- 1Henan Province Hospital of Traditional Chinese Medicine (The Second Affiliated Hospital of Henan University of Chinese Medicine), Zhengzhou, China
- 2The Second Clinical Medical College, Henan University of Chinese Medicine, Zhengzhou, China
- 3The Key Laboratory of Pathogenic Microbes and Antimicrobial Resistance Surveillance of Zhengzhou, Henan Province Hospital of Traditional Chinese Medicine, Zhengzhou, China
- 4Henan Engineering Research Center for Identification of Pathogenic Microbes, Henan Province Hospital of Traditional Chinese Medicine, Zhengzhou, China
- 5Henan Provincial Key Laboratory of Antibiotics-Resistant Bacterial Infection Prevention and Therapy with Traditional Chinese Medicine, Henan Province Hospital of Traditional Chinese Medicine, Zhengzhou, China
Carbapenem-resistant Klebsiella pneumoniae (CRKP) has emerged as a critical global public health threat, characterized by high infection rates, elevated mortality, and limited therapeutic options. In this study, we isolated and characterized a novel bacteriophage (phage), designated as HZJ31, which exhibited potent lytic activity against CRKP strains. Phylogenetic and genomic analyses revealed that phage HZJ31 belongs to the order Caudovirales and lacks virulence factors, antibiotic resistance genes, and lysogeny-related elements, supporting its suitability for therapeutic applications. Phage HZJ31 exhibits remarkable anti-biofilm activity by preventing biofilm formation and disrupting established biofilms, with bacterial reduction rates exceeding 70% (P<0.05). In combination with Tigecycline, it significantly enhanced bactericidal efficacy, delayed the emergence of phage resistant mutants, and improved survival rates in Galleria mellonella larvae infection models. Compared to the bacterial-infected group, which had 80% larval mortality at 96 h, treatment with HZJ31 or TGC alone led to 50% and 60% survival, while their combination improved survival to 70% (P < 0.05). Notably, the phage-resistant mutant, which emerged due to capsule loss, resulted in reduced growth and virulence, while regaining sensitivity to certain antibiotics (such as gentamicin), indicating a fitness cost associated with phage resistance. Collectively, these findings provide valuable insights into phage-antibiotic synergy and underscore the promising clinical potential of phage HZJ31 as a therapeutic agent against CRKP infections.
1 Introduction
Antibiotic resistance is acknowledged as a critical threat to public health. Carbapenems, including imipenem and meropenem, are highly effective broad-spectrum β-lactam antibiotics used as a last resort against multidrug-resistant Gram-negative infections, particularly those caused by ESBL- and AmpC-producing Klebsiella pneumoniae (KP) strains (Bologna et al., 2024; Naghavi et al., 2024). The extensive clinical utilization of carbapenems has led to the global rise of carbapenem-resistant Klebsiella pneumoniae (CRKP) (Ding et al., 2023; Hu et al., 2024a). In 2024, according to the China Antimicrobial Surveillance Network (CHINET), KP strains ranked second among all clinical isolates, accounting for 13.9% (n=458271), second only to Escherichia coli at 18.2%. From 2014 to 2024, clinical KP isolates showed increased resistance to imipenem and meropenem, rising from 11.0% and 14.1% to 22.6% and 23.4%, respectively, indicating a concerning trend (Karampatakis et al., 2023). Infections with CRKP endure a significant fatality rate and are challenging to treat (Lou et al., 2022; Hu et al., 2024a). In 2017, WHO published the WHO Priority Pathogens List for R&D of New Antibiotics, where carbapenem-resistant Enterobacteriaceae (including CRKP) was categorized as a “Critical” priority level (Miller and Arias, 2024). Treatment options for CRKP infections are extremely restricted, emphasizing the urgent need for novel antibacterial strategies (Hu et al., 2024a; Xing et al., 2025).
Bacteriophages (phages) are viruses that specifically infect bacteria and cause lysis, characterized by their ubiquity, abundance, diversity, structural simplicity, and amenability to genetic manipulation. In light of the escalating global antimicrobial resistance crisis, phage therapy has garnered renewed interest from the scientific community (Goodridge, 2010; Jault et al., 2019; Patpatia et al., 2021; Uyttebroek et al., 2022). Phages are classified into lytic and lysogenic phages based on their life cycle and replication characteristics. Lytic phages exhibit specific adsorption to bacterial cells and release progeny phages through a five-stage process: adsorption, penetration, biosynthesis, maturation, and release (Suttle, 2007). Bacteriophages, whether administered as monotherapy or in combination regimens, have demonstrated potent bactericidal activity against KP strains (Martins et al., 2022; Mulani et al., 2022), emerging as a promising therapeutic modality for CRKP infections (Baqer et al., 2022; Gorodnichev et al., 2023; Kou et al., 2024). Phage therapy offers distinct advantages over antibiotics, such as effectiveness against multidrug-resistant bacteria, anti-biofilm capabilities, high specificity, potent bactericidal activity, and the potential for genetic engineering (Kortright et al., 2019; Xing et al., 2025). Phage therapy faces multiple challenges, including a restricted host range, the rapid emergence of phage-resistant mutants, possible clearance by the host immune system, the absence of standardized manufacturing protocols and robust clinical trial data (Strathdee et al., 2023) (Uyttebroek et al., 2022). Moreover, recent studies have highlighted the potential risk of horizontal gene transfer (HGT) mediated by phages, which may unintentionally contribute to the dissemination of virulence factors and antibiotic resistance genes among pathogenic bacteria (Colavecchio et al., 2017; Cui et al., 2024; Cook and Hynes, 2025; She et al., 2025). These limitations significantly impede its broader clinical application.
This study identified a newly isolated lytic phage, HZJ31, targeting CRKP, and evaluated its biology, genome, anti-biofilm activity, and antibacterial efficacy in vitro and in vivo, highlighting its potential as a novel antibacterial strategy for CRKP infections. Notably, the analysis of the phage-resistant strain sheds light on the dynamic interactions between phages and bacteria, while also offering valuable insights into the fitness costs associated with phage resistance.
2 Materials and methods
2.1 Bacterial strains and culture conditions
Clinical KP strains, including KPZ2 and its phage-resistant mutant KPZ2-R, were isolated from the clinical laboratory at Henan Provincial Hospital of Traditional Chinese Medicine and preserved in 25% glycerol at -80°C. The strains were identified using the IVD MALDI Biotyper mass spectrometer (Bruker Daltonik, Germany). Antimicrobial susceptibility testing was performed using the VITEK 2 Compact system (bioMérieux, France), and results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. The strains were revived on blood agar plates and incubated overnight at 37°C with 5% CO2. After that, one colony was added to Luria-Bertani broth and cultured at 37°C with shaking at 180 rpm until the OD600 reached 0.6–1.0, indicating the logarithmic growth phase.
2.2 Phage isolation and purification
Phages were isolated from untreated hospital wastewater using a modified enrichment protocol (Wang et al., 2024). To screen for lytic phages, several clinical KP isolates, including KPZ2, were randomly selected as candidate host strains. After being stored at 4°C for 2 h, untreated sewage samples were concentrated at 8000 rpm for 10 min and filtered through a 0.22 μm filter. The filtered sewage was then mixed with an equal volume of 2× LB broth and inoculated with a clinical isolate (OD600 = 0.5). The mixture was incubated overnight at 37°C with shaking at 120 rpm. The phage stock was obtained by collecting the supernatant after centrifuging and filtering the supernatant through a 0.22 μm filter. The double-layer agar plate approach was used to purify phages until a single type of phage morphology was observed.
2.3 Transmission electron microscopy
The agarose gel blocks containing phages were incubated in PBS at 4°C for 6 h, then centrifuged and filtered through a 0.22 μm filter to obtain phage suspension. The phage suspension was concentrated with a 30 kDa ultrafiltration tube at 3000 × g and 4°C for 15 min. Phages were adsorbed onto the 200-mesh carbon-coated copper grid and stained with 2% phosphotungstic acid. Phage morphology was observed using a Hitachi HT7800.
2.4 Optimal multiplicity of infection
The optimal MOI for phage HZJ31 was determined with a modified protocol (Wang et al., 2024) by incubating it with KPZ2 at MOI values of 0.001, 0.01, 0.1, and 1 at 37°C with shaking at 200 rpm for 4 h. The mixture of cultures was centrifuged at 6000 rpm for 15 min and the supernatant passed through a 0.22 μm filter to remove bacterial cells. Phage titers were determined using the double-layer agar method.
2.5 One-step growth curve
The one-step growth curve of phage HZJ31 was evaluated using the method (Fei et al., 2023). Briefly, phages HZJ31(MOI=0.001) and KPZ2 were incubated at 37°C for 15 min to allow adsorption. The mixture was then centrifuged at 12,000 rpm for 5 min at 4°C, and the supernatant was removed. The pellet was resuspended in LB broth and incubated at 37°C with shaking at 200 rpm for 100 min. The samples were collected at 10-minute intervals, and phage titers were quantified to generate the growth curve. The burst size = (Final phage titer - Initial free phage titer)/Number of infected bacterial cells.
2.6 Host range of phage
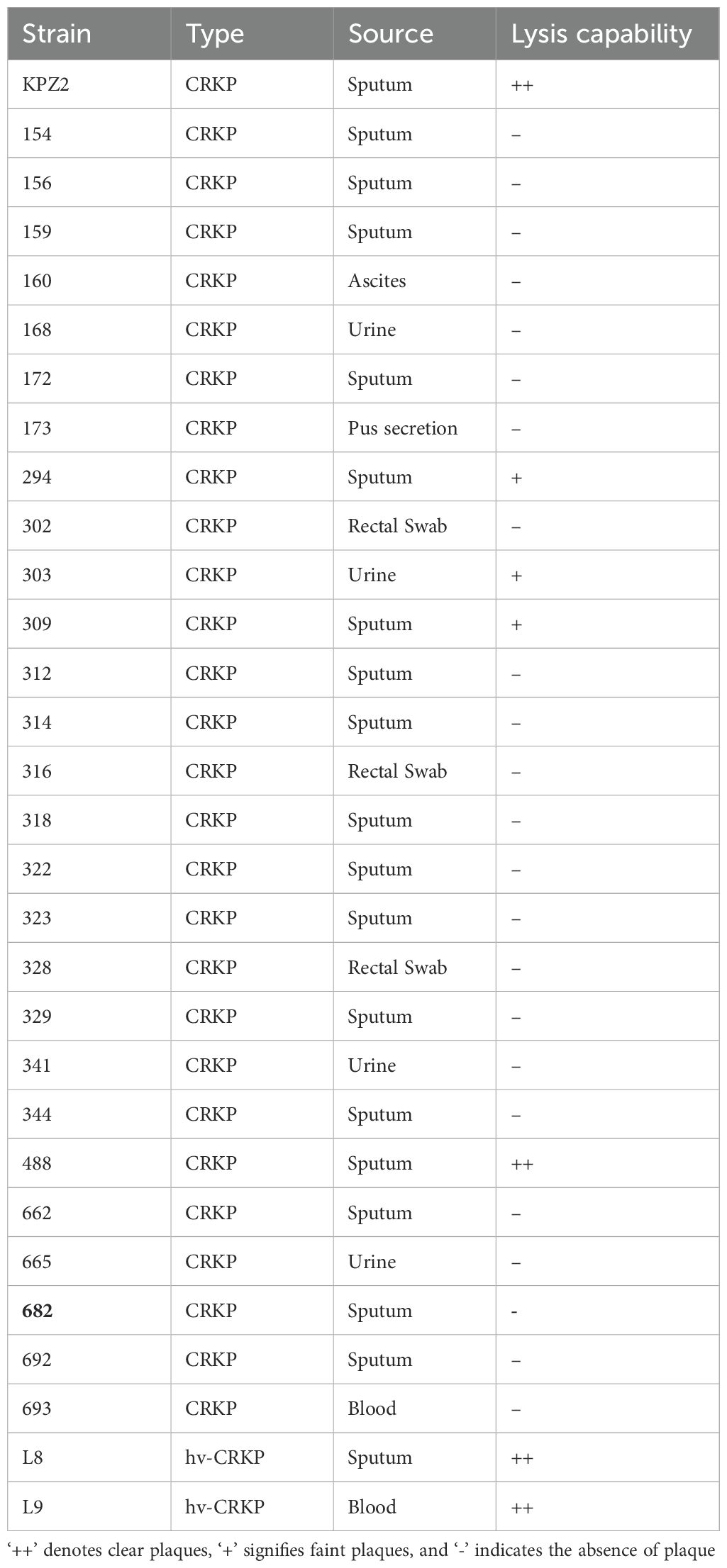
The host range of phage HZJ31 was determined by the spot assay according to the protocol described previously (Wang et al., 2024). Clinical CRKP isolates were adjusted to an OD600 of 0.5 using PBS, and the bacterial suspensions were evenly spread on LB agar plates. Then, 10 μL of phage HZJ31 was immediately spotted onto the surface, followed by overnight incubation at 37°C. The host range was evaluated through plaque formation: ++ denotes clear plaques, + signifies faint plaques, and - indicates the absence of plaque. All experiments were conducted in triplicate for consistency.
