- 1Department of Laboratory Medicine, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Laboratory Medicine, Zhongshan Hospital, Fudan University, Shanghai, China
Methicillin-resistant Staphylococcus aureus (MRSA) stands as a pervasive and important pathogen in the co-infections during the COVID-19 pandemic given its high morbidity and mortality. This study aimed to characterize MRSA isolates obtained from COVID-19 pneumonia with MRSA co-infection patients during the COVID-19 pandemic wave in China from 2022 to 2024. Fifty MRSA isolates collected form COVID-19 pneumonia with MRSA co-infection patients (MRSA-COC) and 50 MRSA isolates collected from MRSA pneumonia patients without COVID-19 (MRSA-CON) were enrolled in this study. Whole-genome sequencing, genomic epidemiology and comparative analysis were conducted to explore differences of MRSA isolates between two groups. Patients with MRSA-COC were significantly older (P=0.0492), had higher rates of severe pneumonia progression (P=0.0006), and carried greater comorbidity burdens. The time from hospital admission to MRSA detected was significantly shorter in MRSA-COC patients than in MRSA-CON patients (P=0.0141). Furthermore, MRSA-COC patients demonstrated significantly higher mortality rates compared with MRSA-CON patients (44% vs. 18%, P=0.0049). However, genomic analysis revealed no statistically significant differences in antimicrobial resistance gene or virulence factor genes between the MRSA-COC and MRSA-CON isolates. CC5 emerged as the predominant clone in both groups, with significantly higher prevalence in MRSA-COC isolates (88% vs. 66%, P=0.0090). The tst-positive ST5-MRSA-II strain was associated with concerning mortality rates in both MRSA-COC (50%) and MRSA-CON (20%) patients, underscoring the critical need for enhanced surveillance in MRSA pneumonia.
Introduction
Staphylococcus aureus is an important and widespread bacterial pathogen, responsible for numerous uncomplicated skin infections and more severe, invasive infections worldwide each year. It is also a main causative agent of pneumonia leading to critical illness and death (He and Wunderink, 2020; Cheung et al., 2021). The pandemic coronavirus disease 2019 (COVID-19) is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has affected hundreds of thousands to millions of people resulting in a profound and detrimental effect on healthcare systems all over the world (Mirzaei et al., 2020). Co-infections and superinfections are common and frequently happened in respiratory viral infections, and secondary or bacterial co-infections with other viruses are known to substantially increase the mortality rate among viral-infected patients as previously documented (Mirzaei et al., 2020). Historically, S. aureus has been a major contributor to secondary bacterial infections in past viral pandemics and has significantly increased the mortality rates of infected patients (Cusumano et al., 2020). S. aureus co-infection and bacteremia were associated with nearly 50% mortality rates in patients during past influenza outbreaks, in contrast to the 1.4% morality rates in those infected with influenza alone (Leung et al., 2014).
There is a growing number of reports highlighting bacterial infections acquired by severe COVID-19 patients after hospital admission. In particular, nosocomial outbreaks in COVID-19 settings have been linked to methicillin-resistant S. aureus (MRSA), carbapenem-resistant Acinetobacter baumannii, and carbapenem-resistant Klebsiella pneumoniae (Segala et al., 2021). Antimicrobial-resistant pathogens including MRSA have been observed to cause healthcare-associated infections (HAIs) in COVID-19 patients, often leading to poorer clinical outcomes (O’Toole, 2021). A systematic review on the impact of COVID-19 pandemic on multidrug-resistant pathogens revealed significant changes in the rate of multidrug-resistant bacteria, and over half of the studies (54.5%) reported an increase in MRSA infection or colonization during the pandemic with the increase ranging from 4.6 to 170.6% (Abubakar et al., 2023). COVID-19 patients with S. aureus co-infection experience notably higher mortality rate during hospital admission compared with patients infected solely with COVID-19. S. aureus co-infection in COVID-19 patients is primarily associated with healthcare, and common hospital interventions such as intubation with mechanical ventilation, central venous catheter, and corticosteroids for severe COVID-19 patients heighten the risk of secondary bacterial infections (Adalbert et al., 2021).
China has experienced a dramatic COVID-19 wave since December 2022, with widespread primary SARS-CoV-2 infections across the population. A substantial proportion of these patients progressed to COVID-19 pneumonia requiring hospitalization, where bacterial co-infections were frequently observed and often associated with poor outcomes. Although the prevalence of COVID-19 in China has since stabilized at low levels, the large population and the extensive outbreak provided a unique context for studying secondary bacterial co-infections. To address the lack of research on MRSA co-infections in COVID-19 pneumonia, particularly in China, we conducted this study in hospitals across Shanghai, China. This study aims to shed light on the dynamics of MRSA co-infection in the context of COVID-19 pneumonia, filling a critical gap in the understanding of this clinical challenge.
Materials and methods
Study design
The patients diagnosed with MRSA co-infection following COVID-19 pneumonia from December 2022 to January 2024 in Ruijin hospital and Zhongshan hospital in Shanghai were enrolled in this study. For comparative analysis, MRSA pneumonia patients without COVID-19 during the same period were included as well. SARS-CoV-2 nucleic acid detection was used for COVID-19 diagnosis in Ruijin Hospital, and SARS-CoV-2 nucleic acid and/or antigen detections were used for COVID-19 diagnosis in Zhongshan Hospital. Ruijin Hospital (Shanghai Jiao Tong University School of Medicine) and Zhongshan Hospital (Fudan University) are both tertiary teaching hospitals in Shanghai, with bed capacities exceeding 3,000 and 2,000 beds respectively, providing comprehensive healthcare services to patients across China.
