- 1School of Biological and Environmental Sciences, Shoolini University of Biotechnology and Management Sciences, Solan, Himachal Pradesh, India
- 2School of Advanced Chemical Sciences, Shoolini University of Biotechnology and Management Sciences, Solan, Himachal Pradesh, India
- 3Crustacean Culture Division, Indian Council of Agricultural Research (ICAR)-Central Institute of Brackish-water Aquaculture, Chennai, Tamil Nadu, India
- 4College of Life Sciences, Department of Aquaculture, National Taiwan Ocean University, Keelung, Taiwan
The gastrointestinal microbiota is crucial for the health and physiology of aquatic organisms, influencing their nutrition, metabolism, and immune responses. This review compares the diversity and function of gut microbial communities in finfish and shellfish, highlighting differences between freshwater and marine species as well as variations within shellfish taxa. We examine how these microbes aid in digesting complex dietary substrates, assimilating nutrients, and synthesizing essential metabolites, all of which are vital for host health. The structure of these microbial communities is shaped by a complex interplay of environmental factors, such as water temperature, salinity, and pH, and host-specific factors, including genetics and diet. A comprehensive understanding of these interactions is key to improving gut health and nutrient use in aquaculture. This review also identifies future research directions, focusing on the use of probiotics, prebiotics, and dietary interventions. These strategies, combined with multi-omics approaches, have great potential to enhance the sustainability of aquaculture by improving growth performance, feed conversion efficiency, and disease resistance in farmed aquatic species.
1 Introduction
Diverse polymicrobial communities—including bacteria, archaea, viruses, yeasts, and protists are ubiquitously associated with aquatic organisms, colonizing niches such as the gastrointestinal tract, skin, gills, and muscle tissues (Li et al., 2022). Their composition and abundance are shaped by environmental factors and host-specific traits like genetics, developmental stage, sex, and diet. Despite this variability, a core gut microbiota often persists across conspecifics, reflecting adaptation to host-specific selective pressures (Diwan et al., 2021). These microbial communities are essential for maintaining physiological homeostasis, supporting nutrient assimilation, immune function, and defense against pathogens. The development of the digestive system and immune modulation are closely tied to the presence of these microbes (Li et al., 2022; Merrifield and Rodiles, 2015). Recent research, including single-cell analyses, highlights the molecular complexity of host microbe interactions and their critical roles in health and disease outcomes (Sharma and Thaiss, 2020).
The study understanding of the complicated internal workings of organisms has been considerably improved by breakthroughs in single-cell analytical tools, particularly those that examine genes, RNA messages, and the spatial organization of cells (Sharma and Thaiss, 2020). While the profound influence of host-microbiota interactions on host development, immunity, metabolism, and associated signalling pathways is now well-established, specific knowledge regarding these interactions in fish and shellfish remains comparatively limited (Sharma and Thaiss, 2020). However, recent research endeavours have begun to elucidate the nature of host-microbiota interactions in the context of various physiological functions within these aquatic organisms. One study investigated these interactions in hybrid fish derived from parental lineages exhibiting contrasting herbivorous and carnivorous dietary adaptations (Pérez et al., 2010). Their findings indicated comparable growth trajectories during early ontogeny and minimal divergence in microbial composition at this initial developmental stage. However, significant alterations in microbiota structure were observed during subsequent developmental phases, coinciding with shifts in dietary regimes. Subsequent analyses revealed a predominance of microbial species associated with metabolic processes and growth in both dietary groups. Furthermore, differentially expressed homologous genes within the intestine, linked to cellular proliferation, immune responses, and metabolic pathways, exhibited correlation with the dominant gut microbiota taxa, suggesting host genes and gut microbes likely work together to help these hybrid fish adapt to their diet (Pérez et al., 2010). Investigations have also focused on the microbial populations within fish digestive systems and their relationships with the mucosal layers (Merrifield and Rodiles, 2015). The research team led by Nerea Arias Jayo also investigated how zebrafish’s gut bacteria changed when the fish ate a diet high in saturated fats with added fish oil. Their work emphasized how important what an animal eats is for the relationship between the animal and the microbes living in its gut (Arias-Jayo et al., 2019). Moreover, research analyzing the gut bacteria of five marine fish species reared together suggested that this intestinal microbial community functions like a second set of genes, influencing various vital physiological processes (Nikouli et al., 2021). The study also reported that a fish gut bacterial community develops in response to both internal factors and external environmental conditions, including diet (Nikouli et al., 2021).
Many studies have elucidated the functional roles of gut microbiota and their interactions with fish and shellfish hosts, including their involvement in digestive enzyme production (Ray et al., 2012) and processes like vitamin synthesis, short-chain fatty acid production, biofilm formation, and iron metabolism in freshwater and marine fish (Romero et al., 2014; Tsuchiya et al., 2008; Xing et al., 2013). The study thoroughly investigated the connection between gut microbiota and growth performance in hybrid fish (Li et al., 2022). While fewer studies have assessed phenotypic variations in fish and shellfish in correlation with gut microbiota, “the species composition of the gut microbiota has been analyzed to determine the involvement of microbial genomes in the selection of core microbiota members (Mushegian et al., 2019). Studies have also examined how the relationship between fish and their gut microbes helps manage stress in different types of fish (Cui et al., 2022; Mohanta et al., 2020). In the context of shellfish, particularly penaeid shrimp, the available information regarding host-microbiota interactions is comparatively limited. Thorough investigation has shown that disease-causing microbes exist in different tissues of penaeid shrimp, negatively impacting their health and the output of aquaculture (Chaiyapechara et al., 2021). However, the burgeoning appreciation for the microbiota’s role in bolstering physiological functions has highlighted its contributions to enhanced immunity and healthy growth. Therefore, to effectively use the shrimp gut microbiota for improving overall health and quality, it’s essential to grasp how external and internal elements affect its composition. Moreover, the study explored how the community of microbes present in the gut of black tiger shrimp (Penaeus monodon) changes, and how their gene activity varies when the shrimp are exposed to different salinities (Chaiyapechara et al., 2021). Their findings indicated that shrimp acclimatized to higher salinities exhibited a gut microbiota dominated by the phylum Proteobacteria, followed by Bacteroidetes and Planctomycetes. The most prevalent genus was Vibrio, belonging to the Harveyi species. Furthermore, they reported differential expression of genes associated with stress and immunity at higher salinities, as the abundance of pathogenic Vibrio increases, expression of genes related to the host’s innate immune response also increases. Other studies have also noted comparable effects of salinity on the bacteria in shrimp intestines (Angthong et al., 2020; Fan et al., 2019; Huang et al., 2016; Rungrassamee et al., 2013). This study looked more closely at the part gut bacteria play in keeping shrimp healthy and controlling diseases. It proposed that changing the gut’s microbial balance by giving shrimp beneficial microbes can have a positive impact on their growth and survival rates (Holt et al., 2020). Recognizing the crucial roles of gut microbiota in shrimp health and immunity (Ma et al., 2018; Tarnecki et al., 2017), an alternative approach for preventing diseases and enhancing shrimp well-being involves strategically modifying the gut microbial community to encourage the growth of beneficial bacteria. Given that factors like culture conditions, developmental phases, and health status can alter shrimp gut microbial composition (Angthong et al., 2020; Fan et al., 2019; Huang et al., 2016; Zheng et al., 2017), a well-defined research strategy is essential to understand these intricate relationships. Researcher investigated the relationship between growth performance in P. monodon and gut microbiota composition, transcriptome, and metabolites, reporting a relative surfeit of bacteria such as Brevibacillus and Spongiimonas in the majority of shrimp guts (Uengwetwanit et al., 2020). Researchers have also identified distinct gene activity patterns in the intestines and specific immune-related genes in shrimp showing different growth rates. Recent progress in studying the microbiota and gene expression in shrimp after probiotic feeding has revealed a strong connection between the types of gut bacteria present and the host’s gene activity related to immunity, digestion, and programmed cell death (Duan et al., 2018, 2019b, 2019a). Researchers investigating the gut bacteria of different penaeid shrimp species have emphasized the crucial part played by the relationships between the host and its microbes in elucidating the basic mechanisms that underpin a wide range of bodily functions (Imaizumi et al., 2021; Dai et al., 2018; Zoqratt et al., 2018).
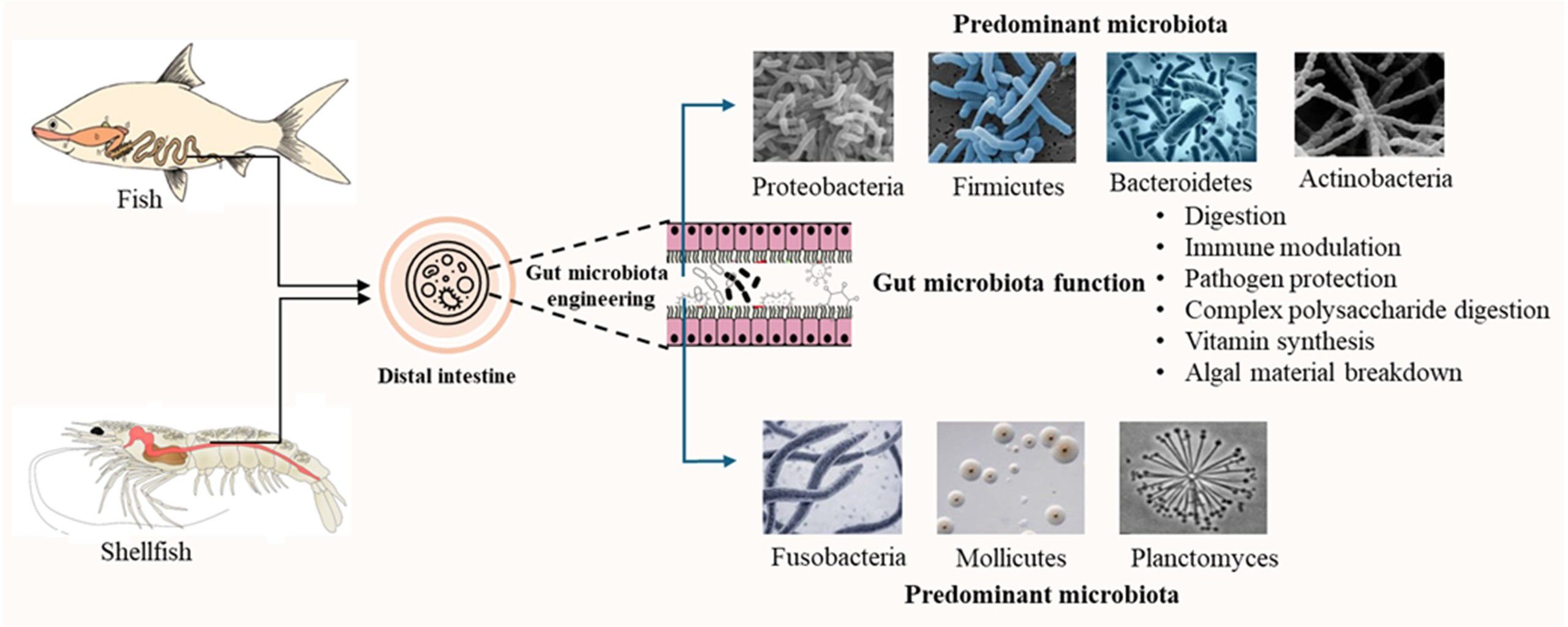
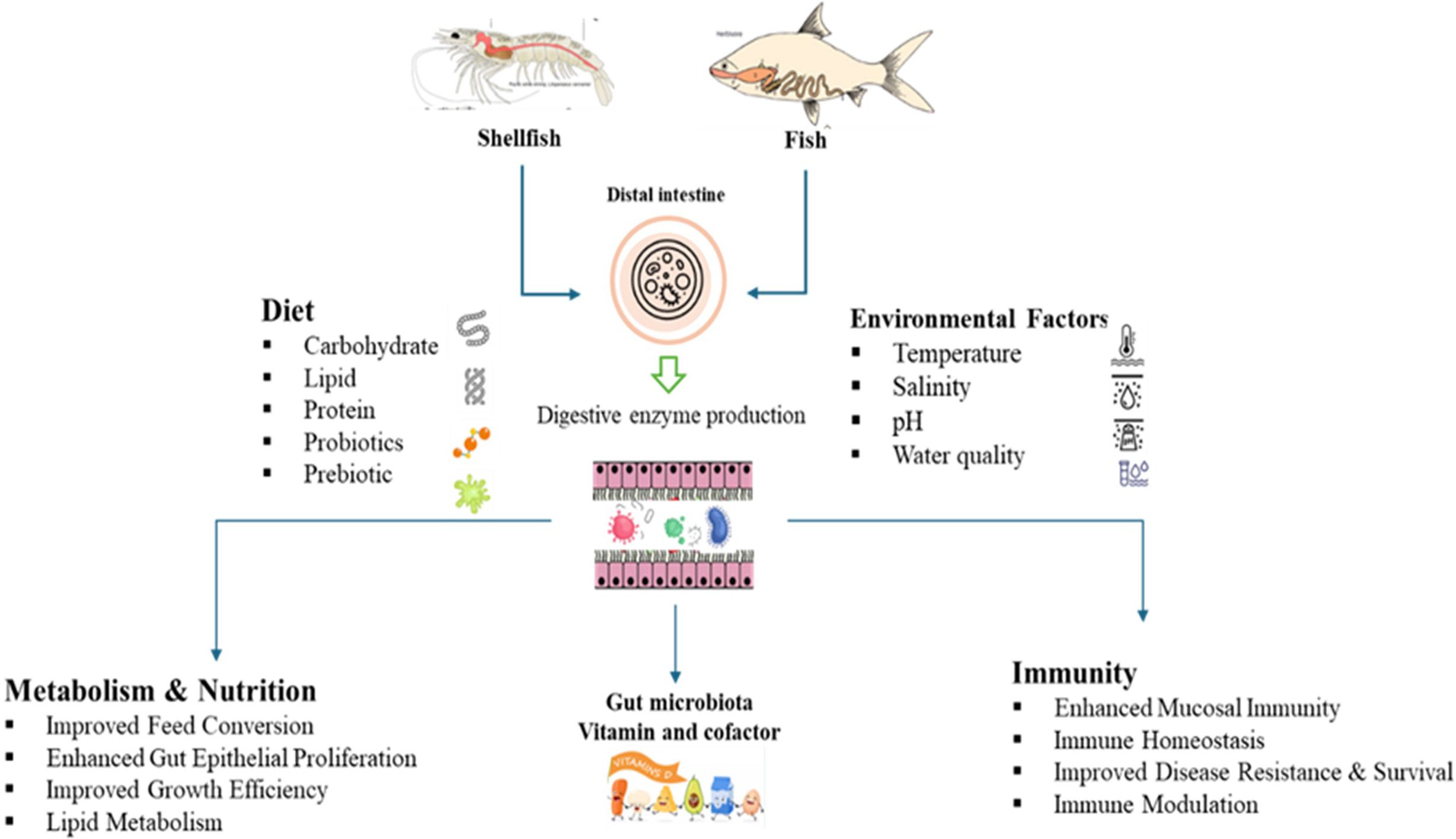
This review synthesizes current knowledge on the complex interactions between host organisms and their gut microbiota in commercially important fish and shellfish species. It critically evaluates the role of gut microbial communities in key physiological processes, with a focus on digestive efficiency, nutrient assimilation, and metabolic regulation. The review first explores the diversity and composition of gut microbiota across freshwater and marine fish, as well as various shellfish species. It then examines the functional contributions of these microbes to host metabolism and analyses the environmental, genetic, and dietary factors that shape microbial diversity and function. Adopting a multidisciplinary perspective, this review highlights the potential of microbiota-driven strategies to enhance aquaculture productivity, support host health, and promote sustainable aquatic food systems (Figure 1).