2.7 The stability for temperature and pH
The stability of phage was carried out according to the published paper (Fei et al., 2023). The thermal stability of phage HZJ31 was tested by incubating the phage (1×1010 PFU/mL) at temperatures of 4, 25, 37, 50, 60, 70, and 80°C for 1 h. For pH stability, phage HZJ31 was mixed with SM buffer at pH values of 3, 5, 7, 9, 10, 11, and 13 in a 1:10 (v/v) ratio and incubated at 37°C for 1 h. Then, phage titers were determined.
2.8 Cytotoxicity of phage by CCK-8 assay
The procedures were performed according to the previous study with minor modifications (Wang et al., 2024). The cells were plated at a density of 1×104 cells per well in a 96-well plate and differentiated with 200 ng/mL of phorbol 12-myristate 13-acetate (PMA) overnight at 37°C with 5% CO2. Phage HZJ31 was diluted from 108 to 1014 PFU/mL in RPMI-1640 with 10% FBS and incubated with cells at 37°C, 5% CO2 for 24, 48, and 72 h. Following incubation, 100 μL of a CCK-8 premix (Beijing Solarbio, China) with 90% PBS was added to each well after the cells were washed twice with PBS. After an additional incubation for 3 h, absorbance at OD450 nm was detected.
2.9 Genomic extraction of phages
The Phage DNA was extracted utilizing the Viral Gene Extraction Kit (Takara Bio, China). A solution containing 200 μL Buffer VGB, 20 μL Proteinase K, one μL Carrier RNA, and 200 μL phage suspension (108 PFU/mL) was incubated at 56°C for 10 min.200 μL of anhydrous ethanol was added and mixed before transferring to a spin column. The mixture was centrifuged at 12,000 rpm for 2 min, and the filtrate was discarded. Subsequently, 500 μL of Buffer RWA and 700 μL of Buffer RWB were added in sequence, centrifuged, and discarded, with Buffer RWB applied twice, followed by centrifugation at 12,000 rpm for 3 min. The spin column was then transferred to a new RNase-free tube, and 60 μL of RNase-free ddH2O was added to elute the DNA. This mixture was incubated for 5 min and centrifuged at 12,000 rpm for 5 min to collect the phage DNA.
2.10 Phage genome sequencing and annotation
The phage genome was sequenced using the Illumina HiSeq 2500 platform, yielding a high average sequencing depth of 24,068×. Raw reads were filtered and quality-checked using fastp, and de novo assembly was performed with SPAdes. The quality of the assembly was evaluated using QUAST, resulting in an N50 value of 40,624 bp, indicating good assembly continuity. The genome was uploaded to the National Center for Biotechnology Information (NCBI). It was then analyzed with BLASTn to identify similar phages in the GenBank database. The tRNA genes encoded by the phage were identified using tRNAscan-SE. Phage-encoded genes were annotated using the Prokaryotic Genome Annotation System (Prokka). Functional characterization of the predicted amino acid sequences of these genes was conducted using NCBI’s BLASTp. Virulence factors and antibiotic resistance genes were identified using the VFDB and ARDB databases (thresholds of ≥70% identity and ≥70% coverage). The genomic map was visualized with CGview.
2.11 Phylogenetic analysis of phage HZJ31
The terminase large subunit sequences were used for phylogenetic analysis. The amino acid sequence of terminase large subunit of phage HZJ31 was entered into NCBI’s BLASTp, resulting in the identification of ten homologous sequences with notable similarity. The basis for generating a phylogenetic tree using the ClustalW alignment tool within the MEGA 11.0 platform. Furthermore, the genome sequences of phage HZJ31 and the selected phage were analyzed by Easyfig.
2.12 Determination of minimum inhibitory concentration
The MICs of tigecycline (TGC) against KPZ2 were conducted by the broth dilution method. TGC was serially diluted in LB broth in twofold dilutions.100 μL of each antibiotic dilution was added to the corresponding wells of a 96-well plate. Negative control wells contained only medium without bacteria, while positive control wells contained bacteria without antibiotics. The KPZ2 bacterial suspension was diluted in LB broth to 5 × 105 CFU/mL from an initial McFarland 0.5 suspension. Then, 100 μL of the bacteria was added to each well. The plate was incubated for 18 h at 37°C. The MIC was determined as the lowest concentration of TGC that inhibited visible bacterial growth.
2.13 Bactericidal kinetics of the combined phage HZJ31 and tigecycline
The assay was performed according to a previously reported method (Wang et al., 2024), with minor modifications. The experimental design included the following groups: The KPZ2 group, The phage HZJ31 group (MOI = 0.001), The 1/2 MIC TGC group, and the phage HZJ31 with 1/2 MIC TGC group. The KPZ2 was standardized to 1×106 CFU/mL. 100 μL of the bacterial culture was added to the 96-well plate, followed by the addition of phage HZJ31 and/or TGC, according to group assignment. The plate was incubated at 37°C for 36 h, with OD600 measurements taken hourly to construct bactericidal kinetics curves to evaluate the inhibitory effects of phage and/or TGC on bacterial growth. Synergy was defined as a statistically significant enhancement of antibacterial activity in the combination group (phage + TGC) compared to each monotherapy group, based on OD600 values measured over time. Statistical analysis was performed using two-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
2.14 Antibacterial efficacy of phage HZJ31 and TGC in the Galleria mellonella infection model
The experiments were evaluated using a Galleria mellonella(G.mellonella)larvae infection model using a protocol modified from a previous study (Peng et al., 2025). The larvae, approximately 2–3 cm in length and weighing 250–300 mg, with a uniform milky white color and no gray spots, exhibiting high vitality, were selected for the experiments (purchased from Tianjin Huiyude Biotechnology Co., Ltd., China). To determine the optimal bacterial infection dose, we selected the concentration that resulted in approximately 80% larval mortality at 24 h post-infection as the target inoculum. Before experimentation, larvae were acclimated at 37°C for 1–2 h, and KPZ2 was cultured in LB broth to reach an OD600 of 0.8-1.0. The culture underwent centrifugation at 7,000 rpm for 10 min, followed by PBS washing and adjustment to concentrations of 105 to 109 CFU/mL. 10 µL bacterial suspension or PBS (as control) was administered into the last left proleg of the larvae. Larvae were incubated at 37°C with survival rates monitored every 12 h.
To evaluate the combined antibacterial efficacy of phage HZJ31 and TGC in the G.mellonella infection model, larvae were randomly assigned to five groups: the PBS control group, the KPZ2 group, the phage HZJ31 group, the TGC group, and the phage HZJ31 + TGC group. Except for the PBS control group (injected with an equal volume of PBS), each larva received 10 μL of KPZ2 suspension (at a concentration determined by preliminary experiments) into the last left proleg. After 1 h, 10 μL of the respective treatment was injected into the last right proleg of each larva: PBS (for the PBS control and KPZ2 group), phage HZJ31 (MOI = 0.001, HZJ31 group), 1/2 MIC TGC (TGC group), and a mixture of phage HZJ31 and 1/2 MIC TGC (combined treatment group). All larvae were incubated at 37°C and monitored every 12 h for 4 consecutive days. Survival status was recorded, and survival rates were calculated for each group to evaluate therapeutic efficacy.
2.15 Biofilm quantification by crystal violet assay
The biofilm quantification was performed using a modified protocol
(Wang et al., 2024). A 96-well plate was incubated at 37°C for 24 h, 48 h, and 72 h with KPZ2 suspension (106 CFU/mL) and phage HZJ31 (MOI =0.001). For biofilm disruption, another set of KPZ2 cultures was incubated for 48 h to form biofilms, which were then treated with phage HZJ31 (MOI = 0.01) for 6 h. After washing with PBS to remove planktonic cells, biofilms were fixed with methanol for 30 min and stained with 0.1% crystal violet solution (Shanghai Baisai, China) for 15 min. The residual stain was removed by PBS washing, and biofilm biomass was quantified by dissolving crystal violet in 200 μL of 30% glacial acetic acid. Absorbance at 595 nm was measured to assess biofilm biomass. All experiments were performed in triplicate.
2.16 Bactericidal activity in biofilms by XTT assay
The experiment was conducted with suitable modifications (Fei et al., 2023). KPZ2 suspension (106 CFU/mL) and phage HZJ31 (MOI = 0.001) were co-incubated in a 96-well plate at 37°C for 24 h, 48 h, and 72 h. For biofilm disruption, preformed 48 h biofilms were treated with HZJ31 (MOI = 0.01) for 6 h. After incubation, PBS was used to wash the plate to remove planktonic bacteria. Subsequently, 200 μL of LB broth and 20 μL of XTT solution (Shanghai Yuanye, China) were introduced into each well. The plate was then incubated at 37°C in the dark for 3 h, followed by absorbance measurement at OD490 nm. The inhibitory rate was calculated using the following formula:
The experiment was repeated three times.
2.17 Screening of phage-resistant bacteria
The experiment was performed with some modifications based on a previous study (Wang et al., 2024). KPZ2 and phage HZJ31 (MOI =0.001) were incubated at 37°C with shaking at 200 rpm for 24 h. Cultures were streaked onto blood agar plates to isolate single colonies. The isolates were identified by the mass spectrometer. Phage resistance was confirmed by spot testing and double-layer agar assays. Resistant strains were designated KPZ2-R.
2.18 Scanning electron microscopy
KPZ2 and KPZ2-R were incubated at 37°C with shaking at 220 rpm for 6 h. The cultures were then centrifuged at 8000 rpm for 10 min to collect the pellets, which were subsequently washed with PBS. Pellets were fixed in 2.5% glutaraldehyde at room temperature for 5 h, followed by PBS washing and 15-minute dehydration using graded ethanol concentrations (30%, 50%, 70%, 90%, 100%). After dehydration, the samples were dried, gold-coated, and observed under the SEM Hitachi SU8100 to capture morphology.
2.19 Antibiotic susceptibility testing
Antibiotic susceptibility of KPZ2 and KPZ2-R was determined using the VITEK-2 Compact system (BioMérieux, France). The KPZ2 and KPZ2-R were standardized to a 0.5 McFarland scale. The prepared bacterial suspension was loaded along with the AST-GN card into the VITEK-2 Compact system for automatic inoculation and determination. The results were analyzed following the guidelines of the Clinical and Laboratory Standards Institute (CLSI).
2.20 Phage adsorption efficiency
The assay was performed using a modified protocol (Wang et al., 2024). KPZ2 and KPZ2-R were incubated with phage HZJ31 (MOI =0.001) at 37°C, with samples collected at 0, 5, 10, 15, and 20 min. The samples were centrifuged at 11000 rpm for 8 min at 4°C. The supernatants were filtered (0.22 μm), and then the phage titer was determined. The Adsorption Efficiency = [(Initial Phage Titer - Phage Titer in Supernatant)/Initial Phage Titer] × 100%.
