- 1Department of Medical Laboratory, The Second Affiliated Hospital of Guangxi Medical University, Nanning, China
- 2Department of Laboratory Medicine, The People’s Hospital of Guangxi Zhuang Autonomous Region (Guangxi Academy of Medical Sciences), Nanning, China
Objective: Pseudomonas aeruginosa is an opportunistic pathogen responsible for nosocomial infections in critically ill and immunocompromised patients. To elucidate genomic-phenotypic-clinical correlations in P. aeruginosa infections, this study integrates targeted next-generation sequencing (tNGS) with conventional diagnostics.
Methods: A retrospective study was conducted between September, 2023 and December, 2024 at The Second Affiliated Hospital of Guangxi Medical University on patient specimens that were subjected to both conventional culture and tNGS testing. Only a single Gram-negative bacterium, P. aeruginosa, was detected by tNGS, and these isolates were subsequently analyzed in order to compare their genomic profiling (virulence/resistance genes), antimicrobial susceptibility testing (AST), and clinical characteristics.
Results: A total of 105 samples were included in the study. Exoenzyme Y gene (exoY) and O-antigen polymerase gene (wzy) were detected in 77.1% and 12.4% of the samples, respectively. Meanwhile, aminoglycoside acetyltransferase gene (aac(6’)aac(3’)), aminoglycoside resistance methyltransferase gene (armA), and chloram phenicol resistance genes (cmlA) were detected in 6.7%, 4.8%, and 12.4% of the samples, respectively. P. aeruginosa isolates detected the exoY gene exhibit a higher level of drug resistance, particularly to cefepime (32.1% vs 4.2%, p<0.05) and piperacillin-tazobactam (33.3% vs 8.3%, p<0.05). aac(6’)aac(3’) and armA genes were statistically associated with tobramycin resistance (p<0.05). The use of antibiotics before hospitalization (within 90 days), hospital-acquired infections, ICU admission, as well as pre-P. aeruginosa detection interventions including invasive procedures, catheterization, mechanical ventilation, duration of antibiotic use time≥14 days, and antibiotic use type ≥3 types (p<0.05) is associated with multidrug-resistant (MDR) and carbapenem-resistant P. aeruginosa (CRPA) infections (p<0.05). Furthermore, the detection of MDR and CRPA strains appears to lead to an increase in the duration and variety of antibiotic use, as well as prolonged of hospital stay (p<0.05).
Conclusion: Our study highlights the importance of integrating tNGS results, which provide insights into pathogen identification, resistance, and virulence genes, with phenotypic and clinical data in order to enhance the accuracy of diagnosis and guide treatment strategies.
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium commonly found in many different environments, including soil, water, plants, and animals, as part of the natural bacterial microbiota, but it is also known to be an opportunistic pathogen that is a frequent cause of nosocomial infections, such as pneumonia, surgical site infections, urinary tract infections, and bacteremia (Sommer et al., 2020). Apart from its high intrinsic resistance, P. aeruginosa exhibits an exceptional ability to acquire resistance to almost all available antibiotics (Botelho et al., 2019). Infections caused by multidrug-resistant (MDR) or extensively drug-resistant (XDR) P. aeruginosa are linked to a significant increase in morbidity and mortality (Del Barrio-Tofiño et al., 2020; Reynolds and Kollef, 2021). Carbapenems are the preferred treatment for severe P. aeruginosa infections, but resistance to this class of antibiotics has recently emerged as well in the eponymous carbapenem-resistant P. aeruginosa (CRPA) (Teo et al., 2021).
The infection potential of P. aeruginosa is partly attributable to the presence of virulence factors and partly to its ability to metabolize a wide range of antibiotics, which is encoded by genes that are collectively organized within genomic islands, making it harder for any single antibiotic to be effective (Botelho et al., 2019). P. aeruginosa utilizes a type III secretion system (T3SS) to inject cytotoxic effector proteins into host cells, and the promiscuous nucleotidyl cyclase, exoenzyme Y (ExoY), is one of the most common effectors found in clinical P. aeruginosa isolates (Mancl et al., 2020). Phosphorylating (APH), adenylylating (ANT), and acetylating (AAC) enzymes compose the three classes of aminoglycoside-modifying enzymes. The acetyltransferases are a particularly important class of resistance enzymes because of their ability to inactivate many of the medically useful aminoglycosides, such as gentamicin, tobramycin, amikacin, and netilmicin (Schwocho et al., 1995). Both ArmA and RmtB, which belong to the 16S rRNA methyltransferase (16S RMTase), have been shown to contribute to aminoglycoside resistance (O’Hara et al., 2013).
In terms of methods of genomic analysis, metagenomic next-generation sequencing (mNGS) work-flows sequence as much DNA and/or RNA as possible in a given sample, whereas targeted next-generation sequencing (tNGS) workflows enrich specific genetic targets for sequencing and therefore have the advantage of enriching genetic targets for specific pathogens or pathogen groups, as well as other genes of interest (Gaston et al., 2022; Ma et al., 2024). Additionally, tNGS can detect drug resistance and virulence genes. However, the clinical application of pathogens identified through tNGS, along with their associated drug resistance and virulence genes, remains inadequately studied. This article therefore compiles data on sequence counts and resistance and virulence genes of P. aeruginosa from tNGS tests conducted at The Second Affiliated Hospital of Guangxi Medical University. By integrating antimicrobial susceptibility test results with patient clinical characteristics, we sought to examine the relationship between the virulence and resistance genes of P. aeruginosa and antibiotic resistance as thoroughly as possible.
Materials and methods
Patient and sample collection
A total of 105 patients admitted to The Second Affiliated Hospital of Guangxi Medical University from September, 2023 to December, 2024 who had P. aeruginosa detected in their tNGS results were retrospectively enrolled. The inclusion criteria were as follows: (1) P. aeruginosa being the sole Gram-negative bacterium detected with tNGS; (2) Specimens being subjected to both conventional culture and tNGS in parallel; and (3) complete clinical data. The exclusion criterion was: (1) Patients from potential transplant units.
Targeted next-generation sequencing
The tNGS assay employs 2,320 specific primers to detect 276 pathogens. Within the pathogen detection module, Mycobacterium tuberculosis complex receives the highest primer allocation (n=73), while P. aeruginosa is targeted by seven specifically configured primer pairs. Additionally, 269 primers were designed for detecting resistance and virulence genes. The primers related to P. aeruginosa are shown in Table 1. Application and practice of tNGS were followed the guidelines of expert consensus (Healthcare, 2024).
tNGS detection was completed by the Department of Medical Laboratory, Second Affiliated Hospital of Guangxi Medical University and Guangxi Huayin Medical Laboratory Co., Ltd. A commercial assay kit was employed for this procedure, in which the manufacturer had pre-optimized the target gene panel and primer sequences. For tNGS, sputum or viscous bronchoalveolar lavage fluid (BALF) samples were mixed with DTT liquefaction reagent in a 1.5 mL tube and centrifuged. After cell lysis, DNA was extracted from 500 μL of the homogenate using a BayBiopure Magnetic Pathogenic Microorganisms Nucleic Acid Kit (Guangzhou Bay Area Biotechnology Co., Ltd., China, Lot number: PMNM-LQ64-Magmix) or a Blood/Cell/Tissue Genomic DNA Extraction Kit (Shanghai Jiachu Biotechnology Co., Ltd., China, Lot number: YDP304-02), following the manufacturer’s protocols. After reverse transcription, the cDNA products were enriched by targeting specific regions with a PCR cycle set at 95°C for 3 minutes, followed by 23 cycles of 95°C for 25 seconds, 63°C for 120 seconds, and 72°C for 120 seconds, with a final extension at 72°C for 5 minutes, before being held at 4°C. Magnetic beads were used to purify the targeted PCR products. Then, 10 μL of the PCR product was used for secondary amplification and purified using magnetic beads once again. Sequencing of the purified libraries was performed using the MGISEQ-200RS High Throughput (Fast) Sequencing Kit (GI).
Antibiotic resistant test
The identification and antimicrobial susceptibility testing (AST) of P. aeruginosa isolated from clinical specimens were performed using the Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and the BD Phoenix M50 automatic microbial identification and drug susceptibility system. Susceptibility testing was done against the following antibiotics: ceftazidime, cefepime, meropenem, imipenem, piperacillin-tazobactam, aztreonam, tobramycin, ciprofloxacin, levofloxacin, ceftazidime-avibactam, cefoperazone-sulbactam and colistin. Antimicrobial sensitivity testing followed the guidelines established by the Clinical and Laboratory Standards Institute v33.0 (CLSI, 2023) and v34.0 (CLSI, 2024). The disk diffusion method was additionally employed for supplementary testing of relevant antibiotics.
