- 1Department of General and Visceral Surgery, Medical Center – University of Freiburg, Freiburg, Germany
- 2Institute of Medical Microbiology and Hygiene, Medical Center – University of Freiburg, Freiburg, Germany
Background: The patient´s microbiome has become a focal point in cancer research. Even for pancreatic cancer, alterations in the microbiome appear to influence cancer formation and progression. The aim of our single-center analysis was the examination of microbiological colonization of pancreas tissue at the time of surgery and its potential influence on complications and outcome.
Methods: We prospectively evaluated patients undergoing pancreatic surgery over a three-year period from June 2018 to June 2021. We focused on the microbiological colonization of pancreatic tissue which was acquired during pancreatic surgery. Tissue samples were cultivated at our institute of microbiology. Patients´ characteristics, complications and postoperative outcome were analyzed using a prospectively maintained SPSS database.
Results: Between June 2018 and June 2021, we collected pancreatic tissue samples of a total of 178 patients undergoing pancreas resections, mostly due to ductal adenocarcinoma (PDAC; 50.6%). We could cultivate bacterial or fungal species in pancreatic tissue samples of 50 of our patients (28.1%). The majority of cases were characterized by the presence of a single microbial species, but 20 patients (11.2%) showed colonization with up to four different species. Among the bacterial species detected were Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Staphylococcus aureus, Enterobacter cloacae and Klebsiella pneumonia. We found significantly more microbiological culture growth in patients with a preoperative biliary stent (74.0% vs. 15.6%, p < 0.001). Concerning postoperative complications, we found no difference concerning pancreatic fistula, but colonization with E. coli was associated with a significantly higher rate of postpancreatectomy hemorrhage (30.0% vs. 8.9%, p = 0.032). Interestingly, survival of PDAC patients seems to be negatively affected by positive microbiological findings at the time of surgery, but without reaching statistical significance (p = 0.770).
Conclusion: In this first analysis of our patient cohort, we could show a microbiological colonization of pancreatic tumor tissue in almost a third of our patients. There seems to be only a minor impact on postoperative complications, but long-term outcome seems to be worse in patients with a positive pancreas microbiome. Further observation is needed to evaluate the influence of the tumor microbiome on the long-term oncological outcome in PDAC patients.
Introduction
The human body is the habitat of diverse microorganisms which – taken together as microbiome – have important metabolic functions (Frost et al., 2022). However, shifts in the composition of the body´s microbiome may lead to disease development and progression (Frost et al., 2022). During recent years, the patient´s microbiome turned in the focus of cancer research (Picardo et al., 2019), as microbiota reside on or within about 20% of malignancies (de Martel et al., 2012; Wei et al., 2019). Even for pancreatic adenocarcinoma – still one of the deadliest malignancies – alterations in the microbiome seem to influence cancer formation and progression. It could be shown that the pancreatic cancer tissue comprises a more abundant microbiome compared to normal pancreatic tissue both in humans as well as in mice (Pushalkar et al., 2018) and that selected bacteria are differentially increased in pancreatic cancer tissue, compared to the gut microbiome (Pushalkar et al., 2018). In mice, ablation of the microbiome seems to protect against pre-invasive and invasive pancreatic ductal adenocarcinoma (PDAC) (Pushalkar et al., 2018). Clinically, intratumoral microbes may influence carcinogenesis and treatment response via different mechanisms (McAllister et al., 2019). A study could show that Fusobacterium species were present in some patients with pancreatic cancer and that their presence within the pancreatic tumor was associated with a worse prognosis of these patients (Mitsuhashi et al., 2015). Geller et al. showed that specific bacteria from the Gammaproteobacteria class, including the Enterobacteriaceae and Pseudomonadaceae families, present in pancreatic tumor tissue, play a role in conferring resistance to gemcitabine, a commonly used chemotherapy drug for pancreatic cancer (Geller et al., 2017). Moreover, the tumor microbiome composition in PDAC-patients may play a role in promoting long-term survival by influencing the host´s immune response (Riquelme et al., 2019) and the presence of intratumoral microbes in long-term survivors was associated with enhanced immune infiltrates (Balachandran et al., 2017). In addition to bacterial colonization, the presence of fungi in pancreatic tumor tissue also affects the course of pancreatic cancer (Aykut et al., 2019). A high abundance and distinct composition of fungal infection was detected in both murine and human pancreatic tumor tissue when compared to normal pancreatic tissue. Moreover, antifungal therapy with oral amphotericin B led to delayed tumorigenesis and tumor growth in the mouse model and potentiated the effect of gemcitabine (Aykut et al., 2019).
The detection of microbes by polymerase chain reaction (PCR), which is performed by the trials mentioned above, is very sensitive; however, it is not able to differentiate between vital and non-vital bacteria. Even in studies evaluating the influence of different risk factors, e.g. smoking, on the pancreatic tumor microbiome, only sequencing methods are used to detect potential bacteria (Liang et al., 2024). So far, PCR and sequencing methods are the predominant methods in order to evaluate the intratumoral microbiota of different tumor entities (Xue et al., 2023), but analyses using conventional microbiological cultures of the tumor tissue are still lacking. Therefore, the aim of our single center study was a first evaluation of the vital microbiome of pancreatic tumors by analyzing bacterial and fungal colonization of patients´ tumor specimens using conventional microbiological cultures. Moreover, our aim was to analyze the potential impact of the pancreatic tumor microbiome on postoperative complications and long-term outcome of patients. In addition, it will be examined whether different microbial species correlate with specific complications after pancreas resections.
Methods
Patient collective and data collection
We prospectively evaluated our patients undergoing pancreatic surgery at the University Hospital Freiburg over a three-year period from June 2018 to June 2021 concerning microbiological colonization of pancreatic tissue. Pancreatic tumor tissue was extracted intraoperatively via Tru-Cut biopsy needles after the resection of the pancreas, so that there was no risk of lacerating neighboring structures. Tissue samples were cultivated for bacterial and fungal species at our institute of microbiology, including testing for bacterial or fungal resistances against special antibiotics or antimycotics. Patients´ characteristics, complications and postoperative outcome were analyzed using a prospectively maintained SPSS database. Postoperative complications such as pancreatic fistula (POPF), postpancreatectomy hemorrhage (PPH) or delayed gastric emptying (DGE) were graded by current international definitions of the International Study Group of Pancreatic Surgery (ISGPS) (Bassi et al., 2005; Wente et al., 2007b; Wente et al., 2007a; Bassi et al., 2017).
Microbiological culture-based methods and microscopy
Pancreatic tissue samples were examined microscopically using Gram staining to detect granulocytes and bacteria. Samples were also plated on various cultural media, including Columbia blood agar (Thermo ScientificTM OxoidTM, Wesel, Germany), chocolate blood agar and MacConkey agar, followed by incubation for at least 48 h under aerobic conditions (36°C, 5% CO2). For the cultivation of strict anaerobic bacteria, yeast cysteine blood agar (HCB; in-house) was used under anaerobic conditions in a jar or plastic bags with either the Genbox ANAER (bioMérieux, Marcy-l’Étoile, France) or the Anaerocult (Merck, Darmstadt, Germany) system. Brain heart infusion broth containing 0.093% (w/v) agar was inoculated and incubated for five days.
Microorganisms were identified using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF, Bruker Daltonics, Bremen, Germany). Antimicrobial susceptibility testing of the detected organisms was conducted using the VITEK®2 system (bioMérieux, Nürtingen, Germany) and interpreted according to EUCAST resistance breakpoints (http://www.eucast.org/).
Microbial genomic DNA preparation and sequencing using illumina 16S rDNA sequencing
Besides conventional microbiological culture, twenty of our PDAC tumor samples were analyzed via 16S rDNA sequencing as well. These samples were processed and analyzed using the ZymoBIOMICS® Targeted Sequencing Service (Zymo Research, Irvine, CA). DNA was extracted using either the ZymoBIOMICS®-96 MagBead DNA Kit on an automated platform or the ZymoBIOMICS® DNA Miniprep Kit (Zymo Research, Irvine, CA). Bacterial 16S ribosomal RNA gene-targeted sequencing was conducted with the Quick-16S™ NGS Library Prep Kit (Zymo Research, Irvine, CA), using primers that amplified the V1-V2 region of the 16S rRNA gene. Final PCR products were quantified using qPCR fluorescence readings and pooled based on equal molarity. The pooled library was cleaned with the Select-a-Size DNA Clean & Concentrator™ (Zymo Research, Irvine, CA), and subsequently quantified with TapeStation® (Agilent Technologies, Santa Clara, CA) and Qubit® (Thermo Fisher Scientific, Waltham, WA). The final library was sequenced on the Illumina® MiSeq™ using a v3 reagent kit (600 cycles) with a 10% PhiX spike-in.
Sequencing analysis pipeline
Raw fastq files were analyzed as previously described (Wetzel et al., 2023; Lichtenegger et al., 2024). In short, raw fastq files’ read quality was assessed using FastQC (Babraham Bioinformatics - FastQC) and MultiQC (Ewels et al., 2016). Further quality control measures, trimming, and analysis of Illumina short-reads were done using the DADA2 analysis pipeline (Callahan et al., 2016). Reads were trimmed after 230 base pairs and filtered with 2 and 5 maximum expected errors in the forward and reverse reads respectively, apart from the default filtering parameters. Amplicon sequence variants (ASVs) were extracted from DADA2 and were assigned to taxonomy ranks using the Genome Taxonomy Database (Parks et al., 2020) release 207. Rarefaction curves were used to estimate sequencing depth.
Bacterial diversity and taxonomy analysis
Further analysis concerning bacterial diversity was carried out using the R programming language. The phyloseq R package (McMurdie and Holmes, 2013) was used to calculate bacterial diversity. Observed, Shannon, and inverse Simpson (InvSimpson) were used as alpha diversity indices. For beta diversity and taxonomy analysis, ASVs with fewer than ten occurrences in all samples were excluded. The microbial Bray-Curtis distance between samples was calculated with the phyloseq R package and visualized using Principal Coordinates Analysis (PCoA). Statistical analyses were conducted with the stats R package (R Core Team. R, 2013). For statistical differences in taxonomy, the Kruskal-Wallis or Wilcoxon rank-sum tests were applied to non-normally distributed variables. Categorical variables were analyzed using the Fisher exact test. Cumulative Sum Scaling (CSS) was performed using the metagenomeSeq package (Paulson et al., 2013) and applied for heatmap visualization. Visualization of samples was achieved using functions from the R packages phyloseq (McMurdie and Holmes, 2013), microViz (Barnett et al., 2021), ggplot2 (Wickham, 2009) and pheatmap (Kolde). The code for this analysis is publicly available at the following link: https://github.com/S-Posadas/Pancreatic_tumor_microbial_colonization.
