Introduction
The intervertebral disc (IVD), serving as a critical load-bearing structure connecting vertebral bodies, maintains spinal mechanical stability and function through the integrated health of its nucleus pulposus, annulus fibrosus, and cartilaginous endplates (Knezevic et al., 2021). However, intervertebral disc degeneration (IVDD) has emerged as a global health challenge, with its prevalence increasing markedly with age. As the leading cause of chronic low back pain, it imposes a substantial healthcare burden on society. Current clinical strategies for managing IVDD primarily focus on symptomatic relief—including physical therapy, pain management, and spinal fusion surgeries aimed at mechanical stabilization—none of which fundamentally reverse the degenerative process or achieve functional tissue regeneration (Copeland, 2007; Binch et al., 2021). With the expanding integration of artificial intelligence (AI) into medical science, particularly its growing capacity to uncover profound patterns within complex biomedical data, a new paradigm has emerged to address this challenge (Zhang et al., 2025). The application of AI in elucidating the mechanisms of IVDD and designing corresponding repair materials offers novel perspectives for regenerative rehabilitation of degenerative discs.
AI for IVD repair mechanisms discovery
Artificial intelligence and machine learning are rapidly transforming intervertebral-disc research by integrating multi-omics data, single-cell profiles, and in-silico drug screening to decode the molecular logic of disc degeneration and repair. These algorithms outperform conventional statistics in ranking non-coding RNAs, programmed-cell-death regulators, or immune–matrix crosstalk genes, providing clinicians with quantitative biomarkers and druggable targets that can be validated in vitro and in rodent models (Liao et al., 2025).
The application of AI now extends to automating the diagnosis and grading of disc degeneration from lumbar MR images, where deep learning models demonstrate high precision in identifying pathological changes. Furthermore, single-cell RNA sequencing (scRNA-seq) has become a pivotal tool, revealing transcriptional shifts in disc resident and infiltrating cell populations following injury, and uncovering novel cellular targets for repair strategies. For instance, specific mesenchymal stem cell (MSC) populations were identified whose differentiation shifts with injury, offering potential for regenerative therapies (Lin et al., 2024; Stirnimann et al., 2025).
Li et al. introduced an RNA-seq-driven competing-endogenous-RNA strategy that couples differential lncRNAs, miRNAs and mRNAs in degenerated versus traumatic discs; they experimentally confirmed the XIST–miR-4775-PLA2G7 and XIST–miR-424-AMOT/TGFBR3 axes as pro-inflammatory circuits that disrupt extracellular-matrix homeostasis (Li et al., 2021). Zhang et al. presented a bioinformatics-plus-machine-learning pipeline that merges four GEO microarray sets, performs WGCNA–LASSO screening and constructs a protein–protein interaction network, identifying IL1R1 and TCF7L2 as central transcriptional hubs whose elevated expression distinguishes advanced Pfirrmann-grade discs with an AUC ≈ 0.7 (Zhang et al., 2024). Lv et al. delineated a comprehensive programmed-cell-death atlas of IVDD by integrating bulk and single-cell transcriptomes; machine-learning models selected PDCD6 and UBE2K as apoptosis-specific drivers, and in vivo administration of the repositioned drug Glibenclamide attenuated caspase-3 activity, preserved disc height and validated the predictive value of their ridge-regression score (Lv et al., 2025).
AI for IVD repair biomaterials design
AI-guided single-cell multi-omics is advancing the rational design of biomaterials for intervertebral disc repair. By mapping cellular heterogeneity, cell death pathways, and fibrotic signatures at single-cell resolution, machine learning models enable the identification of therapeutic targets that are inaccessible via bulk profiling. These insights inform the engineering of mitochondria-targeting carriers, siRNA nanomotors, and immunomodulatory scaffolds—each mathematically optimized to maximize on-site efficacy while minimizing off-target effects. Convolutional neural networks cluster cell populations into distinct transcriptional states, whereas trajectory inference and random forest models predict lineage plasticity and gene significance. Meanwhile, reinforcement learning simulates the spatiotemporal behavior of biomaterials within a reconstructed disc microenvironment, iteratively refining their physical and biochemical properties to achieve an optimal therapeutic profile. This AI-driven approach transforms traditional “make-and-test” cycles into an in-silico “predict-design-validate” workflow, accelerating development, reducing animal use, and enabling patient-specific implants (Gao et al., 2022; Hu et al., 2023).