Based on resistance patterns, P. aeruginosa was categorized into susceptible P. aeruginosa (SPA), MDR, and XDR groups (Magiorakos et al., 2012; Teo et al., 2021; Yaping Xu et al., 2017). In this study, MDR was defined as resistance to at least three classes of the following antibiotics: ceftazidime, cefepime, meropenem, imipenem, piperacillin-tazobactam, aztreonam, tobramycin, ciprofloxacin and levofloxacin. XDR was defined as resistance to all antibiotics except colistin. Additionally, P. aeruginosa was further classified into CRPA and carbapenem-sensitive P. aeruginosa (CSPA) (Teo et al., 2021).
Statistical analysis
Data analysis was performed using SPSS software (IBM, USA), version 19.0. Enumeration data were expressed as cases/percentage [n (%)], and comparisons between groups were made via Chi-squared tests, Continuity-corrected Chi-square Test or Fisher’s exact test. Normally distributed continuous data were presented as mean ± standard deviation (SD). Comparisons between the two groups were conducted using independent samples t-tests. For nonnormally distributed continuous data, results were expressed as median (interquartile range), M (Q1, Q3), and the Wilcoxon rank-sum test was used for group comparisons. A p-value <0.05 was considered to indicate statistically significant test results.
Results
Patient characteristics
The clinical samples included 61 males and 44 females, aged between 7 to 86 years. Isolates were derived from BALF (n=91), sputum (n=5), blood cultures (n=2), cerebrospinal fluid (n=2), tissue (n=2), and other origins (n=3). 55 isolates were collected from the pneumology department, 33 from intensive care unit (ICU), 4 from the rehabilitation department, 3 from the orthopedics department, 3 from cardiovascular thoracic surgery, 2 from emergency ward observation, and 5 from other departments.
Antimicrobial susceptibility profile of P. aeruginosa
In terms of drug resistance, 55 isolates were classified as SPA, 50 as MDR, and 6 as XDR. Among the MDR, 72% exhibited additional resistance to aztreonam, 56% to cefoperazone-sulbactam, 28% to tobramycin, and 30% to ceftazidime-avibactam. All isolates remained susceptible to colistin. Additionally, there were 61 cases of CSPA and 44 cases of CRPA, and among the CRPA isolates, resistance rate was significantly higher than that of CSPA (p<0.05) (Figure 1). All of these isolates were also susceptible to colistin.
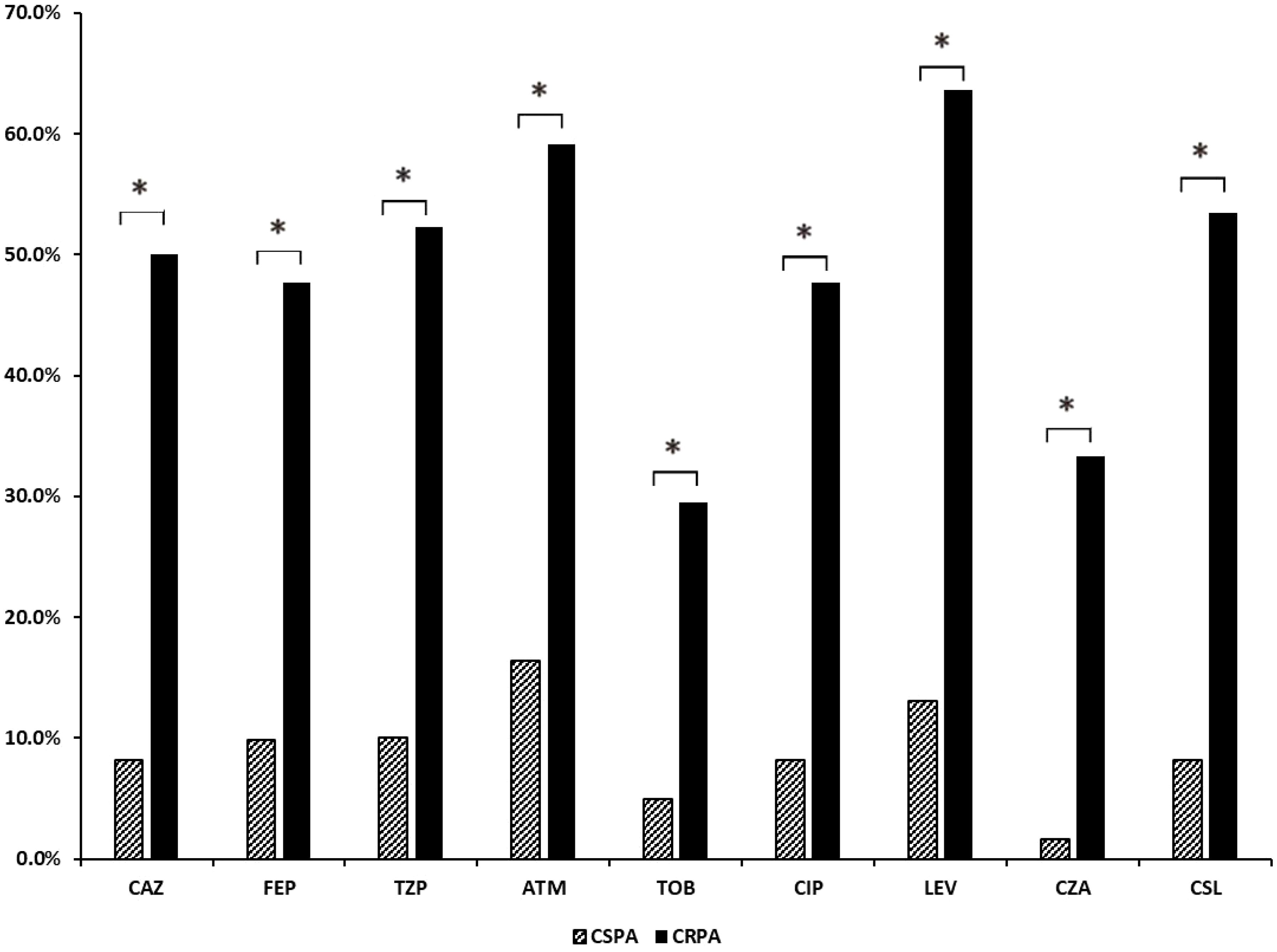
Figure 1. Analysis of drug resistance rates in CSPA and CRPA groups. CAZ, ceftazidime; FEP, cefepime; MEM, meropenem; IPM, imipenem; TZP, piperacillin-tazobactam; ATM, aztreonam; TOB, tobramycin; CIP, ciprofloxacin; LEV, levofloxacin; CZA, ceftazidime-avibactam; CSL, cefoperazone-sulbactam. *, The results indicate a statistically significant difference between the two groups.
Analysis of virulence gene exoY and antimicrobial resistance
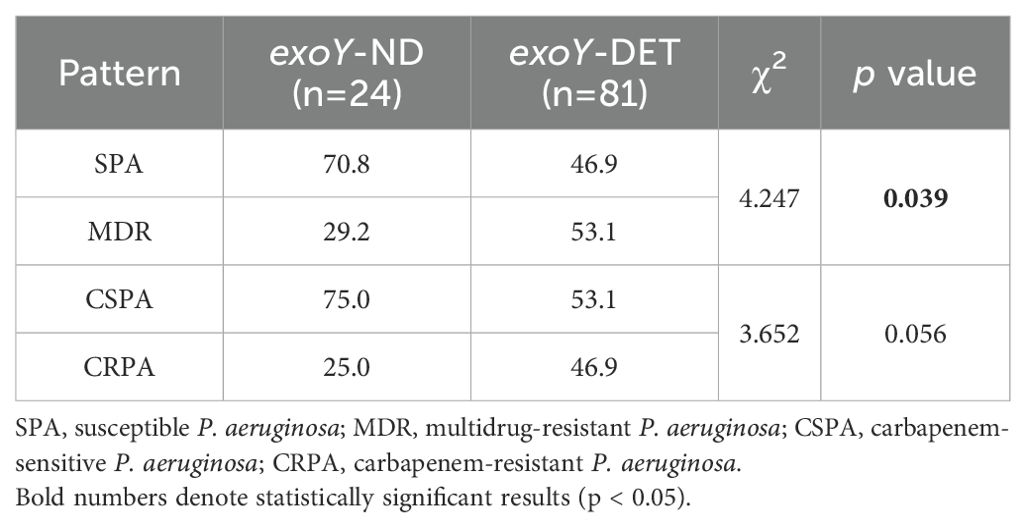
Among the 105 P. aeruginosa isolates, 77.1% (81/105) were exoY detected (exoY-DET), and 22.9% (24/105) were exoY undetected (exoY-ND). The resistance rates of exoY-DET strains to cefepime and piperacillin-tazobactam were significantly higher than those of the exoY-ND strains (p<0.05) (Figure 2). In terms of different resistance patterns, the proportion of exoY-DET strains in MDR isolates was significantly higher than in SPA isolates (p<0.05). However, there was no significant difference in the prevalence of exoY between CSPA and CRPA (Table 2).