Statistical analysis
Statistical analysis concerning patient data was performed using SPSS (IBM SPSS Statistics for Windows, Version 29.0. IBM Corp., Armonk, NY, USA). After performing explorative analysis and descriptive statistics, statistical significance was examined by using chi-square tests and Fisher´s exact tests for categorical variables and ANOVA for continuous variables. Survival estimates were calculated using Kaplan-Meier curves and log-rank tests. Results with a p-value < 0.05 were considered statistically significant.
Ethics
Data collection and analysis were performed in accordance with the Declaration of Helsinki and were approved by the local ethics committee (Ethics Committee of Albert-Ludwigs-University Freiburg, Germany, EK-No. 23-1416-S1-retro).
Results
Baseline characteristics and intraoperative parameters
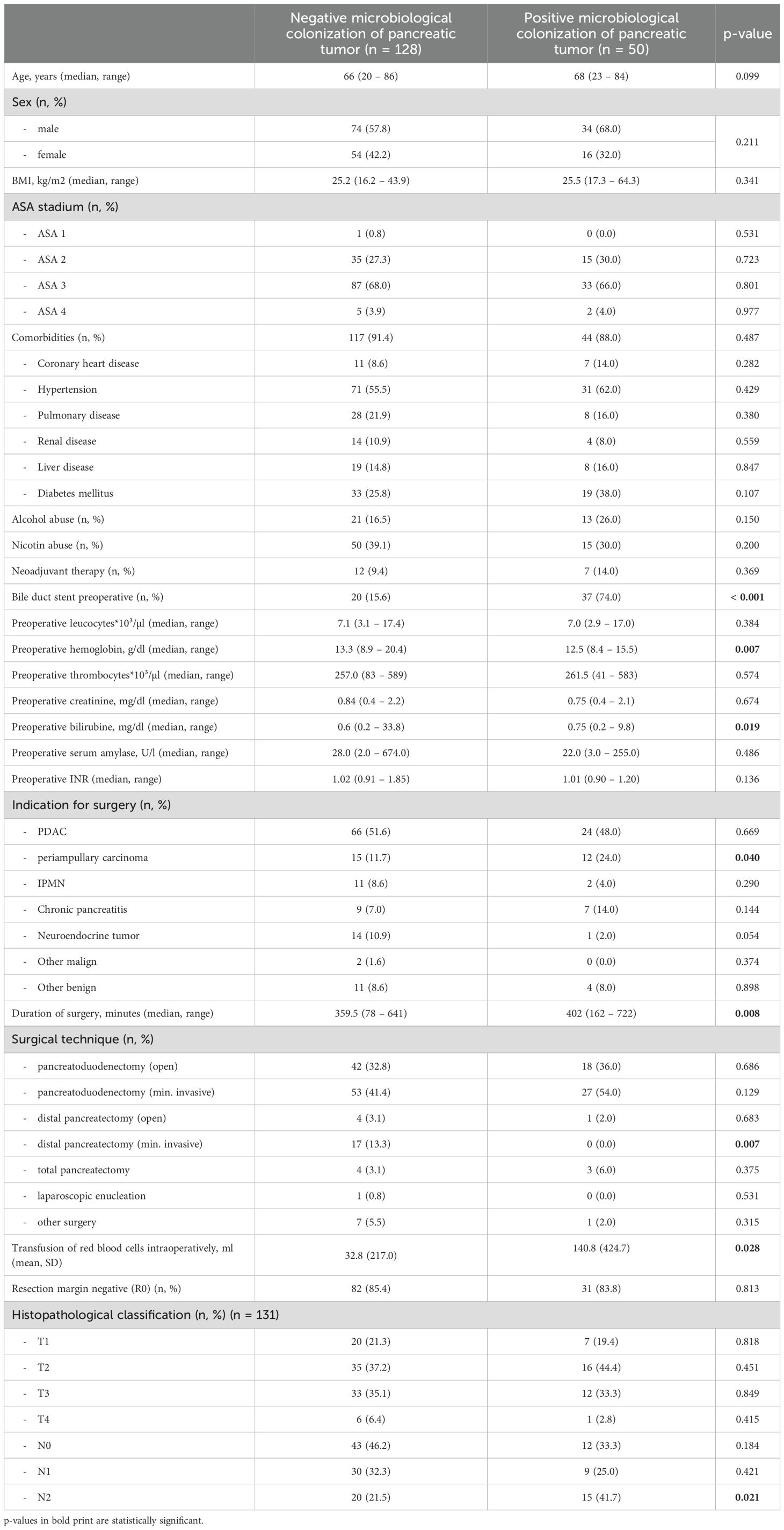
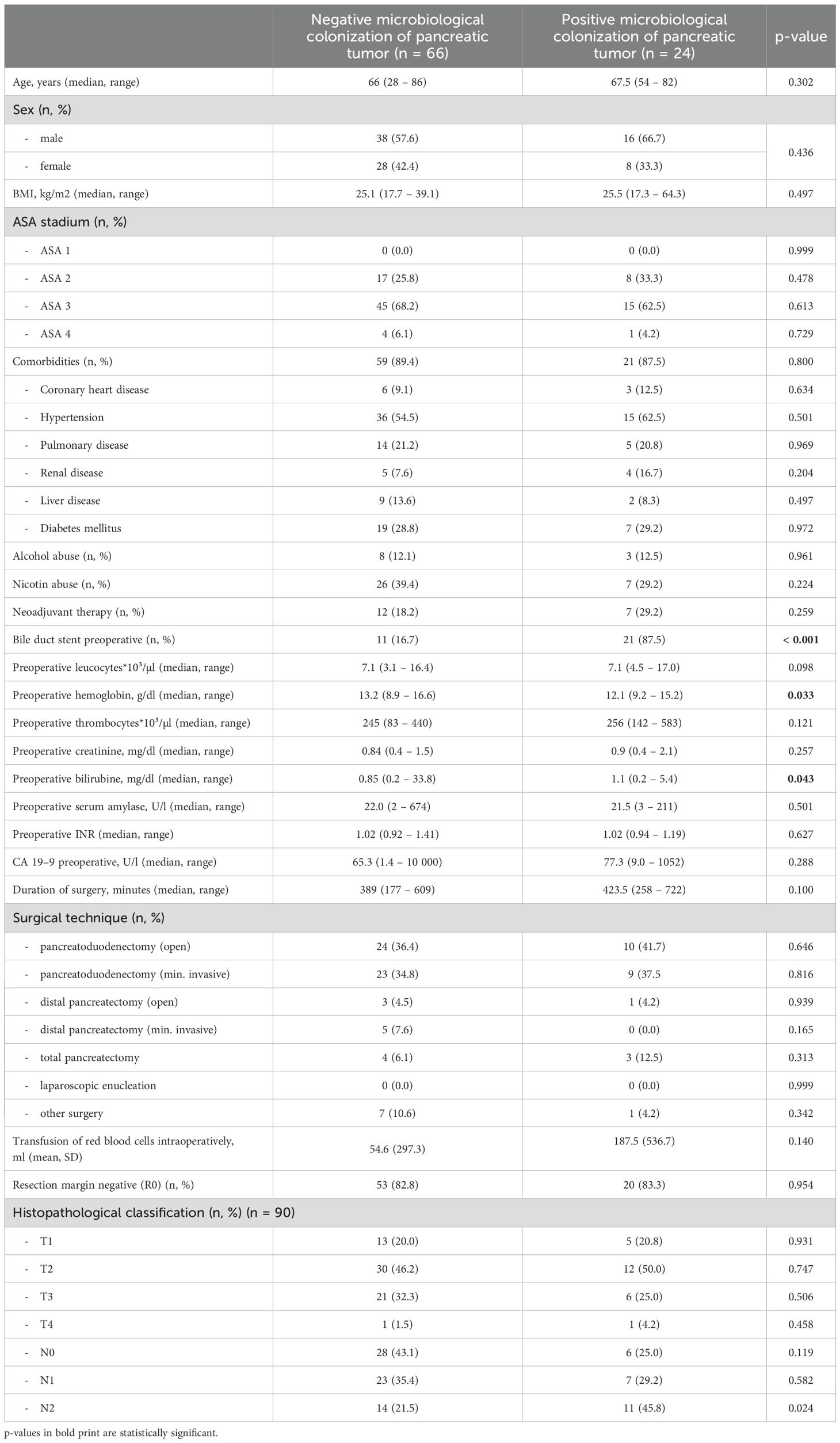
Between June 2018 and June 2021, we collected intraoperative tissue samples of a total of 178 patients undergoing pancreas resections at the University Hospital Freiburg. Most pancreas resections were performed due to pancreatic ductal adenocarcinoma (PDAC; 50.6%). In the majority of cases, patients underwent pancreatoduodenectomies (140 patients, 78.7%). In 50 of our patients (28.1%), we found microbiological colonization of the pancreas tissue at the time of surgery; the remaining 128 samples remained sterile. Dividing the patients in two groups depending on negative (neg) or positive (pos) microbiological findings, we could find no difference concerning age (66 years vs. 68 years, P = 0.099), sex (female 42.2% vs. 32.0%, P = 0.211), preoperative ASA stadium (ASA II 27.3% vs. 30.0%, P = 0.723; ASA III 68.0% vs. 66.0%, P = 0.801) or comorbidities (91.4% vs. 88.0%, P = 0.487) between the groups. Moreover, we could find no difference concerning alcohol (16.5% vs. 26.0%, P = 0.150) or nicotine consumption (39.1% vs. 30.0%, P = 0.200). The rate of neoadjuvant treatment was similar in both groups (9.4% vs. 14.0%, P = 0.369). Preoperative parameters such as preoperative leucocytes (7100/µl vs. 7000/µl, P = 0.384), creatinine (0.84 mg/dl vs. 0.75 mg/dl, P = 0.674), international normalized ratio (INR; 1.02 vs. 1.01, P = 0.136) and serum amylase (28.0 U/l vs. 22.0 U/l, P = 0.486) did not differ between the groups, but we found a significantly lower hemoglobin (12.5 g/dl vs. 13.3 g/dl, P = 0.007) as well as a higher median bilirubin (0.75 mg/dl vs. 0.60 mg/dl, P = 0.019) in patients with a positive microbiological colonization. Furthermore, patients with a positive microbiological colonization showed a significantly longer duration of surgery (402 min vs. 359.5 min, P = 0.008) and needed a higher amount of intraoperative blood transfusions (140.8 ml vs. 32.8 ml, P = 0.028). Interestingly, the rate of advanced lymph node metastasis (N2 stadium) was significantly higher in patients with a positive microbiological colonization of the pancreatic tumor (41.7% vs. 21.5%, P = 0.021). For details concerning baseline characteristics and intraoperative parameters of the entire cohort see Table 1, for the cohort of PDAC patients see Table 2.