2 Diversity and composition of gut microbiota in fish and shellfish
The complex and dynamic microbiota inhabiting the fish gastrointestinal tract exerts a significant influence (Zhang et al., 2021). A stable state characterized by a harmonious interaction between the host and gut microorganisms is vital for healthy intestinal function (Banerjee and Ray, 2017). In fact, these resident bacterial populations significantly influence numerous physiological processes, such as maintaining equilibrium, growth, nutrient processing, reproduction, and immune responses (Butt and Volkoff, 2019). Acknowledging the significance of gut flora for the health of fish, including their immune system, physical functions, and overall well-being (Yang et al., 2021), and given that an unbalanced gut microbiota is associated with internal instability and disease development (Wang A, et al., 2018), current scientific research is focused on the potential of using prebiotics, probiotics, and symbiotic to alter the gut microbiota as a way to improve fish health (Nguyen et al., 2018).
Studies have shown that the Bacteroidetes phylum is important for boosting the natural defences system of fish (Gómez and Balcázar, 2008) and for changing the host immune responses, which helps protect them from diseases (Talwar et al., 2018). Furthermore, the Bacteroidetes phylum exhibits a notable metabolic capability to catabolize complex polysaccharides into simpler monosaccharide units (Xu et al., 2003). The development of fast and affordable high-throughput sequencing has dramatically changed how we measure and describe the makeup of microbial communities. This has greatly improved our understanding of the microbiota and made it possible to thoroughly study the bacteria living in fish guts (Chen et al., 2022). Recent advancements in ichthyic microbiota research have garnered considerable attention, recognizing the microbiota a complex consortium encompassing bacteria, fungi, and viruses (Morshed and Lee, 2023) as a critical determinant of host health and homeostasis. Correspondingly, a growing understanding of the microbiota vital function within the host physiology, representing a significant area of application within holobiont research, which focuses on the prevention and therapeutic intervention of diseases through targeted modulation aimed at restoring dysbiotic microbial communities (Diwan et al., 2023). The phylum Actinobacteria, the most extensive prokaryotic group, predominantly comprises Gram-positive bacterial species exhibiting a diverse array of morphological and developmental characteristics (Bhatti et al., 2017). Notably, comparative analyses have identified Bacteroidetes and Firmicutes as potential biomarkers for assessing lipid metabolism in Cyprinus carpio (Meng et al., 2018). A specific study (Talwar et al., 2018) indicated a positive correlation between the phylum Firmicutes and accelerated growth rates in fish compared to the phylum Bacteroidetes. Geerlings et al., 2021 research suggests that bacteria belonging to the phylum verrucomicrobia play a role in breaking down mucus in fish digestion. Recent scientific discoveries highlight the significant influence of gut microbes on the proper development of reproductive systems and subsequent successful reproduction in fish. When zebrafish (Danio rerio) were given Lactobacillus rhamnosus, a type of bacteria in the Firmicutes phylum, from hatching until they reached sexual maturity, their gut bacteria changed, and they developed faster. This was likely due to enhanced growth and the processes that determine their sex (Avella et al., 2012; Carnevali et al., 2013). Conversely, Bozzi et al., 2021 identified Tenacibaculum dicentrarchi as a pathogenic bacterial strain and observed a distinct microbial profile in the distal gastrointestinal tract (GIT) of diseased fish compared to their healthy counterparts. Early exposure of newly hatched fish larvae to commensal microbiota present in the aquatic environment likely confers a protective advantage against opportunistic pathogenic bacteria such as Aeromonas hydrophila (Brugman et al., 2018).
2.1 Freshwater fish gut microbiota
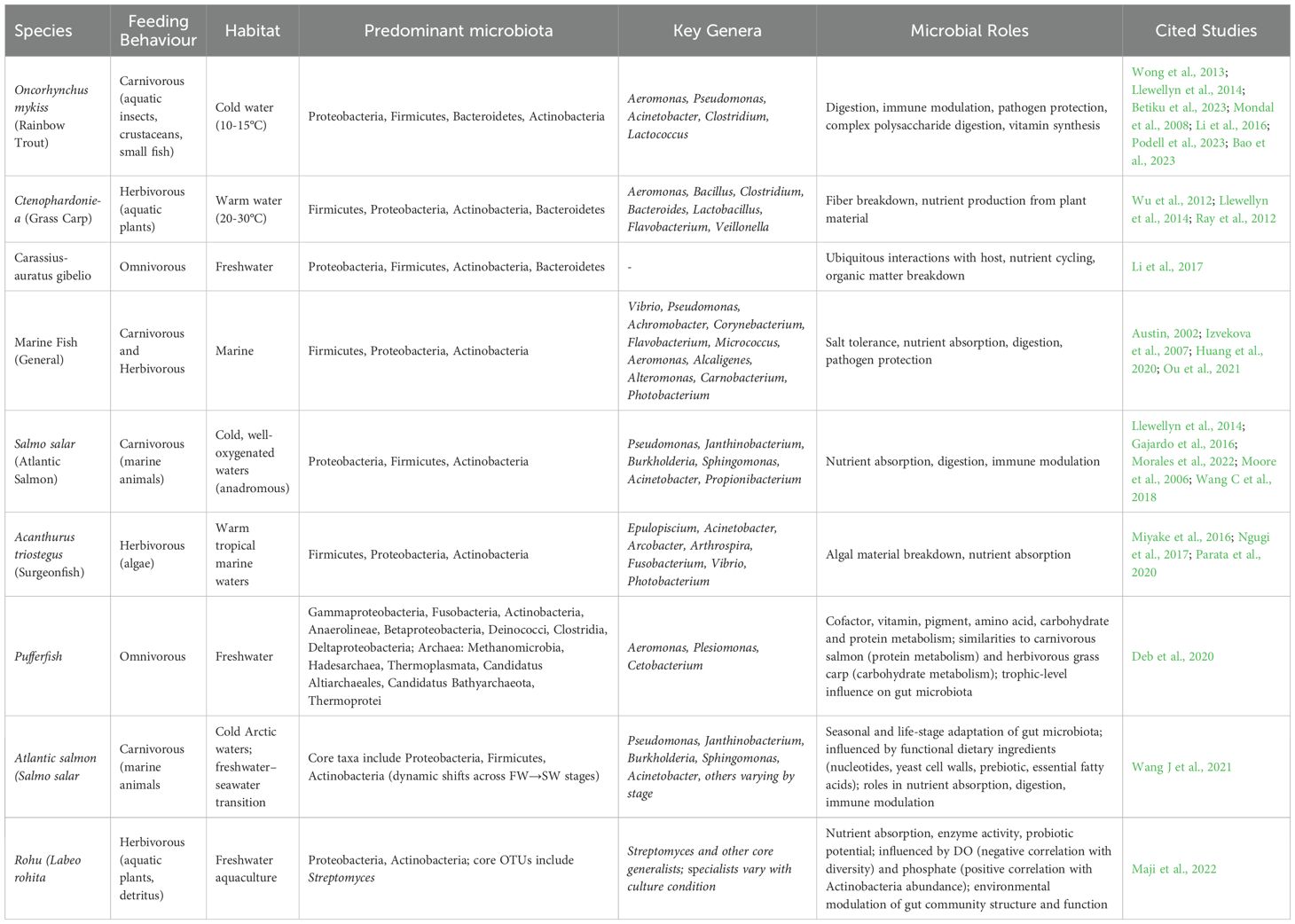
Freshwater investigations have revealed that the core gut bacteria in Oncorhynchus mykiss are resilient to environmental shifts such as different diets and varying stocking densities (Wong et al., 2013), suggesting a stable microbial community. Nevertheless, alterations in diet can still impact the host’s health status (Wong et al., 2013). In herbivorous and omnivorous fish, the enhanced breakdown of cellulose has been linked to the presence of specific bacterial genera, namely Bacillus circulans and B. megaterium (Saha et al., 2006). Investigations into the gut microbiota ontogeny of Carassius auratus gibelio have identified Proteobacteria as the initially dominant phylum colonizing the gastrointestinal tract (Li et al., 2017); this early dominance may be due to their widespread distribution in aquatic environments, facilitating early host–microbe interactions. Beyond Proteobacteria, other commonly found bacterial phyla in freshwater ecosystems include Actinobacteria, Bacteroidetes, Cyanobacteria, and Firmicutes (Cottrell et al., 2005; Jordaan and Bezuidenhout, 2013; Savio et al., 2015; Brar et al., 2023). According to Wu et al. (2012), the most prevalent bacterial group in the gut of freshwater fish is typically Proteobacteria, followed by Firmicutes, Actinobacteria, and Bacteroidetes. Actinobacteria are known for producing a diverse array of secondary metabolites, including hydrolytic enzymes that break down complex molecules, and for contributing to the fermentation of oligosaccharides (Ventura et al., 2007). Members of the phylum Fusobacteria are also frequently detected in freshwater fish (Kim et al., 2021). Common genera and species found include Enterobacter, Aeromonas, Acinetobacter, Escherichia, Klebsiella, Proteus, Serratia, Alcaligenes, Listeria, Bacillus, Bacteroides, Staphylococcus, and Pseudomonas (Austin, 2002; Brown et al., 2019; Hernández et al., 2021; Singh et al., 2021) (Table 1).
A comparative analysis of O. mykiss (rainbow trout) and Ctenopharyngodon idella (grass carp) underscores the influence of trophic ecology and habitat on microbial composition. Rainbow trout, a carnivorous freshwater teleost that consumes aquatic insects, shellfish, and small fish (Huyben et al., 2018), prefers cooler waters (10–15 °C) and harbors gut microbiota dominated by Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, with Aeromonas, Pseudomonas, Acinetobacter, Clostridium, and Lactococcus prevalent at the genus level (Llewellyn et al., 2014; Betiku et al., 2023). These bacteria contribute to digestion, immune modulation (Mondal et al., 2008), pathogen defense, carbohydrate breakdown, and vitamin synthesis (Li et al., 2016; Podell et al., 2023). In contrast, grass carp—a herbivorous fish preferring warmer waters (20–30 °C; Ray et al., 2012)—hosts gut microbiota rich in Aeromonas, Bacillus, Clostridium, Bacteroides, and Leuconostoc (Wu et al., 2012; Llewellyn et al., 2014), reflecting adaptation to plant fiber degradation and plant-derived nutrient assimilation.
2.2 Marine water fish gut microbiota
The elevated salinity of marine environments imposes selective pressures on microbial communities, leading to greater heterogeneity in the gut microbiota of marine fish. Proteobacteria and Actinobacteria are the predominant phyla, although subgroups vary among species. Commonly occurring genera include Vibrio, Flavobacterium, Micrococcus, Aeromonas, Alcaligenes, Alteromonas, Carnobacterium, and Photobacterium (Izvekova et al., 2007; Huang et al., 2020; Ou et al., 2021). Comparisons between Atlantic salmon (Salmo salar) and surgeonfish highlight how habitat and diet shape microbial populations. Atlantic salmon, an anadromous carnivore thriving in cold, oxygen-rich waters, exhibits gut microbiota dominated by Pseudomonas, Janthinobacterium, Burkholderia, Sphingomonas, Acinetobacter, and Propionibacterium (Llewellyn et al., 2014; Gajardo et al., 2016). These microbes aid nutrient absorption and food breakdown (Moore et al., 2006; Wang AR, et al., 2018).
Surgeonfish are herbivorous, inhabiting warm tropical seas and feeding primarily on algae. Their gut microbiota includes Acinetobacter, Arcobacter, Fusobacterium, Vibrio, and Photobacterium (Miyake et al., 2016; Ngugi et al., 2017; Parata et al., 2020), with specialized metabolic capacities for degrading algal polysaccharides, essential for their digestion and nutrient acquisition (Table 1).
