- 1National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, National Center for Tuberculosis Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Beijing Key Laboratory of Pediatric Respiratory Infection Diseases, Key Laboratory of Major Diseases in Children, Ministry of Education, National Clinical Research Center for Respiratory Diseases, Laboratory of Respiratory Diseases, Beijing Pediatric Research Institute, Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health, Beijing, China
- 3Microbial Inspection Department, Changping District Center for Disease Control and Prevention, Beijing, China
- 4Center for Accurate Detection of Tuberculosis, Tianjin Haihe Hospital, Tianjin, China
Objective: Given the increase of treatment failure, relapse and acquired resistance observed in isoniazid (INH) resistance, there is an urgent to improve rifampin (RIF) -priority based diagnostic strategies. Therefore, we evaluated the performance of Innovo GenMax MTB-RIF/INH (GenMax), a moderate- complexity automated nucleic acid amplification test (NAAT), for detecting Mycobacterium tuberculosis complex (MTBC) and resistance to RIF and INH.
Methods: Analytical sensitivity (limit of detection, LOD) was determined using serial dilutions of Mycobacterium tuberculosis H37Rv (ATCC 27249) strains. Diagnostic accuracy was assessed in clinical sputum specimens against microbiological reference standards (MRS: positive by smear microscopy, culture or Xpert MTB/RIF for diagnosis of TB) and phenotypic drug susceptibility testing (DST). Discordant results were resolved by sequencing resistance genes (IS6110, rpoB, katG, inhA, ahpC) and follow-up diagnosis results.
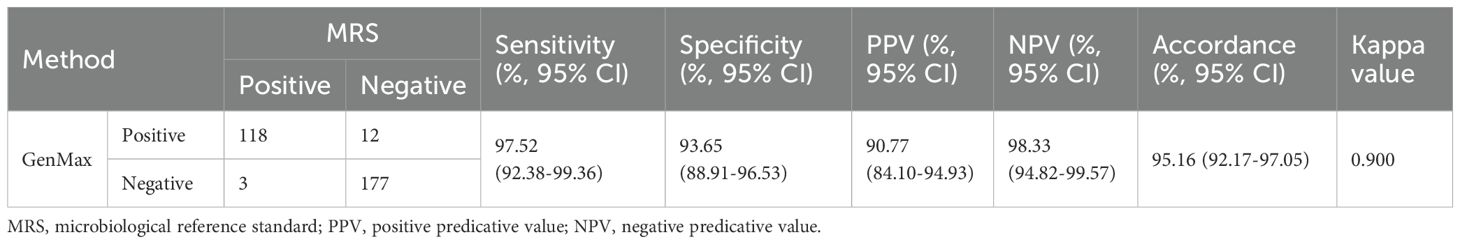
Results: GenMax demonstrated a calculated LOD of 8.8 CFU/mL (95% CI: 7.4-11.4) for MTBC, 674.1 CFU/mL (95% CI: 578.8-923.5) for RIF resistance, and 747.3 CFU/mL (95% CI: 613.7-1081.3) for INH resistance. In clinical evaluation, the sensitivity and specificity for MTBC detection were 97.52% (95% CI: 92.38–99.36) and 93.65% (95% CI: 88.91–96.53), respectively. For RIF and INH resistance, sensitivities were 88.46% (95% CI: 68.72–96.97) and 85.19% (95% CI: 65.39–95.14), with specificity of 92.42% (95% CI: 82.50-97.18) and 94.12% (95% CI: 84.86-98.10).
Conclusion: Innovo GenMax MTB-RIF/INH is a rapid and automated assay with high sensitivity for MTBC detection, suitable for decentralized settings. While its performance for RIF/INH resistance detection is competitive with existing assays, its sensitivity remains gaps relative to WHO targets. Further optimization, particularly through expanded probe coverage, is needed to bridge this gap and ensure reliable detection in clinical settings.
1 Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis complex (MTBC), remains a significant public health challenge in China, which is among the 30 high-burden TB countries globally. In 2023, China was estimated with approximately 741,000 incident TB cases, including 29,000 cases of multidrug-resistant or rifampicin-resistant TB (MDR/RR-TB) (World Health Organization, 2024b). However, only 52.9% MDR/RR-TB cases were identified by genotypic or phenotypic methods in 2023 (World Health Organization, 2024b). Moreover, isoniazid (INH) resistance testing is often restricted to rifampicin-resistant cases in China, leaving isoniazid mono-resistant TB under-diagnosed and mismanaged (Liu et al., 2024). Previous reports revealed that isoniazid resistance affects approximately 11% TB patients nationally (Zhao et al., 2012), while 7.8% culture positive cases were isoniazid-resistant/rifampin-susceptible (Hr-Rs) (Liu et al., 2024). As isoniazid resistance is associated with an increase of treatment failure, relapse and acquired resistance (Gegia et al., 2017; Menzies et al., 2009), it is crucial to improve rifampin-priority based diagnostic strategies, requiring decentralized, rapid diagnostic tools capable of detecting both rifampin and isoniazid resistance, which is critical for clinical decisions making and improved treatment outcomes in Hr-Rs TB patients.
The InnowaveDX MTB/RIF (InnowaveDX company, Suzhou, China), a real-time PCR assay, targeting MTB and rifampicin resistance-determining region (RRDR), demonstrated high accuracy for MTBC and rifampin resistance detection (Deng et al., 2023; Fan et al., 2023). Innovo GenMax MTB-RIF/INH (GenMax), an updated version, expands diagnostic capabilities by incorporating isoniazid resistance with mutations in the promoter of inhA (-18–5), the promoter of ahpC (-15–2), and the 315 codon of katG. This study aimed to evaluate the performance of Innovo GenMax MTB-RIF/INH, which are designed to be implemented in peripheral laboratories with limited infrastructure.
