- Department of Laboratory Medicine, The First Affiliated Hospital of Wannan Medical College (Yijishan Hospital of Wannan Medical College), Wuhu, Anhui, China
Objective: To analyze the identification, antifungal resistance, and genomic characteristics of a Candida auris (C. auris) strain isolated from a urine specimen of an ICU patient at Yijishan Hospital, Wannan Medical College, Anhui Province.
Methods: The isolate was identified by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). The susceptibility of the isolates to fungi was determined by measuring the Minimum Inhibitory Concentration (MIC) values using the VITEK 2 COMPACT system. Whole-genome sequencing (WGS) was performed using high-throughput technology. Resistance and virulence genes were annotated using public databases, including NCBI (https://www.ncbi.nlm.nih.gov/, version 2.2.28), DFVF (http://sysbio.unl.edu/DFVF/, version 1.0), PHI-base (http://www.phi-base.org, version 5.0) and KEGG (https://www.kegg.jp/, version 89.1). A phylogenetic tree was constructed through analysis of the 18S rRNA nucleotide sequence.
Results: The isolate named CAS20503 was identified as C. auris. Antifungal susceptibility testing showed resistance to fluconazole and amphotericin B. Genomic analysis identified resistance genes including ERG11 (azole), FKS1 (echinocandin), ERG3 (polyene), and efflux pumps CDR1, MDR1. Resistance mutations were detected. The virulence genes analyzed based on the DFVF database included CaNik1, CHS2, DUR1,2, HSP90, ICL1, PMT1, PMT2, PMT4, SSD1, TPS2. The host pathogenic genes identified by comparison with the PHI-base database included CaCHS1, ADE2, FAS2, PMR1, CaTPS2, Tfp1. KEGG annotation showed enrichment in infectious disease pathways. The phylogenetic tree constncted based on the nudeotide sequence analysis of 18S rRNA indicated that this strain (Genome accession number: JBPYFS000000000) exhibited a high degree of genomic similarity to the C. auris strain (Genome accession number: CP157510.1), which was isolated in ltaly in 2024 and belonged to clade l (a subset of the South Asian clade).
Conclusion: Through an in-depth analysis of strain CAS20503, which was isolated from a urinary tract infection specimen at a tertiary public hospital in Anhui Province (Yijishan Hospital, Wannan Medical College), this study elucidated the drug resistance profiles and genomic characteristics of C. auris. The findings have provided critical evidence for the early identification, diagnosis, and optimization of antifungal therapeutic regimens for infections caused by this pathogen in clinical practice.
1 Introduction
C. auris is an emerging global microorganism. Through nosocomial transmission, it has spread to over 50 countries across six continents, rapidly garnering attention from global health authorities and emerging as a worldwide threat (Kim et al., 2024). The distinctive characteristics of C. auris have resulted in its inclusion in the “critical priority” category of the World Health Organization (WHO) Fungal Priority Pathogens List (Kriegl et al., 2025). This pathogen is characterized by high-level resistance to multiple antifungal agents and is associated with mortality rates ranging from 29% to 53% (World Health Organization, 2022). C. auris infection can cause severe invasive infections (Khatamzas et al., 2019). Global data indicate that among cases of fungal infections caused by C. auris, the axilla, groin, and nostril are the primary isolation sources, accounting for 29.7% of all isolated strains. The next most frequent sources are blood (17.0%), urine/urinary catheters (14.8%), and skin (13.6%). Notably, from 2022 to 2024, the proportion of isolates from urine/urinary catheters worldwide increased, making it the second most common isolation source in 2024 (19.3%) (La Bella et al., 2025). In recent years, C. auris has been recognized as a pathogen responsible for urinary tract infections (UTIs) on a global scale (Fazeli et al., 2019). The first public report on C. auris in urine dates back to 2010–2014, during which researchers collected urine isolates from a population in India (Kathuria et al., 2015). This study analyzed the drug resistance profiles and genetic characteristics of C. auris isolated from a urinary tract infection specimen of an elderly male patient, thereby providing insights into the diagnosis and treatment of fungal infections in the urinary system.
2 Information and methods
2.1 Patient information
The C. auris strain (CAS20503) utilized in the experiment was isolated from a urine sample of a patient with a urinary tract infection. A 62-year-old male patient presented with persistent right knee pain for over two months, accompanied by limb weakness for one week. He had a long-standing history of hypertension, diabetes mellitus, renal insufficiency, and hemodialysis. Following admission, the patient was transferred to the ICU due to respiratory failure. Five days later, given symptoms of fever and oliguria, a urine sample was collected for relevant examinations. Routine urinalysis revealed 3+ white blood cells. A midstream morning urine specimen was submitted for pathogen culture, which detected C. auris. Following a multidisciplinary consultation, the patient was administered intravenous caspofungin (70 mg as a loading dose, followed by 50 mg daily) in combination with flucytosine (100 mg/kg daily) based on fungal susceptibility testing results. One week after initiation of treatment, no leukocytes were detected in the urine. By the third week of treatment, urine fungal culture yielded negative results.
2.2 Instruments and reagents
Microbial identification and susceptibility testing were performed using the MALDI-TOF MS and VITEK 2 COMPACT systems (bioMérieux, France). Sabouraud dextrose agar medium and CHROMagar Candida chromogenic media were purchased from Jinan Baibo (China). Gram staining reagents were obtained from Zhuhai DL Biotech (China). Additional materials included target plates, formic acid, matrix solution, and fungal susceptibility panels (AST-YST) (bioMérieux, France). Genomic DNA was extracted using the Rapid Fungi Genomic DNA Isolation Kit (Shanghai Sangon Biotech, China). The Hieff NGS Ultima Pro DNA Library Prep Kit was used for library preparation (Yeasen Biotechnology Co., Ltd, China).
2.3 Culture and identification
Midstream urine was aseptically collected and immediately processed. Samples were inoculated onto Sabouraud dextrose agar medium and CHROMagar Candida chromogenic media and incubated at 35°C for 48 hours. A fungal smear was prepared from the colony, followed by Gram staining of the smear, followed by observation of fungal morphology under a microscope. For MALDI-TOF MS identification, colonies were applied to a target plate, treated with formic acid to lyse fungal cell walls, overlaid with matrix solution, and subjected to mass spectrometric analysis.
2.4 Antifungal susceptibility testing
Fungal colonies cultured for 48 hours were collected and adjusted to a 2.0 McFarland standard suspension. Disposable plastic tubes were placed in a rack, each containing 3 ml of 0.45% sodium chloride solution. Subsequently, 280 μl of the 2.0 McFarland suspension was added to each tube and mixed thoroughly. The AST-YST antifungal susceptibility cards were retrieved from the refrigerator, allowed to equilibrate to room temperature for 15–20 minutes, and then inserted into the inoculated tubes. The tubes were transferred to the VITEK 2 COMPACT system for automated determination of MIC.
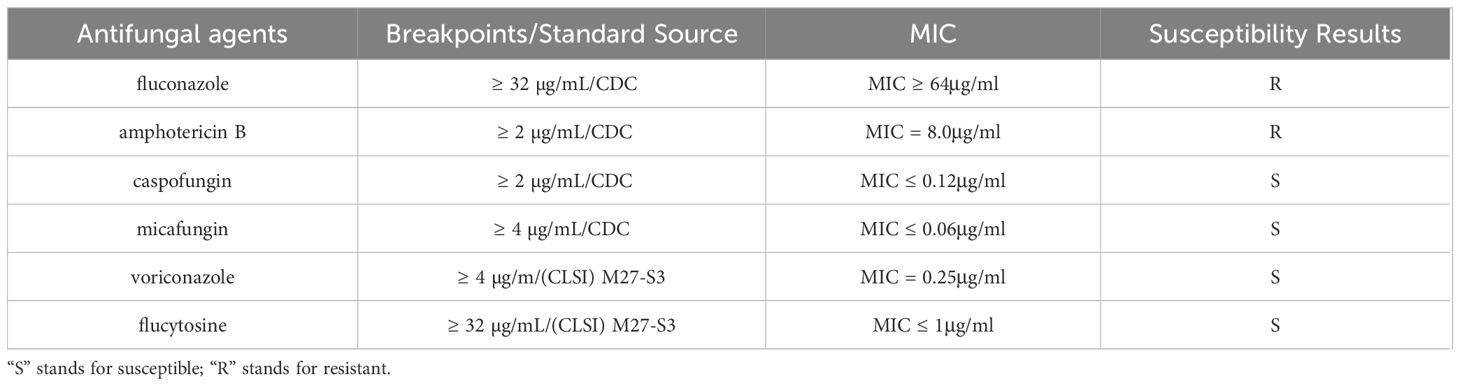
To date, no specific susceptibility breakpoints have been established for C. auris. Therefore, this experiment utilized the tentative MIC breakpoints recommended by the U.S. Centers for Disease Control and Prevention (CDC) as reference standards (Centers for Disease Control and Prevention): fluconazole (FLC) ≥ 32 µg/mL; amphotericin B (AMB) ≥ 2 µg/mL; caspofungin (CSF) ≥ 2 µg/mL; micafungin (MFG) ≥ 4 µg/mL. For antifungal agents lacking CDC breakpoints (voriconazole (VRC) and flucytosine (5-FC), the breakpoints for the genus Candida specified in the Clinical and Laboratory Standards Institute (CLSI) M27-S3 protocol were adopted (voriconazole ≥ 4 µg/mL; flucytosine ≥ 32 µg/mL), which are applicable to C. auris (Ramos et al., 2025) (Table 1).
