- Capital University of Physical Education and Sports, Beijing, China
Objective: This systematic review aimed to examine the relationship between the oral microbiota and the onset and progression of type 2 diabetes mellitus (T2DM).
Methods: A systematic review was conducted in accordance with PRISMA guidelines. Three independent reviewers searched relevant literature across multiple databases, including PubMed/Medline, Web of Science, and Scopus, covering publications from April 2000 to April 2025.
Results: A total of 1,438 publications were initially identified, of which 34 studies met the inclusion criteria after screening, namely 23 cross-sectional studies and 11 case-control studies. These studies involved 2,062 patients with T2DM and 1,445 non-diabetic controls. All included studies reported a correlation or potential association between the oral microbiota and T2DM. Fifteen studies analyzed alpha diversity, revealing heterogeneous findings: three reported increased diversity in T2DM patients, two reported decreased diversity, and the remainder showed either no significant differences or inconsistent trends. At the phylum level, Firmicutes was consistently elevated in T2DM patients (14 studies), whereas Proteobacteria was often reduced, and findings on Bacteroidetes varied. At the genus level, Streptococcus, Porphyromonas, and Treponema were most frequently enriched in T2DM populations, with Streptococcus significantly elevated in 22 studies. Notably, Porphyromonas gingivalis was repeatedly identified as a potential contributor to systemic inflammation and insulin resistance, indicating a potential pathogenic role in the metabolic dysregulation of T2DM. Species-level analyses further revealed increased abundance of Streptococcus mutans, P. gingivalis, and T. denticola, supporting the hypothesis that oral dysbiosis is linked to T2DM pathogenesis.
Conclusion: There is a significant association between oral microbiota composition and T2DM. These findings highlight the potential importance of oral health monitoring as part of preventive and therapeutic strategies in the management of T2DM.
1 Introduction
Diabetes mellitus (DM) is a chronic metabolic disorder characterized by persistent hyperglycemia resulting from insufficient insulin secretion or impaired insulin action, often accompanied by disturbances in glucose, lipid, and protein metabolism (Dale Abel et al., 2024; Yameny, 2024). According to the latest report by the International Diabetes Federation (IDF), 537 million adults worldwide are living with diabetes, a number projected to rise to 783 million by 2045. Type 2 diabetes mellitus (T2DM) is the most prevalent form, accounting for over 90% of all diabetes cases (Schulze and Hu, 2022).T2DM is closely related to obesity (Chandrasekaran and Weiskirchen, 2024), insulin resistance (IR) (Penno et al., 2021), chronic inflammation (Rohm et al., 2022), and oxidative stress (Andreadi et al., 2022; Caturano et al., 2023), which, if not effectively controlled, can lead to complications such as cardiovascular and cerebrovascular diseases, nephropathy, retinopathy, and neuropathy, posing a serious threat to human health (Ali et al., 2022).
The human microbiota plays a significant role in the development and progression of T2DM, involving not only changes in the composition and function of the gut microbiota (Zhou et al., 2022) but also dysbiosis of the oral microbiome (Kumari and Gnanasundaram, 2021). The oral microbiota, as the second most abundant and diverse microbial community in the human body after the gut, comprises approximately 700 microbial species and forms a complex ecological network (Caselli et al., 2020). As a major gateway to the body, the oral microbiota influences both local and systemic health. Its dysbiosis may provoke oral inflammation, compromise mucosal barriers, and allow microbial products into circulation, fueling chronic inflammation and immune imbalance that contribute to diabetes and its complications (Suarez et al., 2020).
In recent years, an increasing number of studies have focused on the association between oral microbiota and diabetes mellitus (Kumari and Gnanasundaram, 2021). For instance, endotoxemia caused by Porphyromonas gingivalis infection has been shown to significantly increase the risk of insulin resistance and diabetes in animal models (Li et al., 2024), suggesting that oral microbiota dysbiosis may directly contribute to the development of diabetes. Additionally, several epidemiological studies have demonstrated that the composition of the oral microbiota is closely associated with glycemic control and systemic inflammation in patients with diabetes (Negrini et al., 2021; Zeng et al., 2024). These microbial profiles are further influenced by lifestyle factors, including diet, smoking, oral hygiene practices, and metabolic status (Shaalan et al., 2022; Mohammed et al., 2024).
Given the complex and dynamic nature of the oral microbiota under diabetic conditions, a systematic review is needed to summarize current evidence. This review investigates differences in oral microbiota composition and diversity in individuals with T2DM and explores potential mechanisms by which oral dysbiosis may influence disease onset and progression.
2 Materials and methods
2.1 Protocol and registration
This systematic review was registered with the International Prospective Register of Systematic Reviews and reported in accordance with the PRISMA statement (Stewart et al., 2015; Chandler et al., 2019)(http://www.crd.york.ac.uk/prospero/, registration number:CRD420251053253).
2.2 Eligibility criteria
The literature search strategy was based on the PICOS framework. Population (P): adult individuals diagnosed with T2DM. Intervention (I): assessment of oral microbiota composition in individuals with T2DM. Comparison (C): adult individuals without T2DM. Outcome (O): the association between oral microbiota composition and the presence of T2DM. Study design (S): observational studies examining the association between oral microbiota and T2DM prevalence, including case-control, cohort, and cross-sectional studies. Exclusion criteria: reviews, conference abstracts, case reports, and other publication types that did not provide original data suitable for analyzing the relationship between oral microbiota and T2DM.
2.3 Information sources and search strategy
Three independent researchers searched PubMed/MEDLINE, Web of Science, Scopus, and the Cochrane Library using the following keywords: “oral microorganism,” “oral microbiota,” “oral microbiome,” “diabetes,” “saliva microbiota,” “type 2 diabetes,” “cross-sectional study,” and “cohort study.” The search was performed up to April 10, 2025. To ensure research quality, all studies were independently screened and extracted by two reviewers, with disagreements resolved through discussion or adjudication by a third reviewer. The search strategy is shown in Table 1.
2.4 Data collection process
The included studies were analyzed, and data were extracted by two independent researchers. After removing duplicates from the literature search, the selected studies were imported into NoteExpress software. The first round of screening was performed by reviewing titles and abstracts, followed by a full-text review to complete the second round. Extracted data included the first author’s name, country, journal, publication year, sample size, patient age, oral diagnosis, microbiome analysis type, sample extraction, detection methods, and key findings. In cases of disagreement, a third researcher was involved in the decision-making process.
2.5 Quality assessment of included studies
The quality of the included studies was assessed using the Joanna Briggs Institute (JBI) critical appraisal checklist, a validated tool for evaluating methodological quality across various study types, including observational and cross-sectional designs. This checklist examines key aspects such as study design relevance, sample selection and size, representativeness, and clarity of data collection procedures.
We also assessed the use of validated measurement tools, potential biases and confounding factors, and the strategies used to control them. The appropriateness and transparency of statistical analyses were evaluated, along with whether ethical approval and informed consent procedures were clearly reported. Based on the number of criteria met, each study was rated as low, moderate, or high quality. These ratings informed the overall strength of the evidence and guided the interpretation of the findings in this review.
3 Results
3.1 Literature search
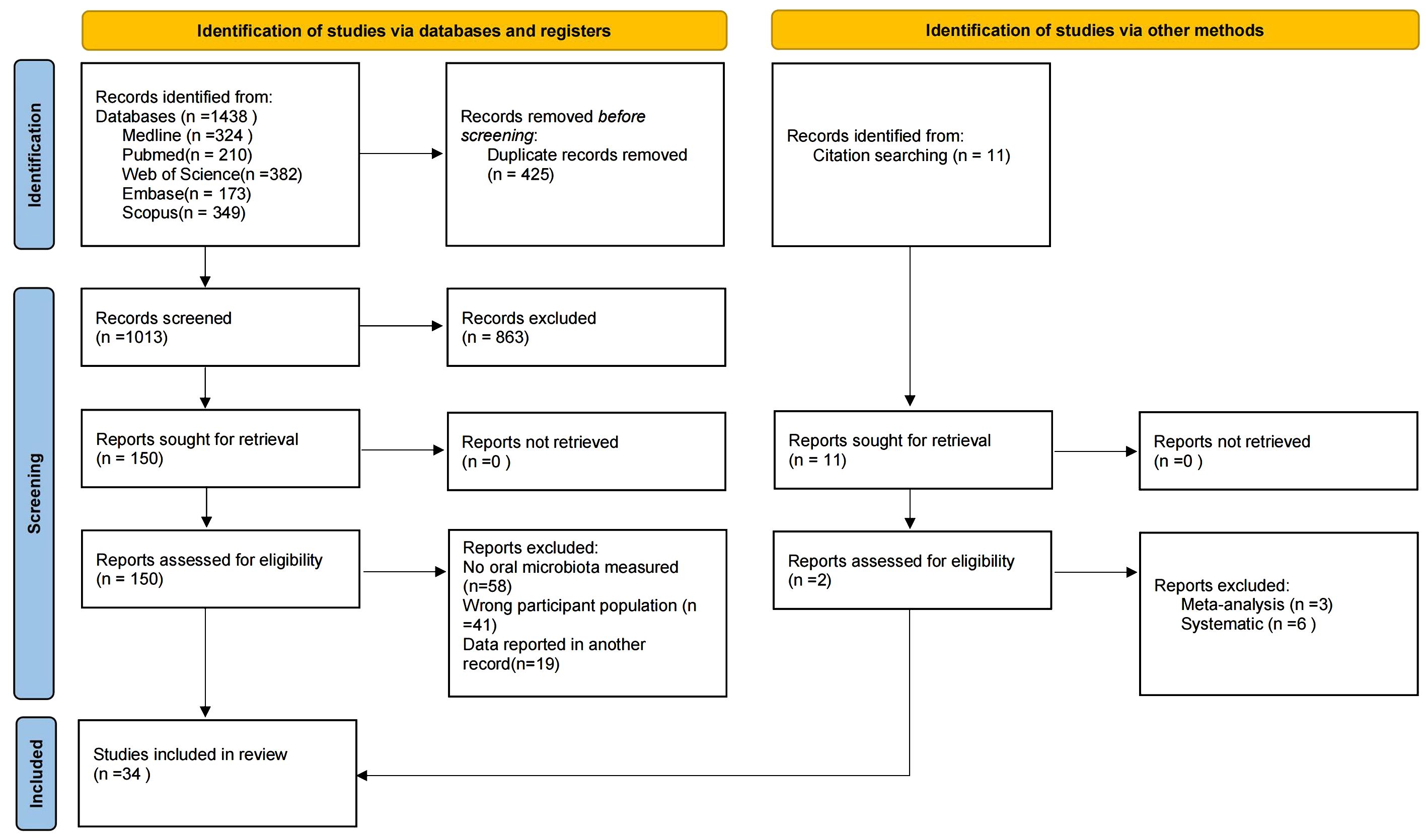
The search strategy is shown in Figure 1. After the initial search, a total of 1,438 studies were retrieved from MEDLINE (n = 324), PubMed (n = 210), Web of Science (n = 382), Embase (n = 173), and Scopus (n = 349), with an additional 11 studies retrieved through other methods. After removing duplicates, 1,013 studies remained. Screening of titles and abstracts yielded 150 studies that met the inclusion and exclusion criteria. Following full-text review, 34 studies were finally included (Hintao et al., 2007; Kamaraj et al., 2011; Adeyemi et al., 2019; Kumar et al., 2012; Al Mubarak et al., 2013; Zhou et al., 2013; Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Shenoy et al., 2014; Mohammadi et al., 2016; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Hsaine et al., 2018; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Matsha et al., 2020; Shi et al., 2020; Sun et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024; Tang et al., 2025).
