- 1Department of Laboratory Medicine, Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, China
- 2Department of Obstetrics and Gynecology, Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, China
- 3Department of Bioinformatics, Guangzhou Forevergen Medical Laboratory Co., Ltd, Guangzhou, Guangdong, China
Background: Detecting microbes in amniotic fluids via amniocentesis represents the standard method for diagnosing intrauterine infections. Given its similarity to metagenomic next-generation sequencing, copy number variation sequencing (CNV-seq) data may also contain microbial sequences. This exploratory study aimed to investigate the feasibility of prenatal CNV-seq for detecting Ureaplasma parvum (U. parvum) in amniotic fluids and to evaluate the pregnancy outcomes in U. parvum-positive cases.
Methods: This retrospective study enrolled 2419 singleton pregnant women who underwent genetic amniocentesis for fetal CNV-seq testing and completed the follow-up with documented pregnancy outcomes. The CNV-seq data were reanalyzed to extract the read counts of U. parvum from each sample’s raw data, and reads per million (RPM) was used to quantify its relative abundance.
Results: The prevalence of asymptomatic intrauterine U. parvum positivity in this cohort was 1.4% (33/2419), with read counts ranging 1 to 30423 and RPM from 0.09 to 3580.65 by reanalysis of CNV-seq data. There was a statistically significantly higher risk for early spontaneous preterm labor (<32 gestational weeks; P<0.001) and preterm premature rupture of the membranes (P<0.001) in women with positive U. parvum compared to negative cases. Among U. parvum positive cases, six cases (6/33, 18.2%) had relatively higher read counts ranging from 2483 to 30423, with corresponding RPM of 406.45 to 3580.65. Adverse pregnancy outcomes were exclusively observed among women with high reads of U. parvum as opposed to those with low reads. Four cases with high U. parvum reads in amniotic fluids, not treated with antibiotics, showed a latency period of 6 to 10 weeks from positive detection to the onset of clinical manifestations.
Conclusions: CNV-seq may be a feasible method for detecting intraamniotic U. parvum infection. High abundance of asymptomatic U. parvum in amniotic fluids are statistically associated with adverse pregnancy outcomes, highlighting its importance in preliminary screening.
Introduction
Intrauterine infection is associated with a variety of adverse consequences, including spontaneous abortion, preterm premature rupture of the membranes (pPROM), early spontaneous preterm labor (SPB), stillbirth, postpartum hemorrhage, and high rates of neonatal morbidity and mortality (Goldenberg et al., 2000, 2008; Kim et al., 2015; Tita and Andrews, 2010). Microbiological studies have found that vaginal organisms ascending from the cervix to the uterus is the primary route of intrauterine infection, and Ureaplasma species (U.spp) are the most frequent organisms detected from the amniotic cavity and vagina of women with preterm birth and pPROM (Goldenberg et al., 2008, 2000; Kim et al., 2015; Rittenschober-Böhm et al., 2021, 2019, 2018; Tita and Andrews, 2010). However, the extensive vaginal colonization of U.spp among healthy pregnant women without overt clinical manifestations of infection or adverse outcomes has cast doubts on the virulence of U.spp, which was even considered as commensal bacteria (Cassell et al., 1993). Consequently, numerous studies based on both animal and human data have been conducted aimed to clarify the pathogenicity of U.spp, but yielding mixed and disparate results (Senthamaraikannan et al., 2016; Tantengco et al., 2021; Noda-Nicolau et al., 2022; Bento et al., 2023).
A recent systematic review elucidated several factors contributing to the inconsistent association of U.spp with poor pregnancy prognosis, such as microbial biovars, colonization sites, microbial loads, and the methods used for microbial identification (Noda-Nicolau et al., 2022). Notably, two predominate species of U.spp in humans are Ureaplasma parvum (U. parvum) and Ureaplasma urealyticum. Many studies have demonstrated that U. parvum has more significant correlation with adverse perinatal outcomes than Ureaplasma urealyticum (Kataoka et al., 2006; Uchida et al., 2013; Prince et al., 2016; Payne et al., 2016; Pavlidis et al., 2020). However, some studies found that ascending infection of U. parvum alone could not produce massive inflammation leading to preterm labor (Tantengco et al., 2021; Bento et al., 2023; Tripathy et al., 2023). Conversely, direct intra-amniotic inoculation of U. parvum in animal models has demonstrated that intrauterine invasion was a major clinical concern than genital colonization (Motomura et al., 2020). Traditional methods like culture and polymerase chain reaction (PCR) can identify U.spp colonization, but their sensitivity relies on the microbial loads and they are unable to distinguish between biovars and genotypes. With the application of new molecular microbiological techniques in the field of obstetric infections, such as real-time quantitative PCR (qPCR), 16S ribosomal RNA (rRNA) sequencing, whole genome sequencing (WGS), and metagenomic next-generation sequencing (mNGS), which has become an alternative approach for simultaneous screening of specific and various microbial pathogens with a strain- or species- or genotype-level resolution (Cox et al., 2016; Prince et al., 2016; Payne et al., 2016; Otgonjargal et al., 2018; Rittenschober-Böhm et al., 2019; Elovitz et al., 2019; Chiu and Miller, 2019). However, despite its potential, the high cost and susceptibility to contamination have limited 16S rRNA and mNGS extensive clinical utility for routine screening. Therefore, it is still worth to explore detection methods for high sensitivity, mature technology, and the relative abundance of microbial loads capacity.
As an important diagnostic tool for microdeletion and microduplication syndromes, copy number variation sequencing (CNV‐seq), a next-generation sequencing (NGS) method, has been widely used in prenatal testing for congenital genetic disorders (Liang et al., 2014; Dong et al., 2016; Zhu et al., 2016; Wang et al., 2018b). Given the similar experimental procedure between CNV-seq and mNGS, CNV-seq data may also contain the sequences of microorganisms. Therefore, in this study, we reanalyzed prenatal CNV-seq data in an attempt to detect U. parvum, a prevalent pathogenic organism associated with adverse pregnancy outcomes. The primary objective is to evaluate the feasibility and efficacy of detecting U. parvum using CNV-seq data. The secondary aim is to explore the nature course and pregnancy outcomes of asymptomatic pregnant women who tested positive for intraamniotic U. parvum at the time of genetic amniocentesis.
Materials and methods
Study design and populations
This retrospective study enrolled singleton pregnant women who underwent genetic amniocentesis at the Third Affiliated Hospital of Sun Yat-sen University from January 2021 to December 2022. The inclusion criteria for participants were that their collected amniotic fluids were sent for fetal chromosomal karyotyping and CNV-seq, with or without whole exome sequencing under the consent of the pregnant women. The exclusion criteria were as follows: (1) maternal age ≥ 45 years; (2) multiple pregnancies; (3) symptoms of spontaneous miscarriage or preterm labor before amniocentesis, such as vaginal bleeding, abdominal pain, or a shortened cervix; (4) fever or other symptoms of systemic inflammatory response before amniocentesis; (5) infectious diseases, including infections with hepatitis viruses, human immunodeficiency virus, or syphilis; and (6) long-term use of glucocorticoids or immunomodulators. Ethical approval for the study was obtained from the Ethics Committee of our hospital (approval number: II2025-130-01). Prior to amniocentesis, all participants provided written informed consents for the further reanalysis of their experimental and clinical data.
Clinical data collection
At our hospital, all pregnant women who underwent amniocentesis were prospectively followed up at least three times, including two weeks post-procedure, four weeks post-procedure, and three to six months postpartum. These follow-ups were conducted through a combination of inpatient and outpatient visits, as well as telephone interviews. Follow-ups mainly focused on collecting data on post-procedure complications, pregnancy outcomes, birth defects, and infant growth, all of which were meticulously documented within prenatal diagnosis records. Clinical data of the enrolled pregnant women were obtained from electronic medical records and prenatal diagnosis files. Maternal prenatal information included age, parity, previous obstetrics history, gestational age at the time of amniocentesis, indication for genetic diagnosis, the color of the amniotic fluid, and the results of genetic tests. Pregnancy outcomes included gestational age at delivery, any obstetric complications, and neonatal outcomes.
CNV sequencing
The experimental steps were as follows: The amniotic fluids were centrifuged at 1500 g for 10 minutes and the resulting cell pellet was used to extract genomic DNA (gDNA) using the QIAamp DNA Mini Kit (QIAGEN, Germany). The gDNA was then fragmented into 200 bp fragments using restriction endonucleases. Libraries were prepared using the CNV-seq Library Preparation Kit (BerryGenomics, China). Library sequencing was performed on the NextSeq CN500 platform (Illumina, USA) using SE36 sequencing mode.
