- 1Department of Orthodontics, School and Hospital of Stomatology, China Medical University, Shenyang, Liaoning, China
- 2Shenyang Clinical Medical Research Center of Orthodontic Disease, Shenyang, China
Under oxygen-limited conditions, the adaptability and underlying mechanisms of bacterial biofilms have become key areas of interest in microbiology and clinical infection research. Within biofilms—composed of bacterial communities and extracellular matrix—an oxygen gradient commonly forms, resulting in hypoxic or even anoxic microenvironments. Such conditions substantially increase biofilm antibiotic resistance and facilitate the persistence of chronic infections. This review systematically summarizes the adaptive strategies employed by biofilms in hypoxic environments, including anaerobic metabolism, phenazine-mediated electron shuttling, and virulence factor regulation. These adaptive responses are governed by genes involved in anaerobic metabolism, quorum sensing systems, and the secondary messenger 3,5-cyclic diguanylic acid (c-di-GMP), which collectively influence biofilm formation. Key transcriptional regulators such as Anr and Dnr, the two-component system NarXL, along with specific functional genes, form an intricate regulatory network. This article aims to provide a comprehensive overview of the adaptive mechanisms of Pseudomonas aeruginosa biofilms under oxygen-limited conditions, providing a theoretical foundation for the development of novel anti-infective therapies, targeting the biofilm infection microenvironment in cystic fibrosis and chronic wounds.
1 Introduction
Pseudomonas aeruginosa is a widely distributed Gram-negative bacterium known to cause nosocomial infections and potentially fatal infections in immunocompromised patients (Gale et al., 2015; Gomila et al., 2018). It is also a persistent colonizer of the lungs in cystic fibrosis (CF) patients, where it is notoriously difficult to eradicate (Singh et al., 2000). During chronic infections, P. aeruginosa predominantly exists in biofilm form. Bacterial biofilms adhere to biological or abiotic surfaces and consist of bacterial communities embedded within an extracellular matrix (ECM), comprising proteins, polysaccharides, and extracellular DNA (eDNA), among other components (Vestby et al., 2020). These molecules provide structural integrity and facilitate intercellular adhesion (Schilcher and Horswill, 2020; Chiba et al., 2022). Unlike planktonic bacteria, biofilms exhibit altered growth rates, metabolism, and gene expression profiles (Donlan, 2002). Bacteria in the inner layers of biofilms adapt to low metabolic activity and undergo anaerobic respiration. Due to the protective barrier of the ECM, biofilm-associated bacteria demonstrate increased antibiotic resistance by approximately 10- to 1000-fold (Wagner and Iglewski, 2008; Stokes et al., 2019). Consequently, biofilms are inherently difficult to treat, leading to persistent and chronic infections that pose significant clinical challenges (Zhao et al., 2023a). It is estimated that 65% to 80% of human bacterial infections are associated with biofilms (Zhao et al., 2023a; Römling and Balsalobre, 2012), underscoring the critical need to eradicate pathogenic biofilms for effective management of chronic infections.
Biofilm formation is a complex process influenced by various external factors, including nutrients, osmotic pressure, temperature, and oxygen availability (Lin et al., 2012). The human body presents numerous low-oxygen or anaerobic niches. For example, P. aeruginosa grows as biofilms in the lungs of CF patients, where chronic infection leads to hypoxic or even anaerobic conditions (Martin et al., 2023; Hall-Stoodley and McCoy, 2022). Similarly, Salmonella colonizes anaerobic niches within the intestinal tract (Ehrhardt et al., 2023), and oxygen concentrations in infected or necrotic tissues and wounds are notably low (Carreau et al., 2011). In vitro studies have confirmed that oxygen gradients are commonly present in bacterial biofilms, including those formed by P. aeruginosa (Papa et al., 2023), Staphylococcus aureus (Zamudio-Chávez et al., 2023), and Enterococcus faecalis (James et al., 2016), particularly in mature biofilms (Borriello et al., 2004). This oxygen limitation enhances pathogen virulence and survival (Wang et al., 2022). Using oxygen microelectrode technology, localized hypoxia within P. aeruginosa biofilms has been confirmed, which restricts protein synthesis in mature biofilm interiors and contributes to antibiotic resistance (Høiby et al., 2024). Oxygen deficiency accounts for at least 70% of antibiotic resistance in mature P. aeruginosa biofilm cells, highlighting oxygen concentration as a critical factor in biofilm formation and multidrug resistance stability. Understanding how bacteria rapidly sense and respond to oxygen-limited environments is essential for improving infection treatments. This review summarizes the adaptations and mechanisms of P. aeruginosa biofilms under oxygen-limited conditions, aiming to provide a novel approach for eradicating biofilms through modulation of the infection microenvironment.