2 Methods
2.1 Participants
Sputum samples were collected from 310 patients presenting with TB symptoms or abnormal chest x-ray results at Tuberculosis Prevention and Control Institute of Changping District and Tianjin Haihe Hospital from February to July in 2024, which were stored in biobank in National Tuberculosis Reference Laboratory. Patients who have received ≥ 2 weeks of anti-tuberculosis treatment at enrollment were excluded to avoid selective pressure induced new resistance mutations, ensure reliable phenotype-genotype correlation, avoid false positives from non-viable bacterial nucleic acids, and patients lacked sputum production capacity were also excluded.
2.2 Microbiological reference standard
All sputum samples were subjected to Ziehl-Neelsen staining directly to confirm acid-fast bacilli rapidly (Steingart et al., 2006). Then the specimens were digested in N-acetyl-L-cysteine NaOH-Na citrate (1.5% final concentration) and neutralized with phosphate buffer (PBS, 0.067 mol/L, pH = 7.4), followed by incubation into the Bactec MGIT 960 system for 6 weeks mycobacterial culture to improve diagnostic accuracy (Hanna et al., 1999). Positive cultures were further subjected to MPT64 antigen detection (Park et al., 2009). For Xpert MTB/RIF assay, a rapid nucleic acid amplification test recommended by the WHO for initial diagnosis of TB with drug resistance, 1 mL of the processed specimen was mixed with 2 mL sample reagent, incubated at room temperature for 10 min, and then transferred into cartridges for analysis using the GeneXpert instrument (Boehme et al., 2010). Microbiological reference standard (MRS) was defined as positive by smear microscopy, culture or Xpert MTB/RIF for the diagnosis of TB (World Health Organization, 2024a).
2.3 Phenotypic drug susceptibility testing
The 1% proportion method on solid L-J medium was used for testing of susceptibility to rifampin and isoniazid. Critical concentration was 40 μg/mL for rifampin and 0.2 μg/mL for isoniazid as recommended by WHO (WHO, 2008).
2.4 Innovo GenMax MTB-RIF/INH
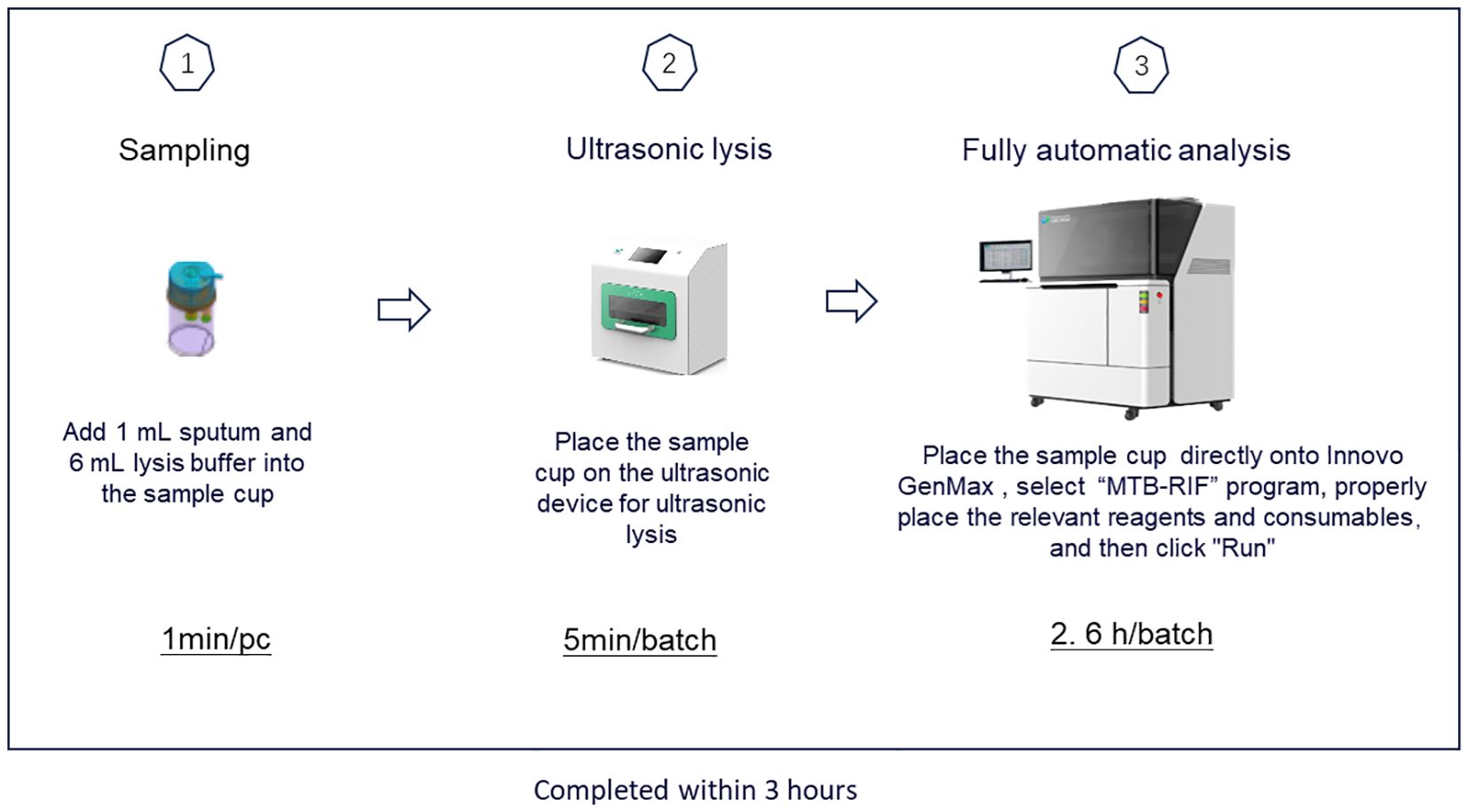
GenMax was operated according to the manufacture’s instruction. Briefly, 1mL sputum was added into the pretreatment tube with 6 mL lysis solution. The pretreated mixed sputum was proceeding with ultrasonic instrument for 5 minutes for DNA extraction, and then uploaded into the GenMax instrument installed with specific software. The results were read and interpreted according to the manual. The whole procedure takes around 3 hours (Figure 1).