3.2 Description of the studies
An overview of study characteristics is provided in Table 2. Thirty-four papers published between 2007 and 2025 were included, comprising 23 cross-sectional studies and 11 case-control studies. These studies were conducted in the following countries or regions: China (n=9), India (n=7), United States (n=4), Iran (n=2), Sweden (n=1), Japan (n=1), Thailand (n=1), Germany (n=1), Saudi Arabia (n=1), South Africa (n=1), Portugal (n=1), Turkey (n=1), Pakistan (n=1), Brazil (n=1), and Morocco (n=1).
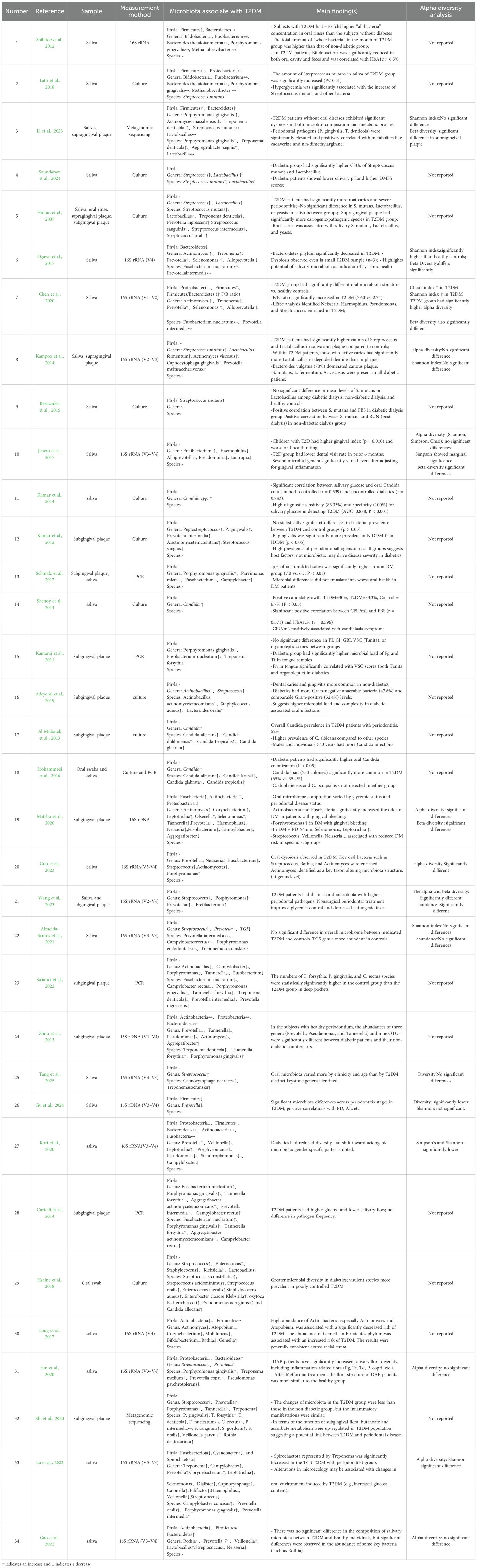
Table 2. Outcomes of the selected studies investigating the association between oral microbiota and T2DM.
Regarding oral microbial detection methods, most studies (n=17) used 16S rRNA high-throughput sequencing technology (Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Rezazadeh et al., 2016; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Sun et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Tang et al., 2025), including Illumina or other high-throughput sequencing platforms; 10 studies used traditional microbial culture methods for strain identification (Hintao et al., 2007; Kamaraj et al., 2011; Kumar et al., 2012; Al Mubarak et al., 2013; Mohammadi et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Li et al., 2023); 5 studies used PCR-related techniques (Adeyemi et al., 2019; Zhou et al., 2013; Shenoy et al., 2014; Hsaine et al., 2018; Matsha et al., 2020); and 2 studies used macro-genome sequencing techniques (Lu et al., 2022; Soundaram et al., 2024). Across all studies, a total of 3,507 subjects were included, comprising 2,062 patients with T2DM patients and 1,445 non-diabetic controls. Sample types included saliva (n=18) (Hintao et al., 2007; Kamaraj et al., 2011; Kumar et al., 2012; Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Janem et al., 2017; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Sun et al., 2020; Gao et al., 2022; Sabancı et al., 2022; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024), subgingival plaque (n=9) (Adeyemi et al., 2019; Al Mubarak et al., 2013; Zhou et al., 2013; Mohammadi et al., 2016; Schmalz et al., 2017; Hsaine et al., 2018; Lu et al., 2022; Guo et al., 2023; Tang et al., 2025), oral swab (n=1) (Long et al., 2017), and six studies using multiple sample types (Shenoy et al., 2014; Rezazadeh et al., 2016; Ogawa et al., 2017; Matsha et al., 2020; Almeida-Santos et al., 2021; Soundaram et al., 2024).
