- State Key Laboratory of Pathogen and Biosecurity, Academy of Military Medical Sciences, Beijing, China
Chlamydia are Gram-negative, obligate intracellular bacterial pathogens that infect eukaryotic cells and reside within a host-derived vacuole known as the inclusion. To facilitate intracellular replication, these bacteria must engage in host-pathogen interactions to obtain nutrients and membranes required for the growth of the inclusion, thereby sustaining prolonged bacterial colonization. Autophagy is a highly conserved process that delivers cytoplasmic substrates to the lysosome for degradation. Pathogens have developed strategies to manipulate and/or exploit autophagy to promote their replication and persistence. This review delineates recent advances in elucidating the interplay between Chlamydia trachomatis infection and autophagy in recent years, emphasizing the intricate strategies employed by both the Chlamydia pathogens and host cells. Gaining a deeper understanding of these interactions could unveil novel strategies for the prevention and treatment of Chlamydia infection.
1 Introduction
Chlamydia are Gram-negative, obligate intracellular bacterial pathogens and symbionts that inhabit a wide range of hosts, from humans to amoebae. The Chlamydiaceae family includes at least 13 species that are pathogenic to humans or animals (Bachmann et al., 2014). Some species, like Chlamydia psittaci and Chlamydia abortus, are known as zoonotic pathogens, while Chlamydia trachomatis is endemic to humans, primarily infecting the female genital tract (Jury et al., 2023). Additionally, Chlamydia pneumoniae is a prevalent pathogen in both humans and animals, commonly associated with respiratory tract infections (Cheong et al., 2019).
As obligate intracellular bacteria, Chlamydia spp. invade eukaryotic cells and reside within a cytoplasmic membrane-bound vacuole known as the inclusion (Stelzner et al., 2023). These bacteria exhibit a biphasic developmental cycle that hinges on specific host-cell interactions for nutrient acquisition necessary for intracellular proliferation (Stephens et al., 1998). The infection process is initiated by the non-replicative, infectious elementary bodies (EBs). Following host cell entry, EBs differentiate into the noninfectious, and replicative reticulate bodies (RBs) (Omsland et al., 2012). Upon completing their developmental cycle, RBs revert to EBs, which are then released upon host cell lysis to propagate subsequent infections. Additionally, some Chlamydia spp. can also exit their host by extrusion, without causing cell lysis.
Chlamydia trachomatis, a critical species within this genus, is the foremost pathogen causing trachoma and sexually transmitted infection (STI), and will be the focus of this review (Liang et al., 2017). Trachoma is the leading infectious cause of blindness globally, with the World Health Organization (WHO) estimating that approximately 190 million people are at risk of trachoma blindness (Woodhall et al., 2018). Most STIs caused by C. trachomatis are asymptomatic, but untreated infections can result in pelvic inflammatory disease (Poston et al., 2019). C. trachomatis strains are classified into two major biovars and further subtyped by serovars based on the chlamydial major outer membrane protein (MOMP) (Miyairi et al., 2006). The lymphogranuloma venereum (LGV) biovar (serovars L1, L2, and L3) targets genital tissues and macrophages, causing the invasive STI known as ‘LGV’ (Abdelsamed et al., 2013; Hepler et al., 2018). Among the non-LGV biovar, serovars A-C infect the ocular tissue and cause trachoma. In contrast, serovars D-K primarily cause inflammatory diseases such as pelvic inflammatory disease and tubal factor infertility (Hepler et al., 2018).
2 Molecular pathogenesis
As an obligate intracellular organism, C. trachomatis employs its virulence factors intricately engage with host cellular mechanisms, eliciting host immune responses and triggering a cascade of cellular pathological alterations (Chen et al., 2019). The Type III secretion system (T3SS) consisting of a needle-like injectosome complex, facilitates the translocation of effector proteins from the chlamydial cytoplasm into the host cell’s cytoplasm and inclusion’s lumen (Deng et al., 2017). It is hypothesized that there are over 80 anti-host T3SS-secreted effectors participating in diverse biological processes, including but not limited to the invasion, cytoskeletal structure reconstruction, modulation of vesicular interactions, apoptosis, signal transduction, promoting the bacterial survival and replication (Betts-Hampikian and Fields, 2010). To date, many proposed effectors remain unverified and even more are yet to be characterized. The advent of genetic manipulation techniques has significantly expanded the research on elucidating the function of T3SS effectors (Wan et al., 2023).
