- 1Department of Laboratory Medicine, The Third Affiliated Hospital of Zunyi Medical University (The First People’s Hospital of Zunyi), Guizhou, China
- 2Department of Pediatrics and Child Health, The Third Affiliated Hospital of Zunyi Medical University (The First People’s Hospital of Zunyi), Guizhou, China
- 3Department of Clinical Laboratory, Fudan University Huashan Affiliated Hospital, Shanghai, China
- 4Scientific Research Center, The Third Affiliated Hospital of Zunyi Medical University (The First People’s Hospital of Zunyi), Guizhou, China
Coxsackievirus A6 (CVA6) ranks as a primary enterovirus associated with hand-foot-mouth disease (HFMD) and herpangina (HA). Given its significant role in these diseases, there is an urgent need for an efficient identification method. This study presents a novel visual approach based on the reverse transcription polymerase spiral reaction (RT-PSR) for the rapid detection of CVA6. We designed an RT-PSR assay that targets and amplifies a segment of the VP1 gene. Hydroxy naphthol blue (HNB) is incorporated as the detection agent in this assay. To evaluate the performance of the RT-PSR assay, we analyzed 142 clinical throat swab samples. The results were benchmarked against those obtained using quantitative reverse transcription - polymerase chain reaction (qRT - PCR). The RT-PSR assay operates at 65°C for 60 minutes and exhibits a detection limit of 10 copies/μL. When tested against other viruses, it consistently yielded negative results, demonstrating its high specificity. Moreover, the RT - PSR assay showed excellent agreement with a commercial qRT - PCR kit. In conclusion, by using HNB as an indicator, the RT - PSR assay emerges as a straightforward and highly sensitive method for detecting CVA6 in symptomatic throat samples. This approach holds great potential for improving the diagnosis and surveillance of CVA6 - related diseases.
1 Introduction
Hand, foot, and mouth disease (HFMD), caused by enteroviruses, is a global public health issue, especially affecting infants and children. Once dominated by Coxsackievirus A16 (CVA16) and Enterovirus 71 (EV71), recent years have seen a surge in CVA6 cases, triggering pandemics in multiple regions (Zhang et al., 2021; Lim et al., 2016; Gopalkrishna and Ganorkar, 2021; Kamau et al., 2021; Ai et al., 2021). CVA6 is also associated with HFMD in adults (Ramirez-Fort et al., 2014). Currently, there are no specific drugs or vaccines for CVA6, and diagnosis mainly relies on enterovirus nucleic acid testing, crucial for CVA6 - related atypical HFMD (Second et al., 2017).
The gold standard for enterovirus detection, combining virus isolation and serological testing, has drawbacks (Bentley et al., 2021). Virus isolation is complex and time - consuming, and serological screening may yield false negatives, particularly in early infection (Zhou et al., 2022). Nucleic acid amplification tests are sensitive, but conventional methods need sophisticated instruments or complex procedures (Grammatikos et al., 2023). Existing RT - PCR - based enterovirus test kits mainly target EV71 and CVA16, with few of them covering CVA6 (Liu et al., 2014; Zhu et al., 2023). As the epidemiology shifts, there’s an urgent need for a simple, sensitive, and affordable CVA6 diagnostic method, especially in resource - poor areas.
Isothermal nucleic acid amplification like reverse transcription-polymerase spiral reaction (RT-PSR) occurs at a fixed temperature, avoiding costly devices (Huang et al., 2020; He et al., 2020). In comparison with reverse transcription loop-mediated isothermal amplification (RT - LAMP), RT - PSR demands fewer primers, thereby reducing costs (Yadav et al., 2023). The VP1 protein of CVA6, key for host cell infection, is variable and a prime target for molecular typing (Lu et al., 2024a). Hydroxy naphthol blue (HNB), a metal ion indicator, can show amplification in PCR through color change with magnesium ions (Thoraneenitiyan et al., 2022).
In this study, we developed a rapid CVA6 detection method using RT - PSR, optimized reaction conditions, evaluated performance, and validated it with clinical samples, comparing results with qRT - PCR.
2 Materials and methods
2.1 Statement on Ethics and Biosafety
The study received ethical approval from the Ethics Committee of the Third Affiliated Hospital of Zunyi Medical University (The First People’s Hospital of Zunyi City; approval number: 2022-202). All experiments were conducted in our biosafety level 2 clinical laboratory.
2.2 Virus strains and RNA extraction
The CVA6 viral nucleic acids utilized in this study were isolated and preserved within our laboratory, and their authenticity was confirmed through VP1 sequencing. Other viruses employed as non-target samples in this study, including respiratory syncytial virus, hepatitis C virus, hepatitis B virus, rotavirus, cytomegalovirus, CVA16, EV71 and CVA10 underwent clinically standardized tests. Viral RNA was extracted using the RNA Kit for virus detection (Ref# DP315-R; Tiangen, China). The concentrations of RNA were determined utilizing the NanoDrop™ Lite Spectrophotometer (Thermo Scientific, USA), and subsequently stored at -80°C freezer (Aucma, China) prior to utilization.
2.3 The design of primer sets and the construction of a plasmid containing a partial VP1 sequence
The VP1 sequences of CVA6 which were retrieved from the NCBI GenBank database, together with the sequences of CVA6 strains isolated by our group in Zunyi City, were analyzed using the multiple sequence alignment tool Geneious Prime to identify the conserved region of the gene, which served as a template for primer selection. Primers for RT-PSR assay were designed using Oligo 7 software. Three pairs of forward and reverse primers were selected, and an unrelated sequence from a plant source was added to obtain an RT-PSR primer (Table 1). To determine the detection limit, a 270 bp fragment containing the target sequence within VP1 coding region was cloned into pGEM-3Zf vectors to generate recombinant plasmids carrying CVA6 (Table 1).
2.4 Optimization of RT-PSR for the detection of CVA6
The CVA6 RNA, verified by sequencing, was utilized for the optimization of the RT-PSR assay. To determine the occurrence of RT-PSR with these primers, an initial test was conducted on all 6 primers to select those that are suitable. The reaction was conducted in a 25 µl volume comprising of 5 µl RNA templates, 2.5 µl isothermal amplification buffer (10 ×, Ref#M0537S; New England BioLabs, United States), 8.0 mM MgSO4 (Ref# M0537S; New England BioLabs, United States), 0.8 M betaine (Ref#B0300; Sigma, United States), 1.4 mM dNTP each (Ref#4019; Takara, Japan), 8 U BcaBEST ® DNA Polymerase ver.2.0 (Ref#RR380A; Takara, Japan), and 7.5 U WarmStart RTx reverse transcriptase (Ref#M0380S; New England BioLabs, United States) along with PSR-F and PSR-R at a concentration of 2.0 µM. The reaction conditions were maintained at a constant temperature of 63°C for 60 minutes. The resulting reaction products were separated through electrophoresis utilizing 1.5% agarose gels.