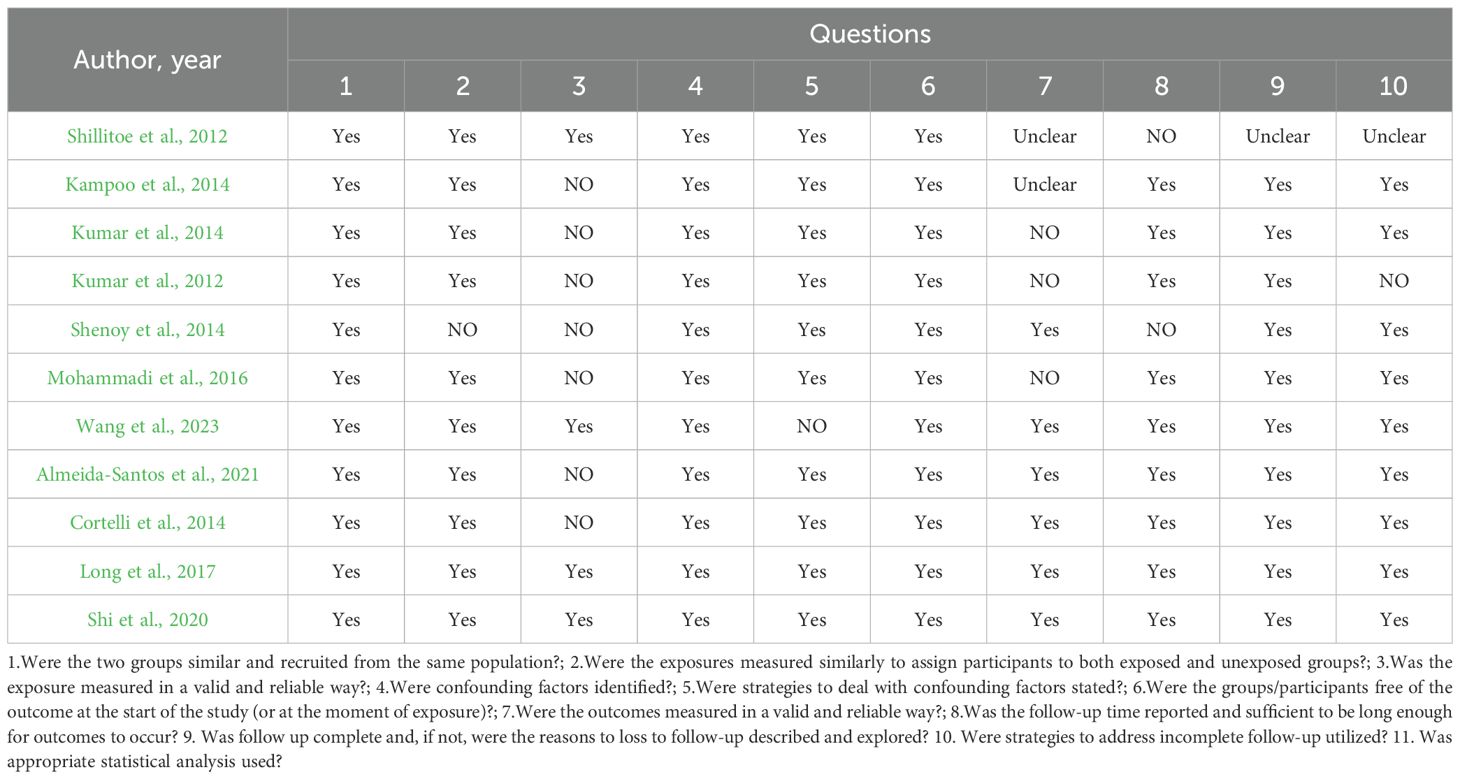
3.3 Quality assessment and risk of bias of included studies
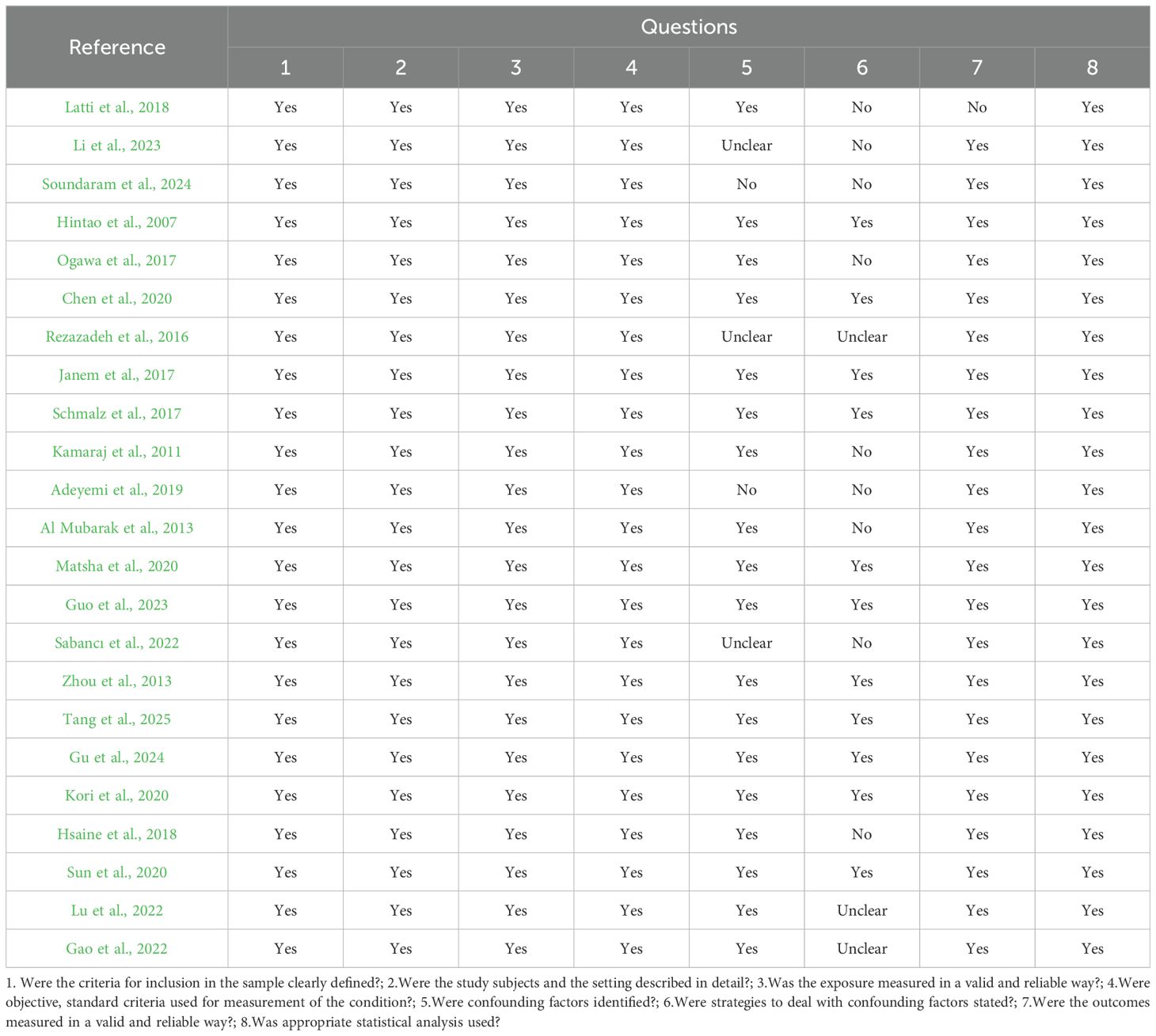
A total of 34 studies were included in this review, and their methodological quality was assessed using the appropriate Joanna Briggs Institute (JBI) critical appraisal checklists according to study design. Among the included studies, 23 were cross-sectional and 11 were case control. Study designs were determined based on the temporal relationship between exposure and outcome, the presence or absence of follow-up, and the comparative group structure.
According to the JBI quality assessment, 13 studies were rated as high quality (Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Shenoy et al., 2014; Ogawa et al., 2017; Kori et al., 2020; Shi et al., 2020; Sun et al., 2020; Lu et al., 2022; Guo et al., 2023; Wang et al., 2023; Gu et al., 2024; Tang et al., 2025). These were characterized by clearly defined inclusion criteria, representative samples, valid and consistent exposure and outcome measurements, identification and control of confounding factors, and appropriate statistical analyses. The remaining 21 studies were rated as moderate quality (Hintao et al., 2007; Kamaraj et al., 2011; Adeyemi et al., 2019; Kumar et al., 2012; Al Mubarak et al., 2013; Zhou et al., 2013; Mohammadi et al., 2016; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Schmalz et al., 2017; Hsaine et al., 2018; Latti et al., 2018; Chen et al., 2020; Matsha et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Sabancı et al., 2022; Li et al., 2023; Rasouli et al., 2023; Soundaram et al., 2024), typically due to limitations in sample size, partial or absent control of confounders, or insufficient reporting of statistical adjustment methods, although most used valid diagnostic and microbiological procedures.
Importantly, study quality appeared to influence the reported findings. High-quality studies tended to show more consistent associations between oral microbiota composition and T2DM, particularly regarding taxa linked with glycemic status. In contrast, several moderate-quality studies yielded heterogeneous or attenuated results, which may be attributable to weaker confounder control, smaller sample sizes, or incomplete adjustment for medication use and comorbidities. This divergence suggests that methodological rigor strengthens the reliability of the evidence base, while limitations in lower-quality studies may partly explain inconsistencies across the literature.
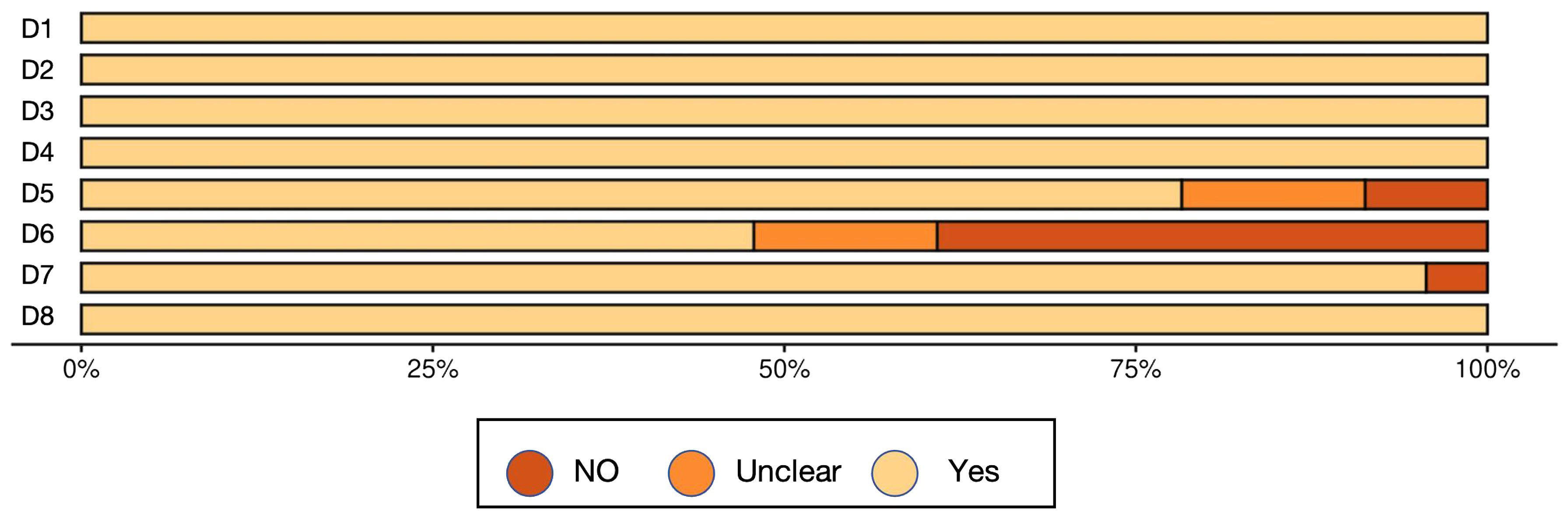
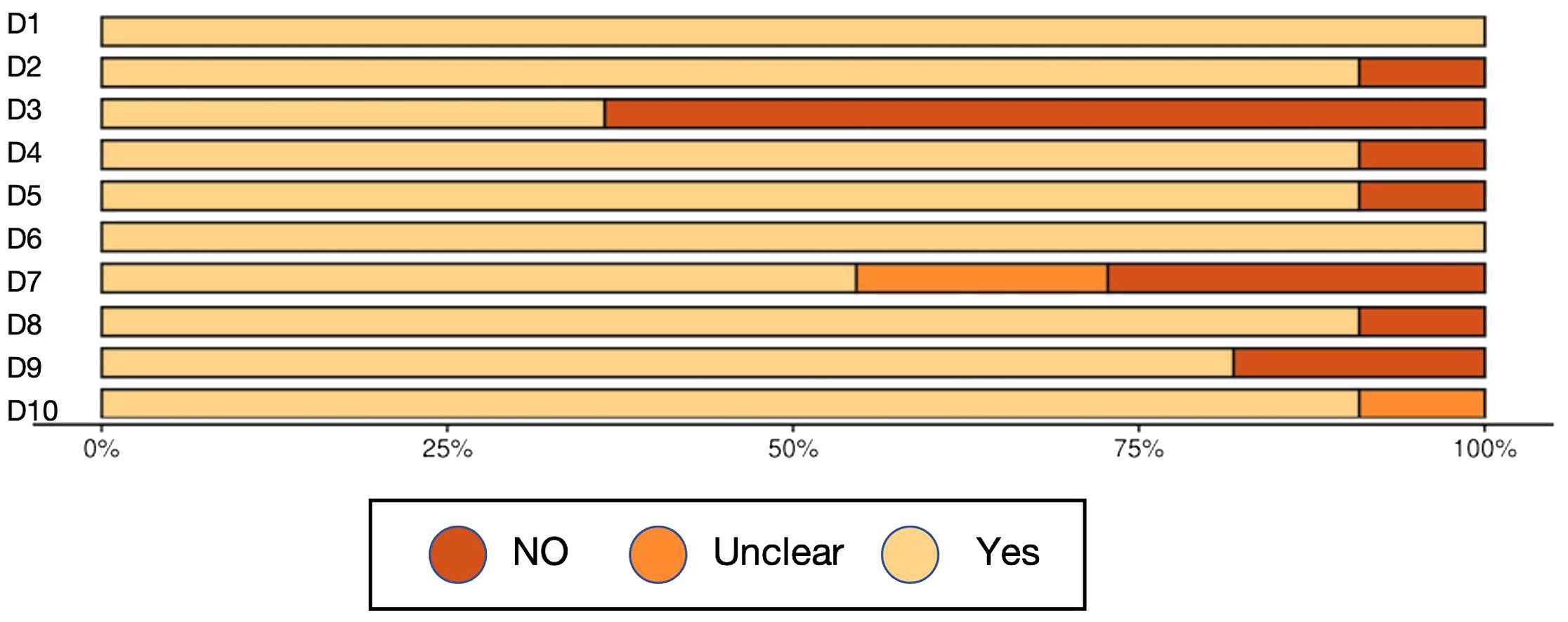
Overall, the methodological quality of the included studies was acceptable, supporting a cautious but meaningful interpretation of the synthesized findings (Table 3, Figure 2, Figure 3, Table 4).
3.4 Oral microbiota and T2DM
A total of 34 studies were included in this systematic review, spanning 2007 to 2025 and covering findings from multiple countries. These studies revealed common features of oral microecological disorders in patients with T2DM. Most reported that the total bacterial load in the oral cavity of patients with T2DM was significantly higher than that in non-diabetic controls. In particular, flora associated with dental caries and yeast infections, such as Streptococcus mutans, Lactobacillus spp., and Candida albicans, showed significant enrichment in the T2DM population. In addition, salivary acidification and decreased pH often showed a synergistic trend with changes in microbial community structure.
