- Institute of Hygiene, University of Münster, Münster, Germany
Autoinducer-2 is a signaling molecule involved in quorum sensing in Escherichia coli. Quorum sensing enables coordinated behavior based on cell density and helps bacteria adapt to their environment. The luxS gene and the lsr locus are involved in the biosynthesis, transport, and intracellular phosphorylation of autoinducer-2. Disruption of autoinducer-2 biosynthesis or transport can reduce biofilm formation, chemotaxis, and the expression of genes relevant for the uropathogenicity of E. coli. Interestingly, most isolates of E. coli phylogroup B2, in which uropathogenic and other extraintestinal pathogenic strains are overrepresented, lack the lsr operon. We show that autoinducer-2-dependent quorum sensing is not fundamentally beneficial for efficient and prolonged urinary bladder colonization. We demonstrate that the lsr-negative asymptomatic bacteriuria isolate 83972 has a higher fitness than its lsr-complemented variant. Using transcriptome analyses, competitive growth assays, and comparisons of selected fitness properties, we show that restoration of the lsr operon in this strain background results in growth retardation, loss of competitiveness, and higher sensitivity to oxidative stress. Our results illustrate that the lack of autoinducer-2-dependent quorum sensing contributes to the well-known fitness and competitiveness of E. coli 83972, on which its effective use for bacterial interference in the urinary bladder relies. It is vital to delve deeper to fully understand the fitness and competitiveness of the ABU strain 83972 if we are to optimize its use in therapeutic colonization. The key is to unravel the underlying molecular mechanisms, thus ensuring the efficacy and safety of this treatment as an alternative to antibiotic therapy.
1 Introduction
Extraintestinal pathogenic Escherichia coli (E. coli) strains (ExPEC) are usually classified as facultative pathogens since they often colonize the gut without causing symptoms (Wiles et al., 2008; Köhler and Dobrindt, 2011). However, they harbor accessory traits that enable them to invade and colonize extraintestinal niches, eventually leading to symptomatic diseases (Vila et al., 2016; Sora et al., 2021). E. coli strains that have characteristic genetic markers that indicate the potential to cause extraintestinal disease, or that were directly isolated from infections outside the gastrointestinal system, are typically classified as ExPEC. ExPEC are usually divided into sepsis-causing (SEPEC), neonatal meningitis-associated (NMEC), and uropathogenic E. coli (UPEC), as well as strains causing systemic disease in animals such as avian pathogenic (APEC) or mammary pathogenic (MPEC) E. coli (Köhler and Dobrindt, 2011; Sora et al., 2021). Several virulence factors are described to promote extraintestinal pathogenicity in ExPEC, including adhesins, invasins, iron uptake systems, or toxins (Sora et al., 2021). Another bacterial trait that is involved in extraintestinal pathogenicity is quorum sensing (QS). Quorum sensing is a process in which bacteria sense and adapt to changing cell densities through signaling molecules called autoinducers (AI). Several AIs that are specific for certain bacterial strains or families have been described (Rutherford and Bassler, 2012). AI-2, on the other hand, serves as an interspecies signaling molecule that is produced and sensed by both Gram-positive and Gram-negative bacteria (Sun et al., 2004; Pereira et al., 2013). Even eukaryotic cells were proposed to participate in AI-2-mediated QS since it was found that Saccharomyces cerevisiae secretes molecules that mimic AI-2, as well as the intestinal epithelial cell (IEC) line Caco-2 in response to bacteria or a tight-junction disruption (Ismail et al., 2016; Valastyan et al., 2021). Moreover, the inflammatory interleukin IL-8 was found to be upregulated in response to external AI-2 in the IEC line HCT-8 (Zargar et al., 2015).
In bacteria, the precursor of AI-2, (S)-4,5-dihydroxy-2,3-pentanedione (DPD) (Xavier and Bassler, 2005), is produced as a byproduct of L-homocysteine synthesis by the synthase LuxS, a widely distributed and conserved enzyme that is part of the activated methyl cycle (Schauder et al., 2001; Vendeville et al., 2005). DPD is a hydrophilic molecule that is actively transported out of the bacterial cell after production (Khera et al., 2022). Outside of the cell, DPD spontaneously circularizes into either boron-containing (S)-2-methyl2,3,3,4-tetrahydroxytetrahydrofuran-borate or boron-free (R)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF), resulting in two different types of active AI-2 (Chen et al., 2002; Miller et al., 2004). Depending on the environmental status, DPD can spontaneously convert between the two active forms and thus be sensed by mixed bacterial communities since both AI-2 forms are perceived by different receptors (Zhang et al., 2020). R-THMF, from now on referred to as AI-2, is an intracellular signaling molecule (Xavier and Bassler, 2005). Different receptors for AI-2 are known, but the best-studied is LsrB. LsrB was first described in Salmonella enterica serovar Typhimurium (S. Typhimurium) (Taga et al., 2001) and is part of a specialized transporter encoded in the lsr (LuxS-regulated) locus (Taga et al., 2003). The lsr locus, or homologs, have been found in various bacterial species, including E. coli (Pereira et al., 2009), and comprises two transcription units. The first transcription unit, lsrRK, encodes the kinase LsrK that phosphorylates the incorporated AI-2 and the repressor LsrR that represses transcription of both lsr operons (Xue et al., 2009), as well as of several other genes in E. coli (Li et al., 2007) and S. Typhimurium (Choi et al., 2012). The binding of phosphorylated AI-2 to LsrR abolishes the transcriptional repression, resulting in AI-2-dependent gene expression (Xavier and Bassler, 2005). The second transcription unit, lsrACDBFG, encodes structural proteins of the AI-2 transporter, i.e., LsrA, LsrC, LsrD, and LsrB as the receptor, as well as the isomerase LsrG that degrades the phosphorylated AI-2 and the thiolase LsrF that catalyzes the terminal step in processing phosphorylated AI-2 (Marques et al., 2014) (Figure 1).
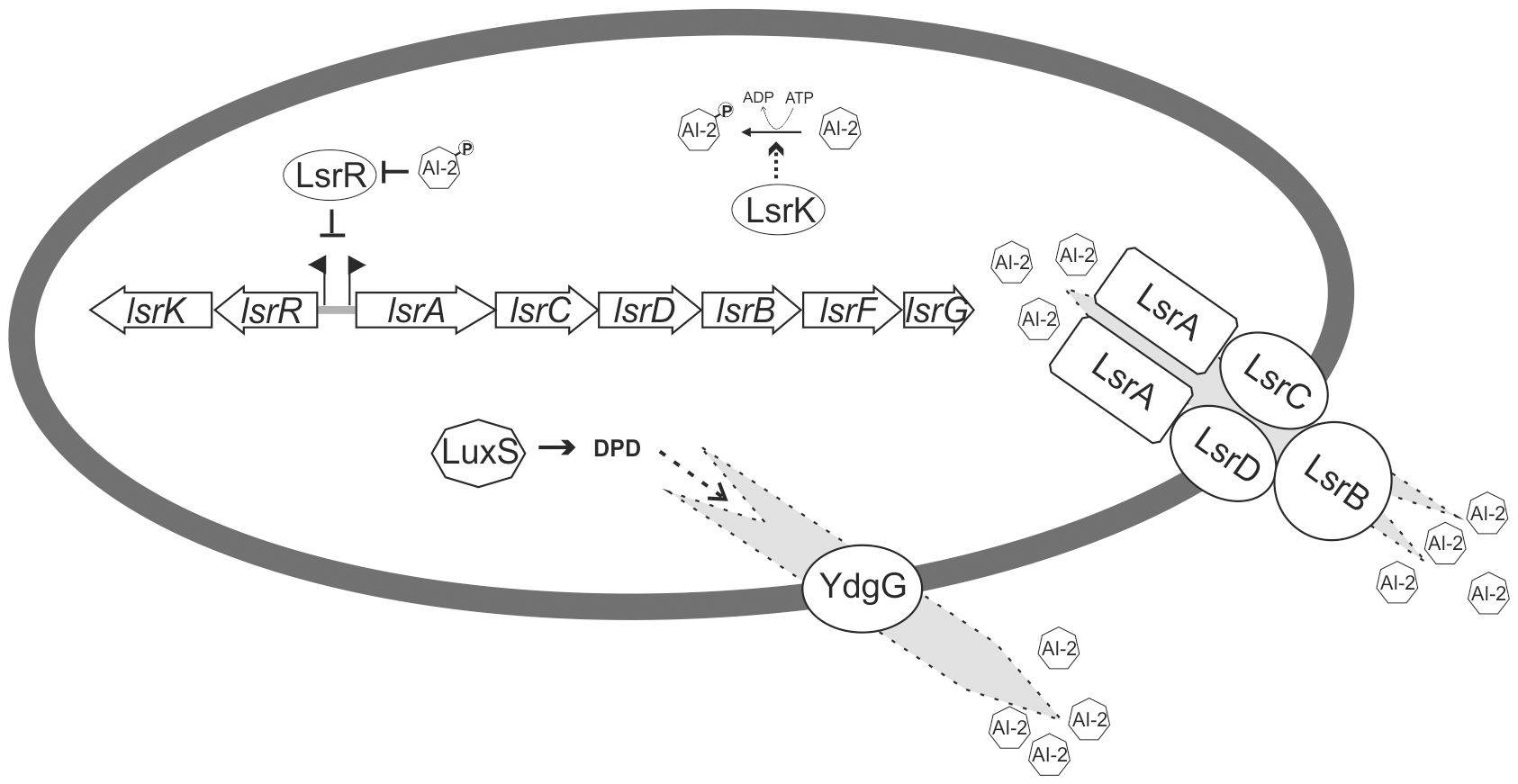
Figure 1. Regulatory circuit and Lsr-mediated transport and modification of AI-2 in E. coli. The AI-2 precursor DPD is produced by LuxS. AI-2 is then exported from the bacterial cell by YdgG. Extracellular AI-2 is imported by the ABC transporter LsrACDB. Intracellular AI-2 will be phosphorylated by the kinase LsrK and remains in the cytoplasm. The transcriptional repressor LsrR represses the expression of the lsrACDBFG and lsrRK transcriptional units by binding to their promoter region. Phosphorylated AI-2 can bind to the repressor LsrR and thereby relieve the transcriptional blockade. The subsequent expression of the LsrACDB transporter facilitates additional uptake of extracellular AI-2. Phosphorylated AI-2 is further processed by LsrG and LsrF.
Interfering with AI-2-dependent QS, either by using QS inhibitors (Helmy et al., 2022) or gene mutations and deletions, led to altered behavior and lower virulence in ExPEC strains. For example, the deletion of luxS has led to a significant reduction of virulence in the APEC O78:K80:H9 strain χ7122 (Palaniyandi et al., 2013) and an attenuated virulence of the APEC strain DE17ΔaroA (Han et al., 2015). Also, the deletion of lsrACD in APEC94, which leads to an impaired AI-2 uptake and decreased bacterial motility, has led to lower bacterial loads and reduced virulence in a duckling infection model (Zuo et al., 2019). Additionally, it was shown that AI-2-dependent QS is associated with hydrogen peroxide (H2O2) resistance in the MPEC strain DCM5 (Wang et al., 2021) and antibiotic sensitivity in APEC strain APECX40 and the MPEC isolate DCM1 (Xue et al., 2016; Yu et al., 2020). Although AI-2-dependent QS has significant effects on APEC and MPEC, little is known about its effects on the virulence and fitness traits of UPEC, which are often genetically very similar to APEC and MPEC (Rodriguez-Siek et al., 2005; Tivendale et al., 2010). UPEC are the leading cause of urinary tract infections (UTI), one of the most prevalent bacterial infections worldwide, accounting for at least 75% of all complicated and uncomplicated cases (Flores-Mireles et al., 2015). Despite several host defense mechanisms, including the physicochemical composition of urine, the expression of antimicrobial peptides, or other innate immune response mechanisms (Abraham and Miao, 2015; Loubet et al., 2020), more than 50% of all women (Medina and Castillo-Pino, 2019) and about 20% of all men (Farrell et al., 2021) suffer at least from one symptomatic UTI episode during their lifetime. This indicates that UPEC strains are well adapted to the bladder environment and can combat the host’s defense strategies (Flores-Mireles et al., 2015; Mann et al., 2017).
The asymptomatic bacteriuria (ABU) E. coli isolate 83972 evolved from UPEC but has lost the ability to express many functional virulence factors (Zdziarski et al., 2008). This strain is well adapted to long-term bladder colonization without causing inflammation or other UTI-related symptoms (Hull et al., 1999). Additionally, E. coli 83972 has a better antioxidant defense than UPEC strains (Aubron et al., 2012) and can outcompete UPEC strains in human urine, amongst others, probably due to fast growth and nutritional adaptation (Ipe et al., 2016; George et al., 2024). Due to these characteristics, deliberate bladder colonization by E. coli 83972 has been suggested as a therapeutic approach to prevent recurring UTI by bacterial interference (Sundén et al., 2010; Köves et al., 2014; Wullt and Svanborg, 2016). Against the background of the increasing spread of multidrug-resistant bacteria, especially in the context of UTI, the use of antibiotics must be reduced (Cook and Wright, 2022). Bacterial interference by bladder colonization with ABU strain 83972 is an alternative way to prevent permanent colonization of the urinary bladder by symptomatic uropathogens (Wullt and Svanborg, 2016; Stork et al., 2018; Kenneally et al., 2022). Deeper insights into the importance of QS for the fitness and competitiveness of ABU strain 83972 may help to optimize the use of this bacterial strain for the deliberate therapeutic colonization of patients by uncovering underlying molecular mechanisms and thus improving the efficacy and safety of this form of treatment as an interesting alternative to antibiotic therapy. Interestingly, E. coli 83972 lacks the complete lsr locus but carries the luxS gene. We hypothesized that the absence of AI-2-dependent QS contributes to the strain’s fitness during bladder colonization. Therefore, we integrated the full-length lsr locus from the E. coli K-12 strain MG1655 into the chromosome of E. coli 83972 and analyzed the transcriptome of the lsr complemented strain to screen for differentially expressed genes that are affected by AI-2-dependent QS and may be relevant during bladder colonization. Additionally, we tested for relevant bacterial phenotypes that may increase this strain`s fitness in the urinary bladder.
2 Material and methods
Bacterial strains and culture conditions. The bacterial strains used in this study are listed in Supplementary Table S5. The asymptomatic bacteriuria E. coli isolate 83972 has been obtained as a gift from C. Svanborg (Lund, Sweden). Bacterial cultures were either cultivated in lysogeny broth (LB) (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) or pooled human urine (four male and six female voluntary individuals, sterile filtered and mixed in a 1:1 male/female ratio (v/v)). When appropriate, antibiotics were added in the following concentrations: kanamycin (25 µg/mL), chloramphenicol (12.5 µg/mL), ampicillin (100 µg/mL), or zeocin (50 µg/mL). Bacterial strains were grown overnight at 37°C on LB agar plates (containing 1.5% agar (w/v)) with the appropriate antibiotics when needed. For overnight cultures, single colonies were picked and incubated in 2 mL of LB at 37°C and with orbital shaking at 180 rpm. E. coli DH5α was used as a host for plasmid construction. Genome manipulation was done by recombineering (Datsenko and Wanner, 2000). When indicated, cultures were supplemented with H2O2 (stabilized, Merck Millipore, Darmstadt, Germany) or synthetic DPD purchased from Rita Ventura’s research group at ITQB-UNL (Oeiras, Portugal) (https://itqb.unl.pt/research/chemistry/bioorganic-chemistry). Synthetic DPD was synthesized as published before (Ascenso et al., 2011), and the concentration was 16.8 mM. Both chemicals were subdiluted to working concentrations using sterile ddH2O.
Cloning methods. DNA amplification for cloning and genetic manipulation was done using the Q5 High-Fidelity DNA Polymerase (New England Biolabs, Frankfurt/Main, Germany). Colony-PCR was done using the GoTaq Green Master Mix (Promega GmbH, Walldorf, Germany). After amplification, the correct size of amplicons was verified using agarose gel electrophoresis in 1x Tris-acetate-EDTA (TAE) buffer with 1-2% (w/v) agarose. The gels were run at 110–130 V for 30–60 min. Amplicons were purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany) or the Wizard SV Gel and PCR Clean-Up System kit (Promega, Walldorf, Germany). Plasmids were isolated using the NucleoSpin Plasmid Mini kit (Macherey-Nagel). Genomic DNA (gDNA) was isolated using the QIAamp DNA Mini kit (QIAGEN, Hilden, Germany). Purified PCR products were quantified using a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Schwerte, Germany). The restriction enzymes and T4 DNA ligase used for cloning were purchased from New England Biolabs (Frankfurt/Main, Germany).
Oligonucleotides and plasmids used. All oligonucleotides and plasmids used are listed in Supplementary Tables S6, S7.
Construction of pMK2. The plasmid pMK2 was constructed by integrating the lsrA promoter-yfp fusion reporter cassette coupled to lsrR (lsrR:PlsrA:yfp fusion) from the previously described E. coli K-12 strain MG1655 (lsrA-G::yfp-cat) (Keizers et al., 2022) into the low-copy number plasmid pWKS30 (Wang and Kushner, 1991). For that, the lsrR:PlsrA:yfp fusion was amplified from the gDNA of E. coli K-12 strain MG1655 (lsrA-G::yfp-cat) using the oligonucleotides pKD3_lsr_seq_4 and LZP50 and introduced into the SmaI-digested pWKS30 by using DNA T4 ligase. The correct insertion of the PCR product into pWKS30 was confirmed by plasmid preparation and subsequent Sanger sequencing.
