- 1Institute of Clinical Medicine, Jiangxi Provincial People’s Hospital, The First Affiliated Hospital of Nanchang Medical College, Nanchang, China
- 2Jiangxi Key Laboratory for Excavation and Utilization of Agricultural Microorganisms, Jiangxi Agricultural University, Nanchang, China
Introduction: Candida auris, known as the "super fungus", is commonly existed in hospital. The treatment of C. auris infection is difficult for its multi-drug resistance and difficult to accurately detect. The use of synthetic antibacterial agents has caused major problems such as drug-resistance and environment pollution and negatively affects non-target species. Microbial biocontrol agents (probiotics) are needed for fungal infection. Bacillus and related genera produce a variety of bioactive substances. As probiotics, it has been widely studied in the field of medicine and is a novel microbial factor for biological control.
Methods: B. velezensis NC-B4 was isolated using gradient dilution method. Then it was identified by phylogenetic analysis and physiological and biochemical characteristics. The antibacterial mechanism of NC-B4 was explored by detecting cellulase, protease and genomic analysis. Then antimicrobial effects were analyzed by detecting the growth and biofilm of C. auris BJCA001. Finally, the cytotoxicity and the protective effect on mice were analyzed by cell line and mouse systemic infection models.
Results: We isolated B. velezensis NC-B4, which showed cellulase, protease enzyme activity and antimicrobial effects against human pathogenic fungi by inhibiting the growth of Candida auris, Cryptococcus neoformans, Candida albicans and mycelial fungus. B. velezensis NC-B4 inhibited the biofilm formation and growth of C. auris. B. velezensis NC-B4 has a protective effect against the toxicity of Candida auris in A549 cell line and mouse systemic infection models. The complete genome of B. velezensis NC-B4 was 3.93 Mb with a 46.5% G+C content and possessed the macrolactin H, bacillaene, fengycin, difficidin, bacillibactin and bacilysin biosynthesis cluster, which known as key factors in biological control.
Discussion: The results of the present study indicated that B. velezensis NC-B4 has antimicrobial properties for its cellulase, protease and antibacterial secondary metabolites, thereby inhibiting the growth of pathogenic bacteria and the formation of biofilms. B. velezensis NC-B4 is expected to be developed as a source for probiotics or new antibiotics.
Introduction
Candida auris, an invasive fungal pathogen commonly colonized in skin, the respiratory tract, and urinary tract, has emerged globally as a multidrug-resistant fungal pathogen (Abdolrasouli et al., 2017; Welsh et al., 2017). There are six distinct clades of C. auris based on genetic and genomic information and locations of first isolates: Clade I (South Asian), Clade II (East Asian), Clade III (South African), Clade IV (South American), Clade V (Iran), and Clade VI (Singapore) (Bhargava, 2025). Ninety percent of clinical isolates of C. auris are resistant to fluconazole, and their sensitivity to other azole antifungal drugs, 5-fluorocytosine, amphotericin B, and echinocandins is also changeable, which often leads to blood infection with high mortality (Chowdhary et al., 2018). At present, antifungal drugs are still the main measures to treat C. auris infection, but the problems of drug resistance and environmental pollution caused by long-term use of antifungal drugs have attracted more and more attention (Osei Sekyere, 2018). Therefore, it is urgent to search for new antifungal methods to solve the drug resistance problem.
Bacillus is the most abundant group of bacteria in the rhizosphere of plants (Choudhary and Johri, 2009; Shao et al., 2022). The bioactive substances produced by Bacillus are harmless to livestock and poultry and can kill bacteria, some fungi, parasites, some viruses, and tumor cells, including drug-resistant strains, and are widely used in industry, agriculture, and medical production (Elshaghabee et al., 2017; Zalila-Kolsi et al., 2023). In addition, the microecological preparation prepared by Bacillus has played an important role in the treatment of intestinal flora imbalance, candida infection, and prevention of wound surface infection (Garvey et al., 2022; Zou et al., 2022; Ramesh and Roy, 2023; Kizhakkekalam, 2022). As a probiotic, it has been widely studied in the medical field and is an ideal new biological control microbial factor. For example, Bacillus licheniformis can inhibit Staphylococcus, Candida albicans, yeast, and Escherichia coli, and was made into capsules and oral liquids with living strains to treat intestinal diseases (Ramirez-Olea et al., 2022); the combination of intestinal ecological preparation of Bifidobacterium and Bacillus licheniformis and chemotherapy drugs cannot only kill and promote apoptosis of H22 ascites cancer cells but also prolong the life cycle of tumor mice and improve the effect of chemotherapy, which laid the foundation for clinical trials (Hirozawa et al., 2023).
There are some reports on the study of B. velezensis as a biological control microbial factor, the possible mechanisms of B. velezensis exert the antifungal effects are as follows: There are many genes responsible for the biosynthesis of antifungal compounds; it was reported that B. velezensis KTA01 can produce lipopeptide, which displayed prominent antifungal activity against B. dothidea KACC45481 (Kang et al., 2024). The research on the mechanism of B. velezensis HeN-7 CFS antifungal action demonstrated that HeN-7 CFS induced the membrane lipid peroxidation in B. sorokiniana, leading to the disruption of cell membrane integrity and resulting in the leakage of cell contents (Lin et al., 2024). B. velezensis CFS may inhibit C. fioriniae through interference with ribosomes, genetic information processing, cell membrane metabolism, and energy metabolism (Fu et al., 2024).
In this study, we aimed to screen isolates and identify functional characteristics of B. velezensis NC-B4 for developing biological control agents against C. auris, which is called super fungus. The antifungal effect of B. velezensis NC-B4 was detected against yeast and filamentous human pathogenic fungus by measuring the antifungal zone, then the protection of B. velezensis NC-B4 on mouse systemic infection with C. auris was detected by measuring the fungal burden [colony-forming unit (CFU)] in each organ after C. auris infection. We provide an understanding of the antibacterial mechanism of action of B. velezensis NC-B4 by detecting extracellular enzyme activity such as cellulase and protease and analyzing the secondary metabolite genes. We also evaluated the toxicity of B. velezensis NC-B4 by detecting the killing of NC-B4 on A549 cells. Based on the effective properties of NC-B4 for its antifungal action, we propose NC-B4 be suggested as a useful biological control agent (probiotic) for the medical and health industry.
Materials and methods
Strains and culture conditions
All the strains used in this study are listed in Table 1. Bacterial strains were grown at 35°C with shaking at 200 rpm in Lurai-Bertani broth (10 g tryptone, 5 g yeast extract, and 10 g NaCl in 1 L, pH 7.0). Fungi were grown at 28°C in Yeast Extract Peptone Dextrose Medium (10 g yeast extract, 20 g peptone, 20 g dextrose, and 20 g agar in 1 L). Candida auris BJCA001, Cryptococcus neoformans H99, Candida albicans SC5314, and four mycelial fungi were obtained from our laboratory. Bacterial growth was determined by measuring optical density at a wavelength of 600 nm.
Isolation, screening, and identification of bacterial strains
B. velezensis NC-B4 was collected from plant rhizosphere soil (Nanchang, China) and isolated using the gradient dilution method. In detail, plant rhizosphere soil was collected from a hospital, park, and mountain and prepared into a 10% soil suspension (10−1), then the soil suspension was diluted by a 10-fold gradient (10−2, 10−3, 10−4, 10−5). Soil suspension (100 µl) of each concentration was spread on the LB plates, and the plates were incubated at 35°C until single colonies grew. Then pick up and inoculate the colonies on the YPD plates that contain C. auris BJCA001 (108 CFU/ml). Finally, we selected the single colony that produced the inhibition zone for further identification. The detection method of physiological and biochemical characteristics refers to Burgey’s Manual of Determinative Bacteriology or the instruction of the kit (Hopebio, HBIG14). Molecular identification was performed by using primers (27F/1492R: AGAGTTTGATCCTGGCTCAG/AGAGTTTGATCCTGGCTCAG; gyrA-F/gyrA-R: CAGTCAGGAAATGCGTACGTCCTT/CAAGGTAATGCTCCAGGCATTGCT; rpoB-F/rpoB-R: AGGTCAACTAGTTCAGTATGGAC/AAGAACCGTAACCGGCAACTT) to amplify and sequence the fragments of 16S rRNA, gyrA, and rpoB, respectively (Lu et al., 2021).
Phylogenetic and statistical analysis
For phylogenetic analysis, 16S rDNA, gyrB, and rpoB sequences closely related to our sequences were retrieved from GenBank based on BLAST results from the National Center for Biotechnology Information. Maximum likelihood (ML) phylogenies were constructed using the ML method in IQTREE v1.6.12 (http://iqtree.cibiv.univie.ac.at/). A bootstrap based on 1,000 replicates was analyzed, the confidence of the nodes was evaluated, and all parameters were kept at the default setting (Nguyen et al., 2015). The trees were visualized using FigureTree v1.4.3 and Adobe Illustrator CC 2018.
