- Department of Microbiology, School of Medicine, Kyungpook National University, Daegu, South Korea
Acinetobacter baumannii is an important nosocomial pathogen that can survive in different environmental conditions and poses a severe threat to public health due to its multidrug resistance properties. Research on transcriptional regulators, which play an essential role in adjusting to new environments, could provide new insights into A. baumannii pathogenesis. LysR-type transcriptional regulators (LTTRs) are structurally conserved among bacterial species and regulate virulence in many pathogens. We identified a novel LTTR, designated as LeuO encoded in the A. baumannii genome. After construction of LeuO mutant strain, transcriptome analysis showed that LeuO regulates the expression of 194 upregulated genes and 108 downregulated genes responsible for various functions and our qPCR validation of several differentially expressed genes support transcriptome data. Our results demonstrated that disruption of LeuO led to increased biofilm formation and increased pathogenicity in an animal model. However, the adherence and surface motility of the LeuO mutant were reduced compared with those of the wild-type strain. We observed some mutations on amino acids sequence of LeuO in clinical isolates. These mutations in the A. baumannii biofilm regulator LeuO may cause hyper-biofilm in the tested clinical isolates. This study is the first to demonstrate the association between the LTTR member LeuO and virulence traits of A. baumannii.
Introduction
Acinetobacter baumannii is a member of ESKAPE pathogens that primarily affect patients with compromised defense in hospitals (Rice, 2008). Hospital-acquired A. baumannii infection can cause bacteremia, urinary tract infection, traumatic skin, and pneumonia (McConnell et al., 2013). Because of the nature of multidrug resistance (MDR), the World Health Organization (WHO) has listed A. baumannii as the “top priority” pathogen that requires new therapeutic options (WHO, 2017). A. baumanni can survive and persist in harsh environmental conditions in hospital settings, an ability that helps prolong outbreaks of nosocomial infection (Jawad et al., 1998). Numerous virulence factors contribute to successful A. baumannii infection, including biofilm formation on biological and innate surfaces (Longo et al., 2014), adherence to and invasion of host cells (Lee et al., 2006), efflux pumps that extrude different molecules and antibiotics (Kumar and Schweizer, 2005), outer membrane protein A (OmpA) that mediates interaction with epithelial cells (Lee et al., 2010), iron acquisition system (Gaddy et al., 2012), and capsular polysaccharide (Russo et al., 2010). A. baumannii pili are a key factor for biofilm formation, and csuA/BABCDE chaperone-usher secretion system-mediated pili help planktonic bacteria to adhere onto abiotic surfaces for biofilm formation (Tomaras et al., 2003). Besides abiotic surfaces, A. baumannii can attach onto biotic surfaces such as respiratory epithelial cells, which is another important virulence factor for infection (Lee et al., 2008). Quorum sensing in A. baumannii is another important pathway by which the pathogen senses extracellular signals and regulates biofilm formation and virulence (Bhargava et al., 2015). However, understanding the molecular mechanisms of virulence factors would help develop novel strategies to prevent multidrug-resistant A. baumannii infection.
Transcriptional regulatory proteins help prokaryotes to communicate between environmental conditions and DNA transcription to survive in different habitats (Santos et al., 2009). Bacterial genomes encode several transcriptional regulatory proteins required for adaptive cellular responses belonging to different families, such as ArsR, AsnC, DeoR, GntR, IclR, LacI, LuxR, XylS, MarR, MerR, NtrC, TetR, YedF, and YhdG. Among these, the family of LysR-type transcriptional regulators (LTTRs) is the largest and resemble approximately 16% of the overall transcriptional factors in bacteria (Srinivasan et al., 2013). A typical LTTR comprises an N-terminal DNA-binding helix-turn-helix (HTH) domain and a C-terminal coinducer-binding domain (also known as a regulatory domain). LTTRs can function as either an activator or repressor of single or operonic gene expression, which is why they have been recently termed as global transcriptional regulators (Schell, 1993). LTTRs are associated with the control of various cellular processes. For instance, VirR in Rhodococcus equi and MvfR and PA2206 in Pseudomonas aeruginosa are involved in virulence and quorum sensing (Russell et al., 2004; Deziel et al., 2005). Moreover, the proteins CidR in Staphylococcus aureus and OxyR in Klebsiella pneumoniae are involved in antibiotic resistance (Rice et al., 2005; Yang et al., 2005). In Yersinia pseudotuberculosis, RovM controls cell invasion, motility, and virulence (Heroven and Dersch, 2006).
LeuO is a member of the LysR family of transcriptional regulators, and members of this family have been investigated in several bacteria, including Escherichia coli, Salmonella enterica, Vibrio cholerae, Yersinia enterocolitica, and Enterobacter cloacae (Guadarrama et al., 2014). LeuO controls several biological functions such as biofilm formation and virulence in V. cholerae and E. coli, regulates the expression of OmpS1 and OmpS2, and downregulates the expression of OmpX, which alter the transport of hydrophobic compounds and virulence in S. enterica (Moorthy and Watnick, 2005; Hernández-Lucas et al., 2008; Shimada et al., 2011). LeuO regulates a wide variety of genes that are involved in amino acid biosynthesis, nitrogen fixation, quorum sensing, and virulence (Schell, 1993). It also regulates bile tolerance, antibiotic resistance, and promoter binding in V. cholera (Bina et al., 2016). However, LeuO has not yet been characterized in A. baumannii and its functions also remain unclear.
In this study, to further understand the role of LeuO in A. baumannii, we generated a knockout mutant of LeuO and conducted transcriptome analysis to compare the differentially expressed genes between ΔLeuO and wild-type strains. Transcriptome analysis showed that several biological and metabolic pathways are altered after the deletion of LeuO. Our experiments on biofilm formation, surface motility and adherence to epithelial cell suggested that some genes related to these features are directly or indirectly regulated by LeuO. Overall, our study results provide novel understanding about the regulatory role of LeuO and the pathogenesis of A. baumannii. This identification of the role of transcriptional regulators may help in the development of novel therapeutics against MDR A. baumannii strains.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
A. baumannii ΔLeuO and complementation strains were constructed using the homologous recombination method as described in the supplementary data. A. baumannii strains were grown in Luria–Bertani (LB) media at 37°C or 30°C, and agar was added at the indicated concentrations obtained from Difco or Eiken (Eiken Chemical, Tokyo, Japan). Chloramphenicol (20 µg/mL), kanamycin (50 µg/mL), and ampicillin (100 µg/mL) were added to LB broth or LB agar plates to maintain the plasmids in E. coli. The bacterial strains, plasmids, and primers used in this study are listed in Supplementary Tables S3 and S4.
Isolation of Bacterial mRNA and RNA Sequencing
Overnight bacterial cultures of both A. baumannii ATCC 17978 wild-type and ΔLeuO strains were subcultured in 10 mL of LB by 1:100 dilution and grown at 37°C until OD600 reached 1.00 under shaking condition. Total RNA was extracted using Qiagen RNeasy Mini kits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The total RNA concentration was measured using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Two biological replicates of each were sent to Macrogen Inc. (Seoul, Republic of Korea), where mRNA quality control (QC), cDNA library preparation, library QC, template preparation, template QC, and RNA sequencing were performed on the Illumina NovoSeq 6000 platform. RNA-sequencing reads were aligned to the A. baumanni strain ATCC 17978 (GCF_ 0000 15 425.1 _ A S M1542v1). Bowtie 1.1.2 (http://bowtie-bio.sourceforge.net/index.shtml) and HTSeq version 0.10.0 software (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) were used for analyzing the sequencing data. Any of the sequencing reads with a fold change of <2 and a p value of >0.05 were eliminated.
Quantitative Real-Time PCR for RNA-seq Data Validation
Total RNA from A. baumannii strains was isolated as described earlier, and cDNA was synthesized using the M-MLV cDNA Synthesis kit (Enzynomics). Real-time PCR amplification of cDNA was performed using the ABI Step One Plus Real-Time System (Applied Biosystems), and TOPreal™ q-PCR 2X PreMIX (SYBR Green with high ROX, Enzynomics) was used. The internal forward and reverse primers used in this study for each gene are listed in Supplementary Table S4. The expression level was standardized relative to the transcription level of 16S rRNA expression level. The fold change was determined using the ΔΔCt method. Experiments were performed in three independent replicates.
Biofilm Formation Assay
Biofilm formation by A. baumannii strains was evaluated according to the method described by (Stepanovic et al. (2000) with some modifications using a crystal violet staining assay. Briefly, bacterial strains were cultured overnight, resuspended in fresh LB broth without salt, and adjusted to a turbidity of 1.0 at 600 nm using a spectrophotometer. After dilution, the bacterial suspensions were aliquoted as 2 ml each into 5-mL polystyrene tubes and incubated for 24 h at 30°C under static conditions in a dark room. After the removal of supernatants, the tubes were washed twice with 2 ml distilled water to remove planktonic or loosely adherent cells. After air-drying the tubes for 10 min, 2 mL of crystal violet (0.1% v/v) was added to each tube to stain the inner wall with biofilm for 15 min. The stained biofilms were solubilized with 2 mL of 95% ethanol for 10 min, and 200 μL of each sample was transferred to a 96-well plate to measure turbidity at 570 nm using a microplate reader (Molecular Devices, Sunnyvale, USA.). To compensate for growth differences, turbidity was also measured at 600 nm before staining the biofilm. Three independent experiments were performed, each in triplicate.
Biofilm formation assays using A. baumannii 17978 wild-type strain, ΔLeuO strain, and clinical isolates of various sequence types (ST-208, ST-229, ST-357, ST-451, ST-552, and ST-784) were also performed as described earlier with two independent experiments.
Pellicle Formation Assay
Pellicle formation assay was performed based on a method described by Martí et al. (2011). Bacterial strains were cultured overnight in LB broth without salt and diluted at 1:40 with the same media. The assay was performed in 5-ml polypropylene tubes, and 2 ml of bacterial suspensions was aliquoted into each tube with incubation at 30°C for 48 h without shaking. The pellicle film was isolated from the tube by adding 1 ml methanol. The pellicle biomass was measured (optical density at 600 nm [OD600]) after resuspending the pellets in 1 ml PBS. Experiments were performed in triplicates.
