- 1Center for Infectious Diseases Research (CIDR) and World Health Organization (WHO) Collaborating Center for Reference and Research on Bacterial Pathogens, American University of Beirut, Beirut, Lebanon
- 2Department of Pediatrics and Adolescent Medicine, American University of Beirut Medical Center, Beirut, Lebanon
- 3Pediatric Infectious Diseases Division, Department of Pediatrics and Adolescent Medicine, American University of Beirut Medical Center, Beirut, Lebanon
- 4Department of Experimental Pathology, Immunology, and Microbiology, Faculty of Medicine, American University of Beirut, Beirut, Lebanon
- 5Clinical Microbiology Laboratory, Department of Pathology and Laboratory Medicine, American University of Beirut Medical Center, Beirut, Lebanon
Introduction: The identification of bacterial pathogens in the clinical setting is essential for providing optimal care and improving outcomes. The primary objective of this study was to assess the performance of the 16S test in bacterial identification from different samples and determine its impact on clinical outcomes.
Methods: This was a retrospective study of patient samples collected from all age-groups at the American University of Beirut Medical Center (AUBMC), from May 2016 to December 2022. Descriptive statistics were conducted to calculate the 16S test positivity rate and to describe the different types of organisms. Univariate analyses were performed to study the clinical impact of the 16S test and its comparison to the conventional bacterial culture among different characteristics. A p ≤ 0.05 was considered statistically significant.
Results: A total of 1489 specimens were submitted for the 16S test during the study period. Of the submitted tests, 395 (26%) had bacteria identified by the 16S test and/or culture. Out of the culture negative/16S positive group, the majority were from specimens collected from the skin and soft tissue system (24 out of 92, 26.1%). This was followed by musculoskeletal specimens (15 out of 92, 16.3%), and central nervous system specimens (14 out of 92, 15.2%). Pus samples had a positivity rate of 66.3% with 5 times higher odds of being positive compared to non-pus samples (25%). Overall, there were 260 identified organisms by 16S test of which the most detected organisms were Staphylococcus spp, Streptococcus spp. and Enterobacterales. The results revealed that 16S testing impacted management in 45.9% of the cases (83/181) showing a change in management. Antibiotic escalation was applied in 31.3% of cases (26/83). Antibiotic de-escalation occurred in 41% of cases (34/83). A change in the treating diagnosis was noted in 26.5% of cases (22/83).
Conclusion: Identification of pathogens using the 16S test in combination with conventional cultures is essential in clinical diagnostics and management of infectious diseases to provide targeted therapy and improve antimicrobial stewardship. Shorter turnaround time, improved patient management, and cost-effectiveness are key factors to consider when advocating for the broader adoption of 16S testing.
Introduction
The identification of bacterial pathogens in the clinical setting is essential for providing optimal care and improving outcomes. Until recently, traditional culture-based identification using appropriate media and Gram stains remained the standard practice. However, some bacterial pathogens are fastidious, slow growing, may not grow unless very special media are used or because they were exposed to antibiotics prior to obtaining samples for culture. Under these circumstances, the use of molecular diagnostic tools becomes advantageous. Sequencing of 16S rRNA gene (16S test) is one of these readily available tools that is becoming more utilized over the past few years. The 16S test can be applied on various clinical specimens such as tissue specimens, pleural, peritoneal, or joint effusions, cerebral spinal fluid (CSF), and blood (Ursenbach et al., 2021a). Universal primers are used to amplify bacterial nucleic acids by targeting the conserved regions; variable regions unique to each bacterial species allow the identification to the species level by PCR followed by sequencing (Cook et al., 2003). Successful identification is reported to be attainable in up to 91% of the cases (Mignard and Flandrois, 2006) in a shorter turnaround time (Mishra et al., 2019) allowing early identification, targeted therapy, favorable clinical outcomes (Fida et al., 2021) and avoiding the use of unnecessary broad antibiotics (Messacar et al., 2017).
Previous studies have demonstrated that the 16S test had both sensitivity and specificity levels above 70%. However, assessment of the impact of the 16S test on clinical outcomes and the influence on antimicrobial prescriptions are limited in the literature. Therefore, it is vital to examine the value of the 16S test on antimicrobial stewardship and cost effectiveness (Mishra et al., 2020). Due to the higher costs and technical challenges of the 16S test, it is usually reserved for patients who may benefit the most from the test to maximize diagnostic yield while taking into consideration cost effectiveness.
The primary objective of this study was to assess the performance of the 16S test in bacterial identification from different samples submitted to our laboratory and determine its impact on clinical outcomes. We specifically examined the role of the 16S test in patients where the results were discordant with conventional cultures, considering the clinical features and impact on clinical outcomes.
Materials and methods
Study design
This was a single center retrospective study of patient samples submitted from different clinical services at the American University of Beirut Medical Center (AUBMC) to our laboratory at the Center for Infectious Diseases Research (CIDR) located at AUBMC.
Study population
The inclusion criteria were 1) all patients (inpatients and outpatients) with clinically suspected infection, 2) all age-groups, with 3) samples received at CIDR for 16S test collected from different body sites from May 2016 to December 2022.
Study procedure
All 16S tests were performed at CIDR at AUBMC without prior knowledge of culture results. For patients where the culture and 16S test results were discordant (culture positive, 16S test negative or culture negative, 16S test positive or those with results showing different microbiological results), clinical data was collected. Patient electronic records were reviewed to assess whether a significant change in management or antimicrobial therapy occurred based on the 16S test result.
Definitions
All specimens with fluids such as joint fluid, pleural fluid, pericardial fluid and abscesses or collections were considered fluid specimens. All other specimens were considered tissue specimens including those from skin and soft tissues, muscles, heart valves, etc. In this study, gastrointestinal (GI) samples referred exclusively to specimens obtained from normally sterile sites, such as peritoneal fluid, liver abscesses, bile, and splenic tissue, collected under aseptic conditions to minimize contamination from commensal flora.
The majority of patients were suspected of having an infection. Prior antimicrobial therapy was defined as any antimicrobial therapy administered in the 2 weeks preceding sample collection.
We reviewed the impact of the 16S test on clinical management, classifying the outcomes into two main categories: ‘Change in Management’ and ‘No Change in Management.’ Among cases with a change in management (n=109), interventions included escalation or de-escalation of antimicrobial therapy, discontinuation of antibiotics, modification of the treatment diagnosis, or withholding antimicrobial therapy due to a negative 16S result. A negative 16S test refers to a result where no bacterial DNA was detected in the specimen by 16S rRNA PCR amplification and subsequent sequencing. This could be due to absence of infection, low bacterial load, or presence of bacteria not detected due to technical limitations of PCR. In cases with no change in management (n=139), factors included confirmation of culture results, severe or critical clinical status, absence of additional pathogens, coverage of identified pathogens by the current regimen, findings of contaminants, or mixed, unidentified bacterial results. We limited our analysis to 250 cases with discordant results between 16S testing and conventional culture, as these cases offered the most insight into the unique clinical contributions of 16S testing. By focusing on discordant cases—where results were either 16S-positive with culture-negative, 16S-negative with culture-positive, or both positive but with different identified organisms—we aimed to understand how 16S testing influences clinical decisions in cases where conventional methods may have limited answers or provide conflicting data. Examining key variables, such as prior antimicrobial therapy, clinical decisions made, and patient outcomes in this subset, allowed us to specifically investigate the added diagnostic and therapeutic value of 16S testing in clinical cases.
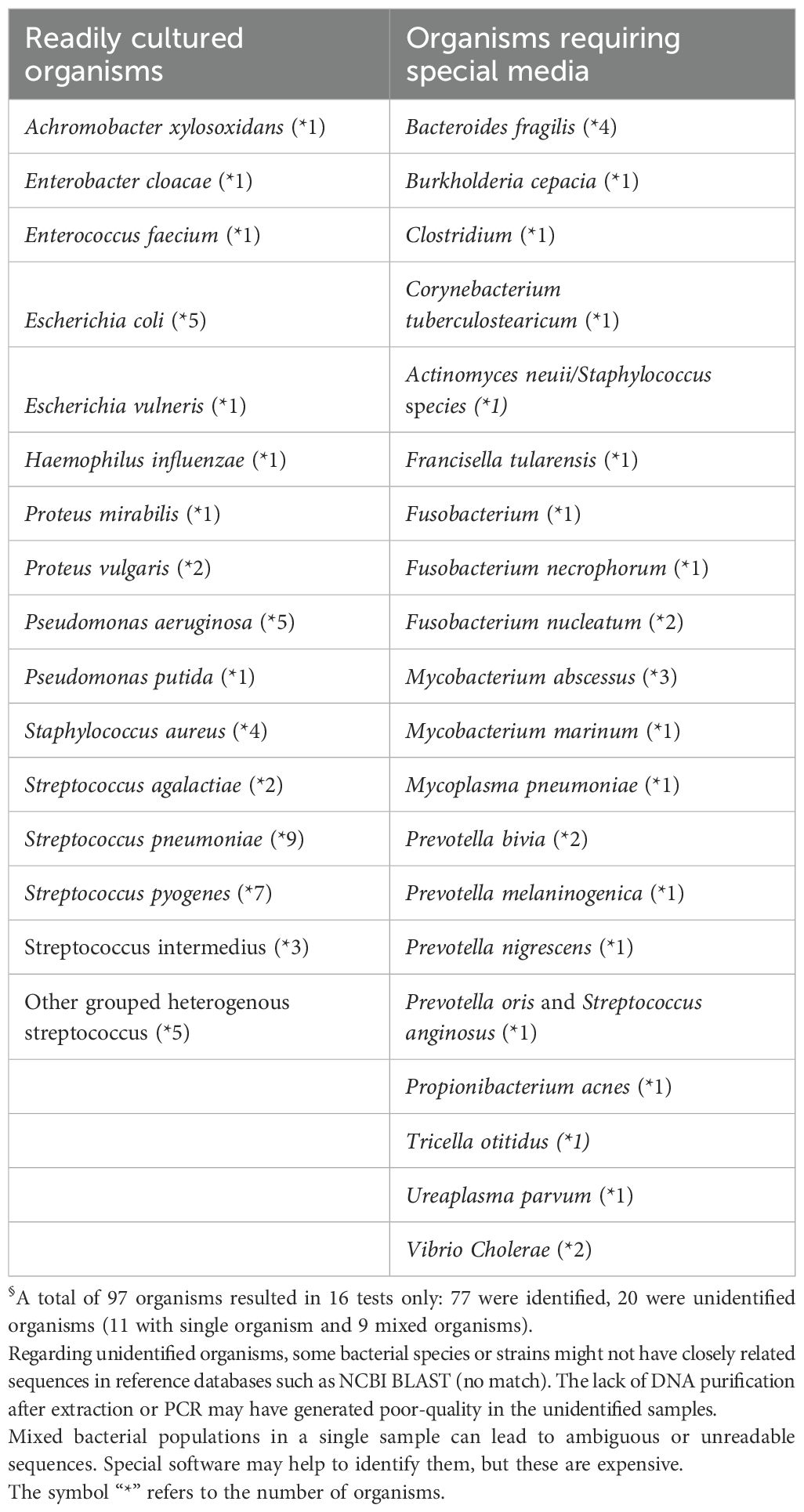
In our study, we divided the organisms detected by 16S with a negative culture into readily cultured organisms and those that require special media. In conventional microbiology, “readily cultured organisms” are those bacteria that can be easily grown using standard laboratory culture techniques and media. These organisms typically do not require special conditions or media to grow and are often identified through conventional culture methods. Conversely, “organisms that require special media to grow” are those that are more fastidious, meaning they have specific nutritional or environmental requirements that are not met by standard culture media. These organisms may require enriched or selective media, specific atmospheric conditions, or other modifications to support their growth. The challenge of culturing these organisms has led to the development of novel cultivation methods, such as the use of dilute nutrient media and simulated natural environmental conditions, to recover previously unculturable bacteria from complex bacterial communities (Vartoukian et al., 2010; Miller et al., 2018).