Enzymatic activity analysis
Cellulase activity was evaluated by cellulase detection plate and DNS (di-nitrosalicylic acid) colorimetry methods. Cellulase detection plate method references (Shen et al., 2020) with minor changes: overnight culture was diluted to an OD600 of 0.01, and 2 µl bacterial suspensions were plotted in cellulase detection medium, then the plates were incubated at 35°C. After 48h, the plates were stained with 0.5% Congo red for 30 min and incubated with 1 M NaCl solution for 10 min at room temperature. Finally, the plates were washed three times by water, and cellulase activity in the plates was assessed by measuring the diameter of the degradation circle. Each treatment was replicated at least three times. The detailed steps of DNS colorimetry are as follows: B. velezensis NC-B4 isolate was grown in LB broth medium for 24h at 37 ± 2°C and then centrifuged at 13,000 rpm for 5 min. 0.5 ml of the supernatant (enzyme solution) was mixed with 1.5 ml of CMC-Na solubilized in phosphate buffer (1%) and incubated at 40°C for 30 min. 1.5 ml of dinitrosalicylic (DNS) acid reagent was added, and the mixture was boiled for 5 min; then cooled down and chilled to 25 ml, and the absorbancy was measured at 520 nm. One unit of enzyme activity was defined as 1 µmol glucose formed per minute.
Protease activity was evaluated by milk plate. In brief, we prepared an LB plate with 5% milk, then added NC-B4 fermentation broth supernatant (OD600 of 2.0, 3.0, 4.0) and incubated at 37°C for 24h. Protease activity was assessed by measuring the diameter of the degradation circle. Another method for analyzing the protease activity was determined by following previously published methods (Wang et al., 2021).
Antifungal analysis
The antifungal ability of B. velezensis NC-B4 was evaluated using the disk diffusion method against several yeasts and filamentous fungi from clinical isolation. For yeast fungi, we prepared the YPD plate with C. auris BJCA001, and 10 µl of 1 × 106 CFUs/ml of the culture suspension was distributed into the hole. After culturing at 35°C for 24h, the diameter of the inhibition zone was measured. For filamentous fungi, a pathogenic agar block was prepared and placed in three corners of the plate, and 10 µl of 1 × 106 CFUs/ml of the culture suspension was distributed in the center. Then culturing at 28°C for 3–5 days, the antifungal activity of NC-B4 was assessed by determining the radial mycelial growth of the fungal pathogen.
Cell growth analysis
For this assay, we firstly prepared NC-B4 fermentation broth supernatant (OD600 of 3.0), which was filtered by a 0.22 µm membrane. C. auris BJCA001 cells were incubated overnight (OD600 of 2.0) and then diluted 1,000 times using YPD medium. Then, 50 µl of diluted cell suspension containing NC-B4 fermentation broth supernatant (5, 10, 20 µl) were added to the 96-well plate in triplicate at 35°C for 2 days, then supplemented with YPD medium to 100 µl. We measured OD600 every six hours then plotted the growth curve.
Biofilm formation assays
Biofilm formation was tested by determining the ability of fungal cells to adhere to the wells of 96-well polypropylene microtiter dishes. C. auris was grown overnight at 35°C and diluted to 1,000 times by using minimal medium (2 g glycerin, 2g mannitol, 10.5 g K2HPO4, 4.5 g KH2PO4, 2 g (NH4)2SO4, 0.2 g MgSO4·7H2O, 0.005 g FeSO4, 0.01 g CaCl2, 0.002 g MnCl2 in 1 L). Then C. auris suspension [with 0, 5, and 10 µl NC-B4 fermentation broth supernatant (OD600 of 3.0)] was added to 96-well polypropylene microtiter plates (100 μl per well) and incubated at 35°C with shaking at 200 rpm for 18h. To remove planktonic cells, we discarded the supernatant and washed twice and stained for 20 min with 1% (wt/vol) crystal violet. Then washed using water, added 200 μl ethanol to the well, and measured the absorbance at 595 nm (Huber et al., 2001).
Cytotoxicity assays
Cytotoxicity was assessed by measuring the release of lactate dehydrogenase (LDH) from A549 cells. The 1 × 104 A549 cells were routinely grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% fetal bovine serum (FBS) in a 96-well plate before infection. C. auris BJCA001 strain was grown in YPD medium at 35°C, then centrifuged and resuspended in DMEM medium (diluted to OD600 = 1), and different concentrations of NC-B4 fermentation broth supernatant (10%, 20%, and 40%) were added. A549 cells were infected with fungi or fungi with NC-B4 fermentation broth supernatant at 109 CFU/ml for 8h. After the 8h incubation, culture supernatants were collected, and LDH in the supernatant was measured following the instruction of the LDH Cytotoxicity Assay Kit (Beyotime Biotechnology, China, C0016). Finally, the cytotoxicity was calculated relative to that of the uninfected control (Yang et al., 2017).
Mouse systemic infection models analysis
All the animal experiments were approved by the Ethics Committee at the Jiangxi Provincial People’s Hospital (approval number KT2023-012). Male BALB/c mice (20–22 g) were used for fungal burden assays; five mice were used for each treated group [phosphate buffered saline (PBS) control, C. auris BJCA001, C. auris BJCA001 + 50% NC-B4 fermentation broth], and 2.5 × 107 cells of BJCA001 in 250 µl PBS were injected into a mouse via tail vein. Mice were humanely killed at 48h after injection. Different organ tissues (liver, kidney, spleen, lung, and brain) of each infected mouse were removed, weighed, homogenized, and diluted in PBS for CFU calculation on YPD medium (Du et al., 2012; Xie et al., 2013).
Whole genome sequencing and analysis
B. velezensis NC-B4 cells were incubated overnight and collected. Genomic DNA was extracted using a bacterial genome extraction kit (Beyotime Biotechnology, China, D0091) according to the manufacturer’s instructions. Whole-genome sequencing was performed using a combination of Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) and Nanopore PromethION platforms. For short reads sequencing on the NovaSeq 6000 platform, a small fragment library was prepared using the VAHTS® Universal Plus DNA Library Prep Kit for MGI V2/for Illumina V2 (Vazyme, China) with an average insertion size of 300 bp. For long-read sequencing, the libraries were prepared using the SQK-LSK110 ligation kit and using the standard protocol.
The assembly of the genome was performed with Unicycler software (0.5.0). Then, Prodigal (v2.6.3), Aragorn (v1.2.38), RNAmmer (v1.2), and Infernal (v1.1) were used for predicting the coding genes, tRNA, rRNA, and mRNA genes, respectively. BLAST software was used for function annotations of genes against Cluster of Orthologous Groups of proteins (COG, http://blast.ncbi.nlm.nih.gov/Blast.cgi), and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) databases.
