- 1Microbiology and Virology, San Gallicano Dermatological Institute IRCCS, Rome, Italy
- 2Hospital Infection Control Committee, Istituti Fisioterapici Ospitalieri (IFO), Rome, Italy
- 3Department of Biology and Biotechnology “C. Darwin”, Sapienza University of Rome, Rome, Italy
Introduction: Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a significant threat in oncology settings due to its multidrug resistance and ability to form biofilms on indwelling medical devices.
Methods: This study investigated the in vitro and in vivo activity of meropenem/vaborbactam (MEV) against two CRKP isolates recovered from catheter-related bloodstream infections in patients undergoing orthopedic oncologic surgery.
Results: Whole-genome sequencing identified the isolates as ST101 and ST307, harboring resistance determinants including blaKPC-3 and blaOXA-1, distributed across IncFII and IncFIB plasmid replicons. Both isolates exhibited extensive resistance to β-lactams, aminoglycosides, and fluoroquinolones but remained susceptible to MEV. Phenotypic assays revealed enhanced biofilm formation and metabolic activity compared to the reference strain Kp ATCC 13883 in the absence of hypervirulence-associated genes. MEV demonstrated bactericidal activity against both planktonic and biofilm-associated cells, with minimum bactericidal concentration (MBC90) and minimum biofilm eradication concentration (MBEC90) values of 0.5/8 μg/ml for CRKP ST101, 0.12/8 μg/ml for CRKP ST307, and 0.25/8 μg/ml for the Kp ATCC 13883 strain. In the Galleria mellonella infection model, MEV significantly improved larval survival following the CRKP challenge.
Discussion: These findings demonstrate that MEV exhibits activity against planktonic and biofilm-associated CRKP cells and highlight the need for further investigation in managing catheter-related bloodstream infections caused by multidrug-resistant K. pneumoniae.
1 Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) represents a critical threat in oncology settings, where immunosuppression, invasive procedures, and prolonged hospital stays increase the risk of life-threatening infections. Among the various carbapenemases, Klebsiella pneumoniae carbapenemase (KPC) enzymes are the most prevalent and clinically significant. Their plasmid-mediated dissemination severely limits treatment options and is associated with high morbidity and mortality rates (Murray et al., 2022).
Catheter-related bloodstream infections (CRBSIs) caused by multidrug-resistant K. pneumoniae are a growing concern in oncology, particularly among neutropenic and immunocompromised patients. Orthopedic oncology settings are particularly high-risk due to the complexity of surgical procedures and the frequent use of indwelling devices, such as central venous catheters (Zimmerli and Moser, 2012). These devices are prone to colonization by biofilm-forming CRKP, which exhibit increased antibiotic tolerance and immune clearance. Biofilm formation contributes to persistent and recurrent infections and has also been identified as a major virulence factor linked to higher mortality rates in this vulnerable population (Gominet et al., 2017; Di Domenico et al., 2021, 2022; Lendak et al., 2021).
Meropenem/vaborbactam (MEV), a novel combination of a carbapenem with a boronic acid β-lactamase inhibitor, has shown potent activity against KPC-producing strains by effectively protecting meropenem from enzymatic degradation (Jorgensen and Rybak, 2018). It has been approved for treating complicated infections caused by multidrug-resistant Gram-negative bacteria, including those involving the bloodstream. Several studies have demonstrated its in vitro efficacy against KPC-producing Enterobacterales, and recent clinical data support its use in real-world settings (Tiseo et al., 2024). Italy represents a region of high endemicity for CRKP, predominantly driven by KPC-type carbapenemases. MEV demonstrated consistent in vitro activity against KPC-producing K. pneumoniae bloodstream isolates, with reported susceptibility rates of 87% (Gaibani et al., 2021). More recent nationwide surveillance data indicate that MEV retains high efficacy, with susceptibility exceeding 95% among KPC producers. When compared with other novel β-lactam/β-lactamase inhibitor combinations, imipenem/relebactam exhibited the highest susceptibility rate (97%), followed by MEV and ceftazidime/avibactam (93.9%) (Bianco et al., 2025). Notably, resistance to MEV and imipenem/relebactam was associated with structural alterations in outer membrane porins, including truncation of OmpK35 and the ins135GD insertion in OmpK36.
This study investigates the efficacy of MEV against CRKP isolates recovered from CRBSI in an orthopedic oncology hospital. We assessed minimum bactericidal concentration (MBC) and minimum biofilm eradication concentrations (MBEC) and characterized relevant resistance and virulence determinants. Additionally, the Galleria mellonella infection model was employed to evaluate the therapeutic potential of MEV in vivo.
By integrating phenotypic susceptibility testing, genotypic profiling, and an experimental infection model, this work aims to provide insight into the effectiveness of MEV in managing CRKP infections in a clinically challenging setting.
2 Methods
2.1 Strain collection and ethical approval
We examined two clinical isolates of carbapenem-resistant K. pneumoniae (CRKP) obtained from hospital-acquired infections in an orthopedic oncology unit. Isolates were obtained from blood cultures collected via central venous access from each patient under sterile conditions and plated on complete and selective media. These strains, part of the Bacterial Strain Collection at the Microbiology and Virology Unit of the San Gallicano Dermatological Institute, Istituti Fisioterapici Ospitalieri (IFO), were collected between January and March 2020. All relevant clinical and microbiological data were recorded in an electronic database.
Catheter-related bloodstream infection (CRBSI) was diagnosed when blood cultures drawn from the catheter hub became positive at least two hours earlier than when peripheral blood cultures of equal volume were simultaneously collected, as measured by an automated blood culture system (Rickard et al., 2021).
This study received approval from the Central Ethics Committee I.R.C.C.S. Lazio (Prot. 5179—18.04.2023, N. 1860/23), and all procedures followed established ethical guidelines and local regulatory requirements.
2.2 Microbiological diagnosis and strain characterization
Carbapenem-resistant K. pneumoniae isolates were recovered from patients with hospital-acquired infections (Di Domenico et al., 2021) and cultured on McConkey (Becton Dickinson, Germany) and blood agar plates (Becton Dickinson, Germany). MALDI-TOF MS (Bruker Daltonik, Germany) was used for bacterial identification, which was subsequently confirmed by 16S rRNA gene sequencing. Antimicrobial susceptibility testing was performed using the BD Phoenix™ automated system (Becton Dickinson Diagnostic Systems, USA) NMIC-474 panel, while colistin susceptibility was determined by broth microdilution (Thermo Scientific, USA). Results were interpreted in accordance with the EUCAST clinical breakpoints (http://www.eucast.org/clinical_breakpoints). The Cepheid Xpert Carba-R assay, integrated with the GeneXpert device (Cepheid, USA), was used to preliminarily detect the presence of blaKPC, blaVIM, blaOXA-48, blaIMP, and blaNDM (Di Domenico et al., 2020). Lateral flow immunoassay was used to detect clinically relevant carbapenemases, specifically KPC, IMP, NDM, VIM, and OXA-48 (Coris BioConcept, Belgium). K. pneumoniae ATCC 13883 was included as a reference strain in all phenotypic tests to provide an internal standard for assay calibration.
2.3 Whole-genome analysis
According to the manufacturer’s protocol, genomic DNA was extracted using the QIAsymphony DSP Virus/Pathogen Kit (Qiagen, Germany). The DNA was sequenced using a hybrid approach combining Illumina MiSeq and Nanopore GridION technologies. Quality control of Illumina reads was performed using FastQC while Nanopore data were assessed with nf-core/nanoseq v3.1.0 (Ewels et al., 2020). Quality metrics were summarized using MultiQC (Ewels et al., 2016). Genome assembly was conducted using Bactopia v.3.2.0 (Petit and Read, 2020) with default options. The pipeline employed the Dragonflye assembler in “short_polish” mode, where long reads were first assembled and subsequently polished using short reads. The assembly process incorporated Medaka (Oxford Nanopore Technologies, 2020) for consensus correction of Nanopore reads and Polypolish (Wick and Holt, 2022) for polishing with Illumina data.
The assembled genomes were characterized using multiple bioinformatic tools. Plasmid replicons were identified using PlasmidFinder v2.1.6 (Carattoli et al., 2014). Antimicrobial resistance determinants, virulence factors, sequence typing (MLST), and capsular loci (KL) were analyzed using Kleborate v3.1.3 (Lam et al., 2021). Kaptive v0.7.3 was used to identify capsule (K-locus) and lipopolysaccharide (O-locus) loci (Lam et al., 2022).
Biosynthetic gene clusters were identified using antiSMASH v8.0 (bacterial version) (Blin et al., 2023). Additional antibiotic resistance gene detection was performed using the Comprehensive Antibiotic Resistance Database (CARD) v4.0 and the Resistance Gene Identifier (RGI) tool v6.0.3 (Alcock et al., 2023), applying “perfect” and “strict” matches with ≥97% identity threshold. Virulence gene profiling was conducted using Kleborate and ABRicate v1.0.1.