Microbiological colonization
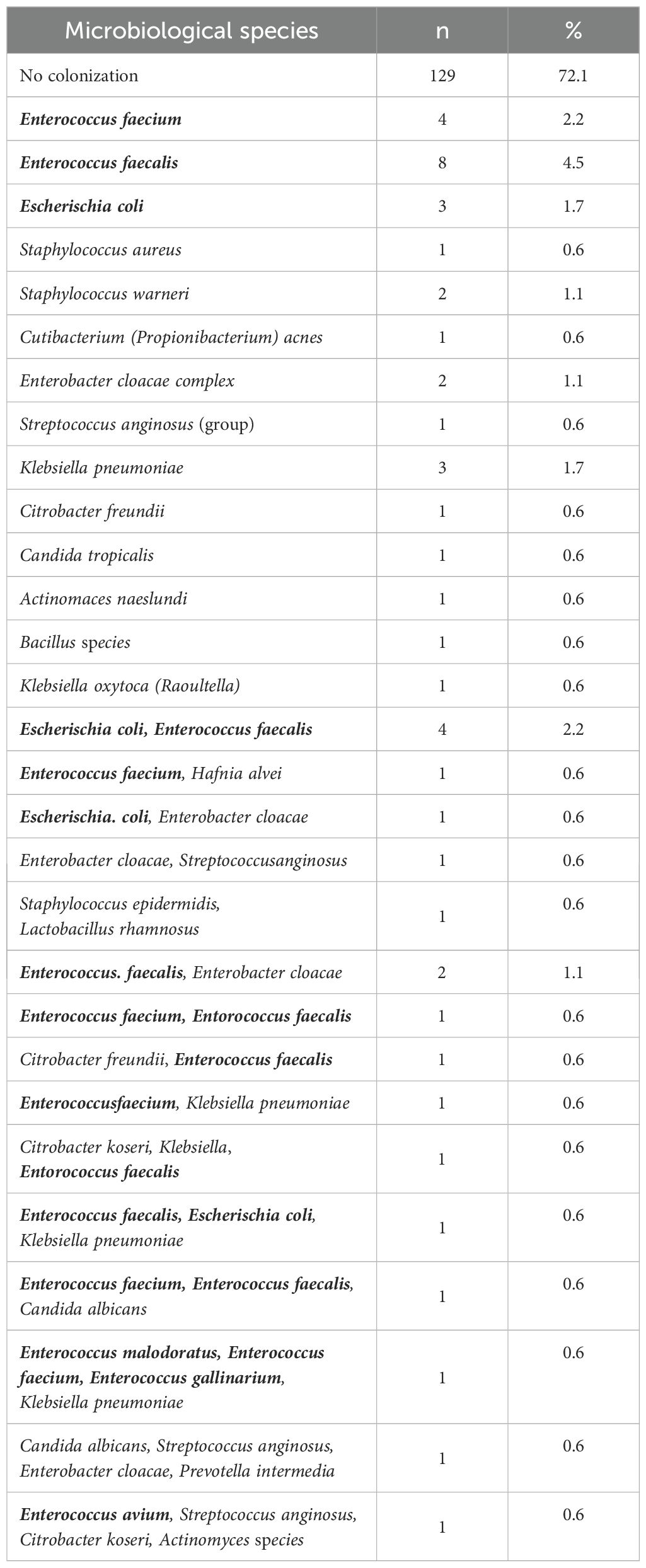
In 50 of our 178 patients, we detected microbiological colonization of the pancreatic tumor at the time of surgery (28.1%). Most of our patients showed only colonization with one microbiological species, but 20 patients (11.2%) revealed colonization with up to four different microbiological species in their tissue samples. Among the bacteria detected were Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Enterobacter cloacae and Klebsiella pneumoniae. An overview of the microbiological findings in our patient collective is given in Table 3. The highest rate of bacterial colonization is found in patients with chronic pancreatitis (7 of 16 patients, 43.8%) and periampullary carcinomas (12 of 27 patients, 44.4%), even reaching statistical significance in the later (P = 0.040). The rate of positive microbiological colonization in the different tumor entities is shown in Figure 1.
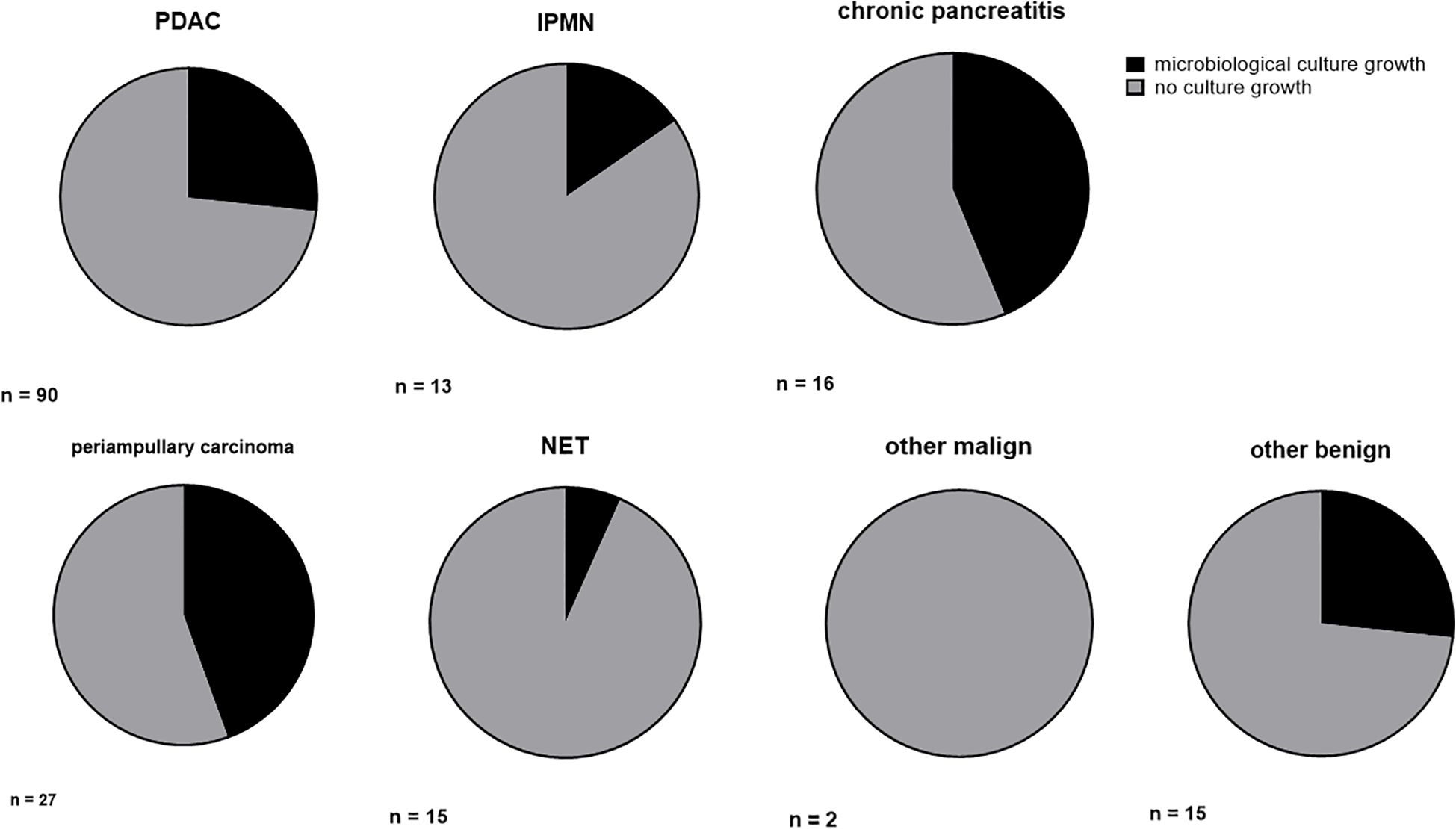
Figure 1. Ratio of microbiological colonization in different tumor entities. Ratio (%) of microbiological colonization in conventional culture growth. PDAC, pancreatic ductal adenocarcinoma; IPMN, intrapapillary mucinous neoplasm; NET, neuroendocrine tumor.
