- 1Centre for Immunology and Infection Control, School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, Brisbane, QLD, Australia
- 2The Ear, Nose and Throat (ENT) Research Group, Centre for Clinical Research, Faculty of Health, Medicine and Behavioral Sciences, The University of Queensland, Brisbane, QLD, Australia
Introduction: Respiratory syncytial virus (RSV) infection in the upper respiratory tract promotes disease progression and transmission, with excessive inflammation contributing to severe lower respiratory tract involvement. This study investigates the immunomodulatory effects of Lactobacillus rhamnosus D3189 on viral kinetics and innate immune responses in well-differentiated nasal epithelial cells (WD-NECs).
Methods: WD-NECs from healthy adult donors (N = 8) were cultured in vitro, treated with L. rhamnosus D3189, and then infected with RSV (strain RS4) 24 hours later. Viral replication and shedding were assessed via RT-qPCR and plaque assays. Cytotoxicity and epithelial integrity were evaluated using LDH release and transepithelial electrical resistance (TEER). Inflammatory and antiviral responses were investigated using multiplex immunoassays, AlphaLISA, and ELISA.
Results: RSV infection induced robust viral replication and shedding, disrupted epithelial barrier integrity, and triggered the release of pro-inflammatory cytokines and type I/III interferons. L. rhamnosus D3189 alone did not induce cytotoxicity or inflammation. While it had no effect on viral replication, TEER, LDH release, or IFN-λ1/3 levels, D3189 significantly enhanced IFN-β production, reduced viral shedding, and attenuated RSV-induced cytokine and chemokine responses.
Discussion: L. rhamnosus D3189 modulates the epithelial immune response to RSV, reducing inflammation and viral shedding without compromising epithelial integrity. These findings support its potential as a novel strategy to limit RSV-associated infection and transmission.
1 Introduction
Human respiratory syncytial virus (RSV), also known as Orthopneumovirus hominis, is an enveloped, negative-sense RNA virus of the Pneumoviridae family that primarily infects and replicates in the airway epithelium (Collins et al., 2013; Duan et al., 2024). RSV initiates infection in the upper respiratory tract (URT) and can progress to the lower respiratory tract (LRT), causing severe diseases such as bronchiolitis and pneumonia, particularly in vulnerable people (Simões et al., 2010; McLaughlin et al., 2022). While RSV infection is often mild, one-third of cases develop severe LRT disease, posing a significant threat to infants under six months, the elderly, and immunocompromised individuals (Piedimonte and Perez, 2014; McLaughlin et al., 2022).
Since the COVID-19 pandemic, RSV epidemiology has shifted, with rising case numbers across all age groups, including older children and adults who were previously less affected (Chuang et al., 2023). While RSV infection in healthy adults is typically mild, they can act as asymptomatic or mildly symptomatic carriers, contributing to virus transmission (Kaler et al., 2023). Given that RSV reinfection occurs throughout life, controlling its spread, particularly amongst high-risk populations, remains a critical public health challenge, yet effective transmission-reduction strategies are underexplored. RSV-induced inflammation plays a key role in disease progression. Viral replication in the URT can trigger an excessive immune response in susceptible individuals, leading to increased production of proinflammatory cytokines and chemokines such as IL-1β, IL-2, IL-6, IL-8, and TNF-α (Garofalo et al., 1996; Jamaluddin et al., 1998; Casola et al., 2001; Garofalo et al., 2013). This inflammation can compromise airway epithelial integrity, disrupt mucociliary clearance, and facilitate viral spread to the LRT (Newton et al., 2016). Consequently, strategies that target RSV at its initial site of infection in the URT, could help limit both transmission and disease severity.
Recent advances in RSV vaccines and therapeutics, such as monoclonal antibodies (Palivizumab, Nirsevimab) and protein subunit vaccines (Arexvy and Abrysvo), have focused on disease prevention in infants (<2 years) and older adults (>60 years), leaving a significant portion of the population unprotected (Gatt et al., 2023; Assad et al., 2024; Moreira et al., 2024; Banooni et al., 2025; Kelleher et al., 2025). While these interventions reduce severe LRT disease, they do not offer complete protection against initial URT infection (Gatt et al., 2023). This highlights the need for approaches that enhance local mucosal immunity in the URT, particularly intranasal strategies that could reduce viral shedding, lower transmission risk, and prevent LRT involvement. Strengthening URT innate immune defenses may also provide protection against other respiratory pathogens.
Recent studies, including our own, have shown that nasal administration of Lactobacillus species can modulate the innate immune response of airway epithelial cells (Gabryszewski et al., 2011; Tomosada et al., 2013; Percopo et al., 2015; Clua et al., 2017; Percopo et al., 2017). In murine models, priming the nasal mucosa with probiotic bacteria has been found to enhance resistance against viral infections, including RSV (Tomosada et al., 2013; Clua et al., 2017), influenza virus (Zelaya et al., 2015), and pneumonia virus of mice (Gabryszewski et al., 2011; Percopo et al., 2015; Percopo et al., 2017). Consistent with these findings, a prospective observational study reported a higher nasopharyngeal abundance of Lactobacillus spp. in healthy infants compared to those with RSV-associated acute respiratory infections (Rosas-Salazar et al., 2016). Additionally, an overrepresentation of Lactobacillus spp. in the respiratory tract of healthy infants has been linked to a reduced risk of developing RSV-associated childhood wheezing illnesses (Rosas-Salazar et al., 2018). Among the Lactobacillus species with immunomodulatory properties, Lactobacillus rhamnosus strains show promise in improving outcomes of respiratory viral infections (Kumova et al., 2019; Yarlagadda et al., 2024). In our work, we identified a novel strain, L. rhamnosus D3189, which significantly reduced the release of rhinovirus (RV)-induced inflammatory markers in an in vitro nasal epithelium model (Yarlagadda et al., 2024), highlighting its potential to mitigate virus-induced upper respiratory tract disease by modulating inflammation.
These findings suggest that lactobacilli, particularly L. rhamnosus strains, may beneficially modulate the respiratory innate immune response against RSV. Building on this and previous studies, we investigated whether L. rhamnosus D3189 influences viral kinetics and the production of cytokines and chemokines in nasal epithelial cells following RSV challenge.
2 Materials and methods
2.1 Human primary nasal epithelial cells
This study was conducted in accordance with the Queensland University of Technology Human Research Ethics Committee (2021000292). Nasal epithelial cells (NECs) were collected from the inferior turbinates of eight healthy adult donors (6 females, 2 males, 28 ± 8.8 years old) in May 2024 using a sterile nasal mucosal curette (Arlington Scientific, USA) with informed consent. NEC cultures were established and expanded as submerged monolayers as previous described (Kicic et al., 2010; Spann et al., 2014; Kicic et al., 2016). Cells were stored in aliquots in freezing media (fetal bovine serum with 10% dimethyl sulfoxide) at passage 1 or 2 in liquid nitrogen until use.
2.2 Air liquid interface culture
Primary NECs were seeded (5 × 105 cells/6.5 mm transwell, 0.4 μm pores; Corning Costar, USA) and cultured in PneumaCult™-Ex Plus Medium (Stemcell Technologies, Canada), as described previously (Yarlagadda et al., 2024). Upon confluence, cells were air-lifted and maintained in PneumaCult™-ALI Medium (Zhu et al., 2022; Yarlagadda et al., 2024). Cultures were differentiated for ≥3 weeks until ciliation, mucus production, and transepithelial electrical resistance (TEER) >250 Ω·cm², measured using an EVOM2 Epithelial Voltohmmeter (World Precision Instruments, USA), were observed. Medium was changed thrice weekly. Supplements were removed 24 h in advance of L. rhamnosus exposure (Yarlagadda et al., 2024).
2.3 Lactobacillus rhamnosus preparation
L. rhamnosus strain D3189, isolated from the URT of healthy children (Coleman et al., 2021), was selected for its in vitro inhibition of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis (Coleman et al., 2022) and its ability to differentially modulate RV-induced innate immune responses in primary NECs from healthy adults (Yarlagadda et al., 2024). D3189 was prepared as previously described (Yarlagadda et al., 2024), grown until the end of the lag phase, and harvested for WD-NEC exposure (Yarlagadda et al., 2024).
