- 1State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
- 2TERRA Teaching and Research Center, Gembloux Agro-Bio Tech, University of Liège, Liège, Belgium
- 3College of Animal Science and Technology, Ningxia University, Yinchuan, China
Schmallenberg virus (SBV) is an emerging orthobunyavirus transmitted by Culicoides midges. It poses a serious global health threat to ruminants, especially during pregnancy, causing abortion, stillbirths, and congenital malformations. Since its first outbreak in 2011, SBV has spread across Europe and other regions. Its transmission has expanded due to global climate change and increased animal trade, resulting in recurrent outbreaks in endemic regions and a growing risk of introduction into non-endemic areas. This situation highlights the urgent need for improved control strategies. This review summarizes the pathogenic and epidemiological characteristics of SBV and provides an overview of recent advancements in diagnostic approaches, vaccine development, and vector control. Diagnostic approaches, such as serological assays and nucleic acid-based tests, have become the primary tools for SBV detection. However, their applicability in clinical settings still requires further optimization. In terms of vaccine development, existing inactivated vaccines have limitations, including the inability to distinguish between vaccinated and infected animals. This has driven the development of next-generation vaccines, such as recombinant protein, viral vector, and mRNA-based platforms. For vector control, integrated approaches combining chemical, ecological, and biological strategies have been proposed to interrupt the transmission of the virus by Culicoides midges. Additionally, this review emphasizes the necessity of region-specific control strategies tailored to the differing epidemiological contexts. In endemic regions, comprehensive measures, including pathogen surveillance, vaccination programs, and Culicoides control, are critical. In non-endemic regions, the focus should be on enhancing border biosecurity, monitoring international trade, and establishing early warning systems. These strategies not only provide a scientific foundation for SBV control but also offer practical guidance for managing the spread of similar vector-borne viruses globally.
1 Introduction
Schmallenberg virus (SBV) was first identified in November 2011 in plasma samples from dairy cows suffering from fever and diarrhea near the town of Schmallenberg, Germany (Hoffmann et al., 2012). The virus was subsequently named after this location. SBV belongs to the Simbu serogroup within the Orthobunyavirus genus and shares a high degree of homology with Akabane virus (Sick et al., 2019). Genomic analysis has revealed frequent recombination events in the S, M, and L genome segments of SBV, which help it adapt to to new hosts and environments (Hughes et al., 2020). The virus is primarily transmitted by Culicoides and is associated with significant reproductive failures in ruminants, including abortion, stillbirth, and arthrogryposis-hydranencephaly syndrome (AHS) (Sibhat et al., 2018). Within a few months, SBV spread across Western Europe, causing significant economic losses to the livestock industry (Stavrou et al., 2017). According to EU statistics, direct losses to the livestock sector exceeded €150 million at the peak of the outbreak in 2012 (Charlier et al., 2020). In recent years, epidemiological data indicate that the virus has spread beyond Europe (Bradshaw et al., 2012; Mason et al., 2013; Rasmussen et al., 2014; Barrett et al., 2015). Combined with the seasonal expansion of Culicoides vectors, this has created a complex “host-vector-environment” transmission network.
The current prevention and control system has three main challenges. First, existing diagnostic methods like pathogen-based, nucleic acid, and serological tests cannot provide rapid on-site detection (Mansfield et al., 2013; Wernike and Beer, 2019). Additionally, serological testing has a high false-positive rate due to cross-reactivity with other Simbu serogroup viruses, such as Akabane virus (Bilk et al., 2012; Loeffen et al., 2012; Bréard et al., 2013). Second, although inactivated vaccines against SBV have demonstrated efficacy, their use remains limited due to the requirement for multiple doses and lack of DIVA capability. Moreover, current research progress on the duration of immunity remains insufficient and needs further investigation. Furthermore, the unpredictable and seasonal circulation of SBV reduces the willingness of farmers to vaccinate, resulting in poor uptake and, in some regions, withdrawal of the vaccine from the market (Wernike and Beer, 2020). Third, traditional vector control strategies are losing effectiveness due to the evolution of insecticide resistance in Culicoides midges (Rasmussen et al., 2014; Naqqash et al., 2016; Sick et al., 2019). Furthermore, studies on the virus’s overwintering mechanisms have indicated that in regions above 45°N latitude, Culicoides midges undergo diapause for up to five months (Purse et al., 2005), yet the virus can still maintain its ecological niche through vertical transmission via the placenta (De Regge et al., 2012; Wernike et al., 2013c; Poskin et al., 2017). This poses a risk for cross-border spread. Moreover, since SBV infections in adult, non-pregnant ruminants are typically asymptomatic or very mild, monitoring efforts are further complicated (Afonso et al., 2014; Wernike et al., 2014). The traditional “one-size-fits-all” approach is no longer sufficient to address the ecological complexity of the virus. There is an urgent need to develop a precise prevention and control system based on geographically stratified transmission risk assessments.
This article provides a comprehensive review of the diagnostic approaches, vaccine development, and vector control strategies for SBV. Additionally, it introduces an innovative “epidemic regions vs. non-epidemic regions” dual-track prevention and control strategy. Based on regional differences, this article proposes scientifically sound and practical control measures aimed at enhancing prevention effectiveness. The goal is to provide theoretical foundations and technical support for global epidemic control efforts.
2 SBV classification and genome
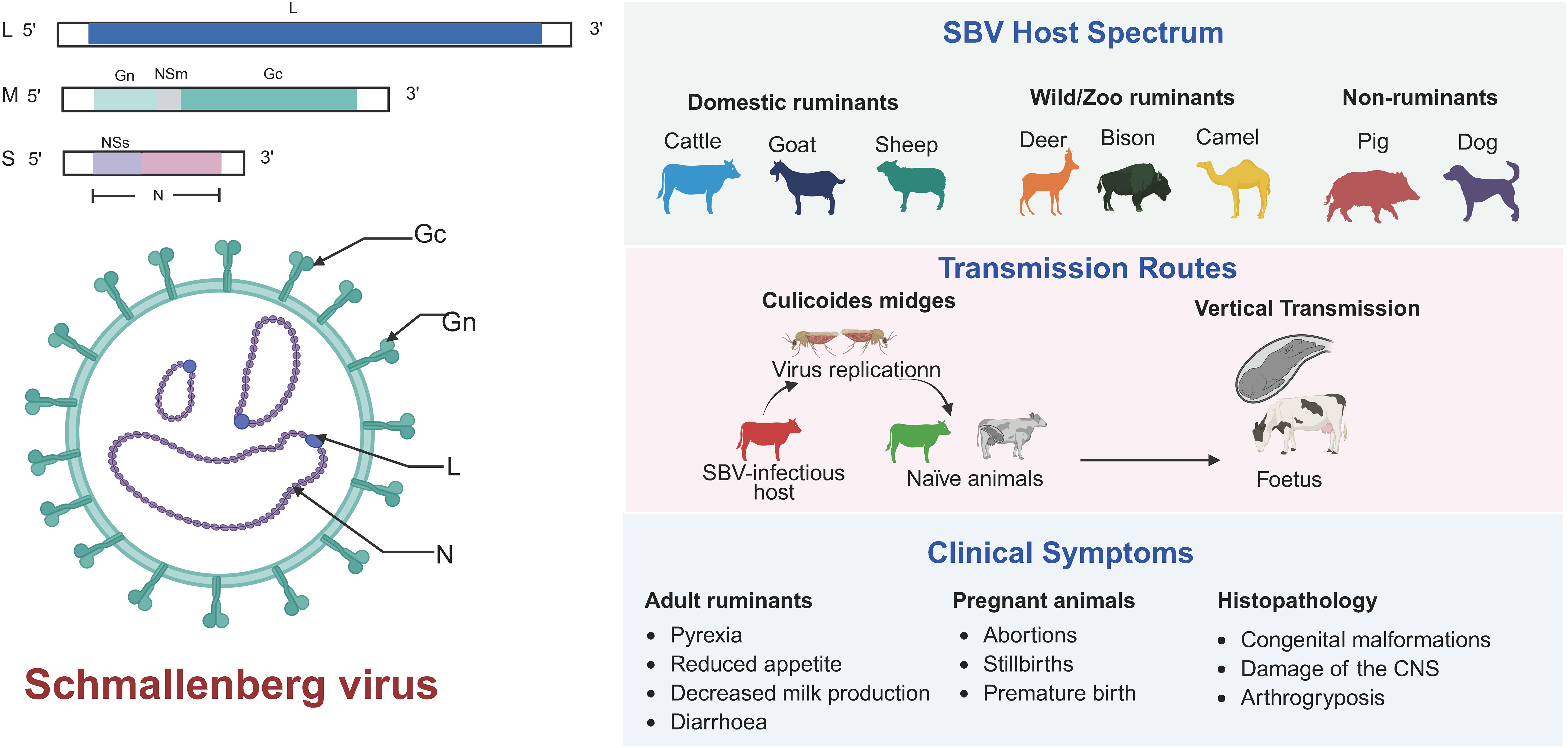
SBV is a Simbu serogroup virus belonging to the genus Orthobunyavirus within the family Peribunyaviridae (Saeed et al., 2001; Rasekh et al., 2018). The genus Orthobunyaviridae contains over 170 species of viruses, including those that cause human diseases (such as Oropouche virus and La Crosse virus) and ruminant diseases (such as Akabane virus, Aino virus, Cache Valley Fever virus) (Abudurexiti et al., 2019). The genome of SBV consists of three negative-strand RNA segments: L segment (large), M segment (medium) and S segment (small) (Figure 1) (Bouloy et al., 1973). The L segment encodes the RNA-dependent RNA polymerase (RdRp) of the virus, which is responsible for viral replication and transcription (Walter and Barr, 2011). The Gn and Gc glycoproteins are essential components of the viral envelope, mediating viral adsorption and membrane fusion (Wernike et al., 2018b). Both glycoproteins serve as the primary targets of the host immune system and can elicit specific antibody responses (Endalew et al., 2019; Wang et al., 2024). Notably, Gc demonstrates particularly strong immunogenicity and has been identified as the predominant target for the production of potent neutralizing antibodies (Wang et al., 2024). Extensive characterization of their antigenic epitopes, especially those on Gc, provides crucial molecular insights for vaccine design, offering strategies to enhance immunogenicity and protective efficacy. The S segment encodes the nucleocapsid protein (N) and the non-structural protein NSs. The N protein is a core component of the viral replication complex, encapsidating viral RNA to form a ribonucleoprotein (RNP) complex, which protects the genome and facilitates viral replication (Ak et al., 2007; Ariza et al., 2013). In vitro studies have shown that deletion of NSs impairs viral replication in interferon-sensitive cells, prevents interferon (IFN) synthesis suppression in infected cells, and disrupts host protein synthesis shutdown (Varela et al., 2016). NSs has been identified as a major virulence factor that downregulates host mRNA synthesis and type I IFN production in mammalian cells, thus enhancing viral replication (Thomas et al., 2004).
The segmented genome characteristic of SBV not only enable the virus to efficiently adapt to different hosts and environments but also significantly enhance its genetic diversity through recombination and reassortment. This provides an evolutionary advantage for the virus in terms of host adaptation (Yanase et al., 2003, 2010; Varela et al., 2016). Due to the segmented genome, the virus can flexibly respond to environmental pressures, such as host immune responses or climate changes, maintaining its infectivity through rapid evolution (Wernike et al., 2021; Sick et al., 2024). Additionally, the segmented genome may facilitate genetic exchange with other Orthobunyavirus species. In the event of co-infection within a host, new recombinant viruses could potentially emerge. This potential genetic compatibility not only influences the ecological adaptability of the virus but also pose public health concerns. Therefore, continuous monitoring of the genetic evolution of SBV, especially interspecies reassortment, is crucial for preventing the emergence of novel pathogens and for informing effective epidemiological surveillance and control strategies. Notably, virus variants isolated from malformed fetuses frequently carry specific mutations, particularly in the S and M genome segments, which impair replication in insect cells and indicate a loss of fitness for vector transmission. Such variants likely do not participate in the natural transmission cycle between mammalian hosts and insect vectors. Studying these mutations is essential to understanding the protein functions that are critical for viral adaptation to different host (Sick et al., 2024).
