- 1School of Traditional Chinese Medicine, Hunan University of Chinese Medicine, Changsha, China
- 2Department of Endocrine, The First Hospital of Hunan University of Chinese Medicine, Changsha, China
- 3Department of Rehabilitation, Hunan Provincial People’s Hospital, Changsha, China
Human microbiota-associated (HMA) animal models have become indispensable tools for investigating microbe-host interactions and disease pathogenesis. However, standardization challenges persist across different research groups when such models are used in fecal microbiota transplantation (FMT) protocols. Establishing a successful HMA model involves multiple stages, including donor screening, fecal suspension preparation, recipient preparation, and FMT. The outcomes of these stages are influenced by donor characteristics, recipient type, microbial viability, and dietary factors. This review examined the critical components of HMA model production, including the inclusion and exclusion criteria for human donors, collection time and processing methodology for fecal samples, recipient animal preparation strategies, and FMT regimens with engraftment validation. The key findings revealed that short-term antibiotic, probiotic, or laxative use constitutes an essential donor exclusion criterion. The time and method of fecal collection should be standardized as much as possible. Fecal samples should be processed as soon as possible, in anaerobic environments, with the addition of suitable protectants if they must be preserved at low temperatures. Microbial community profiling via 16S rRNA gene sequencing represents the primary method for analyzing microbiome composition and verifying microbiota engraftment efficacy throughout FMT procedures. The most commonly used recipients for HMA modeling included germ-free and pseudo-germ-free animals generated through antibiotic-mediated microbiota depletion. Although FMT with a single gavage of fecal suspension proved sufficient for model establishment, multiple frequencies and longer FMT durations significantly improved the efficiency of donor microbiota colonization. Overall, these findings are expected to aid the establishment of a standardized and reproducible protocol for preparing HMA models.
1 Introduction
The microbiota constitutes a complex ecosystem of microorganisms that encompasses bacterial, archaeal, eukaryotic, and viral taxa, each occupying specific ecological niches (Marchesi and Ravel, 2015). These microorganisms demonstrate a ubiquitous natural distribution, with humans serving as one of their primary hosts. Long-term coevolution has cultivated mutualism between humans and their microbiota—particularly within the gastrointestinal tract, where ~95% of endogenous microbes reside. A 2010 metagenomic sequencing analysis revealed that the total human gut microbiome genome exceeds its genomic content by ~150× (Qin et al., 2010). As of 2019, researchers have identified nearly 2,000 novel microbial species in the human intestine (Almeida et al., 2019). Subsequent studies have estimated that the ratio of bacterial to human cells in the adult human body is approximately 1.3:1 (Sender et al., 2016). Recent advancements in multi-omics assay profiling have elucidated the important impact of the microbiome on host health and disease (Integrative HMP (iHMP) Research Network Consortium, 2014). The gut microbial consortium mediates essential physiological functions such as immunological homeostasis, colonization resistance against pathogens, energy metabolism, endocrine regulation, and even certain neurological functions (Lynch and Pedersen, 2016). Dysregulation of the microbial community and abnormalities involving its metabolites have been closely associated with a variety of chronic diseases, including inflammatory bowel disease (Mousa and Al Ali, 2024), certain neuromuscular pathologies (e.g., Alzheimer’s disease (Yang et al., 2024), certain muscular dystrophies (Russo et al., 2024)), metabolic syndromes (e.g., obesity and type 2 diabetes) (Aron-Wisnewsky et al., 2021), and dermatosis (e.g., acne and atopic dermatitis) (Borrego-Ruiz and Borrego, 2024).
The investigation of gut microbe-host interactions offers dual scientific value: elucidating disease mechanisms and pioneering novel diagnostic-therapeutic paradigms. Human microbiota-associated (HMA) animal models have emerged as crucial tools for elucidating the mechanisms underlying microbe-host interactions (Hirayama, 1999; Imaoka et al., 2004; Kibe et al., 2005). Through the transplantation of human microbial communities into recipient animals, HMA models facilitate the longitudinal observation of microbial dynamics or examination of the efficacy of specific therapeutic targets involved in certain interventions (Ridaura et al., 2013). Evidence has demonstrated that HMA models can effectively reconstruct donor microbial signatures and metabolomic profiles (Marcobal et al., 2013). Current applications span four key research domains: the composition of gut microbial consortia, the regulation of gut microbiota in host development, the causal associations between microbes and diseases, and the evaluation of targeted microbiota therapeutic strategies (Sharon et al., 2019). These findings solidify the functional centrality of intestinal microbiomes in terms of maintaining good health. They also provide a scientific basis for microbial interventions that target health benefits across human, animal, and ecological domains.
Despite their scientific utility, HMA animal models derived through fecal microbiota transplantation (FMT) face persistent methodological controversies. The engraftment efficiency of human-derived microbial communities in animal recipients is influenced by several factors. These include the host’s genetic background, gastrointestinal architecture, and behavioral differences—all of which impose certain constraints on HMA animal models (Arrieta et al., 2016). Evidence has indicated that these models risk overestimating the causal associations between microbiomes and disease phenotypes (Walter et al., 2020). Nevertheless, HMA models remain the best choice for investigating host-microbe crosstalk. It remains unclear precisely which methodological refinements in HMA model generation via FMT are required to establish standardized workflows that improve reproducibility and scientific validity. This review highlights key considerations in donor screening, recipient preparation, transplantation protocols, and microbiota validation to enhance HMA model development, experimental reproducibility, and standardization (Figure 1).

Figure 1. The General procedures of human microbiota-associated (HMA) mice models. Using mice as an example, the general procedures of HMA models primarily involves three steps. Donor Preparation: select human donors with balanced diet who meet predefined inclusion and exclusion criteria. Preservation and Processing of Donor Fecal: collect and transport fecal samples and store them under low-temperature conditions. Standardized fecal suspensions are prepared by diluting, homogenizing, filtering, and pooling fecal samples from multiple donors. Fecal Microbiota Transplantation (FMT): recipient mice are adult germ-free animals or antibiotic-induced pseudo-germ-free models. Following FMT, next-generation sequencing (NGS) is utilized as an effective method to quantify microbial engraftment efficiency.
2 Donor preparation
2.1 Inclusion and exclusion criteria for human donors
The 2017 European Consensus Conference established donor inclusion and exclusion criteria for clinical fecal microbiota transplantation (FMT), specifying evaluation parameters that included comprehensive medical histories, same-day donation, clinical signs and symptoms, dietary profiling, and laboratory tests (Cammarota et al., 2017). However, standardized protocols for selecting human fecal donors in animal experiments remain undefined, with significant differences remaining in terms of inclusion and exclusion criteria across studies. Current FMT-based human microbiota-associated (HMA) models predominantly use two donor cohorts: healthy individuals, and patients with the diseases being investigated by the study. The inclusion criteria for healthy individuals reported in existing studies mainly included the following aspects: (1) a minimum of 2–12 months without antibiotic exposure (Cherbuy et al., 2019; Kim et al., 2024; Le Bihan et al., 2015; Aluthge et al., 2020; Chung et al., 2012); (2) the elimination of laxative agents for ≥3 months (Brandi et al., 2024; Gérard et al., 2004; Respondek et al., 2013); (3) a omnivorous diet that includes both vegetarian and meat component (Cherbuy et al., 2019; Dong et al., 2021); and (4) the absence of gastrointestinal disorders (Reygner et al., 2020; Tamura et al., 2019; Gobert et al., 2016; Saint-Cyr et al., 2013), recent pathogen (bacterial or parasitic) infection (Lauko et al., 2023; Nagao-Kitamoto et al., 2020, 2016), and acute or chronic illnesses that can alter gut microbe composition (Salandre et al., 2023; Zabolotneva et al., 2023). The most common exclusion criteria included the following: (1) recent (within 1–2 months) exposure to antimicrobials, prebiotics, or probiotics (Zhang et al., 2023; Chen et al., 2020; Xia et al., 2019); (2) active neuropsychiatric disorders including major depression (Gobert et al., 2016; Kaiser et al., 2021; Zabolotneva et al., 2023; Zhan et al., 2024; Zhang et al., 2020); (3) excessive alcoholism or smoking habits (Zabolotneva et al., 2023; Grabrucker et al., 2023; Gobert et al., 2016); and (4) pregnant or lactating (Zabolotneva et al., 2023; Demir et al., 2022; Zhang et al., 2020; Gobert et al., 2016). The inclusion criteria for disease donors typically add the following requirements: clinical manifestations, laboratory tests, and pathological findings that collectively satisfy the diagnostic criteria for the disease (Zhang et al., 2023; Hsu et al., 2023; Fan et al., 2023; Demir et al., 2022; Zhang et al., 2020; Chen et al., 2020; Duan et al., 2019). Exclusion criteria often include: (1) incomplete information (Zhong et al., 2024); (2) the use of medications that could interfere with the experiment (Hutchison et al., 2024; Zhan et al., 2024; Zhong et al., 2024); and (3) comorbidities of chronic or infectious diseases that could affect the study (Hsu et al., 2023; Fan et al., 2023; Chen et al., 2020; Duan et al., 2019; Xia et al., 2019).
Antibiotic exposure and dietary patterns critically influence gut microbiota composition (Dudek-Wicher et al., 2018). Clinical trials have demonstrated that antibiotic administration reduces microbial diversity. It typically takes ≥1.5 months for the intestinal flora of healthy adults to return to near-baseline levels—with a few common taxa remaining undetectable even after 6 months (Palleja et al., 2018). Diet serves as the substrate for the energy used by microbes, with different microbial species differing in their ability to utilize different foods, resulting in different microbial compositions (Flint et al., 2015). Pharmacological interventions such as laxatives induce clearance of intestinal contents, directly altering the microbial community structure (Drago et al., 2019). Probiotic and prebiotic interventions selectively modulate enteric microbial populations, affecting their health-promoting effects (Sanders et al., 2019). Although evidence regarding the impact of alcohol and tobacco on the gut microbiota remains limited, current findings indicate that excessive alcohol consumption compromises intestinal barrier function and induces dysbiosis (Engen et al., 2015). Cigarette smoking can alter gut microbial composition and diversity through mechanisms involving oxidative stress modulation, the disruption of intestinal tight junctions, and changes in mucin composition (Savin et al., 2018). Current studies report significant variations in donor cohort sizes for FMT, ranging from single donors to multi-donor cohorts (n=1–10) across published protocols (Lauko et al., 2023; Hsu et al., 2023; Hanske et al., 2009; Sánchez-Quintero et al., 2022; Crouzet et al., 2013; Salandre et al., 2023; Mao et al., 2021; Chiu et al., 2017; Xia et al., 2019; Sun et al., 2022; Liu et al., 2023; von Klitzing et al., 2017a; Ye et al., 2023; Zhang et al., 2013; Aluthge et al., 2020; Reygner et al., 2020; Fan et al., 2023; Renu et al., 2022; Zabolotneva et al., 2023; Cherbuy et al., 2019) (Table 1). Research from the Human Microbiome Project has confirmed that significant heterogeneity exists in gut microbial compositions and relative abundances between individuals, even among healthy populations (Human Microbiome Project Consortium, 2012a). Although single-donor FMT ensures traceable microbial origins, it does not adequately address population-level microbial diversity. Conversely, multi-donor strategies enhance ecological validity through sample pooling but increase operational complexity in terms of specimen collection and processing.
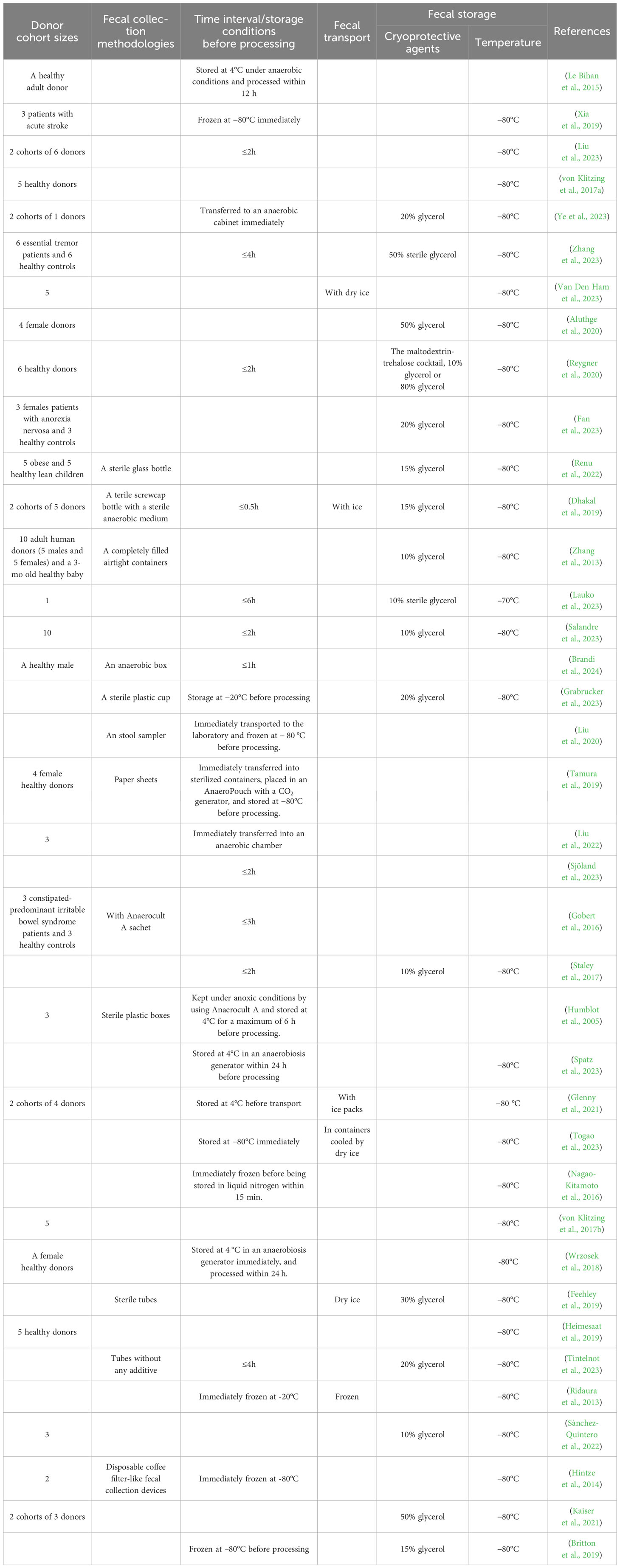
Table 1. Summary of methodological parameters for fecal sample cohort, collection, transport and storage in human microbiota-associated (HMA) studies.
