- 1Animal Husbandry Science Institute of Ganzi Tibetan Autonomous Prefecture, Kangding, China
- 2China Institute of Veterinary Drug Control, Beijing, China
Introduction: Yaks serve as a vital economic and ecological resource in high-altitude regions, but it faces significant health challenges from various pathogens. Among these, Pasteurella multocida and Salmonella are critical pathogens that contribute to severe diseases.
Methods: A duplex real-time fluorescence quantitative PCR assay was developed to simultaneously detect Pasteurella multocida and Salmonella. The species-specific genes kmt1 and invA were selected as target regions for primer and probe design. Following rigorous optimization, a duplex assay was established. Recombinant plasmids were constructed to serve as standards for generating standard curves. The detection thresholds were determined using SPSS statistical analysis and receiver operating characteristic curve methods. Furthermore, the assay’s sensitivity, specificity, stability, and clinical applicability were evaluated.
Results: The established assay demonstrated high sensitivity, with detection limits of 100 and 10 copies for pMD-kmt1 and pMD-invA, respectively. No cross-reactivity was observed with six pathogens, including Mycoplasma bovis, infectious bovine rhinotracheitis virus and others. The standard curves showed strong linearity, with coefficients of determination of 0.995 and 0.998, and amplification efficiencies of 103.37% and 103.47% for pMD-kmt1 and pMD-invA, respectively. No interference was observed between high- and low-concentration templates during simultaneous detection. The intra- and inter-assay coefficients of variation ranged from 0.23% to 1.51%. Detection thresholds were determined to be cycle threshold values of 41.5 for P. multocida and 40.0 for Salmonella. Clinical evaluation was performed on 226 samples collected from yaks in seven counties of Ganzi Prefecture, Sichuan Province, China. The single infection rates of P. multocida and Salmonella were 20.35% (46/226) and 38.50% (87/226), respectively, while the co-infection rate was 6.19% (14/226).
Discussion: This study successfully established a duplex real-time fluorescence PCR assay that enables the simultaneous detection of P. multocida and Salmonella with high sensitivity, specificity, and efficiency. The assay offers a reliable and rapid diagnostic tool that is particularly suited for clinical and epidemiological investigations in yak populations.
1 Introduction
The yak (Bos grunniens), a vital livestock species in high-altitude regions, is widely distributed across the Qinghai-Tibet Plateau and surrounding areas, owing to its exceptional adaptability to cold and hypoxic environments (Tiwari et al., 2024). Yaks serve as a pillar of the local livestock economy and ecosystem, providing essential resources such as meat, milk, and hides to herders (Ai et al., 2024). However, the increasing stocking density of yak populations in recent years has markedly elevated the risk of disease outbreaks and transmission, including bovine pasteurellosis and bovine paratyphoid (Mo et al., 2024; Ran et al., 2024).
Pasteurella multocida (P. multocida), a Gram-negative bacterium, is classified into five capsular serogroups (A, B, D, E, and F) and is a major causative agent of bovine respiratory diseases (serogroup A) and hemorrhagic septicemia (serogroups B and E) (Wang H. et al., 2023). Salmonella, another Gram-negative bacterium with over 2,600 serotypes, is an important pathogen responsible for septicemia and gastroenteritis (Craig et al., 2024; Ran et al., 2024). Commonly present in the intestinal tracts of animals and in environmental reservoirs such as water and soil, Salmonella spreads primarily through contaminated feed, water, or direct contact (Ayuti et al., 2024). P. multocida and Salmonella are opportunistic pathogens, and healthy animals can act as asymptomatic carriers under specific conditions (Mohler et al., 2009; Shehta et al., 2022; Wang H. et al., 2023). In the harsh plateau environment, characterized by hypoxia, extreme cold, and limited forage availability, yaks experience significant physiological stress, which predisposes them to infections by these pathogens (Piorunek et al., 2023a; Wang H. et al., 2023; Ran et al., 2024). Furthermore, P. multocida and Salmonella have zoonotic potential, posing threats to human health (Piorunek et al., 2023a; Piorunek et al., 2023b; Grote et al., 2024).
Current etiology diagnostic methods for P. multocida and Salmonella include bacterial isolation and identification and polymerase chain reaction (PCR) (Islam et al., 2023; Mahboob et al., 2023; Bahr and Abdel, 2024; Shaukat et al., 2024). While bacterial isolation is considered the “gold standard,” it is labor-intensive, time-consuming, and requires specialized laboratory infrastructure (Islam et al., 2023). Traditional PCR is limited by low sensitivity and specificity which reduces its efficiency and accuracy (Bahr and Abdel, 2024).
In contrast, duplex real-time fluorescence quantitative PCR is a highly efficient diagnostic approach that combines speed, sensitivity, specificity, and quantification capabilities (Zheng et al., 2020; Lv et al., 2022). This method enables the simultaneous detection of multiple pathogens in a single assay (Zheng et al., 2020; Lv et al., 2022). Its simplicity and reliability make it an ideal tool for pathogen detection and disease control. Developing a duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of P. multocida and Salmonella would significantly enhance diagnostic sensitivity and specificity, facilitate the rapid identification of infected animals, and provide robust technical support for the effective prevention and control of these diseases in yak populations.
2 Materials and methods
2.1 Nucleic acid of bacteria and viruses
The nucleic acids of Pasteurella multocida (Serotype A), Pasteurella multocida (Serotype B), Pasteurella multocida (Serotype E), Salmonella Dublin, Salmonella typhimurium, Mycoplasma bovis, Escherichia Coli O157, Staphylococcus aureus, Pseudomonas aeruginosa, Infectious Bovine Rhinotracheitis Virus (IBRV), Clostridium perfringens were from the China institute of Veterinary Drug Control.
2.2 Primer and probe design
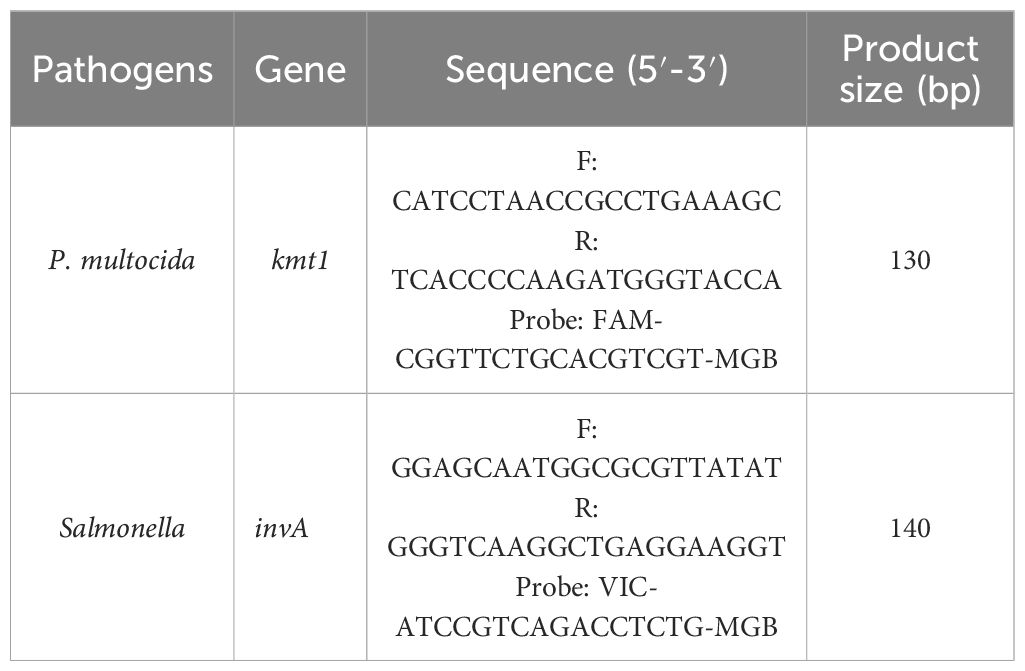
The kmt1 gene of P. multocida (GenBank ID: CP033599.1) and the invA gene of Salmonella (GenBank ID: CP060494.1) were selected as target sequences. Gene sequences were aligned using BLAST, and conserved regions were identified using MegAlign software (Figure 1). Primers and probes were designed with Primer Express 3.0.1 software and synthesized by Tsingke Biotechnology Co., Ltd. Select specific and conserved primers and probes through experiments (Table 1). The 5’ ends of the probes for P. multocida and Salmonella were labeled with FAM and Cy5, respectively, while an MGB quencher was added to the 3’ end. The working concentrations of primers and probes are both 10 μ M.