The diagnosis of community-acquired pneumonia (CAP) relies on the clinical symptoms of lower respiratory tract infection (e.g., dyspnea, productive cough, fever, and chills), combined with radiographic evidence of new pulmonary infiltrates and supportive laboratory findings (e.g., microbiological cultures, inflammatory markers, or molecular testing for pathogens) (Modi and Kovacs, 2020; Cilloniz et al., 2021; Torres et al., 2021). The symptoms of hospital-acquired pneumonia (HAP) may be hidden by either other medications or the cause of admission and HAP diagnosis is believed to be usually delayed (Torres et al., 2021), hence diagnosis of pneumonia can be complicated and requires comprehensive consideration. The pneumonia patients enrolled in this study were diagnosed by MRSA-positive sputum culture combined with clinical symptoms, as well as the presence of new infiltrates on chest radiographs, and/or other laboratory tests. The criteria for defining severe pneumonia comprises: (1) major criteria, including invasive mechanical ventilation, septic shock requiring vasopressor therapy; and (2) minor criteria, such as respiratory rate ≥30 breaths/min, PaO2/FiO2 ratio ≤250, multilobar infiltrates, confusion/disorientation, uremia (blood urea nitrogen level ≥20 mg/dL), leukopenia (white blood cell count <4 ×109/L), thrombocytopenia (platelet count <100×109/L), hypothermia (core temperature <36°C), hypotension requiring aggressive fluid resuscitation (Pletz et al., 2020; Budinger et al., 2021; Cilloniz et al., 2021).
MRSA isolates
A total of 100 MRSA isolates were collected from sputum samples of pneumonia patients and included in this study. Among the 100 MRSA isolates, 50 MRSA isolates were collected from MRSA co-infection with COVID-19 pneumonia patients and named as group MRSA-COC (MRSA co-infection with COVID-19), and the other 50 MRSA isolates were collected from MRSA pneumonia patients without COVID-19 and named as group MRSA-CON (MRSA pneumonia but COVID-19 negative). Group MRSA-COC included 27 MRSA isolates from Ruijin Hospital and 23 MRSA isolates from Zhongshan Hospital. Fifty MRSA isolates of group MRSA-CON (27 from Ruijin Hospital and 23 from Zhongshan Hospital) were randomly selected using the random number generation function of Microsoft Excel 365 (Microsoft Corporation, Redmond, WA, USA).
