. doi: 10.1016/j.matcom.2024.04.002
21. Chukwuma, EC, Okuonghae, D, and Omame, A. Exploring fractional dynamical probes in the context of gender-structured HIV-TB coinfection: a study of control strategies. Chaos, Solitons Fractals. (2023) 175:113929. doi: 10.1016/j.chaos.2023.113929
22. McElreath, R. Statistical rethinking: A Bayesian course with examples in R and Stan. 2nd ed. Boca Raton, FL, USA: CRC Press (2020).
23. Gelman, A, Carlin, JB, Stern, HS, Dunson, DB, Vehtari, A, and Rubin, DB. Bayesian Data Analysis. 3rd ed. Boca Raton, FL, USA: CRC Press (2013).
24. Gelman, A, Hill, J, and Yajima, M. Data analysis using regression and multilevel/hierarchical models. Cambridge, UK: Cambridge University Press (2008).
25. Tian, X, Wang, C, Hao, Z, Chen, J, and Wu, N. Global, regional, and national burden of HIV and tuberculosis and predictions by Bayesian age-period-cohort analysis: a systematic analysis for the global burden of disease study 2021. Front Reprod Health. (2024) 6:1475498. doi: 10.3389/frph.2024.1475498
26. Horton, KC, Mac Pherson, P, Houben, RMGJ, White, RG, and Corbett, EL. Sex differences in tuberculosis burden and notifications in low-and middle-income countries: a systematic review and meta-analysis. PLoS Med. (2016) 13:e1002119. doi: 10.1371/journal.pmed.1002119
27. Medina-Marino, A, de Vos, L, and Daniels, J. Social isolation, social exclusion, and access to mental and tangible resources: mapping the gendered impact of tuberculosis-related stigma among men and women living with tuberculosis in Eastern Cape Province, South Africa. BMC Glob. Public Health. (2025) 3:50. doi: 10.1186/s44263-025-00166-6
28. Wang, W, Liu, A, Liu, X, You, N, Wang, Z, Chen, C, et al. Mycobacterium tuberculosis infection in school contacts of tuberculosis cases: a systematic review and meta-analysis. Am J Trop Med Hyg. (2024) 110:1253–60. doi: 10.4269/ajtmh.23-0038
29. Sibindi, AB, and Ngcobo, L. Migrant remittance patterns in South Africa: a micro-level analysis. J Econ Behav Stud. (2018) 10:109–17. doi: 10.22610/jebs.v10i4(J).2412
30. Adebisi, YA, Amoo, EO, Folorunso, O, Nwaneri, JO, Osungbade, KO, Opiah, MM, et al. Risky sexual behaviour among HIV-infected adults in sub-Saharan Africa: a systematic review and meta-analysis. PLoS One. (2021) 16:e0255676. doi: 10.1371/journal.pone.0255676
31. Vignesh, R, Swathirajan, CR, Solomon, SS, Shankar, EM, and Murugavel, KG. Risk factors and frequency of tuberculosis-associated immune reconstitution inflammatory syndrome among HIV/TB co-infected patients in southern India. Indian J Med Microbiol. (2023) 35:279–81. doi: 10.4103/ijmm.IJMM_16_163
32. Cassim, N, Kimmie, Z, and Govender, N. Socioeconomic status and CD4 count trends in South Africa: a longitudinal study from 2013 to 2023. BMC Public Health. (2024) 24:148. doi: 10.1186/s12889-024-18038-9
33. Buziashvili, M, Djibuti, M, Tukvadze, N, DeHovitz, J, and Baliashvili, D. Incidence rate and risk factors for developing active tuberculosis among people living with HIV in Georgia 2019-2020 cohort. Open forum. Infect Dis. (2024) 11:ofae466. doi: 10.1093/ofid/ofae466
34. Lu, P, Lian, Y, Li, Z, Zhang, J, Chen, W, Zhou, H, et al. Effect of CD4 count on Mycobacterium tuberculosis infection rates in people living with HIV: a comparative study in prison and community. Sci Rep. (2024) 14:26386. doi: 10.1038/s41598-024-77250-8
35. Kouamou, V, Gundidza, P, Ndhlovu, CE, and Makadzange, AT. Factors associated with CD4+ cell count recovery among males and females with advanced HIV disease. AIDS. (2023) 37:2311–8. doi: 10.1097/QAD.0000000000003640
36. Otiende, M, Nyambura, M, Kariuki, D, Kamau, M, and Muchiri, J. Spatial analysis of tuberculosis and HIV co-infection in Nairobi, Kenya: a Bayesian hierarchical model approach. BMC Public Health. (2020) 20:1203. doi: 10.1186/s12889-020-09298-6
37. Bayabil, S, and Seyoum, A. Joint modeling in detecting predictors of CD4+ cell count and status of tuberculosis among people living with HIV/AIDS under HAART at Felege Hiwot teaching and specialized hospital, north-West Ethiopia. HIV AIDS (Auckl). (2021) 13:527–37. doi: 10.2147/HIV.S330914
38. Pilangorgi, SS, Khodakarim, S, Shayan, Z, and Nejat, M. Evaluation of factors related to longitudinal CD4 count and the risk of death among HIV-infected patients using Bayesian joint models. BMC Public Health. (2025) 25:979. doi: 10.1186/s12889-025-22096-6
39. Chattopadhyay, S. (2024). Factor copula models for non-Gaussian longitudinal data: regression for mixed continuous and discrete outcomes with latent variables. arXiv doi: 10.48550/arXiv.2402.00668 [preprint].
Keywords: HIV, TB diagnosis, CD4 cell count, Bayesian joint multivariate model, average marginal effect, latent variable
Citation: Chireshe E, Chifurira R, Batidzirai JM, Chinhamu K and Kharsany ABM (2025) Application of a joint multivariate probit model for mixed outcomes of CD4 cell count and tuberculosis using a Bayesian latent variable approach in KwaZulu-Natal. Front. Appl. Math. Stat. 11:1643745. doi: 10.3389/fams.2025.1643745
Edited by:
Artur Lemonte, Federal University of Rio Grande do Norte, BrazilCopyright © 2025 Chireshe, Chifurira, Batidzirai, Chinhamu and Kharsany. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Exaverio Chireshe, ZWtpY2hpcmVzaGVAZ21haWwuY29t
 Exaverio Chireshe
Exaverio Chireshe Retius Chifurira
Retius Chifurira Jesca Mercy Batidzirai
Jesca Mercy Batidzirai Knowledge Chinhamu
Knowledge Chinhamu Ayesha B. M. Kharsany
Ayesha B. M. Kharsany