2.4 Virus propagation
A clinical isolate of RSV (RS4), generously provided by Professor Paul Young (University of Queensland), was used in this study. RSV was propagated in human epithelial type 2 (HEp-2) cells (ATCC® CCL-23™) and purified as described previously (Spann et al., 2014). Virus stocks were quantified using an immuno-plaque assay (Spann et al., 2014), which was also used to quantify viral shedding in apical washes from experimental NEC cultures.
Briefly, virus stocks were serially diluted 10-fold and used to infect confluent monolayers of HEp-2 cells in duplicate. Two hours post-infection, the cells were overlaid with 0.8% methyl cellulose in OptiMEM/2% fetal bovine serum/1% antibiotic-antimycotic (Thermo Fisher Scientific, USA), and incubated at 37°C with 5% CO2 for 7 days. Cells were then fixed with 60% methanol/40% acetone and blocked with 5% skim milk powder in PBS. RSV plaques were detected using a goat anti-RSV polyclonal antibody (1:500; Sigma-Aldrich, USA), followed by a horseradish peroxidase (HRP)-conjugated anti-goat secondary antibody (1:500; Invitrogen, USA). Both antibody incubations were carried out for 2 hours at 37°C with 5% CO2. Plaques were visualized using a peroxidase substrate with metal enhancer (SigmaFast DAB; Sigma-Aldrich) and counted to calculate viral titers as plaque-forming units (PFU) mL-1 (Yarlagadda et al., 2024).
2.5 Probiotic exposure, viral infection and sample analysis
Duplicate cultures of NECs were exposed to L. rhamnosus D3189 either alone or prior to RSV infection, as described previously (Yarlagadda et al., 2024). Replicate cultures were treated with RSV alone or cell culture media as mock-infection controls. For probiotic pre-treatment, NECs were apically exposed to 80 µL of cell culture media containing either D3189 (2.5 × 107 CFU mL-1) or media alone (virus-only control) for 4 h at 37°C with 5% CO2. Following this exposure, the inoculum was removed to re-establish the air-liquid interface, and the cells were incubated for an additional 20 h. After this incubation period, NECs were either apically treated with 80 µL of cell culture media alone (bacteria-only control) or infected with 80 µL of RSV (7.5 × 106 PFU mL-1) for 4 h at 37°C with 5% CO2. The viral inoculum was then removed, and cultures were maintained at the air-liquid interface for an additional 3 days. Duplicate cultures of NECs from 5 donors were exposed to L. rhamnosus GG (LGG) either alone or prior to RSV infection as above, as a comparison to D3189.
At 3 days post-infection, TEER was measured to assess epithelial barrier integrity. Apical washes (200 µL PBS) were used to quantify shed virus via immuno-plaque assays (Spann et al., 2014), while both apical wash and basal media were analyzed for cytokine and chemokine release using AlphaLISA, ELISA, or magnetic bead assays. Bacterial carriage in the apical wash was quantified using the drop plate technique to determine CFU mL-1. Finally, cells on transwell membranes were lysed in 150 μL of TRIzol® reagent (Thermo Fisher Scientific) for total RNA extraction and reverse transcription quantitative PCR (RT-qPCR).
2.6 Viral gene expression analysis by two-step RT-qPCR
Total RNA was extracted from cell lysates using TRIzol/chloroform phase separation method (Spann et al., 2004), followed by purification and concentration with the PureLink™ RNA Mini Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA quality and quantity were assessed using a Nanodrop Lite Spectrophotometer (Thermo Fisher Scientific). cDNA was synthesised from 200 ng of total RNA using the SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, USA) with Oligo(dT)20 primers, following the manufacturer’s protocol. RSV nucleocapsid (N) mRNA was quantified using the QuantiNova SYBR Green PCR Kit (Qiagen) and RSV N-specific primers (Forward 5’ – 3’: AAG GGA TTT TTG CAG GAT TGT TT, Reverse 5’ – 3’: CTC CCC ACC GTA GCA TTA CTT G). qPCR was performed on the QIAGEN Rotor-gene-Q according to the following cycling conditions: 5˚C for 5 mins, 45 cycles of 95˚C for 5 seconds and 60˚C for 10 seconds. Gene expression was normalized to β-actin mRNA (Forward 5’ – 3’: TAC GCC AAC ACA GTG CTG TCT, Reverse 5’ – 3’: TCT GCA TCC TGT CGG CAA T) (Yarlagadda et al., 2024).
2.7 LDH activity
As an index of cytotoxicity, lactate dehydrogenase (LDH) release was measured using basal media collected 3 days post infection (p.i.) with RSV, using the LDH-Glo Cytotoxicity Assay Kit (Promega) as per the manufacturer’s instructions. The luminescence of the samples was recorded using a CLARIOstar Omega reader. Enzyme activity was expressed as mU/mL.
2.8 Cytokine and chemokine quantification of culture supernatants
Apical wash and basal media were used to quantify IFN-β and IFN-λ1/3 production using an AlphaLISA (Perkin Elmer) and the R&D Systems Human DuoSet ELISA kit, respectively, according to each manufacturer’s instructions. Concentration for apical and basal compartments were combined for total protein production for each individual culture. Inflammatory markers (G-CSF, GM-CSF, IFN-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12, IL-13, IL-17, MCP-1, MIP-1β and TNF-α) secreted in the basal media were quantified using the Bio-Plex Pro Human Cytokine 17-plex Panel (Bio-Rad, USA) on the Bio-Plex®-200 system and concentrations were determined using Bio-Plex Manager Software.
2.9 Statistical analysis
Statistical analyses were conducted using the GraphPad Prism 10.4.1 software. Paired t-tests (Wilcoxon signed-rank test) and one-way ANOVAs (Friedman test and uncorrected Dunn’s post hoc test) were used to assess differences between treatment groups. Statistical significance was defined as p < 0.05 with a 95% confidence interval.
3 Results
3.1 L. rhamnosus D3189 reduced RSV shedding, although not intracellular transcription
To assess the effect of L. rhamnosus D3189 on viral kinetics, we measured shed virus in apical washes and RSV N mRNA expression at 3 days p.i. Pretreatment with D3189 significantly reduced viral shedding (Figure 1A), suggesting a potential impact on viral release or egress. However, there was no significant change in N gene mRNA levels (Figure 1B) indicating no effect on viral transcription.
