- 1Department of Molecular Biology and Microbial Food Safety, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 2Department of Preventive Dentistry, Academic Centre for Dentistry Amsterdam (ACTA), University of Amsterdam and Free University Amsterdam, Amsterdam, Netherlands
- 3RNA Biology Research Group, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 4Laboratory for Mass Spectrometry of Biomolecules, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 5Department of Medical Microbiology and Infection Prevention, Amsterdam Institute for Immunology and Infectious Diseases, Amsterdam University Medical Centers, University of Amsterdam, Amsterdam, Netherlands
- 6Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology, Endocrinology and Metabolism, Amsterdam University Medical Centers, Amsterdam, Netherlands
Background: Co-infections of Candida albicans and Staphylococcus aureus can significantly increase morbidity and mortality. However, the effect of C. albicans–S. aureus co-existence on virulence factor secretion and pro-inflammatory effects remain elusive.
Methods: We systematically investigated the virulence factors released by C. albicans and S. aureus under different culturing conditions using proteomics. We characterized their pro-inflammatory effects in macrophages with transcriptomics and gene set enrichment analysis.
Results and Discussion: We showed that co-culturing of C. albicans and S. aureus promoted the secretion of 7 cytolytic, 11 proteolytic, and 3 lipolytic extracellular virulence factors (ECVFs) and impacted non-ECVFs, owing to Als1/Als3-mediated interactions, the presence of C. albicans, or its pH maintenance. Co-culturing promotes C. albicans hypha formation and β-glucan masking, suggesting that co-culturing enhances both C. albicans invasion and immune evasion. Moreover, the secretome of C. albicans–S. aureus co-culture increased pro-inflammatory pathways including promoting TNF-, NFKB-, and Toll-like receptor signaling pathways, as well as cytokine–cytokine receptor interactions in macrophages. Our findings support that C. albicans and S. aureus reciprocally promote their virulence potential and pro-inflammatory effects, which may provide mechanistic insights into the increased morbidity and mortality during their co-infection in vivo.
Introduction
Candida albicans is a commensal polymorphic fungus of the human oral, urogenital, gastrointestinal, and skin mycobiome (Berman, 2012; Mayer et al., 2013). In immunocompromised individuals, however, the fungus frequently becomes pathogenic (Pasman et al., 2022). Pathogenicity of C. albicans is related to its ability to switch from non-invasive yeast growth to invasive hyphal growth (Talapko et al., 2021). Tissue invasion by hyphae comprises two key processes: the secretion of extracellular virulence factors (ECVFs) that damage the epithelium, facilitating tissue invasion, and invasion by physically forcing the hyphae through epithelial layers (Srikantha et al., 1998; Crampin et al., 2005; Moyes et al., 2016; Desai, 2018). C. albicans hyphal invasion can result in a candidal bloodstream infection (BSI) (McCarty et al., 2021). Interestingly, C. albicans BSIs frequently co-occur with bacterial bloodstream invasion (Klotz et al., 2007), suggesting that the fungus facilitates co-invasion of bacteria. Staphylococcus aureus, an ESKAPE (group of six clinically relevant, highly virulent, and antibiotic-resistant bacterial pathogens) pathogen, is the third most commonly co-isolated bacterium during C. albicans BSIs (Klotz et al., 2007) and is the most common pathogen causing primary bacterial BSIs, i.e., BSIs without an identified portal of entry or associated site of infection (del Rio et al., 2009; Pien et al., 2010). Due to their co-isolation during candidal BSIs, C. albicans has been hypothesized to account for this lacking staphylococcal portal of entry (Schlecht et al., 2015; Allison et al., 2019).
Various in vivo murine studies, using an oral infection model, have shown that C. albicans potently promotes co-invasion and dissemination of S. aureus and significantly increases lethality compared with monomicrobial infection (Carlson, 1982; Carlson, 1983; Carlson, 1988; Nash et al., 2014; Kong et al., 2015; Schlecht et al., 2015; Nash et al., 2016; Allison et al., 2019; Van Dyck et al., 2021). The C. albicans hyphal agglutinin sequence proteins 1 and 3 (Als1, Als3), the proteins responsible for S. aureus binding to hyphae, crucially contribute to S. aureus co-invasion (Kong et al., 2015; Schlecht et al., 2015; Van Dyck et al., 2021). Furthermore, the C. albicans ECVF candidalysin has been shown to significantly contribute to pathogenesis of C. albicans/S. aureus co-infections (Van Dyck et al., 2021). Additional in vitro studies have shown that C. albicans/S. aureus co-culturing also increases the alpha toxin production of the S. aureus secretome by promoting the staphylococcal agr quorum sensing system in a pH-dependent manner (de Carvalho Dias et al., 2017; Todd et al., 2019b; Todd et al., 2019a; Dias et al., 2021). Aside from candidalysin and staphylococcal alpha toxin, C. albicans and S. aureus secrete additional damaging ECVFs (cytolytic, proteolytic, and lipolytic), which could potentially contribute to invasion. Furthermore, non-damaging ECVFs can also indirectly increase pathogenicity of both organisms during co-infection by aiding in immune evasion, adhesion, cell wall biosynthesis, and iron acquisition.
In vivo murine studies using oral co-infection of C. albicans and S. aureus have shown that macrophages and neutrophils, isolated from draining oral lymph nodes, contained viable S. aureus following co-infection with C. albicans but not following any monoculture infections (Allison et al., 2019). Moreover, while low-level immunosuppression crucially contributes to the instigation of invasive candidiasis and the co-invasion/dissemination of S. aureus in mice, high-level immunosuppression significantly reduced S. aureus dissemination during C. albicans co-infection (Van Dyck et al., 2021). This reduced level of S. aureus dissemination was linked to both neutropenia and decreased monocyte production which, together, result in a significantly lower number of neutrophils and macrophages at the site of infection in tissue. In vivo murine studies using intra-abdominal co-infection of C. albicans and S. aureus have shown that co-infection amplified host inflammation, resulting in both a significant increase in neutrophil influx towards the site of infection and prolonged neutrophil residence at the site of infection (Peters and Noverra, 2013; Nash et al., 2014). Together, these studies have highlighted the importance of the immune system in the lethal dissemination of S. aureus during co-infection with C. albicans. The most crucial aspect of immune facilitated dissemination, however, is the fact that S. aureus is notorious for surviving phagocytic killing, allowing S. aureus to disseminate following phagocytosis and phagocyte migration (Kubica et al., 2008; DuMont et al., 2013; Flannagan et al., 2016; Pidwill et al., 2021). While the secretome of C. albicans and S. aureus has been shown to promote the murine macrophage production and secretion of IL-6, NO, and TNF-α during co-culturing (de Carvalho Dias et al., 2017), the effect of C. albicans/S. aureus co-culture secretomes on human macrophages remains unknown. In addition, the impact of co-culturing on the ECVFs in the secretome remain to be elucidated. Therefore, the aim of this study was to identify which ECVFs are secreted by C. albicans and S. aureus and how co-culturing influences the virulence potential. We investigated the contribution of Als1/Als3 binding, biofilm integration, and C. albicans pH maintenance in mediating the changes in ECVF during co-culturing. Additionally, we tested whether co-culturing of C. albicans and S. aureus promoted ECVF cytotoxicity towards human oral squamous cells and inflammation in human macrophages.
Methods
Strains and growth conditions
S. aureus ATCC12600, C. albicans SC5314 wild type, and C. albicans SC5314 als1/als3 (genotype: als1-1Δ::FRT/als1-2Δ::FRTals3-1Δ::FRT/als3-2Δ::FRT) (Van Dyck et al., 2021) were grown as described before (Pasman et al., 2024). In short, C. albicans and S. aureus strains were maintained from glycerol freezer stocks on Sabouraud dextrose/glucose agar supplemented with chloramphenicol (Sigma, 63567) and mannitol salt phenol red agar (MSA, Sigma, 89579), respectively. Single colonies were added to tryptic soy broth (TSB; Brunschwig Chemie, 211825) and cultured overnight at 37°C, 200 rpm. Cultures were rinsed with Dulbecco’s phosphate-buffered saline (DPBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2HPO4) and diluted to ~2 × 106 CFU/mL for C. albicans and ~2 × 107 CFU/mL for S. aureus in Dulbecco’s Modified Eagle’s Medium (Sigma D5030) supplemented with 2.5 g/L dextrose, 1× GlutaMAX (Gibco 35050061), and 1× MEM non-essential amino acids (Gibco 11140050), with a final pH of 7.3 (mDMEM-DMP) (Berman, 2012). Monocultures were generated by further diluting the culture in a 1:1 ratio with mDMEM-DMP, whereas co-cultures were constituted by combining (undiluted) monocultures in a 1:1 ratio. For buffered growth, mDMEM-DMP was supplemented with 100 mM of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Gibco 15630056).
Biofilm growth and assessment
Mono- and co-culture biofilms were grown by inoculating wells of a six-well plate (Corning 3506) with 3 mL of culture, prepared as described above, and incubating it stationarily for 2 h at 37°C. Each well was washed for three times using DPBS to remove non-adherent cells. Finally, fresh mDMEM-DMP was added to the wells and all plates were incubated stationary for 72 h at 37°C in a humidified environment. Concerning Transwell co-cultures, 1.5 mL of C. albicans monoculture was added to the inside of the 0.4 µm Transwell inserts (Thermo Scientific, 140660) whereas 1.5 mL of S. aureus monoculture was added to the inside of the well plate. Transwell co-cultures were further treated identical to normal cultures with half the volume added on either side of the membrane. Medium pH was measured by pH 2.0–9.0 strips (Supelco, 1.09584) inoculated with 40 µL of medium. Culture medium was sampled for downstream analysis, as described below. Biofilms were collected using cell scrapers (Greiner, 391-3010), pelleted, and stored at −80°C until further analysis using qPCR.
Genomic DNA extraction and qPCR
Genomic DNA was extracted from the collected cell pellets using a DNeasy PowerBiofilm Kit (Qiagen, 24000) according to the manufacturer’s protocol. To determine the amount of fungal and bacterial DNA, a quantitative polymerase chain reaction (qPCR) was performed using primers specific to the fungal 28S rRNA gene (forward: GCATATCAATAAGCGGAGGAA AAG; reverse: TTAGCTTTAGATGATTTACCACC; probe: 6FAM-CGGCGAGTG AAGCGGSAARAGCTC-BHQ) (Vollmer et al., 2008) and bacterial 16S rRNA gene (forward: TCCTACGGGAGGCAGCAGT; reverse: GGACTACCAGGGTATCTAATCCTGTT; probe: 6FAM-CGTATTACCGCGGCTGCTGGCAC-BHQ1) (Ciric et al., 2010). Fluorescence was measured using a LightCycler 480 System (Roche) and analyzed using the corresponding system software.