After determining the primers, the subsequent step involved optimizing the conditions for RT-PSR to detect CVA6. Firstly, the primer concentration was optimized by considering concentrations of 1.6 µmol/L, 2.0 µmol/L, 2.4 µmol/L, 2.8 µmol/L, 3.2 µmol/L and 3.6 µmol/L, respectively. DEPC-treated water was utilized as the negative control (NC). To optimize the efficiency of RT-PSR amplification, we conducted the reaction at various temperatures ranging from 59 to 68°C and determined the optimal assay time by testing four different time points (30 min, 40 min, 50 min, and 60 min). The confirmation was obtained through agarose gel electrophoresis using GoldenView staining (Biomed, China) and visualized with a Gel Doc XR+ system (Bio-Rad, United States). As a metal ion indicator, HNB exhibits color transformation in response to the concentration of Mg2+ in the reaction solution.
In order to facilitate visual differentiation between negative and positive results, the optimal concentration of Mg2+ was determined. The color changes were observed by introducing 2 µL of 1.5 mmol/L HNB (Ref#YB335-H; Yubo Biotechnology, China) indicator into the detection system containing varying concentrations of Mg2+ (ranging from 2 mmol/L to 12 mmol/L). A change in color from violet to sky blue indicated a positive result.
2.5 Sensitivity test for RT-PSR assay
To assess the sensitivity of the one-step RT-PSR, we determined the copy number of the standard plasmid using the following formula: (copies/µL = 6.02 × 1023 × DNA concentration, g/µL)/plasmid length × 660. The plasmid was diluted tenfold with TE buffer to obtain concentrations of (1 × 108, 1 × 107, 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, and 1 × 101 copies/µL). Subsequently, we added a volume of five microliters from each dilution to our reaction system. As reverse transcriptase was not required for this sensitivity detection experiment since the plasmid served as our template, DEPC-treated water was used as a negative control. We assessed the sensitivity of the RT-PSR by monitoring changes in color within reaction products and verified the results through subsequent analysis via electrophoresis on 1.5% agarose gels.
2.6 Specificity test for RT-PSR assay
The specificity of the RT-PSR assay was evaluated by testing against respiratory syncytial virus, hepatitis C virus, hepatitis B virus, rotavirus, cytomegalovirus, CVA16, EV71 and CVA10. DEPC-treated water served as the negative control during the experiment.
2.7 Comparison between RT-PSR and TaqMan Real-Time RT-PCR in clinical samples
To assess the clinical applicability of the RT-PSR method, a consecutive series of 142 clinical throat swab samples were selected for examination. Each throat swab was immersed in 1.5 mL of normal saline and subsequently subjected to percussion blending. A supernatant volume of 200 µL was collected and added to the nucleic acid extraction kit (Ref#RT-B-200; Zybio, China). Viral RNA was extracted from the supernatant using an automated nucleic acid extractor following manufacturer’s instructions. Subsequently, both TaqMan Real-Time RT-PCR (Da AN Gene Co., Ltd, China) and RT-PSR were performed on the extracted RNA. The results obtained from these two methods were compared.
2.8 Statistical analysis
Statistical analyses were conducted using in SPSS 18.0 (IBM Corporation, New York, United States). Categorical variables were presented as frequencies and percentages. Kappa statistic was employed to assess the agreement between TaqMan Real-Time RT-PCR and one-step RT-PSR.
3 Results
3.1 Primers
Analysis of the RT-PSR results revealed that when primer sets 1, 3 and 4 were used, a distinct DNA ladder was visible on the agarose gel (Figure 1). This clearly indicated successful nucleic acid amplification under isothermal conditions. Conversely, no amplification was observed with the remaining three primer sets. This could potentially be due to sub-optimal amplification parameters. Among all primer sets, primer set 1 exhibited the highest amplification efficiency. As a result, it was selected for subsequent RT-PSR experiments as detailed in Table 1.
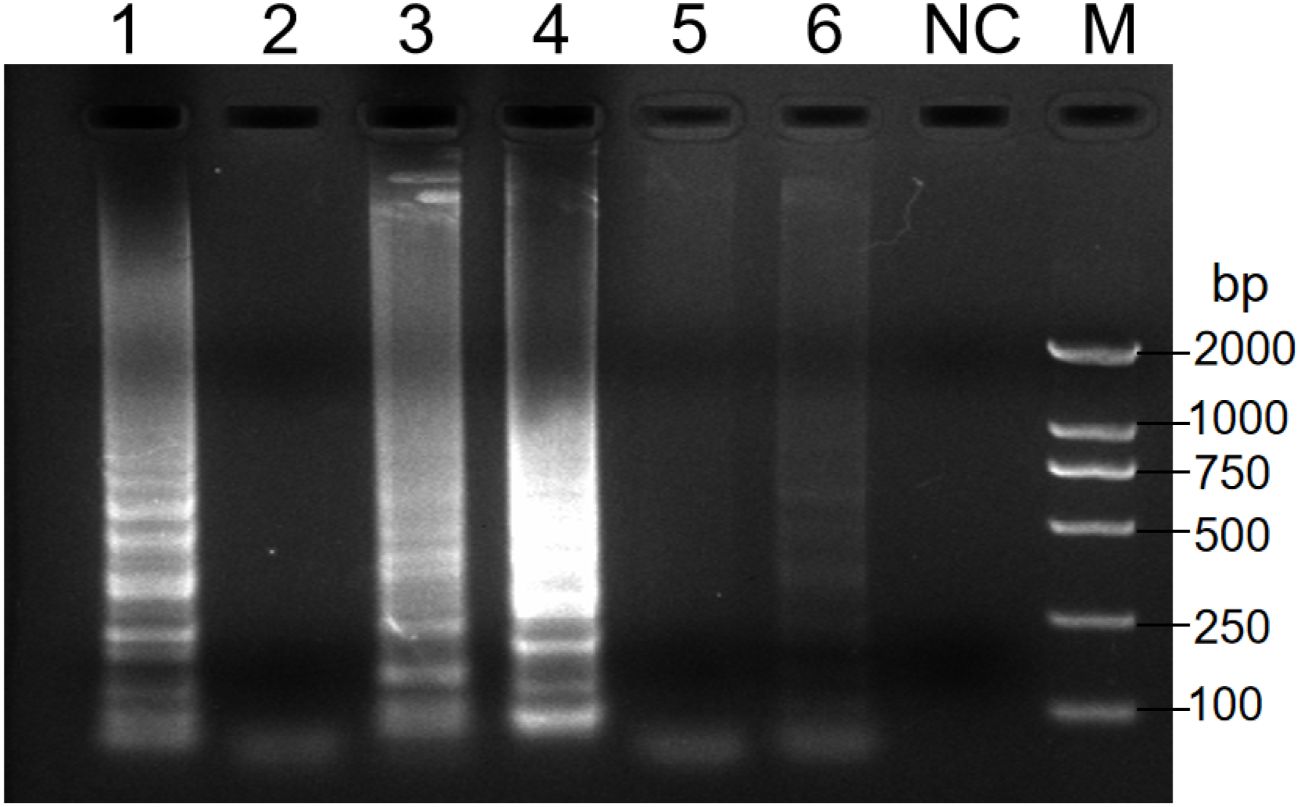
Figure 1. Analysis of the products after amplification with the indicated primers. Electrophoresis was performed on a 1.5% agarose gel to analyze the amplification products of RT-PSR assays with six different sets of primers. Lanes 1-6 indicate primers no. 1 to 6 respectively. Lane NC, negative control (RNase-Free ddH2O); Lane M, DL 2000 DNA marker.