Construction of pLS1 and pLS2. The low-copy-number plasmids pLS1 and pLS2 were constructed by integrating the Pdps-cfp (cfp under the control of the stationary phase-dependent dps promoter) cassette or the Pdps-yfp (yfp under the control of the stationary phase-dependent dps promoter) cassette, respectively, into pWKS30. Both cassettes were amplified using the oligonucleotides bla-TEM-r and MC_185 and pPS1 (Pdps-cfp) or pPS2 (Pdps-yfp) (Schiller et al., 2021) as templates. Amplicons and pWKS30 were cut with KpnI and SmaI. All products were purified, and the digested Pdps-cfp cassette was introduced into digested pWKS30, resulting in pLS1. The digested Pdps-yfp cassette was introduced into digested pWKS30, resulting in pLS2. Ligation was done using DNA T4 ligase. Correct plasmids were confirmed by fluorescence microscopy.
Construction of E. coli strains 83972 ΔluxS and 83972 ΔybhC. For the construction of E. coli 83972 ΔluxS, the luxS gene was replaced by a zeocin resistance cassette (bleR) that was amplified using the oligonucleotides Rec_BleoR_fwd and Rec_BleoR_rev and pEM7/Zeo as a template. Replacement was confirmed by colony PCR using the oligonucleotides CFT073_ΔluxS_CP_fwd and CFT073_ΔluxS_CP_rev and Sanger sequencing of the purified amplicon. For the construction of E. coli strain 83972 ΔybhC, the ybhC gene was replaced by a chloramphenicol resistance cassette (cat) that was amplified using the oligonucleotides del_ybhC_fwd and del_ybhC_rev and pLP2 (Peng et al., 2022) as a template. Replacement was confirmed by colony PCR using the oligonucleotides wt_PybhC and LZP18 and Sanger sequencing of the purified amplicon.
Construction of E. coli strains 83972 attB::lsr and 83972 ΔluxS attB:: lsr. First, the plasmid pWKS30_LSR was generated. The chloramphenicol resistance cassette (cat) was introduced upstream of the lsr locus in the chromosome of E. coli strain MG1655 by recombineering (Datsenko and Wanner, 2000). The resistance cassette was amplified using the oligonucleotides CBL_fwd and CBL_rev and pMB54 (Berger et al., 2016) as a template. The correct chromosomal insertion was confirmed by colony PCR using the oligonucleotides LKRS_fwd and pMB54_lsrRK_CP1_rev. In the same way, the kanamycin resistance cassette (aph) was chromosomally inserted downstream of the lsr locus in the resulting E. coli strain MG1655 cat_lsr. The aph resistance cassette was amplified using the oligonucleotides RBL_fwd and RBL_rev and pKD4 as a template. The correct chromosomal insertion was confirmed by colony PCR using the oligonucleotides pKD3_lsr_seq_12 and LZP50. The full-length lsr locus, flanked by cat and aph, was amplified from gDNA of the resulting E. coli strain MG1655 cat_lsr_aph using the oligonucleotides LKRS_fwd and LZP50, and cloned into the SmaI-digested pWKS30 by using DNA T4 ligase. The relevant parts of the resulting plasmid pWKS30_LSR were afterward confirmed by Sanger sequencing. The complete lsr determinant was then inserted by recombineering (Datsenko and Wanner, 2000) next to the chromosomal attachment site of the bacteriophage λ (attB) in either E. coli 83972 or strain 83972 ΔluxS. For this, the lsr locus flanked by cat and aph (cat_lsr_aph) was amplified using the oligonucleotides pWKS30_attB_fwd and pWKS30_attB_rev and pWKS30_LSR as a template and introduced next to the λ attB site of the strains 83972 and 83972 ΔluxS, respectively. The correct chromosomal insertion was confirmed by colony PCR using the oligonucleotides CFT073_lsrRK_YFP_CP1_fwd and pKD3_lsr_seq_12 or CFT073_lsrRK_YFP_CP1_fwd and CFT073_lsrRK_YFP_CP2_rev and pKD3_lsr_seq_13. Afterward, gDNA was isolated, and the cat_lsr_aph region was amplified using the primer pair CFT073_lsrRK_YFP_CP1_fwd and CFT073_lsrRK_YFP_CP2_rev before the correctness of the DNA sequence was also verified by Sanger amplicon sequencing.
RNA isolation. Total RNA was isolated as previously described (Wallenstein et al., 2020) with the following differences: Overnight cultures of E. coli strains 83972 and 83972 attB::lsr were diluted to a final optical density OD600 = 0.02 in 200 mL LB. At indicated time points, 70 mL of culture (lag phase), 5 mL of culture (exponential phase), or 2 mL of culture (stationary phase) were added to the 0.2x volume of pre-chilled stop solution, and the bacterial pellets were directly lysed. Confirmation of complete gDNA removal was done by qPCR. RNA was extracted from three biological replicates for each strain.
RNAseq and data analysis. Strand-specific cDNA libraries were prepared from the isolated RNA and sequenced (Illumina NextSeq 500, 1 x 75 bp single reads) by vertis Biotechnology AG (Freising, Germany). The obtained raw reads were analyzed as follows: Quality control using FastQC and MultiQC (Bioinformatics; Ewels et al., 2016), adaptor trimming using Cutadapt (Martin, 2011) with subsequent quality control, alignment of reads using burrows-wheeler alignment (Li and Durbin, 2009) to the genome of E. coli 83972 attB::lsr with a subsequent strandedness check using RSeQC (Wang et al., 2012) and quality control, count reading using featureCounts (Liao et al., 2014) and a subsequent differentially expressed gene analysis using DESeq2 (Love et al., 2014) in R. Conversion of formats was done using AGAT (Dainat, 2022). The subsequent data analysis was done in R. The Venn diagram (Figure 2A) was made using venn.diagram (Chen, 2022). The heatmaps (Figures 2B–D) were made using pheatmap (Kolde, 2019). Differentially expressed genes were clustered using dist() and hclust(), and biological replicates were clustered using pheatmap (cluster_cols = T) with the complete linkage method for hierarchial clustering. The scatterplots (Supplementary Figure S2) were computed using pairs.panels() from the package psych (Revelle, 2023).
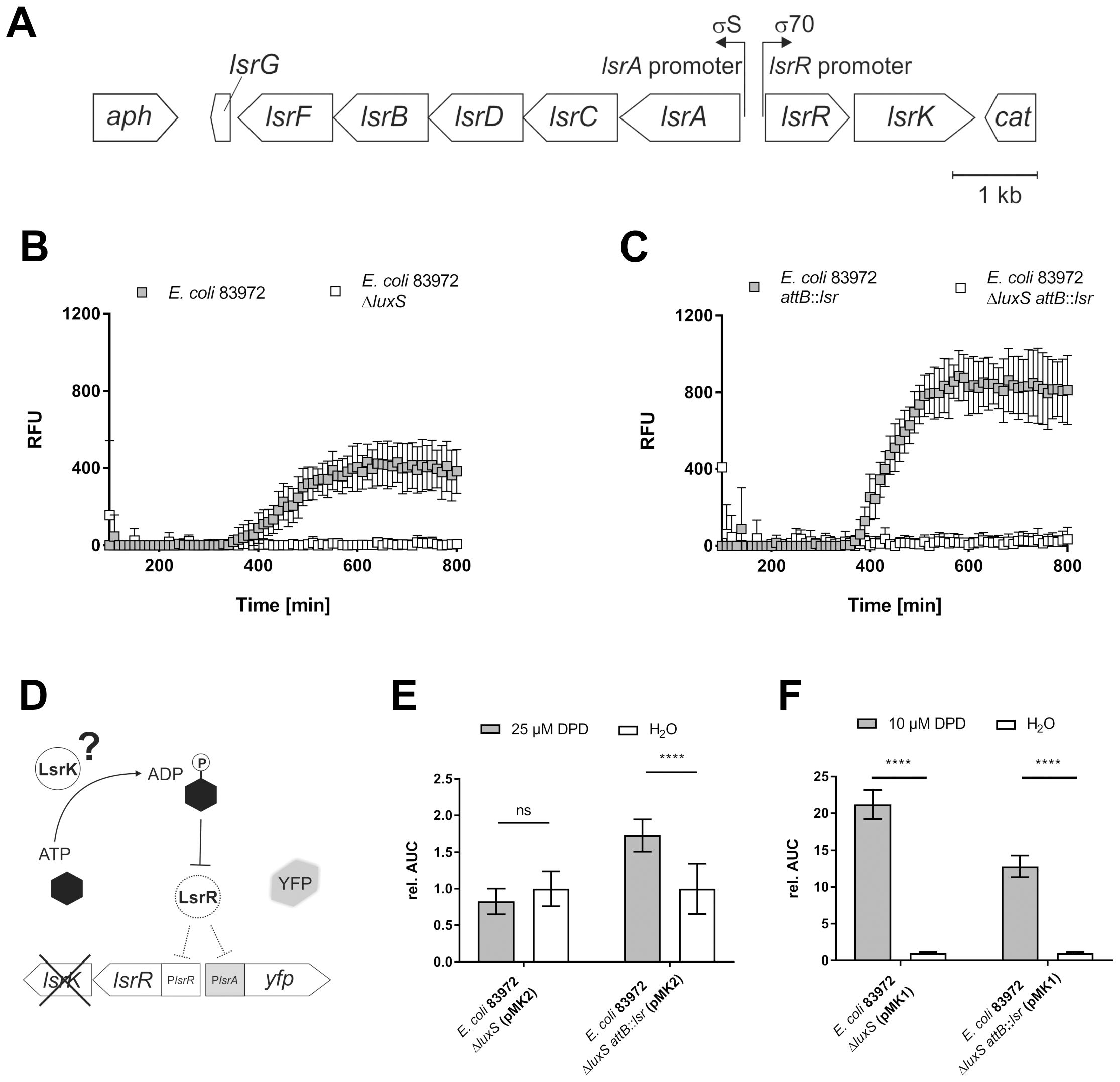
Figure 2. Complementation of E. coli 83972 with the lsr determinant and its impact on AI-2 synthesis, uptake and LsrK phosphorylation. The full-length lsr locus was introduced into E. coli 83972, and subsequent analyses revealed luxS-dependent AI-2 synthesis, the functionality of the complemented gene lsrK and the functionality of AI-2 uptake. (A) The lsr determinant flanked by two resistance cassettes (12,520 bp) was integrated into the chromosomal attB site of E. coli 83972. The luxS-dependent AI-2 synthesis was analyzed by fluorescence kinetics (100–800 min), depicted as RFU, during co-culture with the reporter strain E. coli ΔluxS (attB::PlsrA-yfp) (Keizers et al., 2022). Co-cultures consisting of the reporter strain and (B) either E. coli 83972 (grey squares) or E. coli 83972 ΔluxS (white squares) or (C) E. coli 83972 attB::lsr (grey squares) and E. coli 83972 ΔluxS attB:: lsr (white squares) in a 1:1 ratio in LB. Error bars represent the results of three biological replicates in duplicates each. (D) The functionality of the complemented gene lsrK was analyzed by incorporating the reporter plasmid pMK2 (comprises lsrR and the PlsrA-yfp reporter module) into E. coli strains 83972 ΔluxS and 83972 ΔluxS attB::lsr. YFP expression depends on AI-2 availability and the functional autoinducer-2 kinase LsrK. (E) Relative AUC calculation of PlsrA induction in E. coli strains 83972 ΔluxS (pMK2) and 83972 ΔluxS attB::lsr (pMK2) after the addition of 25 µM DPD compared to water during the late-exponential growth phase, as described before (Keizers et al., 2022). Statistical analysis was performed using the ordinary two-way ANOVA with Tukey’s multiple comparison test; values < 0.05 were considered statistically significant. Error bars represent the results of five biological replicates in duplicates each. (F) Relative AUC calculation of PlsrA induction in E. coli strains 83972 ΔluxS (pMK1) and 83972 ΔluxS attB::lsr (pMK1) after the addition of 10 µM DPD compared to water during the late-exponential growth phase, as described previously (Keizers et al., 2022). Statistical analysis was performed using Welch’s t-test; a value < 0.05 was considered statistically significant. Error bars represent the results of three biological replicates in duplicates each (ns, not significant, **** p<0.0001).
Growth analysis and co-culture assays. For growth analysis, overnight cultures of the bacterial strains were diluted to a final optical density (OD600) of 0.01 in 1 mL of fresh medium with antibiotics when appropriate. 200 µL of freshly diluted cultures were added into one well of a 96-well plate (Thermo Fisher Scientific, Schwerte, Germany) in duplicates for each biological replicate. For the subsequent growth analysis, either an Infinite M NANO+ or an Infinite F200 microplate reader (both from TECAN, Männedorf, Switzerland) was used. The optical density was either measured at 595 nm (± 9 nm) (M NANO+) or 595 nm (± 10 nm) (F200). Signals were measured in ten-minute intervals. The resulting growth curves were analyzed using AMiGA (Analysis of Microbial Growth Assay (Midani et al., 2021)). For the lag phase length analysis, the data was not log-transformed. The co-culture assays were performed as previously described (Keizers et al., 2022). Briefly, overnight cultures of the strains to be analyzed were mixed in a 1:1 ratio with the E. coli K-12 reporter strain MG1655 ΔluxS attB::PlsrA-yfp to a final optical density (OD600) of 0.02. Growth analysis was performed as described above, and the fluorescence signal was either measured using an excitation wavelength of 514 nm (± 9 nm) and an emission wavelength of 550 nm (± 20 nm) (M NANO+) or using an excitation wavelength of 485 nm (± 20 nm) and an emission wavelength of 535 nm (± 25 nm) (F200).
Competition assays. For the competition assays, overnight cultures of E. coli strains 83972 (pLS2) and 83972 attB::lsr (pLS1) were mixed in a 1:1 ratio to a final optical density (OD600) of 0.02 in 25 mL of fresh medium supplemented with ampicillin. The cultures were grown at 37°C with orbital shaking at 180 rpm. After 3 h of growth, the cultures were subdiluted into 25 mL of fresh medium containing ampicillin with an appropriate dilution factor (1:200 for LB and 1:10 for pooled human urine). After subdilution, the cultures were grown for another 3 h. The subdilution procedure was repeated three times with a total assay time of 12 h. For the long-term competition, the cultures were grown at 37°C with orbital shaking at 180 rpm for 72 h. Directly after mixing and at the indicated time points, aliquots were taken from the cultures and microscopically analyzed using a Leica inverted microscope DMi8 with an attached camera at 400 x magnification. For each aliquot, ten microscopic pictures were taken with three channels each – Differential interference contrast (DIC), cyan fluorescent protein CFP (excitation 436 nm (± 20 nm); dichroic mirror 455 nm; emission 480 nm (± 40 nm) and fluorescein isothiocyanate (FITC) (excitation 480 nm (± 40 nm); dichroic mirror 505 nm; emission 527 nm (± 30 nm). Next, the number of bacteria per picture was quantified in the CFP channel and the FITC channel using an ImageJ script. The ratio of bacterial numbers in the FITC channel compared to the total bacterial numbers in both the FITC and CFP channels was calculated for each of the ten pictures, resulting in one mean ratio value with a standard deviation for each time point and biological replicate.
H2O2 resistance assay. For the H2O2 resistance assay, overnight cultures of the bacterial strains were diluted to a final optical density (OD600) of 0.01 in 1 mL of fresh medium. 190 µL of freshly diluted cultures were added into one well of a 96-well plate (Thermo Fisher Scientific) in duplicates for each biological replicate. After adding cultures, 10 µL of H2O2 diluted in ddH2O was added into each well, resulting in the indicated final H2O2 concentrations. The growth was analyzed directly after the addition using an Infinite M NANO+ plate reader (TECAN) or an Infinite F200 (TECAN). The optical density was either measured at 595 nm (± 9 nm) (M NANO+) or 595 nm (± 10 nm) (F200). Signals were measured in ten-minute intervals. The resulting growth curves were analyzed using AMiGA (Analysis of Microbial Growth Assay (Midani et al., 2021)). For the lag phase length analysis, the data was not log-transformed.
Galleria mellonella larvae feeding assay. Galleria mellonella larvae were purchased from Fauna Topics Zoobedarf Zucht und Handels GmbH (Marbach am Neckar, Germany) and reared on an artificial diet (22% maize meal, 22% wheat germ, 11% dry yeast, 17.5% beeswax, 11% honey, and 11% glycerin) as previously described (Mukherjee et al., 2020). To evaluate the fitness of the E. coli wild type strain 83972 and its lsr-complemented mutant in the larval digestive tract, overnight cultures (OD 600 = 1, grown in LB) of both strains were mixed in a 1:1 ratio and force-fed to sixth-instar larvae (weighing approximately 250–300 mg) (Lange et al., 2019). A 10-μL aliquot of this bacterial suspension was gently introduced into the larval mouth using 1-mL disposable syringes fitted with 0.4 x 20-mm needles mounted on a microapplicator. Control larvae received an equivalent volume (10 μL) of sterile LB. Following force-feeding, larvae were incubated at 37°C for 24 hours without food. All larvae survived the incubation period and were subsequently flash-frozen in liquid nitrogen, ground into a fine powder, and homogenized in LB. The homogenates were then plated onto LB agar, and colony-forming units (CFUs) were counted following a 24-hour incubation at 37°C. The survival of the wild type and lsr-complemented mutant strains was analyzed by comparing their initial inoculation ratio to the ratio recovered from larvae after 24 hours of incubation.
