- 1Laboratory of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Science and Peking Union Medical College, Kunming, Yunnan, China
- 2Department of Laboratory Medicine, Fujian Key Clinical Specialty of Laboratory Medicine, Women and Children’s Hospital, School of Medicine, Xiamen University, Xiamen, China
- 3National Health Commission (NHC) Key Laboratory of Research on Quality and Standardization of Biotech Products and National Medical Products Administration (NMPA) Key Laboratory for Quality Research and Evaluation of Biological Products, National Institutes for Food and Drug Control, Beijing, China
- 4Department of Pulmonary and Critical Care Medicine, Xishuangbanna Dai Autonomous Prefecture People’s Hospital, Jinghong, Yunnan, China
- 5Yunnan Key Laboratory of Vaccine Research and Development on Severe Infectious Diseases, Institute of Medical Biology, Chinese Academy of Medical Science & Peking Union Medical College, Kunming, Yunnan, China
Introduction: Various strains emerged in B. pertussis re-emergence, the pathogenic characteristics and mechanisms remain elusive. We aimed to explore the relationship between the in vivo transcriptome and colonization advantage of various pertussis clinical strains during the B. pertussis re-emergence.
Methods: Four pertussis strains were isolated from clinically suspected cases by active surveillance. The phylogenetic relationships of clinical strains and global isolates were compared by a genome-wide SNP-based phylogenetic tree and allele genotyping. LC-MS/MS analysis and binding affinity detection allowed the identification of expression and antigenicity of pertactin. The characteristics of infection and immunity of clinical strains were compared in a BALB/c mouse aerosol challenge model. In vivo RNA-seq analysis was performed in NSG mouse model to describe the transcriptome during infection, and verified by detecting biofilm formation and paraquat tolerance.
Results: The partial pertactin-deficient strain BP-L2 was first reported. It showed significantly enhanced tracheal colonization compared to both CS and BP-L1 strains in naive mice (P < 0.0001 vs. CS) and exhibited superior fitness over BP-L1 in immunized mice (P < 0.001). BP-L1 showed superior lung colonization (P < 0.0001) and tissue-resident memory T cell induction versus BP-L2 and CS (P < 0.001). Colonization dominance of BP-L1 in lungs and BP-L2 in trachea aligned with the pathological injury (P < 0.05) and the inflammatory cytokine enhancement (IL-6 in lungs of BP-L1 group, P < 0.01). In vivo RNA-seq results revealed that BP-L2 significantly upregulated relA (log2FC = 2.1, FDR P-value = 0.019) and sodA (log2FC =2.4, FDR P-value = 8.61E-06) compared to BP-L1, functionally linked to enhanced stringent response and oxidative stress defense. BP-L1 exhibited significant in vivo bipA upregulation over BP-L2 (log2FC = 1.8, FDR P-value = 0.027) without concurrent biofilm enhancement (P = 0.51 vs. BP-L2). Furthermore, the BP-L2 and BP-L3 strains of the same ptxP1-ptxA1-fhaB3 lineage showed significantly higher paraquat tolerance than other strains (P < 0.001), showing extremely high SODs activity.
Conclusion: The emerging pertussis strains exhibit different colonization advantages in the trachea or lungs, which will influence the transmission patterns of the clinical strains. The tracheal colonization advantage of the partial pertactin-deficient strain may be associated with the overexpression of the relA and sodA in vivo infection.
Introduction
Bordetella pertussis (B. pertussis) is the major causative agent of pertussis, a highly contagious, acute respiratory disease in humans. Since the 1980s, there has been a significant increase in the global incidence of pertussis, known as re-emergence of pertussis, which has become a serious public health problem (Burns et al., 2014). China introduced the combined diphtheria-tetanus-whole-cell pertussis (DTwP) vaccine in 1978. In 2007, the combined diphtheria-tetanus-acellular pertussis (DTaP) vaccine was incorporated into the Expanded Program on Immunization (EPI), gradually replacing DTwP until the transition was completed in 2010 (Meng et al., 2018b).The current immunization schedule of China administers the primary DTP series at 2, 4, and 6 months of age, followed by booster doses at 18 months and 6 years of age and the coverage for the three primary doses exceeds 90% (Meng et al., 2018a). However, there has also been an increasing incidence of pertussis in China since 2013, peaking in 2024 (National Disease Control and Prevention Administration, 2025). The re-emergence of pertussis may be related to various factors, including increased awareness, improved diagnostic methods (Belcher and Preston, 2015), suboptimal vaccination (Diavatopoulos and Edwards, 2017), waning vaccine-induced immunity (Klein et al., 2012) and pathogen adaptation (Ma et al., 2021). However, waning immunity of acellular pertussis vaccine (aP) and pathogen adaptation are considered to be important factors affecting the resurgence of B. pertussis.
In recent epidemiological studies of pertussis conducted worldwide, a large number of new B. pertussis epidemic strains were discovered. The latest B. pertussis epidemic strains exhibit multiple antigen alleles with changes, antigen deficiency and other characteristics (Octavia et al., 2012; Ma et al., 2021) under vaccine selection pressure. The re-emergence of pertussis was consistent with the emergence of strains harboring a specific allele of the pertussis toxin operon promoter, ptxP3, which has been reported to be associated with increased pertussis toxin expression (Mooi et al., 2009). Notably, ptxP3 strains have spread globally, as they have emerged in several countries in Europe (Mooi et al., 2009), North America (Schmidtke et al., 2012) and Australia (Octavia et al., 2012). Furthermore, studies have reported a dominance of the bscI3 allele in the ptxP3 lineage, which is associated with decreased type III secretion system and may allow B. pertussis to reduce immune recognition (Xu et al., 2022). The fhaB allele associated with higher mutation rates was also found in the ptxP1 lineage in China, which may be associated with vaccine escape (Xu et al., 2019). However, the ptxP3 strains were still the dominant type in China and most of them were macrolide resistant strains carrying the 23sRNA A2047G mutation (Fu et al., 2023).
In addition, vaccine-associated antigen protein-deficient strains, such as those lacking pertussis toxin (PT), filamentous hemagglutinin (FHA) and pertactin (PRN), have also been identified among the currently dominant isolates. However, pertussis strains lacking additional vaccine antigens have been identified (Bouchez et al., 2015; Williams et al., 2016), though their prevalence remains much lower than that of PRN-deficient isolates (Ma et al., 2021). This fact highlights critical gaps in understanding both the biological role and immunological consequences of PRN during B. pertussis infection.
The ptxP3 allele and PRN-deficient strains are predominant in countries with high vaccination rates (Zhao et al., 2021) and may compromise the protective efficacy of aP vaccines. Although the allele profiles of current epidemic strains differ from historical variants, very few studies have evaluated the infection characteristics and immune protection induced by the current clinical strains (Mooi et al., 2009; Safarchi et al., 2015). The relationship between strain-specific colonization/immune adaptations and transmission efficiency remains unclear. Furthermore, with the development of methods to isolate B. pertussis in the infected state in vivo (Wong et al., 2019), the relative contributions of allelic variation versus infection-phase gene regulation to phenotypic differences require further investigation.
Previous studies have demonstrated that specific gene functions during B. pertussis infection facilitate bacterial colonization within distinct locations of the host respiratory tract (Belcher et al., 2021). For example, the adenylate cyclase toxin-hemolysin (ACT) facilitates early stages of airway colonization by disrupting the phagocytic clearance (Espinosa-Vinals et al., 2025); a study using MyD88-deficient C57BL/6J mice—a model replicating human catarrhal-phase infection—demonstrated that FhaB and Fim are essential for pertussis nasal colonization (Holubova et al., 2022). In addition, a study found the sigma factor RpoN of Bordetella bronchiseptica is involved in bacterial motility and initial biofilm formation, and is essential for tracheal colonization (Ma et al., 2024). The above studies emphasize that the analysis of the expression of key genes during bacterial infection will help discover the mechanisms that cause different infection colonization characteristics of pertussis strains.
In this study, whole-genome sequencing was conducted for all four isolated strains (BP-L1 to BP-L4), which were isolated from 158 clinically suspected cases in the Xishuangbanna region of Yunnan Province, China. Phylogenetic analysis revealed that the four isolates were divided into two major clades, with BP-L2 identified as a partial PRN-deficient strain. Combined analysis of post-infection antibodies, antibody binding affinity and LC-MS/MS confirmed the expression of a partially defective PRN in BP-L2. Subsequently, the pertussis aerosol challenge in mice revealed that BP-L1 and BP-L2 had significantly increased colonization in the lungs and trachea (P < 0.0001 vs. other strains), respectively, and caused pathological injury and elevated inflammatory cytokines at the corresponding locations. BP-L1 infection induced the highest proportion of lung tissue-resident memory T (Trm) cells (P < 0.001), whereas BP-L2 exhibited superior fitness in immunized hosts (P < 0.001 vs. BP-L1). In vivo RNA-seq revealed BP-L2-specific upregulation of stringent response (log2FC = 2.1, FDR P-value = 0.019) and antioxidant stress genes (log2FC =2.4, FDR P-value = 8.61E-06) over the BP-L1. This oxidative stress tolerance was functionally validated through paraquat challenge, with BP-L2 and BP-L3 within the same ptxP1-ptxA1-fhaB3 lineage demonstrating higher survival rates to other strains (P < 0.001). Overall, for the first time, we explored the different infection characteristics and possible underlying mechanisms of various pertussis clinical strains by combining aerosol infection model and in vivo RNA-seq results.
Materials and methods
Ethics approval statement
This study received ethical approval from the People’s Hospital of Xishuangbanna Dai Autonomous Prefecture Ethics Committee (Approval No. 1706001x) with written informed consent from all participants, whose privacy rights were strictly protected. Animal experiments were conducted under specific pathogen-free conditions at the Institute of Medical Biology, Chinese Academy of Medical Sciences (IMBCAMS), in compliance with protocols approved by the Institutional Animal Care and Use Committee (Approval Nos. DWSP202011004 and DWSP201911014).
B. pertussis strains and vaccine
Between October 2018 and October 2021, nasopharyngeal swabs were collected from 158 suspected pertussis cases in Xishuangbanna Dai Autonomous Prefecture. All samples were spread onto Reagan-Lowe agar plates (Oxoid, Thermo Fisher Scientific, Waltham, United States) supplemented with 15% defibrinated sheep blood (Beijing Minhai Biotechnology Co., Ltd, Beijing, China) and Bordetella selective supplement (Oxoid), and cultured at 37°C for 3–7 days. Colonies exhibiting characteristic morphology (diameter 0.5 – 1.0 mm, mercury-drop appearance) were confirmed by the slide agglutination test with B. pertussis and Bordetella parapertussis antisera (Remel Europe Ltd., UK) and subsequent PCR confirmation as stated in the previous study (Wu et al., 2019). Four B. pertussis strains were successfully isolated from158 samples (isolation rate: 2.53%) and designated BP-L1 through BP-L4. Regarding the vaccine, the antigens in the aP vaccine we used were detoxified using glutaraldehyde and subsequently adsorbed onto aluminum hydroxide individually. Each dose of the aP vaccine contains 25 µg of PT, 25 µg of FHA and 8 µg of PRN. The aP vaccine was produced by the Institute of Medical Biology, Chinese Academy of Medical Sciences (IMBCAMS) under good manufacturing practice conditions (Sun et al., 2014) and then adjusted to 1/40 human dose for subsequent use.