3 SBV epidemiological characteristics
Since its first outbreak in Northern Europe in the autumn of 2011, SBV has exhibited a clear trend of cross-border transmission (Figure 2). In 2012, the virus spread rapidly from the British Isles to Scotland and Ireland (Conraths et al., 2013; Dominguez et al., 2014), further expanding into Eastern Europe and the Mediterranean region. Within a year, SBV had spread across Europe and evolved into an endemic pathogen with periodic outbreaks occurring every two to three years. For instance, outbreaks re-emerged in Ireland, the United Kingdom, and Belgium during 2016–2017 (Stokes et al., 2016; Collins et al., 2017; Veldhuis et al., 2017). Notably, in 2022, Germany reported a seroprevalence of 4.92%, which surged to 40.15% in 2023, indicating a significantly rise in viral activity (Wernike et al., 2022). Juvenile ruminants exhibited a seroprevalence of 31.82% in 2023, while the overall adult seroprevalence reached 40.15%, signaling a large-scale outbreak during the summer and autumn months. Although Europe remains the primary endemic region, SBV has expanded to other continents, including Africa and Asia. SBV circulation has been confirmed in Turkey, where it was first detected in 2014 through molecular analysis of aborted ruminant fetuses and showed 29.11% seropositivity and 3.17% PCR positivity in ruminants between 2015 and 2017 in the Eastern Mediterranean region, as well as in Israel in 2019, where genomic detection in both Culicoides midges and affected ruminants demonstrated its presence (Yilmaz et al., 2014; Abutarbush et al., 2017; Behar et al., 2021). Additionally, several countries have also reported serological evidence of SBV, although no pathogen-based evidence has been obtained so far. For example, in 2018, Ethiopia reported seroprevalence rates of 56.6% at the individual level and 82.9% at the herd-level (Sibhat et al., 2018). Meanwhile, serological evidence confirmed its presence in East Asia, specifically in Guangdong Province, China (Zhai et al., 2018). A 2023 study first reported serological evidence of SBV infection in large populations of sheep and goats across multiple states in Peninsular Malaysia. The high seroprevalence of up to 27.8% indicates significant viral circulation among local small ruminants (Jimale et al., 2024). Furthermore, a recent global meta-analysis indicates infection rates of 49% in domestic ruminants and 26% in wild ruminants (Dagnaw et al., 2024).
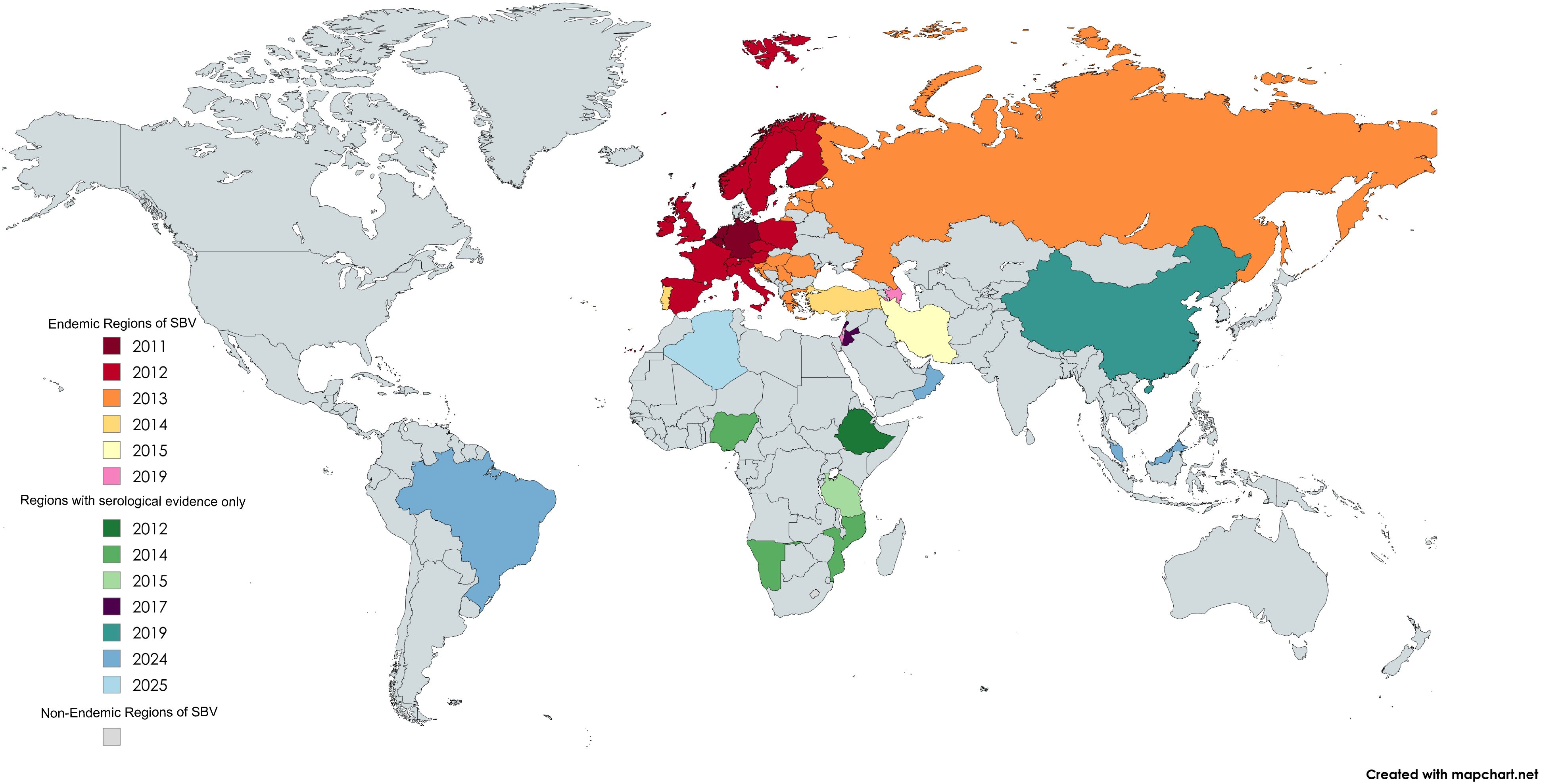
Figure 2. SBV distribution by country and year of first reported case. Map generated using MapChart.net.
SBV is primarily transmitted by the bites of haematophagous midges of the genus Culicoides. These arthropods serve as the principal biological vectors of the virus (Sick et al., 2019). Transmission occurs when an uninfected midge takes a blood meal from an SBV-infected ruminant—typically cattle, sheep, or goats. During the extrinsic incubation period (EIP), the virus replicates within the midge until it reaches transmissible levels. The infected midge can then pass the virus to another host during subsequent feeding. Several Culicoides species have been implicated in SBV transmission, particularly those in the Obsoletus group (Larska et al., 2013). Additionally, vertical (transovarial) transmission within the vector population has been proposed as a mechanism for viral persistence. This is supported by the detection of SBV RNA in nulliparous Culicoides—unfed midges—suggesting that the virus may be transmitted from adult females to their offspring. Such vertical transmission could enable SBV to overwinter within vector populations, thereby facilitating viral re-emergence during subsequent transmission seasons.
SBV is also capable of vertical transmission in ruminant hosts. When a pregnant animal becomes infected, viremia may allow the virus to cross the placenta, infecting the developing fetus (Herder et al., 2012). If infection occurs during critical gestational windows, it can result in congenital malformations, stillbirths, or abortions. Although the exact susceptible period remains undefined, it is estimated to be between gestational days 28–56 in small ruminants and days 80–150 in cattle (Hartley et al., 1977; Parsonson et al., 1988). Detection of SBV RNA in malformed neonates and aborted fetuses has confirmed this route of transmission in field settings (Dogan et al., 2022).
Direct horizontal transmission between animals appears to be unlikely. While experimental subcutaneous inoculation of cattle has resulted in SBV RNA detection in fecal, oral, and nasal swabs, oral or nasal inoculation did not lead to productive infection or seroconversion (Wernike et al., 2013a). These findings indicate that direct transmission through these routes under natural conditions is improbable.
The clinical symptoms of SBV infection vary depending on the host species and age. SBV primarily affects domestic ruminants such as cattle, sheep, and goats, but it has also been detected in wild ruminants, including roe deer and bison (Lievaart-Peterson et al., 2015a; Jiménez-Ruiz et al., 2021, 2022). However, clinical symptoms associated with SBV infection in wild ruminants have not been reported, and further research is needed to elucidate the impact of SBV infection on these species and their role in SBV epidemiology (Schulz et al., 2015; García-Bocanegra et al., 2017). In adult cattle, SBV infection is often asymptomatic or mild, with common symptoms including transient fever, reduced appetite, and decreased milk production, generally resolving within a few days (Wernike et al., 2012). In contrast, adult sheep and goats mainly experience subclinical infections, with only a few acute cases presenting symptoms such as fever, diarrhea, or reduced milk yield (Afonso et al., 2014). However, in pregnant animals, the virus can cross the placenta and infect the fetus, leading to abortion, premature birth, stillbirth, or congenital malformations such as arthrogryposis, hydrocephalus, and porencephaly (De Regge, 2017; Endalew et al., 2018). SBV infection in pregnant animals differ according to the gestational stage at the time of infection. Infection during early pregnancy typically results in embryo loss or abortion, while infection in mid-gestation often causes severe fetal malformations, particularly affecting the central nervous system and musculoskeletal development. Infections occurring late in pregnancy may lead to inflammatory lesions in the fetal brain and neurological symptoms in neonates (O’Connor et al., 2024). There is an inverse correlation between herd immunity levels and the incidence of congenital malformations—higher herd immunity leads to fewer new infections and a reduced rate of congenital defects. Conversely, when the number of susceptible individuals increases, the risk of infection in pregnant animals rises, leading to a higher incidence of fetal malformations. In endemic regions where SBV has circulated for years, most adult animals have acquired natural immunity through prior infection or vaccination. As a result, new infections are relatively rare, and clinical symptoms are usually mild or absent, with recovery occurring within a few days. However, unvaccinated pregnant animals remain at high risk, especially if infection occurs during early to mid-gestation, which significantly increases the likelihood of severe congenital abnormalities, abortion, or stillbirth. Overall, the impact of SBV in endemic regions is relatively limited, but unvaccinated young breeding females remain vulnerable. Both vaccination and naturally acquired herd immunity play a crucial role in reducing new cases. In contrast, animals in non-endemic regions often lack immunity, and newly emerging outbreaks can result in a high rate of fetal malformations, potentially leading to severe economic losses to the livestock industry, including reduced reproductive efficiency and increased farming costs. Therefore, it is essential to implement targeted prevention and control strategies that are tailored to the specific conditions of both endemic and non-endemic regions. This approach will be more effective in controlling the spread of the disease and mitigating its impact.
4 Diagnosis for SBV
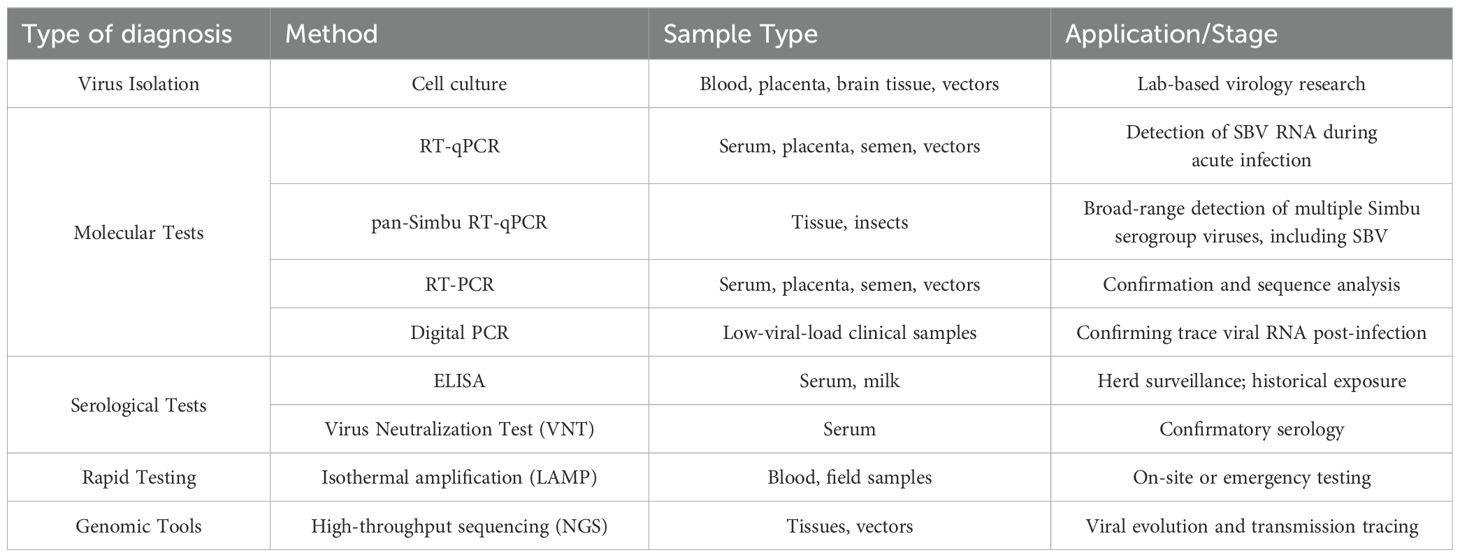
As SBV infection presents symptoms similar to those of other Bunyavirus infections, laboratory diagnostics are essential for accurate confirmation (Garigliany, 2012; Abutarbush et al., 2017). The primary diagnostic methods include virus isolation, molecular detection, and serological testing (Table 1).