Surface Motility Assay
Motility assays were performed as described previously (Clemmer et al., 2011) with some modifications. Modified LB agar containing Eiken agar 3 g/L, tryptone 10 g/L, and yeast extract 5 g/L was autoclaved and cooled at 60°C. Modified LB medium was poured into Petri dishes, dried for 8 h, and used on the same day of preparation. For testing motility, A. baumannii strains were grown overnight and adjusted to the same optical density (OD600 = 1.0) by adding modified LB broth, and then 2 µL of culture was inoculated onto the center of agar plates. The plates were incubated at 37°C for 10 h, after which the diameter of motility zones was measured. The plates were photographed using a digital imaging system. Assays were performed in triplicate with three biological replicates each time.
Adherence Assay
Bacterial adherence to A549 cells was evaluated as described previously (Lee et al., 2006). Briefly, A549 human alveolar epithelial cells were grown in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 100 U/ml penicillin G, and 50 μg/ml streptomycin. Cultures with 80%–90% confluency were trypsinized and seeded at a density of 2 × 105 cells/ml in 6-well culture dishes to obtain a monolayer. After 24-h incubation, the cells were washed twice with PBS and incubated with RPMI 1640 medium without antibiotics. A. baumannii strains were grown to reach an OD A600 of 1.0 and suspended in the same media. Then, 2 × 107 CFU/ml of bacteria were added into each well to obtain a multiplicity of infection (MOI) of 1:100 and incubated for 2 h at 37°C. To determine bacterial adhesion, cells were washed five times with PBS and lysed with 0.1% Triton X-100 at 37°C for 20 min. Colony-forming units were counted after 10-fold serial dilution of lysate samples to determine the number of bacteria that had attached to or invaded the A549 cells. Adherence assays were performed in three independent replicates.
In Vivo Virulence Assay
All animal infection experimental procedures were approved by the Animal Care Committee of Kyungpook National University, South Korea (approval number: KNU-2019-178). Briefly, 8-week-old female BALB/c mice were maintained under conventional conditions at five mice per case and allowed access to food and water throughout the experiment. To promote infection, neutropenic mice were induced by intraperitoneal (IP) injection of cyclophosphamide (150 mg/kg) in PBS before bacterial infection (−4 and −1 day). A. baumannii 17978 WT, ΔLeuO, CP, and A. baumannii 1656-2 WT strains were grown overnight in LB broth at 37°C, washed with PBS, and the concentration was set to 2 × 109 CFU/ml. Mice were injected intraperitoneally with 100 µl PBS (control) and A. baumannii strains (1 × 108 CFU/ml) per mice (n = 5 per group). Animals were monitored every 12 h over a period of 4 days. The number of live and dead animals was input into GraphPad Prism, and survival curve was generated. Statistical analysis was conducted using the Kaplan–Meier test in GraphPad Prism.
Results
A. baumannii A1S_1874 Is the LTTR LeuO
LTTRs are organized as an N-terminal HTH DNA-binding domain and a C-terminal effector-binding domain (EBD) connected by a long linker helix (Muraoka et al., 2003). A. baumannii A1S_1874 constitutes 306 amino acid residues, and an analysis of the amino acid sequence revealed a DNA-binding domain (HTH, 25–79) and a substrate-binding domain (112–304), indicating that A1S_1874 is a member of the LTTR family (Figure 1A). Blast search of the A1S_1874 amino acid sequence (306 aa) was performed to identify the homology sequence of A1S_1874 among other Acinetobacter strains and other bacterial species. The amino acid sequence was conserved among other sequenced A. baumannii strains. Predicted 3D structure of A1S_1874 elucidated using the Phyre2 and PymoL software program clearly displayed two distinct domains (Figure 1B), which support that A1S_1874 is a putative LTTR. Finally, we compared the amino acid sequence of A. baumannii A1S_1874 and other gram-negative bacterial LeuO whose functions have already been characterized. The amino acid sequence of A1S_1874 exhibited 25% sequence homology with E. coli LeuO, 24% sequence homology with S. enterica LeuO, and 25% sequence homology with V. navarrensis LeuO. A. baumannii A1S_1874 demonstrated high sequence homology at the N-terminal region with other bacterial LeuO protein compared with that at the C-terminal region (Figure 1C). LTTRs exhibit low sequence identity among the family members, possibly due to distinct effector recognition. However, the N-terminal region displayed more sequence conservation than the C-terminal region (Schell, 1993). Considering these findings, we predict that A1S_1874 is LeuO, an LTTR in A. baumanni, whose functions must be explored.
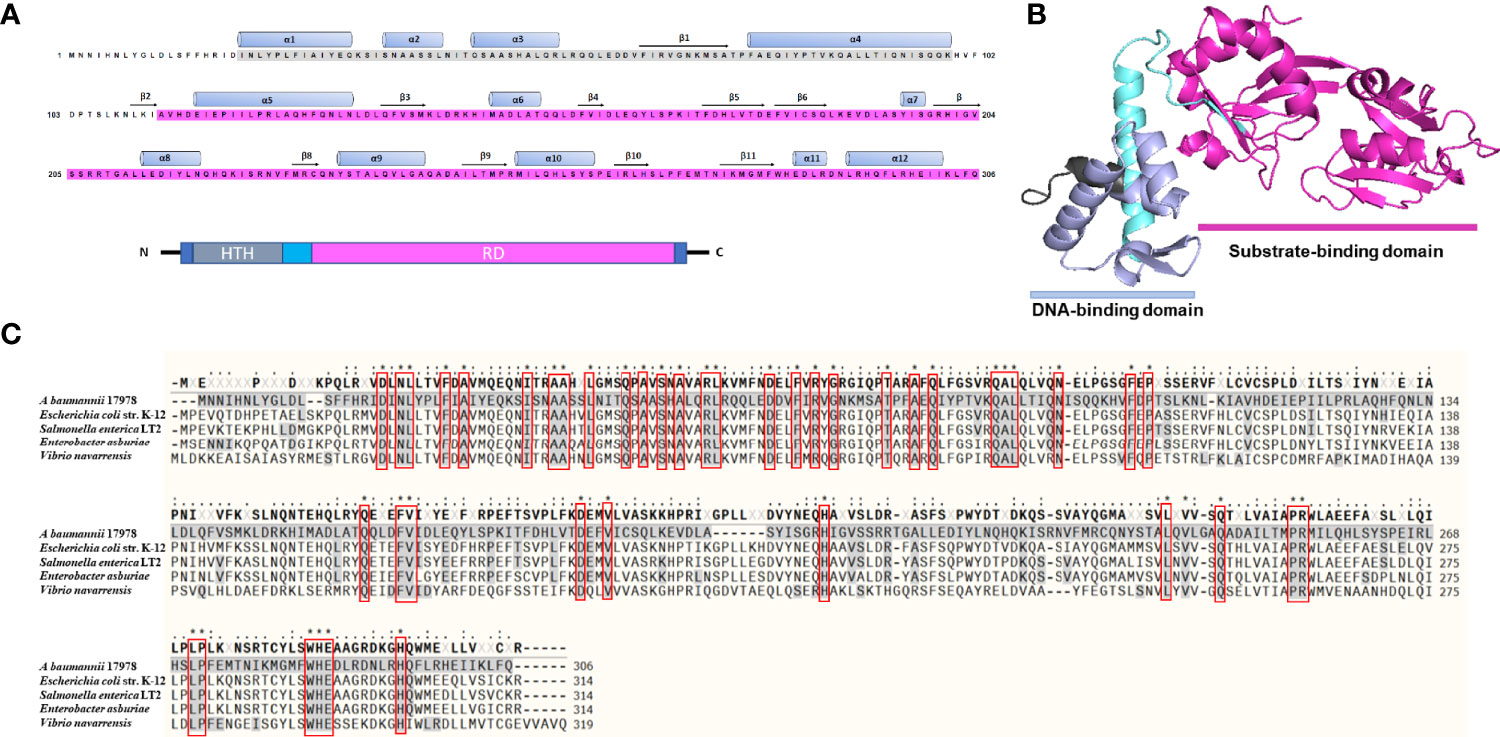
Figure 1 Predicted three-dimensional structure and sequence alignment of LeuO. (A) Deduced amino acid sequence of A1S_1874 where the ash box indicates the helix-turn-helix DNA-binding domain (25–79) and the purple box indicates the LysR family substrate-binding domain (112–304). (B) Predicted 3D structure of A1S_1874 obtained using protein modeling server Phyre2 and PyMoL program. (C) Multiple sequence alignment using COBALT: Multiple Alignment Tool of A. baumannii A1S_1874 with previously characterized LTTR member LeuO from E. coli strain K-12, Salmonella enterica LT2, Enterobacter asburiae, and Vibrio navarrensis. “*”, “.”, “:” indicate most conserved residues and semi-conserved sequence, respectively.