Another important virulence factor is a highly conserved ~7.5 kb plasmid, encoding encodes eight glycoproteins (pGP1-8) that facilitate the ascension of infection, elicit pro-inflammatory responses, and promote extrusion processes (Zhong, 2017; Jury et al., 2023). Under stress conditions, the copy number of C. trachomatis plasmid increases, presumably to upregulate T3SS-secreted substrates and ensure in vivo survival. Nevertheless, the precise role of chlamydial plasmid in its pathogenesis remains to be fully elucidated.
3 Host autophagy in Chlamydia trachomatis infection
To date, three different pathways of autophagy have been described: chaperone-mediated autophagy (CMA), macroautophagy, which is the most extensively studied one, and microphagy (Kim and Lee, 2014). Macroautophagy (hereafter referred to as autophagy) is a fundamental and evolutionary conserved progress that facilitates the delivery of dysfunctional organelles or invading pathogens to lysosome degradation, thereby preserving cellular homeostasis in eukaryotic cells (Yu et al., 2018). Analogous to endocytosis, both pathways converge at the lysosome, where their respective cargos are degraded. Endocytosis is crucial for various cellular processes, including nutrient uptake, cell adhesion and migration, pathogen entry, by internalizing extracellular cargos from the plasma membrane. However, autophagy involves the engulfment of cytoplasmic contents (cargos) into a double membrane vesicle known as the autophagosome. This multifaceted process involves autophagy initiation, autophagosome formation, autophagolysosome formation, and cargo degradation (Hansen and Johansen, 2011). Autophagosomes acquire d lysosomal enzymes necessary for degrading the vacuolar contents, which is subsequently released into the cytosol (Yim and Mizushima, 2020).
Typically, autophagy restricts bacterial proliferation and dissemination, thereby mitigating infection severity. Examples of autophagy-targeted bacteria include Legionella, Salmonella and Mycobacterium (Sharma et al., 2018; Thomas et al., 2020). Correspondingly, these bacteria have developed various strategies to form unique parasitophorous vacuoles and evade the autophagic capture (Deretic et al., 2013). For instance, the cysteine protease RavZ from Legionella pneumophila strain inhibits autophagy by irreversibly deconjugating the ubiquitin-like autophagy-related protein 8 (ATG8) from autophagosome membranes (Choy et al., 2012). Salmonella escaped from Salmonella-containing vacuoles (SCV) into the cytosol, where it is initially ubiquitinated, followed by the recruitment of selective autophagy receptors p62/SQSTM1 and OPTN (optineurin), leading to its phagocytosis and degradation (Gomes and Dikic, 2014). IFN-γ induction of autophagy contributed to the control of Mycobacterium tuberculosis intracellular proliferation (Kim et al., 2011). The role of IFN-γ in autophagy involves the interferon-induced guanylate-binding protein (GBP), which promotes the oxidative killing and the delivery of antimicrobial peptides to autophagolysosome (Gutierrez et al., 2004). However, unlike the aforementioned pathogens, Coxiella burnetii exploits the autophagy for its benefits. The effector protein CvpF of C. burnetii, interacts with the host small GTPase RAB26 to stimulate the autophagy flux, promoting the vacuole biogenesis and its virulence (Siadous et al., 2020) Another effector protein, CpeB, targets Rab11a to promote the accumulation of microtubule-associated protein light chain 3-II (LC3-II), a lipidated form of LC3 that is associated with autophagosomal membranes, thus sustaining the bacteria infection and virulence (Fu et al., 2022).
The role of autophagy in C. trachomatis development is ambiguous. Conflicting results have been reported, depending on experimental conditions, chlamydial serovars, and cell lines used (Witkin et al., 2017). For example, no distinct rim of autophagosomal protein-specific staining was observed around the C. trachomatis’s inclusions (Al-Younes et al., 2004). And the inhibition of inclusion-lysosomes fusion is beneficial for the intracellular survival of C. trachomatis throughout the its developmental cycle, suggesting the evasion of bacteria from the autophagy-mediated destruction (Heinzen et al., 1996). Concurrently, C. trachomatis might modulate interactions between the inclusions with host vesicles and promote nutrient acquisitions from the cytoplasm (McClarty, 1994). Nutrient availability significantly impacts the normal development of C. trachomatis. Thus, it can be inferred that autophagy may serve as a nutrient source of for the replicating bacteria, as demonstrated by the inhibition of the intracellular replication of C. trachomatis serovar L2 upon treatment with autophagy inhibitors (Al-Younes et al., 2004).