Whole-genome sequencing
All four B. pertussis clinical isolates were subsequently subcultured on Bordet-Gengou agar plates supplemented with 15% defibrinated sheep blood and without cephalexin, incubated at 37°C for 72 hours. The resulting colonies were hemolytic and less than 1.0 mm in diameter, resembling mercury droplets and glistening. Then, DNA was isolated using a QIAGEN Genomic-tip 100/G kit (10243, Qiagen, Hilden, Germany) following the manufacturer’s instructions. The whole-genome sequencing process was performed according to the standard protocol provided by Oxford Nanopore Technologies. Bacterial genomic DNA was subjected to rigorous quality assessment using a Nanodrop One spectrophotometer (Thermo Fisher Scientific), a Qubit 4.0 Fluorometer (Thermo Fisher Scientific) and 0.35% agarose gel electrophoresis. Large DNA fragments (>20 kb) were size-selected using the BluePippin automated nucleic acid recovery system (Sage Science) with 0.75% agarose cassettes. Sequencing libraries were prepared with the SQK-LSK109 ligation sequencing kit (Oxford Nanopore Technologies). Final libraries were quantified via Qubit 4.0 Fluorometer and loaded onto FLO-MIN106D flow cells for sequencing on a MinION platform (Oxford Nanopore Technologies) using MinKNOW v22.05.0 software for real-time data acquisition. Raw fast5 data were base-called to fastq format using Guppy v3.2.6 integrated within the MinKNOW software package. Adapter sequences, low-quality reads (Q-score <7), and short fragments (<2000 bp) were filtered out using Porechop v0.2.4 and Filtlong v0.2.1. Filtered subreads were assembled into draft genomes with Canu v1.5. Final assemblies were annotated with Prokka v1.14.6, and all genomes achieved a minimum coverage depth of 50×, validated through alignment statistics and circular completeness checks. The SNPs, insertions and deletions were detected by mapping the contigs to the previously sequenced and annotated B. pertussis Tohama I genome using BLAST and were filtered as described in previous studies (Holt et al., 2008). The complete genomes of 84 pertussis isolates, used to analyze phylogenetic relationships were obtained from databases [PRJNA770762, PRJNA279196, PRJNA1178746, PRJNA695314, PRJNA530108] or recent studies (Bart et al., 2014; Boinett et al., 2015; Alai et al., 2022; Koide et al., 2022; Fu et al., 2023). The detailed strain information is provided in Supplementary Table 1. The assembled whole genome was aligned using snippy v3.1, with Tohama I as the reference genome, and the resulting phylogenetic tree was constructed using tvBOT (Xie et al., 2023). Allelic typing corresponds to the sequence of the Tohama I strain obtained from GenBank. The analysis was conducted using SeqKit v2.9.0, including only the exact sequences. Additionally, IS Finder was utilized to detect Insertion Sequence (IS) elements within the genomes (Park et al., 2012).
PRN purified by anion exchange chromatography
Referring to the previously reported methods (Li et al., 2016), after three subcultures at 36°C and 140 rpm for 48 hours in Stainer and Scholte medium (SS medium), pertactin from B. pertussis strains CS, BP-L1 and BP-L2 was initially extracted through heat treatment. Cells were re-suspended in five times the volume of PBS. The sample was centrifuged to remove the supernatant and other contaminating proteins. The suspension was incubated at 60°C for 3 hours and then centrifuged at 4°C, 8000 rpm for 15 minutes to remove bacterial cells. The supernatant was further concentrated to approximately 1/10 of its initial volume using ultrafiltration. The tangential-flow ultrafiltration module Vivaflow 200 (Sartorius, Germany), which consists of polyethersulfone with a molecular weight cutoff of 3 kDa, was used for ultrafiltration. The concentrated extract solution was dialyzed against 20 mM Tris-HCl, 1 mM EDTA, 1 mM PMSF, pH 7.8 (buffer A) at 4°C. The sample was subsequently centrifuged to eliminate any insoluble impurities prior to chromatographic purification. Anion exchange chromatography was utilized for the capture step, employing Capto adhere (Cytiva, Cat. No. 17544401, USA). Approximately 25 mL of Capto adhere was packed into a column. The column was equilibrated with 10 column volumes (CVs) of buffer A, followed by an additional wash with 2 CVs of the same buffer post-sample loading to remove unbound proteins. A step-gradient concentration of NaCl in buffer A was used, with 0.1 M for eluting the target protein and 1 M for regenerating the column. The flow rate was maintained at 2 mL/min throughout the process. Purification was conducted using the ÄKTA explorer 100 (GE Healthcare, USA), with detection of ultraviolet absorbance at 280 nm. For BP-L1 and CS strains, peak components emerging after approximately 2 CVs were collected, whereas all peak components from BP-L2 strains were recovered and combined as samples for testing.
LC–MS/MS analysis
Load the prepared PRN samples of various B. pertussis strains and protein markers into the wells of a 12% separating gel. Initiate electrophoresis at 80 V for 15 minutes, then increase the voltage to 120 V for an additional hour. Stain the gel with Coomassie Blue dye, excise three gel slices from each gel, and cut them into 1 mm³ cubes. Briefly centrifuge to pellet the slices and wash with 500 µL of 50 mM ammonium bicarbonate/acetonitrile (1:1, v/v) until destained. Remove the supernatant, dehydrate with 500 µL acetonitrile for 10 minutes. Rehydrate the slices in 10 mM DTT/50 mM ammonium bicarbonate at 56°C for 1 hour and repeat the acetonitrile dehydration. Treat the gel slices with 50 mM IAA/50 mM ammonium bicarbonate at room temperature for 1 hour in the dark, and then perform a final acetonitrile dehydration. Add enzyme digestion solution to cover the slices, incubate on ice for 45 minutes, and then add 5-20 µL of digestion solution to maintain hydration during the overnight incubation at 37°C. Collect the supernatant into a fresh tube, extract peptides with 100 µL of 50 mM ammonium bicarbonate/acetonitrile (1:2, v/v, 37°C, 1 hour), combine the extracts, and lyophilize to near dryness. Resuspend the peptides in 10 µL of 0.1% formic acid for LC-MS/MS analysis.
LC-MS/MS was performed on a Q Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer (Thermo Scientific) coupled with an Easy-nLC 1200 system. For each sample, 5 μL was loaded onto a C18 PepMap100 trap column (300 μm × 5 mm) and separated on a Thermo Acclaim PepMap RPLC analytical column (150 μm × 15 cm) using a 66 min gradient: 4-8% B (2 min), 8-28% B (43 min), 28-40% B (10 min), 40-95% B (1 min), 95% B (10 min) (mobile phase A = 0.1% formic acid in water; B = 20% 0.1% formic acid/80% acetonitrile) at a flow rate of 0.6 μL/min. Data-dependent acquisition mode was employed with full MS scans (300–1800 m/z) at 70,000 resolutions (AGC target 3×106 ions, max IT 100 ms) followed by top 15/20 MS/MS scans (HCD, 28% NCE) at 17,500 resolution (AGC target 1×105 ions, max IT 50 ms), using a 3 s cycle time.
The raw MS files were analyzed and searched against target protein database based on the species of the samples using Byonic (v4.2.4 from Protein Metrics, Cupertino, CA). The mass tolerance was set to 20 ppm and 0.02 Da for the precursor and the fragment ion respectively, with up to two missed cleavages allowed. Carbamidomethyl was used as a fixed modification and oxidation (M) (variable), Acetyl (Protein N-term) (variable) were used as a variable modification. Enzyme specificity was defined as chymotrypsin, trypsin, and Glu-C (Junker et al., 2006; Liang et al., 2024).
The binding affinities test by microscale thermophoresis
The isolated strains BP-L1, BP-L2, and CS B. pertussis were harvested in liquid SS medium. The PRN antigen was purified by column chromatography (Cytiva, USA). Rabbit Anti-B. pertussis pertactin antiserum (cat. #PRN11-S, Alpha Diagnostic International, San Antonio, TX) was used as the ligand. The binding affinities between PRN and ligands were determined according to the manufacturer’s protocol for the MO RED-NHS Kit (Sedivy, 2021) (NanoTemper Technologies GmbH, Munich, Germany).
B. pertussis aerosol challenge and vaccines programs
The B. pertussis strains utilized for the aerosol challenge were BP-L1 and BP-L2, which were isolated from clinical cases within this study. Additionally, the B. pertussis strain CS was included as a reference strain (Zhang et al., 2011). B. pertussis infection was induced through aerosol challenge with a concentration of 10^11 CFU/mL, administered via a nebulizer for 30 minutes (Jiang et al., 2021). A total of 132 four-week-old BALB/c mice, used in this study, were procured from Vital River Laboratory Animal Technology Co., Ltd. As previously described, the progression of the infection was monitored by counting colony-forming units (CFU) in lung and tracheal homogenates and nasal lavage at various intervals post-infection (p.i.) (McGuirk et al., 1998). Specifically, plates with CFU counts ranging from 30 to 300 were selected for counting, and the number of viable colonies was calculated based on the dilution factor. Four weeks post B. pertussis challenge, serum PRN-antibody levels were quantified using ELISAs. Furthermore, a total of 52 BALB/c mice, with an equal number of females and males, were vaccinated with a 1/40 human dose of the aforementioned aP vaccine (Zuo et al., 2021). Two doses were administered at 3-week intervals. All vaccinations were conducted under isoflurane anesthesia. Six weeks post-immunization, 48 mice were randomly divided into two groups for respiratory challenge with the BP-L1 strain or BP-L2 strain.
Tissue-resident memory T cell assay
The levels of tissue-resident memory T cells (Trm) in the lungs of mice from different groups were examined 9 weeks following respiratory challenge and 9 weeks post-aP vaccine immunization. The method has been detailed in previous research (Wilk et al., 2017). In brief, we administered an anti-mouse PE-CD45 Ab (eBioscience, San Diego, CA) intravenously to mice 10 minutes before they were euthanized, and the lung lymphocytes were isolated for subsequent flow cytometry, as previously described (Wilk et al., 2017). Flow cytometric analysis was performed on a Beckman Coulter CytoFLEX, and the results were analyzed using CytExpert v2.4.0.28.
B. pertussis in vivo and in vitro RNA-seq
B. pertussis cells from the in vivo infection state were isolated and subjected to in vivo RNA-seq. The specific methods used are detailed in the previous study (Wong et al., 2019). Specifically, 12 NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice, consisting of an equal number of males and females, were purchased from Shanghai Model Organisms Center, Inc., to increase the lung bacterial burden (Wong et al., 2019). The aerosol challenge of NSG mice was conducted using the same procedure as that for BALB/c mice. Specifically, B. pertussis infection was induced via aerosol challenge (10^11 CFU/mL) by administering a nebulizer for 30 minutes. Each of the BP-L1, BP-L2, and CS bacterial strains infected 4 NSG mice, with an equal number of males and females in each group. The lungs and trachea were extracted on the seventh day post-infection. B. pertussis cells were concentrated using a 70-µm-pore-size cell strainer (VWR), and the filtrate was subsequently passed through a 5-µm-pore-size filter (Minisart type 17594 cellulose acetate; Sartorius). The bacteria were then pelleted by final centrifugation at 4°C at 16,100 × g (Wong et al., 2019). For in vitro RNA sequencing analysis, B. pertussis strains were grown in SS liquid medium for 24 hours at 36°C and harvested for transcriptome sequencing.
For the RNA sequencing, rRNA was removed from total RNA using a Ribo-Zero rRNA Removal Kit Bacteria (Illumina, San Diego, CA). RNA-seq was performed on the Illumina NovaSeq 6000 platform. The reads were aligned to the B. pertussis Tohama I genome using Bowtie2 v2.2.8. DEGs were calculated using the edgeR package, with a difference fold threshold of 2 and an FDR-adjusted P (q value) of less than 0.05. The B. pertussis genome contains numerous repeated insertion sequences (IS), and to prevent errors from random mapping, all IS-related transposase reads were excluded from the analysis. Since 99% of the reads in the in vivo RNA sequencing data originated from the host, we increased the sequencing depth to 150 million 2×150-bp paired-end reads to ensure adequate coverage of Bordetella pertussis-derived sequences.
Biofilm formation assay
Following the methods of previous studies (Držmíšek et al., 2023), B. pertussis cells from CS and BP-L1 to BP-L4 were suspended in liquid SS media to achieve a final OD600 of 0.3. Triplicate samples of the diluted cultures (200 µL) were inoculated in parallel into 96-well non-treated tissue culture plates. The plates were incubated at 37°C for 72 hours. Loosely adherent and planktonic bacteria were removed by washing the wells three times with PBS. Adherent cells were stained with a 0.1% crystal violet solution for 45 minutes. The plates were then washed with PBS and dried, after which the adsorbed crystal violet was solubilized in 200 µL of 95% ethanol for 15 minutes. To quantify biofilm formation, the absorbance at A595 of the crystal violet staining was measured using a microplate reader (Thermo Fisher). The readings from the wells were adjusted against the substrate blank well, and each value was the average of the assays, with two replicates per assay. Each experiment was repeated three times.
