- 1Institute of EcoHealth, School of Public Health, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, China
- 2Department of Agricultural and Animal Husbandry Engineering, Cangzhou Technical College, Cangzhou, China
- 3College of Veterinary Medicine, Hebei Agricultural University, Baoding, China
The Respiratory Syncytial Virus (RSV) is a significant agent linked to respiratory infections, representing a considerable health risk for vulnerable populations, including infants, older adults, and those with weakened immune systems. This research successfully introduces an RNA extraction-free rapid detection technique for RSV utilizing real-time reverse transcription recombinase-aided amplification (RT-RAA) technology. Through the crafting of specific primers and probes, this approach enables precise identification of RSV without any interference from other prevalent respiratory viruses. Tests for sensitivity indicated that the detection threshold at a 95% confidence interval was 159 copies per reaction, while the visual detection limit was found to be 1,177 copies per reaction. Testing on clinical samples demonstrated a high degree of consistency with reverse transcription quantitative real-time PCR (RT-qPCR), achieving a Kappa value of 1, which signifies excellent correlation. Furthermore, the amplified products from RT-RAA can be seen with the aid of a portable blue light device, rendering this method appropriate for rapid detection in settings where resources are limited. A total of 265 clinical samples were tested, and the results showed 100% concordance with RT-qPCR. Compared with rapid antigen detection tests (RADTs), RT-RAA exhibited significantly higher sensitivity (100% vs. 93.8%). The rapid detection method for RSV using RT-RAA offers solid technical assistance for the early identification and prevention of RSV.
1 Introduction
Following the COVID-19 pandemic, interest in virology has notably increased, particularly in pathogens that were previously overlooked but have significant impacts on human health. One such pathogen is respiratory syncytial virus (RSV), which significantly contributes to respiratory infections and is the primary viral agent accountable for acute lower respiratory tract infections (ALRTIs) in children younger than five years old (Shi et al., 2017; Bénet et al., 2017). Due to its high transmissibility and associated mortality, infection with RSV has become a major public health issue (Langedijk and Bont, 2023).
RSV belongs to the genus Orthopneumovirus of the family Pneumoviridae, with a single serotype and two antigenic subtypes (A and B). It is an enveloped virus with a non-segmented negative-sense RNA genome, causing severe respiratory diseases in infants, the elderly, and immunocompromised individuals (Rima et al., 2017; Chanock and Finberg, 1957).
RSV is a highly contagious virus and is one of the primary causes of severe respiratory infections in infants and young children, accounting for over 30 million cases globally each year, with approximately 10% of these requiring hospitalization (Nair et al., 2010). Clinically, RSV infection usually manifests as an illness of the upper respiratory tract, with symptoms including a runny nose, nasal blockage, coughing, and sneezing, sometimes accompanied by fever and muscle pain. In youngsters than two years old, bronchiolitis and diseases affecting the lower respiratory tract can also be caused by RSV, often associated with small airway obstruction (Muñoz-Escalante et al., 2019). In severe cases, the infection may progress to pneumonia, respiratory failure, apnea, or even death (Agoti et al., 2014). Among adults, RSV has also been identified as a trigger for hospitalizations due to various clinical syndromes, including pneumonia (11%), chronic obstructive pulmonary disease (11%), asthma (7%), and congestive heart failure (5%) (Falsey et al., 2005). Like other RNA viruses including the influenza virus and coronaviruses, RSV exhibits high genetic variability attributed to the absence of proofreading capabilities in its RNA-dependent RNA polymerase. This results in high mutation rates for RSV-A and RSV-B, estimated at 1.48 × 10-³ and 1.92 × 10-³ substitutions per site annually, respectively (Yu et al., 2021). Consequently, RSV rapidly accumulates single nucleotide polymorphisms (SNPs) and other mutations, leading to continuous genetic and antigenic drift. This high mutation rate significantly undermines the efficacy of vaccines, antiviral drugs, and monoclonal antibodies developed against RSV (Bohmwald et al., 2016).
Because RSV cannot be clinically distinguished from other co-circulating respiratory pathogens based on signs and symptoms alone, it is necessary to conduct laboratory tests on respiratory secretions to verify the presence of infection. Compared to infants, adults tend to have lower viral loads and shorter durations of viral shedding, which limits the sensitivity of current diagnostic methods for detecting RSV in adult patients. Currently, four main diagnostic approaches are available for RSV detection: viral culture, rapid antigen detection tests (RADTs), immunofluorescence (IF) assays, and reverse transcription quantitative real-time PCR (RT-qPCR). Viral culture, once considered the gold standard, is impractical in clinical settings due to its long turnaround time (5–7 days) and low sensitivity (<50%) (Henrickson and Hall, 2007). RADTs and IF assays have replaced culture in routine diagnostics but suffer from low sensitivity (50%-80%) and subjective result interpretation (Chartrand et al., 2015). RT-qPCR, the current gold standard, offers high sensitivity but requires specialized equipment, trained personnel, and high costs, limiting its application in resource-limited settings (Kuypers et al., 2004). RT-qPCR can identify asymptomatic infected individuals, which is an advantage; however, in clinical practice, distinguishing recent infections (requiring intervention) from residual viral shedding (no intervention needed) remains challenging. Approximately 15%-20% of RT-qPCR-positive samples come from individuals with no evidence of acute infection (Lemanske et al., 2005; Kusel et al., 2006; Bulkow et al., 2012), which may affect timely clinical decision-making.
Reverse transcription recombinase-aided amplification (RT-RAA) is an innovative isothermal detection technique that functions at a stable temperature and utilizes four enzymes: recombinase (UvsX, UvsY), single-stranded DNA-binding protein (SSB), and DNA polymerase (Zhang et al., 2017). The underlying principle involves the formation of a complex between UvsX, UvsY, and oligonucleotide primers, which then searches for homologous sequences. Once a homologous region is identified, the corresponding double-stranded DNA is displaced into single strands, allowing the primers to anneal to the complementary sequence. SSB then binds to the resulting single-stranded DNA, and DNA polymerase binds to the primer-DNA complex to initiate amplification. This method is carried out at a stable temperature ranging from 37 to 42 °C, thereby negating the necessity for intricate thermal cycling procedures. As a result, the assay time is significantly reduced and can be completed within 30 minutes. Real-time monitoring of detection results can be achieved with portable fluorescence detectors or blue light emitters. Owing to its exceptional sensitivity, specificity, and rapid response time, RT-RAA has found extensive use in identifying microbial pathogens, establishing itself as a strong asset for point-of-care testing (POCT) (Bai et al., 2020; Wang et al., 2020a, Wang et al., 2020c; Xiong et al., 2020).
In this study, we sought to create an RNA extraction-free RT-RAA assay combined with real-time fluorescence detection for the rapid and convenient identification of RSV. By conducting a comparative analysis with RT-qPCR and RADTs, we assessed the detection capabilities of the RT-RAA approach. Our results demonstrated that the detection time of the RT-RAA assay is similar to that of RADTs, while maintaining the high sensitivity characteristic of RT-qPCR. In summary, this simple, portable, visual, and highly sensitive and specific RSV detection method does not require large or complex equipment, making it highly appropriate for point-of-care applications. This method holds significant promise for practical implementation in the prevention and management of diseases related to RSV.
2 Materials and methods
2.1 Virus and clinical sample sources
In this research, laboratory-preserved RSV strains were used. In 2025, a total of 265 clinical specimens suspected of being infected with RSV were gathered from Hebei Province, including nasopharyngeal swabs, oropharyngeal swabs, and sputum samples. The samples covered both pediatric (<18 years) and adult (≥18 years) populations, with detailed information on age, gender, clinical symptoms, and epidemiological history provided in Supplementary Table S1. Inclusion and exclusion criteria: Samples were included if they met at least one of the following criteria: Acute respiratory infection symptoms (e.g., fever, cough) lasting ≤7 days; Imaging findings suggestive of bronchitis/pneumonia; Epidemiological history (contact with confirmed RSV cases); Clinical suspicion of viral respiratory infection (with exclusion of evidence of bacterial infection). All samples were immediately stored at -80 °C in an ultra-low temperature freezer until further testing.