2.3 Shellfish gut microbiota
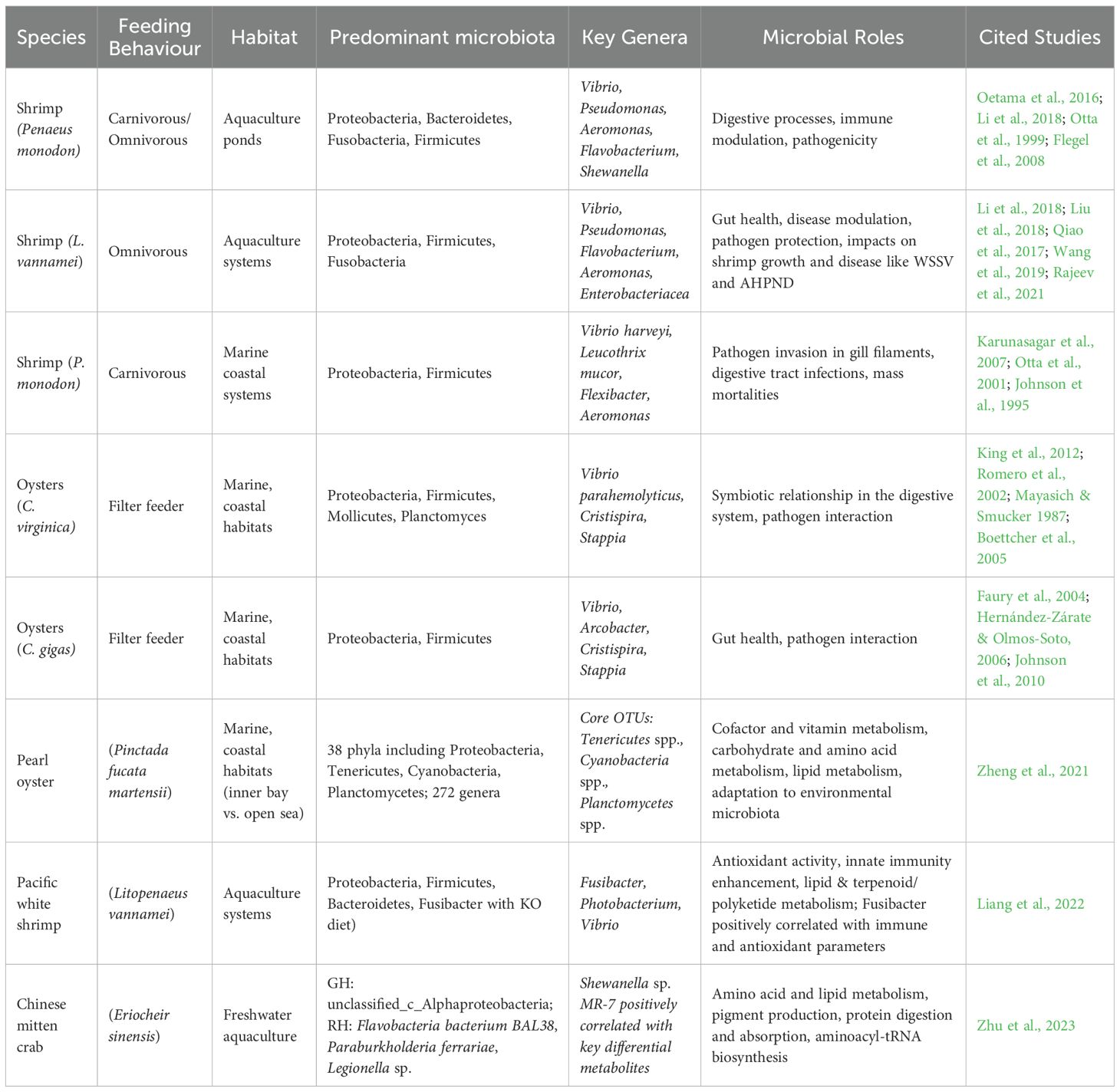
The expansion of commercial shellfish aquaculture, particularly shrimp farming, has coincided with a rise in infectious as well as non-infectious disease etiologies. High prevalence has been observed for filamentous bacteria, peritrich protozoans, invasive bacterial species, and fungi. However, viral agents constitute a critical category of pathogenic microorganisms, having achieved widespread dissemination within shrimp aquaculture facilities. Initially geographically restricted within wild shrimp populations, these viruses have attained global distribution primarily due to the translocation of broodstock and post-larval stages from hatchery systems to geographically distinct locations. A survey by the Global Aquaculture Alliance (GAA) indicated that 60% of shrimp farming losses are attributed to viral infections, while bacterial infections account for 20% of these losses. The first major microbial disease epizootic in shrimp aquaculture occurred in Taiwan during the 1980s, with Monodon baculovirus (MBV) identified as the etiological agent (Flegel et al., 2008). Following this event, outbreaks of infectious hypodermal and hematopoietic necrosis virus (IHHNV) were reported in the United States (Lightner, 1996), yellow head virus (YHV) in Thailand (Flegel, 1997), and Taura syndrome virus (TSV), also in the United States (Brock, 1998). The period from 1993 to 2003 presented further challenges to the shrimp aquaculture sector, characterized by the extensive epizootic of white spot syndrome virus (WSSV), initially documented in China in 1992, which subsequently exhibited rapid pan-Asian spread, resulting in significant economic losses (Flegel et al., 2008). The study documented the presence of six microbial diseases affecting the shrimp Litopenaeus vannamei within Indian aquaculture systems (Gunalan et al., 2014). These included black gill disease, Taura Syndrome Virus (TSV), Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV), white muscle disease (WMD), white gut disease, and muscle cramp disease. While bacterial etiologies, particularly those involving Vibrio spp., have a long-established association with shrimp health and are often correlated with compromised physiological status or suboptimal aquaculture management practices, even immunocompetent individuals can be susceptible to infection under environmental conditions conducive to pathogenic proliferation. The primary sites of bacterial infection are frequently the branchiae and alimentary canal. In severe cases, filamentous bacteria can colonize the branchial lamellae (Johnson et al., 1995). A diverse array of over 20 bacterial species, encompassing human pathogens such as Vibrio cholerae, V. parahaemolyticus, and V. vulnificus, alongside aquatic pathogens like V. harveyi and V. penaeicida, have been implicated in significant shrimp disease outbreaks (Otta et al., 2001). Notably, Vibrio harveyi has been associated with shrimp mortality events and is recognized as the causative agent of brown gill syndrome in Penaeus monodon (Karunasagar et al., 1994; Flegel and Pasharawipas, 1998). Filamentous bacteria, including Leucothrix mucor, Thiothrix sp., Flexibacter sp., Flavobacterium, and Cytophaga sp., have been observed to infect shrimp, particularly during larval ontogeny, manifesting in clinical signs such as branchial discoloration, reduced growth rates, and increased mortality (Karunasagar et al., 2007).
Shrimp farming involving penaeid species faces vulnerability to a wide spectrum of viral pathogens, with over twenty identified as the causative agents of various disease conditions. These viruses are classified within several families, encompassing Parvoviridae, Baculoviridae, Picornaviridae, and Toga-like viruses. The World Organisation for Animal Health (OIE) has identified seven viral pathogens as critically important due to their effects on shrimp aquaculture (Dehaumont, 2004). The alimentary canal of crustaceans, particularly the posterior intestine, offers a favorable milieu for a substantial community of microorganisms (Ceccaldi, 1997). This microbiota is involved in diverse physiological functions, including aiding in nutrient breakdown, synthesizing digestive enzymes, and supplying essential micronutrients such as vitamins (Ceccaldi, 1997). Comparative investigations of shrimp intestinal microbiota across different environmental conditions have indicated variations in microbial composition. Oetama et al. (2016) showed that Proteobacteria represents the dominant bacterial phylum in shrimp originating from both contaminated and less contaminated aquatic environments, followed by less prevalent phyla such as Bacteroidetes, Fusobacteria, and Firmicutes. Of particular note, potentially disease-causing bacteria belonging to the orders Vibrionales and Pseudoaltermonadales were also identified.
Studies examining the intestinal microbial communities of Litopenaeus vannamei have characterized a diverse array of bacteria, exceeding 100 distinct isolates. Within these communities, the genera Photobacterium, Vibrio, Aeromonas, Xanthomonas, Agrobacterium, and Bacillus constitute the predominant taxa (Li et al., 2018). Although the phylum Proteobacteria is generally recognized as the dominant and putatively advantageous component of the healthy shrimp gut microbiota, potentially pathogenic bacteria, including Pseudomonas, Flavobacterium, Escherichia, Aeromonas, Vibrio, Rickettsia, Shewanella, and Desulfovibrio, are also detected at lower relative abundances (Cardona et al., 2016; Qiao et al., 2017).
Research has extensively investigated the modulation of shrimp gastrointestinal microbiota via prebiotic and probiotic administration to influence health outcomes. Numerous studies have assessed their capacity to bolster immune competence and alter the gut microbial profiles (Li et al., 2018; Yukgehnaish et al., 2020). Gut microbial dysbiosis has been implicated in the etiology of various shrimp pathologies. For instance, acute hepatopancreatic necrosis disease (AHPND) in Litopenaeus vannamei is associated with a significant diminution in bacterial richness in affected individuals compared to healthy conspecifics (Liu et al., 2018). Similarly, White Spot Syndrome Virus (WSSV) infection induces a shift in microbiota composition, characterized by an augmentation of Proteobacteria and Fusobacteria and a reduction in Bacteroidetes and Tenericutes (Wang C, et al., 2018). Notably, Huang and Guo reported that microbiota richness in WSSV-infected shrimp showed no significant changes when cultured in biofloc systems, suggesting that this approach may not confer resistance to WSSV (Huang et al., 2018; Guo et al., 2020). White faeces syndrome (WFS) has been correlated with modifications in the gut microbiota, specifically an increase in Tenericutes and Firmicutes and a decrease in Proteobacteria in diseased shrimp (Hou et al., 2018). Investigations into cotton shrimp-like disease (CSL) have indicated an elevation in Tenacibaculum bacteria, although the overall microbiota composition of CSL-affected shrimp demonstrated similarity to that of healthy shrimp (Zhou et al., 2019). Furthermore, Liang et al. (2020) documented significant variations in the microbiota composition of shrimp afflicted with blue body syndrome (BBS) compared to healthy shrimp however, further research is warranted to fully elucidate these relationships. Studies on the microbiota of oysters (Crassostrea virginica and Crassostrea gigas) have concentrated on their interactions with bacteria, including human pathogens such as Vibrio parahaemolyticus and V. vulnificus (Johnson et al., 2010; Sobrinho et al., 2010). Research by King et al. (2012) utilizing high-throughput sequencing methodologies revealed a predominance of Mollicutes and Planctomyctes in the oyster stomach microbiota, whereas the intestinal microbiota exhibited distinct microbial assemblages. These findings indicate the presence of novel microbial communities within oyster digestive systems, although the functional roles of these microbiotas largely remain to be determined (Table 2).
Cross-ecosystem comparisons reveal that dietary ecology and environment are the primary determinants of microbiota structure and function. Freshwater herbivores depend on cellulolytic bacteria for fibre degradation, while carnivorous freshwater and marine fish harbor lipid and protein-degrading microbes, and marine herbivores rely on polysaccharides for algal digestion. In contrast, shellfish exhibit complex host microbe pathogen dynamics, heavily influenced by aquaculture practices and viral disease pressures. Across all groups, the gut microbiota consistently supports three recurring functional themes, nutrient assimilation through the breakdown of carbohydrates, proteins, and lipids, immune modulation and pathogen resistance through commensal protection; and host development and reproduction through microbiota mediated growth, maturation, and sex differentiation. Collectively, these findings underscore that gut microbiota are not passive residents but active contributors to the health, productivity, and resilience of fish and shellfish.
3 Functional roles of fish and shellfish gut microbiota in nutrition and metabolism
The intestinal microbial community significantly influences the nutritional physiology and metabolic homeostasis of fish by actively contributing to the breakdown and absorption of food constituents and modulating the host’s metabolic processes (Figure 2).
Fish rely on their gut microbes to enzymatically digest complex parts of their diet. Numerous bacterial species within the fish gut produce digestive enzymes, such as amylases for carbohydrate breakdown, proteases for protein hydrolysis, and lipases for lipid catabolism, which augment or compensate for the host’s own enzymatic capabilities (Ray et al., 2022; Zhang et al., 2024). This microbial enzymatic activity significantly enhances the efficiency of nutrient digestion in the host (Zhang et al., 2024). Furthermore, the gut microbiota stimulates the proliferation and maturation of the intestinal epithelium, thereby increasing the absorptive surface area available for nutrient uptake (Nayak, 2010). Zebrafish research indicates that gut bacteria promote an elevated count and size of lipid droplets within intestinal epithelial cells, indicating improved lipid absorption (Semova et al., 2012). Cheesman et al. (2011) further elucidated that the microbiota, in conjunction with Wnt signalling pathways, promotes intestinal epithelial cell proliferation by stabilizing β-catenin within gut tissues, potentially contributing to enhanced digestive capacity. Inherent metabolic pathways in fish often demonstrate suboptimal efficiency in processing carbohydrates. However, gut-associated microorganisms enhance carbohydrate digestion through the production of relevant enzymes and by potentiating the activity of the host’s digestive enzymes (Zhang et al., 2024). The microbial fermentation of carbohydrates yields short-chain fatty acids (SCFAs), which are subsequently absorbed by the host and utilized as energy substrates (Pardesi et al., 2022; Hao et al., 2017; Petit et al., 2022). Cetobacterium somerae, a dominant bacterial taxon in the gut of many freshwater fish species, has been shown to improve glucose homeostasis via the production of acetate, which exerts its effects through the activation of parasympathetic pathways in zebrafish (Wang J, et al., 2021). Similarly, the administration of Bacillus amyloliquefaciens SS1 in Nile tilapia fed a high-carbohydrate diet resulted in improved metabolic phenotypes, including reduced fasting glucose levels and decreased lipid accumulation, potentially mediated by an increase in acetate-producing bacteria (Petit et al., 2022). A comparative analysis of the gut microbiota in herbivorous and carnivorous fish by Liu et al. (2016) revealed that cellulase and amylase activities were more closely associated with herbivores, whereas trypsin activity correlated with carnivores. It seems the specific makeup of the microbial community adjusts to the host’s diet and actively participates in breaking down and using carbohydrates and proteins.
The intestinal microbiota significantly enhances host energy extraction via multiple pathways, encompassing the modulation of lipid absorption and the transformation of bile acid profiles and the adjustment of genes involved in maintaining energy balance, Guo and colleagues in 2017 showed that zebrafish consuming diets rich in nucleotides displayed lower basal metabolic rates as a result of changes in their gut bacteria, leading to improved energy storage and growth (Guo et al., 2017). Similarly, Zhang studied that Citrobacter bacteria isolated from Nile tilapia intestines enhanced energy extraction in fish consuming a high-fat diet, highlighting the microbiota’s ability to modulate the adverse outcomes associated with suboptimal nutrient intake (Zhang et al., 2020). Furthermore, De La Torre Canny et al., 2021 indicated that Plesiomonas species reduced fat accumulation in zebrafish, suggesting that specific gut microbes can counteract the obesogenic effects of environmental contaminants like tributyltin (TBT). Bile acids, crucial for the digestion and absorption of lipids, undergo microbial transformation in the gut into secondary bile acids that influence glucose and lipid metabolism. In zebrafish, the primary biliary acids, 5α-cyprinol sulfate (5αCS) and taurocholic acid (TCA), undergo biotransformation mediated by the gut microbiota (Wen et al., 2021). Specifically, an identified Acinetobacter species exhibited the capacity for TCA deconjugation, potentially leading to the activation of farnesoid X receptor (FXR) signaling, a key regulatory axis in lipid and energy homeostasis.