Ethical approval for this retrospective study was granted by the Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The requirement for informed consent was waived, as the research focused solely on bacterial analysis without involving patient interventions.
Identification and MRSA confirmation
Initial species identification of isolates was conducted using MALDI-TOF mass spectrometry (bioMérieux, Marcy-l’Étoile, France). MRSA was screened by the cefoxitin (30 μg) disk diffusion method following the Clinical and Laboratory Standards Institute guidelines (Institute CaLS, 2023). The presence of the mecA was confirmed to validate MRSA identification.
Genome sequencing and assembly
Genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega). DNA samples were fragmented into 400–500 bp segments using the Covaris M220 Focused Acoustic Shearer, following the manufacturer’s protocol. Illumina sequencing libraries were prepared from the sheared fragments with the NEXTflex™ Rapid DNA-Seq Kit. Raw sequencing reads were processed using fastp software (version 0.19.6) for quality filtering and subsequently assembled using the SOPA de novo assembler (version 2.04) (Luo et al., 2012).
Genomic epidemiology analysis
The assembled contigs were analyzed for genomic epidemiology using the Center for Genomic Epidemiology (CGE) platform (http://www.genomicepidemiology.org/). Analyses included phenotyping (antibiotic resistance genes and virulence factor genes) and typing (Multilocus sequence typing [MLST], spa type, and SCCmec type).
Statistical analysis
Statistical analysis was conducted using the t-test, chi-square test, or Fisher’s exact test, as appropriate. A two-sided P value of < 0.05 was considered statistically significant. All analyses were performed using SAS 8.2 software (SAS Institute Inc., Cary, NC, USA).
Results
Clinical data
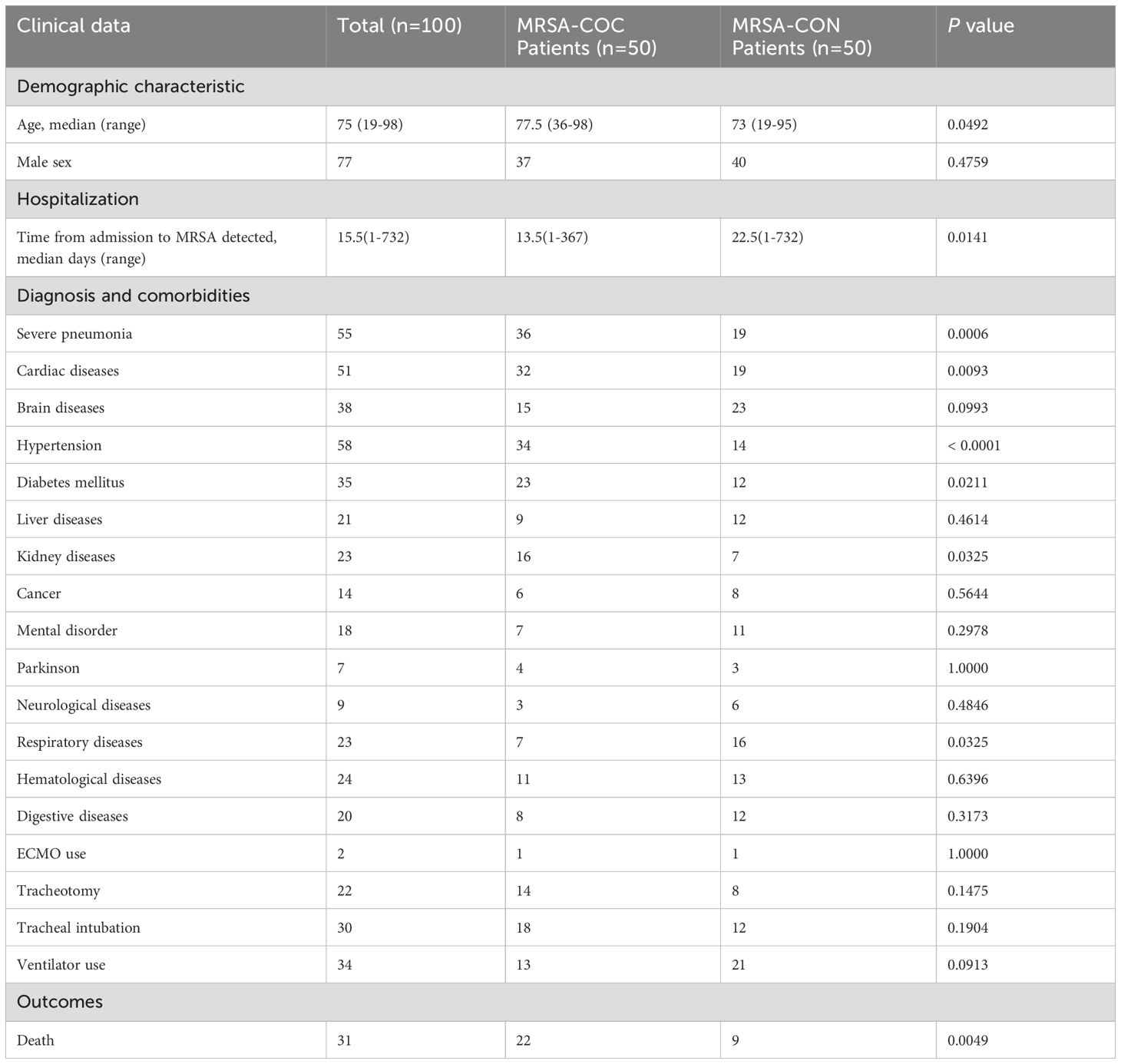
From December 2022 to January 2024, 100 pneumonia patients (54 from Ruijin Hospital, 46 from Zhongshan Hospital) were enrolled in this study. Among them, 50 patients were diagnosed with MRSA co-infection with COVID-19 pneumonia (MRSA-COC), and the other 50 patients were diagnosed with MRSA pneumonia without COVID-19 (MRSA-CON). The median time from COVID-19 (SARS-CoV-2) positive-tested to MRSA detected was 10.5 days (range: 0–69 days). The median time from hospital admission to MRSA detected was 13.5 days (range: 1–367 days) and 22.5 days (range: 1–732 days) respectively in the MRSA-COC