CNV-seq data reanalysis for U. parvum
The CNV-seq data underwent initial processing involving adapter trimming, and reads containing more than three low-quality bases (Q <20) or more than one ambiguous base (N) were removed using an in-house workflow. Subsequently, the remaining clean reads were reanalyzed using the Kraken2 software (version 2.1.2) with Kraken2/Bracken RefSeq PlusPF database (k2_pluspf_20210517) as a reference, established by BenLangmead (https://benlangmead.github.io/aws-indexes/k2). Default parameters were employed for Kraken2 (–minimum-hit-groups 2, –confidence 0.0, -l 31, -k 35). Species of U. parvum (taxid:134821) and the corresponding read counts were extracted from the primary dataset of each sample. The depth-based assessment (relative abundance) of U. parvum was quantified using normalization method based on the ratio of U. parvum read counts to the total reads per million in the CNV-seq dataset, calculated as follows (Qin et al., 2021).
Statistical analysis
The normality of continuous variables was assessed using Shapiro-Wilk test. Continuous data were presented by mean ± standard deviation (SD) or median (interquartile range), with group differences analyzed through Student’s t test or Wilcoxon rank-sum test. Categorical data were expressed as numbers (percentage) and analyzed using chi-square test or Fisher’s exact test. Logistic regression analysis was performed to evaluated risk factors associated with SPB. The cumulative distribution of SPB between U. parvum-positive and U. parvum-negative cases was analyzed using Cox regression. Statistical analysis was performed using SPSS version 27.0 (IBM, USA). A P value <0.05 (two-tailed) was considered statistically significant.
Results
Study population
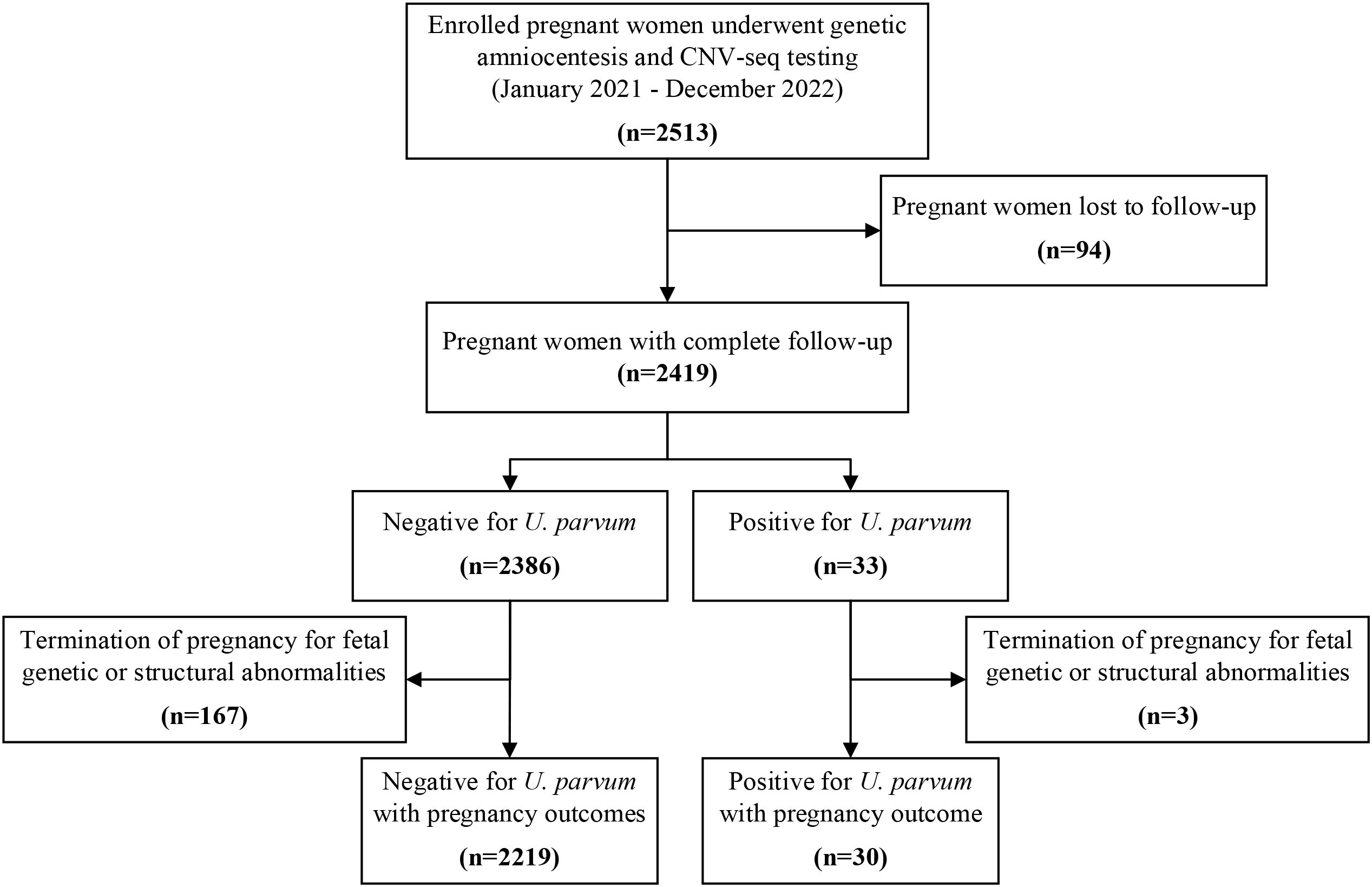
During the study period, amniotic fluid samples from 2513 singleton pregnancies were analyzed for chromosomal karyotyping and CNV-seq analysis, revealing 82 cases with genetic abnormalities. All cases received follow-up evaluations at two weeks and four weeks post-procedure. While 94 (3.7%) were lost to follow-up after delivery, the remaining 2419 women completed the follow-up and their pregnancy outcomes were documented (Figure 1).
Cases positive for U. parvum in amniotic fluids
Upon reanalysis of CNV-seq data from 2419 women, the total sequencing reads per sample ranged from 3.75 million to 20.95 million reads, with an average of 7.08 million reads. Among these, 33 samples (1.4%) showed positive for U. parvum, with read counts ranging from 1 to 30423 and RPM from 0.09 to 3580.65. Within this subset, six cases (6/33, 18.2%) displayed a relative higher number of read counts, ranging from 2483 to 30,423, with corresponding RPM of 406.45 to 3580.65. The remaining 27 women showed low reads of U. parvum, ranging from 1 to 167, with RPM between 0.09 to 19.74. Meanwhile, among the 33 women with positive U. parvum, three underwent mNGS testing, which confirmed that two cases with high reads of U. parvum from CNV-seq data were true positive, whereas one case with a single read of U. parvum was a false positive.
Pregnancy outcome of cases testing positive and negative for U. parvum
Among the 2386 U. parvum-negative cases and 33 U. parvum-positive cases with documented outcomes, 167 and three women, respectively, opted for pregnancy termination due to fetal genetic or structural abnormalities. No miscarriage occurred within two-weeks post-procedure in either group. Of the remaining 2219 women tested negative for U. parvum, the mean gestational age at delivery was significantly earlier than that of the 30 U. parvum-positive cases (P<0.001). SPB and pPROM occurred in 107 (107/2219, 4.8%) and 21 (21/2219, 0.9%) women with U. parvum-negative, respectively. Conversely, among women with positive U. parvum, five (5/30, 16.7%) experienced SPB and four (4/30, 13.3%) suffered pPROM, with rates significantly higher than those in U. parvum-negative cohort (P<0.001). Of note, in the analysis of gestational age for preterm labor, women testing positive for U. parvum were significantly more likely to experience early SPB (<32 weeks gestation) compared to negative cases (P<0.001) (Table 1).