Of the 34 included studies, 15 analyzed the Alpha diversity of oral flora in patients with T2DM (Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Rezazadeh et al., 2016; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024). Overall, α-diversity findings were heterogeneous. Three studies (Kampoo et al., 2014; Chen et al., 2020; Wang et al., 2023) reported significantly higher Alpha diversity in patients with T2DM, suggesting that their oral flora may exhibit a more complex or disorganized structure. In contrast, two studies (Cortelli et al., 2014; Kori et al., 2020) found a significant decrease in Alpha diversity, manifested by reductions in the Shannon and Simpson indices, suggesting impaired richness or homogeneity of the flora. Three additional studies noted statistically significant differences in α-diversity without a clear directional trend, showing complex variations across subgroups (Kumar et al., 2014; Almeida-Santos et al., 2021; Soundaram et al., 2024). No significant differences were reported in the remaining six studies (Kumar et al., 2014; Rezazadeh et al., 2016; Shi et al., 2020; Sabancı et al., 2022; Rasouli et al., 2023; Gu et al., 2024). Overall, T2DM not only alters the number and composition of oral microorganisms but may also disturb oral microecological homeostasis by affecting the local metabolic environment, pH, and salivary flow rate. These alterations may increase the risk of oral diseases such as caries and periodontal disease (Table 5).
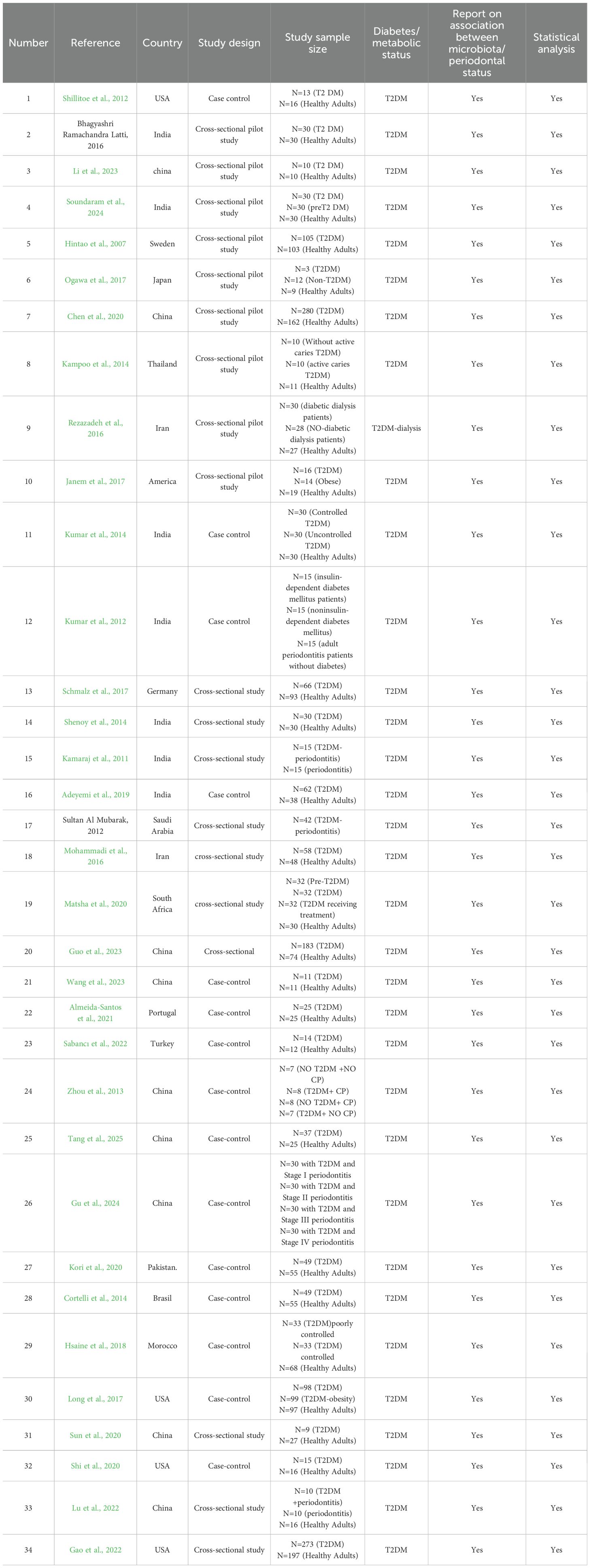
Table 5. General characteristics of the selected studies investigating the association between oral microbiota and T2DM.
3.4.1 Phylum level
A total of 14 studies analyzed phylum-level changes in the oral flora of patients with T2DM (Cortelli et al., 2014; Kampoo et al., 2014; Janem et al., 2017; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Sun et al., 2020; Gao et al., 2022; Guo et al., 2023; Li et al., 2023; Rasouli et al., 2023; Soundaram et al., 2024; Tang et al., 2025). Firmicutes was the phylum most frequently associated with T2DM, reported in all 14 studies, and consistently showed an increasing trend. Bacteroidetes, a common oral phylum, was mentioned in eight studies (Cortelli et al., 2014; Kampoo et al., 2014; Latti et al., 2018; Chen et al., 2020; Shi et al., 2020; Rasouli et al., 2023; Soundaram et al., 2024; Tang et al., 2025). Findings varied, with some studies reporting a decrease in abundance, while others found no significant changes. Proteobacteria were mentioned in five studies, with most reporting a decreasing trend (Cortelli et al., 2014; Kampoo et al., 2014; Shi et al., 2020; Guo et al., 2023; Tang et al., 2025). In addition, Fusobacteria and Actinobacteria were each reported in two studies (Cortelli et al., 2014; Guo et al., 2023), wher eas other phyla, such as Cyanobacteria and Spirochaetota, were only rarely mentioned.