3.2 RT-PSR Optimization
Key reaction parameters for the RT-PSR assay, including primer concentration, reaction temperature, and duration, were optimized using CVA6 RNA. The results of the optimization for primer concentration show that at a final concentration of 1.6 µmol/L, the bands were faint. However, as the concentration increased above 2.0 µmol/L, the amplification bands became stronger, with 2.4 µmol/L identified as the optimal concentration (Figure 2A). Temperature optimization showed that the range of 59-68°C produced minimal variability in band intensity (Figure 2B). A reaction temperature of 65°C was determined to be optimal. Time optimization revealed that no bands formed at 65°C until after 40 minutes, with the bands reaching their best intensity after 60 minutes (Figure 2C). The final optimized conditions for CVA6 detection were 65°C for 60 minutes, with a Mg2+ concentration of 8 mmol/L and 25 µL reaction volumes containing HNB dye for improved differentiation between positive and negative results (Figure 2D).
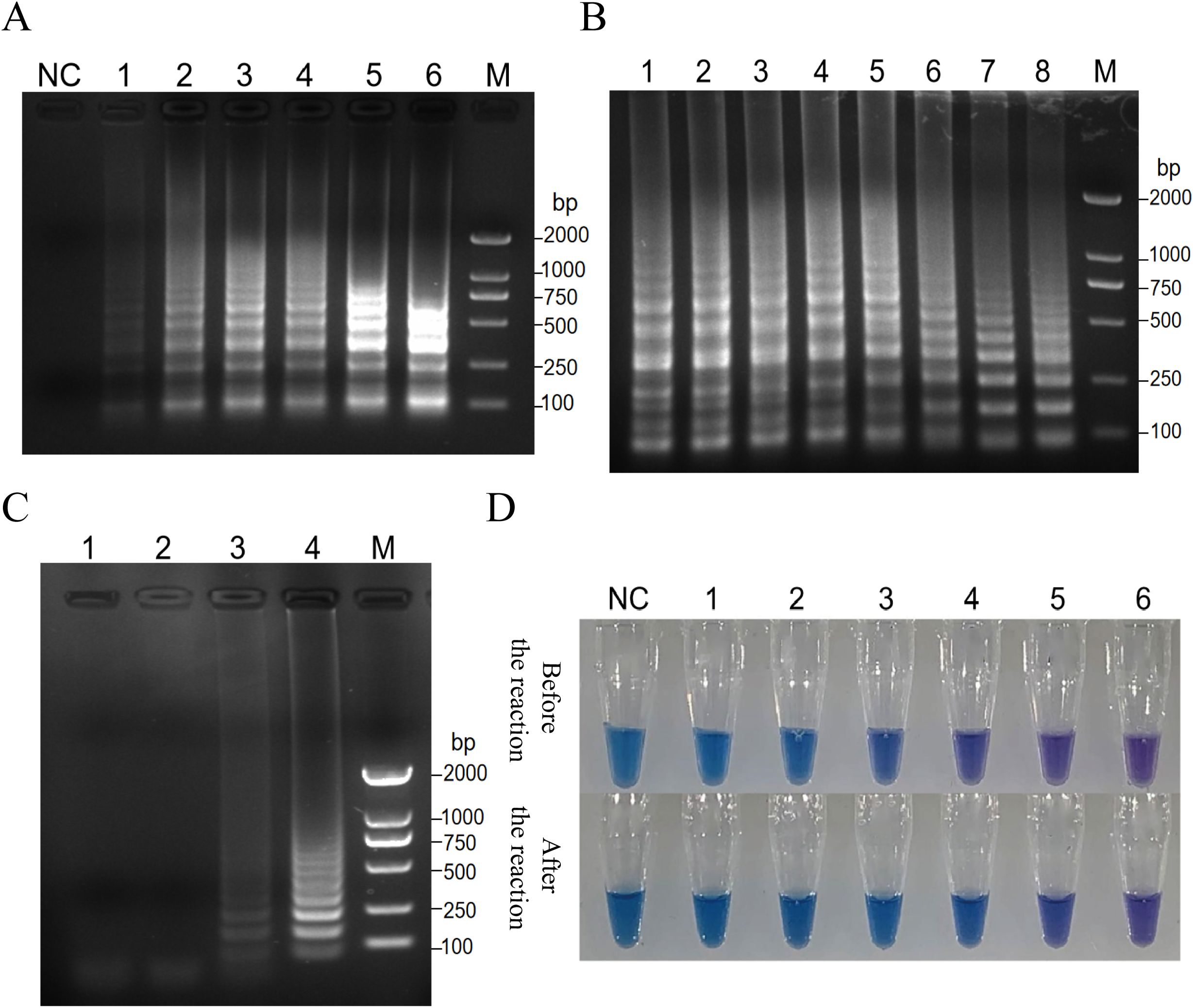
Figure 2. Key reaction parameters for the RT-PSR assay, including primer concentration, reaction temperature, and duration, were optimized using CVA6 RNA. (A) Analysis of the products after amplification with primer 1 at the indicated primer concentrations. Electrophoresis was performed on a 1.5% agarose gel to analyze the amplification products of RT-PSR assays with primer 1 at the primer concentrations of 1.6 µmol/L (Lane 1); 2.0 µmol/L (Lane 2), 2.4 µmol/L (Lane 3), 2.8 µmol/L (Lane 4), 3.2 µmol/L (Lane 5), and 3.6 µmol/L (Lane 6), respectively. Lane NC, negative control (RNase-Free ddH2O); Lane M, DL 2000 DNA marker. (B) Analysis of the product after amplification with primer 1 at the indicated temperatures. RT-PSR amplification was carried out at the temperatures of 59°C (Lane 1), 59.6°C (Lane 2), 60.8°C (Lane 3), 62.4°C (Lane 4), 64.5°C (Lane 5), 66.2°C (Lane 6), 67.3°C (Lane 7), and 68.0°C (Lane 8), respectively. Lane M, DL 2000 DNA marker. (C) Analysis of the product after amplification with primer 1 at 64.5°C for the indicated time periods. RT-PSR amplification was performed for 30 min (Lane 1), 40 min (Lane 2), 50 min (Lane 3), and 60 min (Lane 4), respectively. Lane M, DL 2000 DNA marker. (D) The optimization of Mg2+ concentrations in the reaction. Visual detection of RT-PSR amplification products produced at different concentrations of Mg2+. NC, negative control (DEPC-treated water); Tube 1, 2.0 mmol/L; Tube 2, 4.0 mmol/L; Tube 3, 6.0 mmol/L; Tube 4, 8.0 mmol/L; Tube 5, 10 mmol/L; Tube 6, 12 mmol/L.