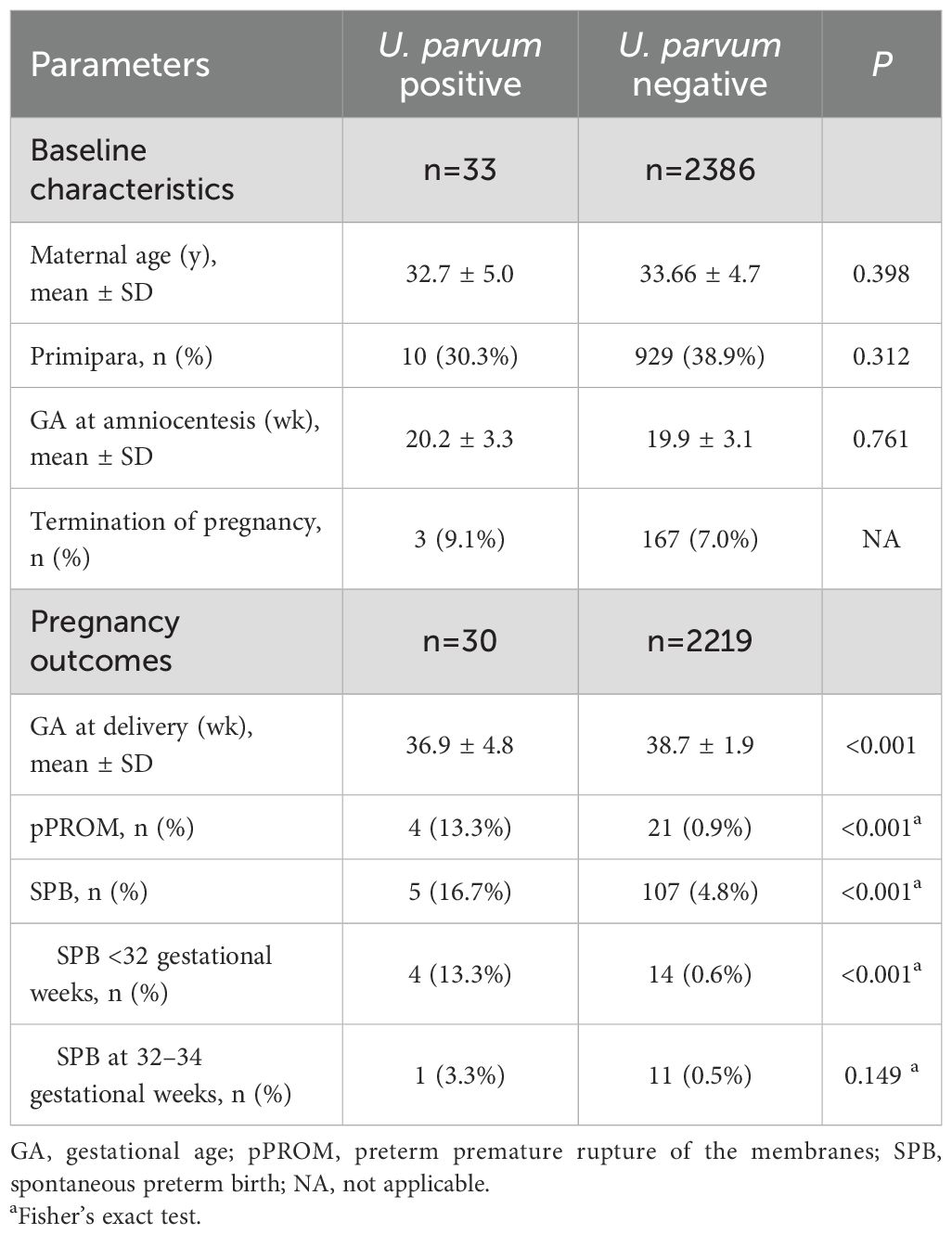
Table 1. Clinical characteristics and outcomes of recruited pregnant women with U. parvum positive and U. parvum negative.
Multivariate logistic analysis revealed that U. parvum positive [adjusted odds ratio (aOR): 3.90, 95% confidence interval (CI): 1.47-10.30, P=0.06] was associated with SPB, while maternal age and gestational weeks at amniocentesis showed no significant correlation (P>0.05). The subsequence of intraamniotic U. parvum infection was further illustrated through the estimated cumulative frequencies of gestational age at delivery among pregnant women who suffered SPB. As depicted in Figure 2, the curve for U. parvum positive was significantly shifted towards earlier gestational weeks and exhibited higher frequencies compared with U. parvum negative. After adjusting for maternal age and gestational weeks at amniocentesis, the presence of U. parvum was identified as an independent risk factor for SPB (adjusted hazard ratio (aHR): 4.03, 95% CI:1.64-9.88, P=0.002).
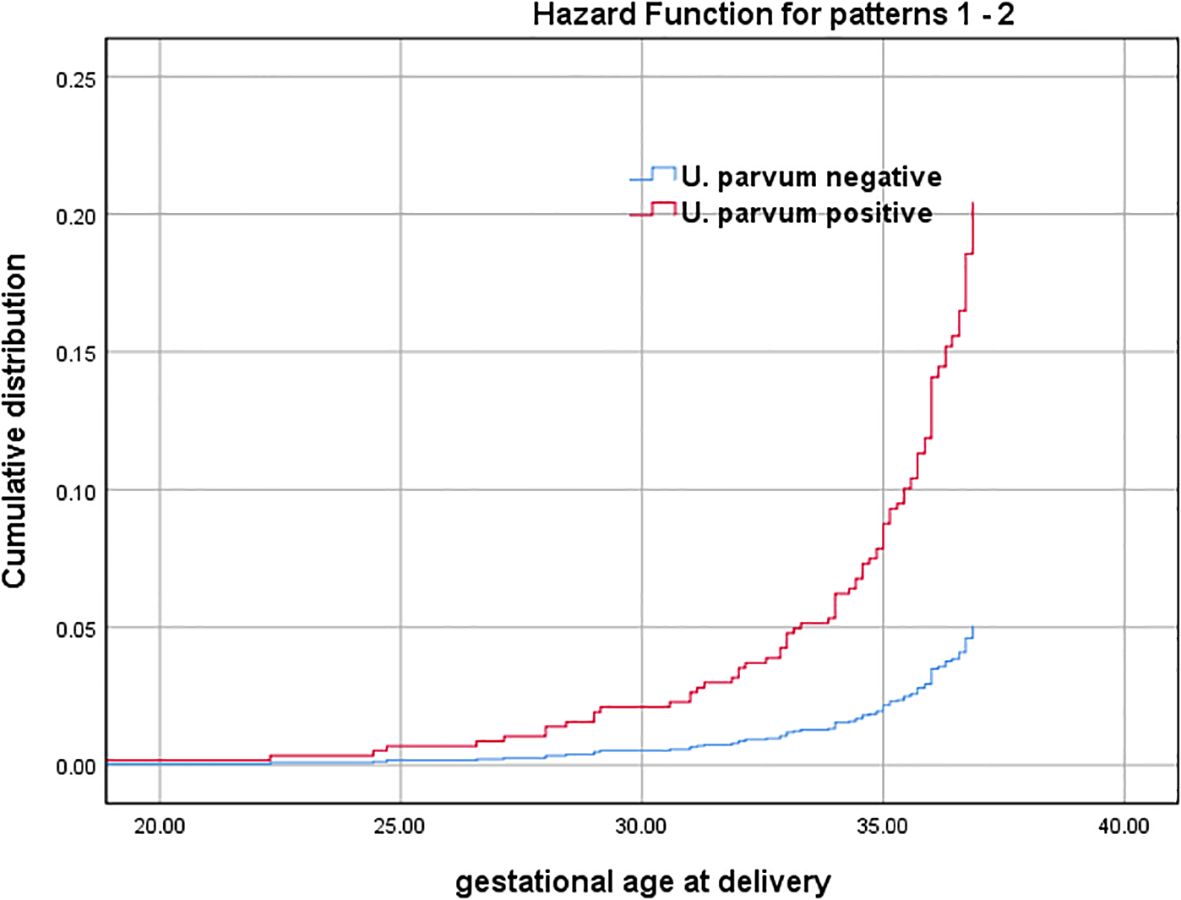
Figure 2. Gestational age at delivery among SPB cases with U. parvum positive and U. parvum negative (Cumulative distribution curves represent estimated cumulative frequencies of gestational age at birth among pregnant women who suffered SPB with U. parvum positive and U. parvum negative). Adjusted for maternal age and gestational weeks at amniocentesis.
When considering the read counts of U. parvum from CNV-seq, five out of six (5/6, 83.3%) pregnancies with relatively higher reads resulted in SPB between 24 and 32 gestational weeks, leading to two (2/6, 33.3%) neonatal deaths and one (1/6, 16.7% intrauterine fetal demise at 27 gestational weeks. Histological examination of the placenta was performed in three cases, revealing the presence of chorioamnionitis in all instances. In contrast, among 27 women with low reads of U. parvum, three chose termination of pregnancy for fetal genetic or structural abnormalities, while the remaining 24 achieved term deliveries. Detailed clinical data for these 33 cases were presented in Tables 2 and 3.