Results
Screening and identification of bacterial strains
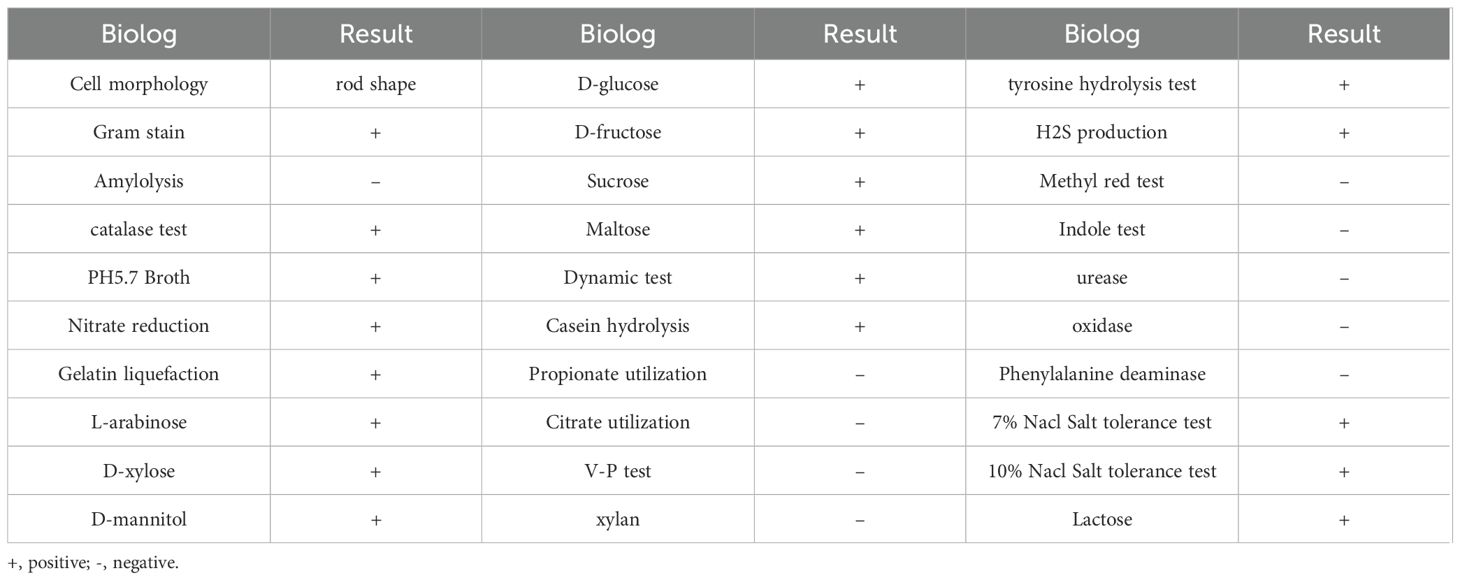
To study the function of microbial biocontrol agents (probiotics) in fungal infection, we collected soil and prepared the soil suspension, then coated the plates. We selected the single clone and inoculated it on a plate that contained C. auris BJCA001. Then, screened strains that can inhibit the growth of C. auris were marked with a red arrow (Figure 1A). Then, we selected the number 4 strain with good antifungal effect; it could form round, milky white, opaque colonies, with dry and wrinkled surfaces, irregular edges, and sunken center on LB agar (Figure 1B). The cells were rod-shaped, single or paired, the spore is nearly round and proximal, and the sporocysts are enlarged (Figure 1C). It was identified as B. velezensis based on physiological and biochemical characteristics (Table 2) and molecular characteristics, including 16S rRNA, gyrA, and rpoB gene sequences, so we named it Nan chang B. velezensis 4(NC-B4). In order to further confirm its classification status, an ML phylogenetic tree was established based on the concatenation of multiple sequences (16S rRNA, gyrA, and rpoB). In the phylogenetic tree, the NC-B4 and isolates of B. velezensis clustered together with 100% bootstrap support (Figure 1D). Therefore, the NC-B4 was identified as B. velezensis.
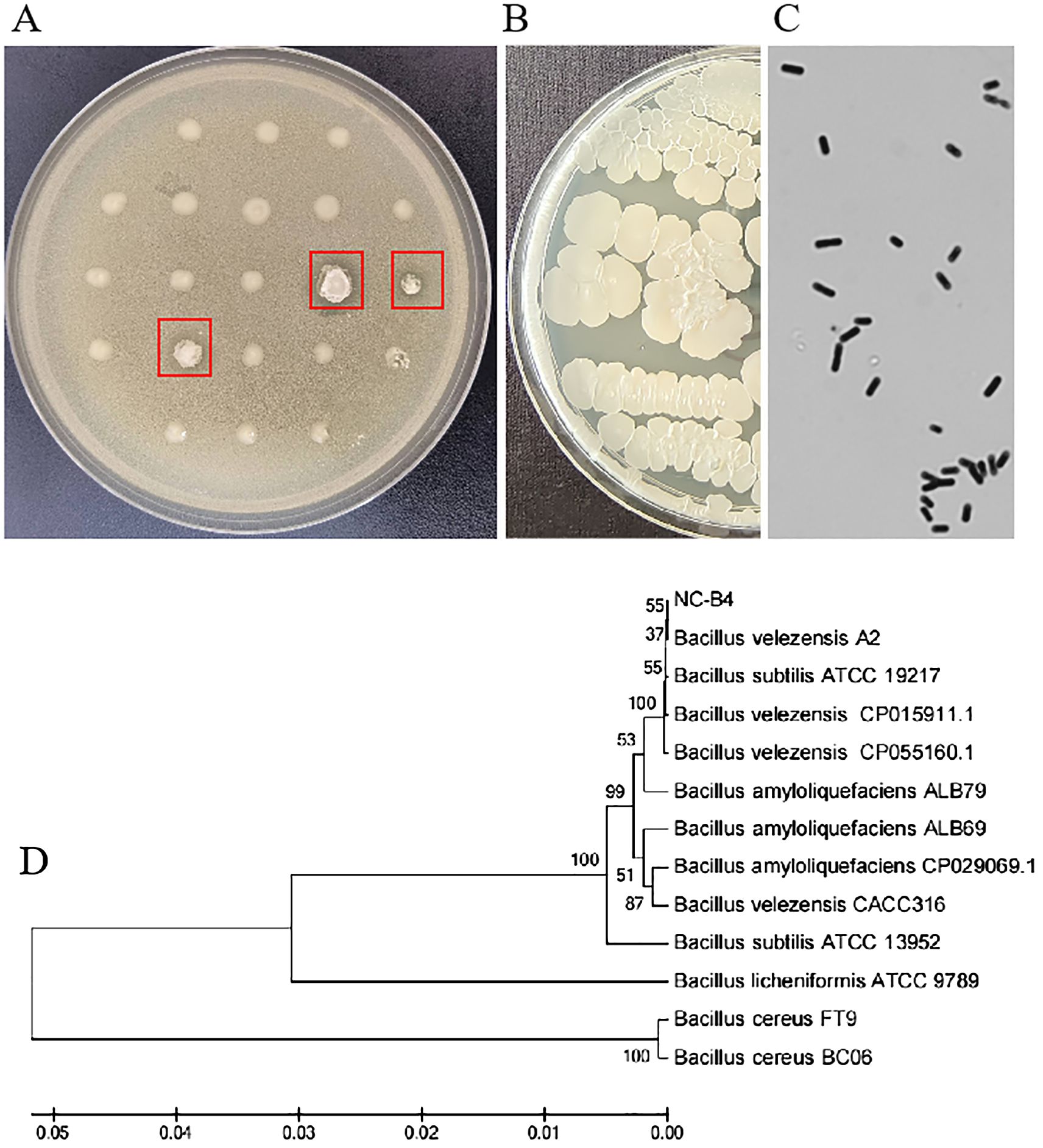
Figure 1. Colony morphology and phylogenetic tree. (A) Preliminary screening picture, strains which can inhibit the growth of C. auris were marked with a red arrow. (B) B. velezensis NC-B4 was inoculated on LB plate at 35°C for 24h, it could form round, milky white, opaque, with dry and wrinkled surface, irregular edge, and sunken center colonies. (C) Observation under microscope, the cells were rod-shaped, single or paired, the spore is nearly round, proximal, and the sporocystis are enlarged. (D) The phylogenetic tree, a maximum likelihood phylogenetic tree was established based on the concatenation of multiple sequences (16S rRNA, gyrA, and rpoB). In the phylogenetic tree, the NC-B4 and isolates of B velezensis clustered together with 100% bootstrap support.
Enzyme activity of B. velezensis NC-B4
To understand the antifungal mechanism of NC-B4, the extracellular enzyme activity of NC-B4 in different growth stages was detected by the plate method and the absorbance method. As shown in Figure 2A, NC-B4 has an obvious protease activity compared with clear LB medium, and the activity of protease increased with the increase of cell concentration and became stable after OD 600 reached 3.0. Another method (5% milk plates) reached the same conclusion (Figure 2B). Similarly, the measured cellulose activity of NC-B4 fermentation broth was 2.7U, 6.8U, and 7.8U at OD 600 2.0, 3.0, and 4.0, respectively, which showed a higher cellulose activity (Figure 2C). It also showed the same result by the cellulase detection plate method (Figure 2D).