2.3 Reaction system optimization
The primer and probe concentrations (ranging from 0.1 μM to 0.5 μM), annealing temperatures (ranging from 56°C to 62°C) and number of cycles (40×, 45×, 50×) for a single-fluorescence quantitative PCR assay were optimized using both the matrix and single-variable methods. The optimal reaction conditions were selected based on criteria of low Ct values and high amplification efficiency. Subsequently, the reaction system and program for the duplex fluorescence quantitative PCR assay were further optimized using the parameters established for the single-fluorescence PCR assay.
2.4 Standard curve and sensitivity testing
The target sequences of P. multocida and Salmonella were synthesized to construct recombinant plasmid standards, pMD-Pm and pMD-SE, respectively. The concentrations of the recombinant plasmid standards were measured and converted to copy numbers using a standard formula. Both plasmid standards were adjusted to a final concentration of 1 × 1010 copies/μl and then mixed in equal volumes to obtain a combined concentration of 0.5 × 1010 copies/μl. A 10-fold serial dilution was performed from 0.5 × 1010 copies/μl to 0.5 × 1010 copies/μl. The optimized duplex fluorescence quantitative PCR method was applied to detect plasmids at various concentrations, generating a standard curve and determining the limit of detection.
2.5 Specificity testing
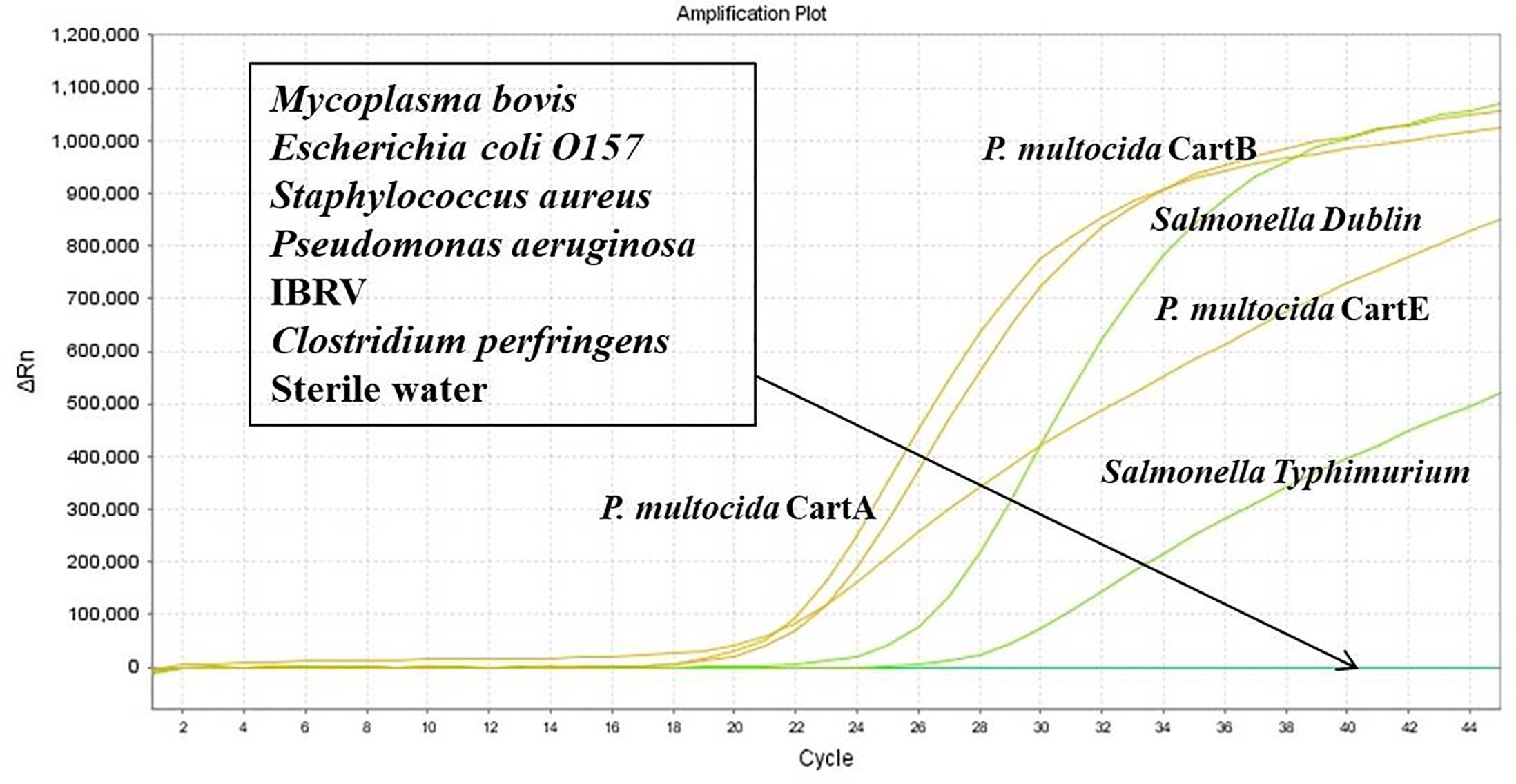
DNA from P. multocida (serotype A, B, and E), Salmonella Dublin, and Salmonella typhimurium was used as positive controls, while sterile water served as the negative control. The optimized dual-fluorescence quantitative PCR method was employed to detect the nucleic acids of Mycoplasma bovis, Escherichia coli O157, Staphylococcus aureus, Pseudomonas aeruginosa, IBRV, and Clostridium perfringens, in order to assess the specificity of the method.