4 Host autophagy manipulation by C. trachomatis
4.1 Initiating autophagy induced by Chlamydia infection
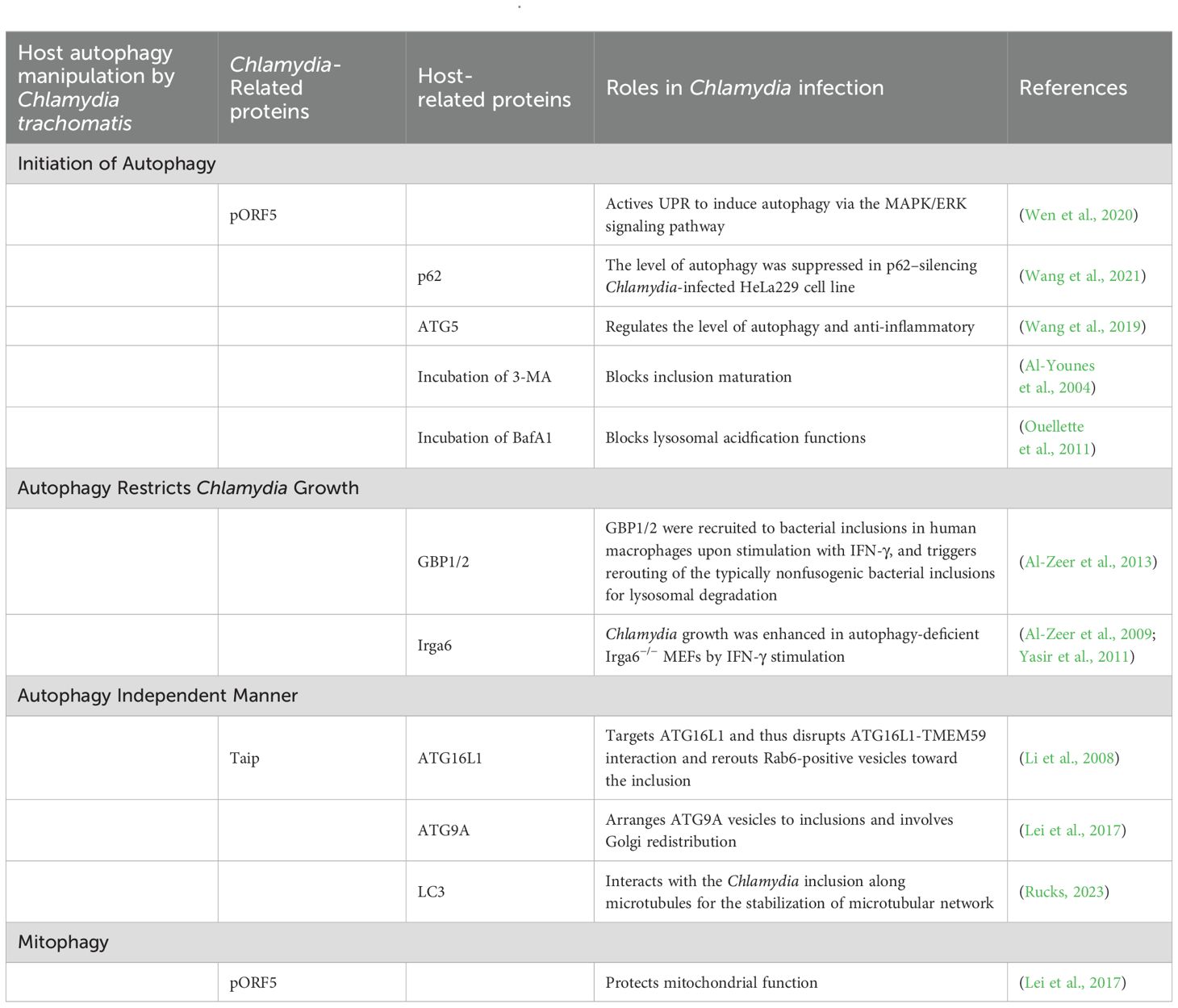
Chlamydia trachomatis has a substantially contracted genome (1.04 Mb encoding 895 open reading frames) that lacks many metabolic enzymes, conferring a restricted biosynthetic capacity that necessitates reliance on eukaryotic host cells for essential metabolites (Stephens et al., 1998). C. trachomatis acquires a variety of nutrients, including amino acids, nucleotides and lipids from the lysosomal-mediated degradation of proteins (Bastidas et al., 2013) and various transporters (Damiani et al., 2014). The active intracellular growth of C. trachomatis results in significant decreases in the nutrient pools in host cells, thereby triggering autophagy around the midpoint of the C. trachomatis developmental cycle, suggesting that Chlamydia infection induces autophagy to secure nutrients for bacterial proliferation (Pachikara et al., 2009). Previous studies have demonstrated that Chlamydia infection could induce autophagy. Autophagosome-associated markers, including microtubule-associated protein light chain 3 (MAP-LC3, autophagosomal markers) and calreticulin, are redistributed to the inclusions of C. trachomatis (Al-Younes et al., 2004). The plasmid-encoded protein pORF5 of C. trachomatis activates the unfolded protein response (UPR) to induce autophagy, with the MAPK/ERK inhibitor PD98059 partially attenuating pORF5-induced autophagy, indicating that pORF5 activates UPR to induce autophagy via the MAPK/ERK signaling pathway (Wen et al., 2020). The infection of mouse embryo fibroblasts (MEFs) by C. trachomatis serovar L2 correlates with upregulated autophagy-related protein production (Pachikara et al., 2009), whereas autophagy is suppressed in p62−silenced Chlamydia-infected HeLa229 cell line (Wang et al., 2021). The numbers of autophagosomes and the expression of LC3-II and Beclin1 were increased, and the expression of p62 was downregulated in C. psittaci-infected human bronchial epithelial cells (HBEs) (Chen et al., 2022). These findings align with other studies showing that Atg5 silencing significantly inhibits the autophagy in the Chlamydia-infected HeLa229 cells (Wang et al., 2019). Furthermore, Atg5 knockdown promotes the secretion of proinflammatory cytokines IL-1β and IFN-γ in C. trachomatis-infected cells, suggesting an anti-inflammatory role for Atg5 in the autophagy induction during chlamydia infection (Wang et al., 2019).