Secretome sample preparation for proteomic analysis, cytotoxicity assay, and macrophage exposure experiments
To concentrate the secreted proteins in the spent media, the medium collected from six wells of the same culturing conditions was pooled and filtered using a 0.2-µm polyethersulfone (PES) filter (Sarstedt, 83.1826.001). The filtrate was further concentrated with a 3-kDa Amicon ultra centrifugal filter (Pall, MAP003C37). The concentrated secretome samples were immediately stored without protease inhibitor supplementation for cytotoxicity assay, or supplemented with protease inhibitors (Roche, 11873580001) according to manufacture manual for proteomic analysis, or supplemented with RPMI 1640 to the original volume before protein extraction for macrophage activation experiments described below. Protein concentrations were determined using a Bradford protein assay, and prior to LC-MS proteomic analysis, protein concentrations were diluted to identical levels across all samples. All secretome samples were stored at −80°C until use.
Sample preparation for LC-MS analysis
Samples were prepared and measured according to Šimkovicová et al. (2024) (Šimkovicová et al., 2024). In short, samples were thawed, reduced, and alkylated by incubation with tris-(2-carboxyethyl)phosphine (10 mM) and chloroacetamide (30 mM) for 30 min, 70°C. Next, samples were prepared for mass spectrometry analysis using the single-pot, solid-phase-enhanced sample preparation (SP3) protocol (Hughes et al., 2014). Soluble protein recovery was optimized by ensuring no detergents were added to the samples and precipitation time was extended to 30 min (room temperature). Beads used for washing were air-dried and resuspended in ammonium bicarbonate (100 mM) after which trypsin (Sequencing Grade Modified) was added at a protease-to-protein ratio of 1:50 (w/w) at 37°C. Formic acid was added to the overnight digestion at a final concentration of 1% and pH of 2. Finally, peptides were recovered using a magnetic separator device.
LC-MS analysis for quantitative proteomics
Samples were separated by reversed-phase chromatography using an UltiMate 3000 RSLCnano UHPLC system (Thermo Scientific, Germeringen, Germany), and peptides were separated using a 75 μm × 250 mm analytical column (C18, 1.6 μm particle size, Aurora, IonOpticks, Australia), maintained at 50°C, and operated at a flow rate of 400 nL/min with 3% solvent B for 3 min (solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile, ULCMS-grade, Biosolve). Next, a multi-stage gradient was applied (17% solvent B at 21 min, 25% solvent B at 29 min, 34% solvent B at 32 min, 99% solvent B at 33 min, kept at 99% solvent B till 40 min). For equilibration, the system was returned to initial conditions (t = 40.1 min) for 58 min. Eluted peptides were electrosprayed by a captive spray source via the column-associated emitter and were analysed by a TIMS-TOF Pro mass spectrometer (Bruker, Bremen, Germany) operated in PASEF mode for standard proteomics acquisition. MS/MS scans were initiated 10× with a total cycle time of 1.16 s, a target intensity of 2 × 104, an intensity threshold of 2.5 × 103, and a charge state range of 0–5. Active exclusion was enabled for a period of 0.4 min and precursors reevaluated when the ratio of current intensity:previous intensity exceeded 4.
Spectral data processing and proteome database search
LC-MS data were processed using MaxQuant software (version 1.16.14.0) using standard settings, i.e., trypsin/p as the enzyme allowing for two missed cleavages with carbamidomethylation at cysteine as a fixed modification and oxidation at methionine as a variable modification searching the proteome databases of: Candida_albicans_UP000000559 and Saureus_UP000008816. MaxQuant outputs were used for subsequent analysis using Perseus (version 2.0.7.0). Proteins that are only identified by peptides carrying one or more modified amino acids as well as reverse and potential contaminant proteins were removed from the dataset. Remaining data were log2 transformed, and proteins that were not measured in both samples of at least one condition were removed. Next, remaining proteins were annotated using the 2019_11 release of the Max Planck Institute of Biochemistry annotation database (C. albicans SC5314 and S. aureus NCTC 8325), after which the dataset was split into two sets: one set containing all C. albicans proteins and one set containing all S. aureus proteins. For both sets, missing values were imputed based on the low end of the corresponding normal distribution (width = 0.3, down shift = 1.8) and all values were subtracted with the most frequent value. Protein differences were tested on significance using ANOVA with a permutation-based FDR of 0.05 and 250 number of randomizations after which non-significant proteins were removed. Principal component analysis was used to identify sample group differences based on the remaining proteins. Finally, remaining data were normalized using Z-score normalization and significantly differing proteins between monoculture and co-culture conditions were identified using volcano plots based on a Pearson correlation with an FDR of 0.05. Using the virulence factor database (Liu et al., 2022), Aureo Wiki (Fuchs et al., 2018), UniProt (UniProt Consortium, 2018), STRING database (Szklarczyk et al., 2023), and various studies (Calderone and Fonzi, 2001; Sorgo et al., 2010; Zecconi and Scali, 2013), extracellular (ECVFs) and non-extracellular (N-ECVFs) virulence factors were identified. Results were visualized using Microsoft Excel (version 2402, build 17328.20282).
Human oral squamous cell culture and cytotoxicity assay
Ca9-22 (gingival squamous carcinoma) and HO1N1 (buccal epithelial carcinoma) cells were cultured in DMEM + 10% foetal bovine serum (FBS; Sigma) at 37°C, 5% CO2. Cells reached confluence within 10 days and were washed using PBS, detached by trypsinization (0.05% trypsin) for 3 min at 37°C, spun down, and diluted to 1 × 105 cells/mL using DMEM/F12 (Gibco) supplemented with 10% FBS. Cells were inoculated (1 × 105 cells/well) into wells of a 24-well plate (Greiner) and incubated for 24 h at 37°C, 5% CO2. Ca 9–22 and HO1N1 cells were exposed to secretome protein isolates (1:10 diluted in mDMEM-DMP, 100 mM HEPES), acquired as described above but without protease inhibitor supplementation, and incubated for 24 h at 37°C, 5% CO2. Medium was collected and stored at −20°C until cytotoxicity assay. Cytotoxicity was determined using a Roche LDH Kit PLUS (Cat. No. 04 744 934 001) according to the manufacturer’s protocol. Medium of cells lysed with 1% Triton-X 100 in mDMEM-DMP (100 mM HEPES) was used as positive control, whereas medium of unexposed macrophages was utilized as negative control.
THP-1 cell culture and differentiation
THP-1 monocytes were maintained in culture in Roswell Park Memorial Institute medium (RPMI 1640, Gibco, 11875093) supplemented with 10% foetal bovine serum (Sigma F9665), 1% GlutaMAX (Gibco, 35050061), and 1% Pen-strep (Gibco, 15140-122). THP-1 monocytes were seeded in either 96- or 24-well plates at a density of 0.5 × 106 cells/mL and differentiated into M0 macrophages by incubating the cells for 72 h at 37°C, 5% CO2 in RPMI 1640 supplemented with 100 nM phorbol 12-myristate 13-acetate (PMA, Millipore 500582). Following a 72-h incubation, cells were washed with Dulbecco’s phosphate-buffered saline (DPBS, Gibco, 14190177, 37°C) and used for downstream application. Differentiated macrophages were imaged at 10× magnification using a Leica DM13000 B coupled to a ZEISS Axiocam MRc.
Secretome cytotoxicity
THP-1 M0 macrophages, differentiated in 96-well plates, were incubated in RPMI 1640 supplemented with various dilutions of the conditioned media (1:25, 1:50, 1:100, and 1:200, diluted in RPMI 1640) for 6 h at 37°C, 5% CO2. Macrophages in RPMI 1640 served as the negative control, whereas macrophages treated with 1% Triton-X 100 for 10 min before medium collection served as the positive control. Following incubation, medium was collected and LDH activity was quantified using an LDH-Glo™ Cytotoxicity Assay (Promega, J2380), measured with a Synergy Mx (Bio Tek). Cytotoxicity was determined as follows: Percentage of cytotoxicity (%) = 100 * ((Experimental LDH release – negative control)/(Positive control LDH release – negative control)).
RNA isolation, sequencing, and data analysis
Macrophages were lysed by incubating in 350 µL of 1% Triton-X 100 (Bio-Rad) in DPBS for 10 min. Subsequently, 350 µL of buffer RLT in the RNeasy Plus Mini Kit (Qiagen, 74134) supplemented with 1% β-mercaptoethanol was added, mixed with pipetting, harvested in 1.5-mL tubes, and stored at −70°C until RNA isolation. The total RNA was then isolated by following the manufacturer’s protocol. The RNA purity was measured using a DeNovix DS-11+ spectrophotometer, with RNA integrity number of all samples 9.9–10.0 except for one at 9.7 due to the high concentration of total RNA.
The mRNA enrichment and library preparation was carried out by the Dutch Genomics Service & Support Provider at the University of Amsterdam. The NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) was used to perform a poly-A enrichment using 1 μg total RNA. RNA-Seq libraries were generated according to the manufacturers’ protocols using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina and NEBNext Multiplex Oligos for Illumina (Unique Dual Index Primer Pairs) (New England Biolabs). The size distribution of the libraries with indexed adapters was assessed using a 2200 TapeStation System with Agilent D1000 ScreenTapes (Agilent Technologies). The libraries were quantified on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) using the NEBNext Library Quant Kit for Illumina (New England Biolabs) according to the instructions of the manufacturer. The libraries were clustered and sequenced (75 bp) on a NextSeq 550 System (Illumina) using a NextSeq 500/550 High Output Kit v2.5 (75 Cycles) (Illumina).