Xia et al. (2023) also observed that bile acids enhance intestinal barrier function through both direct mechanisms and microbiota-dependent routes, potentially impacting nutrient assimilation and metabolic processes. The feed conversion ratio (FCR) represents a crucial economic parameter in aquaculture. Gut microbiota contributes to enhanced FCR by improving digestive and metabolic efficiency. Although more extensively investigated in monogastric animals, accumulating evidence from fish also supports this notion. Specific probiotic strains, such as Lactobacillus acidophilus (Singh et al., 2014), Bacillus coagulans (Adeshina et al., 2020), and Acinetobacter (Amoah et al., 2019), have been associated with improved FCR in fish, although the underlying mechanisms remain to be fully elucidated. Dvergedal et al. (2020) identified three operational taxonomic units (OTUs) that exhibited a positive correlation with enhanced feed efficiency and carbon metabolism in Salmo salar. Similarly, Bozzi et al. (2021) reported a positive association between Mycoplasma abundance and both physiological condition and somatic weight in salmon, indicating a beneficial microbial role in growth and energy utilization. Intestinal microbes can modulate host gene expression related to metabolic pathways. For example, studies in Danio rerio have demonstrated microbiota-mediated regulation of genes involved in lipid uptake, fatty acid metabolism, and energy storage (Sheng et al., 2018). In zebrafish, Semova and colleagues demonstrated in 2012 that the gut microbial community enhanced the uptake of lipids and the accumulation of lipid droplets within the intestinal and hepatic tissues. This observation suggests that the microbially mediated influence on host metabolic pathways particularly lipid processing may represent a conserved biological mechanism across vertebrate species.
The intestinal microbiota of crustaceans, notably shrimp, is fundamental to their nutritional physiology, metabolic homeostasis, immune system function, and capacity to withstand environmental perturbations. Considering the limited capacity of shrimp to digest complex food components such as carbohydrates, proteins, and fats, the gut microbiota plays a vital role in completing these metabolic processes. The dynamic nature of this microbial community’s structure and function allows for rapid adjustments in response to dietary changes, environmental factors, and stressors, with downstream effects on host health and productivity. Shrimp is critically dependent on their gut microbiota for the assimilation of complex feed components. The intestinal microbiota metabolizes unabsorbed nutrients and generates diverse metabolites that positively influence the host digestive processes and general health (Qiao et al., 2017). These micro-organisms express enzymes including proteases, lipases, and amylases that contribute to the catabolism of proteins, lipids, and carbohydrates, respectively. Distinct anatomical sections of the shrimp gastrointestinal tract exhibit specialized microbial metabolic roles. The foregut microbiota is predominantly involved in the biotransformation of amino acids and carbohydrates. The community of microorganisms residing in the insect midgut is crucial for processing fats, polyketide compounds, and terpenoids. The hindgut microbiota is more engaged in vitamin biosynthesis, energy production, and cofactor metabolism (Garibay-Valdez et al., 2021).
The evident functional diversification highlights the adaptive specialization of the gut microbiota, driven by the spatial heterogeneity and nutrient gradients along the host’s digestive tract. Dietary carbohydrates function not only as a principal energy substrate for shrimp but also as crucial substrates supporting the metabolic activities of the gut microbial community. Through fermentative pathways, these microorganisms metabolize carbohydrates, yielding short-chain fatty acids (SCFAs) and other biologically active compounds. The inclusion of various carbohydrates, such as glucose, sucrose, xylooligosaccharides, and starch, in the dietary formulations for shrimp has been shown to positively influence the composition and resilience of their gut microbial communities. This modulation of the intestinal microbiota is associated with enhanced gut health and improved overall growth performance in shrimp aquaculture (Chen et al., 2021; Gyan et al., 2022). Furthermore, modulation of the carbon-to-nitrogen (C/N) ratio in feed has been shown to optimize the gut microbial architecture, increasing microbial efficiency in nutrient catabolism and anabolism, thereby enhancing shrimp productivity (Guo et al., 2020). An optimized C/N ratio fosters the proliferation of beneficial bacterial taxa and supports a conducive environment for microbial fermentation and energy acquisition. Gut microbiota-mediated protein and lipid metabolism is critical for maximizing feed utilization efficiency in shrimp. Modifications in the dietary protein composition or ratio induce shifts in the gut microbial community towards species exhibiting enhanced proteolytic capabilities, consequently improving the digestion and assimilation of amino acids (Gyan et al., 2022).
The nature of dietary lipid sources, such as soybean oil, tallow, or linseed oil, exerts a considerable impact on the structural assembly of the intestinal microbial community. These alterations in microbial composition modulate lipid metabolic pathways and correlate with observed differences in immunological responses and growth performance (Zhang et al., 2014). Furthermore, specific microbial taxa contribute to the biotransformation of bile acids, indirectly facilitating the processes of lipid emulsification and subsequent absorption. The community of microorganisms residing in the shrimp’s gut is essential for regulating its immune system. A balanced and diverse microbial ecosystem can confer protection against pathogenic organisms through mechanisms including competitive exclusion, the enhancement of mucosal immunity, and the production of antimicrobial compounds. Following infection with White Spot Syndrome Virus (WSSV), shrimp demonstrate shifts in their core gut microbial profile. These microbial dysbiosis events are frequently associated with an increased production of antiviral metabolites of microbial origin, which contribute to the maintenance of immune homeostasis and provide defence against the disease (Zhang and Sun, 2022).
The increasing acknowledgement of probiotic supplementation as a more environmentally sustainable and safe approach compared to antibiotic administration in shrimp aquaculture for disease control is significant. Probiotic bacteria, including genera such as Lactobacillus and Bacillus, have demonstrated positive influences on shrimp growth metrics, immune competence, and survivability. These beneficial effects are mediated through the promotion of advantageous microbial communities and the attenuation of pathogenic organism burdens (Holt et al., 2020). Environmental variables, with a particular emphasis on salinity, significantly impact the compositional architecture of the shrimp gastrointestinal microbiota. Research on Penaeus monodon indicated that shrimp exposed to an initial acclimatization at 20 ppt salinity, followed by transfer to environments of 10 ppt and 30 ppt, exhibited modifications in microbial community structure. Specifically, the relative abundance of Vibrio species established sensitivity to salinity shifts (Chaiyapechara et al., 2012). While Vibrio populations increased in correlation with elevated salinity levels, the relative abundance of other genera, remained relatively constant, indicating a degree of stability within the core microbiota.
Alterations in environmental salinity induce shifts in the microbial community structure, which are mirrored at the molecular level. Metatranscriptomic studies show that genes related to stress response and immunity are expressed at varying levels, underscoring the interconnected impact of environmental salinity on the transcriptional profiles of both the host organism and its associated microbiota. However, the precise nature of this interaction whether the shrimp actively regulate these microbial shifts or passively respond to them requires further investigation. Nutrient deprivation, specifically starvation, elicits substantial modifications in the gut microbiota composition and functional attributes. In shrimp experiencing starvation, a downregulation of digestive enzyme activity is observed concurrently with an upregulation of immune response-related genes. Functional pathway analysis reveals a diminished capacity for carbohydrate, protein, lipid, glycan, and enzyme metabolism under conditions of nutrient scarcity (Dai et al., 2020). These physiological alterations heighten the host’s susceptibility to pathogenic invasion, emphasizing the importance of the gut microbiota in maintaining metabolic and immunological homeostasis during periods of stress. Contemporary research has begun to elucidate correlations between the compositional characteristics of the gut microbiota and parameters of shrimp breeding performance. Specific microbial signatures have been linked to enhanced growth rates and weight gain, indicating the potential utility of gut microbial structure as a biomarker for selective breeding initiatives in shrimp aquaculture (Fan & Li, 2019). These findings lend support to the integration of microbiota analysis into aquaculture breeding programs as a strategy to improve sustainability and overall productivity.
4 Sequencing strategies for gut microbiota analysis in fish and shellfish
Advances in next-generation sequencing (NGS) have greatly enhanced the characterization of fish and shellfish gut microbiota, primarily through targeted amplicon sequencing and shotgun metagenomics (Wensel et al., 2022). While differing in scope and resolution, both approaches have transformed microbiome research in aquaculture.
Amplicon sequencing, typically targeting the 16S rRNA gene for bacteria or ITS regions for fungi, offers cost-effective and scalable profiling of microbial communities (Yang et al., 2025). It has been widely applied to assess how gut microbiota shift across diets, developmental stages, and environmental or health conditions (Perez-Bustamante et al., 2024). Although this strategy cannot directly resolve functional genes or capture non-bacterial taxa, it remains well suited for large-scale ecological comparisons.
Shotgun metagenomics, which sequences total genomic DNA without prior amplification, enables comprehensive profiling of both microbial composition and functional capacity (Yang et al., 2025). This approach achieves strain-level resolution across bacteria, archaea, viruses, and microeukaryotes (Talwar et al., 2018), and allows annotation of metabolic pathways, virulence factors, antimicrobial resistance genes, and mobile elements. Such functional insights are particularly valuable for elucidating microbial contributions to nutrient assimilation, immune modulation, pathogen defense, and stress tolerance in aquaculture species (Khurana et al., 2020). For instance, shotgun studies have linked microbial shifts to dietary efficiency, probiotic action, and disease resistance, thereby informing management strategies that enhance productivity and sustainability.
The choice of method depends on study objectives: amplicon sequencing is ideal for high-throughput surveys of microbial dynamics across treatments or time points (Spilsbury et al., 2022), whereas shotgun metagenomics is preferred for mechanistic analyses of host–microbe interactions and functional profiling (Yang et al., 2025). Increasingly, combined approaches are employed to leverage the breadth of amplicon surveys with the depth of shotgun datasets, providing a more holistic understanding of gut microbial ecology (Sheth et al., 2016).
Looking forward, integration of metagenomics with transcriptomics, metabolomics, and metaproteomic will be critical for linking microbial identity with activity. These multi-omics strategies are beginning to reveal not only community composition but also metabolic function within the gut, creating new opportunities to connect microbiome dynamics with growth performance, disease resistance, and sustainability outcomes in aquaculture.
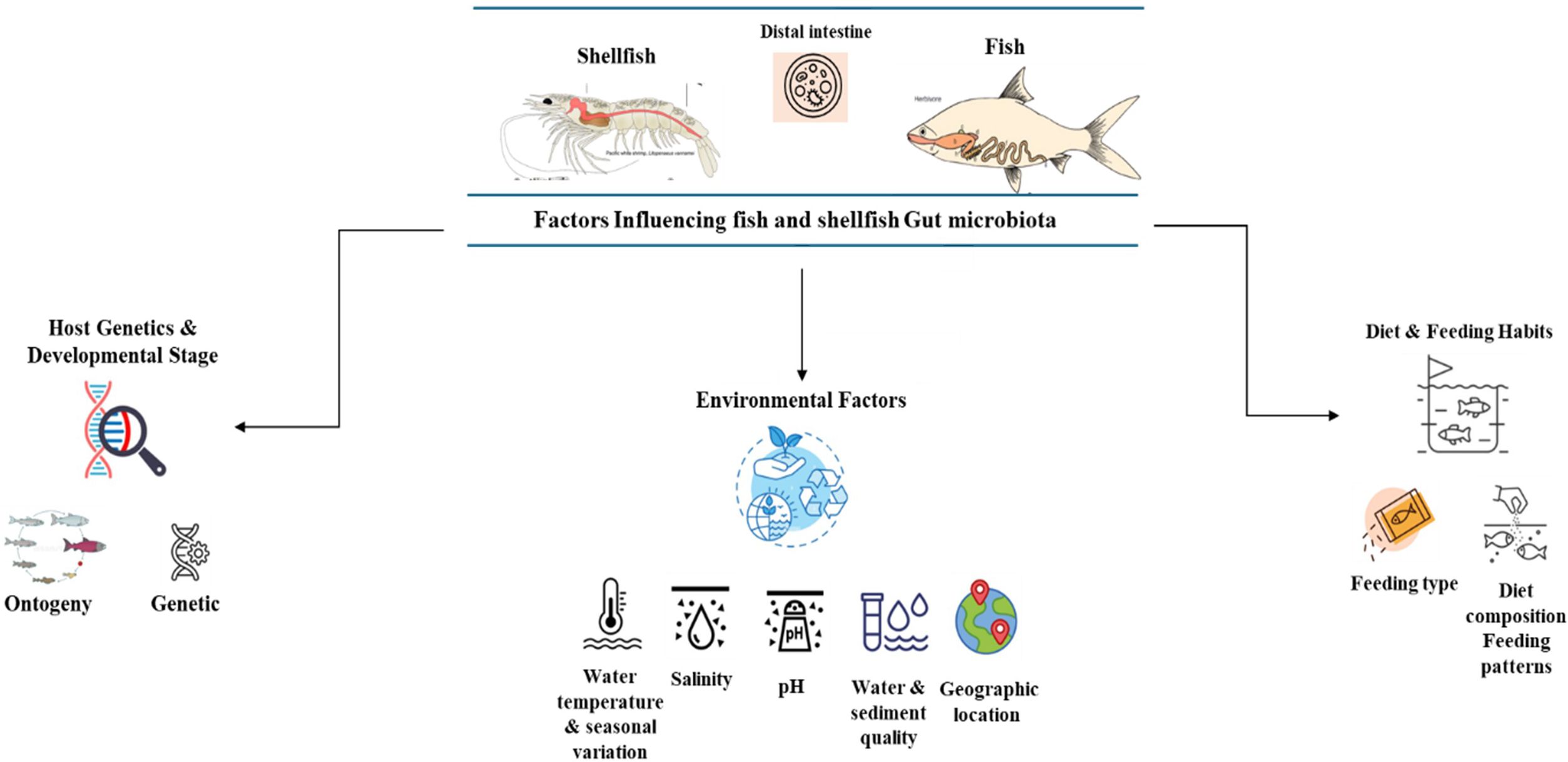
5 Factors influencing the composition of fish and shellfish gut microbiota
The structure and variety of microorganisms inhabiting the fish gut are determined by a complex and ever-changing combination of environmental conditions, inherent biological traits, and nutritional inputs. These factors exert specific effects that vary across the life cycle of the fish and in different habitats. An increasing amount of scientific evidence indicates that comprehending these intricate relationships is essential for refining fish farming techniques, as well as for maintaining fish well-being, strengthening their defense mechanisms, and fostering ecological adaptability (Diwan et al., 2023a; Ou et al., 2021) (Figure 3).
