- 1Department of Laboratory Animal Center, General Hospital of Northern Theater Command, Shenyang, Liaoning, China
- 2College of Life and Health Science, Northeastern University, Shenyang, Liaoning, China
- 3Department of Medical Security Center, General Hospital of Northern Theater Command, Shenyang, Liaoning, China
Introduction: Cryptococcus neoformans is a ubiquitous environmental fungal pathogen whose pathogenicity is closely linked to its ability to adapt to host environments. The RNA-binding protein Puf4 plays a key role in regulating the cell wall of C. neoformans. However, the specific mechanisms by which Puf4 regulates metabolism and virulence remain unclear.
Methods: In this study, we systematically investigated the role of Puf4 in the metabolism and virulence of C. neoformans using RNA sequencing (RNA-seq), biochemical analysis, and in vivo animal experiments.
Results: Colony-spotting assay assays show that Puf4 affects the cell membrane and oxidative stress. RNA-seq analysis revealed that Puf4 was enriched in key metabolic pathways, including carbohydrate metabolism, oxidative phosphorylation, and glycolysis. Biochemical analysis revealed that Puf4 overexpression led to significant increases in both total carbohydrate and glycogen content. Additionally, the Puf4-overexpressing strain showed elevated ROS levels, and reduced resistance to antimycin A, indicating decreased oxidative stress tolerance. Notably, Puf4 overexpression resulted in reduced capsule size and decreased L-DOPA production, accompanied by downregulated expression of the capsule-associated gene CAP10 and the melanin biosynthesis gene LAC1. In vivo animal experiments further confirmed that the virulence of the Puf4-overexpressing strain was significantly attenuated.
Discussion: These findings suggest that Puf4 may serve as a regulator linking metabolic status and pathogenic potential in C. neoformans, although the underlying mechanisms require further elucidation.
1 Introduction
C. neoformans, a conditionally pathogenic fungus ubiquitous in nature, also causes severe pulmonary and central nervous system infections (Fang et al., 2016; El Helou and Hellinger, 2019; Denham et al., 2022), particularly in immunocompromised hosts. It is a major cause of death in human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS) patients globally, being accountable for 19% of AIDS-related deaths, posing a significant public health burden (Rajasingham et al., 2022). The pathogenicity of C. neoformans is closely related to its unique biological characteristics, such as capsule formation, melanin synthesis, and survivability in various environments (Stott et al., 2021; Wake et al., 2023). Its environmental adaptation involves a complex metabolic regulatory network. Studying its metabolic mechanisms and virulence regulation is crucial for understanding its pathogenicity and developing new treatments (Zhao and Lin, 2021; Gao et al., 2022).
Although the research on C. neoformans has been deepening our understanding of this pathogenic fungus in recent years, its pathogenic mechanisms remain incompletely understood. RNA-binding proteins (RBPs) are a class of proteins that regulate gene expression at the post-transcriptional level. They can bind to target mRNAs and influence mRNA stability, translation efficiency, and other processes, thereby affecting cellular physiological functions. RBPs have been shown to play critical roles in fungal physiology (Zhong et al., 2022; Krusche et al., 2025), potentially influencing pathogenicity by regulating gene expression and metabolic pathways (Dallastella et al., 2023; Sankaranarayanan et al., 2023). For example, in filamentous fungi, a novel mechanism has been elucidated, in which the RBP CsdA forms a complex with the global regulatory factor RsdA and regulates the expression of RsdA at the post-transcriptional level, mediating changes in global secondary metabolism of the fungi (Song et al., 2023). In the phytopathogenic fungus Verticillium dahliae, research has revealed that the RBP VdNop12 plays a key role in pathogenicity and low-temperature adaptability (Zhang et al., 2020). The Puf (Pumilio and FBF) family of proteins, a group of conserved RBPs, characterized by multiple Puf domains (Pumilio/FBF homology domains) that specifically bind to the 3’ untranslated regions (3’UTRs) of target mRNAs to repress translation or promote mRNA decay, also warrant further investigation in this context (Wickens et al., 2002; Spassov and Jurecic, 2003; Tam et al., 2010; Wang et al., 2018; Son et al., 2021). In Saccharomyces cerevisiae, Puf4 modulates energy metabolism by regulating gene expression. This finding indicates that Puf family proteins perform both conserved and divergent regulatory functions in fungal adaptation to environmental changes and pathogenesis (Kaeberlein and Guarente, 2002; Russo and Olivas, 2015; Wilinski et al., 2015; Fischer and Olivas, 2018; Sadee et al., 2022).
In C. neoformans, Puf4, a member of the Puf protein family, has been previously studied for its effects on cell wall structure and protein homeostasis. Puf4 knockout mutants show reduced β-glucan content in the cell wall, increasing resistance to echinocandin drugs such as caspofungin. This suggests that Puf4 maintain cell integrity by regulating cell wall synthesis-related genes (e.g., FKS1, CHS3) (Kalem et al., 2021). Proteomic analysis further indicates that Puf4 knockout results in delayed unconventional splicing of HXL1 mRNA and a subsequent delay in the induction of Hxl1 target genes, indicating its role in regulating protein homeostasis networks and influencing fungal adaptation to the host body temperature of 37°C (Glazier et al., 2015). Although Puf4 has been shown to influence cell wall synthesis and protein homeostasis, whether it coordinates metabolic adaptation with virulence regulation remains unclear. This study aims to explore potential links between Puf4-mediated metabolic reprogramming and virulence attenuation in C. neoformans, providing initial evidence and theoretical basis for its multifaceted regulatory role. In addition, Oxidative stress and carbohydrate metabolism play crucial roles in the survival and virulence of C. neoformans. The fungus can encounter oxidative stress from host immune cells during infection, and its ability to respond to oxidative stress is closely linked to virulence. Carbohydrate metabolism provides energy and metabolic precursors for the fungus’s growth and proliferation. Studies have shown that changes in carbohydrate metabolism can affect the virulence of C. neoformans, such as the impact of glycolysis on capsule formation and melanin synthesis.
In this study, we constructed a Puf4-overexpressing strain and used RNA sequencing (RNA-seq), biochemical analysis, and in vivo animal experiments to investigate the role of Puf4 in C. neoformans metabolism and virulence. Given the unique regulatory network of Puf4 in C. neoformans—simultaneously influencing energy metabolism, oxidative stress, and virulence factors—it holds promise as a novel target for antifungal drug development, providing new approaches to address the challenge of drug resistance in C. neoformans infections.