3.3 Sensitivity and specificity of the RT-PSR Assay
The analytical sensitivity of RT-PSR was assessed by performing a 10-fold serial dilution of plasmid templates. The limit of detection for RT-PSR was determined to be 10 copies/µL. When the target template concentration exceeded or equaled 1 × 101 copies/µL, the reaction solution exhibited a sky-blue color under natural light, while RNase-Free ddH2O resulted in a violet coloration of the reaction mixture. These findings were further confirmed by agarose gel electrophoresis (Figures 3A, B). The specificity of the assay was evaluated using a panel of 8 strains. Only CVA6 produced a sky-blue color, while all other strains yielded violet results (Figure 3C). The electrophoretic results clearly showed that amplicons were generated solely in CVA6 samples (Figure 3D). No non - specific amplification occurred in any of the control virus samples, effectively highlighting the high specificity of the assay.
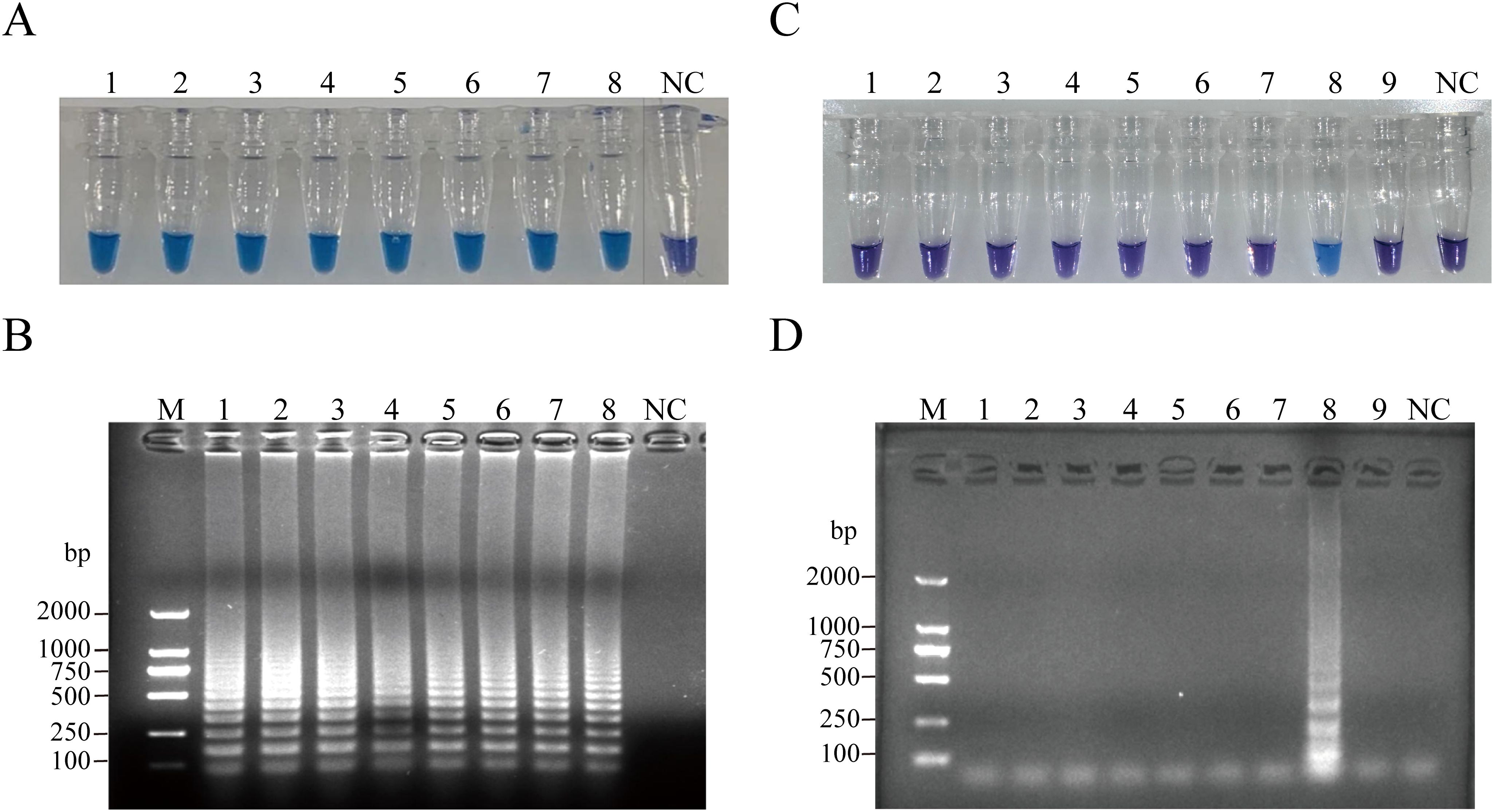
Figure 3. Sensitivity of the RT-PSR amplification for CVA6. (A) Visual detection of RT-PSR amplification products utilizing the HNB indicator. Tube 1, 1 × 108 copies/µL; Tube 2, 1 × 107 copies/µL; Tube 3, 1 × 106 copies/µL; Tube 4, 1 × 105 copies/µL; Tube 5, 1 × 104 copies/µL; Tube 6, 1 × 103 copies/µL; Tube 7, 1 × 102 copies/µL; Tube 8, 1 × 101 copies/µL; NC, negative control (DEPC-treated water). (B) Agarose gel electrophoresis of RT-PSR amplification products. Lane M: DL 1,000 DNA marker. Lane 1, 1 × 108 copies/µL; Lane 2, 1 × 107 copies/µL; Lane 3, 1 × 106 