3.4.2 Genus level
All 34 included studies analyzed changes in the oral flora of patients with T2DM (Hintao et al., 2007; Kamaraj et al., 2011; Adeyemi et al., 2019; Kumar et al., 2012; Al Mubarak et al., 2013; Zhou et al., 2013; Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Shenoy et al., 2014; Mohammadi et al., 2016; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Hsaine et al., 2018; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Matsha et al., 2020; Shi et al., 2020; Sun et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024; Tang et al., 2025) at the genus level. Overall, significant alterations were observed, closely linked to both the pathophysiological characteristics of T2DM and changes in the oral microenvironment. Streptococcus was the genus most significantly affected, reported in 18 publications, with most showing an increased abundance (Hintao et al., 2007; Al Mubarak et al., 2013; Kampoo et al., 2014; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Shi et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024). Prevotella was mentioned in 16 studies, but results were heterogeneous: some reported an increase (Cortelli et al., 2014; Rezazadeh et al., 2016; Ogawa et al., 2017; Schmalz et al., 2017; Chen et al., 2020; Shi et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Rasouli et al., 2023), while others reported a decrease (Kumar et al., 2014; Kori et al., 2020; Tang et al., 2025). Porphyromonas was reported in 10 studies, with most showing increased abundance in patients with T2DM, suggesting a potential role in T2DM-associated oral dysbiosis (Adeyemi et al., 2019; Zhou et al., 2013; Cortelli et al., 2014; Shenoy et al., 2014; Hsaine et al., 2018; Latti et al., 2018; Almeida-Santos et al., 2021; Lu et al., 2022; Wang et al., 2023; Soundaram et al., 2024). Fusobacterium and Treponema were each reported in seven studies (Adeyemi et al., 2019; Zhou et al., 2013; Shenoy et al., 2014; Hsaine et al., 2018; Latti et al., 2018; Guo et al., 2023; Wang et al., 2023), and both were predominantly found in elevated abundance. Overall, the frequent occurrence and altered abundance of these genera suggest that T2DM may drive the oral flora toward increased pathogenicity through changes in the oral environment.
3.4.3 Species level
Of the 17 studies analyzing species-level changes in the oral flora (Hintao et al., 2007; Al Mubarak et al., 2013; Zhou et al., 2013; Mohammadi et al., 2016; Long et al., 2017; Ogawa et al., 2017; Hsaine et al., 2018; Chen et al., 2020; Matsha et al., 2020; Shi et al., 2020; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Li et al., 2023; Gu et al., 2024; Soundaram et al., 2024; Tang et al., 2025), Porphyromonas gingivalis was the most frequently reported species, appearing in seven studies (Zhou et al., 2013; Hsaine et al., 2018; Latti et al., 2018; Shi et al., 2020; Gao et al., 2022; Soundaram et al., 2024; Tang et al., 2025), all of which showed an increasing trend. Streptococcus mutans was the next most frequently reported species, appearing in six studies (Hintao et al., 2007; Rezazadeh et al., 2016; Janem et al., 2017; Ogawa et al., 2017; Latti et al., 2018; Soundaram et al., 2024), and also generally showing an increasing trend. Treponema denticola was mentioned in four studies, mainly with increasing trend (Zhou et al., 2013; Ogawa et al., 2017; Soundaram et al., 2024; Tang et al., 2025). Fusobacterium nucleatum was reported in three studies (Zhou et al., 2013; Hsaine et al., 2018; Chen et al., 2020), but findings varied. Campylobacter rectus was reported in two studies (Hsaine et al., 2018; Sabancı et al., 2022), with inconsistent results: some showed increased abundance, while others reported a decrease or no significant change.
In summary, different taxonomic levels of oral flora showed specific patterns of change in patients with T2DM. Species such as Firmicutes (phylum), Streptococcus spp. (genus), and S. mutans (species) showed a consistent trend of elevation across most studies, suggesting a close relationship with the onset and progression of T2DM. However, some bacterial groups exhibited heterogeneous trends across studies, which may reflect differences in sample characteristics, detection techniques, and study design. Further high-quality studies are needed to confirm these associations.
4 Discussion
T2DM is a rapidly growing chronic metabolic disease worldwide, with more than 500 million patients currently affected by metabolic disease and T2DM accounting for over 90% of these cases (Rasouli et al., 2023). In the human oral cavity, the complex and diverse microbial community has a profound impact on health. Oral flora interacts with host metabolic status through multiple mechanisms, including inflammatory responses, insulin resistance, and immunomodulation (Lee et al., 2021).When oral microorganisms enter the systemic circulation via swallowing or gingival microdamage, they may serve as risk factors for triggering or exacerbating metabolic disorders (Jia et al., 2023).Currently, routine screening for T2DM relies mainly on blood glucose testing, which has limitations, especially during the early asymptomatic stage (Ortiz-Martínez et al., 2022). In recent years, oral microecology, due to its close association with glucose metabolism, has been regarded as an auxiliary screening and early risk prediction tool (Liu et al., 2021). The relative abundance of certain salivary taxa has been shown to significantly differentiate T2DM from nondiabetic individuals with good sensitivity and specificity (Shrivastava et al., 2025). Notably, oral flora is more stable in the short term and easier to sample than intestinal flora, making it a promising source of noninvasive biomarkers. Focusing on oral samples, this review aimed to summarize the characteristic changes of oral flora in patients with T2DM and to deepen the understanding of the possible relationship between T2DM and oral health.
A total of 34 studies with 2,062 patients with T2DM were included. Fourteen studies demonstrated correlations between specific phyla of oral microbiota and T2DM (Cortelli et al., 2014; Kampoo et al., 2014; Janem et al., 2017; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Sun et al., 2020; Gao et al., 2022; Guo et al., 2023; Li et al., 2023; Rasouli et al., 2023; Soundaram et al., 2024; Tang et al., 2025). All of the 34 studies examined associations between specific genera and T2DM (Hintao et al., 2007; Kamaraj et al., 2011; Adeyemi et al., 2019; Kumar et al., 2012; Al Mubarak et al., 2013; Zhou et al., 2013; Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Shenoy et al., 2014; Mohammadi et al., 2016; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Hsaine et al., 2018; Latti et al., 2018; Chen et al., 2020; Kori et al., 2020; Matsha et al., 2020; Shi et al., 2020; Sun et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024; Tang et al., 2025), and 17 analyzed species-level changes in patients with T2DM (Hintao et al., 2007; Al Mubarak et al., 2013; Zhou et al., 2013; Mohammadi et al., 2016; Long et al., 2017; Ogawa et al., 2017; Hsaine et al., 2018; Chen et al., 2020; Matsha et al., 2020; Shi et al., 2020; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Li et al., 2023; Gu et al., 2024; Soundaram et al., 2024; Tang et al., 2025). Notably, 14 studies reported significant differences in oral microbiota composition between patients with T2DM and healthy controls (Hintao et al., 2007; Al Mubarak et al., 2013; Kampoo et al., 2014; Kumar et al., 2014; Shenoy et al., 2014; Mohammadi et al., 2016; Rezazadeh et al., 2016; Ogawa et al., 2017; Hsaine et al., 2018; Latti et al., 2018; Chen et al., 2020; Almeida-Santos et al., 2021; Guo et al., 2023; Wang et al., 2023), suggesting that certain oral microorganisms may be associated with an elevated risk of developing T2DM. In terms of phylum, several studies reported significant differences in the abundance of Firmicutes, Bacteroidetes, and Proteobacteria compared to healthy controls (Cortelli et al., 2014; Shi et al., 2020; Tang et al., 2025). At the genus level, significant differences were reported for Streptococcus, Prevotella, and Porphyromonas in patients with T2DM compared to healthy individuals. In addition, Streptococcus emerged as an important focal point, with 18 studies demonstrating its association with the progression of T2DM (Hintao et al., 2007; Al Mubarak et al., 2013; Kampoo et al., 2014; Rezazadeh et al., 2016; Janem et al., 2017; Long et al., 2017; Ogawa et al., 2017; Schmalz et al., 2017; Shi et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Lu et al., 2022; Sabancı et al., 2022; Li et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024), highlighting its relevance in this area of research. In terms of diversity, 15 studies analyzed the Alpha diversity of oral flora in patients with T2DM (Cortelli et al., 2014; Kampoo et al., 2014; Kumar et al., 2014; Rezazadeh et al., 2016; Chen et al., 2020; Kori et al., 2020; Shi et al., 2020; Almeida-Santos et al., 2021; Gao et al., 2022; Sabancı et al., 2022; Guo et al., 2023; Rasouli et al., 2023; Wang et al., 2023; Gu et al., 2024; Soundaram et al., 2024), with some heterogeneity in the results. Three studies reported significantly higher diversity (Kumar et al., 2014; Almeida-Santos et al., 2021; Soundaram et al., 2024), while no significant differences were observed in six studies (Kumar et al., 2014; Rezazadeh et al., 2016; Shi et al., 2020; Sabancı et al., 2022; Rasouli et al., 2023; Gu et al., 2024). These discrepancies may reflect the influence of multiple underlying factors. Periodontal status may act as an important effect modifier, as active inflammation can alter microbial community richness and evenness in divergent ways depending on disease severity and treatment history (Griffen et al., 2012). Similarly, glycemic control has been shown to shape oral ecological conditions: poorly controlled hyperglycemia favors the dominance of acidogenic taxa and may lead to reduced diversity, whereas in some cases the concurrent colonization of opportunistic species could manifest as apparent diversity gains (Latti et al., 2018). In addition, the use of antidiabetic medications, particularly metformin, has been associated with shifts in microbial composition through both immunomodulatory and metabolic pathways, further contributing to heterogeneity across studies (Gu et al., 2021). Beyond these biological influences, methodological factors such as sampling site, sequencing depth, and the choice of diversity indices may also have contributed to the variability observed.