Optimal donor selection for microbiota studies requires stringent criteria. Based on the above evidence, we believe that healthy donors must demonstrate at least: (1) A ≥3-month abstinence from antibiotics, laxatives, and probiotic or prebiotic supplements (Le Bihan et al., 2015; Aluthge et al., 2020; Chung et al., 2012; Brandi et al., 2024; Gérard et al., 2004; Respondek et al., 2013; Zhang et al., 2023; Chen et al., 2020; Xia et al., 2019); (2) the absence of gastrointestinal disorders or active infections (Reygner et al., 2020; Tamura et al., 2019; Gobert et al., 2016; Saint-Cyr et al., 2013; Lauko et al., 2023; Nagao-Kitamoto et al., 2016, 2020; Salandre et al., 2023; Zabolotneva et al., 2023); (3) adherence to a nutritionally balanced diet (Cherbuy et al., 2019; Dong et al., 2021); (4) A preference for non-smokers and non-drinkers (Zabolotneva et al., 2023; Grabrucker et al., 2023; Gobert et al., 2016); and (5) compliance with fecal collection protocols. Disease cohort donors require additional validation that includes: (1) diagnostic confirmation per established clinical criteria (Zhang et al., 2023; Hsu et al., 2023; Fan et al., 2023; Demir et al., 2022; Zhang et al., 2020; Chen et al., 2020; Duan et al., 2019); (2) the exclusion of confounding comorbidities that could affect gut microbiota (Hsu et al., 2023; Fan et al., 2023; Chen et al., 2020; Duan et al., 2019; Xia et al., 2019); (3) the absence of active infectious diseases (Staley et al., 2017). Fecal samples could be initially collected from multiple donors, after which a suitable number of optimal and representative specimens could be selected for downstream experiments (Fan et al., 2023; Gobert et al., 2016; Xia et al., 2019; Zhang et al., 2023).
2.2 Fecal collection
2.2.1 The time of fecal collection
Both humans and animals, along with their gut microbiotas, are affected by temporal rhythms. Research has demonstrated that 10% of the bacterial operational taxonomic units (OTUs) in humans and 15% of those in mice show significant circadian fluctuations in terms of relative abundance (Thaiss et al., 2014). Reitmeier et al. analyzed fecal samples from 1,943 participants with recorded collection times and revealed that 70% exhibited defecation patterns concentrated between the hours of 5:00–11:00 (Reitmeier et al., 2020). Throughout the day, distinct taxonomic groups dominate the gut microbiota. Firmicutes prevail during daylight hours, for example, whereas Bacteroidetes predominate nocturnally (Reitmeier et al., 2020). Current clinical FMT protocols lack standardized stool collection timing. In the preparation of animal models for FMT-based HMA animal model preparation, certain studies have utilized stool samples obtained from donors’ first morning bowel movements (Zhang et al., 2020; Mao et al., 2021; Sun et al., 2022).
2.2.2 The methodology of fecal collection
Current methodologies for fecal sample collection exhibit significant heterogeneity. Certain protocols require donors to defecate directly into an anaerobic box (Brandi et al., 2024; Gobert et al., 2016), while others use sterile containers or specialized stool collection kits (Hintze et al., 2014; Grabrucker et al., 2023; Liu et al., 2020). Alternative approaches involve paper sheets and immediately transferring them into sterilized containers (Tamura et al., 2019). Standardized collection tools, exemplified by stool collection kits, present three primary advantages. First, they minimize oxygen exposure to protect anaerobic taxa. Second, they prevent environmental contamination, such as from toilet water and urine. Third, they enhance donor compliance through improved hygienic handling and sensory acceptability. The commode kit has gained widespread adoption in large-scale cohort studies such as the Human Microbiome Project, owing to its user-friendly design (Franzosa et al., 2014; Human Microbiome Project Consortium, 2012b). Despite achieving operational simplicity and cost optimization, these systems require detailed instructional protocols and incur additional research expenditures. Conversely, evidence demonstrates that paper-based collection methods preserve fecal microbial diversity and community structure without significant alteration (Al et al., 2018), offering a viable alternative for resource-constrained investigations.
2.2.3 Time interval and storage conditions before processing
For fresh samples, the clinical FMT protocols emphasize that the primary recommendation is to process them within 6 h (Cammarota et al., 2017; Lopetuso et al., 2023). Fecal samples should be stored at a temperature of 20°C–30°C (Cammarota et al., 2017) or at ≤4°C prior to processing (Lopetuso et al., 2023). If feasible, anaerobic storage and processing should be utilized (Cammarota et al., 2017). Similar protocols apply to FMT-based HMA animal model preparation: in some studies, samples were required to be processed in an anaerobic chamber immediately after defecation (Tamura et al., 2019; Ye et al., 2023; Liu et al., 2022; Wrzosek et al., 2018). Consistent with clinical FMT protocols, some studies require microbial slurry extraction and FMT administration to be completed within 2–6 hours post-collection (Sjöland et al., 2023; Gobert et al., 2016; Staley et al., 2017; Zhang et al., 2023; Lauko et al., 2023). When immediate processing is unfeasible, stool samples were stored under anoxic conditions at 4°C for a maximum 6–24 h (Humblot et al., 2005; Wrzosek et al., 2018; Le Bihan et al., 2015; Spatz et al., 2023)(Table 1). These preservation measures aim to maintain donor microbial viability (MV) and protect obligate anaerobes, which outnumber aerobic bacteria by 100–1000× in the human gut (Widjaja and Rietjens, 2023). Lower anaerobe abundance has been reported to correlate with dysbiosis-associated pathologies such as irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) (Rajilić-Stojanović et al., 2011; Sokol et al., 2009). Insufficient anaerobic protection may therefore compromise experimental outcomes through microbial community variation. However, clinical evidence demonstrates comparable efficacy between anaerobic and aerobic FMT preparations when treating Clostridioides difficile infections (Lee et al., 2016). This equivalence may stem from spore-forming bacterial genera, which constitute 50–60% of healthy gut microbiota and exhibit oxygen-resistant sporulation capabilities and thus facilitate inter-host transmission (Browne et al., 2016).
In summary, it is imperative for researchers to meticulously record the precise defecation times of participants and to prioritize the collection of fecal samples from the same timeframe in order to mitigate potential confounding variables associated with circadian rhythms (Thaiss et al., 2014; Reitmeier et al., 2020; Zhang et al., 2020; Mao et al., 2021; Sun et al., 2022). The optimal collection methodology should be selected based on donor cohort size and degree of cooperation, with a standardized sampling methodology maintained to minimize technical variability. Ideally, fresh fecal samples should be processed within 2 h of collection, with a maximum allowable delay of 6 h (Sjöland et al., 2023; Gobert et al., 2016; Staley et al., 2017; Zhang et al., 2023; Lauko et al., 2023). In instances where immediate processing is not feasible, it is advisable to refrigerate the samples at 4°C (Humblot et al., 2005; Spatz et al., 2023). The adoption of anaerobic preservation and processing protocols should be guided by the available laboratory resources and the specific aims of the research. These findings provide preliminary insights into fecal collection and processing methods, but further research is needed for validation.
2.3 Fecal transport and storage protocols
The standardized handling of fresh fecal samples requires predefined transport and storage solutions when immediate processing is not feasible. Current methodologies demonstrate variations in the transportation and preservation of stool samples(Table 1): some studies advocate for ice-based transportation without defined temperature parameters (Dhakal et al., 2019; Glenny et al., 2021), while others recommend using dry ice for cryopreservation prior to shipping (Van Den Ham et al., 2023; Togao et al., 2023; Feehley et al., 2019). A broad consensus exists among researchers regarding –80°C as the optimal long-term storage temperature for fecal specimens (Nagao-Kitamoto et al., 2016; von Klitzing et al., 2017b; Wrzosek et al., 2018; Feehley et al., 2019; Heimesaat et al., 2019; Spatz et al., 2023; Tintelnot et al., 2023; Togao et al., 2023; Zhang et al., 2023; Xia et al., 2019; Liu et al., 2023; Ye et al., 2023; Zhang et al., 2023; Van Den Ham et al., 2023; Aluthge et al., 2020; Reygner et al., 2020; Fan et al., 2023; Renu et al., 2022). Although pragmatic protocols permit short-term preservation at –20°C before inoculum preparation (Grabrucker et al., 2023; Ridaura et al., 2013). Alternatively, storage and transportation at 4°C is permitted within a strict ≤24 h limit (Spatz et al., 2023; Wrzosek et al., 2018).
Current research has not explored how different storage conditions of fecal samples may influence the outcomes of FMT. Nevertheless, multiple studies have reported the finite effects of storage conditions on fecal microbiota. Fouhy et al. observed no significant compositional differences between fresh, dry ice flash-frozen, and –80°C-stored (for 7 days) fecal samples (Fouhy et al., 2015). Tedjo et al. confirmed microbiota stability following 24 h storage at 4°C, and 1-week storage at –20°C preservation whether for healthy, IBS, and IBD cohorts (Tedjo et al., 2015). Similarly, Choo et al. demonstrated that healthy donor fecal samples stored at 4°C for 72 h exhibited no statistically significant differences regarding microbial composition and diversity compared to their –80°C cryopreserved counterparts (Choo et al., 2015). Therefore, 4°C refrigeration and –20°C freezing are recommended as short-term transportation and preservation conditions, while –80°C cryopreservation is reserved for long - term storage.
In the context of cryopreservation, methods encompass direct freezing (Ridaura et al., 2013) as well as the incorporation of various cryoprotective agents like 10–50% glycerol solutions (Lauko et al., 2023; Ye et al., 2023; Feehley et al., 2019; Sánchez-Quintero et al., 2022; Aluthge et al., 2020; Zhang et al., 2023; Fan et al., 2023; Renu et al., 2022; Dhakal et al., 2019; Zhang et al., 2013; Lauko et al., 2023; Salandre et al., 2023; Grabrucker et al., 2023; Feehley et al., 2019; Tintelnot et al., 2023; Kaiser et al., 2021).The academic community remains divided concerning cryoprotectant. Advocates posit that freeze-thaw cycles (FTCs) compromise bacterial viability (Postgate and Hunter, 1961), necessitating the use of protective agents. Due to the uncertainties surrounding the effects of glycerol’s cellular permeation on bacterial viability, novel formulations such as maltodextrin-trehalose have been developed. The maltodextrin-trehalose have been validated through multi-phase assays to optimally preserve fecal microbial vitality during both freezing and thawing (Burz et al., 2019; Reygner et al., 2020). Conversely, some researchers suggest that direct ultra-low-temperature (−80°C) preservation without additives can maintain microbial composition without significant alteration (Tedjo et al., 2015). Three clinical studies provide evidence that the therapeutic effects of fresh and cryopreserved FMT preparations are comparable (Lee et al., 2016; Satokari et al., 2015; Sintes et al., 2024). However, a comparative trial indicated that fecal samples frozen without cryoprotectants showed changes in composition, viability, and cultivability upon thawing compared to fresh feces (Bilinski et al., 2022). Therefore, cryopreservation method should consider the use of cryoprotectants to maintain MV and composition, especially when samples undergo multiple FTCs. For short-term fecal sample storage, direct freezing at ultra-low temperatures without additives may be sufficient for preserving microbial integrity in certain contexts.
