- 1Laboratory of Veterinary Public Health, Joint Graduate School of Veterinary Medicine, Yamaguchi University, Yamaguchi, Japan
- 2Division of Veterinary Public Health, Department of Veterinary Science, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia
- 3Department of Veterinary Science, National Institute of Infectious Diseases, Tokyo, Japan
Background: Francisella tularensis is a highly infectious Gram-negative bacterium that causes tularemia in humans and animals. It has a remarkable ability to survive and replicate within a wide range of host cells. F. novicida shares many characteristics with of F. tularensis. However, it is rarely pathogenic in humans, and its reduced virulence makes it a suitable model organism for studying F. tularensis infection. This study aimed to identify the pathogenic factors of F. novicida.
Methods: Using a novel infection model with HeLa cells expressing FcγRII (HeLa-FcγRII cells), we screened 2,232 transposon mutants of F. novicida pre-treated with antiserum containing F. novicida antibodies to find less cytotoxicity strains. The transposon insertion site was identified by sequencing, leading to the determination of the genes responsible for the attenuated cytotoxicity. Additionally, the intracellular behavior of the mutant was investigated within both HeLa-FcγRII and THP-1 cells.
Results and discussion: A total of thirteen mutants with attenuated cytotoxicity were isolated, and their responsible genes were identified. They are figE, slt, fopA, iglC, igID, iglF, iglI, pdpB, pdpA, ampG, wbtF, and one unnamed gene (FTN_0096). We focused on the wbtF gene. The F. novicida wild-type (WT) strain showed intracellular replication in HeLa-FcγRII and THP-1 cells, but the number of intracellular wbtF mutants decreased. The wbtF mutant could not escape from phagolysosomes in the initial phases of infection and was digested within the lysosome. The wbtF mutant was also detected in the mitochondria and the Golgi complex. The cytokine response induced by wbtF mutant was comparable to that of the WT strain. These findings indicate that wbtF is important for the intracellular replication of F. novicida.
1 Introduction
Francisella tularensis is a highly infectious Gram-negative bacterium that causes tularemia, a zoonosis affecting humans and animals. The genus Francisella comprises several species including F. tularensis, F. philomiragia, F. noatunensis, F. hispaniensis and F. novicida (Kingry and Petersen, 2014; Sjodin et al., 2012). F. tularensis is divided into two main subspecies: Type A (F. tularensis subsp. tularensis [F. tularensis]) and Type B (F. tularensis subsp. holarctica [F. holarctica]). Type A is considerably more virulent than Type B and is commonly associated with severe disease in humans, whereas Type B typically causes milder illness. Pneumonic tularemia, the most dangerous form, is transmitted through aerosols or inhalation. Remarkably, inhaling as few as 25 colony-forming units (CFUs) may result in pulmonary infection, making F. tularensis one of the most infectious bacteria transmitted via the aerosol route (Harrell et al., 2024). Francisella is a significant pathogen associated with high morbidity and mortality rates. The most virulent strain of Francisella induces systemic forms of tularemia (pneumonic, typhoidal), with fatality rates reaching 60% (Yeni et al., 2021; Wawszczak et al., 2022). Due to its high infectivity and potential for airborne spread, F. tularensis has been developed and stockpiled as a biological warfare agent by several nations (Dennis et al., 2001).
F. novicida is classified as a separate species from F. tularensis, but due to the high genomic homology, it is sometimes classified as a subspecies of F. tularensis (Johansson et al., 2010). Because Francisella novicida (F. novicida) is considerably less virulent than F. tularensis, it is commonly used as a surrogate strain for virulent F. tularensis. In contrast to F. tularensis, F. novicida is an opportunistic pathogen that causes illness and even death in debilitated or immunocompromised patients but not in healthy individuals. Due to the rarity of F. novicida infections, effective detection is challenging (Brett et al., 2012; Kingry and Petersen, 2014). F. novicida shares many characteristics with Type A strains of F. tularensis, showing similarities in the genome sequence, intracellular life cycle, and mechanisms of infectivity, including rapid phagosomal escape followed by vigorous cytosolic replication. It also infects macrophages, causing disease in mice. However, due to its lower virulence in humans, it can be safely handled under Biosafety Level 2 laboratory conditions (Kingry and Petersen, 2014; Marecic et al., 2017). F. novicida strain U112 possesses a potent tool in the form of a specific transposon mutant, which has been used to investigate the intracellular life cycle of Francisella and elucidate its virulence mechanisms (Weiss et al., 2007; Gallagher et al., 2007).
Typically, nonpathogenic bacteria taken up by host cells are enclosed in vacuoles or phagosomes, which mature through the endocytic pathway and ultimately fuse with lysosomes, leading to bacterial degradation. Intracellular pathogens have evolved strategies to evade this process within phagocytic cells and resist phagosome–lysosome fusion (Ray et al., 2009). In particular, Francisella species exhibit a complex intracellular life cycle and notable environmental persistence (Rowe and Huntley, 2015). Their survival and replication rely heavily on escaping the initial phagosomal compartment and replicating within the host cell cytosol, making Francisella a model cytosolic pathogen. The ability to persist and multiply inside phagocytes and other host cells is fundamental to its pathogenicity. Disruptions in key mechanisms, such as receptor engagement, resistance to reactive oxygen species (ROS), phagosomal escape, cytosolic replication, and evasion of innate immune recognition, significantly impair F. tularensis survival and reduce its virulence in both in vitro and in vivo models (Celli and Zahrt, 2013).
wbtF is assigned as a potential NAD-dependent epimerase and UDP-glucose 4 epimerase (UGE) (Thomas et al., 2007). It shows sequence similarity with established UGEs, including wbpP from Pseudomonas aeruginosa (Prior et al., 2003). wbpP is necessary for producing UDP-N-acetyl-D-galacturonic acid, a precursor to the galactosaminuronic acid-derived components of the O-antigen (Miller et al., 2008). UGE, part of the short-chain dehydrogenase/reductase enzyme superfamily, catalyzes the final step in the interconversion of UDP-glucose and UDP-galactose during galactose metabolism in bacteria and mammals (Fushinobu, 2021). Thomas et al. (2007) identified wbtF genes as a virulence determinant of F. tularensis, but its function remains unknown.
In this study, we created a transposon mutant library of F. novicida and screened for infection using epithelial cells, HeLa cells expressing the FcγRII (HeLa-FcγRII cells) (Nakamura et al., 2024). We focused our investigation on wbtF and analyzed that the wbtF was associated with the intracellular replication of F. novicida.
2 Materials and methods
2.1 Bacterial strains and culture conditions
F. novicida U112 was obtained from the Pathogenic Microorganism Genetic Resource Stock Center (Gifu University) and cultured aerobically at 37°C in a chemically defined medium (CDM) (Nagle et al., 1960) or on brain–heart infusion broth enriched with cysteine (BHIc) (McGann et al., 2010) and solidified with 1.5% agar (Wako Laboratory Chemicals, Osaka, Japan).
2.2 Cell culture conditions
HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher, Waltham, MA, USA). The cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. Similarly, the human monocytic cell line THP-1 was cultured under the same incubation conditions in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated FBS.
2.3 Transposon mutant library construction
A transposon mutant library was constructed using the Ez-Tn5 Transposome system (Epicentre, Madison, WI, USA). The multiple cloning sites of plasmid pMOD3 were digested with HindIII and EcoRI to construct the transposon. A kanamycin resistance cassette from pKEK1140 (Rodriguez et al., 2008) was inserted into these sites, creating the plasmid pMOD3-FtKm. The transposon region of pMOD3-FtKm was amplified by polymerase chain reaction, purified, and combined with transposase according to the manufacturer’s instructions. This transposon mixture was introduced into F. novicida by cryotransformation. Following transformation, the bacteria were plated on BHIc agar supplemented with 50 μg/mL kanamycin to select for mutants.
2.4 Cytotoxicity assay
HeLa-FcγRII cells (5 × 104 cells/well) were seeded in 48-well tissue culture plates and incubated for 24 h. F. novicida strains preincubated with mouse serum containing anti-F. novicida antibodies were added at a multiplicity of infection (MOI) of 1. The plates were centrifuged at 300 ×g at room temperature for 10 min to promote bacterial contact with the cells and then incubated at 37°C for the designated times. Following incubation, the cells were washed three times with DMEM to remove nonadherent bacteria and treated with gentamicin (50 μg/mL) for 1 h to eliminate extracellular bacteria. Infected cells were monitored microscopically at designated time intervals to assess the relative cytotoxicity of the transposon mutant library. To evaluate cytotoxicity, lactate dehydrogenase (LDH) release was measured after incubating cells in DMEM at 37°C for a defined period. LDH levels in the culture supernatant were then quantified using the LDH Cytotoxicity Detection Kit (Takara Bio, Shiga, Japan). In brief, cell culture supernatants were collected and incubated with the catalyst and dye solutions for 30 min at room temperature in the dark. Absorbance was measured at 490 nm with a reference wavelength of 600 nm using a microplate reader. LDH activity was expressed as a percentage of total LDH release, determined from lysed cell controls.
2.5 Sequence analysis of transposon mutants
The plasmid pMOD3 carries the R6Kγ origin of replication, facilitating propagation in Escherichia coli. Genomic DNA from F. novicida transposon mutants was extracted using a PureLink Genomic DNA Mini Kit (Thermo Fisher) and digested with restriction enzymes (XhoI, BglII, EcoRI, SalI, NotI, BamHI, PstI, and SphI). The resultant DNA fragments were blunted using a DNA Blunting Kit (Takara Bio) and then ligated using Ligation High Ver. 2 (Toyobo). The ligation products were introduced into One Shot PIR1 Chemically Competent E. coli (Thermo Fisher) via transformation. Kanamycin-resistant colonies were selected, and plasmid DNA was isolated. Sequencing was performed using the primers specified in the Ez-Tn5 Transposome system manual to determine transposon insertion sites.
2.6 Green fluorescent protein–expression by F. novicida and complementary strains
GFP-expressing plasmids (pOM5-GFP) were generated as described by Shimizu et al. (2019). Briefly, the plasmids were introduced into the wild-type (WT) and wbtF mutant strains of F. novicida via electroporation. To create pOM5-wbtF or complementary strains, wbtF of F. novicida, along with its native promoter region (300 bp upstream), was cloned into the pOM5 vector. The resultant plasmid (pOM5-wbtF) was used to transform the wbtF mutant strains of F. novicida via electroporation for complementation studies.
2.7 Intracellular replication assay
THP-1 cells (4 × 105 cells/well) were seeded in 24-well tissue culture plates and differentiated by treatment with 200 nM phorbol 12-myristate 13-acetate (PMA) for 48 h. HeLa-FcγRII cells (1 × 105 cells/well) were cultured overnight on 12-mm glass coverslips placed in 24-well plates. F. novicida strains were added to the THP-1 cells and F. novicida strains were opsonized with mouse serum added to the HeLa-FcγRII cells at an MOI of 1. After infection, the THP-1 and HeLa-FcγRII cells were washed three times with RPMI 1640 medium and DMEM, respectively. To remove extracellular bacteria, all wells were treated with gentamicin (50 μg/mL) for 1 h. Then, the cells were incubated in fresh medium at 37°C for the designated times. To analyze intracellular replication, infected cells were washed with phosphate-buffered saline and lysed using 0.1% Triton X-100 in chemically defined medium (CDM). The lysates underwent serial dilution and were plated on BHIc agar to determine the number of CFUs.