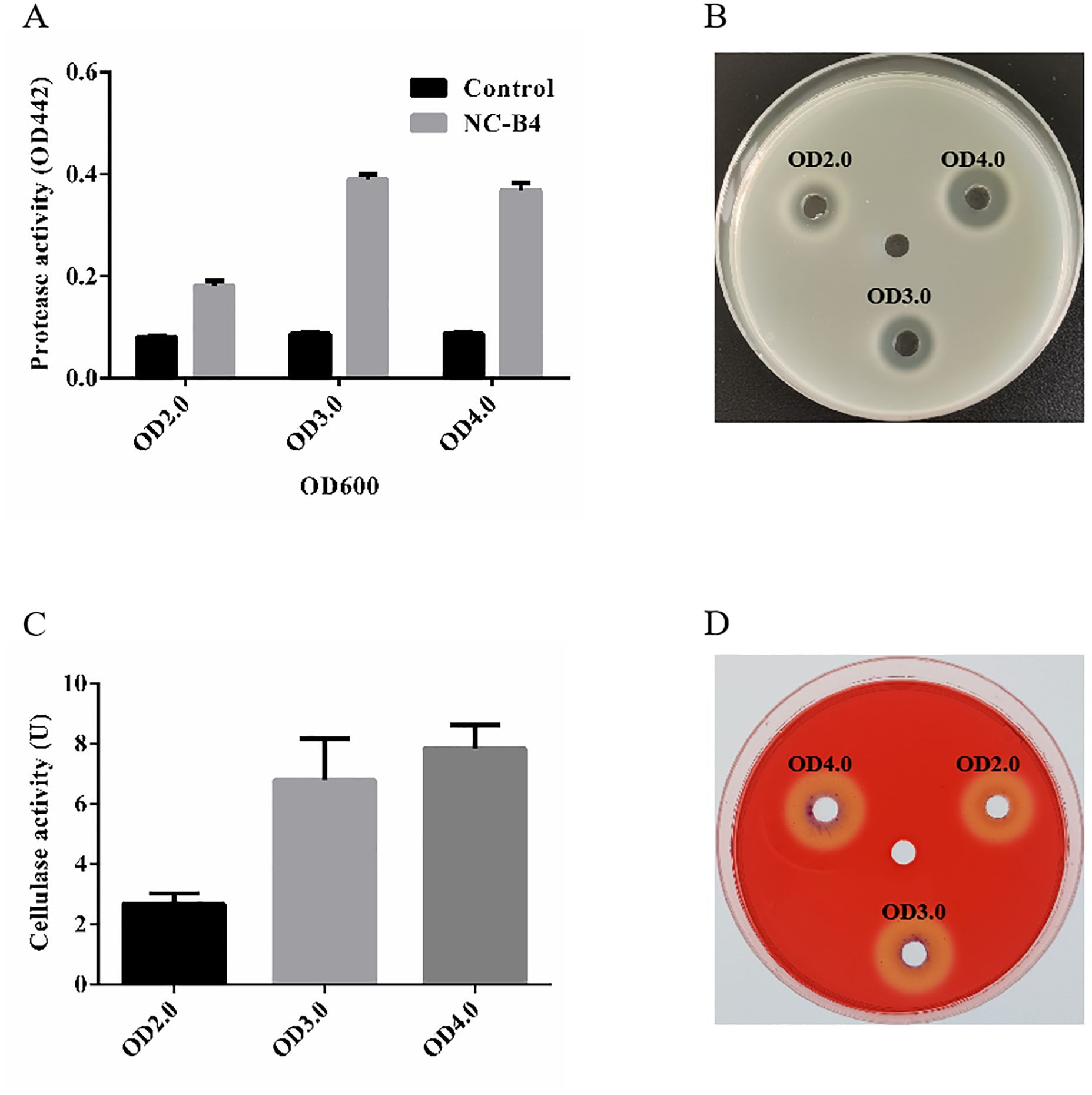
Figure 2. Enzyme activity of B. velezensis NC-B4. The protease (A, B) and cellulase (C, D) activities in fermentation broth were detected at different OD values (2.0, 3.0, and 4.0). (A) OD442 represents the strength of protease activity, NC-B4 has an obvious protease activity compared with clear LB medium (control) at different OD values. (B) NC-B4 fermentation broth can form a decomposition ring on the 5% milk plate, the larger the degradation ring, the stronger the protease activity. The cellulose activity of NC-B4 fermentation broth at OD2.0, 3.0, and 4.0. by absorbance method (C) and cellulase detection plate method (D), the higher the enzyme activity value (U) and the larger the degradation circle are, and the stronger of cellulase activity will be. Each experiment was repeated three times.
In-vitro antifungal effects against human pathogenic fungi
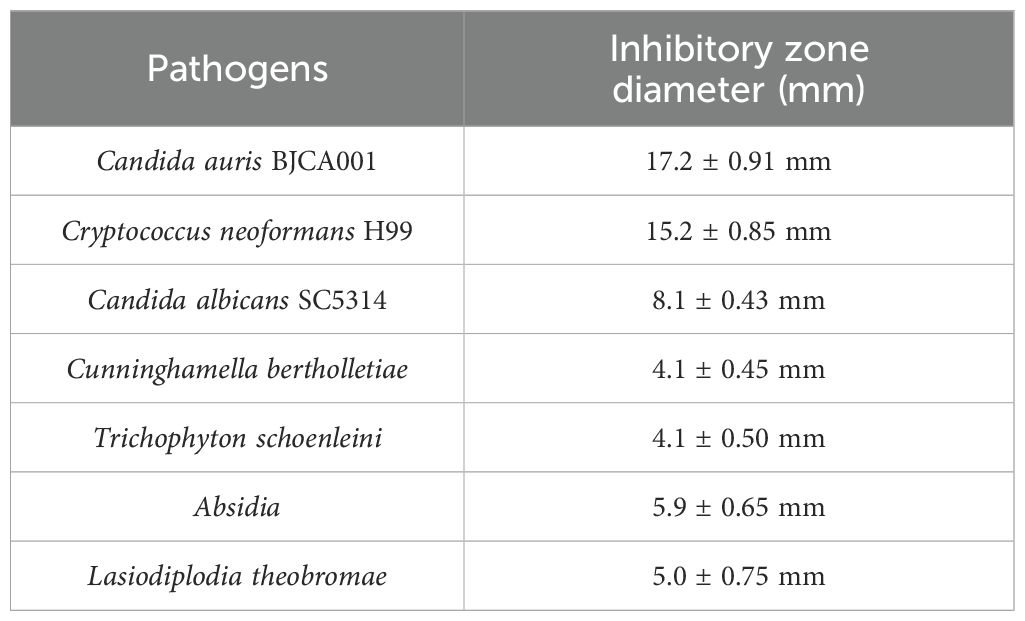
In order to detect whether B. velezensis NC-B4 has an antagonistic effect on other human pathogenic fungi, we selected yeast and filamentous fungi for the antagonistic activity assay. As shown in Figure 3, NC-B4 inhibited the growth of all seven human pathogenic fungi. However, the antagonistic effects on yeast and filamentous fungi were significantly different. NC-B4 strongly inhibited yeast fungi growth of C. auris BJCA001, Cryptococcus neoformans H99, and Candida albicans SC5314, while weakly inhibiting mycelial growth of Lasiodiplodia theobromae, Absidia, Cunninghamella bertholletiae, and Trichophyton schoenleini. The detailed antagonistic effect of strain NC-B4 on seven human pathogenic fungi was listed in Table 3; the diameter (mm) of the inhibitory zone or inhibition rate(%)represents antagonistic effects.
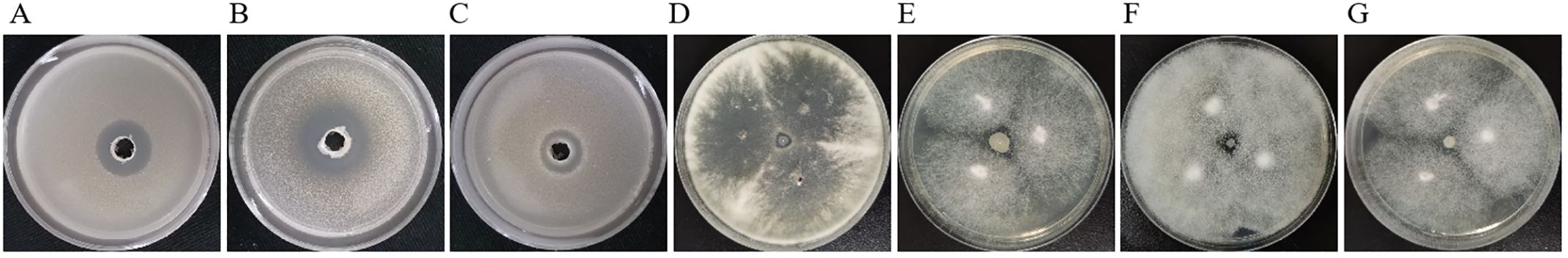
Figure 3. Antifungal activity of test strains against human pathogenic fungi. Inhibition effect of C. auris BJCA001 (A), C. neoformans H99 (B), C. albicans SC5314 (C), L. theobromae (D), Absidia (E), C. bertholletiae (F), T. schoenleini (G). Each experiment was repeated three times. The diameter of inhibitory zone represents antagonistic effects, NC-B4 strongly inhibited yeast fungi growth of C. auris BJCA001, Cryptococcus neoformans H99, Candida albicans SC5314, while weakly inhibited mycelial growth of Lasiodiplodia theobromae, Absidia, Cunninghamella bertholletiae, and Trichophyton schoenleini.