SBV can be isolated and cultured in a variety of insect and mammalian cell types, such as BHK-21, Vero, and KC cells (Hoffmann et al., 2012; Wernike et al., 2013b). However, due to the low viral load in most clinical samples, not all samples are successful in virus isolation, which limits its sensitivity.
Real-time fluorescence quantitative PCR (RT-qPCR) is the most commonly used molecular detection method (De Regge et al., 2013). It amplifies the S gene and L gene of the SBV genome to detect minute amounts of viral RNA with high sensitivity and specificity, making it especially suitable for early detection of acute infections and high-throughput testing. Samples from the placenta, serum, semen, and insect vectors can be used for RT-qPCR testing (Bilk et al., 2012; De Regge et al., 2013; Poel et al., 2014). However, since viremia is short-lived, the timing of detection is crucial. Some adult animals may have cleared the viral RNA by the time of testing, reducing the detection sensitivity. To address this limitation, digital PCR (dPCR) has been introduced, which increases sensitivity for low-load samples and reduces nonspecific interference, making it especially suitable for detecting trace amounts of RNA after virus clearance. Additionally, a broad-spectrum Simbu RT-qPCR detection method has been developed, which can be used to detect multiple Simbu serogroup viruses (Fischer et al., 2013; Golender et al., 2018; Camarão et al., 2019).
SBV-specific antibodies are typically produced 1–3 weeks after infection and can persist for several years. Therefore, serological testing is an effective means of diagnosing SBV infection. Virus neutralization tests (VNT), and enzyme-linked immunosorbent assays (ELISA) are the main serological tests (Loeffen et al., 2012; Beer et al., 2013). VNT is considered the “gold standard” due to its high specificity, but it is time-consuming and operationally complex, typically used only for confirmation (Loeffen et al., 2012). In contrast, ELISA is an efficient screening method that can detect antibodies not only in serum but also in milk, making it suitable for large-scale herd immunity screening and assessing infection history (Humphries and Burr, 2012; Bréard et al., 2013; Daly et al., 2015). However, in regions where other Simbu serogroup viruses co-circulate, the specificity of ELISA may be challenged (Blacksell et al., 1997). In a recent study conducted in Turkey’s Eastern Mediterranean region, the seroprevalence of SBV-specific antibodies detected by ELISA reached 29.11%, whereas only 3.17% of virological samples tested positive by RT-PCR, and no viral RNA was detected in vector samples. While this discrepancy may reflect the temporal gap between exposure and sampling, it may also suggest potential cross-reactivity or false positives inherent in serological assays (Dogan et al., 2022). In such settings, confirmatory testing by VNT becomes particularly essential to ensure diagnostic accuracy. This two-tiered diagnostic strategy has proven feasible in practice; for instance, a sero-epidemiological study in Spain (2006–2015) used ELISA for preliminary screening of wild ruminants, followed by VNT confirmation—demonstrating the reliability of this approach in both domestic and wildlife surveillance programs (García-Bocanegra et al., 2017).
To meet the demand for on-site rapid detection, various portable and rapid testing technologies have been developed in recent years. For example, isothermal amplification technologies, such as LAMP, have proven to perform effective in resource-limited environments due to their simplicity and low equipment requirements (Aebischer et al., 2014). Furthermore, novel detection platforms combining the CRISPR-Cas system have shown great potential, with sensitivity and specificity comparable to laboratory diagnostic technologies, significantly reducing detection times (Padmanaban and Ranganathan, 2022). The development and application of these rapid testing technologies provide crucial support for SBV screening in remote areas and emergency situations. Genome sequencing can track viral mutations and transmission chains, enhancing the epidemic monitoring system. As sequencing technology advances, the collection and analysis of genomic data have become essential tools for monitoring viral evolution and predicting transmission trends.
The epidemiological characteristics and transmission patterns of SBV vary significantly across regions, resulting in different requirements for diagnostic technologies. Diagnostic methods must be adjusted and optimized to the specific conditions of both endemic and non-endemic areas to ensure early detection, accurate assessment, and effective outbreak control.
4.1 Diagnosis in endemic regions
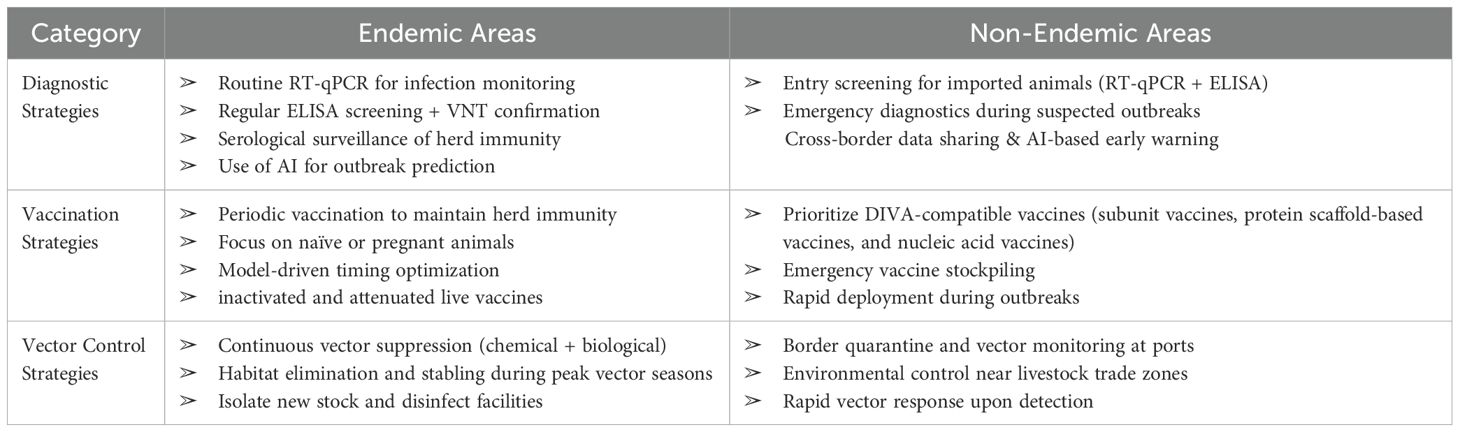
In European regions, where SBV is highly prevalent, the virus causes periodic outbreaks, typically recurring every 2–3 years (Cuéllar et al., 2018; Bayrou et al., 2022). During low prevalence periods, the virus can still be transmitted at low levels, making early detection crucial to prevent undiagnosed cases that could result in uncontrolled outbreaks. Therefore, Molecular diagnostic methods like RT-qPCR and digital PCR are prioritized during acute infection phases. They enable large-scale screening of samples, such as placenta and vector insects, within 24 hours. Additionally, serological monitoring of host immunity dynamics crucial for predicting the next wave of outbreaks. Most European countries have established comprehensive surveillance systems that regularly sample livestock populations and use RT-qPCR and serological tests to track virus transmission in real time (Roberts et al., 2014). To improve monitoring efficiency, some countries are exploring the use of machine learning-based epidemiological models to dynamically predict the virus spread and identify high-risk areas (Ali, 2024; Ekundayo, 2024; Liu et al., 2025).
The livestock industry in Europe is highly concentrated, especially in countries like Germany, the Netherlands, and France. Due to the large livestock populations, rapid testing and screening technologies are needed to reduce the risk of large-scale outbreaks. ELISA technology is widely used for large-scale serological screening due to its high throughput and cost-effectiveness, meeting this demand, while the VNT is used for high-precision confirmation testing of specific samples (Nurtop et al., 2018). Additionally, monitoring post-vaccination responses requires complementary tools to optimize the Differentiating Infected from Vaccinated Animals (DIVA) strategy, accurately distinguishing between naturally infected and vaccinated individuals, and assessing herd immunity levels and the potential for new outbreaks. To maintain high diagnostic standards, laboratories should regularly participate in inter-laboratory proficiency testing (Wernike and Beer, 2019).
In Asia and Africa, SBV outbreaks tend to be smaller in scale, but the potential risk of virus transmission should not be ignored (Zhai et al., 2018; Nadeem et al., 2024). The livestock industry in Asia is highly concentrated, particularly in countries like India and China, where dairy cattle and goat farming are widespread. While SBV outbreaks are currently rare, the active cross-border trade and livestock transport increase the risk of virus spread (Collins et al., 2019; Senf et al., 2020). In these regions, promoting portable diagnostic tools and establishing regional monitoring networks is highly recommended. In Africa, efforts to diagnose SBV face greater challenges due to limited laboratory facilities and diagnostic capabilities. Many remote areas lack basic laboratory infrastructure and specialized personnel, making a strong need for low-cost, easy-to-use field detection technologies. Diagnostics in these regions should focus on developing cost-effective tools, such as rapid test strip devices, which can provide results in just minutes, offering a significant advantage for rapid on-site screening. CRISPR-based diagnostic technologies, characterized by high sensitivity and specificity, hold great promise for improving field testing accuracy, particularly in rural livestock farming areas and during emergency outbreaks (Chertow, 2018; Ramachandran et al., 2020; Kaminski et al., 2021). One example is a CRISPR-Cas12 system combined with electric field control and microfluidics, which can detect SARS-CoV-2 RNA from raw samples in about 35 minutes (Ramachandran et al., 2020). This shows that CRISPR-based methods can be adapted quickly for different pathogens and could be a useful diagnostic tool for SBV. Additionally, the host range of SBV in African wildlife is still unclear, and the conditions for sample collection vary significantly among different species (Al-Busaidy et al., 1987; Mouchantat et al., 2015). Therefore, there is an urgent need to develop detection tools suitable for multiple sample types.
4.2 Diagnosis in non-endemic regions
Although SBV has not spread widely in non-endemic regions, the risk of virus introduction and spread has increased due to the rise in international trade and livestock transportation, especially through the importation of breeding livestock, frozen semen, and embryos, in which subclinically infected individuals may introduce the virus (Gibbens, 2012; Wernike et al., 2022). To address this challenge, a comprehensive prevention and control system must be established, focusing on a “blocking entry - early warning - rapid response” strategy. For imported livestock and reproductive products, RT-qPCR screening combined with serological ELISA/VNT tests to exclude subclinical infections and cross-reaction interference (Aebischer et al., 2014; Golender et al., 2018; Goto et al., 2023).
However, traditional detection methods have limitations in terms of screening speed and costs. To improve early diagnosis and monitoring efficiency, future efforts could focus on developing automated high-throughput screening technologies that enhance detection efficiency and reduce screening costs. Additionally, non-endemic regions should develop real-time monitoring systems based on data analysis and smart algorithms, establishing intelligent networks that integrate climate data (temperature, humidity), vector insect distribution models, and livestock immunization profiles (Robertson et al., 2010; Kapetas et al., 2025). AI algorithms can be used to predict high-risk areas. Drawing on cross-border monitoring experience from African swine fever, sharing virus gene sequences and epidemic dynamics with SBV-endemic countries is essential for timely updates to detection targets (Nadeem et al., 2024; Nie et al., 2024). A rapid response system should be implemented, with emergency protocols (e.g., isolation - re-testing - tracing) to ensure preliminary diagnosis is completed within 24 hours (Aebischer et al., 2014; Wernike and Beer, 2019). In border areas adjacent to endemic regions, pilot vector insect trapping and pathogen monitoring should be conducted to prevent the epidemic from infiltrating.