2.2 Design of RT-RAA primers and probe
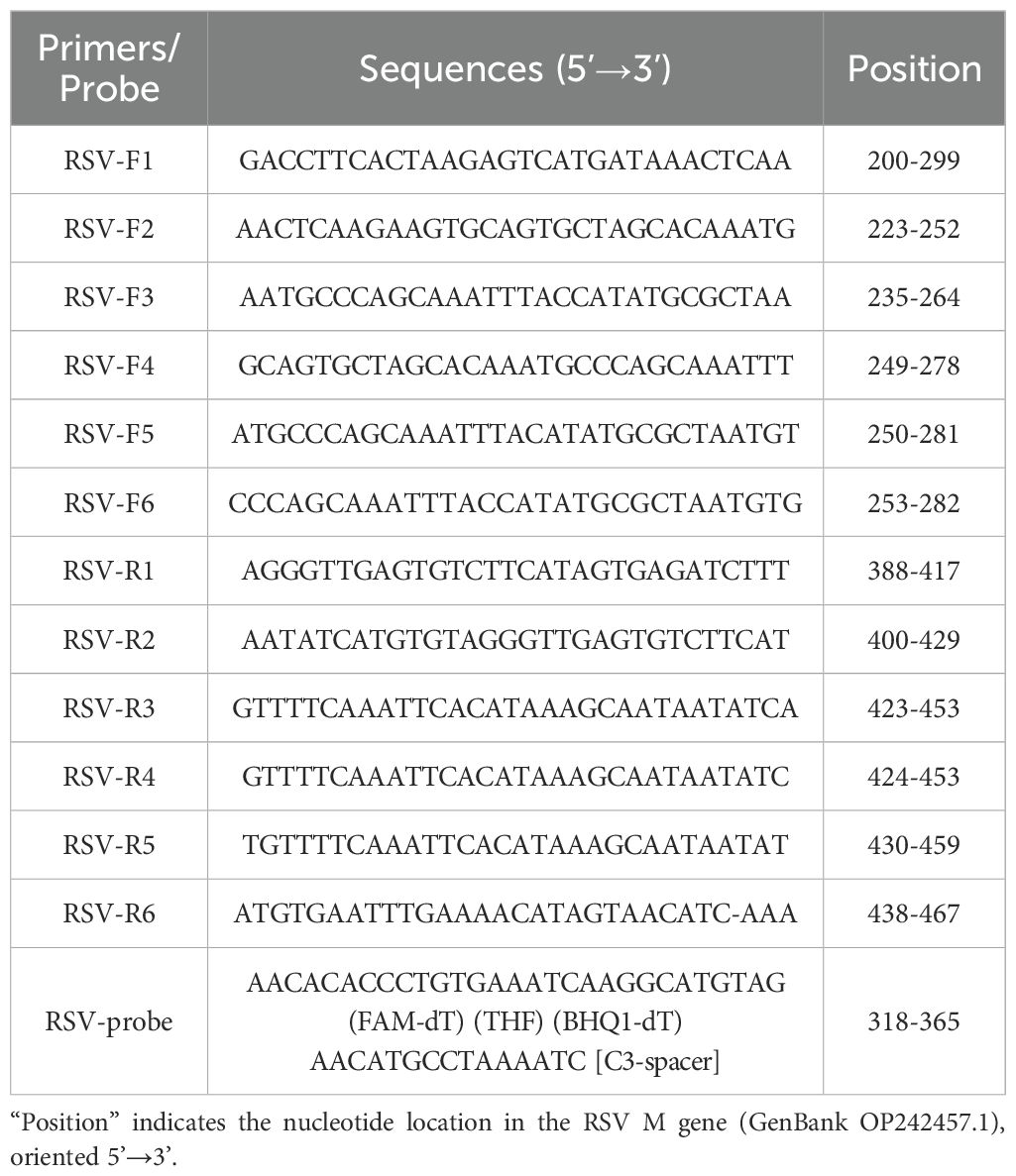
The M gene sequence of RSV (GenBank accession No. OP242457.1) was down-loaded from the GenBank database and subjected to sequence alignment analysis using DNA STAR software to identify conserved regions. Primers and probes were created utilizing SnapGene software, and the best combinations of primers and probes were chosen based on initial screening techniques. The synthesis of the primers and probes was carried out by Shanghai Biotechnology Co., Ltd. Their sequences and positions are listed in Table 1.
2.3 Nucleic acid extraction
Nucleic acids were obtained from the 265 collected clinical samples following the guidelines of the Qiagen RNA Isolation Kit (Hilden, Germany). During the extraction process, RNase-free water was used as a negative control. All isolated nucleic acids were preserved at -80°C for no longer than 24 hours before further analysis.
2.4 Pyrolysis (RNA extraction-free) of clinical samples
The 265 collected clinical samples were processed according to the instructions of the Rapid Nucleic Acid Releaser (RNA)-Type II Kit (AmpFuture, Changzhou, China). RNase-free water was used as the negative control. All extracted nucleic acids were stored at -80 °C for no longer than 24 hours before further analysis.
2.5 RT-RAA amplification
The RNA constant-temperature rapid amplification kit (#WLRE8208KIT) was purchased from AmpFuture Biotechnology Co., Ltd. (Weifang, China). Following the guidelines provided in the kit, a reaction mixture of 25 μL was formulated. This mixture included 14.7 μL Buffer A, 4.75 μL RNase-free water, 1.0 μL forward primer (10 μM), 1.0 μL reverse primer (10 μM), 0.3 μL probe (10 μM), 2.0 μL nucleic acid template, and 1.25 μL Buffer B. The reaction tubes were placed in a 7500 Real-time PCR System (Applied Biosystems) and incubated at a temperature of 42 °C for 30 minutes, cycling once per minute while real-time monitoring of fluorescence signals occurred. Simultaneously, the RT-RAA amplification products were visually observed using a portable blue light imaging device (TGreen, Tiangen Biotech Co., Ltd., Beijing, China) with an excitation wavelength of 480 nm.
2.6 RT-qPCR detection
Detection via RT-qPCR was conducted using the One Step PrimeScript III RT-qPCR Mix (RR600A, Takara), adhering to the guidelines provided by the manufacturer. The reaction mixture, totaling 25 μL, included 12.5 μL of 2× One Step U+ Mix, 1.5 μL of One Step U+ Enzyme Mix, 1.0 μL of forward primer (10 μM), 1.0 μL of reverse primer (10 μM), 0.5 μL of probe (10 μM), 2.0 μL of nucleic acid template, and 6.5 μL of RNase-free water. This reaction was executed on a 7500 Real-time PCR System (Applied Biosystems) following these cycling parameters: reverse transcription at 55°C for 15 minutes, an initial denaturation step at 95°C for 30 seconds, and subsequently 40 cycles at 95°C for 10 seconds and 60°C for 30 seconds, while continuously monitoring the real-time fluorescence signal.
2.7 Rapid antigen detection tests
RADTs were performed using a commercially available colloidal gold-based RSV antigen detection kit (CRIUS: H940-000013) according to the manufacturer’s instructions. Briefly, 100 μL of sample lysate was added to the test cassette, and results were read visually within 15 minutes.
2.8 Specificity analysis
Nucleic acids from the following pathogens were utilized as templates to assess the specificity of the assay: Influenza A H1N1, Influenza A H3N2, Influenza A H9N2, Influenza B Victoria lineage, Influenza B Yamagata lineage, Klebsiella pneumoniae, Parainfluenza virus, Rhinovirus, Adenovirus, Human metapneumovirus, Streptococcus pneumoniae, Chlamydia pneumoniae, and Mycoplasma pneumoniae. The assay’s specificity was confirmed through real-time fluorescent RT-RAA amplification employing the chosen specific primers and probe. All pathogen nucleic acids were obtained from Hebei Houqi Biotechnology Co., Ltd. (Baoding, Hebei, China).