2.5 Sequencing of fragments of IS6110, rpoB, ahpC, inhA, and katG gene
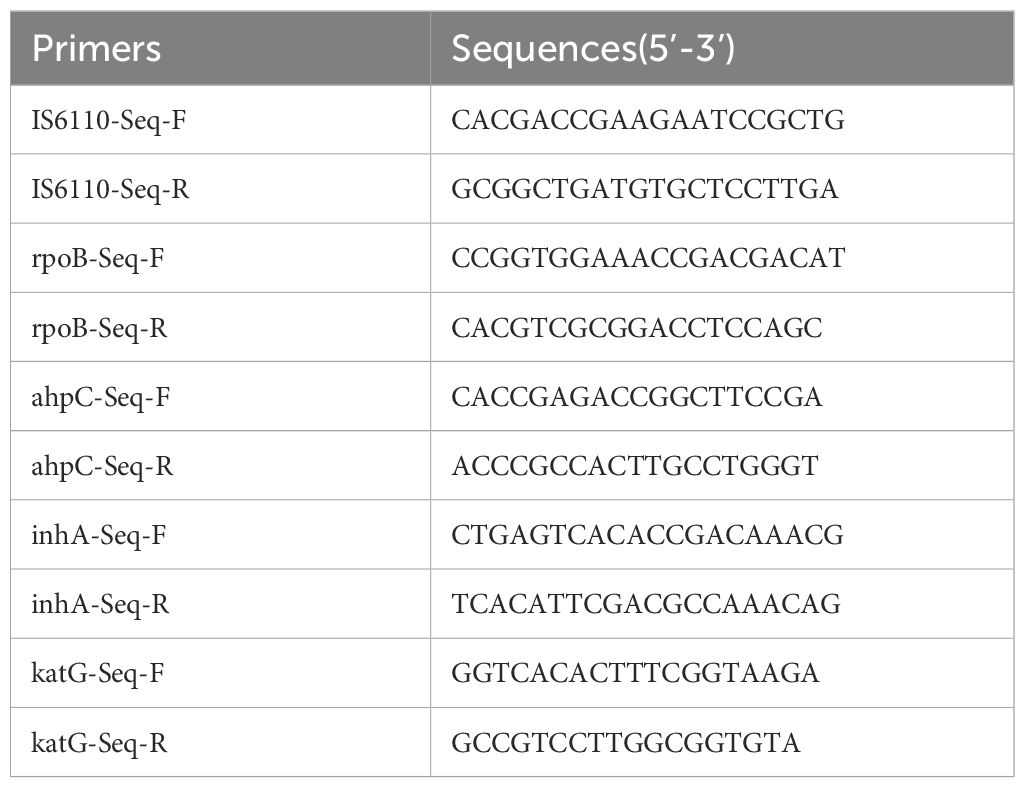
Boiling method was used for crude DNA extraction from sputum for sequencing. The DNA was amplified with primers shown in Table 1 and subjected to sequencing for IS6110, rpoB, ahpC, inhA, and katG gene fragments. The sequencing results were compared with the H37Rv sequence.
2.6 Limitation of detection and analytic specificity
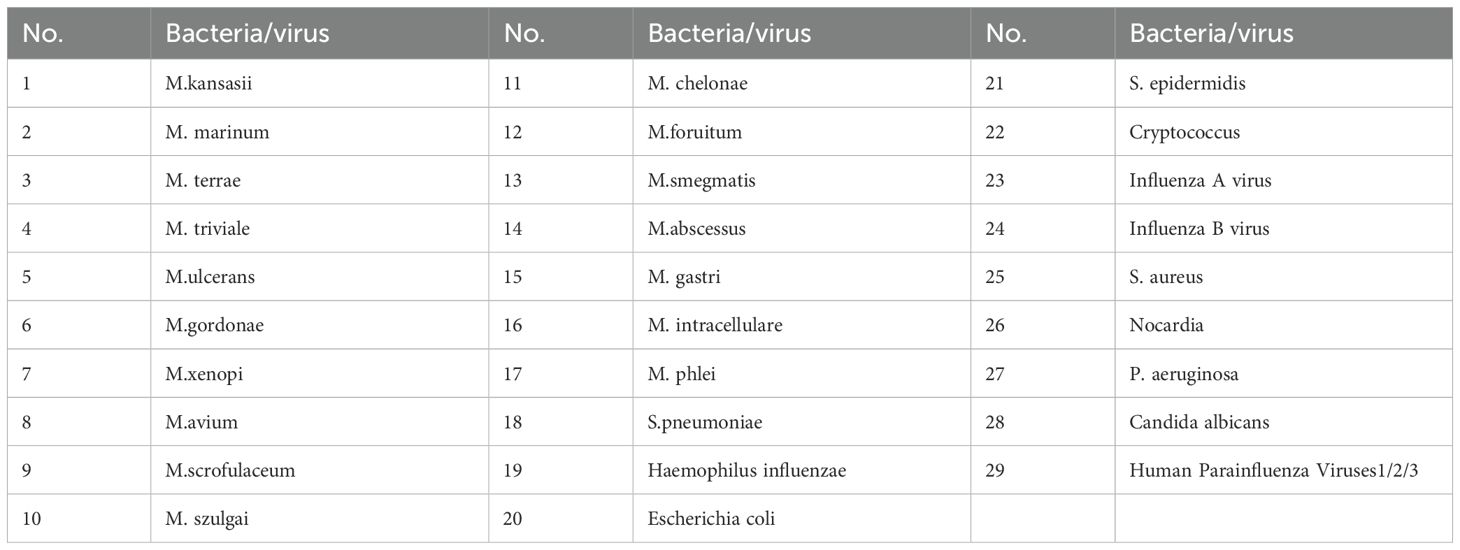
The limit of detection (LOD) was determined by diluted Mycobacterium tuberculosis H37Rv (ATCC 27249) strain with 1×103 CFU/mL at a series of concentrations (0.125 CFU/mL, 1.25 CFU/mL, 2.5 CFU/mL, 5 CFU/mL, 10 CFU/mL, 20 CFU/mL). The LOD for rifampicin and isoniazid resistance was performed by diluted with mono-rifampin resistant or mono-isoniazid resistant MTB strain with 1 × 105 CFU/mL at a series of concentrations (2000 CFU/mL, 1000 CFU/mL, 500 CFU/mL, 250 CFU/mL, 25 CFU/mL). Each sample with a defined dilution was tested 20 replicates. In addition, the analytical specificity was tested using (approximately 104 CFU/ml) strains of 17 different species of nontuberculous mycobacteria (NTM) and 12 other common bacteria or virus (Table 2).