2.5 Whole genome sequencing
The sequencing platform used was DNBSEQ-T7, with library preparation performed using the Hieff NGS Ultima Pro DNA Library Prep Kit; paired-end sequencing was employed, with an average read length of 150 bp, generating raw data containing 31,911,388 total reads (4,786,708,200 bp in total) and achieving 99.997% genome coverage. To ensure sequencing data quality, multiple quality control methods were implemented: Fastp software was used to analyze raw data quality, including base quality values (Q20 ratio 98.63%, Q30 ratio 95.87%) and GC content (45.15%); adapter-containing sequences were removed, with adapters accurately identified via read overlap regions; global trimming and sliding window quality trimming were performed (window length 4, with windows of quality below Q20 trimmed); base inconsistencies in overlap regions were corrected; low-quality sequences (reads containing more than 40% of bases with quality below Q15) and paired-end reads shorter than 35 nt were filtered out; and sequence trimming was conducted using Trimmomatic (version 0.36). Additionally, 10,000 reads were randomly selected and aligned with the NT database for species proportion counting, revealing that 99.52% of reads matched C. auris (indicating an extremely low contamination rate). Post-quality control, the data retention rate reached 99.94%, with the proportion of low-quality sequences at 0.0%, sequences containing excessive Ns at 0.057%, and short sequences at 0.003%, results that indicated high data purity and good integrity.
2.6 Bioinformatics analysis
2.6.1 Drug resistance gene alignment
The target genes included ERG11, FKS1, ERG3, CDR1, and MDR1. Reference sequences and their accession numbers were retrieved from the NCBI database as follows: ERG11 (Gene ID: 40026303), FKS1 (Gene ID: 40026130), ERG3 (Gene ID: 40028956), CDR1 (Gene ID: 40025314), and MDR1 (Gene ID: 40029187). Sequence alignment was performed using NCBI BLAST (version 2.2.28) and SnapGene (Version 8.0.3). For the alignment process, the whole-genome nucleic acid sequences of the test strains were aligned against the aforementioned NCBI reference sequences, and sequence matching patterns were analyzed, including completely matched genes, genes with insertion/deletion variations, and genes with point mutations.
2.6.2 Virulence gene alignment
First, the gene prediction tool Prokka (version 1.10) was used to identify coding sequences (CDS) from the assembled genome, and the corresponding protein sequences were obtained. NCBI BLAST (version 2.2.28) was then employed to align these gene-encoded protein sequences against the DFVF V1.0 database. The screening criteria for virulence gene results were set as follows: E-value = 0 and query coverage > 60%.
2.6.3 Pathogenic gene alignment
NCBI BLAST was used to align the gene-encoded protein sequences against the PHI-base V5.0 database. The screening criteria for pathogenic gene results were defined as E-value = 0 and query coverage > 95%.
2.6.4 Database metabolic pathway classification
The protein sequences predicted in the above steps were converted to FASTA format (a total of 6530 CDS were predicted in this project). The KEGG Automatic Annotation Server (KAAS, version 2.1) was used as the core tool to associate genes with KEGG (version 89.1) data. Standardized protein sequences were batch-uploaded to the KAAS server (https://www.genome.jp/kaas-bin/kaas_main), with the corresponding species classification (fungi) and reference genome database (KEGG Orthology, ko) selected. Using BLASTP alignment, KAAS conducted homology searches between the input protein sequences and KEGG’s KO database to determine the KO number for each gene. The KO number serves as a key identifier linking genes to metabolic pathways. Output results included each gene’s KO number, corresponding metabolic pathway information (Pathway ID), and functional descriptions. The number of genes annotated in each metabolic pathway was counted, and a bar chart was generated.
2.7 Method for constructing the phylogenetic tree
Phylogenetic analysis was performed based on the 18S rRNA homologous sequence of the CAS20503 strain. This sequence was aligned against the NCBI BLAST database with the parameter set to identity > 90%, yielding a total of 19 included sequences. No outgroup was used for rooting the phylogenetic tree, as the strain had already been identified as C. auris by mass spectrometry. Using MEGA12 software (Version 12.0.9), a phylogenetic tree was constructed from these sequences via the maximum likelihood method with 1000 bootstrap replications.
2.8 Ethical approval
The fungal strain used in the experiment was isolated and cultured from a urine specimen of a clinical patient. Informed consent has been obtained from the subject for the use of the experimental specimen.
3 Experimental results
3.1 Morphological characteristics
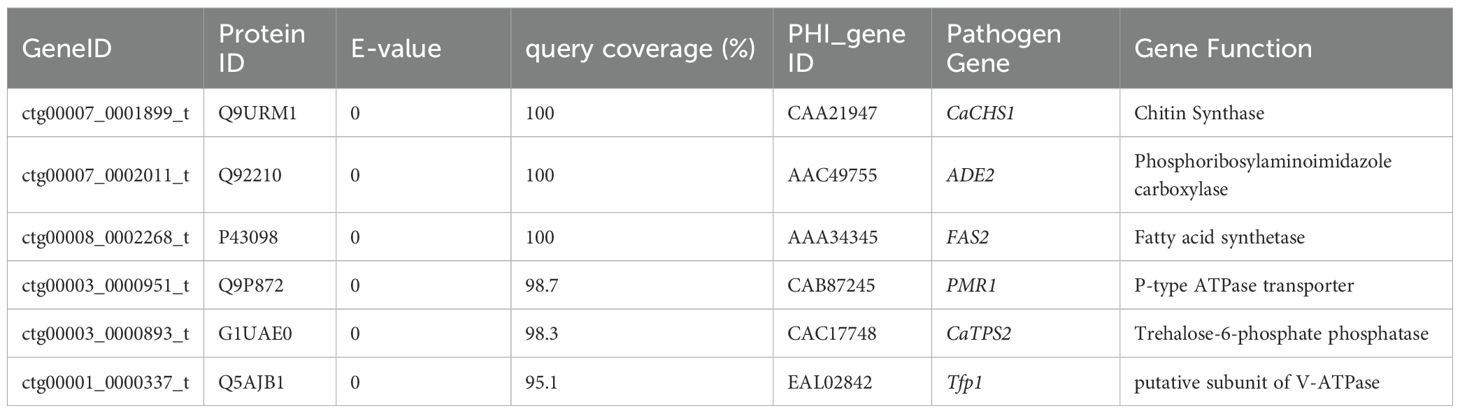
After 48 hours of incubation, on CHROMagar Candida chromogenic medium, colonies were initially white, gradually transitioning to a pinkish-purple hue with prolonged incubation (Figure 1a). Colonies on Sabouraud dextrose agar medium appeared creamy white (Figure 1b). With Gram staining, under the microscope, the fungus could be observed as purple, oval yeast-like cells with a diameter of 3-6 μm (Figure 2a). Wet mount microscopic examination of the fungal suspension prepared with 10% potassium hydroxide (KOH) revealed the presence of budding spores in the fungus (Figure 2b).
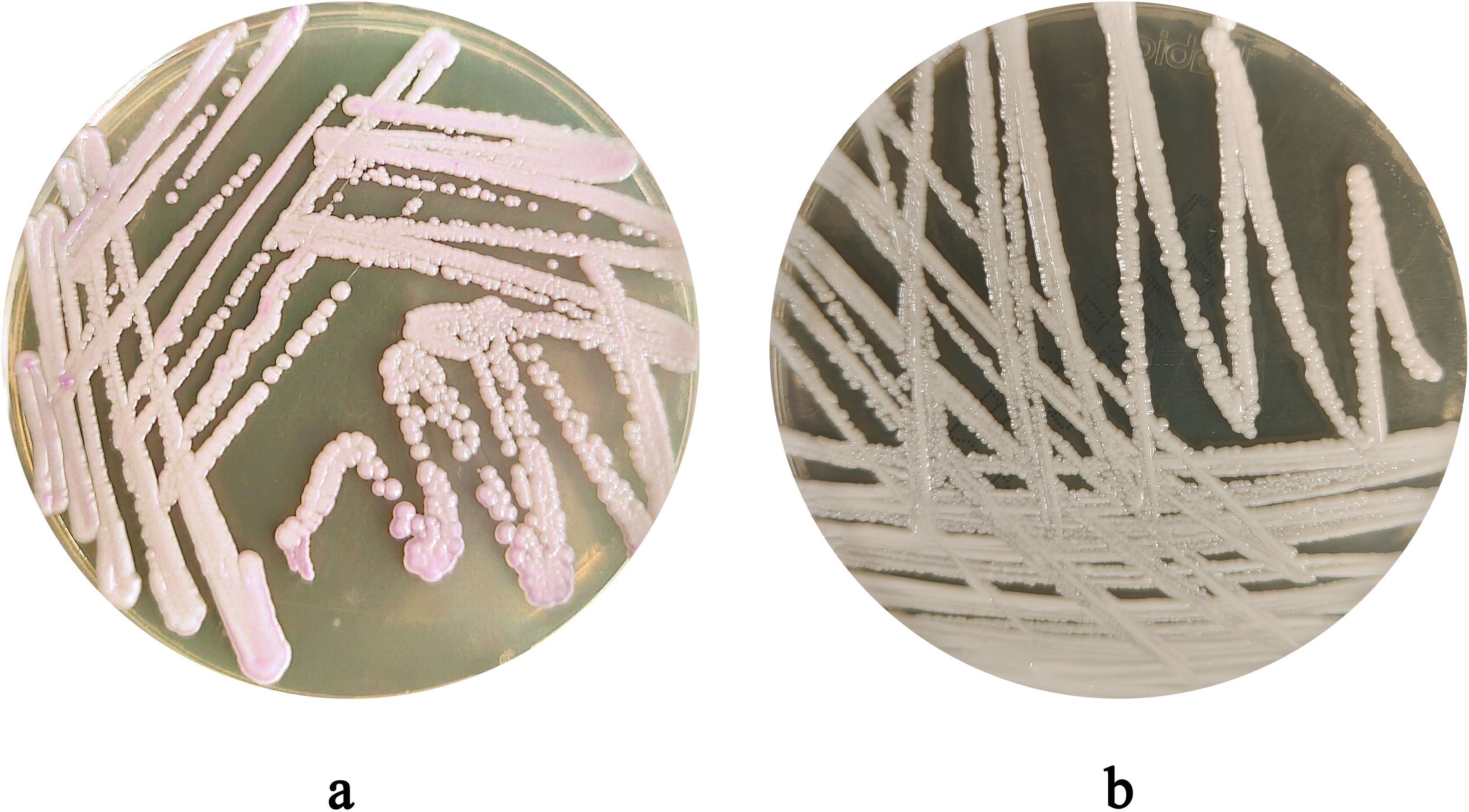
Figure 1. (a) Colony Morphology of C. auris strain CAS20503 Incubated on Sabouraud dextrose agar medium for 48 hours. (b) Colony Morphology of C. auris strain CAS20503 Incubated on CHROMagar Candida chromogenic media for 48 hours.