The raw sequencing data were subjected to quality control with FastQC v0.11.9 (Andrews, 2010) and MultiQC version 1.21 (Ewels et al., 2016). Subsequently, the reads were trimmed with Trimmomatic v0.39 (Bolger et al., 2014). Post-trimming quality control indicated that all samples were of similar high quality. The reads were aligned (without soft clipping) to the human reference genome (GRCh38.111) using HISAT2 version 2.2.1 (Kim et al., 2015). HTSeq-count version 2.0.5 (Anders et al., 2015) was used to count the amount of reads per gene and determine the gene expression values. The gene expression data were checked for quality and subsequently normalized and analysed with DESeq2 (Love et al., 2014). The technical replicates were collapsed and statistical tests controlling for batch effects were performed. The differences between the monocyte control and the medium control were assessed by comparing differentially expressed genes (DEGs), i.e., genes with a FDR-adjusted p-value < 0.01 and a log2 fold change above 1 or below −1, with previously identified DEGs following THP-1 monocyte differentiation (Liu et al., 2023). DEGs were identified between all conditions and visualized using Microsoft Excel (version 2403). Expression of M1 and M2 polarization related genes was identified according to previously published gene sets for differential gene expression (Mills and Ley, 2014; Murray, 2017; Shapouri-Moghaddam et al., 2018; Rynikova et al., 2023). Finally, to identify pathways containing overrepresented genes, a Gene Set Enrichment Analysis (GSEA) was performed using WebGestalt 2019 (Liao et al., 2019) with a KEGG-based pathway analysis, a weighted set cover redundancy removal, and a significance level cutoff of FDR-adjusted p-value < 0.01. WebGestalt output pathways were visualized using both Microsoft Excel (version 2403) and Python (version 3.8.5). Gene ratios were determined by dividing the number of enriched genes in a pathway by the total number of genes of that pathway.
Statistical analysis
All data were tabulated and visualized using Microsoft Excel (version 2403). When applied, data normality was tested using a Shapiro–Wilk test, and group comparison was performed using either a one-way ANOVA (normally distributed data) or Kruskal–Wallis test (non-normally distributed data) combined with a Tukey post analysis using Prism graph pad (8.3.0). All conditions were tested in at least three biological replicates and technical duplicates.
Results
Co-culturing of C. albicans and S. aureus increases the ECVF and N-ECVF secretion by C. albicans
To investigate the effect of C. albicans and S. aureus co-culturing on the secreted proteins, we measured the levels of the proteins in the spent media collected from different co-culture and monoculture conditions (see Methods). Following mass spectrometry analysis, we detected 183 C. albicans proteins in both samples of at least one culturing condition. Principal component analysis (PCA) revealed a clear separation between co-cultures and monocultures (Figure 1A), indicating that C. albicans-secreted protein profiles were changed by the presence of S. aureus. Of the 21 known C. albicans ECVFs (Supplementary Table S1), 14 ECVFs were detected in both samples of at least one culturing condition and 12 ECVFs were significantly increased in co-culture compared with monoculture (Figure 1B; Supplementary Table S1). Among these, five ECVFs, i.e., Plb1, Sap4, Sap6, Sod5, and Csa2, were significantly increased only in wild-type co-culture compared with monoculture, but this increase was abolished in ALS1/ALS3 ΔΔ/ΔΔ or Transwell setting (Supplementary Table S1), suggesting that their secretion was strongly dependent on Als1/Als3-mediated binding, whereas secreted aspartic protease 5 (Sap5) and glycosidase Crh11 was independent of Als1/Als3-mediated binding and other types of physical interaction (Supplementary Table S1). Interestingly, the remaining five ECVFs, i.e., Sap9, glycosidase Utr2, glucan 1,3-beta-glucosidase Xog1, PR-1 protein homolog Rbe1, and Rbt4, were significantly increased during both regular and Transwell-separated co-culturing with wild-type C. albicans but were not changed following co-culturing with C. albicans ALS1/ALS3 ΔΔ/ΔΔ (Supplementary Table S1), indicating that their release was strongly dependent on soluble factors and independent of Als1/Als3-mediated binding (Figure 1B; Supplementary Table S1). These significantly increased C. albicans ECVFs contribute to damaging functions such as proteolysis, immune evasion, cell wall modelling, and iron acquisition, suggesting that co-culturing C. albicans with S. aureus promotes virulence of C. albicans. It is worth noting that C. albicans candidalysin, a known virulence factor, does not contribute to the virulence increase in co-cultures since C. albicans candidalysin was detected but did not significantly change following co-culturing with S. aureus (Supplementary Table S1).
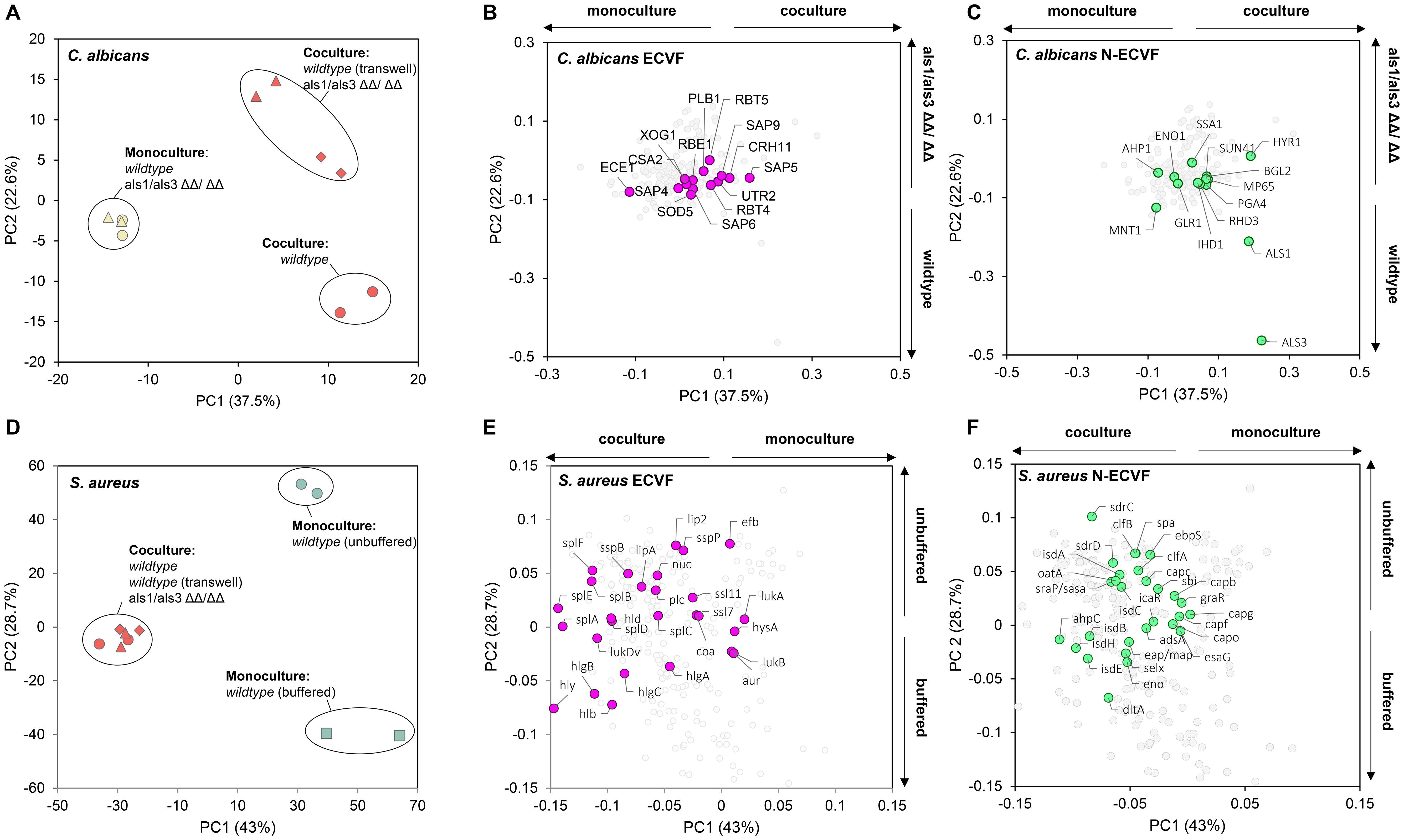
Figure 1. Co-culturing changes extracellular virulence factors and non-extracellular virulence factors released by C. albicans and S. aureus. (A) Principal component analysis and (B, C) corresponding loading plot of Candida albicans spent medium proteins that were detectable in at least one of the samples. Extracellular virulence factors (ECVF) and non-ECVF (N-ECVF) are highlighted in magenta in (B) and in green in (C), respectively. Culture conditions in (A) yellow filled circle: C. albicans wild-type, yellow filled triangle: C. albicans ALS1/ALS3 ΔΔ/ΔΔ, red filled circle: co-culture wild-type, red filled triangle: co-culture ALS1/ALS3 ΔΔ/ΔΔ, red filled diamond: co-culture Transwell. C. albicans ECVFs are colored in magenta in (B). (D) Principal component analysis and (E, F) corresponding loading plot of Staphylococcus aureus spent medium proteins that were detectable in at least one of the samples. ECVF and N-ECVF are highlighted in magenta in (E) and in green in (F), respectively. Culture conditions in (C): red filled circle: co-culture wild-type, red filled triangle: co-culture ALS1/ALS3 ΔΔ/ΔΔ, red filled diamond: co-culture Transwell, green filled square: S. aureus monoculture buffered, green filled circle: S. aureus monoculture unbuffered. S. aureus ECVFs are colored in pink in (E).
Next, we expanded our investigation to non-extracellular virulence factors (N-ECVFs), as cytoplasmic proteins in C. albicans were showed to be released extracellularly via extracellular vescicles (Gil-Bona et al., 2015). We identified 42 N-ECVFs, of which 14 C. albicans N-ECVFs were significantly changed and mainly contributed to co-culture-based (PC1, 37.5%) and ALS1/ALS3 deletion-based (PC2, 22.6%) separation (Figure 1C). Most of the 14 significantly changed C. albicans N-ECVFs were related to adhesion or cell wall remodeling. For instance, hyphae-regulated cell wall protein 1 (Hyr1) and heat shock protein 70 (Ssa1) were significantly increased in wild-type co-culture versus C. albicans monoculture, but this change was attenuated in als1/als3 ΔΔ/ΔΔ co-culture and Transwell settings (Supplementary Table S2). Additionally, thioredoxin peroxidase (Ahp1) and glycolipid 2-alpha-mannosyltransferase 1 (Mnt1), involved in the reduction of hydrogen peroxide and O-glycosylation of cell wall, respectively, were significantly decreased in wild-type and Transwell co-culture, and this decrease was enhanced in ALS1/ALS3 ΔΔ/ΔΔ co-culture. These results suggest that Hyr1, Ssa1, Ahp1, and Mnt1 are influenced by both Als1/Als3-dependent C. albicans–S. aureus interaction and other S. aureus factors. In addition, cell wall protein (Rhd3) and cell wall remodeling-related proteins Ihd1, Sun41, Pga4, Als1, Als3, Mp65, and Bgl2 were significantly increased in wild-type co-culture (Supplementary Table S2), which was attenuated by the physical separation and completely abolished in ALS1/ALS3 ΔΔ/ΔΔ co-culture (Supplementary Table S2). Moreover, the enolase (Eno1) level was not altered during wild-type co-culturing, whereas its level was decreased in ALS1/ALS3 ΔΔ/ΔΔ and Transwell co-cultures (Supplementary Table S2). Together, these results suggest that the Als1/Als3-dependent and independent physical interaction between C. albicans and S. aureus promotes the levels of non-extracellular virulence factors.