2.7 Data analysis
SPSS version 20.0 (IBM, Chicago, IL) software was used. The diagnostic accuracy of the GenMax assay was described as point estimates and 95% confidence intervals (95% CIs). The consistency between GenMax and MRS for MTB detection, and accordance between GenMax and phenotypic DST for rifampin and isoniazid resistance was conducted with Kappa analysis. For calculation of the LOD values, the percentages of the replicates resulting in successful TB detection and rifampin/isoniazid resistance were calculated at each input CFU concentration in suspensions. Probit analysis was used to generate the curve through the tested concentrations, and lower and upper 95% confidence intervals (95% CIs).
3 Results
3.1 The limit of detection and analytic specificity
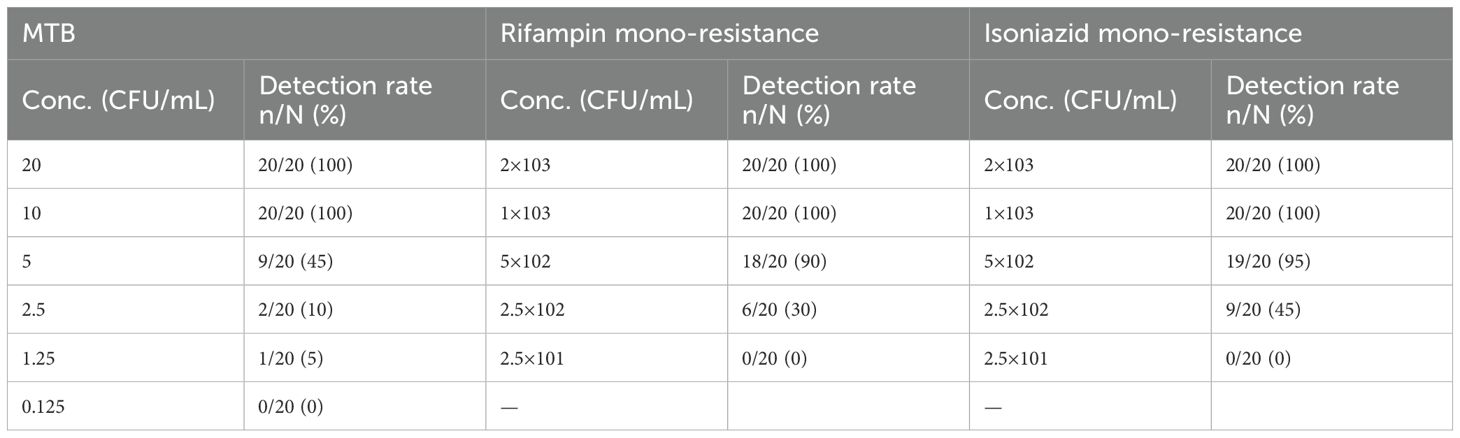
GenMax demonstrated 100% accuracy in detecting the target MTB strain across all tested samples, with a detection limit as low as 10 CFU/mL. At dilutions below 10 CFU/mL, correct detection rate decreased to 45% for 5 CFU/mL, 10% for 2.5 CFU/mL, and 5% for 1.25 CFU/mL (Table 3). The calculated limitation of detection (LOD) was 8.8 CFU/mL (95% CI 7.4-11.4). Correct detection of rifampin mono-resistant strains by GenMax was 100% down to 1×103 CFU/mL, which decreased to 90% for 5×102 CFU/mL, and 30% for 2.5×102 CFU/mL. The calculated rifampin resistance detection limit was 674.1 CFU/mL (95% CI: 578.8- 923.5). As was the case with isoniazid resistance detection limit, 100% corrective detection was down to 1×103 CFU/mL and 95% for 5×102 CFU/mL. The calculated detection limit of isoniazid resistance was 747.3 CFU/mL (95% CI: 613.7-1081.3) (Figure 2). For specificity, none of the tested NTM, bacteria or virus strains were detected as Mycobacterium tuberculosis.