Phylogenetic analysis of chromosomes was performed using kSNP v4.0 (Gardner et al., 2015) to identify core single-nucleotide polymorphisms. The maximum likelihood tree generated by kSNP was selected for phylogenetic inference. Plasmid phylogeny was assessed using Mash v2.3 (Ondov et al., 2016) for sequence similarity estimation. Phylogenetic trees were visualized using Interactive Tree Of Life (iTOL) (Letunic and Bork, 2024). Circular representations of chromosomal features were generated using Proksee (Grant et al., 2023), highlighting antimicrobial resistance genes, biosynthetic gene clusters, and other relevant genomic features.
2.4 Determination of meropenem/vaborbactam minimum bactericidal concentration (MBC90)
The bactericidal activity of MEV was assessed using the broth microdilution method in cation-adjusted Mueller-Hinton broth (CAMHB), following standardized procedures. MEV was tested in a concentration range of 0.008–16 μg/ml for meropenem, combined with a fixed 8 μg/ml of vaborbactam. Bacterial inocula were prepared by suspending overnight cultures grown on MacConkey agar in 0.45% saline and adjusted to a turbidity equivalent to a 0.5 McFarland standard (approximately 108 CFU/ml). The suspension was diluted to approximately 105 CFU/ml, and 100 μl was added to each well of a sterile 96-well flat-bottom polystyrene microtiter plate containing 100 μl of CAMHB. After 20 h of incubation at 37°C, minimum inhibitory concentrations (MICs) were recorded as the lowest concentration inhibiting visible bacterial growth. These MIC values confirmed susceptibility to MEV (≤2/8 μg/ml) for both CRKP isolates and the reference strain, and were in full agreement with results obtained using the automated BD Phoenix™ system. For determination of MBC90, viable bacterial counts were determined by serial dilution and plating on MacConkey agar, and results were expressed as CFU/ml. The MBC90 was defined as the lowest antibiotic concentration, resulting in a ≥90% reduction in CFU/ml relative to the untreated control.
2.5 Determination of meropenem/vaborbactam minimum biofilm eradication concentration (MBEC90)
Biofilm eradication assays were performed as previously described, with modifications. Overnight cultures grown on MacConkey agar were suspended in 0.45% saline and adjusted to 0.5 McFarland standard. The suspension was diluted to ~105 CFU/ml, and 100 μl aliquots were added to sterile 96-well flat-bottom polystyrene plates containing 100 μl of CAMHB. The plates were incubated at 37°C for five hours to allow biofilm formation. Wells were then rinsed with 0.45% saline to remove planktonic cells and filled with 100 μl of CAMHB containing meropenem at 0.008–16 μg/ml combined with a fixed 8 μg/ml of vaborbactam. The plates were incubated for an additional 20 h at 37°C. Untreated wells served as growth controls. Following treatment, wells were washed twice with saline, and biofilms were mechanically disrupted by scraping. Cells were resuspended in 100 μl of saline, serially diluted, and plated on MacConkey agar for CFU enumeration. K. pneumoniae ATCC 13883 was included as a reference strain to standardize biofilm quantification procedures. The MBEC90 was defined as the lowest antibiotic concentration, resulting in a ≥90% reduction in viable biofilm-associated cells relative to the untreated control.
2.6 Phenotypic characterization of virulence-associated traits
Biofilm formation was assessed using a static microtiter plate assay. Strains were cultured in CAMHB at 37°C with agitation (150 rpm) for 24 hours, washed, adjusted to a 0.5 McFarland standard, and inoculated (100 μl) into 96-well flat-bottom polystyrene plates. Following 24 hours of static incubation at 37°C, wells were washed with phosphate-buffered saline (PBS), air-dried for 45 minutes, stained with 0.1% crystal violet (CV), and destained with ethanol-acetone, 4:1. Biofilm biomass was quantified by measuring absorbance at 570 nm (Sivori et al., 2024).
Planktonic growth was evaluated using a resazurin-based fluorescence assay. Bacterial suspensions (~1×105 CFU/ml) were inoculated into 96-well plates containing CAMHB and 0.01% resazurin. Plates were incubated at 37°C, and fluorescence (excitation/emission: 560/590 nm) was measured every 30 minutes for 24 hours. Growth was expressed as the area under the fluorescence curve (AUC) relative to untreated controls.
Overnight bacterial cultures were harvested by centrifugation at 9,000 × g and resuspended in phosphate-buffered saline (PBS) to an optical density at 600 nm (OD600) of 1.0. Cell suspensions were centrifuged at 1,000 × g for 5 minutes to separate capsular material released into the supernatant. The OD600 of each supernatant was then measured and normalized to the initial OD600 of the corresponding bacterial suspension prior to centrifugation (Di Domenico et al., 2020).
Siderophore production was measured using the chrome azurol S (CAS) assay. Cultures (~1×105 CFU/ml) were grown in CAMHB, centrifuged, and 100 μl of supernatant was mixed with 100 μl of CAS reagent in a 96-well plate. After 20 minutes at room temperature, absorbance was recorded at 630 nm. Siderophore activity was calculated as percent siderophore units (PSU) using the formula: PSU (%) = [(Ar − As)/Ar] × 100, where Ar is the absorbance of the reference (uninoculated control) and As the absorbance of the sample (Guembe et al., 2025).
The hypermucoviscosity phenotype was evaluated by the string test. A bacterial colony was touched with a standard inoculation loop and lifted vertically; a string length ≥5 mm was interpreted as positive. All assays were performed in biological triplicate (Di Domenico et al., 2020).
2.7 In vivo infection model using Galleria mellonella
The Galleria mellonella infection model was used to evaluate the in vivo efficacy of MEV against CRKP. Larvae (250–320 mg; Bigserpens, Paliano, Italy) were selected based on size and lack of melanization. Bacterial strains were cultured in CAMHB to mid-logarithmic phase (OD600 = 0.4–0.6), harvested by centrifugation, washed in PBS, and adjusted to a final concentration of ~1×105 CFU per larva. Infection was established by injecting 10 μl of the bacterial suspension into the hemocoel via the last right proleg using a Hamilton syringe (10 μl volume, 26s gauge needle). Specifically, 30 minutes post-infection, treatment was administered by injecting 10 μl of MEV (Sigma–Aldrich) at 4 mg/kg. Control groups included untreated infected larvae and uninfected larvae injected with PBS. For each condition, 10 larvae were used. Infected larvae were incubated at 37°C in the dark, and their survival was monitored over 48 hours. Survival data were analyzed using the Mantel-Cox log-rank test (GraphPad Prism v10.0, GraphPad Software, San Diego, CA, USA). Experimental procedures were adapted from published protocols (Ignasiak and Maxwell, 2017; Guembe et al., 2025).
2.8 Statistical analysis
Statistical analysis was performed using GraphPad Prism v10.0 (GraphPad Software, San Diego, CA, USA) and IBM SPSS Statistics for Windows, Version 21.0 (IBM Corp., Armonk, NY, USA). Continuous variables are presented as mean ± standard deviation (SD) or as median with interquartile range (IQR), as appropriate. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was used to evaluate differences among groups for virulence-associated phenotypic traits. Kaplan–Meier survival curves for the G. mellonella model were analyzed using the log-rank (Mantel-Cox) test. A p-value < 0.05 was considered statistically significant for all comparisons. All experiments were conducted in biological triplicate unless otherwise stated.
3 Results
3.1 Isolation of carbapenem-resistant K. pneumoniae from catheter-related bloodstream infections
Two hospitalized patients in an orthopedic oncology unit were diagnosed with CRBSIs. The isolates were obtained from blood cultures collected via central venous access.
The antibiotic susceptibility testing of K. pneumoniae CRKP ST101 and CRKP ST307 was reported in Figure 1A. The K. pneumoniae ATCC 13883 (Kp ATCC 13883) was included as a reference strain (Figure 1A). CRKP ST101 and CRKP ST307 displayed extensive drug resistance, with high MICs to β-lactams, aminoglycosides, ciprofloxacin, and meropenem (MIC >16 μg/ml).