2.8 Fluorescence microscopy
THP-1 cells (4 × 105 cells/well) were seeded on 12-mm glass coverslips in 24-well tissue culture plates and differentiated with 100 nM phorbol 12-myristate 13-acetate (PMA) for 48 h. HeLa-FcγRII cells (1 × 105 cells/well) were cultured overnight on 12-mm glass coverslips placed in 24-well plates. The THP-1 cells were infected with GFP-expressing F. novicida strains and incubated for the designated times. For HeLa-FcγRII cells, GFP-expressing F. novicida strains were preincubated with mouse serum and added at an MOI of 1, followed by incubation for the designated times. The cells were stained with LysoTracker Red DND-99, MitoTracker Deep Red FM, and BODIPY TR Ceramide (Thermo Fisher) to visualize lysosomes, mitochondria, and the Golgi complex, respectively. To detect lysosomal-associated membrane protein 1 (LAMP-1), cells were fixed using a PLP Solution Set (FUJIFILM Wako Chemicals) containing 5% sucrose for 1 h at 37°C and then permeabilized using ice-cold methanol for 10 s. The fixed cells were incubated with anti-LAMP-1 antibody (ab25245, 1:100; Abcam), followed by staining with TRITC-conjugated anti-rat IgG secondary antibody (ab150158, 1:1000; Abcam). Confocal images were captured using a FluoView FV1000 laser scanning microscope (Olympus, Tokyo, Japan).
2.9 ELISA
THP-1 cells (4 × 105 cells per well) were seeded into a 48-well culture plate and treated with 100 nM PMA for 48 hours. Following this, they were infected with F. novicida strains, including the wbtF mutant and its complementary strains. After 2, 6, 12, and 24 h of incubation, the levels of tumor necrosis factor (TNF)-α and interleukin (IL)-6 in the culture supernatants were quantified using the ELISA MAX Standard Kit (Biolegend, San Diego, CA), following the manufacturer’s protocol.
2.10 Statistical analysis
Differences between groups were evaluated using Welch’s t-test or multiple comparison tests, including the Bonferroni and Dunnett methods, as appropriate. Before these analyses, data normality was assessed using the Shapiro–Wilk test. P < 0.01 was considered statistically significant.
3 Results
3.1 Thirteen genes in the F. novicida transposon mutant library showed less cytotoxicity to HeLa-FcγRII cells
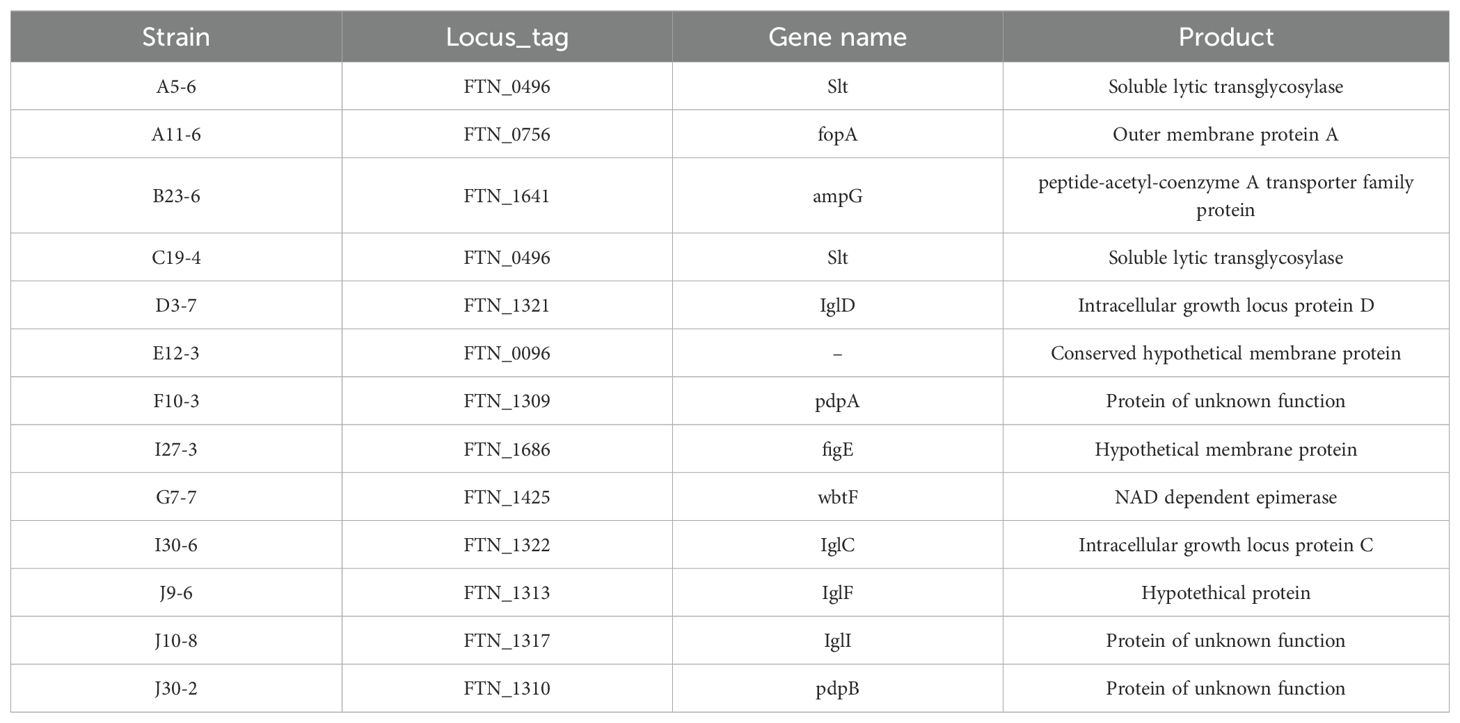
We constructed an F. novicida transposon mutant library to identify the virulence factors of F. novicida. We previously developed a novel infection model of Francisella using HeLa cells expressing mouse FcγRII (HeLa-FcγRII) cells (Nakamura et al., 2024). In this cell line, cellular uptake occurs via antibodies and FcγRII. Therefore, it was expected that screening with this cell line would allow us to identify novel virulence factors crucial for pathogenesis after cellular uptake. We screened the F. novicida transposon mutant library using HeLa-FcγRII cells. F. novicida exhibits cytotoxicity toward epithelial cells, including HeLa-FcγRII cells, leading to cell death. We microscopically screened the transposon mutant library for strains lacking cytotoxic activity to identify genes associated with this cytotoxic effect. Of the 2,232 transposon mutants screened, 13 were identified as less cytotoxic. Significant differences in host cell condition were evident, while wild-type infection caused pronounced disruption and cell loss, monolayers exposed to the 13 mutants retained their structure and remained largely intact. To confirm these results, LDH assays were performed. The mutant strains caused less LDH release from HeLa-FcγRII cells than the WT strain, indicating reduced cytotoxicity (Figure 1). The insertion sites in these mutant strains were sequenced to determine the disrupted genes (Table 1). We focused on mutants with the code number G7-7, which encode wbtF, and analyzed the gene function.
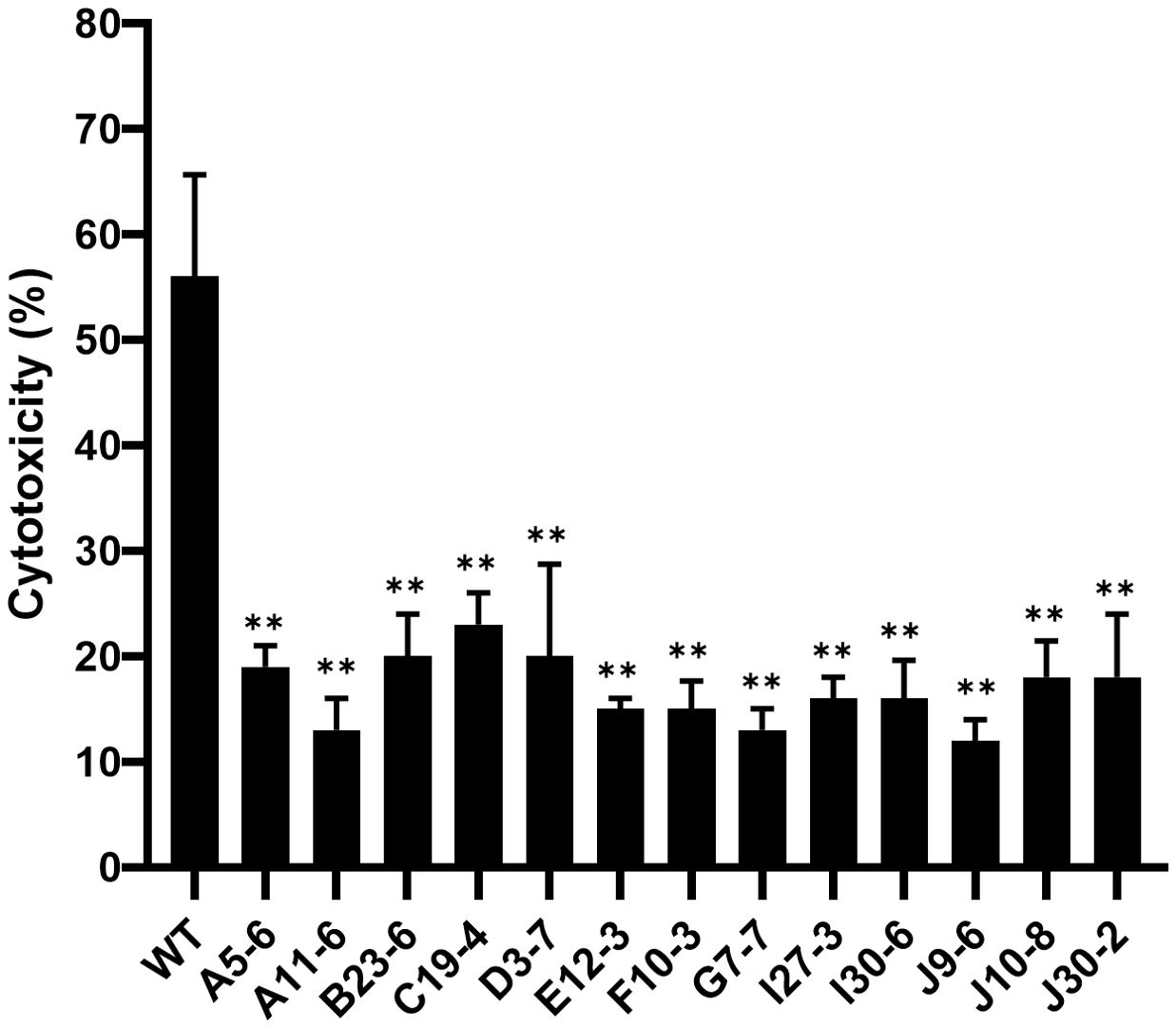
Figure 1. HeLa-FcγRII cells were infected with transposon mutants of F. novicida preincubated with mouse serum containing anti-F. novicida antibodies at an MOI of 1 and incubated at 37°C for 24 h. LDH release was measured as an indicator of cytotoxicity. The graph presents the mean and standard deviations (SD) of the LDH release at 24 h from three identical experiments. Statistical significance relative to the WT strain was determined using multiple comparisons and is indicated by asterisks. **P < 0.01.