and MRSA-CON patients as shown in Table 1, and the time from admission to MRSA detected was significantly shorter in the MRSA-COC patients compared with MRSA-CON patients (P=0.0141). The age of patients with MRSA-COC was higher than those with MRSA-CON (P=0.0492). The patients with MRSA-COC demonstrated significantly higher rates of severe pneumonia progression (72% vs. 38%, P=0.0006) and greater comorbidity burdens such as cardiac diseases (64% vs. 38%, P=0.0093), hypertension (68% vs. 28%, P<0.0001), diabetes mellitus (46% vs. 24%, P=0.0211) and kidney diseases (32% vs. 14%, P=0.0325). Conversely, the patients with MRSA-CON were more frequently associated with underlying respiratory conditions (14% vs. 32%, P=0.0325). The mortality rate was significantly higher in MRSA-COC patients compared with MRSA-CON patients (44% vs. 18%, P=0.0049).
Antimicrobial resistance genes
No genes or mutations associated with resistance to oxazolidinones, glycopeptides or lipoglycopeptides were identified in this study. Aminoglycosides resistance genes including aac(6’)-aph(2’’), aph(2’’)-Ia, aph(3’)-III, aadD were detected in 71 isolates. Genes conferring resistance to MLS-B (macrolides, lincosamides, ketolides, and streptogramin B) such as ermA, ermB or ermC were found in 84 isolates. Tetracycline resistance genes tetM and/or tetK were detected in 80 isolates. High-level mupirocin resistance gene mupA was present in 50 isolates. No significant differences in antimicrobial resistance genes were observed between MRSA-COC and MRSA-CON groups as presented in Table 2.
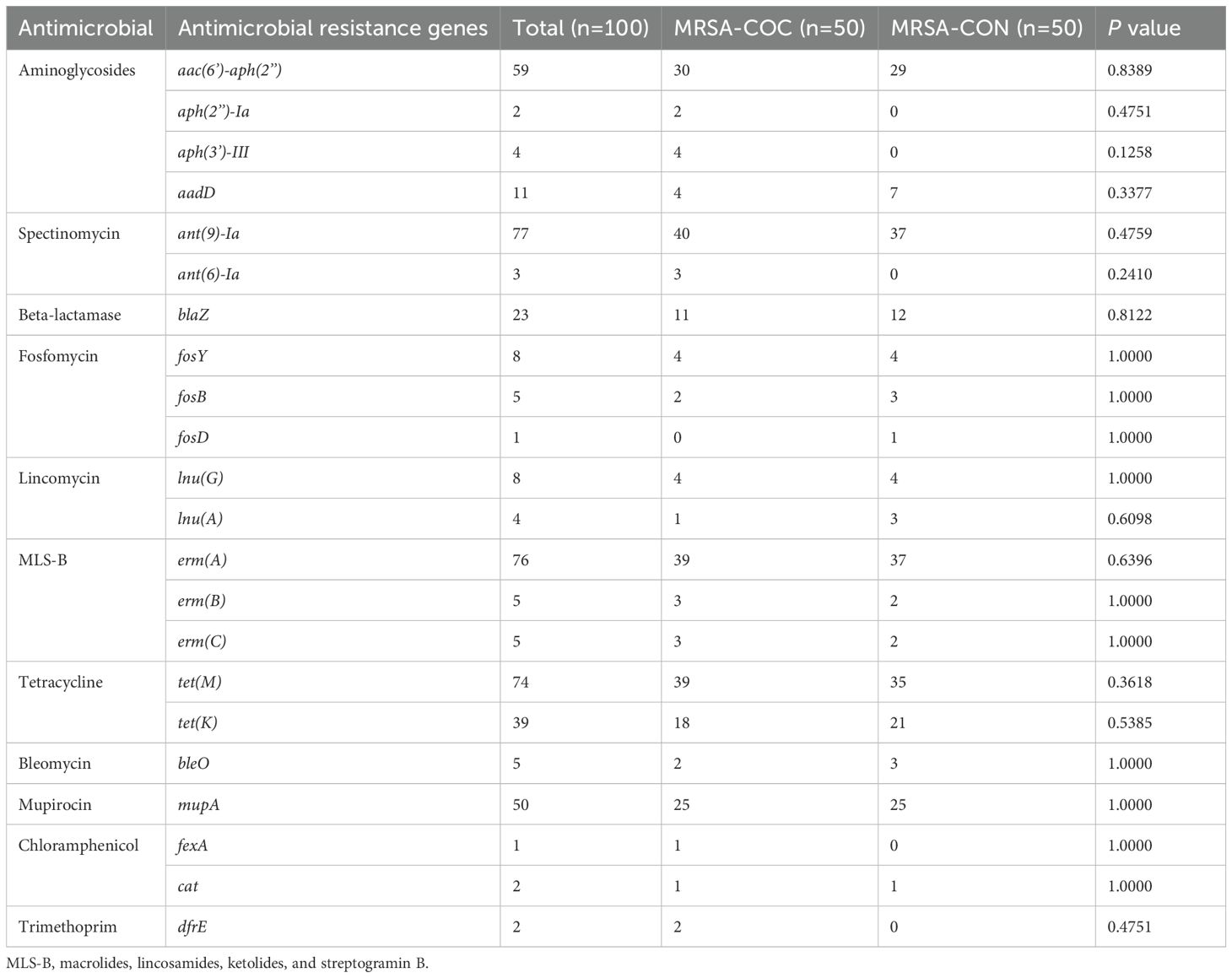
Table 2. The antimicrobial resistance genes of MRSA isolates from MRSA pneumonia patients with/without COVID-19.
Virulence factor genes
All MRSA isolates in this study carried genes encoding aureolysin (aur) and hemolysin (hlgA, hlgB, hlgC), as detailed in Table 3. The tst gene (encoding toxic shock syndrome toxin-1) was identified in 13 isolates overall (8 from MRSA-COC group and 5 from MRSA-CON group). The gene lukS/F-PV encoding Panton-Valentine leukocidin was found in only one MRSA isolate in the MRSA-COC group. No significant differences in the distribution of virulence factor genes were observed between two groups as shown in Table 3.
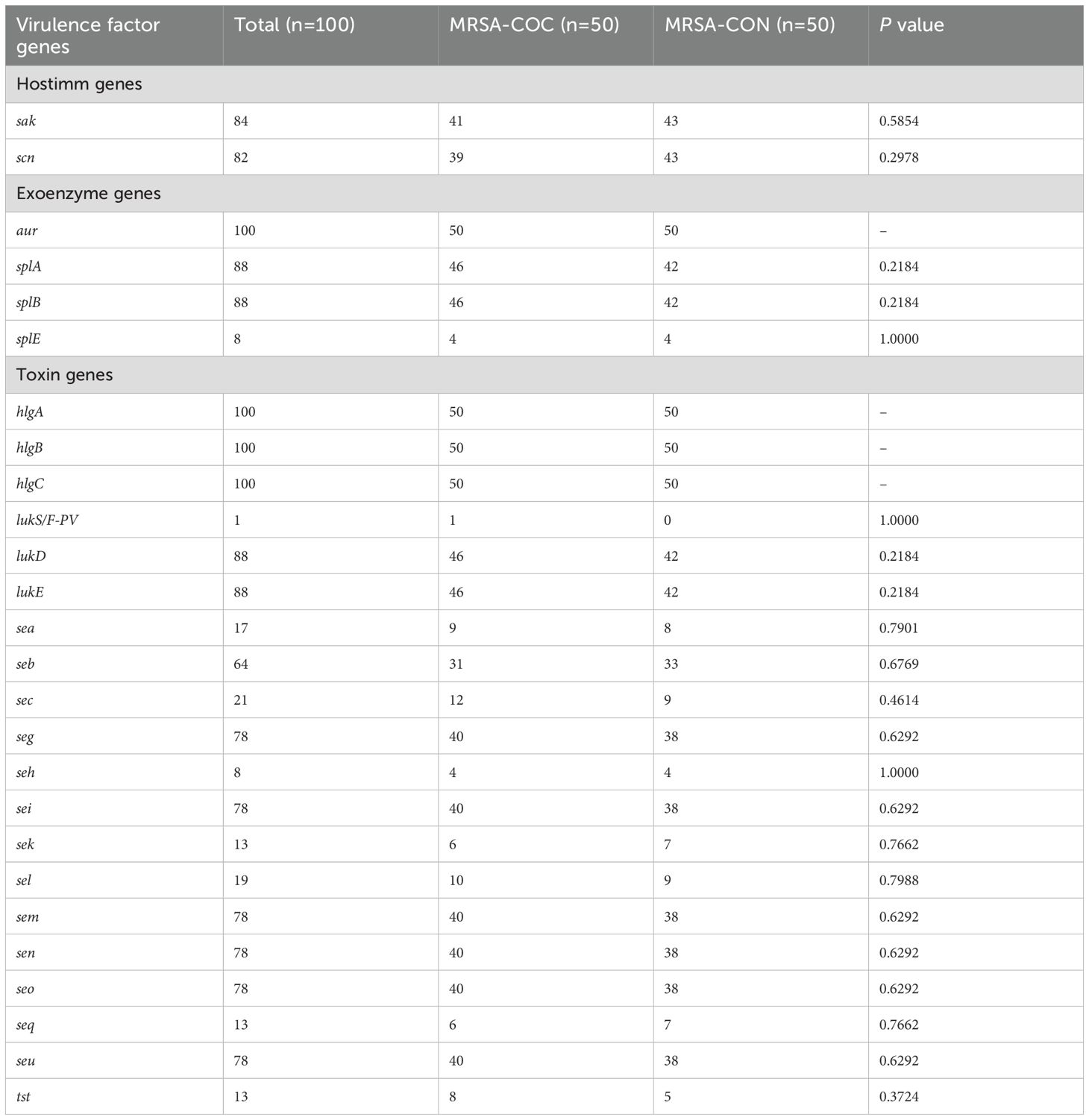
Table 3. The virulence factor genes of MRSA isolates from MRSA pneumonia patients with/without COVID-19.