Transcriptome Analysis of LeuO Mutant Strain
To characterize LeuO regulation in A. baumannii ATCC 17978, we conducted transcriptome profiling of LeuO mutant strain and compared with wild-type strain to obtain insights into the global transcriptomic changes caused by LeuO deletion. The LeuO mutant strain was constructed as described by Jung et al. (2020) (Supplementary data section), and it was confirmed using PCR analysis (Supplementary Figure S1). For RNA sequencing, we extracted RNA from cells growing up to an OD600 = 1. Differentially expressed genes were selected when the fold change in expression was ≥2 with p values <0.05 (Figure 2A). We found that a total of 302 genes were differentially expressed, among which 194 were upregulated and 108 were downregulated compared with those of A. baumannii WT and LeuO mutant. The complete list of differentially expressed genes is shown in Supplementary Table S1. An acyl carrier protein (locus tag A1S_0114) was the highest upregulated differentially expressed gene (fold change 201.60), and a hypothetical protein (locus tag A1S_0645) was the highest downregulated gene (fold change −16.02). In A. baumannii ATCC 17978, the A1S_0112-A1S_0119 cluster is the polycistronic operon responsible for biofilm formation and virulence. Our transcriptome data indicated that the A1S_0112-A1S_0119 cluster was highly upregulated (9.5- to 201.60-fold) in the absence of LeuO. Csu operon (CsuA/BABCDE) genes (A1S_2213-2218) were also upregulated in the LeuO mutant strain (10.40- to 64.16-fold). Proteins encoded by the csu operon have been identified in pellicle and biofilm formation. Quorum sensing–related genes (A1S_0109-A1S_0111) were also upregulated after LeuO mutation. Several genes related to iron ion binding and transport such as A1S_0242, A1S_0243, A1S_0980, A1S_0981, A1S_1063, and A1S_3369 were also upregulated after LeuO mutation. Some efflux pump–related genes such as the MFS family transporters A1S_1440 and A1S_1772 and the RND family transporters A1S_1649 and A1S_1773 were also upregulated. Transcriptional regulators of different families such as the GntR family (A1S_0072), TetR family (A1S_0548), and AsnC family (A1S_1090) and another transcriptional regulator (A1S_1256) were also overexpressed. Interestingly, the gene cluster A1S_0640-A1S_0647 (putative hypothetical protein) was highly downregulated (−3.53- to −16.02-fold) in the LeuO mutant strain. VGR-like proteins are putative T6SS effectors in A. baumannii that regulate cell invasion. The expression of several VGR-like protein genes (A1S_1288, A1S_1289, and A1S_3364) was downregulated. Iron-storing bacterioferritin (A1S_0800 and A1S_3175) and several efflux pump transporters, especially RND efflux, were downregulated. Numerous genes involved in metabolism such as dehydrogenase, hydrolase, and hydratase were also downregulated. Several genes were classified as hypothetical proteins whose functions are unknown in A. baumannii.
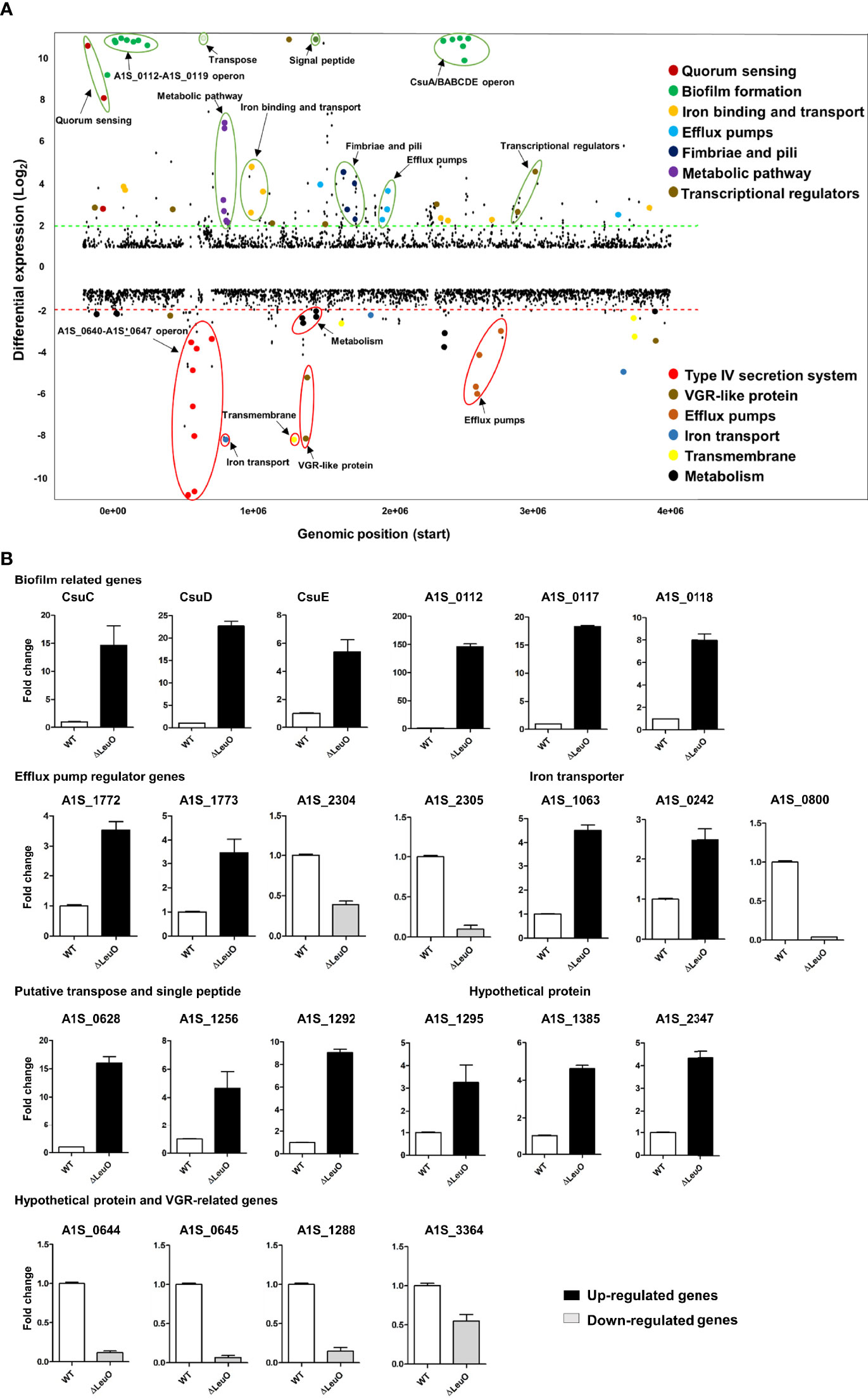
Figure 2 Overview of transcriptional differences between ΔLeuO and ATCC 17978 wild-type A. baumannii strains and qRT-PCR validation. (A) Comparative transcriptomics of A. baumannii ATCC 17978 and ΔLeuO strain are displayed as the differential expression. Differential expression levels are presented as fold change (mutant/wild-type) in the Y-axis, and more than 10-fold expression levels are presented above the 10-scale. Each dot indicates the differential expression levels of all predicted open reading frames of the ATCC 17978 genome and sorted according to the locus tag on the X-axis. The dash lines indicate 2-fold differential expression; upregulated and downregulated genes are located above the green line and below the red line, respectively. Gene names or functions of various highly differentially expressed genes are indicated as colored dots and circles and grouped according to functions on the right side. (B) qRT-PCR analysis of selected differentially expressed genes categorized as different functional groups such as biofilm-related genes, efflux pump regulator genes, iron transporter, putative transpose and signal peptide, hypothetical protein genes and VGR-related genes. Upregulated genes are presented as black-colored bars, and ash-colored bars represent downregulated genes. The data represent mean ± standard deviation from three biological replicates.
We performed qPCR using the same RNA sample to validate differential gene expression levels obtained from RNA-seq. A total of 23 genes from different functional groups were selected for qPCR, including members of A1S_0112-A1S-0119 operon, csu operon, efflux pump regulator, iron transporter, putative transpose and signal peptide, hypothetical proteins and VGR-like proteins (Figure 2B). The expression profiles were found to be consistent with data obtained from RNA-seq experiments. In some cases, there was a fold change difference in qPCR and RNA-seq data, which may be due to differences in sensitivity and specificity between the two technologies.
Contribution of LeuO to Biofilm Formation and Pellicle Formation
Biofilm formation is an important virulence factor for A. baumannii persistent infection. We conducted biofilm formation assay using the crystal violet staining method to determine the effect of LeuO deletion on the biofilm formation ability of A. baumannii on abiotic surfaces. The ΔLeuO strain produced significantly more biofilm than that produced by the wild-type strain. The LeuO-complemented (CP) strain yielded less biofilm than the mutant strain (Figure 3A). The biofilm mass formed by each strain was measured using absorbance at 570 nm of retained crystal violet, which was normalized relative to the growth of each strain using absorbance at 600 nm. The ΔLeuO strain demonstrated approximately 6-fold more biofilm than that of the wild-type strain, and CP restored the biofilm formation (Figure 3B). A. baumannii can form pellicles at the air–liquid interface. We analyzed the role of LeuO in pellicle formation using the A. baumannii strains in modified Luria-Bertani (LB) broth. During static culture, the ΔLeuO strain formed significantly more pellicles than the WT strain as photographed using a digital imaging system (Figure 3C). The pellicle biomass was measured at OD600 for quantification, wherein it was 0.51 for the ΔLeuO strain and 0.14 for the WT strain (Figure 3D). The complementation strain restored the pellicle formation to almost the same level as that of the wild-type strain.