2.4 Fecal suspensions preparation
Fresh fecal specimens are typically reconstituted using phosphate-buffered saline (PBS) (Mao et al., 2021; Chiu et al., 2017; Dong et al., 2021; Sun et al., 2022; Sánchez-Quintero et al., 2022, 2023; Wahlström et al., 2017) or brain heart infusion (BHI) culture medium (Spatz et al., 2023; Wrzosek et al., 2018) before FMT administration, as shown in Table 2. During the post-thaw processing of cryopreserved fecal samples, common dilution vehicles include sterile saline (Hintze et al., 2014; Huang et al., 2020; Van Den Ham et al., 2023; Zabolotneva et al., 2023), PBS buffer (Grabrucker et al., 2023; Kaiser et al., 2021; Xia et al., 2019; Ye et al., 2023), media contain glycerol (Britton et al., 2019), and BHI medium (Tintelnot et al., 2023). The standard dilution ratios range from 1:10 to 1:1000 (w/v) (Le Bihan et al., 2015; Nagao-Kitamoto et al., 2016; Wrzosek et al., 2018; Crouzet et al., 2013; Togao et al., 2023; Zabolotneva et al., 2023; Wahlström et al., 2017)(Table 2). Sample preparation strategies include donor-specific retention through individual processing (Nagao-Kitamoto et al., 2016) and homogenized aliquots via pooled sample blending (Sánchez-Quintero et al., 2023). Clinical guidelines explicitly discourage the pooling of fecal samples from multiple donors during processing, to maintain donor traceability and mitigate the potential for pathogen dissemination (Keller et al., 2021). However, HMA model development strategies often involve compositing donor material to achieve a uniform distribution of human-derived gut microbiota across recipient animals (Sánchez-Quintero et al., 2023), thereby minimizing inter-individual variability. Fecal homogenization tools include traditional mortar-pestle grinding (Turnbaugh et al., 2009), dedicated blenders (Lauko et al., 2023; Reygner et al., 2020), the Ultra-Turrax blender (Humblot et al., 2005), and the Nanogenizer-Titanium High-Pressure Homogenizer (Staley et al., 2017; Kaiser et al., 2021). At present, there is a deficiency of comparative research examining the effects of various homogenization instruments on FMT. The available evidence suggests that following the blending process using either a blender or a pneumatic mixer, high-throughput DNA sequencing reveals a notable decrease in intra-sample heterogeneity (Hsieh et al., 2016). Dilution and filtration are common procedures during suspension preparation (Grabrucker et al., 2023; Zabolotneva et al., 2023; Zhang et al., 2023; Han et al., 2021; Reygner et al., 2020; Zhang et al., 2020; Staley et al., 2017; Zhang et al., 2013), which may help remove food debris, reduce the viscosity of the suspension, and prevent catheter occlusion during administration. Drawing from the aforementioned information, we recommend blending fecal samples followed by sequential dilution, homogenization, and filtration to obtain representative suspensions. Researchers should explicitly document their procedural details during such experiments—particularly the diluent composition and dilution ratio—to enhance experimental reproducibility.
2.5 Fecal microbiota assessment methodologies
Before FMT implementation, fecal suspensions are typically assessed via culturing-based methods (Bereswill et al., 2011), flow cytometry (Bilinski et al., 2020), 16S rRNA sequencing (Bereswill et al., 2011), shotgun metagenomic sequencing (Liu et al., 2023), or agar spot assays (Reygner et al., 2020). These analytical modalities collectively evaluate MV, composition, quantitation, and antagonistic capacity against specific bacterial strains. Conventional culturing methods typically detect only ~30–50% of viable gut microbes (Adak and Khan, 2019). Bilinski et al. demonstrated that flow cytometry with fluorochromes provides superior bacterial viability validation (Bilinski et al., 2020). The next-generation sequencing (NGS)—including 16S rRNA gene sequencing and shotgun metagenomics represent the common methodologies used in microbial studies, both of which carry distinct advantages. The 16S rRNA gene sequencing is well-suited to large-scale cohort analyses. However, it suffers from reduced accuracy in terms of species-level classification and functional profiling capacity—thus precluding the detection of strain-level variations (Jovel et al., 2016; Wensel et al., 2022). Conversely, the shotgun metagenomics facilitates strain identification and functional prediction but carries substantially higher costs (Jovel et al., 2016; Wensel et al., 2022). The agar spot test serves as a simple and effective preliminary screening tool for selecting antagonistic fecal samples in FMT-bacterial infection therapy, thus reducing downstream experimental expenditures (Salandre et al., 2023). In summary, shotgun metagenomic sequencing and agar spot assays are considered more suitable analytical methods for conducting detailed characterizations of specific bacterial strains or for selecting functionally specialized samples. However, the viable microbial number and 16S rRNA gene sequencing are recommended for initial community profiling due to its cost-effectiveness, ease of use, and suitability for large-scale or routine analyses.
3 Recipient selection
3.1 Recipient types
3.1.1 Germ-free animals
Germ-free (GF) animals are born and maintained in isolators throughout their lifespans, thus having minimal or no microbial exposure (Dremova et al., 2023). GF mice are still the most extensively used model organisms of this type—although axenic pig, dog, and chicken systems have been successfully generated through the progressive development of various technologies (Dhakal et al., 2019; Uzbay, 2019; Zhang et al., 2013). The establishment of gnotobiotic models through the colonization of GF animals with defined microbial consortia can provide controllable platforms for investigating host-microbe interactions (Dremova et al., 2023). Excluding the confounding effects of indigenous microbiota and antibiotics, this approach is widely regarded as an optimal strategy for generating human microbiota-associated (HMA) models. The applications of GF animals primarily include the following aspects: (1) elucidating the relationship between microbes and diseases to explore pathogenic mechanisms (Huang et al., 2020); (2) investigating the protective roles of microbes, such as resistance to the pathogen Clostridioides difficile (Sulaiman et al., 2025, 2024), mitigation of obesity (Mao et al., 2021), and alleviation of gastrointestinal discomfort (Rocha Martin et al., 2022); (3) studying metabolites produced by gut microbial communities, such as short-chain fatty acids (Liu et al., 2025), bile acids (Xue et al., 2025), and lactate (Li et al., 2022); (4) examining factors influencing microbial communities, including responses and functional outputs to dietary fibers and different types of diets (Feng et al., 2022; Turnbaugh et al., 2009); and (5) exploring the mechanisms by which drugs target the gut microbiota for therapeutic effects (Li et al., 2023). However, the utility of axenic models is constrained by three intrinsic barriers: first, the operational costs of isolator-based husbandry and sterile maintenance are prohibitive (Kennedy et al., 2018); second, open-environment behavioral assays and coinfection studies cannot be implemented (Kennedy et al., 2018); and third, immuno-developmental deficits inevitably arise because of the absence of gut microbiota (Kennedy et al., 2018). Collectively, these limitations have reduced the applicability of such models in terms of sophisticated pathophysiological research.
3.1.2 Altered Schaedler’s flora animals
To circumvent the immunological and developmental deficits of GF animals while maintaining controlled microbial status, altered Schaedler’s flora (ASF) animals were developed as well-defined microbiota models. Originating from Schaedler’s 1965 longitudinal tracking of gut microbiota succession in Nelson-Collins Swiss mice from birth to weaning, this model incorporates a standardized bacterial consortium that has been designated Schaedler’s flora (Schaedler et al., 1965). In 1978, Orcutt et al. refined and standardized this microbial consortium for stable intestinal colonization in murine hosts, and formally designated it ASF (Trexler and Orcutt, 1999). ASF serves as a representation of conventional murine gut microbiota (Deloris Alexander et al., 2006), demonstrating heritable stability through transgenerational propagation after colonization (Sarma-Rupavtarm et al., 2004). Compared to GF mice, ASF mice exhibit normal gastrointestinal architecture and physiological functions, along with fully developed immune systems (Proctor et al., 2022; Sarma-Rupavtarm et al., 2004). These animals are preferentially used to investigate specific microbial influences, intestinal mucosal responses, and the development of the enteric nervous system (Wymore Brand et al., 2015). However, the use of ASF mice in HMA studies remains scarce. Staley et al. demonstrated separate human donor microbiota transferability to ASF mice but revealed divergent outcomes. One cage exhibited significant microbial divergence from the donors (P=0.002), while another maintained no detectable divergence (P=0.012) (Staley et al., 2017). This heterogeneity suggests a potential niche competition between native ASF and humanized microbiomes (Staley et al., 2017), though the specific mechanisms underlying this phenomenon merit further investigation. The current evidence in the field is insufficient in terms of clearly defining the utility of ASF systems regarding humanized microbiota transfer, thus demanding expanded experimental validation.
3.1.3 Antibiotic administration-induced pseudo-germ-free animals
Although rodent and human gut microbiomes share taxonomic similarities, 85% of the microbial genera present in rodents are absent in humans (Ley et al., 2005). Thus, pre-FMT preparation must maximize the depletion of native microbiota to enhance the engraftment efficiency of transplanted communities. Specific pathogen-free (SPF) animals are those maintained in barrier-controlled environments, with certification confirming the absence of a defined set of common pathogens to which the species is typically exposed in a natural setting (Dobson et al., 2019; Lane-Petter, 1962). The establishment of pseudo-GF animals using various antibiotic regimens (Table 3) constitutes the primary preparatory phase for establishing HMAs based on SPF animals. This strategy originated in 1954 with Bohnhoff’s seminal discovery that the oral administration of high-dose streptomycin (50 mg) significantly increased the susceptibility to Salmonella enteritidis infection in mice (Bohnhoff et al., 1954). This discovery revealed that antibiotics disrupt gut microbiota homeostasis. It also established an experimental approach that leverages the antimicrobial suppression of native microbiota to enhance colonization potential. Subsequent studies demonstrated a 10× reduction in fecal 16S rDNA load and drastic structural alterations in microbial communities by day 10 of antibiotic treatment (Hill et al., 2010). This significantly increased the probability of effective donor microbiota colonization via FMT.
The administration routes include ad libitum antibiotic solution (Amorim et al., 2022; Bereswill et al., 2011; Grabrucker et al., 2023; Heimesaat et al., 2024, 2018; Kaiser et al., 2021; Salandre et al., 2023; Shayya et al., 2023), oral gavage (Chen et al., 2020; Kong et al., 2023; Liang et al., 2020; Sun et al., 2022; Xia et al., 2019), and injection (Lauko et al., 2023). Among these, drinking antibiotic solutions offers maximal technical simplicity.However, it carries a risk of dehydration, which may result from animals avoiding water due to the taste of the antibiotic or from antibiotic-associated diarrhea caused by prolonged exposure to the solution (Hill et al., 2010; Reikvam et al., 2011; Xu et al., 2023). Modified regimens, such as removing gentamicin or supplementing with sweeteners, have failed to mitigate this issue (Hill et al., 2010; Reikvam et al., 2011). By contrast, gavage delivery circumvents the dehydration trap while displaying microbiota depletion-associated phenotypes (Hill et al., 2010; Reikvam et al., 2011).Furthermore, several investigators have combined various delivery modalities like “oral gavage + subcutaneous injection” (Lauko et al., 2023), “ad libitumantibiotic solution + intraperitoneal injection” (Reygner et al., 2020), and “ad libitumantibiotic solution + oral gavage” (Sánchez-Quintero et al., 2022) to achieve superior methodological outcomes.
Different antimicrobial agents exhibit different targeting mechanisms. Metronidazole selectively impacts anaerobes, vancomycin targets gram-positive bacteria, and ampicillin and ciprofloxacin act against both gram-positive and gram-negative species (Schubert et al., 2015; Zackular et al., 2016). Consequently, antibiotic cocktails (including dual or multiple antibiotics and antifungals) are essential for comprehensive microbial eradication (von Klitzing et al., 2017b, 2017a; Zhan et al., 2024).
In pseudo-GF animal models generation, different types of antibiotics exhibit varying dosages depending on the administration route. For example, the commonly used oral gavage dose of vancomycin is 100 mg/kg (Chen et al., 2020; Kong et al., 2023; Liang et al., 2020), while the dose via drinking water is 500 mg/L (Grabrucker et al., 2023; Amorim et al., 2022; Kløve et al., 2020; Heimesaat et al., 2018; von Klitzing et al., 2017b, 2017a; Bereswill et al., 2011). The typical gavage dose of ampicillin is 200 mg/kg (Kong et al., 2023; Chen et al., 2020; Liang et al., 2020), whereas the dose in drinking water is 1 g/L (Grabrucker et al., 2023; Amorim et al., 2022; Kaiser et al., 2021; Kløve et al., 2020; Heimesaat et al., 2018; von Klitzing et al., 2017b, 2017a; Bereswill et al., 2011). For metronidazole, the gavage dose is 200 mg/kg (Kong et al., 2023; Sun et al., 2022; Chen et al., 2020; Liang et al., 2020), while the drinking water concentration is 1 g/L (Amorim et al., 2022; Kløve et al., 2020; Heimesaat et al., 2018; von Klitzing et al., 2017b, 2017a; Bereswill et al., 2011). Treatment timeframes also vary significantly. Gavage persists for 3–21 days (Liang et al., 2020; Sun et al., 2022), whereas aqueous delivery via the drinking of antibiotic solutions lasts between 3–56 days (Heimesaat et al., 2024, 2018; Liu et al., 2023; Reygner et al., 2020; Shayya et al., 2023; Wos-Oxley et al., 2012). Amorim et al. administered broad-spectrum antibiotics (ampicillin 1 g/L, vancomycin 0.5 g/L, neomycin 1 g/L, and metronidazole 1 g/L) through drinking an antibiotic solution and subsequently quantified the depletion of gut microbiota (Amorim et al., 2022). They demonstrated a 96% reduction by day 3, progressive declines through days 7–14, and stabilization by day 21 (Amorim et al., 2022). Tirelle et al. compared administration routes across temporal fecal bacterial density profiles and reported that twice-daily gavage (amphotericin-B 0.1 g/L, ampicillin 10 g/L, neomycin trisulfate salt hydrate 10 g/L, metronidazole 10 g/L, and vancomycin hydrochloride 5 g/L) achieved a bacterial depletion efficiency comparable to that of drinking water (amphotericin-B 0.01 g/L, ampicillin 1 g/L, neomycin trisulfate salt hydrate 1 g/L, metronidazole 1 g/L, and vancomycin hydrochloride 0.5 g/L). They demonstrated significant depletion by day 4, which was sustained until day 12 without additional clearance effects (Tirelle et al., 2020). These findings indicate that 3-day administration achieves fundamental microbiota eradication regardless of the delivery method, whereas optimized durations maintain persistent effects. Prolonged treatment regimens risk inducing antibiotic-resistant strains and compromising the health of the model animals, altering their phenotypes and increasing their mortality rates (Hill et al., 2010; Tirelle et al., 2020).