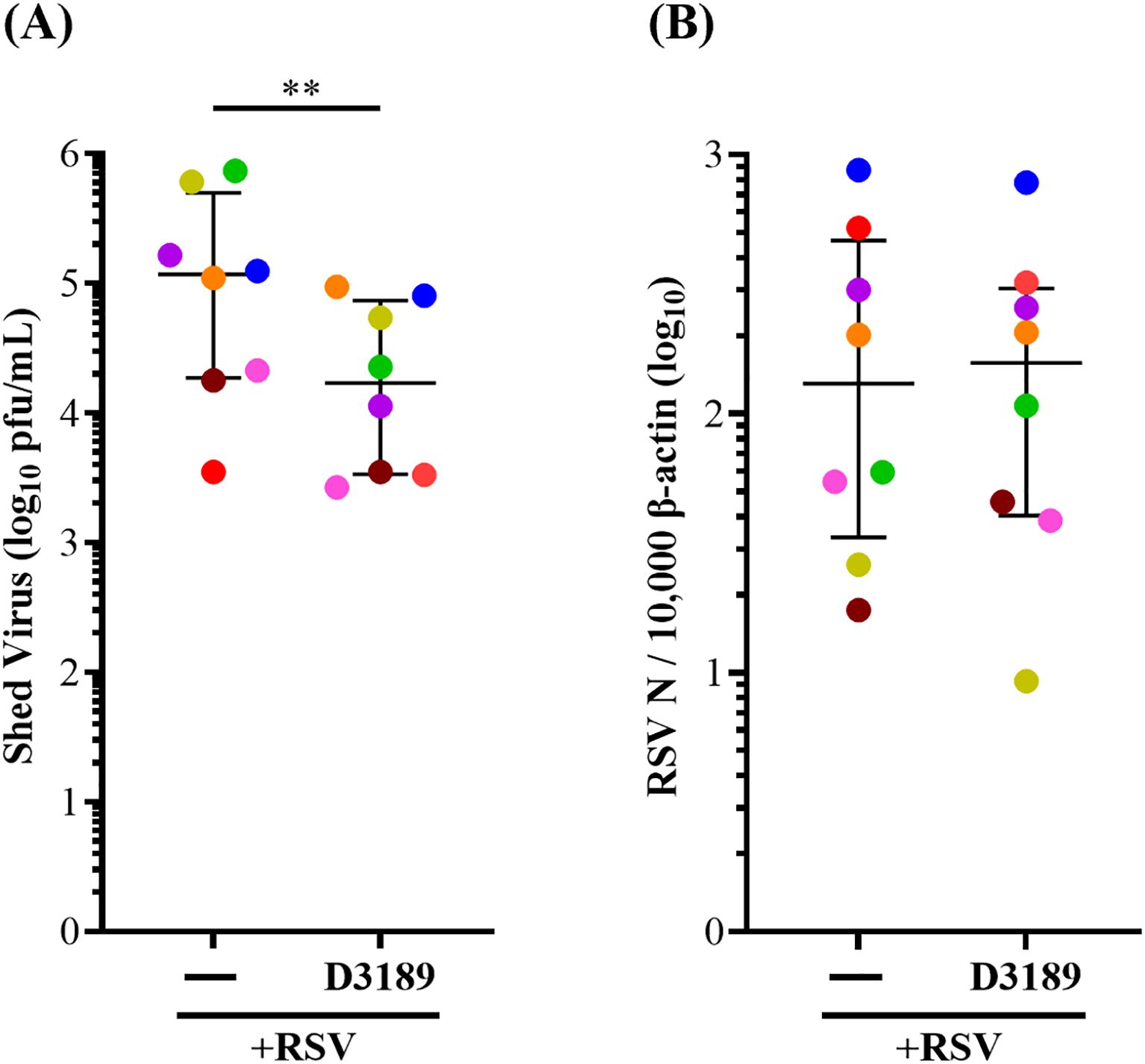
Figure 1. Effect of L. rhamnosus D3189 on RSV load 3 days post-infection. WD-NECs were pretreated with D3189 (80 µL of 2.5 × 107 CFU mL-1) or media (negative control) for 24 h before RSV infection (80 µL of 7.5 × 106 PFU mL-1) or mock exposure (virus-negative control). At 3 days post-infection, apical washes were collected. (A) Shed virus was quantified by immuno-plaque assay. (B) Viral transcription was assessed by RT-qPCR using RSV N-specific primers, normalised to β-actin expression. Data are presented as median and interquartile range, analysed by Wilcoxon signed-rank test. Each data point represents the mean of duplicate cultures per donor, with donors distinguished by colour (n = 8). **, p < 0.01.
Bacterial carriage was evaluated four days post-exposure to assess potential variations in lactobacilli levels among donors at the time of sampling and to confirm colonization by D3189. All cultures exhibited comparable bacterial loads in the apical wash, measured as CFU mL-1, confirming successful colonization (Supplementary Figure 1).
3.2 L. rhamnosus D3189 does not alter cytotoxicity or barrier integrity during RSV infection
LDH, a cytosolic enzyme released upon damage to the plasma membrane, was measured as an indicator of cytotoxicity. Although LDH release varied among donors, D3189 alone did not induce cytotoxicity, consistent with our previous findings (Yarlagadda et al., 2024). However, RSV infection with and without D3189 pretreatment significantly elevated LDH release compared to the uninfected or D3189 only controls, indicating that D3189 did not mitigate RSV-induced cytotoxicity (Figure 2A). Similarly, TEER, an indicator of tight junction integrity, was unaffected by D3189 alone but significantly reduced by RSV infection, regardless of probiotic pretreatment, suggesting that D3189 did not protect against virus-induced barrier disruption (Figure 2B).
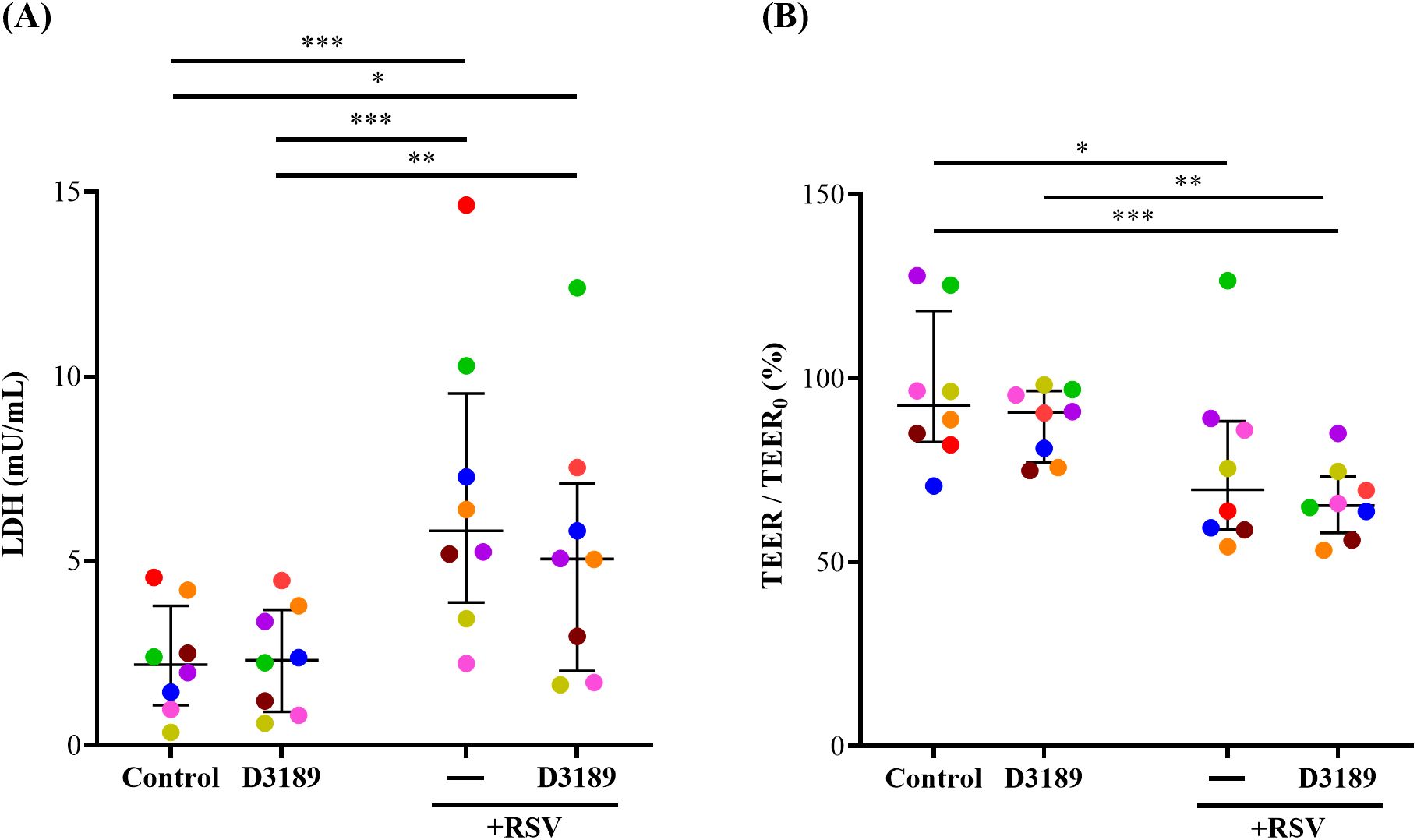
Figure 2. Effect of L. rhamnosus D3189 on LDH release and TEER 3 days post-infection with RSV. WD-NECs were pretreated with D3189 (80 µL of 2.5 × 107 CFU mL-1) or media (negative control) for 24 h before RSV infection (80 µL of 7.5 × 106 PFU mL-1) or mock exposure (virus-negative control). At 3 days post-infection, (A) LDH release in the basal media was quantified by LDH-Glo Cytotoxicity Assay Kit, and (B) TEER was measured using the EVOM2 Epithelial Voltohmmeter and is presented as a percentage fold change relative to baseline TEER. Data are presented as median and interquartile range, analysed by the Friedman test and uncorrected Dunn’s post hoc test. Each data point represents the mean of duplicate cultures per donor, with donors distinguished by colour (n = 8). *, p < 0.05; **, p < 0.01; ***, p < 0.005.