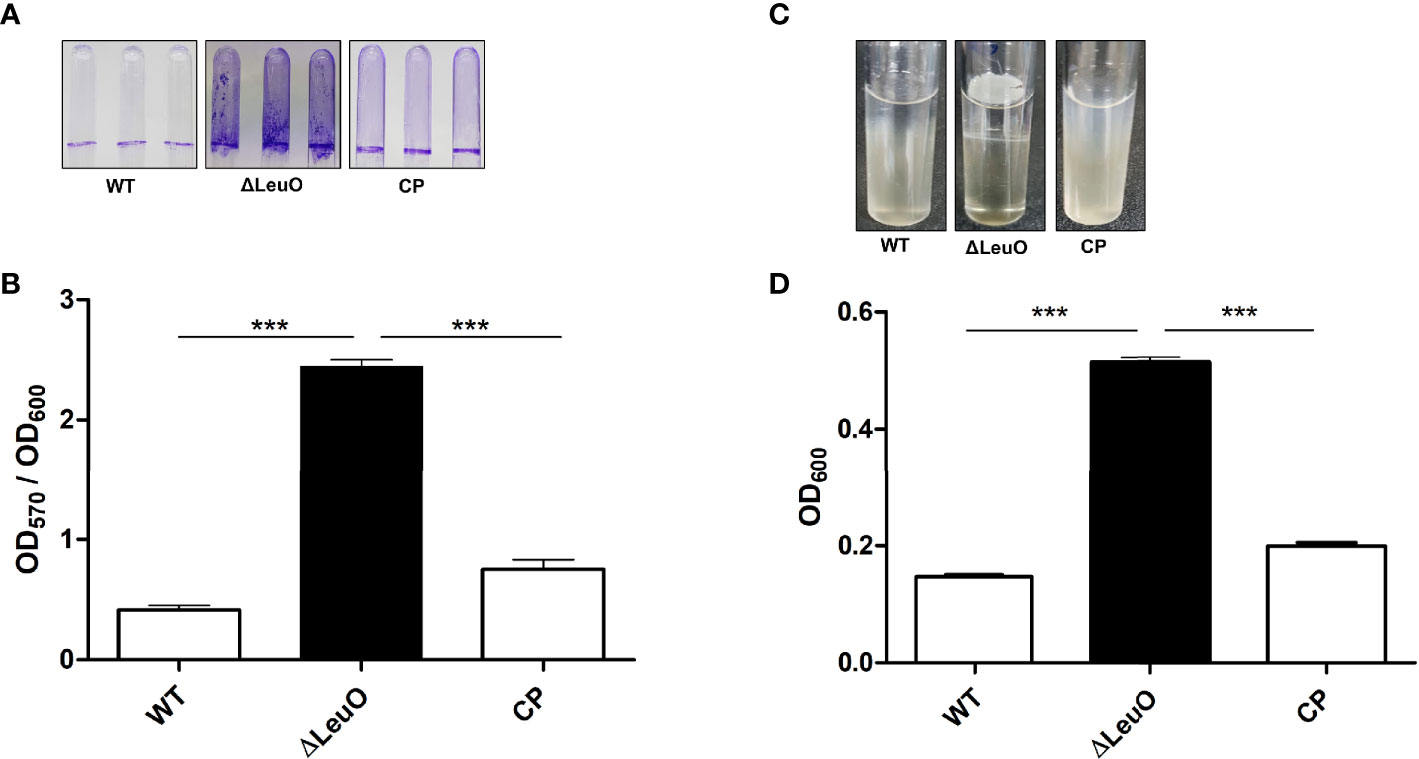
Figure 3 Biofilm and air–liquid interface pellicle formed by A. baumannii 17978 WT, ΔLeuO, and complementation strains. A. baumannii ATCC 17978 WT, ΔLeuO, and its CP strains were cultured in 5-ml polystyrene tubes at 30°C for 24 h in LB broth without salt. (A) Biofilm formation on polystyrene tubes was photographed after staining with 0.1% crystal violet. (B) Biofilm values (OD570) were normalized by growth levels (OD600) to compensate for the levels of biofilm formation on the polystyrene surface. Biofilm formation assays were conducted in triplicate, and average values of three replicates were plotted with standard deviation. (C) A. baumannii strains were cultured at 30°C for 48 h in LB broth without salt, and the pellicle formed by each strain was photographed. (D) Pellicles were separated by adding methanol, and pellicle biomass was measured as OD600. This experiment was performed in triplicate. Data are expressed as average values of three replicates with standard deviation. ***P < 0.001, significantly higher than that of the wild-type strain.
Biofilm formation in A. baumannii is regulated by several specific genes, including csuA/BABCDE, ompA, abaI, and pgaABCD (Longo et al., 2014). We performed qPCR using the WT and ΔLeuO strains to determine the role of LeuO in the expression of Csu operon genes and thus in biofilm formation. Our results revealed that CsuC, CsuD, and CsuE showed 14-, 22-, and 5-fold higher expression levels, respectively, in the LeuO mutant strain than those in the wild-type strain (Figure 2B). We also conducted qPCR of several genes from the A1S_0112-A1S_0119 cluster, which are responsible for biofilm formation. We observed that A1S_0112, A1S_0117, and A1S_0118 exhibited 146-, 18-, and 8-fold higher expression levels, respectively, in the LeuO mutant strain than those in the wild-type strain (Figure 2B). These findings suggest that LeuO regulates the expression of biofilm-related genes and LeuO mutation results in high biofilm formation in A. baumannii.
Point Mutations in LeuO Contribute to Biofilm Formation of A. baumannii Clinical Isolates
Considering the significant regulatory role of LeuO in the biofilm formation of A. baumannii ATCC 17978 strain, we conducted biofilm formation assay using different clones of A. baumannii clinical isolates (ST-208, ST-229, ST-357, ST-451, ST-552, and ST-784). We observed that clinical A. baumannii isolates formed much more biofilm than ATCC 17978 strain and almost similar biofilm to that formed by the ΔLeuO strain in some cases (Figure 4A and Supplementary Figure S3B). We focused on the genetic analysis of LeuO locus (A1S_1874) in our tested strains to identify the cause of high biofilm formation. Sequencing analysis of LeuO locus revealed that each of the clinical isolates had 306 amino acid residues identical to those in the ATCC 17978 strain but carried several point mutations in the linker helix or in the EBD (Supplementary Figure S3A). In ST-208, ST-451, ST-357 and ST-784 isolates, D-to-E, K-to-R, and N-to-S point mutation changed the amino acid aspartic acid to glutamic acid, lysine to arginine, and asparagine to serine at positions 63, 99, and 109, respectively (Figure 4B). The isolate ST-229 exhibited a seven-point mutation at positions 109, 187, 194, 195, 198, 264, and 303 compared with that in the ATCC 17978 strain. The clinical isolate ST-552 shared two-point mutations in the regulatory domain at positions 198 and 264, which changed serine to asparagine and glutamic acid to lysine, respectively (Figure 4B and Supplementary Figure S3A). These mutations in the A. baumannii biofilm regulator LeuO may modulate LeuO stability and cause hyper-biofilm formation in the tested clinical isolates.
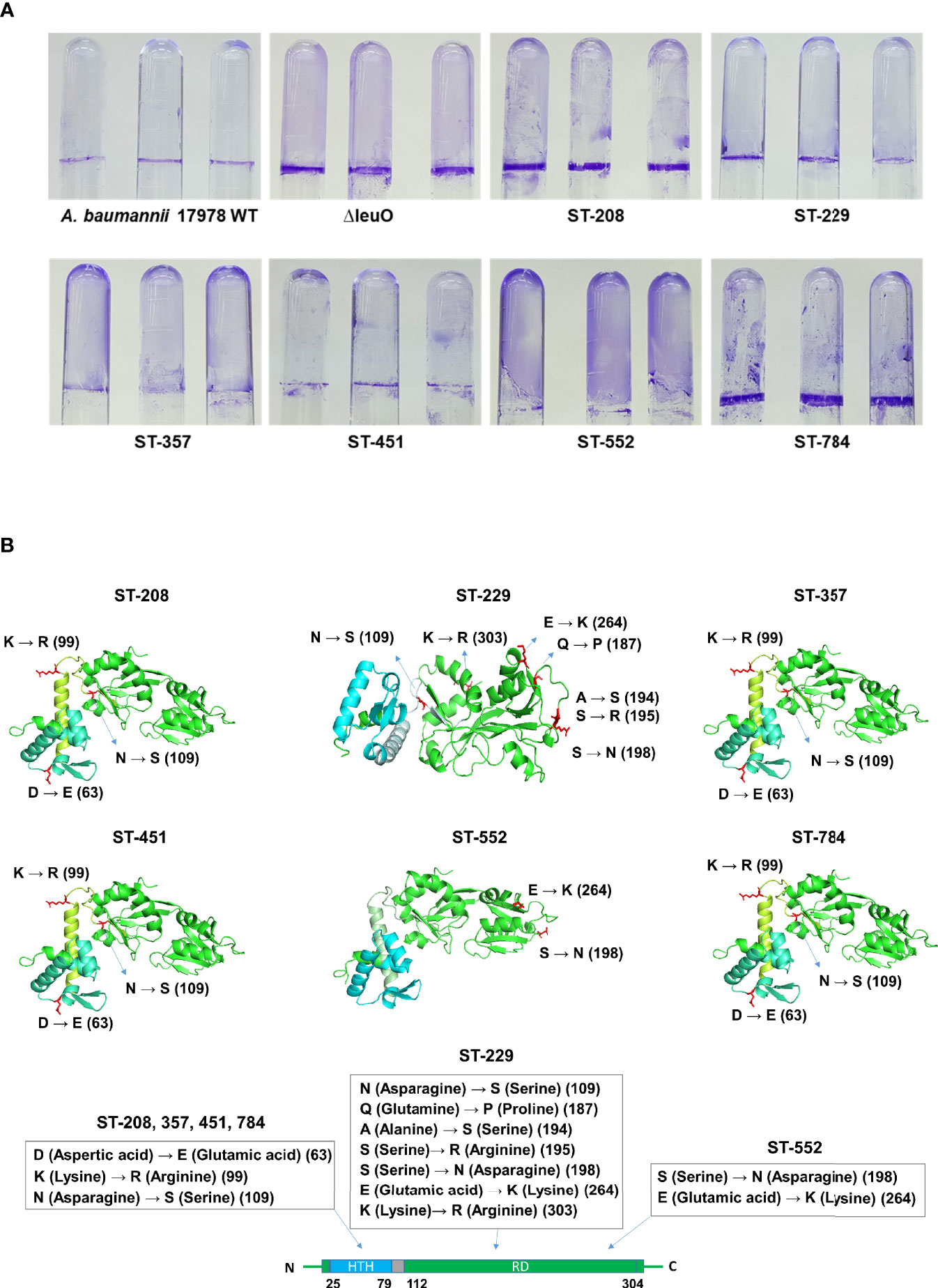
Figure 4 Hyper-biofilm-forming clinical strains have point mutation in the LeuO gene locus. (A) A. baumannii ATCC 17978 WT, ΔLeuO, and clinical strains of different sequence type ST-208(011), ST-229(079), ST-357(004), ST-451(001), ST-552(015), and ST-784(001) were cultured in 5-ml polystyrene tubes at 30°C for 24 h in LB broth without salt. (A) Biofilm formation on polystyrene tubes was photographed after staining with 0.1% crystal violet. (B) Cartoon representations of predicted LeuO gene structures of the clinical strains ST-208, ST-229, ST-357, ST-451, ST-552, and ST-784, respectively, obtained using the PyMoL software. Blue color indicates the N-terminal HTH domain, and green color indicates the C-terminal regulatory domain. Mutated residues are shown in red sticks at different positions such as 63, 99, 109, 187, 194, 195, 198, 264, and 303 and are marked beside.