2 Materials and methods
2.1 Fungal strains and growth conditions
The wild-type strain H99 was used to generate Puf4-overexpressing mutants. Mutants were generated by homologous recombination and transformed using biolistic transformation as previously described (Toffaletti et al., 1993). The sequences of the primers used are listed in Supplementary Table S1. In order to create a Puf4-overexpressing strain, the GFP open reading frame (primer pair 44/45) was amplified, the PUF4 open reading frame (primer pair 46/47) was amplified, Subsequently, PUF4-GFP fusion sequence was generated using primers 44 and 47, and the resulting polymerase chain reaction (PCR) products were cloned into the PacI site in the pXL-HYG plasmid. The correct plasmid was then transformed into H99, and transformants were analyzed by PCR and quantitative real-time PCR (qRT-PCR). Fungal cells were grown in Yeast Peptone Dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% glucose) at 30°C. The transformant selection was performed using YPD agar supplemented hygromycin B (200 U/mL). The GFP strain was primarily used to verify the successful construction of the Puf4-overexpressing strain and to monitor Puf4 expression levels. The empty vector was introduced into H99 to generate the control strain.
2.2 Spotting assay
Control and Puf4-overexpressing (Puf4 OE) strains were independently grown in YPD liquid medium overnight, then washed twice with phosphate-buffered saline (PBS), Control and Puf4-overexpressing (Puf4 OE) cells were diluted in PBS to an OD600 of 1.0. Ten-fold serial cell dilutions were spotted onto spotted onto YPD agar with or without 0.02% SDS, 0.8% Congo red, 4% H2O2. and YPD agar with 1M Sorb. Plates were incubated at 30°C or 37°C for 2 days before photographs were taken.
2.3 RNA-seq analysis
Control and Puf4 OE strains were independently grown in YPD medium overnight, then inoculated into 50 ml fresh YPD medium at an OD600 of 0.2 and were incubated for additional 4 hr before RNA was isolated, After RNA isolation, the samples for RNA-seq analysis(Personal Biotechnology Co., Ltd. Shanghai, China). RNA-seq was performed by Personal Biotechnology Co., Ltd. (Shanghai, China). Briefly, total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and sequencing was performed on an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Differential gene expression analysis was performed using the DESeq2 software package, and genes with a fold change ≥2 and a p-value <0.05 were considered differentially expressed. Three biological replicates were carried out.
2.4 Total RNA preparation and qRT-PCR analysis
Control and mutant strains were inoculated into 10 mL YPD medium and incubated overnight at 30°C. The samples were diluted to an OD600 of 0.2 with YPD medium and then incubated at 30°C for 4 h. Cells were harvested at 3,000 × g for 5 min at 4°C and then washed twice with ice-cold phosphate-buffered saline (PBS). Cells were lysed using a bead beater, and total RNA was extracted using the E.Z.N.A. Total RNA Kit I (Omega Bio-tek Inc., Norcross, GA, USA),The quality of RNA was assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) by measuring the A260/A280 ratio, which was maintained between 1.8 and 2.0, and cDNA was synthesized using a Reverse Transcription All-in-one Mix (Mona Biotech Co., Ltd., Suzhou, China). The qRT-PCR analysis was performed using a CFX Connect Thermocycler (Bio-Rad Laboratories Inc., Hercules, CA, USA). The qRT-PCR conditions were as follows: initial denaturation at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds and annealing/extension at 55°C for 30 seconds The sequences of primers used are listed in Supplementary Table S1.
2.5 Reactive oxygen species detection
Control and Puf4-overexpressing (Puf4 OE) strains were independently grown in YPD medium overnight, then inoculated into 10 ml fresh YPD medium at an OD600 of 0.2 and shaken in an orbital shaker until OD600 reached 1. Cells were harvested at 3,000 × g for 5 min at 4 °C, washed twice with ice-cold ddH2O, and then disrupted using glass beads. ROS levels were measured using the Reactive Oxygen Species Assay Kit (Beyotime Biotechnology, Shanghai, China) according to manufacturer’s instructions. The luminescent signal was measured using a Synergy H4 microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
2.6 Glycogen staining
Cell were collected at an OD600 of 1 and glycogen was detected by staining with the Periodic Acid-Schiff Staining Kit (Gefan Biotechnology Co. Ltd, Shanghai, China) following the manufacturer’s instructions. Briefly, cells were fixed with Cornoy’s fluid for 15 min, and then washed with distilled water for 1–2 min. Then, carbohydrates were oxidized with periodic acid solution for 5–10 min followed by three 5-min washes with distilled water. Subsequently, cells were stained with Schiff’s reagent for 8 min, then washed three times with distilled water for 5 min each. Finally, slides were prepared and observed under a microscope(Nikon DS-Ri-1, Japan) with a 100× objective lens was used for observation.
2.7 MitoSOX staining
Cell were collected at an OD600 of 1 and glycogen was detected by staining with the MitoSOX™ (Thermo Fisher Scientific Inc., Waltham, MA, USA) following the manufacturer’s instructions. Briefly, wash the cells with PBS and resuspend them in a solution of MitoSOX™ Red at a working concentration of 5 μM. Incubate the cells at 37°C in the dark for 10 minutes to allow the dye to penetrate the cells and specifically stain the mitochondria. After the incubation period, wash the cells with PBS to remove any unbound MitoSOX™ Red. Finally, analyze the fluorescence intensity using flow cytometer.
2.8 PIR-PCR
Yeast cells were cultured in YPD medium to the logarithmic growth phase, typically reaching an OD600 of 0.5–0.8. Crosslinking was performed by adding formaldehyde to a final concentration of 0.3% and incubating at room temperature for 30 minutes. The reaction was quenched by adding glycine, followed by 5 minutes of mixing. Cells were harvested by centrifugation at 3000 rpm for 5 minutes at 4 °C and washed twice with ice-cold PBS. Cell pellets were resuspended in 500 μL of RIPA lysis buffer. The lysate was sonicated using the following parameters: 5% power, 5 seconds on, 5seconds off, for a total of 5 minutes. After sonication, the lysate was centrifuged at 10000 rpm for 10 minutes at 4 °C, and the supernatant was transferred to a fresh tube containing 20 μL of anti-GFP magnetic bead. The mixture was incubated overnight at 4°C. Following incubation, the beads were washed three times with lysis buffer. Subsequently, 100 μL of elution buffer was added, and the mixture was incubated at 65 °C for 50 minutes. RNA was then extracted from the eluate. The purified RNA was subsequently used for cDNA synthesis and qPCR analysis.
2.9 Carbohydrate measurement
Inducing cells using ROS detection method, 2×108 number of cells of the strain were collected and their carbohydrate content was determined using a Total Carbohydrate Assay Kit (Absin Biotechnology, Shanghai, China), following the manufacturer’s instructions. Briefly, after homogenizing with Buffer I, Cell homogenate was boiled at 100°C for 30 min, cooled, and then the Detection Solution II was added. Subsequently, after centrifuging at 8,000 × g for 10 min, the supernatant was collected, and the Dye Detection Solution was added. Then, after heating at 98°C for 10 min, the samples were cooled on ice, and the absorbance was measured at 540 nm.