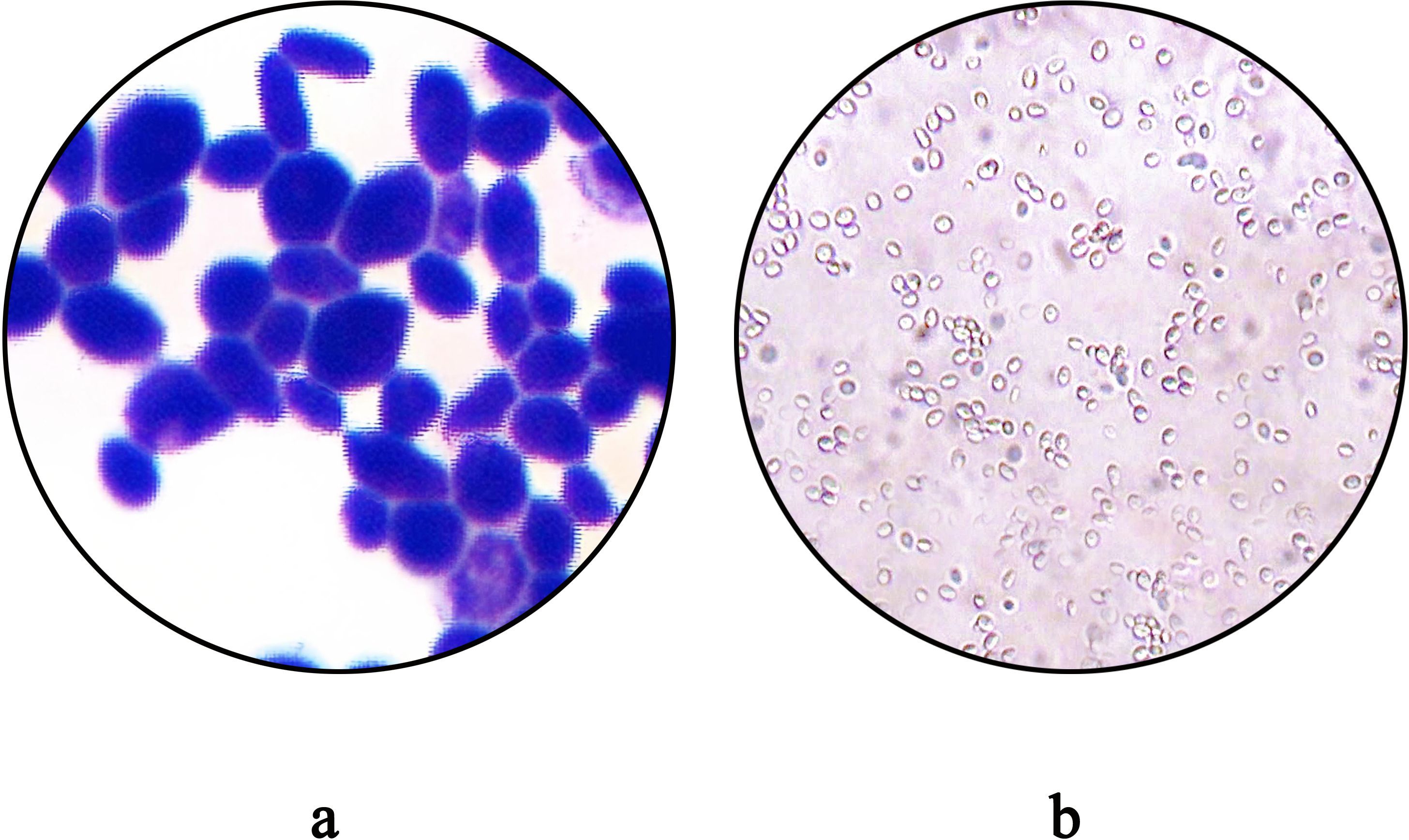
Figure 2. (a) Morphological Characteristics of C. auris strain CAS20503 Observed by Gram Staining Microscopy. (b) The microscopic characteristics of the wet mount of C. auris strain CAS20503 fungal colony suspension treated with 10% potassium hydroxide (KOH) were as follows.
3.2 Strain identification
MALDI-TOF MS analysis identified the isolate as C. auris with a high confidence score (99.9%) (Figure 3).
Figure 3. Identification of the Protein Fingerprint of C. auris strain CAS20503 Using MALDI-TOF MS. Peaks are visible across 2,000 to 12,000 m/z (Da) range, with significant peaks around 3,000 and 7,000 m/z, indicating protein masses.
3.3 Antifungal susceptibility profile
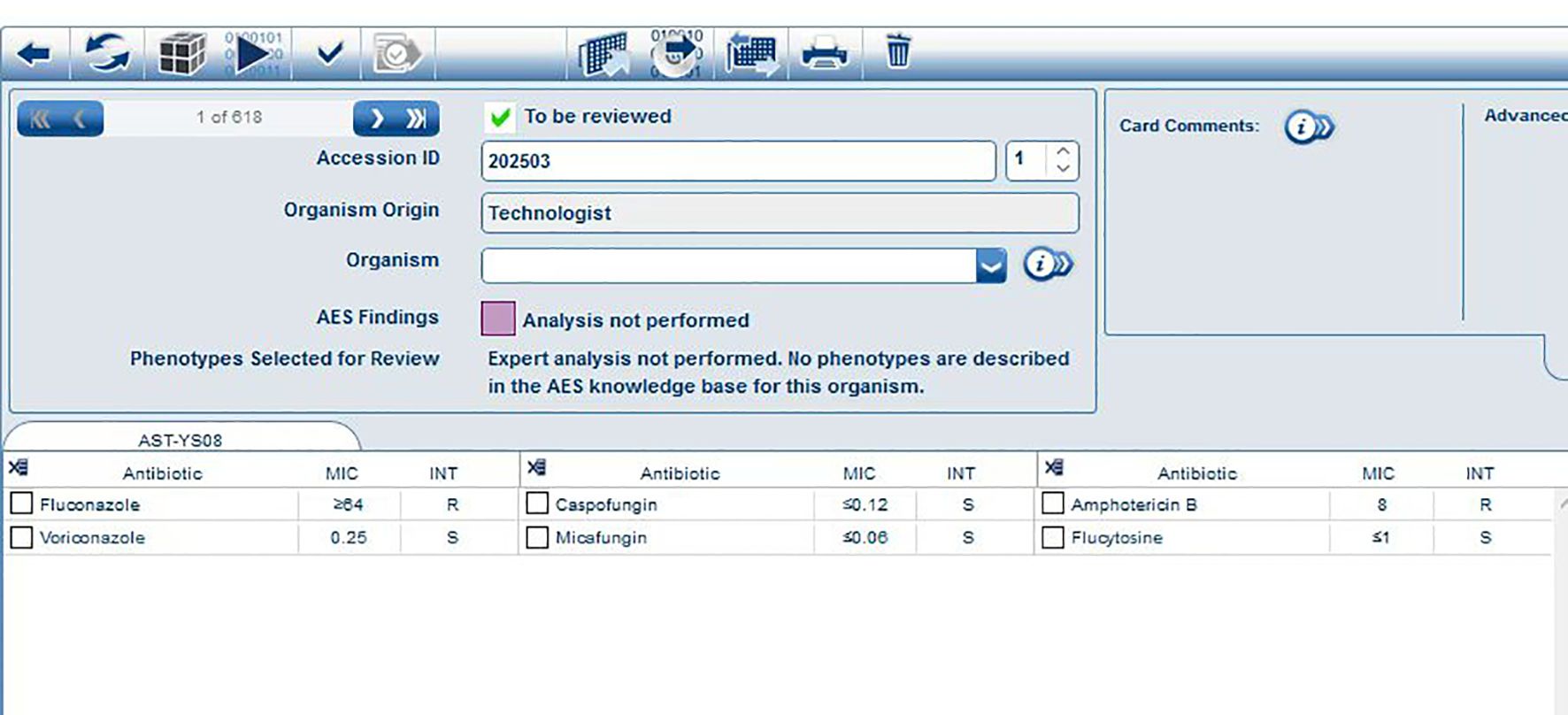
The fungal susceptibility test results demonstrated that the strain was resistant to fluconazole (MIC ≥ 64 μg/ml) and amphotericin B (MIC = 8.0 μg/ml), while being sensitive to voriconazole (MIC = 0.25 μg/ml), caspofungin (MIC ≤ 0.12 μg/ml), micafungin (MIC ≤ 0.06 μg/ml), and flucytosine (MIC ≤ 1 μg/ml) (Table 1). The results of the MIC determined automatically by the VITEK 2 COMPACT system were shown in Figure 4.