Tu et al. presented an atlas of human nucleus pulposus at single-cell resolution that resolves six NPC sub-states, fibro-progenitor trajectories and CD90+ progenitors with tri-lineage potency; bioinformatic deconvolution of immune infiltration revealed G-MDSC-mediated immunosuppression, inspiring a progenitor-enriched hydrogel that arrests fibrosis in rat IVDD (Tu et al., 2022). Zhou et al. introduced Motor@TA-siRNA, an H2O2-propelled nanomotor whose trajectory, siRNA load and tannic-acid shield were optimized by single-cell evidence of STING-driven pyroptosis; the carrier self-enriches within fibrotic NP, silences STING and simultaneously scavenges ROS, doubling disc height retention relative to passive vectors (Zhou et al., 2025) (Figure 1). Yang et al. described a deep-learning-assisted mitochondrial therapy in which scRNA-seq pinpointed OXPHOS-deficient fibrochondrocytes as the prime driver of fibrosis; exogenous mitochondria engineered with a mitochondria-targeting macromolecule PSP were shown to restore respiration, block mtDNA leakage and disrupt the SPARC-STING axis, achieving near-native NP architecture in a rat puncture model (Yang et al., 2025).
FIGURE 1
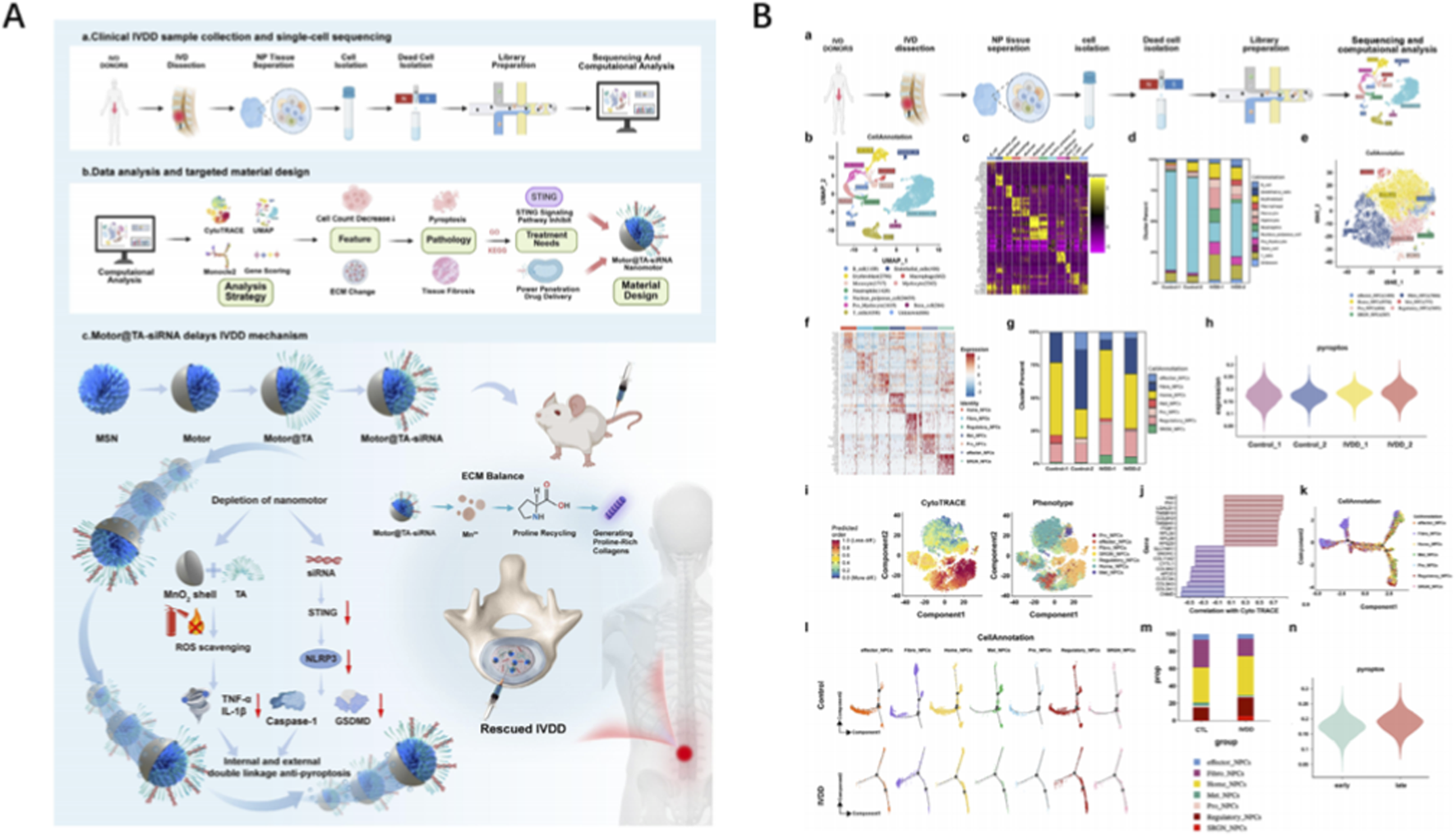
Application of AI in IVD repair biomaterials design (A) Schematic illustration of a single-cell-inspired self-enrichment pneumatic nanocarrier that inhibits pyroptosis to delay IVDD through intracellular and extracellular synergisms. (B) Single-cell analysis of nucleus pulposus tissues. Reproduced with permission from Copyright 2025 Wiley.
Conclusion
In summary, AI is revolutionizing the study and treatment of intervertebral disc degeneration by enabling a shift from descriptive observation to predictive, mechanism-based intervention. It excels in decoding complex molecular mechanisms underlying IVDD, identifying critical biomarkers and cellular pathways through integrated multi-omics and single-cell analyses. In biomaterial design, AI accelerates the development of intelligent implants—such as targeted nanocarriers and mitochondrial therapies—by optimizing material properties and predicting therapeutic behavior through in silico modeling, thereby streamlining the traditional development pipeline.