2.10 Animal infection
Female 4-6-week old C57BL/6 mice were purchased from Liaoning Changsheng Biotech Co., Ltd. (Benxi, China)., and housed at 22°C with a 12 h light-dark cycle. Mice were anesthetized and intranasally administered with 105 fungal cells in 50 μl of PBS. The mice were monitored daily and sacrificed when moribund (defined as hunched posture, minimal motor activity and weight loss of 15% of their initial body weight). For the fungal burden analysis, infected mice at 14 days post infection were killed by exposure to CO2. Lung and brain tissues were collected, weighed, and homogenized in 5 ml of PBS. The suspensions were diluted with PBS, and samples of the lysates were plated onto YPD agar. Fungal colony-forming units (CFU) were counted after incubation at 30°C for 2 days.
2.11 Melanin formation and capsule induction
Melanin was quantified using L-DOPA agar medium. Overnight cell cultures were diluted to an OD600 of 1.0 and spotted onto L-DOPA plates. The plates were incubated at 37°C until black pigment appeared, and photographs were then taken. For the induction of capsule formation, Cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), in an incubator at 37°C and 5% CO2, for 3 days. Following incubation, the cells were harvested and stained with India ink to visualize the capsule formation, Capsule measurement was performed using India ink staining. Cells were stained with India ink and observed under a microscope. The capsule thickness was measured using image analysis software, with at least 100 cells measured per sample to ensure statistical significance.
2.12 Data availability
The transcriptome (RNA-seq) data are deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (PRJNA1267736). Any other data necessary to support the conclusions of this study are available from the authors upon request.
2.13 Quantification and statistical analyses
All statistical analyses were performed using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA). Shpiro-Wilk test was used to assess the normality of the data. We have specified which statistical methods were used for which data, including the use of non-parametric statistical methods for non-normally distributed data The statistical test Kaplan Meier was used for the animal survival tests. Statistically significant differences between the two groups were determined using an unpaired two-tailed Student’s t-test. Statistical analyses across two or more groups were performed using one-way analysis of variance (ANOVA) test or two-way ANOVA test. Significant differences were defined as p < 0.05. p values are indicated in the plots or represented as *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001. All experiments were performed using three biological replicates to ensure the reproducibility.
3 Results
3.1 Puf4 affects cellular stress responses and membrane homeostasis
In order to explore the role of Puf4 in the environmental adaptation of C. neoformans, we constructed a Puf4-overexpressing strain (Puf4 OE). After evaluating its growth capacity under various stressful conditions, we found that under SDS and Congo red treatment, the growth of Puf4 OE cells was weaker than that of the control cells, which indicates that the cell membrane integrity may be compromised. In addition, under hydrogen peroxide treatment and at high temperatures of 37°C, the growth rate of the Puf4 OE strain was lower than that of the control cells, which implies that the antioxidant capacity and thermostress tolerance of the Puf4 OE strain were reduced (Figure 1A).To verify whether the above phenotypes are related to cell wall stress, we added 1 M Sorbitol (Sorb) to the medium to maintain osmotic pressure. The results showed that sorbitol treatment could partially restore the growth defect of the Puf4 OE strain under high temperature conditions (Figure 1B). To sum up, overexpression of Puf4 may disrupt the homeostasis of the cell membrane and cell wall, thereby reducing the tolerance of C. neoformans to chemical and thermal stresses. At the same time, these results also suggest that Puf4 plays a key role in the antioxidant stress response.
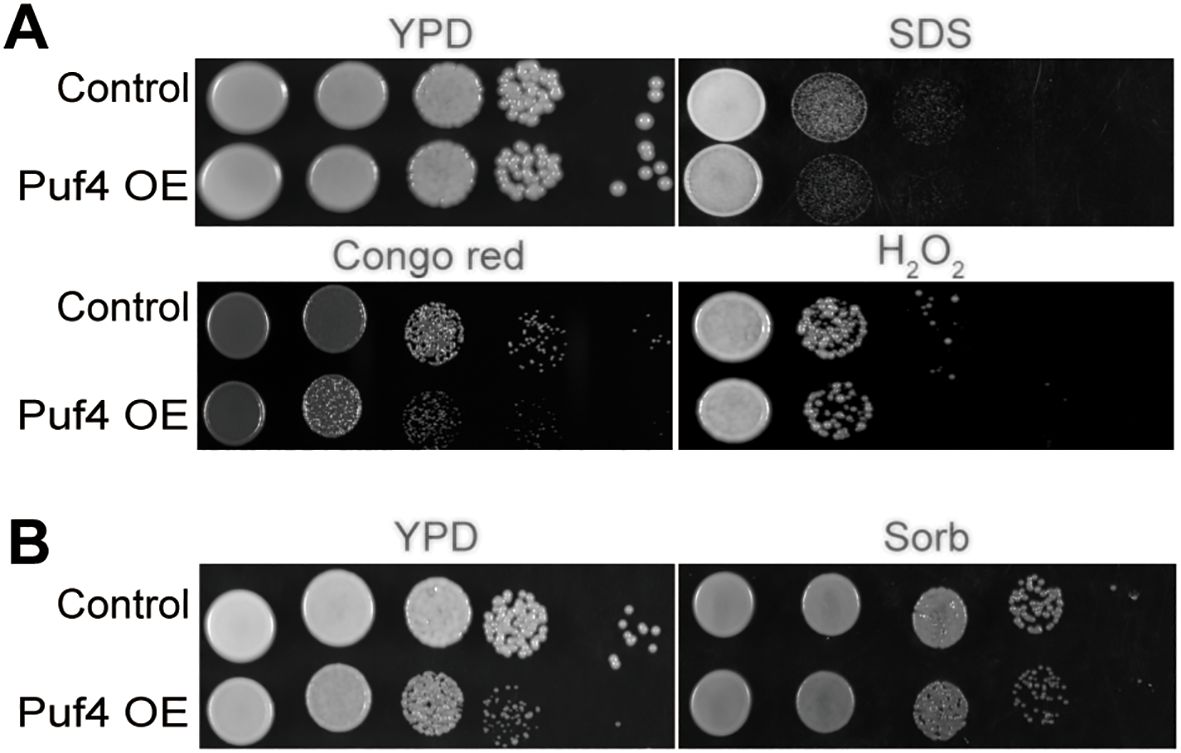
Figure 1. Overexpression of Puf4 damages cell membrane integrity and reduces oxidative stress tolerance. (A) Spotting assay of the Puf4 overexpressing strain. This strain was spotted onto YPD agar with or without 0.02% SDS, 0.8% Congo red and 4% H2O2, then was incubated at 30°C for 2 days. (B) Treatment of Puf4 OE with Sorb. Control and Puf4 OE cells were spotted onto YPD agar with or without 1M Sorb, then were incubated at 37°C for 2 days.
3.2 Puf4 overexpression alters metabolism and redox state
RNA-seq analysis identified 517 differentially expressed genes (DEGs) in the Puf4 OE strain compared to the control, which were grouped into 9 gene clusters (149 upregulated and 368 downregulated) (Figures 2A, B). Gene Ontology (GO) term enrichment analysis revealed that Puf4 primarily regulates processes such as carbohydrate metabolism, ribosomal metabolism, and oxidoreductase and peroxidase activities (Figure 2C). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that DEGs in the Puf4 OE strain were significantly enriched in carbohydrate metabolism pathways, including glycolysis, sucrose metabolism, and pentose and glucuronate interconversions (Figure 2D). The results of gene set enrichment analysis (GSEA) further supported the role of Puf4 in regulating carbohydrate metabolism and glycolysis (Figures 2E, F), suggesting that Puf4 Overexpression Alters Carbon Metabolism and Redox State.