3.2 Effect of wbtF on intracellular replication in HeLa-FcγRII cells
To determine the effect of wbtF on intracellular replication in HeLa-FcγRII cells in detail, we infected them with F. novicida WT and the wbtF mutant. Through a fluorescence microscope, we discovered intracellular bacteria using GFP-expressing F. novicida strains. We analyzed the intracellular of the WT and wbtF mutant 12h and 24h after infection in HeLa-FcγRII cells (Figure 2A). From 12 to 48 h postinfection, the rate of intracellular replication of the WT strain increased, while that of the wbtF mutant decreased (Figure 2B). Complementation of the wbtF mutant restored replication to the WT level (Figure 2B). These results suggested wbtF involvement in the intracellular replication of F. novicida in HeLa-FcγRII cells.
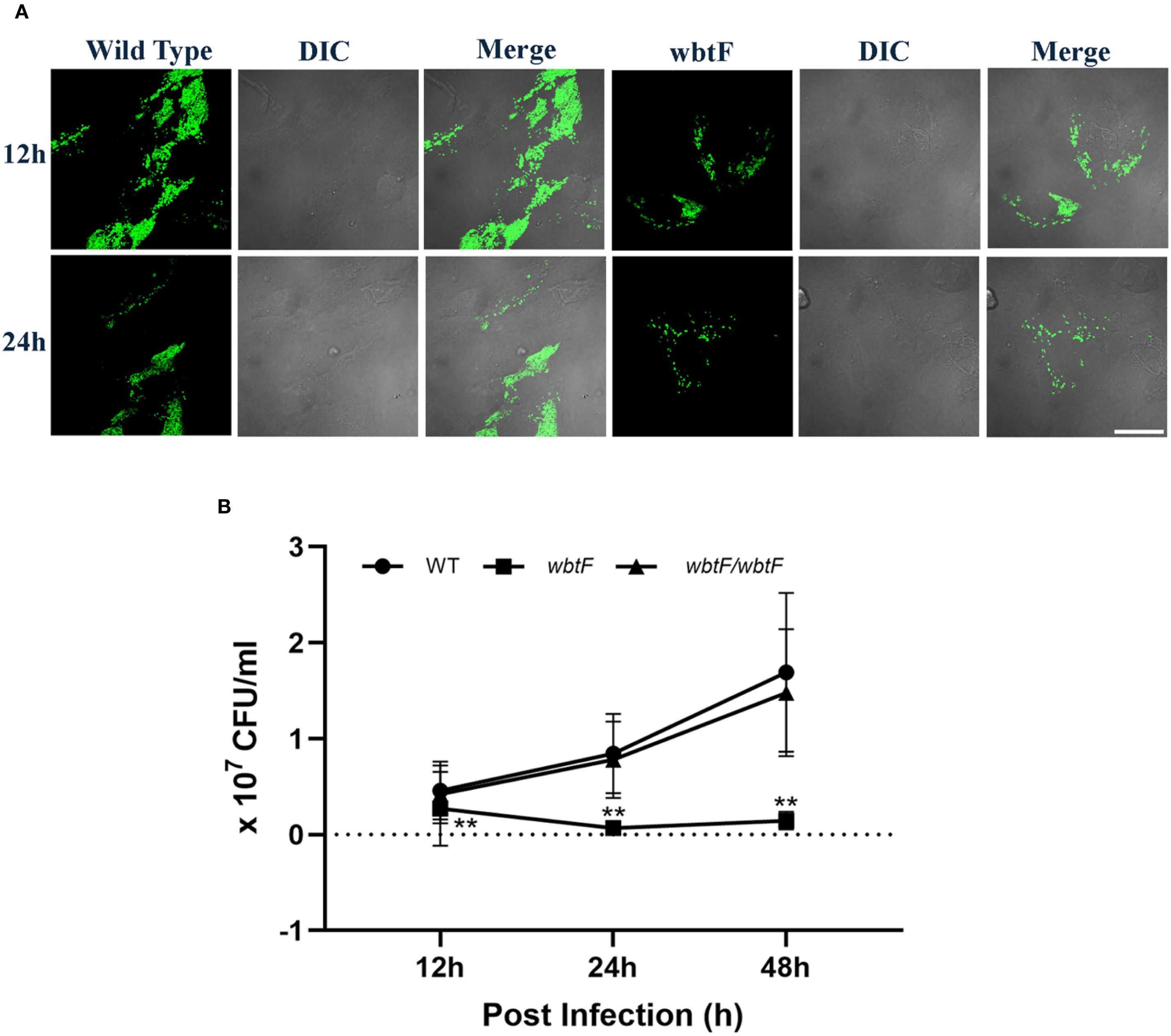
Figure 2. HeLa-FcγRII cells were infected with F. novicida strains preincubated with mouse serum containing anti-F. novicida antibodies at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h (A) The infected HeLa-FcγRII cells were fixed and observed at 12 and 24 h postinfection. Scale bar = 20 μm. (B) At 12, 24, and 48 h postinfection, the infected HeLa-FcγRII cells were lysed with 0.1% Triton X-100, and the lysates were plated on BHIc agar. The graph presents the mean and SD of the number of CFUs at the designated time points from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using multiple comparisons and is indicated by asterisks. **P < 0.01.
3.3 Role of wbtF in avoiding phagolysosomes in HeLa-FcγRII cells
We studied how the wbtF mutant replicates intracellularly in HeLa-FcγRII cells to clarify the role of wbtF in the intracellular replication of Francisella and its mechanism of escape from phagolysosomal digestion during the early stages of infection. HeLa-FcγRII cells were infected with GFP-expressing F. novicida strains. At 2 and 6 h postinfection, the cells were stained with LysoTracker and examined using confocal microscopy. The F. novicida WT bacteria did not exhibit LysoTracker colocalization. In contrast, the wbtF mutant strain exhibited noticeable colocalization with LysoTracker-positive compartments, suggesting defective phagolysosomal evasion (Figure 3A). A low percentage of WT bacteria showed lysosomal colocalization in HeLa-FcγRII cells. In contrast, a significantly higher percentage of wbtF mutant bacteria showed lysosomal colocalization (Figure 3B). This suggested that the WT bacteria, but not the wbtF mutant bacteria, escaped from the phagolysosomes in HeLa-FcγRII cells.
Figure 3. (A) HeLa-FcγRII cells were infected with F. novicida strains preincubated with mouse serum containing anti-F. novicida antibodies at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. The infected HeLa-FcγRII cells were treated with LysoTracker Red DND-99 to visualize the acidic compartments at 2 and 6 h postinfection. Scale bar = 20 μm. (B) Percentage of F. novicida colocalization with LysoTracker-labeled lysosomes. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (C) Percentage of colocalization between F. novicida and LAMP-1. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (D) HeLa-FcγRII cells were similarly infected then treated with an anti-LAMP-1 antibody and stained with a TRITC-conjugated anti-rat IgG for 2–24 h postinfection. Scale bar = 20 μm.
3.4 F. novicida escape from lysosomes in HeLa-FcγRII cells
HeLa-FcγRII cells were infected with GFP-expressing F. novicida WT and wbtF mutant strains to determine whether they underwent lysosomal digestion. Lysosomes were visualized using an anti-LAMP-1 antibody (Figure 3D). At 2–24 h postinfection, the WT strain replicated intracellularly, with bacterial numbers increasing over time. The wbtF mutant showed intracellular replication for up to 12 h postinfection. However, replication was decreased at 24 h postinfection, suggesting impaired long-term survival or replication within the host cells (Figure 3C). Compared with the wbtF mutant, a significantly lower percentage of WT strains that infected the HeLa-FcγRII cells colocalized with LAMP-1 at 2, 6, 12, and 24 h postinfection (Figure 3C). These results indicated that F. novicida WT escaped lysosomal degradation in the HeLa-FcγRII cells compared to the wbtF mutant, which exhibits reduced escape capacity.
3.5 Impact of wbtF in the mitochondria of HeLa-FcγRII cells
We infected HeLa-FcγRII cells with WT and wbtF mutant strains to determine the role of wbtF in the mitochondria. We visualized the mitochondria using MitoTracker Deep Red FM (Figure 4A) to observe whether the WT and the wbtF mutant strains colocalized with the mitochondria. At 2 and 6 h postinfection, a low percentage of the HeLa-FcγRII cells infected with the WT strain colocalized with the mitochondria (Figure 4B). In contrast, HeLa-FcγRII cells infected with the wbtF mutant showed 61%–76% colocalization with the mitochondria (Figure 4B). These results demonstrated that F. novicida WT effectively modified mitochondrial function and sustained replication, while the wbtF mutant could not.
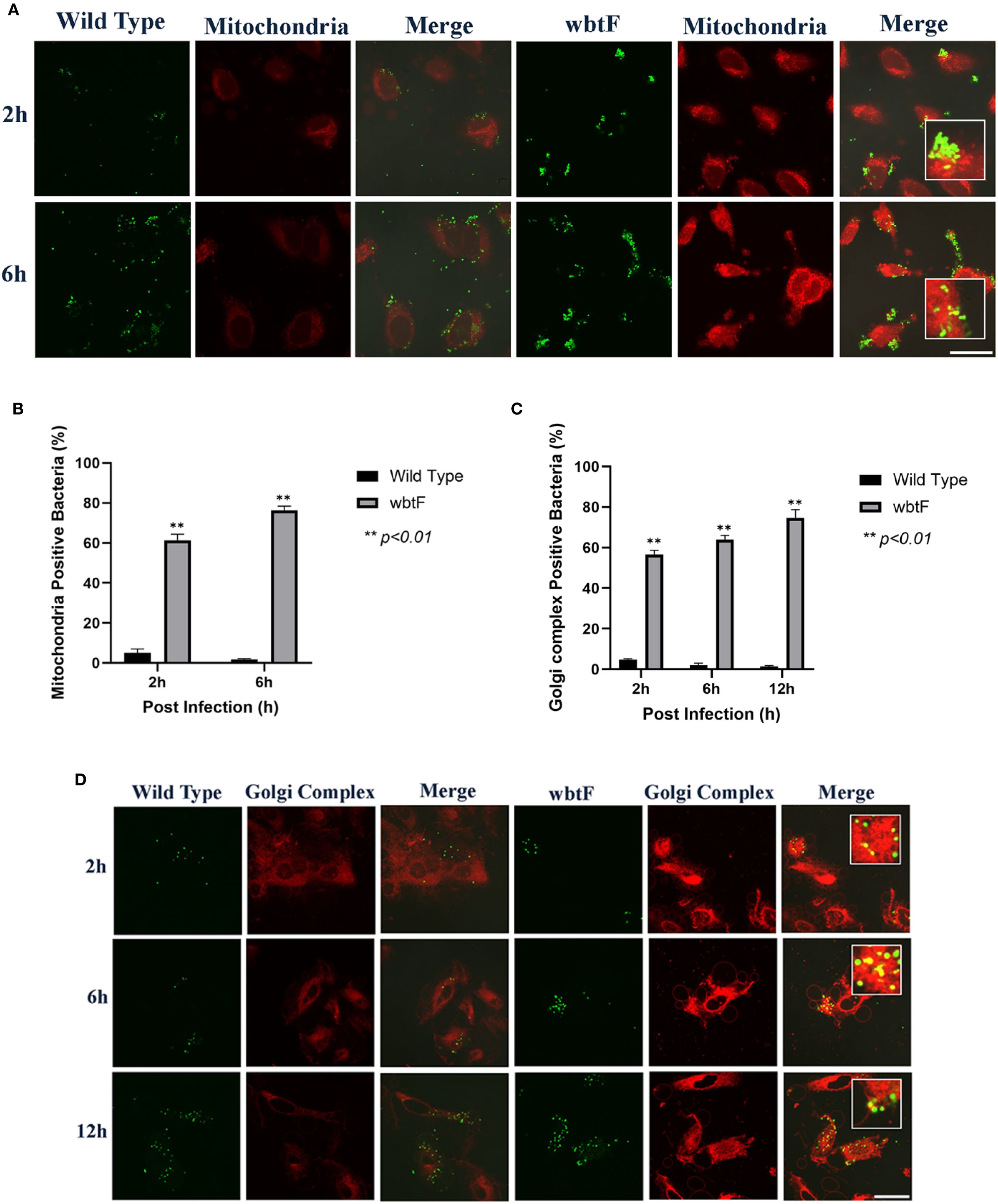
Figure 4. HeLa-FcγRII cells were infected with F. novicida strains preincubated with mouse serum containing anti-F. novicida antibodies at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. (A) The infected HeLa-FcγRII cells were stained with MitoTracker Deep Red FM at 2 and 6 h postinfection. Scale bar = 20 μm. (B) Percentage of F. novicida colocalized with the mitochondria. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (C) Percentage of F. novicida colocalized with the Golgi complex. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (D) The infected HeLa-FcγRII cells were stained with BODIPY TR ceramide at 2, 6, and 12 h postinfection. Scale bar = 20 μm.