Conventional microbial testing
Bacterial identification was performed using standard microbiological and biochemical techniques. Clinical specimens were first cultured on appropriate selective and differential media to allow for bacterial isolation and preliminary differentiation based on colony morphology, pigmentation, and hemolytic activity. Gram staining was subsequently conducted to classify isolates as Gram-positive or Gram-negative. Further identification was achieved through a series of biochemical assays, including catalase, coagulase, oxidase, urease, indole, and carbohydrate fermentation tests, as appropriate. These tests were interpreted according to standard bacteriological protocols to determine the species-level identity of the isolates.
DNA extraction and 16 rRNA PCR protocol
DNA extraction was performed using the NucleoSpin Bloodkit (Macherey-Nagel, Germany). Briefly, the specimens were incubated with lysozyme (10837059001; Sigma-Aldrich, Germany) for 20 minutes at 37°C followed by incubation with Proteinase K for 30 minutes at 70°C. The DNA was bound to the silica membrane and washed twice then eluted. Extraction kit buffers and polymerase chain reaction (PCR) reagents were routinely tested for bacterial DNA contamination. PCR was performed using the 27F/519R primers (Hassan et al., 2014).
Each PCR reaction (20 μl) consisted of 1 × HOT FIREPOL BLEND Master Mix supplemented with 7.5 mM Mgcl2 (Solis BioDyne), DNA template and nuclease free water. Escherichia coli chromosomal DNA was used as a positive control. The 16S PCR was performed using the C1000 Thermal Cycler (BioRad, USA). The cycling conditions were as follows: 95°C for 12 minutes corresponding to an initial denaturation, followed by denaturation over 30 cycles at 95°C for 30 seconds, annealing at 54°C for 30 seconds, elongation at 72°C for 1 minute, with a final elongation at 72°C for 5 minutes. The amplified PCR products were analyzed by gel electrophoresis through a 1% agarose gel in Tris-borate-EDTA buffer. Bands were detected by ethidium bromide staining and UV transillumination. Amplified PCR products were then purified with ExoSAP-IT PCR Product Cleanup Reagent (78201.1.ML; ThermoFisher Scientific, USA) as per the manufacturer’s instructions. Sequencing reactions were performed using the Sanger method on an ABI 3730 XL system (Applied Biosystems). Sequences were analyzed using the blast tool against the NCBI core nucleotide database with a similarity threshold of ≥90% for species-level identification.
Statistical analysis
Statistical analysis was done using the Statistical Package for the Social Sciences (SPSS) V.28 (SPSS™ Inc., Chicago, IL USA). Descriptive statistics were conducted to identify the demographic characteristics of subjects, to calculate the 16S test positivity rate and to describe the different types of organisms. Afterward, univariate analyses using the binomial logistic regression were performed to study the clinical impact of the 16S test and its comparison to the conventional bacterial culture among different characteristics. The strength of association was interpreted using the unadjusted odds ratio (UOR) with 95% confidence interval (CI). A p ≤ 0.05 was considered statistically significant.
Ethical considerations
This study was approved by the AUB Institutional Review Board (IRB) (Protocol number: BIO-2021-0205). The enrollment of patients occurred under complete confidentiality. The patient’s name or other identifying information was not used, each sample and its data had a specific study code. Data was collected from the patient’s chart and was coded rather than deidentified. The SPSS was secured by a username and password accessible to study team members only and was stored in password-protected computers. The data will remain and be secured for only five years after the end of the study. After that period, and after its publication, all the data will be permanently deleted.
Results
A total of 1,489 specimens were submitted for 16S rRNA testing during the study period. Of these, 395 specimens (26%) were positive by either the 16S test and/or conventional culture. Specifically, 225 samples (15.1%) were positive by both methods, while 97 samples (6.5%) were positive by the 16S test only—92 were culture-negative (6.2%), and 5 were not cultured (0.3%) (Supplementary Figure S1, Supplementary Table S1). Among the 322 samples that were PCR-positive, 79.5% (n = 256) yielded specific bacterial identification by sequencing, including 3.7% with polymicrobial findings. Species identification was not successful in 20.5% (n = 66) of PCR-positive samples. The overall distribution of organisms detected by 16S testing and conventional culture is summarized in Supplementary Table S2.
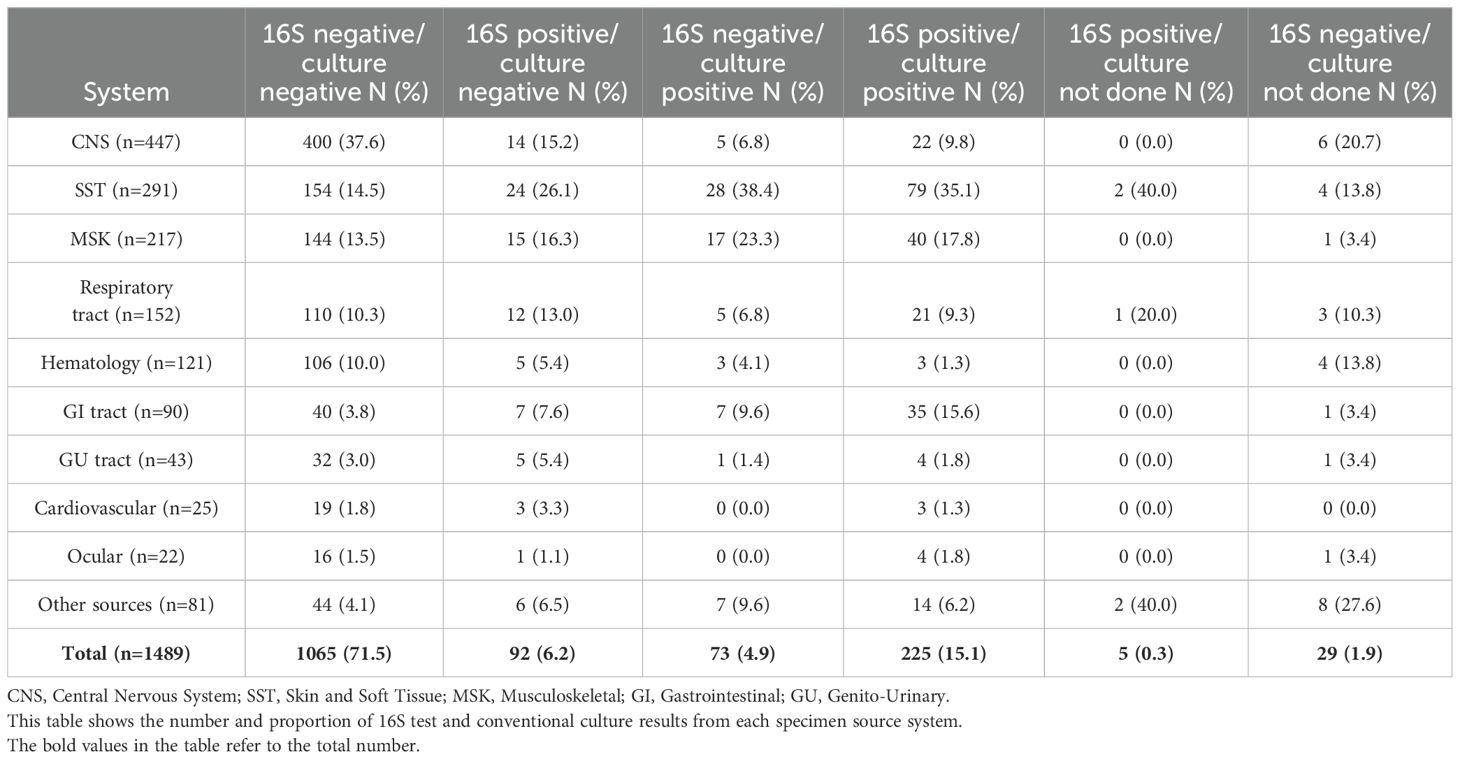
Among the 1,455 specimens tested by both 16S and culture, high concordance between the two methods was observed in 1,185 cases (81.4%) (Table 1). Discordant results were observed in 270 cases (18.6%), with 92 (34.1%) being culture-negative but 16S-positive, and 73 (27%) being culture-positive but 16S-negative. Within the culture-negative/16S-positive group, most specimens were derived from the skin and soft tissue system (26.1%), followed by musculoskeletal (16.3%) and central nervous system (15.2%) samples (Table 2).