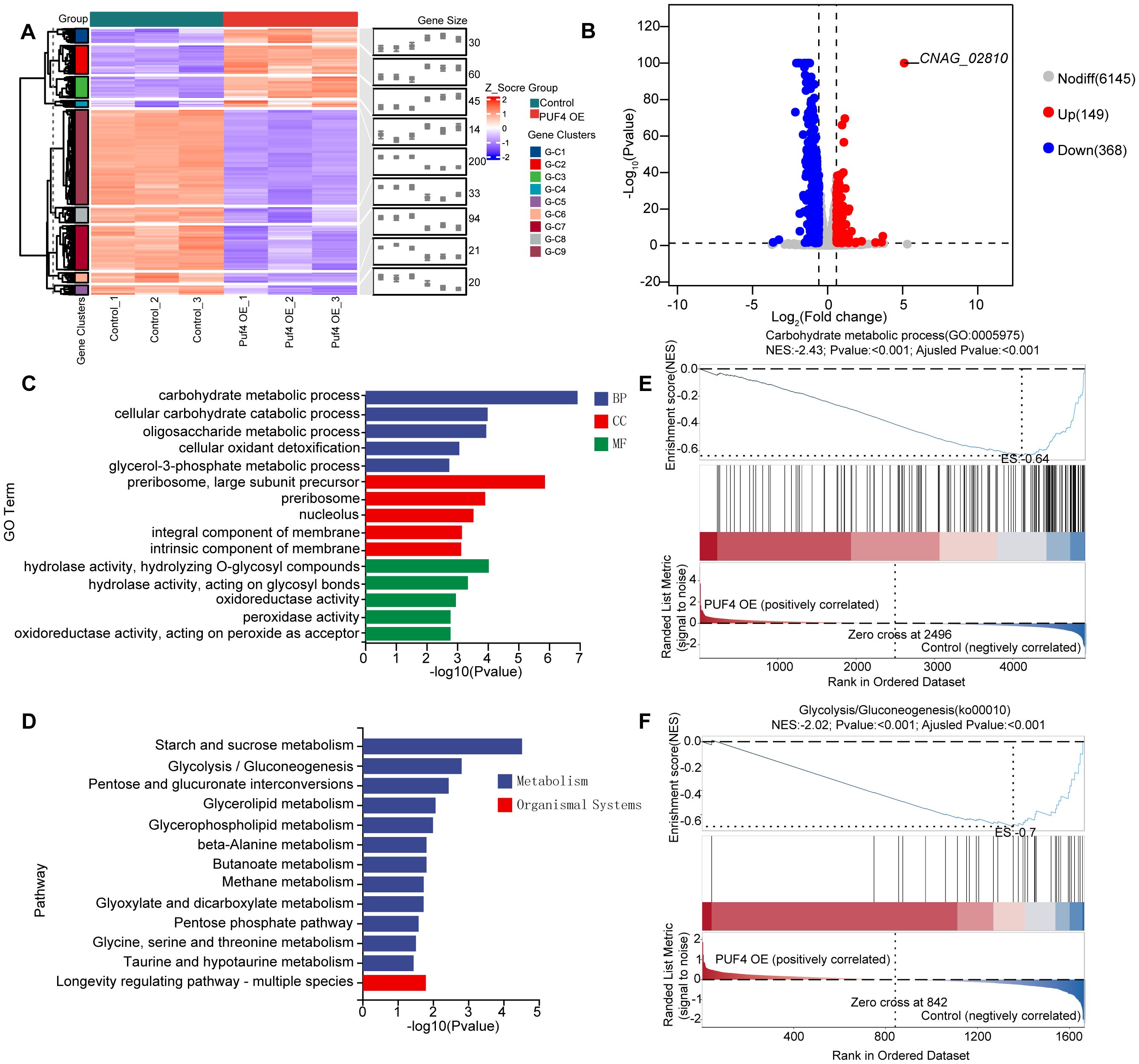
Figure 2. RNA-seq Analysis and Functional Enrichment of Puf4 OE Strains. (A) Clustered heatmap of DEGs. comparing the transcriptome data from the Control and Puf4 OE strains, Clustered heatmap of differentially expressed genes (DEGs) in the Puf4-overexpressing strain compared to the control strain. The heatmap was generated based on the expression levels of DEGs, with red indicating upregulated genes and blue indicating downregulated genes. The DEGs were grouped into 9 gene clusters, with 149 upregulated genes and 368 downregulated genes. (B) Volcano plot of DEGs. genes with p-values less than 0.05 and fold changes greater than 1.5 or less than 0.667 were considered to be differentially expressed. (C) Gene ontology (GO). GO term enrichment analyses were performed using the RNA-seq data from Control and Puf4 OE strains. (D) Kyoto Encyclopedia of Genes and Genomes (KEGG). KEGG pathway enrichment analyses were performed using RNA-seq data from Control and Puf4 OE strains. (E, F) Gene Set Enrichment Analysis (GSEA). comparing the transcriptome data from the Control and Puf4 OE strains.
3.3 Puf4 promotes carbohydrate accumulation and reduces oxidative stress efficiency
Biochemical analysis revealed that total carbohydrate content and glycogen reserves increased by 40 and 50%, respectively, in the Puf4 OE strain compared to the control cells (Figures 3A-C). The expression of the key glycolytic gene HXK1 was significantly downregulated (Figure 3D), the downregulation of HXK1 expression in Puf4 OE strains, along with increased glycogen accumulation, suggests a potential shift in carbon flux from glycolysis toward energy storage pathways. RNA-binding proteins primarily regulate RNA activity by interacting with the untranslated regions (UTRs) of target transcripts. To investigate this mechanism, we performed RNA immunoprecipitation (RIP) followed by qPCR. Our results showed that the UTR regions of HXK1 mRNA, along with those of other top differentially expressed genes, were significantly enriched for Puf4 binding (Figure 3E). Energy metabolism assays revealed a 10 - fold increase in ROS levels in the Puf4 OE strain (Figure 3G), MitoSOX staining also yielded consistent results (Figure 3F). Meanwhile, the Puf4 OE strain displayed enhanced sensitivity to the respiratory chain inhibitor antimycin A (Figure 3H). Although increased ROS levels and antimycin A sensitivity imply mitochondrial dysfunction, the specific targets or pathways through which Puf4 mediates this effect remain to be determined.