3.6 Effect of wbtF on the Golgi complex of infected HeLa-FcγRII cells
We infected HeLa-FcγRII cells with WT and wbtF mutant strains to investigate the function of wbtF in the Golgi complex of infected cells. The Golgi complex was stained using BODIPY TR ceramide at 2, 6, and 12 h postinfection. A low percentage of WT bacteria colocalized with the Golgi complex (Figure 4C). In contrast, 57%–75% of the wbtF mutant bacteria colocalized with the Golgi complex at 2, 6, and 12 h postinfection (Figure 4C). Visualization of intracellular colocalization of the bacteria with the Golgi complex 2, 6, and 12 h postinfection can be seen in Figure 4D.
3.7 Effect of wbtF on intracellular replication in THP-1 cells
THP-1 cells were infected with the WT and wbtF mutant of F. novicida to evaluate the role of wbtF in intracellular replication. Fluorescence microscopy using GFP-expressing strains confirmed the presence and localization of intracellular bacteria. Our analysis of the intracellular levels of WT and wbtF mutant bacteria at 12, 24, and 48 h postinfection in THP-1 cells further validated the reduced replication capacity of the wbtF mutant compared with the WT strain (Figure 5A). The WT strain showed a steady increase in intracellular replication, while the number of wbtF mutants was significantly decreased between 12 and 48 h postinfection (Figure 5B). Complementation of the wbtF mutant restored replication to the WT level, confirming the role of wbtF in intracellular survival (Figure 5B). These findings suggested wbtF involvement in F. novicida intracellular replication in THP-1 cells.
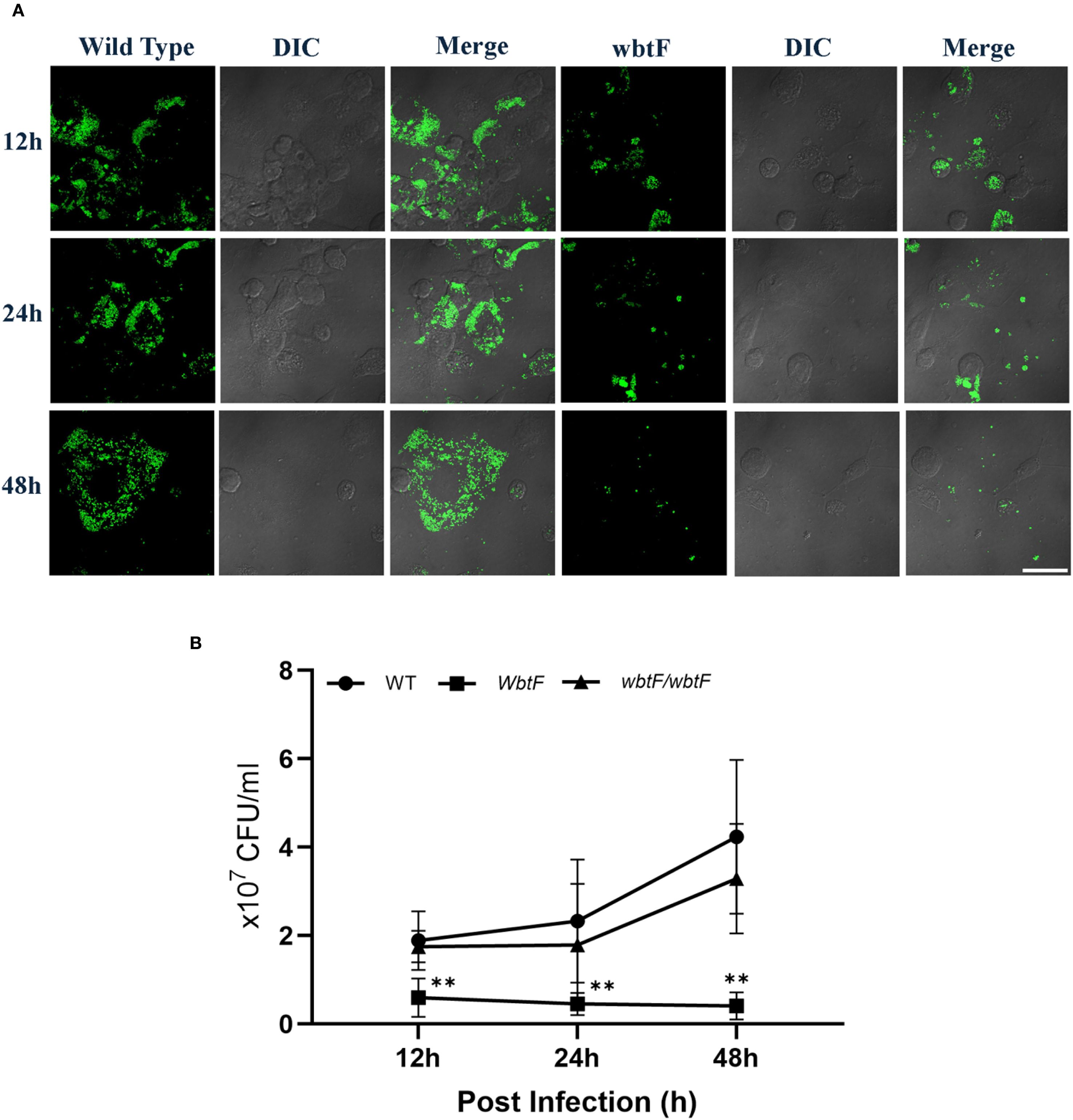
Figure 5. THP-1 cells were infected with F. novicida at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. (A) The infected THP-1 cells were fixed and observed at 12, 24, and 48 h after infection. Scale bar = 20 μm. (B) The infected THP-1 cells were lysed with 0.1% Triton X-100 at 12, 24, and 48 h postinfection, and the lysates were plated on BHIc agar. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using multiple comparisons and is indicated by asterisks. **P < 0.01.
3.8 Role of wbtF in avoiding phagolysosomes in THP-1 cells
We infected THP-1 cells with GFP-expressing F. novicida strains to investigate the role of wbtF in intracellular survival, specifically its involvement in phagolysosomal escape. We assessed the ability of F. novicida to evade phagolysosomal fusion during the initial stage of infection (2 and 6 h postinfection) using LysoTracker staining visualized under confocal microscopy. F. novicida WT showed low levels of colocalization with LysoTracker, indicating efficient evasion of phagolysosomal fusion. In contrast, the wbtF mutant colocalized with LysoTracker-positive lysosomes (Figure 6A). Only a limited number of WT bacteria were colocalized with the lysosomes, while a significantly higher number of wbtF mutant bacteria were detected within the lysosomal compartments (Figure 6B). This suggested that the WT strain escaped from the phagolysosomes in THP-1 cells, but the wbtF mutant could not.
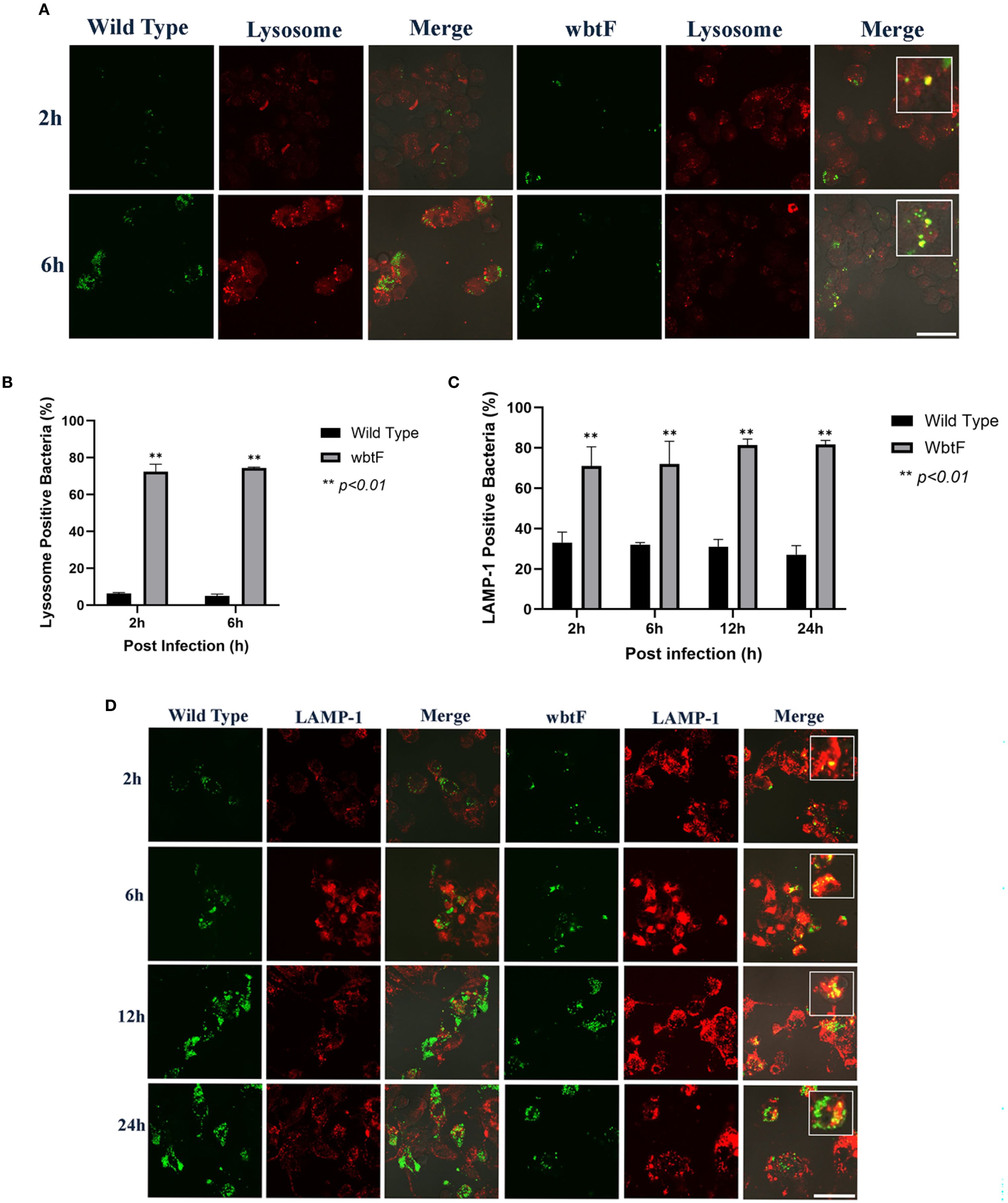
Figure 6. THP-1 cells were infected with F. novicida strains at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. (A) The infected THP-1 cells were treated with LysoTracker Red DND-99 for 2–6 h postinfection. Scale bar = 20 μm. (B) Percentage of F. novicida colocalized with lysosomes. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (C) Percentage of F. novicida colocalized with LAMP-1. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (D) The infected THP-1 cells were treated with anti-LAMP-1 antibody and stained with TRITC-conjugated anti-rat IgG for 2–24 h postinfection. Scale bar = 20 μm.