5.1 Environmental factors
5.1.1 Water temperature and seasonal variation
Fish gut microbiota composition is strongly affected by temperature, driving the proliferation or decline of specific microbial taxa in response to thermal conditions (Ghosh et al., 2022; Hassenrück et al., 2020; Sepúlveda-Quiroz et al, 2021). In young milkfish (Chanos chanos), alterations in ambient temperature between 26 and 33 °C elicited notable modifications in the intestinal microbiota composition, thereby supporting the host’s physiological acclimatization to variable thermal conditions (Hassenrück et al., 2020). In a parallel observation within Oncorhynchus tshawytscha, differing temperature regimes induced a shift in the dominant microbial communities, characterized by the displacement of families like Vibrionaceae and their subsequent substitution with Fusobacteriaceae and Brevenemataceae (Steiner et al., 2022; Dulski et al., 2020; Hovda et al., 2012).
5.1.2 Salinity
Salinity represents a pivotal environmental determinant shaping the gut microbial architecture, particularly for species undergoing transitions between marine and freshwater habitats. In Salmo salar, the migration from saltwater to freshwater ecosystems induced alterations in the prevailing bacterial phyla. Notably, the genera Escherichia and Shigella exhibited increased relative abundance in populations inhabiting seawater (Morales et al., 2022). Conversely, certain taxa, including the phylum Proteobacteria and the genus Lactobacillus, demonstrated a capacity to withstand fluctuations in salinity in aquaculture settings involving Oncorhynchus tschawytscha (Zhao et al., 2020; Liu et al., 2023).
5.1.3 pH
The acidity of aquatic habitats significantly impacts the equilibrium of gut microorganisms. Deviations from a neutral pH, whether towards acidic or alkaline conditions, can disrupt this microbial balance (homeostasis) and favor the proliferation of disease-causing microbes (pathogens). Fonseca et al. (2019) observed a decrease in beneficial lactic acid bacteria in sea bream when pH was lowered. Conversely, Kong et al. (2022) and Shang et al. (2024) describes an increase in detrimental Vibrio species in common carp under alkaline conditions. Exposure of the Pacific oyster, Crassostrea gigas, and other consumable oyster species to conditions of ocean acidification, characterized by decreased pH levels, has been observed to induce shifts in their associated microbial communities (Kong et al., 2022).
5.1.4 Water and sediment quality
Aquatic microbial communities, residing in the water column and sediment, exert a considerable influence on fish gut microbiota via ongoing exposure. In shrimp species like Litopenaeus vannamei and Penaeus japonicus, studies have evidenced a notable similarity between the microbial compositions of the ambient water and the host’s intestinal tract (Huang et al., 2018; Song et al., 2020). These environmental microbial assemblages function as microbial reservoirs, facilitating the ingestion and subsequent colonization of the gut by microorganisms (Huang et al., 2021).
5.1.5 Geographic location
Geographic partitioning results in environmental heterogeneity, characterized by variations in salinity, temperature, and the spectrum of microorganisms. These variations, in turn, influence the structural makeup of the gut microbial communities within organisms. For instance, research employing high throughput 16S rRNA sequencing by Liu et al. (2022b); Liu et al. (2022a) investigations revealed significant variations in the gut microbial community structure across Megalobrama terminalis populations residing in the Pearl, Moyang, and Wanquan River systems. This phenotypic divergence is attributed to allopatric speciation driven by genetic drift and adaptation to distinct environmental pressures. Conversely, studies by Noman et al. (2024) indicated a negligible impact of geographic location on the gut microbiota of gilthead seabream (Sparus aurata) and European seabass (Dicentrarchus labrax), suggesting that certain species may maintain relatively stable microbial communities irrespective of their geographic distribution.
5.2 Host genetics and developmental stage
Genetic traits inherited from a host significantly impact the development of their microbial communities, the way their immune system functions, and their enzyme production, ultimately determining the makeup of the gut microbiota (Naya-Català et al., 2022). Studies in Gasterosteus aculeatus revealed that genetically differentiated populations harbored divergent gut microbial assemblages (Smith et al., 2015). Conversely, in Ictalurus punctatus and Ictalurus furcatus, environmental variables appeared to be the dominant drivers of gut microbial composition when ontogenetic trajectories were comparable (Bledsoe et al., 2018). Furthermore, the host’s developmental phase represents a crucial determinant of microbial community structure, the microbiota undergoes temporal shifts in relation to host maturation., Sparus aurata in later life stages exhibited greater microbial richness compared to younger individuals (Parata et al., 2020). Analogous ontogenetic patterns of microbial succession have been documented in Acipenseridae species and Silurus meridionalis, where microbial community dynamics correlated with host developmental transitions (Parata et al., 2020).
5.3 Diet and feeding habits
The interplay between piscian alimentary regimes and foraging habits significantly modulates the constitution of their intestinal microbial consortia. Herbivorous generally possess comparatively longer alimentary canals and harbor distinct gut microbial communities compared to carnivorous species. The impact of nutritional factors on the composition and function of these microbial consortia is well-established in scientific publications, for instance, in Atlantic salmon, diets rich in carbohydrates have been shown to diminish overall bacterial load while fostering the proliferation of taxa specialized in carbohydrate metabolism (Villasante et al., 2019). Ontogenetic shifts in feeding patterns, as observed in Megalobrama amblycephala, result in microbial community reorganization and modifications in enzymatic activities (Wei et al., 2018). Furthermore, periods of nutritional deprivation also exert influence on gut microbial assemblages; in Plectropomus leopardus, Firmicutes was the predominant phylum during periods of feeding, whereas Proteobacteria became dominant during fasting (Mekuchi et al., 2018).
6 Conclusion
Gut-associated microbial communities in fish and shellfish are now widely regarded as a functional “accessory organ” that exerts significant influence on host physiology, nutrition, and metabolic efficiency. These microbial assemblages differ considerably among freshwater and marine species, as well as across various groups of shellfish, reflecting the combined effects of host traits, ecological roles, and environmental conditions. Through processes such as enzymatic degradation of complex substrates, vitamin biosynthesis, and enhancement of nutrient absorption, the gut microbiota directly contributes to host growth and health. The composition of these microbial communities is not static but dynamically shaped by multiple internal and external drivers, including environmental parameters (temperature, salinity, pH, sediment quality, and geography) and host-related factors (genetics, development, and diet). Collectively, this host microbe environment interaction forms a regulatory network essential for maintaining intestinal balance and overall organismal health. Despite notable advances, the precise cellular, molecular, and metabolic mechanisms that govern these interactions remain insufficiently understood. Future research should focus on unravelling the influence of gut microbiota on host gene expression, immune regulation, and metabolic pathways, using integrative multi-omics and controlled experimental approaches. In addition, the microbial communities of aquatic organisms harbor an untapped reservoir of bioactive molecules with promising applications in pharmaceuticals, nutraceuticals, and sustainable aquaculture practices. A more comprehensive mechanistic understanding will not only optimize health and productivity in aquaculture but also open new frontiers in microbial biotechnology and environmentally responsible cultivation of fish and shellfish.
7 Future perspectives and recommendation
To significantly propel gut microbiota research forward, foundational experimental methodologies necessitate enhancement. The integration of sophisticated multi-omics analyses offers deeper understanding into the gut microbiota’s role, function, and composition, facilitating the optimization of the intestinal ecosystem for future applications (Ou et al., 2021). Gnotobiotic piscine models, such as zebrafish and stickleback, present valuable systems for these investigations (Zhang et al., 2021). Diverse bioengineering strategies can be employed to cultivate a beneficial microbiota or to remediate dysbiosis in animal models. However, a thorough understanding of the interplay between host disease states and the intestinal microbial community is crucial for effective implementation (Filardo et al., 2024). A synergistic application of various engineering methodologies, targeted at gut microbiota modulation, represents a primary focus for future research endeavours (Nath et al., 2022). Expanding beyond conventional engineering approaches, metabolic engineering strategies hold promises for improved outcomes in gut microbiota modulation (Li et al., 2023). CRISPR-based technologies offer the potential to significantly advance engineering techniques with enhanced precision and efficiency (Careaga, 2024). Beyond single-strain manipulation, the application of synthetic microbial communities (SynComs) provides an emerging strategy. These rationally designed consortia can deliver consistent functions such as nutrient assimilation, pathogen resistance, and stress tolerance, thereby enabling more predictable and stable outcomes in aquaculture systems. Another promising direction is precision aquaculture microbiome engineering, where microbial interventions are tailored to host species, developmental stage, or environmental conditions. Supported by artificial intelligence (AI) and machine learning, such targeted approaches could optimize microbial community dynamics in real time, thereby improving feed conversion efficiency, growth, and disease resilience. In addition, integrating microbiome engineering with climate change resilience strategies represents a crucial future perspective. Microbial communities may buffer fish and shellfish against stressors such as ocean acidification, salinity fluctuations, and rising water temperatures. Designing microbiomes with adaptive traits could thus strengthen the robustness of aquaculture systems under global climate variability.
The development of natural bacteria-derived therapeutics for various human diseases represents a burgeoning field with potential future applications in aquaculture. Further advancements in gut microbiota engineering techniques, coupled with artificial intelligence (AI) and synthetic biology, can drive substantial progress in sustainable aquaculture (Kumar et al., 2022). The establishment of comprehensive protocols and rigorous training programs encompassing advanced strategies and addressing safety considerations is paramount for the successful translation of microbiota engineering to real world applications. Given its capacity to bolster aquaculture sustainability through enhanced productivity, disease resilience, and ecological equilibrium, microbiota engineering emerges as a resilient and forward-looking strategy for advancing sustainable aquaculture practices.
Author contributions
NR: Conceptualization, Data curation, Methodology, Visualization, Writing – original draft, Writing – review & editing. AK: Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. JJ: Conceptualization, Validation, Writing – original draft, Writing – review & editing. AP: Investigation, Writing – original draft, Writing – review & editing. SD: Conceptualization, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. F-HN: Investigation, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
The authors are thankful to Shoolini University of Biotechnology and Management Sciences, Solan, Himachal Pradesh for helping in conducting this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeshina, I., Abubakar, M. I., and Ajala, B. E. (2020). Dietary supplementation with Lactobacillus acidophilus enhanced the growth, gut morphometry, antioxidant capacity, and the immune response in juveniles of the common carp, Cyprinus carpio. Fish Physiol. Biochem. 46, 1375–1385. doi: 10.1007/s10695-020-00796-7
Amoah, K., Huang, Q., Tan, B., Zhang, S., Chi, S., Yang, Q., et al. (2019). Dietary supplementation of probiotic Bacillus coagulans ATCC 7050 improves the growth performance, intestinal morphology, microflora, immune response and disease confrontation of Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 87, 796–808. doi: 10.1016/j.fsi.2019.02.029