Paraquat sensitivity assay
Adopting the methods from previous studies (Graeff-Wohlleben et al., 1997; Martins et al., 2018), the B. pertussis strains CS and BP-L1 to BP-L4 were cultured overnight in liquid SS medium. The bacterial cultures were then diluted in PBS to an optical density of OD590 = 0.1, and the CFU for each strain were determined on Reagan-Lowe agar plates supplemented with 15% defibrinated sheep blood. The entire 5 g of paraquat powder (Sigma) was dissolved in 50 mL of PBS, followed by sterile filtration and the transfer of 1,285 µL of the paraquat solution into 10 mL of bacterial suspension to achieve a final concentration of 50 mM. The samples were subsequently shaken at 37°C. After 30 minutes, the samples were diluted again, and the CFU was determined on the same Reagan-Lowe plates. The percentage of surviving bacteria for each strain was calculated, with each experiment being repeated three times.
Statistical analysis
All data are expressed as the mean ± SEM and were analyzed using GraphPad Prism v9.0 (GraphPad, San Diego, CA, USA). One-way analysis of variance (ANOVA) was utilized to assess the statistical significance of differences among three or more groups. A value of P < 0.05 was considered to indicate statistical significance. For transcriptome analysis, Bonferroni-corrected q values of less than 0.05 were deemed statistically significant.
Results
Whole-genome sequence characteristics of clinical strains
Whole-genome sequencing was conducted on all four clinical isolates in this study. Following rigorous quality assessments and data cleaning, between 60,262 and 111,052 sequence reads were obtained for BP-L1 to BP-L4, with mean quality scores ranging from 8.98 to 9.27 (refer to Supplementary Table 2). Allelic genotyping indicated that BP-L1 and BP-L4 possessed the ptxP3/ptxA1/prn2/fhaB1/bscI3 genotype; BP-L3 had the ptxP1/ptxA1/prn1/fhaB3/bscI1 genotype, and BP-L2 exhibited the ptxP1/ptxA1/fhaB3/bscI1 genotype. The phylogenetic relationships among 82 recent B. pertussis isolates and 2 reference strains are depicted in Figure 1. These strains were sourced from China, the USA, Australia, Japan, India, and the Netherlands, and were isolated between the years 2000 and 2024. The phylogenetic tree was constructed using whole-genome SNPs, with Tohama I (NC_002929.2) serving as the reference strain. The SNPs for BP-L1 to BP-L4 were detailed in Supplementary Table 3. The analysis also included multiple allelic types and macrolide resistance mutations, such as the 23sRNA A2047G. The phylogenetic tree identified two main lineages characterized by ptxP/fhaB/bscI allele profiles and the A2047G mutation. The ptxP1 cluster primarily consists of Chinese isolates (32 out of 35 strains), including BP-L2/L3 strains with the A2047G mutation and fhaB3 alleles. The ptxP3 cluster includes both Chinese and Western strains, with BP-L1/L4 strains featuring bscI3 alleles. The copy numbers of IS1002, IS481, and IS1663 in strains BP-L1 to BP-L4, CS, and the reference strain Tohama I were compared. All four clinical isolates exhibited nearly identical insertion sequence (IS) copy numbers (249 to 253 copies for IS481, 6 to 7 copies for IS1663, and 6 copies for IS1002), whereas CS and Tohama I had only 236 and 230 copies of IS481 in their genomes, respectively (see Supplementary Table 4 for details). In addition, IS481 showed consistent insertion in the gene coding regions and promoters of the four isolates. However, no insertion occurred in the known antigen gene coding regions or promoters. The specific insertion positions and functional annotation results are shown in Supplementary Table 5. More importantly, the PRN coding sequence of the BP-L2 strain was only 1,629 bp. It exhibited a substantial base deletion spanning positions 52 to 1156, corresponding to the deletion of amino acids at positions 18 to 391. The deleted segment is situated within the region encoding the P.69 protein, specifically the PRN antigen. The N-terminal signal sequence and the C-terminal self-transport element P.30 remained intact and matched the reference strain (Junker et al., 2006). Based on the sequencing results, the PRN protein encoded by BP-L2 comprises 174 amino acids, with a theoretical monomer molecular weight of approximately 19 kDa. The PRN sequence of BP-L2 is presented in Supplementary Material 1.
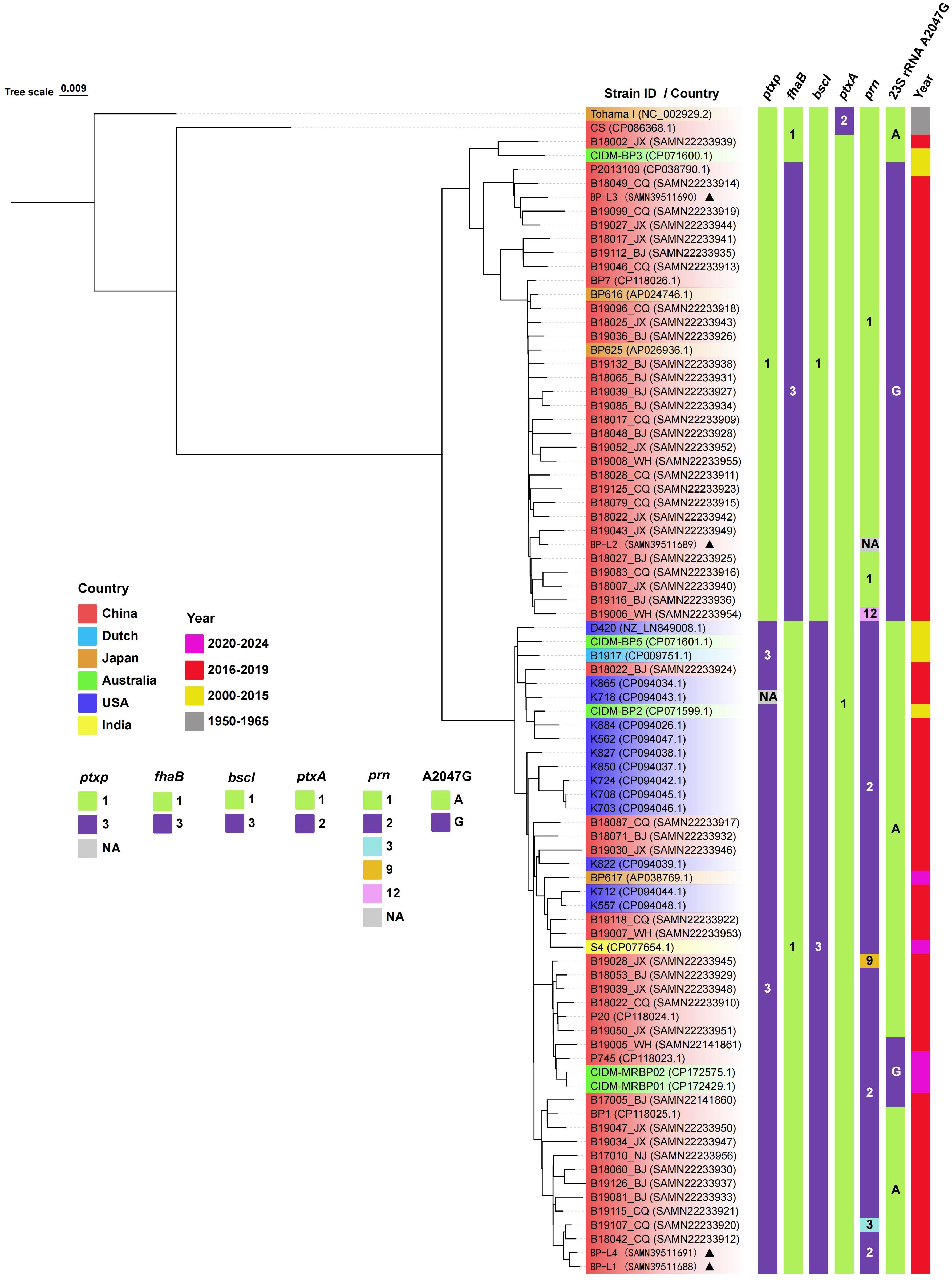
Figure 1. Phylogenetic relationship of recent B. pertussis isolates. A phylogenetic tree of 84 B. pertussis isolates is constructed using the maximum likelihood method based on the whole-genome SNPs. The tree is rooted using the B. pertussis reference genome Tohama I as the outgroup. The leaf labels are formatted as ‘Strain ID (accession number)’, and the background color represents the geographic source of the isolates. The strains in this study are marked with triangles. The vertical color blocks on the right represent the allele types of ptxP, fhaB, bscI, ptxA, prn, 23S rRNA A2047G, and the isolation year for each strain. Complete strain information is provided in Supplementary Table 1.
BP-L2 expresses partial PRN antigen
The samples of CS, BP-L1, and BP-L2 containing PRN were analyzed using SDS–PAGE and western blotting (Supplementary Figure 1). Consistent results were found for BP-L1 and CS, while the PRN components of BP-L2 formed complex bands, with the main band located at approximately 72 kDa. The N-terminal amino acid sequencing results (Figure 2) were consistent with the amino acid sequence at positions 65–83 in the PRN coding region of BP-L2 (score 490.7), demonstrating that BP-L2 was capable of expressing a partial PRN antigen.
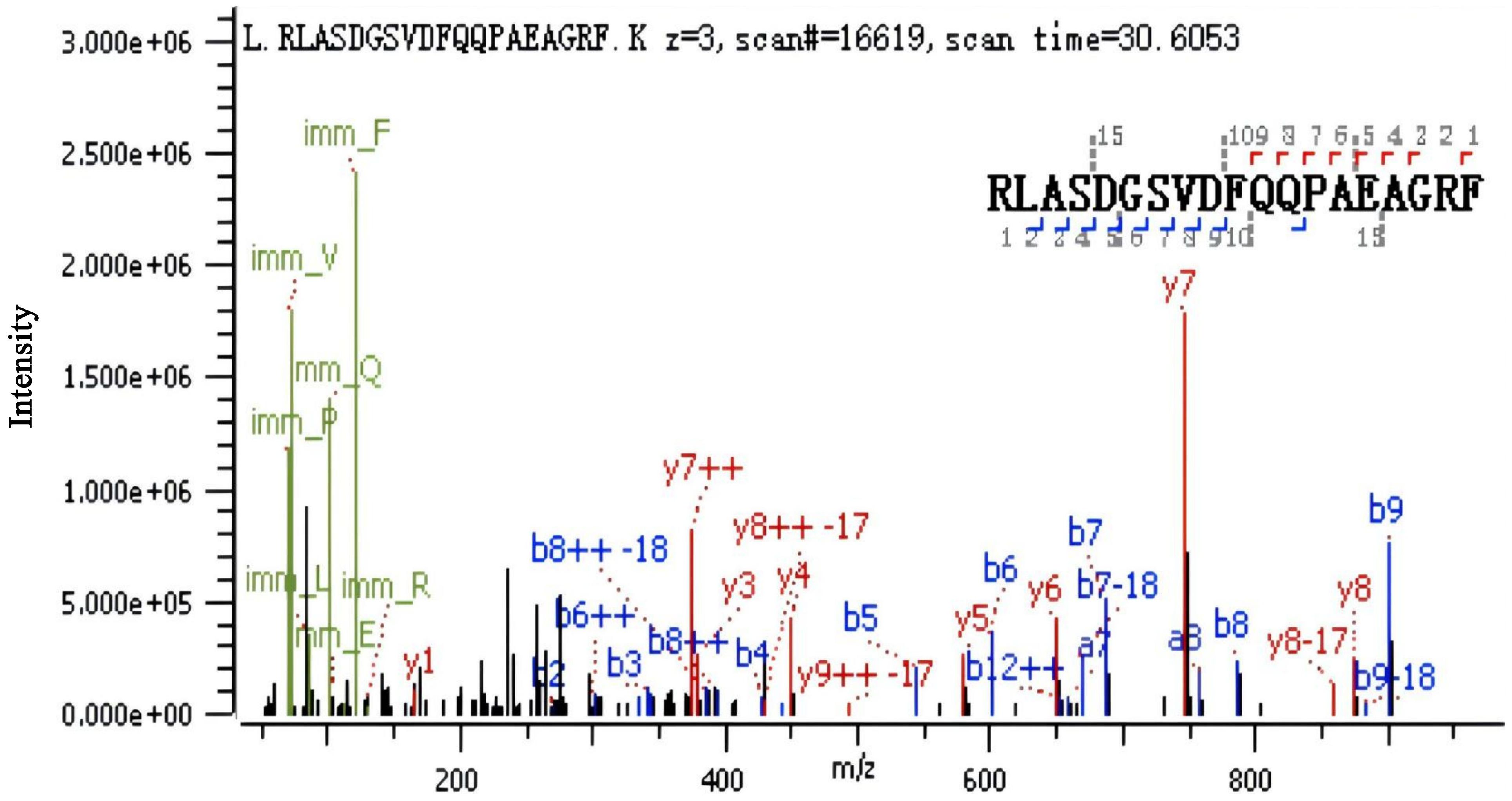
Figure 2. Pertactin expression in the BP-L2 strain confirmed using N-terminal peptide sequencing. LC-MS/MS analysis identified the peptide ‘RLASDGSVDFQQPAEAGRF’, corresponding to amino acids 65–83 in the pertactin coding region, confirming protein expression in BP-L2.