3.9 F. novicida escape from lysosomes in THP-1 cells
THP-1 cells were infected with the GFP-expressing F. novicida WT and wbtF mutant strains to investigate whether lysosomes contributed to F. novicida degradation. Lysosomes were visualized using an anti-LAMP-1 antibody (Figure 6D). The WT strain exhibited consistent intracellular replication at 2–24 h postinfection, as indicated by the increasing number of bacteria. In contrast, the wbtF mutant replicated up to 12 h postinfection, but the intracellular bacterial load declined markedly by 24 h postinfection. Compared with the wbtF mutant, a significantly lower percentage of WT strains colocalized with LAMP-1 at 2, 6, 12, and 24 h postinfection (Figure 6C). These results indicated that F. novicida WT escaped lysosomal degradation in the THP-1 cells, whereas the wbtF mutant could not.
3.10 Impact of wbtF in the mitochondria of THP-1 cells
THP-1 cells were infected with WT and wbtF mutant strains to determine the role of wbtF in the mitochondria. We visualized the mitochondria using MitoTracker Deep Red FM (Figure 7A) to observe whether the WT and the wbtF mutant bacteria colocalized with the mitochondria. A low percentage of the WT bacteria colocalized with the mitochondria (Figure 7B). In contrast, 69%–78% of the wbtF mutant bacteria colocalized with the mitochondria at 2 and 6 h postinfection (Figure 7B). These results indicated that F. novicida WT modified mitochondrial function and sustained replication in the THP-1 cells, while the wbtF mutant could not.
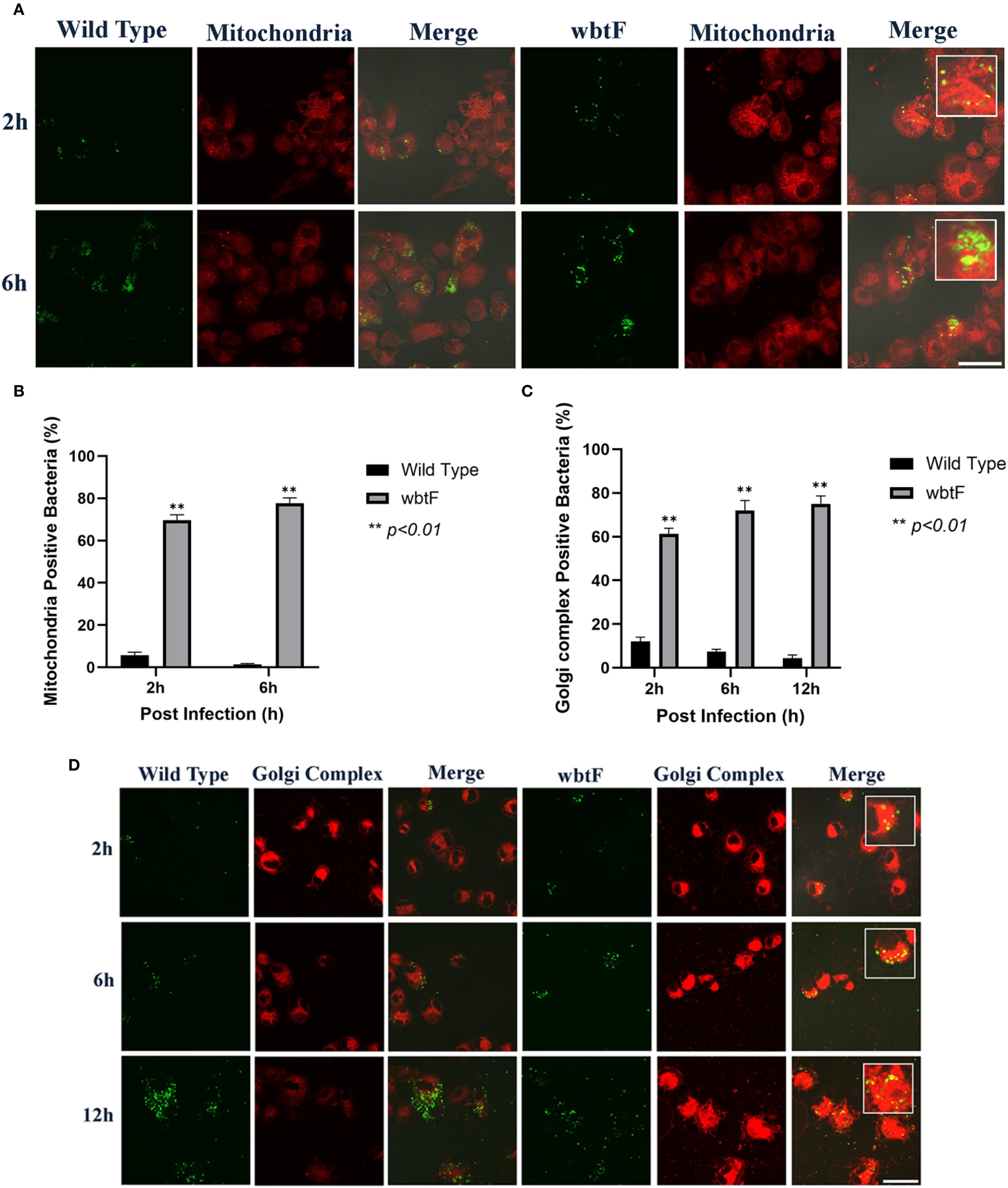
Figure 7. THP-1 cells were infected with F. novicida at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. (A) The infected THP-1 cells were treated with MitoTracker Deep Red FM at 2 and 6 h postinfection. Scale bar = 20 μm. (B) Percentage of F. novicida colocalized with the mitochondria. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (C) Percentage of F. novicida colocalized with the Golgi complex. Data represent the mean and SD from three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using Welch’s t-test and is indicated by asterisks. **P < 0.01. (D) The infected THP-1 cells were treated with BODIPY TR ceramide at 2, 6, and 12 h after infection. Scale bar = 20 μm.
3.11 Effect of wbtF on the Golgi complex of infected THP-1 cells
We infected THP-1 cells with the WT and wbtF mutant strains to analyze the function of wbtF in the Golgi complex. The Golgi complex was stained using BODIPY TR ceramide to determine if the Golgi complex could colocalize the WT strain and the wbtF mutant. The intracellular bacteria were examined at 2, 6, and 12 h after infection. A low percentage of the WT strain colocalized with the Golgi complex (Figure 7C). In contrast, 61%–75% of the wbtF mutant bacteria were colocalized with the Golgi complex at 2, 6, and 12 h after infection (Figure 7C). Visualization of intracellular colocalization of the bacteria with the Golgi complex 2, 6, and 12 h postinfection can be seen in Figure 7D.
3.12 Effect of wbtF on cytokine production
To evaluate the effect of wbtF in cytokine responses, we measured the induction of cytokines produced by THP-1 cells that were infected with WT, wbtF mutant and complementary strains. The induction of TNF-α (Figure 8A) and IL-6 (Figure 8B) did not differ significantly among the WT, wbtF mutant, and complementary strains.
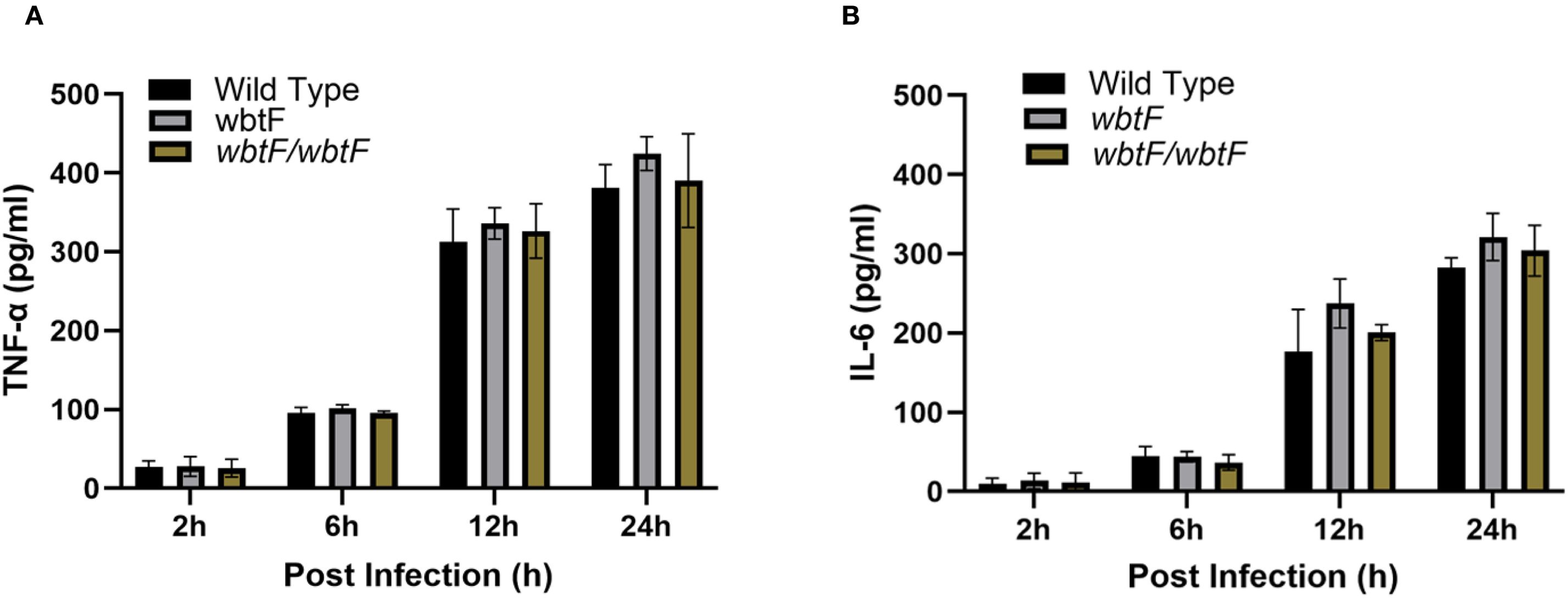
Figure 8. THP-1 cells were infected with F. novicida at an MOI of 1, followed by gentamicin treatment (50 μg/mL) for 1 h to eliminate extracellular bacteria. After 2 h, 6 h, 12 h, and 24 h of incubation, cell supernatants were collected, and the levels of TNF-α (A) and IL-6 (B) were determined using ELISA. The data represent the averages and standard deviations of three identical experiments. Statistical significance relative to the WT strain at each designated time point was determined using multiple comparisons and no significant difference was observed.