5 Vaccine development for SBV
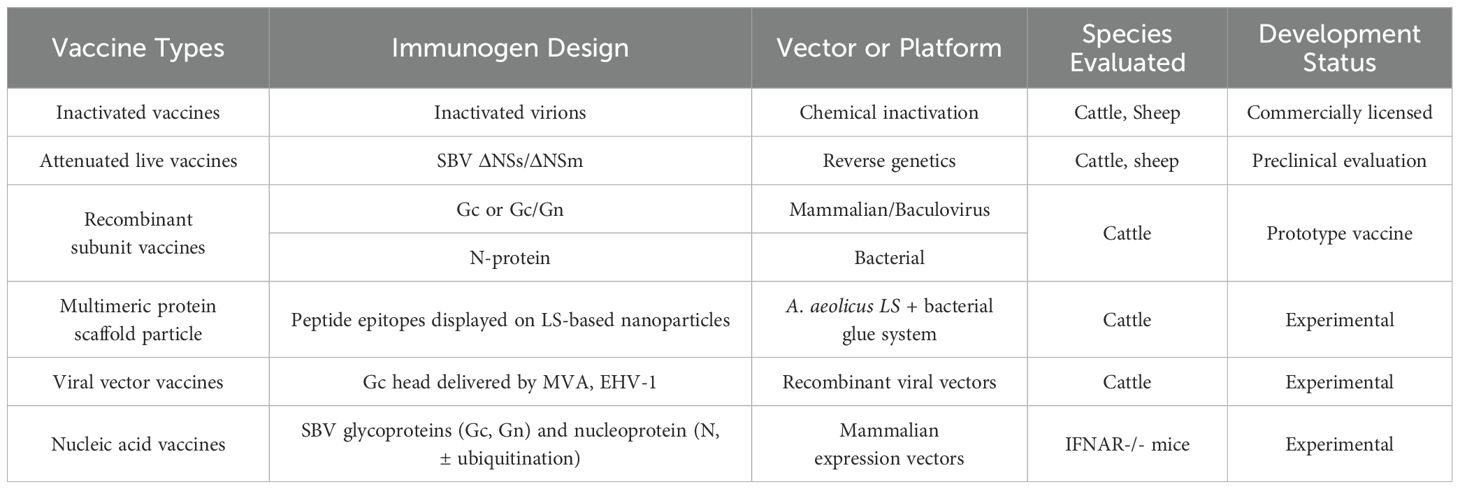
The development of vaccines for SBV is a key strategy for controlling the virus. Currently, there are several types of vaccines under development, including inactivated vaccines, attenuated live vaccines, recombinant subunit vaccines, viral vector vaccines, and nucleic acid vaccines (Table 2) (Hechinger et al., 2014; Kraatz et al., 2015; Wernike et al., 2018b; Wernike and Beer, 2020). Among these, inactivated vaccines are the only vaccines currently approved for market use. Examples of approved vaccines include Bovilis SBV (MSD Animal Health), Zulvac SBV (Zoetis), and SBVvax (Merial). These vaccines function by chemically inactivating the viral particles, which triggers an immune response. Initially licensed in the United Kingdom and France in 2013, they subsequently received European Union-wide marketing authorization in May 2015. Inactivated vaccines have demonstrated high efficacy in preventing SBV infection, thereby significantly reducing the risk of fetal abnormalities and miscarriage in pregnant dams (Wernike et al., 2013d; Hechinger et al., 2014). A study showed that a single dose of an inactivated vaccine completely inhibited viral replication in all vaccinated sheep (5/5), as confirmed by competitive ELISA, microneutralization tests, and SBV-specific real-time RT-PCR (Hechinger et al., 2014). However, these vaccines come with some limitations, including high production costs, and the inability to differentiate between vaccinated and naturally infected animals.
Attenuated live vaccines are commonly developed through targeted deletion of viral genes, such as NSm or NSs, aiming to attenuate viral pathogenicity while preserving immunogenic properties (Bird et al., 2011; Kraatz et al., 2015). SBV mutants with deletions in the NSs and NSm genes were evaluated in vitro, in IFNAR−/− mice, and in cattle. The double gene-deletion mutant demonstrated no detectable viral replication in cattle and conferred complete protection in all vaccinated animals following immunization. Importantly, this mutant also shows potential for DIVA capability (Kraatz et al., 2015; Varela et al., 2016). However, live vaccines may pose a risk of reversion to virulence, and further research and safety evaluations are needed before they can be widely used.
Recombinant subunit vaccines produce antigens through expression systems and induce immune responses with adjuvants, and are a key focus in SBV vaccine development. The envelope glycoproteins Gn and Gc mediate viral entry, with Gc identified as the main target of neutralizing antibodies. Sera from SBV-infected animals react strongly with the full-length Gc and its N-terminal domain, indicating that this region contains key neutralizing epitopes (Roman-Sosa et al., 2016). Several studies have evaluated the immunogenicity of Gc-based subunit vaccines. Kerstin et al. expressed the N-terminal domain of Gc using both prokaryotic and mammalian systems and assessed their protective efficacy in IFNAR−/− mice and cattle (Wernike et al., 2017). Prokaryotically expressed forms generally failed to induce protective immunity, whereas mammalian-expressed proteins conferred partial protection in both models. A multivalent vaccine combining Gc domains from SBV and Akabane virus achieved full protection in animal models. Additionally, vaccinated animals lacked antibodies against the viral N-protein, allowing differentiation from natural infection, which supports a marker vaccine approach (Wernike et al., 2017). Subunit vaccines based on the Gc head–stalk construct have demonstrated strong protective efficacy, inducing sterilizing immunity in animal models. Compared to the head domain alone, the inclusion of the stalk enhances immune responses and protection. Moreover, the conserved structure of Gc across Orthobunyaviruses suggests potential for broad cross-protection (Roman-Sosa et al., 2016; Hellert et al., 2019). In addition to humoral responses, cellular immunity may also contribute to protection against SBV. Recent studies have explored the nucleoprotein (SBV-N) as an alternative immunogen. Bacterially expressed SBV-N, when combined with a veterinary-grade saponin adjuvant, reduced viremia and clinical signs in mice despite not inducing neutralizing antibodies, suggesting a role for T-cell–mediated immunity (Boshra et al., 2020). Further research showed that the C4 fragment of SBV-N elicited strong cellular responses and exhibited high sequence similarity with other Simbu viruses, supporting its potential as a broad-spectrum vaccine candidate (Guerra et al., 2023) While recombinant protein vaccines offer advantages in safety and DIVA compatibility, their immunogenicity depends on the stability and properties of the antigen, as well as the compatibility with adjuvants. Additionally, their relatively high production costs may limit their use in resource-limited regions.
Vaccines based on nanoparticles and protein structures are emerging as promising options in SBV vaccine development. Recently, the multimeric protein scaffold particle (MPSP) platform, derived from lumazine synthase (LS) of Aquifex aeolicus, has introduced a new strategy for vaccine design (Aebischer et al., 2021). This platform allows peptide epitopes to be presented via genetic fusion, while large antigens are conjugated to pre-assembled particles using bacterial “superglue”, enhancing the vaccine’s polyvalency and immunogenicity. In the study using SBV as a model, the MPSPs presented the key immunogens of the virus and showed strong protective effects in both mouse and cattle models. Compared to monomeric subunit vaccines, the multivalent antigens on the MPSPs significantly enhanced the immune response, with a single dose protecting 80% of mice from a lethal dose of SBV and inducing nearly sterile immunity in cattle (Aebischer et al., 2021).
Live viral vectors can serve as an effective delivery system for SBV Gc antigens, enabling efficient expression of the Gc protein in vivo and enhancing its immunogenicity. Kerstin et al. used modified Ankara vaccinia virus (MVA) and equine herpesvirus type 1 (EHV-1) attenuated strains as viral vectors, inserting the N-terminal of the SBV Gc glycoprotein to develop SBV live viral vector vaccines (Wernike et al., 2018b). The results showed that cattle vaccinated with the recombinant EHV-1 vector vaccine achieved 50% protection after challenge, while all cattle vaccinated with the recombinant MVA vector vaccine received complete immune protection, confirming that the MVA-delivered SBV Gc domain live viral vector vaccine exhibited higher immunogenicity (Wernike et al., 2018b). Due to the replication deficiency of MVA in mammals, it has a safety advantage over traditional vaccinia virus. Furthermore, since the recombinant MVA vector vaccine based on SBV Gc protein does not generate antibodies against the N protein in vaccinated animals, the use of N protein serological testing allows for DIVA compatibility of the vaccine.
Nucleic acid vaccines express viral antigens in host cells, activating both humoral and cellular immune responses, making them a promising vaccine development direction. DNA vaccines, which encode SBV Gc and N proteins, have demonstrated the ability to reduce viremia and provide partial protection, though they have only been tested in small animal models (Boshra et al., 2017). However, DNA vaccines face challenges such as low delivery efficiency, requiring optimization of delivery systems like electroporation or nanoparticle-based methods. mRNA vaccines have emerged a new direction for SBV vaccine research due to their short development cycles, high immunogenicity, and DIVA compatibility (Iavarone et al., 2017; Maruggi et al., 2019). The success of BNT162b2 and mRNA-1273 during the COVID-19 pandemic illustrates how quickly effective vaccines can be developed once the delivery platform is established (Baden et al., 2021; Khoury et al., 2021). Additionally, in recent years, mRNA has made significant progress in optimizing antigen design, improving immune efficacy, and enhancing delivery systems (Maruggi et al., 2019; Clemente et al., 2023; Mochida and Uchida, 2024). It is believed that mRNA vaccines for SBV will play an important role in disease prevention and control in the future.
Although significant progress has been made in the research of various types of SBV vaccines, vaccine development and vaccination strategies must fully consider factors such as the intensity of outbreaks, geographic conditions, economic constraints, and technical accessibility. These strategies should be tailored to the specific characteristics of the epidemic in different regions to ensure maximum effectiveness and feasibility of vaccine implementation.
5.1 Vaccination strategies in endemic regions
In Europe, the main endemic region for SBV, vaccination strategies focus on establishing and maintaining herd immunity to prevent viral infections that cause fetal deformities and miscarriages (Hoffmann et al., 2012; Veldhuis et al., 2017). Studies show a high seroprevalence of SBV in these regions, with many adult animals acquiring immunity through natural infection. However, first-time pregnant dams remain at significant risk of infection. Additionally, due to changes in livestock populations, older animals with natural antibodies are gradually replaced by younger, susceptible animals, leading to a decline in herd immunity levels and promoting virus re-circulation (Bessell et al., 2014; Wernike et al., 2018a). Therefore, regular vaccination, particularly targeting susceptible young dams, may be an effective control strategy.
Inactivated vaccines have been widely used and, when combined with seasonal monitoring, help reduce the risk of outbreaks. Studies show that a single dose of the vaccine can induce sufficient antibody levels within two weeks (Wernike et al., 2013d). Vaccination is recommended to be completed prior to mating or during early gestation to confer protective immunity throughout pregnancy and to ensure that maternal antibody levels reach a protective threshold during the critical period of fetal susceptibility (Martinelle et al., 2015; König et al., 2019). Due to the uncertainty of SBV transmission, some regions may use emergency vaccination after an outbreak. However, it should be noted that emergency vaccination may be less effective in pregnant animals (Græsbøll et al., 2014).
The demand for new vaccines in European endemic areas is gradually increasing, especially those that support DIVA diagnostic strategies. These vaccines not only optimize epidemic dynamic monitoring but also ensure smooth health certification in livestock trade, thus preventing economic losses caused by trade restrictions. Additionally, by integrating epidemiological modeling technology, predictive tools can be used to dynamically adjust the vaccination timing and coverage, enabling more precise control. Vaccination in endemic regions should be integrated with molecular and serological diagnostic techniques to create a dual defense system for both immunity and monitoring (Wernike and Beer, 2020).
SBV prevalence in Asia and Africa is lower than in Europe, but cross-border trade and natural transmission risks are increasingly evident. Vaccination strategies should focus on key breeding livestock populations, combined with vector control measures to reduce the risk of virus transmission. Studies show that genetically engineered attenuated live vaccines can provide long-lasting immunity with a single dose and reduce cold chain transportation requirements. Moreover, research is exploring oral or spray vaccine delivery methods to lower vaccination costs and improve herd immunity coverage. In regions with dense wild animal populations or complex breeding environments in Africa, vaccine distribution should be adapted to local ecological conditions to ensure coverage of a sufficiently wide host population (Songane, 2018).
In summary, SBV vaccination strategies in endemic regions extend beyond vaccination alone, encompassing rigorous surveillance, ongoing vaccine development, and informed policy implementation to effectively mitigate the impact of the virus on the livestock industry.
5.2 Vaccination strategies in non-endemic regions
In non-endemic areas such as North America and Australia, vaccination strategies should focus on preventing virus introduction and enabling rapid response in the event of an outbreak. The main risks arise from international trade and cross-border livestock transportation, especially through the import of breeding animals, frozen semen, and embryos, through which the virus may enter local herds via subclinical infections. Therefore, the core task of vaccination is not large-scale distribution, but rather the establishment of emergency immunization reserves. Vaccine stockpiles should be set up in major livestock trade hubs and breeding centers to ensure rapid deployment in case of an outbreak. Since inactivated vaccines cannot differentiate between infected and immune animals, and attenuated live vaccines pose risks of reversion to virulence and environmental leakage, non-endemic areas are more likely to choose new vaccines that support DIVA strategies, such as subunit vaccines, protein scaffold-based vaccines, and nucleic acid vaccines. Particularly, protein scaffold-based vaccines are promising for providing rapid protection, reducing the need for frequent vaccinations, and should be prioritized for emergency reserves (Aebischer et al., 2021). Furthermore, mRNA vaccines, with their ability to induce a comprehensive immune response and their rapid production advantages, will also be suitable for emergency immunization once successfully developed (Maruggi et al., 2019). Notably, emergency vaccination should be integrated with cross-border quarantine systems. Once an imported case is detected, emergency vaccination should be carried out for susceptible animals around the outbreak point to ensure immunity coverage. To address potential viral mutations, vaccine stockpiles should be regularly evaluated for efficacy and updated according to the standards of the World Organisation for Animal Health (WOAH). In summary, vaccination strategies in non-endemic regions should focus on establishing emergency immunization reserves and rapid response mechanisms, prioritizing the use of innovative vaccines that support DIVA strategies to manage the risk of virus introduction and improve immunity coverage. By combining cross-border quarantine and dynamically adjusted vaccine reserves, the risk of virus transmission can be effectively reduced, ensuring the sustainability of control measures.