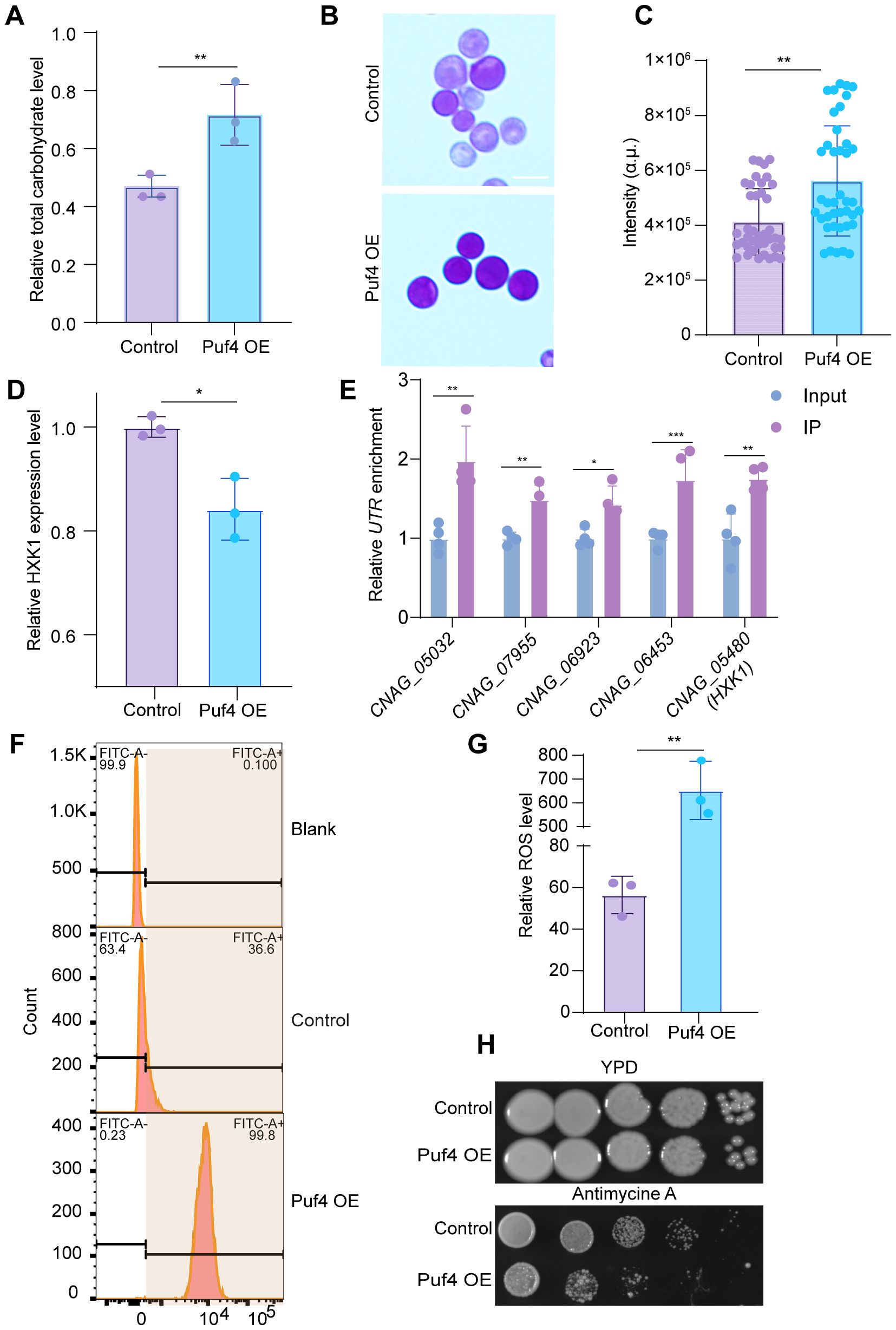
Figure 3. Puf4 Overexpression reprograms C. neoformans metabolism and energy generation pathways. (A) Quantification of Total carbohydrate. Control and Puf4 OE cells were grown at 30°C for 12 h and subsequently used for the determination of total carbohydrate content. (B) Glycogen staining detection. glycogen reserve content determination in Puf4 OE and Control strains. Bar = 5 μm. (C) Quantification of glycogen formation. glycogen formation from Figure 2B was measured and plotted; at least 50 cells were measured and quantified. (D) Analysist of HXK1 gene expression level in Puf4 OE strains. measurements were performed using RNA samples from Puf4 OE cells and Control cells (n = 3). (E) RNA-immunoprecipitation (RNA-IP) PCR analyses. Cell lyses (n=4) were isolated from the Puf4 OE strain, and RNA-IP-PCR was performed to determine the binding enrichment of Puf4 at the UTRs. (F) Cell population analysis of MitoSOX-stained Puf4 OE strains. Wild type and Puf4 OE strains were incubated at 30°C for 30 min, then MitoSOXbased flow cytometry was performed. (G) Analysis of ROS levels. Puf4 OE and Control strains were incubated at 30°C for 12 h, then ROS levels were measured. (H) Spotting assay of the Puf4 OE strain. this strain was spotted onto YPD agar plates with or without 15 μM antimycin A, and then incubated at 30°C for 2 days. *p < 0.05, **p < 0.01 (Two-tailed unpaired t tests were used for statistical analyses.). Data are expressed as the mean ± SD.
3.4 Puf4 OE reduces the virulence of C. neoformans
In vitro assays showed that the capsule area of the Puf4 OE strain was reduced by approximately 50% compared to the control cells, along with reduced melanin formation (Figures 4A, B). The qRT-PCR analysis revealed that the mRNA levels of the capsule synthesis gene CAP10 and the melanin synthesis gene LAC1 were downregulated by 20 and 40%, respectively (Figure 4C), suggesting that Puf4 reduces the pathogenic potential of C. neoformans by suppressing virulence factor-related genes. In a mouse model of intravenous infection, the survival rate of mice infected with the Puf4 OE strain was significantly higher than that of the control cells (Figure 4D). By day 14 post-infection, the CFUs in the lung and brain tissues of mice in the Puf4 OE group were reduced compared to the control group (Figures 4E, F). Histological staining further confirmed that the fungal burden in the lung tissue of mice in the Puf4 OE group was significantly lower than that in the control cells (Figure 4G), indicating that Puf4 OE significantly reduces the virulence of C. neoformans in vivo, which is consistent with the in vitro virulence factor phenotypes. These findings indicate that overexpression of Puf4 attenuates the virulence of C. neoformans.