2 Formation of oxygen-limited microenvironment
The oxygen concentration gradient is an important characteristic of bacterial biofilms. The oxygen concentration in the environment is mostly around 19.95%. Generally, an oxygen concentration ranging from 11% to 1% is regarded as hypoxic (Carreau et al., 2011), while an environment with less than 1% oxygen or completely devoid of oxygen is an anoxic environment (Mashruwala et al., 2017). In CF lungs, P. aeruginosa forms biofilms where oxygen penetration is severely restricted. Microelectrode measurements reveal that oxygen levels decline progressively with depth and penetrate only 50 μm from the biofilm surface (Werner et al., 2004), whereas the average biofilm thickness can reach 210 μm (Borriello et al., 2004). Consequently, cells deep within the biofilm experience hypoxic or anoxic conditions (Werner et al., 2004; Dietrich et al., 2013). Active protein synthesis is confined to a zone roughly 30 μm above the base (Xu et al., 1998). This oxygen limitation arises from both physical and biological factors: the dense biofilm matrix and viscous CF mucus impede diffusion, while host inflammatory responses recruit neutrophils whose respiratory burst consumes oxygen (Kolpen et al., 2010). Additionally, oxygen is consumed directly by host and bacteria respiration at the biofilm periphery, maintaining a persistent hypoxic gradient (Kolpen et al., 2010), as shown in Figure 1. Sputum from CF patients is predominantly micro-aerobic to anaerobic (Yoon et al., 2002; Alvarez-Ortega and Harwood, 2007a; Hassett et al., 2009). The extent of anoxic regions correlates with bacterial load and mucus thickness, potentially occupying large portions of mucus volume (Cowley et al., 2015). Notably, multidrug resistance protein (MexA) is more abundant in anoxic zones, suggesting enhanced drug tolerance under hypoxia in P. aeruginosa (Schaible et al., 2012; Martin et al., 2023; Pulukkody et al., 2021). Despite being a facultative anaerobe, P. aeruginosa can maintain growth under oxygen-limited conditions (Jackson et al., 2014), which in turn promotes robust biofilm formation (Rossi et al., 2021).
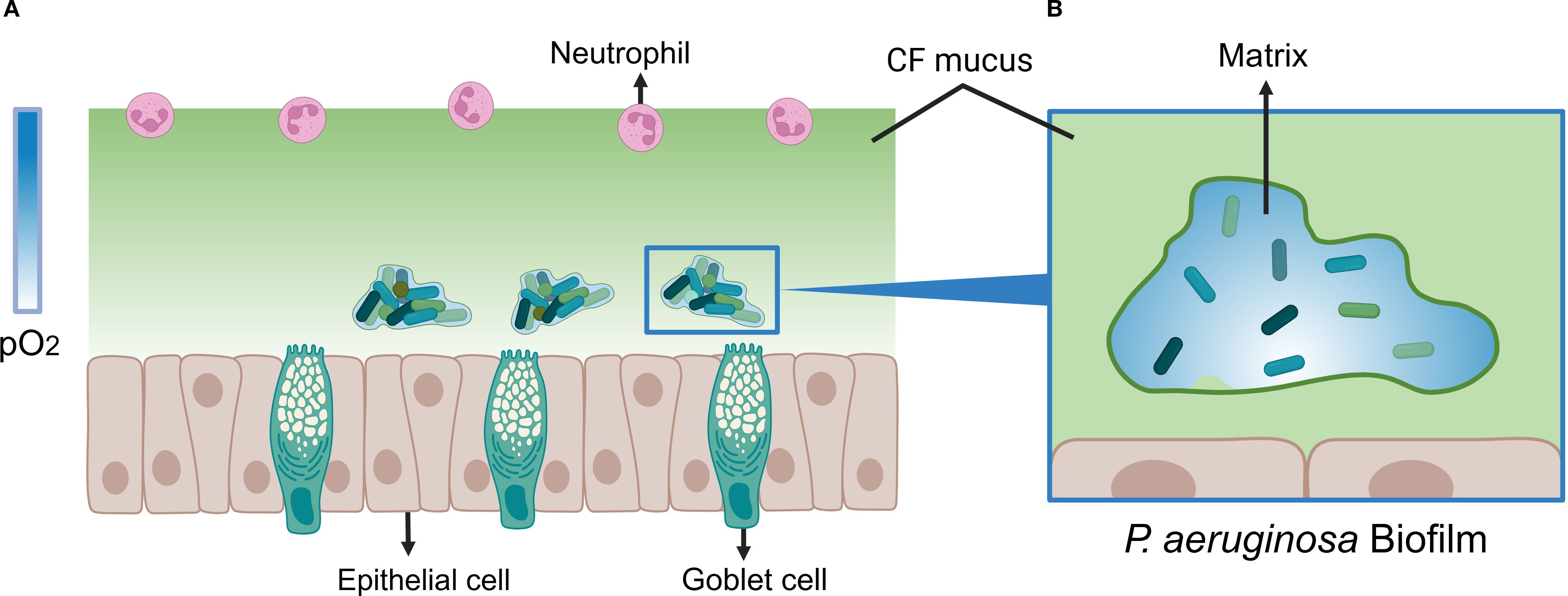
Figure 1. Model of P. aeruginosa Biofilm Under Oxygen-Limited Conditions. (A) P. aeruginosa biofilm model in cystic fibrosis (CF) mucus under oxygen-limited conditions. The color bar on the left side of the figure represents the partial pressure of oxygen (pO2). The gradient from deep to light blue indicates an increasing anaerobic state within the mucus. Goblet cells continuously secrete mucus, and combined with elevated oxygen consumption by host cells, a steep oxygen gradient forms within the thickened CF mucus (depicted in light green). P. aeruginosa forms biofilms within the hypoxic regions of the mucus, provoking a chronic inflammatory response in the host. This response leads to neutrophil infiltration and a respiratory burst, which further depletes oxygen levels within the mucus. (B) Oxygen-Limited Microenvironment within P. aeruginosa Biofilm. The transition from deep to light blue illustrates the gradual decrease in oxygen concentration observed inside the biofilm. Oxygen depletion occurs predominantly in the deep regions (white area). This gradient arises because oxygen is directly consumed by host cells and bacterial respiration at the biofilm surface, leading to a progressive oxygen decline with increasing distance from the surface. Created with BioRender.com.