To confirm the expression of the PRN antigen, we determined serum IgG antibody levels 6 weeks after the challenge (Figure 3). The geometric mean of antibody titers was 476 for BP-L1, 141 for BP-L2, and 673 for CS, respectively. Although the PRN antibody levels of the BP-L2 group were lower than those in the other two groups, the differences were not significant (P > 0.05). The results indicated that a partial PRN was expressed and that some degree of antibody response was induced during BP-L2 infection.
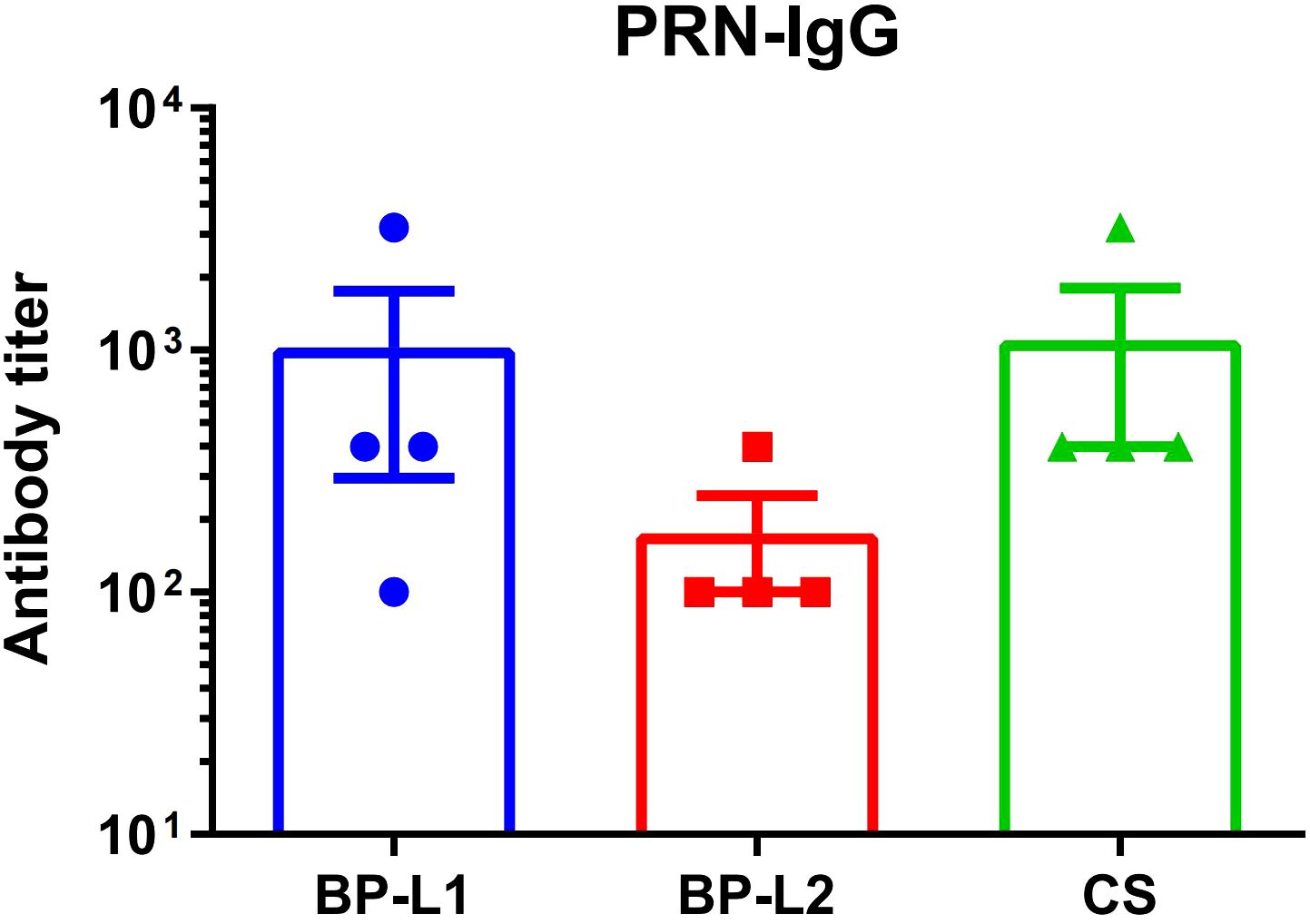
Figure 3. Serum antibody levels of PRN in the 6 weeks after infection using ELISA, the results are shown as the anti-PRN IgG antibody titer of the mean ± SEM (n=4).
The binding affinities of the antiserum to the PRN of the CS, BP-L1, and BP-L2 strains were 8.8193 nM, 9.0359 nM, and 9.0448 nM by Kd values, respectively. Since Kd values and binding affinities are inversely correlated, the antigenicity ranking of PRN is CS, BP-L1, BP-L2. However, they are very similar according to the Kd values.
BP-L1 and BP-L2 exhibit different infection and immune characteristics in a mice infection model
In a mice aerosol infection model, the bacterial burden of BP-L1 was significantly greater in the lungs (P < 0.0001 on the 3rd and 7th days post-challenge) and significantly lower in the nasal cavity compared to the BP-L2 and CS strains (P < 0.0001 on the 7th and 14th days post-challenge). However, the tracheal colonization ability of the BP-L2 strain was significantly greater (P < 0.0001 on the 10th days post challenge) than that of the other two strains (Figure 4). These differences in colonization in the lungs of BP-L1 and the trachea of BP-L2 were also validated by pathological scores (Supplementary Figure 2) and cytokine test of lungs or nasal associated lymphoid tissues lymphocytes (Supplementary Figures 3, 4). Pathological injury or the increase of inflammatory cytokines were observed in the lungs of BP-L1 and the trachea of BP-L2 correspondingly. Specifically, the pathological results showed that BP-L1 infection caused significant alveolar exudate (P < 0.05 compared to CS and BP-L2), while BP-L2 infection caused significant exfoliation of tracheal epithelium (P<0.05 compared to BP-L1, P < 0.001 compared to CS). Four weeks after BP-L1 infection, the level of pulmonary IL-6 was significantly higher than that of BP-L2 (P < 0.01) and CS (P < 0.001). However, nasal cytokines of BP-L2 also showed an overall increasing trend compared with the BP-L1 and CS strains (P > 0.05).
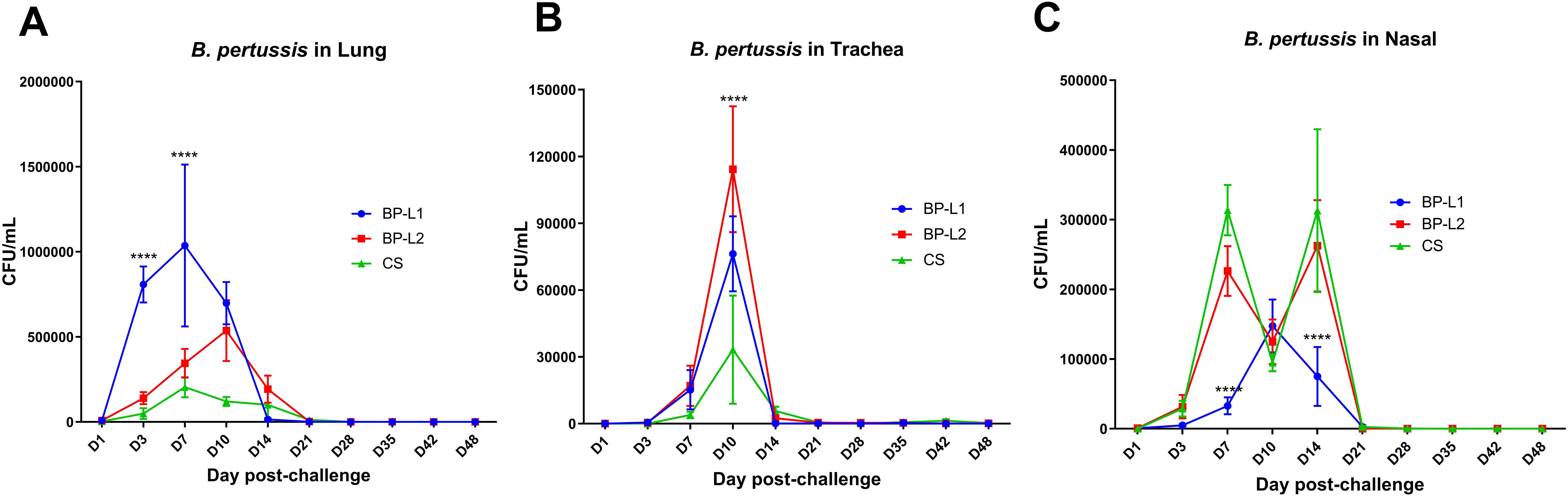
Figure 4. Colonization of bacteria after aerosol challenge of the mice by isolates and reference strains; (A) B. pertussis in Lung; (B) B. pertussis in Trachea; (C) B. pertussis in Nasal, results shown as CFU/mL of the mean ± SEM, P values are determined by one way analysis of variance, ****P<0.0001 (n=4).
Trm levels were examined at 9 weeks after challenge with the different strains or initial immunization with the aP vaccine (Figure 5). The results showed that the level of tissue-resident CD4+ T cells after BP-L1 infection was significantly higher than that after vaccine immunization (P < 0.0001), BP-L2 (P < 0.01) and CS (P < 0.001) challenge. The Trm-specific subtyping results also showed that BP-L1 infection caused a significant increase in the IFN-γ (P < 0.05 vs. CS) and IL-17a (P < 0.01 vs. CS) subtypes. Trm is regarded as a more effective immune indicator for B. pertussis infection. These cells can persist long-term at the infection site and rapidly proliferate and differentiate after reinfection (Wilk et al., 2017).