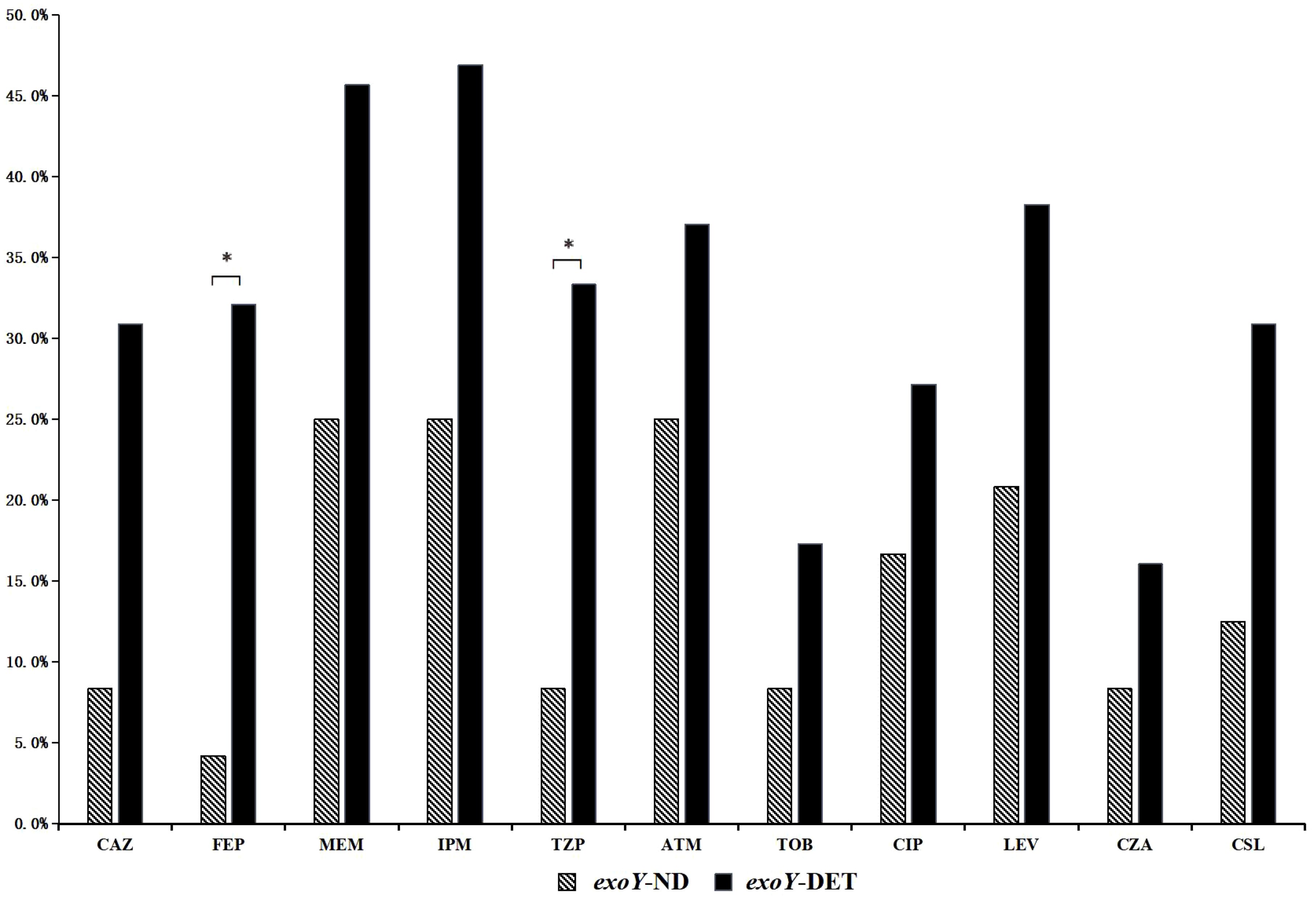
Figure 2. Resistance rates to common antibiotics among exoY-ND and exoY-DET strains. *, The results indicate a statistically significant difference between the two groups.
Associations between exoY and patient clinical characteristics
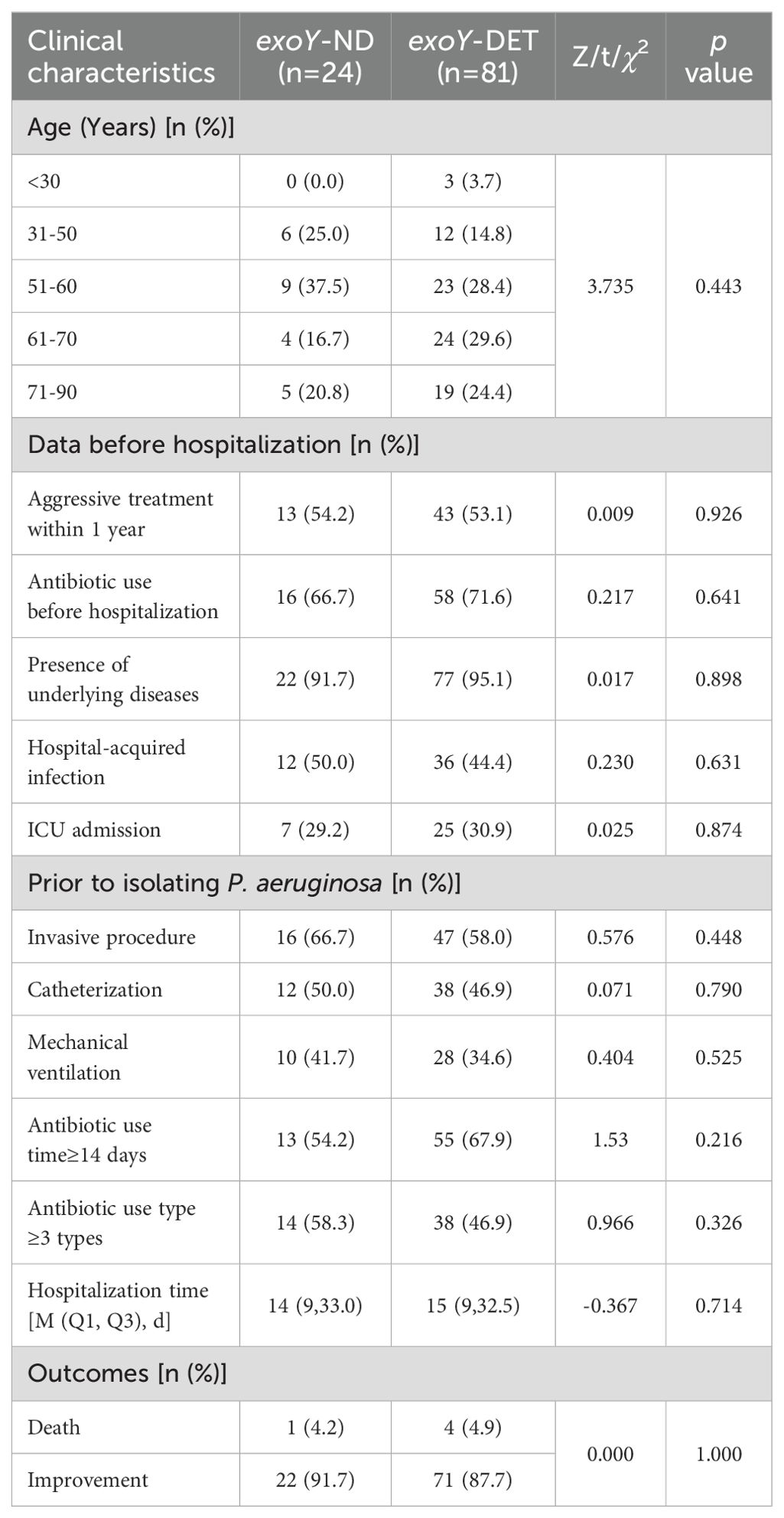
There were no significant differences between the exoY-ND group and the exoY-DET group in terms of age, history of aggressive treatment within the past year, antibiotic use before hospitalization (within 90 days), presence of underlying diseases, hospital-acquired infection, ICU admission, as well as pre-P. aeruginosa detection intervention including invasive procedures, catheterization, mechanical ventilation, duration of antibiotic use time≥14 days, the number of antibiotic use type ≥3 types, and metrics including hospitalization time and outcomes (Table 3).