3 The effect of oxygen-limited conditions on P. aeruginosa biofilms
3.1 Anaerobic metabolism
In human host environments, pathogens encounter fluctuating oxygen levels, with adaptive responses to such fluctuations primarily occurring at the metabolic level (Ferreira et al., 2013). Metabolism reprogramming is central to initiating bacterial tolerance mechanisms and reactivating transitions from a non-replicating to an actively growing state. P. aeruginosa exhibits remarkable metabolic versatility, utilizing diverse catabolic and anabolic pathways encoded in its genome to thrive in harsh environments (La Rosa et al., 2018; Crabbé et al., 2019). While aerobic metabolism dominates under oxygen-replete conditions, biofilm-embedded bacteria frequently reside in oxygen-limited niches. Under such constraints, P. aeruginosa shifts to aerobic metabolism—enabling sustained growth and metabolic activity without oxygen (Yoon et al., 2002). This adaptation encompasses anaerobic respiration and fermentation, as illustrated in Figure 2. Critically, this metabolic switch confers enhanced antibiotic tolerance and specific molecule resistance (Peng et al., 2017).
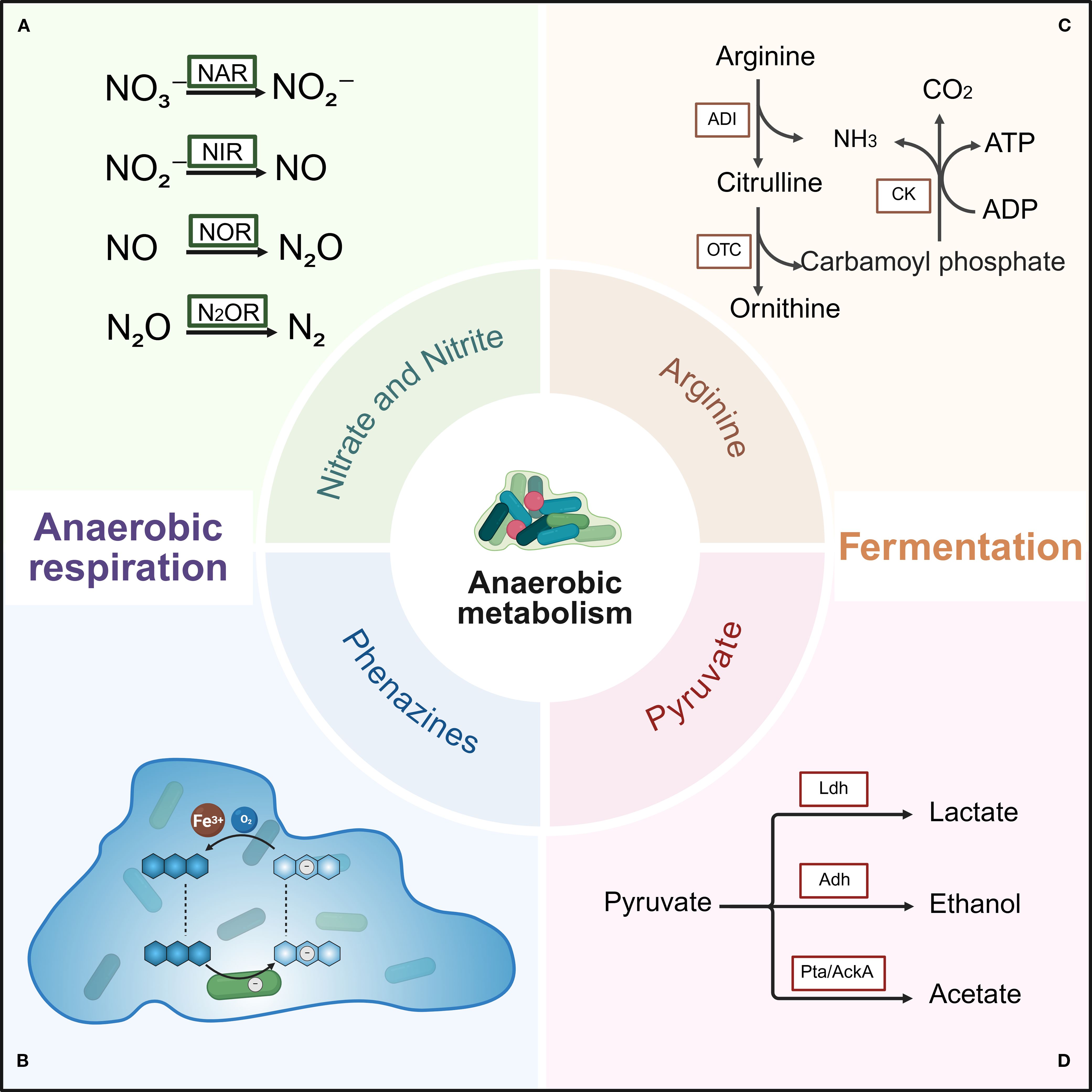
Figure 2. Anaerobic metabolism of P. aeruginosa biofilms under oxygen-limited conditions. Two primary anaerobic metabolic pathways are shown: anaerobic respiration and fermentation. Anaerobic respiration mainly involves denitrification pathways utilizing nitrate and phenazine to maintain cellular redox balance. Fermentation includes the arginine and pyruvate fermentation pathways, which generate ATP to sustain bacterial survival. (A) Reduction of nitrate, nitrite, nitric oxide, and nitrous oxide by nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase, respectively. (B) Electron transfer cycle of phenazine within biofilm. Cells are represented as rods, phenazine as blue hexagons, electrons as white circles, and oxygen concentration is depicted by the blue background. (C) Arginine deiminase (ADI) pathway. Key enzymes include arginine deiminase (ADI), ornithine carbamoyltransferase (OTC), and carbamate kinase (CK). (D) Pyruvate fermentation pathway. Key enzymes include lactate dehydrogenase (Ldh), alcohol dehydrogenase (Adh), phosphate acetyltransferase (Pta), and acetate kinase (AckA). Created with BioRender.com.
For energy generation via respiratory, P. aeruginosa can utilize oxygen, nitrogen compounds, and potentially other electron acceptors such as thiosulfate (Rossi et al., 2021). Under oxygen-limited conditions, P. aeruginosa performs anaerobic respiration using nitrate, nitrite, or nitrous oxide as terminal electron acceptors, facilitating rapid growth and energy production. In the absence of nitrate and nitrite, P. aeruginosa employs two alternative fermentation pathways to support slow growth or survival. The first involves substrate-level phosphorylation through arginine utilization, resulting in very slow growth. The second fermentation pathway uses pyruvate as a substrate, converting it into acetate, lactate, and small amounts of succinate.