copies/µL; Lane 4, 1 × 105 copies/µL; Lane 5, 1 × 104 copies/µL; Lane 6, 1 × 103 copies/µL; Lane 7, 1 × 102 copies/µL; Lane 8, 1 × 101 copies/µL; NC, negative control (RNase-Free ddH2O). (C) Visual detection of RT-PSR amplification products. Tube 1, hepatitis B virus; Tube 2, hepatitis C virus; Tube 3, respiratory syncytial virus; Tube 4, cytomegalovirus; Tube 5, EV71; Tube 6, CVA10; Tube 7, CVA16; Tube 8, CVA6; Tube 9, rotavirus; NC, negative control (RNase-Free ddH2O). (D) Agarose gel electrophoretic profile of RT-PSR amplification products from 9 viral strains. Lane M: DL 2,000 DNA marker. Lane 1, hepatitis B virus; Lane 2, hepatitis C virus; Lane 3, respiratory syncytial virus; Lane 4, cytomegalovirus; Lane 5, EV71; Lane 6, CVA10; Lane 7, CVA16; Lane 8, CVA6; Lane 9, rotavirus; Lane NC, negative control (RNase-Free ddH2O).
3.4 Clinical specimens testing
RT-PSR and TaqMan real-time RT-PCR were used to test 142 clinical samples. Both methods yielded 50/142 positive results (Figure 4). Using TaqMan real-time RT-PCR as a reference, the RT-PSR assay demonstrated a 100% positive and negative agreement, with a total agreement of 100%, a Youden index of 1, and a Kappa coefficient of 1 (Table 2). This indicates that the RT-PSR method is highly consistent with qRT-PCR results.
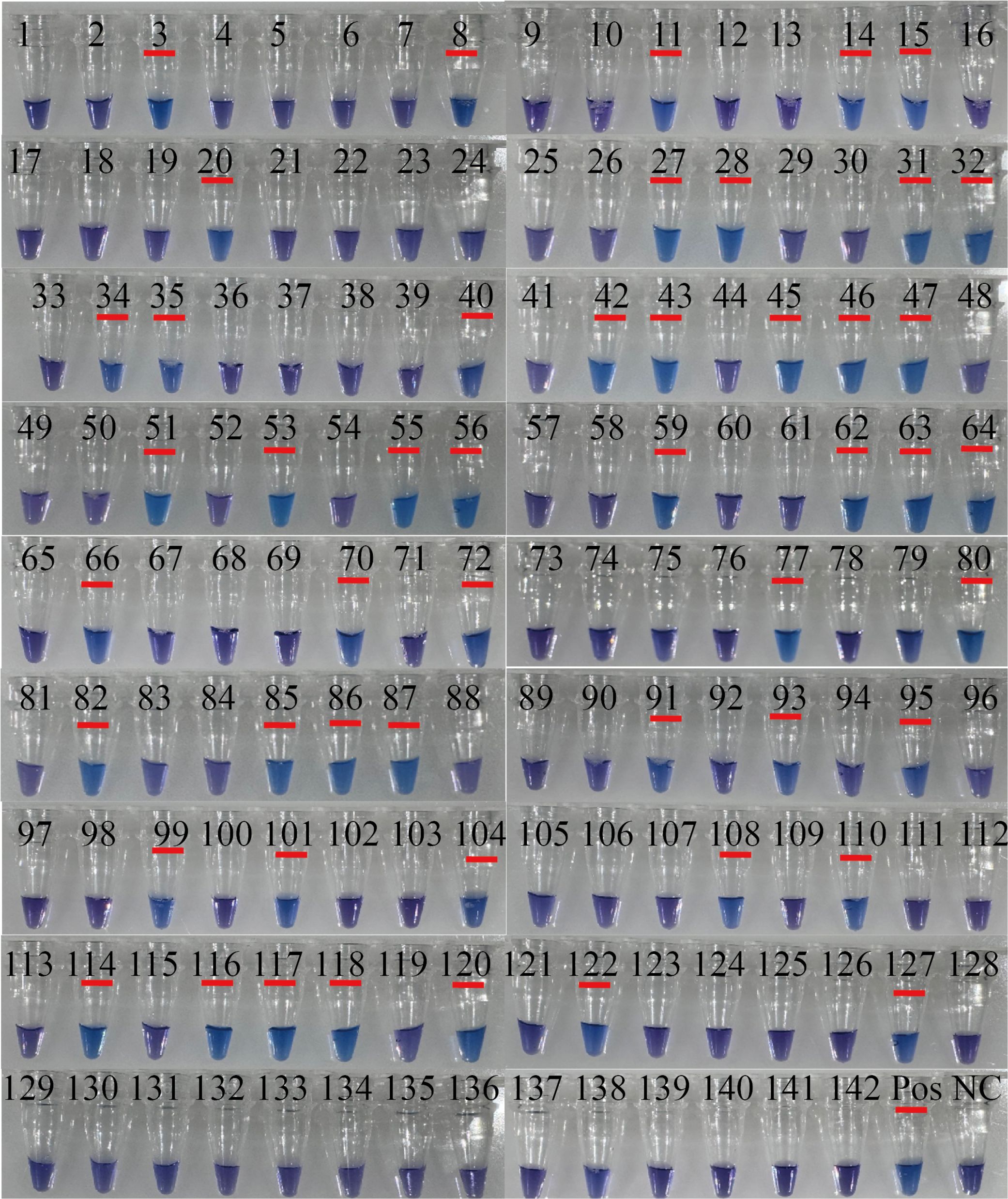
Figure 4. The results of visualized RT-PSR to detect clinical samples. Visual detection of RT-PSR amplification products from clinical samples (tube no. 1 to 142). NC, negative control (RNase-FreeH2O); Pos, 108 copies/µL the standard plasmid.