3.4 Resistance gene profiling
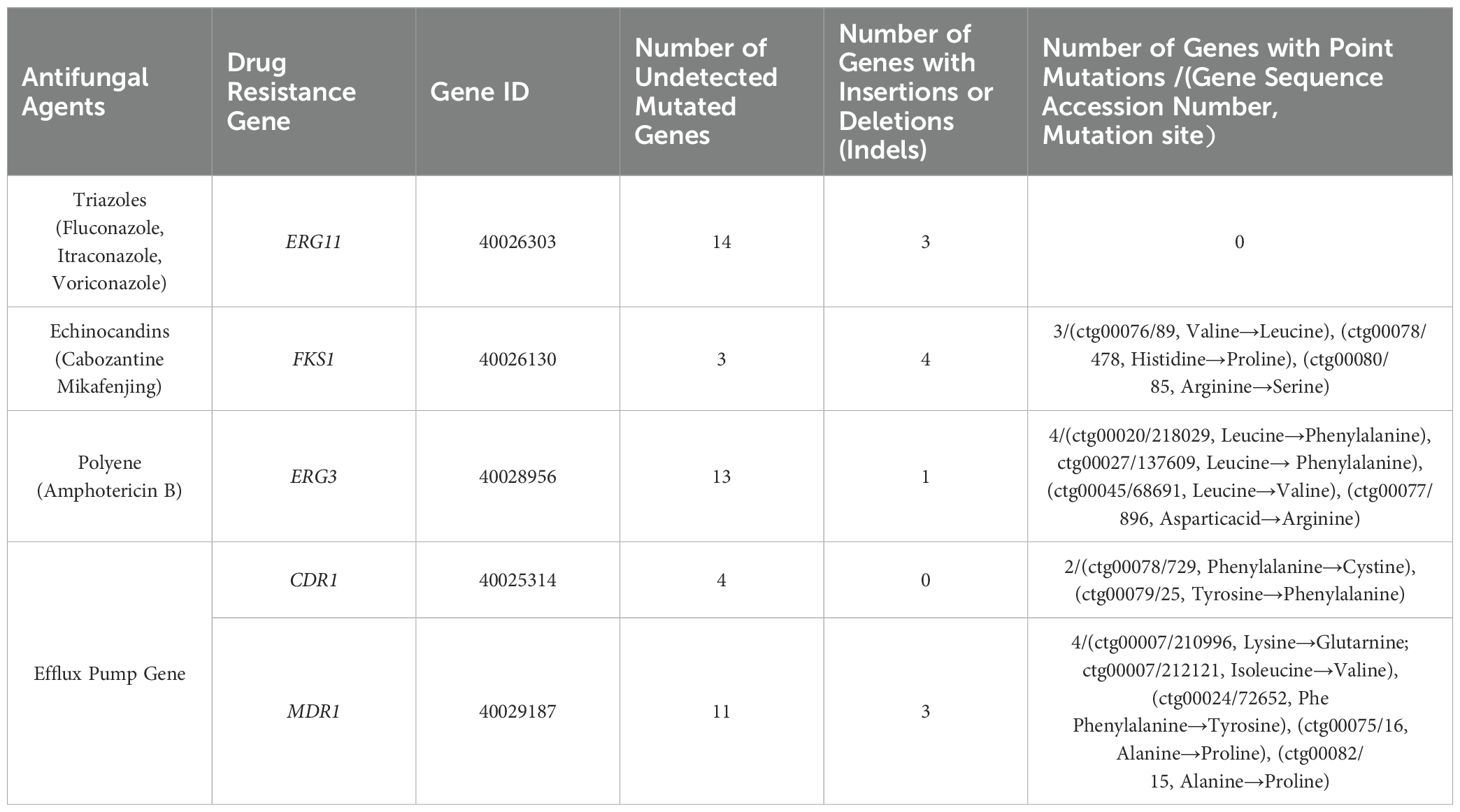
For ERG11, 14 genes showed complete matches with the reference gene’s nucleic acid sequence, while 3 exhibited insertion or deletion variations. For FKS1, 3 genes perfectly matched the reference sequence, 4 had insertion or deletion variations, and 3 contained point mutations. For ERG3, 13 genes perfectly matched the reference sequence, 1 showed insertion or deletion variations, and 4 had point mutations. For CDR1, 4 genes perfectly matched the reference sequence, with 2 containing point mutations. For MDR1, 11 genes perfectly matched the reference sequence, 3 exhibited insertion or deletion variations, and 4 contained point mutations (Table 2).
3.5 Virulence gene prediction
Through gene prediction and alignment analysis, this study identified 10 virulence genes that met the screening criteria (E-value = 0 and query coverage > 60%). Details of their encoded proteins and functions are as follows:
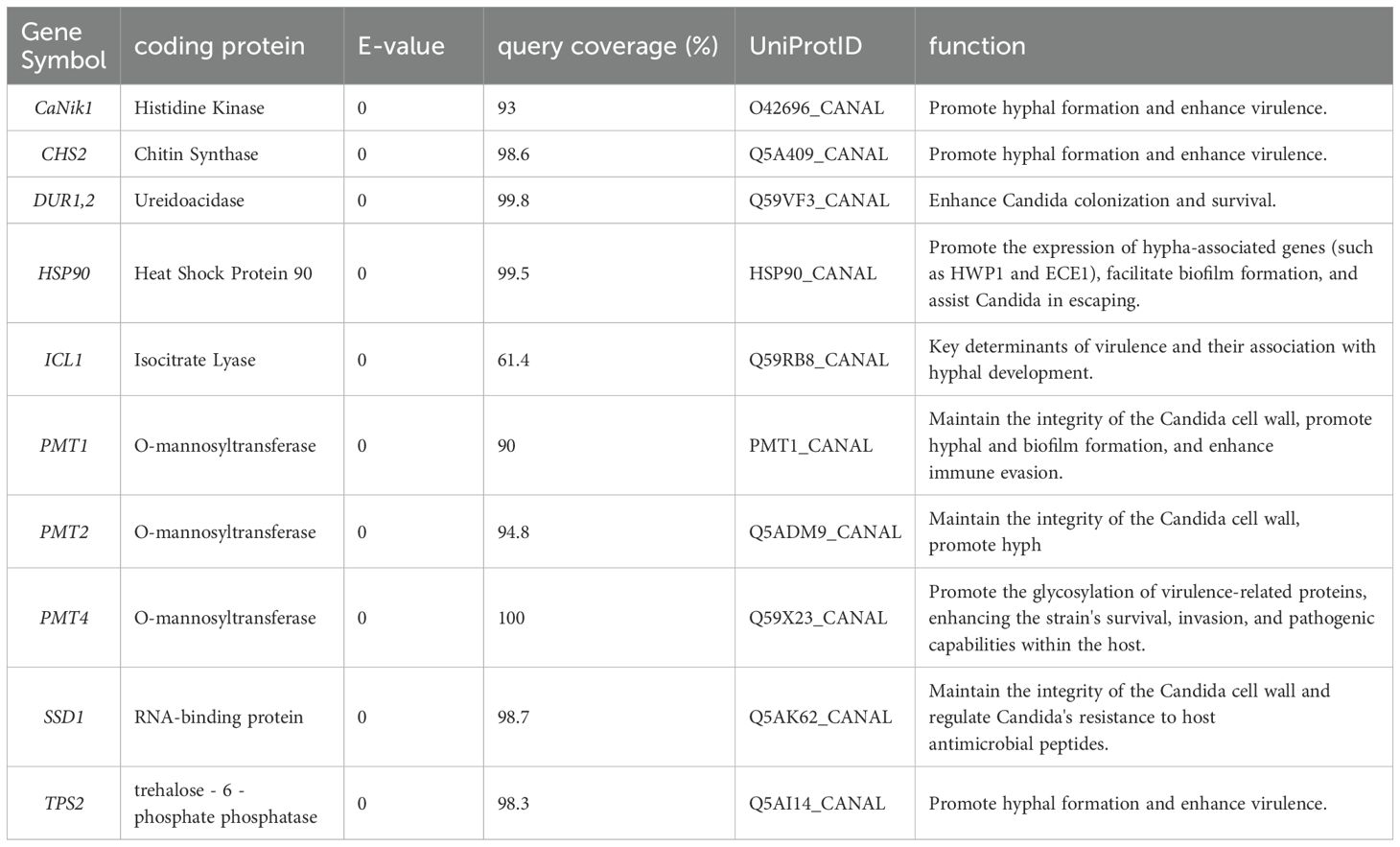
Genes related to pathogenicity: CaNik1 encoded histidine kinase, whose function was to promote hyphal formation and enhance virulence. CHS2 encoded chitin synthase, which could promote hyphal formation and enhance virulence. DUR1,2 encoded ureidoacidase, enabling Candida to enhance its colonization and survival abilities. HSP90 encoded heat shock protein 90, which could promote the expression of hypha-related genes (e.g., HWP1 and ECE1), contribute to biofilm formation, and assist Candida in immune evasion. ICL1 encoded isocitrate lyase, which was a key determinant of virulence and associated with hyphal development. PMT1 and PMT2 encoded O-mannosyltransferase, which could maintain the integrity of the Candida cell wall, promote the formation of hyphae and biofilms, and enhance immune evasion ability. PMT4 encoded O-mannosyltransferase, which could promote the glycosylation of virulence-related proteins and enhance the strain’s survival, invasion, and pathogenic abilities in the host. SSD1 encoded an RNA-binding protein, which could maintain the integrity of the Candida cell wall and regulate Candida’s resistance to host antimicrobial peptides. TPS2 encoded trehalose-6-phosphate phosphatase, which could promote hyphal formation and enhance virulence. The E-values, query coverage, and UniProt IDs of the above gene alignments were shown in Table 3.