3.1.1 Anaerobic respiration
Nitrogen metabolism under anaerobic and biofilm conditions promotes the virulence and tolerance of pathogens under hypoxic stress. Denitrification, a continuous four-step, eight-electron reduction process converting nitrate to nitrogen, is a key pathway enabling P. aeruginosa to respire anaerobically under oxygen-limited conditions (Yoon et al., 2002; Stoodley et al., 2024). In this process, nitrate (NO3-) or nitrite (NO2-) serves as the terminal electron acceptor (TEA), replacing oxygen generate energy. Denitrification also serves as an important cellular redox balancing mechanism within biofilms, provided these electron acceptors are available in sufficient concentrations (Line et al., 2014; Palmer et al., 2007). Using an alginate-encapsulated P. aeruginosa chronic infection model, it has been demonstrated that oxygen depletion limits bacterial growth. Supplementing nitrate as an alternative electron acceptor sustains the growth of P. aeruginosa microcolonies under oxygen-limited conditions, although the overall respiration rate decreases (Sønderholm et al., 2017). Consistent with these findings, lower nitrate and nitrite levels have been observed in infected wounds compared to non-infected controls, reflecting bacterial consumption via denitrification (Debats et al., 2006). Clinically, the ability of P. aeruginosa to perform anaerobic respiration under hypoxic conditions contributes to enhanced biofilm formation (Balasubramanian et al., 2017), aligning with model predictions (Aristotelous, 2022).
During hypoxic growth, nitric oxide (NO) can be produced endogenously as a denitrification intermediate or derived from exogenous NO donors. NO exhibits potent bactericidal properties (Fang, 1997). Accumulation of NO under anaerobic conditions acts as a stress signal, ultimately promoting biofilm formation as a defense mechanism (Yoon et al., 2011; Cutruzzolà and Frankenberg-Dinkel, 2016). NO detoxification proteins, including NO reductase (NOR, encoded by norVW) and flavohemoglobin (Hmp), help mitigate NO toxicity during denitrification (Gardner et al., 2003). Interestingly, nitrite reductase (NIR) also has NO-independent functions; NO can indirectly induce NIR expression and regulate flagella biosynthesis and swimming motility by forming a ternary complex with the molecular chaperone DnaK and flagellin FliC in the periplasm, serving as a scaffold (Borrero-de Acuña et al., 2015).