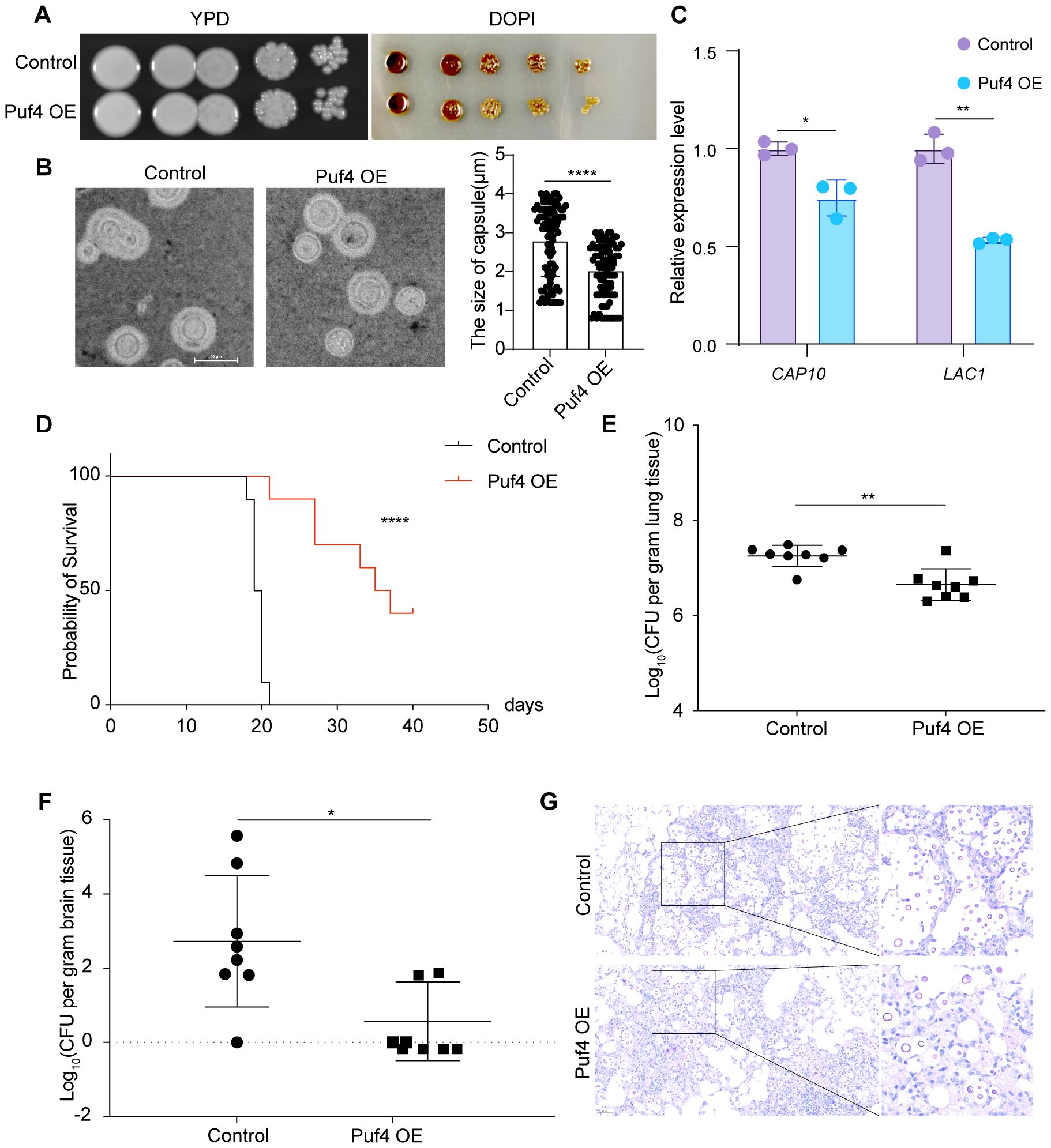
Figure 4. Puf4 Overexpression reduces C. neoformans virulence. (A) Melanin formation in Control and Puf4 OE strains. Overnight cell cultures were diluted to an OD600 of 1.0, and then spotted onto L-DOPA plates, melanin production was induced and photographs were taken after three days of incubation. (B) Comparison of capsule size between the Control and Puf4 OE strains. Capsule formation was induced in DMEM supplemented with 10% FBS, and cells were imaged using light microscopy, scale bar = 10 μm, capsule thickness was measured and plotted, at least 100 cells were measured and quantified. (C) Analysis of the mRNA expression levels of virulence-associated genes by qRT-PCR. mRNA samples (n = 3) were isolated from the indicated strains, and cDNAs were synthesized And used for qRT-PCR analysis (n = 3) of indicated genes. (D) Survival analysis of the Control and Puf4 OE strains. overnight Control and Puf4 OE cell cultures were washed twice with PBS and diluted in YPD medium. Then, mice were infected with a PBS suspension containing 105 cells of the Control or Puf4 OE strain and animal survival rates were recorded. (n = 10) (E) Fungal burden analysis in the lungs. mice were infected with Control and Puf4 OE strains as described for (D). Then, 2 weeks days post infection, mice were killed, and CFUs from lung tissues were counted. (n = 8) (F) Brain fungal burden analysis. fungal burden tests were performed as described in (E). (G) Histopathological analysis of C. neoformans-infected lung tissues. lung tissues from Control and Puf4 OE infected mice were isolated, fixed, and stained with mucicarmine or hematoxylin and eosin (H&E) and examined under a microscope; scale bar: 50 µm; *p < 0.05, **p < 0.01, ****p < 0.0001 (Two-tailed unpaired t tests were used for statistical analyses.); data are expressed as the mean ± SD.
4 Discussion
This study delves into the role of Puf4 in the metabolism and virulence of C. neoformans, revealing its significant impact on cell wall integrity, antioxidant stress capacity, and key metabolic pathways. The research shows that Puf4 influences carbon metabolism, oxidative phosphorylation, sucrose metabolism, and glycolysis. Moreover, Puf4 overexpression leads to substantial increases in total carbohydrate and glycogen content, highlighting its key role in regulating intracellular carbohydrate metabolism. Our findings collectively indicate that Puf4 acts as a pivotal regulator linking cellular metabolism to pathogenic adaptation, thereby providing a novel understanding of cryptococcal pathogenesis.
Compared to Puf4 knockout strains, Puf4 - overexpressing strains exhibit distinct differences in cell membrane integrity, antioxidant capacity, and carbon metabolism. Puf4 knockout strains show reduced cell wall β-glucan content and increased echinocandin drug resistance, whereas Puf4 - overexpressing strains display compromised cell membrane integrity, weakened antioxidant capacity, and reprogrammed carbohydrate metabolism. These differences likely reflect the dose - dependent role of Puf4 in regulating C. neoformans metabolism and virulence. Carbohydrate metabolism is crucial for maintaining cell wall integrity. The results of this study show that Puf4 overexpression leads to increased total carbohydrate and glycogen content, which may affect the synthesis and composition of cell wall components. This could, in turn, impact cell wall integrity and the fungus’s ability to withstand external stress. Cell wall integrity is closely related to the virulence of C. neoformans. A compromised cell wall can affect the fungus’s ability to adhere to host cells and resist host immune defenses. The reduced cell wall integrity caused by Puf4 overexpression may decrease the virulence of C. neoformans, making it easier for the host immune system to clear the fungus. However, the specific mechanisms by which Puf4 regulates these processes and the exact impact of these regulations on pathogenicity remain to be further investigated in future studies.