Additionally, animal studies from rat donors have demonstrated that transplantation of homologous microbiota on the second day following antibiotic administration leads to novel microbial reorganization (Manichanh et al., 2010). This phenomenon may be attributed to collateral damage caused by antibiotic residues, which can adversely affect both native and transplanted microbial communities (Manichanh et al., 2010). Therefore, restoring sterile water for a certain period prior to FMT could help mitigate the interference caused by antibiotic residues. According to existing evidence, this period typically ranges from 48 to 72 hours (Grabrucker et al., 2023; Heimesaat et al., 2024, 2018; Kaiser et al., 2021; Kløve et al., 2020; Shayya et al., 2023; Staley et al., 2017; von Klitzing et al., 2017b, 2017a; Zhan et al., 2024; Zhang et al., 2023).
3.1.4 Bowel cleansing-induced pseudo-germ-free animals
Laxative-based bowel-cleansing agents provide another effective microbiota-depleting strategy. Polyethylene glycol (PEG), a standard colonic preparation agent for colonoscopy procedures, has been used in many clinical studies to reduce microbial biomass and diversity when administered via split-dose regimens (Harrell et al., 2012; Jalanka et al., 2015; Zhou et al., 2025). Current clinical FMT guidelines rank PEG enemas as the optimal secondary preparatory intervention following antibiotic pretreatment (Cammarota et al., 2017). Wrzosek et al. demonstrated the applicability of PEG in animal bowel preparation protocols (Wrzosek et al., 2018). Murine models that received four cycles of intragastric 425 g/L PEG 4000 (200 µL per dose at 20 min intervals) achieved complete gastrointestinal evacuation with 90% reductions in microbial biomass (Wrzosek et al., 2018). Experimental data from mouse donor studies also indicated that 4-week regimens of PEG 400 or PEG 4000 (40% concentration, 100 µL oral gavage delivered 5 times weekly) significantly reduced gut microbial diversity in mice, with the 40% PEG 4000 cohort showing superior efficacy (Ishibashi et al., 2023). This approach preserves intestinal immune function and gut microbiome stability vs antibiotic-mediated depletion protocols (Wrzosek et al., 2018). In complex HMA models that require concurrent antibiotic therapy because of coinfection (Spatz et al., 2023), PEG lavage prevents antibiotic-associated carryover effects. However, as an osmotic cathartic, PEG requires elevated concentrations and substantial dosages to achieve effective intestinal clearance (Ishibashi et al., 2023; Le Roy et al., 2018)—which can induce electrolyte disturbances and dehydration. PEG-induced osmotic diarrhea disrupts the protective colonic mucus barrier, potentially influencing host immunocompetence (Tropini et al., 2018).
Overall, since antibiotic administration and bowel cleansing-induced pseudo-germ-free animals both retain residual native microbiota, these microbes may compete with the transplanted microbes or potentially develop into new ecological structures. Such models may not accurately represent a truly germ-free environment (Amorim et al., 2022; Hill et al., 2010; Tirelle et al., 2020; Wrzosek et al., 2018). Therefore, GF animals may be the optimal research model for exploring the causal relationships between microbiota and phenotypes. However, antibiotic-mediated pseudo-axenic models exhibit methodological superiority in studies focused on immunological regulation, developmental research, or targeted pathogen challenges. PEG bowel-cleansing protocols merit primary consideration if required to circumvent antibiotic-induced microbiota remodeling or residual impacts.
3.2 Recipient age
Human microbiota-associated (HMA) animal models common receptor types and ages include: (1) Fischer 344 rat, 8 -week-old (Crouzet et al., 2013); (2) Sprague dawley (SD) Rat, with various starting ages including 8 -week-old (Mao et al., 2021), 10 -week-old (Hanske et al., 2009), and 13-week-old (Grabrucker et al., 2023); (3) C57BL/6 mouse, with a range of starting ages from 3 to 8-week-old (Chiu et al., 2017; Hutchison et al., 2024; Hsu et al., 2023; Xia et al., 2019; Sun et al., 2022; Spatz et al., 2023; Salandre et al., 2023; Liu et al., 2023; Wrzosek et al., 2018; Huang et al., 2020; von Klitzing et al., 2017a); (4) BALB/c mouse, with various starting ages including 4,6,8-week-old (Kibe et al., 2005; Lin et al., 2021; Togao et al., 2023; Zabolotneva et al., 2023); as shown in Table 4. Due to the lack of humanized microbiota animal studies across different age groups, a study describing FMT from animal donors to same-species recipients of varying ages was selected as an indirect reference for analysis. In this study, age significantly influenced the efficacy of gut microbiota colonization (Le Roy et al., 2018).Comparative analyses by Le Roy demonstrated superior donor microbiota engraftment in 3-week-old weaned SPF micecompared to 8-week-old adults (Le Roy et al., 2018). This may be because animals with low gut microbiota richness exhibit superior engraftment efficacy (Ericsson et al., 2017), as microbial diversity naturally increases with age (Zhang et al., 2015). By contrast, the dietary transition to solid food during weaning generates transient microbial instability (Zhang et al., 2015) that requires 11–15 days to achieve full community stabilization (Schloss et al., 2012). Other compelling evidence has demonstrated that microbiota alterations established during juvenile stages are sustained into adulthood and induce phenotypic convergence between host organisms and donor profiles (Cox et al., 2014). Collectively, these findings suggest that 3-week-old or weaning-stage juvenile animals may represent the optimal candidates for FMT selection. However, given the critical role of microbiota-immune crosstalk in host immunological maturation (Al Nabhani et al., 2019), studies advocate using 6–8-week-old adult animals with fully developed immune systems (Crouzet et al., 2013; von Klitzing et al., 2017b; Wrzosek et al., 2018; Daharsh et al., 2019; Xia et al., 2019; Aluthge et al., 2020; Basson et al., 2020; Zhang et al., 2020; Huang et al., 2020; Lin et al., 2021; Mao et al., 2021; Han et al., 2021; Sun et al., 2022; Salandre et al., 2023; Spatz et al., 2023; Liu et al., 2023; Togao et al., 2023; Fan et al., 2023). Although this age-specific model better recapitulates microbiota-mature immune system interactions, it may compromise the efficiency of colonization. Therefore, in HMA model, we recommend strategic selection based on research priorities: juvenile models for microbiota colonization dynamics, and adult animals when investigating immunomodulatory mechanisms.
3.3 Dietary impact
Dietary modulation plays a pivotal role in shaping the gut microbiome (Zmora et al., 2019). Empirical evidence has confirmed that different diets influence both the composition and function of intestinal microorganisms in humans as well as animals (Beam et al., 2021). This principle is equally applicable to HMA animals. Turnbaugh et al. proved that high-fat high-sugar diets rapidly remodeled the microbiota architectures of HMAs, impaired donor microbiota engraftment, and induced phenotypes associated with metabolic obesity (Turnbaugh et al., 2009). Dietary heterogeneity constitutes a critical determinant that prevents HMA animals from fully replicating the gut microbial profiles of their donors (Silley, 2009). Comparative studies have revealed that donor-matched diets fail to enhance gut microbial engraftment efficiency in HMA mice vs fixed-formula grain-based chows (Van Den Ham et al., 2023). By contrast, Schoeler et al. demonstrated superior microbiota transfer success rates in HMA mice that received analog diets identical to those of their human donors (Schoeler et al., 2024). In a 28-day dietary intervention study, Dong et al. observed equivalent microbial colonization rates between coarse-feed diet (CFD) and purified-feed diet groups (70.00% vs. 72.69%) in HMA mice (Dong et al., 2021). In particular, the CFD-fed mice exhibited gut microbial diversity profiles and functional signatures that demonstrated close proximities to those of their human donors (Dong et al., 2021). Although the effects of standardized feeds on HMA animals remain unclear, current evidence demonstrates that donor-aligned dietary formulations may reduce enteric microbiota discrepancies between donor and recipient ecosystems.
4 Experimental administration protocols and treatment duration
Common methods for administering fecal microbiota transplantation (FMT) include rectal enema, co-housing, and oral-gastric gavage. Rectal administration requires anesthetizing the animals, gently inserting a tube into the colon, and slowly injecting a fecal bacteria suspension (Zhou et al., 2019). Nevertheless, this procedure presents technical challenges such as mucosal damage, infection, and uncontrollable absorption efficacy (Bokoliya et al., 2021). Co-housing protocols, which let germ-free (GF) mice be co-housed with colonized mice, are effective for microbiota transfer between conspecifics (Hansen et al., 2012; Seedorf et al., 2014). However, it is not suitable for the establishment of human microbiota-associated (HMA) models (Bokoliya et al., 2021). Oral gastric gavage is a method that involves using a stainless-steel or flexible cannula to a syringe to deliver the fecal suspension directly into the stomach (Bokoliya et al., 2021). This method carries potential complications, including respiratory tract injury, gastric rupture, and weight loss (Bokoliya et al., 2021). Nonetheless, empirical evidence has confirmed that single-dose FMT delivery via gavage reliably induces human microbial colonization in experimental animals (Hanske et al., 2009; Reygner et al., 2020). This approach therefore remains the preferred methodology for establishing HMA models.
Notably, emerging nanotechnology applications have introduced single-cell nanocapsules as a novel delivery vehicle for FMT (Hou et al., 2025). This innovative approach utilizes silk fibroin and phosphatidylcholine to form reinforced nanoshells around intestinal microbiota within 1 hour, achieving microbial encapsulation without compromising viability. Experimental trials involving oral administration of these nanocapsules to GF mice and colitis murine models demonstrated superior performance compared to conventional FMT through three key advantages: (1) protecting microbial communities from gastric acid and pepsin degradation; (2) significantly enhancing microbial engraftment efficiency; and (3) providing additional anti-inflammatory benefits while preserving intestinal epithelial integrity (Hou et al., 2025).
Another critical aspect that merits attention is the dosage and frequency of fecal suspension administration (Table 1). The typical standard gavage volumes are 1–2 mL for rats (Hanske et al., 2009; Crouzet et al., 2013; Le Bihan et al., 2015) and 0.1–0.5 mL for mice (Chiu et al., 2017; Han et al., 2021; Liu et al., 2022). For developing pig HMA models, the sparse existing literature on the subject suggests an ideal inoculum volume of 1 mL (Dhakal et al., 2019; Renu et al., 2022; Zhang et al., 2013). Furthermore, some studies have characterized the total number of cells administered within these volumes, reporting, for instance, 1 mL (2.7–5.5 × 109 cells) for SD rats (Hanske et al., 2009), 0.2 mL (109 CFU/ml) for C57BL/6 mice (Ye et al., 2023), 0.1 mL (107 bacteria) for C3H/HeN mice (Reygner et al., 2020), and 0.20 mL/10 g (108–9 CFU/mouse) for SAMP mice (Basson et al., 2020). Administration frequencies vary widely, ranging from single-bolus delivery to daily regimens (1–3 doses/day) spanning 2–60 days, or periodic administration at 2–7-day intervals (Basson et al., 2020; Daharsh et al., 2019; Fan et al., 2023; Wahlström et al., 2017; Zabolotneva et al., 2023; Zhan et al., 2024). Hanke et al. demonstrated that HMA rats exhibited 55.8–64.5% gut microbial similarity to their human donors at 2–12 weeks post-FMT, with no significant differences observed between time points (Hanske et al., 2009). Despite the variations present in murine strains, studies by (Liu et al., 2022) (Ye et al., 2023), and (Dong et al., 2021) demonstrated that fecal suspension doses of 0.1, 0.2, and 0.3 mL achieved colonization efficiencies of 59–81% (operational taxonomic unit, OTU level), 65–66% (genus level), and 67.5–85.96% (OTU level), respectively. The relationship between dosage and engraftment efficiency has yet to be elucidated. Nevertheless, current studies consistently demonstrate ≥50% donor microbiota retention in HMA models following single-dose FMT following adequate intestinal preparation, regardless of the volume administered (Dong et al., 2021; Hanske et al., 2009; Liu et al., 2022; Ye et al., 2023).
Thus, the question has arisen of whether chronic FMT protocols with increased frequency can optimize colonization success has garnered significant attention. Experimental protocols by Aluthge et al. revealed that a secondary 0.2 mL fecal transplant in C3H/HeN mice (delivered at a 2-week interval) induced >96% amplicon sequence variant (ASV)-level microbial retention (Aluthge et al., 2020). By contrast, Hutchison et al.’s cohort of C57BL/6 mice, who received multiple 0.1 mL doses at 7-day intervals, exhibited 49–52% ASV and 58–68% genus-level colonization fidelity (Hutchison et al., 2024). The twice-weekly administration of a 0.2 mL fecal suspension to BALB/c mice revealed 70% genus-level colonization efficiency via 16S rRNA gene sequencing (Lin et al., 2021). Although FMT protocols with increased administration frequency appear to improve colonization success, experimental outcomes varied substantially across the above study. A comprehensive study by Van Den Ham et al. evaluated three fecal transplant schedules (single-dose, 4-day consecutively, and once a week for 4 weeks) using 0.2 mL inocula administered to GF mice (Van Den Ham et al., 2023). The once a week for 4 weeks protocol demonstrated superior colonization efficiency vs the other interventions, which was attributed to its capacity to establish a stabilized intestinal condition that minimized pre-engraftment microbial fluctuations, thereby narrowing the donor-recipient microbiota divergence (Van Den Ham et al., 2023). Another comparative study evaluated four FMT strategies in Polyethylene glycol (PEG)-cleansed C57BL/6J mice: (1) a single round during the first week; (2) two rounds of FMT in the first week; (3) once a week for four weeks; and (4) twice a week for four weeks (Wrzosek et al., 2018). After four weeks, the results showed that: (1) a single FMT enabled the observation of human-derived microorganisms; (2) two rounds of FMT in the first week allowed for the engraftment of sub-dominant human bacteria; (3) once-weekly regimen for four weeks was sufficient to establish dominant bacterial populations; (4) in contrast, FMT twice weekly for four weeks disrupted the stability of the newly established microbial ecosystem (Wrzosek et al., 2018). Therefore, the above evidence supports administering multiple FMT doses (cumulatively ≥2 doses) over a 2–4weeks period during the HMA model preparation to optimize the efficacy of donor microbiota engraftment.