Contribution of LeuO to Surface Motility
We next determined whether there was any influence of LeuO on surface motility by comparing motility with that of wild-type parent strain, ΔLeuO strain, and complementation strain on semisolid motility agar plates. Bacterial migration from the center of agar plates was measured at a point of time. Migration distance of the ΔLeuO strain from its inoculating point was smaller than that of the wild-type strain (68 mm in the mutant and 89 mm in the wild-type strain; (Figures 5A, B). The impaired motility of the ΔLeuO strain was restored in the complementation strain. Comparative transcriptome analysis revealed that several type VI pili genes (VGR-like proteins) were downregulated in the LeuO mutant strain compared with those in the wild-type strain. VGR-like protein genes were validated by qPCR (Figure 2B), which illustrated decreased expression of those genes in the LeuO mutant strain. These results imply that LeuO is an important regulator of surface motility.
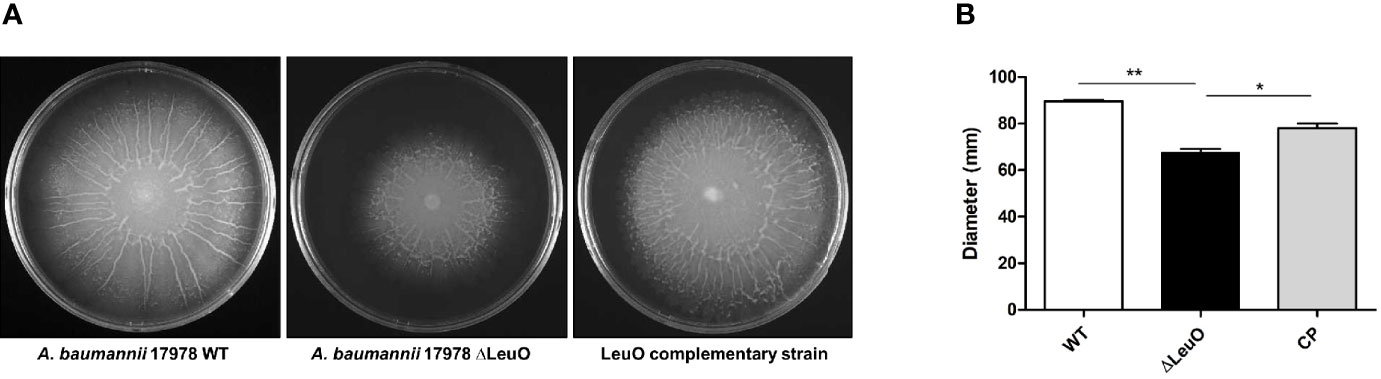
Figure 5 Comparison of surface motility of A. baumannii 17978 WT, ΔLeuO, and complementation strains. (A) 0.3% Eiken soft agar plates were used to test the surface motility of A. baumannii strains. Migration of bacteria from the inoculation point was photographed after culturing at 37°C for 10 h. (B) Average area of migration (diameter in mm) of three biological replicates is presented with standard deviation. The experiment was performed in triplicates. *P < 0.05, **P < 0.01, significantly lower than that of the wild-type strain.
LeuO Contributes to A. baumannii Adherence and Invasion Onto A549 Cell Line
A. baumannii pathogenesis largely depends on cellular adhesion and invasion. For determining the importance of LeuO in adherence to human alveolar epithelial cells, we performed an adherence assay on A549 alveolar epithelial cells using A. baumanni ATCC 17978 wild-type strain, ΔLeuO strain, and its complementation strain. In contrast to the result of biofilm formation assay on the polystyrene surface, the ΔLeuO strain exhibited approximately 10-fold reduction in attachment compared with that in the wild-type strain (Figure 6). This difference in adherence to epithelial cells was statistically significant. The complementation strain exhibited partial recovery of adhesion properties similar to the wild-type strain.
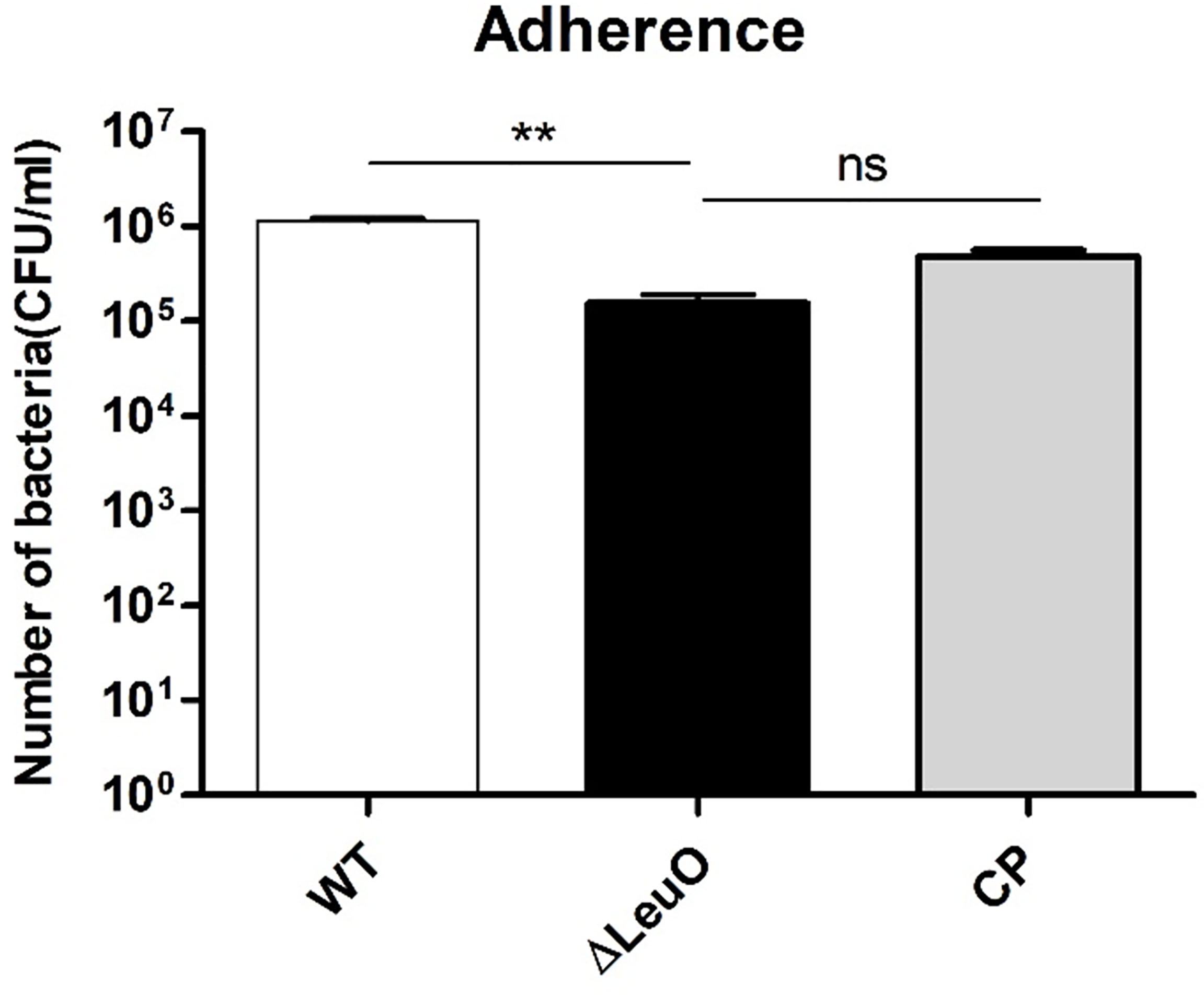
Figure 6 Adherence of A. baumannii strains to A549 cells. A549 alveolar epithelial cells were infected with A. baumannii wild-type, ΔLeuO, .and CP strains at a MOI of 1:100 for 2 h. After lysis with Triton-X, cell lysates were diluted and plated onto LB agar plates for CFU counting. The experiment was performed with three replicates. Data are presented as mean ± SD. ns, non-significant; **P < 0.01, significantly lower than that of the wild-type strain.
LeuO Regulates Virulence in Murine Infection
Because of the differences in some virulence-related traits such as biofilm formation and surface motility, we investigated the in vivo virulence of A. baumannii WT strain, ΔLeuO strain, and CP strain in a mice infection model. Because A. baumannii commonly infected immunocompromised patients, neutropenic mice were infected intraperitonially (108 CFU/mouse) and monitored for 96 h postinfection. After bacterial challenge with the LeuO mutant strain, all of mice succumbed to infection within 18 h (Figure 7). However, mice exposed to wild-type strain infection survived at 40% till the end of experimental period and all mice (100%) injected with PBS survived during the experimental period. Complementation of LeuO disruption restored the ability of A. baumannii to survive during the experimental period. The highly virulent A. baumannii 1656-2 strain (Park et al., 2011) showed almost the same result as that of the LeuO mutant strain. The Kaplan–Meier survival curve of the tested strains revealed statistical significance between the strains. Altogether, these data suggested that LeuO was responsible for the virulence of A. baumannii in the mice model.