Beyond anaerobic respiration, P. aeruginosa in the mucus layer of CF lungs can perform microaerobic respiration. This process rapidly consumes oxygen, creating an oxygen gradient (Alvarez-Ortega and Harwood, 2007b). In the CF lung microenvironment, microaerobic respiration can occur simultaneously with nitrate respiration when both oxygen and nitrate are present (Chen et al., 2006). Three high-affinity terminal oxidases—the cbb3-1, cbb3-2, and cyanide-insensitive oxidases—enable growth at low oxygen concentrations and facilitate microaerobic growth. Notably, the genes encoding cbb3–2 oxidase and the cyanide-insensitive oxidase (cioAB) are highly expressed under oxygen-limited conditions (Alvarez-Ortega and Harwood, 2007b). The cyanide-insensitive oxidases not only increase under microaerobic conditions but may also protect cells against hydrogen cyanide toxicity during growth. The cbb3–1 oxidase is consistently expressed across various oxygen levels, suggesting P. aeruginosa maintains preparedness for sudden oxygen depletion without needing to trigger a transcriptionally regulated hypoxic response (Alvarez-Ortega and Harwood, 2007b).
3.1.2 Fermentation
Under oxygen-limited and nitrate-depleted conditions, P. aeruginosa biofilms sustain other energy supply through fermentation, primarily by activating the arginine fermentation pathway (Scribani Rossi et al., 2022) and pyruvate fermentation (Eschbach et al., 2004), which moderately support anaerobic growth and survival. L-arginine serves as a substrate for ATP production, enabling bacterial persistence under these conditions (Scribani Rossi et al., 2022). Specifically, P. aeruginosa utilizes the arginine deiminase (ADI) pathway to generate energy under oxygen-limited conditions, producing 1 mole of ATP per mole of L-arginine consumed. When arginine concentrations are sufficiently high, substrate-level phosphorylation can yield enough ATP to maintain bacterial growth. Thus, denitrification and arginine fermentation represent core metabolic processes in P. aeruginosa under oxygen-limited conditions.
Additionally, under anaerobic conditions, P. aeruginosa ferments pyruvate into lactate, acetate, and succinate. Although pyruvate fermentation does not support substantial anaerobic growth, it promotes long-term bacterial viability without contributing directly to proliferation (Eschbach et al., 2004). Proteomic analyses of hypoxic biofilm regions further suggest that cells produce fermentation by-products such as acetate (Pulukkody et al., 2021). Notably, nitrate respiration inhibits pyruvate fermentation, whereas arginine fermentation proceeds independently of pyruvate metabolism (Eschbach et al., 2004).
3.2 Phenazines as an electron shuttle
P. aeruginosa is well known for producing colored, redox-active metabolites called phenazines (Glasser et al., 2017). These phenazine compounds produced vary in structure and chemical properties (Mavrodi et al., 2010), functioning as electron-cycling molecules. The redox cycling of phenazine involves alternating reduction and oxidation reactions, which reoxidize accumulated NADH, thereby facilitating the transfer of electrons from reducing agents such as NAD(P)H to oxidizing agents like oxygen. This process promotes adenosine triphosphate (ATP) production and the generation of proton-motive force, allowing cells to survive in hypoxic regions and supporting colony growth (Dietrich et al., 2013; Ciemniecki and Newman, 2020). The redox potential of phenazines enables their reduction by bacterial cells and subsequent reaction with higher-potential oxidizing agents outside the cells, such as ferric iron and oxygen (Glasser et al., 2014). Acting as electron shuttles between bacteria and external substrates (Wang and Newman, 2008), phenazines alleviate limitations posed by scarce electron acceptors (Dietrich et al., 2013). Effectively, within the deep layers of the biofilm community, electrons accumulated for ATP synthesis can be accepted by oxidized phenazines and transferred to extracellular oxidants like oxygen. The cbb3-type terminal oxidases Cco1 and Cco2 of P. aeruginosa, key components of the respiratory chain, participate in phenazine reduction (Jo et al., 2017).
Phenazines and the cellular redox state directly influence biofilm morphogenesis via the regulatory protein RmcA, which modulates matrix components responsible for wrinkle formation (Dietrich et al., 2008; Okegbe et al., 2017). Phenazine-producing colonies tend to grow smoothly, whereas phenazine-deficient strains exhibit rougher, highly wrinkled biofilms that maximize oxygen contact (Kempes et al., 2014). Wrinkling serves as an adaptive mechanism to optimize oxygen accessibility and maintain metabolic homeostasis. Beyond their redox roles, phenazines act as signaling molecules that promote biofilm formation. P. aeruginosa synthesizes phenazine pigments such as pyocyanin, which intercalate into DNA base-pair regions, enhancing electron transfer, causing structural perturbations, and increasing DNA viscosity. The interaction is crucial for biofilm development (Das et al., 2015). Disrupting the pyocyanin—DNA interaction—via antioxidants or other inhibitors—can impede biofilm formation and associated infections (Das et al., 2015). The redox cycling of pyocyanin also generates reactive oxygen species (ROS), which damage host cells and pathogen cells, releasing eDNA (Das and Manefield, 2012; Rada et al., 2013). Phenazine compounds also mediate efficient extracellular electron transfer (EET) by interacting with eDNA in P. aeruginosa biofilms (Saunders et al., 2020). Together with the anaerobic stress responses, phenazines promote antibiotic tolerance and contribute to disease progression (Schiessl et al., 2019; Jiménez Otero et al., 2023). Notably, phenazines enhance biofilm tolerance to antibiotics such as ciprofloxacin (Schiessl et al., 2019).