Drug resistance genes and drug resistance
The resistance genes that could be detected in this study including β-lactamase, vancomycins, methicillins, aminoglycosides, macrolides, sulfonamides, quinolones, chloramphenicols, tetracyclines, polymyxins and efflux pump class. For aminoglycoside resistance genes, we designed primers for: aac, ant, aph, armA and rmtB (Supplementary Table 1). But our study only detected aac(3’), aac(6’), armA, cmlA, blaTEM, floR, blaGES and tetC genes in P. aeruginosa.
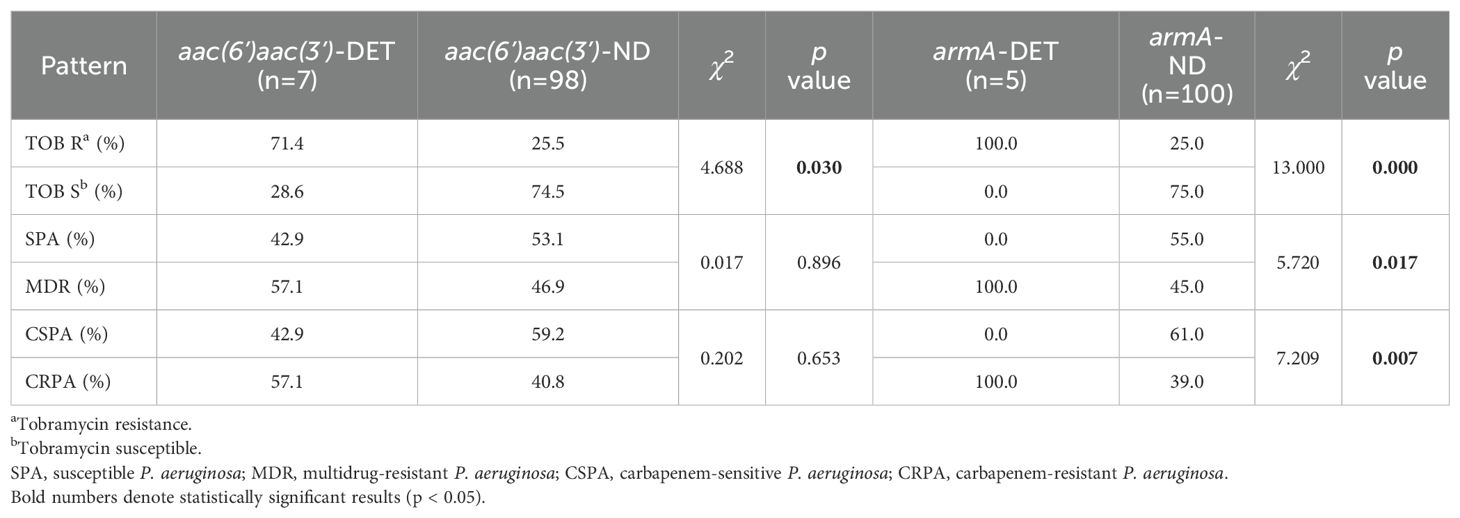
Among the 105 isolates, 7 were detected the aac(6’) and aac(3’) resistance gene, and these exhibited significantly higher resistance to tobramycin compared to isolates undetected the gene (p<0.05). No significant difference was observed in the detection rate of the aac(6’)aac(3’) across the SPA and MDR, or CSPA and CRPA groups (Table 4). 5 isolates detected the armA resistance gene, and these isolates demonstrated significantly higher resistance to tobramycin compared to the armA-ND group as well (p<0.05). The armA resistance gene was detected exclusively in MDR and CRPA isolates (Table 4), and only one armA-DET isolate was classified as XDR (data not shown). In addition, the resistance genes cmlA, blaTEM, floR, blaGES, and tetC were detected in 13, 3, 2, 1, and 1 isolate(s), respectively.
Correlation between different resistance phenotypes and patient clinical characteristics
There were no statistically significant differences between SPA and MDR or between CSPA and CRPA in terms of age, history of aggressive treatment within the past year, and underlying diseases (p>0.05). However, significant differences were observed between groups regarding the use of antibiotics prior to hospitalization (within 90 days), hospital-acquired infections, ICU admission, as well as pre-P. aeruginosa detection interventions including invasive procedures, catheterization, mechanical ventilation, duration of antibiotic use time≥14 days, and the number of antibiotic use type ≥3 types (p<0.05). Following the detection of P. aeruginosa, the duration of antibiotic use time≥14 days, and the number of antibiotic use type ≥3 types, and the length of hospital stay were also significantly different between groups (p<0.05). These findings suggest that the detection of MDR and CRPA strains is associated with prolonged antibiotic use, a greater diversity of antibiotics, and extended hospital stays (Table 5).
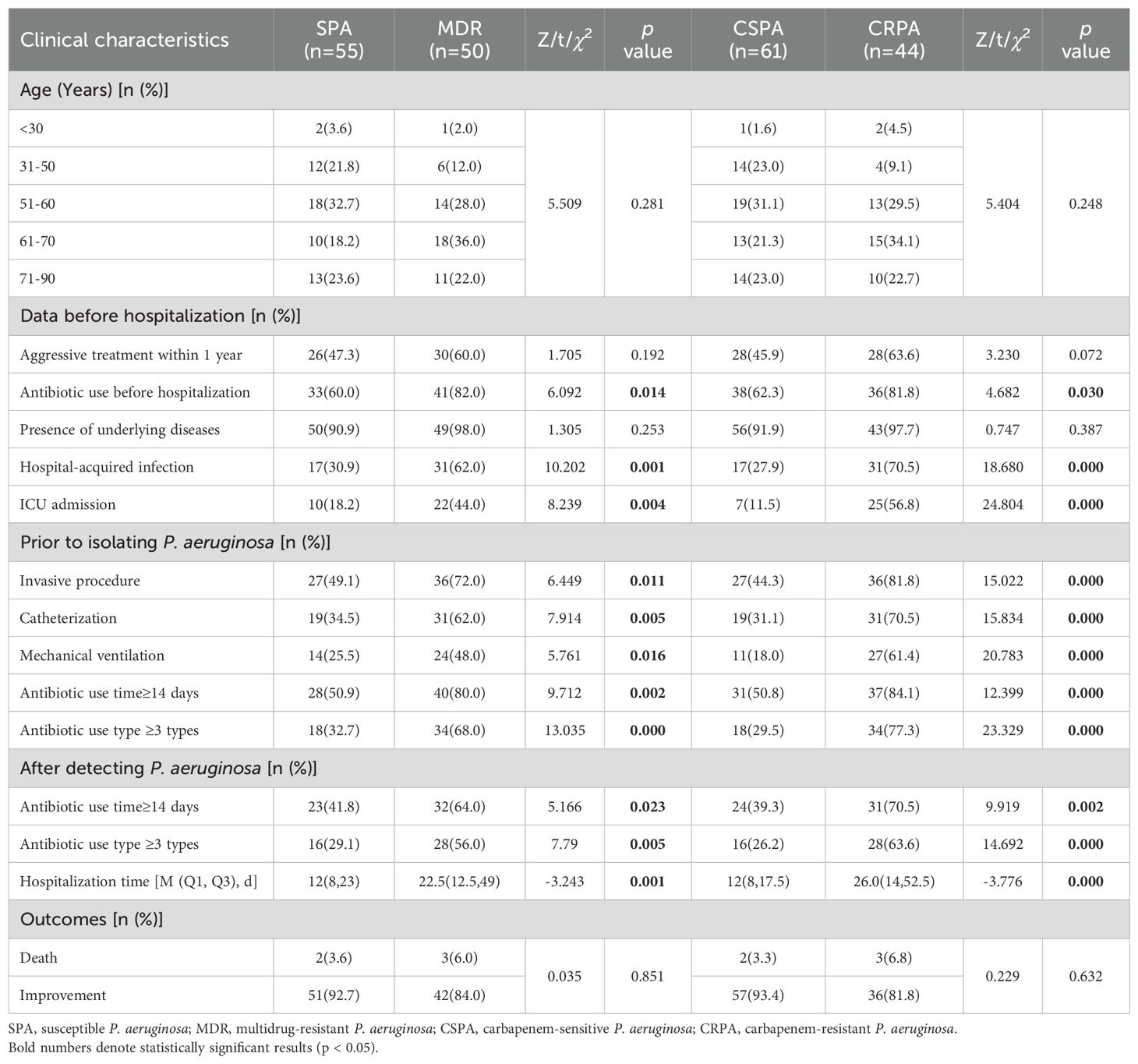
Table 5. Clinical characteristics of patients with P. aeruginosa infections with different resistance patterns.