At the phylum level, 14 studies analyzed changes in patients with T2DM. Firmicutes consistently showed a higher abundance and was the phylum most closely associated with T2DM. In contrast, Proteobacteria showed a significant decrease in several studies, while Bacteroidetes displayed inconsistent trends, with some studies reporting reduced abundance and others finding no significant differences. These results are consistent with previous findings; for example, Tokman et al. reported that an increase in Firmicutes may be strongly associated with chronic low-grade inflammation and metabolic disturbances in patients with T2DM (Bahar-Tokman et al., 2022).Some strains in this phylum can produce metabolites such as short-chain fatty acids during carbohydrate metabolism. These metabolites may not only alter oral ecology by affecting the mucosal barrier and local pH but also modulate host immune responses and insulin sensitivity, thereby promoting diabetes progression (Molinsky et al., 2025).
At the genus level, the oral flora of patients with T2DM showed changes in the abundance of several key taxa. Notably, Streptococcus was repeatedly reported to be elevated in most studies. This genus is widely present in normal oral ecology and has a strong ability to metabolize sugar, rapidly proliferating and producing acidic metabolites in high-sugar environments, thereby lowering oral pH and promoting the development of dental caries and periodontal disease (Ali et al., 2021).Patients with T2DM provide favorable conditions for Streptococcus enrichment due to decreased salivary flow rate and altered salivary composition, which may constitute an important mechanism driving the oral flora toward pathogenicity. In addition, the genus Porphyromonas (particularly P. gingivalis) also showed a trend of increased abundance in several studies (Gu et al., 2024). P. gingivalis is a key causative agent of periodontal disease and represents the core of the “oral–systemic inflammatory axis.” It secretes virulence factors such as lipopolysaccharides (LPS) and proteases that activate the host immune system and induce systemic inflammatory responses, which in turn may promote insulin resistance (Reyes, 2021; Murugaiyan et al., 2024). Its enrichment in the T2DM population suggests that this genus may not only contribute to oral disease but may also participate in the systemic regulation of T2DM progression. Prevotella is another genus frequently reported in T2DM studies, though its trends vary. Some studies found an increase in its abundance in the oral or intestinal flora of patients with T2DM, potentially linked to its ability to ferment carbohydrates and produce butyrate (Zhang et al., 2021). In contrast, other studies reported a decrease. These differences may reflect ecological roles of Prevotella in different oral sites, disease stages, or comorbid contexts and may also be influenced by factors such as diet and oral hygiene.
At the species level, changes in specific bacteria provide more precise insights into the relationship between oral flora and T2DM. Streptococcus mutans, the main causative agent of dental caries, was significantly increased in patients with T2DM. The hyperglycemic environment favors the growth of S. mutans, and the acidic substances it produces further damage tooth enamel, creating a vicious cycle (Brito et al., 2021). Similarly, periodontal pathogens such as Porphyromonas gingivalis and Treponema denticola were also increased in most studies. These species exacerbate systemic inflammatory responses by triggering periodontal inflammation and releasing inflammatory mediators, which in turn impair glycemic control and insulin sensitivity (Liu et al., 2024). Although trends for other species such as Fusobacterium nucleatum were less consistent, their role in oral dysbiosis should not be overlooked. F. nucleatum may aggravate oral disease progression by promoting biofilm formation and enhancing the invasiveness of other pathogenic bacteria.
Research indicates that metabolic dysregulation in diabetes mellitus (DM) exacerbates inflammation and promotes microbial dysbiosis in the subgingival microbiome, which is a key factor in the progression of periodontitis in diabetic patients. Hyperglycemia elevates glucose levels in saliva, providing a nutrient-rich environment for cariogenic bacteria in the dental biofilm. Studies have shown that saliva in patients with DM contains higher levels of glucose, urea, and total protein, while exhibiting lower calcium levels and acidic pH (Verhulst et al., 2019), which further support the growth of pathogenic bacteria. These dysbiotic shifts in the oral microbiome are not only associated with local tissue destruction but also contribute to systemic inflammation. Cytokines such as interleukin-1e (IL-1leu), tumor necrosis factor-s (TNF-r-s), and the receptor activator of nuclear factor κa ligand (RANKL) have been implicated in mediating periodontitis in diabetic patients. Additionally, interactions between advanced glycation end products (AGEs) and their receptor (RAGE) exacerbate inflammation and periodontal tissue destruction (Mealey and Oates, 2006; Taylor et al., 2013). Recent studies suggest that diabetes may enhance the pathogenicity of the oral microbiome through IL-17-mediated pro-inflammatory mechanisms. These immune disruptions in DM lead to dysbiosis in the subgingival microbiome, predisposing individuals to periodontitis. Moreover, these dysbiotic shifts may also affect the gut microbiome via the oral–gut pathway, contributing to systemic inflammation and insulin resistance, thereby linking oral health to broader metabolic dysfunction in diabetic patients (Li et al., 2023).
Emerging evidence suggests that antidiabetic medications, particularly metformin, may influence hostuence,lys.r interactions beyond glucose regulation. Metformin has been shown to modulate the gut microbiota by enriching beneficial taxa such as Akkermansia muciniphila and other short-chain fatty acid producers, while reducing potentially pathogenic bacteria. Although direct evidence of its impact on the oral microbiota is limited, preliminary findings indicate distinct microbial signatures in patients with T2DM receiving metformin therapy, possibly mediated by reduced systemic inflammation, improved immune balance, and altered salivary metabolic profiles. In contrast, evidence regarding the microbiome-related effects of other antidiabetic drugs, such as DPP-4 and SGLT2 inhibitors, remains scarce. Longitudinal and interventional studies are needed to clarify whether these agents exert protective, neutral, or adverse effects on the oral microbial ecosystem and metabolic outcomes (Hung and Hung, 2020).
This study has several methodological limitations. Different detection methods (e.g., PCR, 16S rRNA gene sequencing) significantly influence the interpretation of results, as each technique has distinct advantages and inherent biases. PCR can precisely amplify target microbial DNA but is restricted to known species and may overrepresent certain taxa, whereas 16S rRNA sequencing provides broader community profiles but with limited resolution and lower sensitivity for low-abundance microbes. These methodological differences may contribute to inconsistencies in reported abundance and limit comparability across studies. In addition, most included studies were cross-sectional in design, which restricts causal inference. Another important limitation is the inconsistent handling of key confounders, including oral hygiene, diet, smoking, and metformin use. Quality assessment revealed that 21 studies did not adequately control for these factors, which may introduce systematic bias and obscure whether observed microbial changes are attributable to T2DM itself or to external influences. To address these issues, future studies should adopt prospective cohort designs, use standardized microbiome sequencing technologies, and apply rigorous statistical methods to control for confounders. Considering lifestyle, metabolic status, and oral environmental factors will further strengthen the validity and reliability of conclusions.
5 Conclusions
This systematic review identified significant changes in the oral flora of patients with T2DM across 34 studies. These changes were observed at the phylum, genus, and species levels, with the most consistent increases reported for the phylum Firmicutes, the genus Streptococcus, and the species Porphyromonas gingivalis. Total oral bacterial load was generally higher in patients with T2DM, while bacterial diversity showed heterogeneous patterns across studies. Given the strong association between oral flora and T2DM, future research should prioritize clarifying causal relationships. In addition, maintaining good oral hygiene may contribute to both the prevention and management of diabetes.
Author contributions
MH: Methodology, Writing – original draft. XZ: Data curation, Writing – review & editing. LD: Data curation, Writing – review & editing. ZY: Conceptualization, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the “Internal mechanism, key dimensions and practice path of the construction of “Master Inheritance Studio” in physical training (145124038/016)” grant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeyemi, B., Abimbola, O., and Kolude, B.. (2019). A comparative study of oral health status in diabetic and non-diabetic patients. Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiol. 128 (1), e68–e69.
Ali, T., Rumnaz, A., Urmi, U. L., Nahar, S., Rana, M., Sultana, F., et al. (2021). Type-2 diabetes mellitus individuals carry different periodontal bacteria. Pesquisa Bras. em Odontopediatria e Clínica Integrada 21, e0107. doi: 10.1590/pboci.2021.049
Ali, M. K., Pearson-Stuttard, J., Selvin, E., and Gregg, E. W. (2022). Interpreting global trends in type 2 diabetes complications and mortality. Diabetologia 65, 3–13. doi: 10.1007/s00125-021-05585-2
Almeida-Santos, A., Martins-Mendes, D., Gayà-Vidal, M., Pérez-Pardal, L., Beja-Pereira, A., Almeida-Santos, A., et al. (2021). Characterization of the oral microbiome of medicated type-2 diabetes patients. Front. Microbiol. 12, 610370. doi: 10.3389/fmicb.2021.610370
Al Mubarak, S., Robert, A. A., Baskaradoss, J. K., Al-Zoman, K., Al Sohail, A., Alsuwyed, A., et al. (2013). The prevalence of oral Candida infections in periodontitis patients with type 2 diabetes mellitus. J. infection Public Health 6, 296–301. doi: 10.1016/j.jiph.2012.12.007
Andreadi, A., Bellia, A., Di Daniele, N., Meloni, M., Lauro, R., Della-Morte, D., et al. (2022). The molecular link between oxidative stress, insulin resistance, and type 2 diabetes: A target for new therapies against cardiovascular diseases. Curr. Opin. Pharmacol. 62, 85–96. doi: 10.1016/j.coph.2021.11.010
Bahar-Tokman, H., Demirci, M., Keskin, F., Cagatay, P., Taner, Z., Ozturk-Bakar, Y., et al. (2022). Firmicutes/bacteroidetes ratio in the gut microbiota and IL-1β, IL-6, IL-8, TLR2, TLR4, TLR5 gene expressions in type 2 diabetes. Clin. Lab. 68 (9). doi: 10.7754/Clin.Lab.2022.211244
Brito, A. C. M., Bezerra, I. M., Borges de, M. H. S., Cavalcanti, Y. W., Almeida de de, L. F. D., Brito, A. C. M., et al. (2021). Effect of different salivary glucose concentrations on dual-species biofilms of Candida albicans and Streptococcus mutans. Biofouling 37, 615–625. doi: 10.1080/08927014.2021.1946519
Caselli, E., Fabbri, C., D’Accolti, M., Soffritti, I., Bassi, C., Mazzacane, S., et al. (2020). Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 20, 120. doi: 10.1186/s12866-020-01801-y
Caturano, A., D’Angelo, M., Mormone, A., Russo, V., Mollica, M. P., Salvatore, T., et al. (2023). Oxidative stress in type 2 diabetes: impacts from pathogenesis to lifestyle modifications. Curr. Issues Mol. Biol. 45, 6651–6666. doi: 10.3390/cimb45080420
Chandler, J., Cumpston, M., Li, T., Page, M. J., and Welch, V. (2019). Cochrane handbook for systematic reviews of interventions (Hoboken: Wiley).