Molecular types
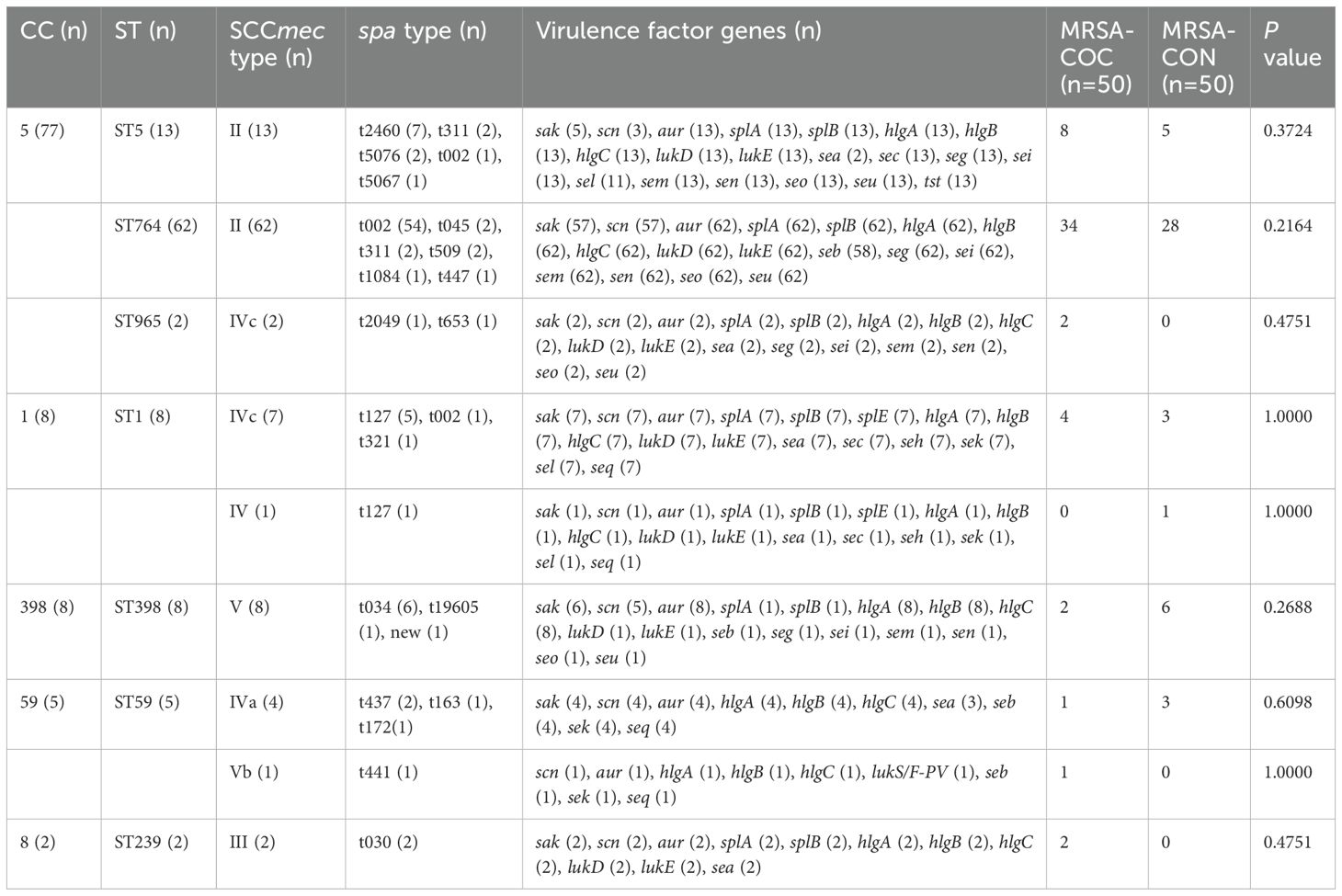
As presented in Table 4, the clonal complex CC5 was the most prevalent (77/100, 77%), found in 88% (44/50) of MRSA-COC isolates and 66% (33/50) of MRSA-CON isolates, and CC5 was much more frequently found in MRSA-COC group (88% vs. 66%, P=0.0090). All 13 tst-positive isolates exclusively comprised ST5-MRSA (CC5), with mortality rates of 50% (4/8) in MRSA-COC patients and 20% (1/5) in MRSA-CON patients. Overall, the most common sequence type (ST) and spa type were ST764 (62/100, 62%) and t002 (56/100, 56%) respectively. SCCmecII (75%) was the most frequent SCCmec type, followed by SCCmecIV (14%), SCCmecV (9%), and SCCmecIII (2%). Among MRSA-COC isolates, ST764 (30/50, 60%), t002 (30/50, 60%), and SCCmecII (38/50, 76%) were the most common molecular types.