Detection of the lsr determinant in E. coli genomes. To screen for the presence and conservation of the lsr determinant in E. coli, all published E. coli genomes that were available at the time point of the analysis were downloaded from NCBI Assembly (Kitts et al., 2016) (time point of download: 15.03.2023) in FASTA format (Status: latest RefSeq). In total, 32,594 genomes were downloaded. Next, the phylogroup was determined using ClermonTyping (Beghain et al., 2018) for each genome. The analysis was continued with 32,404 genomes belonging to the phylogroups A, B1, B2, C, D, E, F, and G. Next, each genome was analyzed using the basic local alignment search tool (BLAST+ (Camacho et al., 2009)) to screen for the nucleotide sequence (blastn) of the full-length lsr locus (lsrRK-lsrACDBFG), the luxS gene and the tam gene (reference sequences from the E. coli K-12 strain MG1655). Output possibilities were “complete” (blastn hit length equal query length), “incomplete” (blastn hit length inequal query length), “no match” (no blastn hit) or “multiple matches” (multiple blastn hits). The output “< 100 bp” is a summary of a blastn hit length of below 100 bp and the “no match” hits. Please note that our analysis only focuses on gene length, whereas the functionality of the encoded gene product was not part of the analysis.
Statistical analysis. Statistical analysis was performed using GraphPad Prism v8.0.2 (San Diego, USA).
3 Results
3.1 Complementation of E. coli 83972 with the lsr locus: impact on AI-2 uptake, synthesis and secretion
We complemented E. coli strain 83972 with the lsr locus by integrating the full-length lsr determinant (lsrACDBFG and lsrRK operons), flanked by two resistance markers, into the chromosomal attachment site (attB) of the bacteriophage λ (Figure 2A). The resulting E. coli strain 83972 attB::lsr, the wild type strain 83972, as well as their isogenic luxS deletion mutants (83972 ΔluxS and 83972 ΔluxS attB::lsr), were used for further analysis. The ability of strains 83972 and 83972 attB::lsr to secrete AI-2 was evaluated in co-cultures with the reporter strain E. coli ΔluxS (attB::PlsrA-yfp), as described previously (Keizers et al., 2022). We detected AI-2-dependent YFP-expression in the co-cultures with E. coli 83972 and E. coli 83972 attB::lsr, whereas we observed no YFP-expression in co-cultures with the isogenic luxS deletion strains. Both strains 83972 and 83972 attB::lsr synthesized AI-2in a luxS-dependent manner and exported it, which can be sensed by the PlsrA reporter strain in the co-culture. In the presence of the lsr determinant, E. coli 83972 can accumulate additional AI-2, leading to a stronger lsrA promoter induction in the reporter strain (Figures 2B, C).
To test if the lsr-complemented strain was capable of taking up and phosphorylating AI-2, we introduced pMK2, a derivative of the previously described reporter plasmid pMK1 that lacks lsrK (Keizers et al., 2022), into the strains 83972 ΔluxS and 83972 ΔluxS attB::lsr (Figure 2D). The functionality of LsrK was analyzed by supplementing the growth medium with 25 µM synthetic DPD in the late logarithmic growth phase and comparing the AI-2-dependent YFP expression to a water control. There was no significant difference in YFP expression between E. coli 83972 ΔluxS (pMK2) supplemented with 25 µM DPD and the water control. The expression of the AI-2 kinase LsrK in E. coli 83972 ΔluxS attB::lsr (pMK1) led to a significantly higher YFP expression when the strain was supplemented with 25 µM DPD as compared to the water control (p < 0.001) (Figure 2E). To determine if the observed differences were due to an overall defect in AI-2 uptake in the lsr-negative 83972 wild type strain, we introduced the reporter plasmid pMK1 into E. coli 83972 ΔluxS and E. coli 83972 ΔluxS attB::lsr. Again, AI-2 uptake was analyzed by supplementing the medium with 10 µM synthetic DPD in the late logarithmic growth phase and comparing the AI-2-dependent YFP expression to a water control. In both strains harboring pMK1, we detected a significantly higher YFP expression after the addition of 10 µM DPD relative to the corresponding water controls (p < 0.001). This confirmed that external AI-2 can be taken up by E. coli 83972 in an LsrACBD-independent way (Figure 2F).
3.2 Influence of the lsr determinant on global gene expression in E. coli 83972
To analyze the effect of AI-2-dependent quorum sensing on the transcriptome, we performed an RNA-seq analysis of E. coli strains 83972 and 83972 attB::lsr. Both strains were cultivated in LB, and samples were taken in the lag phase, during mid-exponential growth (exp. phase), and during the transition to the stationary phase (stat. phase) (Supplementary Figure S1). We isolated total RNA from three biological replicates of each strain and time point for RNAseq analysis. For each library, we obtained ~ 11.5 x 106 reads (± 2 x 106 reads), from which, on average, ~ 89% (± 3-5%) were mapped to the reference genome (E. coli 83972 attB::lsr). Data analysis was done using DESeq2 in R (Love et al., 2014), and the resulting expression data was used for further analysis (Supplementary Figure S2). The transcriptome analysis revealed 492 significantly differentially expressed genes (DEG; adjusted p-value < 0.05) in E. coli 83972 attB::lsr compared to the wild type strain 83972 in the three growth phases. A complete list of all DEGs is provided in the Supplementary Table S1. Of the 492 DEGs, 199 genes had a log2-fold change (L2FC) in expression of at least 1 (DEG (± 1 L2FC)). Among all DEG (± 1 L2FC), nine genes were differentially expressed during all three growth phases (Figure 3A). We found 60 DEG (± 1 L2FC) in the lag phase, 67 DEG (± 1 L2FC) during mid-exponential growth, and 72 DEG (± 1 L2FC) during the transition to the stationary phase (Figures 3B–D). Besides lsrR, lsrK and the two resistance cassettes aph and cat that were used to complement E. coli 83972 attB::lsr, genes related to the isoleucine/valine biosynthesis pathway, namely ilvNBAG, were upregulated, and the gene ybhC, which encodes an uncharacterized protein that was suggested to be involved in H2O2 resistance, was downregulated in all three growth phases. The DEG (± 1 L2FC) can be functionally clustered into four main groups, i.e., metabolism, motility/biofilm formation/adhesion, growth and stress response, and some remaining genes (Tables 1–5).
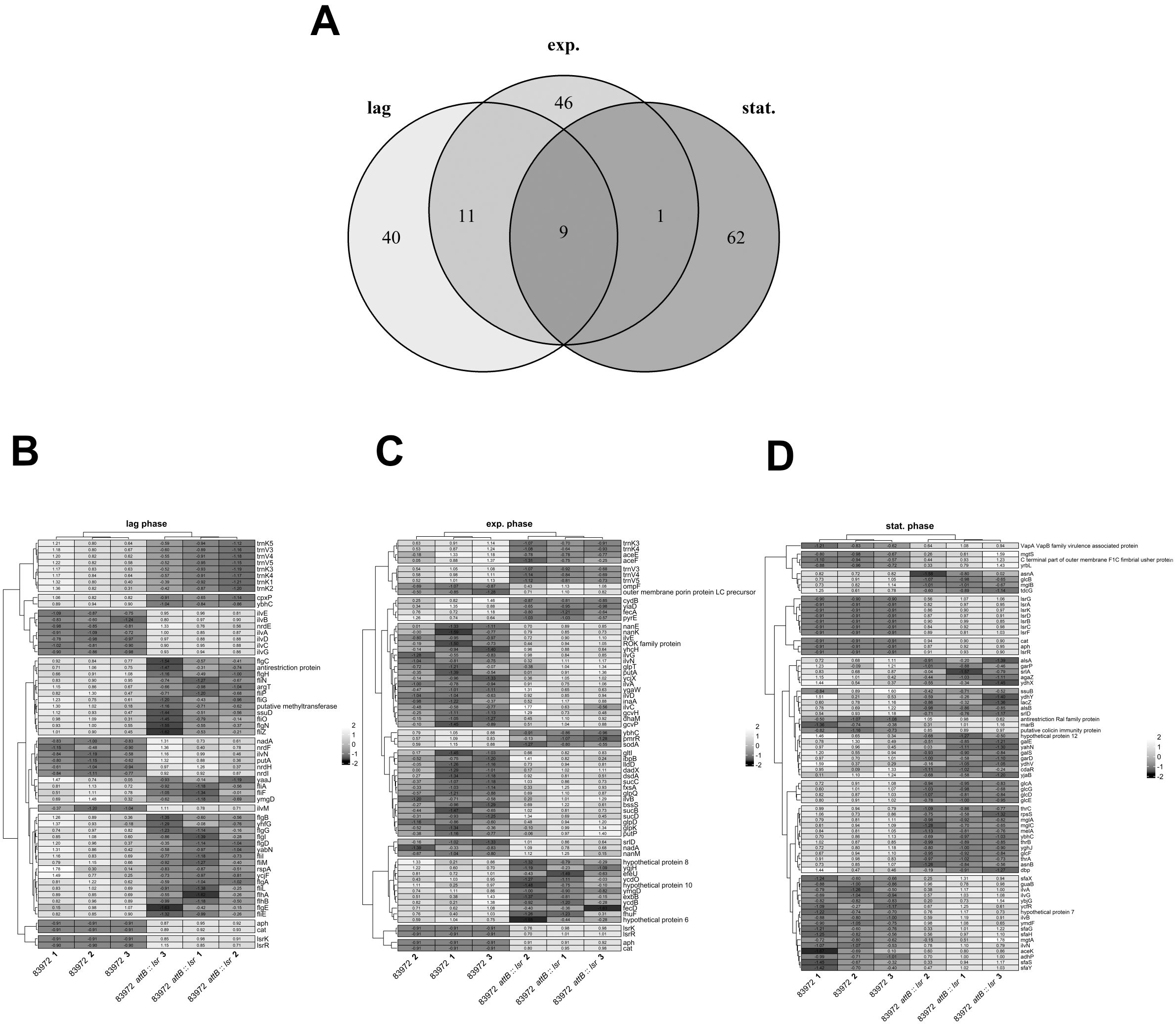
Figure 3. Differentially expressed genes in E. coli 83972 attB::lsr compared to its lsr-negative parental strain 83972. (A) The Venn diagram indicates the number of differentially expressed genes (DEG) in the lag phase (lag), mid-exponential phase (exp.) and stationary phase (stat.). The heatmaps depict the groups of genes which are differentially expressed in an lsr-dependent manner in (B) lag phase, (C) exp. phase and (D) stat. phase.
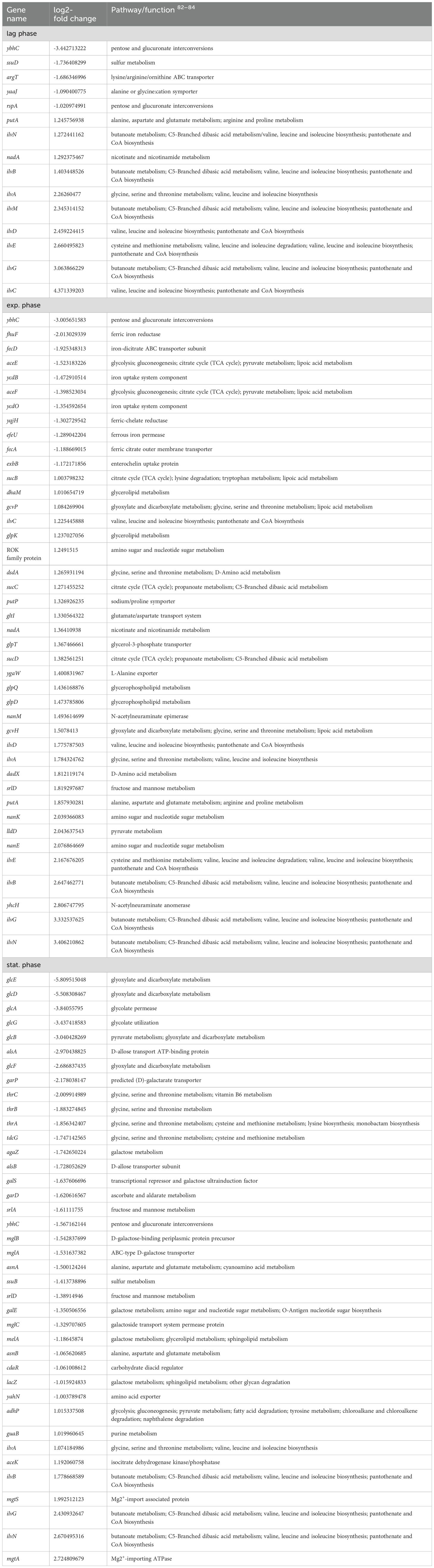
Table 1. Differentially expressed genes in E. coli 83972 attB::lsr relative to E. coli 83972 associated with metabolism.
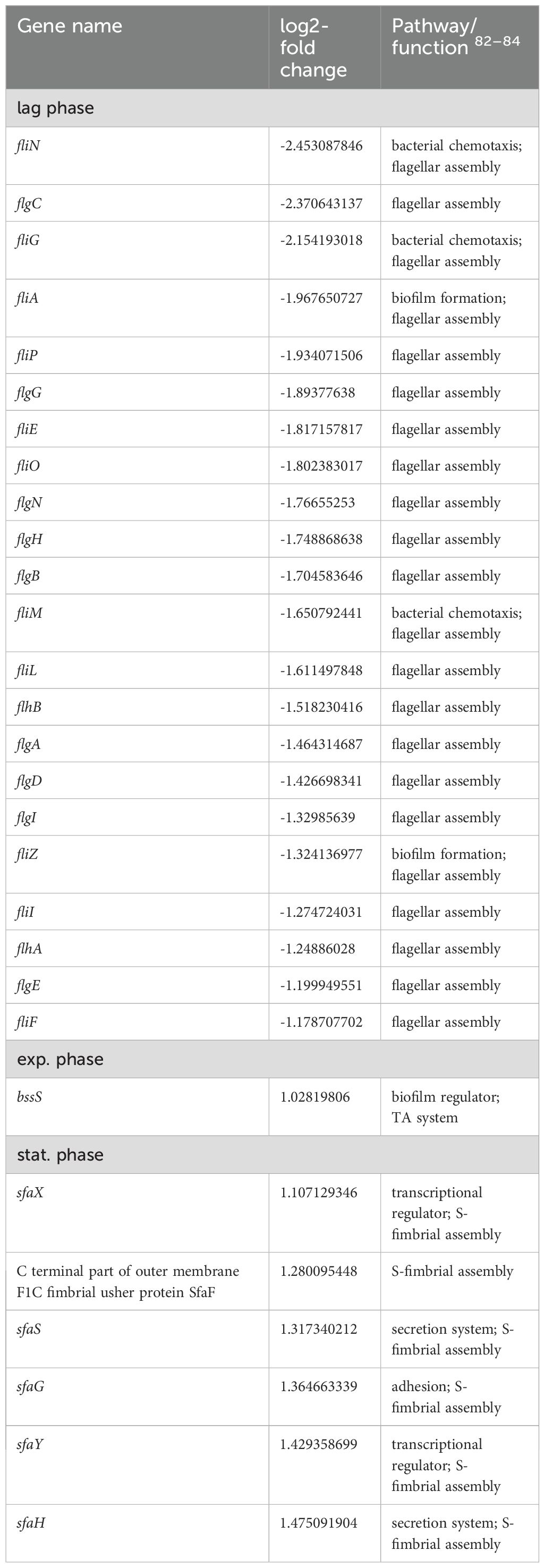
Table 2. Differentially expressed genes in E. coli 83972 attB::lsr relative to E. coli 83972 associated with motility, biofilm formation and adhesion.