Co-culturing of C. albicans and S. aureus promotes ECVF and N-ECVF secretion by S. aureus
Co-culturing also influenced the release of virulence factors by S. aureus. We detected 930 S. aureus proteins in at least one culturing condition. Principal component analysis indicates a clear separation between monocultures and co-cultures (Figure 1D), suggesting that the presence of C. albicans has substantial impact on the proteins released by S. aureus. It is worth noting that no separation was observed among three co-culture conditions (Figure 1D), indicating that deletion of ALS1/ALS3 or lack of physical interaction with C. albicans did not influence the S. aureus secretome compared with wild-type co-culture. Of the 50 known S. aureus ECVFs, 27 were detected in both samples of at least one culture condition and 20 were found statistically significantly changed (Figure 1E, Supplementary Table S3). Of the 20 changed ECVFs, hemolysins including alpha hemolysin (Hly/Hla), Hlb, and HlgA-C were increased in all conditions (Supplementary Table S3). During the co-culture, C. albicans can maintain the pH (Supplementary Figure S1A) from decreasing caused by S. aureus. Here, we observed that buffering the pH alone increased the secretion of these hemolysins to a similar extent as the C. albicans, and this increase was maintained despite the lack of physical contact or Als1/Als3-mediated binding (Figure 1E; Supplementary Table S3). This result, together with the maintained pH during C. albicans–S. aureus co-culture (Supplementary Figure S1A), suggests that C. albicans-maintained pH likely contributed to the increased release of the hemolysins. On the other hand, other S. aureus ECVFs were not influenced by pH but by other factors during co-culturing. For instance, cysteine proteinase staphopain B (SspB), serine protease-like protein A-F (SplA-F), lipase (Lip), phospholipase C (Plc), delta hemolysin (Hld), and leukotoxin D (LukD) were significantly more present in all co-culture conditions, but significantly less present or unaltered during buffered monoculture (Supplementary Table S3). Because ALS1/ALS3 deletion and separated growth did not deviate from wild-type co-culture results, these results are independent of Als1/Als3 and physical binding.
Similar to C. albicans, we also detected S. aureus N-ECVFs. Of the 28 detected N-ECVFs, 17 were significantly changed (Figure 1F). Buffering strongly impacted N-ECVF levels, evidenced by the fact that 11 and 2 N-ECVFs in buffered monoculture were significantly higher and lower than those in unbuffered monoculture, respectively (Figure 1F). This pH-mediated effect seemed to be divergently interfered with other factors from C. albicans. For example, the buffering increase of d-alanine-d-alanyl carrier protein ligase (DltA) and enolase (Eno) was further enhanced by the presence of C. albicans, and this enhancement is independent of Als1/Als3 or physical proximity (Figure 1F; Supplementary Table S4). For those proteins that were decreased by buffering, i.e., ClfB, Ebp, Oata, Srap/Sasa, IsdA, IsdC, IsdE, SpA, SdrC, and ClfA, the decrease was attenuated or even reserved by the presence of C. albicans (Supplementary Table S4), despite that C. albicans maintained pH comparably with HEPES buffering (Supplementary Figure S1A). These results suggest that other C. albicans-derived soluble factors have stronger effects in influencing these proteins compared with pH. Together, these results suggest that both C. albicans-derived pH maintenance has a broader impact than that of Als1/Als3-mediated binding on N-ECVFs of S. aureus.
Co-culturing promotes cytotoxicity to oral squamous cells
Previous in vivo murine studies have reported that oral inoculation of C. albicans potently promotes co-invasion and dissemination of co-inoculated S. aureus (Carlson, 1982; Carlson, 1983; Carlson, 1988; Nash et al., 2014; Kong et al., 2015; Schlecht et al., 2015; Nash et al., 2016; Allison et al., 2019; Van Dyck et al., 2021), suggesting that epithelium in the oral cavity is disrupted by co-infection. Reasoning that the increase in ECVF and N-ECVF in C. albicans–S. aureus co-culture would increase the damaging potential, we next sought to test whether the co-culture induces higher cytotoxicity to oral squamous cells. To test this, we exposed human gingival squamous Ca 9–22 and human buccal mucosa squamous HO1N1 cells to undiluted mono- and co-culture secretomes of C. albicans wild-type, C. albicans ALS1/ALS3 ΔΔ/ΔΔ, and/or S. aureus (cultured in either buffered or unbuffered medium) for 24 h and determined cytotoxicity by measuring lactate dehydrogenase activity in the medium. All monoculture secretomes, i.e., C. albicans wild-type, C. albicans ALS1/ALS3 ΔΔ/ΔΔ, and S. aureus (unbuffered), induce similar levels of cytotoxicity as the negative control (~20%–30%; Figure 2). In contrast, all tested co-culture secretomes and S. aureus buffered induced higher cytotoxicity to both Ca 9–22 cells and HO1N1 cells (Figure 2). These results are in agreement with the observed elevated levels of ECVFs in co-culture secretomes compared with monoculture. Interestingly, the secretome of buffered S. aureus monocultures showed higher cytotoxicity to Ca 9-22 (92%) and HO1N1 (79%) (Figure 2), despite that the levels of ECVFs in buffered S. aureus were comparable or lower than that in co-cultures (Supplementary Table S3).
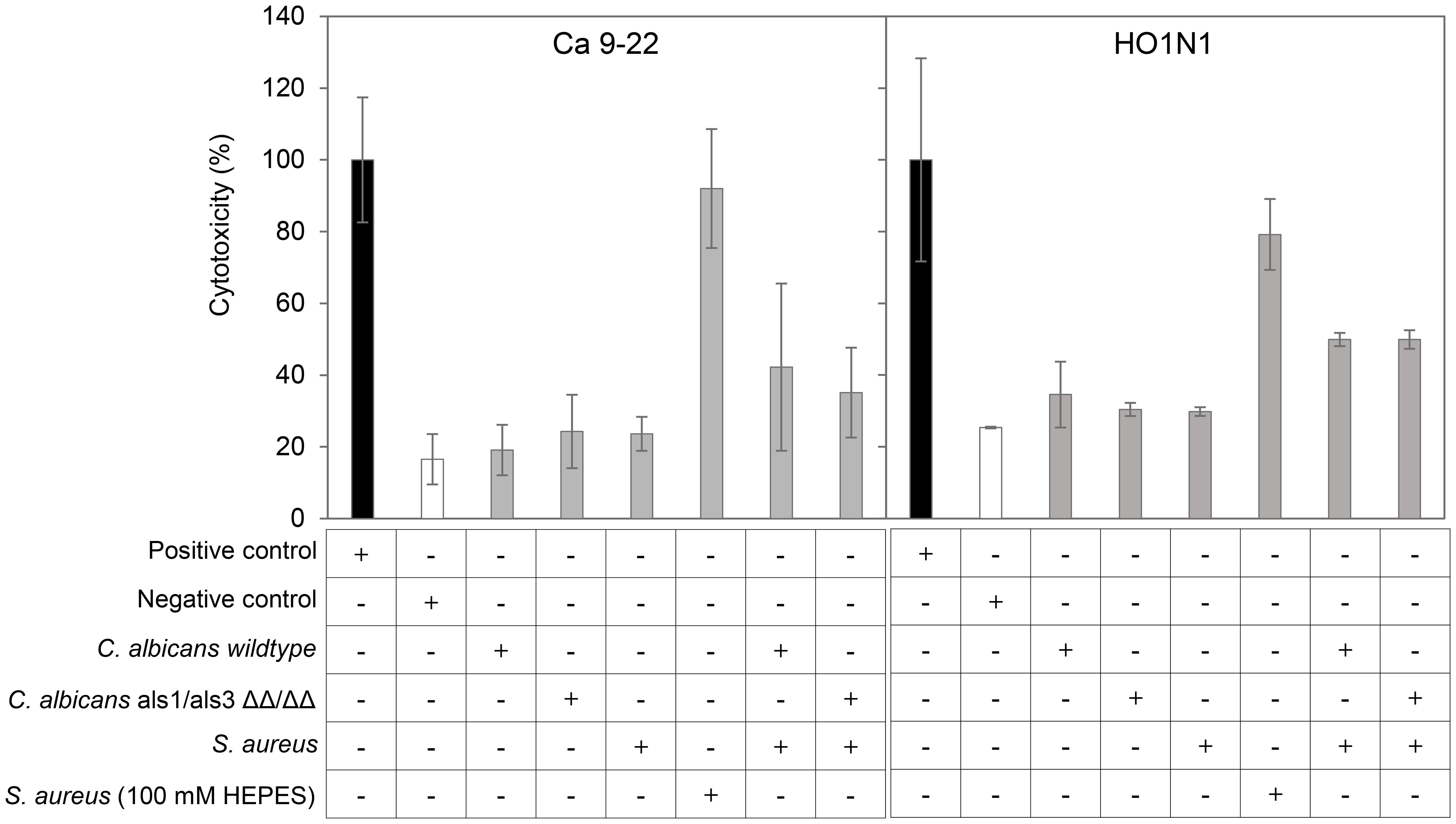
Figure 2. Co-culturing C. albicans and S. aureus induces slightly higher cytotoxicity on human oral epithelial cells. Cytotoxicity, expressed as the percentage of LDH activity in the medium relative to positive control 1% Triton X-100, after a 24-h exposure to secretomes from different culturing conditions on human gingival/oral squamous carcinoma Ca 9–22 and HO1N1 cells. Error bars represent standard variation of three wells in the same experiment. Negative control: corresponding medium (mDMEM-DMP).