B. velezensis NC-B4 inhibited the biofilm formation and growth of C. auris
Biofilm is an important virulence factor, which is related to antibiotic resistance, escape of microbes from the body’s immune system, recalcitrant infections, and biofilm-associated deaths. So we evaluated the capacity of B. velezensis NC-B4 to inhibit the biofilm formation of C. auris. We obtained supernatant of NC-B4 fermentation broth then tested the effects on the growth curve and biofilm formation of C. auris BJCA001. We found NC-B4 fermentation broth can inhibit the growth of C. auris when added 5, 10, and 20 µl of the supernatant in 100 µl, respectively (Figure 4A). The biofilm production reduced to 67% and 33% when 5 and 10 µl of the supernatant were to 100 µl, respectively (Figure 4B).
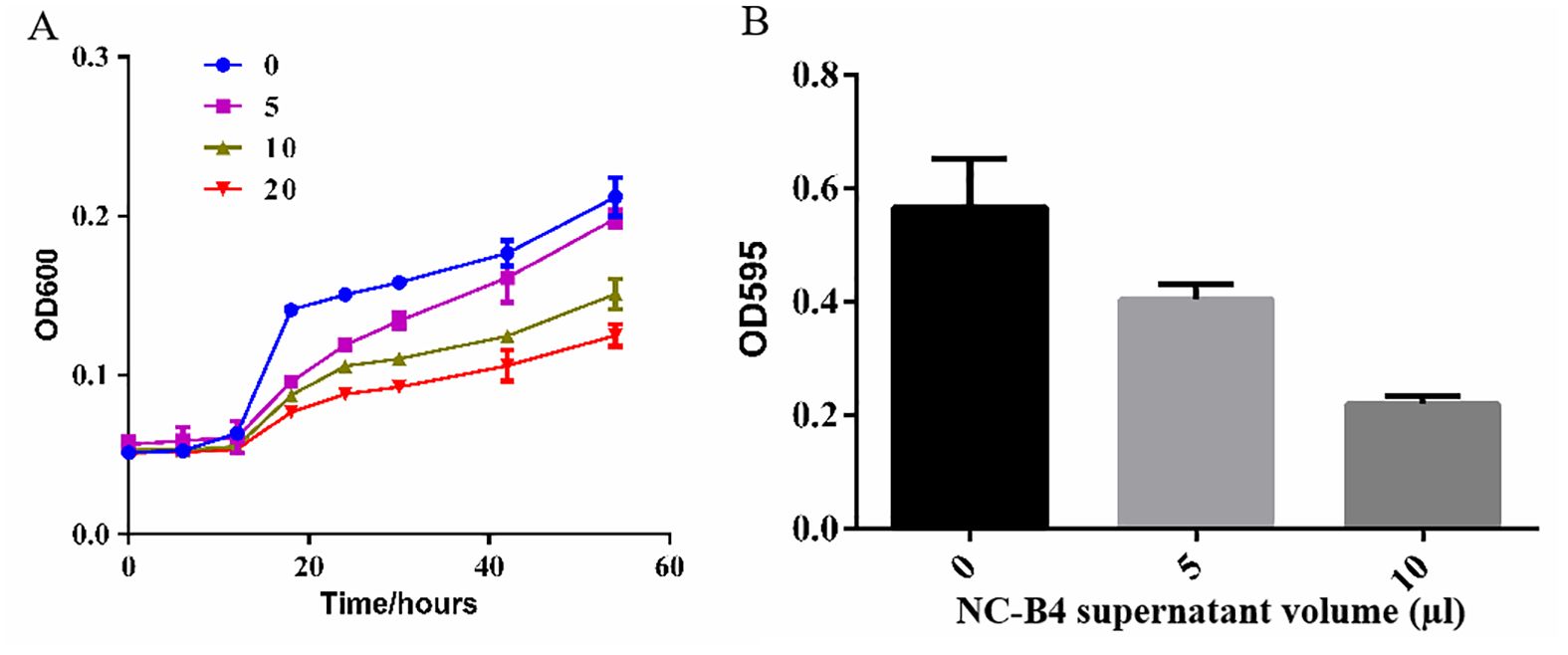
Figure 4. The supernatant B. velezensis NC-B4 controls the phenotypes in C. auris. Growth curve (A), OD600 represents the density of strain, NC-B4 fermentation broth can inhabit the growth of C. auris when added 5, 10, and 20 µl of the supernatant in 100 µl at different cultivation periods, respectively; effect on C. auris biofilm formation (B), OD595 represents the biofilm production, it was reduced when added 5, 10 µl of the supernatant in 100 µl, respectively. The data shown are the mean of three replicates, and error bars indicate the standard deviation. The experiment was repeated three times.
The effects of B. velezensis NC-B4 fermentation broth on the pathogenicity of C. auris
To detect the effects of fermentation broth on the cytotoxicity of C. auris, we measured the cytotoxicity by quantifying the release of LDH into the supernatant of a human cell line, A549. The result showed that the NC-B4 pellet (labeled P in Figure 5A) and supernatant (labeled B in Figure 5A) did not exhibit toxicity when added at 40 µl/100 µl; however exogenous addition of the NC-B4 supernatant at 10, 20, and 40 µl/100 µl (10%, 20%, and 40% B) reduced C. auris virulence by 33%, 70%, and 90%, respectively (Figure 5A). NC-B4 supernatant also reduced C. auris infection in the mouse systemic infection models. Measurement of the fungus CFUs in different tissues of the mouse revealed that the addition of 125 µl/250 µl of NC-B4 supernatant decreased the fungal burden (CFU) in the spleen and brain, while there was no significant difference in the kidney, lung, and liver (Figure 5B).
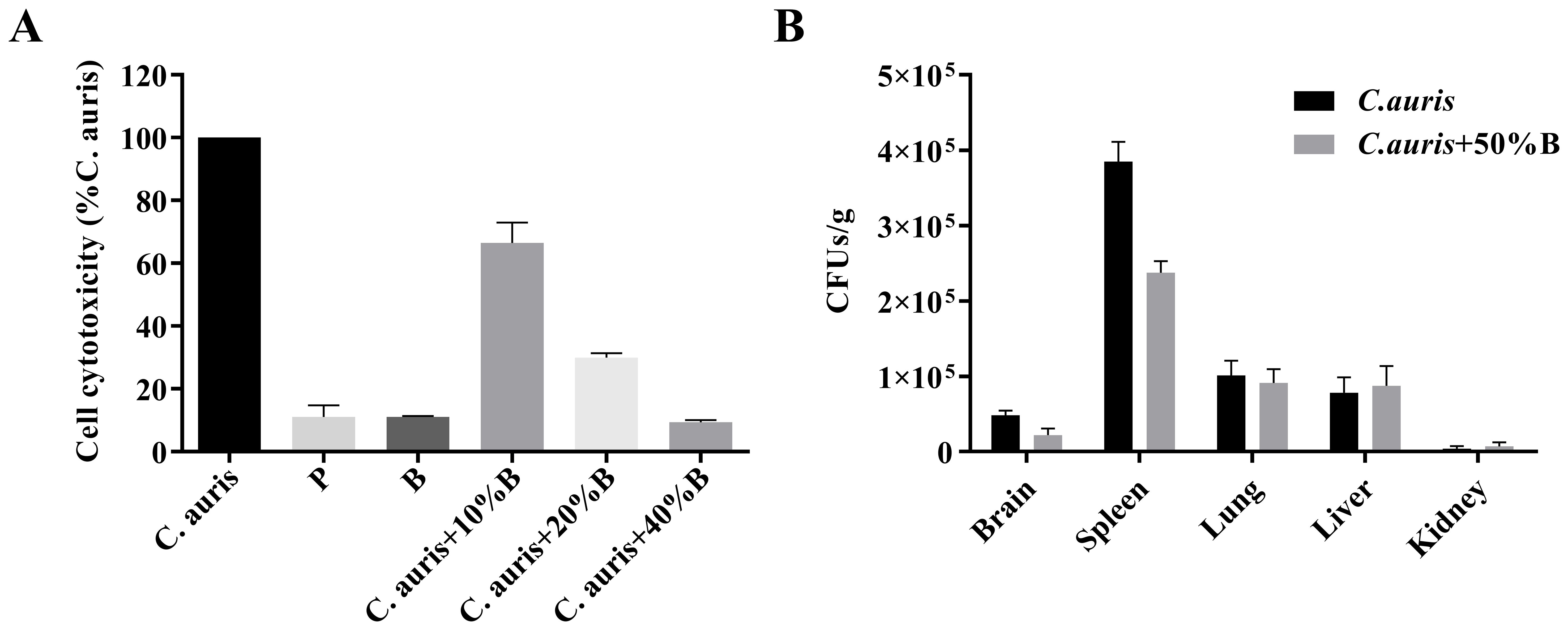
Figure 5. The effects of B. velezensis NC-B4 extraction on the pathogenicity of C. auris were determined using the A549 cell line (A) and mouse model (B). They were repeated three times. (A) Cell cytotoxicity was detected and measured as LDH release. P and B represent adding 40 µl of NC-B4 pellet (1 × 109 CFU/ml) and NC-B4 supernatant. LDH released by C. auris BJCA001 was arbitrarily defined as 100% and used to normalize the LDH release ratios of B. velezensis NC-B4 fermentation broth and the BJCA001 with different gradient fermentation broth. The data shown are the mean of three replicates, and error bars indicate the standard deviations. (B) Measurement of the fungus colony-forming units (CFUs) in different tissues of mouse after 48h of infection, it revealed that the addition of 125 µl/250 µl of NC-B4 supernatant (50% B) decreased the fungal burden (CFU) in spleen and brain, while there are no significant difference in kidney, lung, and liver.