2.6 Anti-interference and stability testing
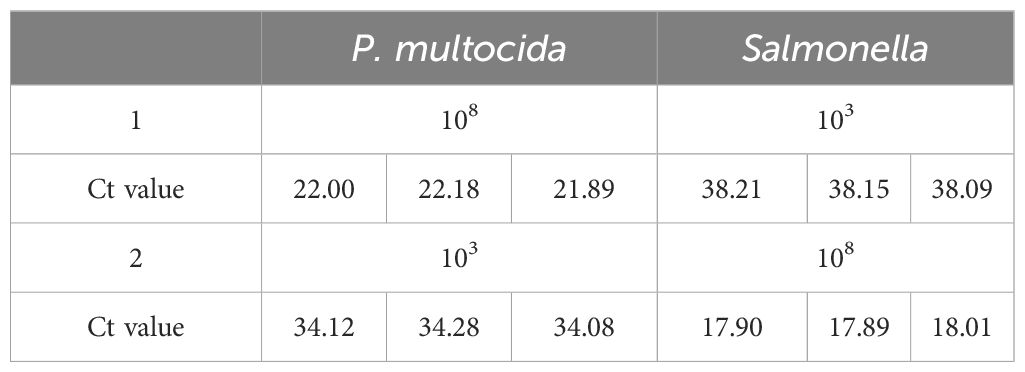
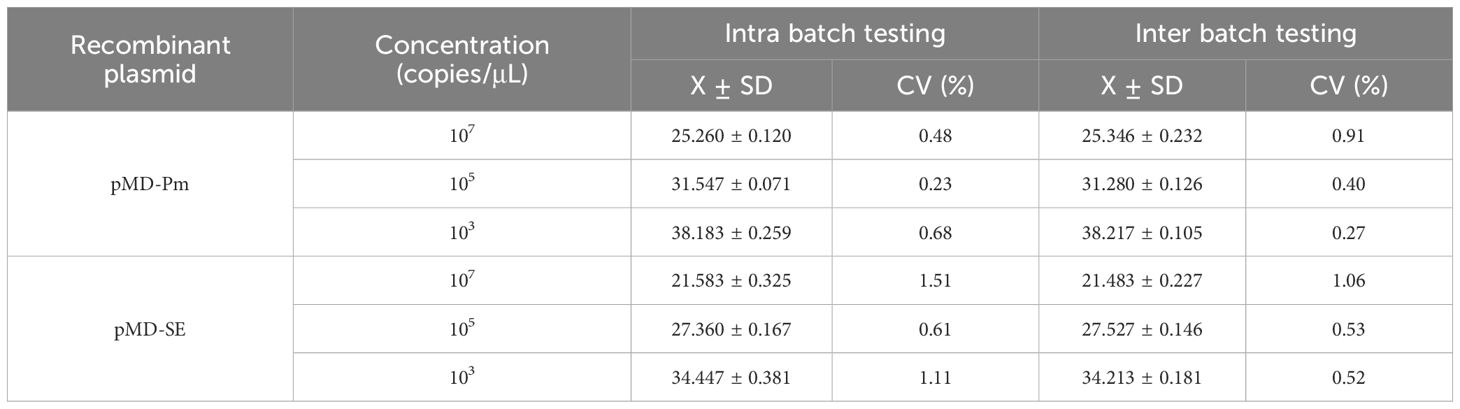
After cross-mixing the plasmid standards at 108 and 103 copies/µL, the mixture was used as the template for detection using the optimized dual-fluorescence quantitative PCR method. To evaluate whether high-concentration plasmids could interfere with the amplification of low-concentration plasmids. The reaction components, including enzymes, buffers, primers, and probes, were assembled under optimal conditions. Three gradient-positive control plasmid standards (107, 105, and 10³ copies/µL) were stored at -20°C. To assess the method’s reproducibility and stability, these controls were tested every two weeks.
2.7 Critical value analysis
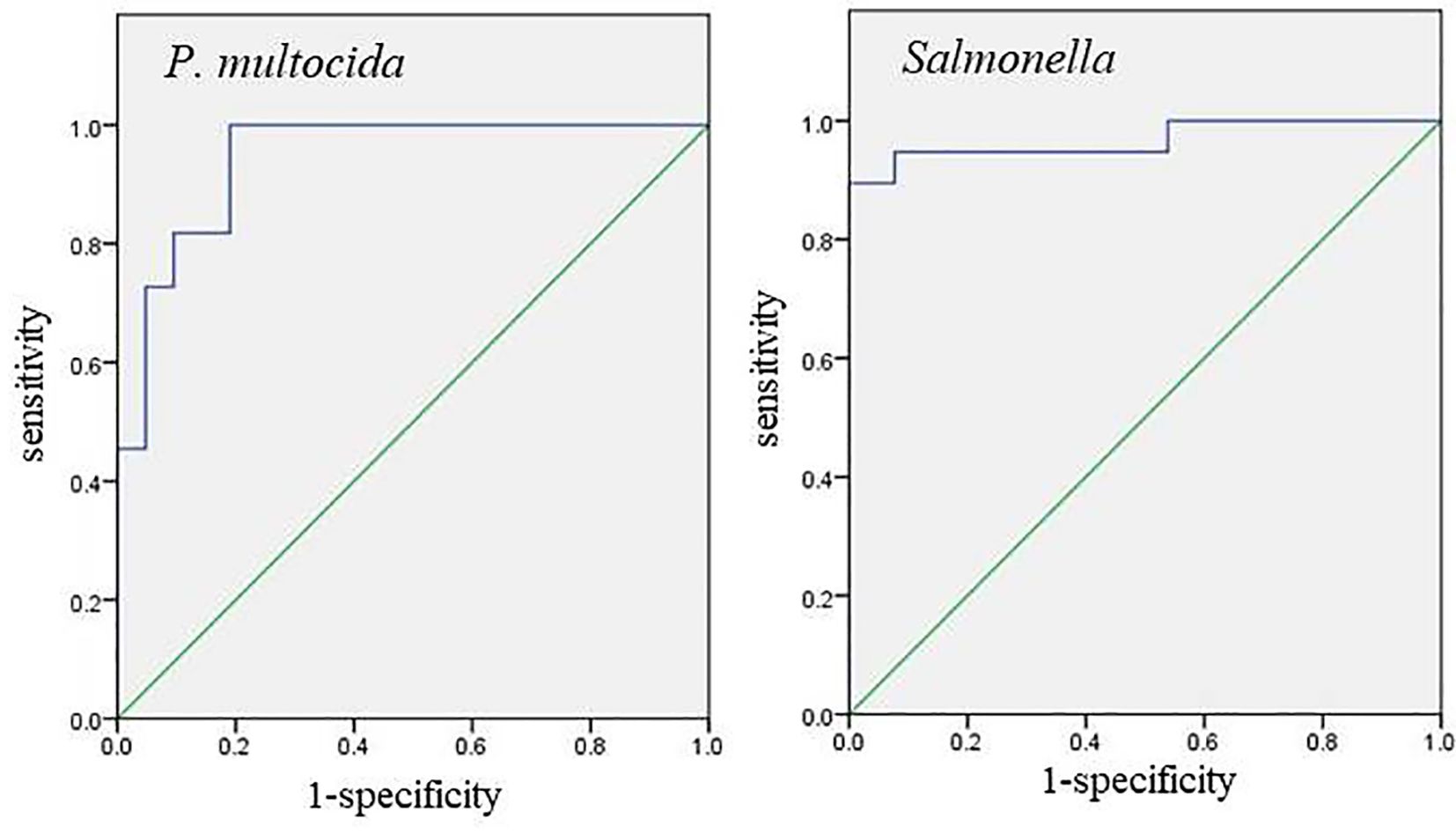
This method was applied to test 50 negative samples and 50 positive samples. These 100 samples were tested using the standards issued by Sichuan Province, China “Diagnostic Technical Specification for Yak Pasteurellosis Disease” (Standard number: DB51/T 2298-2016) and “Technical Specification for Isolation and Identification of Salmonella from Yak Sources” (Standard number: DB51/T 1836-2014) recommend methods for testing to determine the background of the sample. Statistical analysis was performed using SPSS, and a Receiver Operating Characteristic (ROC) curve was generated to determine the sensitivity (Se), specificity (Sp), and area under the curve (AUC) of the detection method. The Youden index (Se+Sp−1) was calculated, with the detection threshold corresponding to the value yielding the highest Youden index (Barraclough, 2012; Krupinski, 2021; Pantoja-Galicia et al., 2021).
2.8 Clinical sample testing
A total of 226 clinical samples (nasal swabs, rectal swabs, feces, and serum) of yaks were collected from various regions of Garze Tibetan Autonomous Prefecture, Sichuan Province from July 2023 to December 2024, including 22 samples from Jiulong County, 15 samples from Garze County, 59 samples from Daofu County, 31 samples from Liuhe County, 28 samples from Litang County, 37 samples from Yajian County, and 34 samples from Seda County. These samples were collected from healthy yaks, yaks with respiratory symptoms, and calves with diarrhea symptoms. Nucleic acids were extracted following the manufacturer’s instructions for commercial kits (TIANGEN, Beijing). The nucleic acids of these samples were then simultaneously detected using the dual-fluorescence quantitative PCR method established in this study and a previously reported single-fluorescence quantitative PCR method (Demirci et al., 2019; Otten et al., 2024). The concordance between the two methods was compared.