Despite its promise, AI integration into disc research faces key challenges. The “black-box” nature of complex models obscures prediction interpretability, potentially limiting clinical adoption. Moreover, algorithm performance depends inherently on the quality, volume and diversity of training data; biases or noise in datasets compromise generalizability. Finally, translating in silico predictions and optimized biomaterial designs into safe, effective clinical applications remains a major hurdle requiring rigorous validation.
In our view, the full integration of AI into future intervertebral disc repair strategies is not only highly valuable but also an indispensable component. Conventional approaches struggle to address the complex etiological factors and significant inter-individual variability of IVDD. In contrast, AI provides a robust, data-driven framework for developing personalized therapeutic regimens grounded in solid mechanistic foundations. We should strive to build multimodal AI systems that integrate biomechanical, imaging, and clinical data to simulate the processes of disc degeneration and repair within a holistic digital twin framework. On the basis of interdisciplinary collaboration, we will advance these technological breakthroughs from theoretical concepts to clinical practice.
Statements
Author contributions
YS: Writing – review and editing, Writing – original draft. YW: Visualization, Writing – original draft, Formal Analysis, Project administration, Funding acquisition, Methodology, Data curation, Software, Investigation, Supervision, Conceptualization, Validation, Writing – review and editing, Resources.
Funding
The author(s) declared that financial support was not received for this work and/or its publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declared that generative AI was not used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Binch A. L. A. Fitzgerald J. C. Growney E. A. Barry F. (2021). Cell-based strategies for IVD repair: clinical progress and translational obstacles. Nat. Rev. Rheumatol.17 (3), 158–175. 10.1038/s41584-020-00568-w
2
Copeland B. (2007). Surgical versus nonsurgical treatment for back pain. N. Engl. J. Med.357 (12), 1255–1256.