3.6 Genes related to pathogenicity annotation
Based on the PHI-base database, a total of 6 pathogen-related genes were identified through screening with the criteria of E-value = 0 and query coverage > 95%. Among them, CaCHS1 encoded chitin synthase, ADE2 encoded phosphoribosylaminoimidazole carboxylase, FAS2 encoded fatty acid synthetase, PMR1 encoded a P-type ATPase transporter, CaTPS2 encoded trehalose-6-phosphate phosphatase, and Tfp1 encoded a putative subunit of V-ATPase. The coverage, PHI gene, and Protein ID corresponding to each gene were shown in Table 4.
3.7 Metabolic pathway annotation
The pathways of the CAS20503 isolate annotated by the KEGG database consisted of 6 branches, namely: Cellular Processes, Environmental Information Processing, Genetic Information Processing, Human Diseases, Metabolism, and Organic Systems. KEGG pathway analysis indicated that among the annotated human disease categories, genes associated with infectious disease pathways were the most prominently expressed (Figure 5).
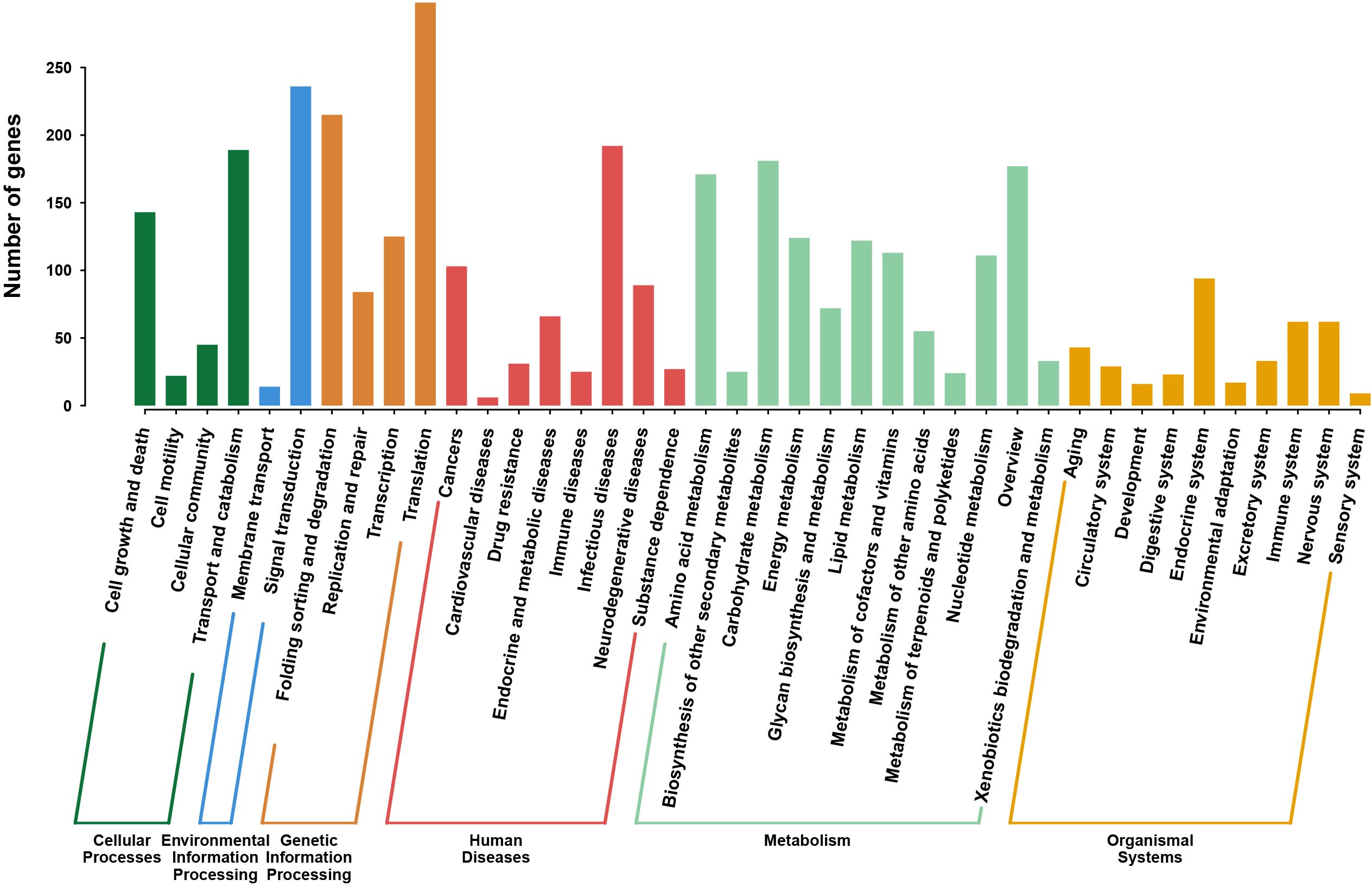
Figure 5. KEGG Pathway Classification of C. auris strain CAS20503. The pathway was composed of six branches. The vertical axis denotes the number of genes.The horizontal axis was categorized into six major functional branches in humans. Green: Cellular Processes, Dark Blue: Environmental Information Processing, Orange: Genetic Information Processing, Red: Human Diseases, Light Blue: Metabolism, Yellow: Organic Systems.
4 Phylogenetic analysis
Phylogenetic reconstruction based on 18S rRNA homologous sequences from the core genome revealed that the strain clustered closely with other C. auris strains. Notably, it showed the highest sequence similarity to the C. auris strain belonging to clade I (a subset of the South Asian clade) isolated in Italy in 2024 (GenBank accession number: CP157510.1) (Figure 6).