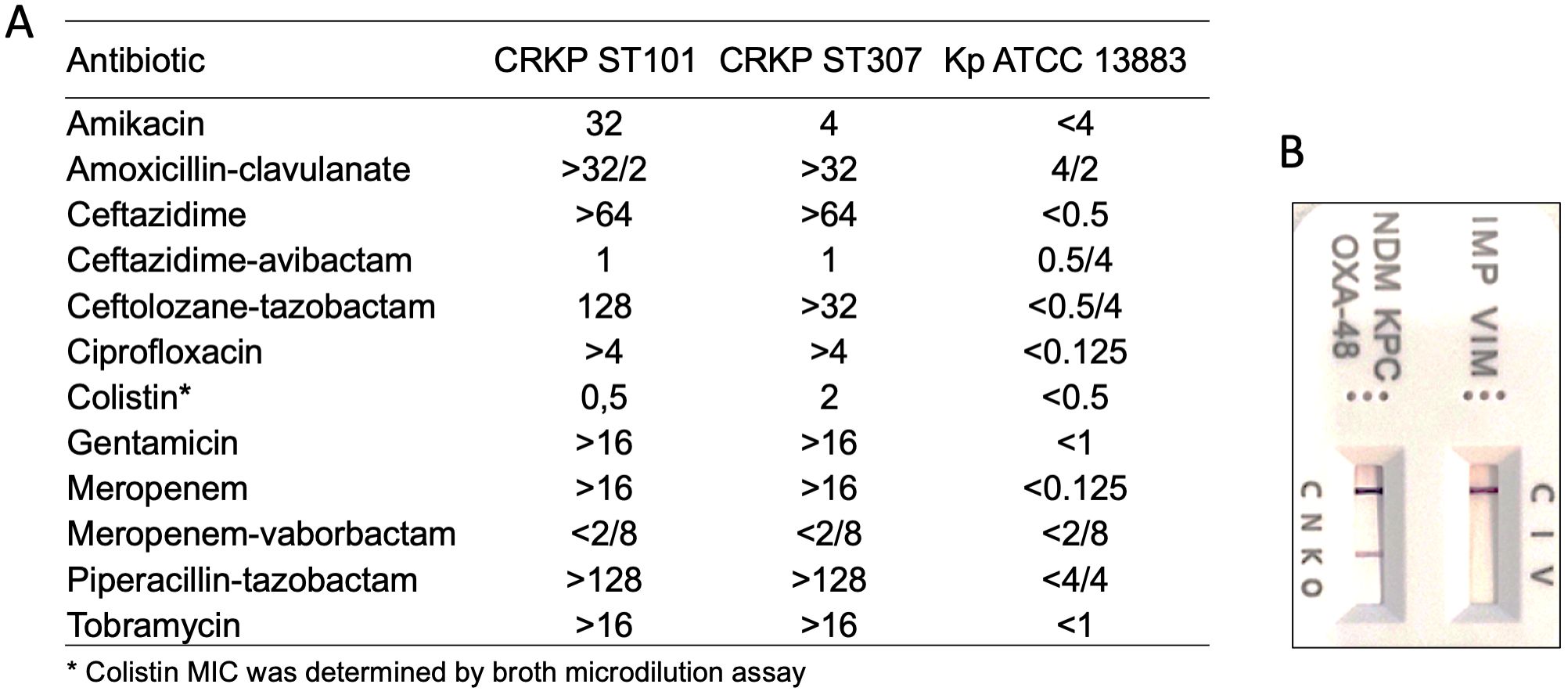
Figure 1. Antimicrobial susceptibility profiles and carbapenemase detection in Klebsiella pneumoniae strains. (A) Minimum inhibitory concentrations (MICs, μg/ml) for a panel of antibiotics were determined for CRKP ST101, CRKP ST307, and the reference strain K. pneumoniae ATCC 13883 using broth microdilution or automated systems, as appropriate. (B) Detection of carbapenemase enzymes was performed using a lateral flow immunoassay. Test bands indicate the presence of specific carbapenemases: IMP (I), VIM (V), NDM (N), KPC (K), and OXA-48-like enzymes (O). The control (C) band serves as an internal validity marker for test performance.
In contrast, KP ATCC 13883 remained susceptible to all tested agents, with low MIC values across all antibiotic classes. Notably, both CRKP ST101 and CRKP ST307 isolates were susceptible to MEV (MIC ≤ 2/8 μg/ml) despite their resistance to meropenem alone. The presence of carbapenemase enzymes was further verified by lateral flow immunoassay (Figure 1B). Colistin retained activity against CRKP ST101 (MIC = 0.5 μg/ml) but showed reduced efficacy against CRKP ST307 (MIC = 2 μg/ml), as determined by broth microdilution.
3.2 Genetic determinants and plasmid composition of two clinical CRKP isolates
Whole-genome sequencing of the K. pneumoniae isolates identified two distinct multilocus sequence types (STs), ST101 and ST307 (Figure 2A), corresponding to capsule loci KL17 and KL102, respectively. The genome of the ST101 isolate (CRKP ST101) comprised a 5.44 Mb chromosome and seven plasmids ranging in size from 6,972 bp to 180,962 bp. The ST307 isolate (CRKP ST307) possessed a 5.32 Mb chromosome and two plasmids of 147,570 bp and 103,862 bp (Figure 2B). A maximum-likelihood phylogenetic tree was constructed based on core single-nucleotide polymorphisms (SNPs) from whole-genome sequences to investigate the relatedness of two CRKP isolates (CRKP ST101 and CRKP ST307) to reference genomes available in the NCBI Pathogens database. The inclusion criteria comprised genome assemblies of high quality or complete status, the presence of blaKPC, blood as the source of isolation, and origin from European countries. The analysis shows that both isolates cluster more closely with a strain from England than other Italian isolates (Figure 2A).
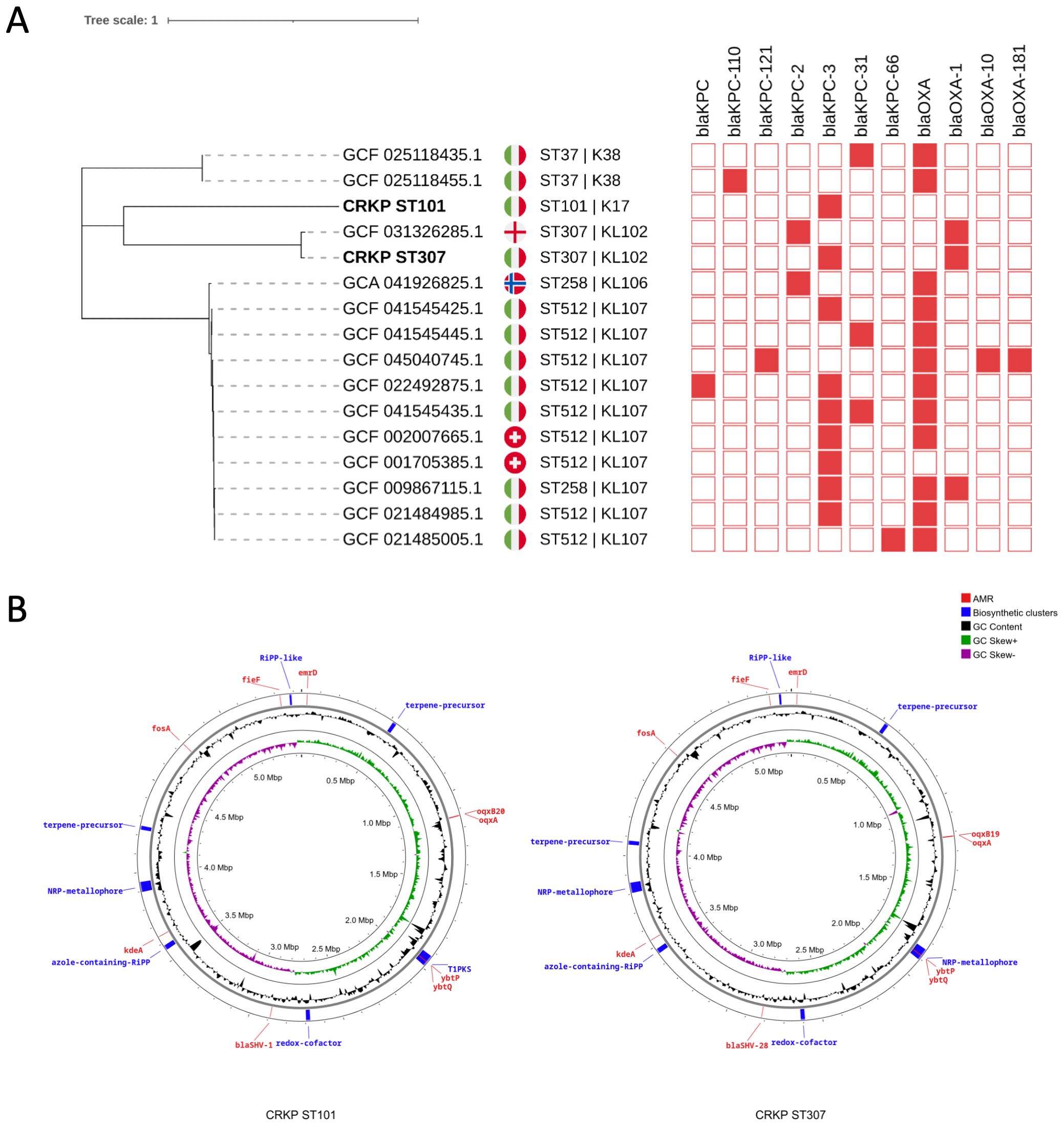
Figure 2. Phylogenetic analysis and genomic features of K. pneumoniae isolates. (A) A maximum-likelihood phylogenetic tree was constructed based on whole-genome sequences to assess the relatedness among K. pneumoniae strains. Isolates CRKP ST101 and CRKP ST307 (bold) are highlighted among reference genomes, revealing their phylogenetic placement. Genomes included in the analysis were filtered for high-quality completeness, and the presence of blaKPC with metadata restricted to blood-derived isolates from European countries was confirmed. National flags indicate the country of origin. Sequence types (ST) and capsular loci (KL) are shown next to each genome. The presence or absence of blaKPC or blaOXA carbapenemase genes (red) are displayed in a binary heatmap. (B) Circular chromosomal maps of Klebsiella pneumoniae isolates CRKP ST101 and CRKP ST307. Chromosomal representations of CRKP ST101 (left) and CRKP ST307 (right) display annotated genomic features. Outer rings highlight predicted antimicrobial resistance (AMR) genes (red), biosynthetic gene clusters (blue), and relevant virulence or metabolic determinants. Inner rings display GC content (black), GC skew+ (green), and GC skew– (purple). Biosynthetic clusters include RIPP-like, nonribosomal peptide synthetase (NRP)-metallophore, azole-containing RIPP, and terpene precursor regions.