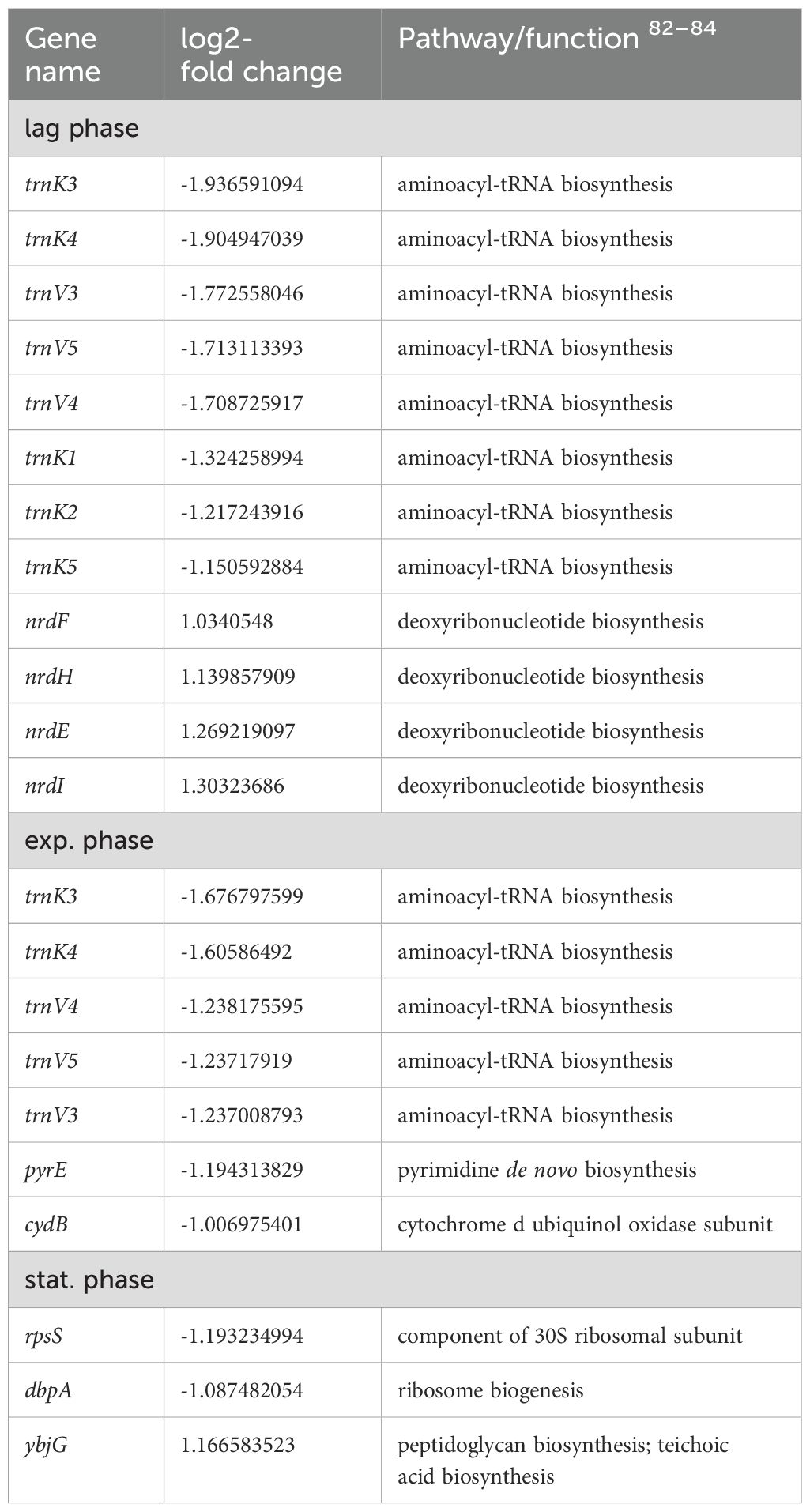
Table 3. Differentially expressed genes in E. coli 83972 attB::lsr relative to E. coli 83972 associated with growth.
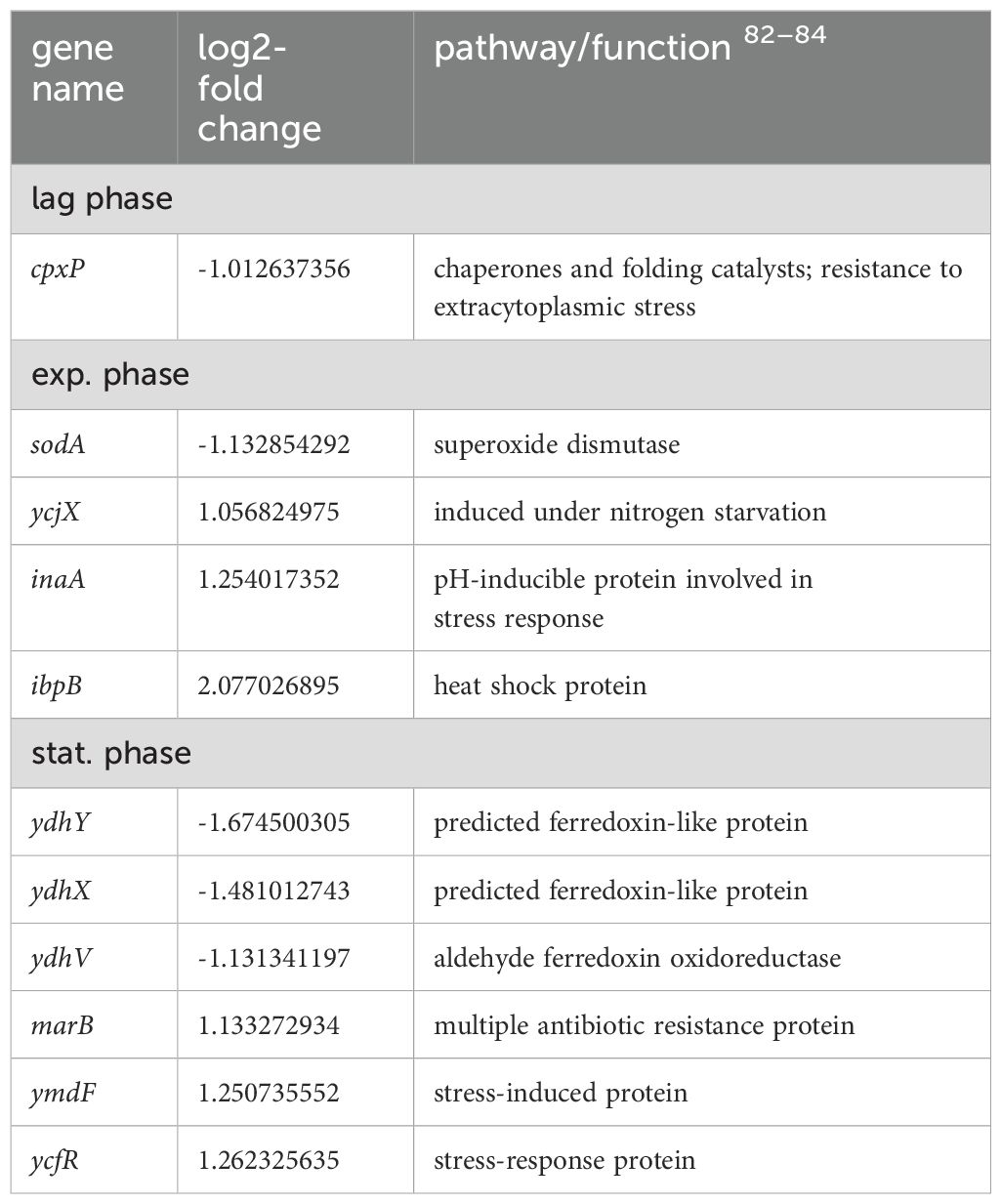
Table 4. Differentially expressed genes in E. coli 83972 attB::lsr relative to E. coli 83972 associated with stress response.
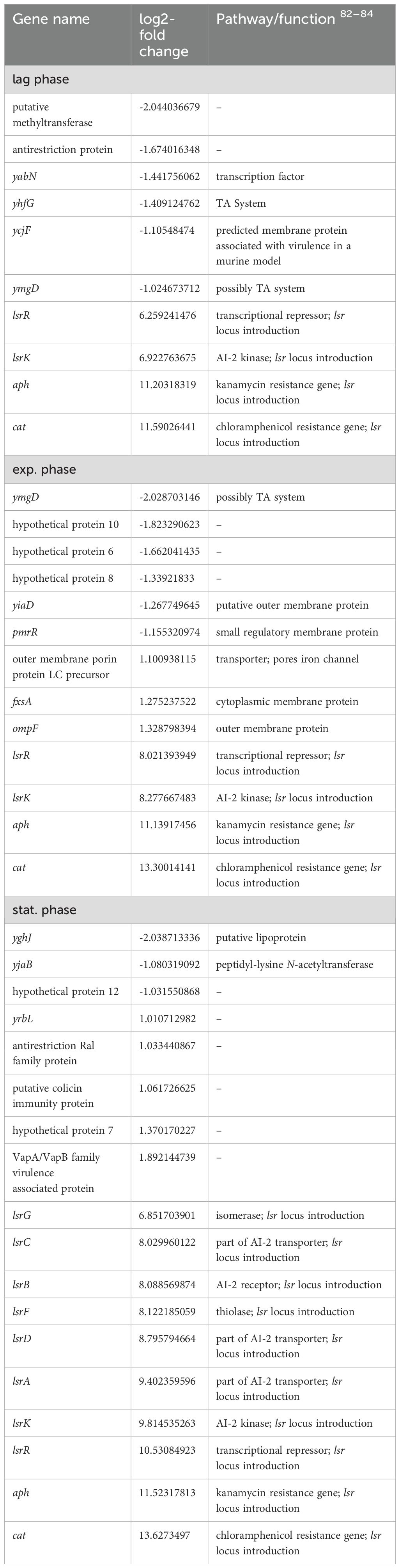
Table 5. Remaining differentially expressed genes in E. coli 83972 attB::lsr relative to E. coli 83972.
In the lag phase, mainly biosynthesis genes were upregulated. Together with the genes of the valine, leucine, and isoleucine biosynthesis pathway (ilvNBAMDEGC), also genes of the nrd operon, encoding for a ribonucleotide reductase (nrdHIEF) were significantly upregulated. On the contrary, several metabolic genes were downregulated. We found that genes involved in arginine uptake and metabolism (argT), sulfur utilization (ssuD), or coding for an alanine/glycine:cation symporter (yaaJ), as well as a mannonate dehydratase (rspA) were downregulated. Also, the expression of genes associated with the aminoacyl-tRNA biosynthesis of lysine (trnK1-5) and valine (trnV3-5) or with resistance against extracytoplasmic stress (cpxP) was downregulated. Amongst all genes that were downregulated in strain 83972 in the presence of the lsr determinant in the lag phase, more than 50% code for components of the flagellar assembly apparatus. In addition to the flagellar sigma factor σ28-encoding gene fliA, the transcriptional regulator gene fliZ, and the chaperone gene flgN, nearly all flagellar genes belonging to class 2 were downregulated (fliE, fliFG, fliI, fliLMNOP, flhBA, flgBCDE, flgGHI, and flgA).
During mid-exponential growth, we predominantly found genes associated with metabolic pathways upregulated in E. coli 83972 attB::lsr relative to E. coli strain 83972. The encoded gene products are involved in the metabolism of D-serine (dsdA), glutamate (gltI), glycine (gcvPH), L-lactate (lldD), sorbitol (srlD), L-alanine (dadX, ygaW), glycerol (glpDTQK), proline (putAP), N-acetylneuraminate (nanMKE-yhcH), dihydroxyacetone (dhaM) as well as the TCA cycle (sucBCD). Also, three genes encoding outer membrane proteins (ompF and the outer membrane porin protein LC precursor) were upregulated. The genes coding for the valine, leucine, and isoleucine biosynthesis pathway (ilvNBADEGC) were upregulated. Interestingly, we also found upregulated genes involved in stress response, including the nitrogen starvation gene ycjX, the heat-shock protein gene ibpB, and the pH-stress response protein gene inaA. On the contrary, the gene cluster aceEF, encoding a pyruvate dehydrogenase, was downregulated during mid-exponential growth. Of all downregulated genes during mid-exponential growth, 36% are associated with iron sensing and utilization, including the genes coding for the ferric siderophore reductase (fhuF), the ferric chelate reductase (yqjH), the ferric citrate transporter genes (fecA) and (fecD), the ferrous iron permease and transport genes efeU, ycdB, and ycdO, the energy transducing Ton complex subunit gene exbB and pmrR, which is associated with the iron and acidic pH sensing BasSR complex. Additionally, apart from ybhC, the superoxide dismutase SodA-encoding gene was downregulated. The latter two proteins have been described to protect against oxidative stress.
As expected, we observed that the genes of the lsr locus were highly expressed during the transition to the stationary phase. Also, the expression of valine, leucine, and isoleucine biosynthesis pathway genes (ilvGABN) and genes contributing to magnesium sensing and utilization were upregulated. These included the import-associated genes mgtA and mgtS and the Mg2+-induced kinase gene yrbL. Interestingly, several genes associated with general stress response were upregulated. These included marB, encoding the protein MarB that reduces the transcription rate of marA, which codes for a pleiotropic regulator also involved in general stress response. The expression of ycfR, which is involved in stress response and outer membrane permeability was also upregulated. We also detected that the transcript levels of several genes (focDFGHYX) of the F1C fimbrial operon were significantly increased in E. coli 83972 attB::lsr in comparison to E. coli strain 83972. The main proportion of downregulated genes, was related to metabolism. The yghJ (sslE) gene, for example, codes for a metalloprotease contributing to mucin degradation in the intestinal tract or the bladder (Nesta et al., 2014). The most strongly downregulated genes were linked to glycolate utilization (glcDEFGBA). Additionally, genes that are involved in D-allose transport (alsAB), galactarate metabolism (garPD and cdaR), galactose metabolism (mglABC, lacZ, melA, agaZ and galES), sorbitol utilization (srlAD), D-serine metabolism (tdcG) and sulfur utilization (ssuB) were significantly downregulated, too. Furthermore, genes associated with L-threonine (thrABC) and asparagine (asnAB) synthesis, as well as three genes that code for components of a putative oxidoreductase (ydhYVX) were also downregulated.
3.3 Reintroduction of the lsr locus reduces competitiveness in vitro
As the majority of differentially regulated genes affect the bacterial metabolism, we hypothesized that E. coli 83972 attB::lsr might have a growth defect when compared to the wild type strain 83972. Therefore, we analyzed and compared the growth behavior of both strains under aerobic and anaerobic conditions in LB (Figure 4) and pooled human urine (Supplementary Figure S3). In all conditions tested, E. coli 83972 attB::lsr had a significantly longer lag phase (Figures 4B, E; Supplementary Figures S3B, E) and a significantly higher doubling time than parental strain 83972 (Figures 4C, F; Supplementary Figures S3C, F). Next, we performed a growth competition experiment with E. coli 83972 and E. coli 83972 attB::lsr to test this phenotype for its biological relevance. To be able to differentiate between both strains by fluorescence microscopy, we introduced the plasmid pLS1 (cfp under the control of the stationary phase-dependent dps promoter) into E. coli 83972 attB::lsr and the plasmid pLS2 (identical to pLS1 on the nucleotide level, but cfp is replaced by yfp) into E. coli 83972. First, we mimicked bottleneck situations for which we mixed these two strains in a 1:1 ratio and grew the competing strains for 3 h, followed by a subsequent dilution into fresh medium. This cycle was repeated three times. After 3 h of growth, the proportion of E. coli 83972 (pLS2) increased from 52 ± 3.7% to 72 ± 13.3% while further increasing to 97 ± 0.8% after three transfers (Figure 5A). After 3 h of growth in pooled human urine, the proportion of E. coli 83972 (pLS2) rose from 49 ± 6.1% to 61 ± 2.1% while making up 79 ± 1.1% after three serial passages (Supplementary Figure S4A). We also tested for the long-term competitiveness of both strains. We observed that the proportion of E. coli 83972 (pLS2) significantly rose from 51 ± 1.7% to 75 ± 0.3% in LB after 24 h of direct competition (Figure 5B). Within the following 48 h, the proportion of E. coli 83972 (pLS2) remained unchanged. In pooled human urine, we saw that the proportion of the wild type strain significantly rose from 48 ± 2.8% to 67 ± 2.7% after 72 h of competition (Supplementary Figure S4B).