2.9 Sensitivity analysis
Sensitivity analysis was performed using the laboratory-preserved RSV M gene plasmid (GenBank accession No. OP242457.1) (pMD18-T-M). The plasmid underwent a serial 10-fold dilution to create template concentrations that varied from 105 to 100 copies in a 2 μL volume. A volume of 2 μL from each dilution was used to evaluate the sensitivity of the real-time RT-RAA assay, which was conducted in parallel with the RT-qPCR method. To accurately determine the detection limits, both assays were independently repeated eight times. Probit regression analysis was executed using SPSS software (version 22.0).
2.10 Reproducibility and stability analysis
High, medium, and low concentrations of RSV M gene plasmid (GenBank accession No. OP242457.1) (107, 105, and 103 copies/reaction) and clinical sample nucleic acids were selected for testing. Each concentration was tested in triplicate within the same batch and in three independent experiments conducted at different times. To assess the intra-batch and inter-batch reproducibility of the RT-RAA assay, the threshold time’s coefficient of variation (CV) was computed.
2.11 Clinical sample testing
A total of 265 clinical samples were tested using the real-time RT-RAA method in parallel with the RT-qPCR assay and RADTs. The concordance between the two methods’ results was compared and analyzed.
2.12 Statistical analysis
Probit regression analysis was conducted with SPSS software, utilizing a confidence level of 95% to establish the detection limit. To assess the agreement between the results obtained from real-time RT-RAA, RT-qPCR and RADTs, the Kappa statistic was employed. Chi-square tests were used to compare the sensitivity of RT-RAA and RADTs, with P<0.05 considered statistically significant.
3 Results
3.1 Optimal primer screening for RT-RAA
An Exo-probe targeting positions 318–365 was first designed (Figure 1A), based on which six upstream candidate primers (F1-F6) and six downstream candidate primers (R1-R6) were generated (Figure 1A). To begin the screening process, the upstream primer F1 was chosen at random and combined with every one of the six downstream primers. The results showed that the F1/R3 pair exhibited the most robust amplification (Figure 1B). Subsequently, using R3 as the fixed downstream primer, all six upstream primers were evaluated in various combinations. Among these, the F5/R3 pair demonstrated the best amplification performance (Figure 1C). Therefore, the primer pair F5/R3, along with the Exo-probe (targeting positions 318-365), was ultimately selected for real-time RT-RAA detection.
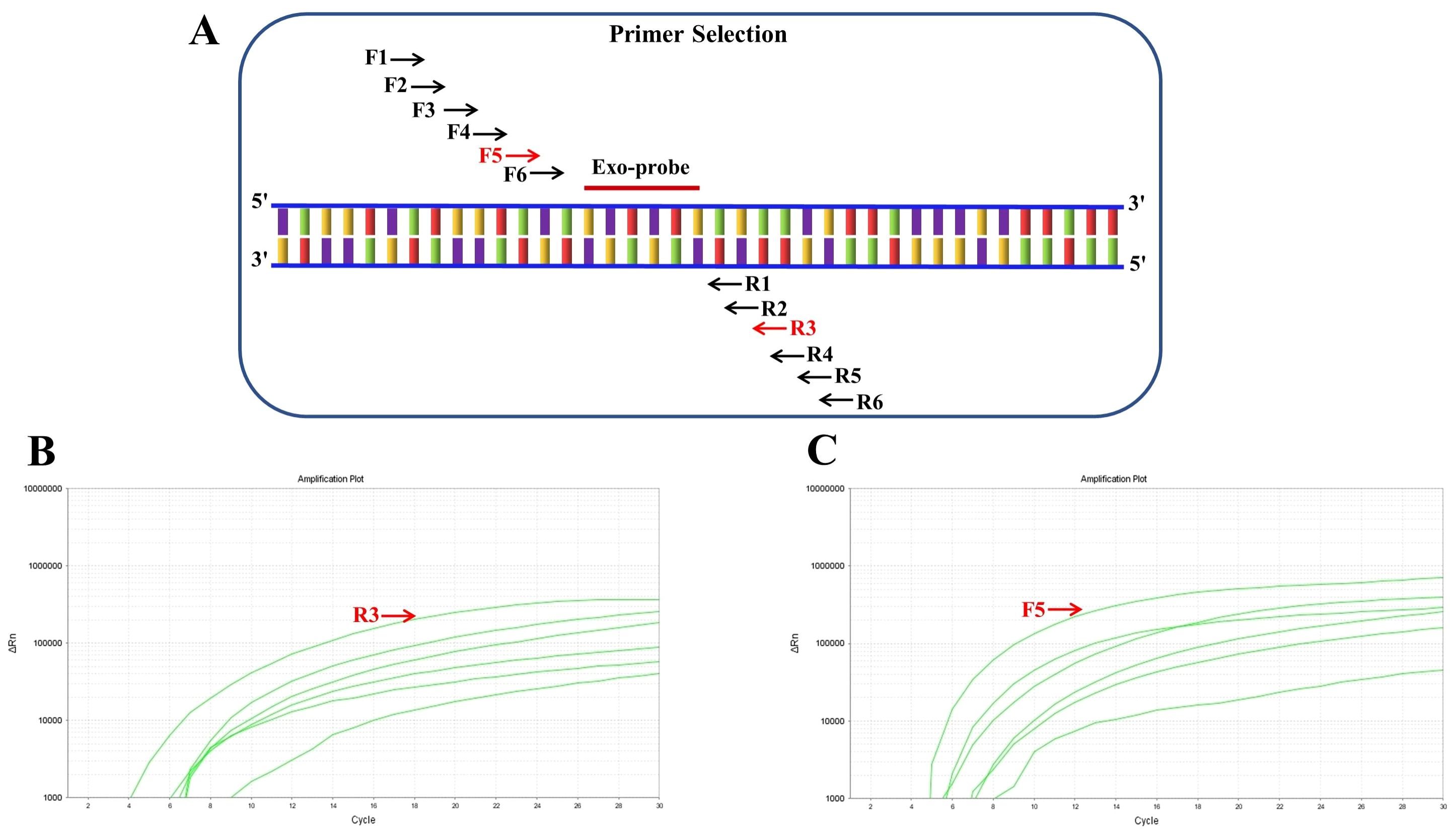
Figure 1. Screening of RT-RAA primers for respiratory syncytial virus detection. (A) Schematic representation of the positions of six upstream candidate primers (F1-F6) and six downstream candidate primers (R1-R6) designed based on the Exo-probe. (B) Screening results of randomly paired upstream primer F1 with six downstream primers, showing the most significant amplification when paired with R3. (C) Screening results using R3 as the fixed downstream primer paired with six upstream primers, indicating that the F5/R3 pair yields the best amplification efficiency.
3.2 Specific analysis
The specificity analysis results demonstrated that only RSV samples (RSV-A and RSV-B) exhibited positive reactions, while samples of H1N1 influenza A, H3N2 influenza A, H9N2 influenza A, Victoria and Yamagata strains of influenza B, Klebsiella pneumoniae, parainfluenza virus, rhinovirus, adenovirus, human metapneumovirus, Streptococcus pneumoniae, Chlamydia pneumoniae, Mycoplasma pneumoniae, and the negative control group all tested negative (Figure 2A). Additionally, the products resulting from the RT-RAA assay were directly visible using a portable blue light device, further validating the high specificity of this technique for detecting RSV (Figure 2B).