Postoperative complications and length of hospital stay
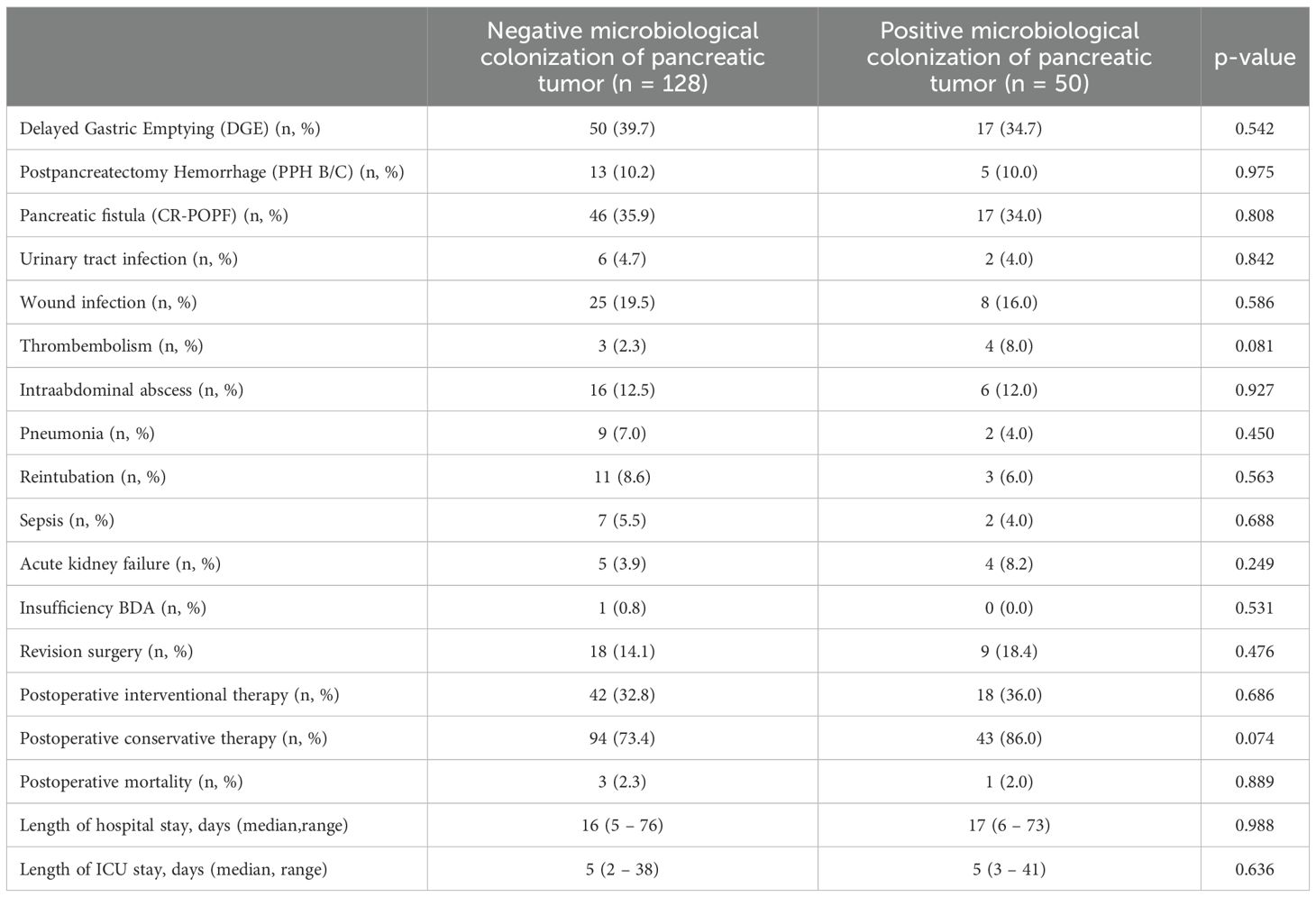
In our patient collective, we found a similar distribution of delayed gastric emptying (DGE), postpancreatectomy hemorrhage (PPH) and clinically relevant pancreatic fistula (CR-POPF) between patients with and without microbiological findings in the pancreatic tumor (DGE neg 39.7% vs. pos 34.7%, P = 0.542; PPH B/C neg 10.2% vs. pos 10.0%, P = 0.975; CR-POPF neg 35.9% vs. pos 34.0%, P = 0.808) (Table 4). Moreover, we found no difference between both groups concerning urinary tract infection (neg 4.7% vs. pos 4.0%, P = 0.842), wound infections (19.5% vs. 16.0%, P = 0.586), intraabdominal abscesses (12.5% vs. 12.0%, P = 0.927), pneumonia (7.0% vs. 4.0%, P = 0.450) and acute kidney failure (3.9% vs. 8.2%, P = 0.249) following surgery. Even concerning postoperative sepsis, we found no difference between patients with negative and positive microbiological colonization (neg 5.5% vs. pos 4.0%, P = 0.688), but there seemed to be a trend towards more thromboembolic complications in patients with a microbiological colonization of the pancreatic tumor (pos 8.0% vs. neg 2.3%, P = 0.081). There was no significant difference concerning revision surgery (neg 14.1% % vs. pos 18.4%, P = 0.476) or the need of postoperative interventional therapies (neg 32.8% vs. pos 36.0%, P = 0.686), but patients with a positive microbiological colonization showed a trend towards receiving additional conservative treatment more frequently (pos 86.0% vs. neg 73.4%, P = 0.074). The latter was mainly due to the preoperatively inserted bile duct stent, which leads to a routine postoperative antibiotic therapy following our hospital standards. The rate of postoperative mortality was similar between both groups (neg 2.3% vs. pos 2.0%, P = 0.889) as well as the length of stay of the intensive care unit (ICU) (neg median 5 days (2–38 days) vs. pos 5 days (3–41 days), P = 0.636) and the length of hospital stay (16 days (5–76 days) vs. 17 days (6–73 days), P = 0.988). Details on postoperative complications and hospital stay are summarized in Table 4.
Association of specific bacteria and postoperative complications
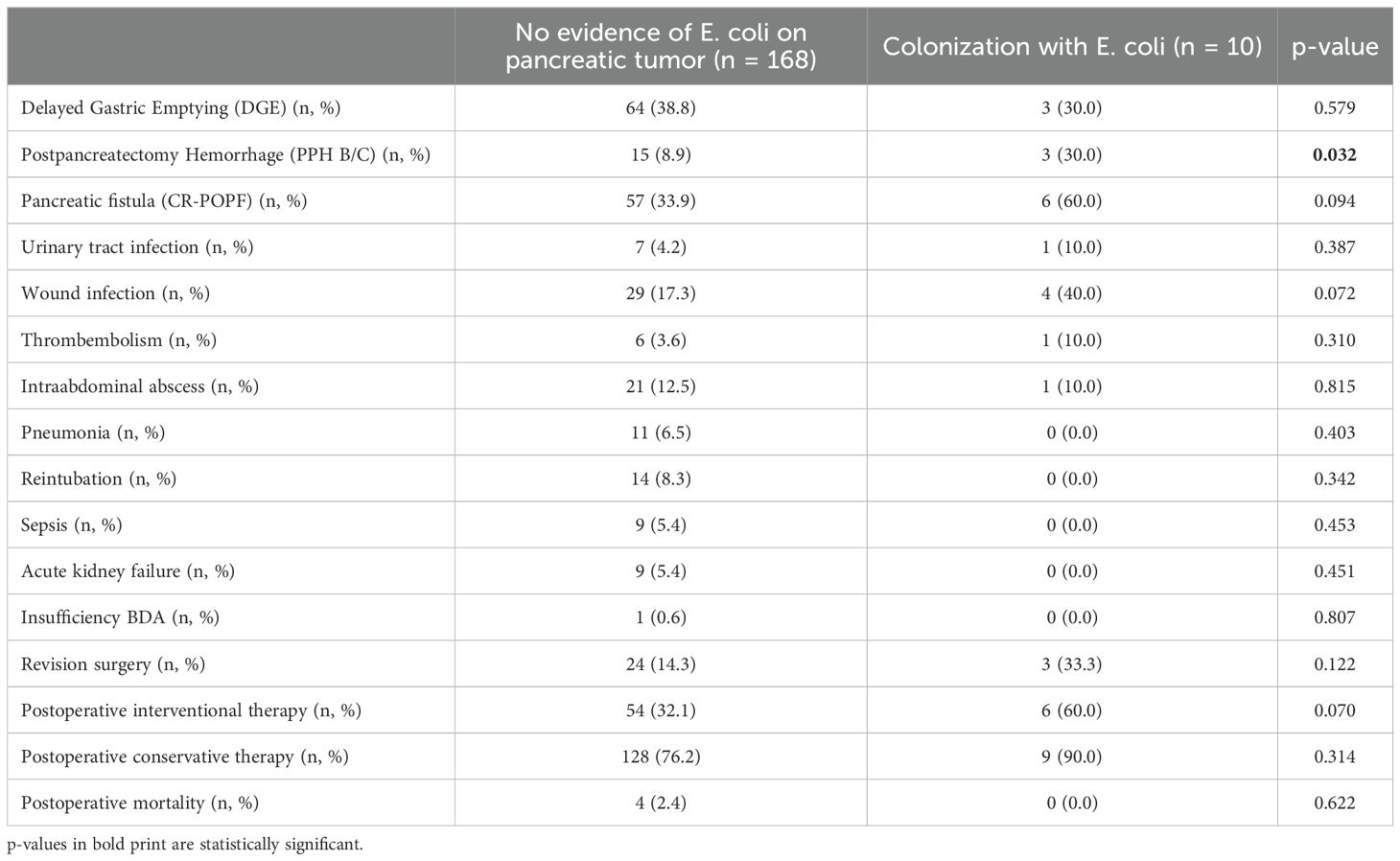
In a next step, we analyzed if a specific microbiological colonization of the pancreatic tumor was associated with postoperative complications. Here, we could find no influence of enterococcus species (neither E. faecium nor E. faecalis nor both) on postoperative complications (Table 5). In case of infections with E. coli species in the pancreatic tumor, significantly more cases of postpancreatectomy hemorrhage grade B/C (30.0% vs. 8.9%, p = 0.032) and a trend towards more clinically relevant pancreatic fistula (60.0% vs. 33.9%, p = 0.094) and wound infections (40.0% vs. 17.3%, p = 0.072) were observed (Table 6).
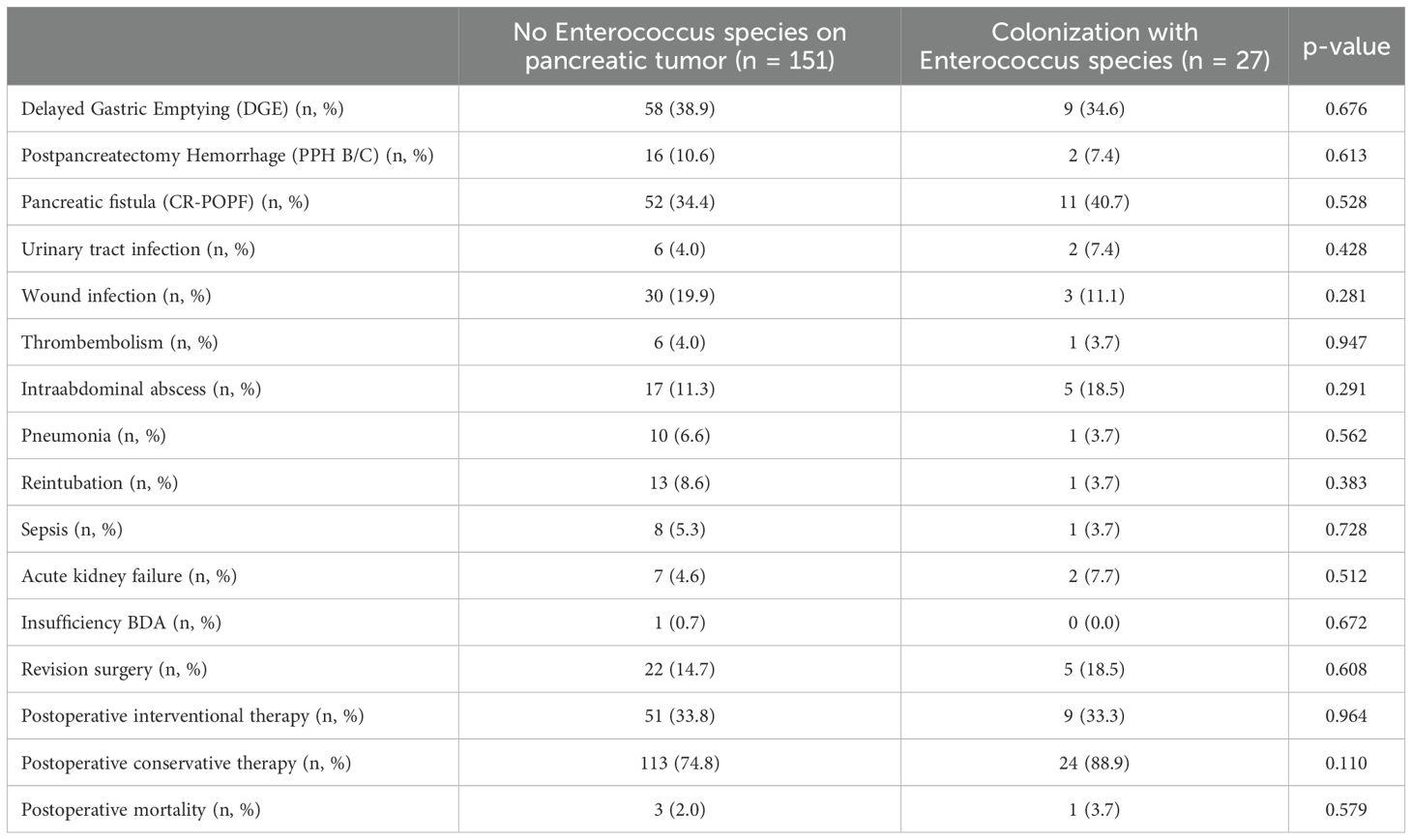
Table 5. Complications in association with colonization with Enterococcus species at the time of surgery.