Angthong, P., Uengwetwanit, T., Arayamethakorn, S., Chaitongsakul, P., Karoonuthaisiri, N., and Rungrassamee, W. (2020). Bacterial analysis in the early developmental stages of the black tiger shrimp (Penaeus monodon). Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-61559-1
Arias-Jayo, N., Abecia, L., Lavín, J. L., Itziar, T., Arranz, S., Ramírez, A., et al. (2019). Host-microbiota interactions in response to a high-saturated fat diet and fish-oil supplementation in zebrafish adult. J. Funct. Foods 60, 103416. doi: 10.1016/j.jff.2019.103416
Austin, B. (2002). The bacterial microflora of fish. Sci. World J. 2, 558–572. doi: 10.1100/tsw.2002.137
Avella, M. A., Place, A., Du, S.-J., Williams, E., Silvi, S., Zohar, Y., et al. (2012). Lactobacillus rhamnosus accelerates zebrafish backbone calcification and gonadal differentiation through effects on the GnRH and IGF systems. 7 (9), e45572 doi: 10.1371/journal.pone.0045572
Bao, S., Zhuo, L., Qi, D., Tian, H., Wang, D., Zhu, B., et al (2023). Comparative study on the fillet nutritional quality of diploid and triploid rainbow trout (Oncorhynchus mykiss). Aquac. Rep. 28, 101431. doi: 10.1016/j.aqrep.2022.101431
Banerjee, G. and Ray, A. K. (2017). Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 72, 1–11. doi: 10.1007/s13199-016-0441-8
Betiku, O. C., Yeoman, C. J., Gaylord, T. G., Ishaq, S. L., Duff, G. C., Sealey, W. M., et al. (2023). Evidence of a divided nutritive function in rainbow trout (Oncorhynchus mykiss) midgut and hindgut microbiotas by whole shotgun metagenomic approach. Aquac. Rep. 30, 101601. doi: 10.1016/j.aqrep.2023.101601
Bhatti, A. A., Haq, S., and Bhat, R. A. (2017). Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 111, 458–467. doi: 10.1016/j.micpath.2017.09.036
Bledsoe, J. W., Waldbieser, G. C., Swanson, K. S., Peterson, B. C., and Small, B. C. (2018). Comparison of channel catfish and blue catfish gut microbiota assemblages shows minimal effects of host genetics on microbial structure and inferred function. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01073
Boettcher, M. I., Schettgen, T., Kütting, B., Pischetsrieder, M., and Angerer, J. (2005). Mercapturic acids of acrylamide and glycidamide as biomarkers of the internal exposure to acrylamide in the general population. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 580, 167–176. doi: 10.1016/j.mrgentox.2004.11.010
Bozzi, D., Rasmussen, J. A., Carøe, C., Sveier, H., Nordøy, K., Gilbert, M. T. P., et al. (2021). Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Anim. Microbiota 3, 30. doi: 10.1186/s42523-021-00096-2
Brar, B., Thakur, K., and Mahajan, D. (2023). Mapping water quality parameters in Baner and Gaj rivulets: insights into the potential impact on river Beas in Himachal Pradesh using ArcGIS. World Water Policy 9, 550–590. doi: 10.1002/wwp2.12122
Brock, G. J. (1998). Foreign direct investment in Russia's regions 1993–95: Why so little and where has it gone? Econ. Transit. 6, 349–360. doi: 10.1111/j.1468-0351.1998.tb00053.x
Brown, R., Wiens, G., and Salinas, I. (2019). Analysis of the gut and gill microbiota of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 86, 497–506. doi: 10.1016/j.fsi.2018.11.079
Brugman, S., Ikeda-Ohtsubo, W., Braber, S., Folkerts, G., Pieterse, C. M. J., and Bakker, P. A. H. M. (2018). A comparative review on microbiota manipulation: lessons from fish, plants, livestock, and human research. Front. Nutr. 5. doi: 10.3389/fnut.2018.00080
Butt, R. L. and Volkoff, H. (2019). Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00009
Cardona, E., Gueguen, Y., Magré, K., Lorgeoux, B., Piquemal, D., Pierrat, F., et al. (2016). Bacterial community characterization of water and intestine of the shrimp Litopenaeus stylirostris in a biofloc system. BMC Microbiol. 16, 157. doi: 10.1186/s12866-016-0770-z
Carnevali, O., Avella, M. A., and Gioacchini, G. (2013). Effects of probiotic administration on zebrafish development and reproduction. Gen. Comp. Endocrinol. 188, 297–302. doi: 10.1016/j.ygcen.2013.04.020
Ceccaldi, H. J. (1997). Anatomy and physiology of the digestive system of crustacean. In: Crustacean Nutrition. L. D’Abramo and D. Conklin (eds.). Adv. World Aquacult. 6, 261–291. doi: 10.1016/j.aquaculture.2004.05.032
Chaiyapechara, S., Rungrassamee, W., Suriyachay, I., Kuncharin, Y., Klanchui, A., Karoonuthaisiri, N., et al. (2012). Bacterial community associated with the intestinal tract of Penaeus monodon in commercial farms. Microb. Ecol. 63, 938–953. doi: 10.1007/s00248-011-9936-2
Chaiyapechara, S., Uengwetwanit, T., Arayamethakorn, S., Bunphimpapha, P., and Rungrassamee, W. (2021). Understanding the host-microbe-environment interactions: intestinal microbiota and transcriptomes of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture 546, 737371. doi: 10.1016/j.aquaculture.2021.737371
Cheesman, S. E., Neal, J. T., Mittge, E., Seredick, B. M., and Guillemin, K. (2011). Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci. U.S.A. 108, 4570–4577. doi: 10.1073/pnas.1000072107
Chen, Y., Chi, S., Zhang, S., Dong, X., Yang, Q., Liu, H., et al. (2021). Replacement of fish meal with methanotroph (Methylococcus capsulatus, Bath) bacteria meal in the diets of Pacific white shrimp (Litopenaeus vannamei). Aquaculture 541, 736801. doi: 10.1016/j.aquaculture.2021.736801
Chen, C.-Z., Li, P., Liu, L., and Li, Z.-H. (2022). Exploring the interactions between the gut microbiota and the shifting surrounding aquatic environment in fisheries and aquaculture: a review. Environ. Res. 214, 114202. doi: 10.1016/j.envres.2022.114202
Cottrell, M. T., Waidner, L. A., and Yu, L. (2005). Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ. Microbiol. 7, 1883–1895. doi: 10.1111/j.1462-2920.2005.00882.x
Cui, X., Zhang, Q., Zhang, Q., Zhang, Y., Chen, H., Liu, G., et al. (2022). Research progress of the gut microbiota in hybrid fish. Microorganisms 10, 891. doi: 10.3390/microorganisms10050891
Dai, W.-F., Zhang, J.-J., Qiu, Q.-F., Chen, J., Yang, W., Ni, S., et al. (2018). Starvation stress affects the interplay among shrimp gut microbiota, digestion, and immune activities. Fish Shellfish Immunol. 80, 191–199. doi: 10.1016/j.fsi.2018.05.040
Dai, W., Sheng, Z., Chen, J., and Xiong, J. (2020). Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture 517, 734802. doi: 10.1016/j.aquaculture.2019.734802
Deb, S., Das, L., and Das, S. K. (2020). Composition and functional characterization of the gut microbiome of freshwater pufferfish (Tetraodon cutcutia). Arch. Microbiol. 202, 2761–2770. doi: 10.1007/s00203-020-01997-7
Dehaumont, P. (2004). OIE international standards on antimicrobial resistance. Journal of Veterinary Medicine, Series B, 51 (8-9), 411–414. doi: 10.1111/j.1439-0450.2004.00784.x
De La Torre Canny, S. G., Mueller, O., Craciunescu, C. V., Blumberg, B., and Rawls, J. F. (2021). Tributyltin exposure leads to increased adiposity and reduced abundance of leptogenic bacteria in the zebrafish intestine. 2021–07. doi: 10.1101/2021.07.09.451869
Diwan, A., Harke, S. N., and Panche, A. (2023). “Impact of climate change on the gut microbiota of fish and shellfish,” in Microbiota of Finfish and Shellfish. Ed. Diwan, A., et al (Springer Nature Singapore, Singapore), 255–294. doi: 10.1007/978-981-99-3217-9_12
Diwan, A., Harke, S. N., and Panche, A (2023a). Host-microbiota interaction in fish and shellfish: an overview. Fish Shellfish Immunol. Rep. 4, 100091. doi: 10.1016/j.fsirep.2023.100091
Diwan, A. D., Harke, S. N., Panche, G., and Archana, N. (2021). Aquaculture industry perspective from gut microbiota of fish and shellfish: an overview. J. Anim. Physiol. Anim. Nutr. 106, 441–469. doi: 10.1111/jpn.13619
Duan, Y., Liu, Q., Wang, Y., Zhang, J., and Xiong, D. (2018). Impairment of the intestine barrier function in Litopenaeus vannamei exposed to ammonia and nitrite stress. Fish Shellfish Immunol. 78, 279–288. doi: 10.1016/j.fsi.2017.12.016
Duan, Y., Wang, Y., Liu, Q., Zhang, J., and Xiong, D. (2019b). Changes in the intestine barrier function of Litopenaeus vannamei in response to pH stress. Fish Shellfish Immunol. 88, 142–149. doi: 10.1016/j.fsi.2018.09.028
Duan, Y., Wang, Y., Liu, Q., Xiong, D., and Zhang, J. (2019a). Transcriptomic and microbiota response on Litopenaeus vannamei intestine subjected to acute sulphide exposure. Fish Shellfish Immunol. 88, 335–343. doi: 10.1016/j.fsi.2018.10.027
Dulski, T., Kozłowski, K., and Ciesielski, S. (2020). Habitat and seasonality shape the structure of tench (Tinca L.) gut microbiota. Sci. Rep. 10, 4460. doi: 10.1038/s41598-020-61395-3
Dvergedal, H., Sandve, S. R., Angell, I. L., Klemetsdal, G., and Rudi, K. (2020). Association of gut microbiota with metabolism in juvenile Atlantic salmon. Microbiota 8, 160. doi: 10.1186/s40168-020-00913-4
Fan, L. and Li, Q. (2019). Characteristics of intestinal microbiota in the Pacific white shrimp Litopenaeus vannamei differing growth performances in the marine cultured environment. Rev. Aquacult. 13, 505–520. doi: 10.1016/j.aquaculture.2019.02.075
Fan, L., Wang, Z., Chen, M., Qu, Y., Li, J., Zhou, A., et al. (2019). Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci. Total Environ. 657, 1194–1204. doi: 10.1016/j.scitotenv.2018.12.139
Faury, N., Saulnier, D., Thompson, F. L., Gay, M., Swings, J., Le Roux, F., et al. (2004). Vibrio crassostreae sp. nov., isolated from the haemolymph of oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 54, 2137–2140. doi: 10.1099/ijs.0.63232-0
Filardo, S., Di Pietro, M., and Sessa, R. (2024). Current progresses and challenges for microbiota research in human health: a perspective. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1377012
Flegel, T. W. (1997). Major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand. World J. Microbiol. Biotechnol. 13, 433–442. doi: 10.1023/A:1018580301578
Flegel, T. W., Lightner, D. V., Lo, C. F., and Owens, L. (2008). Shrimp disease control: past, present and future. In: Diseases in Asian Aquaculture VI. Fish Health Section, Asian Fisheries Society, Manila, Philippines. 355–378.
Flegel, T. W. and Pasharawipas, T. (1998). Advances in shrimp biotechnology. In: Proceedings of the National Centre for Genetic Engineering and Biotechnology, Thailand, 245–250.
Fonseca, F., Cerqueira, R., and Fuentes, J. (2019). Impact of ocean acidification on the intestinal microbiota of the marine sea bream (Sparus aurata L.). Front. Physiol. 10. doi: 10.3389/fphys.2019.01446
Gajardo, K., Rodiles, A., Kortner, T. M., Krogdahl, Å., and Bakke, A. M. (2016). A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci. Rep. 6, 30893. doi: 10.1038/srep30893
Garibay-Valdez, E., Cicala, F., Martinez-Porchas, M., Gómez-Reyes, R., Vargas-Albores, F., Gollas-Galván, T., et al. (2021). Longitudinal variations in the gastrointestinal microbiota of the white shrimp, Litopenaeus vannamei. PeerJ 9, e11827. doi: 10.7717/peerj.11827
Geerlings, S. Y., Ouwerkerk, J. P., Koehorst, J. J., Ritari, J., Aalvink, S., Stecher, B., et al. (2021). Genomic convergence between Akkermansia muciniphila in different mammalian hosts. BMC Microbiol. 21, 1–13. doi: 10.1186/s12866-021-02190-3
Ghosh, S. K., Wong, M. K. S., Hyodo, S., Goto, S., and Hamasaki, K. (2022). Temperature modulation alters the gut and skin microbial profiles of chum salmon (Oncorhynchus keta). Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1027621
Gómez, G. D. and Balcázar, J. L. (2008). A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52, 145–154. doi: 10.1111/j.1574-695X.2007.00343.x
Gunalan, B., Soundarapandian, P., Anand, T., Kotiya, A. S., and Simon, N. T. (2014). Disease occurrence in Litopenaeus vannamei shrimp culture systems in different geographical regions of India. Int. J. Aquac. 4, 1–6. doi: 10.5376/ija.2014.04.0004
Guo, H., Huang, L., Hu, S., Chen, C., and Zhang, D. (2020). Effects of carbon/nitrogen ratio on growth, intestinal microbiota and metabolome of shrimp (Litopenaeus vannamei). Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00652
Guo, X., Ran, C., Zhang, Z., He, S., Jin, M., and Zhou, Z. (2017). The growth-promoting effect of dietary nucleotides in fish is associated with an intestinal microbiota-mediated reduction in energy expenditure. J. Nutr. 147, 781–788. doi: 10.3945/jn.116.245506
Gyan, R. W., Yang, Q., Tan, B., Dong, X., Chi, S., Liu, H., et al. (2022). Effects of replacing fish meal with distillers’ dried grains with solubles on the growth performance and gut microbiota in juvenile Pacific whiteleg shrimp Litopenaeus vannamei. N. Am. J. Aquacult. 84, 191–205. doi: 10.1002/naaq.10236
Hao, Y. T., Wu, S. G., Jakovlić, I., Zou, H., Li, W. X., and Wang, G. T. (2017). Impacts of diet on hindgut microbiota and short-chain fatty acids in grass carp (Ctenopharyngodon idella). Aquacult. Res. 48, 5595–5605. doi: 10.1111/are.13381
Hassenrück, C., Reinwald, H., Kunzmann, A., Tiedemann, I., and Gärdes, A. (2020). Effects of thermal stress on the gut microbiota of juvenile milkfish (Chanos chanos). Microorganisms 9, 5. doi: 10.3390/microorganisms9010005
Hernández-Zárate, G. and Olmos-Soto, J. (2006). Identification of bacterial diversity in the oyster Crassostrea gigas by fluorescent in situ hybridization and polymerase chain reaction. J. Appl. Microbiol. 100, 664–672. doi: 10.1111/j.1365-2672.2005.02800.x
Hernández‐Zárate, G. and Olmos‐Soto, J. (2021). Dietary filamentous fungi and duration of feeding modulate gut microbial composition in rainbow trout (Oncorhynchus mykiss). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.728569