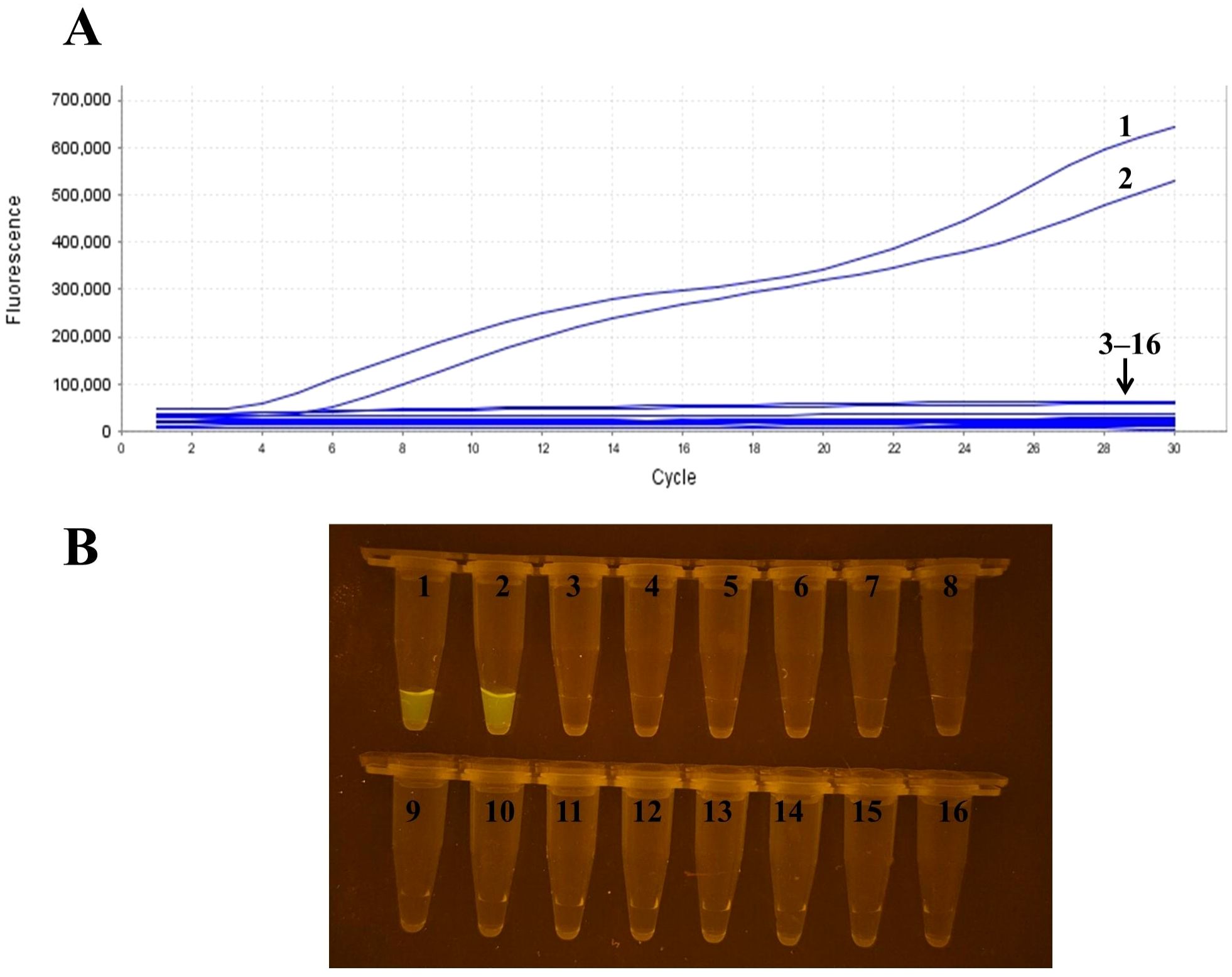
Figure 2. Specificity analysis of RSV RT-RAA detection. (A) Real-time fluorescence detection results showing positive amplification only in RSV samples (RSV-A, RSV-B), while other virus samples including influenza A H1N1, H3N2, etc., and the negative control group showed no amplification. (B) Visualization of RT-RAA amplification products under a portable blue light de-vice, further confirming the high specificity of the method for RSV. Sample numbers correspond to different templates: samples 1–2 are RSV, samples 3–15 represent influenza A H1N1, H3N2, H9N2, influenza B Victoria lineage, influenza B Yamagata lineage, Klebsiella pneumoniae, parainfluenza virus, rhinovirus, adenovirus, human metapneumovirus, Streptococcus pneumoniae, Chlamydia pneumoniae, and Mycoplasma pneumoniae, respectively; sample 16 is the negative control.
3.3 Sensitivity analysis
The evaluation of the sensitivity for RT-RAA and RT-qPCR was conducted utilizing serial dilutions of plasmid templates containing the RSV M gene. Based on re-al-time fluorescence readouts from both methods, the limit of detection (LOD) for RT-RAA was established at 159 copies per reaction at a 95% confidence interval (Figures 3A, B). For visual detection using RT-RAA, the LOD was 1177 copies per reaction (Figure 3C). In comparison, for RT-qPCR, the LOD was determined to be 140 copies per reaction.

Figure 3. Sensitivity analysis of RSV RT-RAA detection. samples 1–6 in (C) represent RSV plasmid templates with 105, 104, 103, 102, 101, and 100 copies per reaction, respectively; sample 7 is the negative control (RNase-free water). All dilutions were prepared with TE buffer (pH 8.0) and calibrated by RT-qPCR. (A) Real-time fluorescence readout of RT-RAA assays using serial dilutions of RSV plasmid templates, showing fluorescence signal changes at different template concentrations. The detection limit of RT-RAA was determined to be 159 copies per reaction at a 95% confidence interval. (B) Real-time fluorescence readout of RT-qPCR assays using the same serially diluted RSV plasmid templates for sensitivity comparison. (C) Visualization results of RT-RAA, indicating a detection limit of 1177 copies per reaction. The different curves or sample numbers correspond to varying template concentrations, with samples 1–6 representing RSV plasmid templates from 105 to 100 copies, and sample 7 as the negative control (RNase-free water). All dilutions were prepared with TE buffer (pH 8.0) and calibrated by RT-qPCR.
3.4 Repeatability and stability analysis
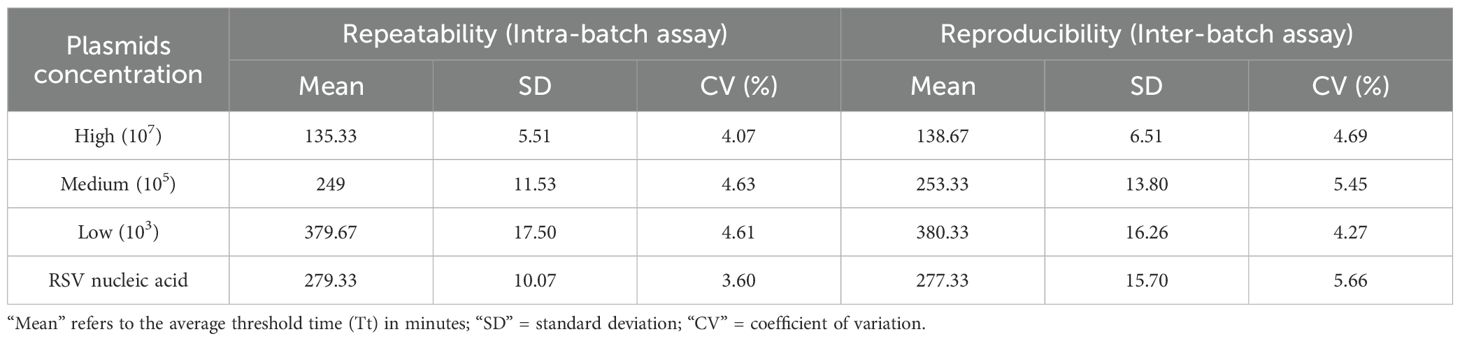
Reproducibility testing was performed using RSV M gene plasmids and clinical sample nucleic acids at different concentrations. The intra-assay coefficients of variation (CVs) were 4.07%, 4.63%, and 4.61%, while the inter-assay CVs were 4.69%, 5.45%, and 4.27%. All intra- and inter-assay CVs were below 5.66%, indicating that the RNA extraction-free RT-RAA method possesses good repeatability and stability (Table 2).