3 Results
3.1 Optimization of dual fluorescence quantitative PCR conditions
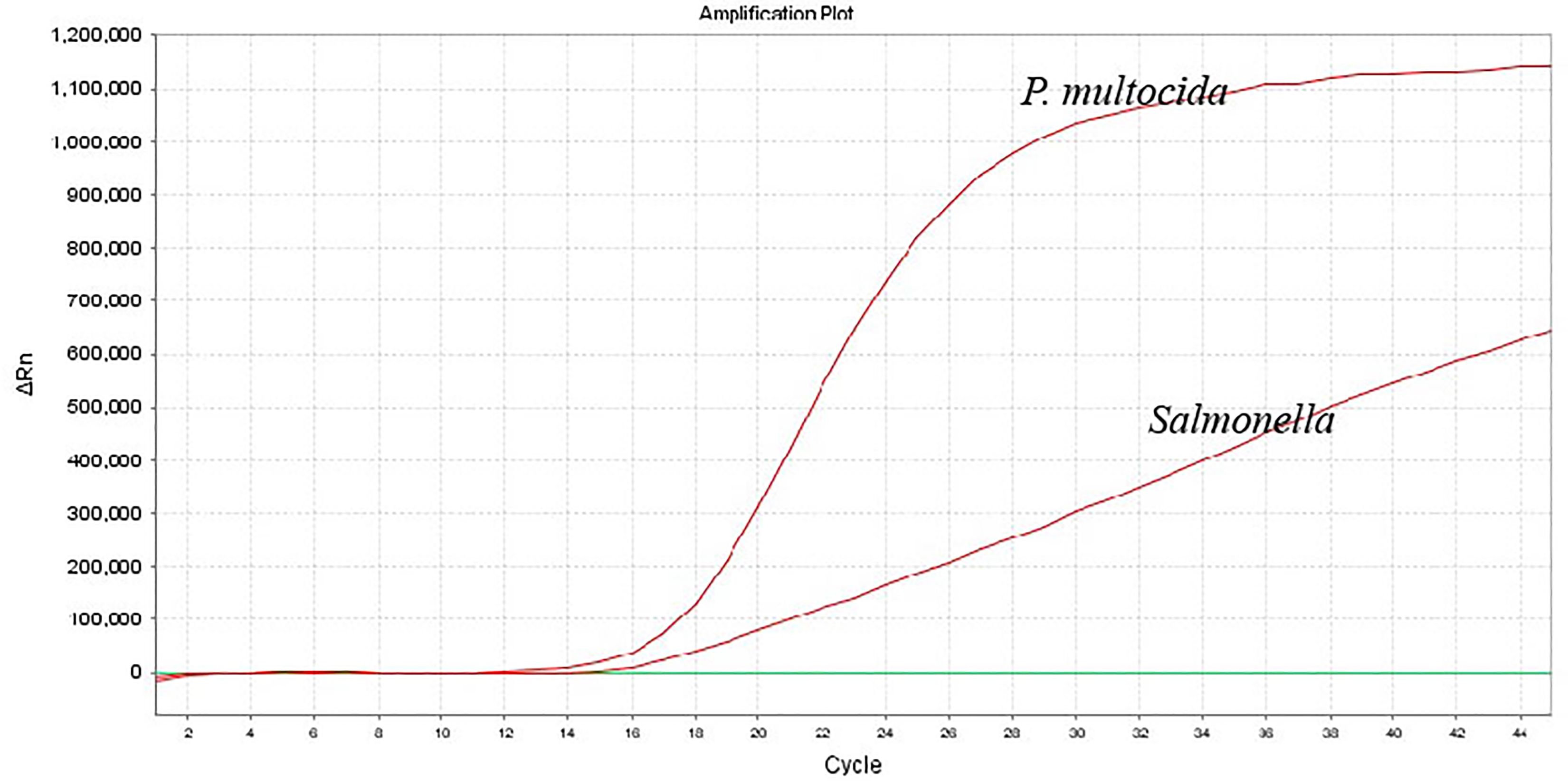
To enhance the amplification efficiency of the dual-fluorescence quantitative PCR, the annealing temperature, number of cycles, as well as the primer and probe concentrations, were optimized using the single-variable control method. The dual fluorescence quantitative PCR assay utilized 25 μL reaction mixture, consisting of 12.5 μL of 2 × Probe qPCR Mix (Takara, Dalian), 0.5 μM of kmt1-forward primer and kmt1-reverse primer, 0.2 μM of kmt1-probe, 0.45 μM of invA-forward primer and invA-reverse primer, 0.3 μM of invA-probe, 2.0 μL of template, add ddH2O to a final volume of 25 µL. When the number of cycles is 45, the amplification efficiency of this method is high and there are no non-specific reactions. The amplification parameters were 95°C for 30 s, and then 45 cycles of 95°C for 5 s and 60°C (annealing temperature and extension temperature) for 34 s. It can specifically amplify P. multocida and Salmonella (Figure 2).
3.2 Standard curves and sensitivity
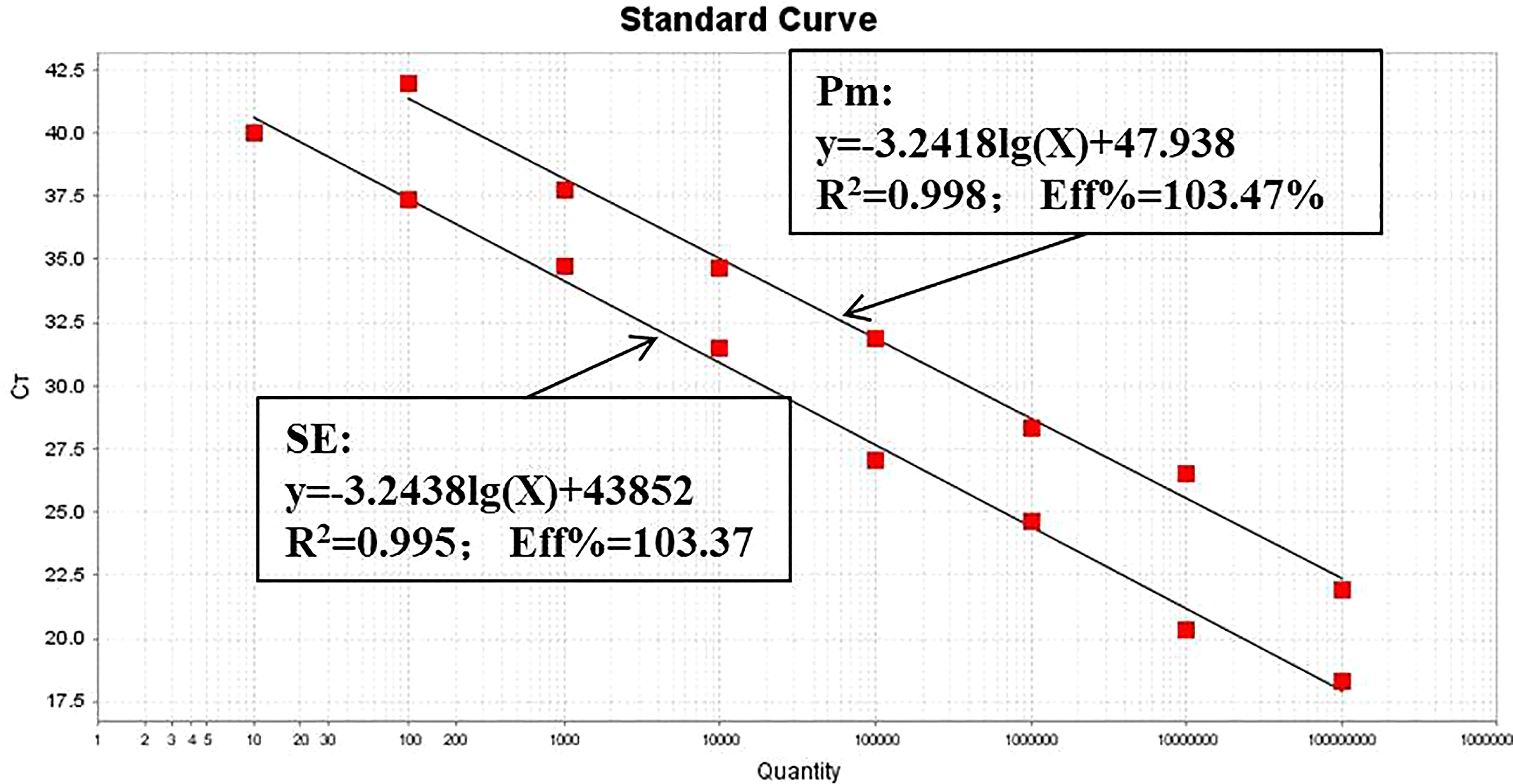
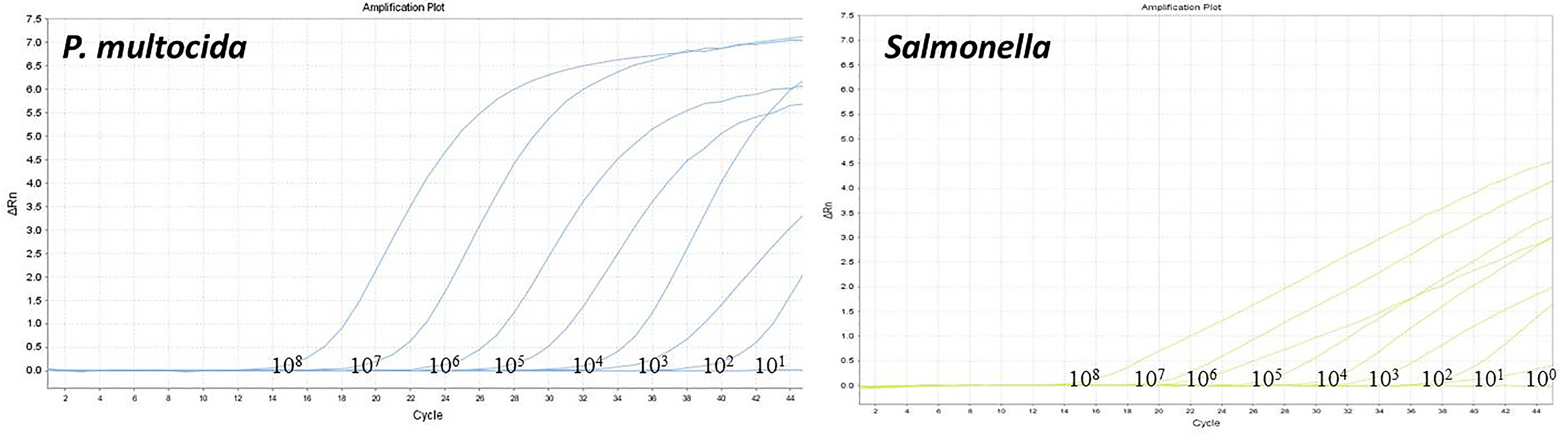
The optimized dual-fluorescence quantitative PCR method was used to amplify standard plasmids at different gradient concentrations. The standard curve of P. multocida was established using 108 to 102 copies, the standard curve of Salmonella was constructed using 108 to 101 copies. Both standard curves demonstrated strong linear relationships, with R² values of 0.995 and 0.996, respectively, both exceeding 0.990. The amplification efficiencies (Eff) were high, at 103.47% and 103.37%, respectively (Figure 3). The limit of detection of the dual-fluorescence quantitative PCR method for the recombinant plasmid standards of P. multocida and Salmonella were 100 copies and 10 copies, respectively (Figure 4).
3.3 Results of specificity
As shown in Figure 5, using the nucleic acids of P. multocida (serotype A, B, and E), Salmonella Dublin, and Salmonella Typhimurium as templates, fluorescence signals and amplification curves were observed in both the FAM and VIC channels. In contrast, no amplification curves or fluorescence signals were detected for the nucleic acids of non-target pathogens such as Mycoplasma bovis, Escherichia coli O157, Staphylococcus aureus, Pseudomonas aeruginosa, IBRV, and Clostridium perfringens.