Comparative analysis of tNGS and whole genome sequencing results
As a newly developed platform for detecting virulence and resistance genes, the validity of tNGS needs to be confirmed. To address this issue, we conducted whole genome sequencing (WGS) on 10 enriched bacterial samples from the included studies and compared the results with those from the tNGS (Supplementary Tables 1, 2). The results demonstrated a 100% concordance rate between WGS and tNGS for exoY, cmlA, and armA. However, although WGS achieved 100% detection rates for mucA, fliC/D, flgC/D/E/G/H/I/J, flhB, fliI/J/M/P/Q/R, fleN, and aph(3’)-IIb, 20% for wzz and 10% for ant(2’’), tNGS failed to detect these genes. It should be noted that both aac(6’) and wzy were detected in Case 9 by WGS but were missed by tNGS, suggesting potential false negatives in tNGS (Table 6). These findings suggest that WGS outperforms tNGS in detecting virulence and resistance genes. Moreover, among 4,206 historical tNGS reports, aph(3’)-IIb was detected in 108 cases, mainly associated with Staphylococcus aureus. This observation implies that the tNGS primers targeting the aph resistance gene may have strain specificity and thus may not be suitable for detecting in P. aeruginosa.
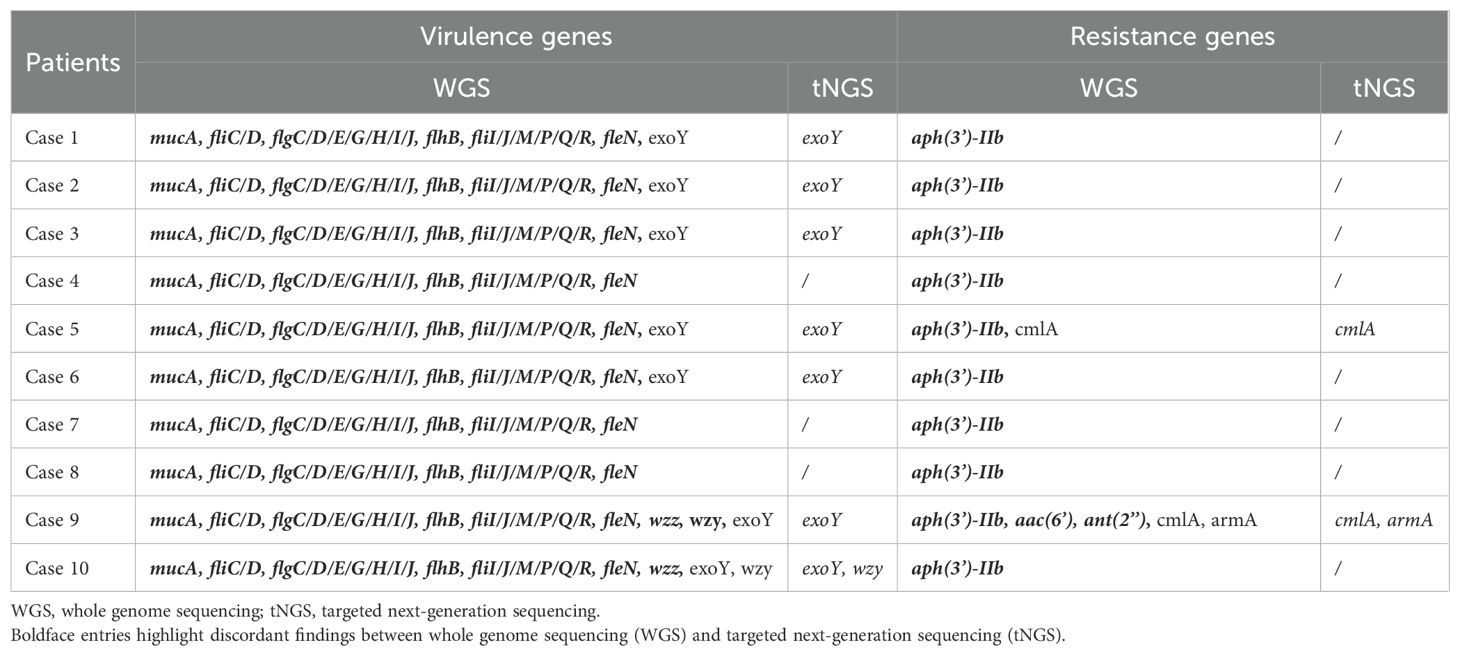
Table 6. Comparative detection of virulence and resistance genes by WGS and tNGS in clinical samples.
Discussion
The infection potential of P. aeruginosa is partly attributable to its virulence factors and partly to its ability to metabolize a wide range of antibiotics, which collectively enhance its infectivity potential (Botelho et al., 2019), and it is crucial to understand the genetic mechanisms that underpin both of these sources of drug resistance. tNGS has the advantage of enriching genetic targets for specific pathogens or pathogen groups as well as other genes of interest (Gaston et al., 2022; Ma et al., 2024). Due to its high sensitivity, affordability, and strong concordance rate, tNGS has begun to emerge as a valuable tool in the clinical detection of infectious diseases (Liu et al., 2024). In this study, a commercial assay kit was used where genes and primers tested were preselected by the manufacturer. By December 20, 2024, a total of 4,206 specimens had been tested using tNGS. Among these, respiratory samples accounted for 60.0%, and blood samples accounted for 32.9%. Non-sterile body fluid samples frequently contain mixed microbes, including diverse Gram-negative and Gram-positive bacteria, viruses, and other pathogens, resulting in complex detected resistance and virulence genes. To minimize interference from resistance and virulence genes of other bacteria, we exclusively selected samples in which Gram-negative bacteria consisted solely of P. aeruginosa and for which both microbial culture and AST data were available.
Of the 105 cases, the sequence count for P. aeruginosa totaling 92,857(55,184-122,118), and the minimum sequence count was 949. This suggests that when the sequence count for P. aeruginosa is below a certain threshold, its colony count in culture may be low or even undetectable. Since culture is still the gold standard for bacterial confirmation, clinicians should carefully consider this diagnostic limitation when making antibiotic treatment strategies. And detecting substantial amounts of P. aeruginosa in respiratory tract specimens doesn’t mean an infection, as this opportunistic pathogen may colonize alongside polymicrobial flora. In such cases, the hospital’s microbial staff, after reviewing both culture and smear results, considered it may belong to typical pharyngeal flora and ultimately did not conduct drug susceptibility testing, or the sample was deemed unqualified and required additional inspection.
Virulence gene selection criteria prioritize clinical relevance, functional representativeness and detection feasibility. For clinical relevance, genes should be linked to infection severity or therapeutic decisions. For example: Based on clinical phenotype correlations and detection positivity rates, exoY-DET strains demonstrate higher prevalence in burn, bacteremia, urine, and respiratory samples (51.7-95.0%), whereas exoT exhibits substantial inter-study variability in detection rates (5-100%). Although exoU and exoS exhibit potent virulence, their limited carriage rates (approximately 13.3-64.5% and 20.0-38.46%, respectively) and poor representativeness across clinical cohorts restrict their broader applicability (Azimi et al., 2016; Jabalameli et al., 2012; Ullah et al., 2023). Regarding biofilm-related genes, the mucA mutation directly induces alginate overproduction, which is strongly associated with chronic pulmonary infections and mucoid colony formation in cystic fibrosis (CF) patients (Schofield et al., 2021). In contrast, Psl and Pel, while contributing to biofilm formation, are primarily linked to acute-phase infections or environmental adaptation, lacking well-defined clinical biomarkers (de Sousa et al., 2021). For functional representativeness, the Wzy/Wzz system regulates the synthesis of the O-antigen of lipopolysaccharide (LPS), determining the serotype of P. aeruginosa (e.g., O1–O20). This regulation impacts host immune recognition and antibody neutralization efficiency (Islam et al., 2010). Meanwhile, type IV pili (encoded by pilA, pilB, pilC, pilQ) are involved in adhesion and motility, but their functions can be partially compensated by flagella or other adhesins (e.g., CupA) (de Sousa et al., 2021).
In this study, the virulence factors were classified into nine functional categories: toxins, adhesion and invasion systems, secretion systems, biofilm and polysaccharide production, iron uptake mechanisms, LPS biosynthesis, genotoxins, flagella and motility, and heme uptake and utilization systems (Supplementary Table 2). Among them, the virulence genes specific to P. aeruginosa include exoY and mucA, while those shared with other bacteria comprise wzy, wzz, lpxA/B/C/D/L/M, fleN, flgC/D/E/G/H/I/J, and fliC/D/I/J/M/P/Q/R. However, only the exoY and wzy genes were detected in this study. Regarding resistance genes, only aac(6’), aac(3’), armA, cmlA, blaTEM, floR, blaGES, and tetC were detected.