Comparison of colonization of PDAC tissue versus benign tumors
Comparing PDAC with other tumor entities, we could find no difference in the total amount of a positive microbiological culture growth (26.7% vs. 29.5%, p = 0.669). Interestingly, we found significantly more cases of colonization with E. faecium in patients with PDAC compared to other tumor entities (8.9% vs. 1.1%, p = 0.018), but a trend towards less colonization with E. faecalis in PDAC patients (6.7% vs. 14.8%, p = 0.080). Concerning E. coli, we found no difference between PDAC and other tumor entities (p = 0.181). When comparing PDAC tumors of the pancreatic head with tumors of corpus or tail, we found no difference in the amount of microbiological colonization (27.9% vs. 22.7%, p = 0.631).
Survival
Concerning the 90 PDAC patients, we found a trend towards a worse survival in patients with microbiological colonization of the pancreatic tumor (17.5 months vs. 25.5 months), but without reaching statistical significance (p = 0.770). This can be observed in the Kaplan-Meier curves, which indicate a trend toward reduced survival in patients with positive microbiological findings, especially during the first 36 months following tumor resection. Subsequently, both survival curves run in parallel, suggesting that in long-term PDAC survivors, factors beyond microbial colonization may contribute to outcomes (Figure 2A). Comparing the survival curves of patients with and without preoperative bile duct stenting, we could find similar curves with stented patients tending to have a poorer survival (20.5 months vs. 25.5 months, p = 0.520) (Figure 2B).

Figure 2. Kaplan Meier curves of PDAC patients. (A) negative vs. positive microbiome (B) stented vs. non-stented patients.
Comparison between conventional microbiological culture and 16S rDNA-sequencing
As our study is one of the first studies analyzing the vital microbiome of pancreatic tumors via conventional microbiological culture of tumor tissue, we performed an analysis of 20 of our PDAC patient samples (10 with positive and 10 with negative microbial colonization in the conventional microbial culture) via 16S-rDNA sequencing in order to evaluate if the results in our cohort differ between conventional culture und 16S-rDNA sequencing. In this first comparison of conventional microbiological culture and 16S-rDNA sequencing in our cohort, there was a strong correlation between cultural growth of staphylococci and enterococci and the identification of these bacteria via sequencing. On the other hand, especially for Enterobacteriaceae, the most frequently found species in sequencing do not match the species growing in conventional culture. Moreover, even in tumor specimens without growth of bacteria in the conventional culture, we could detect several bacteria via sequencing. In contrast, with the exception of one tumor sample, all bacteria identified via cultural growth could be identified via sequencing too. The results concerning abundant phyla and genera according to microbial growth in culture are shown in Figure 3 and grouped by the family of the cultured bacteria (Enterobacteriaceae, Enterococcaceae, Staphylococcaceae) or by no growth in Figures 4A, B. Most of the samples (4 out of 5) in which Enterococcus spp. were identified through 16S rDNA sequencing also exhibited growth of Enterococcus in culture. Conversely, multiple taxa from Enterobacteriaceae and Staphylococcaceae identified by 16S-rDNA sequencing were not detected in culture (Figure 4C). The broader microbial community structure was not substantially influenced by the cultured bacteria, as shown by the minimal impact on beta diversity (Figure 5). We further investigated the bacterial community structure in pancreatic tumor samples by examining the alpha diversity, which captures both the richness (variety of bacterial taxa) and evenness (the distribution of their abundances). Alpha diversity was assessed using the observed species, Shannon index and inverse Simpson (InvSimpson) index. Here, we found no difference between patients with positive or negative microbiological growth in the conventional culture (Observed P = 0.6, Shannon P = 0.91, InvSimpson P = 0.8). However, in patients with preoperatively inserted bile duct stents, there seems to be a trend towards a reduced alpha diversity in comparison to patients without bile duct stents, but without reaching statistical significance (Observed P = 0.3, Shannon P = 0.097, InvSimpson P = 0.11). We found no differences in alpha-diversity between patients with and without postoperative sepsis (Observed P = 0.38, Shannon P = 0.26, InvSimpson P = 0.32) or between different Clavien-Dindo stages (Observed P = 0.46, Shannon P = 0.84, InvSimpson P = 0.65). Results concerning alpha diversity are shown in Figure 6. Moreover, via 16S-rDNA sequencing, we could find a shift towards an increase in Cutibacterium in patients with bile duct stents. Cutibacterium represent typically stent-associated bacteria that grow hardly in conventional culture, so that we couldn´t find them via conventional culture, but verify them via sequencing especially in the stented patients. An overview of the abundant phyla and genera according to stent presence is given in Figure 7.
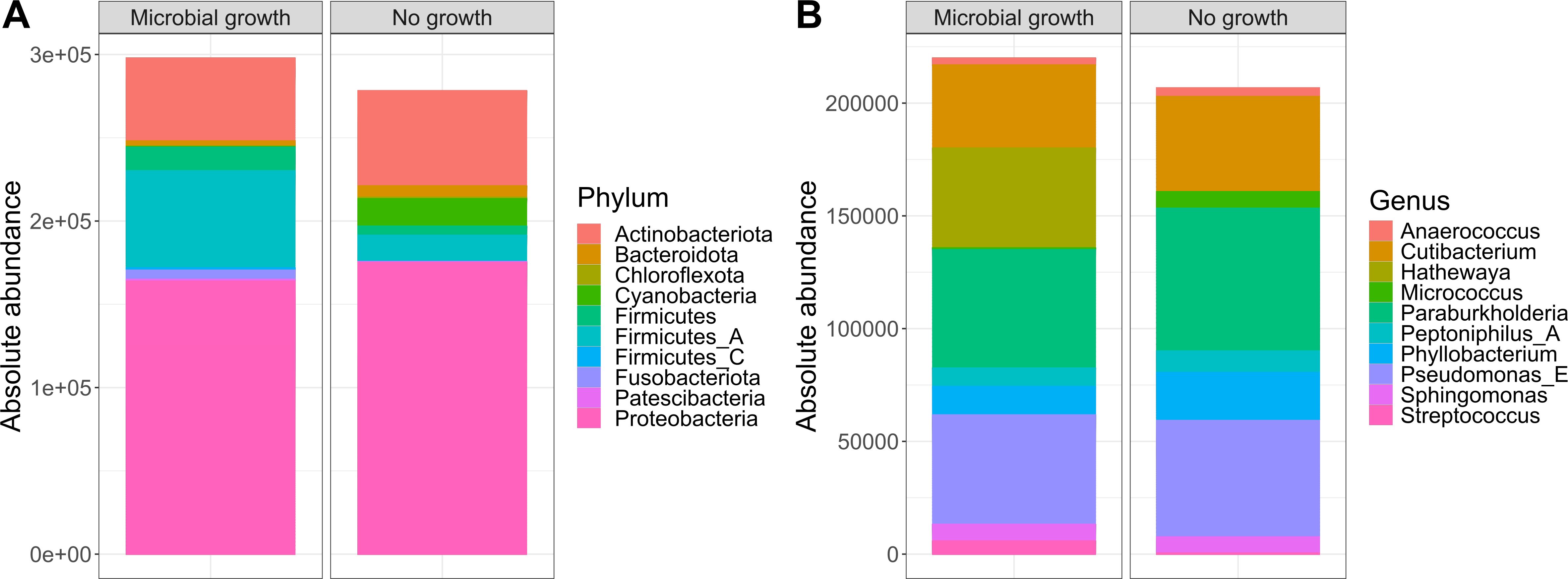
Figure 3. Abundant phyla and genera according to microbial growth. Ten most abundant phyla (A) and genera (B) from 16S rDNA-sequencing grouped by the presence of microbial growth in culture (n=10 per group). The y-axis represents absolute abundance.
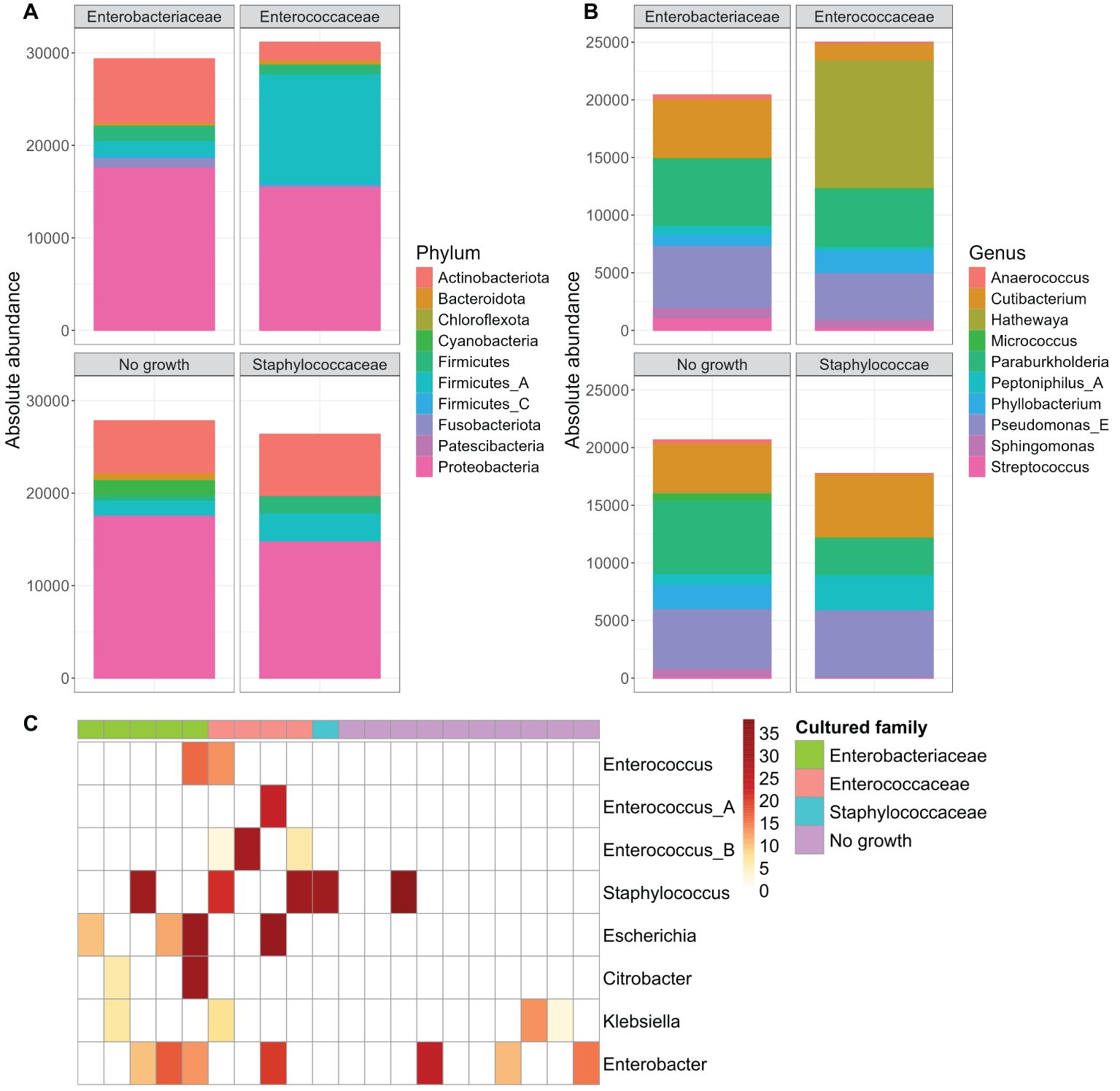
Figure 4. Abundant phyla and genera according to microbial growth in culture, grouped by family. Ten most abundant phyla (A) and genera (B) from 16S rDNA-sequencing grouped by the family of the cultured bacteria (Enterobacteriaceae n=5, Enterococcaceae n=4, Staphylococcaceae n=1) or by no observed growth (n=10). Read counts are normalized to the sample size (n) of each group. The y-axis represents absolute abundance. Heatmap of genera abundances that were cultured in the samples. Sample abundances were normalized using Cumulative Sum Scaling (CSS) (C).