RNA-seq and biochemical analyses revealed that Puf4 overexpression substantially reprograms carbohydrate metabolism and decreases oxidative stress tolerance in C. neoformans. Specifically, total carbohydrate and glycogen levels, ROS production are increased, and resistance to oxidative stress are significantly decreased. These findings indicate that Puf4 redirects carbon flux toward energy storage by inhibiting glycolysis and promoting glycogen synthesis, mechanistically, The observed downregulation of HXK1 suggests that Puf4 may indirectly influence carbon flux through modulation of glycolytic gene expression, although direct regulatory mechanisms remain to be determined. In addition, decreased oxidative phosphorylation efficiency—as evidenced by elevated ROS levels and reduced antimycin A resistance—suggests that Puf4 may regulate mitochondrial complex III assembly genes (e.g., QCR2, QCR9), changing the electron transport chain function. Mitochondria, as energy - producing and oxidative - stress - related organelles, may have their functions affected by Puf4, potentially disrupting redox balance and impacting C. neoformans normal physiology. These findings offer new insights into C. neoformans metabolic regulation and oxidative - stress response in diverse environments.
Puf4 overexpression led to decreased capsule size and melanin production, ultimately weakening its virulence, which may result from reprogramming of metabolic pathways and oxidative stress response. These effects may arise from multiple interconnected mechanisms. First, Puf4 may modulate metabolic allocation in response to stress, potentially prioritizing survival over virulence. Alternatively, Puf4 may influence virulence-related gene expression via post-transcriptional mechanisms unrelated to metabolic flux, such as mRNA stability or translation efficiency. Future studies using RNA immunoprecipitation (RIP-seq) are needed to determine whether CAP10 or LAC1 mRNAs are direct targets of Puf4. Second, the observed “carbon flux bias toward storage” may directly suppress capsular polysaccharide synthesis. Glycogen accumulation consumes metabolic intermediates (e.g., UDP-glucose) that are critical precursors for capsule biosynthesis (Wolfe et al., 2021), thereby limiting the availability of resources for virulence factor production. Third, elevated ROS levels in the Puf4 OE strain may inhibit melanin synthesis by destabilizing LAC1 mRNA or impairing Lac1 enzyme activity. ROS-mediated post-transcriptional regulation of virulence genes is well-documented (Lee et al., 2013; Samarasinghe et al., 2021; Xue et al., 2024); for instance, in Candida albicans, oxidative stress suppresses hyphal formation by downregulating EFG1, a key transcriptional regulator of morphogenesis (Glazier, 2022, Prasad et al., 2010). Additionally, ROS can directly oxidize phenolic substrates like L-DOPA, reducing melanin polymerization efficiency (Lee et al., 2013; Samarasinghe et al., 2021; Xue et al., 2024).
Although our data reveals that Puf4 plays a key regulatory role in C. neoformans metabolism and virulence, it remains unclear whether the metabolic pathways altered by Puf4 overexpression are truly downregulated or modulated during host infection. Future research needs to further explore the dynamic effects of Puf4 on metabolic pathways during host infection and how these metabolic changes interact with the host immune response to influence the pathogenic process of C. neoformans. Future studies could further explore the specific molecular mechanisms by which Puf4 regulates metabolic reprogramming and virulence in C. neoformans. This could involve identifying the direct mRNA targets of Puf4 using RNA immunoprecipitation sequencing (RIP-seq) technology and investigating the role of Puf4 in regulating metabolic flux. Additionally, the potential of Puf4 as a target for antifungal drug development could be explored to provide new strategies for the treatment of C. neoformans infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Animal Welfare & Ethics Committee of Northern Theater Command General Hospital (2025-47). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CS: Project administration, Visualization, Methodology, Conceptualization, Funding acquisition, Validation, Formal Analysis, Writing – original draft, Supervision, Software, Data curation, Resources, Investigation. JL: Writing – review & editing, Conceptualization, Writing – original draft, Data curation. WZ: Writing – review & editing, Writing – original draft. LJ: Writing – review & editing, Data curation, Methodology. HZ: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the Liaoning Province Science and Technology Plan Joint Project (2023JH2/101700140 to C.S).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1628448/full#supplementary-material
References
Dallastella, M., De Oliveira, W. K., Rodrigues, M. L., Goldenberg, S., and Alves, L. R. (2023). The characterization of RNA-binding proteins and RNA metabolism-related proteins in fungal extracellular vesicles. Front. Cell Infect. Microbiol. 13, 1247329. doi: 10.3389/fcimb.2023.1247329, PMID: 37780856
Denham, S. T., Brammer, B., Chung, K. Y., Wambaugh, M. A., Bednarek, J. M., Guo, L., et al. (2022). A dissemination-prone morphotype enhances extrapulmonary organ entry by Cryptococcus neoformans. Cell Host Microbe 30, 1382–1400 e1388. doi: 10.1016/j.chom.2022.08.017, PMID: 36099922
El Helou, G. and Hellinger, W. (2019). Cryptococcus neoformans pericarditis in a lung transplant recipient: Case report, literature review and pearls. Transpl Infect. Dis. 21, e13137. doi: 10.1111/tid.13137, PMID: 31267603
Fang, W., Chen, M., Liu, J., Hagen, F., Al-Hatmi, M. A., Zhang, P., et al. (2016). Cryptococcal meningitis in systemic lupus erythematosus patients: pooled analysis and systematic review. Emerg. Microbes Infect. 5, e95. doi: 10.1038/emi.2016.93, PMID: 27599471
Fischer, A. D. and Olivas, W. M. (2018). Multiple Puf proteins regulate the stability of ribosome biogenesis transcripts. RNA Biol. 15, 1228–1243. doi: 10.1080/15476286.2018.1521211, PMID: 30251908
Gao, X., Fu, Y., Sun, S., Gu, T., Li, Y., Sun, T., et al. (2022). Cryptococcal Hsf3 controls intramitochondrial ROS homeostasis by regulating the respiratory process. Nat. Commun. 13, 5407. doi: 10.1038/s41467-022-33168-1, PMID: 36109512
Glazier, V. E. (2022). EFG1, everyone's favorite gene in candida albicans: A comprehensive literature review. Front. Cell Infect. Microbiol. 12, 855229. doi: 10.3389/fcimb.2022.855229, PMID: 35392604