3.5 Clinical sample testing
A total of 265 clinical samples were assessed utilizing both real-time RT-RAA, RT-qPCR and RADTs. The real-time fluorescence-based RT-RAA assay identified 112 positive samples, which were in complete agreement with the RT-qPCR results. The real-time RT-RAA method exhibited a sensitivity of 100%, specificity of 100%, and overall accuracy of 100% (265/265), with a Kappa value of 1, indicating excellent concordance between the two methods. In comparison, RADTs detected 105 positive samples among the 112 RT-qPCR-positive samples, showing a sensitivity of 93.8% (105/112) and a specificity of 89.5% (137/153 negative samples), with a Kappa value of 0.820. These results confirm that RT-RAA is significantly more sensitive than RADTs. Additionally, among the 112 samples that tested positive via real-time RT-RAA, 107 were concurrently classified as positive through visual RT-RAA detection. The visual RT-RAA assay showed a sensitivity of 95.5% (107/112), specificity of 100%, and an overall accuracy of approximately 98.5% [(107 + 153)/265], with a Kappa value of 0.961.
Five samples that were positive by real-time RT-RAA but negative by visual detection had RT-qPCR CT values indicating viral loads close to the visual detection limit: Sample S089 (CT = 34.2, ~1300 copies/reaction), S156 (CT = 35.5, ~1250 copies/reaction), S192 (CT = 36.1, ~1200 copies/reaction), S215 (CT = 36.3, ~1190 copies/reaction), and S253 (CT = 37.8, ~1180 copies/reaction). These results further confirm the strong correlation between the two detection approaches (Table 3).
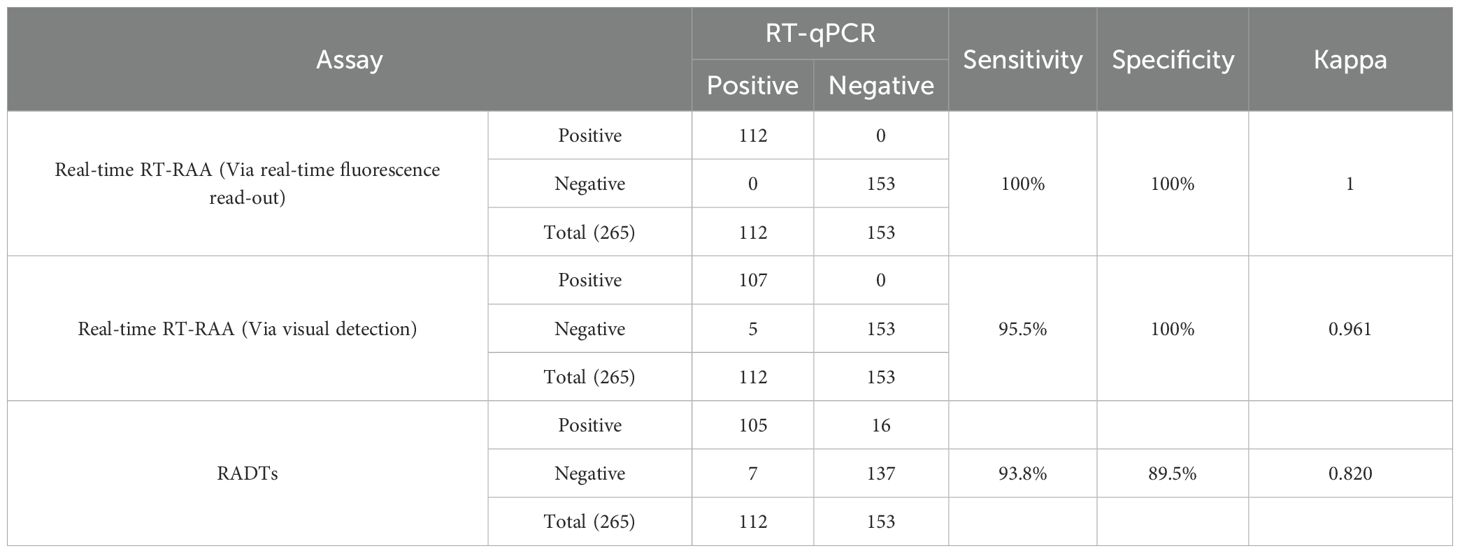
Table 3. Comparison of the RSV real-time RT-RAA with the RT-qPCR assay and RADTs on clinical samples.
To confirm the specificity of RT-RAA amplification, 20 positive products (covering high, medium, and low viral loads) were randomly selected for Sanger sequencing. The sequences showed 100% identity with the RSV M gene (GenBank OP242457.1), with no non-specific amplification.
4 Discussion
In 1956, Morris and colleagues first identified this pathogen while studying acute respiratory disease in chimpanzees. It was officially named respiratory syncytial virus (RSV) in 1957 due to its distinctive cytopathic effects. RSV accounts for the primary reason behind acute respiratory infections that lead to outpatient consultations, emergency room visits, and hospital admissions for children globally under the age of five. During seasonal outbreaks, the number of RSV infections often exceeds those caused by influenza and other viruses. The epidemic season typically spans from autumn to spring, with cases emerging earlier in temperate regions (Hall et al., 2009; Papenburg et al., 2012; Zhou et al., 2012). Preterm infants and immunocompromised individuals are at an elevated risk for serious infections (Papenburg and Boivin, 2010), while RSV notably affects the rates of morbidity and mortality among adults with pre-existing health conditions and the elderly demographic (Falsey et al., 2005). RSV is transmitted both through direct contact with respiratory droplets released by coughing or sneezing patients and indirectly via contact with contaminated secretions on environmental surfaces. Given the absence of distinctive clinical signs and symptoms that set RSV infection apart from other simultaneously circulating respiratory pathogens, a precise laboratory diagnosis of RSV in respiratory samples is essential. Currently, treatment for RSV-associated acute respiratory infections mainly relies on supportive care. Early and precise diagnosis can reduce empirical antibiotic use and allow timely adjustment of therapy in patients already receiving antibiotics, thereby avoiding unnecessary medication—an especially important consideration for infants with severe bacterial coinfections (Adcock et al., 1997; Woo et al., 1997; Barenfanger et al., 2000; Byington et al., 2002; Purcell and Fergie, 2002; Ferronato et al., 2012; Rogers et al., 2015). Moreover, RSV is a common pathogen responsible for nosocomial outbreaks. Rapid detection facilitates timely isolation measures to interrupt transmission chains, optimize bed management, and improve both infection control efficiency and healthcare quality (Madge et al., 1992; Mills et al., 2011).
The identification of RSV through microbiological diagnosis mainly relies on successfully detecting the virus in respiratory secretions from individuals. Prior to the development of innovative rapid molecular diagnostic tests, four main diagnostic methods were commonly used: cell culture, RT-qPCR, IF, and RADTs. Cell culture was previously deemed the gold standard due to its high specificity, but it suffers from low sensitivity, complex procedures, and lengthy turnaround times. RT-qPCR has become the new gold standard because of its high sensitivity, specificity, and rapid turnaround; however, it requires specialized personnel and equipment, making it primarily suitable for large centralized laboratories. Additionally, interpreting RT-qPCR results can be challenging—for example, prolonged viral shedding in recovered patients may lead to false-positive interpretations. IF offers a relatively quick diagnostic option but has lower sensitivity compared to RT-qPCR and relies on skilled personnel and specific equipment. Traditional RADTs depend on visual interpretation of results, which can be subjective, and their relatively low sensitivity makes them unreliable for ruling out RSV cases during outbreaks. Serological testing, while capable of identifying infections based on antibody titers, is limited in its application since some children do not undergo seroconversion after infection. With advancements in molecular biology, novel automated immunoassays and molecular diagnostic methods have emerged. Although these techniques offer standardized result interpretation, they require auxiliary equipment and are costly, making them unsuitable for point-of-care or large-scale screening (Queiróz et al., 2002; Henrickson and Hall, 2007).