Virulence-associated genes typically linked to hypervirulent K. pneumoniae (hvKP), including iucA, iroB, rmpA, and rmpA2, were undetected in either isolate. CRKP ST101 and CRKP ST307 carried antimicrobial resistance determinants conferring resistance to three or more antibiotic classes (Figure 2B).
Plasmid phylogeny (Figure 3) revealed that the blaKPC-3 gene in CRKP ST101 was located on plasmid p4, which clustered with a group of blaKPC-carrying plasmids, including GCF_009867115.2, GCF_045040745.1, GCF_021848955.1, and GCF_001053585.1.3. These plasmids formed a distinct clade within the blaKPC-containing group and carried replicons associated with the IncFII incompatibility group. In CRKP ST307, plasmid p2 carried the blaOXA-1 gene and grouped with blaOXA-containing plasmids such as GCF_031362851.2 and GCF_045040745.1.3. This plasmid encoded an IncFIB replicon. Plasmid p3 harbored the blaKPC-3 gene and clustered with blaKPC-only plasmids, including GCF_001053585.1.3, and carried both IncFIB and IncFII replicons.
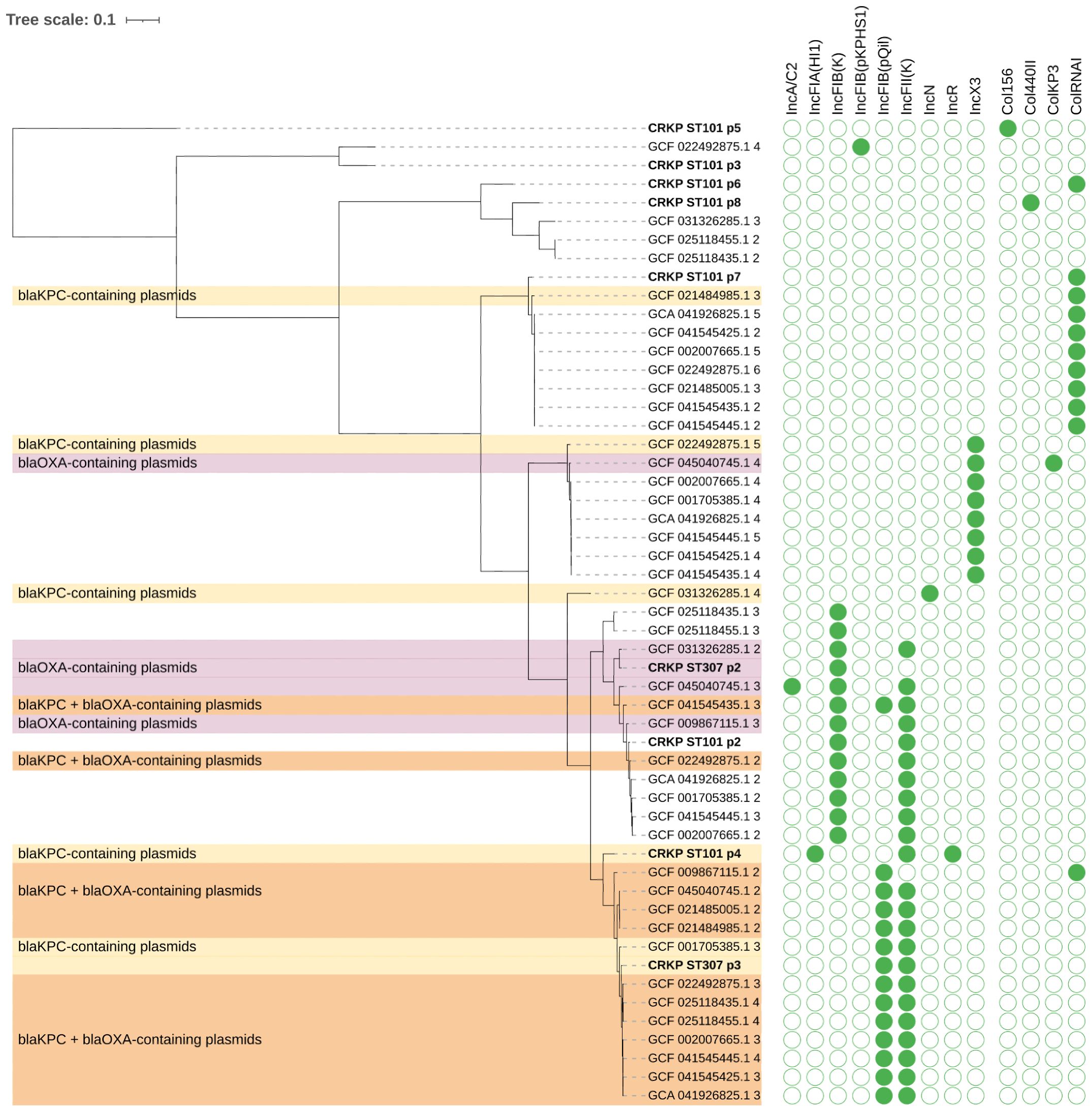
Figure 3. Phylogenetic tree of plasmids carrying carbapenemase genes blaKPC and/or blaOXA in K. pneumoniae. Plasmid sequences were clustered based on sequence similarity and colored according to the presence of blaKPC (yellow), blaOXA (purple), or both genes (orange). Plasmids (p) from CRKP ST101 and CRKP ST307 are highlighted (bold). The matrix on the right displays the presence (green) or absence (white) of plasmid replicons across plasmid genomes.
3.3 Phenotypic characterization of virulence-associated traits in CRKP isolates
Biofilm formation, metabolic activity, capsule production, and siderophore secretion in CRKP ST101, CRKP ST307, and the reference strain Kp ATCC 13883 were analyzed to investigate the virulence-associated traits. CRKP ST307 exhibited significantly higher biofilm biomass than CRKP ST101 (p < 0.0292) and Kp ATCC 13883 (p < 0.0001) (Figure 4A). Metabolic activity, assessed using a resazurin-based assay, was lower in Kp ATCC 13883 compared to CRKP ST101 (p = 0.0074) and CRKP ST307 (p = 0.0148) (Figure 4B). Capsule production was significantly higher in Kp ATCC 13883 than in CRKP ST307 (p = 0.0002) (Figure 4C), while siderophore secretion was also greater in Kp ATCC 13883 (p = 0.0003) (Figure 4D). A positive string test identified Kp ATCC 13883 as the only strain exhibiting a hypermucoviscous (HMV) phenotype (Figure 4E).