Discussion
COVID-19, characterized by high transmission rates, poses severe complications including acute respiratory distress syndrome (ARDS), thromboembolic events, septic shock, and multi-organ failure. Bacterial co-infection in COVID-19 patients often exacerbates the immunosuppressive effects of the viral infection, worsening clinical outcomes (Adalbert et al., 2021). According to the results we studied, the mortality rate among the COVID-19 pneumonia patients with MRSA co-infection was significantly higher than that among the MRSA pneumonia patients without COVID-19 (44% vs. 18%, P=0.0049). This mortality rate was also much higher than previously we have studied for S. aureus bloodstream infections in Shanghai in recent years (Gu et al., 2020). These findings align with previous reviews, which demonstrate that S. aureus co-infection in COVID-19 pneumonia patients leads to worse outcomes compared with those with COVID-19 or MRSA solely during hospital admission (Adalbert et al., 2021; O’Toole, 2021; Abubakar et al., 2023). MRSA co-infection in COVID-19 patients is predominantly associated with healthcare, and common hospital interventions for severe COVID-19 patients, such as prolonged hospitalization and invasive procedures, may increase the risk of bacterial co-infection. To mitigate the risks, preventive measures such as COVID-19 vaccination and outpatient care may be key initiatives to reduce hospital admissions and subsequent MRSA co-infections. Infection prevention, control programs, and antimicrobial stewardship must be prioritized, particularly in light of the long-term burden of HAIs during the COVID- 19 pandemic (O’Toole, 2021).
Although no significant differences in antimicrobial resistance profiles were found between MRSA-COC and MRSA-CON isolates, antimicrobial resistance (AMR) problem remains a critical concern in MRSA co-infection in COVID-19. Many antibiotics such as azithromycin have been used to prevent and treat bacterial co-infection and secondary bacterial infections in patients with viral respiratory infections such as SARS-CoV-2. Although antibiotics do not directly affect SARS-CoV-2, viral respiratory infections often lead to bacterial pneumonia (Mirzaei et al., 2020). The high prevalence of MLS-B antibiotics resistance observed in this study (overall: 84/100, 84%; MRSA-COC isolates: 43/50, 86%; MRSA-CON isolates: 41/50, 82%) may significantly compromise therapeutic efficacy in both MRSA co-infected COVID-19 pneumonia and MRSA pneumonia cases. The overuse of broad-spectrum antibiotics during the COVID-19 pandemic may have inadvertently accelerated the emergence of antimicrobial-resistant pathogens, emphasizing the need for robust surveillance and antimicrobial stewardship programs. As demonstrated by our findings, no resistance to oxazolidinones, glycopeptides or lipoglycopeptides was detected, suggesting these antibiotic classes may remain viable for MRSA pneumonia treatment regardless of COVID-19 infection status. However, continuous vigilance against emerging resistant strains is deeply imperative.
In acute and intensive care units, inappropriate antimicrobial exposure and disrupted infection control measures have contributed to the selection and spread of antimicrobial-resistant pathogens. The COVID-19 pandemic is likely to play an important role in the emergence and transmission of resistant pathogens in hospital settings, hence there is a need to enhance infection prevention and antimicrobial management, as well as robust and consistent surveillance for antimicrobial resistance as part of the pandemic response and recovery (Langford et al., 2023). Antimicrobial resistance will continue to pose a substantial threat to healthcare systems in the coming years. To mitigate the potential long-term impact of COVID-19 on antimicrobial resistance, it is necessary to integrate both infection prevention and control strategies and antimicrobial stewardship activities into the pandemic response. Management strategies to reduce the emergence of AMR should therefore be investigated and implemented at local and global levels (Rehman, 2023). Proper prescription and optimized use of antimicrobials, coupled with high-quality diagnostics and aggressive infection control measures, may prevent the onset of multidrug-resistant pathogens during the pandemic (Lai et al., 2021). Research into new therapeutic approaches and improved diagnostic tools can help reduce reliance on broad-spectrum antibiotics and prevent the diffusion of multidrug-resistant pathogens.
Consistent with the findings of antimicrobial resistance genes in this study, no statistically significant differences in virulence factor genes were observed between MRSA-COC and MRSA-CON groups. These findings suggest that MRSA strains exhibit comparable levels of antimicrobial resistance and pathogenicity in both MRSA co-infection with COVID-19 pneumonia and MRSA pneumonia cases, warranting equal clinical attention. Consequently, similar antibiotic treatment and prevention strategies may be adopted for MRSA pneumonia patients with/without COVID-19. However, the higher mortality rate observed in MRSA co-infected COVID-19 pneumonia cases compared with MRSA pneumonia cases may be attributed to the underlying comorbidities and COVID-19-related pathological effects, which require specific clinical interventions and targeted therapies.
CC5 is one of the five major global MRSA clonal complexes (CC5, CC8, CC22, CC30, CC45) and has been frequently reported worldwide including in the USA, South America, Europe, Asia, Africa, and Australia (Lakhundi and Zhang, 2018). In this study, CC5 was the dominant clonal complex, present in 77% of MRSA isolates, with significantly higher prevalence in MRSA-COC group than in MRSA-CON isolates (88% vs. 66%, P=0.0090). This aligns with reports identifying CC5 as a major global MRSA clonal complex associated with endemic outbreaks in regions with high MRSA morbidity rates (Wang et al., 2022). As presented in Table 4 in our current study, it was revealed that all 13 MRSA isolates carrying the tst gene pertained to the ST5-MRSA-II lineage. Notably, this tst-positive ST5-MRSA-II strain exhibited high mortality rates in both MRSA-COC patients (50%) and MRSA-CON patients (20%). The close binding-like relationship between tst gene and CC5 (ST5) has been discovered in our precious studies about S. aureus bloodstream infections and MRSA burn wound infection (Gu et al., 2020; Gu et al., 2023). The toxin gene tst encodes Toxic Shock Syndrome Toxin-1 (TSST-1) which is a 22 kDa extracellular protein toxin secreted by S. aureus, a virulence factor that triggers immune dysregulation, fever, rash, vascular disorders, toxic shock syndrome vascular disorders, and multi-organ failure (Zhu et al., 2023). The TSST-1 gene (tst) has also been found predominantly in CC5 MRSA isolates in Suzhou, China associated with higher mortality (Wang et al., 2017) and tst-positive ST5-MRSA-II has been proved to be represented one dominated clone in China as reported (Zhao et al., 2019). These findings highlight the need for strict infection prevention and control measures and innovative therapeutic strategies to prevent further spread of the tst-positive CC5 MRSA strains in China, particularly in Shanghai and Zhejiang province. Some protein synthesis inhibitory antibiotics targeting TSST-1 or herbal extracts inhibiting the production of the TSST-1 toxin can be considered as an potential alternative strategy for managing tst-positive S. aureus infection because toxic shock syndrome (TSS) caused by TSST-1 is frequently fatal without prompt and targeted therapeutic intervention (Qiu et al., 2011; Hodille et al., 2017; Katahira et al., 2019). In addition, it has been discovered that the expression of tst potentially associated with the mutation of its promoter and variations in specific virulence regulators expression (Zhao et al., 2019). Therefore, sequencing of the tst promoter and quantitative analysis of major virulence regulators expression may provide important information for investigation of MRSA infection.