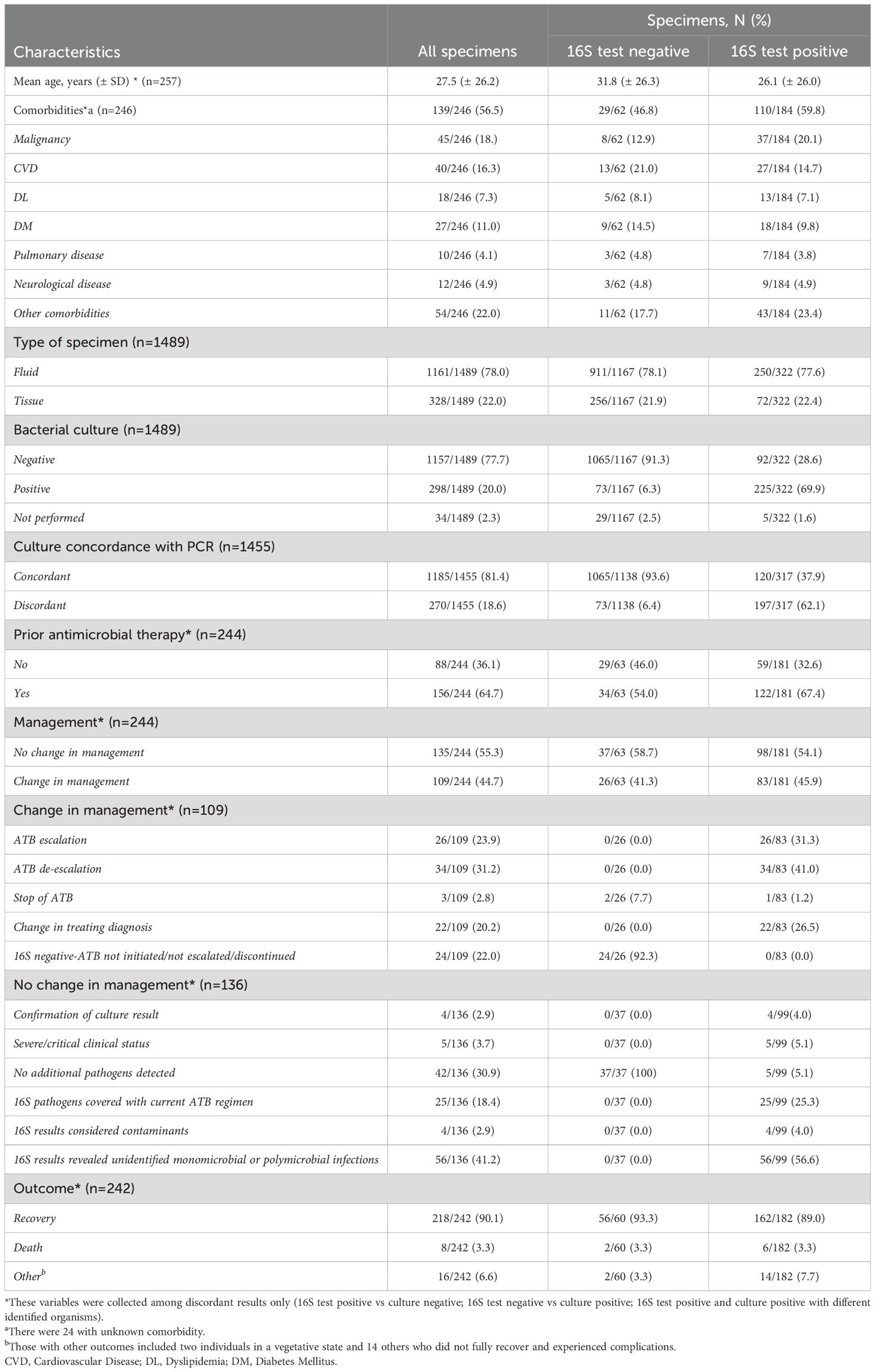
Table 1. Characteristics of specimens and comparison between 16S test negative and positive results.
The overall positivity rate of the 16S test remained relatively stable throughout the study period, ranging from 20.6% to 29.9%, except for the year 2021, which showed a significant drop to 10.7% (p = 0.003, OR = 0.402, 95% CI: 0.222–0.726) (Figure 1). The likelihood of a positive 16S result was significantly higher in gastrointestinal (GI) specimens, with a positivity rate of 46.7% (42/90) (p < 0.001). GI specimens primarily included peritoneal fluid (61.8%), liver tissue (27%), splenic tissue (5.6%), abdominal wall (4.5%), and bile tree fluid (1.1%). Skin and soft tissue (SST) specimens followed, with a positivity rate of 36.1% (105/291) (p < 0.001) (Figure 2). Among SST specimens, 16S positivity was higher in non-surgical samples compared to surgical ones (45.9% vs. 27.6%, p = 0.005, OR = 0.449, 95% CI: 0.257–0.786) (Table 3). Additionally, 16S positivity in cardiovascular system specimens (24%) was significantly higher than in CNS specimens (8.1%) (p = 0.010, OR = 3.605, 95% CI: 1.354–9.597) (Figure 3). Similarly, culture positivity was significantly higher among GI (47.2%) and SST specimens (37.5%) compared to CNS specimens (p < 0.001).
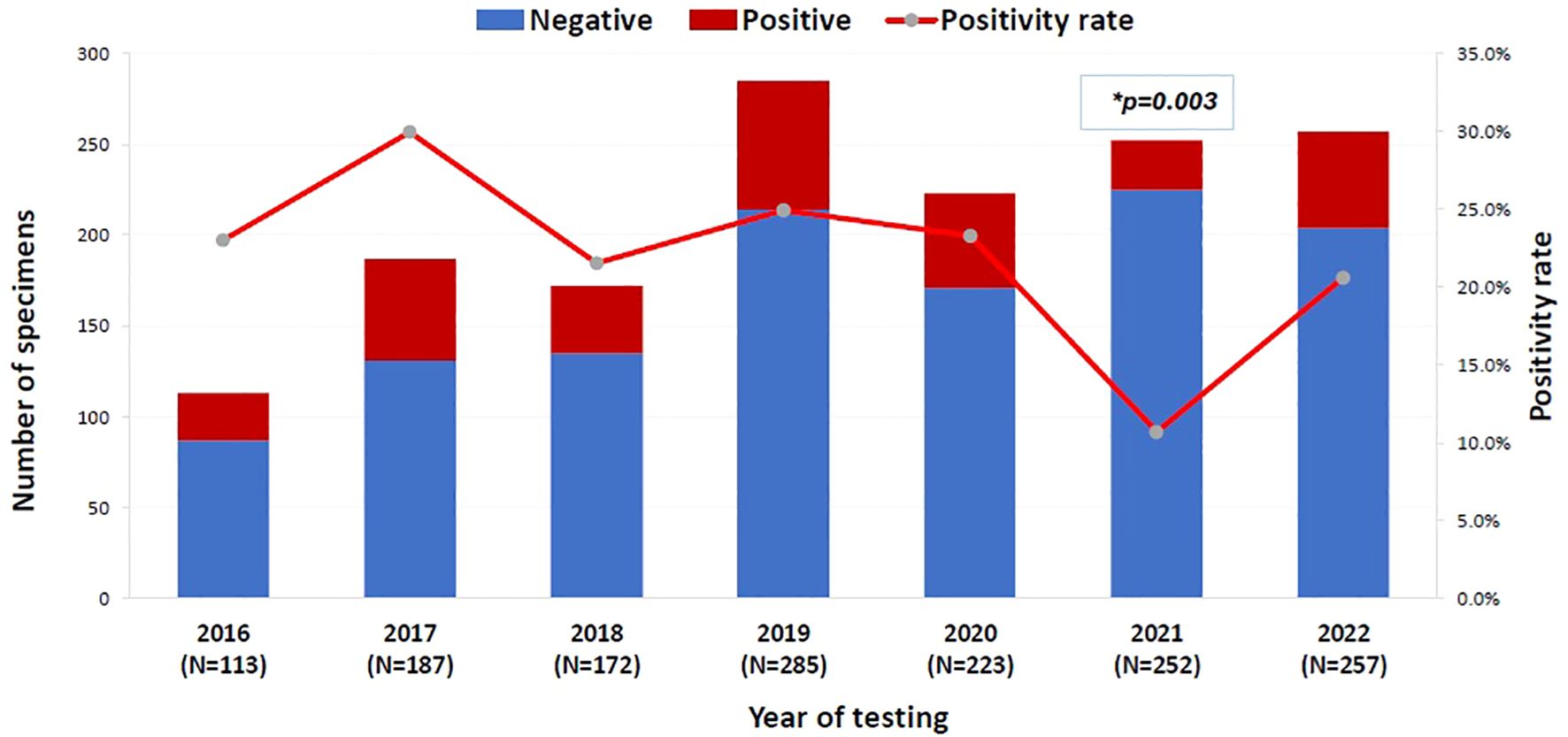
Figure 1. The positivity rate of 16S test across the years (N=1489). 16S rRNA PCR, 16S ribosomal Ribonucleic Acid Polymerase Chain Reaction. Binomial logistic regression analysis was used to compare the 16S rRNA PCR positivity rate across years. The year 2016 was the reference category. *P-value= 0.003, OR= 0.402, 95% CI= [0.222-0.726]. The x-axis represents the years of 16S testing from 2016 to 2022. The left y-axis is the number of 16S test negative (blue bar) and 16S test positive (red bar) specimens per year and the right y-axis is the 16S test positivity rate per year (red line).

Figure 2. 16S test positivity rate per sample source. CNS, Central Nervous System; GU, Genito-Urinary; MSK, Musculoskeletal; SST, Skin and Soft Tissue; GI, Gastrointestinal. Binomial logistic regression analysis was used to compare the 16S rRNA PCR positivity rate among sample sources by system. The CNS was the reference category. Sample sources: GI tract: peritoneal fluid, liver, spleen, abdominal wall, bile tree; SST: leg tissues, neck abscess, lymph nodes, back abscess, thigh tissues and fluids, toe swab, perianal abscess; Cardiovascular: pericardial fluid, valve, pericardial tissue, mediastinal fluid, aortic tissue, pus surrounding pacemaker; MSK: knee tissues and fluids, bone tissues, joint fluids, synovial fluids, shoulder fluids. Respiratory tract: BAL, Pleural fluid, DTA, lung tissues; Ocular: eye discharge, eye swab; GU tract: urine, urethral discharge, vaginal swab, cervical tissue; CNS: CSF, brain abscess, brain tissue; Hematology: blood, bone marrow aspirate. The x-axis represents the 16S test positivity rate. The y-axis represents the rate in each sample source system.