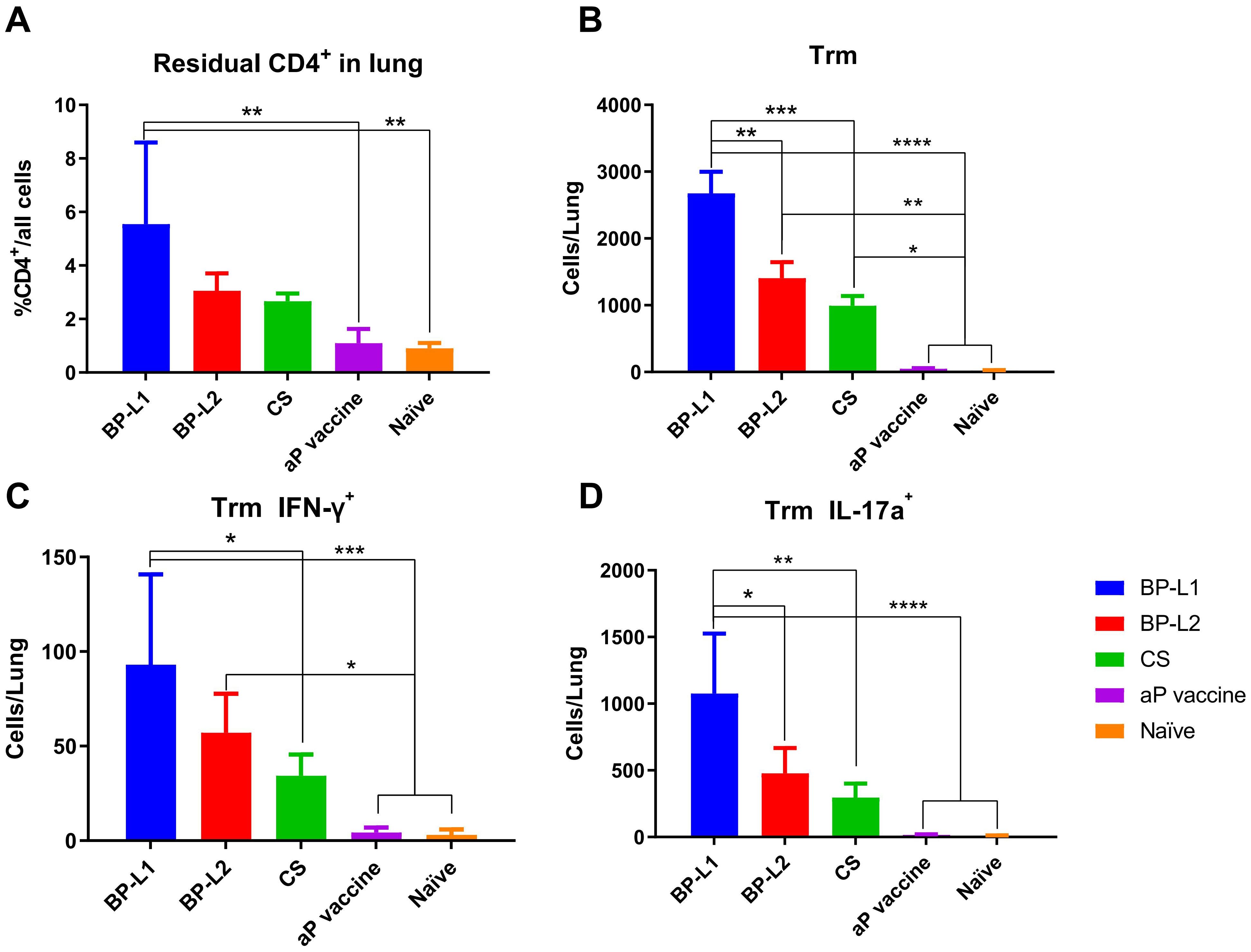
Figure 5. Lung tissue-resident memory T cell levels and typing results; (A) tissue-resident CD4+ T cells; (B) Trm; (C) IFN-γ+ Trm; (D) IL-17a+ Trm, the results shown as (A) the proportion of CD4+ resident cells; (B–D) the number of target cells in the total lung of the mean ± SEM, P values are determined by one way analysis of variance, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (n=4).
The colonization ability of BP-L1 in immunized mice was significantly lower than that in naïve mice (Figure 6). The colonization ability of BP-L2 was also affected by aP immunization, but an obvious colonization peak and clearance trend were still observed. Bacterial colonization in the BP-L2 group was greater than that in the BP-L1 group in multiple results (P < 0.001 in the lungs on the 3rd day post challenge; P < 0.01 in nasal on the 10th day post challenge).
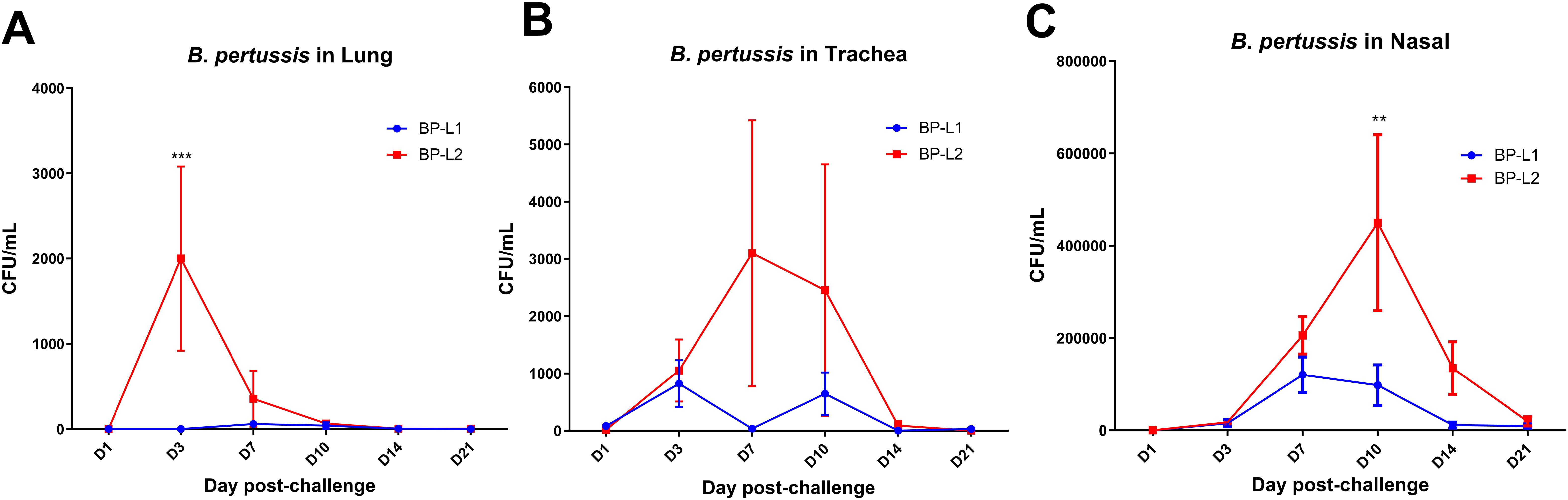
Figure 6. Colonization of B. pertussis in various regions of aP-immunized mice post-infection; (A) B. pertussis in Lung; (B) B. pertussis in Trachea; (C) B. pertussis in Nasal, results shown as CFU/mL of the mean ± SEM, P values are determined by one way analysis of variance, **P<0.01, ***P<0.001 (n=4).
In vivo and in vitro transcriptome sequencing
Compared to BALB/c mice, aerosol infection results indicated a significantly higher overall bacterial load in the lungs and trachea of NSG mice, with the highest load in the CS group, followed by BP-L2, and the lowest in BP-L1 (Supplementary Figure 5). For the in vivo study, total RNA of B. pertussis was isolated from infected mice and transcriptome sequencing analysis was conducted (Figure 7). Overall, an average of 155,463 2-by 150-bp reads from in vivo samples mapped to the reference genome, resulting in 1% of the reads corresponding to the pathogen. These mapped reads cover an average of 63% of the reference genome sequences (Supplementary Table 6). Subsequent differential analysis of the BP-L1, BP-L2 and CS strains revealed only 104 gene expression differences across all comparisons. Among these, expressions of relA (log2FC = 2.1, FDR P-value = 0.019), fim3 (log2FC =5.7, FDR P-value = 6.02E-28) and sodA (log2FC =2.4, FDR P-value = 8.61E-06) in BP-L2 were significantly upregulated compared to BP-L1. All the above genes are involved in the bacterial stringent response (Sugisaki et al., 2013). For the ptxP3 strain BP-L1, the gene bipA is upregulated in vivo as a major component of biofilms, with a log2 fold change of 1.8 and an FDR P-value of 0.027 when compared to BP-L2. However, no genes associated with the PT antigen were found to be significantly highly expressed during in vivo infection. Subsequently, we reduced the fold change analysis parameter to 1.5, yet the conclusion remained unchanged. The in vivo RNA-seq results for gene expression differences in all comparisons are presented in Supplementary Table 7.
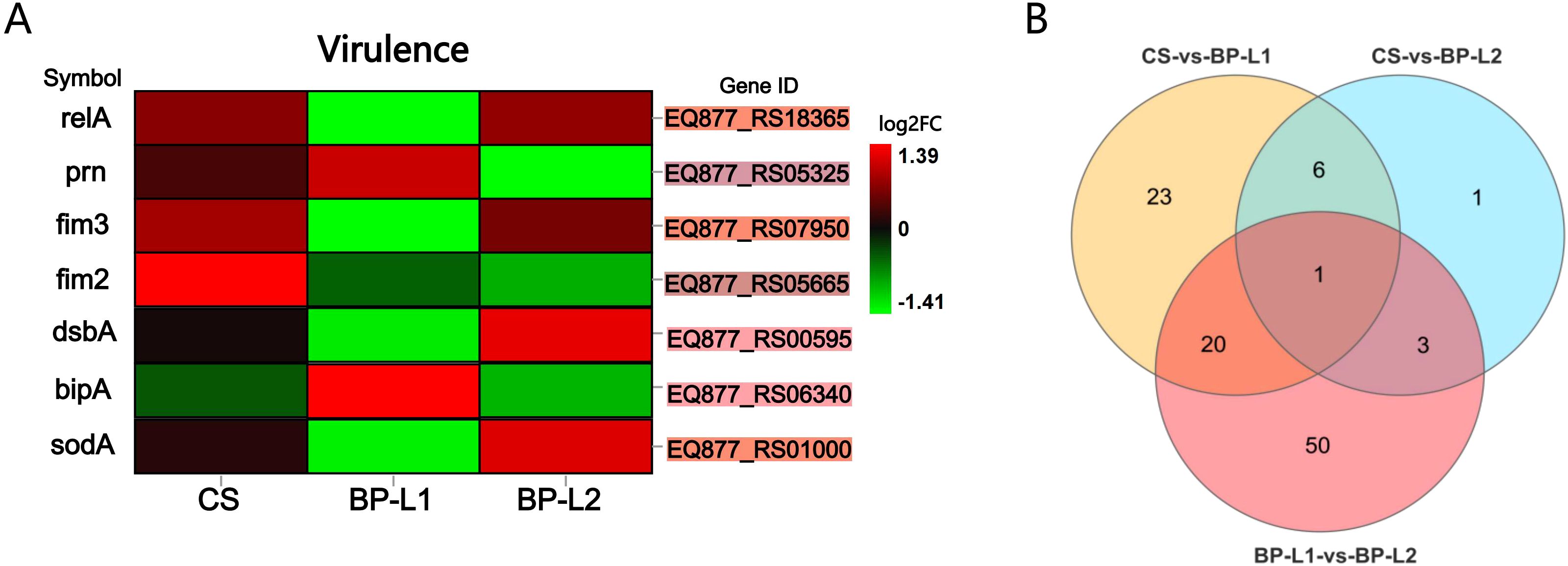
Figure 7. Gene expression levels of pertussis-infected strains in vivo; (A) Heatmap of virulence-related gene expression levels in DEGs. The background color of the ‘Gene ID’ represents the location in the Venn diagram; (B) Venn diagram of differentially expressed genes between the BP-L1, BP-L2 and CS group. Details of the differentially expressed genes is provided in Supplementary Table 4 marked in the corresponding color. [fold change > 2, Q value < 0.05, corrected by the Bonferroni test] (n=4).
The results of in vitro transcriptome sequencing indicated that genes associated with PT and adenylate cyclase toxin were downregulated in BP-L1 and upregulated in BP-L2. This variation may be associated with the differential expression of the virulence regulatory gene bvgA/S (Chen and Stibitz, 2019) (Supplementary Figure 6). The sodB was significantly downregulated in BP-L1 (log2FC = -0.76, FDR P-value = 4.37E-11) and BP-L2 (log2FC = -0.60, FDR P-value = 8.13E-08) compared to CS, which was associated with antioxidant stress (DeShazer et al., 1994). The in vitro RNA-seq results for gene expression differences in all comparisons are shown in Supplementary Table 8.
BP-L2 and BP-L3 have higher paraquat tolerance than BP-L1, BP-L4 and CS strains
Because of the increased sodA expression, we predicted that BP-L2 and even BP-L3 strains of the same lineage would exhibit greater tolerance to superoxide effects. To compare the ability of different clinical strains to counter oxidative stress, we assessed their sensitivities to paraquat, which induces oxidative stress by generating O2-. For this, CS and BP-L1 to BP-L4 cells were exposed to 50 mM paraquat for 30 minutes. The CFU of the bacteria before and after paraquat treatment were then counted on R-L agar plates, and the results were presented as the calculated proportion of surviving bacteria. The results indicated that the paraquat challenge caused a significant decrease (approximately 30%) in the viable counts of BP-L1, BP-L4, and CS strains, whereas the viability of BP-L2 and BP-L3 was only slightly impaired (approximately 1%) (Figure 8). These outcomes demonstrate that BP-L2 has a higher tolerance to oxidative stress (P < 0.001), which may be attributed to its elevated superoxide dismutases (SODs) activity. Moreover, the similar tolerance exhibited by BP-L3 within the same ptxP1-ptxA1-fhaB3 lineage suggests that this characteristic may be lineage-specific.