Given the close association between CC5-MRSA and key virulence factors such as tst as reported, enhanced global and regional surveillance of CC5-MRSA lineage is critically needed. Further genetic investigations into the tst promoter region and the regulatory mechanisms of virulence factor expression may yield insights into its pathogenesis and resistance. Implementing integrated infection control strategies alongside antimicrobial stewardship programs is vital to curbing the dissemination of hypervirulent and highly transmissible CC5-MRSA strains. Additionally, advancing diagnostic precision and developing targeted therapeutic approaches are imperative to mitigate the escalating threat of antimicrobial resistance and optimize clinical outcomes.
This study is limited by the inclusion of only two hospitals in Shanghai, which may affect its generalizability and introduce potential bias. The findings should therefore be interpreted with caution, as they represent only a subset of MRSA pneumonia patients in Shanghai. Future multicenter studies incorporating a broader range of hospitals are needed to enhance epidemiological representativeness and yield more robust findings.
In conclusion, this study revealed that the patients with MRSA-COC were significantly older, exhibited higher rates of severe pneumonia progression with greater comorbidity burdens, and showed shorter time from admission to MRSA infection compared with MRSA-CON patients. The mortality rate of MRSA-COC patients was significantly higher than that of MRSA-CON patients. However, genomic analysis revealed no significant differences in antimicrobial resistance genes or virulence factors genes between two groups, suggesting comparable antimicrobial resistant and pathogenic potential between MRSA strains causing MRSA co-infections in COVID-19 pneumonia and conventional MRSA pneumonia. Clonal distribution analysis identified CC5 as the dominant clone in both groups, with significantly higher prevalence among MRSA-COC isolates. Most critically, tst-positive ST5-MRSA-II strains demonstrated concerning mortality rates in both MRSA-COC and MRSA-CON pneumonia patients, highlighting the urgent need for enhanced surveillance and tailored management strategies for MRSA pneumonia especially in COVID-19 pneumonia.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
FG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Writing – original draft. YZ: Data curation, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft. JS: Conceptualization, Funding acquisition, Investigation, Writing – review & editing. WG: Conceptualization, Investigation, Resources, Writing – review & editing. LH: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by National Natural Science Foundation of China (grant numbers 82272372 and 82102452). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abubakar, U., Al-Anazi, M., Alanazi, Z., and Rodriguez-Bano, J. (2023). Impact of COVID-19 pandemic on multidrug resistant gram positive and gram negative pathogens: A systematic review. J. Infect. Public Health 16, 320–331. doi: 10.1016/j.jiph.2022.12.022
Adalbert, J. R., Varshney, K., Tobin, R., and Pajaro, R. (2021). Clinical outcomes in patients co-infected with COVID-19 and Staphylococcus aureus: a scoping review. BMC Infect. Dis. 21, 985. doi: 10.1186/s12879-021-06616-4
Budinger, G. R. S., Misharin, A. V., Ridge, K. M., Singer, B. D., and Wunderink, R. G. (2021). Distinctive features of severe SARS-CoV-2 pneumonia. J. Clin. Invest. 131, e149412. doi: 10.1172/JCI149412
Cheung, G. Y. C., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569. doi: 10.1080/21505594.2021.1878688
Cilloniz, C., Torres, A., and Niederman, M. S. (2021). Management of pneumonia in critically ill patients. BMJ 375, e065871. doi: 10.1136/bmj-2021-065871
Cusumano, J. A., Dupper, A. C., Malik, Y., Gavioli, E. M., Banga, J., Berbel Caban, A., et al. (2020). Staphylococcus aureus bacteremia in patients infected with COVID-19: A case series. Open Forum Infect. Dis. 7, ofaa518. doi: 10.1093/ofid/ofaa518
Gu, F., He, W., Xiao, S., Wang, S., Li, X., Zeng, Q., et al. (2020). Antimicrobial Resistance and Molecular Epidemiology of Staphylococcus aureus Causing Bloodstream Infections at Ruijin Hospital in Shanghai from 2013 to 2018. Sci. Rep. 10, 6019. doi: 10.1038/s41598-020-63248-5
Gu, F., He, W., Zhu, D., Yang, P., Sun, J., and Han, L. (2023). A 10-year retrospective study of methicillin-resistant Staphylococcus aureus from burn wound infection in southeast China from 2013 to 2022. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1301744
He, H. and Wunderink, R. G. (2020). Staphylococcus aureus pneumonia in the community. Semin. respiratory Crit. Care Med. 41, 470–479. doi: 10.1055/s-0040-1709992
Hodille, E., Rose, W., Diep, B. A., Goutelle, S., Lina, G., and Dumitrescu, O. (2017). The role of antibiotics in modulating virulence in staphylococcus aureus. Clin. Microbiol. Rev. 30, 887–917. doi: 10.1128/cmr.00120-16
Institute CaLS (2023). Performance Standards for Antimicrobial Susceptibility Testing Vol. 43 (Wayne, PA, USA: CLSI document M100-S29).