2.21 Bacterial growth rates
KPZ2 and KPZ2-R were adjusted to 1.0 × 106 CFU/mL. The 96-well plate was prepared with 200 μL of bacterial suspension in each test well and 200 μL of LB broth in the control wells. The plate was incubated at 37°C for 24 h with hourly OD600 measurements.
2.22 Comparison of virulence using the G.mellonella infection model
The concentrations of KPZ2 and KPZ2-R were adjusted to 106 and 107 CFU/mL, respectively. Each G.mellonella larvae received an injection of 10 µL bacterial suspension (PBS as a control) into the last left proleg. Larvae were incubated at 37°C with survival monitored every 12 h over 4 days.
2.23 Bacterial whole-genome sequencing
Genomic DNA of strains KPZ2 and KPZ2-R was extracted using a whole-genome extraction kit (Sangon Biotech, China) according to the manufacturer’s instructions. Briefly, 5 mL of overnight bacterial culture was centrifuged to collect the cells, followed by the addition of lysis buffer and incubation at 65 °C for 1 h to lyse the cells. Subsequently, Buffer PB was added, mixed thoroughly, and briefly incubated at −20 °C. After centrifugation, the supernatant was transferred, isopropanol was added, mixed well, and centrifuged again to pellet the DNA. The DNA pellet was washed with 75% ethanol, dried after removing the ethanol, and then dissolved in TE buffer. DNA concentration and integrity were assessed using a Qubit 4.0 fluorometer (Thermo Fisher Scientific, China). The extracted DNA was either used for subsequent experiments or stored at −20°C.
The genomes of strains KPZ2 and KPZ2-R were sequenced using the BGI MGISEQ 2000 platform. The extracted DNA was enzymatically fragmented, end-repaired, ligated with adaptors, and amplified by PCR to construct sequencing libraries. Library concentration was quantified using a Qubit fluorometer, and fragment size distribution was assessed using the Agilent 2100 Bioanalyzer; libraries with a main peak around 500 bp were considered qualified. The libraries were then denatured, circularized, and purified before sequencing. Raw sequencing data were subjected to quality control to remove adaptors and low-quality reads. The filtered reads were assembled using SPAdes (v3.13.0). Based on alignment results, ANGSD (v0.940) was used to reconstruct the genome sequences, and genome completeness and contamination were evaluated using CheckM (v1.2.2).
The K-locus types of KPZ2 and KPZ2-R were identified using the Kaptive Web tool (https://kaptive-web.erc.monash.edu/). Genome assemblies in FASTA format were uploaded, and typing was based on sequence coverage, identity, and completeness. Confidence levels were recorded for each result.
2.24 Statistical analysis
All statistical analyses were conducted using GraphPad Prism 9.2. One-way or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was employed to undertake multiple group comparisons. Survival curves for G.mellonella were analyzed using the log-rank (Mantel-Cox) test. Statistical significance was indicated by *P <0.05, **P <0.01, ***P < 0.001, and ****P <0.0001.
3 Results
3.1 Isolation and characterization of phage HZJ31
Phage HZJ31 was isolated using strain KPZ2 as the host bacterium from wastewater. TEM revealed the phage had an icosahedral head approximately 80 nm in diameter and a short tail measuring 10 nm (Figure 1A). The plaques were large, clear, and uniform, reaching up to 4.0 mm in diameter, with a halo of approximately 7.2 mm, which indicates the high efficiency (Figure 1B). Phage HZJ31 was classified within the Caudovirales based on its morphological characteristics.
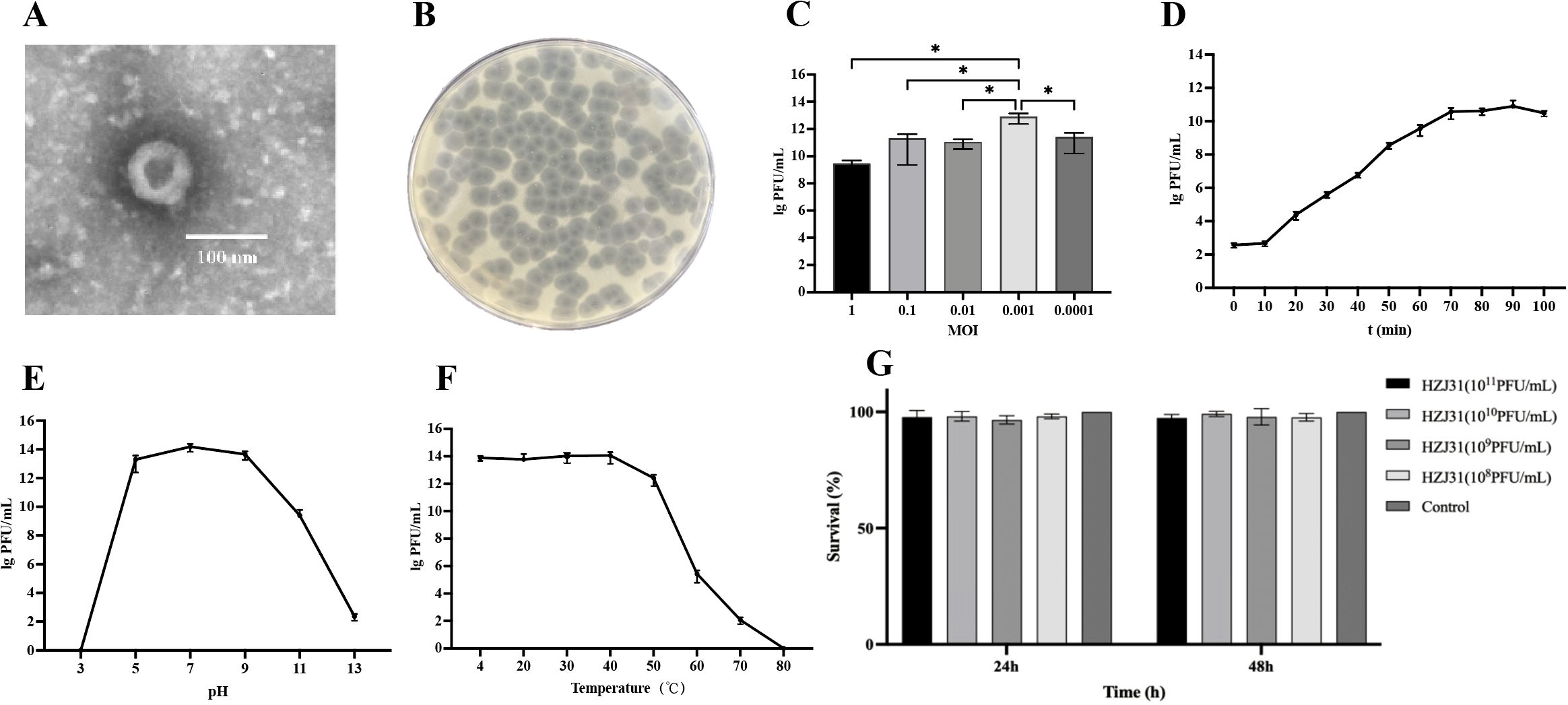
Figure 1. Biological characteristics, stability and cytotoxicity of phage HZJ31. (A) TEM morphology of phage HZJ31, revealing an icosahedral head (~80 nm) and short tail (~10 nm), consistent with Caudovirales. (B) Plaque morphology of phage HZJ31 on a double-layer agar plate. (C) The optimal MOI of phage HZJ31, the highest titer was observed at MOI = 0.001, mean ± SD. (D)The one-step growth curve of phage HZJ31, mean ± SD. (E) The pH stability of phage HZJ31, the phage remained stable between pH 5–9 but was inactivated under extreme acidic or alkaline conditions. mean ± SD. (F) Thermal stability of phage HZJ31. phage activity was maintained up to 50 °C and declined at higher temperatures, with complete inactivation at 80 °C. mean ± SD. (G) Cytotoxicity assessment of phage HZJ31 on THP-1 cells by CCK-8 assay. no significant cytotoxicity was observed after 24 h and 48 h exposure to phage HZJ31 at various concentrations (108 to 1011 PFU/mL). mean ± SD. *P < 0.05.
The double-layer agar plate method was employed to determine the optimal MOI, one-step growth, and stability of phage HZJ31. Phage titer of HZJ31 remained largely stable across MOIs from 0.1 to 0.0001, peaking at an MOI of 0.001, which was chosen for further experiments (Figure 1C). The one-step growth curve showed a 10-minute latent period, followed by a rapid exponential rise in phage population between 10 and 70 min, culminating in a plateau phase. The estimated burst size was 374 PFU/cell, suggesting that HZJ31 possesses a highly efficient host cell lysis capability (Figure 1D).
The environmental stability of phage HZJ31 was further assessed under varying pH and temperature conditions. Phage HZJ31 exhibited stable titers between pH 5 and 9, with the highest stability observed around neutral pH. In contrast, the phage titer drastically decreased under highly alkaline (pH > 9) and acidic (pH < 5) conditions, becoming nearly undetectable at pH 3 (Figure 1E). The thermal stability of HZJ31 was robust between 4°C and 50°C, but its titer began to decrease beyond 50°C, and complete inactivation occurred at 80°C (Figure 1F). The stability of HZJ31 indicates its potential suitability for phage therapy applications. Phage HZJ31 demonstrates strong host specificity, with a lysis rate of 23.33% (7/30) against clinical CRKP isolates and only 13.33% (4/30) achieving complete lysis (Table 1). Furthermore, CCK-8 assays showed that co-culturing THP-1 cells with HZJ31 at concentrations ranging from 108 to 1011 PFU/mL for 24 h and 48 h did not significantly affect cell viability compared to the untreated control (mean ± SD, n=6, Figure 1G), confirming the cellular safety of HZJ31 for therapeutic applications.