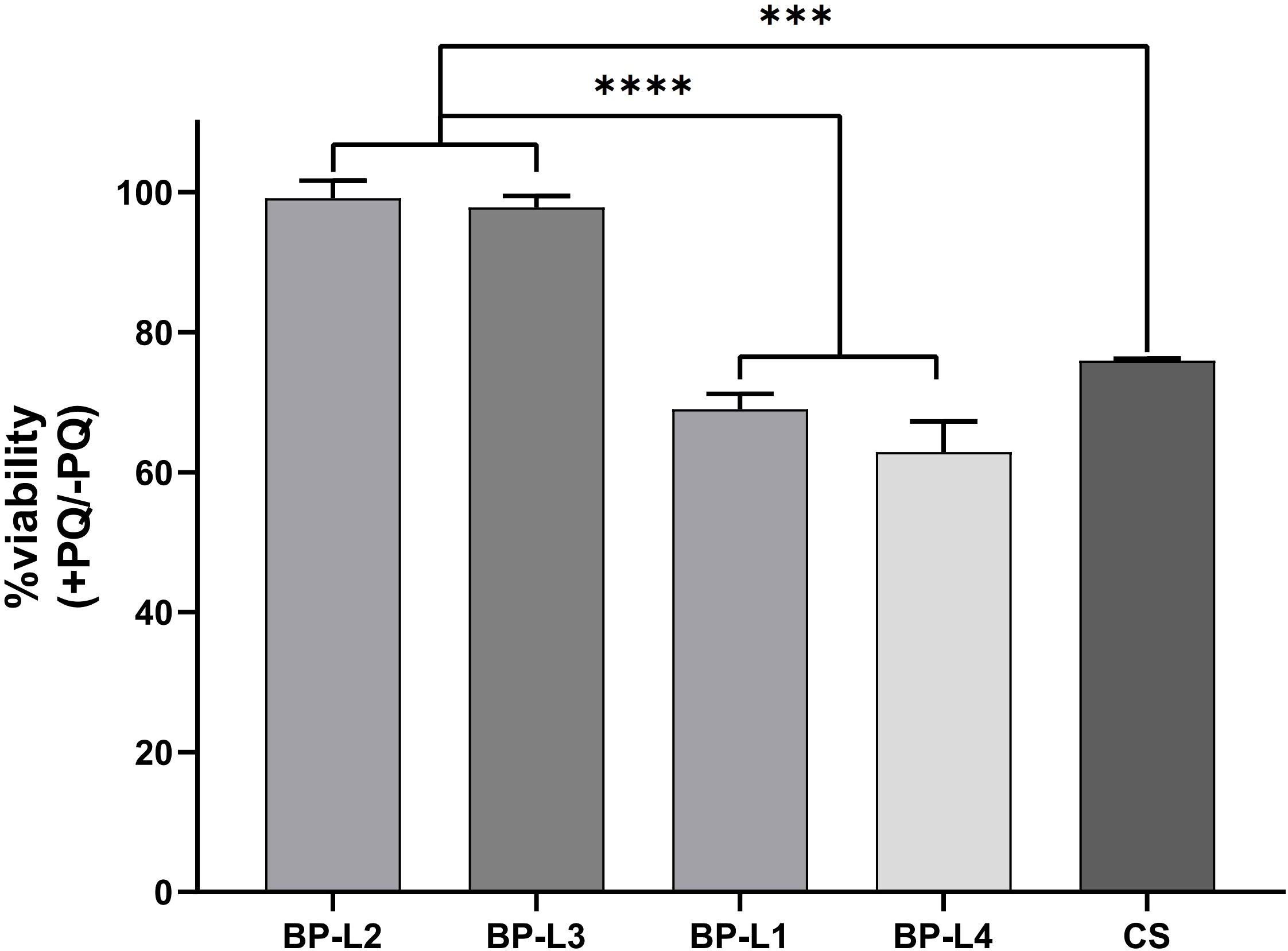
Figure 8. Tolerance of clinical strains BP-L1 to L4 and CS strains to paraquat. The results are calculated as percent bacterial survival after the paraquat challenge. Results are shown as mean ± SEM (n=3). ***P < 0.001 vs. CS, ****P < 0.0001 vs. BP-L1 and BP-L4.
Additionally, as the major component of the biofilm, bipA was found to have increased expression in BP-L1 infections in vivo. We compared the biofilm formation ability of all clinical strains of the aforementioned species using the crystal violet method, and only observed a statistical difference between BP-L2 and BP-L3 (Supplementary Figure 7). However, there was no significant difference in biofilm counts between the other strains, including BP-L1 (P = 0.51 compared to BP-L2).
Discussion
The resurgence of B. pertussis, characterized by various epidemic strains exhibiting antigenic drift and deficiencies, has facilitated the transmission of the bacterium. This study involved active monitoring and strain isolation of B. pertussis, screening 158 clinically suspected cases. From these, four clinical strains, designated BP-L1 to BP-L4, were successfully isolated. Notably, the partial PRN-deficient strain BP-L2 was reported for the first time, demonstrating a unique colonization advantage in the tracheas of naïve mice and a fitness advantage in immunized mice. In vivo RNA-seq results indicated that the high expression of genes associated with the stringent response and antioxidant stress during infection might contribute to BP-L2’s colonization in the trachea. In line with these findings, strains BP-L2 and BP-L3, from the same lineage, exhibited significantly higher tolerance to oxidative stress, with notably elevated SODs activity. Furthermore, strain BP-L1 displayed a pronounced advantage in lung colonization and overexpressed bipA in vivo, although its biofilm formation was not enhanced.
Although the allele profiles correspond with Western strains, the ptxP3 strains BP-L1/L4 formed a distinct clade with Chinese isolates in 2019. This suggests that the ptxP3 strains isolated in this study may have originated from epidemic strains already prevalent in China. Furthermore, all strains isolated after 2020 in the phylogenetic tree were bscI3 strains, including ptxP3/fhaB1/bscI3 strains carrying macrolide resistance mutations, aligning with a recent report from China (Fu et al., 2024). This indicates that attention should be given to the macrolide resistance and bscI typing of ptxP3 strains and the extent of their dissemination in China. The ptxP3 allele has been the most frequently isolated allele in recent pertussis outbreaks (Cai et al., 2023). Mooi et al. (2009) showed by ELISAs that the PT expression of the ptxP3 strains was modestly greater (1.62-fold) than that of the ptxP1 strains in Bordet-Gengou media. In mice, PT plays a central role in immune suppression, facilitating early lung colonization and inhibiting neutrophil recruitment to sites of infection (Kirimanjeswara et al., 2005). Consistent with mentioned studies, we observed that the ptxP3 strain BP-L1 had a significantly greater ability to colonize the lungs of naïve mice.
However, our in vivo results suggest that the colonization ability of BP-L1 in the lungs may not be directly related to the elevated PT expression, indicating that the pulmonary colonization ability of the BP-L1 strain could be associated with other mutations. Bart et al. (2010) hypothesized that ptxP3 might be a “hitchhiker” mutation, resulting from advantageous mutations selected elsewhere in the genome, and subsequent research supports this hypothesis (Schmidtke et al., 2012). King et al. (2013) reported that ptxP3 strains represented a lineage with a fitness advantage over other strains. In fact, previous study has also reported that no significant differences in the PT expression levels between ptxP3 and ptxP1 strains in THIJS medium (Luu et al., 2018), suggesting that ptxP3 strains may represent a lineage with a series of other mutations in the genome. Our study reinforces this perspective from an in vivo viewpoint.
BipA, the major antigen of biofilms, has been found to be significantly upregulated during BP-L1 infection in our study. Research has indicated that biofilm formation involving bipA can enhance early colonization of the lungs (de Gouw et al., 2014; Hiramatsu et al., 2016). However, we verified that the biofilm formation capacity of BP-L1 was not significantly increased in the SS liquid medium. Previous studies have reported that bipA is expressed maximally under the Bvg-intermediate (Bvg i) phase and was also the first identified Bvg i phase gene (Williams et al., 2005). Indeed, the in vitro Bvg phases were defined based on specific culture conditions (Hoo et al., 2014), which may explain why the biofilm verification results are inconsistent with the bipA expression in vivo. Alternatively, biofilm formation may not be responsible for the colonization advantage of BP-L1 in the lungs, which necessitates further experimental confirmation. Nonetheless, a significant Trm response induced by BP-L1 was observed at 9 weeks post-aerosol challenge. As a significantly upregulated antigen during BP-L1 infection, bipA may still be a candidate antigen for enhancing the immune persistence of an aP vaccine.
RN-deficient strains have been consistently reported in countries with high vaccine coverages (Otsuka et al., 2012; Pawloski et al., 2014). Nao Otsuka found that PRN-deficient strains exhibited greater growth advantages in vitro (Otsuka et al., 2012), which helps maintain transmissibility between hosts. A retrospective study (Bodilis and Guiso, 2013) showed that a longer interval between the onset of pertussis symptoms and hospitalization was observed in infants infected with PRN-deficient strains, indicating an increased chance of transmission. In addition, Marjolein van Gent et al. (Salaün et al., 2003) demonstrated that PRN deletion significantly weakened lung and tracheal colonization through the PRN knockout strain. However, there appears to be no major loss of in vitro or in vivo fitness in the natural PRN-deficient strain, which raises the question of whether the contributions of PRN to pathogenesis might be redundant or complementary functions. Unlike the currently widely isolated PRN-deficient strains (Ma et al., 2021), BP-L2 was able to express the partial PRN antigen. Whole genome sequencing showed that the PRN coding region of BP-L2 was missing from amino acids 18 to 391 and the theoretical molecular weight was about 19kDa. The missing region contains the RGD sequence, which is crucial for PRN’s adhesive role, and the highly variable region 1 (Junker et al., 2006). However, the N-terminal signal sequence and C-terminal transport element P.30 remained intact and identical to the reference strain. Consequently, we hypothesize that this segment of the PRN antigen can still be expressed and transported to the outer membrane. Subsequent ELISA, binding affinity tests, and LC-MS/MS analyses confirmed the expression of the partial PRN. We observed a distinct respiratory colonization advantage for BP-L2, as well as characteristics of immune escape. Thus, this pattern of partial expression may represent an adaptive evolution that maximally retains the ability to infect, highlighting the significant role of the missing part in immune protection (the RGD sequence) or the crucial role of the remaining part in the colonization and infection of B. pertussis (Hijnen et al., 2004).
Compared to conventional RNA sequencing, in vivo RNA-seq revealed a remarkably limited number of DEGs, highlighting the association between the colonization advantage and transcriptional regulation in this study. The in vivo expression level of PRN in BP-L2 was significantly lower than that of the other two strains. However, whether this is related to the partial deletion of the PRN coding region remains to be further studied. During BP-L2 infection in vivo, the relA and sodA genes were significantly overexpressed, which was related to the stringent response and antioxidant stress (Martins et al., 2018). As the major synthetase of the stringent response, relA plays important roles in the regulation of survival during host invasion (Pacios et al., 2020). The stringent response resists intracellular oxidative stress by regulating the production of the antioxidant enzyme soda (Martins et al., 2018). Superoxide radicals are the main source of bacterial oxidative stress within the host and are the primary mechanism of sterilization for some antibiotics and macrophages. SODs can rapidly eliminate superoxide radicals, which is crucial for the long-term colonization of pertussis (Nguyen et al., 2011; Schatzman and Culotta, 2018). Then,we simulated in vivo oxidative stress using paraquat and observed elevated SOD activity in BP-L2, which could contribute to the survival of B. pertussis within macrophages (Mitchell et al., 2016) and provide an advantage in tracheal colonization for BP-L2. More importantly, the same SODs activity shown by BP-L3 in the same ptxP1-ptxA1-fhaB3 lineage suggests that this character may be lineage specific. The fhaB3 allele has been recently identified exclusively in Chinese ptxP1 isolates, and these strains exhibit a higher mutation rate compared to the global ptxP1-ptxA1 B. pertussis population (Xu et al., 2019). Considering the potential impact of increased tracheal colonization on the transmission capacity of B. pertussis, these findings suggest that enhanced surveillance of BP-L2 and other ptxP1-ptxA1-fhaB3 strains is warranted. Additionally, the use of a stringent response inhibitor (Léger et al., 2021) should be contemplated for the treatment and prevention of pertussis infection.