5 Microbial colonization assessment strategies
The assessment of donor microbiota engraftment efficiency is performed through diverse detection modalities. These modalities include conventional culturing (Hirayama et al., 1995), next-generation sequencing (NGS) (Kong et al., 2023; Wensel et al., 2022), selective culturing (Chiu et al., 2017), fluorescence in situ hybridization (Gérard et al., 2004), and temporal-temperature gradient gel electrophoresis (Respondek et al., 2013). Cultivation and isolation represent conventional approaches wherein microbiota are taxonomically enumerated post-development on selective media. Intrinsic limitations persist as slow-growing or fastidious bacteria, which are subject to microbial competition and stringent nutrient requirements, often resist in vitro isolation and cultivation (Kato et al., 2018). Strategies such as oligotrophic media, extended incubation periods, and anaerobic culturing conditions have been implemented to address these limitations (Goodman et al., 2011; Pulschen et al., 2017). Nevertheless, their high level of technical demand exacerbates in the challenges inherent to culturing gut microbiota. The advent of culture-independent NGS has resolved these obstacles by facilitating the direct sequencing of microbial DNA and RNA, thereby facilitating the detection of unculturable bacterial taxa. The emergence of culture-independent NGS has addressed these challenges by enabling the amplification and direct sequencing of microbial DNA and RNA, which in turn enhances the identification of unculturable bacterial taxa (Wensel et al., 2022). Microbial colonization efficacy can be quantified using three principal approaches derived from sequencing data. The first is donor-specific retention percentages calculated using operational taxonomic unit (OTU) (Knights et al., 2011) or amplicon sequence variant (ASV) (Gray et al., 2024) classification systems. Studies have revealed that OTU-based calculations systematically overestimate colonization efficiency vs ASV-resolution analyses (Gray et al., 2024). This might be because OTU analysis provides more spurious taxa (Reitmeier et al., 2021). Consequently, the assessment of colonization efficiency at this tier remains contentious, warranting genus-level analysis (Ye et al., 2023) or the implementation of alternative assessment methodologies. The second approach comprises monitoring the emergence of donor-enriched or species-specific bacterial taxa in recipient microbiota (Xia et al., 2019). This approach faces validity challenges related to interspecies microbial overlap (e.g., Prevotella, Bacteroides, Clostridium, and Eubacterium-dominant genera across human, murine, and porcine gut communities) (Li et al., 2018), rendering FMT-dependent colonization indistinguishable from native microbiota. The third approach involves assessing microbial transfer via abundance ratios (e.g., the Bacteroidetes/Firmicutes ratio) (Sun et al., 2022). However, such evaluations lack diagnostic precision, owing to multifactorial influences on microbial abundance and pre-existing microbial overlap between donors and recipients.
6 Conclusion and prospects
HMA animal models serve as indispensable tools for deciphering the roles of microbes in states of both health and disease, by simulating humanized gut microbiomes. The core technical aspects underlying the construction of HMA models remain under investigation. This review of the critical elements involved in the development of HMA models has delineated the following key findings(Figure 2): (1) Donor screening necessitates rigorous interviews regarding dietary habits, medication history, and pre-existing pathologies to eliminate a host of factors that can influence gut microbial composition. (2) Fecal preservation mandates immediate refrigeration within a 2–6 h window after collection. (3) Fecal suspension preparation should employ multi-donor blending strategies coupled with 16S rRNA sequencing to verify microbial composition. (4) Recipient selection should preferentially utilize adult germ-free (GF) or antibiotic-induced pseudo-GF animals that are fed diets matching their human donors. (5) Oral gavage represents the ideal route for FMT, with protocols utilizing high administration frequencies (cumulatively ≥2 doses) and extended durations (2–4 weeks) demonstrating significantly higher engraftment rates. (6) Next-generation sequencing (NGS) represents an efficient methodology for quantifying microbial engraftment. Metrics used include retention rates of operational taxonomic unit (OTU)/amplicon sequence variant (ASV) between donor and recipient microbiomes. Other metrics involve the detection of donor-specific bacterial strains, and phylum-level abundance ratios. These findings establish a methodological foundation for standardizing HMA model generation protocols. The development of HMA models faces persistent challenges that include objective microbial disparities between donors, unstable colonization of human microbiota in animal recipients, methodological variations in recipient animal preparations, and various dietary influences on microbial colonization. Moreover, a rational method for assessing the efficiency of colonization is needed to maximize the preparation of reproducible and representative HMA models.
Author contributions
XH: Conceptualization, Writing – original draft. YY: Supervision, Writing – original draft. NT: Investigation, Writing – original draft. JH: Writing – original draft. XZ: Writing – original draft. RY: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (U21A20411), Natural Science Foundation of Hunan Province of China (2024JJ1007), and Hunan University of Chinese Medicine Disciplinary Construction “Revealing the List and Appointing Leaders” Project (22JBZ002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adak, A. and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Al, K. F., Bisanz, J. E., Gloor, G. B., Reid, G., and Burton, J. P. (2018). Evaluation of sampling and storage procedures on preserving the community structure of stool microbiota: A simple at-home toilet-paper collection method. J. Microbiol. Methods 144, 117–121. doi: 10.1016/j.mimet.2017.11.014
Almeida, A., Mitchell, A. L., Boland, M., Forster, S. C., Gloor, G. B., Tarkowska, A., et al. (2019). A new genomic blueprint of the human gut microbiota. Nature 568, 499–504. doi: 10.1038/s41586-019-0965-1
Al Nabhani, Z., Dulauroy, S., Marques, R., Cousu, C., Al Bounny, S., Déjardin, F., et al. (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014
Aluthge, N. D., Tom, W. A., Bartenslager, A. C., Burkey, T. E., Miller, P. S., Heath, K. D., et al. (2020). Differential longitudinal establishment of human fecal bacterial communities in germ-free porcine and murine models. Commun. Biol. 3, 760. doi: 10.1038/s42003-020-01477-0
Amorim, N., McGovern, E., Raposo, A., Khatiwada, S., Shen, S., Koentgen, S., et al. (2022). Refining a protocol for faecal microbiota engraftment in animal models after successful antibiotic-induced gut decontamination. Front. Med. (Lausanne) 9. doi: 10.3389/fmed.2022.770017
Aron-Wisnewsky, J., Warmbrunn, M. V., Nieuwdorp, M., and Clément, K. (2021). Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health-pathophysiology and therapeutic strategies. Gastroenterology 160, 573–599. doi: 10.1053/j.gastro.2020.10.057
Arrieta, M.-C., Walter, J., and Finlay, B. B. (2016). Human microbiota-associated mice: A model with challenges. Cell Host Microbe 19, 575–578. doi: 10.1016/j.chom.2016.04.014
Basson, A. R., Gomez-Nguyen, A., Menghini, P., Buttó, L. F., Di Martino, L., Aladyshkina, N., et al. (2020). Human gut microbiome transplantation in ileitis prone mice: A tool for the functional characterization of the microbiota in inflammatory bowel disease patients. Inflammation Bowel Dis. 26, 347–359. doi: 10.1093/ibd/izz242
Beam, A., Clinger, E., and Hao, L. (2021). Effect of diet and dietary components on the composition of the gut microbiota. Nutrients 13, 2795. doi: 10.3390/nu13082795
Bereswill, S., Fischer, A., Plickert, R., Haag, L.-M., Otto, B., Kühl, A. A., et al. (2011). Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6, e20953. doi: 10.1371/journal.pone.0020953
Bilinski, J., Dziurzynski, M., Grzesiowski, P., Podsiadly, E., Stelmaszczyk-Emmel, A., Dzieciatkowski, T., et al. (2020). Multimodal approach to assessment of fecal microbiota donors based on three complementary methods. J. Clin. Med. 9, 2036. doi: 10.3390/jcm9072036
Bilinski, J., Dziurzynski, M., Grzesiowski, P., Podsiadly, E., Stelmaszczyk-Emmel, A., Dzieciatkowski, T., et al. (2022). Fresh versus frozen stool for fecal microbiota transplantation—assessment by multimethod approach combining culturing, flow cytometry, and next-generation sequencing. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.872735
Bohnhoff, M., Drake, B. L., and Miller, C. P. (1954). Effect of streptomycin on susceptibility of intestinal tract to experimental salmonella infection. Exp. Biol. Med. 86, 132–137. doi: 10.3181/00379727-86-21030
Bokoliya, S. C., Dorsett, Y., Panier, H., and Zhou, Y. (2021). Procedures for fecal microbiota transplantation in murine microbiome studies. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.711055
Borrego-Ruiz, A. and Borrego, J. J. (2024). Nutritional and microbial strategies for treating acne, alopecia, and atopic dermatitis. Nutrients 16, 3559. doi: 10.3390/nu16203559
Brandi, G., Calabrese, C., Tavolari, S., Bridonneau, C., Raibaud, P., Liguori, G., et al. (2024). Intestinal microbiota increases cell proliferation of colonic mucosa in human-flora-associated (HFA) mice. IJMS 25, 6182. doi: 10.3390/ijms25116182
Britton, G. J., Contijoch, E. J., Mogno, I., Vennaro, O. H., Llewellyn, S. R., Ng, R., et al. (2019). Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity 50, 212–224.e4. doi: 10.1016/j.immuni.2018.12.015
Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., et al. (2016). Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. doi: 10.1038/nature17645
Burz, S. D., Abraham, A.-L., Fonseca, F., David, O., Chapron, A., Béguet-Crespel, F., et al. (2019). A guide for ex vivo handling and storage of stool samples intended for fecal microbiota transplantation. Sci. Rep. 9, 8897. doi: 10.1038/s41598-019-45173-4
Cammarota, G., Ianiro, G., Tilg, H., Rajilić-Stojanović, M., Kump, P., Satokari, R., et al. (2017). European consensus conference on faecal microbiota transplantation in clinical practice. Gut 66, 569–580. doi: 10.1136/gutjnl-2016-313017
Chen, X., Li, P., Liu, M., Zheng, H., He, Y., Chen, M.-X., et al. (2020). Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut 69, 513–522. doi: 10.1136/gutjnl-2019-319101
Cherbuy, C., Bellet, D., Robert, V., Mayeur, C., Schwiertz, A., and Langella, P. (2019). Modulation of the caecal gut microbiota of mice by dietary supplement containing resistant starch: impact is donor-dependent. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01234
Chiu, C.-C., Ching, Y.-H., Li, Y.-P., Liu, J.-Y., Huang, Y.-T., Huang, Y.-W., et al. (2017). Nonalcoholic fatty liver disease is exacerbated in high-fat diet-fed gnotobiotic mice by colonization with the gut microbiota from patients with nonalcoholic steatohepatitis. Nutrients 9, 1220. doi: 10.3390/nu9111220
Choo, J. M., Leong, L. E. X., and Rogers, G. B. (2015). Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 5, 16350. doi: 10.1038/srep16350
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. doi: 10.1016/j.cell.2012.04.037
Cox, L. M., Yamanishi, S., Sohn, J., Alekseyenko, A. V., Leung, J. M., Cho, I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721. doi: 10.1016/j.cell.2014.05.052
Crouzet, L., Gaultier, E., Del’Homme, C., Cartier, C., Delmas, E., Dapoigny, M., et al. (2013). The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil. 25, e272–e282. doi: 10.1111/nmo.12103
Daharsh, L., Zhang, J., Ramer-Tait, A., and Li, Q. (2019). A double humanized BLT-mice model featuring a stable human-like gut microbiome and human immune system. J. Vis. Exp. 150, 1–16. doi: 10.3791/59773
Deloris Alexander, A., Orcutt, R. P., Henry, J. C., Baker, J., Bissahoyo, A. C., and Threadgill, D. W. (2006). Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome 17, 1093–1104. doi: 10.1007/s00335-006-0063-1
Demir, M., Lang, S., Hartmann, P., Duan, Y., Martin, A., Miyamoto, Y., et al. (2022). The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 76, 788–799. doi: 10.1016/j.jhep.2021.11.029
Dhakal, S., Wang, L., Antony, L., Rank, J., Bernardo, P., Ghimire, S., et al. (2019). Amish (Rural) vs. non-Amish (Urban) Infant Fecal Microbiotas Are Highly Diverse and Their Transplantation Lead to Differences in Mucosal Immune Maturation in a Humanized Germfree Piglet Model. Front. Immunol. 10. doi: 10.3389/fimmu.2019.01509
Dobson, G. P., Letson, H. L., Biros, E., and Morris, J. (2019). Specific pathogen-free (SPF) animal status as a variable in biomedical research: have we come full circle? Ebiomedicine 41, 42–43. doi: 10.1016/j.ebiom.2019.02.038
Dong, S., Zeng, B., Hu, L., Zhang, Y., Xiong, J., Deng, J., et al. (2021). Effect of a humanized diet profile on colonization efficiency and gut microbial diversity in human flora-associated mice. Front. Nutr. 8. doi: 10.3389/fnut.2021.633738
Drago, L., Valentina, C., and Fabio, P. (2019). Gut microbiota, dysbiosis and colon lavage. Dig Liver Dis. 51, 1209–1213. doi: 10.1016/j.dld.2019.06.012
Dremova, O., Mimmler, M., Paeslack, N., Khuu, M. P., Gao, Z., Bosmann, M., et al. (2023). Sterility testing of germ-free mouse colonies. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1275109
Duan, Y., Llorente, C., Lang, S., Brandl, K., Chu, H., Jiang, L., et al. (2019). Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511. doi: 10.1038/s41586-019-1742-x
Dudek-Wicher, R. K., Junka, A., and Bartoszewicz, M. (2018). The influence of antibiotics and dietary components on gut microbiota. Prz Gastroenterol. 13, 85–92. doi: 10.5114/pg.2018.76005
Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B., and Keshavarzian, A. (2015). The gastrointestinal microbiome: alcohol effects on the composition of intestinal microbiota. Alcohol Res. 37, 223–236.