Additionally, autophagy inhibitors impede the intracellular replication of Chlamydia. Hep-2 cells infected with C. trachomatis Serovar L2 subsequently treated with 3-methyladenine (3-MA) exhibits a decreased expression LC3-II/LC3-I ratio, abnormal inclusion maturation, and diminished progeny, indicating that 3-MA inhibits of C. trachomatis growth by reducing autophagy (Al-Younes et al., 2004). Bafilomycin A1 (BafA1), a vacuolar H+/ATPase inhibitor that blocks lysosomal acidification and functions, is one of the widely used to inhibit the last stage of autophagy and impair the growth of C. trachomatis (Ouellette et al., 2011).
4.2 Autophagy restricts Chlamydia growth
One of the host cell strategies to block intracellular bacterial proliferation involves phagolysosomal degradation, wherein the host cell directs internalized bacteria into vesicles delivered to the acidic lysosome for degradation. Early-stage C. trachomatis infected RAW macrophages exhibit impaired intracellular proliferation, while inclusions of C. trachomatis were positive for the autophagy marker LC3 during the late stage of infection. Sun et al. also reported that C. trachomatis is rapidly transported to lysosomes, with significantly augmented bacterial growth in RAW macrophages pretreated with BafA1 (Sun et al., 2012). These findings underscore the crucial role of lysosomal inclusion fusion in modulating C. trachomatis infection.
Interferon γ (IFN-γ), a cytokine involved in immune regulation plays a pivotal role in cell-intrinsic immunity against various intracellular pathogens, including Chlamydia (Vollmuth et al., 2022). Exogenous IFN-γ diminished chlamydial infectivity by 50% in wild-type human macrophages. Al-Zeer and colleagues demonstrated that the human GBP1/2 are recruited to bacterial inclusions in human macrophages upon IFN-γ stimulation, prompting the rerouting of the typically nonfusogenic bacterial inclusions for lysosomal degradation. This suggests that autophagy confines C. trachomatis proliferation in human macrophages via IFN-γ-inducible GBPs (Al-Zeer et al., 2013).
Subsequent studies identified immunity-related GTPases (IRGs), such as Irga6, as critical in IFN-γ-induced autophagy for C. trachomatis eradication (Al-Zeer et al., 2009). Enhanced C. trachomatis growth has been observed in autophagy-deficient Atg5−/− or Irga6−/− MEFs upon IFN-γ stimulation compared to that in wild-type MEFs. IFN-γ treatment inhibits the growth of C. trachomatis inclusions and formation of infectious EBs via lysosomal fusion with early inclusions, a process reversible with BafA1 treatment (Al-Zeer et al., 2009; Yasir et al., 2011).
Additionally, autophagy pathway component deficiency in MEFs or macrophages leads to increased C. pneumoniae proliferation, indicating autophagy’s bactericidal role (Crother et al., 2019). The inflammasome, a crucial innate immune component, fight against pathogenic microorganisms (Biasizzo and Kopitar-Jerala, 2020). Autophagy negatively regulates the nucleotide binding domain and leucine-rich repeat (NLR) pyrin domain containing 3 (NLRP3) inflammasome and subsequent IL-1β secretion (Crother et al., 2019). Recent studies have revealed that Chlamydia induces inflammasome priming and activation signals (Webster and Goodall, 2018), and the autophagy deficiency results in increased pulmonary inflammasome activity and IL-1β production in response to C. pneumoniae infection in mice (Crother et al., 2019).