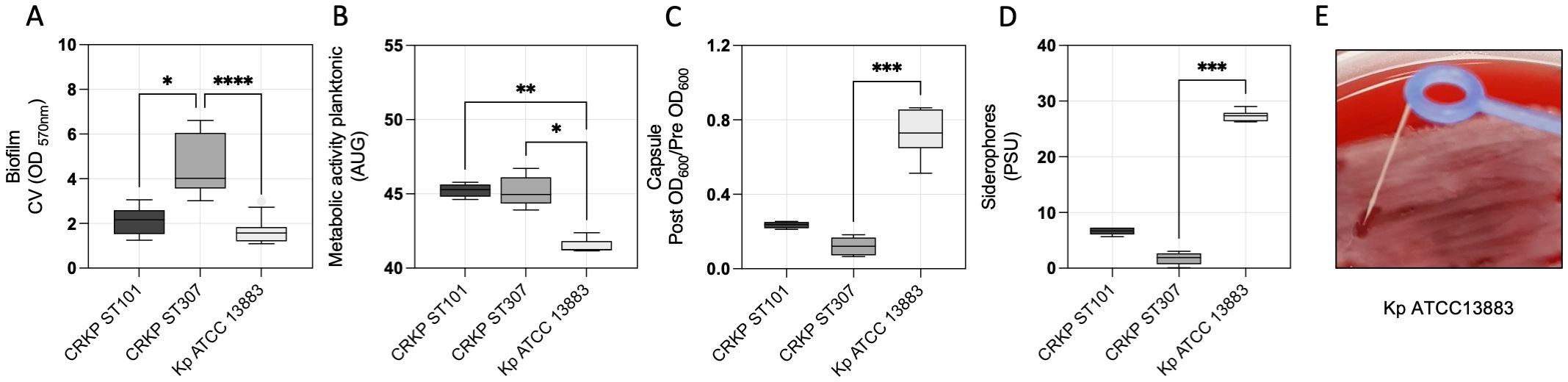
Figure 4. Virulence-associated traits in K. pneumoniae clinical isolates. (A) Quantitative analysis of biofilm biomass, (B) metabolic activity of planktonic cells, (C) capsule production, (D) and siderophore secretion in CRKP ST101, CRKP ST307, and Kp ATCC 13883. Statistical significance was determined using a one-way ANOVA followed by Tukey’s multiple comparisons test (p < 0.05). (E) A representative image of the string test performed on Kp ATCC 13883 to assess the hypermucoviscous phenotype. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
3.4 Antibacterial Activity of MEV against planktonic and biofilm-associated CRKP Isolates
The antibacterial activity of MEV was assessed against K. pneumoniae isolates CRKP ST101, CRKP ST307, and the reference strain Kp ATCC 13883 under both planktonic and biofilm-associated conditions (Figure 5). Bacterial viability was determined following treatment with increasing concentrations of MEV, and survival was expressed as the percentage of CFU/ml relative to untreated controls. For planktonic cells, MEV induced a concentration-dependent reduction in bacterial survival across all tested strains. The minimum bactericidal concentration (MBC90), defined as the lowest concentration of MEV resulting in ≥90% reduction in CFU, was 0.5/8 μg/ml for CRKP ST101, 0.12/8 μg/ml for CRKP ST307, and 0.25/8 μg/ml for Kp ATCC 13883. Similar concentrations were required for biofilm-associated cells to achieve ≥90% reduction in viability. Minimum biofilm eradication concentration (MBEC90) values were 0.5/8 μg/ml for CRKP ST101, 0.12/8 μg/ml for CRKP ST307, and 0.25/8 μg/ml for the reference strain. MEV showed negligible activity at concentrations below 0.06/8 μg/ml across all strains and conditions.
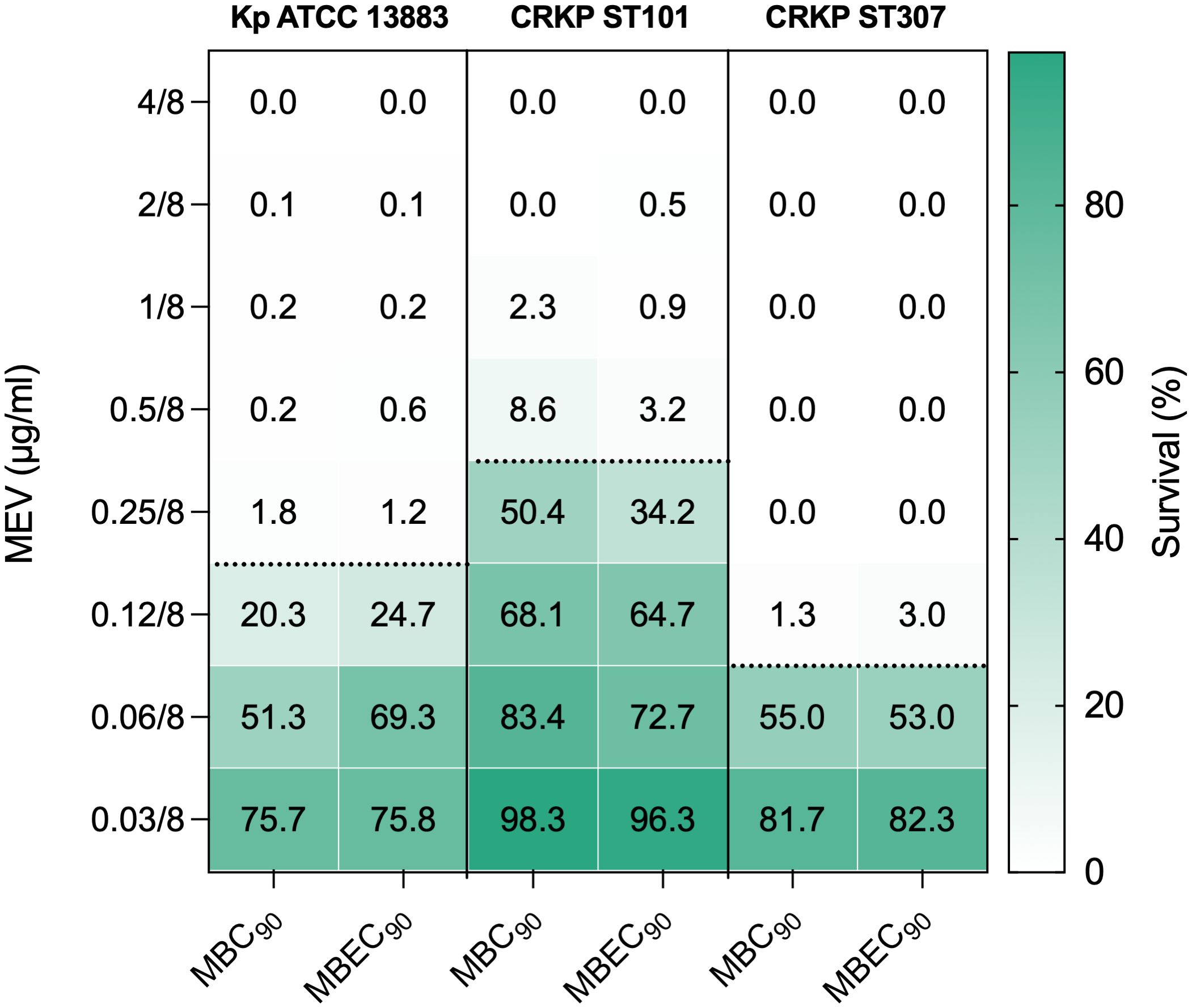
Figure 5. Antibacterial activity of MEV against K. pneumoniae planktonic and biofilm-associated cells. Bacterial survival was quantified following treatment with increasing concentrations of MEV and expressed as the percentage of CFU/ml relative to untreated controls. Data are shown for CRKP ST101, CRKP ST307, and the reference strain K. pneumoniae ATCC 13883 under planktonic and biofilm-associated conditions. The color gradient indicates growth compared to the untreated controls. The dashed black horizontal line indicates the 90% reduction threshold used to define the minimum bactericidal concentration (MBC90) for planktonic cells and the minimum biofilm eradication concentration (MBEC90) for biofilm-associated cells.
3.5 In vivo assessment of virulence and therapeutic efficacy of MEV in the Galleria mellonella infection model
Survival assays in the Galleria mellonella infection model were conducted to evaluate the virulence of CRKP isolates and the in vivo efficacy of MEV (Figure 6A). A dose-dependent mortality was observed following infection with Kp ATCC 13883, with 100% lethality reached at an inoculum of 1 × 105 CFU/larva within 24 hours (Figure 6B). This concentration was used to test the activity of MEV in the control Kp ATCC 13883, CRKP ST101, and CRKP ST307. Comparative analysis of larvae infected with 1×105 CFU/larva revealed significantly increased mortality in larvae infected with Kp ATCC 13883 (p = 0.0052) compared to both CRKP ST101 and CRKP ST307 (Figure 6C). At 48 hours post-infection, survival rates were 40.0% for CRKP ST101, 20.0% for CRKP ST307, and 0% for Kp ATCC 13883. Treatment with MEV (2 g meropenem + 2 g vaborbactam equivalent), administered 30 minutes post-infection, significantly improved survival in infected larvae (Figures 6D-F). Larvae infected with K. pneumoniae and treated with a single dose of MEV exhibited a significantly higher survival rate compared to untreated Kp ATCC 13883 (p = 0.0019), CRKP ST101 (p = 0.0204), and CRKP ST307 (p = 0.0058), at 24 hours.