Ericsson, A. C., Personett, A. R., Turner, G., Dorfmeyer, R. A., and Franklin, C. L. (2017). Variable colonization after reciprocal fecal microbiota transfer between mice with low and high richness microbiota. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00196
Fan, Y., Støving, R. K., Berreira Ibraim, S., Hyötyläinen, T., Thirion, F., Arora, T., et al. (2023). The gut microbiota contributes to the pathogenesis of anorexia nervosa in humans and mice. Nat. Microbiol. 8, 787–802. doi: 10.1038/s41564-023-01355-5
Feehley, T., Plunkett, C. H., Bao, R., Choi Hong, S. M., Culleen, E., Belda-Ferre, P., et al. (2019). Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 25, 448–453. doi: 10.1038/s41591-018-0324-z
Feng, J., Qian, Y., Zhou, Z., Ertmer, S., Vivas, E. I., Lan, F., et al. (2022). Polysaccharide utilization loci in bacteroides determine population fitness and community-level interactions. Cell Host Microbe 30, 200–215.e12. doi: 10.1016/j.chom.2021.12.006
Flint, H. J., Duncan, S. H., Scott, K. P., and Louis, P. (2015). Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 74, 13–22. doi: 10.1017/S0029665114001463
Fouhy, F., Deane, J., Rea, M. C., O’Sullivan, Ó., Ross, R. P., O’Callaghan, G., et al. (2015). The effects of freezing on faecal microbiota as determined using miSeq sequencing and culture-based investigations. PLoS One 10, e0119355. doi: 10.1371/journal.pone.0119355
Franzosa, E. A., Morgan, X. C., Segata, N., Waldron, L., Reyes, J., Earl, A. M., et al. (2014). Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. U.S.A. 111, E2329–E2338. doi: 10.1073/pnas.1319284111
Gérard, P., Béguet, F., Lepercq, P., Rigottier-Gois, L., Rochet, V., Andrieux, C., et al. (2004). Gnotobiotic rats harboring human intestinal microbiota as a model for studying cholesterol-to-coprostanol conversion. FEMS Microbiol. Ecol. 47, 337–343. doi: 10.1016/S0168-6496(03)00285-X
Glenny, E. M., Fouladi, F., Thomas, S. A., Bulik-Sullivan, E. C., Tang, Q., Djukic, Z., et al. (2021). Gut microbial communities from patients with anorexia nervosa do not influence body weight in recipient germ-free mice. Gut Microbes 13 (1), 1–15. doi: 10.1080/19490976.2021.1897216
Gobert, A. P., Sagrestani, G., Delmas, E., Wilson, K. T., Verriere, T. G., Dapoigny, M., et al. (2016). The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci. Rep. 6, 39399. doi: 10.1038/srep39399
Goodman, A. L., Kallstrom, G., Faith, J. J., Reyes, A., Moore, A., Dantas, G., et al. (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 6252–6257. doi: 10.1073/pnas.1102938108
Grabrucker, S., Marizzoni, M., Lopizzo, N., Mombelli, E., Nicolas, S., Dohm-Hansen, S., et al. (2023). Microbiota from Alzheimer’s patients induce deficits in cognition and hippocampal neurogenesis. Brain 146, 4916–4934. doi: 10.1093/brain/awad303
Gray, S. M., Moss, A. D., Herzog, J. W., Kashiwagi, S., Liu, B., Young, J. B., et al. (2024). Mouse adaptation of human inflammatory bowel diseases microbiota enhances colonization efficiency and alters microbiome aggressiveness depending on the recipient colonic inflammatory environment. Microbiome 12, 147. doi: 10.1186/s40168-024-01857-2
Han, X., Wang, Y., Zhang, P., Zhu, M., Li, L., Mao, X., et al. (2021). Kazak faecal microbiota transplantation induces short-chain fatty acids that promote glucagon-like peptide-1 secretion by regulating gut microbiota in db/db mice. Pharm. Biol. 59, 1077–1087. doi: 10.1080/13880209.2021.1954667
Hansen, C. H. F., Nielsen, D. S., Kverka, M., Zakostelska, Z., Klimesova, K., Hudcovic, T., et al. (2012). Patterns of early gut colonization shape future immune responses of the host. PLoS One 7, e34043. doi: 10.1371/journal.pone.0034043
Hanske, L., Loh, G., Sczesny, S., Blaut, M., and Braune, A. (2009). The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. J. Nutr. 139, 1095–1102. doi: 10.3945/jn.108.102814
Harrell, L., Wang, Y., Antonopoulos, D., Young, V., Lichtenstein, L., Huang, Y., et al. (2012). Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS One 7, e32545. doi: 10.1371/journal.pone.0032545
Heimesaat, M. M., Escher, U., Grunau, A., Fiebiger, U., and Bereswill, S. (2018). Peroral low-dose toxoplasma gondii infection of human microbiota-associated mice - A subacute ileitis model to unravel pathogen-host interactions. Eur. J. Microbiol. Immunol. (Bp) 8, 53–61. doi: 10.1556/1886.2018.00005
Heimesaat, M. M., Escher, U., Grunau, A., Kühl, A. A., and Bereswill, S. (2019). Multidrug-resistant pseudomonas aeruginosa accelerate intestinal, extra-intestinal, and systemic inflammatory responses in human microbiota-associated mice with subacute ileitis. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00049
Heimesaat, M. M., Langfeld, L. Q., Schabbel, N., Mousavi, S., and Bereswill, S. (2024). Carvacrol prophylaxis improves clinical outcome and dampens apoptotic and pro-inflammatory immune responses upon Campylobacter jejuni infection of human microbiota-associated IL-10–/– mice. EuJMI 14, 166–179. doi: 10.1556/1886.2024.00009
Hill, D. A., Hoffmann, C., Abt, M. C., Du, Y., Kobuley, D., Kirn, T. J., et al. (2010). Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158. doi: 10.1038/mi.2009.132
Hintze, K. J., Cox, J. E., Rompato, G., Benninghoff, A. D., Ward, R. E., Broadbent, J., et al. (2014). Broad scope method for creating humanized animal models for animal health and disease research through antibiotic treatment and human fecal transfer. Gut Microbes 5, 183–191. doi: 10.4161/gmic.28403
Hirayama, K. (1999). Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp. Anim. 48, 219–227. doi: 10.1538/expanim.48.219
Hirayama, K., Miyaji, K., Kawamura, S., Itoh, K., Takahashi, E., and Mitsuoka, T. (1995). Development of intestinal flora of human-flora-associated (HFA) mice in the intestine of their offspring. Exp. Anim. 44, 219–222. doi: 10.1538/expanim.44.219
Hou, W., Cao, Y., Wang, J., Yin, F., Wang, J., Guo, N., et al. (2025). Single-cell nanocapsules of gut microbiota facilitate fecal microbiota transplantation. Theranostics 15, 2069–2084. doi: 10.7150/thno.104852
Hsieh, Y.-H., Peterson, C. M., Raggio, A., Keenan, M. J., Martin, R. J., Ravussin, E., et al. (2016). Impact of different fecal processing methods on assessments of bacterial diversity in the human intestine. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01643
Hsu, C. L., Wang, Y., Duan, Y., Chu, H., Hartmann, P., Llorente, C., et al. (2023). Differences in bacterial translocation and liver injury in ethanol versus diet-induced liver disease. Dig Dis. Sci. 68, 3059–3069. doi: 10.1007/s10620-023-07860-1
Huang, Z., Chen, J., Li, B., Zeng, B., Chou, C.-H., Zheng, X., et al. (2020). Faecal microbiota transplantation from metabolically compromised human donors accelerates osteoarthritis in mice. Ann. Rheum Dis. 79, 646–656. doi: 10.1136/annrheumdis-2019-216471
Human Microbiome Project Consortium (2012a). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Human Microbiome Project Consortium (2012b). A framework for human microbiome research. Nature 486, 215–221. doi: 10.1038/nature11209
Humblot, C., Combourieu, B., Väisänen, M.-L., Furet, J.-P., Delort, A.-M., and Rabot, S. (2005). 1H nuclear magnetic resonance spectroscopy-based studies of the metabolism of food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline by human intestinal microbiota. Appl. Environ. Microbiol. 71, 5116–5123. doi: 10.1128/AEM.71.9.5116-5123.2005
Hutchison, E. R., Yen, M.-I., Peng, H. W., Davis, C. R., Vivas, E. I., Tallon, M. M., et al. (2024). The gut microbiome modulates the impact of Anaerobutyricum soehngenii supplementation on glucose homeostasis in mice. doi: 10.21203/rs.3.rs-4324489/v1
Imaoka, A., Setoyama, H., Takagi, A., Matsumoto, S., and Umesaki, Y. (2004). Improvement of human faecal flora-associated mouse model for evaluation of the functional foods. J. Appl. Microbiol. 96, 656–663. doi: 10.1111/j.1365-2672.2004.02189.x
Integrative HMP (iHMP) Research Network Consortium (2014). The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289. doi: 10.1016/j.chom.2014.08.014
Ishibashi, R., Matsuhisa, R., Nomoto, M., Chudan, S., Nishikawa, M., Tabuchi, Y., et al. (2023). Effect of oral administration of polyethylene glycol 400 on gut microbiota composition and diet-induced obesity in mice. Microorganisms 11, 1882. doi: 10.3390/microorganisms11081882
Jalanka, J., Salonen, A., Salojärvi, J., Ritari, J., Immonen, O., Marciani, L., et al. (2015). Effects of bowel cleansing on the intestinal microbiota. Gut 64, 1562–1568. doi: 10.1136/gutjnl-2014-307240
Jovel, J., Patterson, J., Wang, W., Hotte, N., O’Keefe, S., Mitchel, T., et al. (2016). Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00459
Kaiser, T., Nalluri, H., Zhu, Z., and Staley, C. (2021). Donor microbiota composition and housing affect recapitulation of obese phenotypes in a human microbiota-associated murine model. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.614218
Kato, S., Yamagishi, A., Daimon, S., Kawasaki, K., Tamaki, H., Kitagawa, W., et al. (2018). Isolation of previously uncultured slow-growing bacteria by using a simple modification in the preparation of agar media. Appl. Environ. Microbiol. 84, e00807–e00818. doi: 10.1128/AEM.00807-18
Keller, J. J., Ooijevaar, R. E., Hvas, C. L., Terveer, E. M., Lieberknecht, S. C., Högenauer, C., et al. (2021). A standardised model for stool banking for faecal microbiota transplantation: a consensus report from a multidisciplinary UEG working group. United Eur. Gastroenterol. J. 9, 229–247. doi: 10.1177/2050640620967898
Kennedy, E. A., King, K. Y., and Baldridge, M. T. (2018). Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9. doi: 10.3389/fphys.2018.01534
Kibe, R., Sakamoto, M., Yokota, H., Ishikawa, H., Aiba, Y., Koga, Y., et al. (2005). Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl. Environ. Microbiol. 71, 3171–3178. doi: 10.1128/AEM.71.6.3171-3178.2005
Kim, H.-N., Cheong, H. S., Kim, B., Sohn, W., Cho, Y. K., Kwon, M.-J., et al. (2024). Human gut microbiota from hepatitis B virus-infected individuals is associated with reduced triglyceride level in mice: faecal transplantation study. Microbes Infection 26, 105281. doi: 10.1016/j.micinf.2023.105281
Kløve, S., Genger, C., Mousavi, S., Weschka, D., Bereswill, S., and Heimesaat, M. M. (2020). Toll-like receptor-4 dependent intestinal and systemic sequelae following peroral campylobacter coli infection of IL10 deficient mice harboring a human gut microbiota. Pathogens 9, 386. doi: 10.3390/pathogens9050386
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. doi: 10.1038/nmeth.1650
Kong, C.-Y., Yang, Y.-Q., Han, B., Chen, H.-L., Mao, Y.-Q., Huang, J.-T., et al. (2023). Fecal microbiome transplant from patients with lactation mastitis promotes mastitis in conventional lactating mice. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1123444
Lauko, S., Gancarcikova, S., Hrckova, G., Hajduckova, V., Andrejcakova, Z., Fecskeova, L. K., et al. (2023). Beneficial effect of faecal microbiota transplantation on mild, moderate and severe dextran sodium sulphate-induced ulcerative colitis in a pseudo germ-free animal model. Biomedicines 12, 43. doi: 10.3390/biomedicines12010043
Le Bihan, G., Jubelin, G., Garneau, P., Bernalier-Donadille, A., Martin, C., Beaudry, F., et al. (2015). Transcriptome analysis of Escherichia coli O157:H7 grown in vitro in the sterile-filtrated cecal content of human gut microbiota associated rats reveals an adaptive expression of metabolic and virulence genes. Microbes Infect. 17, 23–33. doi: 10.1016/j.micinf.2014.09.008