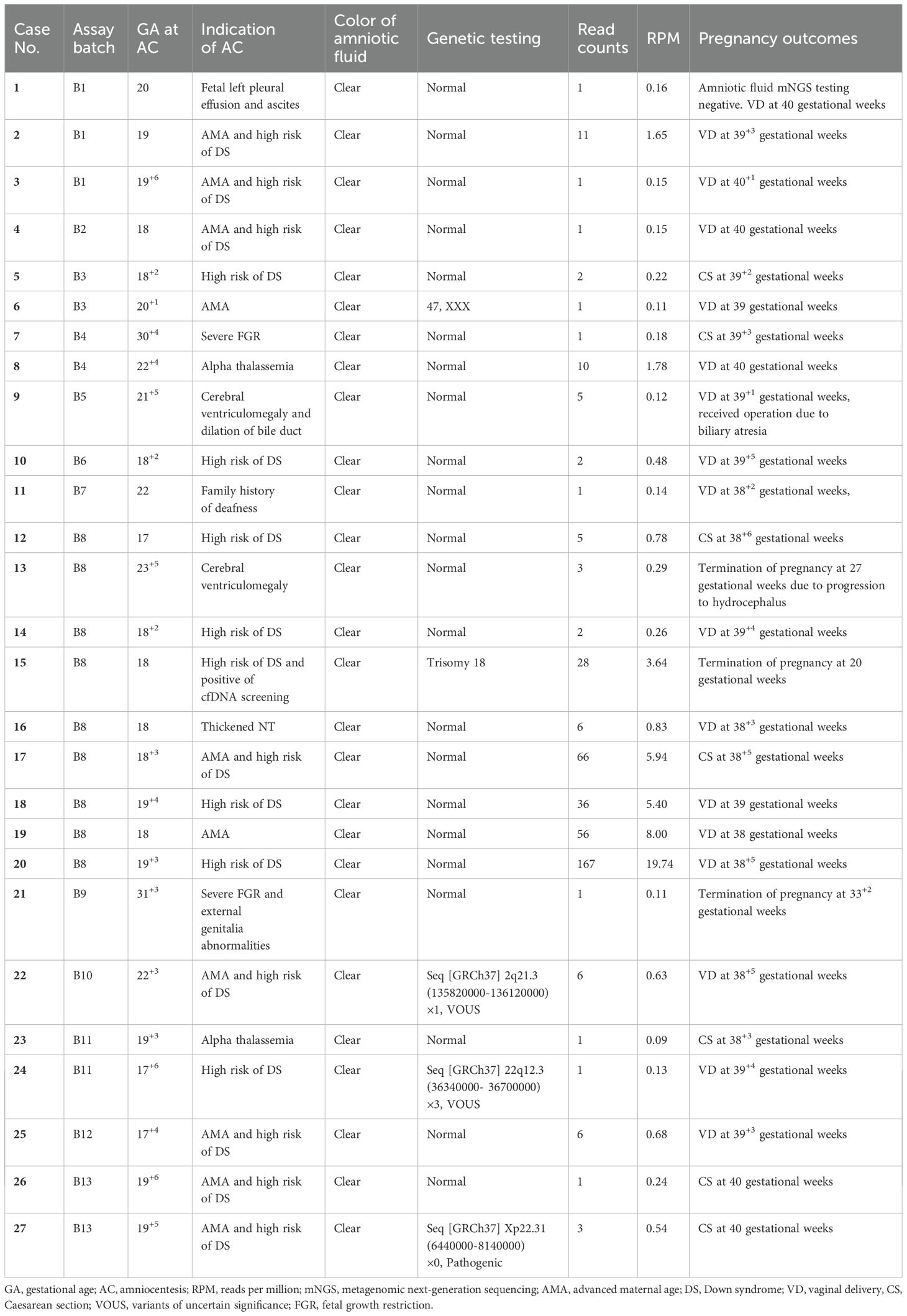
Table 2. Detailed clinical data and pregnancy outcomes of 27 cases with low reads of positive U. parvum.
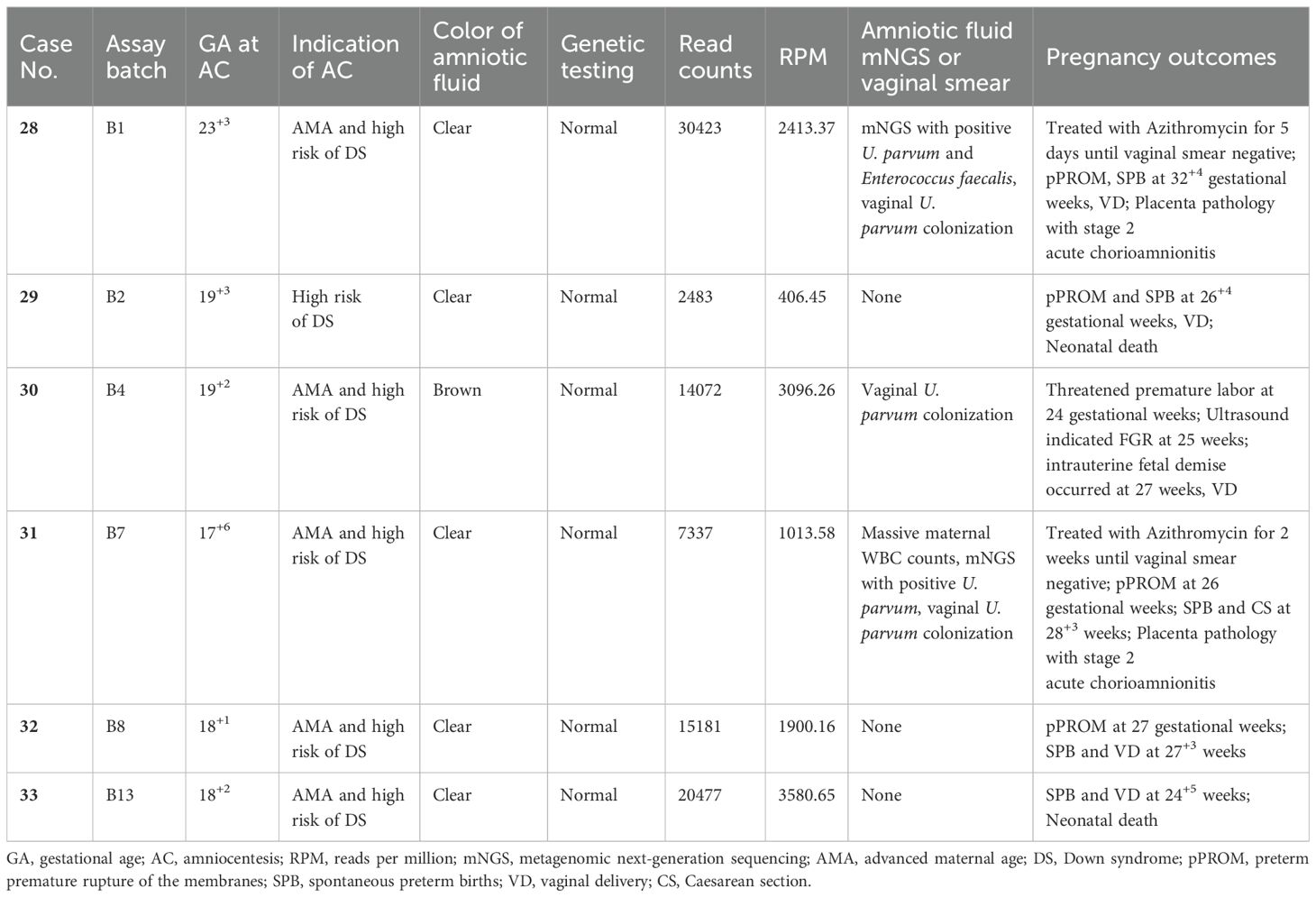
Table 3. Detailed clinical data and pregnancy outcomes of six cases with high reads of positive U. parvum.
Regarding the latency period from detection of high reads of U. Parvum in amniotic fluids to the onset of overt clinical manifestation, our four cases (case 29, 30, 32 and 33) without antibiotics treatment showed a relative long interval ranging from 6 to 10 weeks. In other two cases (case 28 and 31), Azithromycin was administrated until the vaginal swabs turned negative; however, both cases experienced SPB and pPROM approximately 9 weeks after amniocentesis.
Discussion
Our reanalysis of prenatal amniotic CNV-seq data showed that the prevalence of U. parvum positive in amniotic fluids was 1.4%, with only 0.2% (6/2419) exhibiting high read counts of U. parvum. Meanwhile our clinical data indicated a statistically significant association between asymptomatic high reads of U. Parvum with subsequent adverse pregnancy outcomes, such as SPB, pPROM, stillbirth, and histological chorioamnionitis. Conversely, the presence of low reads of U. Parvum in amniotic fluid did not increase the risk of poor pregnancy outcomes. These findings underscore the potential for detecting and quantifying the relative abundance of U. Parvum using prenatal amniotic CNV-seq data without the need for additional tests and associated costs. Additionally, our clinical data showed a relatively long latency period from the detection of subclinical high reads of U. Parvum in amniotic fluids to the onset of SPB or pPROM. This observation signifies a nature course of intrauterine U. Parvum infection and highlights its chronic progression from infection to subsequent clinical symptoms.