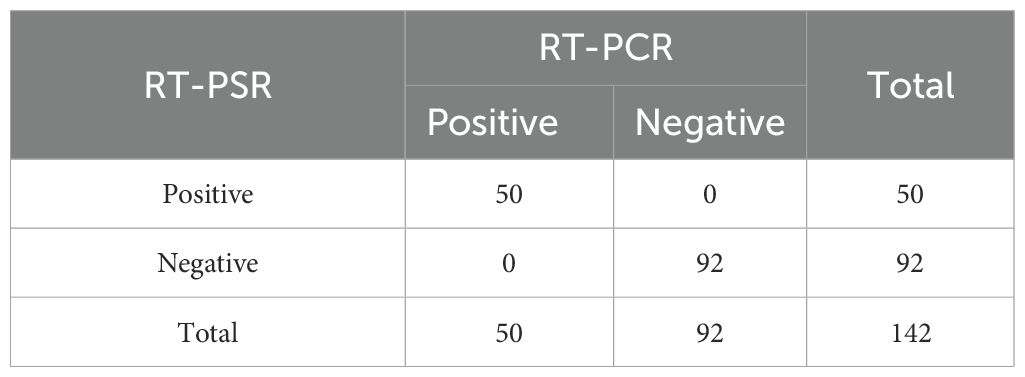
Table 2. The RT-PSR assay was comparatively evaluated with the commercial CVA6 kits for the detection of CVA6 in throat swab samples from human patients.
4 Discussion
This study successfully developed a highly sensitive and specific RT - PSR assay for rapid visual detection of CVA6, a primary causative agent of HFMD and HA. By targeting the VP1 gene and employing HNB as a visual indicator, this assay is capable of detecting as few as 10 copies/μL within 60 minutes at 65°C. When evaluated using 142 clinical throat swab samples, the RT-PSR method demonstrated complete concordance with quantitative RT-PCR (qRT-PCR), validating its efficacy for on-site clinical diagnostics and highlighting its potential utility in resource-constrained environments.
In recent years, the extensive administration of EV71 vaccines has notably reduced the incidence of EV71-associated HFMD cases. Research has revealed that CVA6 has become the primary pathogen in severe HFMD cases across diverse regions in China (Lu et al., 2024b). The growing prevalence of CVA6 in recent years has contributed to an elevated incidence of HFMD among children (Noisumdaeng and Puthavathana, 2023). However, the clinical manifestations of CVA6-related HFMD differ from those of other types. Besides typical symptoms like fever, sore throat, and herpes simplex on the skin of the hands, feet, and mouth, atypical features such as herpetic lesions and nail abnormalities are more frequently observed (Horsten et al., 2018). This situation underscores the insufficiency of infection control strategies that solely target typically symptomatic patients in preventing viral transmission, which may lead to misdiagnoses (Chen et al., 2023). Thus, performing CVA6 enterovirus detection in patients with atypical rashes is of great clinical significance for improving disease diagnosis and identification. In this study, we have developed a novel isothermal nucleic acid amplification method by designing specific primers targeting the conserved sequence of VP1 gene in CVA6. The established method exhibited excellent performance in terms of sensitivity and specificity. In addition, this method is well-suited for POCT (point-of-care testing) and can save approximately 1 hour in testing time compared to qRT-PCR.
The primers were designed on the basis of VP1 sequences. The VP1 capsid protein gene of enterovirus is genetically correlated with the serotype of enterovirus (Liu et al., 2024). In this study, these designed primers demonstrated remarkable specificity for the CVA6 genotype. No cross-reactivity was detected with CVA16, EV71, and CVA10 genotypes. Compared to LAMP, recombinase polymerase amplification, and helicase-dependent amplification, RT-PSR presents several advantages. RT-PSR requires fewer primers than LAMP, streamlining primer design and cutting costs. Similar to LAMP and HDA, it functions at a single temperature. However, it attains high sensitivity and specificity comparable to RPA without the need for recombinase or helicase proteins, which can complicate reaction setup and increase costs. Additionally, RT-PSR adopts a visual detection method with HNB, enabling straightforward result interpretation without complex equipment. This makes it especially suitable for point-of-care testing and resource-limited settings (Dixit et al., 2024).
Several indicators have been utilized to report the amplification process. One approach is to use a pH indicator such as phenol red and conduct the reaction in a weakly buffered environment. As the chain reaction progresses, the pH decreases, causing a visible color change from red to yellow. However, it should be noted that samples do not alter the PH of the reaction (Dao Thi et al., 2020). SYBR green is another common indicator. Positive reactions fluoresce brightly green, while negative samples maintain an orange color. Nevertheless, as more and more double-stranded DNA chimeras are formed due to continuous DNA amplification product generation in the reaction, SYBR Green inhibits the strand displacement activity of Bst DNA polymerase, thus impeding the reaction rate (Xu et al., 2019). In this study, we selected HNB because it has minimal interference. HNB is a metal ion - binding indicator dye that binds to Mg2+ ions and changes color according to the Mg2+ concentration in the reaction mixture. When the Mg2+ concentration is 6 mM or higher, HNB appears purple; below 6 mM, it turns sky blue (Pauly et al., 2023).
The assay demonstrated a high sensitivity in CVA6 detection, with a minimum limit of detection (LOD) of 1 × 101 copies/µL, which is approximately equivalent to the sensitivity of conventional RT-PCR of 0.9 × 101 copies/µl. RT-PSR is an isothermal amplification method. Although it may sacrifice some sensitivity compared to traditional methods, it offers substantial advantages in terms of convenience, specificity, and speed. In 2020, He et al (He et al., 2020). established an RT-PSR for CVA16, achieving a sensitivity range of 2.4 × 102 and 2.4 × 101 copies/µL. Combined with our established method will greatly increase the diagnostic rate of HFMD. Only three previous studies of PSR estimated diagnostic indicators such as sensitivity, specificity and LOD. The reported specificities of PSR were 92%, 93.7% and 100% respectively (Sharma et al., 2020; Milton et al., 2021; Tomar et al., 2020). Our study showed higher specificity, despite having slightly lower sensitivity compared to qRT-PCR-based assays. This closed-tube real-time detection method allows for easy visual result interpretation, making it more convenient and appropriate for primary hospitals and on-site detection in resource-limited regions.