3.2 Genomic analysis of phage HZJ31
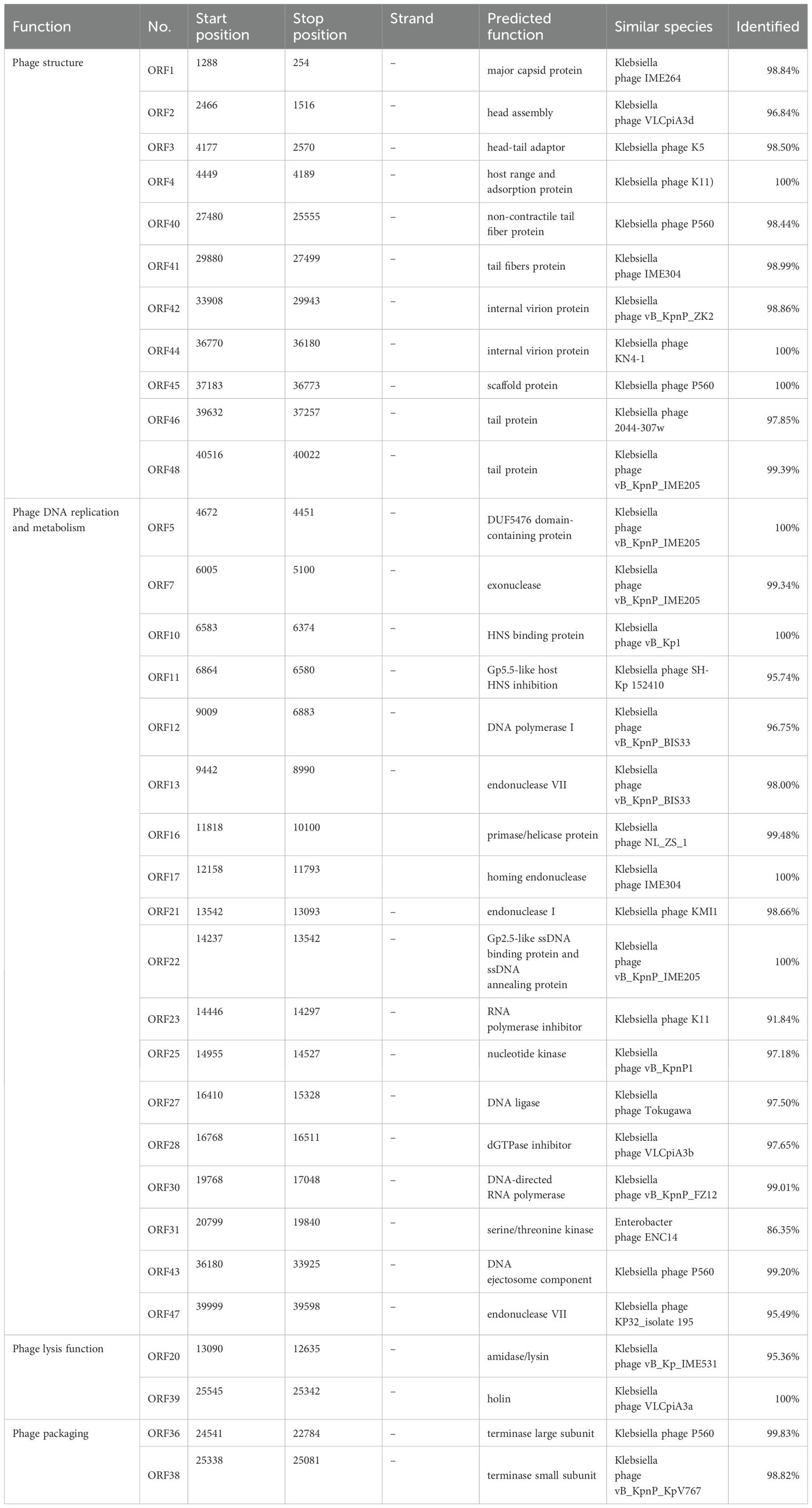
The genome of Phage HZJ31 (GenBank: OR050820.1) was sequenced using Illumina HiSeq 2500 platform, identifying a circular dsDNA molecule of 40,624 bp with a GC content of 53.16%. The genomic map was constructed using GCview (Figure 2A). Annotation using Prokka identified 48 ORFs, of which 33 were functionally annotated and classified into four major categories: structural proteins, DNA replication and metabolism, lysis mechanisms, and genome packaging (Table 2). In the structural module, ORF1 encodes a major capsid protein (98.84% identity with phage IME264), and ORF4 encodes a host adsorption protein (100% identity with phage K11), both essential for host recognition and infection. Tail-associated proteins such as ORF40 and ORF41 (tail fiber proteins) contribute to phage structural assembly. Replication-related genes include ORF12 (DNA polymerase I) and ORF16 (primase/helicase), while ORF30 encodes a DNA-directed RNA polymerase, suggesting the phage can replicate and transcribe independently of the host. Lysis functions are attributed to ORF20, encoding an amidase/lysin, and ORF39, encoding a holin, which together facilitates host cell wall degradation and phage release. Packaging proteins ORF36 and ORF38 encode the large and small terminase subunits, respectively, responsible for DNA cleavage and encapsidation. Overall, HZJ31 harbors a complete set of functional modules characteristic of a lytic phage, supporting its potential as a therapeutic agent against CRKP infections.
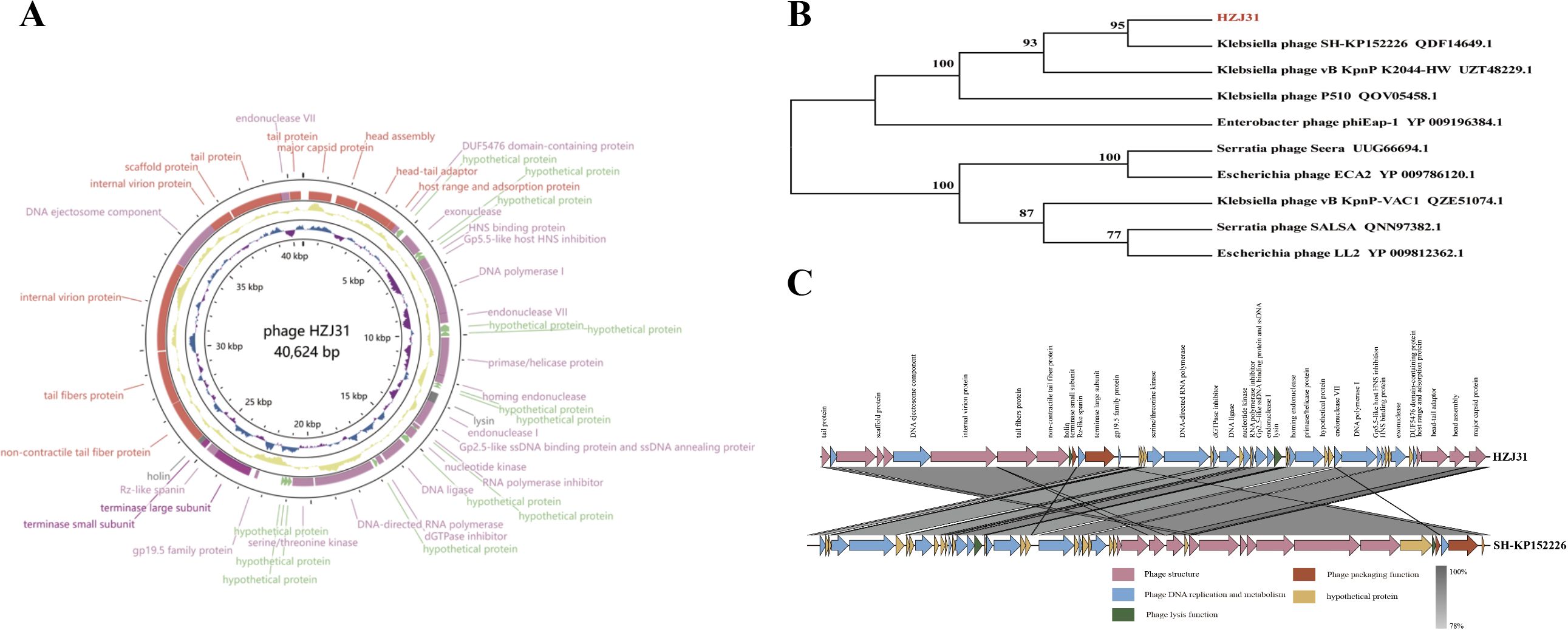
Figure 2. Genomic analysis of phage HZJ31. (A) Gene map of Phage HZJ31. (B) Phylogenetic tree phage HZJ31 based on terminase large subunit. (C) Genomic comparative of phage HZJ31 and Klebsiella phage SH-KP152226.
The absence of virulence or antibiotic resistance genes in the VFDB and ARDB databases highlights the phage’s biosafety for therapeutic use. Genomic analysis using BLASTn revealed that phage HZJ31 exhibits 94.84% sequence identity with Klebsiella phage 066012 (GenBank: MW042787.1) with 94% coverage. The terminase large subunit gene of HZJ31 was found to be highly conserved, and phylogenetic analysis based on this gene positioned HZJ31 in close relation to Klebsiella phage SH-KP152226 (QDF14649.1) (Figures 2B, C). These findings collectively indicate that HZJ31 represents a novel Klebsiella-specific phage, expanding our understanding of phage diversity and potential for therapeutic exploitation.
3.3 Antibacterial efficacy of phage HZJ31 and TGC in vitro and in vivo
The schematic diagrams of the in vitro bactericidal kinetics assay and the G. mellonella infection model are shown in Figures 3A and 3B, respectively. In this study, the combined antibacterial effect of phage HZJ31 and TGC was first evaluated in vitro. Prior to combination testing, the MIC of TGC against strain KPZ2 was determined to be 1.0 μg/mL. Then the bactericidal kinetics of phage HZJ31(MOI = 0.001), TGC (1/2 MIC, 0.5 μg/mL), and their combination against strain KPZ2 were evaluated over a 36-h period (Figure 3C). During the initial 6 h, the bacterial growth among all groups was generally consistent, with no significant differences in OD600. In the HZJ31 group, although the OD600 values gradually increased after 7 h, they remained significantly lower than those of the KPZ2 group throughout the entire experiment. In the 1/2 MIC TGC group, bacterial growth was also delayed, with OD600 increasing at a slower rate than in the HZJ31 group, reaching approximately 0.45 at 36 h. In the HZJ31 combined with the 1/2 MIC TGC group, no significant increase in OD600 was observed during the first 10 h, followed by a slight increase thereafter. However, the OD600 of the HZJ31 with 1/2 MIC TGC group remained at a low level (< 0.3) throughout the 36-h period, significantly lower than those in all other groups (P < 0.05), indicating a synergistic antibacterial effect of the combined treatment.
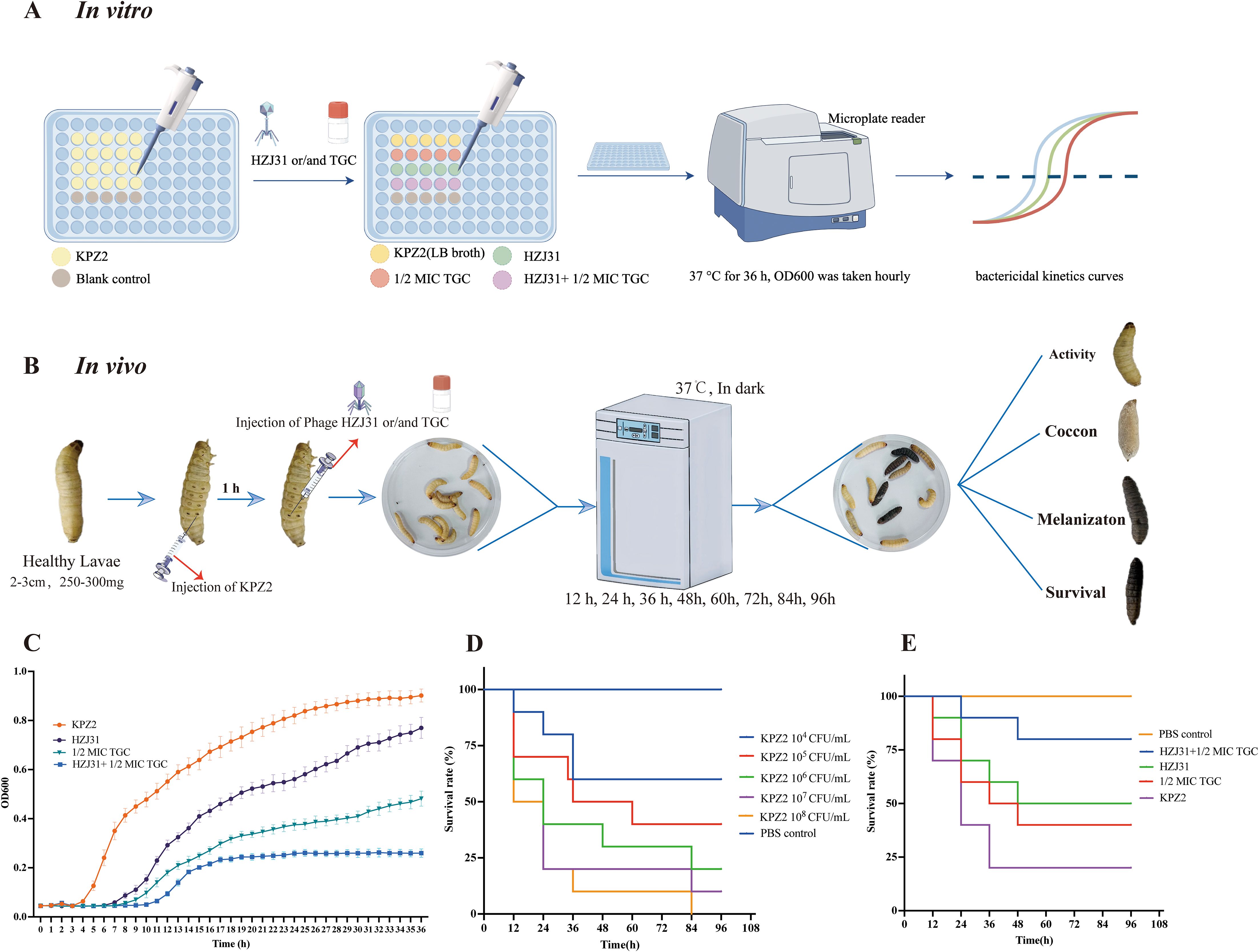
Figure 3. Antibacterial efficacy of phage HZJ31 and TGC against KPZ2 in Vivo and in Vitro. (A) Schematic of the in vitro bactericidal kinetics assay. KPZ2 was cultured in a 96-well plate and treated with phage HZJ31 (MOI = 0.001), 1/2 MIC TGC, or their combination. OD600 values were measured hourly over 36 hours at 37 °C using a microplate reader to generate bactericidal kinetics curves. (B) Schematic of the bactericidal efficacy in the G.mellonella infection model. Larvae were injected with KPZ2 (10 μL) into the last left proleg and incubated for 1 h, followed by injection of phage HZJ31, 1/2 MIC TGC, or their combination into the last right proleg. Larvae were monitored at 12-h intervals for survival over 96 hours. (C) In vitro bactericidal activity of phage HZJ31 and TGC against KPZ2 strain, mean ± SD. (D) Survival curves of G.mellonella larvae injected with different concentrations of KPZ2 to determine the optimal bacterial inoculum. An inoculum of 1×107 CFU/mL resulted in approximately 80% mortality at 24 h, mean ± SD. (E) Therapeutic efficacy of HZJ31 and TGC in G.mellonella infection model, mean ± SD.