Regarding the other in vivo RNA-seq results for BP-L2, there were 127 SNP differences distinguishing BP-L1 from BP-L2 (Supplementary Table 9). However, no related SNPs were identified in the mentioned genes such as bipA, bvgA/S, sodA/B, fim2/3, or relA between BP-L1 and BP-L2. The Fim serotype for BP-L1 is Fim2/3, and for BP-L2, it is Fim3, thus accounting for the observed differences. Gene rearrangement is also a significant mode of adaptive evolution in pertussis. The published results support the expected correlation between IS elements and the observation of rearrangements in B. pertussis (Luu et al., 2018). This study noted similar IS counts in four clinical strains, and collinearity analysis revealed comparable gene rearrangements in BP-L1 and BP-L2 (Supplementary Figure 8). In contrast, the reference strains CS and Tohama I exhibited lower amounts of IS and different rearrangements, suggesting that these clinical strains may have a higher frequency of gene rearrangement.
The aP vaccine elicits a robust antibody response against PRN, which is crucial for complement-mediated killing of B. pertussis (Lesne et al., 2020). Consequently, PRN-deficient strains have been demonstrated to exhibit enhanced fitness in aP-immunized mice (Safarchi et al., 2015) and are more commonly found among B. pertussis isolates from vaccinated individuals (Bodilis and Guiso, 2013), indicating evidence of vaccine-driven evolution. Similarly, significant fitness of BP-L2 was observed in aP-immunized mice in this study. We observed a slight weakening in both the post-infection PRN antibody levels and binding affinities compared with other strains. By comparing with previous a study (Hijnen et al., 2004), it was also found that the residual PRN retained only 3 (epitope-16,17, and 18) out of 18 epitopes. Therefore, the adaptability in immunized hosts of BP-L2 may be the result of a combination of these factors. Furthermore, BP-L2 was isolated from a vaccinated adult patient, and family transmission was not ruled out. These results indicate the limited protection provided by the aP vaccine against current B. pertussis clinical strains.
However, this study has several limitations. Utilizing NSG mice enabled us to achieve sufficient levels of bacterial colonization for in vivo RNA-seq. Nonetheless, this approach also exposed B. pertussis to an immune-deficient environment lacking mature lymphocytes, functional macrophages, and dendritic cells, which could influence the activation and repression of certain virulence factors (Wong et al., 2019). Thus, this constitutes a limitation of the study. In future research, we plan to analyze the differential expression of target genes in normal infection models using RT-PCR, including relA, sodA, bipA, and other associated genes.
In summary, through in vivo functional analysis, animal modeling, and vaccine effectiveness studies on B. pertussis clinical strains, we first reported a partial PRN-deficient strain named BP-L2, which exhibited a distinct colonization advantage in the trachea and fitness in aP-immunized mice. The tracheal colonization advantage of the partial pertactin-deficient strain BP-L2 may be associated with the overexpression of relA and sodA during in vivo infection. Further experiments revealed that the SODs activity of BP-L2 was indeed higher than that of other strains, indicating higher paraquat tolerance. More importantly, the same tolerance displayed by BP-L3 within the same ptxP1-ptxA1-fhaB3 lineage suggests that this characteristic may be lineage-specific. We propose that our results could offer a novel perspective for investigating emerging B. pertussis strains. According to the results of our study, despite Yunnan’s proximity to Laos, Vietnam, and Myanmar in southwestern China, the B. pertussis epidemic trend is consistent with that of China and most countries. These results are crucial not only for expanding our understanding of B. pertussis strains but also for promoting the development of new vaccine formulations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1066958.
Ethics statement
The studies involving humans were approved by The ethics committee of the People’s Hospital of the Xishuangbanna Dai Autonomous Prefecture. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. The animal study was approved by The institutional Experimental Animal Ethics Committee of Institute of Medical Biology, Chinese Academy of Medical Science & Peking Union Medical College. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
WZ: Data curation, Investigation, Funding acquisition, Writing – original draft. CW: Data curation, Investigation, Writing – review & editing. MH: Resources, Writing – review & editing. MZ: Data curation, Investigation, Writing – review & editing. JLL: Investigation, Writing – review & editing. XM: Data curation, Writing – review & editing. NG: Investigation, Writing – review & editing. QG: Investigation, Writing – review & editing. YM: Data curation, Writing – review & editing. JYL: Data curation, Investigation, Writing – review & editing. SL: Data curation, Writing – review & editing. YH: Resources, Writing – review & editing. MS: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. LS: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the Yunnan Provincial Science and Technology Department (Grant number 202002AA10009), the National Health, Family Planning Commission of China (Grant number 2015ZX09101031-002) and the Fujian Provincial Natural Science Foundation of China (Grant number 2023J05267). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We appreciate all the volunteers for participating in this experiment. We also appreciate the contributions of all investigators at Department of Pulmonary and Critical Care Medicine, Xishuangbanna Dai Autonomous Prefecture People’s Hospital for clinic sampling collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1547751/full#supplementary-material
References
Alai, S., Gautam, M., Palkar, S., Oswal, J., Gairola, S., and Dhotre, D. P. (2022). Characterization of Bordetella pertussis strains isolated from India. Pathogens 11, 794. doi: 10.3390/pathogens11070794
Bart, M. J., van Gent, M., van der Heide, H. G., Boekhorst, J., Hermans, P., Parkhill, J., et al. (2010). Comparative genomics of prevaccination and modern Bordetella pertussis strains. BMC Genomics 11, 627. doi: 10.1186/1471-2164-11-627
Bart, M. J., Zeddeman, A., van der Heide, H. G., Heuvelman, K., van Gent, M., and Mooi, F. R. (2014). Complete genome sequences of Bordetella pertussis isolates B1917 and B1920, representing two predominant global lineages. Genome Announc 2, e01301–14. doi: 10.1128/genomeA.01301-14
Belcher, T., Dubois, V., Rivera-Millot, A., Locht, C., and Jacob-Dubuisson, F. (2021). Pathogenicity and virulence of Bordetella pertussis and its adaptation to its strictly human host. Virulence 12, 2608–2632. doi: 10.1080/21505594.2021.1980987
Belcher, T. and Preston, A. (2015). Bordetella pertussis evolution in the (functional) genomics era. Pathog. Dis. 73, ftv064. doi: 10.1093/femspd/ftv064
Bodilis, H. and Guiso, N. (2013). Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg. Infect. Dis. 19, 471–474. doi: 10.3201/1903.121475
Boinett, C. J., Harris, S. R., Langridge, G. C., Trainor, E. A., Merkel, T. J., and Parkhill, J. (2015). Complete genome sequence of Bordetella pertussis D420. Genome Announc 3 (3), e00657-15. doi: 10.1128/genomeA.00657-15
Bouchez, V., Hegerle, N., Strati, F., Njamkepo, E., and Guiso, N. (2015). New data on vaccine antigen deficient Bordetella pertussis isolates. Vaccines (Basel) 3, 751–770. doi: 10.3390/vaccines3030751
Burns, D. L., Meade, B. D., and Messionnier, N. E. (2014). Pertussis resurgence: perspectives from the Working Group Meeting on pertussis on the causes, possible paths forward, and gaps in our knowledge. J. Infect. Dis. 209 Suppl 1, S32–S35. doi: 10.1093/infdis/jit491
Cai, J., Chen, M., Liu, Q., Luo, J., Yuan, L., Chen, Y., et al. (2023). Domination of an emerging erythromycin-resistant ptxP3 Bordetella pertussis clone in Shanghai, China. Int. J. Antimicrob. Agents 62, 106835. doi: 10.1016/j.ijantimicag.2023.106835
Chen, Q. and Stibitz, S. (2019). The BvgASR virulence regulon of Bordetella pertussis. Curr. Opin. Microbiol. 47, 74–81. doi: 10.1016/j.mib.2019.01.002
de Gouw, D., Serra, D. O., de Jonge, M. I., Hermans, P. W., Wessels, H. J., Zomer, A., et al. (2014). The vaccine potential of Bordetella pertussis biofilm-derived membrane proteins. Emerg. Microbes Infect. 3, e58. doi: 10.1038/emi.2014.58
DeShazer, D., Bannan, J. D., Moran, M. J., and Friedman, R. L. (1994). Characterization of the gene encoding superoxide dismutase of Bordetella pertussis and construction of a SOD-deficient mutant. Gene 142, 85–89. doi: 10.1016/0378-1119(94)90359-x
Diavatopoulos, D. A. and Edwards, K. M. (2017). What is wrong with pertussis vaccine immunity? Why immunological memory to pertussis is failing. Cold Spring Harb. Perspect. Biol. 9, a029553. doi: 10.1101/cshperspect.a029553
Držmíšek, J., Petráčková, D., Dienstbier, A., Čurnová, I., and Večerek, B. (2023). T3SS chaperone of the CesT family is required for secretion of the anti-sigma factor BtrA in Bordetella pertussis. Emerg. Microbes Infect. 12, 2272638. doi: 10.1080/22221751.2023.2272638
Espinosa-Vinals, C., Holubova, J., Stanek, O., Osicka, R., Masin, J., Arellano Herencia, F. E., et al. (2025). Intranasal application of a bifunctional pertactin-RTX fusion antigen elicits protection of mouse airway mucosa against Bordetella pertussis colonization. mSphere 10, e0095924. doi: 10.1128/msphere.00959-24
Fu, P., Yan, G., Li, Y., Xie, L., Ke, Y., Qiu, S., et al. (2024). Pertussis upsurge, age shift and vaccine escape post-COVID-19 caused by ptxP3 macrolide-resistant Bordetella pertussis MT28 clone in China. Clin. Microbiol. Infect. 30, 1439–1446. doi: 10.1016/j.cmi.2024.08.016
Fu, P., Zhou, J., Yang, C., Nijiati, Y., Zhou, L., Yan, G., et al. (2023). Molecular evolution and increasing macrolide resistance of Bordetella pertussis, Shanghai, China 2016-2022. Emerg. Infect. Dis. 30, 29–38. doi: 10.3201/eid3001.221588
Graeff-Wohlleben, H., Killat, S., Banemann, A., Guiso, N., and Gross, R. (1997). Cloning and characterization of an Mn-containing superoxide dismutase (SodA) of Bordetella pertussis. J. Bacteriol 179, 2194–2201. doi: 10.1128/jb.179.7.2194-2201.1997
Hijnen, M., Mooi, F. R., van Gageldonk, P. G., Hoogerhout, P., King, A. J., and Berbers, G. A. (2004). Epitope structure of the Bordetella pertussis protein P.69 pertactin, a major vaccine component and protective antigen. Infect. Immun. 72, 3716–3723. doi: 10.1128/iai.72.7.3716-3723.2004
Hiramatsu, Y., Saito, M., Otsuka, N., Suzuki, E., Watanabe, M., Shibayama, K., et al. (2016). BipA is associated with preventing autoagglutination and promoting biofilm formation in Bordetella holmesii. PloS One 11, e0159999. doi: 10.1371/journal.pone.0159999
Holt, K. E., Parkhill, J., Mazzoni, C. J., Roumagnac, P., Weill, F. X., Goodhead, I., et al. (2008). High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40, 987–993. doi: 10.1038/ng.195