Katahira, E. J., Davidson, S. M., Stevens, D. L., and Bolz, D. D. (2019). Subinhibitory concentrations of tedizolid potently inhibit extracellular toxin production by methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 68, 255–262. doi: 10.1099/jmm.0.000905
Lai, C. C., Chen, S. Y., Ko, W. C., and Hsueh, P. R. (2021). Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antimicrob. Agents 57, 106324. doi: 10.1016/j.ijantimicag.2021.106324
Lakhundi, S. and Zhang, K. (2018). Methicillin-resistant staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, e00020–e00018. doi: 10.1128/CMR.00020-18
Langford, B. J., Soucy, J. R., Leung, V., So, M., Kwan, A. T. H., Portnoff, J. S., et al. (2023). Antibiotic resistance associated with the COVID-19 pandemic: a systematic review and meta-analysis. Clin. Microbiol. Infect. 29, 302–309. doi: 10.1016/j.cmi.2022.12.006
Leung, C. H., Tseng, H. K., Wang, W. S., Chiang, H. T., Wu, A. Y., and Liu, C. P. (2014). Clinical characteristics of children and adults hospitalized for influenza virus infection. J. microbiol. immunol. infection = Wei mian yu gan ran za zhi 47, 518–525. doi: 10.1016/j.jmii.2013.06.002
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18. doi: 10.1186/2047-217x-1-18
Mirzaei, R., Goodarzi, P., Asadi, M., Soltani, A., Aljanabi, H. A. A., Jeda, A. S., et al. (2020). Bacterial co-infections with SARS-coV-2. IUBMB Life 72, 2097–2111. doi: 10.1002/iub.2356
Modi, A. R. and Kovacs, C. S. (2020). Hospital-acquired and ventilator-associated pneumonia: Diagnosis, management, and prevention. Cleve Clin. J. Med. 87, 633–639. doi: 10.3949/ccjm.87a.19117
O’Toole, R. F. (2021). The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 27, 1772–1776. doi: 10.1016/j.cmi.2021.06.001
Pletz, M. W., Blasi, F., Chalmers, J. D., Dela Cruz, C. S., Feldman, C., Luna, C. M., et al. (2020). International perspective on the new 2019 american thoracic society/infectious diseases society of america community-acquired pneumonia guideline: A critical appraisal by a global expert panel. Chest 158, 1912–1918. doi: 10.1016/j.chest.2020.07.089
Qiu, J., Wang, J., Luo, H., Du, X., Li, H., Luo, M., et al. (2011). The effects of subinhibitory concentrations of costus oil on virulence factor production in Staphylococcus aureus. J. Appl. Microbiol. 110, 333–340. doi: 10.1111/j.1365-2672.2010.04888.x
Rehman, S. (2023). A parallel and silent emerging pandemic: Antimicrobial resistance (AMR) amid COVID-19 pandemic. J. Infect. Public Health 16, 611–617. doi: 10.1016/j.jiph.2023.02.021
Segala, F. V., Bavaro, D. F., Di Gennaro, F., Salvati, F., Marotta, C., Saracino, A., et al. (2021). Impact of SARS-coV-2 epidemic on antimicrobial resistance: A literature review. Viruses 13, 2110. doi: 10.3390/v13112110
Torres, A., Cilloniz, C., Niederman, M. S., Menéndez, R., Chalmers, J. D., Wunderink, R. G., et al. (2021). Pneumonia. Nat. Rev. Dis. Primers 7, 25. doi: 10.1038/s41572-021-00259-0
Wang, B., Xu, Y., Zhao, H., Wang, X., Rao, L., Guo, Y., et al. (2022). Methicillin-resistant Staphylococcus aureus in China: a multicentre longitudinal study and whole-genome sequencing. Emerging microbes infections 11, 532–542. doi: 10.1080/22221751.2022.2032373
Wang, M., Zheng, Y., Mediavilla, J. R., Chen, L., Kreiswirth, B. N., Song, Y., et al. (2017). Hospital Dissemination of tst-1-Positive Clonal Complex 5 (CC5) Methicillin-Resistant Staphylococcus aureus. Front. Cell. infection Microbiol. 7. doi: 10.3389/fcimb.2017.00101
Zhao, H., Xu, S., Yang, H., He, C., Xu, X., Hu, F., et al. (2019). Molecular Typing and Variations in Amount of tst Gene Expression of TSST-1-Producing Clinical Staphylococcus aureus Isolates. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01388
Keywords: MRSA, Staphylococcus aureus, COVID-19, SARS-CoV-2, co-infection
Citation: Gu F, Zhang Y, Sun J, Guo W and Han L (2025) The characteristics of methicillin-resistant Staphylococcus aureus co-infection in COVID-19 pneumonia. Front. Cell. Infect. Microbiol. 15:1560688. doi: 10.3389/fcimb.2025.1560688
Received: 14 January 2025; Accepted: 25 April 2025;
Published: 16 May 2025.
Edited by:
Elijah Ige Ohimain, Niger Delta University, NigeriaReviewed by:
Gahee Kim, Chosun University Hospital, Republic of KoreaAkira Toga, Keio University, Japan
Copyright © 2025 Gu, Zhang, Sun, Guo and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizhong Han, aGFubGl6aG9uZzExMDdAMTYzLmNvbQ==; Wei Guo, Z3VvLndlaUB6cy1ob3NwaXRhbC5zaC5jbg==
†These authors have contributed equally to this work
 Feifei Gu
Feifei Gu Yuyi Zhang
Yuyi Zhang Jingyong Sun
Jingyong Sun Wei Guo
Wei Guo Lizhong Han
Lizhong Han