6 Vector control strategies for SBV
Effective control of vector-borne viruses such as SBV requires more than just epidemiological surveillance and vaccination (Gubbins et al., 2014; Sumner et al., 2017; Achee et al., 2019). Central to this is the management of Culicoides midges—the primary vectors responsible for transmission. By integrating chemical control, biological control, environmental and farm interventions, and climate monitoring strategies, a comprehensive and multidimensional vector control system can be established, facilitating early warning and rapid response to SBV outbreaks.
Key chemical strategies include using pyrethroid-based adulticides during peak midge activity seasons via ground spraying, indoor residual spraying, or insecticide-treated materials in livestock housing (Gubler, 2005). Larvicidal agents such as insect growth regulators (IGRs)—including pyriproxyfen and methoprene—are used to disrupt the development of immature stages at breeding sites (Baldacchino et al., 2015). While these interventions can rapidly reduce vector abundance, their sustained or indiscriminate use poses challenges such as insecticide resistance, ecological toxicity, and negative impacts on non-target species (Wernike et al., 2013b; Snyder et al., 2016; Matsuo, 2019).
Biological control strategies have emerged as sustainable alternatives (Huang et al., 2017). This approach employs natural agents—such as entomopathogenic fungi, bacterial larvicides, and aquatic predators—to reduce vector populations or impair their transmission capacity. Fungi like Beauveria bassiana and Metarhizium anisopliae have demonstrated efficacy against various arthropods, though their performance is sensitive to environmental conditions including temperature, humidity, and formulation parameters (Ansari et al., 2011; Fernandes et al., 2012). Bacterial larvicides, such as Bacillus thuringiensis var. israelensis and Lysinibacillus sphaericus, produce toxins specifically targeting mosquito larvae, and their combined application can help mitigate resistance development (Silva-Filha et al., 2021).
Environmental and farm interventions are equally crucial. Reducing standing water, improving drainage, and optimizing barn structures can limit Culicoides breeding sites—especially in intensive farming systems (Lievaart-Peterson et al., 2015b; Kohara et al., 2018). Small-scale farms can further minimize vector-host contact using physical barriers like insect-proof screens and nets. Adjusting livestock reproduction schedules to avoid peak vector activity, along with evening stabling and rotational grazing, are practical risk-reduction tactics. International trade and transport pose significant risks for vector and pathogen spread, requiring stringent inspection, disinfection protocols, and vector surveillance at ports, airports, and border crossings to mitigate introductions via cargo, vehicles, and animal movements.
Climate factors—particularly rising temperatures, shifting precipitation patterns, and wind dynamics—play a pivotal role in SBV transmission (De Regge, 2017). Elevated temperatures can extend the seasonal activity of Culicoides midges, while altered precipitation reshapes breeding habitat distribution (Haider et al., 2018). Wind influences midge flight and host-seeking behavior, thus modulating viral spread (Mellor et al., 2000). These drivers collectively determine the spatiotemporal dynamics of SBV, especially in temperate zones where vectors are climate-sensitive. Establishing robust meteorological monitoring systems and integrating climate, vector ecology, and host data into predictive models is essential. The incorporation of real-time satellite data on land surface temperature, vegetation indices, and precipitation enhances early warning accuracy and guides timely control measures (Ceccato et al., 2018; Pley et al., 2021). Additionally, wind conditions should be incorporated into risk assessments to guide livestock movement and housing strategies aimed at reducing vector-host contact.
A One Health approach that fosters collaboration across veterinary, entomological, environmental, and public health disciplines is fundamental to devising effective, climate-resilient, and region-specific SBV control strategies (Socha et al., 2022). Tailoring interventions based on local climate, vector presence, livestock production systems, and trade patterns enhances prevention efficacy and sustainability. While many control measures can be broadly applied, their successful implementation must reflect the distinct epidemiological contexts of endemic versus virus introduction-risk areas.
6.1 Vector control in endemic regions
In SBV-endemic areas with established virus circulation, vector control aims to suppress transmission cycles and reduce disease impact on livestock health and productivity. Control strategies emphasize sustained application of chemical insecticides timed to vector seasonal peaks, often supported by biological control, which helps limit insecticide reliance and resistance. Environmental management is intensive, focusing on eliminating vector breeding habitats within and around farms, combined with physical barriers to minimize vector-host interactions. Livestock management—such as adjusting reproduction to avoid peak vector seasons and housing animals during high-risk periods—is a key component of risk mitigation. Biosecurity within endemic zones centers on preventing virus reintroduction and spread among herds, involving quarantine of new stock and disinfection protocols (Pley et al., 2021). Furthermore, evidence indicates that climate change has extended the active vector season and increased the number of transmission cycles. To address these emerging challenges in endemic regions, monitoring systems should integrate high-resolution satellite data with local vector surveillance to produce real-time SBV risk maps (Fairbanks et al., 2024). Climate-driven surveillance and modeling inform adaptive management strategies, ensuring timely and effective deployment of control measures tailored to shifting vector population dynamics.
6.2 Vector control in non-endemic regions
Non-endemic regions prioritize prevention of virus and vector introduction (Roberts et al., 2014). With no active circulation, control efforts focus on robust border biosecurity, including quarantine and inspection of animals and goods, disinfection of transport vehicles, and surveillance of vectors at entry points such as airports and seaports. Environmental and livestock management aim at reducing potential vector habitats in areas surrounding livestock operations, particularly near international trade hubs. Physical barriers and husbandry practices serve as preventive buffers, limiting any inadvertent contact between vectors and susceptible animals (Narladkar, 2018). Surveillance in non-endemic areas focuses on early detection and rapid response to vector incursions or virus introductions, guided by climate and ecological modeling to anticipate and mitigate invasion risks before local transmission can be established. Predictive models based on satellite climatic data can help identify areas that may become suitable for vector introduction due to climate shifts. Surveillance should prioritize these high-risk zones with sentinel herd monitoring, vector trapping, and environmental assessments.
7 Conclusion and future perspectives
SBV is an orthobunyavirus transmitted by Culicoides midges, with its epidemiological characteristics influenced by multiple factors, including climate change, vector ecology, livestock management, and international trade (Walter and Barr, 2011; Hoffmann et al., 2013; Wernike et al., 2014). This paper systematically reviews pathogen biology, epidemiology, detection methods, vaccine development, and vector control strategies for SBV. Based on the differing needs of endemic and non-endemic regions, differentiated and comprehensive response strategies are proposed (Table 3). In endemic regions, SBV exhibits periodic outbreaks, requiring a comprehensive control approach that includes vaccination, pathogen and serological monitoring, and vector management, in order to minimize economic losses in the livestock industry. In contrast, non-endemic regions should focus on managing border and import risks, including monitoring for pathogens and vectors, and strengthening emergency immunization reserves to prevent cross-border transmission of the virus and reduce the risk of outbreaks.
Although available control technologies have played a significant role in epidemic monitoring in both endemic and non-endemic regions, they still face several key challenges. Currently, RT-qPCR, dPCR, and ELISA are widely used for virus monitoring, while rapid detection technologies such as LAMP also show promising applications (Aebischer et al., 2014; Daly et al., 2015; Nurtop et al., 2018; Goto et al., 2023). However, these methods still have limitations in terms of specificity, sensitivity, and applicability, particularly in resource-limited areas. There is a need to accelerate the development of faster, simpler, and field-appropriate detection technologies to improve monitoring efficiency and accuracy. In vaccine development, the existing inactivated vaccines remain the only approved type, but they are unable to differentiate between vaccinated and naturally infected animals (DIVA incompatibility) (Endalew et al., 2019). Researchers are exploring new vaccine types such as recombinant protein, vector, DNA/mRNA, and VLP vaccines to address these issues. The goal of developing these novel vaccines is not only to enhance immune efficacy but also to ensure biosafety and support disease control and eradication in endemic regions. Furthermore, vaccine development must also take into account regional resource constraints and production capacities, particularly in resource-poor areas, where there is a need to develop low-cost and heat-stable vaccine technologies to ensure broad applicability.
Given the challenges posed by the cross-border transmission of SBV, global cooperation is crucial for establishing a systematic SBV control strategy (Ma et al., 2022). To develop precise control measures, strengthening international data sharing, technology transfer, and cross-national epidemiological research is essential (Chokshi et al., 2006; Lang, 2011). Additionally, establishing a global animal disease monitoring system, optimizing vaccine stockpiling mechanisms, and enhancing cross-border surveillance networks can effectively slow the rapid spread of outbreaks. As climate change increasingly affects virus transmission, integrating real-time meteorological monitoring, climate modeling, and remote sensing technologies will not only improve prediction capabilities for vector-borne viruses but also facilitate more precise cross-border control measures (Davis et al., 2017; Merkord et al., 2017; Wimberly et al., 2022). By establishing a multi-layered global cooperation framework, a solid foundation can be laid for the long-term control of SBV and other emerging viruses, ensuring the sustainable development of livestock industries.
Author contributions
JW: Data curation, Investigation, Software, Writing – original draft. QJ: Data curation, Writing – original draft. HX: Software, Writing – original draft. FW: Data curation, Writing – original draft. SC: Writing – review & editing. JC: Writing – review & editing. ZJ: Funding acquisition, Writing – review & editing. XY: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Key Research and Development Project of China (2022YFD1800500).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