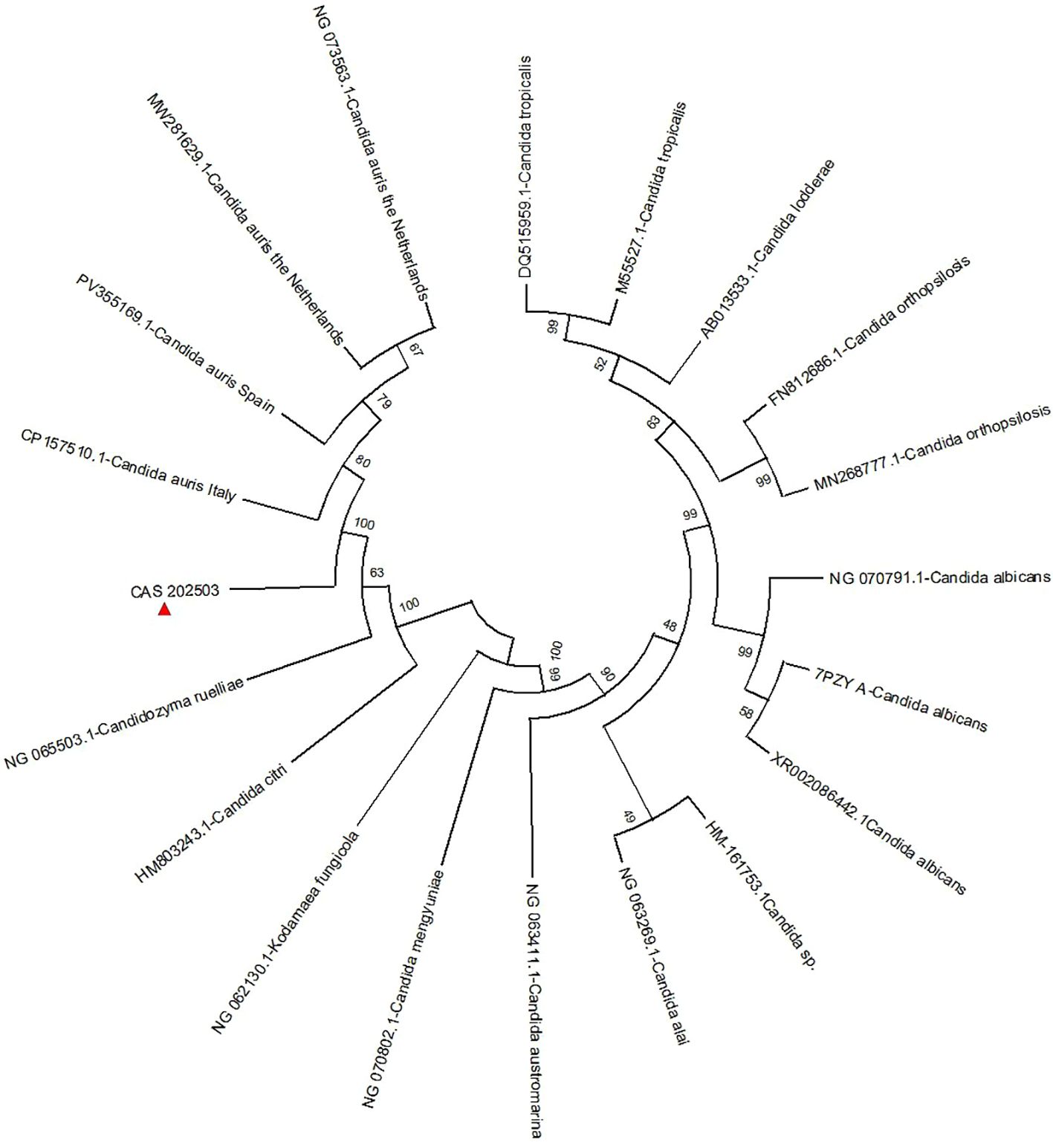
Figure 6. A phylogenetic tree was constructed through analysis of the 18S rRNA nucleotide sequence. The naming convention for strains in the figure follows the format: Assembly Accession ID + fungal name.The naming convention for C. auris strains includes the country of origin appended to the strain designation. The isolate CAS20503 is marked with a red triangle. Bootstrap values indicate branch support..
5 Discussion
An analysis was conducted on the high-risk factors for C. auris infection in the patients involved in this study. Candida species were one of the main pathogens associated with fungal infections, particularly susceptible to infecting immunocompromised patients, as well as those who had been hospitalized for a long time or undergone invasive medical procedures (Halioti et al., 2024). Although C. auris was an emerging Candida species, the risk factors for patients infected with it were similar to those for infections caused by other Candida species (Biagi et al., 2019; Bradley, 2019). These factors included: advanced age; presence of indwelling medical devices (such as central venous catheters or urinary catheters); specific comorbidities (including diabetes mellitus, neoplastic diseases, and chronic kidney disease); receipt of total parenteral nutrition; dependence on mechanical ventilation; and receipt of hemodialysis (Biran et al., 2023). In addition, it had been documented that invasive infections could occur within 48 hours after patients were admitted to the intensive care unit (ICU) (Jeffery-Smith et al., 2017). In this study, the CAS20503 strain was isolated from the urine of an elderly male patient with multiple underlying diseases, including diabetes mellitus, hypertension, and renal insufficiency. The patient was admitted to the hospital due to knee joint effusion, and later transferred to the ICU due to acute respiratory failure. Five days later, C. auris was detected after urine culture was performed due to symptoms of fever and a significant decrease in urine output. The patient had been receiving long-term hemodialysis treatment due to renal insufficiency. Additionally, after being transferred to the ICU, he had a history of invasive procedures such as central venous catheterization, cystocentesis, endotracheal intubation, and ventilator support. These factors were likely the causes of the fungal infection in this patient (Rossow et al., 2021). Our findings highlight that clinicians should enhance fungal culture and surveillance for infected patients with underlying diseases, undergoing invasive treatment, and exhibiting poor response to antifungal agents, so as to avoid missed diagnosis.
In terms of identification, after 48 hours of culture on Sabouraud dextrose agar medium, the CAS20503 isolate formed smooth, milky white colonies. On CHROMagar Candida chromogenic medium, it formed pink, smooth, and moist colonies, which was consistent with literature reports (Cortegiani et al., 2018). However, these morphological characteristics of C. auris were similar to those exhibited by other common Candida species, so in vitro morphological evaluation of colonies alone was insufficient for accurate laboratory identification. Studies have reported that the VITEK 2 COMPACT system may produce false-negative results due to missed detection (Ambaraghassi et al., 2019). Since traditional morphological observation and biochemical identification methods often struggled to accurately identify C. auris, we were able to accurately and definitively identify C. auris through MALDI-TOF MS combined with genome sequencing technology. Therefore, for difficult or rare bacteria, especially in cases where traditional methods could not distinguish them from closely related Candida species, molecular detection technology was recommended (Satoh et al., 2009).
In the fungal drug susceptibility testing, the C. auris strain CAS20503 exhibited resistance to fluconazole and amphotericin B, and susceptibility to voriconazole, caspofungin, micafungin, and flucytosine. This was consistent with the known multidrug-resistant profile of C. auris. In the alignment of whole-genome nucleic acid sequences, key antifungal resistance genes were identified, including ERG11, FKS1, ERG3, CDR1, and MDR1. Among them, ERG11 was associated with resistance to triazole drugs; FKS1 was related to resistance to echinocandin drugs; ERG3 was linked to resistance to polyene antibiotics; and CDR1 and MDR1, which belonged to efflux pump genes, were involved in the active pumping of intracellular substances such as antibiotics out of the cell by pathogens, thereby leading to drug resistance. In addition, these genes harbored insertions, deletions, and point mutations, which might collectively contribute to the development of drug-resistant phenotypes. For instance, in the alignment of the ERG11 gene, 3 insertion or deletion variations were identified. Such mutations altered the structure of lanosterol 14α-demethylase, reduced the binding capacity of triazole drugs, and thereby conferred fluconazole resistance in the strain (Jangir et al., 2023). The research results revealed that the ERG3 gene contained 1 insertion or deletion variation and 4 point mutations; these mutations might affect ergosterol biosynthesis and impair the efficacy of amphotericin B by altering cell membrane integrity (Rybak et al., 2022; Carolus et al., 2024). The FKS1 gene carried 4 insertion or deletion variations and 3 point mutations; these variations and mutations might disrupt the active site of 1,3-β-D-glucan synthase, which was further associated with decreased sensitivity to echinocandin drugs (Frras-De-LeLe et al., 2020; Lee et al., 2021). Moreover, mutations in the efflux pump genes CDR1 (with 2 point mutations) and MDR1 (with 3 insertion or deletion variations and 4 point mutations) might further enhance antifungal resistance (Li et al., 2024a; Li et al., 2024b).
Pathogenic fungi could cause a spectrum of diseases, ranging from superficial cutaneous and mucosal infections to deep-seated tissue and organ infections. In severe instances, such infections could result in multiple organ failure and even death (Denning, 2024). Through analysis of the virulence genes of the CAS20503 strain using the DFVF database, virulence-associated genes were identified. These genes might have exerted a synergistic effect during human infection by regulating key processes of C. auris, including morphological transformation, host colonization, immune evasion, and environmental adaptation, thereby significantly augmenting its pathogenicity and the persistence of infection. Among them, CaNik1, CHS2, TPS2, and ICL1 synergistically promoted hyphal elongation, helping Candida penetrate host epithelial tissues and spread to the urinary system. DUR1,2 combined with PMT4 could jointly promote the colonization of Candida on the surface of the urethral mucosa. HSP90 regulated the expression of biofilm-related genes to promote the synthesis of biofilm matrix and maintain the structural integrity of biofilms. It could synergize with PMT1 and PMT2 to enhance the ability of Candida to form biofilms on the surface of medical devices (such as urinary catheters and central venous catheters).
Based on the PHI-base database, a total of 6 pathogenicity-related genes were identified through the screening criteria of E-value = 0 and query coverage > 95%. They were CaCHS1, ADE2, FAS2, PMR1, CaTPS2, and Tfp1. During the process of Candida infecting the host, these 6 genes synergistically participated in the structural maintenance, substance metabolism, environmental adaptation, and virulence regulation of Candida. They enhanced its abilities of host colonization, tissue invasion, immune evasion, and persistent infection from multiple dimensions such as structural stability, metabolic support, and stress adaptation, and were important molecular bases for the pathogenic mechanism. KEGG pathway analysis showed that genes related to the metabolism of infectious diseases were enriched, indicating that the strain might have enhanced its own survival ability through specific metabolic adaptations during the process of host infection.
The drug resistance genes, virulence genes, and pathogenic genes of C. auris might have acted synergistically through a complex regulatory network to enhance its infectivity. Their drug resistance genes could help the strain resist clinical therapeutic interventions, virulence genes could improve its environmental adaptability and immune evasion ability, and pathogenic genes could provide guarantees for its survival and proliferation in the host. However, existing studies remained limited to the level of gene identification and functional speculation. Therefore, it was necessary to carry out C. auris-specific gene knockout or overexpression experiments to verify the actual functions of drug resistance genes and virulence genes.