Glazier, V. E., Kaur, J. N., Brown, N. T., Rivera, A. A., and Panepinto, J. C. (2015). Puf4 regulates both splicing and decay of HXL1 mRNA encoding the unfolded protein response transcription factor in Cryptococcus neoformans. Eukaryot Cell 14, 385–395. doi: 10.1128/EC.00273-14, PMID: 25681267
Kaeberlein, M. and Guarente, L. (2002). Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160, 83–95. doi: 10.1093/genetics/160.1.83, PMID: 11805047
Kalem, M. C., Subbiah, H., Leipheimer, J., et al. (2021). Puf4 mediates post-transcriptional regulation of cell wall biosynthesis and caspofungin resistance in cryptococcus neoformans. mBio 12. doi: 10.1128/mBio.03225-20, PMID: 33436441
Krusche, J., Beck, C., Lehmann, E., Gerlach, D., Daiber, E., Mayer, C., et al. (2025). Characterization and host range prediction of Staphylococcus aureus phages through receptor-binding protein analysis. Cell Rep. 44, 115369. doi: 10.1016/j.celrep.2025.115369, PMID: 40022731
Lee, I. R., Lui, E. Y., Chow, E. W., Arras, S. D., Morrow, C. A., and Fraser, J. A. (2013). Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics 194, 421–433. doi: 10.1534/genetics.113.150326, PMID: 23564202
Prasad, T., Hameed, S., Manoharlal, R., Biswas, S., Mukhopadhyay, C. K., Goswami, S. K., et al. (2010). Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res. 10, 587–596. doi: 10.1111/j.1567-1364.2010.00639.x, PMID: 20491944
Rajasingham, R., Govender, N. P., Jordan, A., Loyse, A., Shroufi, A., Denning, D. W., et al. (2022). The global burden of HIV-associated cryptococcal infection in adults in 2020: a modelling analysis. Lancet Infect. Dis. 22, 1748–1755. doi: 10.1016/S1473-3099(22)00499-6. Erratum in: Lancet Infect Dis. (2023) 23(1):e1. doi: 10.1016/S1473-3099(22)00597-7, PMID: 36049486
Russo, J. and Olivas, W. M. (2015). Conditional regulation of Puf1p, Puf4p, and Puf5p activity alters YHB1 mRNA stability for a rapid response to toxic nitric oxide stress in yeast. Mol. Biol. Cell 26, 1015–1029. doi: 10.1091/mbc.E14-10-1452, PMID: 25631823
Sadee, C., Hagler, L. D., Becker, W. R., Jarmoskaite, I., Vaidyanathan, P. P., Denny, S. K., et al. (2022). A comprehensive thermodynamic model for RNA binding by the Saccharomyces cerevisiae Pumilio protein PUF4. Nat. Commun. 13, 4522. doi: 10.1038/s41467-022-31968-z, PMID: 35927243
Samarasinghe, H., Aceituno-Caicedo, D., Cogliati, M., Kwon-Chung, K. J., Rickerts, V., Velegraki, A., et al. (2021). Author correction: genetic factors and genotype-environment interactions contribute to variation in melanin production in the fungal pathogen cryptococcus neoformans. Sci. Rep. 11, 12860. doi: 10.1038/s41598-021-91476-w, PMID: 34127698
Sankaranarayanan, S., Haag, C., Petzsch, P., Köhrer, K., Matuszyńska, A., Zarnack, K., et al. (2023). The mRNA stability factor Khd4 defines a specific mRNA regulon for membrane trafficking in the pathogen Ustilago maydis. Proc. Natl. Acad. Sci. U.S.A. 120, e2301731120. doi: 10.1073/pnas.2301731120, PMID: 37590419
Son, S. H., Jang, S. Y., and Park, H. S. (2021). Functions of PUF family RNA-binding proteins in aspergillus nidulans. J. Microbiol. Biotechnol. 31, 676–685. doi: 10.4014/jmb.2101.01011, PMID: 33746193
Song, Z., Zhou, S., Zhang, H., et al. (2023). Fungal secondary metabolism is governed by an RNA-binding protein CsdA/RsdA complex. Nat. Commun. 14, 7351. doi: 10.1038/s41467-023-43205-2, PMID: 37963872
Spassov, D. S. and Jurecic, R. (2003). The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55, 359–366. doi: 10.1080/15216540310001603093, PMID: 14584586
Stott, K. E., Loyse, A., Jarvis, J. N., et al. (2021). Cryptococcal meningoencephalitis: time for action. Lancet Infect. Dis. 21, e259–e271. doi: 10.1016/S1473-3099(20)30771-4, PMID: 33872594
Tam, P. P., Barrette-Ng, I. H., Simon, D. M., et al. (2010). The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10, 44. doi: 10.1186/1471-2229-10-44, PMID: 20214804
Toffaletti, D. L., Rude, T. H., Johnston, S. A., et al. (1993). Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol 175, 1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993, PMID: 8444802
Wake, R. M., Molloy, S. F., Jarvis, J. N., et al. (2023). Cryptococcal antigenemia in advanced human immunodeficiency virus disease: pathophysiology, epidemiology, and clinical implications. Clin. Infect. Dis. 76, 764–770. doi: 10.1093/cid/ciac675, PMID: 35986670
Wang, M., Oge, L., Perez-Garcia, M. D., et al. (2018). The PUF protein family: overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19020410, PMID: 29385744
Wickens, M., Bernstein, D. S., Kimble, J., et al. (2002). A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 18, 150–157. doi: 10.1016/S0168-9525(01)02616-6, PMID: 11858839
Wilinski, D., Qiu, C., Lapointe, C. P., et al. (2015). RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 6, 8213. doi: 10.1038/ncomms9213, PMID: 26364903
Wolfe, A. L., Zhou, Q., Toska, E., et al. (2021). UDP-glucose pyrophosphorylase 2, a regulator of glycogen synthesis and glycosylation, is critical for pancreatic cancer growth. Proc. Natl. Acad. Sci. U.S.A. 118. doi: 10.1073/pnas.2103592118, PMID: 34330832
Xue, P., Sanchez-Leon, E., Hu, G., et al. (2024). The interplay between electron transport chain function and iron regulatory factors influences melanin formation in Cryptococcus neoformans. mSphere 9, e0025024. doi: 10.1128/msphere.00250-24, PMID: 38687055
Zhang, J., Cui, W., Abdul Haseeb, H., et al. (2020). VdNop12, containing two tandem RNA recognition motif domains, is a crucial factor for pathogenicity and cold adaption in Verticillium dahliae. Environ. Microbiol. 22, 5387–5401. doi: 10.1111/1462-2920.15268, PMID: 33000558
Zhao, Y. and Lin, X. (2021). Cryptococcus neoformans: Sex, morphogenesis, and virulence. Infect. Genet. Evol. 89, 104731. doi: 10.1016/j.meegid.2021.104731, PMID: 33497839
Keywords: Cryptococcus neoformans, RNA-binding protein, carbohydrate metabolism, oxidative stress, fungal virulence
Citation: Suo C, Lei J, Zhang W, Jia L and Zhang H (2025) Puf4 -mediated oxidative stress virulence attenuation in Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 15:1628448. doi: 10.3389/fcimb.2025.1628448
Received: 14 May 2025; Accepted: 14 July 2025;
Published: 11 August 2025.
Edited by:
Yuanwei Zhang, Nanjing Normal University, ChinaReviewed by:
Marcos Sergio Toledo, Federal University of São Paulo, BrazilRita Silva-Gomes, University of Minho, Portugal
Rosanna Baker, Johns Hopkins University, United States
Copyright © 2025 Suo, Lei, Zhang, Jia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: He Zhang, YWx3YXlzemhoQDE2My5jb20=; Lihui Jia, NDE4MjExNzg2QHFxLmNvbQ==; Wanli Zhang, MjMwMTUzOUBzdHUubmV1LmVkdS5jbg==
 Chenhao Suo
Chenhao Suo Jianjun Lei1
Jianjun Lei1 He Zhang
He Zhang