In recent years, nucleic acid isothermal amplification technologies have become essential tools for detecting RNA viruses. This category includes various techniques such as loop-mediated isothermal amplification (LAMP) (Fukuta et al., 2003; Meyers et al., 2023), recombinase polymerase amplification (RPA) (Zaghloul, 2014; Lobato and O’Sullivan, 2018), recombinase-aided amplification (RAA) (Chen et al., 2018), nucleic acid sequence-based amplification (NASBA) (Gabrielle et al., 1993; Li et al., 2023a), and helicase-dependent amplification (HDA) (Goldmeyer et al., 2007; Zasada et al., 2022). By amplifying DNA at a constant temperature, these methods eliminate the need for the thermal cycling required by PCR. Due to their simple equipment requirements, rapid detection times, and high feasibility, isothermal amplification techniques are well-suited for point-of-care testing and resource-limited settings (Obande and Banga Singh, 2020). As a novel isothermal amplification method, RT-RAA utilizes DNA polymerase, UvsX, UvsY and SSB to complete amplification within 30 minutes at a constant temperature of 37-42°C (Yan et al., 2018; Li et al., 2023c, Li et al., 2023b). It requires only a single primer pair, making it more efficient than LAMP, which demands 4–6 primers and operates at 65°C (Pang and Long, 2023). When combined with portable devices such as lateral flow devices (LFDs), handheld isothermal fluorescence detectors, or portable blue-light illuminators, RT-RAA effectively meets the needs of on-site detection (Chen et al., 2021; Cui et al., 2022; Li et al., 2023c). It has been widely applied across various pathogen detection scenarios, covering animal, human, and animal-derived food sectors (Wang et al., 2020b; Ding et al., 2022; Feng et al., 2022; Xia et al., 2022, Xia et al., 2024; Li et al., 2024).
The RNA extraction-free RT-RAA method established in this study demonstrated significant advantages in RSV detection, showing better compatibility and innovation compared to previously reported methods. Compared with PCR or RT-qPCR diagnostic methods that rely on purified nucleic acids, our RAA assay eliminates the need for conventional nucleic acid extraction, reducing sample preparation time from 2 hours to just 5 minutes. From a technical perspective, this method is similar to the multiplex RT-RAP technology developed by Fan et al (Fan et al., 2023), enabling rapid nucleic acid amplification at a constant temperature of 42 °C without the thermal cycling required in conventional PCR, thereby simplifying the workflow and reducing the detection time to within 30 minutes. Regarding detection performance, the RT-RAA developed here achieved a detection limit of 159 copies/reaction for RSV (95% confidence interval), surpassing the sensitivity of 162 copies/reaction reported by Zhou et al (Zhou et al., 2025). Notably, this study introduced a visual detection mode, which, although with a detection limit of 1177 copies/reaction, offers practical utility in resource-limited settings through the use of a portable blue-light device. This aligns well with the point-of-care testing requirements emphasized by Hou et al. in bovine RSV detection (Qi et al., 2019). Clinically, the detection results of 265 samples by RT-RAA were in complete agreement with RT-qPCR (Kappa = 1), corroborating the 100% concordance reported by Fan et al. in 252 clinical samples, thus confirming the clinical re-liability of the RT-RAA assay. Moreover, RT-RAA showed significantly higher sensitivity than RADTs (100% vs. 93.8%), addressing the limitation of low sensitivity in traditional antigen-based tests. The establishment of the RT-RAA assay critically depends on the design of primers and probes; the optimal primer pair identified in this study (R3/F5) exhibited high specificity with no cross-reactivity observed against 13 common respiratory pathogens including influenza A/B viruses and Klebsiella pneumoniae. Sequencing verification further confirmed the specificity of the amplified products. Overall, streamlining the procedure reduces the likelihood of errors and minimizing reagent use lowers costs, which facilitates the promotion and clinical application of this assay. Building on the strengths of existing RAA technology, this study further enhanced the field applicability of RSV detection by integrating an RNA extraction-free strategy, optimizing the target gene, and incorporating visual detection. The performance of this method complements similar approaches reported in the literature, providing a diversified technical solution for rapid diagnosis of respiratory viruses.
In summary, the RSV RNA extraction-free RT-RAA assay developed in this study is characterized by high sensitivity, accuracy, rapidity, ease of operation, and cost-effectiveness, making it highly suitable for point-of-care testing. This method provides robust technical support for early surveillance and precise control of RSV infections. It is portable, requires no bulky instrumentation, and enables visual detection under blue light, while maintaining excellent sensitivity and specificity. Clinical evaluation demonstrated 100% concordance with RT-qPCR results, fully validating its re-liability and potential for clinical application.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Cangzhou Technical College (protocol code JYJF0029 and 27 January 2025). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. JZ: Conceptualization, Methodology, Writing – original draft. XH: Conceptualization, Methodology, Writing – original draft. CW: Formal analysis, Investigation, Writing – original draft. ZL: Conceptualization, Methodology, Software, Writing – original draft. ZY: Conceptualization, Investigation, Software, Writing – original draft. CZ: Conceptualization, Funding acquisition, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. TF: Conceptualization, Funding acquisition, Methodology, Software, Writing – original draft, Writing – review & editing. KS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Central government guided local science and technology development fund projects, grant number 246Z6604G and 246Z6605G, basic scientific research funds for universities under Hebei Province, grant number KY2024007 and KY2024006.