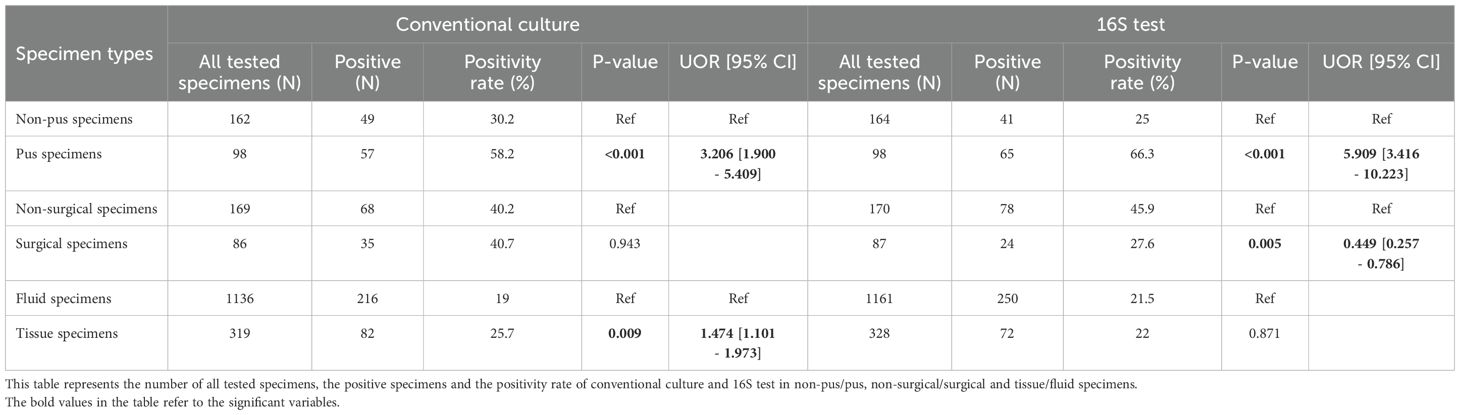
Table 3. Positivity rate of the conventional culture and 16S test among the different specimen types.
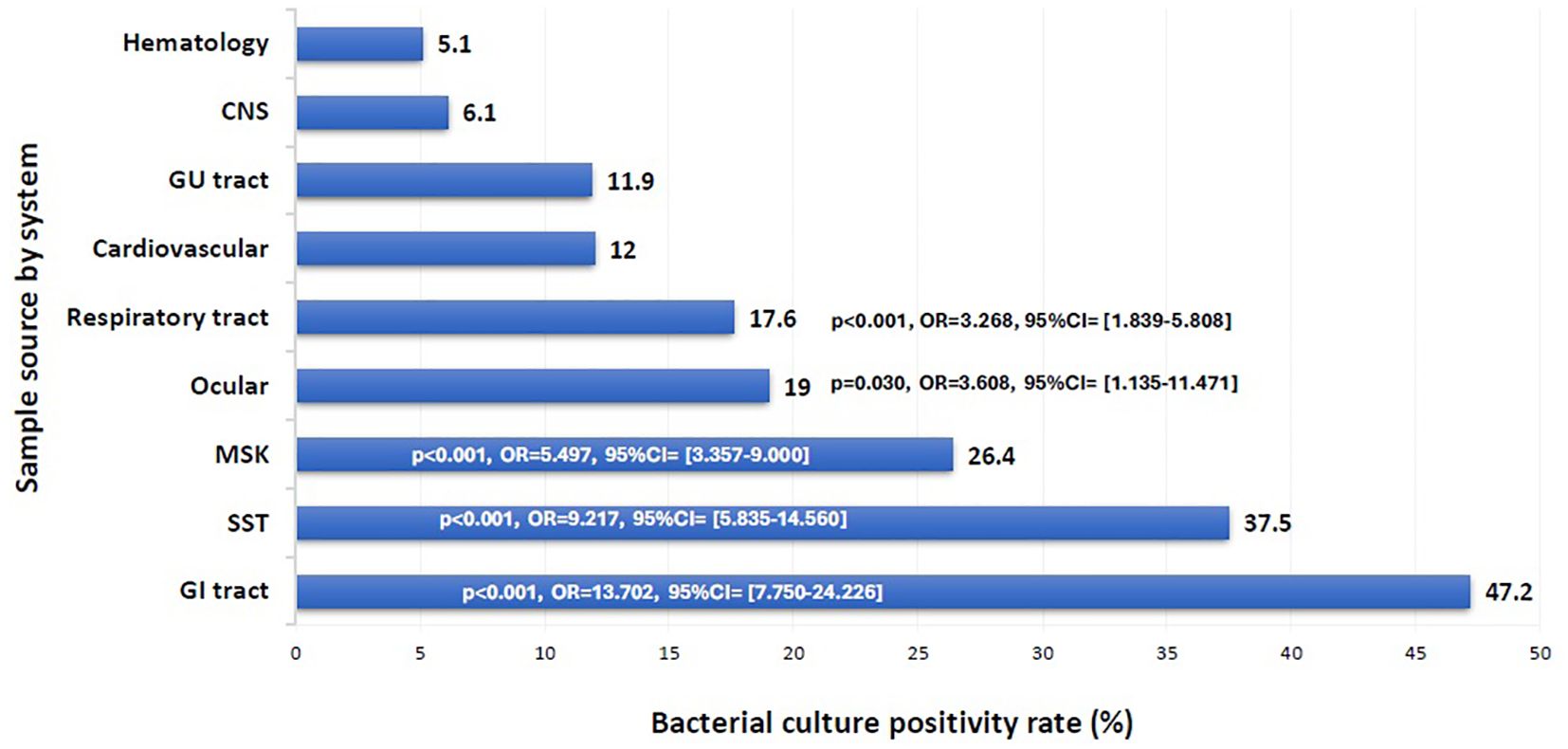
Figure 3. Bacterial culture positivity rate per sample source. CNS, Central Nervous System; GU, Genito-Urinary; MSK, Musculoskeletal; SST, Skin and Soft Tissue; GI, Gastrointestinal. Binomial logistic regression analysis was used to compare the bacterial culture positivity rate among sample sources by system. The CNS was the reference category. Sample sources: GI tract: peritoneal fluid, liver, spleen, abdominal wall, bile tree; SST: leg tissues, neck abscess, lymph nodes, back abscess, thigh tissues and fluids, toe swab, perianal abscess; Cardiovascular: pericardial fluid, valve, pericardial tissue, mediastinal fluid, aortic tissue, pus surrounding pacemaker; MSK: knee tissues and fluids, bone tissues, joint fluids, synovial fluids, shoulder fluids. Respiratory tract: BAL, Pleural fluid, DTA, lung tissues; Ocular: eye discharge, eye swab; GU tract: urine, urethral discharge, vaginal swab, cervical tissue; CNS: CSF, brain abscess, brain tissue; Hematology: blood, bone marrow aspirate. The x-axis represents the bacterial culture positivity rate. The y-axis represents the rate in each sample source system.
Of the 92 culture-negative/16S-positive specimens, the majority (83.7%) were fluids, including 26.1% from SST samples (Tables 1, 2; Supplementary Figure S1). The 16S positivity rate was similar between fluid and tissue samples (21.5% vs. 22%). Notably, pus samples had a substantially higher positivity rate of 66.3%, with an approximately fivefold increased likelihood of positivity compared to non-pus samples (25%) (p < 0.001, OR = 5.909, 95% CI: 3.416–10.223) (Table 3). Conversely, cerebrospinal fluid (CSF) specimens showed the lowest positivity rate by 16S testing (24/414 (5.8%), p = 0.001), despite being the most frequently submitted sample type (Table 4).
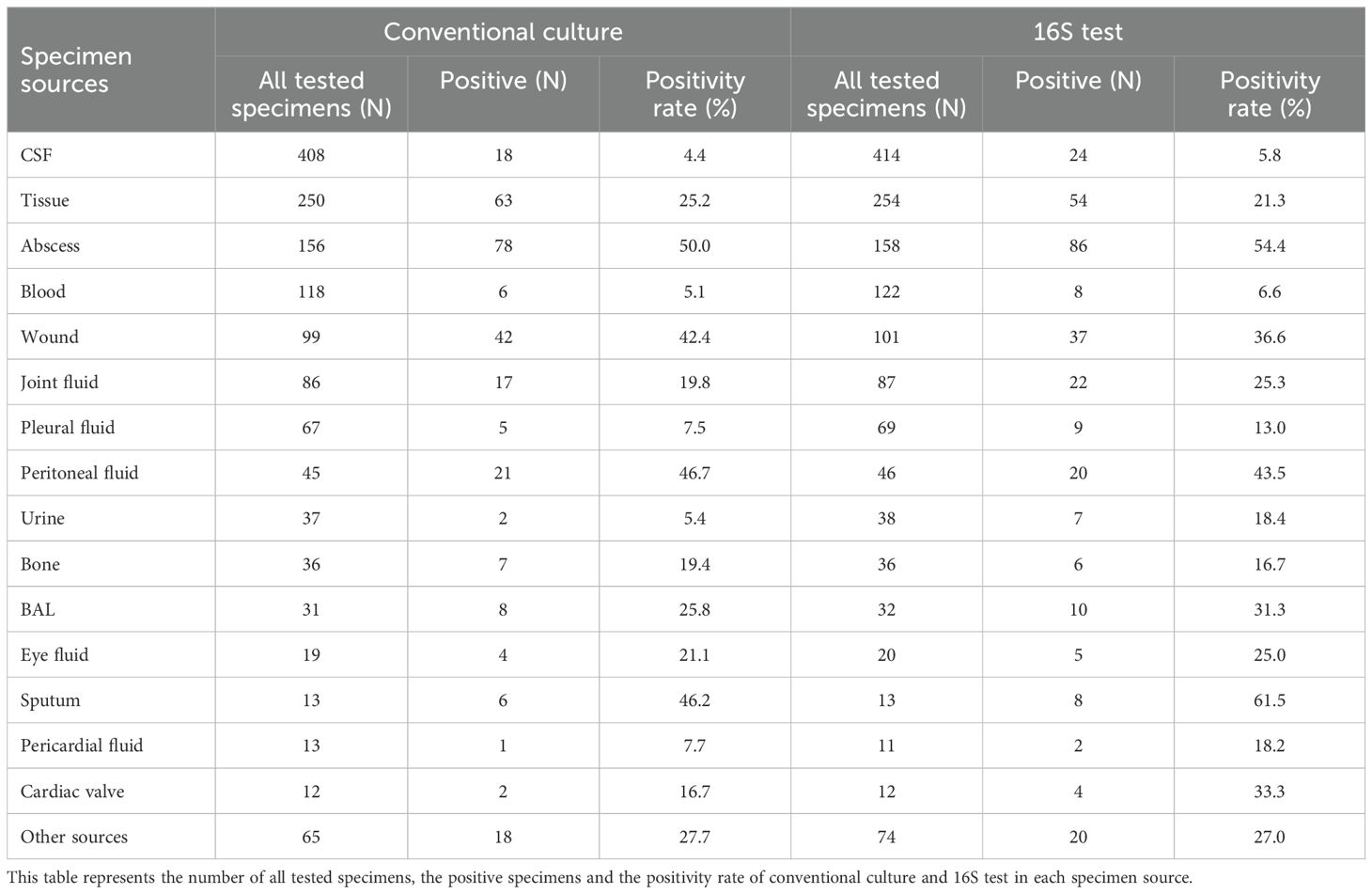
Table 4. Positivity rate of the conventional culture and 16S test among the different specimen sources.