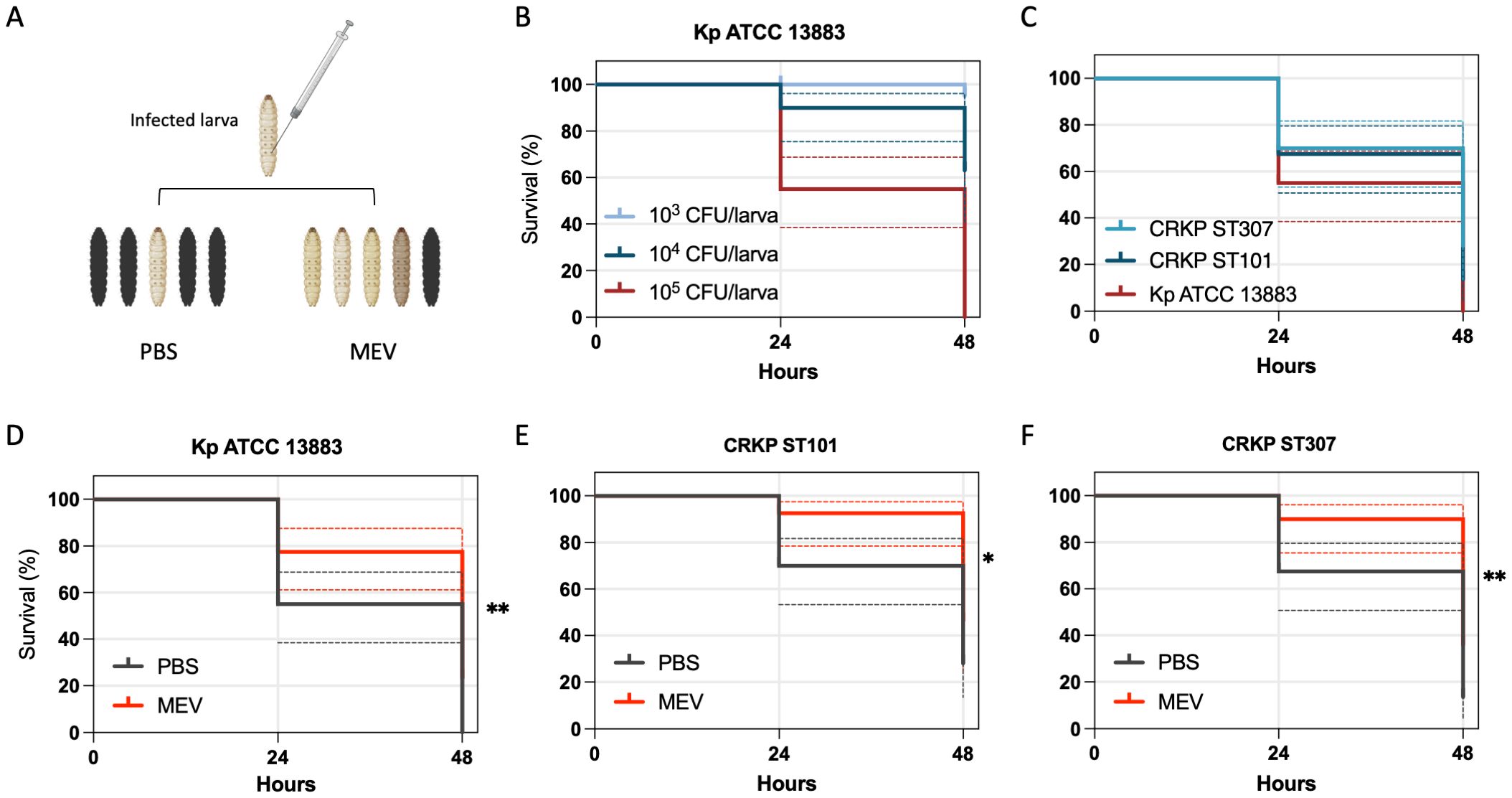
Figure 6. Kaplan-Meier survival curves of G. mellonella infected with K. pneumoniae and treated with MEV. (A) Schematic representation of the G. mellonella infection model, illustrating the injection procedure and representative images of healthy (light-colored) and dead (dark-colored) larvae. (B) Survival of G. mellonella following infection with Kp ATCC 13883 at inocula of 1×10³, 1×104, and 1×105 CFU/larva over 48 hours. (C) Comparative survival analysis of G. mellonella infected with Kp ATCC 13883, CRKP ST101, and CRKP ST307 at 1×105 CFU/larva. (D–F) Kaplan-Meier survival curves of G. mellonella infected with Kp ATCC 13883 (D), CRKP ST101 (E), or CRKP ST307 (F) at 1×105 CFU/larva and treated with a single dose of MEV (2g meropenem + 2g vaborbactam equivalent) administered 30 minutes post-infection. Control groups included larvae treated with either MEV (2 g/2 g equivalent) or PBS (Data not shown). Statistical significance was determined using the Log-rank test. *P < 0.05; **P < 0.01.
4 Discussion
The increasing prevalence of CRKP in oncology settings represents a critical clinical challenge, particularly in patients requiring prolonged use of indwelling devices. In this study, we characterized two CRKP isolates from CRBSIs in an orthopedic oncology unit, integrating genomic, phenotypic, and in vivo data to evaluate the activity of MEV. Whole-genome sequencing revealed that the isolates belonged to high-risk clones ST101 and ST307, both associated with extensive antimicrobial resistance (AMR) and nosocomial transmission. These isolates harbored clinically relevant resistance determinants, including blaKPC-3 and blaOXA, distributed across IncFII and IncFIB plasmid replicons. Both sequence types have been increasingly reported in bloodstream infections across Italy, with ST101 and ST307 representing emerging epidemic clones associated with hospital outbreaks and poor clinical outcomes in southern regions (Loconsole et al., 2020; Di Pilato et al., 2024). These isolates harbored clinically relevant resistance determinants, including blaKPC-3 and blaOXA. This observation aligns with national surveillance studies reporting KPC-3 as the most prevalent carbapenemase variant among K. pneumoniae isolates circulating in Italy (Giani et al., 2013; Bonura et al., 2015; Calia et al., 2017; Fasciana et al., 2019; Guembe et al., 2025). The identified genetic determinants are also consistent with previous reports describing the plasmid-mediated convergence of resistance in CRKP strains circulating in healthcare environments (Wyres et al., 2020; Di Pilato et al., 2024; Zhao et al., 2024). A recent nationwide genomic surveillance in Italy documented the widespread circulation of high-risk clones, particularly those within clonal complexes CC258, CC101, and CC307, among CRKP (Bianco et al., 2025). These lineages were also predominant among strains resistant to novel β-lactam/β-lactamase inhibitor combinations, including MEV and imipenem/relebactam and ceftazidime/avibactam. Although all isolates in our study remained susceptible to MEV, they belonged to clonal backgrounds (ST101 and ST307) that have been previously implicated in cross-resistance to multiple β-lactam/β-lactam inhibitor combinations. This overlap highlights the potential for further resistance development and supports the need for ongoing genomic monitoring to track the dissemination of epidemiologically successful clones and detect early signs of reduced susceptibility.
Phenotypic assays demonstrated that CRKP ST307 exhibited enhanced biofilm production and metabolic activity relative to CRKP ST101 and the reference strain Kp ATCC 13883 in the absence of hypervirulence-associated markers. These findings align with previous observations that classical CRKP lineages can express robust biofilm phenotypes contributing to persistence and therapeutic recalcitrance in the bloodstream and hospital-acquired infections (Gominet et al., 2017; Di Domenico et al., 2020; Fabrizio et al., 2024; Guerra et al., 2025). In contrast, biofilm production did not correlate with increased in vivo mortality in the G. mellonella model. Despite CRKP ST307 exhibiting the highest biofilm biomass in vitro, larvae infected with this strain demonstrated lower mortality rates than those infected with the reference strain Kp ATCC 13883. This dissociation between biofilm-forming capacity and acute virulence in G. mellonella suggests that biofilm-associated traits may primarily contribute to persistence and antibiotic tolerance rather than acute lethality in this model (Vuotto et al., 2017; Di Domenico et al., 2020). The observation supports previous studies indicating that while biofilm formation enhances survival under antimicrobial pressure and host immunity, it does not necessarily translate to increased pathogenicity in short-term infection models lacking complex immune responses (Wand et al., 2012; Benthall et al., 2015; Guembe et al., 2025).
The pronounced virulence of Kp ATCC 13883 in the G. mellonella infection model, as evidenced by complete lethality at 1×105 CFU/larva within 24 hours, can be attributed to its elevated production of capsule and siderophores. Quantitative assays confirmed significantly greater capsule polysaccharide, HMV phenotype, and siderophore secretion in Kp ATCC 13883 compared to CRKP ST101 and CRKP ST307. Both factors have been independently associated with increased immune evasion and enhanced fitness in host environments, including invertebrate models (Paczosa and Mecsas, 2016). Capsule production inhibits phagocytosis and complement activation, while siderophores facilitate iron acquisition under host-imposed nutritional immunity, contributing synergistically to virulence (Paczosa and Mecsas, 2016). These findings are consistent with previous studies demonstrating that hypercapsulated and siderophore-rich K. pneumoniae strains elicit high mortality in G. mellonella independent of classical hypervirulence genotypes (Pu et al., 2023).