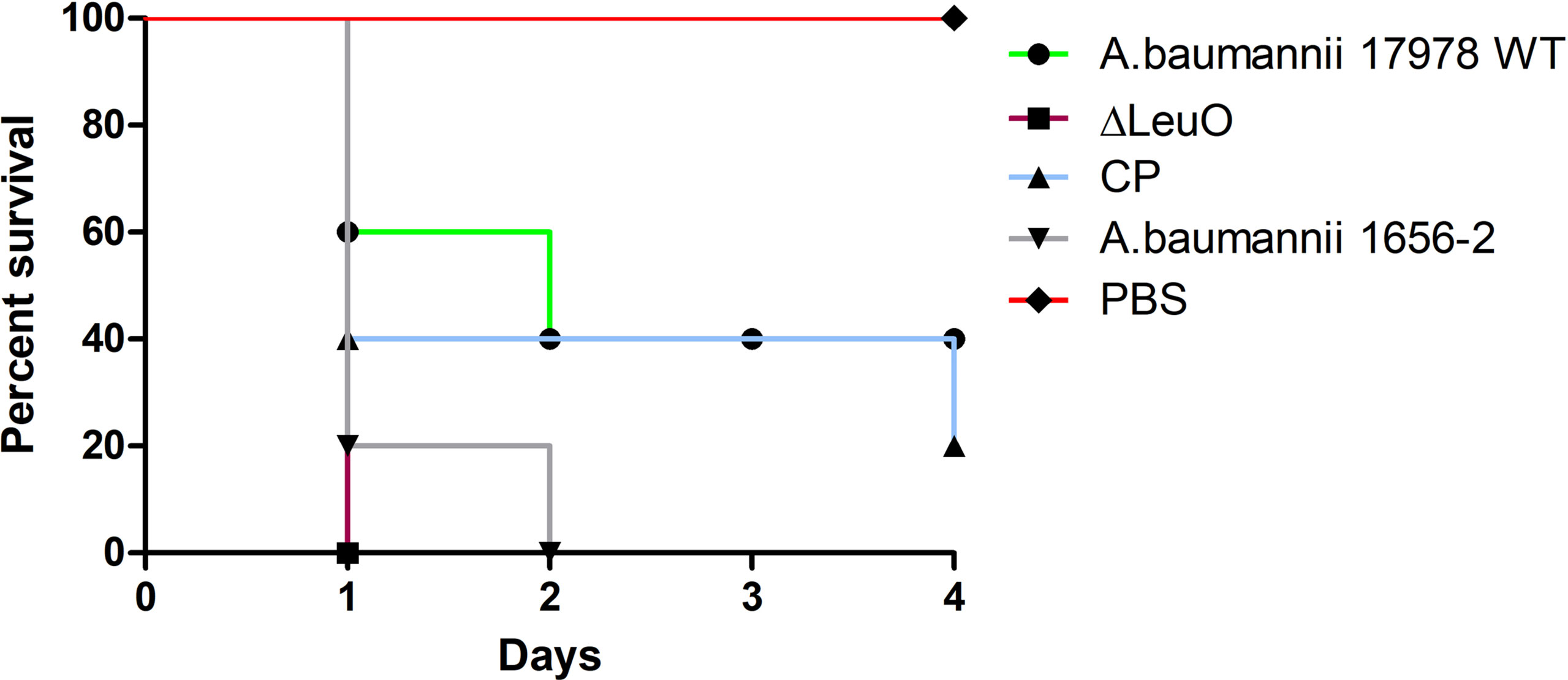
Figure 7 Survival of mice infected with A. baumannii strains. Kaplan-Meier survival curves of the virulence of A. baumannii 17978 wild-type, ΔLeuO, CP strain and A. baumannii 1656-2 in mice were determined up to 4 days of infection. 8-week-old female BALB/c mice were injected intraperitoneally (IP) with 1 × 108 CFU/ml bacterial suspension of tested strains (n = 5 per group). PBS was administered as control group and survival of infected mice were monitored every 12h for 4 days. Survival curve was generated using GraphPad Prism software.
Discussion
Despite the increasing prevalence of multidrug-resistant strains, the molecular mechanism underlying A. baumannii pathogenesis remains poorly defined. Prokaryotes have diverse transcription factors that regulate gene expression to adjust to new environments, among which the LTTR family is highly conserved in bacteria (Rivera-Gómez et al., 2011). In the present study, the predicted 3D structure of A. baumannii A1S_1874 showed two domains, a DNA-binding domain and a substrate-binding domain, suggesting that A1S_1874 belongs to the LTTR family (Figure 1). LeuO showed the highly conserved region at the N-terminal region compared with other bacterial LTTR family member protein. We suggested that A1S_1874 encodes LeuO in A. baumannii and a new global transcriptional regulator controlled different gene expression and biological functions. In S. enterica, LeuO regulates virulence-related genes (Fernández-Mora et al., 2004). In this study, we observed that LeuO is involved in regulating several phenotypes of A. baumannii. Our initial identification of the LeuO mutation indicated that there is also no significant difference in the growth of A. baumannii strains after LeuO deletion (Supplementary Figure S2). This result is consistent with another finding of LTTR deletion in Listeria monocytogenes (Abdelhamed et al., 2020). The RND superfamily efflux pump member AdeABC is responsible for resistance to aminoglycoside antibiotics (Marchand et al., 2004). In this experiment, the LeuO mutant exhibited higher susceptibility to the aminoglycoside antibiotics tobramycin, amikacin and gentamicin and also to the widely used beta-lactam antibiotics imipenem and meropenem (Supplementary Table S2).
Our RNA-seq data showed that 194 genes were upregulated and 108 genes were downregulated in the LeuO mutant compared with those in the wild-type strain (Supplementary Table S1). Differentially expressed genes were further categorized into COG categories to elucidate the potential roles of LeuO in transcriptomic regulation in A. baumannii (Figure 2A). Transcriptomic data showed that the csu operon genes and the A1S_0112-A1S_0119 cluster were highly upregulated, which are responsible for biofilm formation. The quorum sensing–related genes A1S_0109 and A1S_0111 (AbaI and AbaR) were also upregulated. We also observed that some putative transcriptional regulators of a different family were also upregulated. This could potentially affect different regulatory networks, and hence, further investigation would help understand the role of LeuO in these regulatory networks. We observed that the type VI secretion system-related genes were downregulated, which may play a role in surface motility. A large number of downregulated genes were hypothetical proteins with unknown functions. Further research is necessary to decipher the role of these genes in the A. baumannii genome. Previous studies have shown that the members of LTTRs regulate the expression of numerous genes involved in essential bacterial functions, including virulence, motility, metabolism, cell division, and oxidative stress response (Russell et al., 2004; Heroven and Dersch, 2006; Lu et al., 2007; O’Grady et al., 2011). Our transcriptomic data and observation of these LysR family members indicated almost the same effects of gene expression.
Biofilm formation by bacteria provides persistence and protects them from unfavorable conditions, including antimicrobial activity, and it is a major virulence factor (Costerton et al., 1999). A variety of transcriptional regulators have been identified to control biofilm formation in different organisms, for example, the transcriptional regulator LrhA in E. coli, CytR in V. cholerae, Fur in Yersinia pestis, SinR in Bacillus subtilis, and LcrX in Xanthomonas axonopodis (Haugo and Watnick, 2002; Blumer et al., 2005; Kearns et al., 2005; Sun et al., 2012; Park et al., 2020). LeuO, a putative LTTR, was identified as a repressor of biofilm synthesis in A. baumannii for the first time in this study. Our results demonstrated that biofilm formation on abiotic surface was significantly increased in the LeuO mutant strain compared with that in the wild-type strain (Figure 3). The csuA/BABCDE chaperon-usher secretion system-mediated pili play an important role in biofilm formation (Gaddy and Actis, 2009). Our RNA transcriptomic data revealed that csu operon genes were highly upregulated in the LeuO mutant strain. Another gene cluster, A1S_0112-A1S_0119, was also highly upregulated. A previous study showed that the A1S_0112-A1S_0119 cluster is critical for biofilm synthesis (Rumbo-Feal et al., 2017). These findings may explain the cause of high biofilm synthesis in the LeuO mutant strain. LeuO may be a transcriptional regulator that directly or indirectly controls genes that regulate biofilm formation. In contrast, several A. baumannii clinical isolates (ST-208, ST-229, ST-357, ST-451, ST-552, and ST-784) exhibited robust biofilm formation, which was almost similar to that of the LeuO mutant strain (Figure 4A). Sequence analysis of LeuO locus revealed several amino acid mutations in all the tested clinical strains compared with those in the wild-type strain in the linker helix and regulatory domain regions (Figure 4B). These mutations in amino acids may alter LeuO regulation in clinical strains and result in hyper-biofilm formation. It has been reported that mutation in the amino acids of the B. subtilis master biofilm regulator sinR modulated biofilm formation (Leiman et al., 2014).
A. baumannii displays surface-associated motility, which is an important virulence factor (Tomaras et al., 2003). We observed that the surface motility of the LeuO mutant was reduced compared with that in the wild-type strain (Figure 5). Extension and retraction of type IV pilus is required for motility (Clemmer et al., 2011). In our RNA-seq data, we observed that the type IV pilus genes A1S_0646 (IcmB) and VGR-like protein (type VI secretion system) A1S_1288 were downregulated by log 8.29- and 8.3-fold, respectively (Supplementary Table S1). Numerous transcriptional factors are known to be involved in the motility of other bacteria, such as ArcB, QseD, and RvoM (Heroven and Dersch, 2006; Habdas et al., 2010; Zhang et al., 2017). These results suggest that LeuO downregulates type IV and type VI pili genes and attenuates motility in the LeuO mutant strain. Surface motility is also controlled by quorum sensing in A. baumannii. Although abaRI genes were upregulated in LeuO mutant strain, surface motility was reduced. Further research will help to describe this discrepancy.