3.3 L. rhamnosus D3189 does not significantly modulate the IFN response to RSV infection
IFN-β and -λ are principal antiviral proteins induced by RSV infection of airway epithelial cells (AECs) (Bergeron et al., 2023). Given the observed reduction in viral shedding, we investigated whether D3189 influenced the antiviral response by enhancing IFN production. IFN-β and -λ1/3 release were quantified in apical washes and basal media from duplicate cultures and combined to determine secretion. D3189 alone did not induce IFN release by WD-NECs (Figures 3A, B). As expected, RSV infection significantly increased IFN-β and IFN-λ1/3 production compared to uninfected controls, although with large donor variability. While D3189 pretreatment did not statistically significantly alter this response, there was a strong trend toward increased IFN-β levels in co-exposed cultures (Figure 3A). Notably, 6 out of 8 donors (orange, red, yellow, green, brown, blue) exhibited higher IFN-β secretion when pretreated with D3189 compared to no pretreatment, suggesting donor specific variability regarding the effect on RSV-induced IFN-β production.
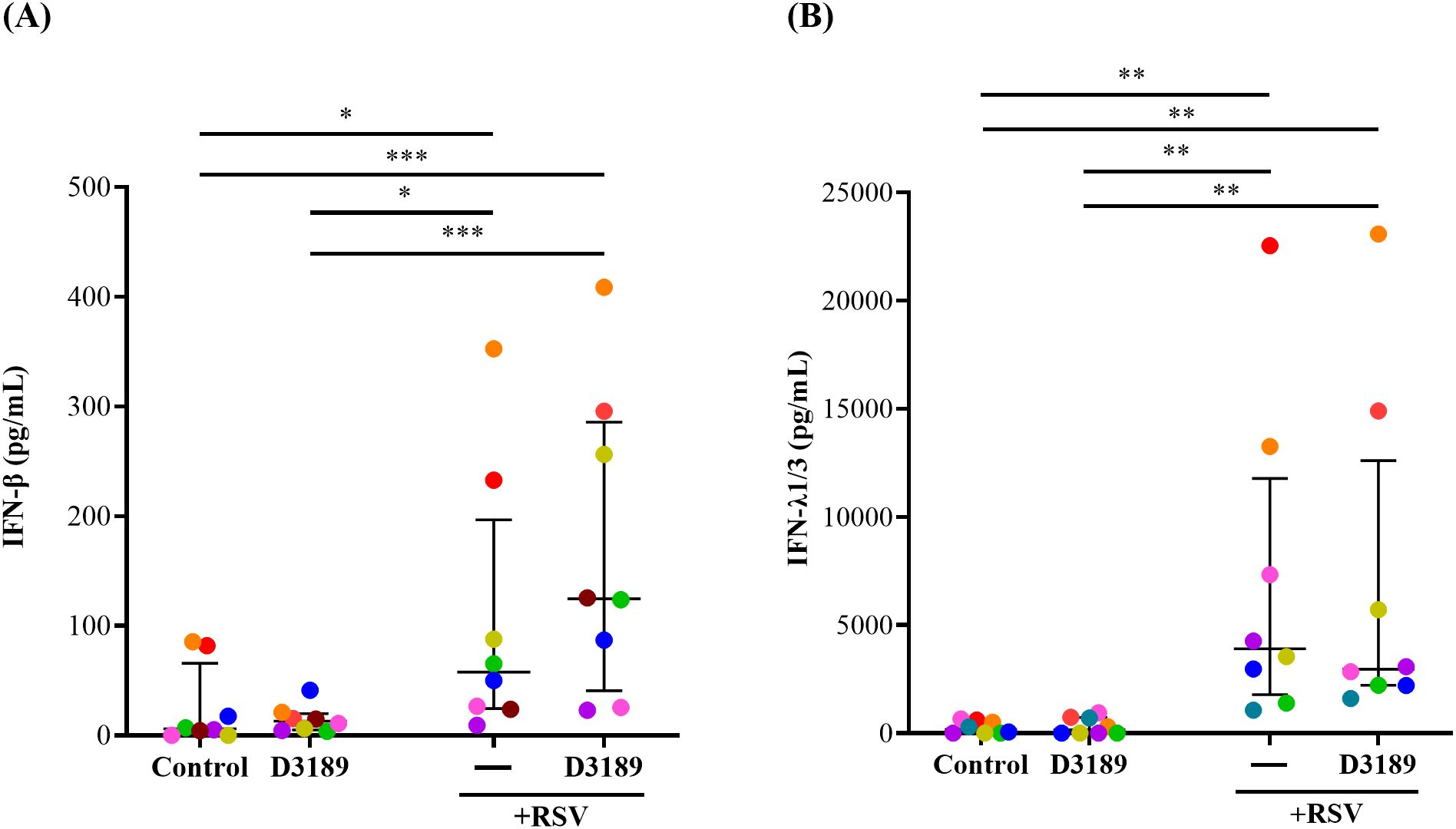
Figure 3. Effect of L. rhamnosus D3189 IFN-β and IFN-λ1/3–3 days post-infection with RSV. WD-NECs were pretreated with D3189 (80 µL of 2.5 × 107 CFU mL-1) or media (negative control) for 24 h before RSV infection (80 µL of 7.5 × 106 PFU mL-1) or mock exposure (virus-negative control). At 3 days post-infection, secreted (A) IFN-β and (B) IFN-λ1/3 were quantified by AlphaLISA and standard ELISA, respectively, using both apical wash and basal media. Data are presented as median and interquartile range, analysed by the Friedman test and uncorrected Dunn’s post hoc test. Each data point represents the mean of duplicate cultures per donor, with donors distinguished by colour (n = 8). *, p < 0.05; **, p < 0.01; ***, p < 0.005.
3.4 L. rhamnosus D3189 attenuates RSV-induced inflammatory responses
Host innate inflammatory responses to RSV infection play a critical role in determining disease severity, as excessive inflammation, characterized by increased production of proinflammatory cytokines and chemokines have been linked to more severe RSV disease, prolonged illness, and heightened risk of secondary infections (Sun and López, 2017; Agac et al., 2023). We investigated whether D3189 could modulate the release of proinflammatory cytokines and chemokines from RSV-infected WD-NECs, particularly to assess whether pretreatment dampens inflammation, as we previously observed in RV-infected WD-NECs (Yarlagadda et al., 2024). A 17-plex bead array was performed using basal medium to quantify key proinflammatory cytokines and chemokines. As expected, RSV infection significantly induced the release of IFN-γ, IL-1β, IL-4, IL-6, IL-17, MCP-1, MIP-1β and TNF-α compared to uninfected controls (Figures 4A–H). D3189 alone did not induce cytokine or chemokine expression, consistent with our previous findings (Yarlagadda et al., 2024). However, D3189 pretreatment prior to RSV infection significantly attenuated IFN-γ, IL-1β and MCP-1 production (Figures 4A, B, G, respectively), with a strong trend toward reduced IL-17 and TNF-α levels (Figures 4E, H, respectively). Notably, this dampening effect was most pronounced in donors exhibiting the highest inflammatory responses to RSV infection (as indicated by color coding). A heatmap (Figure 4I) illustrating fold changes over uninfected controls further demonstrated an overall reduction in inflammatory cytokine and chemokine release following D3189 pretreatment, although not all changes reached statistical significance. These findings suggest that D3189 has the potential to suppress RSV-induced inflammation in WD-NECs.