Given the significant similarities between the innate immune responses of G.mellonella larvae and vertebrates, the G.mellonella model is widely used as an alternative to evaluating bacterial virulence and in vivo antimicrobial efficacy. We then evaluated the bactericidal efficacy of phage HZJ31 and TGC in the G.mellonella infection model. In this study, we established an infection model by injecting G.mellonella larvae with varying concentrations of bacteria, determining that an optimal inoculum of 107 CFU/mL for KPZ2 resulted in an 80% mortality rate at 24 h (Figure 3D). Significant differences were observed in the survival curves among the groups (log-rank test, P < 0.05). Compared to the KPZ2 group, the survival rates at 96 h were 50% and 60% for the phage HZJ31 and TGC treatments, respectively, while the survival rate for larvae treated with the combination of phage HZJ31 and TGC was 70% (Figure 3E). In summary, phage HZJ31 effectively controlled KPZ2 infection in vivo.
3.4 The antibiofilm activity of phage HZJ31
The results showed that phage HZJ31 significantly inhibited KPZ2 biofilm formation at 24 h, 48 h, and 72 h (P < 0.05, Figure 4A). The inhibitory effect of phage HZJ31 on bacterial growth within the KPZ2 biofilm was assessed, showing a 59.45% inhibition rate at 72 h (P < 0.05) (Figure 4C). When phage HZJ31 was co-cultured with preformed KPZ2 biofilm for 6 h, significant disruption of the biofilm was observed (Figure 4B), with bacterial inhibition within the biofilm exceeding 70% (P < 0.05, Figure 4D). These findings demonstrate that phage HZJ31 effectively inhibits the formation of KPZ2 biofilms and disrupts preformed biofilms, exerting a substantial inhibitory effect on the bacteria within the biofilm.
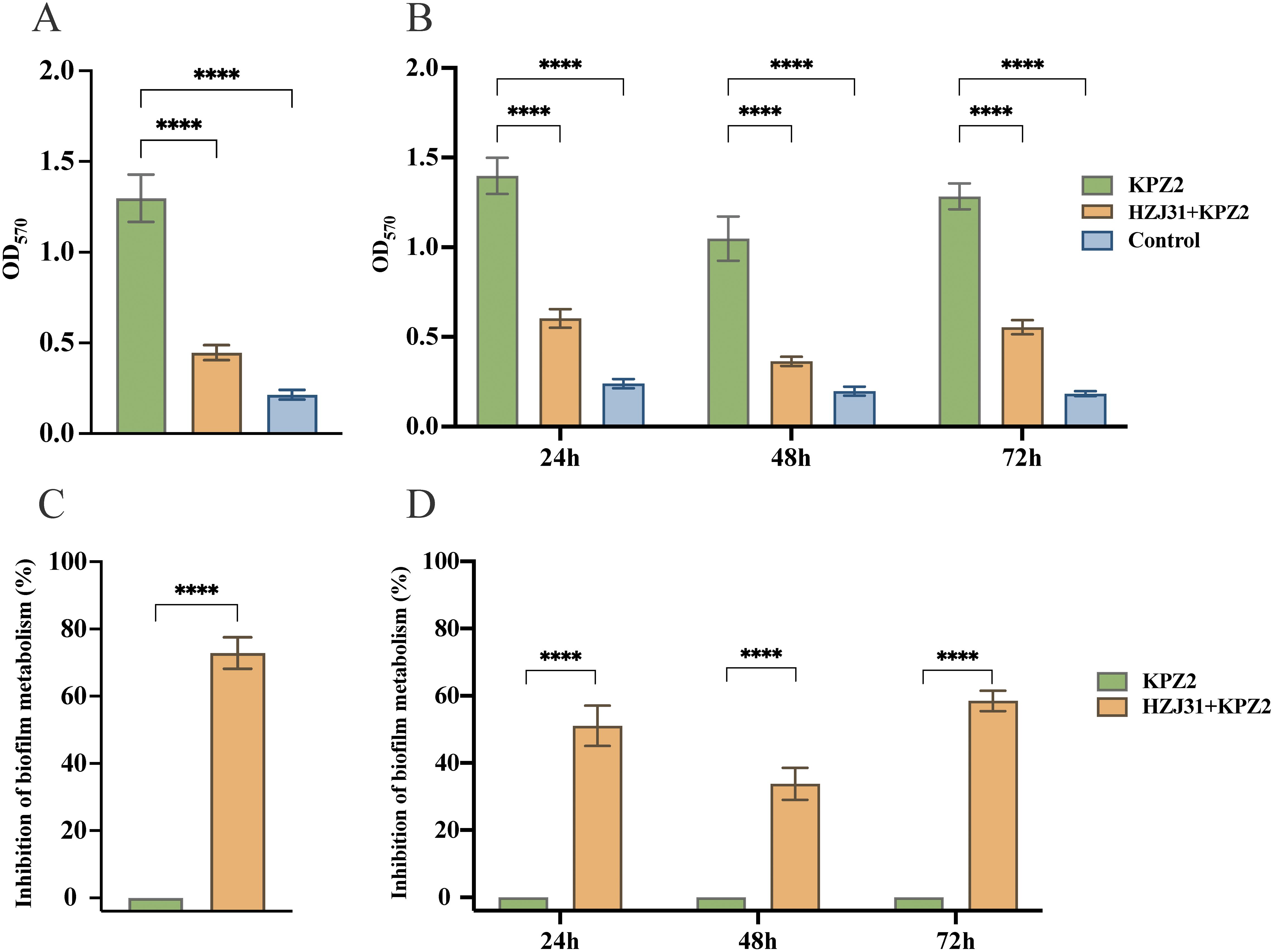
Figure 4. The antibiofilm activity of phage HZJ31.The crystal violet assay was used to quantify biofilm biomass and assess the disruptive effect of phage HZJ31 on pre-formed biofilms (A)and its inhibitory effect on biofilm formation (B). The XTT assay measured bacterial viability within biofilms to assess the disruptive (C) and inhibitory effects (D) of phage HZJ31. All data are shown as mean ± SD. ****P <0.0001.
3.5 Fitness cost of phage resistance in KPZ2-R: impaired capsule, growth, and virulence
After 24 h of co-culturing KPZ2 with phage HZJ31 (MOI = 0.001), the culture appeared visibly turbid, suggesting the emergence of phage-resistant mutants. Subsequent plating on LB agar and purification on blood agar yielded colonies that were no longer susceptible to phage HZJ31, as confirmed by both the double-layer agar method and spot assay, which showed no lysis zones. Mass spectrometry analysis identified all resistant colonies as Klebsiella pneumoniae, excluding the possibility of contamination. One phage-resistant isolate (designated KPZ2-R) was randomly selected from these colonies for further comparison with the wild-type strain. KPZ2 exhibited a slightly faster growth rate in LB broth than KPZ2-R, as measured by OD600 (Figure 5A). The phage adsorption efficiency of KPZ2 was 65.82% at 5 min, significantly higher than that of KPZ2-R (24.24%), and continued to increase over time, reaching 88.00% at 15 min. Conversely, the adsorption efficiency of KPZ2-R remained low throughout the incubation period (Figure 5B).
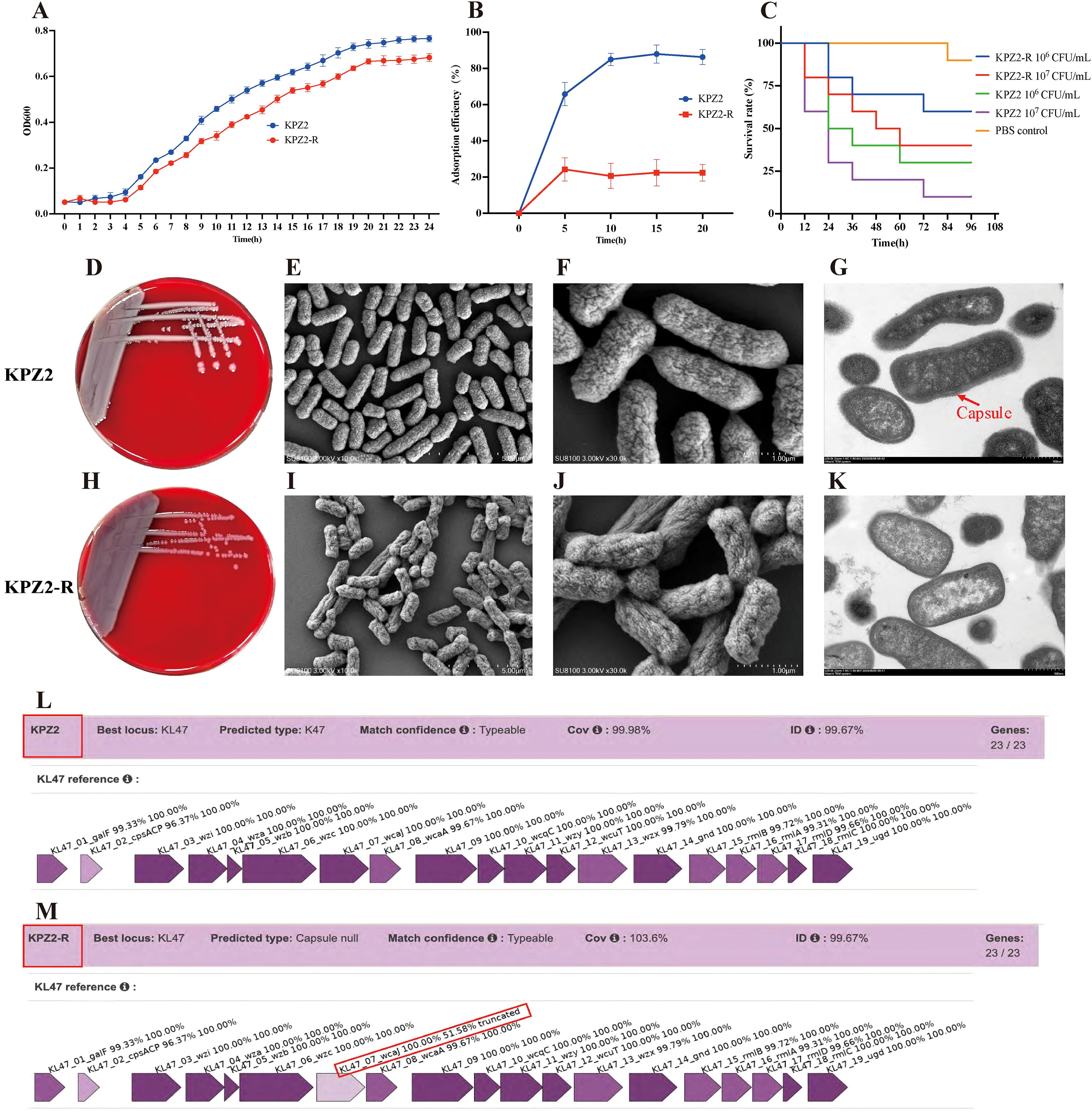
Figure 5. Characterization of phage-resistant strain KPZ2-R. (A) Growth curves of KPZ2 and KPZ2-R in LB broth measured by OD600, mean ± SD. (B) Comparison of adsorption efficiency between KPZ2 and KPZ2-R, mean ± SD. (C) Comparison of virulence between KPZ2 and KPZ2-R in the G.mellonella infection model. (D, H) Colony morphology of the strain KPZ2 and KPZ2-R on blood agar plates. (E, F, I, J) SEM images of KPZ2 (E, F) and KPZ2-R (I , J). KPZ2 showed a smooth, dense, and uniform surface with a thick structure. In contrast, KPZ2-R had a rough, uneven surface with wrinkles or depressions and a thinner structure, along with evident capsule defects. (G, K) TEM images of KPZ2 (G) and KPZ2-R (K). KPZ2 exhibited a well-defined, prominent capsule surrounding the cell, whereas KPZ2-R displayed a significantly reduced and poorly defined capsule. (L, M) Kaptive-based capsular locus analysis. Both strains matched the KL47 capsular type with 99.67% identity. KPZ2 showed complete locus coverage (99.98%) with intact capsule biosynthesis genes, while KPZ2-R was classified as “capsule null” due to a truncated wcaJ gene (51.58% identity).