A total of 260 organisms were identified through 16S sequencing (Supplementary Figure S2). Among the 77 organisms identified in culture-negative samples, 65% (n = 50) were organisms that are typically culturable under standard conditions, while 35% (n = 27) required special media (Figure 4). The commonly culturable organisms included Streptococcus pneumoniae (n = 9), Streptococcus pyogenes (n = 7), Pseudomonas aeruginosa (n = 5), Escherichia coli (n = 5), Staphylococcus aureus (n = 4), and Streptococcus agalactiae (n = 2). The group requiring special media included Fusobacterium spp. (n = 4), Bacteroides fragilis (n = 4), Prevotella spp. (n = 4), Mycobacterium spp. (n = 4), and other less common species such as Turicella otitidis (n = 1) and Vibrio cholerae (n = 2) (Table 5). Notably, 88.9% of the S. pneumoniae cases detected solely by 16S testing had prior antibiotic exposure.
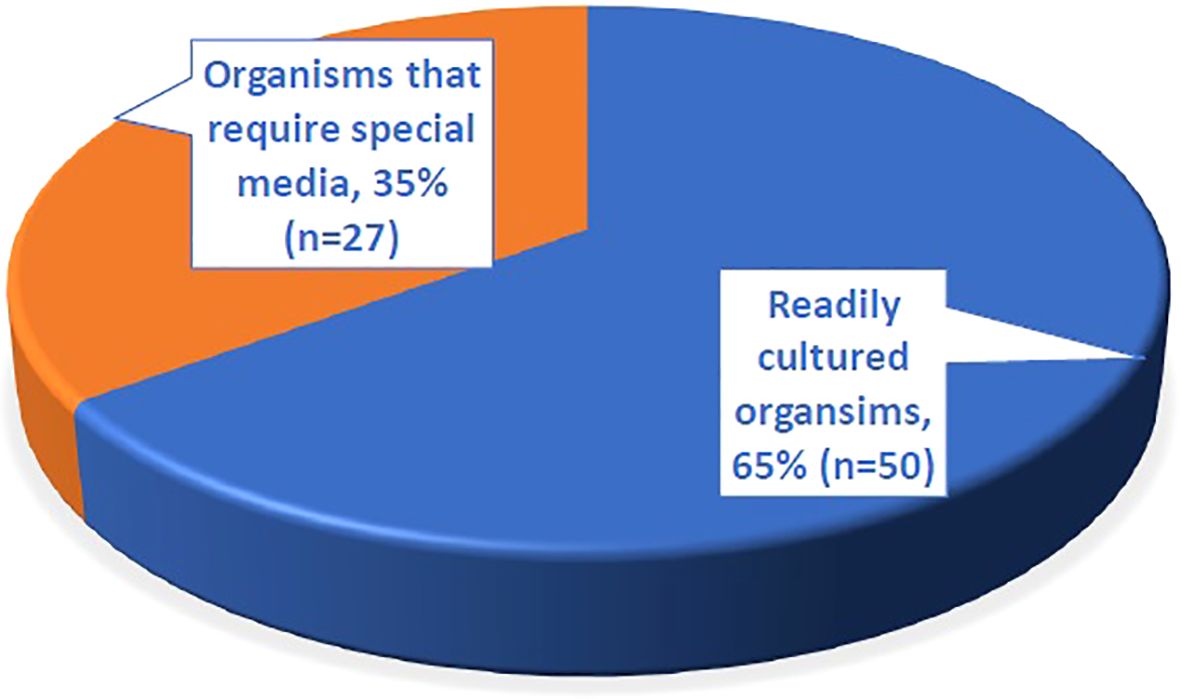
Figure 4. The proportion of different types of organisms identified by 16S testing from 77 culture negative samples.
Of the total discordant results, 64.7% (n = 156) of patients had received antibiotics within two weeks prior to sample collection. Among the culture-negative/16S-positive group, 67.4% had prior antimicrobial exposure, although this finding was not statistically significant (Table 1).
The clinical impact of 16S testing was observed in 45.9% of cases (83/181), resulting in changes in patient management. Among these, antibiotic escalation was implemented in 31.3% (26/83), antibiotic de-escalation in 41% (34/83), and a change in diagnosis in 26.5% (22/83). Additionally, a negative 16S result influenced the decision to withhold or discontinue antibiotic treatment in 24/109 (22%) cases. In the remaining 54.1% (98/181) of cases, no change in management was observed. This was attributed to confirmation of current treatment (4%), severe or critical patient condition necessitating continuation of therapy (5.1%), absence of additional pathogen detection (5.1%), pathogens already covered by current antibiotics (25.3%), detection of likely contaminants (4%), or presence of mixed, unidentified bacteria without specific therapeutic implications (56.6%) (Table 1).
In cases where 16S testing was positive and no culture was requested, the clinical impact varied by sample type. Changes in management were more frequent when specimens originated from cardiac valves or urine (66.7% each), although this was not statistically significant (Table 6). Importantly, among culture-negative/16S-positive cases, a significant change in management was observed in 60.3% (p = 0.001, OR = 1.552–5.268) (Figure 5).
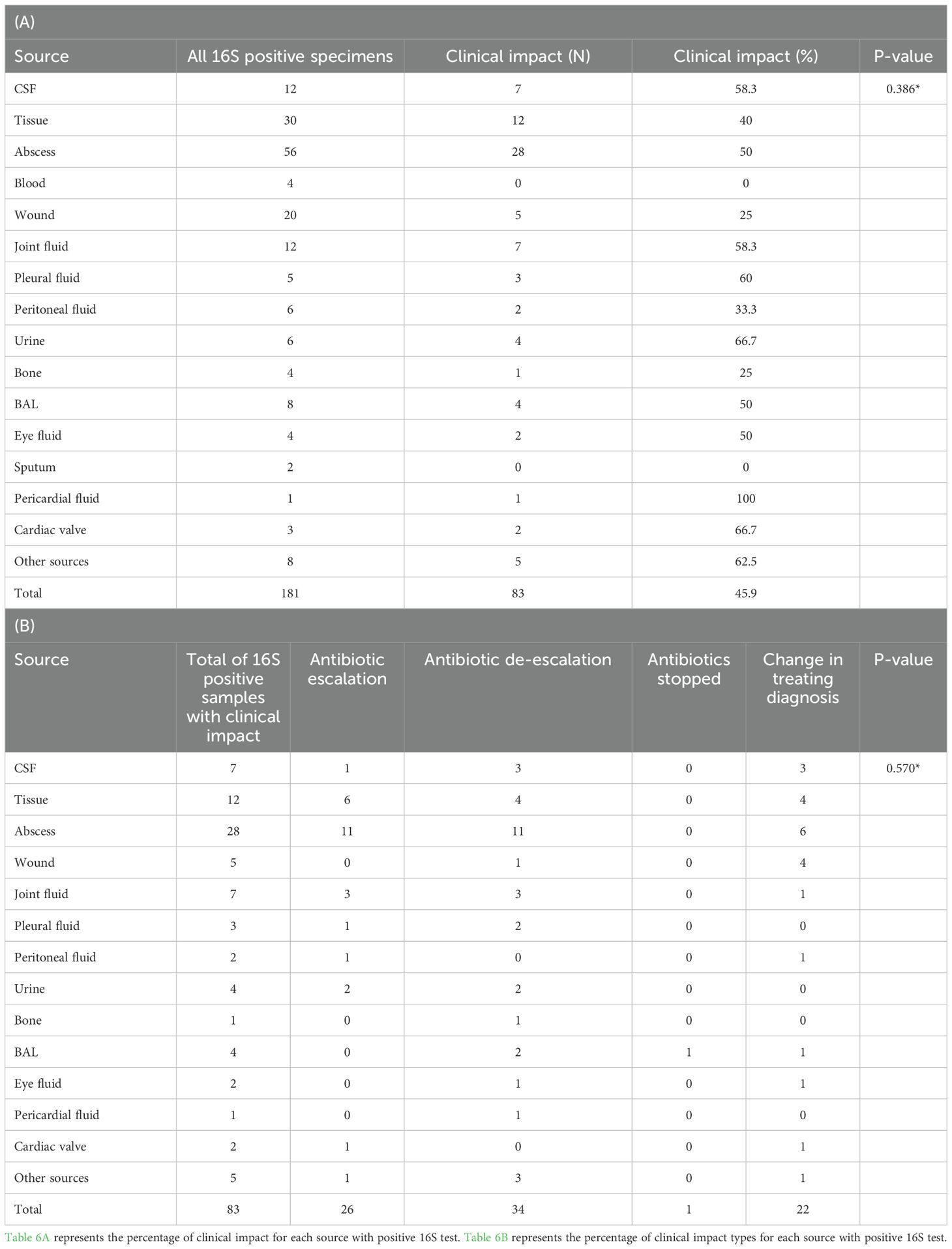
Table 6. (A) Clinical impact and (B) change in management classes among the different sources of the 16S test positive specimens.