4 Discussion
Francisella is a Gram-negative bacterium, and its highly pathogenic species, F. tularensis, is a well-known cause of tularemia in humans and other mammals. F. tularensis outmaneuvers the host’s immune defenses by escaping from the phagosome—a cellular structure meant to destroy invaders. Instead of being degraded, the bacteria multiply inside the host cell’s cytoplasm, where they are better protected from immune responses (Pavlik et al., 2024). Scientists are still piecing together the mechanism of phagosomal escape, and a gene cluster called the Francisella pathogenicity island (FPI) has emerged as a key player. The FPI encodes component proteins of a unique type VI secretion system thought to be critical to the bacterium’s survival and virulence (Clemens et al., 2015; Rigard et al., 2016; Spidlova and Stulik, 2017).
We constructed a transposon mutant library of F. novicida to isolate mutants that were less cytotoxic. We screened the transposon mutants using a new infection model of Francisella, HeLa cells expressing mouse FcγRII (HeLa-FcγRII). HeLa-FcγRII cells are a cell line that expresses the FcγRII receptor, enabling efficient uptake of IgG-opsonized bacteria (Nakamura et al., 2024). Although epithelial cells, like HeLa, are readily amenable to genetic modification, infection with Francisella is rarely effective and significantly lower than the infection efficiency of macrophage-like cells. Francisella infection of epithelial cell lines, such as like HeLa, A549, and HEK293, has been performed at a high MOI (100–10,000) and requires a relatively long time (approximately 8–24 h) (Lo et al., 2013; Amer et al., 2010; Bradburne et al., 2013). The ability of Francisella to infect macrophages largely depends on whether the bacteria are opsonized. The use of unopsonized bacteria leads to rapid disruption of the phagosome. In contrast, opsonizing Francisella with fresh serum facilitates directing the bacteria to various phagocytic receptors, resulting in delayed phagosomal escape (Geier and Celli, 2011). Notably, when HeLa cells engineered to express FcγRII were infected with antibody-coated bacteria, the infection efficiency was approximately 100 times higher compared with normal HeLa cells (Nakamura et al., 2024).
Among the 2,232 transposons mutants library, 13 showed less cytotoxicity and were successfully sequenced. Two mutants involved the same gene: FTN_0496 (slt). FTN_1686 (figE), FTN_0496 (slt), FTN_1322 (IglC), FTN_1321 (IglD), FTN_1309 (pdpA), FTN_1310 (pdpB), FTN_1317 (IglI), FTN_1313 (IglF), and FTN_0096 were previously reported as pathogenic factors of Francisella (Llewellyn et al., 2011; Ozanic et al., 2015; Brodmann et al., 2017; Pérard et al., 2018; Nakamura et al., 2019; Dean et al., 2020). FTN_0756 (fopA) and FTN_1641 (ampG) were previously reported as immunosuppressive factors involved in dampening host immune responses (Nakamura et al., 2022). However, this was the first report of FTN_1425 (wbtF). Therefore, we focused on wbtF. Although wbtF has been reported as a non-essential gene that can be disrupted (Thomas et al., 2007), we were unable remove the gene using a suicide vector (Nakamura et al., 2019) in this study. Therefore, we attempted to directly investigate wbtF gene from a transposon mutant library of F. novicida.
wbtF is one of the O-antigen biosynthetic gene clusters of F. tularensis (Clemens et al., 2012). The O-antigen capsule in Francisella comprises a polysaccharide that is structurally identical to the O-antigen found in lipopolysaccharide (LPS). This LPS O-antigen is a critical pathogenic factor. O-antigen deficiency results in increased sensitivity to serum and a loss of virulence in Type A and B strains of F. tularensis (Catanzaro and Inzana, 2020). Thomas et al. (2007) created knock-out mutants targeting the O-antigen gene cluster in F. novicida and F. tularensis SchuS4, leading to the complete loss of O-antigen production. Notably, the SchuS4 mutant lacking the O-antigen was fully attenuated in mice.
Francisella species are highly pathogenic, largely due to their ability to replicate within the cytosol of phagocytic cells, such as macrophages and dendritic cells. After entering these cells through phagocytosis, the bacteria are initially enclosed within a membrane-bound compartment called the Francisella-containing phagosome. However, they rapidly disrupt the phagosomal membrane and escape into the host cell cytosol, where they replicate. The bacteria adapt their metabolism to the cytosolic environment by relying on gluconeogenesis and amino acid catabolism as carbon and energy sources. Thus, phagosomal escape is critical for Francisella’s ability to replicate inside host cells, leading to disease in vivo. Phagosomal escape also triggers the host’s antimicrobial and innate immune defenses. In many cases, once the infected cell becomes overloaded with bacteria and dies, the bacteria exit and potentially spread to neighboring cells (Degabriel et al., 2023; Chong and Celli, 2010).
In this research, intracellular replication of the F. novicida WT strain in HeLa-FcγRII cells and THP-1 cells increased at 12–48 h after infection. This correlated with previous findings of increased intracellular replication of Francisella WT in HeLa-FcγRII and THP-1 cells (Nakamura et al., 2021, 2024). Compared with the WT strain, the wbtF mutant underwent intracellular replication for up to 12 h after infection in HeLa-FcγRII and THP-1 cells, but then decreased at 24 and 48 h after infection. The rate of intracellular replication of the wbtF mutant was significantly lower than that of the WT strain (P <0.01). Thomas et al. (2007) demonstrated that the O-antigen of F. tularensis was essential for the survival and replication inside host cells.
The acidic organelles were stained with LysoTracker. The wbtF mutant bacteria showed colocalization with LysoTracker in the early stages of infection (2–6 h) of HeLa-FcγRII cells (60%–71%) and THP-1 cells (72%–74%). Furthermore, the wbtF mutant also colocalized with the lysosome marker LAMP-1 at 2, 6, 12, and 48 h postinfection in 78%–88% of HeLa-FcγRII cells and 71%–82% of THP-1 cells.
F. tularensis virulence is closely associated with its ability to survive inside phagocytic cells and escape into the cytosol. Upon host cell entry, the bacterium is initially enclosed in a phagosome that typically matures into a phagolysosome. However, F. tularensis disrupts this maturation by blocking fusion between the phagosome and lysosome. Then, it breaks down the phagosomal membrane and escapes into the cytoplasm, where it replicates. The molecular mechanism underlying the phagosomal escape of Francisella remains poorly understood. However, it has been shown that disrupting genes within the FPI prevents the bacterium from phagosomal escape (Spidlova and Stulik, 2017). Thus, our findings suggest that the wbtF mutant could not escape from the phagosomes, indicating that wbtF is required for intracellular replication of F. novicida.
Mitochondria are double-membraned organelles vital for various cellular processes and cell survival. Their metabolic functions are crucial for maintaining tissue balance across different cell types and organisms. Unsurprisingly, these metabolic pathways often shift in response to stressors, like nutrient deprivation. Beyond their role in energy production, mitochondria are also highly involved in regulating cell death during infection and have been increasingly recognized as important hubs for immune signaling (Spinelli and Haigis, 2018; Mills et al., 2017). We observed that the F. novicida WT strain associates with mitochondria in HeLa–FcγRII cells and THP-1 cells, and underwent intracellular replication. Furthermore, it did not colocalize with mitochondria (MitoTracker) at 2–6 h after infection. In contrast, the wbtF mutant colocalized with the mitochondria during the initial stages of infection (2–6 h) in HeLa-FcγRII cells (61%–76%) and THP-1 cells (69%–78%).
Mitochondria are highly susceptible to damage, and their dysfunction disrupts cellular homeostasis. Such disturbances are closely linked to the development of various diseases, including neurodegenerative and cardiovascular conditions, cancer, metabolic disorders, and infections. Thus, the impaired mitochondria must be promptly isolated and selectively eliminated. Mitophagy (mitochondrial autophagy) is a critical cellular process that eliminates damaged mitochondria, thereby preserving mitochondrial and overall cellular homeostasis. When cells are exposed to stressors such as ROS, nutrient scarcity, or aging, mitochondria become depolarized and dysfunctional, triggering their selective removal through mitophagy (Doblado et al., 2021; Onishi et al., 2021; Xu et al., 2020; Lu et al., 2023).
Pathogens stimulate mitophagy to suppress the host defense mechanism. They likely employ a common mechanism that inhibits innate immune responses and promotes persistent infection by directly or indirectly facilitating mitophagy (Ma et al., 2024). The initiation of mitophagy to eradicate intracellular mitochondrial ROS (mtROS) is an essential survival tactic for intracellular infections. mtROS are essential for innate host defense, functioning as effectors by harming pathogens (Yang et al., 2025; Silwal et al., 2020). Alqahtani et al. (2023) showed that infection with F. tularensis live vaccine strain induced mitophagy. Generally, mitophagy is considered to limit the accumulation of mtROS, a significant source of cellular ROS. Microbial pathogens often evade or suppress the host’s protective mtROS pathway and neutralize bactericidal free radicals (Su et al., 2023; Silwal et al., 2020). Other intracellular bacteria, such as Staphylococcus aureus, promote mitophagy to clear mtROS. S. aureus not only survives within macrophages but also stimulates mitophagy in host cells to clear mtROS. It uses HDAC11 to augment interleukin 10 (IL-10) transcription, promoting mitophagy to clear mtROS, reduce mitochondrial damage, and maintain an ecological environment favorable for bacterial survival. Listeria monocytogenes diminishes mtROS and inhibits cell death by promoting mitophagy through the secretion of its virulence factor, listeriolysin O (Yang et al., 2025; Zhang et al., 2019). These findings suggest that the intracellular survival of Francisella WT depends on mitochondrial modulation and escape. Indeed, the effector protein IglJ has been reported to interact directly with mitochondrial targets (Prokšová et al., 2020). In contrast, the wbtF mutant cannot modulate mitochondria and is therefore detected in association with mitochondria. The wbtF gene might be involved in this kind of mitochondrial modulation.
The Golgi complex is a crucial membrane-bound organelle in the perinuclear region that consists of flattened disk-shaped cisternae connected in a ribbon-like structure. It comprises three distinct subcompartments: the cis-Golgi, medial-Golgi, and trans-Golgi network (Ahat et al., 2019). The Golgi complex is the modification and sorting hub for proteins and lipids. Newly synthesized proteins and lipids are delivered to the Golgi complex, where they undergo posttranslational modifications and are sorted into vesicles for transport to other organelles or extracellular secretion (Makhoul et al., 2019).