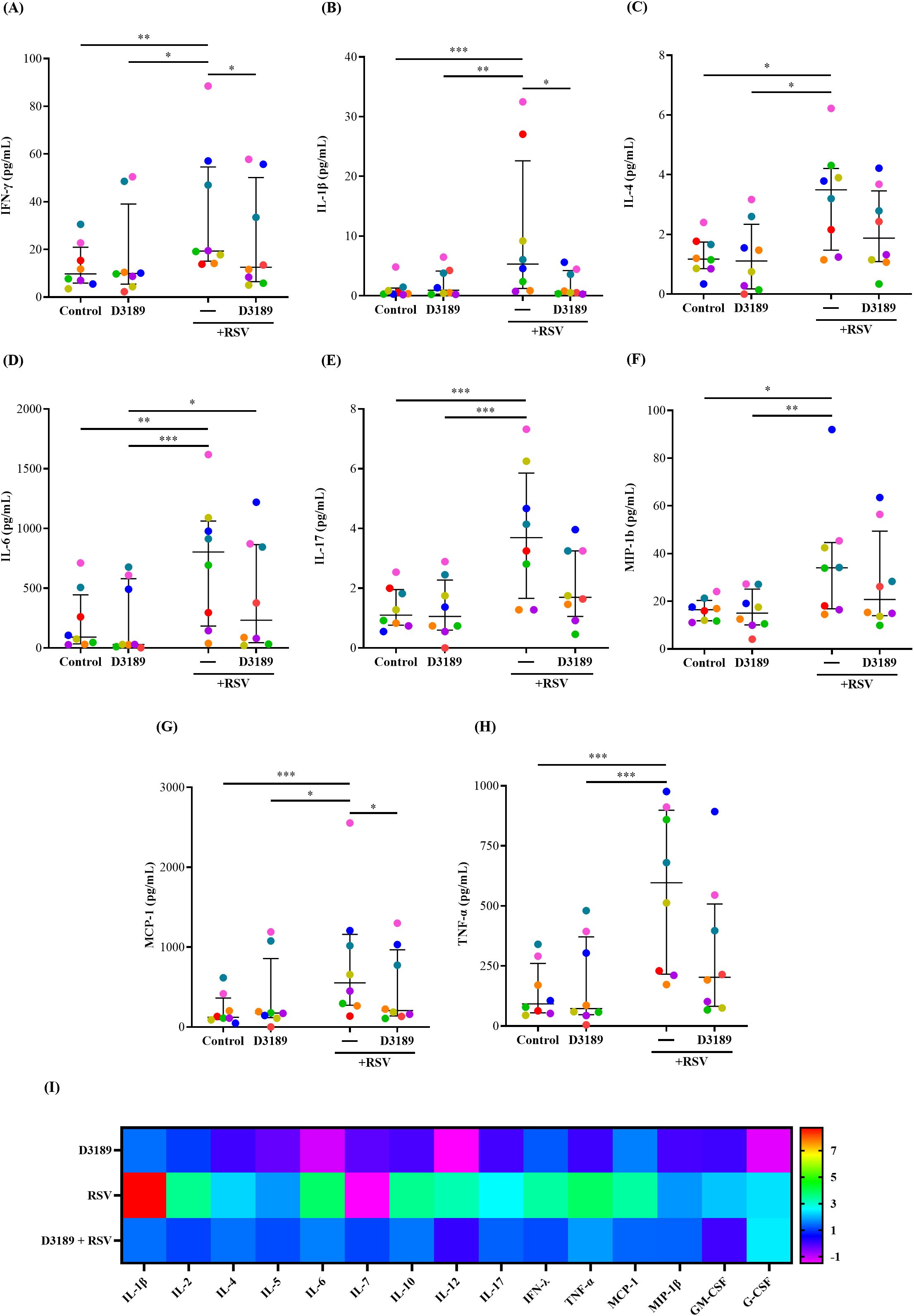
Figure 4. Effect of L. rhamnosus D3189 pro-inflammatory mediator release 3 days post-infection with RSV. WD-NECs were pretreated with D3189 (80 µL of 2.5 × 107 CFU mL-1) or media (negative control) for 24 h before RSV infection (80 µL of 7.5 × 106 PFU mL-1) or mock exposure (virus-negative control). At 3 days post-infection, secreted cytokines and chemokines were quantified in basal media using the Bio-plex pro human cytokine 17-plex assay. Data points represent secreted (A) IFN-γ, (B) IL-1β, (C) IL-4, (D) IL-6, (E) IL-17, (F) MCP-1, (G) MIP-1β and (H) TNF-α from each donor. A (I) heatmap illustrates fold changes in cytokine and chemokine production relative to uninfected controls for all analytes detected with the assay’s limit of detection. Data are presented as median and interquartile range, analysed by the Friedman test and uncorrected Dunn’s post hoc test. Each data point represents the mean of duplicate cultures per donor, with donors distinguished by colour (n = 8). *, p < 0.05; **, p < 0.01; ***, p < 0.005.
4 Discussion
RSV is a major etiological agent of respiratory infections and acute respiratory disease, contributing significantly to morbidity and mortality rates worldwide (Li et al., 2022). Despite extensive research efforts, both prophylactic and therapeutic strategies remain inadequate in reducing infection in the URT. Our previous work demonstrated that pretreatment with specific L. rhamnosus strains reduced RV-induced cytokine and chemokine release in WD-NECs without affecting viral load (Yarlagadda et al., 2024). Building on these findings, we hypothesized that D3189 might confer protection against RSV infection in the same WD-NEC model. Our results reveal that pretreatment with D3189 reduces viral shedding and significantly attenuates pro-inflammatory cytokine and chemokine release following RSV infection. To our knowledge, this is the first study to report the immunomodulatory effects of lactobacilli against RSV infection in primary WD-NECs.
Studies in murine models of IAV, RSV, and pneuomovirus of mice (PVM) infection have demonstrated that intranasal administration of lactobacilli reduces viral load in the respiratory tract and improves survival outcomes (Hori et al., 2001; Gabryszewski et al., 2011; Youn et al., 2012; Park et al., 2013; Tomosada et al., 2013). Although our model of WD-NECs lacks a functional immune system we did also observe reduced RSV viral shedding correlated to pre-exposure to L. rhamnosus D3189. Interestingly, this effect was not observed in our previous study with RV-infected WD-NECs, where D3189 had no impact on viral load (Yarlagadda et al., 2024), indicating virus-specific interactions with D3189. RSV infects AECs by attaching to surface receptors and entering via membrane fusion (Battles and McLellan, 2019). Once inside, the viral RNA-dependent RNA polymerase transcribes genes in a sequential 3’ to 5’ gradient. Full-length genome replication and synthesis of viral proteins support virion assembly and apical release through budding (Shang et al., 2021). This life cycle underpins RSV’s pathogenicity, especially in infants, where epithelial injury and exaggerated inflammatory responses contribute to bronchiolitis. In our study, the observed reduction in RSV shedding without a corresponding decrease in intracellular N gene mRNA suggests that D3189 may interfere with later stages of the virus life cycle, such as assembly, budding, or egress rather than transcription or genome replication. However, the precise mechanisms by which D3189 exerts these effects remain to be elucidated.
We hypothesized that the reduction in RSV virus shedding by pre-exposure to D3189 may correlate with enhanced production of key antiviral cytokines, specifically type I (IFN-α/β) and type III (IFN-λ) interferons, which are critical in the innate antiviral immune response and viral clearance (Yang and Li, 2020). Mouse and airway cell studies have shown that lactobacilli enhance resistance to viral infections, including RSV, by upregulating IFN-β production, which subsequently boosts the expression of viral-sensing receptors such as TLR3 and DDX58, and antiviral factors including Mx1 and OAS1, which limit viral replication (Tomosada et al., 2013; Islam et al., 2021). In our WD-NEC study D3189 alone did not induce either IFN-β or -λ production (Yarlagadda et al., 2025), although similarly to L. rhamnosus-treated mice challenged with RSV (Tomosada et al., 2013), there was a trend for virus-induced IFN-β to be further enhanced by D3189 exposure, at least for 6 of the 8 donors. The boost in IFN may have partially contributed to reduced infectious virus shedding, as RSV both induces IFN-β and is susceptible to IFN-β for viral clearance (Hijano et al., 2019). Unlike RSV, in our previous study using WD-NECs we did not observe any induction of IFN-β by RV, or enhanced IFN-β induction in combination with D3189 exposure. This difference can be attributed to the fact that RV does not rely on IFN-β for viral clearance, and is a poor inducer of IFN-β, unlike RSV (Mueller and Rouse, 2008; Yuan et al., 2022).