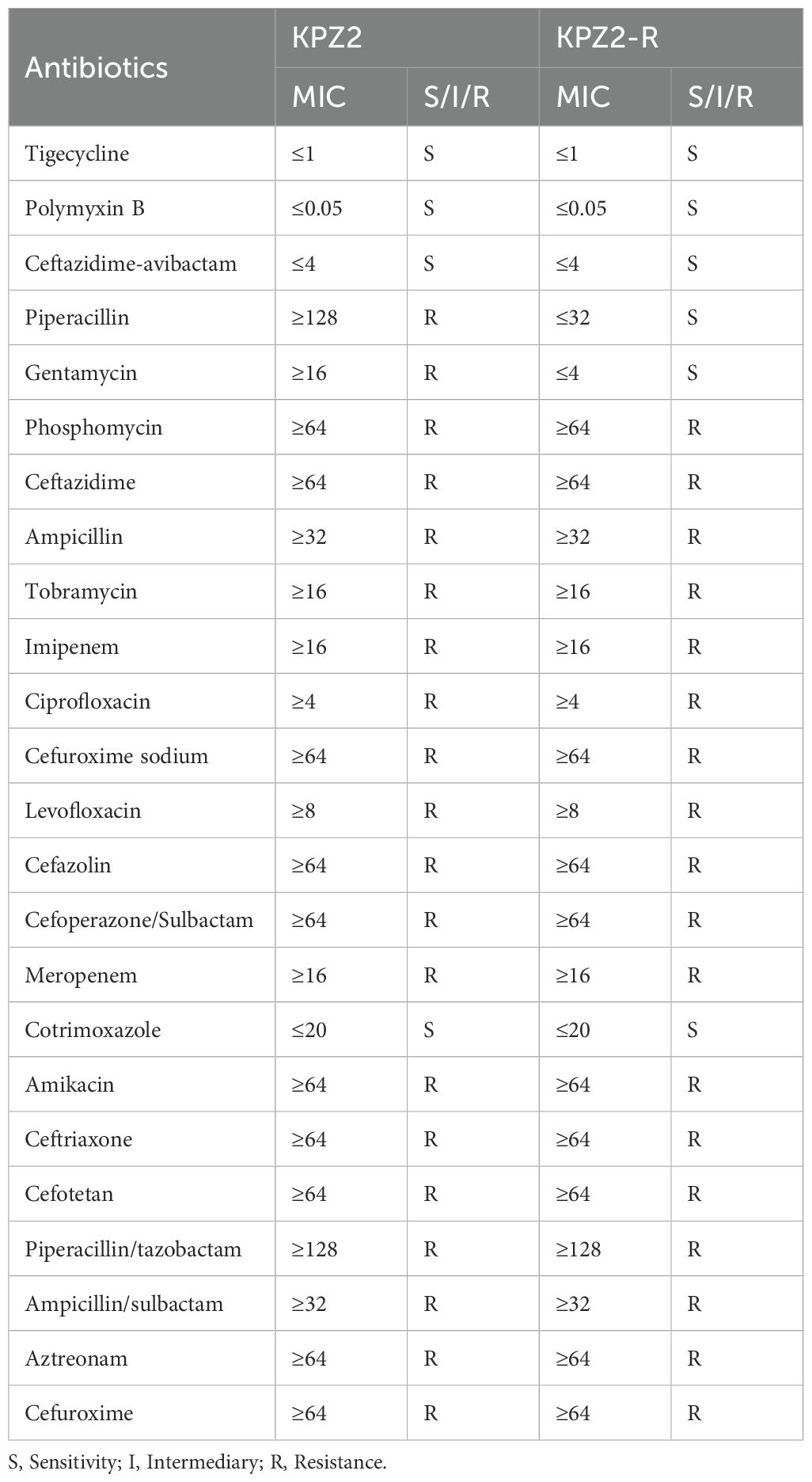
We also compared the virulence of KPZ2 and KPZ2-R using the G.mellonella infection model. At 106 and 107 CFU/mL of KPZ2, the survival rates of larvae at 48 h were 40% and 20%, respectively, whereas larvae injected with the same doses of KPZ2-R had survival rates of 70% and 50% (Figure 5C). Overall, KPZ2-R demonstrated lower virulence than KPZ2 in vivo. We further compared the antibiotic susceptibility of KPZ2 and KPZ2-R (shown in Table 3). Among all antibiotics tested, KPZ2 was only susceptible to TGC, polymyxin B, and ceftazidime/avibactam, while resistant to the others. In contrast, KPZ2-R showed increased susceptibility, shifting from resistance (R) to susceptibility (S) to piperacillin and gentamicin.
On blood agar, KPZ2 formed large, smooth, plump, and moist colonies with a milky white appearance (Figure 5D), while the phage-resistant mutant KPZ2-R formed smaller, gray, semi-transparent colonies, but no obvious difference in diameter (Figure 5H). SEM revealed that the surface of KPZ2 appeared smooth, dense, and uniform, exhibiting a full and thick structure without visible indentations (Figures 5E, F). In contrast, KPZ2-R displayed a rough, uneven surface with wrinkles or depressions, a comparatively thinner structure, and showed evident structural defects in the capsule (Figures 5I, J). TEM further confirmed these differences, revealing a prominent, well-defined capsule surrounding KPZ2 (Figure 5G), while KPZ2-R exhibited a significantly reduced and poorly defined capsule with only a faint or nearly absent surrounding layer (Figure 5K), indicating impaired capsule formation in the resistant mutant.
Consistent with these observations, Kaptive-based analysis revealed that both KPZ2(GenBank: CP178862.1) and its phage-resistant mutant KPZ2-R (GenBank: CP192283.1) matched the KL47 capsular locus with 99.67% sequence identity (Figures 5L, M). KPZ2 exhibited complete locus coverage (99.98%) and intact capsule biosynthesis genes. However, KPZ2-R was classified as “capsule null” due to a truncated wcaJ gene (51.58% identity), suggesting the loss of capsule synthesis capability is associated with phage resistance.
4 Discussion
In recent years, CRKP has rapidly spread worldwide, resulting in high mortality rates and limited treatment options, and has become a serious public health concern (Karampatakis et al., 2023). Therefore, there is an urgent need to explore alternative therapeutic strategies (Chang et al., 2022). Amid the growing challenge of antimicrobial resistance, phage therapy has emerged as a promising antibacterial approach (Hatfull et al., 2022; Kou et al., 2024). Numerous preclinical studies and clinical case reports have demonstrated the therapeutic potential of phages in treating KP infections (Kou et al., 2024; Xing et al., 2025).
Phage HZJ31 demonstrated stability across pH values ranging from 5 to 9 and temperatures from 4°C to 50°C. This robust stability profile facilitates its activity under diverse environmental conditions, simplifying storage and transportation logistics. Following a 48 co-culture of phage HZJ31 with THP-1 cells, the CCK-8 assay yielded no detectable cytotoxicity, indicating biocompatibility. Genomic sequencing further disclosed an absence of genes conferring toxicity or antibiotic resistance, offering initial assurance of its safety for therapeutic applications. These findings underpin the promising clinical potential of phage HZJ31.
Biofilms significantly contribute to recurrent medical device-associated chronic infections and play a pivotal role in bacterial adaptive resistance and evasion of host immune surveillance (Chadha et al., 2022). These complicated microbial communities are composed of bacteria that are embedded within a self-produced matrix of extracellular polymers (EPS) consisting of extracellular polysaccharides, proteins, and extracellular DNA (eDNA) (Flemming et al., 2016). The biofilm-forming ability of KP is closely associated with the expression of virulence factors such as capsular polysaccharides and outer membrane proteins (Sauer et al., 2022) (Li et al., 2024). KP strains with robust biofilm-forming capabilities exhibit heightened virulence, suggesting more severe infections and adverse outcomes (Zheng et al., 2018; Tang et al., 2024). Phage therapy has emerged as a promising strategy to combat biofilm-associated infections. In this study, phage HZJ31 effectively inhibited KPZ2 biofilm formation at 24 h, 48 h, and 72 h and compromised the structural integrity of preformed biofilms, resulting in efficient bacterial lysis within the biofilms.
Phages can encode enzymes, such as depolymerase, endolysin and holin, that degrade biofilm matrices, including polymers and capsular polysaccharides, thus facilitating the disruption of bacterial biofilm integrity, which is a key mechanism of phage action against bacterial biofilms (Lin et al., 2014; Topka-Bielecka et al., 2021; Lendel et al., 2024; Zhao et al., 2024a). The effectiveness of HZJ31 in overcoming bacterial biofilm defenses presents a new avenue for the treatment of refractory infections. A total of 48 ORFs were identified in the genome of phage HZJ31 through Prokka annotation, of which 33 were functionally characterized. Notably, ORF 20 and ORF 39 were predicted to encode a lysin and a holin protein, respectively. In addition, 15 ORFs were annotated as hypothetical proteins, suggesting that some of them may encode previously uncharacterized enzymes involved in biofilm matrix degradation. Further structural and functional analyses are required to elucidate their potential roles.