Genome feature of B. velezensis NC-B4
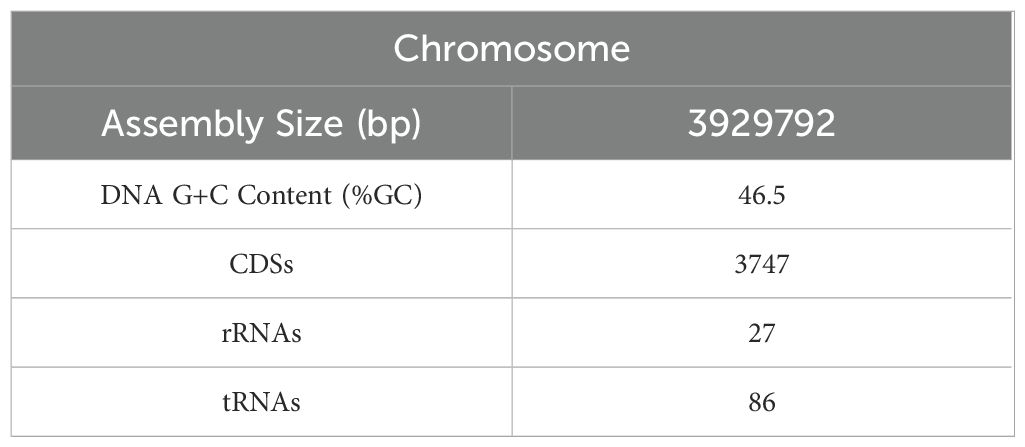
The whole genome of NC-B4 contains a 3,929,792 base pair circular chromosome with a 46.5% G+C content. The genome contains 3,747 protein-coding sequences (CDSs), 27 rRNAs, and 86 tRNAs (Figure 6A and Table 4). Among these coding sequences, 3,618 genes were distributed to 23 orthologous clusters (Figure 6C). More than 250 genes were classified into functional categories for general function prediction only (n = 358), amino acid transport and metabolism (n = 349), transcription (n = 318), and carbohydrate transport and metabolism (n = 293) (Figure 6C). To assess the safety of NC-B4, we analyzed the virulence factors in the genome data of the strain NC-B4. Compared with the virulence factors database (VFDB), 71 genes with pident and Qcovs values greater than 50 were obtained and classed. Most of them belong to metabolism-related enzymes; some of them are transporters, regulators, or motility-related proteins; and there are no exotoxin-related genes (Figure 6B and Table 5). The gene clusters related to secondary metabolite clusters identified in the genome of B. velezensis NC-B4 are listed in Table 6. In detail, NC-B4 possesses six metabolite clusters, five of them have 100% similarity, which are conserved in all B. velezensis members; the remaining one has 82% similarity. This group of six metabolite clusters comprises macrolactin H, bacillaene, fengycin, difficidin, and bacillibactin gene clusters, encoding the antibacterial or antifungal bioactivity (Table 6). The whole genome sequences of the NC-B4 isolate have been deposited in GenBank (Bioproject number: PRJNA995027).
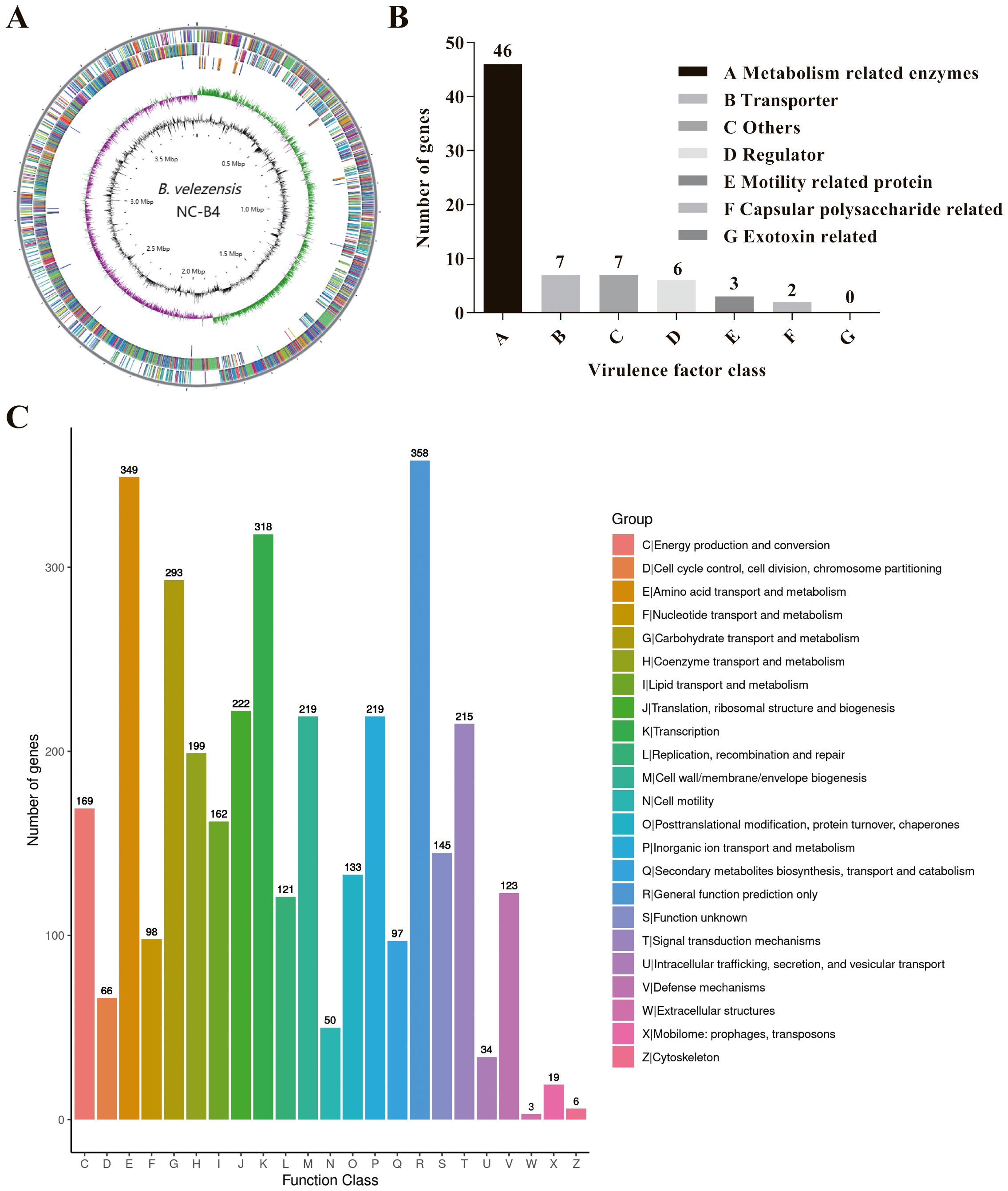
Figure 6. Genome Feature of B. velezensis NC-B4. Circular genome maps of B. velezensis NC-B4 chromosome (A). Circles from the outside to the center denote rRNA and tRNA gene, reverse strand coding sequence, forward strand coding sequence, GC skew, and GC content. Genome number of virulence factor clusters of orthologous groups category (B). Compared with virulence Factors Database (VFDB), 71 genes with pident and Qcovs values greater than 50 were obtained and classed. Most of them belong to metabolism related enzymes, some of them are transporter, regulator, motility related proteins, there is no exotoxin related genes. Genome number of clusters of orthologous groups category (C). Among these coding sequences, 3,618 genes were distributed to 23 orthologous clusters, more than 250 genes were classified into functional categories for general function prediction only (n = 358), amino acid transport and metabolism (n = 349), transcription (n = 318), carbohydrate transport and metabolism (n = 293).