Chandrasekaran, P. and Weiskirchen, R. (2024). The role of obesity in type 2 diabetes mellitus—An overview. Int. J. Mol. Sci. 25, 1882. doi: 10.3390/ijms25031882
Chen, B., Wang, Z., Wang, J., Su, X., Yang, J., Zhang, Q., et al. (2020). The oral microbiome profile and biomarker in Chinese type 2 diabetes mellitus patients. Endocrine 68, 564–572. doi: 10.1007/s12020-020-02269-6
Cortelli, J. T., Pinheiro, R. M. S., Costa de, F. O., Aquino, D. R., Raslan, S. A., Cortelli, S. C., et al. (2014). Salivary and microbiological parameters of chronic periodontitis subjects with and without type 2 diabetes mellitus: a case-control study. Rev. Odontologia da UNESP 43, 196–202. doi: 10.1590/rou.2014.030
Dale, A. E., Gloyn, A. L., Evans-Molina, C., Joseph, J. J., and Misra, S.. (2024). Diabetes mellitus—Progress and opportunities in the evolving epidemic. Cell 187, 3789–3820. doi: 10.1016/j.cell.2024.06.029
Gao, C., Guo, Y., and Chen, F. (2022). Cross-cohort microbiome analysis of salivary biomarkers in patients with type 2 diabetes mellitus. Front. Cell. Infection Microbiol. 12, 816526. doi: 10.3389/fcimb.2022.816526
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. doi: 10.1038/ismej.2011.191
Gu, M., Wang, P., Xiang, S., Xu, D., Jin, C., Jiang, Z., et al. (2021). Effects of type 2 diabetes and metformin on salivary microbiota in patients with chronic periodontitis. Microbial Pathogenesis 161, 105277. doi: 10.1016/j.micpath.2021.105277
Gu, M., Ge, J., Pan, Q., Hu, N., Hua, F., Gu, M., et al. (2024). Salivary microbiome variations in type 2 diabetes mellitus patients with different stages of periodontitis. BMC Oral. Health 24, 1424. doi: 10.1186/s12903-024-05135-3
Guo, X., Dai, S., Lou, J., Ma, X., Hu, X., Tu, L., et al. (2023). Distribution characteristics of oral microbiota and its relationship with intestinal microbiota in patients with type 2 diabetes mellitus. Front. Endocrinol. 14, 1119201. doi: 10.3389/fendo.2023.1119201
Hintao, J., Teanpaisan, R., Chongsuvivatwong, V., Ratarasan, C., Dahlen, G., Hintao, J., et al. (2007). The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral. Microbiol. Immunol. 22, 175–181. doi: 10.1111/j.1399-302X.2007.00341.x
Hsaine, S., Fethi, F. Z., Charof, R., Ounine, K., Hsaine, S., Fethi, F. Z., et al. (2018). Microbiological study of oral flora in diabetic patients with gingivitis. Int. J. Pharm. Pharm. Sci. 10, 113–116. doi: 10.22159/ijpps.2018v10i6.26295
Hung, W.-W. and Hung, W.-C. (2020). How gut microbiota relate to the oral antidiabetic treatment of type 2 diabetes. Med. Microecol. 3, 100007. doi: 10.1016/j.medmic.2020.100007
Janem, W. F., Scannapieco, F. A., Sabharwal, A., Tsompana, M., and Berman, H. A.. (2017). Salivary inflammatory markers and microbiome in normoglycemic lean and obese children compared to obese children with type 2 diabetes. PLoS One 12, e0172647. doi: 10.1371journal.pone.0172647
Jia, S., Li, X., and Du, Q. (2023). Host insulin resistance caused by Porphyromonas gingivalis-review of recent progresses. Front. Cell. Infection Microbiol. 13, 1209381. doi: 10.3389/fcimb.2023.1209381
Kamaraj, D. R., Bhushan, K. S., Laxman, V. K., and Mathew, J.. (2011). Detection of odoriferous subgingival and tongue microbiota in diabetic and nondiabetic patients with oral malodor using polymerase chain reaction. Indian J. Dental Res. 22, 260–265. doi: 10.4103/0970-9290.84301
Kampoo, K., Teanpaisan, R., Ledder, R. G., and McBain, A. J.. (2014). Oral bacterial communities in individuals with type 2 diabetes who live in southern Thailand. Appl. Environ. Microbiol. 80, 662–671. doi: 10.1128/AEM.02821-13
Kori, J. A., Saleem, F., Ullah, S., and Azim, M. K.. (2020). Characterization of Oral bacteriome dysbiosis in type 2 diabetic patients. MedRxiv, 2020–2004. doi: 10.1101/2020.04.09.20052613
Kumar, V. V., Kumar, K. P., Abdul Gafoor, , and Santhosh, V. C.. (2012). Evaluation of subgingival microflora in diabetic and nondiabetic patients. J. Contemp Dent. Pract. 13, 157–162. doi: 10.5005/jp-journals-10024-1113
Kumar, S., Padmashree, S., and Jayalekshmi, R. (2014). Correlation of salivary glucose, blood glucose and oral candidal carriage in the saliva of type 2 diabetics: A case-control study. Contemp. Clin. dentistry 5, 312–317. doi: 10.4103/0976-237X.137925
Kumari, S. and Gnanasundaram, N. (2021). Oral manifestations in diabetes mellitus-a review. J. Indian Acad. Oral. Med. Radiol. 33, 352–356. doi: 10.4103/jiaomr.jiaomr_325_21
Latti, B. R., Kalburge, J. V., Birajdar, S. B., and Latti, R. G.. (2018). Evaluation of relationship between dental caries, diabetes mellitus and oral microbiota in diabetics. J. Oral. Maxillofac. Pathol. 22, 282. doi: 10.4103/jomfp.JOMFP_163_16
Lee, Y.-H., Chung, S. W., Auh, Q.-S., Hong, S.-J., Lee, Y.-A., et al. (2021). Progress in oral microbiome related to oral and systemic diseases: an update. Diagnostics 11, 1283. doi: 10.3390/diagnostics11071283
Li, Y., Qian, F., Cheng, X., Wang, D., Wang, Y., et al. (2023). Dysbiosis of oral microbiota and metabolite profiles associated with type 2 diabetes mellitus. Microbiol. Spectr. 11, e03796–e03722. doi: 10.1128/spectrum.03796-22
Li, X., Yan, F., Chen, W., Huang, M., Mo, Chaolun, et al. (2024). Effects of irrigation of porphyromonas gingivalis on colonic mechanical and immune barriers in type 2 diabetic mice. Chin. Gen. Pract. 27, 2225. doi: 10.12114/j.issn.1007-9572.2023.0386
Li, L., Xie, X., Yun, Wu, et al. (2023). Research progress on the mechanisms of the association between periodontitis and diabetes. J. Sichuan Univ. (Medical Sci. Edition) 54, 71–76. doi: 10.12182/20230160203
Liu, Y.-k., Chen, V., He, J.-z., Zheng, X., Xu, X., et al. (2021). A salivary microbiome-based auxiliary diagnostic model for type 2 diabetes mellitus. Arch. Oral. Biol. 126, 105118. doi: 10.1016/j.archoralbio.2021.105118
Liu, F., Zhu, B., An, Y., Zhou, Z., Xiong, P., Li, X., et al. (2024). Gingipain from Porphyromonas gingivalis causes insulin resistance by degrading insulin receptors through direct proteolytic effects. ” Int. J. Oral. Sci. 16, 53. doi: 10.1038/s41368-024-00313-z
Long, J., Cai, Q., Steinwandel, M., Hargreaves, M. K., Bordenstein, S. R., et al. (2017). Association of oral microbiome with type 2 diabetes risk. J. periodontal Res. 52, 636–643. doi: 10.1111/jre.12432
Lu, C., Zhao, Q., Deng, J., Chen, K., Jiang, X., et al. (2022). Salivary microbiome profile of diabetes and periodontitis in a Chinese population. Front. Cell. Infection Microbiol. 12, 933833. doi: 10.3389/fcimb.2022.933833
Matsha, T. E., Prince, Y., Davids, S., Chikte, U., Erasmus, R. T., Kengne, A. P., et al. (2020). Oral microbiome signatures in diabetes mellitus and periodontal disease. J. Dental Res. 99, 658–665. doi: 10.1177/0022034520913818
Mealey, B. L. and Oates, T. W. (2006). American academy of periodontology. Diabetes mellitus and periodontal diseases. J. Periodontol 77, 1289–1303. doi: 10.1902/jop.2006.050459
Mohammadi, F., Javaheri, M. R., Nekoeian, S., and Dehghan, P.. (2016). Identification of Candida species in the oral cavity of diabetic patients. Curr. Med. mycology 2, 1. doi: 10.18869/acadpub.cmm.2.2.4
Mohammed, L. I., Zakaria, Z. Z., Benslimane, F. M., Al-Asmakh, M., Mohammed, L. I., Zakaria, Z. Z., et al. (2024). Exploring the role of oral microbiome dysbiosis in cardiometabolic syndrome and smoking. Exp. Lung Res. 50, 65–84. doi: 10.1080/01902148.2024.2331185
Molinsky, R. L., Johnson, A. J., Marotz, L., Roy, S., Bohn, B., Goh, C. E., et al. (2025). Association between Dietary Patterns and subgingival microbiota: results from the oral infections, glucose intolerance, and insulin resistance study (ORIGINS). J. Clin. periodontology 52, 2–15. doi: 10.1111/jcpe.14067