3.4 Results of the stability and anti-interference tests
As shown in Table 2, no interference was detected in the amplification of high-concentration pathogen against the another low-concentration pathogens. After statistical analysis of the Ct values of the dual fluorescence quantitative PCR amplification curves, it was found that the coefficient of variation for Ct values of P. multocida and Salmonella ranged from 0.23% to 1.51% (Table 3).
3.5 Determination of cut-off value
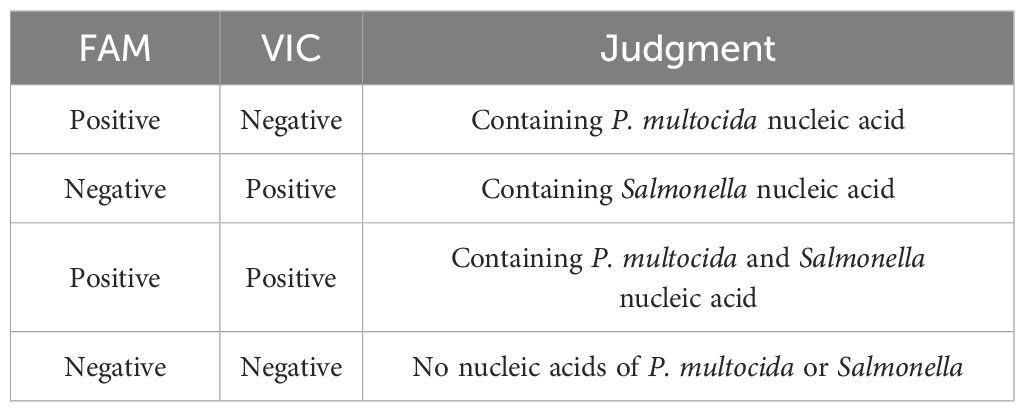
Fifty positive samples and fifty negative samples with known backgrounds were tested. For P. multocida and Salmonella, the sensitivity values were 1.0 and 0.895, respectively, while the specificity values were 0.81 and 1.0, respectively. The areas under the curve (AUC) were 0.944 and 0.968, respectively (Figure 6). The maximum Youden index were 0.81 and 0.895, corresponding to Ct values of 41.0 and 40.5, respectively. The experiment was considered valid when the positive controls for P. multocida and Salmonella (FAM and VIC) displayed typical S-shaped amplification curves, while the negative controls (FAM and VIC) showed no amplification curves and Ct values ≥45 or no values.
3.6 Positive and negative result determination
If a typical S-shaped amplification curve appears in the FAM detection channel and the Ct value is ≤ 41; If the Ct value of the VIC detection channel is ≥ 45 or there is no Ct value, and there is no typical S-shaped amplification curve, it is determined to be P. multocida. If a typical S-shaped amplification curve appears in the VIC detection channel and the Ct value is ≤ 40.5; If the Ct value of the FAM detection channel is ≥ 40 or there is no Ct value, and there is no typical S-shaped amplification curve, it is determined to be Salmonella. If the critical value of the test is ≤ Ct value < 45, it is considered suspicious and it is recommended to perform a double dose retest. If the retest Ct value is<the critical value of the test, it is considered positive. If the retest Ct value is ≥ the critical value of the test, it is considered negative. If the Ct value is ≥ 45 or there is no Ct value and no typical S-type amplification curve, it is judged as negative for P. multocida and Salmonella. The results are summarized in Table 4.