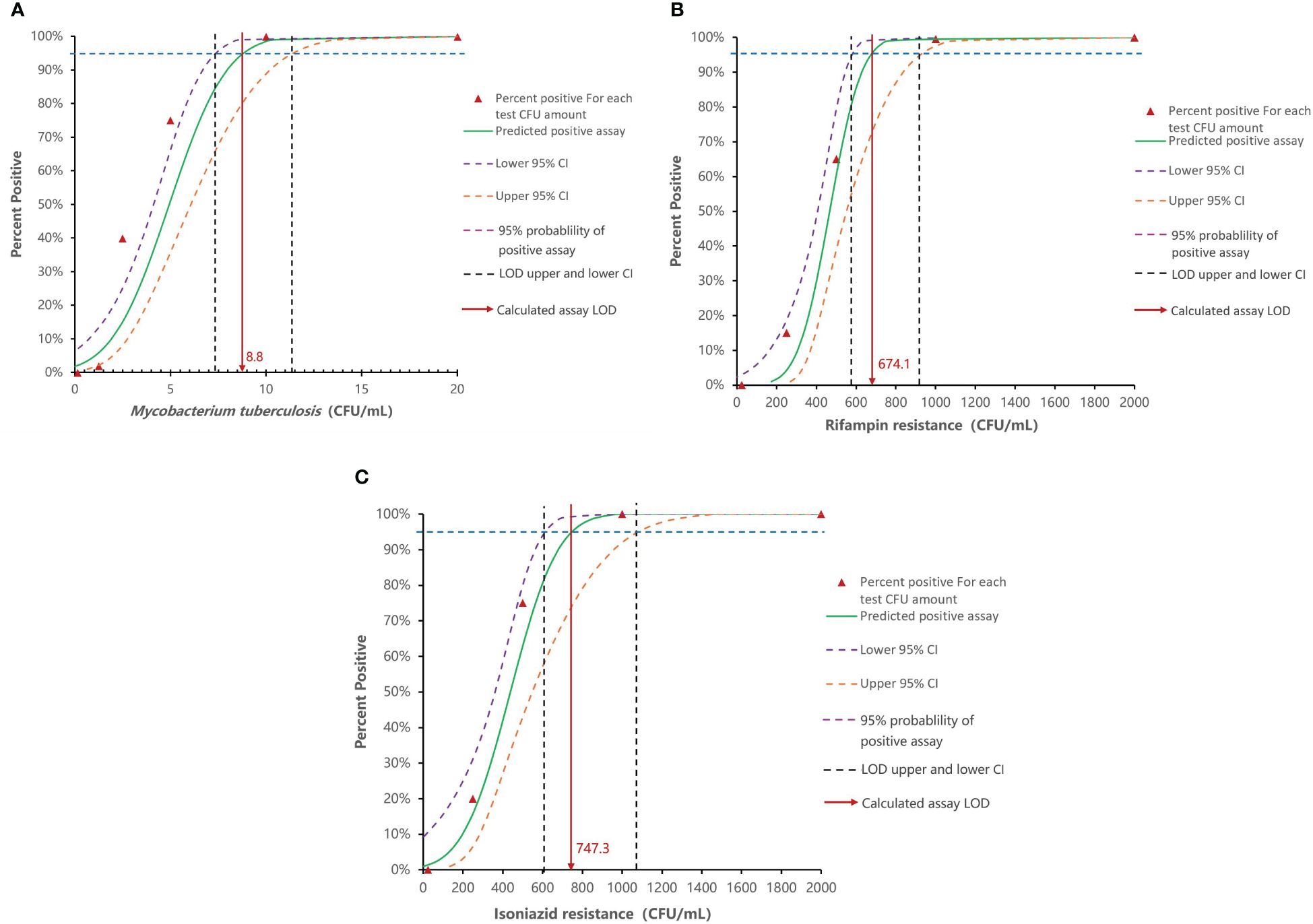
Figure 2. Analytical sensitivity of Innovo GenMax MTB-RIF/INH. (A) MTB detection limit (8.8 CFU/mL, 95% CI: 7.4–11.4). (B) Rifampin resistance detection (674.4 CFU/mL, 95% CI: 578.8- 923.5). (C) Isoniazid resistance detection (747.3 CFU/mL, 95% CI: 613.7-1081.3).
3.2 Diagnostic accuracy of GenMax for pulmonary TB compared with MRS
A total of 310 cases with presumed pulmonary TB were analyzed. 121 cases were microbiological confirmed by smear microscopy, culture or Xpert MTB/RIF. The sensitivity and specificity of GenMax for the detection of MTBC were 97.52% (95% CI: 92.38-99.36) and 93.65% (95% CI: 88.91-96.53) relative to the MRS, respectively. The PPV and NPV of GenMax was 90.77% (95% CI: 84.10-94.93) and 98.33% (95% CI: 94.82-99.57), respectively (Table 4). Among 12 MRS-negative/GenMax-positive samples, IS6110 sequencing successfully confirmed GenMax results in 8 cases (66.7%), while 4 samples failed of sequencing due to insufficient DNA concentration in the specimen. Follow-up data revealed that 6/12 were bacteriologically confirmed cases, 4/12 cases had previous TB history, and 2 cases lacked subsequent clinical results. 3 MRS-positive/GenMax-negative samples were all XpertMTB/RIF positive.
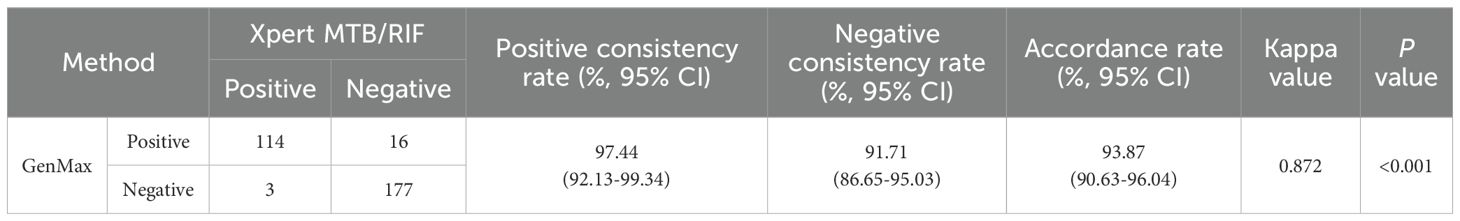
3.3 Diagnostic accuracy of GenMax for pulmonary TB compared with XpertMTB/RIF
When conducting a head-to-head comparison between GenMax and XpertMTB/RIF, the accordance rate was 93.87% (291/310), with kappa value 0.872 (Table 5). Among 16 specimens showing Xpert MTB/RIF-negative/GenMax-positive discordant results, MTBC were confirmed in 4 cases through culture. Subsequent IS6110 sequencing were performed on 12 smear-negative and culture-negative samples, yielding positive results in 8 samples, while sequencing failed in 4 specimens due to insufficient DNA concentration in sputum. Follow-up results showed that 6 of these 12 initially microscopy/culture-negative cases were bacteriologically confirmed during subsequent following-up. 4/12 cases had a previous TB history, while follow-up information was unavailable for 2 cases.