To assess whether D3189 modulates RSV-induced cytotoxicity, we measured LDH release as an indicator of epithelial damage. D3189 alone had no deleterious effect on WD-NECs, consistent with our previous findings (Yarlagadda et al., 2024). However, while our earlier study assessed D3189 exposure over 48 hours, the present study extended this duration to four days post-exposure, further confirming the absence of cytotoxic effects associated with prolonged D3189 exposure. As anticipated, RSV induced LDH release, consistent with multiple studies reporting LDH detection in nasopharyngeal samples as a marker of RSV-induced epithelial injury and a predictor of disease severity (Vázquez et al., 2019). Previous studies in mice have demonstrated that nasal delivery of lactobacilli prior to RSV infection or poly(I:C) challenge significantly reduces LDH release, suggesting a protective effect against virus-induced cytotoxicity (Tomosada et al., 2013). However, despite the significant reduction in viral shedding, D3189 pretreatment did not mitigate RSV-induced LDH release within the 3 days of experimentation. This finding suggests that, unlike the mouse studies, D3189 does not confer similar protection in the context of RSV infection in the WD-NEC model. These results align with our previous study where D3189 and other L. rhamnosus isolates had no effect on RV-induced LDH release (Yarlagadda et al., 2024). The mechanisms underlying this response remain unclear, however they may be influenced by distinct host immune interactions, where factors essential for lactobacilli-enhanced epithelial repair are absent from the WD-NEC model. Further investigations are warranted to delineate the precise factors governing lactobacilli-mediated cytoprotective effects in viral infections.
RSV infection is known to compromise airway epithelial barrier integrity, leading to increased permeability and disruption of tight junctions (Kast et al., 2017; Smallcombe et al., 2019). While the impact of probiotics on epithelial barrier dysfunction in the URT remains largely unexplored, some evidence suggests that lactobacilli can enhance and regulate epithelial barrier function in AECs (De Boeck et al., 2020; De Rudder et al., 2020). In the present study, exposure to D3189 did not compromise tight junction integrity, indicating an absence of deleterious effects. As anticipated, RSV infection led to a significant reduction in TEER, consistent with previous reports demonstrating RSV-mediated disruption of AEC tight junctions (Kast et al., 2017; Smallcombe et al., 2019). However, similarly to LDH release, this barrier dysfunction was not modulated by D3189 pretreatment, suggesting that D3189 does not confer protective effects on tight junction integrity under these conditions.
RSV infection triggers a robust cytokine and chemokine response essential for viral clearance. However, it is often associated with a dysregulated hyper-inflammatory response, particularly in RSV bronchiolitis (Das et al., 2005; Rayavara et al., 2018). RSV is a potent activator of NF-κB signaling pathways in airway epithelial cells, leading to upregulation of inflammatory mediators such as IL-1α/β, IL-6, IL-8, TNF-α, IFN-γ, IL-17, MCP-1, MIP-1α/β, RANTES, and IP-10 (Garofalo et al., 1996; Jamaluddin et al., 1998; Casola et al., 2001; Garofalo et al., 2013). Consistent with this, we observed that RSV infection induced the release of IFN-γ, IL-1β, IL-4, IL-6, IL-17, MCP-1, MIP-1β, and TNF-α from WD-NECs, all of which influence RSV pathogenesis and disease severity. Pre-exposure to D3189 significantly reduced RSV-induced release of IFN-γ, IL-1β, and MCP-1, with additional trends towards reduced IL-17 and TNF-α. While IFN-γ is essential for antiviral immunity, excessive levels contribute to airway obstruction and inflammation (Estripeaut et al., 2008). IL-1β induces epithelial damage and mucus hypersecretion, while IL-17 contributes to neutrophilic damage and post-viral wheezing (Mukherjee et al., 2011). MCP-1 recruits monocytes, intensifying inflammation and viral spread, while TNF-α enhances epithelial apoptosis and mucus production, further compromising airway function (Bergeron and Tripp, 2021). The observed shift in cytokine and chemokine profiles following D3189 treatment suggests an immunomodulatory effect that could mitigate RSV-induced inflammation and epithelial damage. We previously hypothesized that L. rhamnosus D3189 metabolites might inhibit NF-κB signaling to suppress pro-inflammatory cytokine and chemokine production (Yarlagadda et al., 2024). In this study, D3189 may have directly dampened cytokine and chemokine production as we have observed previously (Yarlagadda et al., 2024), although the reduction in virus shedding with D3189 pretreatment may have also indirectly influenced reduced immune activation thereby contributing to lower inflammatory mediator release. Thus, D3189 appears to both directly modulate immune responses and indirectly reduce inflammation by limiting viral shedding, alleviating RSV-associated immune activation.
We previously observed that different L. rhamnosus strains modulate host responses to RV infection in WD-NECs differently (Yarlagadda et al., 2024). To determine whether the observed antiviral effects were unique to L. rhamnosus D3189, we also tested another commonly studied probiotic strain, LGG, in a subset of donors (n = 5). These results, presented in Supplementary Figure 2, showed that LGG did not significantly reduce RSV shedding, enhance IFN-β expression, or attenuate RSV-induced proinflammatory responses. Moreover, LGG co-exposure resulted in greater disruption of epithelial barrier integrity, as reflected by reduced TEER compared to RSV infection alone. These findings suggest that the antiviral and immunomodulatory effects observed in this study are strain-specific and highlight the importance of functional screening when selecting probiotic candidates for respiratory applications.
Although this study offers valuable insights, several limitations should be considered when translating the results of probiotic therapy from the in vitro model to clinical settings. The WD-NEC model lacks immune cell components and other features of a fully functional mucosa, limiting our observations to the responses of structural cells to both RSV infection and probiotic exposure. Additionally, variability in human donor responses, driven by genetic and environmental differences, may influence NEC responses. Expanding the donor cohort in future studies may help clarify the broach applicability of the observed effects.
To build on findings of this study, future research will focus on elucidating the mechanisms underlying the antiviral effects of L. rhamnosus D3189. Specifically, we will investigate whether reduced RSV shedding results from inhibited release of fully assembled and infectious virions from NECs. To determine whether bacterial viability is required, we will compare the effects of live and heat-inactivated bacteria as well as conditioned media, to understand whether secreted factors or direct surface interactions are responsible. Time-course experiments introducing D3189 at different stages of infection will further define the window during which the probiotic is effective. Additionally, we will investigate D3189’s immunomodulatory activity by using inhibition studies to clarify host pathways involved in antiviral defense.