It is well-established that intrauterine infection is considered the main cause of preterm birth and pPROM (Goldenberg et al., 2000; Romero et al., 2007; Goldenberg et al., 2008), with the majority of cases beginning as subclinical (Romero et al., 2014). The most common organism isolated from the amniotic cavity is U. spp, with the reported positive rate ranging from 11.4 to 18.4% among asymptomatic women at mid-trimester (Gerber et al., 2003; Perni et al., 2004), and from 22% to 43.9% in cases complicated with preterm birth or pPROM from early studies (Witt et al., 2005; Kirchner et al., 2007; Kasper et al., 2010). Moreover, both direct and indirect causal relationships between U. spp and preterm birth has been confirmed by numerous human pregnancies and animal models (Gerber et al., 2003; Perni et al., 2004; Witt et al., 2005; Kasper et al., 2010; Viscardi, 2010; Uchida et al., 2013; Yoneda et al., 2016; Motomura et al., 2020). Consistent with these studies, our analysis of prenatal amniotic CNV-seq data revealed that asymptomatic positive of U. Parvum, particularly in high reads of case series, significantly linked to a spectrum of adverse pregnancy outcome including SPB, pPROM, stillbirth, and histological chorioamnionitis. However, a multicenter randomized trial in France tried to assess the effectiveness of antibiotics treatment in asymptomatic second-trimester women with Mycoplasma or U. spp positivity, and found a relatively low rate (3.0%) of amniotic fluid colonization which was not associated with any adverse pregnancy or neonatal outcomes (Kayem et al., 2018). Nevertheless, two recent systematic review and meta-analysis investigating U. spp’s pathogenic potential for early deliveries, but yielded unexpectedly inconsistent results (Noda-Nicolau et al., 2022; Jonduo et al., 2022). An important reason for this may be the heterogeneous bacterial detection methods and differences in sample types among the eligible studies.
Prior to 2010, conventional culture methods were predominantly used for detection of U. spp in the early reported studies (Gerber et al., 2003; Perni et al., 2004; Witt et al., 2005; Kirchner et al., 2007). However, a research team from South Korea found that more than one third of amniotic microbial invasion with U. spp was missed by culture method when compared with PCR technique, and these cases with negative culture but positive through PCR were also associated with high risk of adverse outcomes (Yoon et al., 2000, 2003). Afterwards, many studies had revealed that modern molecular biological methodologies not only can discriminate different biovars and genotypes of U.spp, but also can detect bacterial loads compared to traditional culture-based methods (Kim et al., 2003; Kasper et al., 2010; Xiao et al., 2011; Yoneda et al., 2016; Prince et al., 2016; Payne et al., 2016; Otgonjargal et al., 2018; Rittenschober-Böhm et al., 2019; Stranik et al., 2022). Meanwhile, those findings also showed that intrauterine infection with U. parvum was the predominant biovar and significantly associated with adverse perinatal outcomes (Cox et al., 2016; Prince et al., 2016; Otgonjargal et al., 2018; Motomura et al., 2020). Even with a very low positive rate (1.4%) of U. Parvum from amniotic fluid, the present prenatal data found that all six asymptomatic cases with high reads of this microorganism during second trimester ultimately experienced preterm delivery at very early gestational age between 24 to 32 weeks. Therefore, our data provides additional evidence that the abundant invasion of U. Parvum in the amniotic cavity possesses intensive pathogenic potential for clinical adverse consequences associated with intrauterine infection.
Regarding quantification of bacterial loads within the uterine cavity, several microbiological methods have been developed, including qPCR, 16S rRNA sequencing, and mNGS (Jacobsson et al., 2009; Kasper et al., 2010; DiGiulio et al., 2010; Kacerovsky et al., 2012; Aagaard et al., 2014; Sweeney et al., 2016). qPCR is particularly noted for its rapid and sensitive quantification of specific microbial DNA in clinical samples, also provides valuable insights into microbial burden levels. However, the limitation of qPCR is reliance on predetermined target sequences, potentially missing unrecorded pathogens and lacking a detailed microbial profile. The modern techniques of 16S rRNA sequencing and mNGS, on the other hand, offer a comprehensive assessment of the microbial composition and diversity within a specific compartment, also providing the relative abundance of different microorganisms simultaneously by counting sequenced reads in the sample (Kembel et al., 2012; Bertelli and Greub, 2013; Chiu and Miller, 2019). This capability is essential for assessing the severity of intrauterine infections and predicting their related adverse pregnancy outcomes (Jacobsson et al., 2009; Kasper et al., 2010; Kacerovsky et al., 2011, 2012). Nonetheless, 16S rRNA lacks detailed quantitative data at the species level, while mNGS requires significant computational resources and may be cost-prohibitive for routine clinical use (Deurenberg et al., 2017; Johnson et al., 2019). Similar to the process of mNGS, CNV-seq technique may also detect microbial sequences. Our data confirmed this conception and also demonstrated a close link between the high burden of U. Parvum in the amniotic cavity and adverse outcomes. Conversely, cases with low levels of U. Parvum consistently resulted in uneventful term deliveries. These contrasting results were mainly attributed to different microbial loads in the amniotic cavity. It is well understood that the degree of microbial load directly correlates with the intensity of inflammatory response, and higher microbial burdens tend to trigger a more robust immune response, often leading to significant complications (Jacobsson et al., 2009; Kasper et al., 2010).
Another potential explanation for our observations of low concentrations of intrauterine U. Parvum presence could be false-positive results. Given that modern molecular technologies are highly sensitive and capable of detecting very small amounts of nucleic acid, the testing process is relatively prone to contamination and makes the results susceptible to false-positive (Zinter et al., 2019; Cantalupo and Pipas, 2019; Du et al., 2022). We noted that cases 1-3, 4, 7-8, 11, 12-20, and 26–27 were processed for CNV-seq testing in the same assay batch as one of six samples with high microbial loads, raising suspicious of cross-contaminated from the severely infected samples due to index swapping (Costello et al., 2018). For the other nine cases with low read counts, ranging from 1 to 6, we considered the possibility of non-specific read mapping. However, verifying false-positive through bioinformatics analysis to establish thresholds for sequence reads and RPM is both technique-intensive prior to testing and time-consuming afterwards (Chiu and Miller, 2019). Unfortunately, retrospective data is not feasible for this purpose. Another possible explanation could be a mild intrauterine U. parvum infection that was effectively cleared by the host immune system without causing any clinical symptoms and subsequent complications. This suggests that intrauterine presence of U. parvum is not necessarily indicative of adverse pregnancy outcomes (Rodríguez et al., 2011; Kayem et al., 2018). Therefore, interpreting these low read counts of intrauterine microbial presence demands considerable cautions.
Given the chronicity of intrauterine infections (Goldenberg et al., 2000, 2008), early detection of microbial presence in the amniotic cavity is theoretically crucial for timely and effective antibiotics treatment. Our four clinical cases (case 29, 30, 32 and 33) without antibiotics treatment exhibited latency periods ranging from 6 to 10 weeks from the detection of subclinical high reads of U. Parvum in amniotic fluids to overt clinical manifestation. This finding further highlights the chronic nature of intrauterine infection, from the detection of U. Parvum positive in mid-trimester to the emergence of overt clinical symptoms, thereby providing sufficient time for clinical antibiotic intervention. However, despite the timely administration of antibiotics until the vaginal swabs turned negative, SPB and pPROM still occurred in other two cases (case 28 and 31). These findings suggested that antibiotics treatment may not completely eradicate severe amniotic infections, particularly with drugs like Azithromycin, which have limited ability to cross the placental barrier (Heikkinen et al., 2000; Witt et al., 2003).
As an untargeted microbial pathogen screening tool, mNGS boasts significant advantages over traditional microbial culture and has been increasingly applied in clinic for patients with severe or rare infections (Gu et al., 2019; Sun et al., 2022). It predominantly uses host-cell-free samples such as plasma, cerebrospinal fluid, or bronchoalveolar lavage fluid, and often involves the removal of host DNA to enrich microbial genomes, thereby enhancing its sensitivity (Blauwkamp et al., 2019; Miller et al., 2019; Huang et al., 2020). For optimal sensitivity, mNGS data should consist of no fewer than 10 million sequencing reads, with some studies even up to 36 million reads (Li et al., 2021). In contrast, CNV-seq, designed for detecting CNVs in the human genome, does not intentionally enrich microbial genomes and generally requires only a minimum of 3 million sequencing reads (Zhu et al., 2016; Wang et al., 2018b; Hayes et al., 2013; Wang et al., 2018a; Meng and Jiang, 2022). As such, despite a similar testing process to mNGS, the suitability of CNV-seq data for analyzing microbial pathogens remains uncertain. Notably, in this study cohort, we identified six cases from 2419 prenatal samples with high read counts of intrauterine U. parvum, whose clinical manifestations and outcomes were consistent with previous reported intrauterine U. parvum infection (Gray et al., 1992; Gerber et al., 2003; Perni et al., 2004; Witt et al., 2005; Kirchner et al., 2007; Kasper et al., 2010). Due to the low-depth sequencing of CNV-seq, the definitive connection between U. parvum colonization and adverse outcomes should be interpreted with cautions, and a larger sample size and additional research are needed to further validate the performance of CNV-seq in detecting U. parvum and other microorganisms.