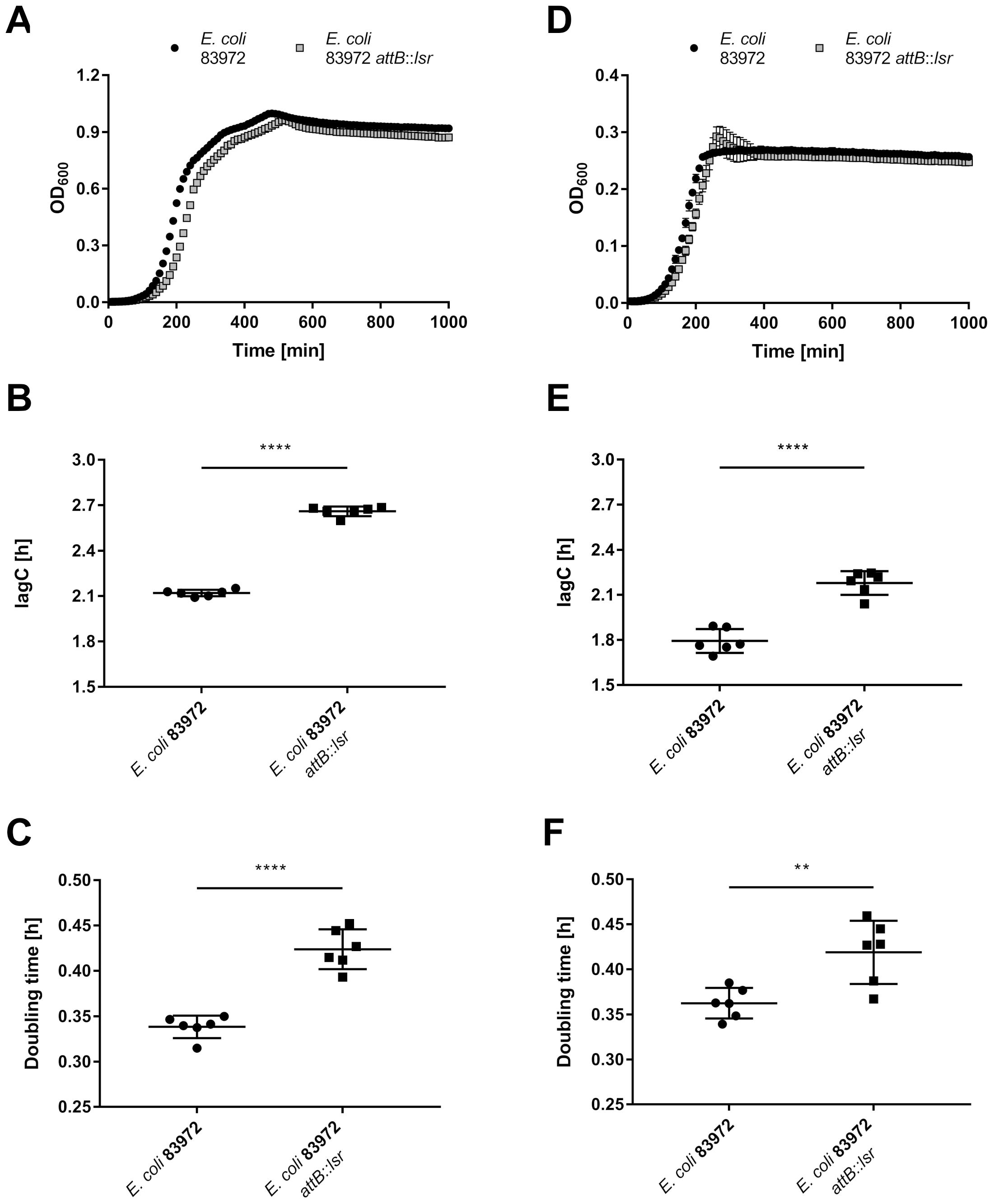
Figure 4. Restoration of the lsr determinant impairs growth of E. coli 83972. Growth analysis of E. coli strains 83972 and 83972 attB::lsr was done in LB under (A-C) aerobic and (D-F) anaerobic conditions. (A, D) Growth curves over a time span of 1000 min with optical density (OD600) measurements every 10 min of E. coli 83972 (black circles) and E. coli 83972 attB::lsr (grey squares). (B, E) Time until the cultures reached the exponential growth phase (lagC). (C, F) Doubling time during the exponential growth phase. Depicted are three biological replicates performed in duplicates each. The starting OD600 was 0.01 for both strains. Growth curve analysis was done using AMiGA (Midani et al., 2021). Statistical analysis was performed using an unpaired t-test; values < 0.05 were considered statistically significant (** p<0.01, **** p<0.0001).
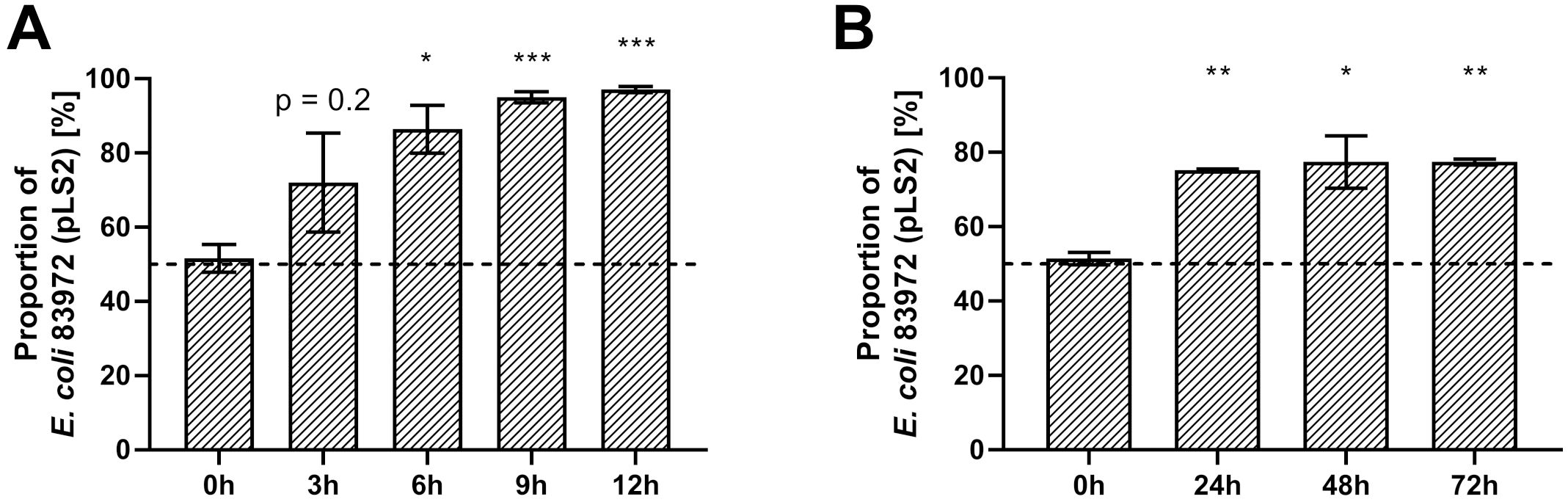
Figure 5. Restoration of the lsr determinant reduces competitiveness in serial passaging of E. coli 83972. Competition assays were done by mixing overnight cultures of strains 83972 (pLS2) and 83972 attB::lsr (pLS1) in a 1:1 ratio in LB. (A) The cultures were grown and subsequently diluted (1:200) into fresh medium every 3 h. Depicted are the mean values and standard deviations of the ratio analysis of ten microscopic pictures for four biological replicates at the indicated time points. (B) The cultures were grown over a total of 72 h. Depicted are the mean values and standard deviations of the ratio analysis of ten microscopic pictures for three biological replicates at the indicated time points. Statistical analysis was performed using RM one-way ANOVA (Geisser-Greenhouse correction) with Dunnett’s multiple comparison test; values < 0.05 were considered statistically significant (* p<0.05, ** p<0.01, *** p<0.001).
3.4 Reintroduction of lsr locus or deletion of ybhC leads to a lower resistance against oxidative stress
To test for resistance to oxidative stress, we added different concentrations of H2O2 to low bacterial numbers of the E. coli strains 83972, 83972 ΔybhC and 83972 attB::lsr and analyzed their growth behavior. We observed that the length of the lag phase was significantly longer for E. coli 83972 after the addition of 160 µM H2O2 as compared to the water control, while the addition of 40 µM or 100 µM did not lead to a significantly longer lag phase. In the case of the E. coli strains 83972 ΔybhC and 83972 attB::lsr, on the other hand, already the addition of 100 µM resulted in significantly longer lag phases as compared to the corresponding water controls (Figure 6). Once the strains entered exponential growth, however, the doubling times were not affected by the initial H2O2 stress (Supplementary Figure S5).
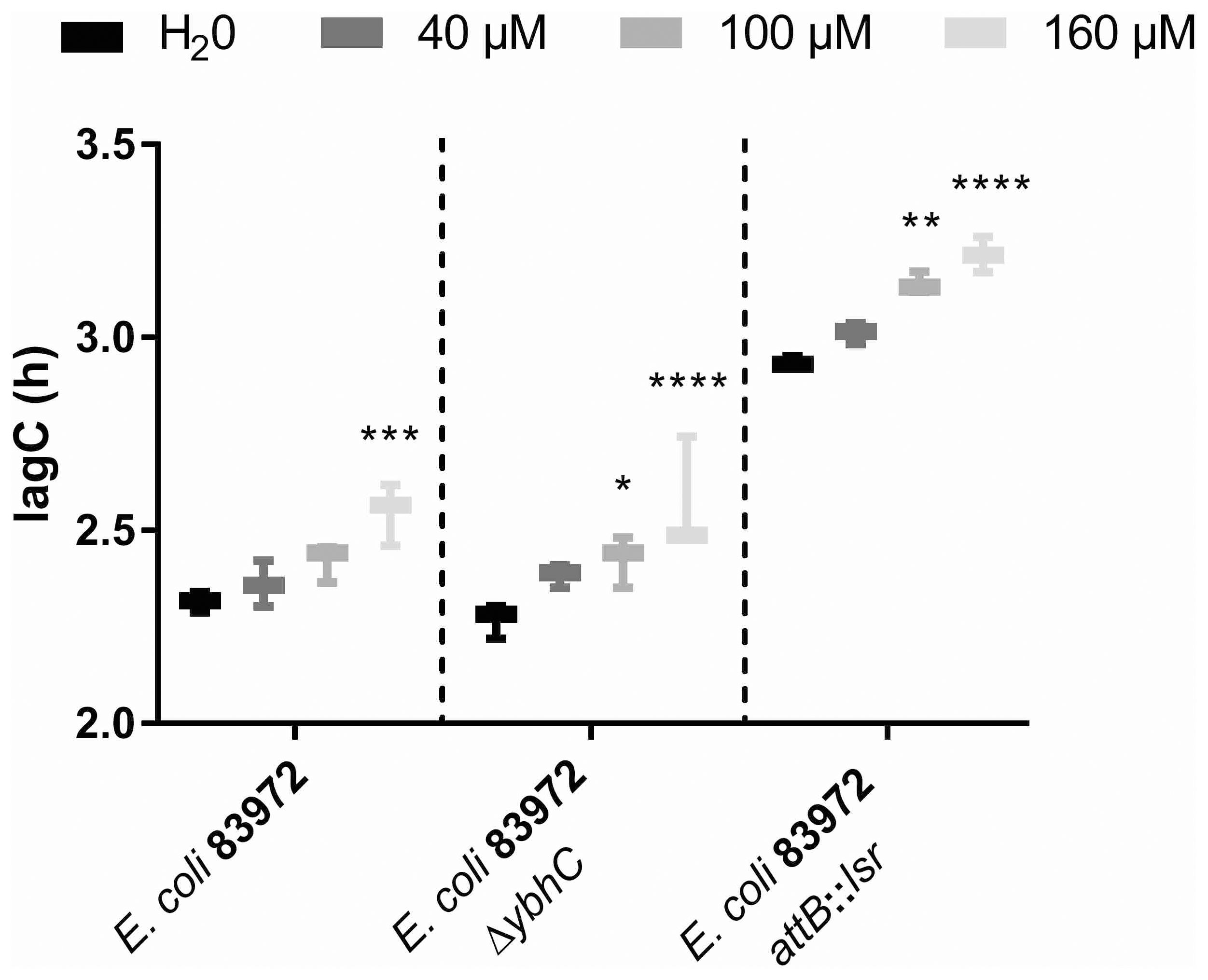
Figure 6. Introduction of the lsr locus or deletion of ybhC negatively affects oxidative stress resistance of E. coli 83972. Shown is the time until the bacteria reach the exponential growth phase (lagC) during growth of E. coli strains 83972, 83972 ΔybhC and 83972 attB::lsr in LB. The starting OD600 was 0.01 for each strain. Before growth analysis, the strains were challenged with a final H2O2 concentration of 40 µM, 100 µM or 160 µM. Water was added as a negative control. LagC was analyzed using AMiGA (Midani et al., 2021). The statistical analysis was performed using ordinary two-way ANOVA with Tukey’s multiple comparison test. Only the effect of different H2O2 concentrations on the growth of the respective E. coli strains was compared (83972, 83972 ΔybhC and 83972 attB::lsr; simple effect within rows); values < 0.05 were considered statistically significant (** p<0.01, *** p<0.001, **** p<0.0001).
3.5 Complementation of E. coli strain 83972 with the lsr operon results in impaired colonization capacity in the digestive tract of Galleria mellonella larvae
Uropathogenic E. coli normally have their reservoir in the densely populated intestinal tract, and urinary tract infections usually occur through smear infection with bacteria originating from the intestinal tract. To test whether the presence of a functional lsr operon affects the ability of E. coli strain 83972, as a degenerate uropathogen, to efficiently colonize a niche that is densely populated by different types of bacteria such as the intestinal tract, we fed G. mellonella larvae with a mixture of E. coli strain 83972 and its lsr-complemented variant in a 1:1 ratio. Quantification after 24 h incubation showed that the lsr-positive E. coli 83972 variant was found in significantly lower numbers in the digestive tract of the G. mellonella larvae than the wild type strain. After 24 hours, the lsr-negative wild type E. coli 83972 outcompeted its lsr-positive complemented counterpart in the digestive tract of G. mellonella larvae (Figure 7).
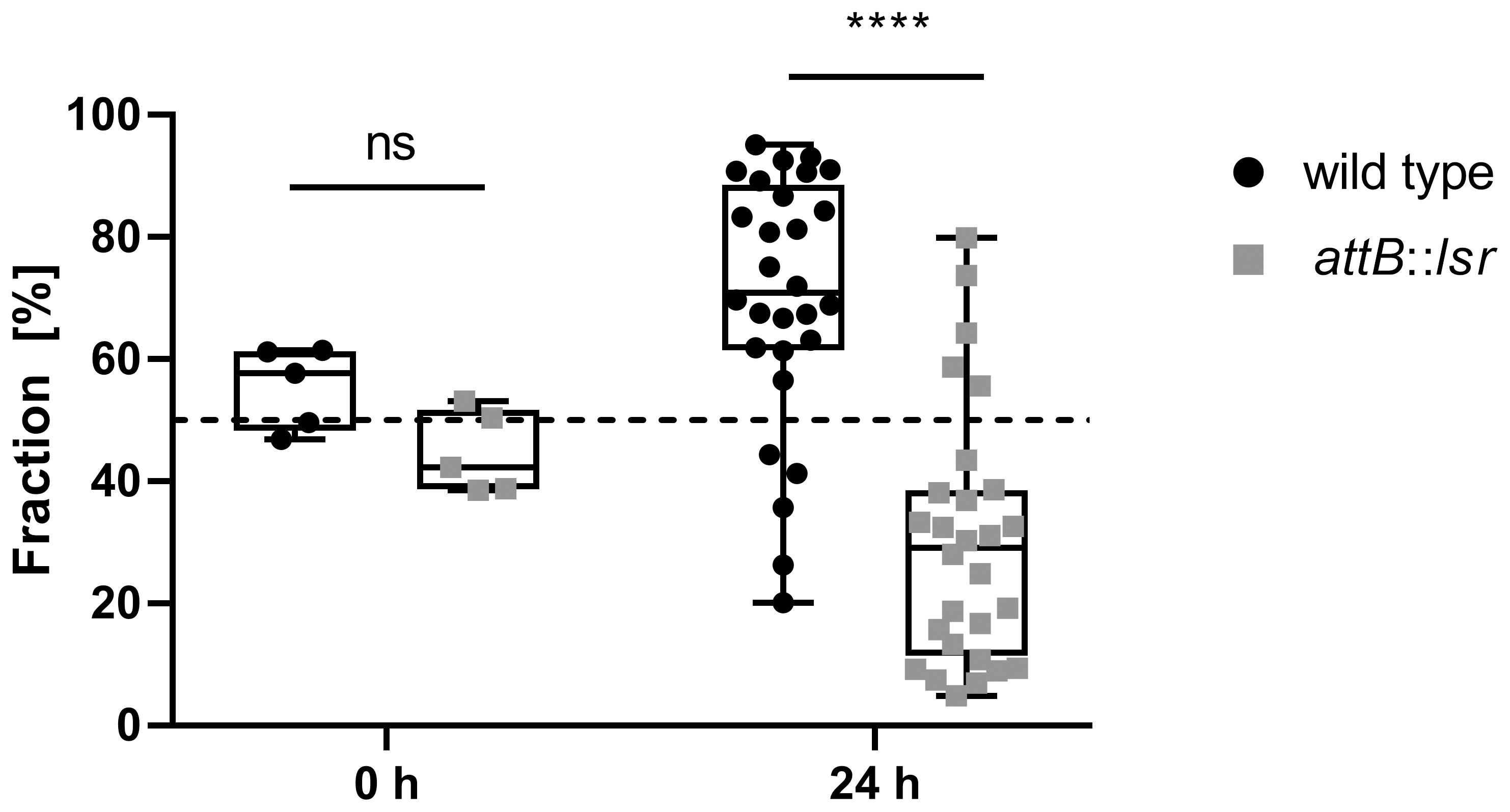
Figure 7. Restoration of the lsr locus negatively affects the competitiveness of E. coli 83972 in the digestive tract of Galleria mellonella larvae. Statistical analysis considering a mixed effects analysis was performed using Sidak’s multiple comparison test; values < 0.05 were considered statistically significant (**** p<0.0001).