3.4 Diagnostic accuracy of GenMax for detection of rifampin and isoniazid resistance
92 phenotypic susceptibility results were available for comparison of the diagnostic accuracy of rifampin resistance (Table 6). The sensitivity of GenMax and Xpert MTB/RIF in detection of rifampin resistance was 88.46% (95% CI: 68.72-96.97) and 84.62% (95% CI: 64.27-94.95), respectively, with the same specificity of 92.42% (95% CI: 82.50-97.18). 3 samples with GenMax-susceptible/phenotypic-resistant were consistently classified as susceptible by both Xpert MTB/RIF and rpoB sequencing. Conversely, among five specimens showing GenMax-resistant/phenotypic-susceptible, sequencing revealed RIF resistance-associated with mutations in all cases. A total of 95 cases underwent parallel phenotypic DST and GenMax test. When compared against phenotypic DST, GenMax demonstrated 85.19% (95% CI: 65.39-95.14) sensitivity (23/27) and 94.12% (95% CI: 84.86-98.10) specificity (64/68) for INH resistance detection. The overall concordance rate between GenMax and phenotypic DST reached 91.58% (95% CI: 83.76-95.52). Notably, genetic sequencing of the 8 discordant cases revealed complete alignment with GenMax results (8/8).
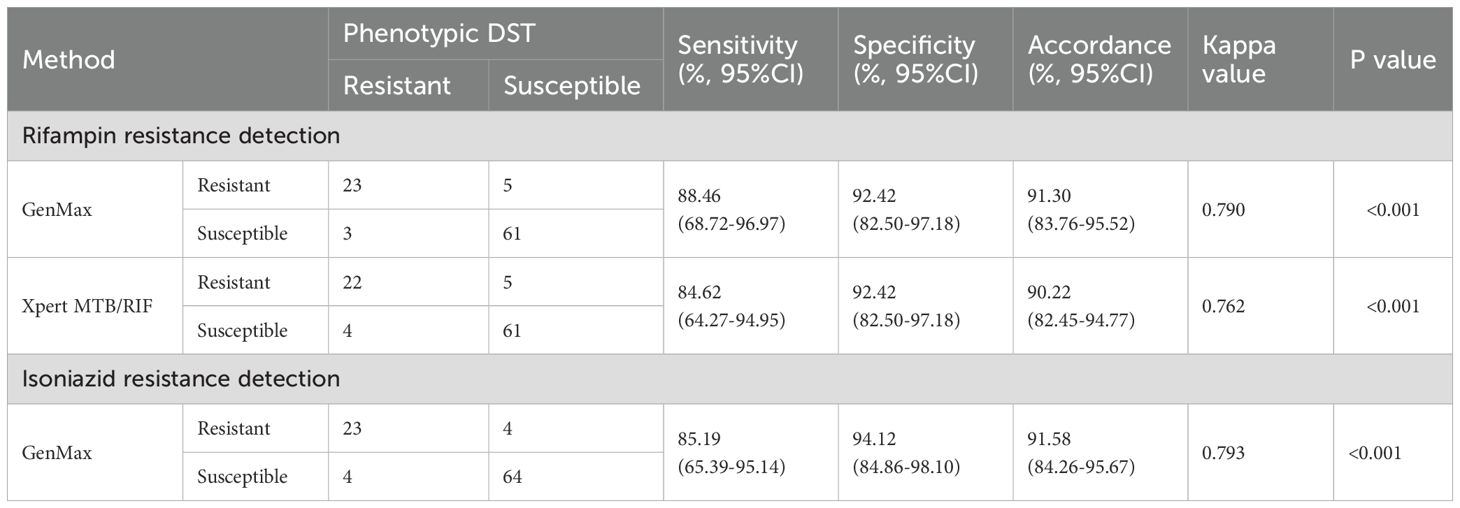
Table 6. Diagnostic accuracy of GenMax and Xpert MTB/RIF for detection of Rifampin and isoniazid resistance.
4 Discussion
To facilitate accurately and rapidly diagnosis of pulmonary tuberculosis, novel diagnostics with improved sensitivity are urgently needed (Seki et al., 2018; MacLean et al., 2020). The GenMax provides an option in automated nuclei acid amplification tests (NAATs) for TB diagnosis and resistance detection, particularly in decentralized settings. This study demonstrated high sensitivity for detecting MTBC and its capacity to simultaneously identify resistance to rifampin and isoniazid, two cornerstone drugs in TB treatment.
GenMax exhibited a limit of detection (LOD) of 8.8 CFU/mL for MTBC, surpassing InnowaveDX MTB/RIF (LOD: 9.6 CFU/mL) (Deng et al., 2023), Xpert MTB/RIF (LOD: 131 CFU/mL) (Helb et al., 2010) and Ultra (LOD: 16 CFU/mL) (Chakravorty et al., 2017), though it should be noted that this value was derived solely from testing diluted bacterial suspensions, which has not been validated in sputum specimens spiked with known bacterial loads. The assay’s sensitivity drops to 45% at 5 CFU/mL emphasized optimizing sample processing steps (e.g., pre-enrichment or centrifugation) to concentrate low-abundance targets could improve sensitivity. Moreover, a high diagnostic sensitivity (97.52%) to some extent also reflects a lower LOD of GenMax, although such sensitivity may be also influenced by factors such as the bacterial load in included cases, the preparation of the specimens, and the amplification efficiency of the assay, etc (Dinnes et al., 2007; Chakravorty et al., 2017). The sensitivity for MTBC detection approached the optimal sensitivity requirements (≥95%) on sputum-based assays outlined in the WHO’s Target Product Profile (TPP) (World Health Organization, 2024c), positioning it as a robust tool for paucibacillary samples, such as those from pediatric or HIV/TB co-infected patients. Future studies should expand clinical validation to include larger cohorts of TB cases to confirm robustness. The diagnostic sensitivity is superior to Xpert MTB/RIF Ultra, which demonstrates ~90% sensitivity (World Health Organization, 2024c). GenMax’s automated workflow and shorter turnaround time (~3 hours) make it particularly advantageous in resource-limited settings where skilled personnel and infrastructure are scarce.