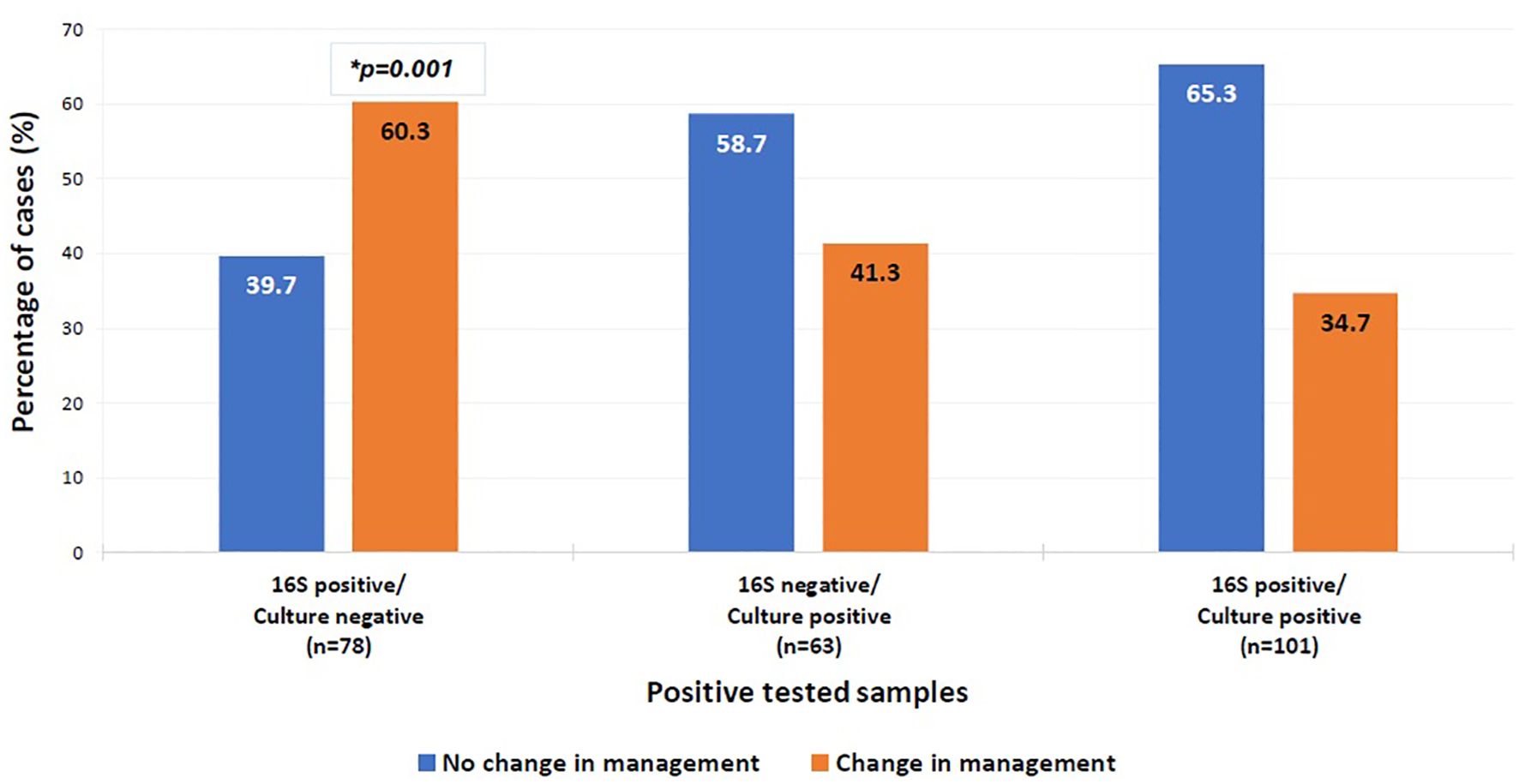
Figure 5. Clinical impact on management of positive tested samples. Multinomial logistic regression analysis was used to compare the clinical impact on management of the different categories of the positive tested samples. The category “16S positive/culture positive” was the reference category. *P-value=0.001, OR=2.859, 95%CI= [1.552-5.268]. The x-axis represents the positive tested samples. The y-axis represents the percentage of positive cases with a change in management (orange bar) versus the percentage of cases without a change in management (blue bar).
Examples of clinically useful 16S test results
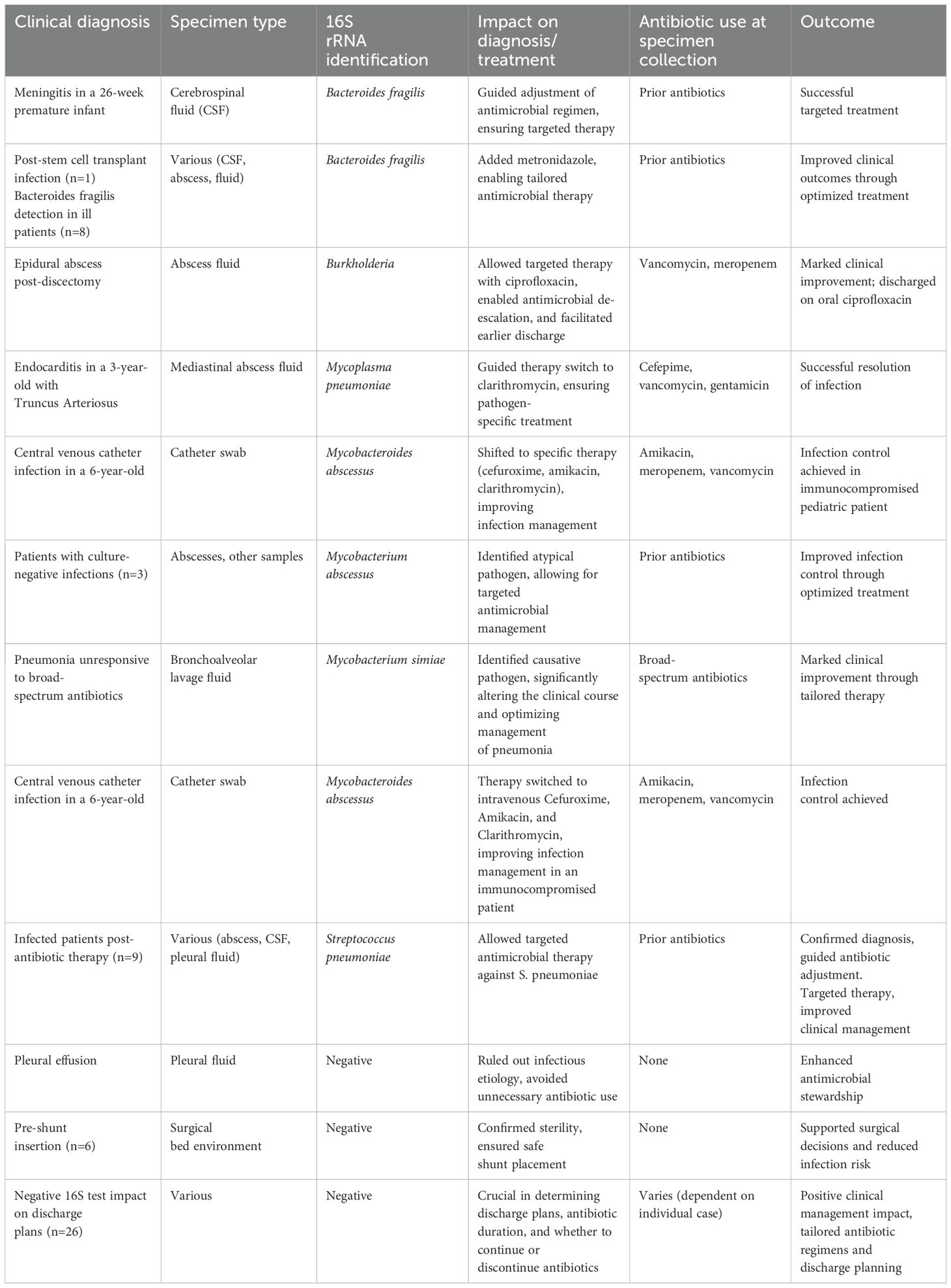
In the following section, several examples of clinically useful 16S test results impacting management will be presented.
In 12 patients with negative cultures likely due to prior antimicrobial use, the 16S test identified Streptococcus pneumoniae from various samples, such as abscesses, CSF, and pleural fluid. This allowed targeted antimicrobial therapy against S. pneumoniae. In several ill patients, Bacteroides fragilis was only detected with the 16S test leading to the addition of metronidazole to the antimicrobial regimen. On the other hand, negative 16S test results confirmed surgical bed environment in at least 6 patients prior to ventriculo-peritoneal shunt insertion. Negative 16S test results enhanced antimicrobial stewardship in cases of suspected non-infectious pleural effusions where the use of antibiotics was avoided. In the majority of cases, negative 16S test results were pivotal in determining discharge plans and the continuing need for antibiotics and their duration, positively impacting clinical management. Some additional illustrative cases follow:
1. A 57-year-old male patient with lumbar discitis was started on broad-spectrum antibiotics with repeatedly negative cultures, the 16S test detected Vibrio cholerae from specimens collected from disc tissue and disc fluid, leading to adjustments in the duration and management of the disease.
2. A 3-year-old patient with Truncus Arteriosus post total repair was admitted for documented endocarditis and initially treated with cefepime, vancomycin, and gentamicin. The 16S test identified Mycoplasma pneumoniae in mediastinal abscess fluid where blood and mediastinal fluid cultures were negative. The patient was switched to intravenous clarithromycin.
3. A 26-week premature infant was admitted to the NICU with meningitis. Despite negative CSF, blood, and urine cultures, 16S testing identified Bacteroides fragilis in the CSF.
4. A 36-year-old male, one month post L4-L5 discectomy, presenting with a four-day history of lower extremity weakness and pain. MRI revealed a right-sided epidural postsurgical collection. The patient was initially started on intravenous vancomycin and meropenem. However, 16S testing identified Burkholderia as the causative organism. This result led to a switch in therapy to intravenous ciprofloxacin, and after two weeks of marked clinical and laboratory improvement, the patient was discharged on oral ciprofloxacin.
5. Identification of Non-Tuberculosis Mycobacterial infections
We encountered a patient with a well-defined severe skin infection that was resistant to several antimicrobial regimens with repeated cultures showing no bacterial growth. 16S test identified Mycobacterium abscessus guiding the treating physician in both choice and duration of specific antimicrobial therapy. Similarly, three additional patients had Mycobacterium abscessus identified with the 16S test while all cultures were negative. Another patient transferred to our center for investigation and management of a pneumonia that was unresponsive to multiple broad-spectrum antibiotics with negative cultures, including bronchioalveolar lavage, the 16S test identified Mycobacterium simiae as the cause of pneumonia. Similarly, a 6-year-old boy with high-risk neuroblastoma, post-autologous stem cell transplant, was admitted with a suspected central venous catheter (CVC) infection. Workup showed negative blood and wound cultures, the patient was empirically started on amikacin, meropenem, and vancomycin. However, CVC swab sent for 16S testing identified Mycobacterium abscessus as the causative pathogen. This finding prompted a switch in therapy to intravenous cefuroxime, amikacin, and clarithromycin.