3.6 E. coli of the phylogroup B2 are predominantly lsr-negative
To obtain an overview of the abundance of the lsr locus in the E. coli population, we screened all E. coli genomes that were available from the NCBI database concerning their phylogroup and the presence of the full-length lsr locus. In total, we analyzed 32,404 genomes of isolates from the phylogroups A, B1, B2, C, D, E, F, and G. Genomes from isolates of phylogroup B1 accounted for the largest share (29.1%), followed by phylogroup A (28.2%) and phylogroup B2 (18.6%) (Supplementary Table S2). We screened these genomes for the presence of the full-length lsr locus (reference DNA sequence from the E. coli K-12 strain MG1655) in all genomes. The search resulted in 22,259 complete, 1,861 incomplete and 2,400 multiple matches. In 5,884 genomes, we detected no match or a blastn hit length that was shorter than 100 bp (Supplementary Table S3). We found that > 81% of E. coli strains belonging to the phylogroups A1, B1, C, D, E, F, or G carry a full-length lsr locus. On the contrary, in > 95% of E. coli isolates belonging to the phylogroup B2, a homologous region smaller than 100 bp was detected (Figure 8, Supplementary Figure S6A). We analyzed the prevalence of the luxS and tam genes in the E. coli population. The search for the luxS genes in the genome collection resulted in 32,301 complete, 22 incomplete, and 62 multiple matches. In 19 genomes, we found no match (Supplementary Table S3). The search for the tam gene, which is located downstream of lsrG resulted in 30,364 complete, 1,679 incomplete, and 181 multiple matches. In 180 genomes, we found no match (Supplementary Table S3). Accordingly, more than 99% of the analyzed E. coli genomes include a full-length luxS gene (Supplementary Figure S6B), and > 88% of the analyzed E. coli genomes possess a full-length tam gene (Supplementary Figure S6C).
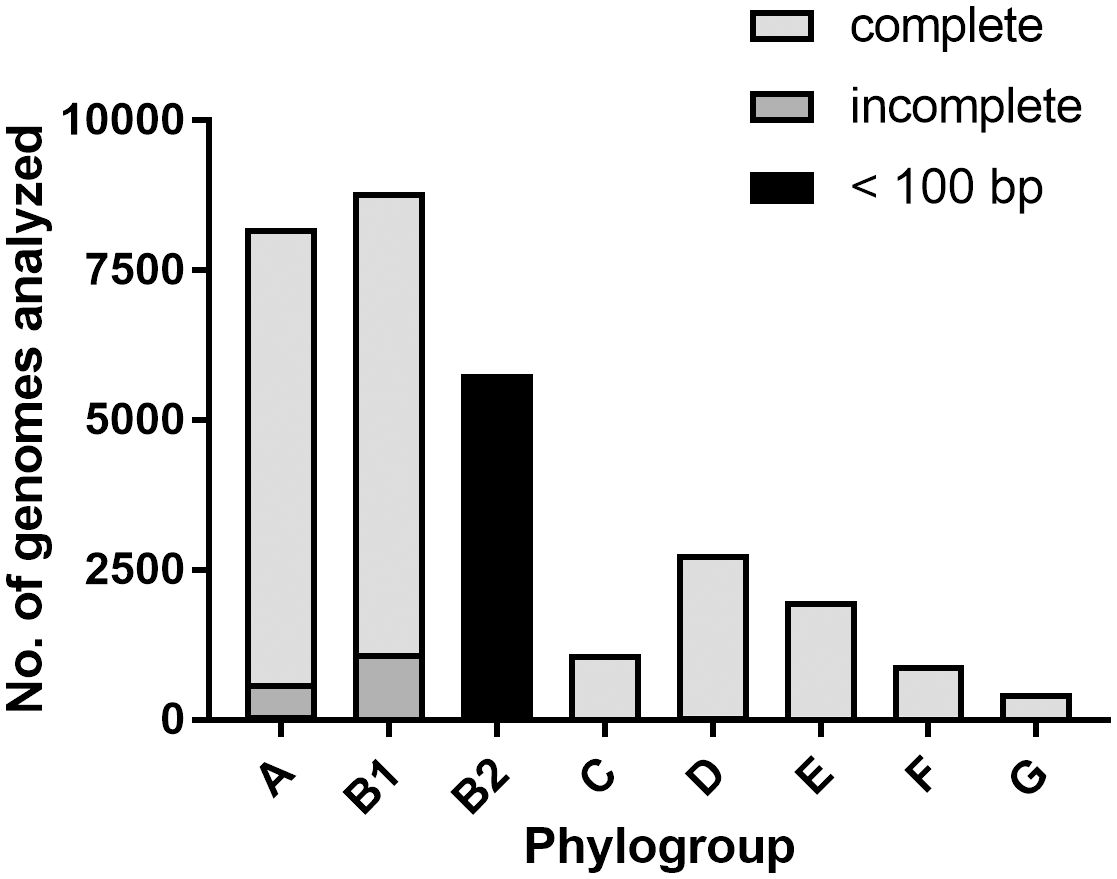
Figure 8. The majority of E. coli isolates of the phylogroup B2 does not encode an lsr locus. Shown is the distribution of 30,004 E. coli genomes with respect to their phylogroup (A, B1, B2, C, D, E, F or G) and the presence of the full-length lsr locus (E. coli K-12 strain MG1655 lsrRK-lsrACDBFG; size: 8,673 bp). “Complete” (light grey) equals a blastn match of 8,673 bp, “incomplete” (dark grey) equals a blastn match < 8,673 bp and “< 100 bp” (black) equals a blastn match < 100 bp.
4 Discussion
AI-2-dependent QS was shown to play an essential role in APEC and MPEC strains for virulence, colonization, or resistance (Palaniyandi et al., 2013; Han et al., 2015; Xue et al., 2016; Zuo et al., 2019; Yu et al., 2020; Wang et al., 2021; Helmy et al., 2022). However, except for knowledge about the role of QS-induced biofilm formation in catheter-associated UTIs (Laganenka and Sourjik, 2018; Henly et al., 2021; Lila et al., 2023), little is known about the function of AI-2-dependent QS in UTIs. E. coli 83972 is naturally lsr-negative but luxS-positive and causes asymptomatic bacteriuria (Medina and Castillo-Pino, 2019). We analyzed this strain’s ability to produce, export, import, and phosphorylate AI-2. We found that E. coli 83972 produced AI-2, which could be sensed by another E. coli strain during co-culture (Figure 2B). The AI-2 production was LuxS-dependent, as judged by the inability to induce AI-2-dependent YFP expression in our reporter strain when luxS was deleted in E. coli 83972 (Figure 2B). However, we observed no AI-2-dependent YFP expression when E. coli 83972 ΔluxS (pMK2) was challenged with extracellular DPD (Figures 2D, E). In comparison, the addition of low concentrations of extracellular DPD to E. coli 83972 ΔluxS (pMK1) has led to a significantly higher YFP expression compared to the water control (Figure 2F). This result was expected since E. coli 83972 does not encode LsrK, which is so far the only described kinase that phosphorylates incorporated AI-2 (Pereira et al., 2012). Since AI-2 is initially taken up in an Lsr-independent way by a phosphoenolpyruvate phosphotransferase system, the lack of an active AI-2 kinase would not lead to an AI-2 sequestration inside the cell because unphosphorylated AI-2 is transported back into the extracellular space (Marques et al., 2011; Pereira et al., 2012; Trappetti et al., 2017). When pMK1, a plasmid encoding LsrK, was introduced into E. coli 83972, externally added AI-2 was incorporated, phosphorylated, and sequestered inside the cell, while the introduction of pMK2, lacking lsrK, did not lead to AI-2 phosphorylation. These results let us conclude that E. coli 83972 produces AI-2, which can be sensed by other bacteria but cannot process AI-2 itself. This is not unusual, as other bacteria are also known to be unable to sense AI-2 and express luxS only because of its role in the activated methyl cycle (Rezzonico and Duffy, 2008). One example is the uropathogenic Proteus mirabilis strain BB2000, which does not possess a luxP or lsrB homolog but does express luxS (Rezzonico and Duffy, 2008). Furthermore, the deletion of luxS did not lead to altered colonization or pathogenicity of this strain during UTI in a mouse model (Schneider et al., 2002), suggesting that AI-2-dependent QS does not play a role during UTI in this strain. To reintroduce the ability to use AI-2 as a signaling molecule, we integrated the lsr locus into the chromosomal attB site of E. coli 83972 (Figure 2A). Indeed, when E. coli 83972 ΔluxS attB::lsr (pMK2) was challenged with extracellular DPD, we observed a significantly higher YFP expression as compared to the water control (Figures 2D, E). This result suggests that lsrK is functionally expressed in E. coli 83972 attB::lsr. Additionally, E. coli 83972 attB::lsr could export AI-2, which could be sensed by another E. coli strain in coculture (Figure 2C).
The observation that global gene expression of UPEC strains in vivo during human UTI (hUTI) or murine UTI (mUTI) was more similar to their in vitro gene expression in LB than to their in vitro gene expression in pooled human urine (Frick-Cheng et al., 2020) allows us to assess the importance of lsr-dependent gene expression for colonization of the urinary bladder using our own in vitro RNA-seq data in LB. Our transcriptome analysis revealed that the transcriptomes of the E. coli strains 83972 and 83972 attB::lsr were not drastically different during the three growth phases (Supplementary Figure S2). Only 60–72 genes were differentially expressed in E. coli strains 83972 and 83972 attB::lsr with a log2-fold change of higher or lower 1 (Figure 3). By comparing the results of Frick-Cheng and colleagues (Frick-Cheng et al., 2020) to our data, we observed that some genes or operons that were differentially expressed in vivo during hUTI/mUTI relative to in vitro growth in LB, also seem to be affected by AI-2-dependent QS. For example, several genes from the flagellar machinery (flgCFGLM and fliS) were downregulated during mUTI/hUTI as compared to growth in LB (Frick-Cheng et al., 2020). More than 50% of all downregulated genes in E. coli 83972 attB::lsr in the lag phase were linked to the flagellar machinery (Table 2). The link between AI-2-dependent QS and flagellar gene expression was also seen in the EHEC strain 86-24, where the deletion of luxS led to a downregulation of flagellar genes (Sperandio et al., 2001), which is contradictory to our results where AI-2-dependent QS seems to repress flagellar gene expression. In UPEC strain CFT073, flagella expression was not crucial for bladder colonization efficiency, whereas the loss of flagellar genes was a disadvantage during bladder colonization (Lane et al., 2005). During UTI, the flagella induce a Toll-like receptor-dependent immune response and, thus, inflammation (Subashchandrabose and Mobley, 2015; Acharya et al., 2019). However, since the flhDC genes coding for the master regulator of the flagellar biosynthesis cascade (Subashchandrabose and Mobley, 2015), and the highly antigenic flagellin, encoded by fliC (Acharya et al., 2019), were not differentially expressed in E. coli 83972 attB::lsr (Table 2, Supplementary Table S1), and since E. coli 83972 expresses only little flagella (Hancock et al., 2008), it appears unlikely that the further downregulation of already weakly expressed flagellar genes is negatively impacting this strain’s bladder colonization ability.
One of the main characteristics of E. coli 83972 is its fast growth in urine and, thus, the outgrowth of competing UPEC strains in direct competition (Roos et al., 2006; Ipe et al., 2016). One factor that might contribute to this higher competitiveness is metabolic adaptation (Ipe et al., 2016). We found several genes associated with metabolism to be differentially expressed in all three growth phases in E. coli 83972 attB::lsr (Table 1). AI-2-dependent QS was already linked to sugar metabolism (Ha et al., 2018), catabolite repression (Wang et al., 2005) and carbon regulation (Mitra et al., 2016). The strong upregulation of genes involved in the valine, leucine, and isoleucine biosynthesis (ilv operon) was interesting since ilvA and ilvC were found to be essential factors during growth in urine (Vejborg et al., 2012), probably because the overall concentrations of the three amino acids are relatively low as compared to other amino acids in urine (Bouatra et al., 2013) (Table 1, Supplementary Table S1). In E. coli strain CFT073, valine overproduction is considered a metabolic adaptation during biofilm formation (Valle et al., 2008). We also found several genes related to biofilm formation and adhesion upregulated in the transition to the stationary phase upon restoration of the lsr operon (Supplementary Table S1, Table 2). Thus, we checked for biofilm formation according to the protocol of (Laganenka et al., 2016). in E. coli strains 83972 and 83972 attB::lsr, but observed no significant differences (Supplementary Figure S7). Thus, the observed lsr-dependent upregulation of the ilv genes had no marked effect on biofilm formation of E. coli strain 83972 attB::lsr.
We wondered whether the differential regulation of metabolic genes in E. coli 83972 attB::lsr might affect growth. Indeed, we observed a growth retardation of this strain compared to the lsr-negative wild type 83972, independent of growth medium or oxygen availability (Figure 4, Supplementary Figure S3). Consistent with the findings of Sperandio and colleagues made in EHEC strain 86-24 (Sperandio et al., 2001), we observed that the deletion of luxS resulted in a significantly longer lag phase of E. coli strain 83972 (Supplementary Figure S8). Interestingly, the restoration of the lsr determinant in E. coli 83972 significantly extended the lag phase as well as the doubling time of E. coli 83972 (Supplementary Figure S8). As we did not observe an additive effect of luxS deletion and lsr restoration on the length of the lag phase in E. coli 83972, and since luxS was not differentially expressed in E. coli 83972 attB::lsr in all three growth phases, the longer lag phase of the strains 83972 ΔluxS and 83972 ΔluxS attB::lsr as compared to the wild type might thus be due to the loss of LuxS as metabolic enzyme (Schauder et al., 2001; Vendeville et al., 2005; Zdziarski et al., 2008) and not due to AI-2 secretion, as E. coli 83972 and E. coli 83972 ΔluxS are not able to sense AI-2. Taken together, these findings showed that the presence of the full-length lsr locus alone was not sufficient to cause the observed slowdown in growth but that it most likely resulted from an AI-2-dependent deregulation of specific gene expression. Because we performed the complementation as a single copy-insertion of the lsr determinant into a neutral site of the chromosome, overexpression effects and artificial phenotypes due to incompatibilities in gene regulation or metabolic stress caused by overexpression should be excluded. To validate our assumption, additional E. coli isolates will have to be tested for their lsr-dependent growth characteristics. In this context, fitness costs from the two cat and aph resistance cassettes used for complementation could be considered as well as non-QS-related functions of the lsr determinant.
In the intestinal tract, where the bacterial composition is diverse and the bacterial load is high, a negative effect of AI-2-dependent QS on growth rate does not necessarily lead to a reduction in fitness as AI-2-dependent QS significantly impacts microbiota composition, which in turn affects the fitness of individual bacterial species (Christiaen et al., 2014; Thompson et al., 2015; Deng et al., 2022). Nevertheless, our feeding experiments of G. mellonella larvae with equal ratios of E. coli 83972 wild type and the lsr-complemented strain impressively showed that the restoration of the lsr operon significantly reduced the colonization ability and competitiveness of E. coli 83972 attB::lsr in the digestive tract compared to the lsr-negative parent strain. We are fully aware that the human intestinal tract and that of G. mellonella larvae differ substantially, not only in their microbiome composition. We applied G. mellonella larvae as a 3R-compliant model to investigate the ability of E. coli 83972 to assert itself in a niche densely populated by a complex microbiota. We view this experiment as a proxy for intestinal colonization and understand that a more relevant colonization model for the human digestive tract must be used in further studies to clarify the impact of AI-2-dependent QS on the intestinal colonization capacity of phylogroup B2 strains. It has recently been described in a mouse model that AI-2 production increases chemotaxis and thus the metabolic trait-dependent fitness of E. coli strain Z1331 in the intestine. AI-2-dependent chemotaxis can also promote the coexistence of different E. coli strains in the intestine through niche segregation (Laganenka et al., 2023). Related to hardly flagellated E. coli strain 83972, we interpret our results as indicating that the importance of AI-2 in bacterial fitness in the digestive tract can be strain-dependent and influenced by individual metabolic and phenotypic characteristics. In the human bladder, however, where bacterial numbers are typically not as high as in the intestinal tract (Perez-Carrasco et al., 2021), regulating growth by monitoring cell density may be a disadvantage for urine isolates, because of rapid changes of the population size in the bladder. The urine void drastically reduces the overall bacterial count and volume of the growth compartment, urging non-voided bacteria to regrow fast to maintain bladder colonization, especially when competing with a second bacterial species (Ipe et al., 2016). QS is known to slow down growth and metabolism as soon as a high cell density is reached (Sperandio et al., 2001; An et al., 2014). When the bacteria encounter a rapid reduction in cell density, this switch leads to a subsequent adaptation of gene expression in quorum sensing strains (Pereira et al., 2013), which likely differs from that in bacteria that cannot sense the bacterial cell density. In our co-cultivation experiment, where the lsr-positive strain 83972 attB::lsr (pLS1) and the lsr-negative strain 83972 (pLS2) were in direct competition, the lsr-positive variant capable of QS was outcompeted by the isogenic lsr-negative wild type, which is unable to sense bacterial cell density but displays a shorter lag phase and doubling time (Figure 4). The observed slowdown in growth and prolonged doubling time of the lsr-positive variant could underlie the displacement of this mutant by the QS-insensitive wild type due to QS-dependent adaptations in gene expression. Especially in the case of frequent changes in population density, here mimicked by serial passage after 3 h of growth (Figure 5A), compared to the long-term competition experiment (Figure 5B), the displacement by the lsr-negative strain is particularly evident. This property benefits E. coli 83972 in bacterial interference in the urinary bladder because it can compensate for bottlenecks due to rapid reductions in population size as a result of repeated voiding. In summary, our results suggest that the differential expression of metabolic genes by AI-2-dependent QS may result in different growth characteristics and reduced competitiveness of E. coli 83972 attB::lsr. The direct evidence, however, is still missing, and the precise regulatory mechanisms remain to be elucidated.