Lee, C. H., Steiner, T., Petrof, E. O., Smieja, M., Roscoe, D., Nematallah, A., et al. (2016). Frozen vs Fresh Fecal Microbiota Transplantation and Clinical Resolution of Diarrhea in Patients With Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA 315, 142–149. doi: 10.1001/jama.2015.18098
Le Roy, T., Debédat, J., Marquet, F., Da-Cunha, C., Ichou, F., Guerre-Millo, M., et al. (2018). Comparative evaluation of microbiota engraftment following fecal microbiota transfer in mice models: age, kinetic and microbial status matter. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03289
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Li, X., Liang, S., Xia, Z., Qu, J., Liu, H., Liu, C., et al. (2018). Establishment of a Macaca fascicularis gut microbiome gene catalog and comparison with the human, pig, and mouse gut microbiomes. Gigascience 7, giy100. doi: 10.1093/gigascience/giy100
Li, H., Xie, J., Guo, X., Yang, G., Cai, B., Liu, J., et al. (2022). Bifidobacterium spp. and their metabolite lactate protect against acute pancreatitis via inhibition of pancreatic and systemic inflammatory responses. Gut Microbes 14, 2127456. doi: 10.1080/19490976.2022.2127456
Li, Z., Zhang, B., Wang, N., Zuo, Z., Wei, H., and Zhao, F. (2023). A novel peptide protects against diet-induced obesity by suppressing appetite and modulating the gut microbiota. Gut 72, 686–698. doi: 10.1136/gutjnl-2022-328035
Liang, W., Zhao, L., Zhang, J., Fang, X., Zhong, Q., Liao, Z., et al. (2020). Colonization potential to reconstitute a microbe community in pseudo germ-free mice after fecal microbe transplant from equol producer. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01221
Lin, L., Song, J., Li, J., Zuo, X., Wei, H., Yang, C., et al. (2021). Imaging the in vivo growth patterns of bacteria in human gut Microbiota. Gut Microbes 13, 1960134. doi: 10.1080/19490976.2021.1960134
Liu, Y., Cheng, Y.-Y., Thompson, J., Zhou, Z., Vivas, E. I., Warren, M. F., et al. (2025). Decoding the role of the arginine dihydrolase pathway in shaping human gut community assembly and health-relevant metabolites. Cell Syst. 16, 101292. doi: 10.1016/j.cels.2025.101292
Liu, D., Gao, X., Huang, X., Fan, Y., Wang, Y.-E., Zhang, Y., et al. (2023). Moderate altitude exposure impacts host fasting blood glucose and serum metabolome by regulation of the intestinal flora. Sci. Total Environ. 905, 167016. doi: 10.1016/j.scitotenv.2023.167016
Liu, L., Kirst, M. E., Zhao, L., Li, E., and Wang, G. P. (2022). Microbiome Resilience despite a Profound Loss of Minority Microbiota following Clindamycin Challenge in Humanized Gnotobiotic Mice. Microbiol. Spectr. 10, e0196021. doi: 10.1128/spectrum.01960-21
Liu, H., Tian, R., Wang, H., Feng, S., Li, H., Xiao, Y., et al. (2020). Gut microbiota from coronary artery disease patients contributes to vascular dysfunction in mice by regulating bile acid metabolism and immune activation. J. Transl. Med. 18, 382. doi: 10.1186/s12967-020-02539-x
Liu, Y., Yang, M., Tang, L., Wang, F., Huang, S., Liu, S., et al. (2022). TLR4 regulates RORγt+ regulatory T-cell responses and susceptibility to colon inflammation through interaction with Akkermansia muciniphila. Microbiome 10, 98. doi: 10.1186/s40168-022-01296-x
Lopetuso, L. R., Deleu, S., Godny, L., Petito, V., Puca, P., Facciotti, F., et al. (2023). The first international Rome consensus conference on gut microbiota and faecal microbiota transplantation in inflammatory bowel disease. Gut 72, 1642–1650. doi: 10.1136/gutjnl-2023-329948
Lynch, S. V. and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Manichanh, C., Reeder, J., Gibert, P., Varela, E., Llopis, M., Antolin, M., et al. (2010). Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 20, 1411–1419. doi: 10.1101/gr.107987.110
Mao, K., Gao, J., Wang, X., Li, X., Geng, S., Zhang, T., et al. (2021). Bifidobacterium animalis subsp. lactis BB-12 Has Effect Against Obesity by Regulating Gut Microbiota in Two Phases in Human Microbiota-Associated Rats. Front. Nutr. 8. doi: 10.3389/fnut.2021.811619
Marchesi, J. R. and Ravel, J. (2015). The vocabulary of microbiome research: a proposal. Microbiome 3, 31. doi: 10.1186/s40168-015-0094-5
Marcobal, A., Kashyap, P. C., Nelson, T. A., Aronov, P. A., Donia, M. S., Spormann, A., et al. (2013). A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 7, 1933–1943. doi: 10.1038/ismej.2013.89
Mousa, W. K. and Al Ali, A. (2024). The gut microbiome advances precision medicine and diagnostics for inflammatory bowel diseases. Int. J. Mol. Sci. 25, 11259. doi: 10.3390/ijms252011259
Nagao-Kitamoto, H., Leslie, J. L., Kitamoto, S., Jin, C., Thomsson, K. A., Gillilland, M. G., et al. (2020). Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 26, 608–617. doi: 10.1038/s41591-020-0764-0
Nagao-Kitamoto, H., Shreiner, A. B., Gillilland, M. G., Kitamoto, S., Ishii, C., Hirayama, A., et al. (2016). Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol. Gastroenterol. Hepatol. 2, 468–481. doi: 10.1016/j.jcmgh.2016.02.003
Palleja, A., Mikkelsen, K. H., Forslund, S. K., Kashani, A., Allin, K. H., Nielsen, T., et al. (2018). Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 3, 1255–1265. doi: 10.1038/s41564-018-0257-9
Postgate, J. R. and Hunter, J. R. (1961). On the survival of frozen bacteria. J. Gen. Microbiol. 26, 367–378. doi: 10.1099/00221287-26-3-367
Proctor, A., Parvinroo, S., Richie, T., Jia, X., Lee, S. T. M., Karp, P. D., et al. (2022). Resources to facilitate use of the altered schaedler flora (ASF) mouse model to study microbiome function. mSystems 7, e0029322. doi: 10.1128/msystems.00293-22
Pulschen, A. A., Bendia, A. G., Fricker, A. D., Pellizari, V. H., Galante, D., and Rodrigues, F. (2017). Isolation of uncultured bacteria from Antarctica using long incubation periods and low nutritional media. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01346
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Rajilić-Stojanović, M., Biagi, E., Heilig, H. G. H. J., Kajander, K., Kekkonen, R. A., Tims, S., et al. (2011). Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141, 1792–1801. doi: 10.1053/j.gastro.2011.07.043
Reikvam, D. H., Erofeev, A., Sandvik, A., Grcic, V., Jahnsen, F. L., Gaustad, P., et al. (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6, e17996. doi: 10.1371/journal.pone.0017996
Reitmeier, S., Hitch, T. C. A., Treichel, N., Fikas, N., Hausmann, B., Ramer-Tait, A. E., et al. (2021). Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. ISME Commun. 1, 31. doi: 10.1038/s43705-021-00033-z
Reitmeier, S., Kiessling, S., Clavel, T., List, M., Almeida, E. L., Ghosh, T. S., et al. (2020). Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28, 258–272.e6. doi: 10.1016/j.chom.2020.06.004
Renu, S., Deblais, L., Patil, V., Schrock, J., Kathayat, D., Srivastava, V., et al. (2022). Gut microbiota of obese children influences inflammatory mucosal immune pathways in the respiratory tract to influenza virus infection: optimization of an ideal duration of microbial colonization in a gnotobiotic pig model. Microbiol. Spectr. 10, e0267421. doi: 10.1128/spectrum.02674-21
Respondek, F., Gerard, P., Bossis, M., Boschat, L., Bruneau, A., Rabot, S., et al. (2013). Short-chain fructo-oligosaccharides modulate intestinal microbiota and metabolic parameters of humanized gnotobiotic diet induced obesity mice. PLoS One 8, e71026. doi: 10.1371/journal.pone.0071026
Reygner, J., Charrueau, C., Delannoy, J., Mayeur, C., Robert, V., Cuinat, C., et al. (2020). Freeze-dried fecal samples are biologically active after long-lasting storage and suited to fecal microbiota transplantation in a preclinical murine model of Clostridioides difficile infection. Gut Microbes 11, 1405–1422. doi: 10.1080/19490976.2020.1759489
Ridaura, V. K., Faith, J. J., Rey, F. E., Cheng, J., Duncan, A. E., Kau, A. L., et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. doi: 10.1126/science.1241214
Rocha Martin, V. N., Del’Homme, C., Chassard, C., Schwab, C., Braegger, C., Bernalier-Donadille, A., et al. (2022). A proof of concept infant-microbiota associated rat model for studying the role of gut microbiota and alleviation potential of cutibacterium avidum in infant colic. Front. Nutr. 9. doi: 10.3389/fnut.2022.902159
Russo, C., Surdo, S., Valle, M. S., and Malaguarnera, L. (2024). The gut microbiota involvement in the panorama of muscular dystrophy pathogenesis. Int. J. Mol. Sci. 25, 11310. doi: 10.3390/ijms252011310
Saint-Cyr, M. J., Perrin-Guyomard, A., Houée, P., Rolland, J.-G., and Laurentie, M. (2013). Evaluation of an oral subchronic exposure of deoxynivalenol on the composition of human gut microbiota in a model of human microbiota-associated rats. PLoS One 8, e80578. doi: 10.1371/journal.pone.0080578
Salandre, A., Delannoy, J., Goudiaby, M. T. B., Barbut, F., Thomas, M., Waligora-Dupriet, A.-J., et al. (2023). A simple in vitro test to select stools for fecal microbiota transplantation to limit intestinal carriage of extensively drug-resistant bacteria. Microorganisms 11, 2753. doi: 10.3390/microorganisms11112753
Sánchez-Quintero, M. J., Delgado, J., Martín Chaves, L., Medina-Vera, D., Murri, M., Becerra-Muñoz, V. M., et al. (2023). Multi-omics approach reveals prebiotic and potential antioxidant effects of essential oils from the mediterranean diet on cardiometabolic disorder using humanized gnotobiotic mice. Antioxidants 12, 1643. doi: 10.3390/antiox12081643
Sánchez-Quintero, M. J., Delgado, J., Medina-Vera, D., Becerra-Muñoz, V. M., Queipo-Ortuño, M. I., Estévez, M., et al. (2022). Beneficial effects of essential oils from the mediterranean diet on gut microbiota and their metabolites in ischemic heart disease and type-2 diabetes mellitus. Nutrients 14, 4650. doi: 10.3390/nu14214650
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Sarma-Rupavtarm, R. B., Ge, Z., Schauer, D. B., Fox, J. G., and Polz, M. F. (2004). Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 70, 2791–2800. doi: 10.1128/AEM.70.5.2791-2800.2004
Satokari, R., Mattila, E., Kainulainen, V., and Arkkila, P. E. T. (2015). Simple faecal preparation and efficacy of frozen inoculum in faecal microbiota transplantation for recurrent Clostridium difficile infection–an observational cohort study. Aliment Pharmacol. Ther. 41, 46–53. doi: 10.1111/apt.13009
Savin, Z., Kivity, S., Yonath, H., and Yehuda, S. (2018). Smoking and the intestinal microbiome. Arch. Microbiol. 200, 677–684. doi: 10.1007/s00203-018-1506-2
Schaedler, R. W., Dubs, R., and Costello, R. (1965). Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122 (1), 77–82. doi: 10.1084/jem.122.1.77
Schloss, P. D., Schubert, A. M., Zackular, J. P., Iverson, K. D., Young, V. B., and Petrosino, J. F. (2012). Stabilization of the murine gut microbiome following weaning. Gut Microbes 3, 383–393. doi: 10.4161/gmic.21008
Schoeler, M., Chakaroun, R., Brolin, H., Larsson, I., Perkins, R., Marschall, H.-U., et al. (2024). Moderate variations in the human diet impact the gut microbiota in humanized mice. Acta Physiol. (Oxf) 240, e14100. doi: 10.1111/apha.14100
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio 6, e00974. doi: 10.1128/mBio.00974-15
Seedorf, H., Griffin, N. W., Ridaura, V. K., Reyes, A., Cheng, J., Rey, F. E., et al. (2014). Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159, 253–266. doi: 10.1016/j.cell.2014.09.008
Sender, R., Fuchs, S., and Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Sharon, G., Cruz, N. J., Kang, D.-W., Gandal, M. J., Wang, B., Kim, Y.-M., et al. (2019). Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600–1618.e17. doi: 10.1016/j.cell.2019.05.004