In the present study, we used alveolar epithelial cells (A549) to determine the role of LeuO in A. baumannii adherence. The LeuO mutant displayed significantly lower adherence to A549 epithelial cells than that shown by the wild-type strain (Figure 6). Our observations of the LeuO-mediated epithelial cell adherence and biofilm formation on abiotic surface are quite opposite, which suggests the presence of different mechanisms of adherence to either biotic or abiotic surface. Although csu operon is essential for biofilm formation, other studies have suggested that A. baumannii adherence to abiotic surfaces is independent of csuA/BABCDE-mediated pili and that csu-knockout strains showed no difference in binding to bronchial cells (de Breij et al., 2009; Gaddy and Actis, 2009). Our data corroborate with these findings. Further research is necessary to understand the exact molecular mechanisms involved in adherence to epithelial cells.
Several LTTRs are known to control the expression of virulence genes to maintain the host–pathogen interaction, such as ShvR in B. cenocepacia, MexT in P. aeruginosa, and LeuO in S. enterica serovar Typhimurium (Tropel and van der Meer, 2004; Tian et al., 2009; Espinosa and Casadesús, 2014). In this study, we observed that LeuO regulated several genes related to virulence. In our study, LeuO deletion increased lethal infection in the intraperitoneal mouse infection model (Figure 7). This increase in pathogenicity may be due to the result of increased virulence-related gene expression. Some previous studies have shown that disruption of LTTR family members such as GigC, MvfR, and ShvR reduced virulence in mammalian and plant models of infection (Cao et al., 2001; Déziel et al., 2005; Gebhardt et al., 2020). This contrasting result requires further analysis to understand the actual role of LTTRs in A. baumannii pathogenicity.
In conclusion, LeuO (A1S_1874) was identified as an LTTR, and transcriptome analysis revealed that LeuO regulated divergent sets of genes with different biological functions that were altered after LeuO deletion. Altogether, LeuO is involved in the regulation of biofilm formation, adherence, motility, and virulence of A. baumannii. This study provides valuable information regarding the role of an LTTR in the pathogenesis of A. baumannii.
Statistical Analysis
All experiments in this study were performed independently, and data are expressed as mean and standard deviations (SDs). All raw data were saved in Excel files and imported to GraphPad Prism for statistical analysis. Statistical differences between groups of data were compared using Student’s t-tests or one-way analysis of variance along with Turkey’s multiple comparisons.
Data Availability Statement
The findings of this study are available within this paper and its Supplementary Material. The transcriptomic data discussed in this publication have been deposited in the NCBI’s gene expression omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed using the accession number GSE173626.
Ethics Statement
Animal experiments were conducted according to experimental procedures approved by the Animal Care Committee of Kyungpook National University, South Korea (approval number: KNU-2019-178).
Author Contributions
MI, KK, and MS designed the study and MI wrote the manuscript. MI, KK, and MS performed the experiments. JL reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by a grant from the Korea Government National Research Foundation Grants 2016R1D1A1B01008960 (to MS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.738706/full#supplementary-material
References
Abdelhamed, H., Ramachandran, R., Narayanan, L., Ozdemir, O., Cooper, A., Olivier, A. K., et al. (2020). Contributions of a LysR Transcriptional Regulator to Listeria Monocytogenes Virulence and Identification of Its Regulons. J. Bacteriol. 202 (10), e00087–20. doi: 10.1128/JB.00087-20
Bhargava, N., Singh, S. P., Sharma, A., Sharma, P., Capalash, N. (2015). Attenuation of Quorum Sensing-Mediated Virulence of Acinetobacter Baumannii by Glycyrrhiza Glabra Flavonoids. Future Microbiol. 10, 1953–1968. doi: 10.2217/fmb.15.107
Bina, X. R., Howard, M. F., Ante, V. M., Bina, J. E. (2016). Vibrio Cholerae LeuO Links the ToxR Regulon to Expression of Lipid A Remodeling Genes. Infect. Immun. 84, 3161–3171. doi: 10.1128/IAI.00445-16
Blumer, C., Kleefeld, A., Lehnen, D., Heintz, M., Dobrindt, U., Nagy, G., et al. (2005). Regulation of Type 1 Fimbriae Synthesis and Biofilm Formation by the Transcriptional Regulator LrhA of Escherichia Coli. Microbiology (Reading) 151, 3287–3298. doi: 10.1099/mic.0.28098-0
Cao, H., Krishnan, G., Goumnerov, B., Tsongalis, J., Tompkins, R., Rahme, L. G. (2001). A Quorum Sensing-Associated Virulence Gene of Pseudomonas Aeruginosa Encodes a LysR-Like Transcription Regulator With a Unique Self-Regulatory Mechanism. Proc. Natl. Acad. Sci. U.S.A. 98, 14613–14618. doi: 10.1073/pnas.251465298
Clemmer, K. M., Bonomo, R. A., Rather, P. N. (2011). Genetic Analysis of Surface Motility in Acinetobacter Baumannii. Microbiology (Reading) 157, 2534–2544. doi: 10.1099/mic.0.049791-0
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial Biofilms: A Common Cause of Persistent Infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
De Breij, A., Gaddy, J., van der Meer, J., Koning, R., Koster, A., Van Den Broek, P., et al. (2009). CsuA/BABCDE-Dependent Pili are Not Involved in the Adherence of Acinetobacter Baumannii ATCC19606(T) to Human Airway Epithelial Cells and Their Inflammatory Response. Res. Microbiol. 160, 213–218. doi: 10.1016/j.resmic.2009.01.002
Deziel, E., Gopalan, S., Tampakaki, A. P., Lepine, F., Padfield, K. E., Saucier, M., et al. (2005). The Contribution of MvfR to Pseudomonas Aeruginosa Pathogenesis and Quorum Sensing Circuitry Regulation: Multiple Quorum Sensing-Regulated Genes are Modulated Without Affecting lasRI, rhlRI or the Production of N-Acyl-L-Homoserine Lactones. Mol. Microbiol. 55, 998–1014. doi: 10.1111/j.1365-2958.2004.04448.x
Espinosa, E., Casadesús, J. (2014). Regulation of Salmonella Enterica Pathogenicity Island 1 (SPI-1) by the LysR-Type Regulator LeuO. Mol. Microbiol. 91, 1057–1069. doi: 10.1111/mmi.12500
Fernández-Mora, M., Puente, J. L., Calva, E. (2004). OmpR and LeuO Positively Regulate the Salmonella Enterica Serovar Typhi Omps2 Porin Gene. J. Bacteriol. 186, 2909–2920. doi: 10.1128/JB.186.10.2909-2920.2004
Gaddy, J. A., Actis, L. A. (2009). Regulation of Acinetobacter Baumannii Biofilm Formation. Future Microbiol. 4, 273–278. doi: 10.2217/fmb.09.5
Gaddy, J. A., Arivett, B. A., Mcconnell, M. J., Lopez-Rojas, R., Pachon, J., Actis, L. A. (2012). Role of Acinetobactin-Mediated Iron Acquisition Functions in the Interaction of Acinetobacter Baumannii Strain ATCC 19606T With Human Lung Epithelial Cells, Galleria Mellonella Caterpillars, and Mice. Infect. Immun. 80, 1015–1024. doi: 10.1128/IAI.06279-11
Gebhardt, M. J., Czyz, D. M., Singh, S., Zurawski, D. V., Becker, L., Shuman, H. A. (2020). GigC, a LysR Family Transcription Regulator, Is Required for Cysteine Metabolism and Virulence in Acinetobacter Baumannii. Infect. Immun. 89 (1), e00180-20. doi: 10.1128/IAI.00180-20
Guadarrama, C., Villaseñor, T., Calva, E. (2014). The Subtleties and Contrasts of the LeuO Regulator in Salmonella Typhi: Implications in the Immune Response. Front. Immunol. 5, 581–581. doi: 10.3389/fimmu.2014.00581
Habdas, B. J., Smart, J., Kaper, J. B., Sperandio, V. (2010). The LysR-Type Transcriptional Regulator QseD Alters Type Three Secretion in Enterohemorrhagic Escherichia Coli and Motility in K-12 Escherichia Coli. J. Bacteriol. 192, 3699–3712. doi: 10.1128/JB.00382-10
Haugo, A. J., Watnick, P. I. (2002). Vibrio Cholerae CytR is a Repressor of Biofilm Development. Mol. Microbiol. 45, 471–483. doi: 10.1046/j.1365-2958.2002.03023.x
Hernández-Lucas, I., Gallego-Hernández, A. L., Encarnación, S., Fernández-Mora, M., Martínez-Batallar, A. G., Salgado, H., et al. (2008). The LysR-Type Transcriptional Regulator LeuO Controls Expression of Several Genes in Salmonella Enterica Serovar Typhi. J. Bacteriol. 190, 1658–1670. doi: 10.1128/JB.01649-07
Heroven, A. K., Dersch, P. (2006). RovM, a Novel LysR-Type Regulator of the Virulence Activator Gene Rova, Controls Cell Invasion, Virulence and Motility of Yersinia Pseudotuberculosis. Mol. Microbiol. 62, 1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x
Jawad, A., Seifert, H., Snelling, A. M., Heritage, J., Hawkey, P. M. (1998). Survival of Acinetobacter Baumannii on Dry Surfaces: Comparison of Outbreak and Sporadic Isolates. J. Clin. Microbiol. 36, 1938–1941. doi: 10.1128/JCM.36.7.1938-1941.1998
Jung, H. W., Kim, K., Islam, M. M., Lee, J. C., Shin, M. (2020). Role of Ppgpp-Regulated Efflux Genes in Acinetobacter Baumannii. J. Antimicrob. Chemother. 75, 1130–1134. doi: 10.1093/jac/dkaa014
Kearns, D. B., Chu, F., Branda, S. S., Kolter, R., Losick, R. (2005). A Master Regulator for Biofilm Formation by Bacillus Subtilis. Mol. Microbiol. 55, 739–749. doi: 10.1111/j.1365-2958.2004.04440.x
Kumar, A., Schweizer, H. P. (2005). Bacterial Resistance to Antibiotics: Active Efflux and Reduced Uptake. Adv. Drug Deliv. Rev. 57, 1486–1513. doi: 10.1016/j.addr.2005.04.004
Lee, J. S., Choi, C. H., Kim, J. W., Lee, J. C. (2010). Acinetobacter Baumannii Outer Membrane Protein A Induces Dendritic Cell Death Through Mitochondrial Targeting. J. Microbiol. 48, 387–392. doi: 10.1007/s12275-010-0155-1
Lee, J. C., Koerten, H., Van Den Broek, P., Beekhuizen, H., Wolterbeek, R., Van Den Barselaar, M., et al. (2006). Adherence of Acinetobacter Baumannii Strains to Human Bronchial Epithelial Cells. Res. Microbiol. 157, 360–366. doi: 10.1016/j.resmic.2005.09.011
Lee, H. W., Koh, Y. M., Kim, J., Lee, J. C., Lee, Y. C., Seol, S. Y., et al. (2008). Capacity of Multidrug-Resistant Clinical Isolates of Acinetobacter Baumannii to Form Biofilm and Adhere to Epithelial Cell Surfaces. Clin. Microbiol. Infect. 14, 49–54. doi: 10.1111/j.1469-0691.2007.01842.x
Leiman, S. A., Arboleda, L. C., Spina, J. S., Mcloon, A. L. (2014). SinR is a Mutational Target for Fine-Tuning Biofilm Formation in Laboratory-Evolved Strains of Bacillus Subtilis. BMC Microbiol. 14, 301–301. doi: 10.1186/s12866-014-0301-8
Longo, F., Vuotto, C., Donelli, G. (2014). Biofilm Formation in Acinetobacter Baumannii. New Microbiol. 37, 119–127.