Overall, this study successfully developed a visual RT-PSR assay for CVA6. The assay exhibits remarkable sensitivity, rapid reaction times, and high specificity. In addition, it enables non-invasive analysis and is user-friendly. Despite these achievements, the study has several limitations. The performance of the RT-PSR assay is highly reliant on the optimized concentration of Mg²⁺ and reaction temperature. Therefore, standardized conditions and strict operational procedures are essential. Currently, the assay has only been validated using throat swab samples. Thus, further research is required to explore its applicability to other sample types. Another shortcoming is the limited number of clinical samples included in this study. As a result, a comprehensive evaluation of the assay’s performance relative to other molecular diagnostic techniques in a wider range of clinical settings is still needed. For future research, a significantly larger and more diverse sample population should be incorporated. Also, under various field conditions, systematic comparisons between the RT - PSR detection method and established diagnostic techniques should be carried out. These steps will help to better assess the reliability and generalizability of the RT - PSR assay. Notwithstanding these limitations, this innovative RT - PSR assay offers practical technical support for real - time, on - site testing. It holds particular promise for resource - constrained rural and suburban areas. As such, it can contribute to the expansion of hand, foot, and mouth disease (HFMD) control measures, as well as management strategies for other viral diseases.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Human Research Ethics Committee of The Third Affiliated Hospital of Zunyi Medical University (The First People’s Hospital of Zunyi). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
KFW: Funding acquisition, Supervision, Writing – original draft. KW: Investigation, Methodology, Writing – original draft. YA: Conceptualization, Supervision, Writing – original draft. JL: Supervision, Writing – review & editing. LL: Supervision, Writing – review & editing. WZ: Data curation, Investigation, Writing – original draft. GC: Data curation, Investigation, Writing – review & editing. HZ: Data curation, Writing – review & editing. JW: Data curation, Writing – review & editing. KL: Data curation, Writing – review & editing. SY: Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was financially supported by the Guizhou High-level (BAI) Innovative Talents Project (QIANKehe Platform & Talents-GCC [2022]042-1), Shanghai-Zunyi Cooperation Project (202150524), Innovation Group Project provided by Education Department of Guizhou Province (QianJiaoheKYzi [2021]019), Key Discipline Project of Clinical Laboratory Diagnostics funded by Guizhou Provincial Health Commission (QianWeijianhan [2021]160), and Key Discipline Project of Clinical Laboratory Diagnostics funded by Zunyi Municipal Health Bureau (2022-1444).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, Y., Zhang, W., Wu, J., Zhang, J., Shen, M., Yao, S., et al. (2021). Molecular epidemiology and clinical features of enteroviruses-associated hand, foot, and mouth disease and herpangina outbreak in zunyi, China 2019. Front. Med. (Lausanne) 8, 656699. doi: 10.3389/fmed.2021.656699
Bentley, K., Tee, H. K., Pearson, A., Lowry, K., Waugh, S., Jones, S., et al. (2021). Isolation and identification of inter-species enterovirus recombinant genomes. Viruses 13, 2390. doi: 10.3390/v13122390
Chen, S., Chen, Y., Ji, W., Wang, F., Zhang, X., Jin, Y., et al. (2023). Emerging concerns of atypical hand foot and mouth disease caused by recombinant Coxsackievirus A6 variants in Henan, China. J. Med. Virol. 95, e29316. doi: 10.1002/jmv.v95.12
Dao Thi, V. L., Herbst, K., Boerner, K., Meurer, M., Kremer, L. P., Kirrmaier, D., et al. (2020). A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12, eabc7075. doi: 10.1126/scitranslmed.abc7075
Dixit, R., Kodali, N. K., Biswal, M., Prakash, J. A. J., Gopalan, N., Das, P., et al. (2024). Polymerase Spiral Reaction (PSR) as a point-of-care diagnostic assay: A systematic review. Expert Rev. Mol. Diagn. 24, 79–88. doi: 10.1080/14737159.2024.2315286
Gopalkrishna, V., Ganorkar, N. (2021). Epidemiological and molecular characteristics of circulating CVA16, CVA6 strains and genotype distribution in hand, foot and mouth disease cases in 2017 to 2018 from Western India. J. Med. Virol. 93, 3572–3580. doi: 10.1002/jmv.26454
Grammatikos, S., Svoliantopoulos, I., Gizeli, E. (2023). Naked-eye detection of LAMP-produced nucleic acids in saliva using chitosan-capped auNPs in a single-tube assay. Analytical Chem. 95, 18514–18521. doi: 10.1021/acs.analchem.3c03878
He, S., Huang, Y., Zhao, Y., Pang, B., Wang, L., Sun, L., et al. (2020). A reverse transcription-polymerase spiral reaction (RT-PSR)-based rapid coxsackievirus A16 detection method and its application in the clinical diagnosis of hand, foot, and mouth disease. Front. Microbiol. 11, 734. doi: 10.3389/fmicb.2020.00734
Horsten, H.-H., Kemp, M., Fischer, T. K., Lindahl, K. H., Bygum, A. (2018). Atypical hand, foot, and mouth disease caused by coxsackievirus A6 in Denmark: A diagnostic mimicker. Acta Dermato-venereologica 98, 350–354. doi: 10.2340/00015555-2853
Huang, W. E., Lim, B., Hsu, C. C., Xiong, D., Wu, W., Yu, Y., et al. (2020). RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 13, 950–961. doi: 10.1111/1751-7915.13586
Kamau, E., Nguyen, D., Celma, C., Blomqvist, S., Horby, P., Simmonds, P., et al. (2021). Seroprevalence and virologic surveillance of enterovirus 71 and coxsackievirus A6, United Kingdom 2006-2017. Emerg. Infect. Dis. 27, 2261–2268. doi: 10.3201/eid2709.204915