GenMax demonstrated sensitivities of 88.46% for RIF resistance and 85.19% for INH resistance. While these values fall short of WHO TPP targets (≥95% for RIF, ≥90% for INH) (World Health Organization, 2024c), they remain competitive with first-generation molecular RIF/INH assays. For instance, in China the GenoType MTBDR assay achieves ~91% sensitivity for RIF resistance but only ~80% for INH resistance (Sun et al., 2019). Similarly, the Genechip shows comparable performance with 88% sensitivity for RIF resistance and 80% for INH resistance detection (Pang et al., 2013). Notably, RIF resistance is predominantly mediated by rpoB mutations, whereas INH resistance involves complex mechanisms. The lower sensitivity for INH resistance likely stems from its genetic complexity, involving mutations in katG, ahpC-inhA promoter and other unknown mechanisms (Liu et al., 2022; WHO, 2023). GenMax’s targeted approach covers some particular regions, such as RRDR for RIF resistance, katG codon 315, promoter region of inhA and ahpC for INH resistance detection, but may miss uncommon variants, necessitating expanded probe coverage or supplemental sequencing. Importantly, sequencing confirmed GenMax results in all discordant cases (8/8), underscoring its reliability in detecting known resistance-associated mutations. However, phenotypic DST discrepancies (e.g., 3 GenMax-susceptible/phenotypic-resistant RIF cases) suggest that rare rpoB mutations outside the rifampicin resistance-determining region (RRDR) or heteroresistance may contribute to false negatives of this GenMax assay. Therefore, in high-risk clinical scenarios (e.g., treatment failure cases or contacts of known DR-TB patients), GenMax should be used in conjunction with phenotypic DST or WHO-recommended molecular tests to minimize the risk of missed resistance detection.
The WHO TPP emphasizes the need for rapid, user-friendly, and cost-effective diagnostics in decentralized settings (WHO, 2024). GenMax meets these criteria through its almost automated system (hands-on time <10 minutes) and minimal infrastructure requirements. Results are typically available within 3 hours. This important feature can potentially result in dramatically reduced turnaround times for MTB and resistance to rifampin and isoniazid. Its ability to concurrently detect MTBC, RIF, and INH resistance in a single test streamlines workflows, reducing delays in initiating appropriate therapy. This is critical in China, where only half of MDR/RR-TB cases are currently detected (WHO, 2024), and INH mono-resistance remains underdiagnosed due to limited testing access and RIF resistance prioritized diagnostic algorithm.
This study has several limitations. Firstly, the sample size for drug resistance testing was relatively small (92 cases for rifampin and 95 cases for isoniazid), necessitating validation through multicenter studies and geographically diverse cohorts to confirm its applicability across varying Mycobacterium tuberculosis strains and resistance patterns. Secondly, the research focused solely on pulmonary tuberculosis cases; future investigations should evaluate the efficacy of GenMax in extrapulmonary and pediatric tuberculosis samples, where bacterial loads are typically lower. Finally, a comprehensive cost-effectiveness analysis is essential to justify its scalability in resource-limited healthcare systems.
5 Conclusion
The Innovo GenMax MTB-RIF/INH assay is a promising tool for rapid, decentralized TB diagnosis and resistance screening, which is critical for clinical decisions making and improved treatment outcomes. While its MTBC detection performance aligns with WHO targets, sensitivity gaps for RIF/INH resistance underscore the need for iterative optimization. Strategic integration with existing technologies and expanded clinical validation will maximize its impact on TB control, particularly in high-burden settings like China.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of China CDC (202201). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
XO: Conceptualization, Writing – original draft. BZ: Methodology, Writing – original draft. HZ: Conceptualization, Writing – original draft. RX: Investigation, Writing – original draft. QS: Methodology, Resources, Writing – original draft. ZQ: Resources, Supervision, Writing – original draft. LZ: Resources, Formal analysis, Writing – original draft. KC: Methodology, Writing – original draft. YS: Investigation, Writing – original draft. YZhe: Data curation, Writing – original draft. YZho: Software, Writing – original draft. SW: Writing – original draft. HX: Methodology, Writing – review & editing. YZha: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key R&D Program of China (2022YFC2305204, 2023YFC2307301), Public Health Personnel Training Support Program(01056), Beijing Natural Science Foundation (7224328), Funding For Reform and Development of Beijing Municipal Health Commission (EYGF-HX-05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Boehme, C. C., Nabeta, P., Hillemann, D., Nicol, M. P., Shenai, S., Krapp, F., et al. (2010). Rapid molecular detection of tuberculosis and rifampin resistance. New. Engl. J. Med. 363, 1005–1015. doi: 10.1056/NEJMoa0907847
Chakravorty, S., Simmons, A. M., Rowneki, M., Parmar, H., Cao, Y., Ryan, J., et al. (2017). The new Xpert MTB/RIF ultra: improving detection of mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 8, e00812-17. doi: 10.1128/mBio.00812-17