MEV exhibited potent bactericidal activity against both planktonic and biofilm-associated cells. MBC90 and MBEC90 values for CRKP ST101, CRKP ST307, and Kp ATCC 13883 were ≤0.5/8 μg/ml, confirming the efficacy of MEV across multiple growth states. This result is consistent with in vitro studies demonstrating MEV’s preserved activity against KPC-producing strains despite resistance to meropenem monotherapy (Tumbarello et al., 2024; Giuliano et al., 2025). The low MBEC90 values further support its potential use in device-associated infections where biofilm formation contributes to therapeutic failure.
In the G. mellonella model, MEV significantly improved survival in larvae infected with CRKP ST101 or CRKP ST307. These findings are in line with prior preclinical studies demonstrating the in vivo efficacy of MEV in bloodstream and device-related infection models (Tiseo et al., 2024). Notably, while both strains were susceptible to MEV in vitro, differential virulence was observed, with CRKP ST307 exhibiting higher biofilm biomass and lower capsule and siderophore production. This phenotypic variation may reflect strain-specific adaptations influencing pathogenesis and treatment response.
The in vivo efficacy of MEV observed in the G. mellonella model must be interpreted within the context of the experimental design, particularly regarding pharmacokinetics and dosage regimens. In clinical settings, MEV is administered as a combination of 2 g meropenem and 2 g vaborbactam every 8 hours by intravenous infusion over 3 hours. This extended infusion strategy ensures sustained plasma concentrations exceeding the MIC, which is critical for optimizing bactericidal activity against K. pneumoniae, particularly in severe infections such as bacteremia. In contrast, the G. mellonella model employed a single-dose administration of MEV (2 g meropenem + 2 g vaborbactam equivalent) delivered 30 minutes post-infection. While this regimen enabled a comparative assessment of strain-specific virulence and drug responsiveness, it does not replicate the pharmacodynamic exposure achieved in human therapy. This discrepancy highlights a key limitation of the study, as the simplified dosing in the invertebrate model does not capture the complexity of drug absorption, distribution, metabolism, and elimination in mammalian systems. Additionally, although G. mellonella offers a cost-effective model for the preliminary evaluation of antimicrobial efficacy and virulence, it lacks the immunological and physiological complexity of vertebrate hosts.
Further limitations include the small number of isolates examined, which restricts the generalizability of our findings. While our two CRKP strains provide valuable mechanistic insights into MEV activity, they may not fully represent the genetic and phenotypic diversity observed among CRKP in clinical settings. For instance, Giuliano et al. (2025) described two KPC-producing CRKP isolates causing ventriculitis, which responded favorably to MEV therapy, highlighting the utility of such studies despite the inherent limitations of small sample sizes. Similarly, Larcher et al. (2023) reported a single case of bloodstream infection due to a KPC-3-producing K. pneumoniae in a critically ill patient with augmented renal clearance. The successful use of continuous infusion MEV in this context illustrates how individualized therapeutic strategies can be informed by detailed isolate characterization even in single-isolate studies. These examples underscore the need for broader datasets to capture the full spectrum of resistance mechanisms, virulence factors, and biofilm phenotypes within CRKP populations. Moreover, the absence of pharmacokinetic/pharmacodynamic analyses in our study limits the translation of in vitro findings into optimized dosing regimens for clinical use. Future studies incorporating dynamic infection models and standard human-equivalent dosing schedules will be crucial for more accurately assessing the translational potential of MEV in treating CRKP-associated bloodstream infections.
Furthermore, caution is warranted in interpreting MEV’s activity against biofilm-associated K. pneumoniae. Although MEV showed in vitro activity against mature biofilms, the extracellular matrix and reduced metabolic activity of biofilm-embedded cells may limit antibiotic penetration and bactericidal efficacy in vivo. These features may limit the clinical utility of in vitro MBEC90 values as predictors of treatment success in device-associated infections.
Collectively, our findings highlight the genomic complexity and phenotypic heterogeneity of CRKP strains causing CRBSI in immunocompromised hosts. MEV demonstrated activity against planktonic and biofilm-associated CRKP and improved survival in an invertebrate infection model. These data support the continued evaluation of MEV as a therapeutic option for CRKP-associated bloodstream infections and underscore the importance of integrating genomic surveillance with functional assays to inform antimicrobial stewardship strategies in high-risk populations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB88190.
Ethics statement
The studies involving humans were approved by Central Ethics Committee I.R.C.C.S. Lazio (Prot. 5179—18.04.2023, N. 1860/23). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
FS: Writing – review & editing, Formal Analysis, Investigation. MF: Writing – review & editing, Investigation, Formal Analysis. MT: Writing – review & editing, Formal Analysis, Data curation, Software. ICa: Formal Analysis, Writing – review & editing, Investigation. CP: Resources, Writing – review & editing. GF: Writing – review & editing, Investigation, Formal Analysis. ICe: Methodology, Writing – review & editing, Data curation. AC: Data curation, Writing – review & editing, Methodology. LF: Data curation, Writing – review & editing, Methodology. FP: Supervision, Writing – original draft. ED: Conceptualization, Supervision, Validation, Data curation, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Italian Ministry of Health (RC 2025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MM declared a shared affiliation with the author GF to the handling editor at the time of review.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AMR, antimicrobial resistance; ATCC, American Type Culture Collection; CARD, Comprehensive Antibiotic Resistance Database; CFU, colony forming units; CRBSI, catheter-related bloodstream infection; CRKP, carbapenem-resistant Klebsiella pneumoniae; HMV, hypermucoviscous; MBEC90, minimum biofilm eradication concentration 90%; MBC90, minimum bactericidal concentration 90%; MEV, meropenem/vaborbactam; MLST, multilocus sequence typing; SNP, single nucleotide polymorphism; ST, sequence type; WGS, whole-genome sequencing.
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Benthall, G., Touzel, R. E., Hind, C. K., Titball, R. W., Sutton, J. M., Thomas, R. J., et al. (2015). Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int. J. Antimicrobial. Agents 46, 538–545. doi: 10.1016/j.ijantimicag.2015.07.014
Bianco, G., Boattini, M., Lupo, L., Ambretti, S., Greco, R., Degl’Innocenti, L., et al. (2025). In vitro activity and genomic characterization of KPC-producing Klebsiella pneumoniae clinical blood culture isolates resistant to ceftazidime/avibactam, meropenem/vaborbactam, imipenem/relebactam: an Italian nationwide multicentre observational study, (2022–23). J. Antimicrobial. Chemother. 80, 583–592. doi: 10.1093/jac/dkae450
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Bonura, C., Giuffrè, M., Aleo, A., Fasciana, T., Di Bernardo, F., Stampone, T., et al. (2015). An Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy 2014: emergence of multiple non-ST258 clones. PloS One 10, e0132936. doi: 10.1371/journal.pone.0132936
Calia, C., Pazzani, C., Oliva, M., Scrascia, M., Lovreglio, P., Capolongo, C., et al. (2017). Carbapenemases-producing Klebsiella pneumoniae in hospitals of two regions of Southern Italy. APMIS 125, 491–498. doi: 10.1111/apm.12666
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Di Domenico, E. G., Cavallo, I., Sivori, F., Marchesi, F., Prignano, G., Pimpinelli, F., et al. (2020). Biofilm production by carbapenem-resistant klebsiella pneumoniae significantly increases the risk of death in oncological patients. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.561741
Di Domenico, E. G., Marchesi, F., Cavallo, I., Toma, L., Sivori, F., Papa, E., et al. (2021). The impact of bacterial biofilms on end-organ disease and mortality in patients with hematologic Malignancies developing a bloodstream infection. Microbiol. Spectr. 9, e00550–e00521. doi: 10.1128/Spectrum.00550-21