4.3 Autophagy-independent manner
Autophagy-related gene (ATG) proteins are pivotal in orchestrating the biogenesis of autophagosomes, facilitating the transport of cytoplasmic constituents to lysosomes or vacuoles (Kirkin and Rogov, 2019). Furthermore, ATG protein activities modulate cell cycle progression through both autophagy-dependent and autophagy-independent pathways (Palikaras et al., 2018).
Research indicates that the proliferation of C. trachomatis is modulated by ATG9A and ATG16L1 via autophagy-independent mechanisms. Hamaoui et al. revealed that the Chlamydia effector protein TaiP (CT622) targets ATG16L, disrupting the ATG16L1-TMEM59 interaction and redirecting Rab6-positive vesicles to the inclusion (Li et al., 2008). Moreover, a recent investigation elucidated that ATG9A organizes ATG9A vesicles to inclusions and mediates Golgi apparatus redistribution to enhance C. trachomatis proliferation (Zou et al., 2018).
LC3 proteins are extensively utilized as biomarkers in autophagy flux studies. The cytosolic non-lipidated form of LC3 (LC3-I) conjugates with phosphatidylethanolamine, forming the LC3-phosphatidylethanolamine conjugate (LC3-II), which is subsequently recruited to autophagosomal membranes. Notably, LC3 has been implicated in stabilizing the microtubule network of the cytoskeleton (Lei et al., 2017). Al-Younes et al. demonstrated that LC3 associates with C. trachomatis inclusions along microtubules, and depletion of LC3 in autophagy-deficient cells significantly impairs chlamydial propagation, highlighting a novel role for LC3, independent of autophagy, in Chlamydia pathogenesis (Rucks, 2023).
4.4 Chlamydia protects mitochondrial function by mitophagy
Autophagy can be either selective or nonselective. Selective autophagy leads to the lysosomal degradation of specific cargoes like mitochondria, known as mitophagy (Kirkin and Rogov, 2019). Mitophagy is crucial for maintaining mitochondrial quality control and homeostasis, as it facilitates the selective degradation of dysfunctional mitochondria (Palikaras et al., 2018).
pORF5 among the eight plasmid-encoded proteins in C. trachomatis, is the sole secreted protein (Li et al., 2008). Previous studies have confirmed that pORF5 modulates the expression of autophagy-related proteins in host cells (Zou et al., 2018). Lei et al. demonstrated that C. trachomatis pORF5 preserves mitochondrial function in HeLa cells carbonyl treated with cyanide 3-chlorophenylhydrazone (CCCP). Subsequent experimental data indicated following pORF5 treatment, LC3 accumulated around mitochondria, with p62 subsequently recruited to the mitochondria via its binding to LC3, implying that pORF5 safeguards mitochondrial function through the mechanism of mitophagy (Lei et al., 2017).
5 Conclusion and future directions
In summary, C. trachomatis has developed a variety of strategies to exploit host cellular mechanisms, thereby facilitating its own intracellular proliferation Autophagy, the host cell’s intrinsic defense mechanism, presents a dual role for C. trachomatis. On one hand, C. trachomatis can induce autophagy to procure nutrients sustain host cell homeostasis. On the other hand, the fusion of autophagosomes and lysosomes leads to acidification and the activation of degradative enzymes that inhibit the proliferation of C. trachomatis (Table 1). However, the exact interplay between C. trachomatis and autophagy remains to be elucidated.
Coxiella burnetii, an obligate intracellular bacterial pathogen, can actively manipulate the autophagic pathway to establish an intracellular niche known as the Coxiella-containing vacuole (CCV). Accumulating evidence indicates that the functional type IV secretion system (T4SS) of C. burnetii is pivotal for the manipulation of host autophagy. Similar to C. burnetii, Chlamydia survives and reproduces within the inclusions that sequester the pathogens with the cytoplasma and can manipulate host cellular processes through the T3SS-secreted effector proteins. Currently, research on C. trachomatis T3SS-secreted effector proteins is limited; many proposed effectors are yet to be confirmed, and even more remain uncharacterized, with only evidence of secretion predominantly derived from heterologous T3SS expression systems (Rucks, 2023). Genetic approaches have made substantial advancements in C. trachomatis, which should be leveraged to further dissect on autophagy (Bastidas and Valdivia, 2023). Ultimately, a deeper understanding of the molecular pathogenesis of C. trachomatis will be of great help for increasing the diagnostic efficiency and providing insights for the development of novel vaccines and therapeutic interventions.
Author contributions
SZ: Writing – original draft. YJ: Writing – original draft. YY: Writing – original draft. XO: Writing – original draft. DZ: Writing – review & editing. YS: Writing – review & editing. JJ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the State Key Laboratory of Pathogen and Biosecurity (Academy of Military Medical Science) (SKLPBS2217).