Murugaiyan, V., Utreja, S., Hovey, K. M., Sun, Y., LaMonte, M. J., Wactawski‑Wende, J., et al. (2024). Defining Porphyromonas gingivalis strains associated with periodontal disease. Sci. Rep. 14, 6222. doi: 10.1038/s41598-024-56849-x
Negrini de, T. C., Carlos, I. Z., Duque, C., Caiaffa, K. S., Arthur, R. A., Negrini de, T. C., et al. (2021). Interplay among the oral microbiome, oral cavity conditions, the host immune response, diabetes mellitus, and its associated-risk factors—An overview. Front. Oral. Health 2, 697428. doi: 10.3389/froh.2021.697428
Ogawa, T., Honda-Ogawa, M., Ikebe, K., Notomi, Y., Iwamoto, Y., Shirobayashi, I., et al. (2017). Characterizations of oral microbiota in elderly nursing home residents with diabetes. J. Oral. Sci. 59, 549–555. doi: 10.2334/josnusd.16-0722
Ortiz-Martínez, M., González-González, M., Martagón, A. J., Hlavinka, V., Willson, R. C., Rito-Palomares, M., et al. (2022). Recent developments in biomarkers for diagnosis and screening of type 2 diabetes mellitus. Curr. Diabetes Rep. 22, 95–115. doi: 10.1007/s11892-022-01453-4
Penno, G., Solini, A., Orsi, E., Bonora, E., Fondelli, C., Trevisan, R., et al. (2021). Insulin resistance, diabetic kidney disease, and all-cause mortality in individuals with type 2 diabetes: a prospective cohort study. BMC Med. 19, 1–13. doi: 10.1186/s12916-021-01936-3
Rasouli, H., Ramalho, T. C., Popović-Djordjević, J. B., Devkota, H. P., Rasouli, H., Ramalho, T. C., et al. (2023). New opportunities in drug design for the management and treatment of type 2 diabetes. Front. Pharmacol. 14, 1187057. doi: 10.3389/fphar.2023.1187057
Rezazadeh, F., Bazargani, A., Roozbeh-Shahroodi, J., Pooladi, A., Arasteh, P., Zamani, K., et al. (2016). Comparison of oral Lactobacillus and Streptococcus mutans between diabetic dialysis patients with non-diabetic dialysis patients and healthy people. J. Renal injury Prev. 5, 148. doi: 10.15171/jrip.2016.31
Rohm, T. V., Meier, D. T., Olefsky, J. M., Donath, M. Y., Rohm, T. V., Meier, D. T., et al. (2022). Inflammation in obesity, diabetes, and related disorders. Immunity 55, 31–55. doi: 10.1016/j.immuni.2021.12.013
Sabancı, A., Eltas, A., Celik, B., Otlu, B., Sabancı, A., Eltas, A., et al. (2022). The influence of diabetes mellitus on the peri-implant microflora: A cross-sectional study. J. Oral. Biol. Craniofacial Res. 12, 405–409. doi: 10.1016/j.jobcr.2022.05.007
Schmalz, G., Schiffers, N., Schwabe, S., Vasko, R., Müller, G. A., Haak, R., et al. (2017). Dental and periodontal health, and microbiological and salivary conditions in patients with or without diabetes undergoing haemodialysis. Int. Dental J. 67, 186–193. doi: 10.1111/idj.12282
Schulze, M. B. and Hu, F. B. (2022). “Epidemiology of diabetes,” in Handbook of epidemiology (Springer New York, New York, NY), 1–49.
Shaalan, A., Lee, S., Feart, C., Garcia-Esquinas, E., Gomez-Cabrero, D., Lopez-Garcia, E., et al. (2022). Alterations in the oral microbiome associated with diabetes, overweight, and dietary components. Front. Nutr. 9, 914715. doi: 10.3389/fnut.2022.914715
Shenoy, M., Puranik, R., Vanaki, S., Puranik, S., Shetty, P., Shenoy, R., et al. (2014). A comparative study of oral candidal species carriage in patients with type1 and type2 diabetes mellitus. J. Oral. Maxillofac. Pathol. 18.Suppl 1, S60–S65. doi: 10.4103/0973-029X.141361
Shi, B., Lux, R., Klokkevold, P., Chang, M., Barnard, E., Haake, S., et al. (2020). The subgingival microbiome associated with periodontitis in type 2 diabetes mellitus. ISME J. 14, 519–530. doi: 10.1038/s41396-019-0544-3
Shillitoe, E., Weinstock, R., Kim, T., Simon, H., Planer, J., Noonan, S., et al. (2012). The oral microflora in obesity and type-2 diabetes. J. Oral. Microbiol. 4, 19013. doi: 10.3402/jom.v4i0.19013
Shrivastava, R., Kumar, S., and Suman, D. (2025). “Role of the oral microbiome in prognosis and diagnosis,” in Oral Microbiome (CRC Press), 288–301.
Soundaram, B., Murali, C. R., Ajithkumar, M., Anjali, A., JothiPriya, D., Vikraman, S., et al. (2024). Evaluation of oral microflora-related to dental caries and salivary pH in type II diabetic patients. J. Pharm. Bioallied Sci. 16.Suppl 2, S1843–S1849. doi: 10.4103/jpbs.jpbs_1291_23
Stewart, L. A., Clarke, M., Rovers, M., Riley, R. D., Simmonds, M., Stewart, G., et al. (2015). Preferred reporting items for a systematic review and meta-analysis of individual participant data: the PRISMA-IPD statement. JAMA 313, 1657–1665. doi: 10.1001/jama.2015.3656
Suarez, L. J., Garzon, H., Arboleda, S., and Rodrıguez, A. (2020). Oral dysbiosis and autoimmunity: from local periodontal responses to an imbalanced systemic immunity. A review. Front. Immunol. 11. doi: 10.3389/fimmu.2020.591255
Sun, X., Li, M., Xia, L., Fang, Z., Yu, S., Gao, J., et al. (2020). Alteration of salivary microbiome in periodontitis with or without type-2 diabetes mellitus and metformin treatment. Sci. Rep. 10, 15363. doi: 10.1038/s41598-020-72035-1
Tang, L., Ding, K., Li, M., Chao, X., Sun, T., Guo, Y., et al. (2025). Differences in oral microbiota associated with type 2 diabetes mellitus between the Dai and Han populations. J. Oral. Microbiol. 17, 2442420. doi: 10.1080/20002297.2024.2442420
Taylor, J. J., Preshaw, P. M., and Lalla, E. (2013). A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Clin. Periodontol. 40, S113–S134. doi: 10.1902/jop.2013.134005
Verhulst, M. J. L., Loos, B. G., Gerdes, V. E. A., and Teeuw, W. J. (2019). Evaluating all potential oral complications of diabetes mellitus. Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00056
Wang, L., Gao, Z., Zhao, Z., Shen, X., Feng, J., Xiong, J., et al. (2023). Oral microbiota in periodontitis patients with and without type 2 diabetes mellitus and their shifts after the nonsurgical periodontal therapy. Heliyon 9, 11. doi: 10.1016/j.heliyon.2023.e22110
Yameny, A. A. (2024). Diabetes mellitus overview 2024. J. Bioscience Appl. Res. 10, 641–645. doi: 10.21608/jbaar.2024.382794
Zeng, X., Huang, S., Ye, X., Song, S., He, J., Hu, L., et al. (2024). Impact of HbA1c control and type 2 diabetes mellitus exposure on the oral microbiome profile in the elderly population. J. Oral. Microbiol. 16, 2345942. doi: 10.1080/20002297.2024.2345942
Zhang, Z., Tian, T., Chen, Z., Liu, L., Luo, T., et al. (2021). Characteristics of the gut microbiome in patients with prediabetes and type 2 diabetes. PeerJ 9, e10952. doi: 10.7717/peerj.10952
Zhou, M., Rong, R., Munro, D., Zhu, C., Gao, X., Zhang, Q., et al. (2013). Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16S rDNA pyrosequencing. PLoS One 8, e61516. doi: 10.1371/journal.pone.0061516
Keywords: oral microbiota, oral microbiology, diabetes, type 2 diabetes mellitus, T2DM, systematic review
Citation: Huang M, Zhang X, Di L and Yi Z (2025) Advances in the study of oral microbiota in association with T2DM: a systematic review. Front. Cell. Infect. Microbiol. 15:1629304. doi: 10.3389/fcimb.2025.1629304
Received: 15 May 2025; Accepted: 08 September 2025;
Published: 01 October 2025.
Edited by:
Soumyadev Sarkar, Arizona State University, United StatesReviewed by:
Mayank Hans, ESIC Medical College (Faridabad), IndiaYanlong Shi, Fuyang Hospital of Anhui Medical University, China
Shimaa Hussein Kotb, Sphinx University, Egypt
Copyright © 2025 Huang, Zhang, Di and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Yi, eWl6aGVuZ0BjdXBlcy5lZHUuY24=
 Mingming Huang
Mingming Huang Xinbi Zhang
Xinbi Zhang Leiming Di
Leiming Di Zheng Yi*
Zheng Yi*