In conclusion, our study highlights the potential of L. rhamnosus D3189 as a promising immunomodulatory agent in the context of RSV infection. D3189 enhances IFN-β production, reduces viral shedding, and mitigates the release of pro-inflammatory cytokines and chemokines, which could collectively reduce the severity of RSV-induced inflammation within the URT. Reduced viral shedding may also indicate a positive influence of nasal probiotics in reducing the risk of transmission. However, detailed mechanistic studies are required to fully understand how D3189 mediates these effects and to inform its optimal use in clinical applications.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Queensland University of Technology Human Research Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
TY: Data curation, Methodology, Validation, Conceptualization, Investigation, Project administration, Writing – review & editing, Formal Analysis, Writing – original draft, Visualization. JS: Data curation, Conceptualization, Writing – review & editing, Investigation. DM-P: Writing – review & editing, Resources. ACo: Resources, Writing – review & editing. ACe: Resources, Writing – review & editing. KS: Writing – original draft, Formal Analysis, Resources, Methodology, Supervision, Project administration, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We would like to acknowledge Dr Yanshan Zhu and Professor Kirsty Short from The University of Queensland and Saeideh Hajighasemi from Queensland University of Technology for their contributions towards sample collection and culturing of nasal epithelial cells.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1625517/full#supplementary-material
References
Agac, A., Kolbe, S. M., Ludlow, M., Osterhaus, A., Meineke, R., and Rimmelzwaan, G. F. (2023). Host responses to respiratory syncytial virus infection. Viruses. 15. doi: 10.3390/v15101999
Assad, Z., Romain, A. S., Aupiais, C., Shum, M., Schrimpf, C., Lorrot, M., et al. (2024). Nirsevimab and hospitalization for RSV bronchiolitis. N Engl. J. Med. 391, 144–154. doi: 10.1056/NEJMoa2314885
Banooni, P., Gonik, B., Epalza, C., Reyes, O., Madhi, S. A., Gomez-Go, G. D., et al. (2025). Efficacy, immunogenicity, and safety of an investigational maternal respiratory syncytial virus prefusion F protein-based vaccine. Clin. Infect. Dis. doi: 10.1093/cid/ciaf033
Battles, M. B. and McLellan, J. S. (2019). Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 17, 233–245. doi: 10.1038/s41579-019-0149-x
Bergeron, H. C., Hansen, M. R., and Tripp, R. A. (2023). Interferons-implications in the immune response to respiratory viruses. Microorganisms. 11 (9), 2179. doi: 10.3390/microorganisms11092179
Bergeron, H. C. and Tripp, R. A. (2021). Immunopathology of RSV: an updated review. Viruses 13 (12), 2478. doi: 10.3390/v13122478
Casola, A., Garofalo, R. P., Haeberle, H., Elliott, T. F., Lin, R., Jamaluddin, M., et al. (2001). Multiple cis regulatory elements control RANTES promoter activity in alveolar epithelial cells infected with respiratory syncytial virus. J. Virol. 75, 6428–6439. doi: 10.1128/JVI.75.14.6428-6439.2001
Chuang, Y. C., Lin, K. P., Wang, L. A., Yeh, T. K., and Liu, P. Y. (2023). The impact of the COVID-19 pandemic on respiratory syncytial virus infection: A narrative review. Infect. Drug Resist. 16, 661–675. doi: 10.2147/IDR.S396434
Clua, P., Kanmani, P., Zelaya, H., Tada, A., Kober, A., Salva, S., et al. (2017). Peptidoglycan from immunobiotic lactobacillus rhamnosus improves resistance of infant mice to respiratory syncytial viral infection and secondary pneumococcal pneumonia. Front. Immunol. 8, 948. doi: 10.3389/fimmu.2017.00948
Coleman, A., Bialasiewicz, S., Marsh, R. L., Grahn Håkansson, E., Cottrell, K., Wood, A., et al. (2021). Upper respiratory microbiota in relation to ear and nose health among Australian aboriginal and torres strait islander children. J. Pediatr. Infect. Dis. Society. 10, 468–476. doi: 10.1093/jpids/piaa141
Coleman, A., Håkansson, A., Grahn Håkansson, E., Cottrell, K., Bialasiewicz, S., Zaugg, J., et al. (2022). In vitro Inhibition of respiratory pathogens by lactobacillus and alpha haemolytic streptococci from Aboriginal and Torres Strait Islander children. J. Appl. Microbiol. 132, 2368–2378. doi: 10.1111/jam.15320
Collins, P. L., Fearns, R., and Graham, B. S. (2013). Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 372, 3–38. doi: 10.1007/978-3-642-38919-1_1
Das, S., Palmer, O. P., Leight, W. D., Surowitz, J. B., Pickles, R. J., Randell, S. H., et al. (2005). Cytokine amplification by respiratory syncytial virus infection in human nasal epithelial cells. Laryngoscope. 115, 764–768. doi: 10.1097/01.MLG.0000159527.76949.93
De Boeck, I., van den Broek, M. F. L., Allonsius, C. N., Spacova, I., Wittouck, S., Martens, K., et al. (2020). Lactobacilli have a niche in the human nose. Cell Rep. 31, 107674. doi: 10.1016/j.celrep.2020.107674
De Rudder, C., Garcia-Tímermans, C., De Boeck, I., Lebeer, S., Van de Wiele, T., and Calatayud Arroyo, M. (2020). Lacticaseibacillus casei AMBR2 modulates the epithelial barrier function and immune response in a donor-derived nasal microbiota manner. Sci. Rep. 10, 16939. doi: 10.1038/s41598-020-73857-9
Duan, Y., Liu, Z., Zang, N., Cong, B., Shi, Y., Xu, L., et al. (2024). Landscape of respiratory syncytial virus. Chin. Med. J. (Engl) 137, 2953–2978. doi: 10.1097/CM9.0000000000003354
Estripeaut, D., Torres, J. P., Somers, C. S., Tagliabue, C., Khokhar, S., Bhoj, V. G., et al. (2008). Respiratory syncytial virus persistence in the lungs correlates with airway hyperreactivity in the mouse model. J. Infect. Dis. 198, 1435–1443. doi: 10.1086/592714
Gabryszewski, S. J., Bachar, O., Dyer, K. D., Percopo, C. M., Killoran, K. E., Domachowske, J. B., et al. (2011). Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J. Immunol. (Baltimore Md: 1950). 186, 1151–1161. doi: 10.4049/jimmunol.1001751
Garofalo, R. P., Kolli, D., and Casola, A. (2013). Respiratory syncytial virus infection: mechanisms of redox control and novel therapeutic opportunities. Antioxid Redox Signal. 18, 186–217. doi: 10.1089/ars.2011.4307
Garofalo, R., Sabry, M., Jamaluddin, M., Yu, R. K., Casola, A., Ogra, P. L., et al. (1996). Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70, 8773–8781. doi: 10.1128/jvi.70.12.8773-8781.1996
Gatt, D., Martin, I., AlFouzan, R., and Moraes, T. J. (2023). Prevention and treatment strategies for respiratory syncytial virus (RSV). Pathogens. 12 (2), 154. doi: 10.3390/pathogens12020154
Hijano, D. R., Vu, L. D., Kauvar, L. M., Tripp, R. A., Polack, F. P., and Cormier, S. A. (2019). Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front. Immunol. 10, 566. doi: 10.3389/fimmu.2019.00566
Hori, T., Kiyoshima, J., Shida, K., and Yasui, H. (2001). Effect of intranasal administration of Lactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin. Diagn. Lab. Immunol. 8, 593–597. doi: 10.1128/CDLI.8.3.593-597.2001
Islam, M. A., Albarracin, L., Tomokiyo, M., Valdez, J. C., Sacur, J., Vizoso-Pinto, M. G., et al. (2021). Immunobiotic lactobacilli improve resistance of respiratory epithelial cells to SARS-CoV-2 infection. Pathog. (Basel Switzerland). 10, 1197. doi: 10.3390/pathogens10091197
Jamaluddin, M., Casola, A., Garofalo, R. P., Han, Y., Elliott, T., Ogra, P. L., et al. (1998). The major component of IkappaBalpha proteolysis occurs independently of the proteasome pathway in respiratory syncytial virus-infected pulmonary epithelial cells. J. Virol. 72, 4849–4857. doi: 10.1128/JVI.72.6.4849-4857.1998
Kaler, J., Hussain, A., Patel, K., Hernandez, T., and Ray, S. (2023). Respiratory syncytial virus: A comprehensive review of transmission, pathophysiology, and manifestation. Cureus. 15, e36342. doi: 10.7759/cureus.36342
Kast, J. I., McFarlane, A. J., Globinska, A., Sokolowska, M., Wawrzyniak, P., Sanak, M., et al. (2017). Respiratory syncytial virus infection influences tight junction integrity. Clin. Exp. Immunol. 190, 351–359. doi: 10.1111/cei.13042
Kelleher, K., Subramaniam, N., and Drysdale, S. B. (2025). The recent landscape of RSV vaccine research. Ther. Adv. Vaccines Immunother. 13, 25151355241310601. doi: 10.1177/25151355241310601