The above cases demonstrate the crucial role of 16S in enabling targeted therapy, antimicrobial de-escalation, and contribution to antimicrobial stewardship. The above illustrative cases are summarized in Table 7.
Discussion
Managing patients with suspected bacterial infections can be particularly challenging in the absence of microbial identification using conventional tests. This study aimed to review the utility of combining the 16S test with traditional microbiological methods for diagnosing infections and guiding targeted antimicrobial treatment. We observed a high concordance between the 16S test and culture results. Among the 16S test-identified organisms, there were almost twice the proportion of readily cultured organisms in comparison to those that need special media. The majority of patients who had received antibiotics prior to collection of specimens had readily cultured organisms detected (71.8%) whereas their cultures were negative. This highlights the utility of the 16S test in complementing the traditional microbiology cultures in the identification of organisms in both groups. This is particularly important for “precious” specimens where sampling is obtained by invasive procedures such as biopsy, or surgery (e.g. heart valve vegetation). The impact of the 16S test is demonstrated in microbial identification in 60.3% of culture-negative samples from patients with high clinical suspicion of infection. This result is superior to a previous observation reported by Rampini et.al, where culture negative/16S positive identified pathogens formed 42.9% of the total specimens (Rampini et al., 2011).
In Lebanon, antibiotics are available over the counter without a prescription resulting in haphazard self-treatment by patients prior to evaluation by a health-care provider. This practice results in frequent negative bacterial cultures for patients who eventually present to the hospital (Reslan et al., 2022). The 16S test is particularly useful in this group of patients as positive results allow for an accurate diagnosis and tailored management. Our findings echo other studies conducted on the importance of 16S test utilization where successful de-escalation of therapy was evident after the introduction of 16S test in clinical diagnostics (O’Donnell et al., 2018). Ursenbach et al. also reported a 32% impact on antimicrobial regimen change in those with positive PCR results (Ursenbach et al., 2021a).
Overall, the 16S test was more likely to be positive with GI samples [peritoneum (61.8%), liver (27%), spleen (5.6%), abdominal wall (4.5%), and bile tree (1.1%)] in comparison to other specimen sources. This contrasts with what was previously reported in the literature with cardiovascular specimens being the samples with the most positive 16S test results (Fida et al., 2021) probably reflecting differing patient populations in these studies. Our data indicates that pus samples had the highest positivity rate for 16S testing at 66.3% (p < 0.0001), whereas CSF samples had the lowest detection rate at 5.8% (p = 0.003). Additionally, non-surgical samples had almost twice the positivity detection rate compared to surgical samples (66.3% vs. 27.6%).
This finding aligns with the literature, which consistently shows that 16S testing has a higher diagnostic yield in certain sample types. Harris and Brown reported that abscess, pus, and empyema samples had the highest diagnostic yield, with 44.9% positivity by 16S testing (Harris and Brown, 2022).
Aggarwal et al. found that pus samples were most frequently positive (34.5%; p<0.0001), while CSF samples had the lowest positivity rate (5.4%; p=0.003) (Aggarwal et al., 2020).
The low detection rate in CSF samples, despite being the most commonly referred sample type, underscores the challenges in diagnosing central nervous system infections. This is further validated by Ursenbach et al., who noted that 16S results on CSF had a significant clinical impact, particularly in patients with previous antibiotic treatment, where traditional cultures often fail (Ursenbach et al., 2021b).
The findings from our study align with and expand upon the existing literature on the impact of the 16S test on clinical management, with both similarities and notable differences in key areas such as changes in antimicrobial therapy, escalation, de-escalation, and the proportion of cases showing no impact on outcome. Our analysis showed that 45.9% of cases were impacted with a change in management as a result of 16S testing, which is notably higher compared to the 2.2% of cases impacted in Teoh et al.’s study (Teoh et al., 2021). This suggests that, in our cohort, 16S testing may have played a more significant role in guiding treatment decisions, potentially due to differences in patient demographics, infection types, or clinical practices between studies. However, it is noteworthy to mention that our analysis was solely done on discordant results which would affect the findings in comparison to other studies. Despite being limited to 45.9% of cases, the impact of 16S testing on clinical management remains substantial. In clinical practice, a diagnostic tool that leads to a change in management for nearly half of patients is valuable. While 16S testing did not significantly influence cases concordant with culture results, it still contributed to improved clinical outcomes by enabling faster pathogen identification and more targeted treatments. This may reduce reliance on broad-spectrum antibiotics, lowering the risk of antimicrobial resistance while enhancing infection treatment with more appropriate antimicrobial agents. The high rate of management change observed in our findings aligns with Ursenbach et al., who reported a substantial effect, with 50% of cases showing a change in antimicrobial regimen (Ursenbach et al., 2021a).
When exploring management changes resulting from 16S test results in our study, escalation of antibiotics occurred in 31.3% of cases where a change in management occurred, closely matching the 39% escalation rate in Ursenbach et al.’s findings (Ursenbach et al., 2021b). This consistency suggests that in specific cases, 16S testing reliably supports the decision to intensify therapy based on identified pathogens, underscoring its role in addressing previously unidentified or unexpected infections. On the other hand, de-escalation of antibiotics was observed at a higher rate than the 15.6% in Akram et al. and the 21% noted by O’Donnell et al. (Akram et al., 2017). This discrepancy may reflect differences in clinical thresholds for reducing therapy intensity or the types of pathogens commonly encountered in our study compared to others. The high de-escalation rate in the current study indicates that 16S testing provided sufficient information to support more targeted therapy and reduce unnecessary antibiotic exposure, aligning with antimicrobial stewardship goals.
Our study also captured the impact of 16S testing in situations where antibiotics were discontinued or where a change in the diagnosis prompted treatment adjustments. Ursenbach et al. similarly reported significant rationalization of antibiotic treatment in 50% of cases, suggesting that the 16S test consistently enables tailored adjustments in diverse clinical settings (Ursenbach et al., 2021b).
Lastly, our study revealed that 56% of cases had no change in management, compared to the majority of cases in Teoh et al.’s findings (Teoh et al., 2021). This outcome indicates that while the 16S test frequently supports antimicrobial decisions, a substantial proportion of cases still benefit from confirmation of adequate management without requiring alterations. In our cohort, factors such as confirmation of culture results, severe clinical status, or contaminants identified by 16S testing contributed to this lack of change, reinforcing its utility as a complementary diagnostic tool.
In this study, the 16S sequencing success rate in PCR-positive cases was 79.5%. In the remaining cases (20.5%), species-level identification may have been hindered by the presence of mixed bacterial populations. The latter may contribute to the generation of ambiguous or unreadable sequences. Additionally, some bacterial species or strains might not have closely related sequences in reference databases such as NCBI BLAST. Furthermore, lower DNA purity in some unidentified samples may have generated poor quality. Similar to our findings, a study by Fida et al. reported that 22% of samples with positive 16S PCR were not identified by Sanger sequencing (Fida et al., 2021). Another study demonstrated that 2.8% of samples with a positive 16s amplification product indicated suspected mixed infections (Eamsakulrat et al., 2022). The authors attributed the failure of species identification in additional 12.3% of specimens to unsuccessful Sanger sequencing.
Furthermore, negative 16S test results led to no initiation, escalation, or discontinuation of antibiotics in a quarter of the cases reported in the current study. This finding aligns with the literature, which supports the utility of negative 16S results in guiding clinical decisions, particularly in antimicrobial stewardship. Gilbert et al. demonstrated that negative 16S results were associated with shorter lengths of antibiotic therapy and higher rates of antibiotic discontinuation in culture-negative patients (Gilbert et al., 2017). Similarly, O’Donnell et al. found that negative 16S results contributed to the discontinuation of antimicrobials in 8% of cases, highlighting its role in preventing unnecessary antibiotic use (O’Donnell et al., 2018). These studies underscore the importance of negative 16S results in clinical decision-making, particularly in avoiding unnecessary antimicrobial therapy, which is crucial for antimicrobial stewardship (Gilbert et al., 2017; O’Donnell et al., 2018).
The samples that were negative by both 16S and culture were not included in the discordant analysis but were part of the total cohort to provide a comprehensive overview of test performance. The double negatives are important in measuring the specificity of the 16S test. This group includes patients with no bacterial infection, cases with prior effective antibiotic treatment, and non-bacterial etiologies such as viral, autoimmune, or sterile inflammatory conditions. Importantly, in some cases, these double-negative results served a crucial confirmatory role, for example, confirming the sterility of the surgical field before ventriculoperitoneal shunt placement or other invasive procedures. Such findings provided reassurance to clinicians and directly influenced management decisions by supporting the safety of proceeding without antimicrobial coverage or delaying interventions. However, careful clinical correlation would be essential in interpreting such negative microbiological findings, as molecular diagnostics alone cannot exclude infection in the appropriate context.
Limitations
One of the limitations of this study is its retrospective nature, where some of the cases had missing data due to incomplete notes. Importantly, the decision-making process is not always documented clearly in medical records, possibly leading to under or overestimation of the impact of 16S test results on clinical management. Moreover, due to the heterogeneous sources of samples submitted, comparisons regarding sensitivity of the test and its impact on outcome according to sample source were not possible. Including cost-effectiveness as an essential factor associated with the routine use of 16S testing is warranted in future studies. Additionally, sequencing analysis could have been improved to enhance species identification in polymicrobial infections. A key limitation of our approach was the inability to resolve polymicrobial infections using Sanger sequencing. In cases where multiple bacterial templates were present, the resulting chromatograms frequently produced overlapping signals, rendering species-level identification inconclusive. These findings were reported as mixed bacterial populations and, in the absence of definitive identification, did not influence clinical decision-making. The integration of advanced sequencing technologies such as next-generation sequencing (NGS) or Whole Genome Sequencing (WGS), along with specialized bioinformatics tools, may facilitate more accurate characterization of polymicrobial samples and enhance the clinical applicability of 16S testing in future investigations.
While this case of lumbar discitis associated with Vibrio cholerae is unusual, and the patient’s clinical response supported its role as the causative pathogen, we cannot exclude the possibility of misidentification or sample contamination as an alternative explanation, highlighting the need for careful interpretation of molecular findings in rare presentations.