Acknowledgments
We apologize to researchers whose work was not cited here due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelsamed, H., Peters, J., Byrne, G. I. (2013). Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol. 8, 1129–1146. doi: 10.2217/fmb.13.80
Al-Younes, H. M., Brinkmann, V., Meyer, T. F. (2004). Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect. Immun. 72, 4751–4762. doi: 10.1128/IAI.72.8.4751-4762.2004
Al-Zeer, M. A., Al-Younes, H. M., Braun, P. R., Zerrahn, J., Meyer, T. F. (2009). IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PloS One 4, e4588. doi: 10.1371/journal.pone.0004588
Al-Zeer, M. A., Al-Younes, H. M., Lauster, D., Lubad, M.A., Meyer, T. F. (2013). Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins 9 (1), 50–62. doi: 10.4161/auto.22482
Bachmann, N. L., Polkinghorne, A., Timms, P. (2014). Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22, 464–472. doi: 10.1016/j.tim.2014.04.013
Bastidas, R. J., Elwell, C. A., Engel, J. N., Valdivia, R. H. (2013). Chlamydial intracellular survival strategies. Cold Spring Harb. Perspect. Med. 3, a010256. doi: 10.1101/cshperspect.a010256
Bastidas, R. J., Valdivia, R. H. (2023). The emerging complexity of Chlamydia trachomatis interactions with host cells as revealed by molecular genetic approaches. Curr. Opin. Microbiol. 74, 102330. doi: 10.1016/j.mib.2023.102330
Betts-Hampikian, H. J., Fields, K. A. (2010). The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front. Microbiol. 1, 114. doi: 10.3389/fmicb.2010.00114
Biasizzo, M., Kopitar-Jerala, N. (2020). Interplay between NLRP3 inflammasome and autophagy. Front. Immunol. 11, 591803. doi: 10.3389/fimmu.2020.591803
Chen, H., Wen, Y., Li, Z. (2019). Clear victory for chlamydia: the subversion of host innate immunity. Front. Microbiol. 10, 1412. doi: 10.3389/fmicb.2019.01412
Chen, L., Huang, Q., Bai, Q., Tong, T., Zhou, Y., Li, Z., et al. (2022). Chlamydia psittaci Induces Autophagy in Human Bronchial Epithelial Cells via PERK and IRE1alpha, but Not ATF6 Pathway. Infect. Immun. 90, e0007922. doi: 10.1128/iai.00079-22
Cheong, H. C., Lee, C. Y. Q., Cheok, Y. Y., Tan, G. M. Y., Looi, C. Y., Wong, W. F. (2019). Chlamydiaceae: diseases in primary hosts and zoonosis. Microorganisms 7 (5), 146. doi: 10.3390/microorganisms7050146
Choy, A., Dancourt, J., Mugo, B., O'Connor, T. J., Isberg, R. R., Melia, T. J., et al. (2012). The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076. doi: 10.1126/science.1227026
Crother, T. R., Porritt, R. A., Dagvadorj, J., Tumurkhuu, G., Slepenkin, A. V., Peterson, E. M., et al. (2019). Autophagy limits inflammasome during chlamydia pneumoniae infection. Front. Immunol. 10, 754. doi: 10.3389/fimmu.2019.00754
Damiani, M. T., Gambarte Tudela, J., Capmany, A. (2014). Targeting eukaryotic Rab proteins: a smart strategy for chlamydial survival and replication. Cell. Microbiol. 16, 1329–1338. doi: 10.1111/cmi.2014.16.issue-9
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 15 (6), 323–337. doi: 10.1038/nrmicro.2017.20
Deretic, V., Saitoh, T., Akira, S. (2013). Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 13 (10), 722–737. doi: 10.1038/nri3532.