The primary strength of our study lies in our innovative reanalysis of prenatal CNV-seq data to detect U. parvum intrauterine infection, providing a novel method for identifying early infection beyond prenatal genetic diagnosis. To our knowledge, this study represents the first attempt to evaluate the feasibility and efficacy of using CNV-seq data for detecting U. parvum in amniotic fluids. Give these results, this approach could theoretically be extended as an initial screening tool for other latent intrauterine microbial pathogens after amniocentesis, particularly in cases presenting with fetal growth restriction, ventriculomegaly, and fetal hydrops. In such scenarios, reanalyzing existing CNV-seq data might offer valuable microbial insights without the need for universal mNGS screening, thereby reducing the medical burden on these pregnant women.
However, our study had several limitations that should be acknowledged. First, CNV-seq testing does not involve microbial enrichment, and the number of sequencing reads is approximately one-third that of mNGS, which would inevitably reduce its sensitivity in detecting microbial infections. Second, due to the absence of orthogonal clinical testing and external no-template control samples, current retrospective data could not conclusively determine whether the detection of low abundance U. parvum represented a true or false positive, nor confirm the thresholds for relatively high and low read counts. Third, given the limited numbers of cases with positive U. parvum detection and wide variations in read counts and corresponding RPM values, it is not feasible to perform correlation analysis or determine predictive values for adverse pregnancy outcomes. Lastly, the absence of standard methods such as qPCR, 16S rRNA, and mNGS as controls hinders our ability to accurately determine the precise intrauterine infection rate of U. parvum in the study population.
Conclusion
It is feasible to detect and quantify the relative abundance of U. parvum in amniotic fluids using CNV-seq data. Asymptomatic high intrauterine U. parvum loads are statistically linked to adverse pregnancy outcomes, particularly an increased risk of early SPB. Additionally, CNV-seq has potential as a preliminary screening approach for other microbes, particularly beneficial for pregnant women who are at high risk of intrauterine infections and require prenatal genetic amniocentesis. Important directions for future research include the following: (1) Optimization of U. parvum detection methods through incorporation of independent validation techniques, such as qPCR, culture, or targeted metagenomic sequencing, to confirm the presence of U. parvum and assess potential false positives or negatives in CNV-seq data. (2) Enhancement of abundance quantification accuracy by utilizing tools like Bracken2 or other abundance refinement methods in conjunction with Kraken2. (3) Implementation large-scale prospective longitudinal studies addressing additional confounding factors, including maternal health conditions, co-infections, antibiotic use, gestational age, and inflammatory markers, to investigate how initial U. parvum abundance and its temporal changes influence the progression and severity of adverse pregnancy outcomes.
Data availability statement
The CNV-seq data from all cases have been deposited in the National Genomics Data Center (NGDC), with the submission accession number HRA011914 (bioProject accession: PRJCA041124; https://bigd.big.ac.cn/gsa-human/browse/HRA011914). Datasheets generated by Kraken2 are available from the corresponding author upon reasonable request.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University (approval number: II2025-130-01). Prior to amniocentesis, all participants provided written informed consents for the further reanalysis of their experimental and clinical data.
Author contributions
GW: Data curation, Methodology, Software, Writing – original draft. WC: Methodology, Software, Validation, Writing – original draft. XC: Investigation, Writing – original draft. HH: Resources, Supervision, Writing – review & editing. JZ: Conceptualization, Project administration, Writing – review & editing. ZH: Formal analysis, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors declare financial support from the Natural Science Foundation of Guangdong Province (Grant Number 2024A1515012206) for this research and publication.
Acknowledgments
We thank all the subjects for cooperating with our study.
Conflict of interest
Author WC is employed by the company Guangzhou Forevergen Medical Laboratory Co., Ltd.
The remaining authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., and Versalovic, J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65. doi: 10.1126/scitranslmed.3008599
Bento, G. F. C., Richardson, L. S., Da Silva, M. G., Tantengco, O. A. G., and Menon, R. (2023). Modeling an ascending infection by Ureaplasma parvum and its cell signaling and inflammatory response at the feto-maternal interface. Am. J. Reprod. Immunol. 90, e13770. doi: 10.1111/aji.13770
Bertelli, C. and Greub, G. (2013). Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin. Microbiol. Infect. 19, 803–813. doi: 10.1111/1469-0691.12217
Blauwkamp, T. A., Thair, S., Rosen, M. J., Blair, L., Lindner, M. S., Vilfan, I. D., et al. (2019). Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674. doi: 10.1038/s41564-018-0349-6
Cantalupo, P. G. and Pipas, J. M. (2019). Detecting viral sequences in NGS data. Curr. Opin. Virol. 39, 41–48. doi: 10.1016/j.coviro.2019.07.010
Cassell, G. H., Waites, K. B., Watson, H. L., Crouse, D. T., and Harasawa, R. (1993). Ureaplasma urealyticum intrauterine infection: role in prematurity and disease in newborns. Clin. Microbiol. Rev. 6, 69–87. doi: 10.1128/CMR.6.1.69
Chiu, C. Y. and Miller, S. A. (2019). Clinical metagenomics. Nat. Rev. Genet. 20, 341–355. doi: 10.1038/s41576-019-0113-7
Costello, M., Fleharty, M., Abreu, J., Farjoun, Y., Ferriera, S., Holmes, L., et al. (2018). Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics 19, 332. doi: 10.1186/s12864-018-4703-0
Cox, C., Saxena, N., Watt, A. P., Gannon, C., Mckenna, J. P., Fairley, D. J., et al. (2016). The common vaginal commensal bacterium Ureaplasma parvum is associated with chorioamnionitis in extreme preterm labor. J. Matern Fetal Neonatal Med. 29, 3646–3651. doi: 10.3109/14767058.2016.1140734
Deurenberg, R. H., Bathoorn, E., Chlebowicz, M. A., Couto, N., Ferdous, M., García-Cobos, S., et al. (2017). Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 243, 16–24. doi: 10.1016/j.jbiotec.2016.12.022
DiGiulio, D. B., Romero, R., Kusanovic, J. P., Gómez, R., Kim, C. J., Seok, K. S., et al. (2010). Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 64, 38–57. doi: 10.1111/j.1600-0897.2010.00830.x
Dong, Z., Zhang, J., Hu, P., Chen, H., Xu, J., Tian, Q., et al. (2016). Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet. Med. 18, 940–948. doi: 10.1038/gim.2015.199
Du, J., Zhang, J., Zhang, D., Zhou, Y., Wu, P., Ding, W., et al. (2022). Background Filtering of Clinical Metagenomic Sequencing with a Library Concentration-Normalized Model. Microbiol. Spectr. 10, e0177922. doi: 10.1128/spectrum.01779-22
Elovitz, M. A., Gajer, P., Riis, V., Brown, A. G., Humphrys, M. S., Holm, J. B., et al. (2019). Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat. Commun. 10, 1305. doi: 10.1038/s41467-019-09285-9
Gerber, S., Vial, Y., Hohlfeld, P., and Witkin, S. S. (2003). Detection of Ureaplasma urealyticum in second-trimester amniotic fluid by polymerase chain reaction correlates with subsequent preterm labor and delivery. J. Infect. Dis. 187, 518–521. doi: 10.1086/368205