Shayya, N. W., Foote, M. S., Langfeld, L. Q., Du, K., Bandick, R., Mousavi, S., et al. (2023). Human microbiota associated IL-10-/- mice: A valuable enterocolitis model to dissect the interactions of Campylobacter jejuni with host immunity and gut microbiota. Eur. J. Microbiol. Immunol. (Bp) 12, 107–122. doi: 10.1556/1886.2022.00024
Silley, P. (2009). Human flora-associated rodents–does the data support the assumptions? Microb. Biotechnol. 2, 6–14. doi: 10.1111/j.1751-7915.2008.00069.x
Sintes, R., McLellan, P., Navelli, G., Landman, C., Delage, S., Truong, S., et al. (2024). Use of frozen native feces for fecal microbiota transplantation in recurrent clostridioides difficile infection: a simple way to improve the efficiency of donor feces preparation. Antimicrob. Agents Chemother. 68 (10), e0073424. doi: 10.1128/aac.00734-24
Sjöland, W., Wahlström, A., Makki, K., Schöler, M., Molinaro, A., Olsson, L., et al. (2023). Absence of gut microbiota reduces neonatal survival and exacerbates liver disease in Cyp2c70 -deficient mice with a human-like bile acid composition. Clin. Sci. 137, 995–1011. doi: 10.1042/CS20230413
Sokol, H., Seksik, P., Furet, J. P., Firmesse, O., Nion-Larmurier, I., Beaugerie, L., et al. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflammation Bowel Dis. 15, 1183–1189. doi: 10.1002/ibd.20903
Spatz, M., Da Costa, G., Ventin-Holmberg, R., Planchais, J., Michaudel, C., Wang, Y., et al. (2023). Antibiotic treatment using amoxicillin-clavulanic acid impairs gut mycobiota development through modification of the bacterial ecosystem. Microbiome 11, 73. doi: 10.1186/s40168-023-01516-y
Staley, C., Kaiser, T., Beura, L. K., Hamilton, M. J., Weingarden, A. R., Bobr, A., et al. (2017). Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5, 87. doi: 10.1186/s40168-017-0306-2
Sulaiman, J. E., Thompson, J., Cheung, P. L. K., Qian, Y., Mill, J., James, I., et al. (2025). Phocaeicola vulgatus shapes the long-term growth dynamics and evolutionary adaptations of clostridioides difficile. Cell Host Microbe 33, 42–58.e10. doi: 10.1016/j.chom.2024.12.001
Sulaiman, J. E., Thompson, J., Qian, Y., Vivas, E. I., Diener, C., Gibbons, S. M., et al. (2024). Elucidating human gut microbiota interactions that robustly inhibit diverse clostridioides difficile strains across different nutrient landscapes. Nat. Commun. 15, 7416. doi: 10.1038/s41467-024-51062-w
Sun, D., Wang, C., Sun, L., Hu, L., Fang, Z., Deng, Q., et al. (2022). Preliminary report on intestinal flora disorder, faecal short-chain fatty acid level decline and intestinal mucosal tissue weakening caused by litchi extract to induce systemic inflammation in HFA mice. Nutrients 14, 776. doi: 10.3390/nu14040776
Tamura, M., Nakagawa, H., Hori, S., Suzuki, T., and Hirayama, K. (2019). Plasma quercetin metabolites are affected by intestinal microbiota of human microbiota-associated mice fed with a quercetin-containing diet. J. Clin. Biochem. Nutr. 65, 232–239. doi: 10.3164/jcbn.19-45
Tedjo, D. I., Jonkers, D. M. A. E., Savelkoul, P. H., Masclee, A. A., van Best, N., Pierik, M. J., et al. (2015). The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One 10, e0126685. doi: 10.1371/journal.pone.0126685
Thaiss, C. A., Zeevi, D., Levy, M., Zilberman-Schapira, G., Suez, J., Tengeler, A. C., et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529. doi: 10.1016/j.cell.2014.09.048
Tintelnot, J., Xu, Y., Lesker, T. R., Schönlein, M., Konczalla, L., Giannou, A. D., et al. (2023). Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 615, 168–174. doi: 10.1038/s41586-023-05728-y
Tirelle, P., Breton, J., Riou, G., Déchelotte, P., Coëffier, M., and Ribet, D. (2020). Comparison of different modes of antibiotic delivery on gut microbiota depletion efficiency and body composition in mouse. BMC Microbiol. 20, 340. doi: 10.1186/s12866-020-02018-9
Togao, M., Kurakawa, T., Tajima, S., Wagai, G., Ohta-Takada, Y., Otsuka, J., et al. (2023). Human gut microbiota influences drug-metabolizing enzyme hepatic Cyp3a: A human flora-associated mice study. J. Toxicol. Sci. 48, 333–343. doi: 10.2131/jts.48.333
Trexler, P. C. and Orcutt, R. P. (1999). Development of gnotobiotics and contamination con- trol in laboratory animal science. In: Macpherson, C. W. and Mattingly, S. F.. eds. 50 Years of Laboratory Animal Science. Memphis: American Association for Laboratory Animal Science. p. 121–128.
Tropini, C., Moss, E. L., Merrill, B. D., Ng, K. M., Higginbottom, S. K., Casavant, E. P., et al. (2018). Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 173, 1742–1754.e17. doi: 10.1016/j.cell.2018.05.008
Turnbaugh, P. J., Ridaura, V. K., Faith, J. J., Rey, F. E., Knight, R., and Gordon, J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14. doi: 10.1126/scitranslmed.3000322
Uzbay, T. (2019). Germ-free animal experiments in the gut microbiota studies. Curr. Opin. Pharmacol. 49, 6–10. doi: 10.1016/j.coph.2019.03.016
Van Den Ham, K. M., Little, M. R., Bednarski, O. J., Fusco, E. M., Mandal, R. K., Mitra, R., et al. (2023). Creation of a non-Western humanized gnotobiotic mouse model through the transplantation of rural African fecal microbiota. Microbiol. Spectr. 11, e0155423. doi: 10.1128/spectrum.01554-23
von Klitzing, E., Ekmekciu, I., Bereswill, S., and Heimesaat, M. M. (2017a). Intestinal and Systemic Immune Responses upon Multi-drug Resistant Pseudomonas aeruginosa Colonization of Mice Harboring a Human Gut Microbiota. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02590
von Klitzing, E., Ekmekciu, I., Bereswill, S., and Heimesaat, M. M. (2017b). Acute ileitis facilitates infection with multidrug resistant Pseudomonas aeruginosa in human microbiota-associated mice. Gut Pathog. 9, 4. doi: 10.1186/s13099-017-0154-4
Wahlström, A., Kovatcheva-Datchary, P., Ståhlman, M., Khan, M.-T., Bäckhed, F., and Marschall, H.-U. (2017). Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota. J. Lipid Res. 58, 412–419. doi: 10.1194/jlr.M072819
Walter, J., Armet, A. M., Finlay, B. B., and Shanahan, F. (2020). Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232. doi: 10.1016/j.cell.2019.12.025
Wensel, C. R., Pluznick, J. L., Salzberg, S. L., and Sears, C. L. (2022). Next-generation sequencing: insights to advance clinical investigations of the microbiome. J. Clin. Invest. 132, e154944. doi: 10.1172/JCI154944
Widjaja, F. and Rietjens, I. M. C. M. (2023). From-toilet-to-freezer: A review on requirements for an automatic protocol to collect and store human fecal samples for research purposes. Biomedicines 11, 2658. doi: 10.3390/biomedicines11102658
Wos-Oxley, M., Bleich, A., Oxley, A. P. A., Kahl, S., Janus, L. M., Smoczek, A., et al. (2012). Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 3, 234–249. doi: 10.4161/gmic.19934
Wrzosek, L., Ciocan, D., Borentain, P., Spatz, M., Puchois, V., Hugot, C., et al. (2018). Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Sci. Rep. 8, 6854. doi: 10.1038/s41598-018-25300-3
Wymore Brand, M., Wannemuehler, M. J., Phillips, G. J., Proctor, A., Overstreet, A.-M., Jergens, A. E., et al. (2015). The altered schaedler flora: continued applications of a defined murine microbial community. ILAR J. 56, 169–178. doi: 10.1093/ilar/ilv012
Xia, G.-H., You, C., Gao, X.-X., Zeng, X.-L., Zhu, J.-J., Xu, K.-Y., et al. (2019). Stroke dysbiosis index (SDI) in gut microbiome are associated with brain injury and prognosis of stroke. Front. Neurol. 10. doi: 10.3389/fneur.2019.00397
Xu, H., Wang, S., Jiang, Y., Wu, J., Chen, L., Ding, Y., et al. (2023). Poria cocos polysaccharide ameliorated antibiotic-associated diarrhea in mice via regulating the homeostasis of the gut microbiota and intestinal mucosal barrier. Int. J. Mol. Sci. 24, 1423. doi: 10.3390/ijms24021423
Xue, J., Allaband, C., Zuffa, S., Poulsen, O., Meadows, J., Zhou, D., et al. (2025). Gut microbiota and derived metabolites mediate obstructive sleep apnea induced atherosclerosis. Gut Microbes 17 (1), 2474142. doi: 10.1080/19490976.2025.2474142
Yang, J., Liang, J., Hu, N., He, N., Liu, B., Liu, G., et al. (2024). The gut microbiota modulates neuroinflammation in alzheimer’s disease: elucidating crucial factors and mechanistic underpinnings. CNS Neurosci. Ther. 30, e70091. doi: 10.1111/cns.70091
Ye, H., Ghosh, T. S., Hueston, C. M., Vlckova, K., Golubeva, A. V., Hyland, N. P., et al. (2023). Engraftment of aging-related human gut microbiota and the effect of a seven-species consortium in a pre-clinical model. Gut Microbes 15, 2282796. doi: 10.1080/19490976.2023.2282796
Zabolotneva, A. A., Gaponov, A. M., Roumiantsev, S. A., Vasiliev, I. Y., Grigoryeva, T. V., Kit, O. I., et al. (2023). Alkylresorcinols as new modulators of the metabolic activity of the gut microbiota. IJMS 24, 14206. doi: 10.3390/ijms241814206
Zackular, J. P., Baxter, N. T., Chen, G. Y., and Schloss, P. D. (2016). Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere 1, e00001–e00015. doi: 10.1128/mSphere.00001-15
Zhan, K., Wu, H., Xu, Y., Rao, K., Zheng, H., Qin, S., et al. (2024). The function of the gut microbiota–bile acid–TGR5 axis in diarrhea-predominant irritable bowel syndrome. mSystems 9, e01299–e01223. doi: 10.1128/msystems.01299-23
Zhang, P.-P., Li, L.-L., Han, X., Li, Q.-W., Zhang, X.-H., Liu, J. J., et al. (2020). Fecal microbiota transplantation improves metabolism and gut microbiome composition in db/db mice. Acta Pharmacol. Sin. 41, 678–685. doi: 10.1038/s41401-019-0330-9
Zhang, H., Sparks, J. B., Karyala, S. V., Settlage, R., and Luo, X. M. (2015). Host adaptive immunity alters gut microbiota. ISME J. 9, 770–781. doi: 10.1038/ismej.2014.165
Zhang, Q., Widmer, G., and Tzipori, S. (2013). A pig model of the human gastrointestinal tract. Gut Microbes 4, 193–200. doi: 10.4161/gmic.23867
Zhang, R.-X., Xu, J.-T., Zhong, H.-J., Cai, Y.-L., Zhuang, Y.-P., Xie, Y.-T., et al. (2023). Gut microbiota from essential tremor patients aggravates tremors in mice. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1252795
Zhong, H.-J., Zhuang, Y.-P., Xie, X., Song, J.-Y., Wang, S.-Q., Wu, L., et al. (2024). Washed microbiota transplantation promotes homing of group 3 innate lymphoid cells to the liver via the CXCL16/CXCR6 axis: a potential treatment for metabolic-associated fatty liver disease. Gut Microbes 16, 2372881. doi: 10.1080/19490976.2024.2372881
Zhou, Y., Ji, H., Zhang, S., Zhang, X., Zhang, J., Wang, Y., et al. (2025). Effects of different bowel preparation regimens and age factors on the gut microbiota: a prospective randomized controlled study. J. Gastroenterol. Hepatol. 40, 599–608. doi: 10.1111/jgh.16868
Zhou, J., Zhou, Z., Ji, P., Ma, M., Guo, J., and Jiang, S. (2019). Effect of fecal microbiota transplantation on experimental colitis in mice. Exp. Ther. Med. 17, 2581–2586. doi: 10.3892/etm.2019.7263
Keywords: human microbiota-associated animal models, fecal microbiota transplantation, gut microbe-host interactions, microbiome, engraftment, procedure
Citation: Huang X, Yu Y, Tian N, Huang J, Zhang X and Yu R (2025) Human microbiota-associated animal models: a review. Front. Cell. Infect. Microbiol. 15:1644187. doi: 10.3389/fcimb.2025.1644187
Received: 10 June 2025; Accepted: 14 August 2025;
Published: 27 August 2025.
Edited by:
Aabid Hussain, Cleveland Clinic, United StatesReviewed by:
Jordy Evan Sulaiman, Hong Kong Polytechnic University, Hong Kong SAR, ChinaAwatif Abid Al-Judaibi, Jeddah University, Saudi Arabia
Qiwen Cheng, Shandong University, China
Copyright © 2025 Huang, Yu, Tian, Huang, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Yu, eXVyb25nMTk2OTA1QDE2My5jb20=
†These authors have contributed equally to this work
 Xiangning Huang
Xiangning Huang Yunfeng Yu
Yunfeng Yu Na Tian3
Na Tian3 Rong Yu
Rong Yu