3.7 Detection results of clinical samples
A total of 226 samples were tested using the dual-fluorescence quantitative PCR method established in this study and a previously reported method. The results showed an overall positivity rate of 52.65% (119/226), with a positivity rate of 20.35% (46/226) for P. multocida, 38.50% (87/226) for Salmonella, and a co-infection rate of 6.19% (14/226). The detailed infection status is presented in Figures 7A, B. Additionally, the concordance rate with the previously reported method was 100%.
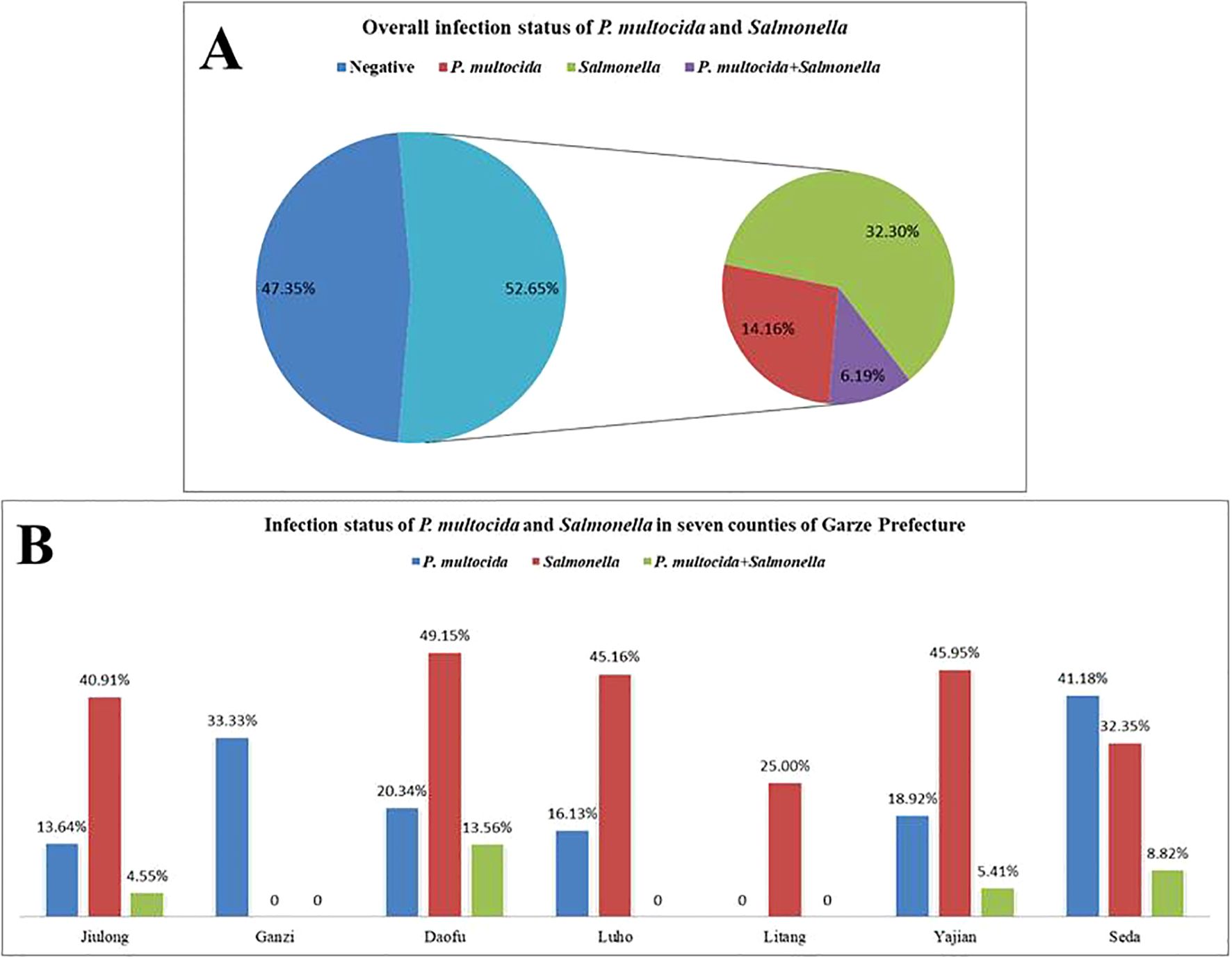
Figure 7. Detection results of clinical samples from seven counties in Ganzi Prefecture. (A) Overall infection status of P.multocida and Salmonella; (B) Infection status of P.multocida and Salmonella in seven counties of Garze Prefecture.
4 Discussion
The yak, a vital economic and ecological resource in high-altitude regions, is essential for local livelihoods and regional sustainability. However, yaks face significant health threats from pathogens like Pasteurella multocida and Salmonella, which cause respiratory diseases, diarrhea, and hemorrhagic septicemia, resulting in substantial economic losses. Additionally, Salmonella poses zoonotic risks, while P. multocida may also threaten human health through environmental exposure. Traditional diagnostic methods, such as pathogen isolation and PCR, are limited by low sensitivity, specificity, and throughput. This underscores the need for a sensitive, specific, and high-throughput duplex real-time fluorescence quantitative PCR (qPCR) assay to improve disease diagnosis and control.
In this study, we developed a duplex qPCR assay targeting the species-specific genes kmt1 of P. multocida and invA of Salmonella. Conserved and specific regions were identified through sequence alignment, and primers and probes were designed accordingly. After optimizing the assay conditions, the assay demonstrated strong specificity, with no cross-reactivity observed with other pathogens, such as Mycoplasma bovis, infectious bovine rhinotracheitis virus and others. The assay exhibited high sensitivity, with detection limits of 100 copies for P. multocida and 10 copies for Salmonella. Compared to the single-plex qPCR assays for P. multocida (50 copies) and Salmonella (100 copies) reported by Pansri et al (Pansri et al., 2020; Pansri et al., 2022), our method achieved equivalent or superior sensitivity. The assay also displayed excellent repeatability and stability, with intra- and inter-assay variations below 2%. Statistical analysis using SPSS and Receiver Operating Characteristic (ROC) curves determined the critical Ct values for detecting P. multocida and Salmonella to be 41.0 and 40.5, respectively. Unlike conventional methods, which rely solely on the presence of amplification curves or Ct values to define positive results potentially leading to false positives due to non-specific amplification (Xin et al., 2023; Mengoli et al., 2009; Cruciani et al., 2023). We used statistical methods to ensure greater accuracy in setting positive and negative thresholds.
Epidemiological studies on P. multocida and Salmonella have been widely reported both domestically and internationally. The prevalence of P. multocida in cattle ranges from 8.35% to 33.01% in regions such as Northeast China and Xinjiang (Almoheer et al., 2022; Wang H. et al., 2023; Zhou et al., 2023), with a prevalence of 38.4% reported in Denmark (Goecke et al., 2021). Salmonella infection rates in livestock vary from 13% to 33.3%, with S. Dublin accounting for 20% of cases (Liu et al., 2022; Wang et al., 2023a; Wang et al., 2023b). In Henan Province, Salmonella was recovered from 21.09% (89/422) of raw milk samples (Liu et al., 2022). In California, USA, the infection rate among cattle was reported to be 6.6%, with Salmonella isolates derived from 60% of water buffalo and 64.28% of dairy cattle (Chen et al., 2019; Fatima et al., 2023). In Ganzi Prefecture, Sichuan Province, yak farming is gradually becoming more intensive and large-scale; however, data on the prevalence of P. multocida and Salmonella in yak populations are limited. In this study, samples were collected from seven counties in Ganzi Prefecture to assess the prevalence of these pathogens. The results showed that the single infection rates for P. multocida and Salmonella were 20.35% (46/226) and 38.50% (87/226), respectively, with a co-infection rate of 6.19% (14/226). These pathogens were found to be prevalent to varying degrees across all counties. Importantly, the results obtained using the developed duplex qPCR assay were consistent with those from previously reported methods, confirming the reliability and applicability of this assay.