Goldenberg, R. L., Culhane, J. F., Iams, J. D., and Romero, R. (2008). Epidemiology and causes of preterm birth. Lancet 371, 75–84. doi: 10.1016/S0140-6736(08)60074-4
Goldenberg, R. L., Hauth, J. C., and Andrews, W. W. (2000). Intrauterine infection and preterm delivery. N Engl. J. Med. 342, 1500–1507. doi: 10.1056/NEJM200005183422007
Gray, D. J., Robinson, H. B., Malone, J., and Thomson, R. B., Jr (1992). Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat. Diagn. 12, 111–117. doi: 10.1002/pd.1970120206
Gu, W., Miller, S., and Chiu, C. Y. (2019). Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Hayes, J. L., Tzika, A., Thygesen, H., Berri, S., Wood, H. M., Hewitt, S., et al. (2013). Diagnosis of copy number variation by Illumina next generation sequencing is comparable in performance to oligonucleotide array comparative genomic hybridisation. Genomics 102, 174–181. doi: 10.1016/j.ygeno.2013.04.006
Heikkinen, T., Laine, K., Neuvonen, P. J., and Ekblad, U. (2000). The transplacental transfer of the macrolide antibiotics erythromycin, roxithromycin and azithromycin. Bjog 107, 770–775. doi: 10.1111/j.1471-0528.2000.tb13339.x
Huang, J., Jiang, E., Yang, D., Wei, J., Zhao, M., Feng, J., et al. (2020). Metagenomic Next-Generation Sequencing versus Traditional Pathogen Detection in the Diagnosis of Peripheral Pulmonary Infectious Lesions. Infect. Drug Resist. 13, 567–576. doi: 10.2147/IDR.S235182
Jacobsson, B., Aaltonen, R., Rantakokko-Jalava, K., Morken, N. H., and Alanen, A. (2009). Quantification of Ureaplasma urealyticum DNA in the amniotic fluid from patients in PTL and pPROM and its relation to inflammatory cytokine levels. Acta Obstet Gynecol Scand. 88, 63–70. doi: 10.1080/00016340802572646
Johnson, J. S., Spakowicz, D. J., Hong, B. Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029. doi: 10.1038/s41467-019-13036-1
Jonduo, M. E., Vallely, L. M., Wand, H., Sweeney, E. L., Egli-Gany, D., Kaldor, J., et al. (2022). Adverse pregnancy and birth outcomes associated with Mycoplasma hominis, Ureaplasma urealyticum and Ureaplasma parvum: a systematic review and meta-analysis. BMJ Open 12, e062990. doi: 10.1136/bmjopen-2022-062990
Kacerovsky, M., Pliskova, L., Bolehovska, R., Musilova, I., Hornychova, H., Tambor, V., et al. (2011). The microbial load with genital mycoplasmas correlates with the degree of histologic chorioamnionitis in preterm PROM. Am. J. Obstet Gynecol 205, 213.e1–213.e7. doi: 10.1016/j.ajog.2011.04.028
Kacerovsky, M., Pliskova, L., Bolehovska, R., Skogstrand, K., Hougaard, D. M., Tsiartas, P., et al. (2012). The impact of the microbial load of genital mycoplasmas and gestational age on the intensity of intraamniotic inflammation. Am. J. Obstet Gynecol 206, 342.e1–342.e8. doi: 10.1016/j.ajog.2012.01.004
Kasper, D. C., Mechtler, T. P., Reischer, G. H., Witt, A., Langgartner, M., Pollak, A., et al. (2010). The bacterial load of Ureaplasma parvum in amniotic fluid is correlated with an increased intrauterine inflammatory response. Diagn. Microbiol. Infect. Dis. 67, 117–121. doi: 10.1016/j.diagmicrobio.2009.12.023
Kataoka, S., Yamada, T., Chou, K., Nishida, R., Morikawa, M., Minami, M., et al. (2006). Association between preterm birth and vaginal colonization by mycoplasmas in early pregnancy. J. Clin. Microbiol. 44, 51–55. doi: 10.1128/JCM.44.1.51-55.2006
Kayem, G., Doloy, A., Schmitz, T., Chitrit, Y., Bouhanna, P., Carbonne, B., et al. (2018). Antibiotics for amniotic-fluid colonization by Ureaplasma and/or Mycoplasma spp. to prevent preterm birth: A randomized trial. PloS One 13, e0206290. doi: 10.1371/journal.pone.0206290
Kembel, S. W., Wu, M., Eisen, J. A., and Green, J. L. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PloS Comput. Biol. 8, e1002743. doi: 10.1371/journal.pcbi.1002743
Kim, M., Kim, G., Romero, R., Shim, S. S., Kim, E. C., and Yoon, B. H. (2003). Biovar diversity of Ureaplasma urealyticum in amniotic fluid: distribution, intrauterine inflammatory response and pregnancy outcomes. J. Perinat Med. 31, 146–152. doi: 10.1515/JPM.2003.020
Kim, C. J., Romero, R., Chaemsaithong, P., Chaiyasit, N., Yoon, B. H., and Kim, Y. M. (2015). Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am. J. Obstet Gynecol 213, S29–S52. doi: 10.1016/j.ajog.2015.08.040
Kirchner, L., Helmer, H., Heinze, G., Wald, M., Brunbauer, M., Weninger, M., et al. (2007). Amnionitis with Ureaplasma urealyticum or other microbes leads to increased morbidity and prolonged hospitalization in very low birth weight infants. Eur. J. Obstet Gynecol Reprod. Biol. 134, 44–50. doi: 10.1016/j.ejogrb.2006.09.013
Li, N., Cai, Q., Miao, Q., Song, Z., Fang, Y., and Hu, B. (2021). High-Throughput Metagenomics for Identification of Pathogens in the Clinical Settings. Small Methods 5, 2000792. doi: 10.1002/smtd.202000792
Liang, D., Peng, Y., Lv, W., Deng, L., Zhang, Y., Li, H., et al. (2014). Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J. Mol. Diagn. 16, 519–526. doi: 10.1016/j.jmoldx.2014.05.002
Meng, X. and Jiang, L. (2022). Prenatal detection of chromosomal abnormalities and copy number variants in fetuses with congenital gastrointestinal obstruction. BMC Pregnancy Childbirth 22, 50. doi: 10.1186/s12884-022-04401-y
Miller, S., Naccache, S. N., Samayoa, E., Messacar, K., Arevalo, S., Federman, S., et al. (2019). Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 29, 831–842. doi: 10.1101/gr.238170.118
Motomura, K., Romero, R., Xu, Y., Theis, K. R., Galaz, J., Winters, A. D., et al. (2020). Intra-Amniotic Infection with Ureaplasma parvum Causes Preterm Birth and Neonatal Mortality That Are Prevented by Treatment with Clarithromycin. mBio 11, e00797-20. doi: 10.1128/mBio.00797-20
Noda-Nicolau, N. M., Tantengco, O. A. G., Polettini, J., Silva, M. C., Bento, G. F. C., Cursino, G. C., et al. (2022). Genital Mycoplasmas and Biomarkers of Inflammation and Their Association with Spontaneous Preterm Birth and Preterm Prelabor Rupture of Membranes: A Systematic Review and Meta-Analysis. Front. Microbiol. 13, 859732. doi: 10.3389/fmicb.2022.859732
Otgonjargal, B., Batbaatar, G., Pfeffer, K., Bruhn, T., Battogtokh, C., and Henrich, B. (2018). A novel mba-based Real time PCR approach for genotyping of Ureaplasma parvum validated in a cohort of Mongolian mothers and offspring. Int. J. Med. Microbiol. 308, 865–871. doi: 10.1016/j.ijmm.2018.08.001
Pavlidis, I., Spiller, O. B., Sammut Demarco, G., Macpherson, H., Howie, S. E. M., Norman, J. E., et al. (2020). Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice. Nat. Commun. 11, 199. doi: 10.1038/s41467-019-14089-y
Payne, M. S., Ireland, D. J., Watts, R., Nathan, E. A., Furfaro, L. L., Kemp, M. W., et al. (2016). Ureaplasma parvum genotype, combined vaginal colonisation with Candida albicans, and spontaneous preterm birth in an Australian cohort of pregnant women. BMC Pregnancy Childbirth 16, 312. doi: 10.1186/s12884-016-1110-x
Perni, S. C., Vardhana, S., Korneeva, I., Tuttle, S. L., Paraskevas, L. R., Chasen, S. T., et al. (2004). Mycoplasma hominis and Ureaplasma urealyticum in midtrimester amniotic fluid: association with amniotic fluid cytokine levels and pregnancy outcome. Am. J. Obstet Gynecol 191, 1382–1386. doi: 10.1016/j.ajog.2004.05.070
Prince, A. L., Ma, J., Kannan, P. S., Alvarez, M., Gisslen, T., Harris, R. A., et al. (2016). The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am. J. Obstet Gynecol 214, 627.e1–627.e16. doi: 10.1016/j.ajog.2016.01.193