To explore the activities of the F. novicida WT and wbtF mutant strains within the Golgi complex, infected HeLa-FcγRII and THP-1 cells were stained with BODIPY TR ceramide. The Francisella WT strain only showed colocalization with the Golgi complex in 2%–5% of HeLa-FcγRII cells and 4%–12% of THP-1 cells. However, the wbtF mutant was colocalized with the Golgi complex in 57%–75% of HeLa-FcγRII cells and 61%–75% of THP-1 cells.
The Golgi complex is a crucial organelle in the host defense systems against intracellular infections. Additionally, recent research indicates that the Golgi complex plays a critical role in immunological signaling (Tao et al., 2020). Disruption of the structure of the Golgi complex catalyzes the pathogenesis of infectious diseases. Various intracellular bacterial, viral, and protozoan infections trigger Golgi complex destabilization. This evolutionarily conserved strategy enables pathogens to access nutrients, hijack organelle membranes for replication, and subvert host immune defenses (Read et al., 2024). Type I interferons (IFNs) are induced by the Golgi complex in response to various stimuli, including Francisella infection (Gregory et al., 2020). Type I IFNs are a group of cytokines crucial for shaping innate and adaptive immune responses during viral and bacterial infections. However, they also suppress antibacterial immunity by promoting IL-10 production, inhibiting proinflammatory cytokines and chemokines, and promoting immune cell apoptosis, thereby exacerbating Francisella infections (Xia et al., 2025; Li et al., 2024). The translocation of the signaling intermediate stimulator of IFN genes (STING, TMEM173) from the endoplasmic reticulum to the cis-Golgi is initiated by cyclic GMP-AMP synthase or interferon-γ inducible protein 16 following the identification of cytosolic exogenous DNA (Storek et al., 2015; Meunier et al., 2015; Man et al., 2015). STING activates TANK-binding kinase 1 (TBK1) at the Golgi complex by inducing its autophosphorylation at serine 172. Following activation, TBK1 phosphorylates interferon regulatory factor 3, facilitating dimerization and translocation to the nucleus, which drives the expression of immune-related genes, including type I IFNs (Barber, 2011). Interestingly, structural disruption of the Golgi complex using golgicide A blocks TBK1 phosphorylation in response to F. tularensis infection and LPS (Gregory et al., 2020). These results suggest that F. novicida WT escapes from the Golgi complex, but wbtF, which was detected in the Golgi complex, cannot escape from the immune response.
Since wbtF is a gene involved in O-antigen synthesis, we investigated the cytokine production of the wbtF deletion mutant to confirm whether the attenuated cytotoxicity and intracellular proliferation of the mutant were due to the LPS-mediated activation of immune responses. Infection of THP-1 cells with F. novicida WT, wbtF mutant, and the complemented strain produced similar TNF-α and IL-6 responses, indicating that wbtF does not significantly affect cytokine production. The wbtF gene, a consecutive gene located within the O-antigen-biosynthetic gene clusters spanning a 17-kb chromosomal region of F. tularensis, is directly implicated in LPS biosynthesis. The O-antigen constitutes one of the three components of LPS, along with the lipid A and the oligosaccharide core (Clemens et al., 2012; Krzyżewska-Dudek et al., 2024). Francisella LPS does not function as an antagonist of TLR4-mediated innate immune signaling in either humans or mice. Francisella LPS is tetraacylated, and it elicits only weak activation of TLR4 and demonstrates a markedly reduced ability to induce the secretion of inflammatory cytokines like TNF-α and IL-6 by murine and human innate immune cells (Hajjar et al., 2006; Gunn and Ernst, 2007; Noah et al., 2010). It seems that Francisella LPS represents, at most, a minor contributor to the induction of proinflammatory responses within the innate immune system. Although deletion of LPS synthesis genes, such as kdsA, have been reported to enhance cytokine production (Nakamura et al., 2022), the absence of wbtF did not impact cytokine production in this study. Taken together, these results imply that the attenuated cytotoxicity and intracellular growth ability observed in wbtF deletion mutant are likely due to mechanisms distinct from LPS-mediated immune activation. Although comprehensive understanding of the detailed mechanisms necessitates further investigation, our results may suggest that wbtF is important for modulating LPS structure, potentially facilitating the evasion of recognition and uptake by cellular organelles, including phagosomes, mitochondria, and the Golgi apparatus.
In summary, our research focused on identifying a virulence factor of F. novicida by screening a transposon mutant library. We used a new infection model of Francisella using HeLa cells expressing mouse FcγRII (HeLa-FcγRII) (Nakamura et al., 2024). We identified wbtF as a pathogenic factor of F. novicida. wbtF is required for escaping from organelles such as phagosomes, mitochondria, and the Golgi apparatus, followed by intracellular replication in HeLa-FcγRII and THP-1 cells. Thus, it may be a potential target for controlling Francisella infection. However, further research is necessary to elucidate the detailed mechanism of wbtF function in Francisella infection.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
DKW: Data curation, Formal analysis, Resources, Validation, Visualization, Writing – original draft. TS: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. KW: Investigation, Methodology, Resources, Writing – review & editing. AU: Methodology, Resources, Writing – review & editing. MW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Number 22K07054 and 21H02360). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahat, E., Li, J., and Wang, Y. (2019). New insights into the Golgi stacking proteins. Front. Cell. Dev. Biol. 7. doi: 10.3389/fcell.2019.00131
Alqahtani, M., Ma, Z., Miller, J., Yu, J., Malik, M., and Bakshi, C. S. (2023). Comparative analysis of absent in melanoma 2-inflammasome activation in Francisella tularensis and Francisella novicida. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1188112
Amer, L. S., Bishop, B. M., and van Hoek, M. L. (2010). Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem. Biophys. Res. Commun. 396, 246–251. doi: 10.1016/j.bbrc.2010.04.073
Barber, G. N. (2011). Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 23, 10–20. doi: 10.1016/j.coi.2010.12.015
Bradburne, C. E., Verhoeven, A. B., Manyam, G. C., Chaudhry, S. A., Chang, E. L., and Thach, D. C. (2013). Temporal transcriptional response during infection of type II alveolar epithelial cells with Francisella tularensis live vaccine strain (LVS) supports a general host suppression and bacterial uptake by macropinocytosis. J. Biol. Chem. 288, 10780–10791. doi: 10.1074/jbc.M112.362178
Brett, M., Doppalapudi, A., Respicio-Kingry, L. B., Myers, D., Husband, B., Pollard, K., et al. (2012). Francisella novicida bacteremia after a near-drowning accident. J. Clin. Microbiol. 50, 2826–2829. doi: 10.1128/JCM.00995-12
Brodmann, M., Dreier, R. F., Broz, P., and Basler, M. (2017). Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat. Commun. 8, 15853. doi: 10.1038/ncomms15853
Catanzaro, K. C. F. and Inzana, T. J. (2020). The francisella tularensis polysaccharides: what is the real capsule? MMBR. 84. doi: 10.1128/mmbr.00065-19
Celli, J. and Zahrt, T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3, a010314. doi: 10.1101/cshperspect.a010314
Chong, A. and Celli, J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1. doi: 10.3389/fmicb.2010.00138
Clemens, D. L., Ge, P., Lee, B.-Y., Horwitz, M. A., and Zhou, Z. H. (2015). Atomic structure of T6SS reveals interlaced array essential to function. Cell. 160, 940–951. doi: 10.1016/j.cell.2015.02.005
Clemens, D. L., Lee, B.-Y., and Horwitz, M. A. (2012). O-antigen-deficient Francisella tularensis live vaccine strain mutants are ingested via an aberrant form of looping phagocytosis and show altered kinetics of intracellular trafficking in human macrophages. Infect. Immun. 80, 952–967. doi: 10.1128/IAI.05221-11
Dean, S. N., Milton, M. E., Cavanagh, J., and van Hoek, M. L. (2020). Francisella novicida Two-Component System Response Regulator BfpR Modulates iglC Gene Expression, Antimicrobial Peptide Resistance, and Biofilm Production. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00082
Degabriel, M., Valeva, S., Boisset, S., and Henry, T. (2023). Pathogenicity and virulence of Francisella tularensis. Virulence. 14. doi: 10.1080/21505594.2023.2274638
Dennis, D. T., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., et al. (2001). Tularemia as a biological weapon: medical and public health management. J. Am. Med. Assoc. 285, 2763–2773. doi: 10.1001/jama.285.21.2763
Doblado, L., Lueck, C., Rey, C., Samhan-Arias, A. K., Prieto, I., and Stacchiotti, A. (2021). Mitophagy in human diseases. Int. J. Mol. Sci. 22, 3903. doi: 10.3390/ijms22083903
Fushinobu, S. (2021). Molecular evolution and functional divergence of UDP-hexose 4-epimerases. Curr. Opin. Chem. Biol. 61, 53–62. doi: 10.1016/j.cbpa.2020.09.007
Gallagher, L. A., Ramage, E., Jacobs, M. A., Kaul, R., Brittnacher, M., and Manoil, C. (2007). A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U.S.A 104, 1009–1014. doi: 10.1073/pnas.0606713104
Geier, H. and Celli, J. (2011). Phagocytic receptors dictate phagosomal escape and intracellular proliferation of Francisella tularensis. Infect. Immun. 79, 2204–2214. doi: 10.1128/IAI.01382-10
Gregory, D. J., DeLoid, G. M., Salmon, S. L., Metzger, D. W., Kramnik, I., and Kobzik, L. (2020). SON DNA-binding protein mediates macrophage autophagy and responses to intracellular infection. FEBS Lett. 594, 2782–2799. doi: 10.1002/1873-3468.13851
Gunn, J. S. and Ernst, R. K. (2007). The structure and function of Francisella lipopolysaccharide. Ann. N. York. Acad. Sci. 1105, 202–218. doi: 10.1196/annals.1409.006
Hajjar, A. M., Harvey, M. D., Shaffer, S. A., Goodlett, D. R., Sjöstedt, A., Edebro, H., et al. (2006). Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect. Immun. 74, 6730–6738. doi: 10.1128/IAI.00934-06
Harrell, J. E., Roy, C. J., Gunn, J. S., and McLachlan, J. B. (2024). Current vaccine strategies and novel approaches to combatting Francisella infection. Vaccine. 42, 2171–2180. doi: 10.1016/j.vaccine.2024.02.086
Johansson, A., Celli, J., Conlan, W., Elkins, K. L., Forsman, M., Keim, P. S., et al. (2010). Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int. J. Syst. Evol. Microbiol. 60, 1717–1718. doi: 10.1099/ijs.0.022830-0