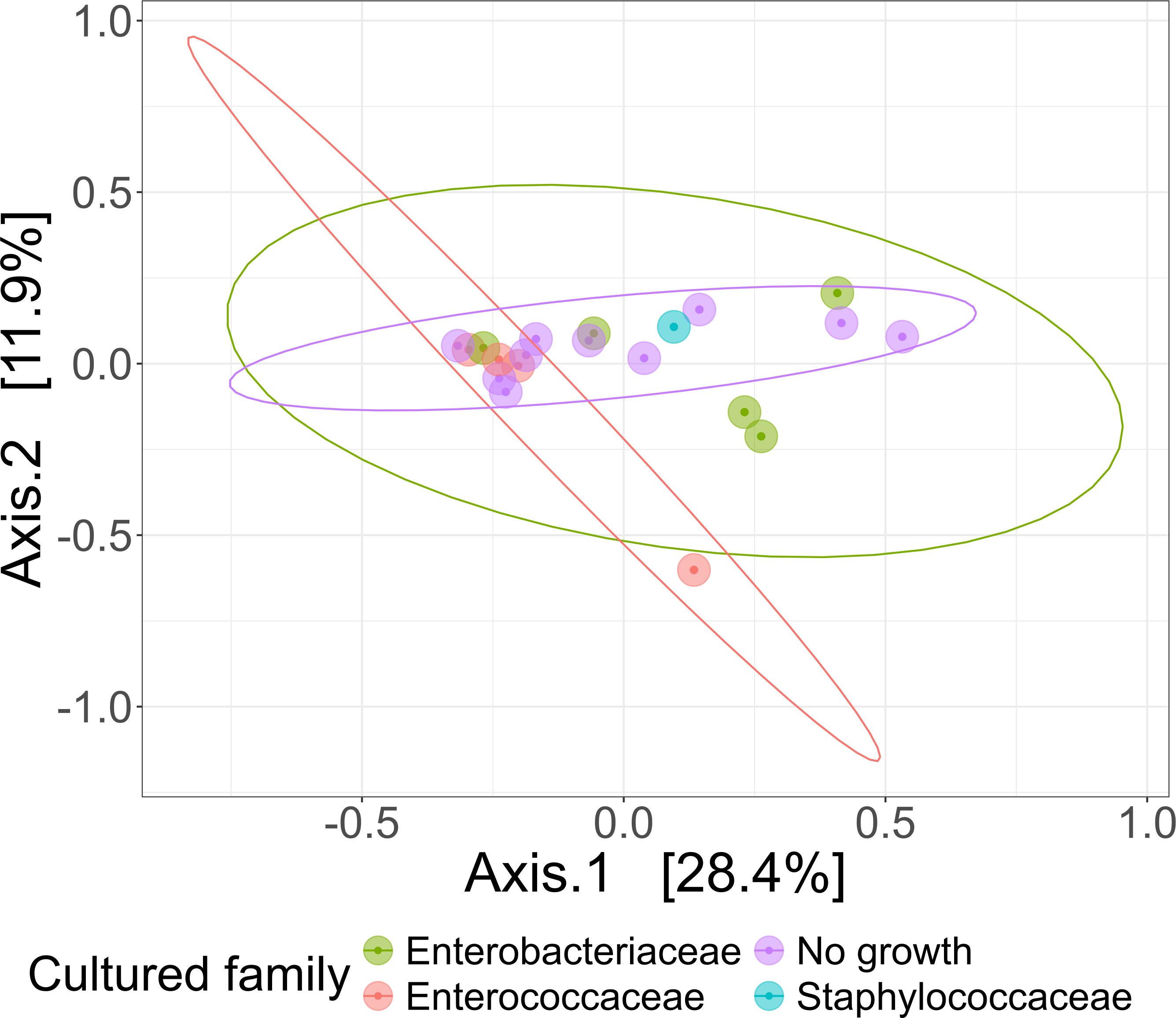
Figure 5. Comparison of beta diversities of microbial communities of patient samples, grouped based on their culture growth. PCoA visualization of bacterial community composition of different bacterial culture growth groups grouped by the family of the cultured bacteria (Enterobacteriaceae n=5, Enterococcaceae n=4, Staphylococcaceae n=1) or by no observed growth (n=10). Beta diversity measured with Bray-Curtis Dissimilarity Distances and visualized using Principal Coordinates Analysis (PCoA) plot.
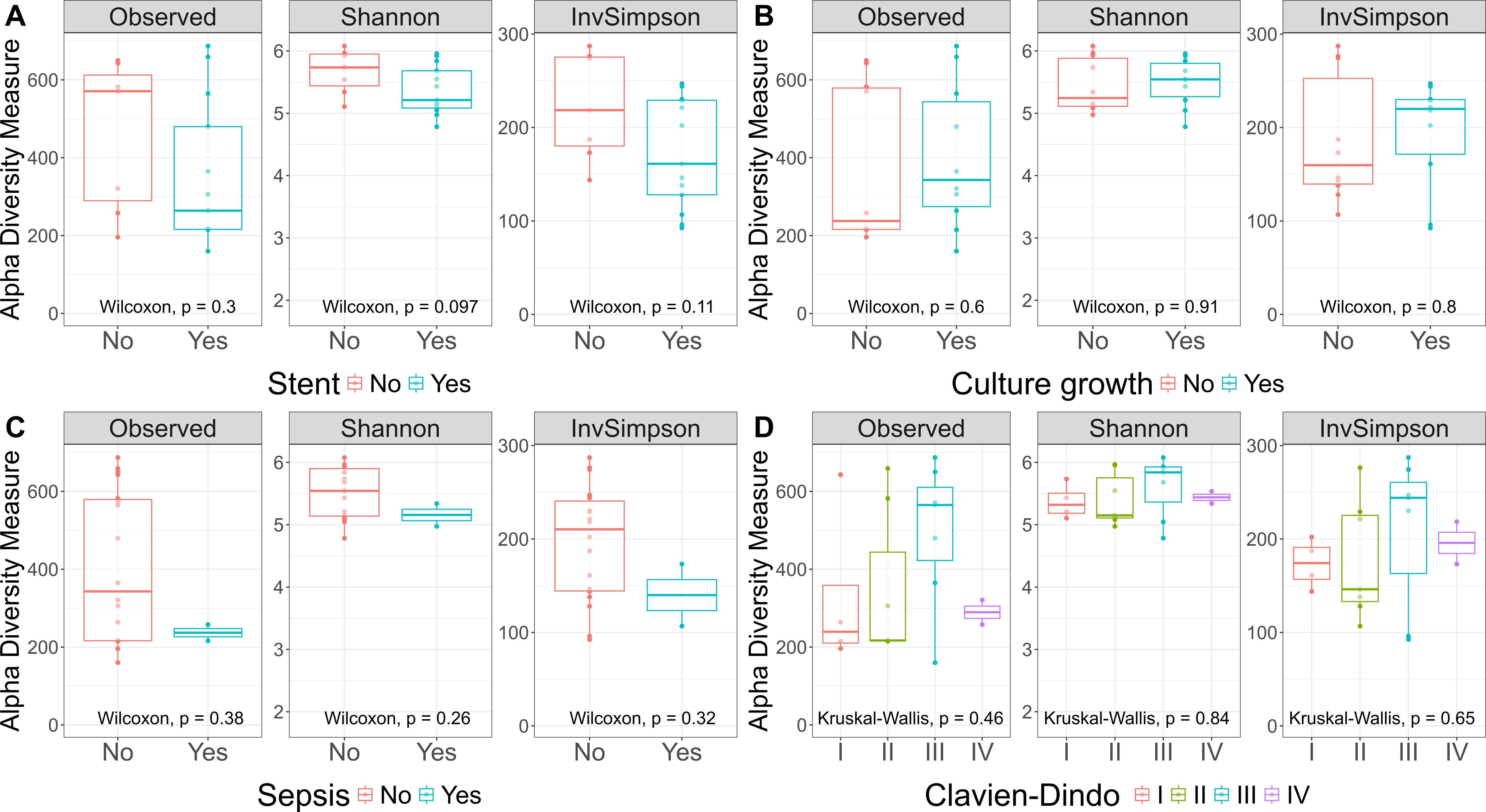
Figure 6. Alpha Diversity. Observed, Shannon, and InvSimpson Alpha Diversity. This boxplot showcases three alpha diversity indices across various patient groups: patients with and without stent (A), patients with and without microbial growth in culture (B), patients with post-surgery sepsis (C), and Clavien-Dindo classification groups (D). The central line within each box represents the median diversity value. The box spans the interquartile range (IQR), from the 25th to the 75th percentile, illustrating the middle 50% of the data. Whiskers extend to the smallest and largest values within 1.5 times the IQR from the quartiles. The Wilcoxon rank-sum test and the Kruskal-Wallis test showed no significant differences between the conditions for any of the alpha diversity indices.