Acknowledgments
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1640503/full#supplementary-material
References
Adcock, P. M., Stout, G. G., Hauck, M. A., and Marshall, G. S. (1997). Effect of rapid viral diagnosis on the management of children hospitalized with lower respiratory tract infection. Pediatr. Infect. Dis. J. 16, 842–846. doi: 10.1097/00006454-199709000-00005
Agoti, C. N., Otieno, J. R., Gitahi, C. W., Cane, P. A., and Nokes, D. J. (2014). Rapid spread and diversification of respiratory syncytial virus genotype ON1, Kenya. Emerg. Infect. Dis. 20, , 950–, 959. doi: 10.3201/eid2006.131438
Bai, X., Ma, X., Li, M., Li, X., Fan, G., Zhang, R., et al. (2020). Field applicable detection of hepatitis B virus using internal controlled duplex recombinase-aided amplification assay and lateral flow dipstick assay. J. Med. Virol. 92, 3344–3353. doi: 10.1002/jmv.25778
Barenfanger, J., Drake, C., Leon, N., Mueller, T., and Troutt, T. (2000). Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J. Clin. Microbiol. 38, 2824–2828. doi: 10.1128/JCM.38.8.2824-2828.2000
Bénet, T., Sánchez Picot, V., Messaoudi, M., Chou, M., Eap, T., Wang, J., et al. (2017). Microorganisms associated with pneumonia in children <5 years of age in developing and emerging countries: the GABRIEL pneumonia multicenter, prospective, case-control study. Clin. Infect. Dis. 65, 604–612. doi: 10.1093/cid/cix378
Bohmwald, K., Espinoza, J., Rey-Jurado, E., Gómez, R., González, P., Bueno, S., et al. (2016). Human respiratory syncytial virus: infection and pathology. Semin. Respir. Crit. Care Med. 37, , 522–, 537. doi: 10.1055/s-0036-1584799
Bulkow, L. R., Singleton, R. J., DeByle, C., Miernyk, K., Redding, G., Hummel, K. B., et al. (2012). Risk factors for hospitalization with lower respiratory tract infections in children in rural Alaska. Pediatrics 129, e1220–e1227. doi: 10.1542/peds.2011-1943
Byington, C. L., Castillo, H., Gerber, K., Daly, J. A., Brimley, L. A., Adams, S., et al. (2002). The effect of rapid respiratory viral diagnostic testing on antibiotic use in a children’s hospital. Arch. Pediatr. Adolesc. Med. 156, 1230. doi: 10.1001/archpedi.156.12.1230
Chanock, R. and Finberg, L. (1957). Recovery from infants with respiratory illness of A virus related to chimpanzee coryza agent (CCA). II. Epide-miologic aspects of infection in infants and young children. Am. J. Hyg 66, 291–300. doi: 10.1093/Oxfordjournals.Aje.A119902
Chartrand, C., Tremblay, N., Renaud, C., and Papenburg, J. (2015). Diagnostic accuracy of rapid antigen detection tests for respiratory syncytial virus infection: systematic review and meta-analysis. J. Clin. Microbiol. 53, 3738–3749. doi: 10.1128/JCM.01816-15
Chen, W., Fan, J., Li, Z., Zhang, Y., Qin, Y., Wu, K., et al. (2021). Development of recombinase aided amplification combined with disposable nucleic acid test strip for rapid detection of porcine circovirus type 2. Front. Vet. Sci. 8. doi: 10.3389/fvets.2021.676294
Chen, C., Li, X., Li, G., Zhao, L., Duan, S., Yan, T., et al. (2018). Use of a rapid reverse-transcription recombinase aided amplification assay for respiratory syncytial virus detection. Diagn. Microbiol. Infect. Dis. 90, 90–95. doi: 10.1016/j.diagmicrobio.2017.10.005
Cui, H., Tu, F., Zhang, C., Zhang, C., Zhao, K., Liu, J., et al. (2022). Real-time reverse transcription recombinase-aided amplification assay for rapid amplification of the N gene of SARS-CoV-2. IJMS 23, 15269. doi: 10.3390/ijms232315269
Ding, X., Wang, H., Cui, M., Cheng, M., Zhao, Q., Bai, Y., et al. (2022). Development of a real-time recombinase-aided amplification method to rapidly detect methicillin-resistant staphylococcus aureus. Microorganisms 10, 2351. doi: 10.3390/microorganisms10122351
Falsey, A. R., Hennessey, P. A., Formica, M. A., Cox, C., and Walsh, E. E. (2005). Respiratory syncytial virus infection in elderly and high-risk adults. N Engl. J. 352, 1749–1759. doi: 10.1056/NEJMoa043951
Fan, G., He, X., Zhang, R., Tian, F., Sun, X., Zhang, M., et al. (2023). A rapid and highly sensitive multiple detection of human adenovirus type 3, type 7 and respiratory syncytial virus by recombinase-aided reverse transcription PCR. Clin. Lab. Anal. 37, e24889. doi: 10.1002/jcla.24889
Feng, X., Zhou, D., Gan, B., Xie, G., and Xu, H. (2022). A combination of novel nucleic acid cross-linking dye and recombinase-aided amplification for the rapid detection of viable salmonella in milk. Foods 11, 2375. doi: 10.3390/foods11152375
Ferronato, Â.E., Gilio, A. E., Ferraro, A. A., De Paulis, M., and Vieira, S. E. (2012). Etiological diagnosis reduces the use of antibiotics in infants with bronchiolitis. Clinics 67, 1001–1006. doi: 10.6061/clinics/2012(09)03
Fukuta, S., Iida, T., Mizukami, Y., Ishida, A., Ueda, J., Kanbe, M., et al. (2003). Detection of Japanese yam mosaic virus by RT-LAMP. Arch. Virol. 148, 1713–1720. doi: 10.1007/s00705-003-0134-5
Gabrielle, M. E., Van Der, V., Schukkink, R. A. F., Van Gemen, B., Schepers, Y., and Klatser, Y. (1993). Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J. Gen. Microbiol. 139, 2423–2429. doi: 10.1099/00221287-139-10-2423
Goldmeyer, J., Kong, H., and Tang, W. (2007). Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid rna detection. J. Mol. Diagnostics 9, 639–644. doi: 10.2353/jmoldx.2007.070012
Hall, C. B., Weinberg, G. A., Iwane, M. K., Blumkin, A. K., Edwards, K. M., Staat, M. A., et al. (2009). The burden of respiratory syncytial virus infection in young children. N Engl. J. Med. 360, 588–598. doi: 10.1056/NEJMoa0804877
Henrickson, K. J. and Hall, C. B. (2007). Diagnostic assays for respiratory syncytial virus disease. Pediatr. Infect. Dis. J. 26, S36–S40. doi: 10.1097/INF.0b013e318157da6f
Kusel, M. M. H., De Klerk, N. H., Holt, P. G., Kebadze, T., Johnston, S. L., and Sly, P. D. (2006). Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr. Infect. Dis. J. 25, 680–686. doi: 10.1097/01.inf.0000226912.88900.a3
Kuypers, J., Wright, N., and Morrow, R. (2004). Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J. Clin. Virol. 31, 123–129. doi: 10.1016/j.jcv.2004.03.018
Langedijk, A. C. and Bont, L. J. (2023). Respiratory syncytial virus infection and novel interventions. Nat. Rev. Microbiol. 21, 734–749. doi: 10.1038/s41579-023-00919-w
Lemanske, R. F., Jr., Jackson, D. J., Gangnon, R. E., Evans, M. D., Li, Z., Shult, P. A., et al. (2005). Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J. Allergy Clin. Immunol. 116, 571–577. doi: 10.1016/j.jaci.2005.06.024
Li, J., Cui, H., Zhang, Y., Wang, X., Liu, H., Mu, Y., et al. (2024). A rapid detection method for H3 avian influenza viruses based on RT-RAA. Animals 14, 2601. doi: 10.3390/ani14172601
Li, H., Song, W., Li, H., Cui, J., Xie, Y., Wu, B., et al. (2023a). Advances in isothermal nucleic acid amplification methods for hepatitis B virus detection. Analyst 1483708–3718. doi: 10.1039/D3AN00700F
Li, R., Su, N., Ren, X., Sun, X., Li, W., Li, Y., et al. (2023b). Centrifugal microfluidic-based multiplex recombinase polymerase amplification assay for rapid detection of SARS-CoV-2. iScience 26, 106245. doi: 10.1016/j.isci.2023.106245
Li, X., Zhu, S., Zhang, X., Ren, Y., He, J., Zhou, J., et al. (2023c). Advances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1215466
Lobato, I. M. and O’Sullivan, C. K. (2018). Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Analytical Chem. 98, 19–35. doi: 10.1016/j.trac.2017.10.015
Madge, P., Paton, J. Y., McColl, J. H., and Mackie, P. L. K. (1992). Prospective controlled study of four infection-control procedures to prevent nosocomial infection with respiratory syncytial virus. Lancet 340, 1079–1083. doi: 10.1016/0140-6736(92)93088-5