5 Conclusion
In this study, we successfully developed a highly sensitive, specific, rapid, duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of P. multocida and Salmonella in yaks. This method offers robust technical support for the diagnosis and control of P. multocida-induced pasteurellosis and Salmonella-associated diseases. Furthermore, the application of this detection method to clinical samples from various counties in Ganzi Prefecture has supplemented the epidemiological data on the prevalence of Pasteurella multocida and Salmonella in local yak populations. These findings are of significant value for the prevention and control of these diseases, contributing to the sustainable development of yak farming in high-altitude regions.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
YP: Writing – review & editing, Project administration, Methodology, Writing – original draft, Data curation, Conceptualization. QY: Resources, Conceptualization, Project administration, Data curation, Writing – original draft, Software. QW: Supervision, Data curation, Investigation, Writing – original draft, Formal Analysis. QL: Investigation, Writing – original draft, Conceptualization, Data curation. WT: Data curation, Investigation, Methodology, Writing – original draft, Software. LX: Data curation, Methodology, Conceptualization, Software, Validation, Writing – original draft. XH: Writing – original draft, Supervision, Investigation, Project administration, Data curation. HX: Project administration, Writing – original draft, Data curation, Investigation. YL: Investigation, Formal Analysis, Validation, Methodology, Writing – original draft. LD: Resources, Data curation, Project administration, Investigation, Writing – original draft. LL: Visualization, Formal Analysis, Writing – original draft, Methodology, Investigation, Writing – review & editing. LZ: Conceptualization, Validation, Methodology, Data curation, Supervision, Writing – review & editing, Writing – original draft. JW: Writing – review & editing, Writing – original draft, Formal Analysis, Resources, Data curation, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was supported by grants from the Southwest University for Nationalities and Ganzi Prefecture Cooperation Project - Key Technologies and Integrated Application Research for Efficient Yak Breeding, Sichuan Province Transfer Payment Science and Technology Plan Project (220022), the National Key Research and Development Program of China 2022YFD1800703), and the Third Batch of Public Welfare Special Projects of China Veterinary Drug Administration (GY202403).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, Y., Yuan, R., Jin, S., Lin, W., and Zhang, Y. (2024). Understanding consumer preferences for attributes of yak meat: implications for economic growth and resource efficiency in pastoral areas. Meat Sci. 216, 109586. doi: 10.1016/j.meatsci.2024.109586
Almoheer, R., Abd Wahid, M. E., Zakaria, H. A., Jonet, M. A. B., Al-Shaibani, M. M., Al-Gheethi, A., et al. (2022). Spatial, temporal, and demographic patterns in the prevalence of hemorrhagic septicemia in 41 countries in 2005-2019: A systematic analysis with special focus on the potential development of a new-generation vaccine. Vaccines (Basel) 10. doi: 10.3390/vaccines10020315
Ayuti, S. R., Khairullah, A. R., Al-Arif, M. A., Lamid, M., Warsito, S. H., Moses, I. B., et al. (2024). Tackling salmonellosis: A comprehensive exploration of risks factors, impacts, and solutions. Open Vet J. 14, 1313–1329. doi: 10.5455/OVJ.2024.v14.i6.1
Bahr, A. D. and Abdel, M. E. (2024). Susceptibility of Pasteurella multocida isolated from cattle in Egypt to antibiotics, silver, chitosan and curcumin nanoparticles. Vet Res. Forum 15, 455–462. doi: 10.30466/vrf.2024.2017140.4090
Barraclough, K. (2012). Diagnosis: shifting the ROC curve. Br. J. Gen. Pract. 62, 452–453. doi: 10.3399/bjgp12X653796
Chen, Z., Biswas, S., Aminabadi, P., Stackhouse, J. W., Jay-Russell, M. T., and Pandey, P. K. (2019). Prevalence of Escherichia coli O157 and Salmonella spp. in solid bovine manure in California using real-time quantitative PCR. Lett. Appl. Microbiol 69, 23–29. doi: 10.1111/lam.13156
Craig, M. J., Cummings, K. J., Aprea, M. S., Franklin-Guild, R. J., and Altier, C. (2024). Serotype and anti-microbial resistance trends among bovine Salmonella isolates from samples submitted to a veterinary diagnostic laboratory in central New York, 2007-2021. Zoonoses Public Health 71, 359–368. doi: 10.1111/zph.13108
Cruciani, M., White, P. L., Barnes, R. A., Loeffler, J, Donnelly, J. P., Rogers, T. R., et al. (2023). An overview of systematic reviews of polymerase chain reaction (PCR) for the diagnosis of invasive aspergillosis in immunocompromised people: A report of the fungal PCR initiative (FPCRI)-an ISHAM working group. J. Fungi (Basel) 9. doi: 10.3390/jof9100967
Demirci, M., Yigin, A., Altun, S. K., Uysal, H. K., Saribas, S., and Kocazeybek, B. S. (2019). Salmonella Spp. and Shigella Spp. detection via multiplex real-time PCR and discrimination via MALDI-TOF MS in different animal raw milk samples. Niger J. Clin. Pract. 22, 1083–1090. doi: 10.4103/njcp.njcp_596_18
Fatima, A., Saleem, M., Nawaz, S., Khalid, L., Riaz, S., and Sajid, I. (2023). Prevalence and antibiotics resistance status of Salmonella in raw meat consumed in various areas of Lahore, Pakistan. Sci. Rep. 13, 22205. doi: 10.1038/s41598-023-49487-2
Goecke, N. B., Nielsen, B. H., Petersen, M. B., and Larsen, L. E. (2021). Design of a high-throughput real-time PCR system for detection of bovine respiratory and enteric pathogens. Front. Vet Sci. 8, 677993. doi: 10.3389/fvets.2021.677993
Grote, A., Piscon, B., Manson, A. L., Adani, B., Cohen, H., and Livny, J.. (2024). Persistent Salmonella infections in humans are associated with mutations in the BarA/SirA regulatory pathway. Cell Host Microbe 32, 79–92.e7. doi: 10.1016/j.chom.2023.12.001
Islam, E. A., Fegan, J. E., Tefera, T. A., Curran, D. M., Waeckerlin, R. C., Ng, D., et al. (2023). Reverse vaccinology-based identification of a novel surface lipoprotein that is an effective vaccine antigen against bovine infections caused by Pasteurella multocida. PloS Pathog 19, e1011249. doi: 10.1371/journal.ppat.1011249
Krupinski, E. A. (2021). Evaluating AI clinically-it’s not just ROC AUC! Radiology 298, 47–48. doi: 10.1148/radiol.2020203782
