- 1Department of Biochemistry, Bahauddin Zakariya University, Multan, Pakistan
- 2Institute of Molecular Biology and Biotechnology, The University of Lahore, Lahore, Pakistan
- 3Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Jouf University, Sakaka, Saudi Arabia
- 4Department of Biochemistry and Molecular Biology, Federal University of Ceara, Fortaleza, Brazil
- 5Department of Chemistry, Kohat University of Science and Technology, Kohat, Pakistan
- 6College of Engineering and Technology, American University of the Middle East, Egaila 54200, Kuwait
- 7Department of Pharmacognosy, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 8Department of Infectious Diseases, The Affiliated Hospital of Southwest Medical University, Luzhou, China
A Corrigendum on
Computational analysis of Ayurvedic metabolites for potential treatment of drug-resistant Candida auris
By Shah M, Zia M, Ahmad I, Umer Khan M, Ejaz H, Alam M, Aziz S, Nishan U, Dib H, Ullah R and Ojha SC (2025). Front. Cell. Infect. Microbiol. 15:1537872. doi: 10.3389/fcimb.2025.1537872
In the published article, there was an error in Figure 8 as published. [A part of the figure was not labeled]. The corrected Figure 8 and its caption appear below.
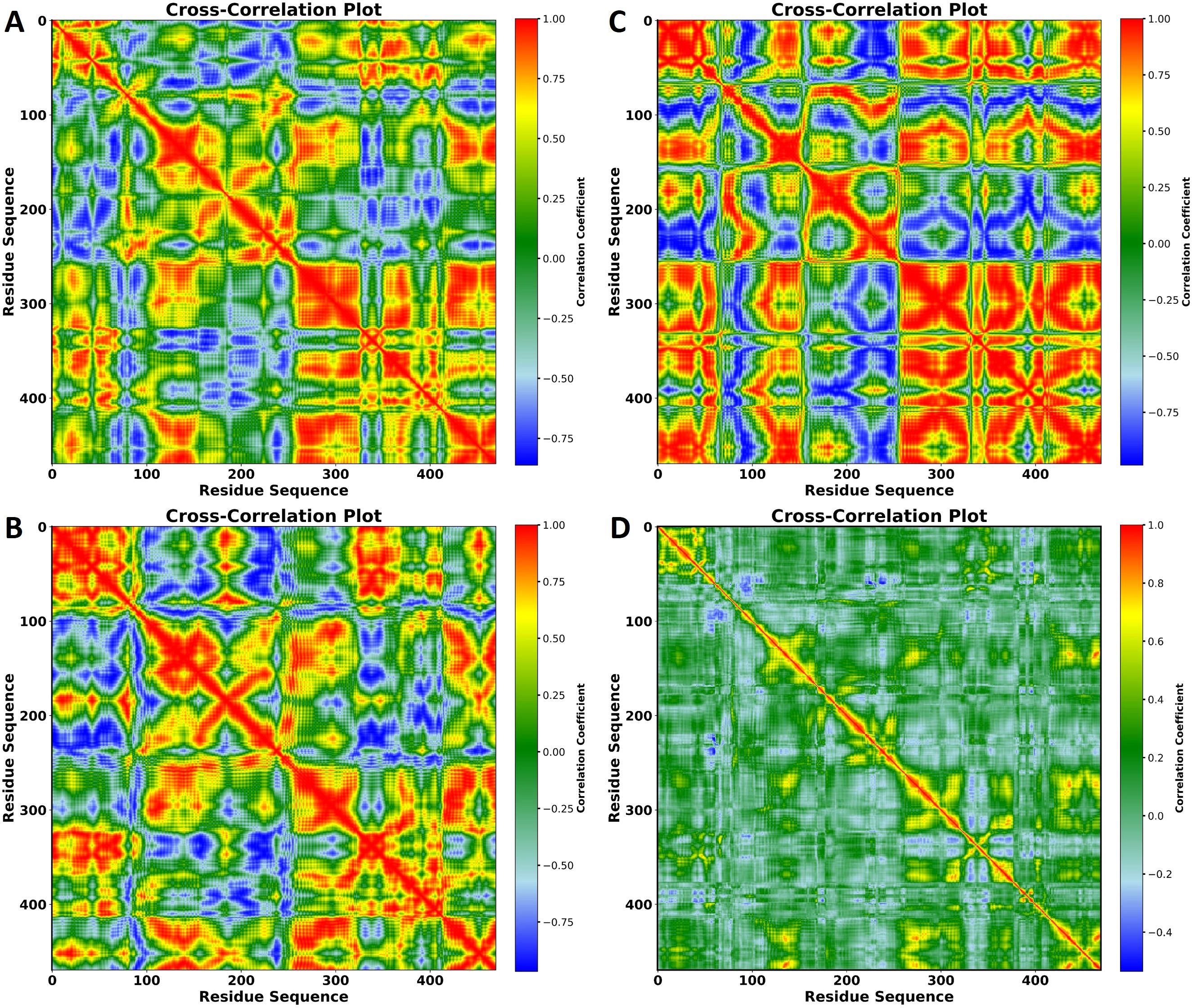
Figure 8. Cross-correlation plots of residue motions in the protein in complexes with (A) VNI, (B) trans-p-coumaric acid, (C) MCPHB, and (D) Cross-correlation plot of residue motions in the apoprotein. The x- and y-axes represent the residue sequence, and the color scale on the right indicates the correlation coefficients, ranging from -1 (anti-correlated, blue) to +1 (positively correlated, red). These plots show the degree of correlated and anti-correlated motions between residues throughout the 100 ns MD simulation.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: fungal infections, Candida auris, computational chemistry, plants, molecular docking
Citation: Shah M, Zia M, Ahmad I, Khan MU, Ejaz H, Alam M, Aziz S, Nishan U, Dib H, Ullah R and Ojha SC (2025) Corrigendum: Computational analysis of Ayurvedic metabolites for potential treatment of drug-resistant Candida auris. Front. Cell. Infect. Microbiol. 15:1595290. doi: 10.3389/fcimb.2025.1595290
Received: 17 March 2025; Accepted: 21 April 2025;
Published: 08 May 2025.
Edited and Reviewed by:
Maryam Roudbary, The University of Sydney, AustraliaCopyright © 2025 Shah, Zia, Ahmad, Khan, Ejaz, Alam, Aziz, Nishan, Dib, Ullah and Ojha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohibullah Shah, bW9oaWJAYnp1LmVkdS5waw==; bW9oaWJ1c2JAZ21haWwuY29t; Suvash Chandra Ojha, c3V2YXNoX29qaGFAc3dtdS5lZHUuY24=
†These authors have contributed equally to this work
‡ORCID: Mohibullah Shah, orcid.org/0000-0001-6126-7102
 Mohibullah Shah
Mohibullah Shah Mahnoor Zia1†
Mahnoor Zia1†