ExoY is one of the effectors injected by the T3SS of P. aeruginosa into host cells. Inside eukaryotic cells, ExoY interacts with F-actin, which stimulates its potent nucleotidyl cyclase activity to produce cyclic nucleotide monophosphates (cNMPs), which disrupts the actin cytoskeleton and increases endothelial permeability (Galle et al., 2012; Mancl et al., 2020; Reynolds and Kollef, 2021; Silistre et al., 2021). Among the 105 P. aeruginosa isolates in this study, 81 (77.1%) were detected the exoY virulence gene, which is inconsistent with previous findings (Azimi et al., 2016; Ullah et al., 2023), possibly due to variations in specimen types. This study primarily used respiratory tract specimens, whereas other studies have focused on wound or secretion samples. The resistance rate of exoY-DET strains to testing drugs was higher than that of exoY-ND strains, though with statistically significant differences observed only for cefepime and piperacillin-tazobactam (p<0.05). Additionally, the proportion of exoY-DET strains in the MDR group was significantly higher than that in SPA isolates (p<0.05). These findings suggest that P. aeruginosa isolates that carry the exoY gene exhibit a more serious degree of drug resistance.
No significant differences were observed between the exoY-DET and exoY-ND groups in clinical features such as the duration and type of antibiotics used, hospitalization time and outcome. This is possibly because ExoY counteracts the overall cytotoxicity of P. aeruginosa toward human epithelial cells and may exert a protective role at certain stages of bacterial infection, potentially facilitating host colonization or contributing to the establishment and/or maintenance of chronic infection (Silistre et al., 2021). This tNGS panel balances the detection of antimicrobial resistance and virulence genes across multiple bacterial pathogens (Supplementary Table 1, 2), which limits the proportion of P. aeruginosa-specific gene targets. However, exoS or exoU may hold greater direct diagnostic relevance when prioritizing strongly virulent factors. Future tNGS protocols could incorporate multiplex detection of exoS, exoU, and exoT alongside integrated analysis of exoY with other T3SS effector proteins to comprehensively evaluate their synergistic contributions in polymicrobial infections.
Biosynthesis of B-band LPS in P. aeruginosa follows the Wzy-dependent pathway, which requires the integral inner membrane proteins Wzx O-antigen [O-Ag] flippase), Wzy (O-Ag polymerase), and WaaL (O-Ag ligase), the O-Ag chain length is determined by Wzz, a polysaccharide copolymerase type 1 protein (Islam et al., 2010). Many important bacterial virulence factors are encoded by prophage or have a phage origin. Due to this, Wzy may greatly contribute to P. aeruginosa virulence (Kaluzny et al., 2007). However, no significant differences were observed in drug sensitivity or clinical characteristics between the wzy-DET and wzy-ND groups (Supplementary Table 3, Supplementary Figure 1).
The lpx series is involved in lipid A biosynthesis and is conserved across Gram-negative bacteria. Currently, the best-characterized lipid A synthesis pathway is the Raetz pathway in Escherichia coli (Romano and Hung, 2023). Regarding flagellar assembly, the fleN, flgC/D/E/G/H/I/J, and fliC/D/I/J/M/P/Q/R genes are widely distributed across Gram-negative bacteria, including P. aeruginosa, Salmonella enterica, Yersinia spp., and Shigella spp (Erhardt et al., 2010; Oladosu et al., 2024). However, none of these genes were detected in our current study.
As a highly adaptable opportunistic pathogen, P. aeruginosa relies on the synergistic effects of multifaceted virulence mechanisms to establish infections. Beyond the well-characterized T3SS and LPS, it secretes a variety of exotoxins and proteases to directly disrupt host tissues. For example, Elastase (LasB, LasA) specifically degrade elastin-rich connective tissues in pulmonary structures (Casilag et al., 2016), while phospholipases and lipases disrupt surfactant function and modulate local immune responses (Korotkov et al., 2012; Ostroff et al., 1990). Exotoxin A inhibits protein synthesis by ADP-ribosylation of cell elongation factor-2 in the host cell, consequently leading to cell death (Allured et al., 1986). Further proteolytic activity is demonstrated by PrpL, a serine protease targeting multiple host substrates including casein, lactoferrin, transferrin, elastase, and decorin (Wilderman et al., 2001). Simultaneously, biosurfactant molecules like rhamnolipids serve dual functions, facilitating biofilm maturation while impairing host immune cell function (Alhede et al., 2009; Pearson et al., 1997).
The pathogen’s iron acquisition systems constitute another critical virulence dimension. P. aeruginosa employs high-affinity siderophores (pyoverdine and pyochelin) and direct heme-iron uptake via PhuR/HasR transporters to overcome iron-deficient environment (Ochsner et al., 2000; Schalk and Guillon, 2013). This efficient iron-capturing ability not only supports bacterial proliferation but also modulates virulence gene expression through iron-responsive regulatory networks (Minandri et al., 2016). Chronic infection strategies center on sophisticated biofilm development mediated by exopolysaccharides (Psl, Pel, and alginate) that establish protective matrices, and dynamically regulates virulence factor expression through quorum-sensing (QS) systems (Las, Rhl) (de Sousa et al., 2021). Moreover, the type V secretion system (T5SS) strengthens its ecological niche competitiveness by promoting biofilm formation and auto-aggregation, and influencing the host’s response to bacterial infection (Borlee et al., 2010; Salacha et al., 2010). The interplay of these virulence mechanisms poses significant clinical challenges. Future tNGS design could consider incorporating more virulence factor detection.
AAC enzymes are a subset of aminoglycoside-modifying enzymes. Acetyltransferases, in particular, play a crucial role in bacterial resistance due to their ability to inactivate several medically important aminoglycosides, including gentamicin, tobramycin and amikacin (Dolgusevs et al., 2024; Schwocho et al., 1995). In this study, the aac(6’)aac(3’) resistance gene was detected in 7 of 105 P. aeruginosa isolates, a detected rate of 6.7%. The resistance rate to tobramycin among aac(6’)aac(3’)-DET strains was 71.4% (5/7), which was significantly higher than that of aac(6’)aac(3’)-ND strains (p<0.05) (Table 3). This suggests that the presence of aac(6’)aac(3’) were highly correlated with tobramycin resistance.
16S RMTases represent one of the most concerning mechanisms of resistance to aminoglycosides. Among the G1405 16S RMTases, ArmA and AmtB are the most widely distributed, having been found in various species of Enterobacteriaceae, as well as in P. aeruginosa and Acinetobacter baumannii (O’Hara et al., 2013). In this study, armA resistance genes were detected in 5 cases, and the tobramycin resistance rate was 100% in the armA-DET group, significantly higher than that in the armA-ND group (p<0.05). armA was present only in MDR and CRPA strains, and the degree of drug resistance was severe. In addition to colistin, only penicillins with b-lactamase inhibitors, ceftazidime, ceftazidime-avibactam, cefoperazone-sulbactam and/or aztreonam remained sensitive. Strains detected both the aac(6’)aac(3’) and armA resistance genes exhibited higher resistance to aminoglycosides; however, 68.7% of tobramycin-resistant strains did not detect either the aac(6’)aac(3’) or armA genes. cmlA was associated with chloramphenicol resistance, and P. aeruginosa is naturally resistant to chloramphenicol (Osei Sekyere and Reta, 2020), but only 12.4% percent of P. aeruginosa had the gene detected. Additionally, resistance genes for β-lactamase, chloramphenicol, and tetracyclines were detected in a subset of samples. These findings raise concerns about the reliability of the use of genomic diagnostics as the sole tool for resistance prediction, as dependence on genomics alone may lead to erroneous diagnoses and inappropriate therapeutic decisions (Sommer et al., 2020).
tNGS and drug susceptibility tests were repeated in six patients within six months. One patient who was diagnosed with severe pneumonia caused by P. aeruginosa was admitted to the ICU and the infection could not be controlled. After six days, a second BALF was collected for tNGS analysis. The results revealed that P. aeruginosa had detected the exoY, armA and cmlA genes. The abrupt detection of exoY, armA and cmlA genes in P. aeruginosa may reflect a polymicrobial infection with population dynamics. Prolonged antimicrobial selection drives resistant strain dominance through competitive advantage, achieving tNGS-detectable thresholds. As a result, the drug susceptibility profile shifted from full sensitivity to multiple drug resistance, with susceptibility remaining only to cefoperazone-sulbactam and colistin. Unfortunately, treatment ultimately failed. This case highlights the utility of tNGS not only in identifying the primary pathogens of infection but also in predicting changes in drug resistance through the detection of resistance and virulence genes. Such early identification of resistance mechanisms can guide timely adjustments in antimicrobial therapy, even before traditional drug susceptibility results are available. But the mechanisms of resistance to carbapenems or other drugs among P. aeruginosa strains are multifactorial, such as the fact that repression or inactivation of the carbapenem porin OprD and the hyperexpression of the chromosomal cephalosporinase AmpC are associated with reduced susceptibility to carbapenems (Hu et al., 2021). This suggests that, in addition to genetic factors, other mechanisms may contribute to resistance.