Liu, B. G., Xie, M., Gong, Y. T., Dong, Y., Zheng, G. M., Wu, H., et al. (2022). Prevalence, resistance phenotypes, and fluoroquinolone resistance genes of Salmonella isolates from raw milk of healthy dairy cows in Henan province, China. Eur. Rev. Med. Pharmacol. Sci. 26, 6837–6844. doi: 10.26355/eurrev_202209_29786
Lv, K., Zhang, Y., Yang, Y., Liu, Z., and Deng, L. (2022). Development of Nested PCR and Duplex Real-Time Fluorescence Quantitative PCR Assay for the Simultaneous Detection of Theileria equi and Babesia caballi. Front. Vet Sci. 9, 873190. doi: 10.3389/fvets.2022.873190
Mahboob, S., Iqbal, M., and Rahman, M. (2023). Development of ELISA-based diagnostic methods for the detection of haemorrhagic septicaemia in animals. J. Microbiol Methods 204, 106652. doi: 10.1016/j.mimet.2022.106652
Mengoli, C., Cruciani, M., Barnes, R. A., Loeffler, J., and Donnelly, J. P. (2009). Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9, 89–96. doi: 10.1016/S1473-3099(09)70019-2
Mo, Q., Nawaz, S., Kulyar, M. F., Li, K., Li, Y., Zhang, Z., et al. (2024). Exploring the intricacies of Pasteurella multocida dynamics in high-altitude livestock and its consequences for bovine health: A personal exploration of the yak paradox. Microb Pathog 194, 106799. doi: 10.1016/j.micpath.2024.106799
Mohler, V. L., Izzo, M. M., and House, J. K. (2009). Salmonella in calves. Vet Clin. North Am. Food Anim Pract. 25, 37–54. doi: 10.1016/j.cvfa.2008.10.009
Otten, N. D., Goecke, N. B., Michelsen, A. M., Nielsen, L. R., Capion, N., Martin, H. L., et al. (2024). Comparing occurrence of bovine respiratory pathogens detected by high-throughput real-time PCR in nasal swabs and non-endoscopic bronchoalveolar lavage samples from dairy and veal calves. Pathogens 13. doi: 10.3390/pathogens13060479
Pansri, P., Katholm, J., Krogh, K. M., Aagaard, A. K., Schmidt, L. M. B., Kudirkiene, E., et al. (2020). Evaluation of novel multiplex qPCR assays for diagnosis of pathogens associated with the bovine respiratory disease complex. Vet J. 256, 105425. doi: 10.1016/j.tvjl.2020.105425
Pansri, P., Svensmark, B., Liu, G., Thamsborg, S. M., Kudirkiene, E., Nielsen, H. V., et al. (2022). Evaluation of a novel multiplex qPCR method for rapid detection and quantification of pathogens associated with calf diarrhoea. J. Appl. Microbiol 133, 2516–2527. doi: 10.1111/jam.15722
Pantoja-Galicia, N., Okereke, O. I., Blacker, D., and Betensky, R. A. (2021). Concordance measures and time-dependent ROC methods. Biostat Epidemiol. 5, 232–249. doi: 10.1080/24709360.2021.1926189
Piorunek, M., Brajer-Luftmann, B., and Walkowiak, J. (2023a). Pasteurella multocida infection in humans. Pathogens 12. doi: 10.3390/pathogens12101210
Piorunek, M., Brajer-Luftmann, B., Trafas, T., Schneider, A., and Walkowiak, J. (2023b). Lower respiratory infection in humans caused by pasteurella multocida. Respir. Physiol. Neurobiol. 315, 104091. doi: 10.1016/j.resp.2023.104091
Ran, X., Li, X., Xie, X., Lei, J., Yang, F., Chen, D., et al. (2024). Effects of probiotic enterococcus faecium from yak on the intestinal microflora and metabolomics of mice with salmonella infection. Probiotics Antimicrob Proteins 16, 1036–1051. doi: 10.1007/s12602-023-10102-5
Shaukat, W., de Jong, E., McCubbin, K. D., Biesheuvel, M. M., van der Meer, F. J. U. M., De Buck, J., et al. (2024). Herd-level prevalence of bovine leukemia virus, Salmonella Dublin, and Neospora caninum in Alberta, Canada, dairy herds using ELISA on bulk tank milk samples. J. Dairy Sci. 107, 8313–8328. doi: 10.3168/jds.2023-24611
Shehta, A., El-Zahar, H., Mansour, A., Mustafa, B., and Shety, T. (2022). Clinical, hematological and some biochemical alterations during diarrhea in Friesian calves naturally infected with E. coli and Salmonella. Beni Suef Univ J. Basic Appl. Sci. 11, 128. doi: 10.1186/s43088-022-00309-w
Tiwari, M., Gujar, G., Shashank, C. G., and Ponsuksili, S. (2024). Selection signatures for high altitude adaptation in livestock: A review. Gene 927, 148757. doi: 10.1016/j.gene.2024.148757
Wang, H., Xin, L., Wu, Y., Liu, Y., Yao, W., Zhang, H., et al. (2023). Construction of a one-step multiplex real-time PCR assay for the detection of serogroups A, B, and E of Pasteurella multocida associated with bovine pasteurellosis. Front. Vet Sci. 10, 1193162. doi: 10.3389/fvets.2023.1193162
Wang, J., Zhu, X., Zhao, Y., Liu, H., Zhang, Z., Yan, L., et al. (2023a). Prevalence and antimicrobial resistance of Salmonella and ESBL E. coli isolated from dairy cattle in Henan Province, China. Prev. Vet Med. 213, 105856. doi: 10.1016/j.prevetmed.2023.105856
Wang, J., Zhu, X., Wang, Z., Chen, Y., Robertson, I. D., Guo, A., et al. (2023b). Prevalence and antimicrobial resistance of Salmonella and the enumeration of ESBL E. coli in dairy farms in Hubei Province. China. Prev. Vet Med. 212, 105822. doi: 10.1016/j.prevetmed.2022.105822
Xin, L., Wang, H., Hu, Y., Liu, Y., Yao, W., Wang, X., et al. (2023). The establishment and application of a one-step multiplex real‐time polymerase chain reaction assay for the detection of Streptococcus suis, Streptococcus suis serotype 2, and Glaesserella parasuis. Animal Research and One Health, 1–12. doi: 10.1002/aro2.37
Zheng, H. H., Zhang, S. J., Cui, J. T., Zhang, J., Wang, L., Liu, F., et al. (2020). Simultaneous detection of classical swine fever virus and porcine circovirus 3 by SYBR green I-based duplex real-time fluorescence quantitative PCR. Mol. Cell Probes 50, 101524. doi: 10.1016/j.mcp.2020.101524
Keywords: yak, Pasteurella multocida, Salmonella, duplex real-time fluorescence PCR, detection
Citation: Pan Y, Yu Q, Wang Q, Li Q, Tian W, Xin L, Hu X, Xiao H, Liu Y, Zhu LRD, Lan L, Zhu L and Wu J (2025) Establishment and epidemiological investigation of a dual fluorescent qPCR assay for Pasteurella multocida and Salmonella in yaks in the Tibetan Autonomous Prefecture of Garzê, China. Front. Cell. Infect. Microbiol. 15:1599817. doi: 10.3389/fcimb.2025.1599817
Received: 25 March 2025; Accepted: 14 May 2025;
Published: 04 June 2025.
Edited by:
Qiang Zhang, Huazhong Agricultural University, ChinaReviewed by:
Zhang Yu, South China Agricultural University, ChinaZhiyu Zhou, National Institutes for Food and Drug Control, China
Copyright © 2025 Pan, Yu, Wang, Li, Tian, Xin, Hu, Xiao, Liu, Zhu, Lan, Zhu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Wu, MjU4NjI4OTU1QHFxLmNvbQ==; Liangquan Zhu, MTM2NzM5MTg5NEBxcS5jb20=; Lan Lan, ODgyNDAyNzNAcXEuY29t
 Yao Pan
Yao Pan Qingting Yu1
Qingting Yu1 Qiang Li
Qiang Li Liangquan Zhu
Liangquan Zhu