Fu, M., Zhang, J., Zhao, M., Zhang, S., Dai, L., Ouyang, X., et al. (2022). Coxiella burnetii plasmid effector B promotes LC3-II accumulation and contributes to bacterial virulence in a SCID mouse model. Infect. Immun. 90, e0001622. doi: 10.1128/iai.00016-22
Gomes, L. C., Dikic, I. (2014). Autophagy in antimicrobial immunity. Mol. Cell 54, 224–233. doi: 10.1016/j.molcel.2014.03.009
Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., Deretic, V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766. doi: 10.1016/j.cell.2004.11.038
Hansen, T. E., Johansen, T. (2011). Following autophagy step by step. BMC Biol. 9, 39. doi: 10.1186/1741-7007-9-39
Heinzen, R. A., Scidmore, M. A., Rockey, D. D., Hackstadt, T. (1996). Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64, 796–809. doi: 10.1128/iai.64.3.796-809.1996
Hepler, R. W., Nahas, D. D., Lucas, B., Kaufhold, R., Flynn, J. A., Galli, J. D., et al. (2018). Spectroscopic analysis of chlamydial major outer membrane protein in support of structure elucidation. Protein Sci. 27, 1923–1941. doi: 10.1002/pro.3501
Jury, B., Fleming, C., Huston, W. M., Luu, L. D. W. (2023). Molecular pathogenesis of Chlamydia trachomatis. Front. Cell Infect. Microbiol. 13, 1281823. doi: 10.3389/fcimb.2023.1281823
Kim, K. H., Lee, M. S. (2014). Autophagy–a key player in cellular and body metabolism. Nat. Rev. Endocrinol. 10, 322–337. doi: 10.1038/nrendo.2014.35
Kim, B. H., Shenoy, A. R., Kumar, P., Das, R., Tiwari, S., MacMicking, J. D. (2011). A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721. doi: 10.1126/science.1201711
Kirkin, V., Rogov, V. V. (2019). A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 76, 268–285. doi: 10.1016/j.molcel.2019.09.005
Lei, W., Li, Q., Su, S., Bu, J., Huang, Q., Li, Z. (2017). Chlamydia trachomatis plasmid-encoded protein pORF5 protects mitochondrial function by inducing mitophagy and increasing HMGB1 expression. Pathog. Dis. 75(9). doi: 10.1093/femspd/ftx111
Li, Z., Chen, D., Zhong, Y., Wang, S., Zhong, G. (2008). The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76, 3415–3428. doi: 10.1128/IAI.01377-07
Liang, S., Bulir, D., Kaushic, C., Mahony, J. (2017). Considerations for the rational design of a Chlamydia vaccine. Hum. Vaccin. Immunother. 13, 831–835. doi: 10.1080/21645515.2016.1252886
McClarty, G. (1994). Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2, 157–164. doi: 10.1016/0966-842X(94)90665-3
Miyairi, I., Mahdi, O. S., Ouellette, S. P., Belland, R. J., Byrne, G. I. (2006). Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J. Infect. Dis. 194, 350–357. doi: 10.1086/505432
Omsland, A., Sager, J., Nair, V., Sturdevant, D. E., Hackstadt, T. (2012). Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. U. S. A 109, 19781–19785. doi: 10.1073/pnas.1212831109
Ouellette, S. P., Dorsey, F. C., Moshiach, S., Cleveland, J. L., Carabeo, R. A. (2011). Chlamydia species-dependent differences in the growth requirement for lysosomes. PloS One 6, e16783. doi: 10.1371/journal.pone.0016783
Pachikara, N., Zhang, H., Pan, Z., Jin, S., Fan, H. (2009). Productive Chlamydia trachomatis lymphogranuloma venereum 434 infection in cells with augmented or inactivated autophagic activities. FEMS Microbiol. Lett. 292, 240–249. doi: 10.1111/fml.2009.292.issue-2
Palikaras, K., Lionaki, E., Tavernarakis, N. (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022. doi: 10.1038/s41556-018-0176-2
Poston, T. B., Gottlieb, S. L., Darville, T. (2019). Status of vaccine research and development of vaccines for Chlamydia trachomatis infection. Vaccine 37, 7289–7294. doi: 10.1016/j.vaccine.2017.01.023
Rucks, E. A. (2023). Type III secretion in chlamydia. Microbiol. Mol. Biol. Rev. 87, e0003423. doi: 10.1128/mmbr.00034-23
Sharma, V., Verma, S., Seranova, E., Sarkar, S., Kumar, D. (2018). Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 6, 147. doi: 10.3389/fcell.2018.00147
Siadous, F. A., Cantet, F., Van Schaik, E., Burette, M., Allombert, J., Lakhani, A., et al. (2020). Coxiella effector protein CvpF subverts RAB26-dependent autophagy to promote vacuole biogenesis and virulence. Autophagy. 17 (3), 706–722. doi: 10.1080/15548627.2020.1728098