Phenazines also stimulate pyruvate fermentation under anoxic conditions by mediating expression of the ackA and pta genes required for this pathway (Glasser et al., 2014). Through their redox cycling, phenazines enable P. aeruginosa to oxidize pyruvate to acetate and couple acetate metabolism with ATP synthesis via acetate kinase, thereby enhancing survival. In pyruvate fermentation, ATP generation is tightly linked to redox balance, a contrast to the arginine fermentation pathway where this connection is absent (Glasser et al., 2014).
3.3 Virulence expression
Mathematical modeling studies have demonstrated that bacterial biofilms growing anaerobically in anoxic environments secrete elevated levels of toxins. These toxins diffuse through the environment and lyse neutrophils, helping the biofilm resist neutrophil-mediated attacks. Consequently, bacterial adaptability and biofilm formation are enhanced under these conditions (Aristotelous, 2022). P. aeruginosa biofilms express a highly regulated protein secretion apparatus known as the type III secretion system (T3SS), which planktonic cells cannot deploy (Mikkelsen et al., 2009). The T3SS directly translocates a specific subset of exotoxin effector proteins–including ExoS, ExoU, ExoT, and ExoY–into host cells, driving P. aeruginosa pathogenicity (Cowell et al., 2005; Taylor and Winter, 2020). Multiple studies indicate that bacteria mount adaptive responses to diverse environments by modulating gene expression and protein production, thereby regulating the expression of virulence factors (Cullen and McClean, 2015; Teng et al., 2018) and inducing virulence protein synthesis (Fang et al., 2016). Oxygen limitation is a critical regulator of T3SS expression, a major virulence determinant in P. aeruginosa. Exposure to hypoxic conditions activates the T3SS (O’Callaghan et al., 2011; Chung et al., 2013). This activation strongly depends on the glyoxylate shunt enzyme isocitrate lyase (ICL, encoded by aceA), which is highly expressed in cystic fibrosis isolates specifically under oxygen-limited conditions (Chung et al., 2013). ICL-dependent regulation influences the expression of the T3 structural proteins, effectors, and regulatory proteins (ExsC, ExsD, and ExsE). Additionally, aceA modulates biofilm formation by affecting the expression of pslA, a gene involved in the biosynthesis of an extracellular polysaccharide (Chung et al., 2013). Notably, aceA mutants display enhanced biofilm formation during anaerobic growth.
The RetS/LadS signaling pathway reciprocally regulates T3SS expression and biofilm formation through a complex mechanism involving the GacS/GacA two-component system, the small regulatory RNAs RsmZ and RsmY, and the translational repressor RsmA (Goodman et al., 2004; Goodman et al., 2009; Kay et al., 2006; Ventre et al., 2006; O’Callaghan et al., 2011). Activation of LadS—or downregulation of RetS—promotes GacS homodimer formation, resulting in GacA phosphorylation and activation, accompanied by increased production of rsmZ and rsmY. These small RNAs sequester RsmA, relieving its repression and thereby activating biofilm formation (Kay et al., 2006; Brencic et al., 2009). Conversely, RetS activation induces heterodimer formation with GacS, inhibiting the GacS/GacA pathway. Free RsmA then binds specific mRNA targets, modulating their stability and indirectly activating exsA-dependent T3SS expression (Brencic et al., 2009). The regulation of AceA may be mediated by the RetS/LadS signaling pathway (Chung et al., 2013).
Moreover, the anaerobic regulator Anr senses oxygen limitation and induces expression of narL within the NarL/NarX two-component system. NarL represses the RsmA-antagonistic RNAs rsmZ and rsmY, resulting in increased levels of free RsmA, which stimulates T3SS expression. Free RsmA positively regulates T3SS and possibly other virulence determinants under its control, serving as a convergence point for P. aeruginosa’s response to various environmental cues (O’Callaghan et al., 2011).
Finally, P. aeruginosa OprG, an outer membrane protein belonging to the OmpW family of eight β-barrel porins, is broadly distributed. In iron-rich anaerobic environments, ANR significantly upregulates oprG transcription. Purified OprG forms cation-selective channels and substantially enhances cytotoxicity (McPhee et al., 2009).
4 Regulatory mechanisms under oxygen-limited conditions
P. aeruginosa biofilms adapt to oxygen-limited conditions through coordinated regulatory mechanisms. Anaerobic metabolism, including denitrification, arginine fermentation, and pyruvate fermentation, is activated under oxygen-limited conditions. Additionally, quorum sensing systems and the secondary messenger cyclic di-GMP (c-di-GMP) play crucial roles in modulating biofilm formation in response. Transcriptional regulatory networks involving the transcription factors ANR and DNR, and the NarXL two-component system, orchestrate the biofilm’s adaptive responses under oxygen-limited environments. Moreover, recent studies have also identified specific genes that further support biofilms growth and development under such conditions.