The nrdHIEF operon was upregulated during mUTI/hUTI as compared to growth in LB (Frick-Cheng et al., 2020), and we found nrdHIEF upregulated in E. coli 83972 attB::lsr as compared to E. coli 83972 in the lag phase (Table 3). Though the operon encodes a ribonucleotide reductase that is required for dNTP synthesis and is thus associated with DNA synthesis, nrdHIEF expression was found to be stimulated upon oxidative stress, particularly in mutants that miss major antioxidant defenses (Monje-Casas et al., 2001). It is thought that E. coli overexpresses several reductases and electron donors, including nrdHIEF, to cope with oxidative stress (Monje-Casas et al., 2001; Abdelwahed et al., 2022). Interestingly, the gene ybhC, which leads to a significantly lower minimal inhibitory concentration against H2O2 when deleted in the E. coli strain BW25113 (Chen et al., 2021), was the most strongly downregulated gene during the lag and exponential growth phase and was also strongly downregulated during the transition to the stationary phase in E. coli 83972 attB::lsr (Supplementary Table S1). We wondered whether this strong downregulation might contribute to a higher sensitivity to H2O2. Therefore, we challenged E. coli 83972, its isogenic ybhC deletion mutant 83972 ΔybhC, and E. coli 83972 attB::lsr with H2O2 concentrations that can be physiologically measured in human urine (Varma and Devamanoharan, 1990). Indeed, we observed that E. coli 83972 was more resistant to H2O2 than the strains 83972 ΔybhC and 83972 attB::lsr (Figure 6). We observed that the ybhC deletion did not affect the length of the lag phase as compared to E. coli 83972 (unpaired t-test; p = 0.19), whereas E. coli 83972 attB::lsr had a significantly longer lag phase than both, E. coli 83972 and E. coli 83972 ΔybhC (Figure 6). Thus, the overall longer lag phase of the lsr-positive variant of strain 83972 does not explain its higher sensitivity to H2O2, while the strong downregulation of ybhC in this strain might be one factor responsible for this phenotype. Notably, the increased H2O2 sensitivity of E. coli 83972 attB::lsr is in agreement with a previous report of increased H2O2 sensitivity in the MPEC strain DCM5 expressing a functional LsrR (Wang et al., 2021). However, in this MPEC strain, the higher sensitivity was explained by LsrR-mediated repression of H2O2 scavenging enzyme-encoding genes, including ahpCF. As opposed to that, we found that ahpCF was slightly upregulated in strain 83972 attB::lsr in the lag phase (Supplementary Table S1). Therefore, the decreased expression of ahpCF is unlikely to explain the lsr-dependent sensitivity of E. coli 83972 to H2O2. E. coli 83972 has an increased level of endogenous reactive oxygen species while growing in urine compared to other ABU and UPEC strains, but correspondingly, it also has a more active antioxidant defense system (Aubron et al., 2012). We found several differentially expressed genes in E. coli 83972 attB::lsr that have been associated with oxidative stress in other studies (Supplementary Table S4) or other stress-related responses (Table 4). Thus, the overall H2O2 detoxification or stress response seems to be imbalanced in the presence of functional AI-2-dependent QS in E. coli 83972 attB::lsr, resulting in a higher sensitivity against oxidative stress. During ABU, however, increased sensitivity to H2O2 is a disadvantage since the infiltration of activated neutrophils into the bladder represents one host defense mechanism to kill bacteria with reactive oxygen species during UTI (Demirel et al., 2020). Especially in a competition situation in the bladder, a significantly prolonged lag phase and doubling time of E. coli 83972 attB::lsr in addition to the increased H2O2 sensitivity could constitute a clear colonization disadvantage relative to the lsr-negative wild type strain (Supplementary Figure S5).
AI-2-dependent QS in ExPEC has pleiotropic effects since it contributes to virulence, colonization, and antibiotic resistance but also H2O2 sensitivity, metabolism, and growth defects, as can also be seen in our study (Xue et al., 2009; Kathayat et al., 2021; Wang et al., 2021). Epidemiological studies have shown that ExPEC frequently belong to the phylogroup B2 (Picard et al., 1999; Micenková et al., 2016). However, in the collection of E. coli genomes we examined, genomes of phylogroup A and B1 isolates were predominant (Supplementary Table S2). Intriguingly, it was already suggested that E. coli strains of the phylogroup B2 might have lost the lsr locus during evolution (Quan and Bentley, 2012; Brito et al., 2013). However, all B2 strains analyzed by Brito and colleagues encoded a functional LuxS synthase and thus should be capable of producing AI-2 (Brito et al., 2013). Following Brito and colleagues, we found that the vast majority of E. coli belonging to the phylogroup B2 lack a full-length lsr locus (Figure 8, Supplementary Figure S6A), while the presence of luxS seems to be remarkably conserved (Supplementary Figure S6B). The high prevalence of luxS in E. coli and also over a long time in bacteria (Santiago-Rodriguez et al., 2014) underlines its crucial role as a metabolic enzyme in the activated methyl cycle (Vendeville et al., 2005). The tam gene, which is located downstream of the lsrACDBFG operon, was also widely distributed (> 88%) in the different phylogroups (Supplementary Figure S6C). Since E. coli 83972 encodes Tam and its role in AI-2-dependent QS remains unclear, we omitted tam from the complemented lsr locus. Indeed, we observed that tam was upregulated during the transition to the stationary phase (data not shown) but was not differentially expressed in E. coli 83972 attB::lsr as compared to E. coli 83972. Evolutionary studies suggest that the lsr locus was passed on via horizontal gene transfer (Rezzonico et al., 2012), and lateral gene transfer events were also described (Pereira et al., 2009). Thus, it is interesting that the operon was not reintroduced into the phylogroup B2 (Manges and Johnson, 2015). These observations and our data suggest that the loss of AI-2-dependent QS could be an evolutionary advantage for E. coli 83972, which efficiently colonizes the urinary bladder as an extraintestinal body niche. Whether the absence of the lsr determinant in almost all phylogroup B2 isolates investigated so far may be correlated, at least in part, with generally increased fitness properties in extraintestinal niches or superior colonization properties in densely populated niches such as the intestinal tract will require more detailed future analyses of the effect of lsr complementation in a diverse collection of phylogroup B2 strains.
5 Conclusion
It is imperative that we explore non-antibiotic solutions to prevent UTI, with bacterial interference being a promising avenue for treating symptomatic episodes of uncomplicated cystitis. Our study provides new information on gene regulation in E. coli 83972. The more we understand which bacterial characteristics improve fitness and colonization properties in the urinary bladder, the better we could rationally improve the engineering of E. coli 83972 or predict outcomes in therapeutic colonization. The impact of AI-2-mediated QS for pathogenicity or fitness of ExPEC is not uniform and is probably dependent on the individual strain background. Whereas interference with AI-2-dependent QS resulted in downregulation of ExPEC virulence traits and attenuation in some isolates, increased fitness due to enhanced resistance to oxidative stress was observed in others. It is tempting to speculate that the absence of the lsr operon in the majority of phylogroup B2 isolates may potentially suggest that the loss of AI-2-dependent QS could be associated with a fitness advantage in extraintestinal niches or their reservoir, i.e., the intestinal tract. However, this must be further analyzed in detail in the future. In this study, we investigated the impact of AI-2-mediated QS on fitness traits of asymptomatic bacteriuria E. coli isolate 83972, a strain that has already been used for therapeutic bladder colonization, thereby interfering with bladder infections caused by uropathogens. We found that the reintroduction of AI-2-dependent QS in E. coli 83972, which is optimally adapted to the growth conditions in the urinary bladder, has led to a phenotype that is disadvantageous for efficient and long-term bladder colonization, especially in a competitive situation. Restoration of the lsr determinant in E. coli 83972 resulted in growth retardation, loss of competitiveness and increased susceptibility to oxidative stress, all characteristics relevant to this strain’s colonization ability in the bladder. Thus, our findings indicate that the absence of AI-2-dependent QS in E. coli 83972 may be an advantage during the colonization of the urinary bladder. Further studies on the benefits of the lack of the lsr determinant in a broad spectrum of ExPEC isolates with different phylogenetic backgrounds and genome content are needed to gain deeper insights into the general importance of AI-2-dependent QS for E. coli fitness and pathogenicity in extraintestinal niches and or the colonization of the intestinal tract.
Data availability statement
The datasets presented in this study are publicly accessible at NCBI GEO (SRA accession number GSE300954). This data can be found here: (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE300954).
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
MK: Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. KM: Data curation, Investigation, Methodology, Writing – review & editing. MB: Conceptualization, Formal analysis, Supervision, Validation, Writing – review & editing. UD: Conceptualization, Formal analysis, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. German Research Foundation grant no. 281125614/GRK2220 (EvoPAD project A3)German Research Foundation grant no. CRC1009/3, project B05.
Acknowledgments
Data reported in this study appear in part in the PhD thesis of M. Keizers. The authors thank H. Wami (Münster) and P. Berger (Münster) for helpful discussions and O. Mantel (Münster) for excellent technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1603759/full#supplementary-material
References
Abdelwahed, E. K., Hussein, N. A., Moustafa, A., Moneib, N. A., and Aziz, R. K. (2022). Gene networks and pathways involved in Escherichia coli response to multiple stressors. Microorganisms 10. doi: 10.3390/microorganisms10091793
Abraham, S. N. and Miao, Y. (2015). The nature of immune responses to urinary tract infections. Nat. Rev. Immunol. 15, 655–663. doi: 10.1038/nri3887
Acharya, D., Sullivan, M. J., Duell, B. L., Goh, K. G. K., Katupitiya, L., Gosling, D., et al. (2019). Rapid bladder interleukin-10 synthesis in response to uropathogenic Escherichia coli is part of a defense strategy triggered by the major bacterial flagellar filament fliC and contingent on TLR5. mSphere 4, e00545–e00519. doi: 10.1128/mSphere.00545-19
An, J. H., Goo, E., Kim, H., Seo, Y. S., and Hwang, I. (2014). Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc. Natl. Acad. Sci. U.S.A. 111, 14912–14917. doi: 10.1073/pnas.1412431111
Ascenso, O. S., Marques, J. C., Santos, A. R., Xavier, K. B., Ventura, M. R., and Maycock, C. D. (2011). An efficient synthesis of the precursor of AI-2, the signalling molecule for inter-species quorum sensing. Bioorg Med. Chem. 19, 1236–1241. doi: 10.1016/j.bmc.2010.12.036
Aubron, C., Glodt, J., Matar, C., Huet, O., Borderie, D., Dobrindt, U., et al. (2012). Variation in endogenous oxidative stress in Escherichia coli natural isolates during growth in urine. BMC Microbiol 12, 120. doi: 10.1186/1471-2180-12-120
Babraham Bioinformatics FastQC A Quality Control tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed May 21, 2025).
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4. doi: 10.1099/mgen.0.000192
Berger, M., Gerganova, V., Berger, P., Rapiteanu, R., Lisicovas, V., and Dobrindt, U. (2016). Genes on a wire: the nucleoid-associated protein HU insulates transcription units in Escherichia coli. Sci. Rep. 6, 31512. doi: 10.1038/srep31512
Bouatra, S., Aziat, F., Mandal, R., Guo, A. C., Wilson, M. R., Knox, C., et al. (2013). The human urine metabolome. PloS One 8, e73076. doi: 10.1371/journal.pone.0073076
Brito, P. H., Rocha, E. P., Xavier, K. B., and Gordo, I. (2013). Natural genome diversity of AI-2 quorum sensing in Escherichia coli: conserved signal production but labile signal reception. Genome Biol. Evol. 5, 16–30. doi: 10.1093/gbe/evs122
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Chen, H. (2022). VennDiagram: Generate High-Resolution Venn and Euler Plots. Available online at: https://cloud.r-project.org/web/packages/VennDiagram/index.html (Accessed May 21, 2025).
Chen, H., Wilson, J., Ercanbrack, C., Smith, H., Gan, Q., and Fan, C. (2021). Genome-wide screening of oxidizing agent resistance genes in Escherichia coli. Antioxidants 10, 861. doi: 10.3390/antiox10060861
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Choi, J., Shin, D., Kim, M., Park, J., Lim, S., and Ryu, S. (2012). LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PloS One 7, e37059. doi: 10.1371/journal.pone.0037059
Christiaen, S. E., O'connell Motherway, M., Bottacini, F., Lanigan, N., Casey, P. G., Huys, G., et al. (2014). Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PloS One 9, e98111. doi: 10.1371/journal.pone.0098111
Cook, M. A. and Wright, G. D. (2022). The past, present, and future of antibiotics. Sci. Transl. Med. 14, eabo7793. doi: 10.1126/scitranslmed.abo7793
Dainat, J. (2022). “Another Gtf/Gff Analysis Toolkit (AGAT): Resolve interoperability issues and accomplish more with your annotations. Available at: https://nbisweden.github.io/AGAT/ (Accessed May 21, 2025).
Datsenko, K. A. and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Demirel, I., Persson, A., Brauner, A., Sarndahl, E., Kruse, R., and Persson, K. (2020). Activation of NLRP3 by uropathogenic Escherichia coli is associated with IL-1beta release and regulation of antimicrobial properties in human neutrophils. Sci. Rep. 10, 21837. doi: 10.1038/s41598-020-78651-1
Deng, Z., Hou, K., Valencak, T. G., Luo, X. M., Liu, J., and Wang, H. (2022). AI-2/LuxS Quorum Sensing System Promotes Biofilm Formation of Lactobacillus rhamnosus GG and Enhances the Resistance to Enterotoxigenic Escherichia coli in Germ-Free Zebrafish. Microbiol Spectr. 10, e0061022. doi: 10.1128/spectrum.00610-22
Ewels, P., Magnusson, M., Lundin, S., and Kaller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Farrell, K., Tandan, M., Hernandez Santiago, V., Gagyor, I., Braend, A. M., Skow, M., et al. (2021). Treatment of uncomplicated UTI in males: a systematic review of the literature. BJGP Open 5, bjgpopen20X101140. doi: 10.3399/bjgpopen20X101140
Flores-Mireles, A. L., Walker, J. N., Caparon, M., and Hultgren, S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol 13, 269–284. doi: 10.1038/nrmicro3432
Frick-Cheng, A. E., Sintsova, A., Smith, S. N., Krauthammer, M., Eaton, K. A., and Mobley, H. L. T. (2020). The gene expression profile of uropathogenic Escherichia coli in women with uncomplicated urinary tract infections is recapitulated in the mouse model. mBio 11, e01412–e01420. doi: 10.1128/mBio.01412-20
George, I., Kalairaj, M. S., Zimmern, P. E., Ware, T. H., and Subashchandrabose, S. (2024). Competitive fitness of asymptomatic bacteriuria E. coli strain 83972 against uropathogens in human urine. Infect. Immun. 92, e0017324. doi: 10.1128/iai.00173-24
Ha, J. H., Hauk, P., Cho, K., Eo, Y., Ma, X., Stephens, K., et al. (2018). Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci. Adv. 4, eaar7063. doi: 10.1126/sciadv.aar7063
Han, X., Bai, H., Tu, J., Yang, L., Xu, D., Wang, S., et al. (2015). Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb Pathog 88, 39–47. doi: 10.1016/j.micpath.2015.08.003
Hancock, V., Seshasayee, A. S., Ussery, D. W., Luscombe, N. M., and Klemm, P. (2008). Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972. Mol. Genet. Genomics 279, 523–534. doi: 10.1007/s00438-008-0330-9
Helmy, Y. A., Kathayat, D., Deblais, L., Srivastava, V., Closs, G., Jr., Tokarski, R. J., 2nd, et al. (2022). Evaluation of novel quorum sensing inhibitors targeting auto-inducer 2 (AI-2) for the control of avian pathogenic Escherichia coli infections in chickens. Microbiol Spectr. 10, e0028622. doi: 10.1128/spectrum.00286-22
Henly, E. L., Norris, K., Rawson, K., Zoulias, N., Jaques, L., Chirila, P. G., et al. (2021). Impact of long-term quorum sensing inhibition on uropathogenic Escherichia coli. J. Antimicrob Chemother. 76, 909–919. doi: 10.1093/jac/dkaa517
Hull, R. A., Rudy, D. C., Donovan, W. H., Wieser, I. E., Stewart, C., and Darouiche, R. O. (1999). Virulence properties of Escherichia coli 83972, a prototype strain associated with asymptomatic bacteriuria. Infect. Immun. 67, 429–432. doi: 10.1128/IAI.67.1.429-432.1999
Ipe, D. S., Horton, E., and Ulett, G. C. (2016). The basics of bacteriuria: strategies of microbes for persistence in urine. Front. Cell Infect. Microbiol 6, 14. doi: 10.3389/fcimb.2016.00014
Ismail, A. S., Valastyan, J. S., and Bassler, B. L. (2016). A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480. doi: 10.1016/j.chom.2016.02.020
Kathayat, D., Lokesh, D., Ranjit, S., and Rajashekara, G. (2021). Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens 10, 467. doi: 10.3390/pathogens10040467
Keizers, M., Dobrindt, U., and Berger, M. (2022). A simple biosensor-based assay for quantitative autoinducer-2 analysis. ACS Synth Biol. 11, 747–759. doi: 10.1021/acssynbio.1c00459
Kenneally, C., Murphy, C. P., Sleator, R. D., and Culligan, E. P. (2022). The urinary microbiome and biological therapeutics: Novel therapies for urinary tract infections. Microbiol Res. 259, 127010. doi: 10.1016/j.micres.2022.127010
Khera, R., Mehdipour, A. R., Bolla, J. R., Kahnt, J., Welsch, S., Ermler, U., et al. (2022). Cryo-EM structures of pentameric autoinducer-2 exporter from Escherichia coli reveal its transport mechanism. EMBO J. 41, e109990. doi: 10.15252/embj.2021109990
Kitts, P. A., Church, D. M., Thibaud-Nissen, F., Choi, J., Hem, V., Sapojnikov, V., et al. (2016). Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 44, D73–D80. doi: 10.1093/nar/gkv1226
Köhler, C. D. and Dobrindt, U. (2011). What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol 301, 642–647. doi: 10.1016/j.ijmm.2011.09.006
Kolde, R. (2019). Package ‘pheatmap’. Available online at: https://cran.r-project.org/web//packages/pheatmap/pheatmap.pdf (Accessed May 21, 2025).