Stelzner, K., Vollmuth, N., Rudel, T. (2023). Intracellular lifestyle of Chlamydia trachomatis and host-pathogen interactions. Nat. Rev. Microbiol. 21, 448–462. doi: 10.1038/s41579-023-00860-y
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Sun, H. S., Eng, E. W., Jeganathan, S., Sin, A. T., Patel, P. C., Gracey, E., et al. (2012). Chlamydia trachomatis vacuole maturation in infected macrophages. J. Leukocyte Biol. 92, 815–827. doi: 10.1189/jlb.0711336
Thomas, D. R., Newton, P., Lau, N., Newton, H. J. (2020). Interfering with Autophagy: The Opposing Strategies Deployed by Legionella pneumophila and Coxiella burnetii Effector Proteins. Front. Cell Infect. Microbiol. 10, 599762. doi: 10.3389/fcimb.2020.599762
Vollmuth, N., Schlicker, L., Guo, Y., Hovhannisyan, P., Janaki-Raman, S., Kurmasheva, N., et al. (2022). c-Myc plays a key role in IFN-gamma-induced persistence of Chlamydia trachomatis. Elife 11, e76721. doi: 10.7554/eLife.76721.sa2
Wan, W., Li, D., Li, D., Jiao, J. (2023). Advances in genetic manipulation of Chlamydia trachomatis. Front. Immunol. 14, 1209879. doi: 10.3389/fimmu.2023.1209879
Wang, F., Zhang, L., Lu, X., Zhu, Q., Shi, T., Lu, R., et al. (2019). Inflammatory mechanism of Chlamydia trachomatis-infected HeLa229 cells regulated by Atg5. Biochem. Biophys. Res. Commun. 520, 205–210. doi: 10.1016/j.bbrc.2019.09.132
Wang, F., Zhang, H., Lu, X., Zhu, Q., Shi, T., Lu, R., et al. (2021). Chlamydia trachomatis induces autophagy by p62 in HeLa cell. World J. Microbiol. Biotechnol. 37, 50. doi: 10.1007/s11274-021-03014-5
Webster, S. J., Goodall, J. C. (2018). New concepts in Chlamydia induced inflammasome responses. Microbes Infect. 20, 424–431. doi: 10.1016/j.micinf.2017.11.011
Wen, Y., Luo, F., Zhao, Y., Su, S., Shu, M., Li, Z. (2020). Chlamydia trachomatis plasmid-encoded protein pORF5 activates unfolded protein response to induce autophagy via MAPK/ERK signaling pathway. Biochem. Biophys. Res. Commun. 527, 805–810. doi: 10.1016/j.bbrc.2020.04.117
Witkin, S. S., Minis, E., Athanasiou, A., Leizer, J., Linhares, I. M. (2017). Chlamydia trachomatis: the persistent pathogen. Clin. Vaccine Immunol. 24 (10), e00203-17. doi: 10.1128/CVI.00203-17
Woodhall, S. C., Gorwitz, R. J., Migchelsen, S. J., Gottlieb, S. L., Horner, P. J., Geisler, W. M., et al. (2018). Advancing the public health applications of Chlamydia trachomatis serology. Lancet Infect. Dis. 18, e399–e407. doi: 10.1016/S1473-3099(18)30159-2
Yasir, M., Pachikara, N. D., Bao, X., Pan, Z., Fan, H. (2011). Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect. Immun. 79, 4019–4028. doi: 10.1128/IAI.05308-11
Yim, W. W., Mizushima, N. (2020). Lysosome biology in autophagy. Cell Discovery 6, 6. doi: 10.1038/s41421-020-0141-7
Yu, L., Chen, Y., Tooze, S. A. (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 14 (2), 207–215. doi: 10.1080/15548627.2017.1378838
Zhong, G. (2017). Chlamydial plasmid-dependent pathogenicity. Trends Microbiol. 25, 141–152. doi: 10.1016/j.tim.2016.09.006
Keywords: Chlamydia trachomatis, inclusion, autophagy, mitophagy, LC3
Citation: Zhang S, Jiang Y, Yu Y, Ouyang X, Zhou D, Song Y and Jiao J (2024) Autophagy: the misty lands of Chlamydia trachomatis infection. Front. Cell. Infect. Microbiol. 14:1442995. doi: 10.3389/fcimb.2024.1442995
Received: 03 June 2024; Accepted: 21 August 2024;
Published: 06 September 2024.
Edited by:
Hua Niu, Affiliated Hospital of Guilin Medical University, ChinaReviewed by:
Kun Liu, Air Force Medical University, ChinaNicolas Jacquier, Centre Hospitalier Universitaire Vaudois (CHUV), Switzerland
Copyright © 2024 Zhang, Jiang, Yu, Ouyang, Zhou, Song and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Jiao, amlhb2p1bjUxOTIwQHNpbmEuY29t; Yajun Song, c29uZ3lqQGJtaS5hYy5jbg==
 Shan Zhang
Shan Zhang Yufei Jiang
Yufei Jiang Yonghui Yu
Yonghui Yu Xuan Ouyang
Xuan Ouyang Dongsheng Zhou
Dongsheng Zhou Yajun Song
Yajun Song Jun Jiao
Jun Jiao