4.1 Anaerobic metabolism-related genes
4.1.1 Denitrification regulatory genes
The expression of narI and nirS genes in P. aeruginosa is upregulated under anaerobic conditions, playing key roles in anaerobic respiration (Martin et al., 2023). Notably, P. aeruginosa may induce denitrification genes in response to low oxygen levels regardless of nitrate availability. During anaerobic growth, two nitrate reductase gene clusters are expressed, one encoding a membrane-bound enzyme complex (narGHJI) associated with the cytoplasmic membrane, and another encoding a periplasmic enzyme complex (napAB) (Palmer et al., 2007). The membrane-bound nitrate reductase is essential for anaerobic growth, as P. aeruginosa depends on it for energy generation when nitrate is present (Palmer et al., 2007). Enzymes involved in nitrate respiration, including nitrate reductases NapA and NarG, accumulate in biofilms (Palmer et al., 2007). Furthermore, antibodies against NapA and NarG have been detected in the serum of CF patients, confirming in vivo production of these respiratory enzymes by (Palmer et al., 2007). The periplasmic nitrate reductase is not essential for anaerobic energy generation. However, it may balance intracellular redox states under low oxygen, a role reported in other microorganisms (Richardson et al., 2001). It is plausible that P. aeruginosa utilizes the periplasmic nitrate reductase similarly. The Nar complex, located in the cytoplasmic membrane, drives proton-motive force generation and ATP synthesis, whereas the periplasmic Nap complex mainly balances intracellular redox states without contributing directly to the transmembrane electrochemical gradient (Richardson et al., 2001; Williams et al., 2007; Borrero-de Acuña et al., 2016). This view contrasts with earlier findings where the periplasmic nitrate reductase and nitrate transport genes (e.g., narK2) were downregulated during anaerobic nitrate growth, with no observed differential regulation of the membrane-bound reductase (Filiatrault et al., 2005; Sharma et al., 2006). The discrepancy may stem from methodological differences, as the prior study lacked saturating mutagenesis and thus might have missed mutants in the nar operon. The narG gene is critical for anaerobic growth across varying nitrate concentrations. The narG operon also includes two homologs of the nitrate/nitrite antiporter gene narK, as well as PA3871 (nifM) and moaA1, which encode molybdopterin cofactor synthesis enzymes, and narHJI, encoding additional subunits of the membrane-bound nitrate reductase (Palmer et al., 2007).
4.1.2 Arginine fermentation regulatory genes
The arc operon encodes three key enzymes in the ADI pathway, arcA (arginine deiminase), arcB (catabolic ornithine carbamoyltransferase), and arcC (carbamate kinase), which is induced under oxygen-limited conditions (Vander Wauven et al., 1984). Subsequent studies revealed that the arc operon also includes the arcD, encoding a hydrophobic membrane-associated protein involved in the ADI process (Lüthi et al., 1990). Proteomic analyses of hypoxic P. aeruginosa biofilm regions reveal increased proteins linked to L-arginine and polyamine metabolism, with elevated ArcA and ArcB indicating active arginine-based energy production. Concurrently, the abundance of the cytosolic aminopeptidase PepA is approximately threefold higher in hypoxic compared to aerobic regions. PepA likely contributes to cellular protein degradation, recycling amino acids for stress responses such as pH homeostasis and energy generation (Pulukkody et al., 2021). Furthermore, the DNA-binding protein HupB is eight times more abundant in hypoxic zones relative to aerobic zones (Pulukkody et al., 2021). HupB, a small histone-like protein also known as heat-unstable (HU) protein, protects DNA from oxidative damage and facilitates adaptation to stress (Stojkova et al., 2019). In mammalian hosts, chronic P. aeruginosa infections are modulated by L-arginine metabolism and its derivative NO, with enhanced arginine catabolism observed at chronic wound sites (Debats et al., 2006). Microaerobic environments typical of chronic infection sites favor L-arginine fermentation, leading to NO deficiency, a hallmark of diminished host defense (Grasemann and Ratjen, 2012). Interestingly, L-arginine also supports phenazine production, and arginine metabolism remains largely unexplored (Ha et al., 2011).
4.1.3 Pyruvate fermentation regulatory genes
A genome-wide analysis of anaerobic metabolism in P. aeruginosa identified several key pyruvate-metabolizing genes, including NADH-dependent lactate dehydrogenase (ldhA), phosphotransacetylase (pta), and acetate kinase (ackA) (Eschbach et al., 2004). The conversion of pyruvate to lactate and acetate relies on the intact ldhA and ackA-pta gene clusters, respectively (Eschbach et al., 2004). The anaerobic induction of the ackA-pta promoter is modulated by oxygen tension through the transcriptional regulators Anr and the integration host factor (IHF) (Eschbach et al., 2004). IHF, potentially encoded by the anr gene, is believed to contain a 4Fe-4S cluster that functions as an oxygen tension sensor (Galimand et al., 1991; Sawers, 1991; Green et al., 2001). The ihfA locus encodes a subunit of the DNA-bending IHF protein, which plays a role in transcriptional regulation (Delic-Attree et al., 1995, 1996). Additionally, the gacS-ldhA operon (PA0926 and PA0927) was identified, encoding the sensor kinase GacS of the GacA/GacS two-component regulatory system and a putative fermentative lactate dehydrogenase (LdhA), respectively (Eschbach et al., 2004). The ackA-pta locus (PA0835 and PA0836) likely encodes acetate kinase and Pta, respectively, while the adhA locus (PA5427) encodes a putative alcohol dehydrogenase (AdhA) (Eschbach et al., 2004). Under oxygen-limited conditions, adhA is induced to facilitate ethanol catabolism, contributing to anaerobic energy metabolism (Crocker et al., 2019).