The reviewer JD declared a shared parent affiliation with the author(s) JW, QJ, FW, SC, JC, ZJ, XY to the handling editor at the time of review.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abudurexiti, A., Adkins, S., Alioto, D., Alkhovsky, S. V., Avšič-Županc, T., Ballinger, M. J., et al. (2019). Taxonomy of the order Bunyavirales: update 2019. Arch. Virol. 164, 1949–1965. doi: 10.1007/s00705-019-04253-6
Abutarbush, S. M., La Rocca, A., Wernike, K., Beer, M., Al Zuraikat, K., Al Sheyab, O. M., et al. (2017). Circulation of a simbu serogroup virus, causing schmallenberg virus-like clinical signs in northern Jordan. Transbound Emerg. Dis. 64, 1095–1099. doi: 10.1111/tbed.12468
Achee, N. L., Grieco, J. P., Vatandoost, H., Seixas, G., Pinto, J., Ching-Ng, L., et al. (2019). Alternative strategies for mosquito-borne arbovirus control. PloS Negl. Trop. Dis. 13, e0006822. doi: 10.1371/journal.pntd.0006822
Aebischer, A., Wernike, K., Hoffmann, B., and Beer, M. (2014). Rapid genome detection of Schmallenberg virus and bovine viral diarrhea virus by use of isothermal amplification methods and high-speed real-time reverse transcriptase PCR. J. Clin. Microbiol. 52, 1883–1892. doi: 10.1128/JCM.00167-14
Aebischer, A., Wernike, K., König, P., Franzke, K., Wichgers Schreur, P. J., Kortekaas, J., et al. (2021). Development of a modular vaccine platform for multimeric antigen display using an orthobunyavirus model. Vaccines (Basel) 9, 651. doi: 10.3390/vaccines9060651
Afonso, A., Abrahantes, J. C., Conraths, F., Veldhuis, A., Elbers, A., Roberts, H., et al. (2014). The Schmallenberg virus epidemic in Europe-2011-2013. Prev. veterinary Med. 116, 391–403. doi: 10.1016/j.prevetmed.2014.02.012
Ak, O., Rf, P., and Ep, N. (2007). The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 81, 3198–3205. doi: 10.1128/JVI.02655-06
Al-Busaidy, S., Hamblin, C., and Taylor, W. P. (1987). Neutralising antibodies to Akabane virus in free-living wild animals in Africa. Trop. Anim. Health Prod 19, 197–202. doi: 10.1007/bf02242116
Ali, H. (2024). AI for pandemic preparedness and infectious disease surveillance: predicting outbreaks, modeling transmission, and optimizing public health interventions. Int. J. Res. Publ. Rev. 05, 4605–4619. doi: 10.55248/gengpi.6.0225.0941
Ansari, M. A., Pope, E. C., Carpenter, S., Scholte, E.-J., and Butt, T. M. (2011). Entomopathogenic fungus as a biological control for an important vector of livestock disease: the culicoides biting midge. PloS One 6, e16108. doi: 10.1371/journal.pone.0016108
Ariza, A., Tanner, S. J., Walter, C. T., Dent, K. C., Shepherd, D. A., Wu, W., et al. (2013). Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res. 41, 5912–5926. doi: 10.1093/nar/gkt268
Baden, L. R., El Sahly, H. M., Essink, B., Kotloff, K., Frey, S., Novak, R., et al. (2021). Efficacy and safety of the mRNA-1273 SARS-coV-2 vaccine. N Engl. J. Med. 384, 403–416. doi: 10.1056/NEJMoa2035389
Baldacchino, F., Caputo, B., Chandre, F., Drago, A., della Torre, A., Montarsi, F., et al. (2015). Control methods against invasive Aedes mosquitoes in Europe: a review. Pest Manag Sci. 71, 1471–1485. doi: 10.1002/ps.4044
Barrett, D., More, S. J., O’Neill, R., Bradshaw, B., Casey, M., Keane, M., et al. (2015). Prevalence and distribution of exposure to Schmallenberg virus in Irish cattle during October 2012 to November 2013. BMC Vet. Res. 11, 267. doi: 10.1186/s12917-015-0564-9
Bayrou, C., Lesenfants, C., Paternostre, J., Volpe, R., Moula, N., Coupeau, D., et al. (2022). Schmallenberg virus, cyclical reemergence in the core region: A seroepidemiologic study in wild cervids, Belgium 2012-2017. Transboundary emerging Dis. 69, 1625–1633. doi: 10.1111/tbed.14136
Beer, M., Conraths, F. J., and van der Poel, W. H. M. (2013). ‘Schmallenberg virus’–a novel orthobunyavirus emerging in Europe. Epidemiol. Infect. 141, 1–8. doi: 10.1017/S0950268812002245
Behar, A., Izhaki, O., Rot, A., Benor, T., Yankilevich, M., Leszkowicz-Mazuz, M., et al. (2021). Genomic detection of schmallenberg virus, Israel. Emerg. Infect. Dis. 27, 2197–2200. doi: 10.3201/eid2708.203705
Bessell, P. R., Auty, H. K., Searle, K. R., Handel, I. G., Purse, B. V., and de C. Bronsvoort, B. M. (2014). Impact of temperature, feeding preference and vaccination on Schmallenberg virus transmission in Scotland. Sci. Rep. 4, 5746. doi: 10.1038/srep05746
Bilk, S., Schulze, C., Fischer, M., Beer, M., Hlinak, A., and Hoffmann, B. (2012). Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet. Microbiol. 159, 236–238. doi: 10.1016/j.vetmic.2012.03.035
Bird, B. H., Maartens, L. H., Campbell, S., Erasmus, B. J., Erickson, B. R., Dodd, K. A., et al. (2011). Rift Valley fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J. Virol. 85, 12901–12909. doi: 10.1128/JVI.06046-11
Blacksell, S. D., Lunt, R. A., and White, J. R. (1997). Rapid identification of Australian bunyavirus isolates belonging to the Simbu serogroup using indirect ELISA formats. J. Virological Methods 66, 123–133. doi: 10.1016/S0166-0934(97)00046-3
Boshra, H. Y., Charro, D., Lorenzo, G., Sánchez, I., Lazaro, B., Brun, A., et al. (2017). DNA vaccination regimes against Schmallenberg virus infection in IFNAR-/- mice suggest two targets for immunization. Antiviral Res. 141, 107–115. doi: 10.1016/j.antiviral.2017.02.013
Boshra, H., Lorenzo, G., Charro, D., Moreno, S., Guerra, G. S., Sanchez, I., et al. (2020). A novel Schmallenberg virus subunit vaccine candidate protects IFNAR-/- mice against virulent SBV challenge. Sci. Rep. 10, 18725. doi: 10.1038/s41598-020-73424-2
Bouloy, M., Krams-Ozden, S., Horodniceanu, F., and Hannoun, C. (1973). Three-segment RNA genome of Lumbo virus (Bunyavirus). Intervirology 2, 173–180. doi: 10.1159/000149420
Bradshaw, B., Mooney, J., Ross, P. J., Furphy, C., O’Donovan, J., Sanchez, C., et al. (2012). Schmallenberg virus cases identified in Ireland. Vet. Rec 171, 540–541. doi: 10.1136/vr.e7928
Bréard, E., Lara, E., Comtet, L., Viarouge, C., Doceul, V., Desprat, A., et al. (2013). Validation of a commercially available indirect elisa using a nucleocapside recombinant protein for detection of schmallenberg virus antibodies. PloS One 8, e53446. doi: 10.1371/journal.pone.0053446
Camarão, A. A. R., Swanepoel, R., Boinas, F., and Quan, M. (2019). Development and analytical validation of a group-specific RT-qPCR assay for the detection of the Simbu serogroup orthobunyaviruses. J. Virol. Methods 271, 113685. doi: 10.1016/j.jviromet.2019.113685
Ceccato, P., Ramirez, B., Manyangadze, T., Gwakisa, P., and Thomson, M. C. (2018). Data and tools to integrate climate and environmental information into public health. Infect. Dis. Poverty 7, 126. doi: 10.1186/s40249-018-0501-9
Charlier, J., Rinaldi, L., Musella, V., Ploeger, H. W., Chartier, C., Vineer, H. R., et al. (2020). Initial assessment of the economic burden of major parasitic helminth infections to the ruminant livestock industry in Europe. Prev. Vet. Med. 182, 105103. doi: 10.1016/j.prevetmed.2020.105103
Chertow, D. S. (2018). Next-generation diagnostics with CRISPR. Science 360, 381–382. doi: 10.1126/science.aat4982
Chokshi, D. A., Parker, M., and Kwiatkowski, D. P. (2006). Data sharing and intellectual property in a genomic epidemiology network: policies for large-scale research collaboration. Bull. World Health Organization. 84, 382–387. doi: 10.2471/BLT.06.029843
Clemente, B., Denis, M., Silveira, C. P., Schiavetti, F., Brazzoli, M., and Stranges, D. (2023). Straight to the point: targeted mRNA-delivery to immune cells for improved vaccine design. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1294929
Collins, Á.B., Barrett, D. J., Doherty, M. L., McDonnell, M., and Mee, J. F. (2017). Significant re-emergence and recirculation of Schmallenberg virus in previously exposed dairy herds in Ireland in 2016. Transbound Emerg. Dis. 64, 1359–1363. doi: 10.1111/tbed.12685
Collins, Á.B., Doherty, M. L., Barrett, D. J., and Mee, J. F. (2019). Schmallenberg virus: a systematic international literature review, (2011-2019) from an irish perspective. Ir Vet. J. 72, 9. doi: 10.1186/s13620-019-0147-3
Conraths, F. J., Kämer, D., Teske, K., Hoffmann, B., Mettenleiter, T. C., and Beer, M. (2013). Reemerging Schmallenberg virus infections, Germany 2012. Emerg. Infect. Dis. 19, 513–514. doi: 10.3201/eid1903.121324
Cuéllar, A. C., Kjær, L. J., Kirkeby, C., Skovgard, H., Nielsen, S. A., Stockmarr, A., et al. (2018). Spatial and temporal variation in the abundance of Culicoides biting midges (Diptera: Ceratopogonidae) in nine European countries. Parasites Vectors 11, 112. doi: 10.1186/s13071-018-2706-y
Dagnaw, M., Solomon, A., and Dagnew, B. (2024). Serological prevalence of the Schmallenberg virus in domestic and wild hosts worldwide: a systematic review and meta-analysis. Front. Vet. Sci. 11. doi: 10.3389/fvets.2024.1371495
Daly, J. M., King, B., Tarlinton, R. A., Gough, K. C., Maddison, B. C., and Blowey, R. (2015). Comparison of Schmallenberg virus antibody levels detected in milk and serum from individual cows. BMC Vet. Res. 11, 56. doi: 10.1186/s12917-015-0365-1
Davis, J. K., Vincent, G., Hildreth, M. B., Kightlinger, L., Carlson, C., and Wimberly, M. C. (2017). Integrating environmental monitoring and mosquito surveillance to predict vector-borne disease: prospective forecasts of a west nile virus outbreak. PloS Curr. 9. doi: 10.1371/currents.outbreaks.90e80717c4e67e1a830f17feeaaf85de
De Regge, N. (2017). Akabane, Aino and Schmallenberg virus-where do we stand and what do we know about the role of domestic ruminant hosts and Culicoides vectors in virus transmission and overwintering? Curr. Opin. Virol. 27, 15–30. doi: 10.1016/j.coviro.2017.10.004
De Regge, N., Deblauwe, I., De Deken, R., Vantieghem, P., Madder, M., Geysen, D., et al. (2012). Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound Emerg. Dis. 59, 471–475. doi: 10.1111/tbed.12000
De Regge, N., van den Berg, T., Georges, L., and Cay, B. (2013). Diagnosis of Schmallenberg virus infection in malformed lambs and calves and first indications for virus clearance in the fetus. Vet. Microbiol. 162, 595–600. doi: 10.1016/j.vetmic.2012.11.029
Dogan, F., Dik, B., Bilge-Dagalp, S., Farzani, T. A., Ataseven, V. S., Acar, G., et al. (2022). Prevalance of Schmallenberg orthobunyavirus (SBV) infection in sampled ruminants in Turkey’s Eastern Mediterranean region between 2015 and 2017. Res. Vet. Sci. 145, 63–70. doi: 10.1016/j.rvsc.2022.02.013
Dominguez, M., Gache, K., Touratier, A., Perrin, J.-B., Fediaevsky, A., Collin, E., et al. (2014). Spread and impact of the Schmallenberg virus epidemic in France in 2012-2013. BMC Vet. Res. 10, 248. doi: 10.1186/s12917-014-0248-x
Ekundayo, F. (2024). Using machine learning to predict disease outbreaks and enhance public health surveillance. World J. Adv. Res. Rev. 24, 794–811. doi: 10.30574/wjarr.2024.24.3.3732
Endalew, A. D., Faburay, B., Trujillo, J. D., Gaudreault, N. N., Davis, A. S., Shivanna, V., et al. (2019). Immunogenicity and efficacy of Schmallenberg virus envelope glycoprotein subunit vaccines. J. Vet. Sci. 20, e58. doi: 10.4142/jvs.2019.20.e58
Endalew, A. D., Morozov, I., Davis, A. S., Gaudreault, N. N., Wernike, K., Bawa, B., et al. (2018). Virological and serological responses of sheep and cattle to experimental schmallenberg virus infection. Vector Borne Zoonotic Dis. 18, 697–703. doi: 10.1089/vbz.2018.2297
Fairbanks, E. L., Daly, J. M., and Tildesley, M. J. (2024). Modelling the influence of climate and vector control interventions on arbovirus transmission. Viruses 16, 1221. doi: 10.3390/v16081221
Fernandes, É.K.K., Bittencourt, V. R. E. P., and Roberts, D. W. (2012). Perspectives on the potential of entomopathogenic fungi in biological control of ticks. Exp. Parasitol. 130, 300–305. doi: 10.1016/j.exppara.2011.11.004