Lim, C. T., Jiang, L., Ma, S., James, L., Ang, L. W. (2016). Basic reproduction number of coxsackievirus type A6 and A16 and enterovirus 71: estimates from outbreaks of hand, foot and mouth disease in Singapore, a tropical city-state. Epidemiol. Infect. 144, 1028–1034. doi: 10.1017/S0950268815002137
Liu, W., Wu, S., Xiong, Y., Li, T., Wen, Z., Yan, M., et al. (2014). Co-circulation and genomic recombination of coxsackievirus A16 and enterovirus 71 during a large outbreak of hand, foot, and mouth disease in Central China. PloS One 9, e96051. doi: 10.1371/journal.pone.0096051
Liu, H., Xue, Q., Yang, F., Cao, W., Liu, P., Liu, X., et al. (2024). Foot-and-mouth disease virus VP1 degrades YTHDF2 through autophagy to regulate IRF3 activity for viral replication. Autophagy. 20, 1597–1615. doi: 10.1080/15548627.2024.2330105
Lu, H., Xiao, J., Song, Y., Yan, D., Zhu, S., Yang, Q., et al. (2024a). Evolution, recombination and geographic spreading of global Coxsackievirus A6. J. Clin. Virol. 173, 105691. doi: 10.1016/j.jcv.2024.105691
Lu, H., Xiao, J., Wang, W., Yan, D., Ji, T., Yang, Q., et al. (2024b). Evolutionary diversity of coxsackievirus A6 causing severe hand, foot, and mouth disease - China 2012-2023. China CDC Wkly 6, 442–449. doi: 10.46234/ccdcw2024.086
Milton, A. A. P., Momin, K. M., Ghatak, S., Priya, G. B., Angappan, M., Das, S., et al. (2021). Development of a novel polymerase spiral reaction (PSR) assay for rapid and visual detection of Clostridium perfringens in meat. Heliyon 7, e05941. doi: 10.1016/j.heliyon.2021.e05941
Noisumdaeng, P., Puthavathana, P. (2023). Molecular evolutionary dynamics of enterovirus A71, coxsackievirus A16 and coxsackievirus A6 causing hand, foot and mouth disease in Thailand 2000-2022. Sci. Rep. 13, 17359. doi: 10.1038/s41598-023-44644-z
Pauly, M. D., Weis-Torres, S., Hayden, T. M., Ganova-Raeva, L. M., Kamili, S. (2023). Development of simple, rapid, and sensitive methods for detection of hepatitis C virus RNA from whole blood using reverse transcription loop-mediated isothermal amplification. J. Clin. Microbiol. 61, e0077123. doi: 10.1128/jcm.00771-23
Ramirez-Fort, M. K., Downing, C., Doan, H. Q., Benoist, F., Oberste, M. S., Khan, F., et al. (2014). Coxsackievirus A6 associated hand, foot and mouth disease in adults: clinical presentation and review of the literature. J. Clin. Virol. 60, 381–386. doi: 10.1016/j.jcv.2014.04.023
Second, J., Velter, C., Calès, S., Truchetet, F., Lipsker, D., Cribier, B. (2017). Clinicopathologic analysis of atypical hand, foot, and mouth disease in adult patients. J. Am. Acad. Dermatol. 76, 722–729. doi: 10.1016/j.jaad.2016.10.022
Sharma, S., Pardasani, D., Dash, P. K., Parida, M., Dubey, D. K. (2020). Development of magnetic bead based sample extraction coupled polymerase spiral reaction for rapid on-site detection of Chikungunya virus. Sci. Rep. 10, 11651. doi: 10.1038/s41598-020-68469-2
Thoraneenitiyan, N., Choopara, I., Nuanualsuwan, S., Kokpol, S., Somboonna, N. (2022). Rapid visual Candidatus Liberibacter asiaticus detection (citrus greening disease) using simple alkaline heat DNA lysis followed by loop-mediated isothermal amplification coupled hydroxynaphthol blue (AL-LAMP-HNB) for potential local use. PloS One 17, e0276740. doi: 10.1371/journal.pone.0276740
Tomar, P. S., Kumar, J. S., Patel, S., Sharma, S. (2020). Polymerase spiral reaction assay for rapid and real time detection of west nile virus from clinical samples. Front. Cell Infect. Microbiol. 10, 426. doi: 10.3389/fcimb.2020.00426
Xu, W., Gao, J., Zheng, H., Yuan, C., Hou, J., Zhang, L., et al. (2019). Establishment and application of polymerase spiral reaction amplification for salmonella detection in food. J. Microbiol. Biotechnol. 29, 1543–1552. doi: 10.4014/jmb.1906.06027
Yadav, P., Dhankher, S., Sharma, S. (2023). Simplified visual detection of Kyasanur Forest Disease virus employing Reverse Transcriptase-Polymerase Spiral Reaction (RT-PSR). Virus Res. 335, 199180. doi: 10.1016/j.virusres.2023.199180
Zhang, Z., Liu, Y., Liu, F., Ren, M., Nie, T., Cui, J., et al. (2021). Basic reproduction number of enterovirus 71 and coxsackievirus A16 and A6: evidence from outbreaks of hand, foot, and mouth disease in China between 2011 and 2018. Clin. Infect. Dis. 73, e2552–e2559. doi: 10.1093/cid/ciaa1853
Zhou, Y., Zhou, J., Yang, J., Qiu, Q., Wang, L., Yang, J., et al. (2022). Comparison of neutralizing antibody response kinetics in patients with hand, foot, and mouth disease caused by coxsackievirus A16 or enterovirus A71: A longitudinal cohort study of chinese children 2017-2019. J. Immunol. 209, 280–287. doi: 10.4049/jimmunol.2200143
Keywords: enteroviruses, Coxsackievirus A6, reverse transcription polymerase spiral reaction, visual detection, hand-foot-mouth disease
Citation: Wang K, Ai Y, Luo J, Liang L, Zhang W, Cao G, Zha H, Wu J, Lei K, Yao S and Wu K (2025) Visual detection of Coxsackievirus A6 using a reverse transcription polymerase spiral reaction method. Front. Cell. Infect. Microbiol. 15:1563495. doi: 10.3389/fcimb.2025.1563495
Received: 21 January 2025; Accepted: 21 April 2025;
Published: 13 May 2025.
Edited by:
Xin Ye, The First Affiliated Hospital of Xi’an Jiaotong University, ChinaReviewed by:
A Alwin Prem Anand, University of Tübingen, GermanyAntonio Charlys Da-Costa, University of São Paulo, Brazil
Copyright © 2025 Wang, Ai, Luo, Liang, Zhang, Cao, Zha, Wu, Lei, Yao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shifei Yao, c2hpZmVpeWFvMTAwNUAxNjMuY29t; Kaifeng Wu, a2lwaG9vbnd1QDEyNi5jb20=
†These authors have contributed equally to this work
 Kun Wang
Kun Wang Yuanhang Ai
Yuanhang Ai Juan Luo1†
Juan Luo1† Guojun Cao
Guojun Cao He Zha
He Zha Jie Wu
Jie Wu Kun Lei
Kun Lei Kaifeng Wu
Kaifeng Wu