Meyers, E., Park, J., Coen, A., Raman, L., Heytens, S., Rhee, J., et al. (2023). Evaluation of a smartphone-operated point-of-care device using loop-mediated isothermal amplification technology for rapid and remote detection of SARS-CoV-2. J. Med. Virol. 95, e29158. doi: 10.1002/jmv.29158
Mills, J. M., Harper, J., Broomfield, D., and Templeton, K. E. (2011). Rapid testing for respiratory syncytial virus in a paediatric emergency department: benefits for infection control and bed management. J. Hosp. Infection 77248–251. doi: 10.1016/j.jhin.2010.11.019
Muñoz-Escalante, J. C., Comas-García, A., Bernal-Silva, S., Robles-Espinoza, C. D., Gómez-Leal, G., and Noyola, D. E. (2019). Respiratory syncytial virus A genotype classification based on systematic intergenotypic and intragenotypic sequence analysis. Sci. Rep. 9, 20097. doi: 10.1038/s41598-019-56552-2
Nair, H., Nokes, D. J., Gessner, B. D., Dherani, M., Madhi, S. A., Singleton, R. J., et al. (2010). Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375, 1545–1555. doi: 10.1016/S0140-6736(10)60206-1
Obande, G. A. and Banga Singh, K. K. (2020). Current and future perspectives on isothermal nucleic acid amplification technologies for diagnosing infections. IDR Volume 13, 455–483. doi: 10.2147/IDR.S217571
Pang, F. and Long, Q. (2023). Recent advances in diagnostic approaches for orf virus. Appl. Microbiol. Biotechnol. 107, 1515–1523. doi: 10.1007/s00253-023-12412-8
Papenburg, J. and Boivin, G. (2010). distinguishing features of human metapneumovirus and respiratory syncytial virus. The Rev. Med. Virol. 20, 245–260. doi: 10.1002/rmv.651
Papenburg, J., Hamelin, M.È., Ouhoummane, N., Carbonneau, J., Ouakki, M., Raymond, F., et al. (2012). Comparison of risk factors for human metapneumovirus and respiratory syncytial virus disease severity in young children. J. Infect. Dis. 206, 178–189. doi: 10.1093/infdis/jis333
Purcell, K. and Fergie, J. (2002). Concurrent serious bacterial infections in 2396 infants and children hospitalized with respiratory syncytial virus lower respiratory tract infections. Arch. Pediatr. Adolesc. Med. 156, 322–324. doi: 10.1001/archpedi.156.4.322{/it}
Qi, J., Li, X., Zhang, Y., Shen, X., Song, G., Pan, J., et al. (2019). Development of a duplex reverse transcription recombinase-aided amplification assay for respiratory syncytial virus incorporating an internal control. Arch. Virol. 164, 1843–1850. doi: 10.1007/s00705-019-04230-z
Queiróz, D. A. O., Durigon, E. L., Botosso, V. F., Ejzemberg, B., Vieira, S. E., Mineo, J. R., et al. (2002). Immune response to respiratory syncytial virus in young Brazilian children. Braz. J. Med. Biol. Res. 35, 1183–1193. doi: 10.1590/S0100-879X2002001000011
Rima, B., Collins, P., Easton, A., Fouchier, R., Kurath, G., Lamb, R. A., et al. (2017). ICTV virus taxonomy profile: pneumoviridae. J. Gen. Virol. 98, 2912–2913. doi: 10.1099/jgv.0.000959
Rogers, B. B., Shankar, P., Jerris, R. C., Kotzbauer, D., Anderson, E. J., Watson, J. R., et al. (2015). Impact of a rapid respiratory panel test on patient outcomes. Arch. Pathol. Lab. Med. 139, 636–641. doi: 10.5858/arpa.2014-0257-OA
Shi, T., McAllister, D. A., O’Brien, K. L., Simoes, E. A. F., Madhi, S. A., Gessner, B. D., et al. (2017). Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390, 946–958. doi: 10.1016/S0140-6736(17)30938-8
Wang, J., Cai, K., He, X., Shen, X., Wang, J., Liu, J., et al. (2020a). Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin. Microbiol. Infection 26, 1076–1081. doi: 10.1016/j.cmi.2020.05.007
Wang, W., Wang, C., Bai, Y., Zhang, P., Yao, S., Liu, J., et al. (2020c). Establishment of reverse transcription recombinase-aided amplification-lateral-flow dipstick and real-time fluorescence-based reverse transcription recombinase-aided amplification methods for detection of the Newcastle disease virus in chickens. Poultry Sci. 99, 3393–3401. doi: 10.1016/j.psj.2020.03.018
Wang, L., Zhao, P., Si, X., Li, J., Dai, X., Zhang, K., et al. (2020b). Rapid and specific detection of listeria monocytogenes with an isothermal amplification and lateral flow strip combined method that eliminates false-positive signals from primer-dimers. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02959
Woo, P. C., Chiu, S. S., Seto, W. H., and Peiris, M. (1997). Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 35, 1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997
Xia, W., Chen, K., Liu, W., Yin, Y., Yao, Q., Ban, Y., et al. (2022). Rapid and visual detection of Mycoplasma synoviae by recombinase-aided amplification assay combined with a lateral flow dipstick. Poultry Sci. 101, 101860. doi: 10.1016/j.psj.2022.101860
Xia, W., Yu, S., Huang, J., Li, Y., Wang, P., Shen, S., et al. (2024). Research Note: Real-time fluorescence-based recombinase-aided amplification for rapid detection of Mycoplasma synoviae. Poultry Sci. 103, 103995. doi: 10.1016/j.psj.2024.103995
Xiong, Y., Luo, Y., Li, H., Wu, W., Ruan, X., and Mu, X. (2020). Rapid visual detection of dengue virus by combining reverse transcription recombinase-aided amplification with lateral-flow dipstick assay. Int. J. Infect. Dis. 95, 406–412. doi: 10.1016/j.ijid.2020.03.075
Yan, T., Li, X., Wang, L., Chen, C., Duan, S., Qi, J., et al. (2018). Development of a reverse transcription recombinase-aided amplification assay for the detection of coxsackievirus A10 and coxsackievirus A6 RNA. Arch. Virol. 1631455–1461. doi: 10.1007/s00705-018-3734-9
Yu, J.-M., Fu, Y.-H., Peng, X.-L., Zheng, Y.-P., and He, J.-S. (2021). Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci. Rep. 11, 12941. doi: 10.1038/s41598-021-92435-1
Zaghloul, H. (2014). Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. WJH 6, 916. doi: 10.4254/wjh.v6.i12.916
Zasada, A. A., Mosiej, E., Prygiel, M., Polak, M., Wdowiak, K., Formińska, K., et al. (2022). Detection of SARS-CoV-2 using reverse transcription helicase dependent amplification and reverse transcription loop-mediated amplification combined with lateral flow assay. Biomedicines 10, 2329. doi: 10.3390/biomedicines10092329
Zhang, X., Guo, L., Ma, R., Cong, L., Wu, Z., Wei, Y., et al. (2017). Rapid detection of Salmonella with recombinase aided amplification. J. Microbiological Methods 139, 202–204. doi: 10.1016/j.mimet.2017.06.011
Zhou, Y., Liang, C., Long, Z., Fan, L., Wang, Y., Wang, Z., et al. (2025). Multiplex real-time reverse transcription recombinase-aided amplification assay for the detection of SARS-CoV-2, influenza A virus, and respiratory syncytial virus. Microbiol. Spectr. 13, e02759–e02724. doi: 10.1128/spectrum.02759-24
Keywords: respiratory syncytial virus, real-time reverse transcription recombinase-aided amplification, visual detection, rapid detection, RNA extraction-free
Citation: Wang Z, Zhang J, He X, Wang C, Li Z, Yang Z, Zhang C, Fan T and Su K (2025) Portable, precise, and RNA extraction-free: RT-RAA technology for rapid early RSV identification and prevention. Front. Cell. Infect. Microbiol. 15:1640503. doi: 10.3389/fcimb.2025.1640503
Received: 03 June 2025; Accepted: 18 August 2025;
Published: 02 September 2025.
Edited by:
Pang Yanan, Changhai Hospital, ChinaReviewed by:
Leonardo Sorrentino, Emory University, United StatesKaiqing Zhang, Biocytogen, United States
Copyright © 2025 Wang, Zhang, He, Wang, Li, Yang, Zhang, Fan and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingli Fan, ZnRsMjAxNUAxMjYuY29t; Kai Su, c3VrYWkwMzI0QDEyNi5jb20=
 Zhenfei Wang
Zhenfei Wang Jing Zhang2
Jing Zhang2 Kai Su
Kai Su