Phage-antibiotic combination therapy emerges as a promising strategy for combating antibiotic-resistant infections. This therapeutic approach harnesses the distinct antimicrobial mechanisms of phages and antibiotics to expand the host range, augment antibacterial potency, and retard the development of phage resistance. Furthermore, it has the potential to diminish the necessary antibiotic dosages, alleviate associated side effects, and optimize treatment efficacy in complex infections (Van Nieuwenhuyse et al., 2021; Ashworth, 2024). In our study, tigecycline was selected for combination therapy based on susceptibility testing, which showed that KPZ2 was resistant to most antibiotics but remained sensitive to TGC, polymyxin B, and ceftazidime-avibactam (Table 3). TGC, a tetracycline derivative, manifests broad-spectrum antimicrobial properties and an extended half-life by impeding protein synthesis in a spectrum of pathogens, including antibiotic-resistant bacteria and biofilms (Giamarellou and Poulakou, 2011). Both monotherapy of TGC and its synergistic combinations with other antibiotics have shown promising treatment efficacies against CRKP infections (Park et al., 2024; Xie et al., 2024). In this study, the bactericidal kinetics data revealed that phage HZJ31 and TGC exhibited distinct antimicrobial effects when applied individually or in combination against KPZ2. The phage HZJ31 group demonstrated a moderate inhibitory effect, with OD600 values rising after 7 h but remaining significantly lower than those of the untreated control throughout the 36-h observation period. This indicates that phage HZJ31 can delay bacterial proliferation and partially suppress growth, though phage resistance may emerge over time. Notably, the combination of phage HZJ31 with 1/2 MIC TGC achieved superior bacterial suppression, suggesting a synergistic interaction between phage HZJ31 and sub-inhibitory concentrations of TGC in both augmenting bactericidal efficacy and delaying the emergence of resistant strains. In addition to TGC, other antibiotics such as polymyxin B, colistin, and ceftazidime-avibactam may also be considered for phage-antibiotic combination strategies (Wang et al., 2021; Han et al., 2022; Cristinziano et al., 2024), depending on the antibiotic susceptibility profile and infection context (Diallo and Dublanchet, 2022) (Morrisette et al., 2020; Zhao et al., 2024b).
In the G.mellonella infection model, larvae treated with phage HZJ31 or TGC alone had survival rates of 50% and 60%, respectively, after 96 h, while the HZJ31-TGC combination exhibited a significantly improved survival rate (70%). These results demonstrate that the phage HZJ31-TGC combination enhances antibacterial activity both in vitro and in vivo. Furthermore, the phage-antibiotic combination exhibited synergistic effects in biofilm degradation. The therapeutic efficacy of phage-antibiotic combination therapy is contingent upon a multitude of factors, such as the specific types of phages and antibiotics employed, the order of administration, dosages, and the duration of treatment. It is noteworthy that certain phage-antibiotic combinations may exhibit antagonistic interactions. In light of the absence of standardized protocols for phage-antibiotic therapy, there is an imperative need for systematic experimentation to ascertain the most efficacious combinations. These experiments should also carefully monitor the emergence of resistant strains during treatment. Highly personalized treatment regimens should be designed to maximize therapeutic efficacy while minimizing adverse reactions (Hong et al., 2024) (Fungo et al., 2023).
In monophage therapy, bacteria rapidly develop resistance to phages under selective pressure, which limits the application of phage therapy (Strathdee et al., 2023; Laanto, 2024; Pal et al., 2024). The mechanisms of phage resistance are complex and diverse, including preventing phage adsorption, blocking phage DNA injection, cleaving the phage genome, abortive infection, and quorum sensing, among others (Laanto et al., 2020). Adsorption is the initial step of phage infection of bacteria and a crucial factor determining the success of phage therapy. Bacteria can block phage adsorption through several mechanisms: mutation or modification of the phage receptor, extracellular matrix interference with the receptor, and competitive inhibition by receptor analogs (Laanto et al., 2020). Capsular polysaccharides, outer membrane proteins, and lipopolysaccharides serve as the adsorption receptors for KP phages, and their loss or mutation can lead to phage resistance (Camprubí, 1991; Henrici De Angelis et al., 2021; Cai et al., 2022; Fang and Zong, 2022; Geng et al., 2023). The capsule often masks the bacterial receptors, preventing direct binding between the phage and the receptor (Samson et al., 2013). In this study, we observed that KPZ2 developed phage resistance under selective pressure from phage HZJ31 by losing its capsule, resulting in a significant decrease in the phage’s adsorption efficiency to the resistant mutant KPZ2-R. Moreover, we observed a significant trade-off phenomenon — KPZ2-R exhibited altered colony morphology, slower growth rate, and reduced virulence, which could increase its susceptibility to immune clearance during actual infections. Interestingly, KPZ2-R regained sensitivity to piperacillin and gentamicin, a phenomenon not uncommon in bacterial resistance mechanisms (Hu et al., 2024b). Although bacteria may escape phage infection by altering or shedding their capsule and outer membrane proteins, this adaptation can inadvertently compromise their defense against antibiotics, consequently rendering them susceptible once again (Gordillo Altamirano et al., 2021; Nang et al., 2024). The employment of a phage cocktail comprising both capsule-dependent and -independent phages, or a strategic sequence of phage and antibiotic administration, may yield superior therapeutic outcomes, offering innovative strategies and perspectives for phage therapy.
This study has several limitations that should be acknowledged. First, the host range of phage HZJ31 was assessed using 30 clinical CRKP isolates through spot assays. The limited number of isolates may not fully represent the genetic and phenotypic diversity of CRKP in clinical settings. Moreover, most strains were collected from a single hospital, raising the possibility of strain clustering and potential bias toward nosocomial infection types. Future studies will include a broader and more diverse set of CRKP isolates from multiple geographic and clinical sources to enhance the accuracy and generalizability of the host range evaluation. Second, the antibacterial and antibiofilm effects of phage HZJ31 were evaluated only against a single CRKP strain, without investigating potential cross-species or cross-strain activity. Additionally, the synergistic antibacterial activity was tested only in combination with TGC, to which the tested strain was already susceptible. Expanding the evaluation to include multiple antibiotics and additional strains will provide a more comprehensive understanding of its synergistic potential. The in vivo antibacterial efficacy was limited to the G.mellonella model and has not yet been validated in advanced mammalian models, which restricts the generalizability of the study. Lastly, the biosafety of phage HZJ31 was only preliminarily assessed at the genetic and cellular levels. Further evaluation in mammalian models is necessary to comprehensively assess its safety and therapeutic efficacy.
In conclusion, we successfully isolated a novel lytic phage, HZJ31, which has demonstrated high environmental stability and biocompatibility. Furthermore, the synergistic application of HZJ31 with TGC showed a markedly enhanced antibacterial activity in both in vitro and the G.mellonella infection model. These findings provide theoretical support for the potential clinical application of phage HZJ31.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
RZ: Conceptualization, Data curation, Funding acquisition, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. RW: Investigation, Methodology, Software, Writing – original draft. BF: Investigation, Methodology, Writing – original draft. RL: Data curation, Methodology, Writing – original draft. XY: Investigation, Software, Writing – review & editing. XL: Project administration, Software, Supervision, Writing – original draft. CW: Conceptualization, Resources, Supervision, Writing – review & editing. YL: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was supported by the Henan Provincial Science and Technology Research Project (No. 242102310148), the Natural Science Foundation of Henan Province (No.242300420437), the Key Scientific Research Project of Henan Higher Education Institutions (No.24A310007), and Wu Jieping Medical Foundation (NO.320.6750.2024-03-27).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ashworth, E. A., Wright, R. C. T., Shears, R. K., Wong, J. K. L., Hassan, A., Hall, J. P. J., et al. (2024). Exploiting lung adaptation and phage steering to clear pan-resistant Pseudomonas aeruginosa infections in vivo. Nat. Commun. 15 (1), 1547. doi: 10.1038/s41467-024-45785-z
Baqer, A. A., Fang, K., Mohd-Assaad, N., Adnan, S. N. A., and Md Nor, N. S. (2022). In vitro activity, stability and molecular characterization of eight potent bacteriophages infecting carbapenem-resistant klebsiella pneumoniae. Viruses 15, 117. doi: 10.3390/v15010117
Bologna, E., Licari, L. C., Manfredi, C., Ditonno, F., Cirillo, L., Fusco, G. M., et al. (2024). Carbapenem-resistant enterobacteriaceae in urinary tract infections: from biological insights to emerging therapeutic alternatives. Medicina (Mex.) 60, 214. doi: 10.3390/medicina60020214
Cai, R., Deng, H., Song, J., Zhang, L., Zhao, R., Guo, Z., et al. (2022). Phage resistance mutation triggered by OmpC deficiency in Klebsiella pneumoniae induced limited fitness costs. Microb. Pathog. 167, 105556. doi: 10.1016/j.micpath.2022.105556
Camprubí, S. (1991). Isolation and characterization of bacteriophage FC3–10 from Klebsiella spp. FEMS Microbiol. Lett. 83, 291–297. doi: 10.1016/0378-1097(91)90491-R
Chadha, J., Harjai, K., and Chhibber, S. (2022). Revisiting the virulence hallmarks of Pseudomonas aeruginosa : a chronicle through the perspective of quorum sensing. Environ. Microbiol. 24, 2630–2656. doi: 10.1111/1462-2920.15784
Chang, R. Y. K., Nang, S. C., Chan, H.-K., and Li, J. (2022). Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 187, 114378. doi: 10.1016/j.addr.2022.114378
Colavecchio, A., Cadieux, B., Lo, A., and Goodridge, L. D. (2017). Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the enterobacteriaceae family - A review. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01108
Cook, B. W. M. and Hynes, A. P. (2025). Re-evaluating what makes a phage unsuitab le for therapy. NPJ Antimicrob. Resist. 3, 45. doi: 10.1038/s44259-025-00117-z
Cristinziano, M., Shashkina, E., Chen, L., Xiao, J., Miller, M. B., Doligalski, C., et al. (2024). Use of epigenetically modified bacteriophage and dual beta-lactams to treat a Mycobacterium abscessus sternal wound infection. Nat. Commun. 15, 10360. doi: 10.1038/s41467-024-54666-4
Cui, L., Watanabe, S., Miyanaga, K., Kiga, K., Sasahara, T., Aiba, Y., et al. (2024). A comprehensive review on phage therapy and phage-based drug development. Antibiot. Basel Switz. 13, 870. doi: 10.3390/antibiotics13090870
Diallo, K. and Dublanchet, A. (2022). Benefits of combined phage-antibiotic therapy for the control of antibiotic-resistant bacteria: A literature review. Antibiot. Basel Switz. 11, 839. doi: 10.3390/antibiotics11070839
Ding, L., Shen, S., Chen, J., Tian, Z., Shi, Q., Han, R., et al. (2023). Klebsiella pneumoniae carbapenemase variants: the new threat to global public health. Clin. Microbiol. Rev. 36, e00008–e00023. doi: 10.1128/cmr.00008-23