Discussion
Bacillus spp. have been widely studied as microbial biocontrol agents. As a useful microorganism in the medical industry, microecological preparations prepared by Bacillus play an important role in the treatment of intestinal flora disorders, candida infection, prevention of wound infection, and other medical processes (Garvey et al., 2022; Zou et al., 2022). B. licheniformis has an inhibitory effect on Staphylococcus, Candida albicans, yeast, and Escherichia coli; capsules and oral liquid made from B. licheniformis strains can treat intestinal diseases (Ramirez-Olea et al., 2022). In addition, intestinal ecological preparation of the combination of bifidobacterium and bacillus licheniformis combined with chemotherapy drugs cannot only kill and promote apoptosis of H22 ascites cancer cells but also prolong the life cycle of tumor mice and improve the effect of chemotherapy (H. S. et al., 2023). Here, we isolated and identified a B. velezensis NC-B4 (Figure 1) that has antifungal activity (Figure 3 and Table 3).
Cell-wall lytic enzymes exert antifungal effects by destroying the fungal cell wall, cytoplasmic membrane or affect fungal growth and differentiation (Aimanianda et al., 2017; Choudhary et al., 2014). For example, cellulase enzymes degrade cell walls by cleaving the β-1,4-D-glycosidic bonds that connect the glucose units containing cellulose (Harrison and Bonning, 2010), and proteolytic enzymes capable of hydrolyzing polysaccharides adversely affect fungal growth and differentiation by dissolving or disturbing polymers in the cell wall of pathogenic fungi (Hasan et al., 2013). Here, we detected the potent activity of cellulase and protease in NC-B4 (Figure 2) that supports its correlation with the growth inhibition of various human pathogenic fungi (Figure 3). It has been reported that the enzymes produced by Bacillus spp., such as amylase, cellulase, and protease, are highly associated with antifungal activity against Fusarium oxysporum pathogens (El-Sersawy et al., 2021). Moreover, the synergistic effect of cell-wall lytic enzymes and secondary metabolites may enhance the antifungal effect (Kim et al., 2022).
C. auris is an emerging fungal pathogen that is becoming a serious global health threat. Due to its multidrug-resistant features, invasive infections of C. auris often results in high mortality rates (Du et al., 2020; Spivak and Hanson, 2018). Here, we showed that the antagonistic activity of NC-B4 on human pathogenic fungi revealed a broad capacity to inhibit the growth of fungus, especially on C. auris (Figure 3 and Table 3). Biofilm is an important virulence factor that is related to fungal drug resistance (Rather et al., 2021). Culture broth of NC-B4 not only inhibited the growth and biofilm formation of C. auris (Figure 4) but also can reduce the cytotoxicity of C. auris to A549 cells (Figure 5A). In the mouse systemic infection model, treatment with culture broth of NC-B4 significantly decreased the fungal burden (CFU) in the spleen and brain (Figure 5B). For a good microbial biocontrol agent (probiotic), in addition to the function of inhibiting pathogens, the safety of the probiotic strain is very important. Here, we verified that NC-B4 has no toxicity to A549 cells (Figure 5A). We also analyze virulence factors in the genome data of the strain NC-B4; there are no toxin-related genes (Figure 6B).
Through whole genome sequencing and analysis, we predicted the genes and gene clusters involved in secondary metabolites produced by NC-B4. Bacillus spp. can produce multiple antimicrobials with a variety of chemical structures, including surfactin, fengycin, macrolactin H, bacillaene, difficidin, bacillibactin, bacilysin, and plantazolicin; they have different effects in the medical industry (Salazar et al., 2023; Sansinenea and Ortiz, 2011). In NC-B4, it exhibits a high genetic capacity for synthesizing cyclic lipopeptides (i.e., fengycin, bacillibactin) and polyketides (i.e., macrolactin H, bacillaene, and difficidin) (Table 6). Among them, the biosynthetic gene clusters of macrolactin H, bacillibactin, and bacilysin that have antibacterial activity were detected in the NC-B4 genome and had size of 87.8 kb, 51.8 kb, and 41.4 kb, respectively (Table 4). In addition, the gene cluster bae and pks associated with the biosynthesis of bacillaene were predicted in the NC-B4 genome (100.6 kb), bacillaene is known as a broad-spectrum antibiotic that inhibits bacterial protein. The gene cluster encoding fengycin synthesis was detected with a size of 134.3 kb, which has an antifungal function.
Numerous studies have been conducted to determine the impact of Bacillus secondary metabolite on pathogens (Fira et al., 2018; Hirozawa et al., 2023). It is noteworthy that on average around 5% of the whole genome of the Bacillus spp. is dedicated to the synthesis of secondary metabolites (Salazar et al., 2023). B. subtilis pB2-L produced plipastatin (the fengycin family), which inhibits F. oxysporum mycelium growth (Gao et al., 2017). B. amyloliquefaciens S76–3 produced plipastatin A and iturin A, which have clear antagonistic effects on F. graminearum (Gong et al., 2015). B. velezensis produces secondary metabolites and enzymes such as protease, chitinase, cellulase, and glucanase, and it inhibits B. cinerea growth, and so on (Fazle and Baek, 2020). However, it has only been reported that Bacillus can inhibit plant pathogenic fungi, and its inhibitory effect on human pathogenic fungi has not been reported (Fira et al., 2018). Here, we found B.velezensis NC-B4 also produces a variety of secondary metabolites and enzymes such as protease and cellulase, which have significant antagonistic effects on human pathogenic fungi, especially on C. auris. The whole genome analysis of secondary metabolites and the detection of enzymes provide scientific evidence of the effectiveness of NC-B4 as a biocontrol agent.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical approval was in accordance with the local legislation and institutional requirements. All the animal experiments were approved by the Ethics Committee at the Jiangxi Provincial People’s Hospital (approval number KT2023-012).
Author contributions
CY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. CC: Data curation, Methodology, Supervision, Writing – original draft. YC: Data curation, Formal Analysis, Supervision, Writing – original draft. ZP: Data curation, Formal Analysis, Methodology, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by Jiangxi Provincial Natural Science Foundation (Grant No. 20242BAB25336, 20232BAB213051, 20212BAB216059, 20252BAC200540), the Health Commission of Jiangxi Province (Grant No. 202310199) and the Science and Technology Research Project of Jiangxi Provincial Department of Education (Grant No. GJJ2203554).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdolrasouli, A., Armstrong James, D., Ryan, L., and Schelenz, S. (2017). In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses. 60, 758–763. doi: 10.1111/myc.12699
Aimanianda, V., Simenel, C., Garnaud, C., Clavaud, C., Tada, R., Barbin, L., et al. (2017). The dual activity responsible for the elongation and branching of β-(1,3)-glucan in the fungal cell wall. mBio. 8, e00619-17. doi: 10.1128/mBio.00619-17
Bhargava, A., Klamer, K., Sharma, M., Ortiz, D., and Saravolatz, L. (2025). Candida auris: A continuing threat. Microorganisms. 13, 652. doi: 10.3390/microorganisms13030652
Choudhary, D. K. and Johri, B. N. (2009). Interactions of Bacillus spp. and plants – With special reference to induced systemic resistance (ISR). Microbiol. Res. 164, 493–513. doi: 10.1016/j.micres.2008.08.007
Choudhary, B., Nagpure, A., and Gupta, R. K. (2014). Fungal cell-wall lytic enzymes, antifungal metabolite(s) production, and characterization from Streptomyces exfoliatus MT9 for controlling fruit-rotting fungi. J. Basic Microbiol. 54, 1295–1309. doi: 10.1002/jobm.201400380
Chowdhary, A., Prakash, A., Sharma, C., Kordalewska, M., Kumar, A., Sarma, S., et al. (2018). A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the EGR11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899. doi: 10.1093/jac/dkx480