The S. aureus but not C. albicans monoculture secretome is cytotoxic to macrophages and induces inflammation under non-cytotoxic doses
Next, we sought to investigate the effects of co-culturing on macrophages. Macrophages are first-line defense mechanisms during the infection in oral mucosa and are expected to encounter C. albicans and S. aureus during initial co-invasion of the mucosa. We differentiated THP-1 monocytes into M0 macrophages and verified the differentiation by comparing the macrophage and monocyte transcriptomes, as well as macrophage morphology (Supplementary Figure S2). Secretome of monocultures and co-cultures induced different levels of cytotoxicity to these macrophages (Figure 3A). Surprisingly, C. albicans monoculture showed similar cytotoxicity as the medium control. Similar to oral cells, co-cultures induced higher cytotoxicity than S. aureus and C. albicans monocultures, and the cytotoxicity is dose-dependent, with 1-to-200 dilution showing the same level of cytotoxicity as the medium (Figure 3A). We next sought to identify the molecular response in macrophages that were exposed to a non-cytotoxic level of secretome. Transcriptomic analysis revealed that S. aureus alone significantly increased the expression of 93 and decreased 8 (Figure 3B), with genes related to M1 macrophage polarization were mostly upregulated (Supplementary Figure S3), and M2 polarization-related genes were hardly affected (Supplementary Figure S4). Consistently, multiple proinflammatory pathways were increased in S. aureus secretome-exposed macrophages (Figure 3C). In contrast, C. albicans secretome did not significantly change the transcription of any genes in macrophages (Figure 3D), which is in agreement with the cytotoxicity data (Figure 2).

Figure 3. Effects of S. aureus and C. albicans secretome in macrophages. (A) Cytotoxicity of secretome on THP-1-derived macrophages using LDH assay. Secretome with a 1:200 dilution does not induce apparent cytotoxicity as compared with medium control (RPMI). (B) Volcano plot of gene transcriptional change in macrophages exposed to S. aureus secretome versus no exposure. (C) Volcano plot of gene transcriptional change in macrophages exposed to C. albicans secretome versus no exposure. Genes with a log2 fold change above 1 or below −1 and an FDR-adjusted p-value < 0.01 were considered significant (orange). (D) Gene set enrichment analysis based on the log2 fold change gene expression between macrophages exposed to S. aureus secretome and unexposed macrophages. Represented pathways are displayed. Dot sizes represent the total number of enriched genes found in the corresponding pathway, and color represents the gene ratio.
Secretome of C. albicans and S. aureus co-cultures amplifies inflammatory responses of THP-1 macrophages compared with monoculture secretomes
Compared with S. aureus monoculture secretome, THP-1 M0 macrophages exposed to co-culture secretome showed 467 significantly upregulated genes and 87 downregulated genes (Figure 4A), including higher transcription of genes related to M1 polarization, pro-inflammatory cytokines and chemokines (Supplementary Figure S3). Despite the activation of macrophages by S. aureus monoculture, 29 pro-inflammatory pathways were further enriched in co-culture secretome-exposed macrophages (Figure 4B), with the majority of these pathways (TNF signaling, TLR signaling, NFKB signaling, and cytokine–cytokine receptor interaction) upregulated as a result of exposure to S. aureus secretome versus medium. In agreement, co-culture secretome-exposed macrophages showed enrichment of the NOD2-like receptor signaling pathway, a known activating pathway of these pro-inflammatory pathways. Interestingly, pathways related to ribosomal and oxidative phosphorylation proteins were significantly negatively affected by exposure to co-culture secretome, indicating metabolic downregulation. When compared with C. albicans monoculture secretome, co-culture secretome-exposed macrophages showed 784 upregulated DEGs and 312 downregulated DEGs (Figure 4C) and enriched the proinflammatory pathways that are vastly overlapping with those when co-culture vs. S. aureus monoculture (Figure 4D).
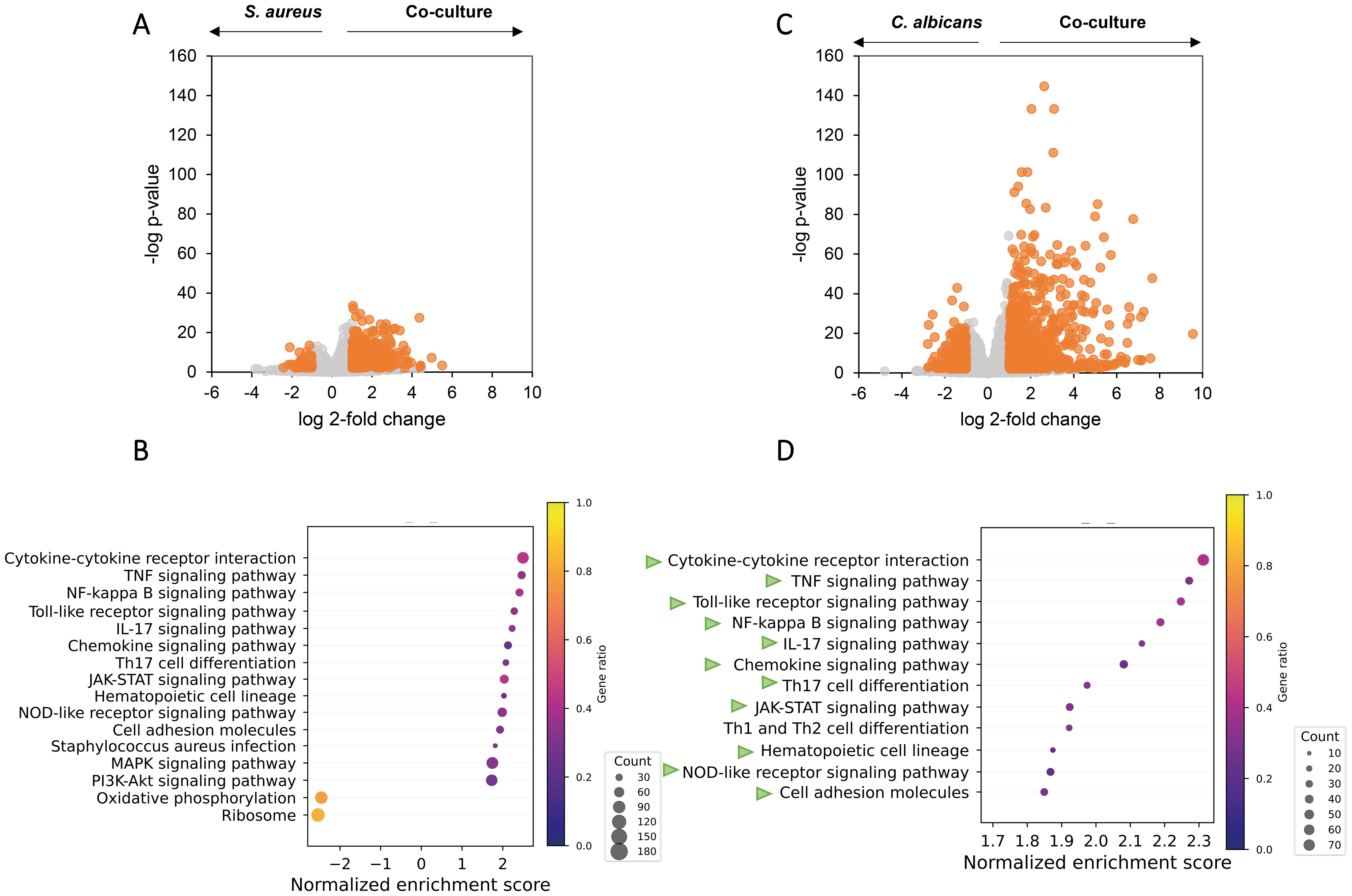
Figure 4. Co-culturing of C. albicans and S. aureus increases pro-inflammatory pathways in macrophages at the transcriptomic level. Volcano plot and overrepresented pathways on the transcriptomes of the macrophages exposed to secretomes from C. albicans wild-type-S. aureus co-culture versus secretomes from monoculture of S. aureus (A, B) or C. albicans (C, D). (A, C) Genes with a Log2 fold change above 1 or below −1 and an FDR-adjusted p-value < 0.01 were considered statistically significant and are highlighted in orange. (B, D) Gene set enrichment analysis based on the Log2 fold change ranking; see more details in Methods. Overrepresented pathways are displayed. Dot sizes represent the total amount of enriched genes found in the corresponding pathway, and color represents the gene ratio. Green arrows in (D) represents the same pathways that are overrepresented in (B).
Als1/Als3 deletion has a marginal impact on the proinflammatory effects of co-culture secretome
Reasoning that Als1/Als3 plays an important role in the virulence factor pattern in C. albicans–S. aureus co-culture, we hypothesize that depletion of ALS1/ALS3 will significantly alleviate the proinflammatory effects induced by wild-type C. albicans–S. aureus co-culture. To test this hypothesis, we compared the transcriptome of macrophages exposed to the wild-type co-culture secretome versus C. albicans ALS1/ALS3 ΔΔ/ΔΔ–S. aureus co-culture secretome. Surprisingly, only two genes were significantly changed (Figure 5A), indicating that ALS1/ALS3 gene deletion has marginal impact on the gene transcription in macrophages. Compared with S. aureus secretome, ALS1/ALS3 ΔΔ/ΔΔ co-culture secretome showed 97 upregulated genes and 2 downregulated genes in macrophages (Figure 5B). Despite that there were less significantly changed genes in ALS1/ALS3 ΔΔ/ΔΔ co-culture versus S. aureus compared with wild-type co-culture versus S. aureus, similar proinflammatory pathways (Figure 5C), including the TLR2 and NOD2-like receptor signaling pathways, were enriched in ALS1/ALS3 ΔΔ/ΔΔ co-culture, affirming that ALS1/ALS3 deletion does not affect the proinflammatory effects of the co-culture.