Fang, Q. and Zong, Z. (2022). Lytic Phages against ST11 K47 Carbapenem-Resistant Klebsiella pneumoniae and the Corresponding Phage Resistance Mechanisms. mSphere 7, e00080–e00022. doi: 10.1128/msphere.00080-22
Fei, B., Li, D., Liu, X., You, X., Guo, M., Ren, Y., et al. (2023). Characterization and genomic analysis of a broad-spectrum lytic phage HZ2201 and its antibiofilm efficacy against Pseudomonas aeruginosa. Virus Res. 335, 199184. doi: 10.1016/j.virusres.2023.199184
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fungo, G. B. N., Uy, J. C. W., Porciuncula, K. L. J., Candelario, C. M. A., Chua, D. P. S., Gutierrez, T. A. D., et al. (2023). Two is better than one”: the multifactorial nature of phage-antibiotic combinatorial treatments against ESKAPE-induced infections. PHAGE 4, 55–67. doi: 10.1089/phage.2023.0007
Geng, H., Song, L., Yang, X., Xing, S., Wang, R., Xu, Y., et al. (2023). Resistance of Klebsiella pneumoniae to Phage hvKpP3 Due to High-Molecular Weight Lipopolysaccharide Synthesis Failure. Microbiol. Spectr. 11, (3), e0438422. doi: 10.1128/spectrum.04384-22
Giamarellou, H. and Poulakou, G. (2011). Pharmacokinetic and pharmacodynamic evaluation of tigecycline. Expert Opin. Drug Metab. Toxicol. 7, 1459–1470. doi: 10.1517/17425255.2011.623126
Goodridge, L. D. (2010). Designing phage therapeutics. Curr. Pharm. Biotechnol. 11, 15–27. doi: 10.2174/138920110790725348
Gordillo Altamirano, F., Forsyth, J. H., Patwa, R., Kostoulias, X., Trim, M., Subedi, D., et al. (2021). Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 6, 157–161. doi: 10.1038/s41564-020-00830-7
Gorodnichev, R. B., Kornienko, M. A., Malakhova, M. V., Bespiatykh, D. A., Manuvera, V. A., Selezneva, O. V., et al. (2023). Isolation and characterization of the first zobellviridae family bacteriophage infecting klebsiella pneumoniae. Int. J. Mol. Sci. 24, 4038. doi: 10.3390/ijms24044038
Han, M.-L., Nang, S. C., Lin, Y.-W., Zhu, Y., Yu, H. H., Wickremasinghe, H., et al. (2022). Comparative metabolomics revealed key pathways associated with the synergistic killing of multidrug-resistant Klebsiella pneumoniae by a bacteriophage-polymyxin combination. Comput. Struct. Biotechnol. J. 20, 485–495. doi: 10.1016/j.csbj.2021.12.039
Hatfull, G. F., Dedrick, R. M., and Schooley, R. T. (2022). Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211. doi: 10.1146/annurev-med-080219-122208
Henrici De Angelis, L., Poerio, N., Di Pilato, V., De Santis, F., Antonelli, A., Thaller, M. C., et al. (2021). Phage resistance is associated with decreased virulence in KPC-producing klebsiella pneumoniae of the clonal group 258 clade II lineage. Microorganisms 9, 762. doi: 10.3390/microorganisms9040762
Hong, Q., Chang, R. Y. K., Assafiri, O., Morales, S., and Chan, H.-K. (2024). Optimizing in vitro phage-ciprofloxacin combination formulation for respiratory therapy of multi-drug resistant Pseudomonas aeruginosa infections. Int. J. Pharm. 652, 123853. doi: 10.1016/j.ijpharm.2024.123853
Hu, F., Pan, Y., Li, H., Han, R., Liu, X., Ma, R., et al. (2024a). Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-centre study. Nat. Microbiol. 9, 814–829. doi: 10.1038/s41564-024-01612-1
Hu, Q., Huang, L., Yang, Y., Xiang, Y., and Liu, J. (2024b). Essential phage component induces resistance of bacterial community. Sci. Adv. 10, eadp5057. doi: 10.1126/sciadv.adp5057
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y.-A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Karampatakis, T., Tsergouli, K., and Behzadi, P. (2023). Carbapenem-resistant klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics 12, 234. doi: 10.3390/antibiotics12020234
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Kou, X., Yang, X., and Zheng, R. (2024). Challenges and opportunities of phage therapy for Klebsiella pneumoniae infections. Appl. Environ. Microbiol. 90, e0135324. doi: 10.1128/aem.01353-24
Laanto, E. (2024). “Overcoming bacteriophage resistance in phage therapy,” in Bacteriophages. Ed. Tumban, E. (Springer US, New York, NY), 401–410. doi: 10.1007/978-1-0716-3549-0_23
Laanto, E., Mäkelä, K., Hoikkala, V., Ravantti, J. J., and Sundberg, L.-R. (2020). Adapting a phage to combat phage resistance. Antibiotics 9, 291. doi: 10.3390/antibiotics9060291
Lendel, A. M., Antonova, N. P., Grigoriev, I. V., Usachev, E. V., Gushchin, V. A., and Vasina, D. V. (2024). Biofilm-disrupting effects of phage endolysins LysAm24, LysAp22, LysECD7, and LysSi3: breakdown the matrix. World J. Microbiol. Biotechnol. 40, 186. doi: 10.1007/s11274-024-03999-9
Li, L., Gao, X., Li, M., Liu, Y., Ma, J., Wang, X., et al. (2024). Relationship between biofilm formation and antibiotic resistance of Klebsiella pneumoniae and updates on antibiofilm therapeutic strategies. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1324895
Lin, T.-L., Hsieh, P.-F., Huang, Y.-T., Lee, W.-C., Tsai, Y.-T., Su, P.-A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210, 1734–1744. doi: 10.1093/infdis/jiu332
Lou, T., Du, X., Zhang, P., Shi, Q., Han, X., Lan, P., et al. (2022). Risk factors for infection and mortality caused by carbapenem-resistant Klebsiella pneumoniae: A large multicentre case–control and cohort study. J. Infect. 84, 637–647. doi: 10.1016/j.jinf.2022.03.010
Martins, W. M. B. S., Li, M., Sands, K., Lenzi, M. H., Portal, E., Mathias, J., et al. (2022). Effective phage cocktail to combat the rising incidence of extensively drug-resistant Klebsiella pneumoniae sequence type 16. Emerg. Microbes Infect. 11, 1015–1023. doi: 10.1080/22221751.2022.2051752
Miller, W. R. and Arias, C. A. (2024). ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 22, 598–616. doi: 10.1038/s41579-024-01054-w
Morrisette, T., Kebriaei, R., Lev, K. L., Morales, S., and Rybak, M. J. (2020). Bacteriophage therapeutics: A primer for clinicians on phage-antibiotic combinations. Pharmacother. J. Hum. Pharmacol. Drug Ther. 40, 153–168. doi: 10.1002/phar.2358
Mulani, M. S., Kumkar, S. N., and Pardesi, K. R. (2022). Characterization of novel klebsiella phage PG14 and its antibiofilm efficacy. Microbiol. Spectr. 10, e01994–e01922. doi: 10.1128/spectrum.01994-22
Naghavi, M., Vollset, S. E., Ikuta, K. S., Swetschinski, L. R., Gray, A. P., Wool, E. E., et al. (2024). Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet 404, 1199–1226. doi: 10.1016/S0140-6736(24)01867-1
Nang, S. C., Lu, J., Yu, H. H., Wickremasinghe, H., Azad, M. A. K., Han, M., et al. (2024). Phage resistance in Klebsiella pneumoniae and bidirectional effects impacting antibiotic susceptibility. Clin. Microbiol. Infec. 30 (6), 787–794. doi: 10.1016/j.cmi.2024.03.015
Pal, N., Sharma, P., Kumawat, M., Singh, S., Verma, V., Tiwari, R. R., et al. (2024). Phage therapy: an alternative treatment modality for MDR bacterial infections. Infect. Dis. 56 (10), 785–817. doi: 10.1080/23744235.2024.2379492
Park, S., Choi, J., Shin, D., Kwon, K. T., Kim, S.-H., Wi, Y. M., et al. (2024). Conversion to colistin susceptibility by tigecycline exposure in colistin-resistant Klebsiella pneumoniae and its implications to combination therapy. Int. J. Antimicrob. Agents 63, 107017. doi: 10.1016/j.ijantimicag.2023.107017
Patpatia, S., Yilmaz, O., Ylänne, M., and Kiljunen, S. (2021). Isolation and Genomic Analysis of the Phage vB_PaeP_fHoPae04 Infecting Pseudomonas aeruginosa. Microbiol. Resour. Announc. 10, e00076–e00021. doi: 10.1128/MRA.00076-21
Peng, X., Chang, J., Zhang, H., Li, X., Zhang, C., Jiao, S., et al. (2025). Isolation, characterization, and genomic analysis of a novel bacteriophage vB_Kp_XP4 targeting hypervirulent and multidrug-resistant Klebsiella pneumoniae. Front. Microbiol. 16. doi: 10.3389/fmicb.2025.1491961
Samson, J. E., Magadán, A. H., Sabri, M., and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096
Sauer, K., Stoodley, P., Goeres, D. M., Hall-Stoodley, L., Burmølle, M., Stewart, P. S., et al. (2022). The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 20, 608–620. doi: 10.1038/s41579-022-00767-0
She, T., Tan, D., Balcazar, J. L., Friman, V., Wang, D., Zhu, D., et al. (2025). Phage-mediated horizontal transfer of Salmonella enterica virulence genes with regulatory feedback from the host. iMeta, e70042. doi: 10.1002/imt2.70042
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K., and Schooley, R. T. (2023). Phage therapy: From biological mechanisms to future directions. Cell 186, 17–31. doi: 10.1016/j.cell.2022.11.017
Suttle, C. A. (2007). Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Tang, M., Zhao, D., Zhang, Y., Qian, C., Chen, H., Chen, L., et al. (2024). Impact of LuxS on virulence and pathogenicity in Klebsiella pneumoniae exhibiting varied mucoid phenotypes. Infect. Immun. 92, e0001224. doi: 10.1128/iai.00012-24
Topka-Bielecka, G., Dydecka, A., Necel, A., Bloch, S., Nejman-Faleńczyk, B., Węgrzyn, G., et al. (2021). Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics 10, 175. doi: 10.3390/antibiotics10020175
Uyttebroek, S., Chen, B., Onsea, J., Ruythooren, F., Debaveye, Y., Devolder, D., et al. (2022). Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect. Dis. 22, e208–e220. doi: 10.1016/S1473-3099(21)00612-5
Van Nieuwenhuyse, B., Galant, C., Brichard, B., Docquier, P.-L., Djebara, S., Pirnay, J.-P., et al. (2021). A Case of In Situ Phage Therapy against Staphylococcus aureus in a Bone Allograft Polymicrobial Biofilm Infection: Outcomes and Phage-Antibiotic Interactions. Viruses 13, 1898. doi: 10.3390/v13101898
Wang, X., Loh, B., Gordillo Altamirano, F., Yu, Y., Hua, X., and Leptihn, S. (2021). Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 10, 2205–2219. doi: 10.1080/22221751.2021.2002671
Wang, R., You, X., Liu, X., Fei, B., Li, Y., Wang, D., et al. (2024). Characterization of phage HZY2308 against Acinetobacter baumannii and identification of phage-resistant bacteria. Virol. J. 21 (1), 283. doi: 10.1186/s12985-024-02556-y
Xie, M., Ye, L., Chen, K., Xu, Q., Yang, C., Chen, X., et al. (2024). Clinical use of tigecycline may contribute to the widespread dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae strains. Emerg. Microbes Infect. 13, 2306957. doi: 10.1080/22221751.2024.2306957
Xing, J., Han, R., Zhao, J., Zhang, Y., Zhang, M., Zhang, Y., et al. (2025). Revisiting therapeutic options against resistant klebsiella pneumoniae infection: Phage therapy is key. Microbiol. Res. 293, 128083. doi: 10.1016/j.micres.2025.128083
Zhao, R., Jiang, S., Ren, S., Yang, L., Han, W., Guo, Z., et al. (2024a). A novel phage putative depolymerase, Depo16, has specific activity against K1 capsular-type Klebsiella pneumoniae. Appl. Environ. Microbiol. 90, e0119723. doi: 10.1128/aem.01197-23
Zhao, M., Li, H., Gan, D., Wang, M., Deng, H., and Yang, Q. E. (2024b). Antibacterial effect of phage cocktails and phage-antibiotic synergy against pathogenic Klebsiella pneumoniae. mSystems. 9, e0060724. doi: 10.1128/msystems.00607-24
Keywords: carbapenem-resistant Klebsiella pneumoniae, phage therapy, biofilm, phage resistance, Galleria mellonella infection model
Citation: Zhu R, Wang R, Fei B, Lu R, You X, Liu X, Wang C and Li Y (2025) In vitro and in vivo antibacterial efficacy of bacteriophage combined with tigecycline against carbapenem-resistant Klebsiella pneumoniae and characterization of phage resistant mutants. Front. Cell. Infect. Microbiol. 15:1610625. doi: 10.3389/fcimb.2025.1610625
Received: 12 April 2025; Accepted: 26 June 2025;
Published: 04 September 2025.
Edited by:
Derry Keith Mercer, INCATE, SwitzerlandReviewed by:
Genyan Liu, Nanjing Medical University, ChinaMansura Sherif Mulani, Savitribai Phule Pune University, India
Jamil Allen Fortaleza, NU Fairview Incorporated, Philippines
Lin Gan, Capital Institute of Pediatrics, China
Copyright © 2025 Zhu, Wang, Fei, Lu, You, Liu, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongwei Li, bHl3QGhhY3RjbS5lZHUuY24=
†These authors share first authorship
 Rui Zhu
Rui Zhu Ruilin Wang2†
Ruilin Wang2† Xiaojuan You
Xiaojuan You Yongwei Li
Yongwei Li