Holt, C. C., David, B., Stentiford, G. D., and Giezen, M. (2020). Understanding the role of the shrimp gut microbiota in health and disease. J. Invertebr. Pathol. 4, 1–16. doi: 10.1016/j.jip.2020.107387
Hou, D., Huang, Z., Zeng, S., Liu, J., Wei, D., Deng, X., et al. (2018). Intestinal bacterial signatures of white feces syndrome in shrimp. Appl. Microbiol. Biotechnol. 102, 3701–3709. doi: 10.1007/s00253-018-8855-2
Hovda, M. B., Fontanillas, R., McGurk, C., Obach, A., and Rosnes, J. T. (2012). Seasonal variations in the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquacult. Res. 43, 154–159. doi: 10.1111/j.1365-2109.2011.02805.x
Huang, Z., Hou, D., Zhou, R., Zeng, S., Xing, C., Wei, D., et al. (2021). Environmental water and sediment microbial communities shape intestine microbiota for host health: the central dogma in an anthropogenic aquaculture ecosystem. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.772149
Huang, Z., Li, X., Wang, L., and Shao, Z. (2016). Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquacult. Res. 47, 1737–1746. doi: 10.1111/are.12628
Huang, F., Pan, L., Song, M., Tian, C., and Gao, S. (2018). Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl. Microbiol. Biotechnol. 102, 8585–8598. doi: 10.1007/s00253-018-9229-5
Huang, Q., Sham, R. C., Deng, Y., Mao, Y., Wang, C., Zhang, T., et al. (2020). Diversity of gut microbiotas in marine fishes is shaped by host-related factors. Mol. Ecol. 29, 5019–5034. doi: 10.1111/mec.15699
Huyben, D., Sun, L., Moccia, R., Kiessling, A., Dicksved, J., Lundh, T. J. J. O. A. M., et al. (2018). Dietary live yeast and increased water temperature influence the gut microbiota of rainbow trout. J. Appl. Microbiol. 124, 1377–1392. doi: 10.1111/jam.13738
Imaizumi, K., Tinwongger, S., Kondo, H., and Hirono, I. (2021). Analysis of microbiota in the stomach and midgut of two penaeid shrimps during probiotic feeding. Sci. Rep. 11, 9936. doi: 10.1038/s41598-021-89415-w
Izvekova, G. I., Izvekov, E. I., and Plotnikov, A. O. (2007). Symbiotic microflora in fishes of different ecological groups. Izv. Akad. Nauk Ser. Biol. 34, 610–618. doi: 10.1134/S106235900706012X
Johnson, C. N., Flowers, A. R., Noriea, N. F. I. I. I., Zimmerman, A. M., Bowers, J. C., DePaola, A., et al. (2010). Relationships between environmental factors and pathogenic vibrios in the northern Gulf of Mexico. Appl. Environ. Microbiol. 76, 7076–7084. doi: 10.1128/AEM.00697-10
Johnson, W. P., Blue, K. A., Logan, B. E., and Arnold, R. G. (1995). Modeling bacterial detachment during transport through porous media as a residence-time-dependent process. Water Resour. Res. 31, 2649–2658. doi: 10.1029/95WR02311
Jordaan, K. and Bezuidenhout, C. C. (2013). The impact of physico-chemical water quality parameters on bacterial diversity in the Vaal River, South Africa. Water SA 39, 385–396. doi: 10.4314/wsa.v39i3.7
Karunasagar, I., Pai, R., Malathi, G. R., and Karunasagar, I. (1994). Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection. Aquaculture 128, 203–209. doi: 10.1016/0044-8486(94)90309-3
Karunasagar, I., Shivu, M. M., Girisha, S. K., Krohne, G., and Karunasagar, I. (2007). Biocontrol of pathogens in shrimp hatcheries using bacteriophages. Aquaculture 268, 288–292. doi: 10.1016/j.aquaculture.2007.04.049
Khurana, H., Singh, D. N., Singh, A., Singh, Y., Lal, R., and Negi, R. K. (2020). Gut microbiota of endangered Tor putitora (Ham.) as a reservoir of antibiotic resistance genes and pathogens associated with fish health. BMC Microbiol. 20, 249. doi: 10.1186/s12866-020-01911-7
Kim, P. S., Shin, N. R., Lee, J. B., Kim, M. S., Whon, T. W., Hyun, D. W., et al. (2021). Host habitat is the major determinant of the gut microbiota of fish. Microbiota 9, 166. doi: 10.1186/s40168-021-01113-x
King, G. M., Judd, C., Kuske, C. R., and Smith, C. (2012). Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS One 7, e51475. doi: 10.1371/journal.pone.0051475
Kong, N., Han, S., Fu, Q., Yu, Z., Wang, L., and Song, L. (2022). Impact of ocean acidification on the intestinal microflora of the Pacific oyster Crassostrea gigas. Aquaculture 546, 737365. doi: 10.1016/j.aquaculture.2021.737365
Kumar, P., Sinha, R., and Shukla, P. (2022). Artificial intelligence and synthetic biology approaches for human gut microbiota. Crit. Rev. Food Sci. Nutr. 62, 2103–2121. doi: 10.1080/10408398.2020.1850415
Li, P., Roos, S., Luo, H., Ji, B., and Nielsen, J. (2023). Metabolic engineering in human gut microbiota: recent developments and future perspectives. Metab. Eng. 79, 1–3. doi: 10.1016/j.ymben.2023.06.006
Li, H., Wu, S., Wirth, S., Hao, Y., Wang, W., Zou, H., et al. (2016). Diversity and activity of cellulolytic bacteria isolated from the gut contents of grass carp (Ctenopharyngodon idellus) fed on Sudan grass (Sorghum Sudanense) or artificial feedstuffs. Aquacult. Res. 47, 153–164. doi: 10.1111/are.12478
Li, L., Wang, P., Zhao, C., and Qiu, L. (2018). The anti-stresses capability of GRP78 in Penaeus monodon: evidence from in vitro and in vivo studies. Fish Shellfish Immunol. 72, 132–142. doi: 10.1016/j.fsi.2017.10.052
Li, W., Zhou, Z., Li, H., Wang, S., Ren, L., Hu, J., et al. (2022). Successional changes of microbial communities and host–microbiota interactions contribute to dietary adaptation in allodiploid hybrid fish. Microb. Ecol. 85(4), 1190–1201. doi: 10.1007/s00248-022-01993-y
Li, X., Zhou, L., Yu, Y., Ni, J., Xu, W., Yan, Q., et al. (2017). Composition of gut microbiota in the gibel carp (Carassius auratus gibelio) varies with host development. Microb. Ecol. 74, 239–249. doi: 10.1007/s00248-016-0924-4
Liang, Q., Li, Z., Ou, M., Wu, X., Qiao, X., Wei, W., et al. (2020). Hypoimmunity and intestinal bacterial imbalance are closely associated with blue body syndrome in cultured Penaeus vannamei. Aquaculture 522, 735118. doi: 10.1016/j.aquaculture.2020.735118
Liang, X., Luo, X., Lin, H., Han, F., Qin, J. G., Chen, L., et al. (2022). Growth, health, and gut microbiota of female Pacific white shrimp, Litopenaeus vannamei broodstock fed different phospholipid sources. Antioxidants 11, 1143. doi: 10.3390/antiox11061143
Lightner, D. V. (1996). Epizootiology, distribution and the impact on international trade of two penaeid shrimp viruses in the Americas. Rev. Sci. Tech. Off. Int. Epiz. 15, 579–601. doi: 10.20506/rst.15.2.944
Liu, D., Zhang, Z., Song, Y., Yang, J., Lu, Y., Lai, W., et al. (2023). Effects of salinity on growth, physiology, biochemistry and gut microbiota of juvenile grass carp (Ctenopharyngodon idella). Aquat. Toxicol. 258, 106482. doi: 10.1016/j.aquatox.2023.106482
Liu, H., Guo, X., Gooneratne, R., Lai, R., Zeng, C., Zhan, F., et al. (2016). The gut microbiota and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci. Rep. 6, 24340. doi: 10.1038/srep24340
Liu, L., Xiao, J., Zhang, M., Zhu, W., Xia, X., Dai, X., et al. (2018). A Vibrio owensii strain as the causative agent of AHPND in cultured shrimp, Litopenaeus vannamei. J. Invertebr. Pathol. 153, 156–164. doi: 10.1016/j.jip.2018.02.005
Liu, R., Han, G., Li, Z., Cun, S., Hao, B., Zhang, J., et al. (2022b). Bacteriophage therapy in aquaculture: status and future challenges. Folia Microbiol. 67, 573–590. doi: 10.1007/s12223-022-00965-6
Liu, Y., Feng, J., Pan, H., Zhang, X., and Zhang, Y. (2022a). Genetically engineered bacterium: principles, practices, and prospects. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.997587
Llewellyn, M. S., Boutin, S., Hoseinifar, S. H., and Derome, N. (2014). Teleost microbiotas: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00207
Ma, T., Suzuki, Y., and Guan, L. L. (2018). Dissect the mode of action of probiotics in affecting host-microbial interactions and immunity in food producing animals. Vet. Immunol. Immunopathol. 205, 35–48. doi: 10.1016/j.vetimm.2018.10.004
Maji, U. J., Mohanty, S., Mahapatra, A. S., Mondal, H. K., Samanta, M., and Maiti, N. K. (2022). Exploring the gut microbiota composition of Indian major carp, rohu (Labeo rohita), under diverse culture conditions. Genomics 114, 110354. doi: 10.1016/j.ygeno.2022.110354
Mayasich, S. A. and Smucker, R. A. (1987). Role of Cristispira sp. and other bacteria in the chitinase and chitobiase activities of the crystalline style of Crassostrea virginica (Gmelin). Microb. Ecol. 14, 157–166. doi: 10.1007/BF02013020
Mekuchi, M., Asakura, T., Sakata, K., Yamaguchi, T., Teruya, K., and Kikuchi, J. (2018). Intestinal microbiota composition is altered according to nutritional biorhythms in the leopard coral grouper (Plectropomus leopardus). PloS One 13, e0197256. doi: 10.1371/journal.pone.0197256
Meng, X.-L., Li, S., Qin, C.-B., Zhu, Z.-X., Hu, W.-P., Yang, L.-P., et al. (2018). Intestinal microbiota and lipid metabolism responses in the common carp (Cyprinus carpio L.) following copper exposure. Ecotoxicol. Environ. Saf. 160, 257–264. doi: 10.1016/j.ecoenv.2018.05.050
Merrifield, D. L. and Rodiles, A. (2015). The fish microbiota and its interactions with mucosal tissues. Mucosal Health Aquacult., 273–295. doi: 10.1016/b978-0-12-417186-2.00010-8
Miyake, S., Ngugi, D. K., and Stingl, U. (2016). Phylogenetic diversity, distribution, and cophylogeny of giant bacteria (Epulopiscium) with their surgeonfish hosts in the Red Sea. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00285
Mohanta, L., Das, B. C., and Patri, M. (2020). Microbial communities modulating brain functioning and behaviors in zebrafish: a mechanistic approach. Microb. Pathog. 145, 104251. doi: 10.1016/j.micpath.2020.104251
Mondal, S., Roy, T., Sen, S. K., and Ray, A. K. (2008). Distribution of enzyme-producing bacteria in the digestive tracts of some freshwater fish. Acta Ichthyol. Piscat. 38, 1–8. doi: 10.3750/AIP2008.38.1.01
Moore, E. R., Tindall, B. J., Martins Dos Santos, V. A., Pieper, D. H., Ramos, J. L., Palleroni, N. J., et al. (2006). “Nonmedical: pseudomonas,” in The Prokaryotes: A Handbook on the Biology of Bacteria, vol. 6 . Eds. Dworkin, M., Falkow, S., Rosenberg, E., et alProteobacteria: Gamma Subclass (Springer, New York, NY), 646–703. doi: 10.1007/0-387-30746-x_21
Morales-Rivera, M. F., Valenzuela-Miranda, D., Nuñez-Acuña, G., Benavente, B. P., Gallardo-Escárate, C., Valenzuela-Muñoz, V., et al. (2022). Atlantic salmon (Salmo salar) transfer to seawater by gradual salinity changes exhibited an increase in the intestinal microbial abundance and richness. Microorganisms 11, 76. doi: 10.3390/microorganisms11010076
Morshed, S. M. and Lee, T.-H. (2023). The role of the microbiota on fish mucosal immunity under changing environments. Fish Shellfish Immunol. 139, 108877. doi: 10.1016/j.fsi.2023.108877
Mushegian, A. A., Arbore, R., Walser, J. C., and Ebert, D. (2019). Environmental sources of bacteria and genetic variation in behavior influence host-associated microbiota. Appl. Environ. Microbiol. 85, e01547–e01518. doi: 10.1128/AEM.01547-18
Nath, A., Bhattacharjee, R., Nandi, A., Sinha, A., Kar, S., Manoharan, N., et al. (2022). Phage-delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiota. Biomed. Pharmacother. 151, 113122. doi: 10.1016/j.biopha.2022.113122
Naya-Català, F., Piazzon, M. C., Torrecillas, S., Toxqui-Rodríguez, S., Calduch-Giner, J.À., Fontanillas, R., et al. (2022). Genetics and nutrition drive the gut microbiota succession and host-transcriptome interactions through the gilthead sea bream (Sparus aurata) production cycle. Biology 11, 1744. doi: 10.3390/biology11121744
Nayak, S. K. (2010). Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x
Ngugi, D. K., Miyake, S., Cahill, M., Vinu, M., Hackmann, T. J., Blom, J., et al. (2017). Genomic diversification of giant enteric symbionts reflects host dietary lifestyles. Proc. Natl. Acad. Sci. U.S.A. 114, E7592–E7601. doi: 10.1073/pnas.1703070114
Nguyen, T. L., Chun, W.-K., Kim, A., Kim, N., Roh, H. J., Lee, Y., et al. (2018). Dietary probiotic effect of Lactococcus lactis WFLU12 on low-molecular-weight metabolites and growth of olive flounder (Paralichthys olivaceus). Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02059
Nikouli, E., Meziti, A., Smeti, E., Antonopoulou, E., and Mente Kormas, E. K. A. (2021). Gut microbiota of five sympatrically farmed marine fish species in the Aegean Sea. Microb. Ecol. 81, 460–470. doi: 10.1007/s00248-020-01580-z
Noman, M., Kazmi, S. S. U. H., Saqib, H. S. A., Fiaz, U., Pastorino, P., Barcelò, D., et al. (2024). Harnessing probiotics and prebiotics as eco-friendly solution for cleaner shrimp aquaculture production: a state of the art scientific consensus. Sci. Total Environ. 915, 169921. doi: 10.1016/j.scitotenv.2024.169921
Oetama, V. S., Hennersdorf, P., Abdul-Aziz, M. A., Mrotzek, G., Haryanti, H., and Saluz, H. P. (2016). Microbiome analysis and detection of pathogenic bacteria of Penaeus monodon from Jakarta Bay and Bali. Mar. Pollut. Bull. 110, 718–725. doi: doi: 10.1016/j.marpolbul.2016.03.043
Otta, S., Karunasagar, I., and Karunasagar, I. (2001). Bacteriological study of shrimp, Penaeus monodon Fabricius, hatcheries in India. J. Appl. Ichthyol. 17, 59–63. doi: 10.1046/j.1439-0426.2001.00249.x
Otta, S. K., Karunasagar, I., and Karunasagar, I. (1999). Bacterial flora associated with shrimp culture ponds growing Penaeus monodon in India. Aquaculture 179, 1–13.