In another case, a patient infected with P. aeruginosa did not detected virulence or drug resistance genes, and the patient’s symptoms failed to improve after five days of treatment. On the sixth day, a follow-up test revealed that the pathogen was still P. aeruginosa, and it still did not detect any virulence or resistance genes. However, the strain had developed MDR, with only carbapenems, tobramycin, and ceftazidime-avibactam remaining effective. A genomic study of 40 nosocomial P. aeruginosa strains demonstrated the organism’s dynamic and highly plastic genome, which enables clinical strains to acquire resistance. These adaptive mechanisms allow P. aeruginosa to respond to selective pressures quickly, such as those from prolonged antibiotic treatment (Liu et al., 2022; Ventero et al., 2023). Therefore, continuous monitoring of P. aeruginosa drug susceptibility is essential, even in the absence of gene acquisition.
In addition, drug sensitivity was continuously monitored in 41 cases, and significant changes were observed in 5. Among these, 2 cases transitioned from full sensitivity to MDR combined with carbapenem resistance, 2 developed into CRPA, and 1 became MDR. However, 10 patients with P. aeruginosa showed no change in their drug sensitivity profiles, and no significant differences in drug use were noted. This suggests that in addition to antibiotic use, the establishment of an infection probably depends much more on adaptation to a range of other selective factors within the human host such as the immune system, altered nutrient availability, fluctuating oxygen concentrations, and the composition of the indigenous microbiota (La Rosa et al., 2018; Pickard et al., 2017; Sommer et al., 2020).
Between the SPA and MDR groups, as well as between the CSPA and CRPA groups, significant differences were observed in several factors, including the use of antibiotics before hospitalization (within 90 days), hospital-acquired infections, ICU admission, as well as pre-P. aeruginosa detection interventions including invasive procedures, catheterization, mechanical ventilation, duration of antibiotic use time≥14 days, and the number of antibiotic use type ≥3 types (p<0.05). After the detection of P. aeruginosa, the duration of antibiotic use time≥14 days, and the number of antibiotic use type ≥3 types, and the length of hospital stay were also significantly different (p<0.05). Furthermore, the detection of MDR and CRPA strains appears to lead to an increase in the duration and variety of antibiotic use, as well as an extended length of hospital stay.
Finally, carbapenem resistance in P. aeruginosa can complicate treatment regimens, given how often P. aeruginosa is resistant to other antimicrobials. In our study, 41.9% of P. aeruginosa isolates were found to be carbapenem-resistant. Notably, over 81.8% of the patients with carbapenem-resistant isolates had prior healthcare exposures, highlighting the significance of nosocomial infections in P. aeruginosa infections. However, these results may be subject to bias, as patients who underwent tNGS testing were primarily from the ICU and respiratory departments, where clinical situations are typically more severe. Additionally, after prolonged treatment, P. aeruginosa is more likely to develop acquired resistance, which may further contribute to the severity of an infection.
tNGS workflows enrich specific genetic targets for sequencing and therefore have the advantage of enriching genetic targets for specific pathogens or pathogen groups. Nevertheless, our analysis revealed that certain virulence and antimicrobial resistance genes identified through WGS were undetectable within the tNGS analytical pipeline. Actually, multiplex PCR scaling to large panels for broad range of most common pathogens causes the nonlinear increase of primer dimer species that reduces tNGS mapping rates (Liu et al., 2024).
A limitation of this study stems from the lack of scientifically substantiated rationale underlying the manufacturer-curated virulence gene panel selection, with future studies benefiting from the inclusion of clinically relevant virulence determinants through expanded genomic profiling.
Conclusions
tNGS is a valuable tool for detecting major pathogens and providing information on virulence and resistance genes. But present detection systems cover only a subset of known virulence/resistance genes, systematic incorporation of newly characterized, medically relevant genes into pipelines warrants urgent development. Although drug resistance genes correlate with resistance phenotypes, they cannot fully replace traditional drug susceptibility tests. A comprehensive approach that integrates both genomic and phenotypic data is thus essential for accurate clinical decision-making.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of the Second Affiliated Hospital of Guangxi Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The study was exempted from requiring informed consent by the Ethical Review Committee as it was a retrospective study and patient data were anonymized.
Author contributions
HW: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. YF: Formal analysis, Investigation, Software, Writing – original draft, Writing – review & editing. WQ: Supervision, Writing – original draft. ZL: Methodology, Writing – original draft. LZ: Validation, Writing – review & editing. JG: Project administration, Writing – review & editing. SH: Funding acquisition, Writing – review & editing. XW: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1563741/full#supplementary-material
References
Alhede, M., Bjarnsholt, T., Jensen, P., Phipps, R. K., Moser, C., Christophersen, L., et al. (2009). Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiol. (Reading) 155, 3500–3508. doi: 10.1099/mic.0.031443-0
Allured, V. S., Collier, R. J., Carroll, S. F., and McKay, D. B. (1986). Structure of exotoxin A of Pseudomonas aeruginosa at 3.0-Angstrom resolution. Proc. Natl. Acad. Sci. U S A 83, 1320–1324. doi: 10.1073/pnas.83.5.1320
Azimi, S., Kafil, H. S., Baghi, H. B., Shokrian, S., Najaf, K., Asgharzadeh, M., et al. (2016). Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect. Control 11, Doc04. doi: 10.3205/dgkh000264
Borlee, B. R., Goldman, A. D., Murakami, K., Samudrala, R., Wozniak, D. J., and Parsek, M. R. (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol 75, 827–842. doi: 10.1111/j.1365-2958.2009.06991.x
Botelho, J., Grosso, F., and Peixe, L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist. Update 44, 100640. doi: 10.1016/j.drup.2019.07.002
Casilag, F., Lorenz, A., Krueger, J., Klawonn, F., Weiss, S., and Häussler, S. (2016). The lasB elastase of pseudomonas aeruginosa acts in concert with alkaline protease aprA to prevent flagellin-mediated immune recognition. Infect. Immun. 84, 162–171. doi: 10.1128/iai.00939-15
CLSI (2023). Performance standards for antimicrobial susceptibility testing,M100. 33st ed (Wayne, PA: Clinical and Laboratory Standards Institute).
CLSI (2024). Performance standards for antimicrobial susceptibility testing,M100. 34st ed (Wayne, PA: Clinical and Laboratory Standards Institute).