Köves, B., Salvador, E., Gronberg-Hernandez, J., Zdziarski, J., Wullt, B., Svanborg, C., et al. (2014). Rare emergence of symptoms during long-term asymptomatic Escherichia coli 83972 carriage without an altered virulence factor repertoire. J. Urol 191, 519–528. doi: 10.1016/j.juro.2013.07.060
Laganenka, L., Colin, R., and Sourjik, V. (2016). Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun. 7, 12984. doi: 10.1038/ncomms12984
Laganenka, L., Lee, J. W., Malfertheiner, L., Dieterich, C. L., Fuchs, L., Piel, J., et al. (2023). Chemotaxis and autoinducer-2 signalling mediate colonization and contribute to co-existence of Escherichia coli strains in the murine gut. Nat. Microbiol 8, 204–217. doi: 10.1038/s41564-022-01286-7
Laganenka, L. and Sourjik, V. (2018). Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species coculture. Appl. Environ. Microbiol 84, e02638–e02617. doi: 10.1128/AEM.02638-17
Lane, M. C., Lockatell, V., Monterosso, G., Lamphier, D., Weinert, J., Hebel, J. R., et al. (2005). Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73, 7644–7656. doi: 10.1128/IAI.73.11.7644-7656.2005
Lange, A., Schäfer, A., and Frick, J. S. (2019). A Galleria mellonella oral administration model to study commensal-induced innate immune responses. J. Vis Exp. 145, e59270. doi: 10.3791/59270
Li, J., Attila, C., Wang, L., Wood, T. K., Valdes, J. J., and Bentley, W. E. (2007). Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol 189, 6011–6020. doi: 10.1128/JB.00014-07
Li, H. and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lila, A. S. A., Rajab, A. A. H., Abdallah, M. H., Rizvi, S. M. D., Moin, A., Khafagy, E. S., et al. (2023). Biofilm lifestyle in recurrent urinary tract infections. Life 13, 148. doi: 10.3390/life13010148
Loubet, P., Ranfaing, J., Dinh, A., Dunyach-Remy, C., Bernard, L., Bruyere, F., et al. (2020). Alternative therapeutic options to antibiotics for the treatment of urinary tract infections. Front. Microbiol 11, 1509. doi: 10.3389/fmicb.2020.01509
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Manges, A. R. and Johnson, J. R. (2015). Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol Spectr. 3. doi: 10.1128/microbiolspec.uti-0006-2012
Mann, R., Mediati, D. G., Duggin, I. G., Harry, E. J., and Bottomley, A. L. (2017). Metabolic adaptations of uropathogenic E. coli in the urinary tract. Front. Cell Infect. Microbiol 7, 241. doi: 10.3389/fcimb.2017.00241
Marques, J. C., Lamosa, P., Russell, C., Ventura, R., Maycock, C., Semmelhack, M. F., et al. (2011). Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): characterization of phospho-(S)-4,5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. J. Biol. Chem. 286, 18331–18343. doi: 10.1074/jbc.M111.230227
Marques, J. C., Oh, I. K., Ly, D. C., Lamosa, P., Ventura, M. R., Miller, S. T., et al. (2014). LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2. Proc. Natl. Acad. Sci. U.S.A. 111, 14235–14240. doi: 10.1073/pnas.1408691111
Martin, M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Medina, M. and Castillo-Pino, E. (2019). An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol 11, 1756287219832172. doi: 10.1177/1756287219832172
Micenková, L., Bosák, J., Vrba, M., Ševčíková, A., and Šmajs, D. (2016). Human extraintestinal pathogenic Escherichia coli strains differ in prevalence of virulence factors, phylogroups, and bacteriocin determinants. BMC Microbiol 16, 218. doi: 10.1186/s12866-016-0835-z
Midani, F. S., Collins, J., and Britton, R. A. (2021). AMiGA: software for automated analysis of microbial growth assays. mSystems 6, e0050821. doi: 10.1128/msystems.00508-21
Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L., et al. (2004). Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15, 677–687. doi: 10.1016/j.molcel.2004.07.020
Mitra, A., Herren, C. D., Patel, I. R., Coleman, A., and Mukhopadhyay, S. (2016). Integration of AI-2 based cell-cell signaling with metabolic cues in Escherichia coli. PloS One 11, e0157532. doi: 10.1371/journal.pone.0157532
Monje-Casas, F., Jurado, J., Prieto-Alamo, M. J., Holmgren, A., and Pueyo, C. (2001). Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 276, 18031–18037. doi: 10.1074/jbc.M011728200
Mukherjee, K., Amsel, D., Kalsy, M., Billion, A., Dobrindt, U., and Vilcinskas, A. (2020). MicroRNAs regulate innate immunity against uropathogenic and commensal-like Escherichia coli infections in the surrogate insect model Galleria mellonella. Sci. Rep. 10, 2570. doi: 10.1038/s41598-020-59407-3
Nesta, B., Valeri, M., Spagnuolo, A., Rosini, R., Mora, M., Donato, P., et al. (2014). SslE elicits functional antibodies that impair in vitro mucinase activity and in vivo colonization by both intestinal and extraintestinal Escherichia coli strains. PloS Pathog 10, e1004124. doi: 10.1371/journal.ppat.1004124
Palaniyandi, S., Mitra, A., Herren, C. D., Zhu, X., and Mukhopadhyay, S. (2013). LuxS contributes to virulence in avian pathogenic Escherichia coli O78:K80:H9. Vet Microbiol 166, 567–575. doi: 10.1016/j.vetmic.2013.07.009
Peng, L., Dumevi, R. M., Chitto, M., Haarmann, N., Berger, P., Koudelka, G., et al. (2022). A robust one-step recombineering system for enterohemorrhagic Escherichia coli. Microorganisms 10, 1689. doi: 10.3390/microorganisms10091689
Pereira, C. S., De Regt, A. K., Brito, P. H., Miller, S. T., and Xavier, K. B. (2009). Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol 191, 6975–6987. doi: 10.1128/JB.00976-09
Pereira, C. S., Santos, A. J., Bejerano-Sagie, M., Correia, P. B., Marques, J. C., and Xavier, K. B. (2012). Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol. Microbiol 84, 93–104. doi: 10.1111/j.1365-2958.2012.08010.x
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Perez-Carrasco, V., Soriano-Lerma, A., Soriano, M., Gutierrez-Fernandez, J., and Garcia-Salcedo, J. A. (2021). Urinary microbiome: yin and yang of the urinary tract. Front. Cell Infect. Microbiol 11, 617002. doi: 10.3389/fcimb.2021.617002
Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67, 546–553. doi: 10.1128/IAI.67.2.546-553.1999
Quan, D. N. and Bentley, W. E. (2012). Gene network homology in prokaryotes using a similarity search approach: queries of quorum sensing signal transduction. PloS Comput. Biol. 8, e1002637. doi: 10.1371/journal.pcbi.1002637
R: The R Project for Statistical Computing. Available online at: https://www.r-project.org/ (Accessed May 21, 2025).
Revelle, W. (2023). Package ‘psych’: Procedures for Psychological, Psychometric, and Personality Research. Available online at: https://cran.r-project.org/web/packages/psych/psych.pdf (Accessed May 21, 2025).
Rezzonico, F. and Duffy, B. (2008). Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol 8, 154. doi: 10.1186/1471-2180-8-154
Rezzonico, F., Smits, T. H., and Duffy, B. (2012). Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors 12, 6645–6665. doi: 10.3390/s120506645
Rodriguez-Siek, K. E., Giddings, C. W., Doetkott, C., Johnson, T. J., Fakhr, M. K., and Nolan, L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151, 2097–2110. doi: 10.1099/mic.0.27499-0
Roos, V., Ulett, G. C., Schembri, M. A., and Klemm, P. (2006). The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74, 615–624. doi: 10.1128/IAI.74.1.615-624.2006
Rutherford, S. T. and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect. Med. 2, a012427. doi: 10.1101/cshperspect.a012427
Santiago-Rodriguez, T. M., Patricio, A. R., Rivera, J. I., Coradin, M., Gonzalez, A., Tirado, G., et al. (2014). luxS in bacteria isolated from 25- to 40-million-year-old amber. FEMS Microbiol Lett. 350, 117–124. doi: 10.1111/fml.2014.350.issue-1
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Schiller, P., Knödler, M., Berger, P., Greune, L., Fruth, A., Mellmann, A., et al. (2021). The superior adherence phenotype of E. coli O104:H4 is directly mediated by the aggregative adherence fimbriae type I. Virulence 12, 346–359. doi: 10.1080/21505594.2020.1868841
Schneider, R., Lockatell, C. V., Johnson, D., and Belas, R. (2002). Detection and mutation of a luxS-encoded autoinducer in Proteus mirabilis. Microbiology 148, 773–782. doi: 10.1099/00221287-148-3-773
Sora, V. M., Meroni, G., Martino, P. A., Soggiu, A., Bonizzi, L., and Zecconi, A. (2021). Extraintestinal pathogenic Escherichia coli: virulence factors and antibiotic resistance. Pathogens 10, 1355. doi: 10.3390/pathogens10111355
Sperandio, V., Torres, A. G., Giron, J. A., and Kaper, J. B. (2001). Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol 183, 5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001
Stork, C., Kovacs, B., Rozsai, B., Putze, J., Kiel, M., Dorn, A., et al. (2018). Characterization of Asymptomatic Bacteriuria Escherichia coli Isolates in Search of Alternative Strains for Efficient Bacterial Interference against Uropathogens. Front. Microbiol 9, 214. doi: 10.3389/fmicb.2018.00214
Subashchandrabose, S. and Mobley, H. L. T. (2015). Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr. 3. doi: 10.1128/microbiolspec.UTI-0015-2012
Sun, J., Daniel, R., Wagner-Döbler, I., and Zeng, A. P. (2004). Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4, 36. doi: 10.1186/1471-2148-4-36
Sundén, F., Håkansson, L., Ljunggren, E., and Wullt, B. (2010). Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J. Urol 184, 179–185. doi: 10.1016/j.juro.2010.03.024
Taga, M. E., Miller, S. T., and Bassler, B. L. (2003). Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol 50, 1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x
Taga, M. E., Semmelhack, J. L., and Bassler, B. L. (2001). The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol 42, 777–793. doi: 10.1046/j.1365-2958.2001.02669.x
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C., and Xavier, K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049
Tivendale, K. A., Logue, C. M., Kariyawasam, S., Jordan, D., Hussein, A., Li, G., et al. (2010). Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78, 3412–3419. doi: 10.1128/IAI.00347-10
Trappetti, C., Mcallister, L. J., Chen, A., Wang, H., Paton, A. W., Oggioni, M. R., et al. (2017). Autoinducer 2 Signaling via the Phosphotransferase FruA Drives Galactose Utilization by Streptococcus pneumoniae, Resulting in Hypervirulence. mBio 8, e02269–e02216. doi: 10.1128/mBio.02269-16
Valastyan, J. S., Kraml, C. M., Pelczer, I., Ferrante, T., and Bassler, B. L. (2021). Saccharomyces cerevisiae requires CFF1 to produce 4-hydroxy-5-methylfuran-3(2H)-one, a mimic of the bacterial quorum-sensing autoinducer AI-2. mBio 12, e03303–e03320. doi: 10.1128/mBio.03303-20
Valle, J., Da Re, S., Schmid, S., Skurnik, D., D'ari, R., and Ghigo, J. M. (2008). The amino acid valine is secreted in continuous-flow bacterial biofilms. J. Bacteriol 190, 264–274. doi: 10.1128/JB.01405-07
Varma, S. D. and Devamanoharan, P. S. (1990). Excretion of hydrogen peroxide in human urine. Free Radic. Res. Commun. 8, 73–78. doi: 10.3109/10715769009087976
Vejborg, R. M., De Evgrafov, M. R., Phan, M. D., Totsika, M., Schembri, M. A., and Hancock, V. (2012). Identification of genes important for growth of asymptomatic bacteriuria Escherichia coli in urine. Infect. Immun. 80, 3179–3188. doi: 10.1128/IAI.00473-12
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol 3, 383–396. doi: 10.1038/nrmicro1146
Vila, J., Saez-Lopez, E., Johnson, J. R., Römling, U., Dobrindt, U., Canton, R., et al. (2016). Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev. 40, 437–463. doi: 10.1093/femsre/fuw005
Wallenstein, A., Rehm, N., Brinkmann, M., Selle, M., Bossuet-Greif, N., Sauer, D., et al. (2020). ClbR is the key transcriptional activator of colibactin gene expression in escherichia coli. mSphere 5, e00591–e00520. doi: 10.1128/msphere.00591-20
Wang, L., Hashimoto, Y., Tsao, C. Y., Valdes, J. J., and Bentley, W. E. (2005). Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol 187, 2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005
Wang, R. F. and Kushner, S. R. (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199. doi: 10.1016/0378-1119(91)90366-J
Wang, H., Shang, F., Shen, J., Xu, J., Chen, X., Ni, J., et al. (2021). LsrR, the effector of AI-2 quorum sensing, is vital for the H(2)O(2) stress response in mammary pathogenic Escherichia coli. Vet Res. 52, 127. doi: 10.1186/s13567-021-00998-8
Wang, L., Wang, S., and Li, W. (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. doi: 10.1093/bioinformatics/bts356
Wiles, T. J., Kulesus, R. R., and Mulvey, M. A. (2008). Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85, 11–19. doi: 10.1016/j.yexmp.2008.03.007
Wullt, B. and Svanborg, C. (2016). Deliberate establishment of asymptomatic bacteriuria-A novel strategy to prevent recurrent UTI. Pathogens 5, 52. doi: 10.3390/pathogens5030052
Xavier, K. B. and Bassler, B. L. (2005). Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol 187, 238–248. doi: 10.1128/JB.187.1.238-248.2005
Xue, T., Yu, L., Shang, F., Li, W., Zhang, M., Ni, J., et al. (2016). Short communication: The role of autoinducer 2 (AI-2) on antibiotic resistance regulation in an Escherichia coli strain isolated from a dairy cow with mastitis. J. Dairy Sci. 99, 4693–4698. doi: 10.3168/jds.2015-10543
Xue, T., Zhao, L., Sun, H., Zhou, X., and Sun, B. (2009). LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Res. 19, 1258–1268. doi: 10.1038/cr.2009.91
Yu, L., Li, W., Li, Q., Chen, X., Ni, J., Shang, F., et al. (2020). Role of LsrR in the regulation of antibiotic sensitivity in avian pathogenic Escherichia coli. Poult Sci. 99, 3675–3687. doi: 10.1016/j.psj.2020.03.064
Zargar, A., Quan, D. N., Carter, K. K., Guo, M., Sintim, H. O., Payne, G. F., et al. (2015). Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. mBio 6, e00025. doi: 10.1128/mBio.00025-15
Zdziarski, J., Svanborg, C., Wullt, B., Hacker, J., and Dobrindt, U. (2008). Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect. Immun. 76, 695–703. doi: 10.1128/IAI.01215-07
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11, 5371. doi: 10.1038/s41467-020-19243-5
Keywords: lsr locus, extraintestinal pathogenic E. coli, fitness, stress response, competitiveness
Citation: Keizers M, Mukherjee K, Berger M and Dobrindt U (2025) Less is more: the lack of autoinducer-2-dependent quorum sensing promotes competitive fitness of Escherichia coli strain 83972. Front. Cell. Infect. Microbiol. 15:1603759. doi: 10.3389/fcimb.2025.1603759
Received: 31 March 2025; Accepted: 07 May 2025;
Published: 02 July 2025.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Ruixue Zhang, Washington University in St. Louis, United StatesWang Zhang, Sir Run Run Shaw Hospital, China
Ignatius Ivan, Siloam Hospitals, Indonesia
Copyright © 2025 Keizers, Mukherjee, Berger and Dobrindt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrich Dobrindt, ZG9icmluZHRAdW5pLW11ZW5zdGVyLmRl
 Marla Keizers
Marla Keizers Krishnendu Mukherjee
Krishnendu Mukherjee Michael Berger
Michael Berger Ulrich Dobrindt
Ulrich Dobrindt