In the treatment of candidal urinary tract infections, fluconazole was the first-choice drug (Escande4 et al., 2019). However, the resistance rate of C. auris to fluconazole was as high as 90%, and to voriconazole was 73% (Rudramurthy et al., 2017), making triazole drugs unsuitable for empirical treatment. Although the resistance rate of C. auris to echinocandins was lower than that to triazoles, resistance remained relatively common (Chowdhary et al., 2018). Literature reported that the resistance rate of C. auris isolates to amphotericin B was as high as 35% (Park et al., 2013). Given that C. auris strains might develop resistance to triazoles, echinocandins, and polyenes, it was necessary to consider alternative drugs in the treatment of urinary tract infections caused by C. auris. flucytosine was an attractive alternative. Compared with commonly used antifungal drugs, it was mainly excreted into urine in its active form, and according to existing studies, primary resistance of C. auris to flucytosine was uncommon (Charlier et al., 2015). The use of flucytosine in multidrug-resistant C. auris strains was consistent with current treatment guidelines for Candida glabrata (Pappas et al., 2016). The C. auris CAS20503 isolate in this study was susceptible to flucytosine, and flucytosine could be used for the treatment of urinary tract infection in this patient. In addition, combination therapy regimens had been used in multiple case reports for the treatment of C. auris, including echinocandins combined with amphotericin B, triazoles combined with echinocandins, or flucytosine combined with amphotericin B (Biagi et al., 2019; Bidaud et al., 2019; Ostrowsky et al., 2020). In the drug susceptibility testing of the CAS20503 isolate, the strain showed resistance to the triazole drug fluconazole and the polyene drug amphotericin B, which was consistent with the literature reports. Although the literature reported that the resistance rate of C. auris to voriconazole reached 73% (Rudramurthy et al., 2017), the isolate in this study was susceptible to voriconazole. Although the literature reported that resistance of C. auris to echinocandins was relatively common (Chowdhary et al., 2018), the CAS20503 isolate was susceptible to this class of antifungal drugs. Based on the results of fungal drug susceptibility testing and the treatment guidelines in the literature, clinicians selected caspofungin combined with flucytosine for treatment. Three weeks after treatment, the fungal culture turned negative, which demonstrated the clinical application value of individualized antifungal susceptibility testing. It had been pointed out in the literature that after the discontinuation of antifungal treatment, weekly cultures should be continued to guide the termination of infection control measures. Two negative culture results at one-week intervals were sufficient as the basis for terminating infection control measures (Griffith and Danziger, 2020). For the treatment of the CAS20503 isolate, clinical antifungal treatment and fungal monitoring were stopped after the fungal culture turned negative detected at three weeks of antifungal treatment. Therefore, there might be deficiencies in the continuous monitoring of fungi in clinical departments.
The genetic relatedness of C. auris strains could be assessed by multiple methods, including multi-locus sequence typing (MLST), proteomic analysis based on MALDI-TOF MS, and WGS (Girard et al., 2016). WGS remained the most advanced method for determining the clonality of C. auris strains (Hata et al., 2020; Kappel et al., 2024). Phylogenetic analysis based on 18S rRNA sequences in this study showed that the CAS20503 strain was genetically close to the C. auris strain with GenBank accession number CP157510.1, which was isolated in Italy in 2024 and belonged to clade I (a subset of the South Asian clade). The Italian isolate was a bloodstream infection isolate. Its drug resistance phenotype was identical to that of the CAS20503 isolate in that it was resistant to fluconazole and amphotericin B, and sensitive to caspofungin and micafungin. The difference was that the Italian isolate was resistant to flucytosine, while the CAS20503 isolate was sensitive to this drug. Both Candida auris isolates had mutations in the resistance genes ERG11, ERG4, and CDR1. The Italian isolate had the FCY1 gene mutation associated with flucytosine resistance (Ben Abid et al., 2023), whereas no such mutated gene was identified in the CAS20503 isolate through alignment. This might have been the main reason why the isolate was sensitive to flucytosine.
Given the high pathogenicity and transmissibility of C. auris, it was advisable to strengthen hospital infection control measures (Aldejohann et al., 2022). After the strain was isolated, as it was the first time such a highly transmissible fungus had been found in the hospital, it attracted strong attention from physicians in the hospital infection control department. They required that the patient be placed in a single room and that standard contact precautions be implemented. All medical staff in contact with the patient were required to strictly follow hand hygiene protocols. Hospital infection control doctors regularly performed swab tests on the patient’s axillae, inguinal regions, and previous infection sites. Various surfaces in the ward were appropriately disinfected, including bedside tables, bed rails, windowsills, blood glucose meters, blood pressure cuff sleeves, crash carts, and nursing carts. In addition, the patient’s previous roommates and medical staff who had long-term contact with the patient also underwent swab tests to prevent the spread of infection to other susceptible patients.
6 Conclusion
An analysis of the C. auris strain CAS20503 in this study showed that its infection was closely associated with high-risk factors such as advanced age, underlying diseases, and invasive procedures, and clinicians needed to strengthen monitoring of such populations. Traditional methods struggled to achieve accurate identification, requiring a combination of MALDI-TOF MS and whole-genome sequencing. The strain was resistant to fluconazole and amphotericin B, while susceptible to caspofungin and others. The drug resistance phenotype was related to mutations in genes such as ERG11 and FKS1, and the combined therapy guided by individualized drug susceptibility testing showed significant efficacy. Virulence and pathogenic genes might have synergistically enhanced infectivity by regulating colonization and biofilm formation, but this speculation still required further verification. The strain was genetically closely related to an Italian strain belonging to the South Asian clade, and they showed differences in susceptibility to flucytosine due to variations in the FCY1 gene mutation. Although infection control measures such as single-room isolation were implemented, there were deficiencies in continuous clinical monitoring after treatment. In summary, this study provided references for the diagnosis, treatment, research on drug resistance mechanisms, and prevention and control of C. auris, highlighting the importance of accurate identification, individualized treatment, and systematic monitoring.
7 Study limitations
This study was based solely on the analysis of a single strain with a limited sample size. Furthermore, the functional speculation about drug resistance, virulence, and pathogenic genes was mainly based on literature reports on studies of Candida albicans. For the homologous genes identified in C. auris that share sequence similarity with these genes, we indeed only confirmed their existence through sequence alignment, and had not yet experimentally verified whether their functions were consistent with those of the homologous genes in Candida albicans. In view of this, it will be necessary for us to expand the sample size in the future and list “functional verification of genes in C. auris” as an important future research direction, so as to clarify their specific roles in the pathogenic process of this strain.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving humans were approved by The Ethics Committee for Scientific Research and New Technologies of the First Affiliated Hospital of Wannan Medical College. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JL: Writing – original draft. ZW: Data curation, Writing – original draft. RZ: Funding acquisition, Writing – original draft. YW: Funding acquisition, Writing – original draft. XG: Formal analysis, Writing – original draft. XL: Conceptualization, Writing – original draft. PZ: Funding acquisition, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by the Postgraduate Education Quality Project of Education Department in Anhui Province (No.2024szsfkc126) and the Youth Foundation of Wannan Medical College (WK2023ZQNZ49 and WK2022F18).
Acknowledgments
We sincerely appreciate the technical support provided by Sangon Biotech (Shanghai, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldejohann, A. M., Wiese-Posselt, M., Gastmeier, P., and Kurzai, O. (2022). Expert recommendations for prevention and management of Candida auris transmission. Mycoses 65, 590–598. doi: 10.1111/myc.13445
Ambaraghassi, G., Dufresne, P. J., Dufresne, S. F., Vallisnea, É, Muñoz, J. F., Cuomo, C. A., et al. (2019). Identification of Candida auris by use of the updated vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J. Clin. Microbiol. 57(11):e00884-19.
Ben Abid, F., Salah, H., Sundararaju, S., Dalil, L., Abdelwahab, A. H., Salameh, S., et al. (2023). Molecular characterization of Candida auris outbreak isolates in Qatar from patients with COVID-19 reveals the emergence of isolates resistant to three classes of antifungal drugs. Clin. Microbiol. Infect. 29, 1083.e1–1083.e7. doi: 10.1016/j.cmi.2023.04.025
Biagi, M. J., Wiederhold, N. P., Gibas, C., Wickes, B. L., Lozano, V., Bleasdale, S. C., et al. (2019). Development of high-level echinocandin resistance in a patient with recurrent Candida auris candidemia secondary to chronic Candiduria. Open Forum Infect. Dis. 6, ofz262. doi: 10.1093/ofid/ofz262
Bidaud, A. L., Botterel, F., Chowdhary, A., and Dannaoui, E. (2019). In vitro antifungal combination of flucytosine with amphotericin B, voriconazole, or micafungin against Candida auris shows no antagonism. Antimicrob. Agents Chemother. 63, e01393–e01319. doi: 10.1128/AAC.01393-19
Biran, R., Cohen, R., Finn, T., Brosh-Nissimov, T., Rahav, G., Yahav, D., et al. (2023). Nationwide outbreak of Candida auris infections driven by COVID-19 hospitalizations, Israel, 2021-2022. Emerg. Infect. Dis. 29, 1297–1301. doi: 10.3201/eid2907.221888
Bradley, S. F. (2019). What is known about Candida auris. JAMA 322, 1510–1511. doi: 10.1001/jama.2019.13843
Carolus, H., Sofras, D., Boccarella, G., Sephton-Clark, P., Biriukov, V., Cauldron, N. C., et al. (2024). Acquired amphotericin B resistance leads to fitness trade-offs that can be mitigated by compensatory evolution in Candida auris. Nat. Microbiol. 9, 3304–3320. doi: 10.1038/s41564-024-01854-z
Centers for Disease Control and Prevention C. auris Screening: Patient Swab Collection. Available online at: https://www.cdc.gov/candida-auris/hcp/screening-hcp/c-auris-screening-patient-swab-collection-1.html (Accessed July 14, 2025).