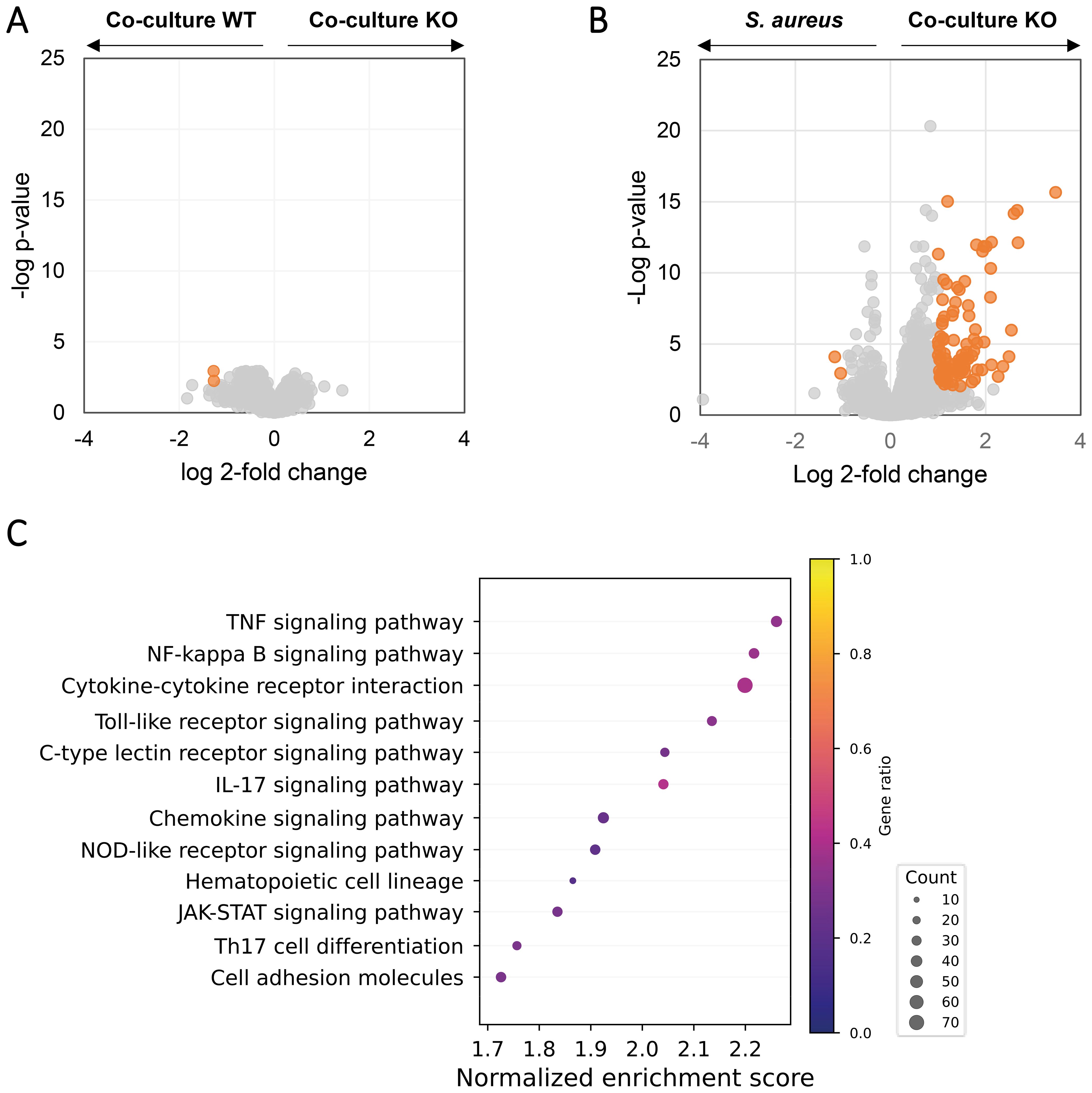
Figure 5. Knockout of als1/als3 in C. albicans does not attenuate the pro-inflammatory potential of the C. albicans–S. aureus co-culture. Volcano plot of gene expression in macrophages exposed to the secretomes from C. albicans ALS1/ALS3 ΔΔ/ΔΔ–S. aureus co-culture versus C. albicans wild-type–S. aureus co-culture (A) or S. aureus monoculture (B). Genes with a Log2 fold change above 1 or below −1 as well as an FDR-adjusted p-value < 0.01 were considered statistically significant and highlighted in orange. (C) Overrepresented pathways in macrophages exposed to the secretome from C. albicans ALS1/ALS3 ΔΔ/ΔΔ–S. aureus co-culture versus S. aureus monoculture based on gene set enrichment analysis. Dot sizes represent the total amount of enriched genes found in the corresponding pathway and color represent the gene ratio.
Discussion
Previous studies have shown that co-infections of C. albicans and S. aureus significantly promote lethality compared with mono infections (Carlson, 1988; Nash et al., 2014; Kong et al., 2016; Nash et al., 2016). Co-invasion and dissemination of S. aureus are crucial to this process and are facilitated by the secretion of damaging ECVFs and hyphal invasion of C. albicans. Our and others’ previous work have shown that C. albicans hyphal formation and invasion is increased by soluble factors of S. aureus (Peters et al., 2010). Using proteomics, we systemically profiled the ECVFs and N-ECVFs released by both C. albicans and S. aureus under various in vivo-relevant conditions. Our results showed that co-culturing significantly increased the levels of ECVFs and N-ECVFs. While Als1/Als3 binding mainly influenced the C. albicans virulence factors, C. albicans-mediated pH maintenance primarily contributed to the increase of S. aureus virulence factors. The increase of these virulence factors promotes cell cytotoxicity and proinflammatory effects.
C. albicans virulence is promoted by S. aureus during co-culturing and mainly attributed to Als binding
ECVFs are essential for various pathogenic processes of C. albicans, such as hyphal formation, damaging host cells, invasion, and immune evasion. Previously, Als3, Csa2, Rbt4, Sap4, and Sap6 proteins were found to be enriched in the secretome of N-acetylglucosamine-induced hyphal growth over yeast and contribute to the virulence of C. albicans pathogenesis, and transcription of SOD5 was increased in hyphal growth (Naglik et al., 2003; Sorgo et al., 2010; Sorgo et al., 2011; Röhm et al., 2013; Okamoto-Shibayama et al., 2014). Similarly, we showed that S. aureus-promoted C. albicans secreted higher levels of Csa2, Rbt4, Sap4-6, and Sod5. Deletion of RBT4 (Naglik et al., 2003; Martchenko et al., 2004; Sorgo et al., 2010; Sorgo et al., 2011; Röhm et al., 2013; Okamoto-Shibayama et al., 2014), SOD5 (Martchenko et al., 2004), SAP4, and SAP6 (Braun et al., 2000) significantly attenuates or completely diminishes C. albicans lethality in animal models, partially due to the depletion of RBT4-mediated resistance to leucocyte attack (Naglik et al., 2003; Martchenko et al., 2004; Sorgo et al., 2010; Sorgo et al., 2011; Röhm et al., 2013; Okamoto-Shibayama et al., 2014), SAP4-6-mediated resistance to macrophage killing (Borg-von Zepelin et al., 1998), and SOD-aided protection against intracellular neutrophil killing (Martchenko et al., 2004). In addition to these proteins, we found that hyphae-related proteins (Bailey et al., 1996; Martin et al., 2013), such as Als1, Als3, Ihd1, and Hyr1, were higher in co-cultures over monoculture. This is in agreement with microscopic/morphological characterization, showing increased hyphae in co-culture (data not shown). Furthermore, the co-culture promotes the level of Xog1 in the secretome. Xog1 is an exo-1,3-beta-glucanase essential for reducing beta-glucan epitope exposure (β-glucan masking) to immune cells, hence reducing the phagocytotic interaction and enhancing immune evasion (Ballou et al., 2016; Childers et al., 2020). Two other cell wall crosslinking enzymes Crh11 and Utr2 were also upregulated during co-culturing. Crh11 and Utr2 were shown to impact β-glucan masking mildly (Ballou et al., 2016). Together, these results support the contribution of these proteins in C. albicans–S. aureus co-culture to the pathogenesis during coinfection.
S. aureus virulence is significantly promoted by C. albicans during co-culturing
Similarly to C. albicans, the co-culturing also promotes the virulence potential of S. aureus by increasing the secretary level of cytolytic, proteolytic, or lipolytic proteins. We found that C. albicans-mediated pH maintenance is a critical factor in regulating these proteins. This is consistent with previous observations, where C. albicans tended to maintain a neutral pH during co-culturing (Supplementary Figure S1A) (Todd et al., 2019b; Pasman et al., 2024) and, thereby, promoted the production and secretion of alpha hemolysin (hla) (Todd et al., 2019b), and hlb and hlg. This is likely due to the C. albicans-mediated activation of the P3 promoter of the S. aureus agr system, which increases the expression of RNAIII that is essential for the production and secretion of these hemolysing toxins (Novick et al., 1993; Dunman et al., 2001). While we and others confirmed the contribution of pH in regulating S. aureus virulence factors, other unknown C. albicans-derived factors also contributed to the elevated virulence potential of S. aureus. For example, the ECVFs were significantly decreased in buffered monocultures over unbuffered monoculture but significantly increased in co-culture over unbuffered monoculture. These results further highlighted the nuanced regulation of the secreted virulence factors and further studies are warranted to identify these C. albicans-derived factors.
As many virulence factors from both C. albicans and S. aureus were substantially increased in co-culturing, it is not surprising that the secretome from co-culture exhibited higher cytotoxicity toward Ca 9–22 and HO1N1 cells compared with that from the monocultures. Consistently, C. albicans–S. aureus co-culturing showed higher cytotoxicity toward keratinocytes NOK-si and HaCat cells, albeit to a different extent (de Carvalho Dias et al., 2017). This discrepancy might be due to the different sensitivity of these cell lines.
C. albicans and S. aureus reciprocally promote the iron acquisition potential of each other
An important strategy for pathogenic microbes is to exploit host heme/iron sources during the infection. The increase in both C. albicans and S. aureus iron acquisition potential may also explain the high lethality of co-infection in vivo in animal models (Carlson, 1983). For C. albicans, Csa2 is critical in the uptake of hemoglobin and heme proteins and their utilization as an iron source (Okamoto-Shibayama et al., 2014) and this process is assisted by Rbt5, Pga7, Frp1, and Frp2 (Kuznets et al., 2014; Nasser et al., 2016; Roy et al., 2022). We found that co-culturing increases the secreted level of Csa2, but not Rbt5, Pga7, Frp1, and Frp2 in C. albicans. For S. aureus, the Isd (iron-regulated surface determinant) system is essential for hemoglobin and heme binding, uptake, and iron release (Caza and Kronstad, 2013). Of all the isd proteins, the levels of cell wall anchored isdA, isdB, isdC, isdE, and isdH, but not intracellular isdI and isdG, were significantly higher in co-culture, supporting the potential contribution of these proteins in the in vivo pathogenesis. Nonetheless, a previous study found that none of isd proteins was significantly changed in co-culture over monoculture (Peters et al., 2010).