Qin, H., Peng, J., Liu, L., Wu, J., Pan, L., Huang, X., et al. (2021). A Retrospective Paired Comparison Between Untargeted Next Generation Sequencing and Conventional Microbiology Tests With Wisely Chosen Metagenomic Sequencing Positive Criteria. Front. Med. (Lausanne) 8, 686247. doi: 10.3389/fmed.2021.686247
Rittenschober-Böhm, J., Habermüller, T., Waldhoer, T., Fuiko, R., Schulz, S. M., Pimpel, B., et al. (2021). Maternal Vaginal Ureaplasma spp. Colonization in Early Pregnancy Is Associated with Adverse Short- and Long-Term Outcome of Very Preterm Infants. Children (Basel) 8, 276. doi: 10.3390/children8040276
Rittenschober-Böhm, J., Waldhoer, T., Schulz, S. M., Pimpel, B., Goeral, K., Kasper, D. C., et al. (2019). Vaginal Ureaplasma parvum serovars and spontaneous preterm birth. Am. J. Obstet Gynecol 220, 594.e1–594.e9. doi: 10.1016/j.ajog.2019.01.237
Rittenschober-Böhm, J., Waldhoer, T., Schulz, S. M., Stihsen, B., Pimpel, B., Goeral, K., et al. (2018). First Trimester Vaginal Ureaplasma Biovar Colonization and Preterm Birth: Results of a Prospective Multicenter Study. Neonatology 113, 1–6. doi: 10.1159/000480065
Rodríguez, N., Fernandez, C., Zamora, Y., Berdasquera, D., and Rivera, J. A. (2011). Detection of Ureaplasma urealyticum and Ureaplasma parvum in amniotic fluid: association with pregnancy outcomes. J. Matern Fetal Neonatal Med. 24, 47–50. doi: 10.3109/14767058.2010.482609
Romero, R., Dey, S. K., and Fisher, S. J. (2014). Preterm labor: one syndrome, many causes. Science 345, 760–765. doi: 10.1126/science.1251816
Romero, R., Espinoza, J., Gonçalves, L. F., Kusanovic, J. P., Friel, L., and Hassan, S. (2007). The role of inflammation and infection in preterm birth. Semin. Reprod. Med. 25, 21–39. doi: 10.1055/s-2006-956773
Senthamaraikannan, P., Presicce, P., Rueda, C. M., Maneenil, G., Schmidt, A. F., Miller, L. A., et al. (2016). Intra-amniotic Ureaplasma parvum-Induced Maternal and Fetal Inflammation and Immune Responses in Rhesus Macaques. J. Infect. Dis. 214, 1597–1604. doi: 10.1093/infdis/jiw408
Stranik, J., Kacerovsky, M., Andrys, C., Soucek, O., Bolehovska, R., Holeckova, M., et al. (2022). Intra-amniotic infection and sterile intra-amniotic inflammation are associated with elevated concentrations of cervical fluid interleukin-6 in women with spontaneous preterm labor with intact membranes. J. Matern Fetal Neonatal Med. 35, 4861–4869. doi: 10.1080/14767058.2020.1869932
Sun, L., Zhang, S., Yang, Z., Yang, F., Wang, Z., Li, H., et al. (2022). Clinical Application and Influencing Factor Analysis of Metagenomic Next-Generation Sequencing (mNGS) in ICU Patients With Sepsis. Front. Cell Infect. Microbiol. 12, 905132. doi: 10.3389/fcimb.2022.905132
Sweeney, E. L., Kallapur, S. G., Gisslen, T., Lambers, D. S., Chougnet, C. A., Stephenson, S. A., et al. (2016). Placental Infection With Ureaplasma species Is Associated With Histologic Chorioamnionitis and Adverse Outcomes in Moderately Preterm and Late-Preterm Infants. J. Infect. Dis. 213, 1340–1347. doi: 10.1093/infdis/jiv587
Tantengco, O. A. G., Kechichian, T., Vincent, K. L., Pyles, R. B., Medina, P. M. B., and Menon, R. (2021). Inflammatory response elicited by Ureaplasma parvum colonization in human cervical epithelial, stromal, and immune cells. Reproduction 163, 1–10. doi: 10.1530/REP-21-0308
Tita, A. T. and Andrews, W. W. (2010). Diagnosis and management of clinical chorioamnionitis. Clin. Perinatol 37, 339–354. doi: 10.1016/j.clp.2010.02.003
Tripathy, S., Burd, I., and Kelleher, M. A. (2023). Membrane Inflammasome Activation by Choriodecidual Ureaplasma parvum Infection without Intra-Amniotic Infection in an NHP Model. Biol. Reprod. 110, 971–984. doi: 10.1093/biolre/ioae027
Uchida, K., Nakahira, K., Mimura, K., Shimizu, T., De Seta, F., Wakimoto, T., et al. (2013). Effects of Ureaplasma parvum lipoprotein multiple-banded antigen on pregnancy outcome in mice. J. Reprod. Immunol. 100, 118–127. doi: 10.1016/j.jri.2013.10.001
Viscardi, R. M. (2010). Ureaplasma species: role in diseases of prematurity. Clin. Perinatol 37, 393–409. doi: 10.1016/j.clp.2009.12.003
Wang, J., Chen, L., Zhou, C., Wang, L., Xie, H., Xiao, Y., et al. (2018a). Identification of copy number variations among fetuses with ultrasound soft markers using next-generation sequencing. Sci. Rep. 8, 8134. doi: 10.1038/s41598-018-26555-6
Wang, J., Chen, L., Zhou, C., Wang, L., Xie, H., Xiao, Y., et al. (2018b). Prospective chromosome analysis of 3429 amniocentesis samples in China using copy number variation sequencing. Am. J. Obstet Gynecol 219, 287.e1–287.e18. doi: 10.1016/j.ajog.2018.05.030
Witt, A., Berger, A., Gruber, C. J., Petricevic, L., Apfalter, P., Worda, C., et al. (2005). Increased intrauterine frequency of Ureaplasma urealyticum in women with preterm labor and preterm premature rupture of the membranes and subsequent cesarean delivery. Am. J. Obstet Gynecol 193, 1663–1669. doi: 10.1016/j.ajog.2005.03.067
Witt, A., Sommer, E. M., Cichna, M., Postlbauer, K., Widhalm, A., Gregor, H., et al. (2003). Placental passage of clarithromycin surpasses other macrolide antibiotics. Am. J. Obstet Gynecol 188, 816–819. doi: 10.1067/mob.2003.171
Xiao, L., Paralanov, V., Glass, J. I., Duffy, L. B., Robertson, J. A., Cassell, G. H., et al. (2011). Extensive horizontal gene transfer in ureaplasmas from humans questions the utility of serotyping for diagnostic purposes. J. Clin. Microbiol. 49, 2818–2826. doi: 10.1128/JCM.00637-11
Yoneda, N., Yoneda, S., Niimi, H., Ueno, T., Hayashi, S., Ito, M., et al. (2016). Polymicrobial Amniotic Fluid Infection with Mycoplasma/Ureaplasma and Other Bacteria Induces Severe Intra-Amniotic Inflammation Associated with Poor Perinatal Prognosis in Preterm Labor. Am. J. Reprod. Immunol. 75, 112–125. doi: 10.1111/aji.12456
Yoon, B. H., Romero, R., Kim, M., Kim, E. C., Kim, T., Park, J. S., et al. (2000). Clinical implications of detection of Ureaplasma urealyticum in the amniotic cavity with the polymerase chain reaction. Am. J. Obstet Gynecol 183, 1130–1137. doi: 10.1067/mob.2000.109036
Yoon, B. H., Romero, R., Lim, J. H., Shim, S. S., Hong, J. S., Shim, J. Y., et al. (2003). The clinical significance of detecting Ureaplasma urealyticum by the polymerase chain reaction in the amniotic fluid of patients with preterm labor. Am. J. Obstet Gynecol 189, 919–924. doi: 10.1067/S0002-9378(03)00839-1
Zhu, X., Li, J., Ru, T., Wang, Y., Xu, Y., Yang, Y., et al. (2016). Identification of copy number variations associated with congenital heart disease by chromosomal microarray analysis and next-generation sequencing. Prenat. Diagn. 36, 321–327. doi: 10.1002/pd.4782
Keywords: intrauterine infection, Ureaplasma parvum, copy number variation sequencing, amniocentesis, pregnancy outcomes
Citation: Wang G, Chen W, Chen X, Hou H, Zhang J and Han Z (2025) Detection of Ureaplasma parvum in amniotic fluids via reanalysis of prenatal copy number variation sequencing data: an exploratory study. Front. Cell. Infect. Microbiol. 15:1579049. doi: 10.3389/fcimb.2025.1579049
Received: 18 February 2025; Accepted: 25 July 2025;
Published: 14 August 2025.
Edited by:
Martin Mueller, Lindenhofspital, SwitzerlandReviewed by:
Andrew Winters, Wayne State University, United StatesTina Perme, University Medical Centre Ljubljana, Slovenia
Copyright © 2025 Wang, Chen, Chen, Hou, Zhang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenyan Han, aGFuemh5YW5AbWFpbC5zeXN1LmVkdS5jbg==; Jun Zhang, emhqdW45QG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
 Guan Wang
Guan Wang Weifen Chen
Weifen Chen Xiaodan Chen1
Xiaodan Chen1 Jun Zhang
Jun Zhang Zhenyan Han
Zhenyan Han