Di Domenico, E. G., Oliva, A., and Guembe, M. (2022). The current knowledge on the pathogenesis of tissue and medical device-related biofilm infections. Microorganisms 10, 1259. doi: 10.3390/microorganisms10071259
Di Pilato, V., Pollini, S., Miriagou, V., Rossolini, G. M., and D’Andrea, M. M. (2024). Carbapenem-resistant Klebsiella pneumoniae : the role of plasmids in emergence, dissemination, and evolution of a major clinical challenge. Expert Rev. Anti-infective Ther. 22, 25–43. doi: 10.1080/14787210.2024.2305854
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Ewels, P. A., Peltzer, A., Fillinger, S., Patel, H., Alneberg, J., Wilm, A., et al. (2020). The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278. doi: 10.1038/s41587-020-0439-x
Fabrizio, G., Sivori, F., Cavallo, I., Truglio, M., Toma, L., Sperati, F., et al. (2024). Efficacy of sodium hypochlorite in overcoming antimicrobial resistance and eradicating biofilms in clinical pathogens from pressure ulcers. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1432883
Fasciana, T., Gentile, B., Aquilina, M., Ciammaruconi, A., Mascarella, C., Anselmo, A., et al. (2019). Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 19, 928. doi: 10.1186/s12879-019-4565-3
Gaibani, P., Lombardo, D., Bussini, L., Bovo, F., Munari, B., Giannella, M., et al. (2021). Epidemiology of meropenem/vaborbactam resistance in KPC-producing klebsiella pneumoniae causing bloodstream infections in northern Italy. Antibiotics 10, 536. doi: 10.3390/antibiotics10050536
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Giani, T., Pini, B., Arena, F., Conte, V., Bracco, S., Migliavacca, R., et al. (2013). Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill 18, 20489. doi: 10.2807/ese.18.22.20489-en
Giuliano, G., Sambo, M., Castellani, P., Benedetti, S., Tarantino, F., and Tumbarello, M. (2025). Meropenem-vaborbactam as intrathecal-sparing therapy for the treatment of carbapenem-resistant K. pneumoniae shunt-related ventriculitis: two case reports and review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 44, 193–196. doi: 10.1007/s10096-024-04986-6
Gominet, M., Compain, F., Beloin, C., and Lebeaux, D. (2017). Central venous catheters and biofilms: where do we stand in 2017? APMIS 125, 365–375. doi: 10.1111/apm.12665
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi: 10.1093/nar/gkad326
Guembe, M., Hafian, R., Díaz-Navarro, M., Visedo, A., De Maio, F., Pimpinelli, F., et al. (2025). Virulence profile of carbapenem-resistant Klebsiella pneumoniae strains by an in vivo model of Galleria mellonella. Microbiol. Spectr., e02215–e02224. doi: 10.1128/spectrum.02215-24
Guerra, M. E. S., Destro, G., Vieira, B., Lima, A. S., Ferraz, L. F. C., Hakansson, A. P., et al. (2025). Corrigendum: Klebsiella pneumoniae biofilms and their role in disease pathogenesis. Front. Cell. Infect. Microbiol. 15. doi: 10.3389/fcimb.2025.1564010
Ignasiak, K. and Maxwell, A. (2017). Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res. Notes 10, 428. doi: 10.1186/s13104-017-2757-8
Jorgensen, S. C. J. and Rybak, M. J. (2018). Meropenem and Vaborbactam: Stepping up the Battle against Carbapenem-resistant Enterobacteriaceae. Pharmacotherapy 38, 444–461. doi: 10.1002/phar.2092
Lam, M. M. C., Wick, R. R., Judd, L. M., Holt, K. E., and Wyres, K. L. (2022). Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microbial. Genomics 8. doi: 10.1099/mgen.0.000800
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188. doi: 10.1038/s41467-021-24448-3
Larcher, R., Laffont-Lozes, P., Naciri, T., Bourgeois, P.-M., Gandon, C., Magnan, C., et al. (2023). Continuous infusion of meropenem–vaborbactam for a KPC-3-producing Klebsiella pneumoniae bloodstream infection in a critically ill patient with augmented renal clearance. Infection 51, 1835–1840. doi: 10.1007/s15010-023-02055-2
Lendak, D., Puerta-Alcalde, P., Moreno-García, E., Chumbita, M., García-Pouton, N., Cardozo, C., et al. (2021). Changing epidemiology of catheter-related bloodstream infections in neutropenic oncohematological patients. PloS One 16, e0251010. doi: 10.1371/journal.pone.0251010
Letunic, I. and Bork, P. (2024). Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
Loconsole, D., Accogli, M., De Robertis, A. L., Capozzi, L., Bianco, A., Morea, A., et al. (2020). Emerging high-risk ST101 and ST307 carbapenem-resistant Klebsiella pneumoniae clones from bloodstream infections in Southern Italy. Ann. Clin. Microbiol. Antimicrob. 19, 24. doi: 10.1186/s12941-020-00366-y
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132. doi: 10.1186/s13059-016-0997-x
Paczosa, M. K. and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Petit, R. A. and Read, T. D. (2020). Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems 5, e00190–e00120. doi: 10.1128/mSystems.00190-20
Pu, D., Zhao, J., Chang, K., Zhuo, X., and Cao, B. (2023). Superbugs” with hypervirulence and carbapenem resistance in Klebsiella pneumoniae: the rise of such emerging nosocomial pathogens in China. Sci. Bull. 68, 2658–2670. doi: 10.1016/j.scib.2023.09.040
Rickard, C. M., Marsh, N. M., Larsen, E. N., McGrail, M. R., Graves, N., Runnegar, N., et al. (2021). Effect of infusion set replacement intervals on catheter-related bloodstream infections (RSVP): a randomised, controlled, equivalence (central venous access device)–non-inferiority (peripheral arterial catheter) trial. Lancet 397, 1447–1458. doi: 10.1016/S0140-6736(21)00351-2
Sivori, F., Cavallo, I., Truglio, M., De Maio, F., Sanguinetti, M., Fabrizio, G., et al. (2024). Staphylococcus aureus colonizing the skin microbiota of adults with severe atopic dermatitis exhibits genomic diversity and convergence in biofilm traits. Biofilm 8, 100222. doi: 10.1016/j.bioflm.2024.100222
Tiseo, G., Galfo, V., Riccardi, N., Suardi, L. R., Pogliaghi, M., Giordano, C., et al. (2024). Real-world experience with meropenem/vaborbactam for the treatment of infections caused by ESBL-producing Enterobacterales and carbapenem-resistant Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. doi: 10.1007/s10096-024-04758-2
Tumbarello, M., Raffaelli, F., Giannella, M., De Pascale, G., Cascio, A., De Rosa, F. G., et al. (2024). Outcomes and predictors of mortality in patients with KPC-kp infections treated with meropenem vaborbactam: an observational multicenter study. Open Forum Infect. Dis. 11, ofae273. doi: 10.1093/ofid/ofae273
Vuotto, C., Longo, F., Pascolini, C., Donelli, G., Balice, M. P., Libori, M. F., et al. (2017). Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol. 123, 1003–1018. doi: 10.1111/jam.13533
Wand, M. E., Bock, L. J., Turton, J. F., Nugent, P. G., and Sutton, J. M. (2012). Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J. Med. Microbiol. 61, 470–477. doi: 10.1099/jmm.0.037523-0
Wick, R. R. and Holt, K. E. (2022). Polypolish: Short-read polishing of long-read bacterial genome assemblies. PloS Comput. Biol. 18, e1009802. doi: 10.1371/journal.pcbi.1009802
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Zhao, Y., Qian, C., Ye, J., Li, Q., Zhao, R., Qin, L., et al. (2024). Convergence of plasmid-mediated Colistin and Tigecycline resistance in Klebsiella pneumoniae. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1221428
Keywords: carbapenem-resistant Klebsiella pneumoniae, multidrug-resistant plasmid, meropenem/vaborbactam, catheter-related bloodstream infections, biofilm-associated infections, surgical site infection
Citation: Sivori F, Francalancia M, Truglio M, Cavallo I, Pronesti C, Fabrizio G, Celesti I, Cazzani A, Furzi L, Pimpinelli F and Di Domenico EG (2025) Meropenem/vaborbactam activity against carbapenem-resistant Klebsiella pneumoniae from catheter-related bloodstream infections. Front. Cell. Infect. Microbiol. 15:1616353. doi: 10.3389/fcimb.2025.1616353
Received: 22 April 2025; Accepted: 14 July 2025;
Published: 31 July 2025.
Edited by:
Luca Cavinato, Washington University in St. Louis, United StatesReviewed by:
Jozsef Soki, University of Szeged, HungaryGabriele Bianco, University Hospital Città della Salute e della Scienza di Torino, Italy
Maria Teresa Mascellino, Sapienza University of Rome, Italy
Copyright © 2025 Sivori, Francalancia, Truglio, Cavallo, Pronesti, Fabrizio, Celesti, Cazzani, Furzi, Pimpinelli and Di Domenico. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mauro Truglio, bWF1cm8udHJ1Z2xpb0BpZm8uaXQ=
†These authors have contributed equally to this work
 Francesca Sivori
Francesca Sivori Massimo Francalancia
Massimo Francalancia Mauro Truglio
Mauro Truglio Ilaria Cavallo
Ilaria Cavallo Carmelina Pronesti2
Carmelina Pronesti2 Giorgia Fabrizio
Giorgia Fabrizio Fulvia Pimpinelli
Fulvia Pimpinelli Enea Gino Di Domenico
Enea Gino Di Domenico