Limitations of the study
While we systemically elucidated the reciprocal effects of C. albicans and S. aureus on their virulence factor secretion, cytotoxicity, and proinflammatory effects, there are several limitations of this study. First, the co-culturing conditions did not perfectly match the in vivo microenvironment, despite our effort to develop a completely defined medium to remove unknown artificial co-founding factors in the culturing media (Pasman et al., 2024). Future investigation may consider more physiologically relevant in vitro systems such as the reconstructed human gingiva–microbe interaction model (Zhang et al., 2022). In addition, all the culturing was performed under an oxygenated environment. Similar to our study, most studies carried out in vitro C. albicans cultures in a regular CO2 incubator that maintains 5% CO2. These conditions do not necessarily represent the in vivo physiology, since commensal C. albicans cells reside in the oral cavity and large intestine, where the oxygen tension can be relatively low. Furthermore, it must be noted that lowered biofilm DNA concentrations were observed regarding the C. albicans ALS1/ALS3 ΔΔ/ΔΔ conditions with respect to its wild-type strain (Supplementary Figure S1). However, no differences were observed between C. albicans wild-type and ALS1/ALS3 ΔΔ/ΔΔ in secretome protein concentrations (Supplementary Figure S1). Finally, we used THP-1-derived macrophages instead of primary macrophages or primary monocyte-derived macrophages. Despite that THP-1 cells are widely used to model human monocytes and macrophages (Mohd Yasin et al., 2022), it has been shown that THP-1 macrophages may show different response patterns as the primary macrophages (Hoppenbrouwers et al., 2022).
Data availability statement
The datasets presented in this study are deposited in the Gene Expression Omnibus repository, accession number GSE289787; the proteomexchange repository, accession number PXD061998.
Author contributions
RP: Conceptualization, Visualization, Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Investigation, Methodology. BK: Conceptualization, Supervision, Writing – review & editing. MJ: Data curation, Formal analysis, Writing – review & editing. Wd: Data curation, Writing – review & editing. GK: Data curation, Writing – review & editing. SB: Writing – review & editing, Conceptualization, Funding acquisition, Supervision. SZ: Conceptualization, Supervision, Writing – review & editing. JZ: Conceptualization, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition, Project administration, Resources, Visualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the University of Amsterdam Research Priority Area Systems Biology Host–Microbiome Interactions. SB is supported by the University of Amsterdam Centre for Urban Mental Health.
Acknowledgments
We thank Selina van Leeuwen (MAD, University of Amsterdam) for providing excellent sequencing services, Carolien Bosch-Tijhof and Caroline de Jongh for the assistance on the Ca 9–22 and HO1N1 cell cultivation, and Winfried Roseboom (University of Amsterdam) for the assistance on LC-MS/MS analysis. We thank Prof. Dr. Patrick van Dyck (KU Leuven) for kindly providing the C. albicans strains. We thank Prof. Dr. Wouter J. de Jonge and Dr. Jurgen Seppen (Amsterdam Gastroenterology, Endocrinology and Metabolism, Amsterdam UMC) for assistance on the THP-1 cells.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1629373/full#supplementary-material
References
Allison, D. L., Scheres, N., Willems, H. M. E., Bode, C. S., Krom, B. P., and Shirtliff, M. E. (2019). The Host Immune System Facilitates Disseminated Staphylococcus aureus Disease Due to Phagocytic Attraction to Candida albicans during Coinfection: a Case of Bait and Switch. Infect. Immun. 87, e00137-19. doi: 10.1128/IAI.00137-19
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data.. Cambridge, United Kingdom. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bailey, D. A., Feldmann, P. J., Bovey, M., Gow, N. A., and Brown, A. J. (1996). The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178, 5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996
Ballou, E. R., Avelar, G. M., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2, 16238. doi: 10.1038/nmicrobiol.2016.238
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borg-von Zepelin, M., Beggah, S., Boggian, K., Sanglard, D., and Monod, M. (1998). The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28, 543–554. doi: 10.1046/j.1365-2958.1998.00815.x
Braun, B. R., Head, W. S., Wang, M. X., and Johnson, A. D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31–44. doi: 10.1093/genetics/156.1.31
Calderone, R. A. and Fonzi, W. A. (2001). Virulence factors of Candida albicans. Trends Microbiol. 9, 327–335. doi: 10.1016/S0966-842X(01)02094-7
Carlson, E. (1982). Synergistic effect of Candida albicans and Staphylococcus aureus on mouse mortality. Infect. Immun. 38, 921–924. doi: 10.1128/iai.38.3.921-924.1982
Carlson, E. (1983). Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect. Immun. 42, 285–292. doi: 10.1128/iai.42.1.285-292.1983
Carlson, E. C. (1988). Synergism of Candida albicans and delta toxin producing Staphylococcus aureus on mouse mortality and morbidity: protection by indomethacin. Zentralblatt für Bakteriol. Mikrobiol. und Hyg. Ser. A Med. Microbiol. Infect. Dis. Virol. Parasitol. 269, 377–386. doi: 10.1016/S0176-6724(88)80181-0
Caza, M. and Kronstad, J. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 3. doi: 10.3389/fcimb.2013.00080
Childers, D. S., Avelar, G. M., Bain, J. M., Pradhan, A., Larcombe, D. E., Netea, M. G., et al. (2020). Epitope shaving promotes fungal immune evasion. MBio 11, 10–1128. doi: 10.1128/mBio.00984-20
Ciric, L., Pratten, J., Wilson, M., and Spratt, D. (2010). Development of a novel multi-triplex qPCR method for the assessment of bacterial community structure in oral populations. Environ. Microbiol. Rep. 2, 770–774. doi: 10.1111/j.1758-2229.2010.00183.x
Crampin, H., Finley, K., Gerami-Nejad, M., Court, H., Gale, C., Berman, J., et al. (2005). Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 118, 2935–2947. doi: 10.1242/jcs.02414
de Carvalho Dias, K., Barbugli, P. A., De Patto, F., Lordello, V. B., de Aquino Penteado, L., Medeiros, A. I., et al. (2017). Soluble factors from biofilm of Candida albicans and Staphylococcus aureus promote cell death and inflammatory response. BMC Microbiol. 17, 1–9. doi: 10.1186/s12866-017-1031-5
del Rio, A., Cervera, C., Moreno, A., Moreillon, P., and Miró, J. M. (2009). Patients at Risk of Complications of Staphylococcus aureus Bloodstream Infection. Clin. Infect. Dis. 48, S246–S253. doi: 10.1086/598187
Desai, J. V. (2018). Candida albicans hyphae: from growth initiation to invasion. J. Fungi 4, 10. doi: 10.3390/jof4010010
Dias, K., de, C., Barbugli, P. A., and Vergani, C. E. (2021). Insights into the activation of oral keratinocyte cell death by Candida albicans and Staphylococcus aureus biofilms. Biofouling 37, 975–983. doi: 10.1080/08927014.2021.1994959
DuMont, A. L., Yoong, P., Surewaard, B. G. J., Benson, M. A., Nijland, R., van Strijp, J. A. G., et al. (2013). Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect. Immun. 81, 1830–1841. doi: 10.1128/IAI.00095-13
Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., et al. (2001). Transcription Profiling-Based Identification ofStaphylococcus aureus Genes Regulated by the agrand/or sarA Loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/jb.183.24.7341-7353.2001
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Kong, E. F., Tsui, C., Kucharíková, S., Andes, D., Van Dijck, P., and Jabra-Rizk, M. A. (2016). Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio 7, e01365-16. doi: 10.1128/mbio.01365-16
Flannagan, R. S., Heit, B., and Heinrichs, D. E. (2016). Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell. Microbiol. 18, 514–535. doi: 10.1111/cmi.12527
Fuchs, S., Mehlan, H., Bernhardt, J., Hennig, A., Michalik, S., Surmann, K., et al. (2018). AureoWiki - The repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 308, 558–568. doi: 10.1016/j.ijmm.2017.11.011
Gil-Bona, A., Llama-Palacios, A., Parra, C. M., Vivanco, F., Nombela, C., Monteoliva, L., et al. (2015). Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J. Proteome Res. 14, 142–153. doi: 10.1021/pr5007944
Hoppenbrouwers, T., Bastiaan-Net, S., Garssen, J., Pellegrini, N., Willemsen, L. E. M., and Wichers, H. J. (2022). Functional differences between primary monocyte-derived and THP-1 macrophages and their response to LCPUFAs. PharmaNutrition 22, 100322. doi: 10.1016/j.phanu.2022.100322
Hughes, C. S., Foehr, S., Garfield, D. A., Furlong, E. E., Steinmetz, L. M., and Krijgsveld, J. (2014). Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757. doi: 10.15252/msb.20145625
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Klotz, S. A., Chasin, B. S., Powell, B., Gaur, N. K., and Lipke, P. N. (2007). Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 59, 401–406. doi: 10.1016/j.diagmicrobio.2007.07.001
Kong, E. F., Kucharíková, S., Van Dijck, P., Peters, B. M., Shirtliff, M. E., and Jabra-Rizk, M. A. (2015). Clinical implications of oral candidiasis: host tissue damage and disseminated bacterial disease. Infect. Immun. 83, 604–613. doi: 10.1128/IAI.02843-14
Kubica, M., Guzik, K., Koziel, J., Zarebski, M., Richter, W., Gajkowska, B., et al. (2008). A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PloS One 3, e1409. doi: 10.1371/journal.pone.0001409