Kingry, L. C. and Petersen, J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00035
Krzyżewska-Dudek, E., Dulipati, V., Kapczyńska, K., Noszka, M., Chen, C., Kotimaa, J., et al. (2024). Lipopolysaccharide with long O-antigen is crucial for Salmonella Enteritidis to evade complement activity and to facilitate bacterial survival in vivo in the Galleria mellonella infection model. Med. Microbiol. Immunol. 213, 8. doi: 10.1007/s00430-024-00790-3
Li, S., Yao, Q., Li, J., Yang, H., Qian, R., Zheng, M., et al. (2024). Inhibition of neutrophil swarming by type I interferon promotes intracellular bacterial evasion. Nat. Commun. 15, 8663. doi: 10.1038/s41467-024-53060-4
Llewellyn, A. C., Jones, C. L., Napier, B. A., Bina, J. E., and Weiss, D. S. (2011). Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PloS One 6. doi: 10.1371/journal.pone.0024201
Lo, K. Y. S., Chua, M. D., Abdulla, S., Law, H. T., and Guttman, J. A. (2013). Examination of in vitro epithelial cell lines as models for Francisella tularensis non-phagocytic infections. J. Microbiol. Methods 93, 153–160. doi: 10.1016/j.mimet.2013.03.004
Lu, Y., Li, Z., Zhang, S., Zhang, T., Liu, Y., and Zhang, L. (2023). Cellular mitophagy: mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics. 13, 736–766. doi: 10.7150/thno.79876
Ma, L., Han, T., and Zhan, Y. A. (2024). Mechanism and role of mitophagy in the development of severe infection. Cell. Death. Discov. 10, 88. doi: 10.1038/s41420-024-01844-4
Makhoul, C., Gosavi, P., and Gleeson, P. A. (2019). Golgi dynamics: the morphology of the mammalian Golgi apparatus in health and disease. Front. Cell. Dev. Biol. 7. doi: 10.3389/fcell.2019.00112
Man, S. M., Karki, R., Malireddi, R. K. S., Neale, G., Vogel, P., Yamamoto, M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475. doi: 10.1038/ni.3118
Marecic, V., Shevchuk, O., Ozanic, M., Mihelcic, M., Steinert, M., Begonja, A. J., et al. (2017). Isolation of F. novicida-containing phagosome from infected human monocyte derived macrophages. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00303
McGann, P., Rozak, D. A., Nikolich, M. P., Bowden, R. A., Lindler, L. E., and Wolcott, M. J. (2010). A novel brain heart infusion broth supports the study of common Francisella tularensis serotypes. J. Microbiol. Methods 80, 164–171. doi: 10.1016/j.mimet.2009.12.005
Meunier, E., Wallet, P., Dreier, R. F., Costanzo, S., Anton, L., Rühl, S., et al. (2015). Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484. doi: 10.1038/ni.3119
Miller, W. L., Matewish, M. J., McNally, D. J., Ishiyama, N., Anderson, E. M., Brewer, D., et al. (2008). Flagellin glycosylation in pseudomonas aeruginosa PAK requires the O-antigen biosynthesis enzyme WbpO. J. Biol. Chem. 283, 3507–3518. doi: 10.1074/jbc.M708894200
Mills, E. L., Kelly, B., and O’Neill, L. A. J. (2017). Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498. doi: 10.1038/ni.3704
Nagle, S. C., Jr., Anderson, R. E., and Gary, N. D. (1960). Chemically defined medium for the growth of Pasteurella tularensis. J. Bacteriol. 79, 566–571. doi: 10.1128/jb.79.4.566-571.1960
Nakamura, T., Shimizu, T., Ikegaya, R., Uda, A., Watanabe, K., and Watarai, M. (2022). Identification of pyrC gene as an immunosuppressive factor in Francisella novicida infection. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1027424
Nakamura, T., Shimizu, T., Inagaki, F., Okazaki, S., Saha, S. S., Uda, A., et al. (2021). Identification of membrane-bound lytic murein transglycosylase A (MltA) as a growth factor for francisella novicida in a silkworm infection model. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.581864
Nakamura, T., Shimizu, T., Nishinakama, N., Takahashi, R., Arasaki, K., Uda, A., et al. (2024). A novel method of Francisella infection of epithelial cells using HeLa cells expressing fc gamma receptor. BMC Infect. Dis. 24. doi: 10.1186/s12879-024-10083-y
Nakamura, T., Shimizu, T., Uda, A., Watanabe, K., and Watarai, M. (2019). Soluble lytic transglycosylase SLT of Francisella novicida is involved in intracellular growth and immune suppression. PloS One 14. doi: 10.1371/journal.pone.0226778
Noah, C. E., Malik, M., Bublitz, D. C., Camenares, D., Sellati, T. J., Benach, J. L., et al. (2010). GroEL and lipopolysaccharide from Francisella tularensis live vaccine strain synergistically activate human macrophages. Infect. Immun. 78, 1797–1806. doi: 10.1128/IAI.01135-09
Onishi, M., Yamano, K., Sato, M., Matsuda, N., and Okamoto, K. (2021). Molecular mechanisms and physiological functions of mitophagy. EMBO J. 40, e104705. doi: 10.15252/embj.2020104705
Ozanic, M., Marecic, V., Kwaik, Y. A., and Santic, M. (2015). The divergent intracellular lifestyle of francisella tularensis in evolutionarily distinct host cells. PloS Pathog. 11, e1005208. doi: 10.1371/journal.ppat.1005208
Pavlik, P., Velecka, E., and Spidlova, P. (2024). Breaking the cellular defense: the role of autophagy evasion in Francisella virulence. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1523597
Pérard, J., Nader, S., Levert, M., Arnaud, L., Carpentier, P., Siebert, C., et al. (2018). Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role in Francisella tularensis virulence. Commun. Biol. 1, 93. doi: 10.1038/s42003-018-0095-6
Prior, J. L., Prior, R. G., Hitchen, P. G., Diaper, H., Griffin, K. F., Morris, H. R., et al. (2003). Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J. Med. Microbiol. 52: 845–851. doi: 10.1099/jmm.0.05184-0
Prokšová, M., Rehulkova, H., Rehulka, P., Lays, C., Lenčo, J., and Stulík, J. (2020). Using proteomics to identify host cell interaction partners for VgrG and IglJ. Sci. Rep. 10. doi: 10.1038/s41598-020-71641-3
Ray, K., Marteyn, B., Sansonetti, P. J., and Tang, C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. 7, 333–340. doi: 10.1038/nrmicro2112
Read, C. B., Ali, A. N., Stephenson, D. J., Macknight, H. P., Maus, K. D., Cockburn, C. L., et al. (2024). Ceramide-1-phosphate is a regulator of Golgi structure and is co-opted by the obligate intracellular bacterial pathogen Anaplasma phagocytophilum. MBio. 15. doi: 10.1128/mbio.00299-24
Rigard, M., Bröms, J. E., Mosnier, A., Hologne, M., Martin, A., Lindgren, L., et al. (2016). Francisella tularensis iglG belongs to a novel family of PAAR-like T6SS proteins and harbors a unique N-terminal extension required for virulence. PloS Pathog. 12, e1005821. doi: 10.1371/journal.ppat.1005821
Rodriguez, S. A., Yu, J. J., Davis, G., Arulanandam, B. P., and Klose, K. E. (2008). Targeted inactivation of Francisella tularensis genes by group II introns. Appl. Environ. Microbiol. 74, 2619–2626. doi: 10.1128/AEM.02905-07
Rowe, H. M. and Huntley, J. F. (2015). From the Outside-In: The francisella tularensis envelope and virulence. Front Cell Infect Microbiol. 5, 94. doi: 10.3389/fcimb.2015.00094
Shimizu, T., Otonari, S., Suzuki, J., Uda, A., Watanabe, K., and Watarai, M. (2019). Expression of Francisella pathogenicity island protein intracellular growth locus E (IglE) in mammalian cells is involved in intracellular trafficking, possibly through microtubule organizing center. Microbiol. Open 8, e00684. doi: 10.1002/mbo3.684
Silwal, P., Kim, J. K., Kim, Y. J., and Jo, E. K. (2020). Mitochondrial reactive oxygen species: double-edged weapon in host defense and pathological inflammation during infection. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01649
Sjodin, A., Svensson, K., Ohrman, C., Ahlinder, J., Lindgren, P., Duodu, S., et al. (2012). Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genom. 13, 268. doi: 10.1186/1471-2164-13-268
Spidlova, P. and Stulik, J. (2017). Francisella tularensis type VI secretion system comes of age. Virulence. 8, 628–631. doi: 10.1080/21505594.2016.1278336
Spinelli, J. B. and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell. Biol. 20, 745–754. doi: 10.1038/s41556-018-0124-1
Storek, K. M., Gertsvolf, N. A., Ohlson, M. B., and Monack, D. M. (2015). cGAS and Ifi204 cooperate to produce type I IFNs in response to francisella infection. J. Immunol. 194, 3226–3245. doi: 10.4049/jimmunol.1402764
Su, L., Zhang, J., Gomez, H., Kellum, J. A., and Peng, Z. (2023). Mitochondria ROS and mitophagy in acute kidney injury. Autophagy. 19, 401–414. doi: 10.1080/15548627.2022.2084862
Tao, Y., Yang, Y., Zhou, R., and Gong, T. (2020). Golgi apparatus: an emerging platform for innate immunity. Trends. Cell. Biol. 30, 467–477. doi: 10.1016/j.tcb.2020.02.008
Thomas, R. M., Titball, R. W., Oyston, P. C. F., Griffin, K., Waters, E., Hitchen, P. G., et al. (2007). The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect. Immun. 75, 371–378. doi: 10.1128/IAI.01241-06
Wawszczak, M., Banaszczak, B., and Rastawicki, W. (2022). Tularaemia-a diagnostic challenge. Ann. Agric. Environ. Med. 29, 12–21. doi: 10.26444/aaem/139242
Weiss, D. S., Henry, T., and Monack, D. M. (2007). Francisella tularensis: activation of the inflammasome. Ann. N Y Acad. Sci. 1105, 219–237. doi: 10.1196/annals.1409.005
Xia, A., Li, X., Zhao, C., Meng, X., Kari, G., and Wang, Y. (2025). For better or worse: type I interferon responses in bacterial infection. Pathogens. 14, 229. doi: 10.3390/pathogens14030229
Xu, Y., Shen, J., and Ran, Z. (2020). Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 18, 1–15. doi: 10.1080/15548627.2019.1603547
Yang, Y., Zhou, H., Li, F., Zhang, Y., Yang, J., Shen, Y., et al. (2025). Staphylococcus aureus induces mitophagy via the HDAC11/IL10 pathway to sustain intracellular survival. J. Transl. Med. 23, 156. doi: 10.1186/s12967-025-06161-7
Yeni, D. K., Büyük, F., Ashraf, A., and Shah, M. S. U. D. (2021). Tularemia: a re-emerging tick-borne infectious disease. Folia Microbiol. 66, 1–14. doi: 10.1007/s12223-020-00827-z
Keywords: F. novicida, wbtF, cytotoxicity, intracellular replication, tularemia
Citation: Wardhana DK, Shimizu T, Watanabe K, Uda A and Watarai M (2025) Identification of the wbtF gene as a cytotoxicity-associated factor in Francisella novicida infection. Front. Cell. Infect. Microbiol. 15:1647652. doi: 10.3389/fcimb.2025.1647652
Received: 16 June 2025; Accepted: 19 September 2025;
Published: 09 October 2025.
Edited by:
Nicolas Jacquier, Centre Hospitalier Universitaire Vaudois (CHUV), SwitzerlandReviewed by:
Petra Spidlova, University of Defence, CzechiaThomas Robert Laws, Defence Science and Technology Laboratory, United Kingdom
Copyright © 2025 Wardhana, Shimizu, Watanabe, Uda and Watarai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Shimizu, c2hpbWl6dXRAeWFtYWd1Y2hpLXUuYWMuanA=
†These authors have contributed equally to this work
 Dhandy Koesoemo Wardhana
Dhandy Koesoemo Wardhana Takashi Shimizu
Takashi Shimizu Kenta Watanabe
Kenta Watanabe Akihiko Uda
Akihiko Uda Masahisa Watarai
Masahisa Watarai