3
Gao X. D. Zhang X. B. Zhang R. H. Yu D. C. Chen X. Y. Hu Y. C. et al (2022). Aggressive strategies for regenerating intervertebral discs: stimulus-responsive composite hydrogels from single to multiscale delivery systems. J. Mater Chem. B10 (30), 5696–5722. 10.1039/d2tb01066f
4
Hu X. Wang Z. Zhang H. Cui P. Li Y. Chen X. et al (2023). Single-cell sequencing: new insights for intervertebral disc degeneration. Biomed. Pharmacother.165, 115224. 10.1016/j.biopha.2023.115224
5
Knezevic N. N. Candido K. D. Vlaeyen J. W. S. Van Zundert J. Cohen S. P. (2021). Low back pain. Lancet398 (10294), 78–92. 10.1016/S0140-6736(21)00733-9
6
Li Z. Sun Y. He M. Liu J. (2021). Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing. Bioengineered12 (1), 1026–1039. 10.1080/21655979.2021.1899533
7
Liao Z. Li M. Chen Z. Yao Z. Wu Y. Tan Z. et al (2025). An integrated multi-algorithm analysis to decipher senescence and metabolic characteristics in nucleus pulposus cells during disc degeneration. Geroscience. 10.1007/s11357-025-01962-6
8
Lin P. Gan Y. B. He J. Lin S. E. Xu J. K. Chang L. et al (2024). Advancing skeletal health and disease research with single-cell RNA sequencing. Mil. Med. Res.11 (1), 33. 10.1186/s40779-024-00538-3
9
Lv Y. Du J. Xiong H. Feng L. Zhang D. Zhou H. et al (2025). Machine learning-based analysis of programmed cell death types and key genes in intervertebral disc degeneration. Apoptosis30 (1-2), 250–266. 10.1007/s10495-024-02047-z
10
Stirnimann A. Schlagenhof L. Gantenbein B. Ille F. (2025). Advancing intervertebral disc biology via omics: implications for nucleus pulposus progenitor cell-based regeneration. JOR Spine8 (4), e70130. 10.1002/jsp2.70130
11
Tu J. Li W. Yang S. Yang P. Yan Q. Wang S. et al (2022). Single-cell transcriptome profiling reveals multicellular ecosystem of nucleus pulposus during degeneration progression. Adv. Sci. (Weinh)9 (3), e2103631. 10.1002/advs.202103631
12
Yang G. Dong C. Wu Z. Wu P. Yang C. Li L. et al (2025). Single-cell RNA sequencing-guided engineering of mitochondrial therapies for intervertebral disc degeneration by regulating mtDNA/SPARC-STING signaling. Bioact. Mater48, 564–582. 10.1016/j.bioactmat.2025.02.036
13
Zhang H. Shi S. Huang X. Gong C. Zhang Z. Zhao Z. et al (2024). Identification of core genes in intervertebral disc degeneration using bioinformatics and machine learning algorithms. Front. Immunol.15, 1401957. 10.3389/fimmu.2024.1401957
14
Zhang J. Li C. Liu H. Wang Q. Xu A. Y. Shah K. et al (2025). Hydrogel delivery systems in intervertebral disc degeneration: current status and future perspectives. J. Control Release386, 114066. 10.1016/j.jconrel.2025.114066
15
Zhou H. Ning H. Liu Q. Tan Q. Zeng D. Feng X. et al (2025). A single-cell-inspired self-enrichment therapeutic strategy delays intervertebral disc degeneration by inhibiting pyroptosis. Adv. Mater, e16405. 10.1002/adma.202516405
Summary
Keywords
artificail intelligence, biomaterials, hydrogel, intervertbral disc, single-cell sequencing
Citation
Shi Y and Wang Y (2025) The AI-driven blueprint: decoding intervertebral disc repair mechanisms for intelligent biomaterial design. Front. Cell Dev. Biol. 13:1751851. doi: 10.3389/fcell.2025.1751851
Received
22 November 2025
Revised
28 November 2025
Accepted
29 November 2025
Published
11 December 2025
Volume
13 - 2025
Edited by
Zhen Yang, Peking University People’s Hospital, China
Reviewed by
Zhang Minglei, Jilin University, China
Updates
Copyright
© 2025 Shi and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifan Wang, wyfspine@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.