Del Barrio-Tofiño, E., López-Causapé, C., and Oliver, A. (2020). Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob Agents 56, 106196. doi: 10.1016/j.ijantimicag.2020.106196
de Sousa, T., Hébraud, M., Dapkevicius, M., Maltez, L., Pereira, J. E., Capita, R., et al. (2021). Genomic and metabolic characteristics of the pathogenicity in pseudomonas aeruginosa. Int. J. Mol. Sci. 22, 12892. doi: 10.3390/ijms222312892
Dolgusevs, M., Jain, N., Savicka, O., Vangravs, R., Bodrenko, J., Bergmanis, E., et al. (2024). Genomic and phenotypic inconsistencies in Pseudomonas aeruginosa resistome among intensive care patients. Front. Cell Infect. Microbiol 14, 1335096. doi: 10.3389/fcimb.2024.1335096
Erhardt, M., Namba, K., and Hughes, K. T. (2010). Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect. Biol. 2, a000299. doi: 10.1101/cshperspect.a000299
Galle, M., Jin, S., Bogaert, P., Haegman, M., Vandenabeele, P., and Beyaert, R. (2012). The Pseudomonas aeruginosa type III secretion system has an exotoxin S/T/Y independent pathogenic role during acute lung infection. PloS One 7, e41547. doi: 10.1371/journal.pone.0041547
Gaston, D. C., Miller, H. B., Fissel, J. A., Jacobs, E., Gough, E., Wu, J., et al. (2022). Evaluation of metagenomic and targeted next-generation sequencing workflows for detection of respiratory pathogens from bronchoalveolar lavage fluid specimens. J. Clin. Microbiol 60, e0052622. doi: 10.1128/jcm.00526-22
Healthcare, S. o. C. M. o. C. I. E. a. P. A. f. M. a (2024). Expert consensus on the application and practice of targeted next-generation sequencing in infectious diseases. Zhonghua Yi Xue Za Zhi 104, 4375–4383. doi: 10.3760/cma.j.cn112137-20240927-02208
Hu, Y., Liu, C., Wang, Q., Zeng, Y., Sun, Q., Shu, L., et al. (2021). Emergence and Expansion of a Carbapenem-Resistant Pseudomonas aeruginosa Clone Are Associated with Plasmid-Borne bla (KPC-2) and Virulence-Related Genes. mSystems 6, e00154–21. doi: 10.1128/mSystems.00154-21
Islam, S. T., Taylor, V. L., Qi, M., and Lam, J. S. (2010). Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 1, e00189–10. doi: 10.1128/mBio.00189-10
Jabalameli, F., Mirsalehian, A., Khoramian, B., Aligholi, M., Khoramrooz, S. S., Asadollahi, P., et al. (2012). Evaluation of biofilm production and characterization of genes encoding type III secretion system among Pseudomonas aeruginosa isolated from burn patients. Burns 38, 1192–1197. doi: 10.1016/j.burns.2012.07.030
Kaluzny, K., Abeyrathne, P. D., and Lam, J. S. (2007). Coexistence of two distinct versions of O-antigen polymerase, Wzy-alpha and Wzy-beta, in Pseudomonas aeruginosa serogroup O2 and their contributions to cell surface diversity. J. Bacteriol 189, 4141–4152. doi: 10.1128/jb.00237-07
Korotkov, K. V., Sandkvist, M., and Hol, W. G. (2012). The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol 10, 336–351. doi: 10.1038/nrmicro2762
La Rosa, R., Johansen, H. K., and Molin, S. (2018). Convergent Metabolic Specialization through Distinct Evolutionary Paths in Pseudomonas aeruginosa. mBio 9, e00269–18. doi: 10.1128/mBio.00269-18
Liu, G., Thomsen, L. E., and Olsen, J. E. (2022). Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: a mini-review. J. Antimicrob Chemother. 77, 556–567. doi: 10.1093/jac/dkab450
Liu, Y., Wu, W., Xiao, Y., Zou, H., Hao, S., and Jiang, Y. (2024). Application of metagenomic next-generation sequencing and targeted metagenomic next-generation sequencing in diagnosing pulmonary infections in immunocompetent and immunocompromised patients. Front. Cell Infect. Microbiol 14, 1439472. doi: 10.3389/fcimb.2024.1439472
Ma, H., Wang, H., Han, X., and Fei, J. (2024). Efficacy of targeted next generation sequencing for pathogen detection in lower respiratory tract infections. Am. J. Transl. Res. 16, 3637–3645. doi: 10.62347/fkwf4589
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mancl, J. M., Suarez, C., Liang, W. G., Kovar, D. R., and Tang, W. J. (2020). Pseudomonas aeruginosa exoenzyme Y directly bundles actin filaments. J. Biol. Chem. 295, 3506–3517. doi: 10.1074/jbc.RA119.012320
Minandri, F., Imperi, F., Frangipani, E., Bonchi, C., Visaggio, D., Facchini, M., et al. (2016). Role of iron uptake systems in pseudomonas aeruginosa virulence and airway infection. Infect. Immun. 84, 2324–2335. doi: 10.1128/iai.00098-16
O’Hara, J. A., McGann, P., Snesrud, E. C., Clifford, R. J., Waterman, P. E., Lesho, E. P., et al. (2013). Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother. 57, 2413–2416. doi: 10.1128/aac.00266-13
Ochsner, U. A., Johnson, Z., and Vasil, M. L. (2000). Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiol. (Reading) 146, 185–198. doi: 10.1099/00221287-146-1-185
Oladosu, V. I., Park, S., and Sauer, K. (2024). Flip the switch: the role of FleQ in modulating the transition between the free-living and sessile mode of growth in Pseudomonas aeruginosa. J. Bacteriol 206, e0036523. doi: 10.1128/jb.00365-23
Osei Sekyere, J. and Reta, M. A. (2020). Genomic and resistance epidemiology of gram-negative bacteria in africa: a systematic review and phylogenomic analyses from a one health perspective. mSystems 5, e00897–20. doi: 10.1128/mSystems.00897-20
Ostroff, R. M., Vasil, A. I., and Vasil, M. L. (1990). Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J. Bacteriol 172, 5915–5923. doi: 10.1128/jb.172.10.5915-5923.1990
Pearson, J. P., Pesci, E. C., and Iglewski, B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol 179, 5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997
Pickard, J. M., Zeng, M. Y., Caruso, R., and Núñez, G. (2017). Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567
Reynolds, D. and Kollef, M. (2021). The epidemiology and pathogenesis and treatment of pseudomonas aeruginosa infections: an update. Drugs 81, 2117–2131. doi: 10.1007/s40265-021-01635-6
Romano, K. P. and Hung, D. T. (2023). Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi-drug resistance. Biochim. Biophys. Acta Mol. Cell Res. 1870, 119407. doi: 10.1016/j.bbamcr.2022.119407
Salacha, R., Kovacić, F., Brochier-Armanet, C., Wilhelm, S., Tommassen, J., Filloux, A., et al. (2010). The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel Type V secretion system. Environ. Microbiol 12, 1498–1512. doi: 10.1111/j.1462-2920.2010.02174.x
Schalk, I. J. and Guillon, L. (2013). Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44, 1267–1277. doi: 10.1007/s00726-013-1468-2
Schofield, M. C., Rodriguez, D. Q., Kidman, A. A., Cassin, E. K., Michaels, L. A., Campbell, E. A., et al. (2021). The anti-sigma factor MucA is required for viability in Pseudomonas aeruginosa. Mol. Microbiol 116, 550–563. doi: 10.1111/mmi.14732
Schwocho, L. R., Schaffner, C. P., Miller, G. H., Hare, R. S., and Shaw, K. J. (1995). Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 39, 1790–1796. doi: 10.1128/aac.39.8.1790
Silistre, H., Raoux-Barbot, D., Mancinelli, F., Sangouard, F., Dupin, A., Belyy, A., et al. (2021). Prevalence of exoY activity in pseudomonas aeruginosa reference panel strains and impact on cytotoxicity in epithelial cells. Front. Microbiol 12, 666097. doi: 10.3389/fmicb.2021.666097
Sommer, L. M., Johansen, H. K., and Molin, S. (2020). Antibiotic resistance in Pseudomonas aeruginosa and adaptation to complex dynamic environments. Microb Genom 6, e000370. doi: 10.1099/mgen.0.000370
Teo, J. Q., Lim, J. C., Tang, C. Y., Lee, S. J., Tan, S. H., Sim, J. H., et al. (2021). Ceftolozane/tazobactam resistance and mechanisms in carbapenem-nonsusceptible pseudomonas aeruginosa. mSphere 6e01026–20. doi: 10.1128/mSphere.01026-20
Ullah, R., Amir, M., Anjum, S., Ur Rehman, M., Noorul Hasan, T., Sajjad Naqvi, S., et al. (2023). Presence of T3SS (exoS, exoT, exoU and exoY), susceptibility pattern and MIC of MDR-Pseudomonas aeruginosa from burn wounds. J. Infect. Dev. Ctries 17, 1130–1137. doi: 10.3855/jidc.17580
Ventero, M. P., Haro-Moreno, J. M., Molina-Pardines, C., Sánchez-Bautista, A., García-Rivera, C., Boix, V., et al. (2023). Role of relebactam in the antibiotic resistance acquisition in pseudomonas aeruginosa: in vitro study. Antibiot (Basel) 12, 1619. doi: 10.3390/antibiotics12111619
Wilderman, P. J., Vasil, A. I., Johnson, Z., Wilson, M. J., Cunliffe, H. E., Lamont, I. L., et al. (2001). Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69, 5385–5394. doi: 10.1128/iai.69.9.5385-5394.2001
Yaping Xu, R. H., Yan, Q., Suo, j., Dong, Z., Xing, Y., Lin, J., et al. (2017). Multidrug-resistant,extensively drug-resistant and pandrug-resistantbacteria:an international expert proposal for interim standard definitionsfor acquired resistance. Chin JNosocomiol 27, 231–240. doi: 10.11816/cn.ni.2016-162935
Keywords: Pseudomonas aeruginosa, resistance genes, virulence genes, targeted next-generation sequencing, multidrug-resistant, carbapenem-resistant Pseudomonas aeruginosa
Citation: Wei H, Fu Y, Qin W, Liang Z, Zhou L, Gao J, He S and Wu X (2025) The relationship between virulence and drug resistance genes in Pseudomonas aeruginosa and antibiotic resistance: a targeted next-generation sequencing approach. Front. Cell. Infect. Microbiol. 15:1563741. doi: 10.3389/fcimb.2025.1563741
Received: 20 January 2025; Accepted: 30 April 2025;
Published: 26 May 2025.
Edited by:
Hong Li, Hainan University, ChinaReviewed by:
Stavros Panagiotou, The University of Manchester, United KingdomDonghoon Alex Kang, Houston Methodist Research Institute, United States
Copyright © 2025 Wei, Fu, Qin, Liang, Zhou, Gao, He and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoning Wu, d3huNTM1QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Huanhuan Wei
Huanhuan Wei Yuanyong Fu2†
Yuanyong Fu2†