Lu, Z., Takeuchi, M., Sato, T. (2007). The LysR-Type Transcriptional Regulator YofA Controls Cell Division Through the Regulation of Expression of ftsW in Bacillus Subtilis. J. Bacteriol. 189, 5642–5651. doi: 10.1128/JB.00467-07
Marchand, I., Damier-Piolle, L., Courvalin, P., Lambert, T. (2004). Expression of the RND-Type Efflux Pump AdeABC in Acinetobacter Baumannii is Regulated by the AdeRS Two-Component System. Antimicrob. Agents Chemother. 48, 3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004
Martí, S., Rodríguez-Baño, J., Catel-Ferreira, M., Jouenne, T., Vila, J., Seifert, H., et al. (2011). Biofilm Formation at the Solid-Liquid and Air-Liquid Interfaces by Acinetobacter Species. BMC Res. Notes 4, 5. doi: 10.1186/1756-0500-4-5
Mcconnell, M. J., Actis, L., Pachon, J. (2013). Acinetobacter Baumannii: Human Infections, Factors Contributing to Pathogenesis and Animal Models. FEMS Microbiol. Rev. 37, 130–155. doi: 10.1111/j.1574-6976.2012.00344.x
Moorthy, S., Watnick, P. I. (2005). Identification of Novel Stage-Specific Genetic Requirements Through Whole Genome Transcription Profiling of Vibrio Cholerae Biofilm Development. Mol. Microbiol. 57, 1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x
Muraoka, S., Okumura, R., Ogawa, N., Nonaka, T., Miyashita, K., Senda, T. (2003). Crystal Structure of a Full-Length LysR-Type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend. J. Mol. Biol. 328, 555–566. doi: 10.1016/S0022-2836(03)00312-7
O’grady, E. P., Nguyen, D. T., Weisskopf, L., Eberl, L., Sokol, P. A. (2011). The Burkholderia Cenocepacia LysR-Type Transcriptional Regulator ShvR Influences Expression of Quorum-Sensing, Protease, Type II Secretion, and Afc Genes. J. Bacteriol. 193, 163–176. doi: 10.1128/JB.00852-10
Park, H., Do, E., Kim, M., Park, H.-J., Lee, J., Han, S.-W. (2020). A LysR-Type Transcriptional Regulator LcrX Is Involved in Virulence, Biofilm Formation, Swimming Motility, Siderophore Secretion, and Growth in Sugar Sources in Xanthomonas Axonopodis Pv. Glycines. Front. Plant Sci. 10, 1657–1657. doi: 10.3389/fpls.2019.01657
Park, J. Y., Kim, S., Kim, S. M., Cha, S. H., Lim, S. K., Kim, J. (2011). Complete Genome Sequence of Multidrug-Resistant Acinetobacter Baumannii Strain 1656-2, Which Forms Sturdy Biofilm. J. Bacteriol. 193, 6393–6394. doi: 10.1128/JB.06109-11
Rice, L. B. (2008). Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Rice, K. C., Nelson, J. B., Patton, T. G., Yang, S. J., Bayles, K. W. (2005). Acetic Acid Induces Expression of the Staphylococcus Aureus cidABC and lrgAB Murein Hydrolase Regulator Operons. J. Bacteriol. 187, 813–821. doi: 10.1128/JB.187.3.813-821.2005
Rivera-Gómez, N., Segovia, L., Pérez-Rueda, E. (2011). Diversity and Distribution of Transcription Factors: Their Partner Domains Play an Important Role in Regulatory Plasticity in Bacteria. Microbiology (Reading) 157, 2308–2318. doi: 10.1099/mic.0.050617-0
Rumbo-Feal, S., Pérez, A., Ramelot, T. A., Ãlvarez-Fraga, L., Vallejo, J. A., Beceiro, A., et al. (2017). Contribution of the A. Baumannii A1S_0114 Gene to the Interaction With Eukaryotic Cells and Virulence. Front. Cell. Infect. Microbiol. 7, 108–108. doi: 10.3389/fcimb.2017.00108
Russell, D. A., Byrne, G. A., O’connell, E. P., Boland, C. A., Meijer, W. G. (2004). The LysR-Type Transcriptional Regulator VirR is Required for Expression of the Virulence Gene vapA of Rhodococcus Equi ATCC 33701. J. Bacteriol. 186, 5576–5584. doi: 10.1128/JB.186.17.5576-5584.2004
Russo, T. A., Luke, N. R., Beanan, J. M., Olson, R., Sauberan, S. L., Macdonald, U., et al. (2010). The K1 Capsular Polysaccharide of Acinetobacter Baumannii Strain 307-0294 is a Major Virulence Factor. Infect. Immun. 78, 3993–4000. doi: 10.1128/IAI.00366-10
Santos, C. L., Tavares, F., Thioulouse, J., Normand, P. (2009). A Phylogenomic Analysis of Bacterial Helix-Turn-Helix Transcription Factors. FEMS Microbiol. Rev. 33, 411–429. doi: 10.1111/j.1574-6976.2008.00154.x
Schell, M. A. (1993). Molecular Biology of the LysR Family of Transcriptional Regulators. Annu. Rev. Microbiol. 47, 597–626. doi: 10.1146/annurev.mi.47.100193.003121
Shimada, T., Bridier, A., Briandet, R., Ishihama, A. (2011). Novel Roles of LeuO in Transcription Regulation of E. coli Genome: Antagonistic Interplay With the Universal Silencer H-NS. Mol. Microbiol. 82, 378–397. doi: 10.1111/j.1365-2958.2011.07818.x
Srinivasan, V. B., Mondal, A., Venkataramaiah, M., Chauhan, N. K., Rajamohan, G. (2013). Role of oxyRKP, a Novel LysR-Family Transcriptional Regulator, in Antimicrobial Resistance and Virulence in Klebsiella Pneumoniae. Microbiology (Reading) 159, 1301–1314. doi: 10.1099/mic.0.065052-0
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., Svabic-Vlahovic, M. (2000). A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
Sun, F., Gao, H., Zhang, Y., Wang, L., Fang, N., Tan, Y., et al. (2012). Fur is a Repressor of Biofilm Formation in Yersinia Pestis. PloS One 7, e52392–e52392. doi: 10.1371/journal.pone.0052392
Tian, Z. X., Fargier, E., Mac Aogáin, M., Adams, C., Wang, Y. P., O’gara, F. (2009). Transcriptome Profiling Defines a Novel Regulon Modulated by the LysR-Type Transcriptional Regulator MexT in Pseudomonas Aeruginosa. Nucleic Acids Res. 37, 7546–7559. doi: 10.1093/nar/gkp828
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., Actis, L. A. (2003). Attachment to and Biofilm Formation on Abiotic Surfaces by Acinetobacter Baumannii: Involvement of a Novel Chaperone-Usher Pili Assembly System. Microbiology (Reading) 149, 3473–3484. doi: 10.1099/mic.0.26541-0
Tropel, D., van der Meer, J. R. (2004). Bacterial Transcriptional Regulators for Degradation Pathways of Aromatic Compounds. Microbiol. Mol. Biol. Rev. 68, 474–500. doi: 10.1128/MMBR.68.3.474-500.2004
WHO (2017). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Saudi Med. J. 38, 444–445.
Yang, S. J., Rice, K. C., Brown, R. J., Patton, T. G., Liou, L. E., Park, Y. H., et al. (2005). A LysR-Type Regulator, CidR, is Required for Induction of the Staphylococcus Aureus cidABC Operon. J. Bacteriol. 187, 5893–5900. doi: 10.1128/JB.187.17.5893-5900.2005
Keywords: Acinetobacter baumannii, LeuO, transcriptome (RNA-seq), biofilm, virulence, adherence
Citation: Islam MM, Kim K, Lee JC and Shin M (2021) LeuO, a LysR-Type Transcriptional Regulator, Is Involved in Biofilm Formation and Virulence of Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 11:738706. doi: 10.3389/fcimb.2021.738706
Received: 09 July 2021; Accepted: 23 September 2021;
Published: 11 October 2021.
Edited by:
Paolo Visca, Roma Tre University, ItalyReviewed by:
Younes Smani, Institute of Biomedicine of Seville (IBIS), SpainBeate Jutta Averhoff, Goethe University Frankfurt, Germany
Copyright © 2021 Islam, Kim, Lee and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minsang Shin, c2hpbm1zQGtudS5hYy5rcg==
†These authors have contributed equally to this work
 Md. Maidul Islam
Md. Maidul Islam Kyeongmin Kim
Kyeongmin Kim Je Chul Lee
Je Chul Lee Minsang Shin
Minsang Shin