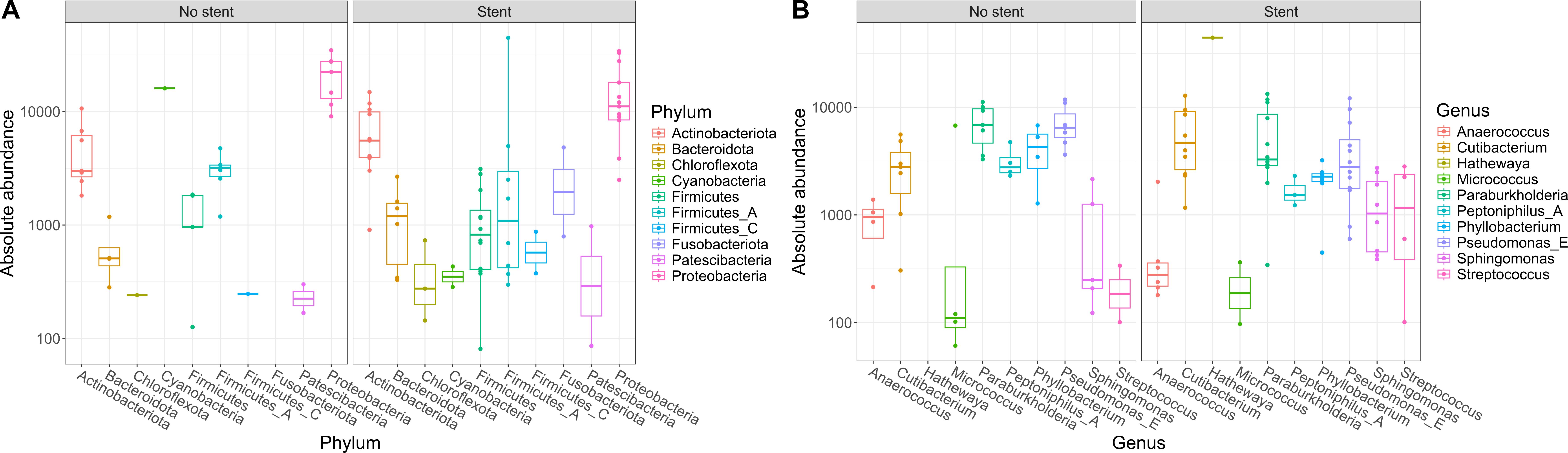
Figure 7. Abundant phyla and genera according to stent presence. Ten most abundant phyla (A) and genera (B) identified in 16S rDNA-sequencing grouped according to stent presence. The y-axis represents absolute abundance. The central line within each box represents the median abundance value. The box spans the interquartile range (IQR), covering the 25th to 75th percentiles, thus illustrating the middle 50% of the data. Whiskers extend to the smallest and largest values within 1.5 times the IQR from the quartiles. Each point on the plot corresponds to one sample.
Discussion
Pancreatic cancer is one of the most common causes of cancer mortality in developed countries (Raimondi et al., 2009). During recent years, the pancreas´ microbiome turned in the focus of cancer research (Picardo et al., 2019), as alterations in the microbiome may lead to disease development and progression (Frost et al., 2022). In cancers not directly linked to known oncogenic microbes (e.g. Helicobacter pylori, HPV, EBV or HBV), accumulating evidence suggests that microbial - particularly bacterial -colonization of tumor tissue actively contributes to the tumor microenvironment (Sepich-Poore et al., 2021). Recent studies employing advanced sequencing technologies, such as those by Galeano Niño et al., demonstrate that the intratumoral microbiota is organized into distinct microniches and functionally impacts tumor biology by activating oncogenic pathways (e.g. JUN/FOS) and immune-suppressive mechanisms (e.g. JAK–STAT), thereby promoting cancer progression (Galeano Niño et al., 2022). This microbial advantage in tumor progression may stem from enhanced survival benefits under fluid shear stress in the circulatory system, as observed in bacterial-colonized tumor cells (Bullman et al., 2017; Fu et al., 2022). In studies examining intratumoral bacteria in pancreatic cancer, Geller et al. demonstrated bacterial colonization in 76% of human PDAC samples, predominantly by Gammaproteobacteria. These bacteria were shown to inactivate the chemotherapy drug gemcitabine through cytidine deaminase (CDDL) activity, thereby promoting treatment resistance, an effect that was reversible with antibiotic administration (Geller et al., 2017). Furthermore, the correlation between metabolic and genetic subtypes in pancreatic cancer highlights the need to further investigate microbiota-metabolism interactions. Notably, early-stage tumors exhibit elevated serum polyamines, a microbial-linked metabolite that could serve as a noninvasive diagnostic biomarker for pancreatic cancer (Mendez et al., 2020; Zhang et al., 2020; Ling and Kalthoff, 2021).
The role of gut microbiota in modulating the efficacy of anticancer treatment and promoting resistance to chemotherapeutic drugs or immune checkpoint inhibitors has been known for several years (Cheng et al., 2020). Recently, it could be shown that pancreatic cancer tissue comprises a more abundant microbiome compared to normal pancreatic tissue both in humans as well as in mice and that selected bacteria are differentially increased in pancreatic cancer tissue, compared to the gut microbiome (Pushalkar et al., 2018). In the first prospective evaluation of our patient cohort over a three-year period, we could prove a microbiological colonization of pancreatic tissue in almost a third of our patients. This microbiological colonization in our collective seems to be promoted by preoperatively inserted bile duct stents as we found significantly more microbiological colonization of the pancreatic tumor in stented patients. The influence of preoperative bile duct stenting on the biliary microbiome was already shown earlier (Scheufele et al., 2017). Alterations of the microbiome in patients undergoing preoperative stent placement were also described by Langheinrich et al (Langheinrich et al., 2020). In this cohort, an increased rate of POPF in stented patients was observed (Langheinrich et al., 2020). We, however, found no difference in fistula rates in our patient cohort although the rate of stented patients was significantly higher in the group with a positive microbiological colonization. Nalluri et al. found a significantly higher rate of positive bacterial colonization of pancreatic tumor tissue in patients with preoperative bile duct stenting, too (Nalluri et al., 2021). Moreover, they observed an association of neoadjuvant chemotherapy with specific alterations of the intra-tumor bacteria in PDAC patients (Nalluri et al., 2021). The alteration of the biliary microbiome by neoadjuvant chemotherapy in PDAC patients was also described by Goel et al., showing significantly more enterococci and Klebsiella in the bile of these patients, but without influence on surgical site infections or POPF (Goel et al., 2019). Similar results, namely an influence of neoadjuvant chemotherapy on the biliary microbiome, but without impact on infectious postoperative outcomes, were found by Nadeem et al (Nadeem et al., 2021). Actually, in our patient collective, we observed an association of neoadjuvant chemotherapy and positive microbiological findings in our evaluation of the first 60 patients after a one-year period. However, in analyzing the entire patient collective after this three-year period, we couldn´t find a significant association between neoadjuvant chemotherapy and microbiological colonization of the pancreatic tumor any more. Another study could show that the bacteria that coexist in the tumor tissues of pancreatic and biliary tract cancer were relatively common to those localized in pancreatic and gastric juice, suggesting that they might originate from these environments (Okuda et al., 2022). Bacterial spread to the pancreas by blood stream, transmurally from the colon or by reflux into the pancreatic duct could already be shown in an animal model in the early nineties of the last century (Widdison et al., 1994). A Chinese review from 2019 showed a summary of microbes influencing tumor development and progression in pancreatic cancer, mentioning amongst others enterococcus species and E. coli as important bacteria leading to the development of PDAC (Wei et al., 2019); these species were also frequent in our patient collective. Especially E. coli seems to be associated with more complications following pancreatic surgery, as we could find a significantly higher rate of postpancreatectomy hemorrhage in these patients. In addition, we observed a trend towards more CR-POPF and towards a higher rate of DGE in the presence of E. coli in the pancreatic tumor. Riquelme et al. could show a different microbiome in resectable PDAC patients with short- and long-term survival by 16S-rRNA sequencing, so that the microbiome seems to influence the hosts immune response against tumor cells and thereby the long-term outcome of PDAC patients (Riquelme et al., 2019). Especially Pseudoxanthomonas, Saccharopolyspora and Streptomyces spp. were associated with long-term survival in this cohort (Riquelme et al., 2019). In our patient cohort, we could find a trends towards a negative influence of a microbiological colonization of the pancreas tumor in PDAC patients on long-term survival in these patients, but without reaching statistical significance. This may be attributed to the relatively small number of PDAC patients with positive microbiological findings in our cohort; therefore, further studies with a larger patient population are needed. Moreover, the RNA-sequencing method used by Riquelme et al. might be more precise in revealing microbiological findings than the standard microbiological culture of tumor tissue used in our study. In our small collective of 20 PDAC patients with additional 16S-rDNA sequencing, we could find a trend towards a reduced alpha diversity in patients with preoperatively inserted bile duct stent. As we could additionally show a trend towards a reduced survival in stented patients, this might support the findings of Riquelme et al. in terms of a higher alpha-diversity in long-term survivors (Riquelme et al., 2019). Furthermore, in our patient cohort, we could find significantly more patients with advanced lymph node involvement (N2-stages) in the group with a positive microbiological culture of the pancreatic tumor, suggesting a more aggressive tumor type in these patients, consequently leading to a poorer survival in this group. A Korean group used extracellular vehicles and 16S-rRNA sequencing to identify the composition and diversity of the microbiome in tissues of pancreatic cancer (Jeong et al., 2020). This group observed differences in the microbiome depending on the rate of lymph node metastasis as well. Moreover, a change in the microbiome depending on the primary tumor size was described (Jeong et al., 2020). In our collective, however, we didn´t observe any correlation between tumor size and the rate of microbiological colonization, at least not by using conventional microbiological culture techniques. Here, further studies with 16S-rRNA sequencing of a larger collective might be warranted.
There are several limitations of our study. First, it is a single center study, even though of a University Hospital. Second, we predominantly performed the standard microbiological cultivation of pancreatic tumor tissue which may have led to less sensitive results concerning bacterial and fungal colonization, compared to 16S-rDNA sequencing methods. However, this study provides an insight of microbiological species associated with pancreatic tumor tissue and their potential influence on patient outcome. Further studies are needed to reveal a closer look on the PDAC microbiome and its influence on oncological long-term outcome.
Data availability statement
The raw sequences generated in this project were deposited in the European Nucleotide Archives (ENA, https://www.ebi.ac.uk/ena) and are available under study number PRJEB89295 and samples accession numbers ERS24440838-ERS24440857.