Kuznets, G., Vigonsky, E., Weissman, Z., Lalli, D., Gildor, T., Kauffman, S. J., et al. (2014). A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PloS Pathog. 10, e1004407. doi: 10.1371/journal.ppat.1004407
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z., and Zhang, B. (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205. doi: 10.1093/nar/gkz401
Liu, T., Huang, T., Li, J., Li, A., Li, C., Huang, X., et al. (2023). Optimization of differentiation and transcriptomic profile of THP-1 cells into macrophage by PMA. PloS One 18, e0286056. doi: 10.1371/journal.pone.0286056
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Martchenko, M., Alarco, A.-M., Harcus, D., and Whiteway, M. (2004). Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15, 456–467. doi: 10.1091/mbc.e03-03-0179
Martin, R., Albrecht-Eckardt, D., Brunke, S., Hube, B., Hünniger, K., and Kurzai, O. (2013). A core filamentation response network in Candida albicans is restricted to eight genes. PloS One 8, e58613. doi: 10.1371/journal.pone.0058613
Mayer, F. L., Wilson, D., and Hube, B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. doi: 10.4161/viru.22913
McCarty, T. P., White, C. M., and Pappas, P. G. (2021). Candidemia and invasive candidiasis. Infect. Dis. Clin. 35, 389–413. doi: 10.1016/j.idc.2021.03.007
Mills, C. D. and Ley, K. (2014). M1 and M2 macrophages: the chicken and the egg of immunity. J. Innate Immun. 6, 716–726. doi: 10.1159/000364945
Mohd Yasin, Z. N., Mohd Idrus, F. N., Hoe, C. H., and Yvonne-Tee, G. B. (2022). Macrophage polarization in THP-1 cell line and primary monocytes: A systematic review. Differentiation 128, 67–82. doi: 10.1016/j.diff.2022.10.001
Moyes, D. L., Wilson, D., Richardson, J. P., Mogavero, S., Tang, S. X., Wernecke, J., et al. (2016). Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68. doi: 10.1038/nature17625
Murray, P. J. (2017). Macrophage polarization. Annu. Rev. Physiol. 79, 541–566. doi: 10.1146/annurev-physiol-022516-034339
Naglik, J. R., Challacombe, S. J., and Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 400–428. doi: 10.1128/MMBR.67.3.400-428.2003
Nash, E. E., Peters, B. M., Fidel, P. L., and Noverr, M. C. (2016). Morphology-independent virulence of Candida species during polymicrobial intra-abdominal infections with Staphylococcus aureus. Infect. Immun. 84, 90–98. doi: 10.1128/IAI.01059-15
Nash, E. E., Peters, B. M., Palmer, G. E., Fidel, P. L., and Noverr, M. C. (2014). Morphogenesis is not required for Candida albicans-Staphylococcus aureus intra-abdominal infection-mediated dissemination and lethal sepsis. Infect. Immun. 82, 3426–3435. doi: 10.1128/IAI.01746-14
Nasser, L., Weissman, Z., Pinsky, M., Amartely, H., Dvir, H., and Kornitzer, D. (2016). Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 1, 16156. doi: 10.1038/nmicrobiol.2016.156
Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B., and Moghazeh, S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x
Okamoto-Shibayama, K., Kikuchi, Y., Kokubu, E., Sato, Y., and Ishihara, K. (2014). Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth. FEMS Yeast Res. 14, 674–677. doi: 10.1111/1567-1364.12160
Pasman, R., Krom, B. P., Zaat, S. A. J., and Brul, S. (2022). The role of the oral immune system in oropharyngeal candidiasis-facilitated invasion and dissemination of Staphylococcus aureus. Front. Oral. Heal. 3. doi: 10.3389/froh.2022.851786
Pasman, R., Zhang, J., Zaat, S. A., Brul, S., and Krom, B. P. (2024). A customizable and defined medium supporting culturing of Candida albicans, Staphylococcus aureus, and human oral epithelial cells. Appl. Environ. Microbiol. 90, e00360–e00324. doi: 10.1128/aem.00360-24
Peters, B. M., Jabra-Rizk, M. A., Scheper, M. A., Leid, J. G., Costerton, J. W., and Shirtliff, M. E. (2010). Microbial interactions and differential protein expression in Staphylococcus aureus–Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 59, 493–503. doi: 10.1111/j.1574-695X.2010.00710.x
Peters, B. M. and Noverra, M. C. (2013). Candida albicans-staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect. Immun. 81, 2178–2189. doi: 10.1128/IAI.00265-13
Pidwill, G. R., Gibson, J. F., Cole, J., Renshaw, S. A., and Foster, S. J. (2021). The role of macrophages in Staphylococcus aureus infection. Front. Immunol. 11, 620339. doi: 10.3389/fimmu.2020.620339
Pien, B. C., Sundaram, P., Raoof, N., Costa, S. F., Mirrett, S., Woods, C. W., et al. (2010). The clinical and prognostic importance of positive blood cultures in adults. Am. J. Med. 123, 819–828. doi: 10.1016/j.amjmed.2010.03.021
Röhm, M., Lindemann, E., Hiller, E., Ermert, D., Lemuth, K., Trkulja, D., et al. (2013). A family of secreted pathogenesis-related proteins in C andida albicans. Mol. Microbiol. 87, 132–151. doi: 10.1111/mmi.12087
Roy, U., Yaish, S., Weissman, Z., Pinsky, M., Dey, S., Horev, G., et al. (2022). Ferric reductase-related proteins mediate fungal heme acquisition. Elife 11, e80604. doi: 10.7554/eLife.80604
Rynikova, M., Adamkova, P., Hradicka, P., Stofilova, J., Harvanova, D., Matejova, J., et al. (2023). Transcriptomic analysis of macrophage polarization protocols: vitamin D3 or IL-4 and IL-13 do not polarize THP-1 monocytes into reliable M2 macrophages. Biomedicines 11, 608. doi: 10.3390/biomedicines11020608
Schlecht, L. M., Peters, B. M., Krom, B. P., Freiberg, J. A., Hänsch, G. M., Filler, S. G., et al. (2015). Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 161, 168–181. doi: 10.1099/mic.0.083485-0
Shapouri-Moghaddam, A., Mohammadian, S., Vazini, H., Taghadosi, M., Esmaeili, S., Mardani, F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. doi: 10.1002/jcp.26429
Šimkovicová, M., Kramer, G., Rep, M., and Takken, F. L. W. (2024). Tomato R-gene-mediated resistance against Fusarium wilt originates in roots and extends to shoots via xylem to limit pathogen colonization. Front. Plant Sci. 15, 1384431. doi: 10.3389/fpls.2024.1384431
Sorgo, A. G., Heilmann, C. J., Dekker, H. L., Bekker, M., Brul, S., de Koster, C. G., et al. (2011). Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell 10, 1071–1081. doi: 10.1128/EC.05011-11
Sorgo, A. G., Heilmann, C. J., Dekker, H. L., Brul, S., de Koster, C. G., and Klis, F. M. (2010). Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27, 661–672. doi: 10.1002/yea.1775
Srikantha, T., Tsai, L., Daniels, K., Enger, L., Highley, K., and Soll, D. R. (1998). The two-component hybrid kinase regulator CaNIKl of Candida albicans. Microbiology 144, 2715–2729. doi: 10.1099/00221287-144-10-2715
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi: 10.1093/nar/gkac1000
Talapko, J., Juzbašić, M., Matijević, T., Pustijanac, E., Bekić, S., Kotris, I., et al. (2021). Candida albicans—The virulence factors and clinical manifestations of infection. J. Fungi 7, 79. doi: 10.3390/jof7020079
Todd, O. A., Fidel, P. L., Jr., Harro, J. M., Hilliard, J. J., Tkaczyk, C., Sellman, B. R., et al. (2019a). Candida albicans augments Staphylococcus aureus virulence by engaging the Staphylococcal agr quorum sensing system. MBio 10, 10–1128. doi: 10.1128/mBio.00910-19
Todd, O. A., Noverr, M. C., and Peters, B. M. (2019b). Candida albicans impacts Staphylococcus aureus alpha-toxin production via extracellular alkalinization. Msphere 4, 10–1128. doi: 10.1128/mSphere.00780-19
UniProt Consortium, T. (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699. doi: 10.1093/nar/gky092
Van Dyck, K., Viela, F., Mathelié-Guinlet, M., Demuyser, L., Hauben, E., Jabra-Rizk, M. A., et al. (2021). Adhesion of Staphylococcus aureus to Candida albicans during co-infection promotes bacterial dissemination through the host immune response. Front. Cell. Infect. Microbiol. 10, 624839. doi: 10.3389/fcimb.2020.624839
Vollmer, T., Störmer, M., Kleesiek, K., and Dreier, J. (2008). Evaluation of novel broad-range real-time PCR assay for rapid detection of human pathogenic fungi in various clinical specimens. J. Clin. Microbiol. 46, 1919–1926. doi: 10.1128/jcm.02178-07
Zecconi, A. and Scali, F. (2013). Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol. Lett. 150, 12–22. doi: 10.1016/j.imlet.2013.01.004
Keywords: Candida albicans (C. albicans), Staphylococcus aureus, inflammation, virulence factors, macrophages, extracellular virulence factors, nonextracellular virulence factors
Citation: Pasman R, Krom BP, Jonker MJ, de Leeuw WC, Kramer G, Brul S, Zaat SAJ and Zhang J (2025) Candida albicans and Staphylococcus aureus reciprocally promote their virulence factor secretion and pro-inflammatory effects. Front. Cell. Infect. Microbiol. 15:1629373. doi: 10.3389/fcimb.2025.1629373
Received: 15 May 2025; Accepted: 24 July 2025;
Published: 22 August 2025.
Edited by:
Justyna Karkowska-Kuleta, Jagiellonian University, PolandReviewed by:
Ashu Sharma, University at Buffalo, United StatesHelena Bujdakova, Comenius University, Slovakia
Copyright © 2025 Pasman, Krom, Jonker, de Leeuw, Kramer, Brul, Zaat and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianbo Zhang, ai56aGFuZzZAdXZhLm5s
†ORCID: Raymond Pasman, orcid.org/0009-0001-8518-5635
Bastiaan P. Krom, orcid.org/0000-0002-1497-1161
Martijs J. Jonker, orcid.org/0000-0003-4304-0486
Wim C. de Leeuw, orcid.org/0000-0001-6707-3894
Gertjan Kramer, orcid.org/0000-0002-8541-0337
Stanley Brul, https://orcid.org/0000-0001-5706-8768
Sebastian A. J. Zaat, orcid.org/0000-0001-9589-186XJianbo Zhang, orcid.org/0000-0003-3526-4586
 Raymond Pasman
Raymond Pasman Bastiaan P. Krom
Bastiaan P. Krom Martijs J. Jonker3†
Martijs J. Jonker3† Gertjan Kramer
Gertjan Kramer Stanley Brul
Stanley Brul Sebastian A. J. Zaat
Sebastian A. J. Zaat Jianbo Zhang
Jianbo Zhang