Fischer, M., Schirrmeier, H., Wernike, K., Wegelt, A., Beer, M., and Hoffmann, B. (2013). Development of a pan-Simbu real-time reverse transcriptase PCR for the detection of Simbu serogroup viruses and comparison with SBV diagnostic PCR systems. Virol. J. 10, 327. doi: 10.1186/1743-422X-10-327
García-Bocanegra, I., Cano-Terriza, D., Vidal, G., Rosell, R., Paniagua, J., Jiménez-Ruiz, S., et al. (2017). Monitoring of Schmallenberg virus in Spanish wild artiodactyls 2006-2015. PloS One 12, e0182212. doi: 10.1371/journal.pone.0182212
Garigliany, M.-M. (2012). Schmallenberg virus: A new Shamonda/Sathuperi-like virus on the rise in Europe. Antiviral Res. 95, 82–87. doi: 10.1016/j.antiviral.2012.05.014
Gibbens, N. (2012). Prevention of schmallenberg virus. Veterinary Rec. 170, 130–130. doi: 10.1136/vr.e816
Golender, N., Bumbarov, V. Y., Erster, O., Beer, M., Khinich, Y., and Wernike, K. (2018). Development and validation of a universal S-segment-based real-time RT-PCR assay for the detection of Simbu serogroup viruses. J. Virol. Methods 261, 80–85. doi: 10.1016/j.jviromet.2018.08.008
Goto, Y., Fukunari, K., and Suzuki, T. (2023). Multiplex RT-qPCR application in early detection of bovine respiratory disease in healthy calves. Viruses 15, 669. doi: 10.3390/v15030669
Græsbøll, K., Enøe, C., Bødker, R., and Christiansen, L. E. (2014). Optimal vaccination strategies against vector-borne diseases. Spat Spatiotemporal Epidemiol. 11, 153–162. doi: 10.1016/j.sste.2014.07.005
Gubbins, S., Richardson, J., Baylis, M., Wilson, A. J., and Abrahantes, J. C. (2014). Modelling the continental-scale spread of Schmallenberg virus in Europe: approaches and challenges. Prev. Vet. Med. 116, 404–411. doi: 10.1016/j.prevetmed.2014.02.004
Gubler, D. (2005). The emergence of epidemic dengue fever and dengue hemorrhagic fever in the Americas: a case of failed public health policy. Rev. Panam Salud Publica 17, 221–224. doi: 10.1590/s1020-49892005000400001
Guerra, G. S., Barriales, D., Lorenzo, G., Moreno, S., Anguita, J., Brun, A., et al. (2023). Immunization with a small fragment of the Schmallenberg virus nucleoprotein highly conserved across the Orthobunyaviruses of the Simbu serogroup reduces viremia in SBV challenged IFNAR-/- mice. Vaccine 41, 3275–3284. doi: 10.1016/j.vaccine.2023.04.027
Haider, N., Cuellar, A. C., Kjær, L. J., Sørensen, J. H., and Bødker, R. (2018). Microclimatic temperatures at Danish cattle farms 2000-2016: quantifying the temporal and spatial variation in the transmission potential of Schmallenberg virus. Parasit Vectors 11, 128. doi: 10.1186/s13071-018-2709-8
Hartley, W. J., De Saram, W. G., Della-Porta, A. J., Snowdon, W. A., and Shepherd, N. C. (1977). Pathology of congenital bovine epizootic arthrogryposis and hydranencephaly and its relationship to Akabane virus. Aust. Vet. J. 53, 319–325. doi: 10.1111/j.1751-0813.1977.tb00240.x
Hechinger, S., Wernike, K., and Beer, M. (2014). Single immunization with an inactivated vaccine protects sheep from Schmallenberg virus infection. Vet. Res. 45, 79. doi: 10.1186/s13567-014-0079-6
Hellert, J., Aebischer, A., Wernike, K., Haouz, A., Brocchi, E., Reiche, S., et al. (2019). Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat. Commun. 10, 879. doi: 10.1038/s41467-019-08832-8
Herder, V., Wohlsein, P., Peters, M., Hansmann, F., and Baumgärtner, W. (2012). Salient lesions in domestic ruminants infected with the emerging so-called Schmallenberg virus in Germany. Vet. Pathol. 49, 588–591. doi: 10.1177/0300985812447831
Hoffmann, B., Scheuch, M., Höper, D., Jungblut, R., Holsteg, M., Schirrmeier, H., et al. (2012). Novel orthobunyavirus in cattle, europe 2011. Emerg. Infect. Dis. 18, 469–472. doi: 10.3201/eid1803.111905
Hoffmann, B., Schulz, C., and Beer, M. (2013). First detection of Schmallenberg virus RNA in bovine semen, Germany 2012. Vet. Microbiol. 167, 289–295. doi: 10.1016/j.vetmic.2013.09.002
Huang, Y.-J. S., Higgs, S., and Vanlandingham, D. L. (2017). Biological control strategies for mosquito vectors of arboviruses. Insects 8, 21. doi: 10.3390/insects8010021
Hughes, H. R., Adkins, S., Alkhovskiy, S., Beer, M., Blair, C., Calisher, C. H., et al. (2020). ICTV virus taxonomy profile: peribunyaviridae. J. Gen. Virol. 101, 1–2. doi: 10.1099/jgv.0.001365
Humphries, D. and Burr, P. (2012). Schmallenberg virus milk antibody ELISA. Vet. Rec 171, 511–512. doi: 10.1136/vr.e7739
Iavarone, C., O'hagan, D. T., Yu, D., Delahaye, N. F., and Ulmer, J. B. (2017). Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines 16, 871–881. doi: 10.1080/14760584.2017.1355245
Jimale, Y. A., Jesse, F. F. A., Paul, B. T., Chung, E. L. T., Zakaria, A., Azhar, N. A., et al. (2024). Seroprevalence and contributing factors of transboundary animal diseases in sheep and goats: a study in Peninsular Malaysia. Trop. Anim. Health Prod 56, 212. doi: 10.1007/s11250-024-04061-4
Jiménez-Ruiz, S., Risalde, M. A., Acevedo, P., Arnal, M. C., Gómez-Guillamón, F., Prieto, P., et al. (2021). Serosurveillance of Schmallenberg virus in wild ruminants in Spain. Transbound Emerg. Dis. 68, 347–354. doi: 10.1111/tbed.13680
Jiménez-Ruiz, S., Vicente, J., Risalde, M. A., Acevedo, P., Cano-Terriza, D., González-Barrio, D., et al. (2022). Survey of Culicoides-borne bluetongue and Schmallenberg viruses at the wildlife-livestock interface in Doñana National Park (Spain). Transbound Emerg. Dis. 69, e1815–e1824. doi: 10.1111/tbed.14516
Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J., Zhang, F., and Collins, J. (2021). CRISPR-based diagnostics. Nat. Biomed. Eng. 5, 643–656. doi: 10.1038/s41551-021-00760-7
Kapetas, D., Christakakis, P., Faliagka, S., Katsoulas, N., and Pechlivani, E. (2025). AI-driven insect detection, real-time monitoring, and population forecasting in greenhouses. AgriEngineering. 7, 29. doi: 10.3390/agriengineering7020029
Khoury, D. S., Cromer, D., Reynaldi, A., Schlub, T. E., Wheatley, A. K., Juno, J. A., et al. (2021). Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211. doi: 10.1038/s41591-021-01377-8
Kohara, J., Takeuchi, M., Hirano, Y., Sakurai, Y., and Takahashi, T. (2018). Vector control efficacy of fly nets on preventing bovine leukemia virus transmission. J. Vet. Med. Sci. 80, 1524–1527. doi: 10.1292/jvms.18-0199
König, P., Wernike, K., Hechinger, S., Tauscher, K., Breithaupt, A., and Beer, M. (2019). Fetal infection with Schmallenberg virus - An experimental pathogenesis study in pregnant cows. Transbound Emerg. Dis. 66, 454–462. doi: 10.1111/tbed.13045
Kraatz, F., Wernike, K., Hechinger, S., König, P., Granzow, H., Reimann, I., et al. (2015). Deletion mutants of Schmallenberg virus are avirulent and protect from virus challenge. J. Virol. 89, 1825–1837. doi: 10.1128/JVI.02729-14
Lang, T. (2011). Advancing global health research through digital technology and sharing data. Science 331, 714–717. doi: 10.1126/science.1199349
Larska, M., Lechowski, L., Grochowska, M., and Żmudziński, J. F. (2013). Detection of the Schmallenberg virus in nulliparous Culicoides obsoletus/scoticus complex and C. punctatus–the possibility of transovarial virus transmission in the midge population and of a new vector. Vet. Microbiol. 166, 467–473. doi: 10.1016/j.vetmic.2013.07.015
Lievaart-Peterson, K., Luttikholt, S., Peperkamp, K., Van den Brom, R., and Vellema, P. (2015a). Schmallenberg disease in sheep or goats: Past, present and future. Vet. Microbiol. 181, 147–153. doi: 10.1016/j.vetmic.2015.08.005
Lievaart-Peterson, K., Luttikholt, S., Peperkamp, K., Van den Brom, R., and Vellema, P. (2015b). Schmallenberg disease in sheep or goats: Past, present and future. Vet. Microbiol. 181, 147–153. doi: 10.1016/j.vetmic.2015.08.005
Liu, M., Liu, Y., and Liu, J. (2025). Machine learning for infectious disease risk prediction: A survey. ACM Comput. Surv. 57, 212:1–212:39. doi: 10.1145/3719663
Loeffen, W., Quak, S., de Boer-Luijtze, E., Hulst, M., van der Poel, W., Bouwstra, R., et al. (2012). Development of a virus neutralisation test to detect antibodies against Schmallenberg virus and serological results in suspect and infected herds. Acta Vet. Scand. 54, 44. doi: 10.1186/1751-0147-54-44
Ma, J., Guo, Y., Gao, J., Tang, H., Xu, K., Liu, Q., et al. (2022). Climate change drives the transmission and spread of vector-borne diseases: an ecological perspective. Biology 11, 1628. doi: 10.3390/biology11111628
Mansfield, K. L., Rocca, S. A. L., Khatri, M., Johnson, N., Steinbach, F., and Fooks, A. R. (2013). Detection of Schmallenberg virus serum neutralising antibodies. J. Virological Methods 188, 139–144. doi: 10.1016/j.jviromet.2012.11.031
Martinelle, L., Poskin, A., Dal Pozzo, F., De Regge, N., Cay, B., and Saegerman, C. (2015). Experimental Infection of Sheep at 45 and 60 Days of Gestation with Schmallenberg Virus Readily Led to Placental Colonization without Causing Congenital Malformations. PloS One 10, e0139375. doi: 10.1371/journal.pone.0139375
Maruggi, G., Zhang, C., Li, J., Ulmer, J. B., and Yu, D. (2019). mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 27, 757–772. doi: 10.1016/j.ymthe.2019.01.020
Mason, C., Stevenson, H., Carty, H., Hosie, B., Caldow, G., and Boyes, G. (2013). SBV in a dairy herd in Scotland. Vet. Rec 172, 403. doi: 10.1136/vr.f2256
Matsuo, N. (2019). Discovery and development of pyrethroid insecticides. Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 95, 378–400. doi: 10.2183/pjab.95.027
Mellor, P. S., Boorman, J., and Baylis, M. (2000). Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol 45, 307–340. doi: 10.1146/annurev.ento.45.1.307
Merkord, C. L., Liu, Y., Mihretie, A., Gebrehiwot, T., Awoke, W., Bayabil, E., et al. (2017). Integrating malaria surveillance with climate data for outbreak detection and forecasting: the EPIDEMIA system. Malar J. 16, 89. doi: 10.1186/s12936-017-1735-x
Mochida, Y. and Uchida, S. (2024). mRNA vaccine designs for optimal adjuvanticity and delivery. RNA Biol. 21, 1–27. doi: 10.1080/15476286.2024.2333123
Mouchantat, S., Wernike, K., Lutz, W., Hoffmann, B., Ulrich, R. G., Börner, K., et al. (2015). A broad spectrum screening of Schmallenberg virus antibodies in wildlife animals in Germany. Vet. Res. 46, 99. doi: 10.1186/s13567-015-0232-x
Nadeem, R. M., Ullah, S., Bajwa, M. T. T., Mahmood, M., Saleem, D. R. M., and Maqbool, M. N. (2024). Machine learning-based prediction of african swine fever (ASF) in pigs. VFAST Trans. Software Eng. 12, 199–216. doi: 10.21015/vtse.v12i3.1909
Naqqash, M. N., Gökçe, A., Bakhsh, A., and Salim, M. (2016). Insecticide resistance and its molecular basis in urban insect pests. Parasitol. Res. 115, 1363–1373. doi: 10.1007/s00436-015-4898-9
Narladkar, B. W. (2018). Projected economic losses due to vector and vector-borne parasitic diseases in livestock of India and its significance in implementing the concept of integrated practices for vector management. Vet. World 11, 151–160. doi: 10.14202/vetworld.2018.151-160
Nie, L., Li, B., Jiao, F., Lu, W., Shi, X., Song, X., et al. (2024). EVIT-YOLOv8: Construction and research on African Swine Fever facial expression recognition. Comput. Electron. Agric. 227, 109575. doi: 10.1016/j.compag.2024.109575
Nurtop, E., Villarroel, P. M. S., Pastorino, B., Ninove, L., Drexler, J. F., Roca, Y., et al. (2018). Combination of ELISA screening and seroneutralisation tests to expedite Zika virus seroprevalence studies. Virol. J. 15, 192. doi: 10.1186/s12985-018-1105-5
O’Connor, T. W., Hick, P. M., Finlaison, D. S., Kirkland, P. D., and Toribio, J.-A. L. M. L. (2024). Revisiting the importance of orthobunyaviruses for animal health: A scoping review of livestock disease, diagnostic tests, and surveillance strategies for the simbu serogroup. Viruses 16, 294. doi: 10.3390/v16020294
Padmanaban, V. and Ranganathan, U. D. K. (2022). CRISPR-Cas system and its use in the diagnosis of infectious diseases. Microbiol. Res. 263, 127100. doi: 10.1016/j.micres.2022.127100