Holubova, J., Stanek, O., Juhasz, A., Hamidou Soumana, I., Makovicky, P., and Sebo, P. (2022). The Fim and FhaB adhesins play a crucial role in nasal cavity infection and Bordetella pertussis transmission in a novel mouse catarrhal infection model. PloS Pathog. 18, e1010402. doi: 10.1371/journal.ppat.1010402
Hoo, R., Lam, J. H., Huot, L., Pant, A., Li, R., Hot, D., et al. (2014). Evidence for a role of the polysaccharide capsule transport proteins in pertussis pathogenesis. PloS One 9, e115243. doi: 10.1371/journal.pone.0115243
Jiang, W., Wei, C., Mou, D., Zuo, W., Liang, J., Ma, X., et al. (2021). Infant rhesus macaques as a non-human primate model of Bordetella pertussis infection. BMC Infect. Dis. 21, 407. doi: 10.1186/s12879-021-06090-y
Junker, M., Schuster, C. C., McDonnell, A. V., Sorg, K. A., Finn, M. C., Berger, B., et al. (2006). Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923. doi: 10.1073/pnas.0507923103
King, A. J., van der Lee, S., Mohangoo, A., van Gent, M., van der Ark, A., and van de Waterbeemd, B. (2013). Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PloS One 8, e66150. doi: 10.1371/journal.pone.0066150
Kirimanjeswara, G. S., Agosto, L. M., Kennett, M. J., Bjornstad, O. N., and Harvill, E. T. (2005). Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J. Clin. Invest. 115, 3594–3601. doi: 10.1172/jci24609
Klein, N. P., Bartlett, J., Rowhani-Rahbar, A., Fireman, B., and Baxter, R. (2012). Waning protection after fifth dose of acellular pertussis vaccine in children. N Engl. J. Med. 367, 1012–1019. doi: 10.1056/NEJMoa1200850
Koide, K., Yamaguchi, T., Katsukawa, C., Otsuka, N., Kenri, T., and Kamachi, K. (2022). Complete genome sequence of a macrolide-resistant Bordetella pertussis isolated in Japan. Microbiol. Resour Announc 11, e0071822. doi: 10.1128/mra.00718-22
Léger, L., Byrne, D., Guiraud, P., Germain, E., and Maisonneuve, E. (2021). NirD curtails the stringent response by inhibiting RelA activity in Escherichia coli. Elife 10, e64092. doi: 10.7554/eLife.64092
Lesne, E., Cavell, B. E., Freire-Martin, I., Persaud, R., Alexander, F., Taylor, S., et al. (2020). Acellular pertussis vaccines induce anti-pertactin bactericidal antibodies which drives the emergence of pertactin-negative strains. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.02108
Li, Z., Zhang, Y., Wang, Q., Li, Z., Liu, Y., Zhang, S., et al. (2016). Purification design and practice for pertactin, the third component of acellular pertussis vaccine, from Bordetella pertussis. Vaccine 34, 4032–4039. doi: 10.1016/j.vaccine.2016.06.029
Liang, Z., Li, C., Gong, X., Ye, G., Jiang, Y., Shi, H., et al. (2024). Development of Glycan-masked SARS-CoV-2 RBD vaccines against SARS-related coronaviruses. PloS Pathog. 20, e1012599. doi: 10.1371/journal.ppat.1012599
Luu, L. D. W., Octavia, S., Zhong, L., Raftery, M. J., Sintchenko, V., and Lan, R. (2018). Proteomic adaptation of Australian epidemic Bordetella pertussis. Proteomics 18, e1700237. doi: 10.1002/pmic.201700237
Ma, L., Caulfield, A., Dewan, K. K., and Harvill, E. T. (2021). Pertactin-deficient Bordetella pertussis, vaccine-driven evolution, and reemergence of pertussis. Emerg. Infect. Dis. 27, 1561–1566. doi: 10.3201/eid2706.203850
Ma, X., Nugraha, D. K., Hiramatsu, Y., and Horiguchi, Y. (2024). RpoN (sigma factor 54) contributes to bacterial fitness during tracheal colonization of Bordetella bronchiseptica. Microbiol. Immunol. 68, 36–46. doi: 10.1111/1348-0421.13109
Martins, D., McKay, G., Sampathkumar, G., Khakimova, M., English, A. M., and Nguyen, D. (2018). Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, 9797–9802. doi: 10.1073/pnas.1804525115
McGuirk, P., Mahon, B. P., Griffin, F., and Mills, K. H. (1998). Compartmentalization of T cell responses following respiratory infection with Bordetella pertussis: hyporesponsiveness of lung T cells is associated with modulated expression of the co-stimulatory molecule CD28. Eur. J. Immunol. 28, 153–163. doi: 10.1002/(SICI)1521-4141(199801)28:01<153::AID-IMMU153>3.0.CO;2-
Meng, Q., Li, L., Shi, W., Wang, Q., Ding, M., Liu, Y., et al. (2018a). Seroprevalence of diphtheria and pertussis immunoglobulin G among children with pneumonia in Ji’nan, China. BMC Pediatr. 18, 383. doi: 10.1186/s12887-018-1337-y
Meng, Q. H., Luo, J., Yang, F., Shen, Y. J., Li, L., Li, L. J., et al. (2018b). A general lack of IgG against pertussis toxin in Chinese pregnant women and newborns. Pediatr. Infect. Dis. J. 37, 934–938. doi: 10.1097/inf.0000000000001933
Mitchell, G., Chen, C., and Portnoy, D. A. (2016). Strategies used by bacteria to grow in macrophages. Microbiol. Spectr. 4, 10.1128/microbiolspec.MCHD-0012-2015. doi: 10.1128/microbiolspec.MCHD-0012-2015
Mooi, F. R., van Loo, I. H., van Gent, M., He, Q., Bart, M. J., Heuvelman, K. J., et al. (2009). Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg. Infect. Dis. 15, 1206–1213. doi: 10.3201/eid1508.081511
National Disease Control and Prevention Administration (2025). National Overview of the Epidemic Situation of Notifiable Infectious Diseases. Available online at: https://www.ndcpa.gov.cn/jbkzzx/c100016/common/list.html (Accessed January 16, 2025).
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Octavia, S., Sintchenko, V., Gilbert, G. L., Lawrence, A., Keil, A. D., Hogg, G., et al. (2012). Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008-2010. J. Infect. Dis. 205, 1220–1224. doi: 10.1093/infdis/jis178
Otsuka, N., Han, H. J., Toyoizumi-Ajisaka, H., Nakamura, Y., Arakawa, Y., Shibayama, K., et al. (2012). Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PloS One 7, e31985. doi: 10.1371/journal.pone.0031985
Pacios, O., Blasco, L., Bleriot, I., Fernandez-Garcia, L., Ambroa, A., López, M., et al. (2020). (p)ppGpp and its role in bacterial persistence: new challenges. Antimicrob. Agents Chemother. 64, e01283–20. doi: 10.1128/aac.01283-20
Park, J., Zhang, Y., Buboltz, A. M., Zhang, X., Schuster, S. C., Ahuja, U., et al. (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13, 545. doi: 10.1186/1471-2164-13-545
Pawloski, L. C., Queenan, A. M., Cassiday, P. K., Lynch, A. S., Harrison, M. J., Shang, W., et al. (2014). Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin. Vaccine Immunol. 21, 119–125. doi: 10.1128/cvi.00717-13
Safarchi, A., Octavia, S., Luu, L. D., Tay, C. Y., Sintchenko, V., Wood, N., et al. (2015). Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine 33, 6277–6281. doi: 10.1016/j.vaccine.2015.09.064
Salaün, L., Snyder, L. A., and Saunders, N. J. (2003). Adaptation by phase variation in pathogenic bacteria. Adv. Appl. Microbiol. 52, 263–301. doi: 10.1016/s0065-2164(03)01011-6
Schatzman, S. S. and Culotta, V. C. (2018). Chemical Warfare at the Microorganismal Level: A Closer Look at the Superoxide Dismutase Enzymes of Pathogens. ACS Infect. Dis. 4, 893–903. doi: 10.1021/acsinfecdis.8b00026
Schmidtke, A. J., Boney, K. O., Martin, S. W., Skoff, T. H., Tondella, M. L., and Tatti, K. M. (2012). Population diversity among Bordetella pertussis isolates, United States 1935-2009. Emerg. Infect. Dis. 18, 1248–1255. doi: 10.3201/eid1808.120082
Sedivy, A. (2021). Standard operating procedure for NanoTemper Monolith measurements. Eur. Biophys. J. 50, 381–387. doi: 10.1007/s00249-021-01534-4
Sugisaki, K., Hanawa, T., Yonezawa, H., Osaki, T., Fukutomi, T., Kawakami, H., et al. (2013). Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiol. (Reading) 159, 1379–1389. doi: 10.1099/mic.0.066597-0
Sun, M., Ma, Y., Xu, Y., Yang, H., Shi, L., Che, Y., et al. (2014). Dynamic profiles of neutralizing antibody responses elicited in rhesus monkeys immunized with a combined tetravalent DTaP-Sabin IPV candidate vaccine. Vaccine 32, 1100–1106. doi: 10.1016/j.vaccine.2013.12.025
Wilk, M. M., Misiak, A., McManus, R. M., Allen, A. C., Lynch, M. A., and Mills, K. H. G. (2017). Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with Bordetella pertussis. J. Immunol. 199, 233–243. doi: 10.4049/jimmunol.1602051
Williams, C. L., Boucher, P. E., Stibitz, S., and Cotter, P. A. (2005). BvgA functions as both an activator and a repressor to control Bvg phase expression of bipA in Bordetella pertussis. Mol. Microbiol. 56, 175–188. doi: 10.1111/j.1365-2958.2004.04526.x
Williams, M. M., Sen, K., Weigand, M. R., Skoff, T. H., Cunningham, V. A., Halse, T. A., et al. (2016). Bordetella pertussis strain lacking pertactin and pertussis toxin. Emerg. Infect. Dis. 22, 319–322. doi: 10.3201/eid2202.151332
Wong, T. Y., Hall, J. M., Nowak, E. S., Boehm, D. T., Gonyar, L. A., Hewlett, E. L., et al. (2019). Analysis of the In Vivo Transcriptome of Bordetella pertussis during Infection of Mice. mSphere 4, e00154–19. doi: 10.1128/mSphereDirect.00154-19
Wu, D. X., Chen, Q., Yao, K. H., Li, L., Shi, W., Ke, J. W., et al. (2019). Pertussis detection in children with cough of any duration. BMC Pediatr. 19, 236. doi: 10.1186/s12887-019-1615-3
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., and Wang, H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–w592. doi: 10.1093/nar/gkad359
Xu, Z., Hu, D., Luu, L. D. W., Octavia, S., Keil, A. D., Sintchenko, V., et al. (2022). Genomic dissection of the microevolution of Australian epidemic Bordetella pertussis. Emerg. Microbes Infect. 11, 1460–1473. doi: 10.1080/22221751.2022.2077129
Xu, Z., Wang, Z., Luan, Y., Li, Y., Liu, X., Peng, X., et al. (2019). Genomic epidemiology of erythromycin-resistant Bordetella pertussis in China. Emerg. Microbes Infect. 8, 461–470. doi: 10.1080/22221751.2019.1587315
Zhang, S., Xu, Y., Zhou, Z., Wang, S., Yang, R., Wang, J., et al. (2011). Complete genome sequence of Bordetella pertussis CS, a Chinese pertussis vaccine strain. J. Bacteriol 193, 4017–4018. doi: 10.1128/jb.05184-11
Zhao, T., Mo, Z., Ying, Z., Huang, T., Che, Y., Li, G., et al. (2021). Post hoc analysis of two clinical trials to compare the immunogenicity and safety of different polio immunization schedules in Chinese infants. Ann. Transl. Med. 9, 253. doi: 10.21037/atm-20-2537
Keywords: Bordetella pertussis, in vivo RNA, clinical strains, tracheal colonization, pertactin deficiency
Citation: Zuo W, Wei C, He M, Zhang M, Liang J, Ma X, Gao N, Gu Q, Ma Y, Li J, Liu S, Huang Y, Sun M and Shi L (2025) In vivo RNA-seq and infection model reveal the different infection and immune characteristics of B. pertussis strains in China. Front. Cell. Infect. Microbiol. 15:1547751. doi: 10.3389/fcimb.2025.1547751
Received: 18 December 2024; Accepted: 14 May 2025;
Published: 11 June 2025.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Laurence Don Wai Luu, University of New South Wales, AustraliaSunil Gairola, Serum Institute of India, India
Jhasketan Badhai, University of South Florida, United States
Copyright © 2025 Zuo, Wei, He, Zhang, Liang, Ma, Gao, Gu, Ma, Li, Liu, Huang, Sun and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Shi, c2hpbGkuaW1iQGdtYWlsLmNvbQ==; Mingbo Sun, c3VubWluZ2JvQHJuYmJpby5jb20=
†These authors have contributed equally to this work and share first authorship
 Weilun Zuo
Weilun Zuo Chen Wei3†
Chen Wei3† Jiangli Liang
Jiangli Liang Jingyan Li
Jingyan Li Shuyuan Liu
Shuyuan Liu Mingbo Sun
Mingbo Sun Li Shi
Li Shi