Deng, Y., Ma, Z., Su, B., Bai, G., Pan, J., Wang, Q., et al. (2023). Accuracy of the InnowaveDX MTB/RIF test for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre study. Emerg. Microbes Infect. 12, 2151382. doi: 10.1080/22221751.2022.2151382
Dinnes, J., Deeks, J. J., Kunst, H., Gibson, A., Cummins, E., Waugh, N., et al. (2007). A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol. Assessment. 11, 1–196. doi: 10.3310/hta11030
Fan, D., Yue, Y., Li, H., Shang, X., Li, H., Xiao, R., et al. (2023). Evaluation of the performances of InnowaveDx MTB-RIF assay in the diagnosis of pulmonary tuberculosis using bronchoalveolar lavage fluid. Tuberculosis 140, 102349. doi: 10.1016/j.tube.2023.102349
Gegia, M., Winters, N., Benedetti, A., van Soolingen, D., and Menzies, D. (2017). Treatment of isoniazid-resistant tuberculosis with first-line drugs: a systematic review and meta-analysis. Lancet Infect. Dis. 17, 223–234. doi: 10.1016/S1473-3099(16)30407-8
Hanna, B. A., Ebrahimzadeh, A., Elliott, L. B., Morgan, M. A., Novak, S. M., Rusch-Gerdes, S., et al. (1999). Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J. Clin. Microbiol. 37, 748–752. doi: 10.1128/JCM.37.3.748-752.1999
Helb, D., Jones, M., Story, E., Boehme, C., Wallace, E., Ho, K., et al. (2010). Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48, 229–237. doi: 10.1128/JCM.01463-09
Liu, D., Huang, F., Zhang, G., He, W., Ou, X., He, P., et al. (2022). Whole-genome sequencing for surveillance of tuberculosis drug resistance and determination of resistance level in China. Clin. Microbiol. Infect. 28, e9–731.e15. doi: 10.1016/j.cmi.2021.09.014
Liu, D., Zhao, B., Zheng, Y., Ou, X., Wang, S., Zhou, Y., et al. (2024). Characterization of isoniazid resistance and genetic mutations in isoniazid-resistant and rifampicin-susceptible Mycobacterium tuberculosis in China. Infect. Med. (Beijing). 3, 100129. doi: 10.1016/j.imj.2024.100129
MacLean, E., Kohli, M., Weber, S. F., Suresh, A., Schumacher, S. G., Denkinger, C. M., et al. (2020). Advances in molecular diagnosis of tuberculosis. J. Clin. Microbiol. 58, e01582-19. doi: 10.1128/JCM.01582-19
Menzies, D., Benedetti, A., Paydar, A., Royce, S., Madhukar, P., Burman, W., et al. (2009). Standardized treatment of active tuberculosis in patients with previous treatment and/or with mono-resistance to isoniazid: a systematic review and meta-analysis. PLoS Med. 6, e1000150. doi: 10.1371/journal.pmed.1000150
Pang, Y., Xia, H., Zhang, Z., Li, J., Dong, Y., Li, Q., et al. (2013). Multicenter evaluation of genechip for detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 51, 1707–1713. doi: 10.1128/JCM.03436-12
Park, M. Y., Kim, Y. J., Hwang, S. H., Kim, H. H., Lee, E. Y., Jeong, S. H., et al. (2009). Evaluation of an immunochromatographic assay kit for rapid identification of Mycobacterium tuberculosis complex in clinical isolates. J. Clin. Microbiol. 47, 481–484. doi: 10.1128/JCM.01253-08
Seki, M., Kim, C. K., Hayakawa, S., and Mitarai, S. (2018). Recent advances in tuberculosis diagnostics in resource-limited settings. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1405–1410. doi: 10.1007/s10096-018-3258-y
Steingart, K. R., Henry, M., Ng, V., Hopewell, P. C., Ramsay, A., Cunningham, J., et al. (2006). Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 6, 570–581. doi: 10.1016/S1473-3099(06)70578-3
Sun, Y., Gao, L., Xia, H., Yang, Z., Deng, S., Yang, J., et al. (2019). Accuracy of molecular diagnostic tests for drug-resistant tuberculosis detection in China: a systematic review. Int. J. Tuberc. Lung. Dis. 23, 931–942. doi: 10.5588/ijtld.18.0550
World Health Organization (2008). Guidelines for the programmatic management of drug-resistant tuberculosis (Geneva: World Health Organization).
World Health Organization (2023). Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. 2nd ed. (Geneva: World Health Organization).
World Health Organization (2024a). “Consolidated guidance on tuberculosis data generation and use,” in Module 1. Tuberculosis surveillance (World Health Organization, Geneva). 2024
World Health Organization (2024b). Global tuberculosis report 2024 (Geneva, Switzerland: World Health Organization).
World Health Organization (2024c). Target product profiles for tuberculosis diagnosis and detection of drug resistance (Geneva, Switzerland: World Health Organization).
Keywords: tuberculosis, rifampin, isoniazid, drug resistance, rapid molecular diagnosis
Citation: Ou X, Zhao B, Zheng H, Xing R, Sun Q, Qin Z, Zhang L, Cui K, Song Y, Zheng Y, Zhou Y, Wang S, Xia H and Zhao Y (2025) Innovo GenMax MTB-RIF/INH: a moderate-complexity automated NAAT for rapid simultaneous detection of Mycobacterium tuberculosis complex and rifampin/isoniazid resistance. Front. Cell. Infect. Microbiol. 15:1600170. doi: 10.3389/fcimb.2025.1600170
Received: 26 March 2025; Accepted: 10 June 2025;
Published: 30 June 2025.
Edited by:
Svetlana Khaiboullina, University of Nevada, United StatesReviewed by:
Samira Tarashi, Pasteur Institute of Iran (PII), IranNtombenhle Gama, University of Pretoria, South Africa
Copyright © 2025 Ou, Zhao, Zheng, Xing, Sun, Qin, Zhang, Cui, Song, Zheng, Zhou, Wang, Xia and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Xia, eGlhaHVpQGNoaW5hY2RjLmNu; Yanlin Zhao, emhhb3lsQGNoaW5hY2RjLmNu
†These authors have contributed equally to this work
 Xichao Ou
Xichao Ou Bing Zhao1†
Bing Zhao1† Huiwen Zheng
Huiwen Zheng Ruida Xing
Ruida Xing Lixia Zhang
Lixia Zhang Yang Zhou
Yang Zhou Shengfen Wang
Shengfen Wang Hui Xia
Hui Xia Yanlin Zhao
Yanlin Zhao