Charlier, C., El Sissy, C., Bachelier-Bassi, S., Scemla, A., Quesne, G., Sitterle, E., et al. (2015). Acquired flucytosine Resistance during Combination Therapy with Caspofungin and flucytosine for Candida glabrata Cystitis. Antimicrob. Agents Chemother. 60, 662–665. doi: 10.1128/AAC.02265-15
Chowdhary, A., Prakash, A., Sharma, C., Kordalewska, M., Kumar, A., Sarma, S., et al. (2018). A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899. doi: 10.1093/jac/dkx480
Cortegiani, A., Misseri, G., Fasciana, T., Giammanco, A., Giarratano, A., and Chowdhary, A. (2018). Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 6, 69. doi: 10.1186/s40560-018-0342-4
Denning, D. W. (2024). Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438. doi: 10.1016/S1473-3099(23)00692-8
Escande4, P., Chow, N. A., Caceres, D. H., Gade, L., Berkow, E. L., Armstrong, P., et al. (2019). Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin. Infect. Dis. 68, 15–21. doi: 10.1093/cid/ciy411
Fazeli, A., Kordbacheh, P., Nazari, A., Daie Ghazvini, R., Mirhendi, H., Safara, M., et al. (2019). Candiduria in hospitalized patients and identification of isolated candida species by morphological and molecular methods in Ilam, Iran. Iran J. Public Health 48, 156–161.
Frías-De-León, M. G., Hernández-Castro, R., Vite-Garín , T., Arenas, R., Bonifaz, A., Castañzr-Olivares, L., et al. (2020). Antifungal resistance in Candida auris: molecular determinants. Antibiotics (Basel) 9(9):568. doi: 10.3390/antibiotics9090568
Girard, V., Mailler, S., Chetry, M., Vidal, C., Durand, G., van Belkum, A., et al. (2016). Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59, 535–538. doi: 10.1111/myc.12519
Griffith, N. and Danziger, L. (2020). Candida auris urinary tract infections and possible treatment. Antibiotics (Basel). 9, 898. doi: 10.3390/antibiotics9120898
Halioti, A., Vrettou, C. S., Neromyliotis, E., Gavrielatou, E., Sarri, A., Psaroudaki, Z., et al. (2024). Cerebrospinal drain infection by Candida auris: A case report and review of the literature. J. Fungi (Basel). 10, 859. doi: 10.3390/jof10120859
Hata, D. J., Humphries, R., and Lockhart, S. R. (2020). Candida auris: an emerging yeast pathogen posing distinct challenges for laboratory diagnostics, treatment, and infection prevention. Arch. Pathol. Lab. Med. 144, 107–114. doi: 10.5858/arpa.2018-0508-RA
Jangir, P., Kalra, S., Tanwar, S., and Bari, V. K. (2023). Azole resistance in Candida auris: mechanisms and combinatorial therapy. Apmis 131, 442–462. doi: 10.1111/apm.13336
Jeffery-Smith, A., Taori, S. K., Schelenz, S., Jeffery, K., Johnson, E. M., Borman, A., et al. (2017). Candida auris: a review of the literature. Clin. Microbiol. Rev. 31(1), e00029–e00017. doi: 10.1128/CMR.00029-17
Kappel, D., Gifford, H., Brackin, A., Abdolrasouli, A., Eyre, D. W., Jeffery, K., et al. (2024). Genomic epidemiology describes introduction and outbreaks of antifungal drug-resistant Candida auris. NPJ Antimicrob. Resist. 2, 26. doi: 10.1038/s44259-024-00043-6
Kathuria, S., Singh, P. K., Sharma, C., Prakash, A., Masih, A., Kumar, A., et al. (2015). Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 53, 1823–1830. doi: 10.1128/JCM.00367-15
Khatamzas, E., Madder, H., and Jeffery, K. (2019). Neurosurgical device-associated infections due to Candida auris - three cases from a single tertiary center. J. Infect. 78, 409–421. doi: 10.1016/j.jinf.2019.02.004
Kim, H. Y., Nguyen, T. A., Kidd, S., Chambers, J., Alastruey-Izquierdo, A., Shin, J. H., et al. (2024). Candida auris-a systematic review to inform the world health organization fungal priority pathogens list. Med. Mycol 62, myae042. doi: 10.1093/mmy/myae042
Kriegl, L., Egger, M., Boyer, J., Hoenigl, M., and Krause, R. (2025). New treatment options for critically important WHO fungal priority pathogens. Clin. Microbiol. Infect. 31, 922–930. doi: 10.1016/j.cmi.2024.03.006
La Bella, A. A., Santiago-Tirado, F. H., and Flores-Mireles, A. L. (2025). Candida auris is emerging as a prevalent urinary pathogen. PloS Pathog. 21, e1013138. doi: 10.1371/journal.ppat.1013138
Lee, Y., Puumala, E., Robbins, N., and Cowen, L. E. (2021). Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem. Rev. 121, 3390–3411. doi: 10.1021/acs.chemrev.0c00199
Li, J., Aubry, L., Brandalise, D., Coste, A. T., Sanglard, D., and Lamoth, F. (2024a). Upc2-mediated mechanisms of azole resistance in Candida auris. Microbiol. Spectr. 12, e0352623. doi: 10.1128/spectrum.03526-23
Li, J., Brandalise, D., Coste, A. T., Sanglard, D., and Lamoth, F. (2024b). Exploration of novel mechanisms of azole resistance in Candida auris. Antimicrob. Agents Chemother. 68, e0126524. doi: 10.1128/aac.01265-24
Ostrowsky, B., Greenko, J., Adams, E., Quinn, M., O’Brien, B., Chaturvedi, V., et al. (2020). C. auris Investigation Work Group. Candida auris Isolates Resistant to Three Classes of Antifungal Medications - New York, 2019. MMWR Morb Mortal Wkly Rep. 69, 6–9. doi: 10.15585/mmwr.mm6901a2
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 62, e1–50. doi: 10.1093/cid/civ933
Park, M., Do, E., and Jung, W. H. (2013). Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 41, 67–72. doi: 10.5941/MYCO.2013.41.2.67
Ramos, L. S., Barbosa, P. F., Lorentino, C. M. A., Lima, J. C., Braga, A. L., Lima, R. V., et al. (2025). The multidrug-resistant Candida auris, Candida haemulonii complex and phylogenetic related species: Insights into antifungal resistance mechanisms. Curr. Res. Microb. Sci. 8, 100354. doi: 10.1016/j.crmicr.2025.100354
Rossow, J., Ostrowsky, B., Adams, E., Greenko, J., McDonald, R., Vallabhaneni, S., et al. (2021). Factors Associated With Candida auris Colonization and Transmission in Skilled Nursing Facilities With Ventilator Units, New York, 2016-2018. Clin. Infect. Dis. 72, e753–e760. doi: 10.1093/cid/ciaa1462
Rudramurthy, S. M., Chakrabarti, A., Paul, R. A., Sood, P., Kaur, H., Capoor, M. R., et al. (2017). Candida auris candidaemia in Indian ICUs: analysis of risk factors. J. Antimicrob. Chemother. 72, 1794–1801. doi: 10.1093/jac/dkx034
Rybak, J. M., Barker, K. S., Muñoz, J. F., Parker, J. E., Ahmad, S., Mokaddas, E., et al. (2022). In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin. Microbiol. Infect. 28, 838–843. doi: 10.1016/j.cmi.2021.11.024
Satoh, K., Makimura, K., Hasumi, Y., Nishiyama, Y., Uchida, K., and Yamaguchi, H. (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53, 41–44. doi: 10.1111/j.1348-0421.2008.00083.x
Keywords: Candida auris, antifungal resistance mechanisms, genomic characterization, database, whole-genome sequencing
Citation: Li J, Wang Z, Zheng R, Wang Y, Guo X, Li X and Zhang P (2025) Identification, antifungal resistance, and genomic characterization of a single Candida auris isolate from urinary tract infection. Front. Cell. Infect. Microbiol. 15:1641542. doi: 10.3389/fcimb.2025.1641542
Received: 05 June 2025; Accepted: 08 August 2025;
Published: 10 September 2025.
Edited by:
Anis Rageh Al-Maleki, University of Malaya, MalaysiaReviewed by:
Andres Herrera Yela, SEK International University, EcuadorNadia Elsayed Ahmed Mohamed Omar Elkady, Ain Shams University, Egypt
Copyright © 2025 Li, Wang, Zheng, Wang, Guo, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhang, enAyMTY5QDE2My5jb20=
†These authors share first authorship
 Jie Li
Jie Li Ziheng Wang
Ziheng Wang Rui Zheng
Rui Zheng Peng Zhang
Peng Zhang