Du, H., Bing, J., Hu, T., Ennis, C. L., Nobile, C. J., Huang, G., et al. (2020). Candida auris: epidemiology, biology, antifungal resistance, and virulence. PloS Pathog. 16, e1008921. doi: 10.1371/journal.ppat.1008921
Du, H., Guan, G., Xie, J., Sun, Y., Tong, Y., Zhang, L., et al. (2012). Roles of Candida albicans GAT2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PloS One 7, e29707. doi: 10.1371/journal.pone.0029707
El-Sersawy, M. M., Hassan, S. E., El-Ghamry, A. A., El-Gwad, A. M. A., and Fouda, A. (2021). Implication of plant growth-promoting rhizobacteria of Bacillus spp. as biocontrol agents against wilt disease caused by Fusarium oxysporum Schlecht. in Vicia faba L. Biomol. Concepts. 12, 197–214. doi: 10.1515/bmc-2021-0020
Elshaghabee, F. M. F., Rokana, N., Gulhane, R. D., Sharma, C., and Panwar, H. (2017). Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 8, 1490. doi: 10.3389/fmicb.2017.01490
Fazle Rabbee, M. and Baek, K. H. (2020). Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules. 25, 4973. doi: 10.3390/molecules25214973
Fira, D., Dimkić, I., Berić, T., Lozo, J., and Stanković, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Fu, C., Wan, S., Yang, P., Zhao, X., Yan, Y., Jiang, S., et al (2024). Identification of the Ilex macrocarpa anthracnose pathogen and the antifungal potential of the cell-free supernatant of Bacillus velezensis against Colletotrichum fioriniae. Front. Microbiol. 15, 1419436. doi: 10.3389/fmicb.2024.1419436
Gao, L., Han, J., Liu, H., Qu, X., Lu, Z., and Bie, X. (2017). Plipastatin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie. Van. Leeuwenhoek. 110, 1007–1018. doi: 10.1007/s10482-017-0874-y
Garvey, S. M., Mah, E., Blonquist, T. M., Kaden, V. N., and Spears, J. L. (2022). The probiotic Bacillus subtilis BS50 decreases gastrointestinal symptoms in healthy adults: a randomized, double-blind, placebo-controlled trial. Gut Microbes 14, 2122668. doi: 10.1080/19490976.2022.2122668
Gong, A., Li, H., Yuan, Q., Song, X., Yao, W., He, W., et al. (2015). Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens s76–3 from wheat spikes against Fusarium graminearum. PloS One 10, e116871. doi: 10.1371/journal.pone.0116871
H, S. R. and Halami, P. M. (2023). The combined effect of potential probiotic Bacillus licheniformis MCC 2514 and Bifidobacterium breve NCIM 5671 towards anti-inflammatory activity on HT-29 cell lines. Probiotics Antimicrob. Proteins. 15, 351–362. doi: 10.1007/s12602-021-09851-y
Harrison, R. L. and Bonning, B. C. (2010). Proteases as insecticidal agents. Toxins. 2, 935–953. doi: 10.3390/toxins2050935
Hasan, S., Ahmad, A., Purwar, A., Khan, N., Kundan, R., and Gupta, G. (2013). Production of extracellular enzymes in the entomopathogenic fungus Verticillium lecanii. Bioinformation. 9, 238–242. doi: 10.6026/97320630009238
Hirozawa, M. T., Ono, M. A., Suguiura, I. M. D. S., Bordini, J. G., and Ono, E. Y. S. (2023). Lactic acid bacteria and Bacillus spp. as fungal biological control agents. J. Appl. Microbiol. 134, lxac083. doi: 10.1093/jambio/lxac083
Huber, B., Riedel, K., Hentzer, M., Heydorn, A., Gotschlich, A., Givskov, M., et al. (2001). The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiol. (Reading). 147, 2517–2528. doi: 10.1099/00221287-147-9-2517
Kang, T. A., Lee, G., Kim, K., Hahn, D., Shin, J. H., and Kim, W. C. (2024). Biocontrol of peach gummosis by Bacillus velezensis KTA01 and its antifungal mechanism. J. Microbiol. Biotechnol. 34, 296–305. doi: 10.4014/jmb.2310.10005
Kim, J., Song, J., Kim, P. I., Kim, D., and Kim, Y. (2022). Bacillus velezensis TSA32–1 as a promising agent for biocontrol of plant pathogenic fungi. J. Fungi. 8, 1053. doi: 10.3390/jof8101053
Kizhakkekalam, V. K., Chakraborty, K., and Krishnan, S. (2022). Antibacterial and wound healing potential of topical formulation of marine symbiotic Bacillus. Arch. Microbiol. 204, 648. doi: 10.1007/s00203-022-03246-5
Lin, X., Wang, J., Hou, Z., Ren, S., Wang, W., Yang, Y., et al (2024). Antifungal potential and mechanism of Bacillus velezensis HeN-7 isolated from tobacco leaves on Bipolaris sorokiniana. Curr. Microbiol. 81, 340. doi: 10.1007/s00284-024-03858-8
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Osei Sekyere, J. (2019). Candida auris: a systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen. 7, e00901. doi: 10.1002/mbo3.901
Ramesh, S., Roy, U., and Roy, S. (2023). The elucidation of the multimodal action of the investigational anti-Candida lipopeptide (AF4) lead from Bacillus subtilis. Front. Mol. Biosci. 10, 1248444. doi: 10.3389/fmolb.2023.1248444
Ramirez-Olea, H., Reyes-Ballesteros, B., and Chavez-Santoscoy, R. A. (2022). Potential application of the probiotic Bacillus licheniformis as an adjuvant in the treatment of diseases in humans and animals: a systematic review. Front. Microbiol. 13, 993451. doi: 10.3389/fmicb.2022.993451
Rather, M. A., Gupta, K., and Mandal, M. (2021). Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 52, 1701–1718. doi: 10.1007/s42770-021-00624-x
Salazar, B., Ortiz, A., Keswani, C., Minkina, T., Mandzhieva, S., Pratap Singh, S., et al. (2023). Bacillus spp. as bio-factories for antifungal secondary metabolites: innovation beyond whole organism formulations. Microb. Ecol. 86, 1–24. doi: 10.1007/s00248-022-02044-2
Sansinenea, E. and Ortiz, A. (2011). Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 33, 1523–1538. doi: 10.1007/s10529-011-0617-5
Shao, J., Liu, Y., Xie, J., Štefanič, P., Lv, Y., Fan, B., et al. (2022). Annulment of bacterial antagonism improves plant beneficial activity of a Bacillus velezensis consortium. Appl. Environ. Microbiol. 88, e0024022. doi: 10.1128/aem.00240-22
Shen, F., Yin, W., Song, S., Zhang, Z., Ye, P., Zhang, Y., et al. (2020). Ralstonia solanacearum promotes pathogenicity by utilizingl -glutamic acid from host plants. Mol. Plant Pathol. 21, 1099–1110. doi: 10.1111/mpp.12963
Spivak, E. S. and Hanson, K. E. (2018). Candida auris: an emerging fungal pathogen. J. Clin. Microbiol. 56, e1588-17. doi: 10.1128/JCM.01588-17
Wang, K., Li, X., Yang, C., Song, S., Cui, C., Zhou, X., et al. (2021). A LysR family transcriptional regulator modulates Burkholderia cenocepacia biofilm formation and protease production. Appl. Environ. Microbiol. 87, e00202-21. doi: 10.1128/AEM.00202-21
Welsh, R. M., Bentz, M. L., Shams, A., Houston, H., Lyons, A., Rose, L. J., et al. (2017). Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 55, 2996–3005. doi: 10.1128/JCM.00921-17
Xie, J., Tao, L., Nobile, C. J., Tong, Y., Guan, G., Sun, Y., et al. (2013). White-opaque switching in natural MTLa/α isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PloS Biol. 11, e1001525. doi: 10.1371/journal.pbio.1001525
Yang, C., Cui, C., Ye, Q., Kan, J., Fu, S., Song, S., et al. (2017). Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc. Natl. Acad. Sci. - Pnas. 114, 13006–13011. doi: 10.1073/pnas.1709048114
Zalila-Kolsi, I., Ben-Mahmoud, A., and Al-Barazie, R. (2023). Bacillus amyloliquefaciens: harnessing its potential for industrial, medical, and agricultural applications—a comprehensive review. Microorganisms. 11, 2215. doi: 10.3390/microorganisms11092215
Keywords: antifungal activity, Bacillus velezensis NC-B4, biological control, Candida auris, genome-sequence analysis
Citation: Yang C, Cui C, Chen Y and Peng Z (2025) Bacillus velezensis NC-B4 as a promising antifungal agent for biocontrol of Candida auris. Front. Cell. Infect. Microbiol. 15:1515537. doi: 10.3389/fcimb.2025.1515537
Received: 23 October 2024; Accepted: 04 August 2025;
Published: 02 September 2025.
Edited by:
María Guadalupe Frías De León, Hospital Regional de Alta Especialidad de Ixtapaluca, MexicoReviewed by:
Ludmila Baltazar, Universidade Federal de Goiás, BrazilSunna Nabeela, Lundquist Institute for Biomedical Innovation, United States
Copyright © 2025 Yang, Cui, Chen and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunxi Yang, Y2h1bnhpeWFuZ0AxMzkuY29t
 Chunxi Yang
Chunxi Yang Chaoyu Cui2
Chaoyu Cui2