Ethics statement
The studies involving humans were approved by Ethics Committee of Albert-Ludwigs-University Freiburg, Germany. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
EB: Conceptualization, Data curation, Visualization, Writing – original draft, Writing – review & editing. JS: Data curation, Writing – review & editing. MB: Formal analysis, Investigation, Visualization, Writing – review & editing. SP-C: Formal analysis, Investigation, Visualization, Writing – review & editing. SC: Resources, Writing – review & editing. SF-F: Resources, Writing – review & editing. UW: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1521952/full#supplementary-material
References
Aykut, B., Pushalkar, S., Chen, R., Li, Q., Abengozar, R., Kim, J. I., et al. (2019). The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. doi: 10.1038/s41586-019-1608-2
Babraham Bioinformatics - FastQC Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed November 1, 2024).
Balachandran, V. P., Łuksza, M., Zhao, J. N., Makarov, V., Moral, J. A., Remark, R., et al. (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. doi: 10.1038/nature24462
Barnett, D. J., Arts, I. C., and Penders, J. (2021). microViz: an R package for microbiome data visualization and statistics. J. Open Source Softw. 6(63), 3201, 1–4. doi: 10.21105/joss.03201
Bassi, C., Dervenis, C., Butturini, G., Fingerhut, A., Yeo, C., Izbicki, J., et al. (2005). Postoperative pancreatic fistula: an international study group (ISGPF) definition. Surgery 138, 8–13. doi: 10.1016/j.surg.2005.05.001
Bassi, C., Marchegiani, G., Dervenis, C., Sarr, M., Abu Hilal, M., Adham, M., et al. (2017). The 2016 update of the International Study Group (ISGPS) definition and grading of postoperative pancreatic fistula: 11 Years After. Surgery 161, 584–591. doi: 10.1016/j.surg.2016.11.014
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1038/nmeth.f.303
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cheng, W. Y., Wu, C.-Y., and Yu, J. (2020). The role of gut microbiota in cancer treatment: friend or foe? Gut 69, 1867–1876. doi: 10.1038/nature09944
de Martel, C., Ferlay, J., Franceschi, S., Vignat, J., Bray, F., Forman, D., et al. (2012). Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 1, 607–615. doi: 10.1016/S1470-2045(12)70137-7
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Frost, F., Weiss, F. U., and Lerch, M. M. (2022). Rolle des Mikrobioms bei Erkrankungen des Pankreas. Internist (Berl) 63, 372–378. doi: 10.1016/j.cell.2019.07.008
Fu, A., Yao, B., Dong, T., Chen, Y., Yao, J., Liu, Y., et al. (2022). Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185, 1356–1372.e26. doi: 10.1016/j.cell.2022.02.027
Galeano Niño, J. L., Wu, H., LaCourse, K. D., Kempchinsky, A. G., Baryiames, A., Barber, B., et al. (2022). Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817. doi: 10.1038/s41467-021-26614-z
Geller, L. T., Barzily-Rokni, M., Danino, T., Jonas, O. H., Shental, N., Nejman, D., et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. doi: 10.1126/science.aah5043
Goel, N., Nadler, A., Reddy, S., Hoffman, J. P., and Pitt, H. A. (2019). Biliary microbiome in pancreatic cancer: alterations with neoadjuvant therapy. HPB (Oxford) 21, 1753–1760. doi: 10.1016/j.hpb.2019.04.005
Jeong, J.-Y., Kim, T.-B., Kim, J., Choi, H. W., Kim, E. J., Yoo, H. J., et al. (2020). Diversity in the extracellular vesicle-derived microbiome of tissues according to tumor progression in pancreatic cancer. Cancers (Basel) 12, 2346. doi: 10.5483/BMBRep.2018.51.9.153
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12. Available online at: https://github.com/raivokolde/pheatmap.
Langheinrich, M., Wirtz, S., Kneis, B., Gittler, M. M., Tyc, O., Schierwagen, R., et al. (2020). Microbiome patterns in matched bile, duodenal, pancreatic tumor tissue, drainage, and stool samples: association with preoperative stenting and postoperative pancreatic fistula development. J. Clin. Med. 9, 2785. doi: 10.1007/s00268-007-9388-5
Liang, X., Zhu, Y., Bu, Y., Dong, M., Zhang, G., Chen, C., et al. (2024). Microbiome and metabolome analysis in smoking and non-smoking pancreatic ductal adenocarcinoma patients. BMC Microbiol. 24, 541. doi: 10.1186/s12866-024-03688-5
Lichtenegger, A. S., Posadas-Cantera, S., Badr, M. T., and Häcker, G. (2024). Comparison of the diversity of anaerobic-cultured gut bacterial communities on different culture media using 16S rDNA sequencing. J. Microbiol. Methods 224, 106988. doi: 10.1016/j.mimet.2024.106988
Ling, Q. and Kalthoff, H. (2021). Transportome malfunctions and the hallmarks of pancreatic cancer. Rev. Physiol. Biochem. Pharmacol. 181, 105–127. doi: 10.1007/112_2020_20
McAllister, F., Khan, M. A. W., Helmink, B., and Wargo, J. A. (2019). The tumor microbiome in pancreatic cancer: bacteria and beyond. Cancer Cell 36, 577–579. doi: 10.1016/j.ccell.2019.11.004
McMurdie, P. J. and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mendez, R., Kesh, K., Arora, N., Di Martino, L., McAllister, F., Merchant, N., et al. (2020). Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis 41, 561–570. doi: 10.1093/carcin/bgz116
Mitsuhashi, K., Nosho, K., Sukawa, Y., Matsunaga, Y., Ito, M., Kurihara, H., et al. (2015). Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 6, 7209–7220. doi: 10.18632/oncotarget.3109
Nadeem, S. O., Jajja, M. R., Maxwell, D. W., Pouch, S. M., and Sarmiento, J. M. (2021). Neoadjuvant chemotherapy for pancreatic cancer and changes in the biliary microbiome. Am. J. Surg. 222, 3–7. doi: 10.1016/j.amjsurg.2020.09.042
Nalluri, H., Jensen, E., and Staley, C. (2021). Role of biliary stent and neoadjuvant chemotherapy in the pancreatic tumor microbiome. BMC Microbiol. 2, 280. doi: 10.1111/j.1442-9993.1993.tb00438.x
Okuda, S., Hirose, Y., Takihara, H., Okuda, A., Ling, Y., Tajima, Y., et al. (2022). Unveiling microbiome profiles in human inner body fluids and tumor tissues with pancreatic or biliary tract cancer. Sci. Rep. 12, 8766. doi: 10.1093/nar/gkp847
Parks, D. H., ChuvoChina, M., Chaumeil, P.-A., Rinke, C., Mussig, A. J., and Hugenholtz, P. (2020). A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086. doi: 10.1038/s41587-020-0501-8
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Picardo, S. L., Coburn, B., and Hansen, A. R. (2019). The microbiome and cancer for clinicians. Crit. Rev. Oncol. Hematol. 141, 1–12. doi: 10.1016/j.critrevonc.2019.06.004
Pushalkar, S., Hundeyin, M., Daley, D., Zambirinis, C. P., Kurz, E., Mishra, A., et al. (2018). The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8, 403–416. doi: 10.1158/2159-8290.CD-17-1134
Raimondi, S., Maisonneuve, P., and Lowenfels, A. B. (2009). Epidemiology of pancreatic cancer: an overview. Nat. Rev. Gastroenterol. Hepatol. 6, 699–708. doi: 10.1038/nrgastro.2009.177
R Core Team. R (2013). A language and environment for statistical computing: reference index (Vienna: R Foundation for Statistical Computing).
Riquelme, E., Zhang, Y., Zhang, L., Montiel, M., Zoltan, M., Dong, W., et al. (2019). Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12. doi: 10.1016/j.cell.2019.07.008
Scheufele, F., Aichinger, L., Jäger, C., Demir, I. E., Schorn, S., Sargut, M., et al. (2017). Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br. J. Surg. 104, e182–e188. doi: 10.1002/bjs.10450
Sepich-Poore, G. D., Zitvogel, L., Straussman, R., Hasty, J., Wargo, J. A., and Knight, R. (2021). The microbiome and human cancer. Science 371. doi: 10.1126/scitranslmed.aax0876
Wei, M.-Y., Shi, S., Liang, C., Meng, Q.-C., Hua, J., Zhang, Y.-Y., et al. (2019). The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol. Cancer 18, 97. doi: 10.1017/S0029665112000158
Wente, M. N., Bassi, C., Dervenis, C., Fingerhut, A., Gouma, D. J., Izbicki, J. R., et al. (2007a). Delayed gastric emptying (DGE) after pancreatic surgery: a suggested definition by the International Study Group of Pancreatic Surgery (ISGPS). Surgery 142, 761–768. doi: 10.1016/j.surg.2007.05.005
Wente, M. N., Veit, J. A., Bassi, C., Dervenis, C., Fingerhut, A., Gouma, D. J., et al. (2007b). Postpancreatectomy hemorrhage (PPH): an International Study Group of Pancreatic Surgery (ISGPS) definition. Surgery 142, 20–25. doi: 10.1016/j.surg.2007.02.001
Wetzel, S., Müller, A., Kohnert, E., Mehrbarzin, N., Huber, R., Häcker, G., et al. (2023). Longitudinal dynamics of gut bacteriome and mycobiome interactions pre- and post-visceral surgery in Crohn’s disease. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1275405
Widdison, A. L., Karanjia, N. D., and Reber, H. A. (1994). Routes of spread of pathogens into the pancreas in a feline model of acute pancreatitis. Gut 35, 1306–1310. doi: 10.1136/gut.35.9.1306
Xue, C., Chu, Q., Zheng, Q., Yuan, X., Su, Y., Bao, Z., et al. (2023). Current understanding of the intratumoral microbiome in various tumors. Cell Rep. Med. 4, 100884. doi: 10.1126/science.abc4552
Keywords: pancreatic tumor, microbiological colonization, postoperative complications, mortality, survival
Citation: Biesel EA, Sundheimer J, Badr MT, Posadas-Cantera S, Chikhladze S, Fichtner-Feigl S and Wittel UA (2025) Microbiological colonization of the pancreatic tumor affects postoperative complications and outcome after pancreatic surgery. Front. Cell. Infect. Microbiol. 15:1521952. doi: 10.3389/fcimb.2025.1521952
Received: 03 November 2024; Accepted: 17 April 2025;
Published: 30 May 2025.
Edited by:
Jitender Monga, Henry Ford Health System, United StatesReviewed by:
Avinash Lomash, Medanta The Medicity Hospital, IndiaVarun Suroliya, Artemis Hospitals, India
Copyright © 2025 Biesel, Sundheimer, Badr, Posadas-Cantera, Chikhladze, Fichtner-Feigl and Wittel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esther Anna Biesel, ZXN0aGVyLmJpZXNlbEB1bmlrbGluaWstZnJlaWJ1cmcuZGU=
 Esther Anna Biesel
Esther Anna Biesel Johanna Sundheimer1
Johanna Sundheimer1 Mohamed Tarek Badr
Mohamed Tarek Badr Sara Posadas-Cantera
Sara Posadas-Cantera Stefan Fichtner-Feigl
Stefan Fichtner-Feigl