Parsonson, I. M., McPhee, D. A., Della-Porta, A. J., McClure, S., and McCullagh, P. (1988). Transmission of Akabane virus from the ewe to the early fetus (32 to 53 days). J. Comp. Pathol. 99, 215–227. doi: 10.1016/0021-9975(88)90073-4
Pley, C., Evans, M., Lowe, R., Montgomery, H., and Yacoub, S. (2021). Digital and technological innovation in vector-borne disease surveillance to predict, detect, and control climate-driven outbreaks. Lancet Planet Health 5, e739–e745. doi: 10.1016/S2542-5196(21)00141-8
Poel, W. H. M. V. D., Parlevliet, J. M., Verstraten, E. R. A. M., Kooi, E. A., Honing, R. H.-V. D., and Stockhofe, N. (2014). Schmallenberg virus detection in bovine semen after experimental infection of bulls. Epidemiol. Infection 142, 1495–1500. doi: 10.1017/S0950268813002574
Poskin, A., Martinelle, L., van der Stede, Y., Saegerman, C., Cay, B., and De Regge, N. (2017). Genetically stable infectious Schmallenberg virus persists in foetal envelopes of pregnant ewes. J. Gen. Virol. 98, 1630–1635. doi: 10.1099/jgv.0.000841
Purse, B. V., Mellor, P. S., Rogers, D. J., Samuel, A. R., Mertens, P. P. C., and Baylis, M. (2005). Climate change and the recent emergence of bluetongue in Europe. Nat. Rev. Microbiol. 3, 171–181. doi: 10.1038/nrmicro1090
Ramachandran, A., Huyke, D. A., Sharma, E., Sahoo, M., Huang, C., Banaei, N., et al. (2020). Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. United States America 117, 29518–29525. doi: 10.1073/pnas.2010254117
Rasekh, M., Sarani, A., and Hashemi, S. H. (2018). Detection of Schmallenberg virus antibody in equine population of Northern and Northeast of Iran. Vet. World 11, 30–33. doi: 10.14202/vetworld.2018.30-33
Rasmussen, L. D., Kirkeby, C., Bødker, R., Kristensen, B., Rasmussen, T. B., Belsham, G. J., et al. (2014). Rapid spread of Schmallenberg virus-infected biting midges (Culicoides spp.) across Denmark in 2012. Transbound Emerg. Dis. 61, 12–16. doi: 10.1111/tbed.12189
Roberts, H. C., Elbers, A. R. W., Conraths, F. J., Holsteg, M., Hoereth-Boentgen, D., Gethmann, J., et al. (2014). Response to an emerging vector-borne disease: surveillance and preparedness for Schmallenberg virus. Prev. Vet. Med. 116, 341–349. doi: 10.1016/j.prevetmed.2014.08.020
Robertson, C., Sawford, K., Daniel, S. L. A., Nelson, T. A., and Stephen, C. (2010). Mobile phone-based infectious disease surveillance system, Sri Lanka. Emerg. Infect. Dis. 16, 1524–1531. doi: 10.3201/eid1610.100249
Roman-Sosa, G., Brocchi, E., Schirrmeier, H., Wernike, K., Schelp, C., and Beer, M. (2016). Analysis of the humoral immune response against the envelope glycoprotein Gc of Schmallenberg virus reveals a domain located at the amino terminus targeted by mAbs with neutralizing activity. J. Gen. Virol. 97, 571–580. doi: 10.1099/jgv.0.000377
Saeed, M. F., Li, L., Wang, H., Weaver, S. C., and Barrett, A. D. T. (2001). Phylogeny of the Simbu serogroup of the genus Bunyavirus. J. Gen. Virol. 82, 2173–2181. doi: 10.1099/0022-1317-82-9-2173
Schulz, C., Beer, M., and Hoffmann, B. (2015). Schmallenberg virus infection in South American camelids: Field and experimental investigations. Vet. Microbiol. 180, 171–179. doi: 10.1016/j.vetmic.2015.08.024
Senf, B., Yeo, W.-H., and Kim, J.-H. (2020). Recent advances in portable biosensors for biomarker detection in body fluids. Biosensors (Basel) 10, 127. doi: 10.3390/bios10090127
Sibhat, B., Ayelet, G., Gebremedhin, E. Z., Skjerve, E., and Asmare, K. (2018). Seroprevalence of Schmallenberg virus in dairy cattle in Ethiopia. Acta Tropica 178, 61–67. doi: 10.1016/j.actatropica.2017.10.024
Sick, F., Beer, M., Kampen, H., and Wernike, K. (2019). Culicoides biting midges—Underestimated vectors for arboviruses of public health and veterinary importance. Viruses 11, 376. doi: 10.3390/v11040376
Sick, F., Zeiske, S., Beer, M., and Wernike, K. (2024). Characterization of a natural “dead-end” variant of Schmallenberg virus. J. Gen. Virol. 105, 376. doi: 10.1099/jgv.0.002005
Silva-Filha, M. H. N. L., Romão, T. P., Rezende, T. M. T., Carvalho, K., da, S., Gouveia de Menezes, H. S., et al. (2021). Bacterial toxins active against mosquitoes: mode of action and resistance. Toxins 13, 523. doi: 10.3390/toxins13080523
Snyder, D., Cernicchiaro, N., Allan, S. A., and Cohnstaedt, L. W. (2016). Insecticidal sugar baits for adult biting midges. Med. Vet. Entomol 30, 209–217. doi: 10.1111/mve.12158
Socha, W., Kwasnik, M., Larska, M., Rola, J., and Rozek, W. (2022). Vector-borne viral diseases as a current threat for human and animal health—One health perspective. J. Clin. Med. 11, 3026. doi: 10.3390/jcm11113026
Songane, M. (2018). Challenges for nationwide vaccine delivery in African countries. Int. J. Health Econ Manag 18, 197–219. doi: 10.1007/s10754-017-9229-5
Stavrou, A., Daly, J. M., Maddison, B., Gough, K., and Tarlinton, R. (2017). How is Europe positioned for a re-emergence of Schmallenberg virus? Veterinary J. 230, 45–51. doi: 10.1016/j.tvjl.2017.04.009
Stokes, J. E., Baylis, M., and Duncan, J. S. (2016). A freedom from disease study: Schmallenberg virus in the south of England in 2015. Vet. Rec 179, 435. doi: 10.1136/vr.103903
Sumner, T., Orton, R. J., Green, D. M., Kao, R. R., and Gubbins, S. (2017). Quantifying the roles of host movement and vector dispersal in the transmission of vector-borne diseases of livestock. PloS Comput. Biol. 13, e1005470. doi: 10.1371/journal.pcbi.1005470
Thomas, D., Blakqori, G., Wagner, V., Banholzer, M., Kessler, N., Elliott, R. M., et al. (2004). Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279, 31471–31477. doi: 10.1074/jbc.M400938200
Varela, M., Pinto, R. M., Caporale, M., Piras, I. M., Taggart, A., Seehusen, F., et al. (2016). Mutations in the schmallenberg virus gc glycoprotein facilitate cellular protein synthesis shutoff and restore pathogenicity of NSs deletion mutants in mice. J. Virol. 90, 5440–5450. doi: 10.1128/JVI.00424-16
Veldhuis, A. M. B., Mars, M. H., Roos, C. A. J., van Wuyckhuise, L., and van Schaik, G. (2017). Two years after the schmallenberg virus epidemic in the Netherlands: does the virus still circulate? Transbound Emerg. Dis. 64, 116–120. doi: 10.1111/tbed.12349
Walter, C. T. and Barr, J. N. (2011). Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 92, 2467–2484. doi: 10.1099/vir.0.035105-0
Wang, J., Chen, D., Wei, F., Yu, R., Xu, S., Lin, X., et al. (2024). Identification of a broadly neutralizing epitope within Gc protein of Akabane virus using newly prepared neutralizing monoclonal antibodies. Veterinary Microbiol. 295, 110123. doi: 10.1016/j.vetmic.2024.110123
Wernike, K., Aebischer, A., Roman-Sosa, G., and Beer, M. (2017). The N-terminal domain of Schmallenberg virus envelope protein Gc is highly immunogenic and can provide protection from infection. Sci. Rep. 7, 42500. doi: 10.1038/srep42500
Wernike, K. and Beer, M. (2019). International proficiency trial demonstrates reliable Schmallenberg virus infection diagnosis in endemic and non-affected countries. PloS One 14, e0219054. doi: 10.1371/journal.pone.0219054
Wernike, K. and Beer, M. (2020). Schmallenberg virus: to vaccinate, or not to vaccinate? Vaccines (Basel) 8, 287. doi: 10.3390/vaccines8020287
Wernike, K., Conraths, F., Zanella, G., Granzow, H., Gache, K., Schirrmeier, H., et al. (2014). Schmallenberg virus-two years of experiences. Prev. Vet. Med. 116, 423–434. doi: 10.1016/j.prevetmed.2014.03.021
Wernike, K., Eschbaumer, M., Breithaupt, A., Hoffmann, B., and Beer, M. (2012). Schmallenberg virus challenge models in cattle: infectious serum or culture-grown virus? Vet. Res. 43, 84. doi: 10.1186/1297-9716-43-84
Wernike, K., Eschbaumer, M., Schirrmeier, H., Blohm, U., Breithaupt, A., Hoffmann, B., et al. (2013a). Oral exposure, reinfection and cellular immunity to Schmallenberg virus in cattle. Vet. Microbiol. 165, 155–159. doi: 10.1016/j.vetmic.2013.01.040
Wernike, K., Fischer, L., Holsteg, M., Aebischer, A., Petrov, A., Marquart, K., et al. (2022). Serological screening in wild ruminants in Germany 2021/2022: No evidence of SARS-CoV-2, bluetongue virus or pestivirus spread but high seroprevalences against Schmallenberg virus. Transbound Emerg. Dis. 10, e3289–e3296. doi: 10.1111/tbed.14600
Wernike, K., Hoffmann, B., Bréard, E., Bøtner, A., Ponsart, C., Zientara, S., et al. (2013b). Schmallenberg virus experimental infection of sheep. Vet. Microbiol. 166, 461–466. doi: 10.1016/j.vetmic.2013.06.030
Wernike, K., Holsteg, M., Szillat, K. P., and Beer, M. (2018a). Development of within-herd immunity and long-term persistence of antibodies against Schmallenberg virus in naturally infected cattle. BMC Vet. Res. 14, 368. doi: 10.1186/s12917-018-1702-y
Wernike, K., Kohn, M., Conraths, F. J., Werner, D., Kameke, D., Hechinger, S., et al. (2013c). Transmission of schmallenberg virus during winter, Germany. Emerg. Infect. Dis. 19, 1701–1703. doi: 10.3201/eid1910.130622
Wernike, K., Mundt, A., Link, E. K., Aebischer, A., Schlotthauer, F., Sutter, G., et al. (2018b). N-terminal domain of Schmallenberg virus envelope protein Gc delivered by recombinant equine herpesvirus type 1 and modified vaccinia virus Ankara: Immunogenicity and protective efficacy in cattle. Vaccine 36, 5116–5123. doi: 10.1016/j.vaccine.2018.07.047
Wernike, K., Nikolin, V. M., Hechinger, S., Hoffmann, B., and Beer, M. (2013d). Inactivated Schmallenberg virus prototype vaccines. Vaccine 31, 3558–3563. doi: 10.1016/j.vaccine.2013.05.062
Wernike, K., Reimann, I., Banyard, A. C., Kraatz, F., La Rocca, S. A., Hoffmann, B., et al. (2021). High genetic variability of Schmallenberg virus M-segment leads to efficient immune escape from neutralizing antibodies. PloS Pathog. 17, e1009247. doi: 10.1371/journal.ppat.1009247
Wimberly, M. C., Davis, J. K., Hildreth, M. B., and Clayton, J. L. (2022). Integrated forecasts based on public health surveillance and meteorological data predict west nile virus in a high-risk region of north america. Environ. Health Perspect. 130, 087006. doi: 10.1289/EHP10287
Yanase, T., Aizawa, M., Kato, T., Yamakawa, M., Shirafuji, H., and Tsuda, T. (2010). Genetic characterization of Aino and Peaton virus field isolates reveals a genetic reassortment between these viruses in nature. Virus Res. 153, 1–7. doi: 10.1016/j.virusres.2010.06.020
Yanase, T., Yoshida, K., Ohashi, S., Kato, T., and Tsuda, T. (2003). Sequence analysis of the medium RNA segment of three Simbu serogroup viruses, Akabane, Aino, and Peaton viruses. Virus Res. 93, 63–69. doi: 10.1016/s0168-1702(03)00066-2
Yilmaz, H., Hoffmann, B., Turan, N., Cizmecigil, U. Y., Richt, J. A., and van der Poel, W. H. M. (2014). Detection and partial sequencing of Schmallenberg virus in cattle and sheep in Turkey. Vector Borne Zoonotic Dis. 14, 223–225. doi: 10.1089/vbz.2013.1451
Keywords: Schmallenberg virus, diagnostic approaches, vaccination strategies, vector control, endemic regions, non-endemic regions
Citation: Wang J, Jia Q, Xiang H, Wang F, Sun C, Chang J, Jiang Z and Yin X (2025) Schmallenberg virus epidemiology and regional control strategies: diagnostics, vaccines, and vector management. Front. Cell. Infect. Microbiol. 15:1633030. doi: 10.3389/fcimb.2025.1633030
Received: 22 May 2025; Accepted: 24 June 2025;
Published: 14 July 2025.
Edited by:
Wang Wei, Inner Mongolia University, ChinaReviewed by:
Junzheng Du, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, ChinaZhanbo Zhu, Heilongjiang Bayi Agricultural University, China
Dongjie Chen, Chinese Academy of Inspection and Quarantine (CAIQ), China
Copyright © 2025 Wang, Jia, Xiang, Wang, Sun, Chang, Jiang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Jiang, amlhbmd6aGlnYW5nQGNhYXMuY24=; Xin Yin, eWlueGluQGNhYXMuY24=
 Jing Wang1,2
Jing Wang1,2 Fang Wang
Fang Wang Chao Sun
Chao Sun Jitao Chang
Jitao Chang Zhigang Jiang
Zhigang Jiang Xin Yin
Xin Yin