Kicic, A., Hallstrand, T. S., Sutanto, E. N., Stevens, P. T., Kobor, M. S., Taplin, C., et al. (2010). Decreased fibronectin production significantly contributes to dysregulated repair of asthmatic epithelium. Am. J. Respir. Crit. Care Med. 181, 889–898. doi: 10.1164/rccm.200907-1071OC
Kicic, A., Stevens, P. T., Sutanto, E. N., Kicic-Starcevich, E., Ling, K. M., Looi, K., et al. (2016). Impaired airway epithelial cell responses from children with asthma to rhinoviral infection. Clin. Exp. Allergy 46, 1441–1455. doi: 10.1111/cea.12767
Kumova, O. K., Fike, A. J., Thayer, J. L., Nguyen, L. T., Mell, J. C., Pascasio, J., et al. (2019). Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLoS Pathog. 15, e1008072. doi: 10.1371/journal.ppat.1008072
Li, Y., Wang, X., Blau, D. M., Caballero, M. T., Feikin, D. R., Gill, C. J., et al. (2022). Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet 399, 2047–2064. doi: 10.1016/S0140-6736(22)00478-0
McLaughlin, J. M., Khan, F., Begier, E., Swerdlow, D. L., Jodar, L., and Falsey, A. R. (2022). Rates of medically attended RSV among US adults: A systematic review and meta-analysis. Open Forum Infect. Diseases. 9. doi: 10.1093/ofid/ofac300
Moreira, A. C., Ribeiro, A. B., Oliveira, I., Sá, M., Lameirão, C., and Marques, P. (2024). Efficacy of anti-RSV vaccination in preventing respiratory syncytial virus disease and severe illness in older adults: a systematic review of randomized controlled trials. Eur. Geriatr. Med. 15, 1215–1229. doi: 10.1007/s41999-024-01066-y
Mueller, S. N. and Rouse, B. T. (2008). Immune responses to viruses. Clin. Immunol., 421–431. doi: 10.1016/B978-0-323-04404-2.10027-2
Mukherjee, S., Lindell, D. M., Berlin, A. A., Morris, S. B., Shanley, T. P., Hershenson, M. B., et al. (2011). IL-17-induced pulmonary pathogenesis during respiratory viral infection and exacerbation of allergic disease. Am. J. Pathol. 179, 248–258. doi: 10.1016/j.ajpath.2011.03.003
Newton, A. H., Cardani, A., and Braciale, T. J. (2016). The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin. Immunopathol. 38, 471–482. doi: 10.1007/s00281-016-0558-0
Park, M. K., Ngo, V., Kwon, Y. M., Lee, Y. T., Yoo, S., Cho, Y. H., et al. (2013). Lactobacillus plantarum DK119 as a probiotic confers protection against influenza virus by modulating innate immunity. PLoS One 8, e75368. doi: 10.1371/journal.pone.0075368
Percopo, C. M., Ma, M., and Rosenberg, H. F. (2017). Administration of immunobiotic Lactobacillus plantarum delays but does not prevent lethal pneumovirus infection in Rag1(-/-) mice. J. Leukoc. Biol. 102, 905–913. doi: 10.1189/jlb.3AB0217-050RR
Percopo, C. M., Rice, T. A., Brenner, T. A., Dyer, K. D., Luo, J. L., Kanakabandi, K., et al. (2015). Immunobiotic Lactobacillus administered post-exposure averts the lethal sequelae of respiratory virus infection. Antiviral Res. 121, 109–119. doi: 10.1016/j.antiviral.2015.07.001
Piedimonte, G. and Perez, M. K. (2014). Respiratory syncytial virus infection and bronchiolitis. Pediatr. Rev. 35, 519–530. doi: 10.1542/pir.35.12.519
Rayavara, K., Kurosky, A., Stafford, S. J., Garg, N. J., Brasier, A. R., Garofalo, R. P., et al. (2018). Proinflammatory effects of respiratory syncytial virus-induced epithelial HMGB1 on human innate immune cell activation. J. Immunol. 201, 2753–2766. doi: 10.4049/jimmunol.1800558
Rosas-Salazar, C., Shilts, M. H., Tovchigrechko, A., Chappell, J. D., Larkin, E. K., Nelson, K. E., et al. (2016). Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am. J. Respir. Crit. Care Med. 193, 1180–1183. doi: 10.1164/rccm.201512-2350LE
Rosas-Salazar, C., Shilts, M. H., Tovchigrechko, A., Schobel, S., Chappell, J. D., Larkin, E. K., et al. (2018). Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J. Allergy Clin. Immunol. 142, 1447–1456. doi: 10.1016/j.jaci.2017.10.049
Shang, Z., Tan, S., and Ma, D. (2021). Respiratory syncytial virus: from pathogenesis to potential therapeutic strategies. Int. J. Biol. Sci. 17, 4073–4091. doi: 10.7150/ijbs.64762
Simões, E. A., Carbonell-Estrany, X., Rieger, C. H., Mitchell, I., Fredrick, L., and Groothuis, J. R. (2010). The effect of respiratory syncytial virus on subsequent recurrent wheezing in atopic and nonatopic children. J. Allergy Clin. Immunol. 126, 256–262. doi: 10.1016/j.jaci.2010.05.026
Smallcombe, C. C., Linfield, D. T., Harford, T. J., Bokun, V., Ivanov, A. I., Piedimonte, G., et al. (2019). Disruption of the airway epithelial barrier in a murine model of respiratory syncytial virus infection. Am. J. Physiol. Lung Cell Mol. Physiol. 316, L358–Ll68. doi: 10.1152/ajplung.00345.2018
Spann, K. M., Baturcam, E., Schagen, J., Jones, C., Straub, C. P., Preston, F. M., et al. (2014). Viral and host factors determine innate immune responses in airway epithelial cells from children with wheeze and atopy. Thorax. 69, 918–925. doi: 10.1136/thoraxjnl-2013-204908
Spann, K. M., Tran, K. C., Chi, B., Rabin, R. L., and Collins, P. L. (2004). Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected. J. Virol. 78, 4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004
Sun, Y. and López, C. B. (2017). The innate immune response to RSV: Advances in our understanding of critical viral and host factors. Vaccine. 35, 481–488. doi: 10.1016/j.vaccine.2016.09.030
Tomosada, Y., Chiba, E., Zelaya, H., Takahashi, T., Tsukida, K., Kitazawa, H., et al. (2013). Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 14, 40. doi: 10.1186/1471-2172-14-40
Vázquez, Y., González, L., Noguera, L., González, P. A., Riedel, C. A., Bertrand, P., et al. (2019). Cytokines in the respiratory airway as biomarkers of severity and prognosis for respiratory syncytial virus infection: an update. Front. Immunol. 10, 1154. doi: 10.3389/fimmu.2019.01154
Yang, E. and Li, M. M. H. (2020). All about the RNA: interferon-stimulated genes that interfere with viral RNA processes. Front. Immunol. 11, 605024. doi: 10.3389/fimmu.2020.605024
Yarlagadda, T., Carey, A., Bryan, E., Huygens, F., Yarlagadda, P., Maresco-Pennisi, D., et al. (2025). The response of nasal epithelial cells exposed to novel Lactobacillus and alpha-haemolytic Streptococcus isolated from the upper respiratory tract of children. J. Appl. Microbiol. 136 (4). doi: 10.1093/jambio/lxaf071
Yarlagadda, T., Zhu, Y., Snape, N., Carey, A., Bryan, E., Maresco-Pennisi, D., et al. (2024). Lactobacillus rhamnosus dampens cytokine and chemokine secretion from primary human nasal epithelial cells infected with rhinovirus. J. Appl. Microbiol. 135. doi: 10.1093/jambio/lxae018
Youn, H. N., Lee, D. H., Lee, Y. N., Park, J. K., Yuk, S. S., Yang, S. Y., et al. (2012). Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res. 93, 138–143. doi: 10.1016/j.antiviral.2011.11.004
Yuan, X.-H., Pang, L.-L., Yang, J., and Jin, Y. (2022). Comparison of immune response to human rhinovirus C and respiratory syncytial virus in highly differentiated human airway epithelial cells. Virol. J. 19, 81. doi: 10.1186/s12985-022-01805-2
Zelaya, H., Tada, A., Vizoso-Pinto, M. G., Salva, S., Kanmani, P., Agüero, G., et al. (2015). Nasal priming with immunobiotic Lactobacillus rhamnosus modulates inflammation-coagulation interactions and reduces influenza virus-associated pulmonary damage. Inflammation Res. 64, 589–602. doi: 10.1007/s00011-015-0837-6
Keywords: lactobacilli, respiratory syncytial virus, nasal epithelium, innate immunity, antiviral, inflammation
Citation: Yarlagadda T, Stedman J, Maresco-Pennisi D, Coleman A, Cervin A and Spann K (2025) Lactobacillus rhamnosus D3189 modulates antiviral and inflammatory responses in primary nasal epithelial cells, reducing respiratory syncytial virus shedding. Front. Cell. Infect. Microbiol. 15:1625517. doi: 10.3389/fcimb.2025.1625517
Received: 09 May 2025; Accepted: 18 June 2025;
Published: 08 July 2025.
Edited by:
Ekta Khattar, SVKM’s Narsee Monjee Institute of Management Studies, IndiaReviewed by:
Smriti Verma, Massachusetts General Hospital and Harvard Medical School, United StatesBhavna Goyal, Achira Labs Pvt. Ltd, India
Copyright © 2025 Yarlagadda, Stedman, Maresco-Pennisi, Coleman, Cervin and Spann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kirsten Spann, a2lyc3Rlbi5zcGFubkBxdXQuZWR1LmF1
 Tejasri Yarlagadda
Tejasri Yarlagadda Jacob Stedman1
Jacob Stedman1 Andrea Coleman
Andrea Coleman Anders Cervin
Anders Cervin Kirsten Spann
Kirsten Spann