Conclusion
This study represents the largest scale study conducted in low- and middle-income countries (LMIC), summarizing findings from 16S rRNA-targeted sequencing conducted on samples suspected of clinical infection. This is especially important because many LMIC have high rates of multi-drug-resistant bacterial infections and lack appropriate antimicrobial stewardship (Khafaja et al., 2024; Zakhour et al., 2025). Identification of pathogens using the 16S test in combination with conventional cultures is essential in clinical diagnostics and management of infectious diseases, particularly those already on antibiotics at the time of sampling to provide targeted therapy and improve antimicrobial stewardship. Shorter turnaround time, improved patient management, and cost-effectiveness are key factors to consider when advocating for the broader adoption of 16S testing. This study provides support for the wider implementation of this test. Further studies are needed on a larger scale to confirm our findings and cover unaddressed factors.
Data availability statement
The datasets presented in this article are not readily available due to privacy and ethical restrictions. Requests to access the datasets should be directed to the corresponding author(s).
Ethics statement
The studies involving humans were approved by Institutional Review Board at the American University of Beirut Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
NY: Writing – review & editing, Writing – original draft. CB: Writing – review & editing, Formal analysis. FD: Writing – review & editing, Data curation. FA: Data curation, Writing – review & editing. LR: Methodology, Writing – review & editing, Validation. MF: Writing – review & editing, Data curation. MM: Data curation, Writing – review & editing. TD: Data curation, Writing – review & editing. ZZ: Writing – review & editing, Data curation. AH: Writing – review & editing, Data curation. RK: Writing – review & editing, Data curation. SK: Writing – review & editing, Data curation. GD: Methodology, Conceptualization, Visualization, Supervision, Validation, Writing – review & editing. GA: Data curation, Resources, Writing – review & editing. GM: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1619640/full#supplementary-material
Abbreviations
16S test, 16S rRNA gene; CSF, Cerebral Spinal Fluid; AUBMC, American University of Beirut Medical Center; CIDR, Center for Infectious Diseases Research; SPSS, Statistical Package for the Social Sciences; UOR, Unadjusted Odds Ratio; CI, Confidence Interval; IRB, Institutional Review Board; GI, Gastrointestinal; SST, Skin and Soft Tissue; LMIC, Low- and Middle-Income Countries.
References
Aggarwal, D., Kanitkar, T., Narouz, M., Azadian, B. S., Moore, L. S. P., and Mughal, N. (2020). Clinical utility and cost-effectiveness of bacterial 16S rRNA and targeted PCR based diagnostic testing in a UK microbiology laboratory network. Sci. Rep. 10, 7965. doi: 10.1038/s41598-020-64739-1
Akram, A., Maley, M., Gosbell, I., Nguyen, T., and Chavada, R. (2017). Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int. J. Infect. Diseases. 57, 144–149. doi: 10.1016/j.ijid.2017.02.006
Cook, V. J., Turenne, C. Y., Wolfe, J., Pauls, R., and Kabani, A. (2003). Conventional methods versus 16S ribosomal DNA sequencing for identification of nontuberculous mycobacteria: cost analysis. J. Clin. Microbiol. 41, 1010–1015. doi: 10.1128/JCM.41.3.1010-1015.2003
Eamsakulrat, P., Santanirand, P., and Phuphuakrat, A. (2022). Diagnostic yield and impact on antimicrobial management of 16S rRNA testing of clinical specimens. Microbiol. Spectrum. 10, e02094–e02022. doi: 10.1128/spectrum.02094-22
Fida, M., Khalil, S., Abu Saleh, O., Challener, D. W., Sohail, M. R., Yang, J. N., et al. (2021). Diagnostic value of 16S ribosomal RNA gene polymerase chain reaction/sanger sequencing in clinical practice. Clin. Infect. Dis. 73, 961–968. doi: 10.1093/cid/ciab167
Gilbert, E. M., Yucebay, F., Malczynski, M., Smith, D., Esterly, J. S., Qi, C., et al. (2017). Use of organism identification by 16S ribosomal RNA polymerase chain reaction to shorten antimicrobial length of therapy. Diagn. Microbiol. Infect. Dis. 88, 163–167. doi: 10.1016/j.diagmicrobio.2017.03.013
Harris, K. A. and Brown, J. R. (2022). Diagnostic yield of broad-range 16s rRNA gene PCR varies by sample type and is improved by the addition of qPCR panels targeting the most common causative organisms. J. Med. Microbiol. 71, 11. doi: 10.1099/jmm.0.001633
Hassan, R. M., El Enany, M. G., and Rizk, H. H. (2014). Evaluation of broad-range 16S rRNA PCR for the diagnosis of bloodstream infections: two years of experience. J. Infect. Dev. Ctries. 8, 1252–1258. doi: 10.3855/jidc.4867
Khafaja, S., Salameh, Y., Boutros, C. F., Awad, C., Faour, K., Tfaily, N., et al. (2024). Increased rate of multidrug-resistant gram-negative bacterial infections in hospitalized immunocompromised pediatric patients. Front. Cell. infection Microbiol. 14, 1382500. doi: 10.3389/fcimb.2024.1382500
Messacar, K., Parker, S. K., Todd, J. K., and Dominguez, S. R. (2017). Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J. Clin. Microbiol. 55, 715–723. doi: 10.1128/JCM.02264-16
Mignard, S. and Flandrois, J. P. (2006). 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J. Microbiol. Methods 67, 574–581. doi: 10.1016/j.mimet.2006.05.009
Miller, J. M., Binnicker, M. J., Campbell, S., Carroll, K. C., Chapin, K. C., Gilligan, P. H., et al. (2018). A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the infectious diseases society of america and the american society for microbiology. Clin. Infect. Dis. 67, e1–e94. doi: 10.1093/cid/ciy381
Mishra, D., Satpathy, G., Chawla, R., Venkatesh, P., Ahmed, N. H., and Panda, S. K. (2019). Utility of broad-range 16S rRNA PCR assay versus conventional methods for laboratory diagnosis of bacterial endophthalmitis in a tertiary care hospital. Br. J. Ophthalmol. 103, 152–156. doi: 10.1136/bjophthalmol-2018-312877
Mishra, D., Satpathy, G., Wig, N., Fazal, F., Ahmed, N. H., and Panda, S. K. (2020). Evaluation of 16S rRNA broad range PCR assay for microbial detection in serum specimens in sepsis patients. J. Infect. Public Health 13, 998–1002. doi: 10.1016/j.jiph.2020.01.007
O’Donnell, S., Gaughan, L., Skally, M., Baker, Z., O’Connell, K., Smyth, E., et al. (2018). The potential contribution of 16S ribosomal RNA polymerase chain reaction to antimicrobial stewardship in culture-negative infection. J. Hosp Infect. 99, 148–152. doi: 10.1016/j.jhin.2017.08.016
Rampini, S. K., Bloemberg, G. V., Keller, P. M., Büchler, A. C., Dollenmaier, G., Speck, R. F., et al. (2011). Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin. Infect. Dis. 53, 1245–1251. doi: 10.1093/cid/cir692
Reslan, L., Youssef, N., Boutros, C. F., Assaf-Casals, A., Fayad, D., Khafaja, S., et al. (2022). The impact of vaccination on the burden of invasive pneumococcal disease from a nationwide surveillance program in Lebanon: an unexpected increase in mortality driven by non-vaccine serotypes. Expert Rev. Vaccines 21, 1905–1921. doi: 10.1080/14760584.2022.2143349
Teoh, T., McNamara, R., Powell, J., O’Connell, N. H., and Dunne, C. P. (2021). A retrospective observational study of the impact of 16s and 18s ribosomal RNA PCR on antimicrobial treatment over seven years: A tertiary hospital experience. PloS One 16, e0258552. doi: 10.1371/journal.pone.0258552
Ursenbach, A., Schramm, F., Séverac, F., Hansmann, Y., Lefebvre, N., Ruch, Y., et al. (2021a). Impact of 16S rDNA sequencing on clinical treatment decisions: a single center retrospective study. BMC Infect. Diseases. 21, 190. doi: 10.1186/s12879-021-05892-4
Ursenbach, A., Schramm, F., Séverac, F., Hansmann, Y., Lefebvre, N., Ruch, Y., et al. (2021b). Revised version (INFD-D-20-00242): impact of 16S rDNA sequencing on clinical treatment decisions: a single center retrospective study. BMC Infect. Dis. 21, 190. doi: 10.1186/s12879-021-05892-4
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. letters. 309, 1–7. doi: 10.1111/j.1574-6968.2010.02000.x
Zakhour, R., Khafaja, S., Korman, R., Boutros, C. F., El Zein, Z., Chmaisse, A., et al. (2025). Rates of multidrug-resistant gram-negative bacterial infections in hospitalized non-immunocompromised pediatric patients: A 9-year retrospective study at a lebanese tertiary medical center. Infection Drug resistance. 18, 363–376. doi: 10.2147/IDR.S488436
Keywords: 16S rRNA, antimicrobial management, clinical specimens, diagnostic yield, clinical impact, bacterial infection, conventional culture, Lebanon
Citation: Youssef N, Boutros CF, Dakroub F, Akl F, Reslan L, Finianos M, Moumneh MBM, Dargham TB, Zein ZE, Haddara A, Korman R, Khafaja S, Matar G, Araj GF and Dbaibo GS (2025) The clinical impact of 16S ribosomal RNA PCR and sequencing in the identification of bacterial infections: a 7-year report from a Lebanese tertiary care center. Front. Cell. Infect. Microbiol. 15:1619640. doi: 10.3389/fcimb.2025.1619640
Received: 28 April 2025; Accepted: 15 July 2025;
Published: 07 August 2025.
Edited by:
Márió Gajdács, University of Szeged, HungaryReviewed by:
Ariadnna Cruz-Córdova, Federico Gómez Children’s Hospital, MexicoSunil Banskar, University of Arizona, United States
Copyright © 2025 Youssef, Boutros, Dakroub, Akl, Reslan, Finianos, Moumneh, Dargham, Zein, Haddara, Korman, Khafaja, Matar, Araj and Dbaibo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ghassan S. Dbaibo, Z2RiYWlib0BhdWIuZWR1Lmxi
 Nour Youssef
Nour Youssef Celina F. Boutros
Celina F. Boutros Fatima Dakroub
Fatima Dakroub Fata Akl
Fata Akl Lina Reslan
Lina Reslan Marc Finianos
Marc Finianos Mohammad Bahij M. Moumneh
Mohammad Bahij M. Moumneh Tarek Bou Dargham
Tarek Bou Dargham Zeinab El Zein1,2,3
Zeinab El Zein1,2,3 Sarah Khafaja
Sarah Khafaja Ghassan Matar
Ghassan Matar George F. Araj
George F. Araj Ghassan S. Dbaibo
Ghassan S. Dbaibo