Ou, W., Yu, G., Zhang, Y., and Mai, K. (2021). Recent progress in the understanding of the gut microbiota of marine fishes. Mar. Life Sci. Technol. 3, 434–448. doi: 10.1007/s42995-021-00094-y
Parata, L., Nielsen, S., Xing, X., Thomas, T., Egan, S., and Vergés, A. (2020). Age, gut location and diet impact the gut microbiota of a tropical herbivorous surgeonfish. FEMS Microbiol. Ecol. 96, fz179. doi: 10.1093/femsec/fiz179
Pardesi, B., Roberton, A. M., Lee, K. C., Angert, E. R., Rosendale, D. I., Boycheva, S., et al. (2022). Distinct microbiota composition and fermentation products indicate functional compartmentalization in the hindgut of a marine herbivorous fish. Mol. Ecol. 31, 2494–2509. doi: 10.1111/mec.16394
Pérez, T., Balcázar, J. L., Ruiz-Zarzuela, I., Halaihel, N., Vendrell, D., de Blas, I., et al. (2010). Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 3, 355–360. doi: 10.1038/mi.2010.12
Perez-Bustamante, I. S., Cruz-Flores, R., López-Carvallo, J. A., and Sánchez-Serrano, S. (2024). Effect of the 16S rRNA gene hypervariable region on the microbiota taxonomic profile and diversity in the endangered fish Totoaba macdonaldi. Microorganisms 12, 2119. doi: 10.3390/microorganisms12112119
Petit, J., de Bruijn, I., Goldman, M. R. G., van den Brink, E., Pellikaan, W. F., Forlenza, M., et al. (2022). β-glucan-induced immuno-modulation: a role for the intestinal microbiota and short-chain fatty acids in common carp. Front. Immunol. 12, 761820. doi: 10.3389/fimmu.2021.761820
Podell, S., Oliver, A., Kelly, L., et al. (2023). Herbivorous fish microbiota adaptations to sulfated dietary polysaccharides. Appl. Environ. Microbiol. 89, e0215422. doi: 10.1128/aem.02154-22
Qiao, F., Liu, Y. K., Sun, Y. H., Wang, X. D., Chen, K., Li, Y. T., et al. (2017). Influence of different dietary carbohydrate sources on the growth and intestinal microbiota of Litopenaeus vannamei at low salinity. Aquacult. Nutr. 23, 444–452. doi: 10.1111/anu.12412
Rajeev, R., Adithya, K. K., Kiran, G. S., and Selvin, J. (2021). Healthy microbiome: A key to successful and sustainable shrimp aquaculture. Rev. Aquac. 13, 238–258. doi: 10.1111/raq.12471
Ray, A. K., Ghosh, K., and Ringø, E. (2012). Enzyme-producing bacteria isolated from fish gut: a review. Aquacult. Nutr. 18, 465–492. doi: 10.1111/j.1365-2095.2012.00943.x
Ray, A. R., Kumar, L., Swain, H. S., and Das, B. K. (2022). Growth, mortality and stock status of three commercially important catfishes from the River Ganga, India. Indian J. Fish. 69, 30–38. doi: 10.21077/ijf.2022.69.2.115676-04
Romero, J., García-Varela, M., Laclette, J. P., and Espejo, R. T. (2002). Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis). Microb. Ecol. 44, 365–371. doi: 10.1007/s00248-002-1063-7
Romero, J., Ringø, E., and Merrifield, D. L. (2014). “The gut microbiota of fish,” in Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics. Eds. Merrifield, D. L. and Ringø, E. (Wiley, London), 75–100. doi: 10.1002/9781118897263.ch4
Rungrassamee, W., Klanchui, A., Chaiyapechara, S., Maibunkaew, S., Tangphatsornruang, S., Jiravanichpaisal, P., et al. (2013). Bacterial population in intestines of the black tiger shrimp (Penaeus monodon) under different growth stages. PloS One 8, e60802. doi: 10.1371/journal.pone.0060802
Saha, S., Roy, R. N., Sen, S. K., et al. (2006). Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac. Res. 37, 380–388. doi: 10.1111/j.1365-2109.2006.01442.x
Savio, D., Sinclair, L., Ijaz, U. Z., et al. (2015). Bacterial diversity along a 2600 km river continuum. Environ. Microbiol. 17, 4994–5007. doi: 10.1111/1462-2920.12886
Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Sepúlveda-Quiroz, C. A., Peña-Marín, E. S., Pérez-Morales, A., Martínez-García, R., Alvarez-Villagomez, C. S., Maytorena-Verdugo, C. I., et al. (2021). Fructooligosaccharide supplementation in diets for tropical gar (Atractosteus tropicus) juvenile: effects on morphophysiology and intestinal barrier function. Aquac. Res. 52, 37–50. doi: 10.1111/are.14867
Shang, X., Geng, L., Wei, H., Che, X., Xing, L., Xing, M., et al. (2024). Selenium-enriched Lactobacillus plantarum alleviates high alkalinity-induced microbiota-gut-blood system effects by improving the gut microbiota. Aquaculture 593, 741294. doi: 10.1016/j.aquaculture.2024.741294
Sharma, P. V. and Thaiss, C. A. (2020). Host-microbiota interactions in the era of single-cell biology. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.569070
Sheng, Y., Ren, H., Limbu, S. M., Sun, Y., Qiao, F., Zhai, W., et al. (2018). The presence or absence of intestinal microbiota affects lipid deposition and related gene expression in zebrafish (Danio rerio). Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01124
Sheth, R. U., Cabral, V., Chen, S. P., and Wang, H. H. (2016). Manipulating bacterial communities by in situ microbiota engineering. Trends Genet. 32, 189–200. doi: 10.1016/j.tig.2016.01.005
Singh, A., Karimi, S., Vidakovic, A., et al. (2021). Dietary filamentous fungi and duration of feeding modulates gut microbial composition in rainbow trout (Oncorhynchus mykiss). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.728569
Singh, K. M., Shah, T. M., Reddy, B., Deshpande, S., Rank, D. N., and Joshi, C. G. (2014). Taxonomic and gene-centric metagenomics of the fecal microbiota of low and high feed conversion ratio (FCR) broilers. J. Appl. Genet. 55, 145–154. doi: 10.1007/s13353-013-0179-4
Smith, C. C., Snowberg, L. K., Caporaso, J. G., Knight, R., and Bolnick, D. I. (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9, 2515–2526. doi: 10.1038/ismej.2015.64
Sobrinho, P. D. S. C., Destro, M. T., Franco, B. D., and Landgraf, M. (2010). Correlation between environmental factors and prevalence of Vibrio parahaemolyticus in oysters harvested in the southern coastal area of Sao Paulo State, Brazil. Appl. Environ. Microbiol. 76, 1290–1293. doi: 10.1128/aem.00861-09
Song, M., Pan, L., Zhang, M., Huang, F., Gao, S., and Tian, C. (2020). Changes of water, sediment, and intestinal bacterial communities in Penaeus japonicus cultivation and their impacts on shrimp physiological health. Aquac. Int. 28, 1847–1865. doi: 10.1007/s10499-020-00562-9
Spilsbury, F., Foysal, M. J., Tay, A., and Gagnon, M. M. (2022). Gut microbiota as a potential biomarker in fish: dietary exposure to petroleum hydrocarbons and metals, metabolic functions and cytokine expression in juvenile Lates calcarifer. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.827371
Steiner, K., Laroche, O., Walker, S. P., and Symonds, J. E. (2022). Effects of water temperature on the gut microbiota and physiology of Chinook salmon (Oncorhynchus tshawytscha) reared in a freshwater recirculating system. Aquaculture 560, 738529. doi: 10.1016/j.aquaculture.2022.738529
Talwar, C., Nagar, S., Lal, R., and Negi, R. K. (2018). Fish gut microbiota: current approaches and future perspectives. Indian J. Microbiol. 58, 397–414. doi: 10.1007/s12088-018-0760-y
Tarnecki, A. M., Burgos, F. A., Ray, C. L., and Arias, C. R. (2017). Fish intestinal microbiome: diversity and symbiosis unravelled by metagenomics. J. Appl. Microbiol. 123, 2–17. doi: 10.1111/jam.13415
Tsuchiya, C., Sakata, T., and Sugita, H. (2008). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 46, 43–48. doi: 10.1111/j.1472-765X.2007.02258.x
Uengwetwanit, T., Uawisetwathana, U., Arayamethakorn, S., Khudet, J., Chaiyapechara, S., Karoonuthaisiri, N., et al. (2020). Multi-omics analysis to examine microbiota, host gene expression and metabolites in the intestine of black tiger shrimp (Penaeus monodon) with different growth performance. PeerJ 8, e9646. doi: 10.7717/peerj.9646
Ventura, M., Canchaya, C., Tauch, A., et al. (2007). Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/mmbr.00005-07
Villasante, A., Ramírez, C., Catalán, N., Opazo, R., Dantagnan, P., and Romero, J. (2019). Effect of dietary carbohydrate-to-protein ratio on gut microbiota in Atlantic salmon (Salmo salar). Animals 9, 89. doi: 10.3390/ani9030089
Wang, J., Jaramillo-Torres, A., Li, Y., Kortner, T. M., Gajardo, K., Brevik, Ø.J., et al. (2021). Microbiota in intestinal digesta of Atlantic salmon (Salmo salar), observed from late freshwater stage until one year in seawater, and effects of functional ingredients: a case study from a commercial sized research site in the Arctic region. Anim. Microbiome 3, 14. doi: 10.1186/s42523-021-00075-7
Wang, A. R., Ran, C., Ringø, E., et al. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquac. 10, 626–640. doi: 10.1111/raq.12191
Wang, C., Sun, G., Li, S., Li, X., and Liu, Y. (2018). Intestinal microbiota of healthy and unhealthy Atlantic salmon Salmo salar L. in a recirculating aquaculture system. J. Oceanol. Limnol. 36, 414–426. doi: 10.1007/s00343-017-6203-5
Wei, J., Guo, X., Liu, H., Chen, Y., and Wang, W. (2018). The variation profile of intestinal microbiota in blunt snout bream (Megalobrama amblycephala) during feeding habit transition. BMC Microbiol. 18, 1–14. doi: 10.1186/s12866-018-1246-0
Wen, J., Mercado, G. P., Volland, A., Doden, H. L., Lickwar, C. R., Crooks, T., et al. (2021). FXR signaling and microbial metabolism of bile salts in the zebrafish intestine. Sci. Adv. 7, eabg1371. doi: 10.1126/sciadv.abg1371
Wensel, C. R., Pluznick, J. L., Salzberg, S. L., and Sears, C. L. (2022). Next-generation sequencing: insights to advance clinical investigations of the microbiota. J. Clin. Invest. 132, e154944. doi: 10.1172/JCI154944
Wong, S., Waldrop, T., Summerfelt, S., Davidson, J., Barrows, F., Kenney, P. B., et al. (2013). Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density. Appl. Environ. Microbiol. 79, 4974–4984. doi: 10.1128/AEM.00924-13
Wu, S., Wang, G., Angert, E. R., and Ringø, E. (2012). Composition, diversity, and origin of the bacterial community in grass carp intestine. PloS One 7, e30440. doi: 10.1371/journal.pone.0030440
Xia, R., Zhang, Q., Xia, D., Hao, Q., Ding, Q., Ran, C., et al. (2023). The direct and gut microbiota-mediated effects of dietary bile acids on the improvement of gut barriers in largemouth bass (Micropterus salmoides). Anim. Nutr. 14, 32–42. doi: 10.1016/j.aninu.2023.03.008
Xing, M., Hou, Z., Yuan, J., Liu, Y., Qu, B., and Liu, B. (2013). Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiota of the farmed adult turbot (Scophthalmus maximus). FEMS Microbiol. Ecol. 86, 432–443. doi: 10.1111/1574-6941.12171
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Yang, T.-T., Liu, Y., Tan, S., Wang, W.-X., and Wang, X. (2021). The role of intestinal microbiota of the marine fish (Acanthopagrus latus) in mercury biotransformation. Environ. pollut. 277, 116768. doi: 10.1016/j.envpol.2021.116768
Yang, J., Qiang, Z., Zhang, D., Hao, H., Wei, J., Hamid, M. S., et al. (2025). Shotgun metagenomics analysis of gut microbiota of three indigenous fish species from the Kizil River, Xinjiang. Front. Microbiol. 16. doi: 10.3389/fmicb.2025.1617701
Yukgehnaish, K., Kumar, P., Sivachandran, P., Marimuthu, K., Arshad, A., Paray, B. A., et al. (2020). Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 12, 1903–1927. doi: 10.1111/raq.12416
Zhang, B. Y., Cai, G. H., Yang, H. L., Nie, Q. J., Liu, Z. Y., and Sun, Y. Z. (2024). New insights on intestinal microorganisms and carbohydrate metabolism in fish. Aquacult. Int. 32, 2151–2170. doi: 10.1007/s10499-024-01009-5
Zhang, M. L., Li, M., Sheng, Y., Tan, F., Chen, L., Cann, I., et al. (2020). Citrobacter species increase energy harvest by modulating intestinal microbiota in fish: nondominant species play important functions. mSystems 5, e00303–e00320. doi: 10.1128/mSystems.00303-20
Zhang, S. and Sun, X. (2022). Core gut microbiota of shrimp function as a regulator to maintain immune homeostasis in response to WSSV. Infect. Microbiol. Spectr. 27, 10.e0246521. doi: 10.1128/spectrum.02465-21
Zhang, M., Sun, Y., Chen, K., Yu, N., Zhou, Z., Chen, L., et al. (2014). Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture 434, 449–455. doi: 10.1016/j.aquaculture.2014.08.019
Zhang, Y., Wen, B., Meng, L.-J., Gao, J.-Z., and Chen, Z.-Z. (2021). Dynamic changes of gut microbiota of discus fish (Symphysodon haraldi) at different feeding stages. Aquaculture 531, 735912. doi: 10.1016/j.aquaculture.2020.735912
Zhao, R., Symonds, J. E., Walker, S. P., Steiner, K., Carter, C. G., Bowman, J. P., et al. (2020). Salinity and fish age affect the gut microbiota of farmed Chinook salmon (Oncorhynchus tshawytscha). Aquaculture 528, 735539. doi: 10.1016/j.aquaculture.2020.735539
Zheng, Z., Liao, Y., Ye, J., Yang, C., Adzigbli, L., Wang, Q., et al. (2021). Microbiota diversity in pearl oyster Pinctada fucata martensii intestine and its aquaculture environment. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.655698
Zheng, Y., Yu, M., Liu, J., Qiao, Y., Wang, L., Li, Z., et al. (2017). Bacterial community associated with healthy and diseased Pacific white shrimp (Litopenaeus vannamei) larvae and rearing water across different growth stages. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01362
Zhou, L., Chen, C., Xie, J., Xu, C., Zhao, Q., Qin, J. G., et al. (2019). Intestinal bacterial signatures of the “cotton shrimp-like” disease explain the change of growth performance and immune responses in Pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 92, 629–636. doi: 10.1016/j.fsi.2019.06.054
Zhu, X., Zhao, Y., Sun, N., Li, C., Jiang, Q., Zhang, Y., et al. (2023). Comparison of the gut microbiota and untargeted gut tissue metabolome of Chinese mitten crabs (Eriocheir sinensis) with different shell colors. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1218152
Keywords: aquaculture sustainability, environmental factors, gut microbiota, innate immunity, metabolism, microbial diversity, nutrition
Citation: Rai N, Kachore A, Julka JM, Panigrahi A, Das SP and Nan F-H (2025) Symbiotic strategies: deciphering the role of gut microbiota in the nutrition and metabolism of fish and shellfish. Front. Cell. Infect. Microbiol. 15:1639426. doi: 10.3389/fcimb.2025.1639426
Received: 02 June 2025; Accepted: 12 September 2025;
Published: 14 October 2025.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Suvra Roy, Central Inland Fisheries Research Institute (ICAR), IndiaSoumya Prasad Panda, Central Inland Fisheries Research Institute (ICAR), India
Eric Amenyogbe, University of Environment and Sustainable Development, Ghana
Copyright © 2025 Rai, Kachore, Julka, Panigrahi, Das and Nan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan-Hua Nan, ZmhuYW5AbWFpbC5udG91LmVkdS50dw==; Sofia Priyadarsani Das, ZGFzLnNvZmlhQGdtYWlsLmNvbQ==
 Nandini Rai
Nandini Rai Ankit Kachore2
Ankit Kachore2 Akshaya Panigrahi
Akshaya Panigrahi Sofia Priyadarsani Das
Sofia Priyadarsani Das Fan-Hua Nan
Fan-Hua Nan