4.2 Quorum sensing
P. aeruginosa utilizes quorum sensing (QS), a cell-density-dependent intercellular communication system, which plays a pivotal role in regulating bacterial virulence and biofilm formation (Lee and Zhang, 2015). Two principal QS signaling molecules, N-butyryl-L-homoserine lactone (C4-HSL) and N-(3-oxododecanoyl)-L-homoserine lactone (3O-C12-HSL), mediate QS through the transcriptional activator pairs lasR/lasI and rhlR/rhlI, respectively. Notably, the expression of lasI and rhlI is significantly upregulated under low oxygen conditions (Alvarez-Ortega and Harwood, 2007b). The QS system modulates genes involved in denitrification, thereby influencing bacterial growth and survival by regulating NO levels within the biofilm. Additionally, QS activates the transcription of the pel gene, partially through the Rhl system. The pel gene product synthesizes the glucose-rich extracellular polysaccharide matrix essential for P. aeruginosa biofilm formation (Friedman and Kolter, 2004; Sakuragi and Kolter, 2007).
In the CF airway environment, optimal expression of the rhl QS component benefits P. aeruginosa persistence. QS controls the expression of the snr-1 gene, which in turn regulates the denitrification rate (Hassett et al., 2002). Specifically, the rhl system acts as an anaerobic repressor of snr-1 transcription. Consequently, in the absence of RhlR, the reducing power provided by Snr-1 increases, thus enhancing NAR activity. However, dysregulation of denitrification genes in rhlR mutants leads to elevated transcription of nar and nir genes, causing accumulation of toxic NO and subsequent self-damage (Yoon et al., 2002). NO, a by-product of anaerobic respiration, accumulates in rhlR mutants despite a modest increase (2-fold) in NOR activity, which is insufficient to mitigate NO toxicity (Yoon et al., 2002). Thus, under anaerobic conditions, P. aeruginosa relies on the rhl QS system and NO reductase to regulate NO levels and sustain robust biofilm formation and survival (Yoon et al., 2002).
4.3 3,5-cyclic diguanylic acid (c-di-GMP)
Another crucial signaling molecule in P. aeruginosa is the second messenger 3,5-cyclic diguanylic acid (c-di-GMP), which facilitates bacterial adaptation to diverse environmental conditions (Ha and O’Toole, 2015; Römling et al., 2013). The c-di-GMP plays a central role in regulating biofilm formation and dispersion (Römling et al., 2013). Elevated intracellular c-di-GMP levels drive the transition from a planktonic to a biofilm lifestyle by repressing motility-related genes, including those regulated by FleQ, and activating genes involved in exopolysaccharide production and biofilm maturation (Baraquet and Harwood, 2013). Conversely, reduced c-di-GMP levels trigger biofilm dispersion by activating motility structures, including flagella and pili (Ha and O’Toole, 2015). The synthesis and degradation of c-di-GMP are catalyzed by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively (Kalia et al., 2013). Although the interplay between QS and c-di-GMP signaling in P. aeruginosa is not fully elucidated, both pathways regulate virulence and biofilm dynamics, suggesting potential crosstalk. Current evidence indicates that the QS system can modulate intracellular c-di-GMP concentrations (Kim et al., 2018). Specifically, the Las-QS system may elevate c-di-GMP levels by stimulating DGC activity, whereas the Rhl-QS system might decrease c-di-GMP by inducing PDE activity. Furthermore, the tyrosine phosphatase TpbA inhibits the activity of the DGC TpbB via dephosphorylation, thereby reducing biofilm formation. Las-QS positively regulates TpbA expression, while Rhl-QS does not influence it (Ueda and Wood, 2009). Interestingly, TpbA also enhances rhl transcription, indicating that QS can negatively regulate c-di-GMP production in P. aeruginosa. However, the exact dynamics of c-di-GMP synthesis under varying QS states remain unclear. Moreover, NO has been reported to modulate DGC and PDE activities (Cutruzzolà and Frankenberg-Dinkel, 2016; Park et al., 2020). NO inhibits biofilm formation by enhancing PDE activity, leading to decreased intracellular c-di-GMP levels (Cutruzzolà and Frankenberg-Dinkel, 2016).
4.4 Transcription factors
TFs play a crucial role in regulating gene expression in response to environmental changes. Bacteria sense oxygen-limited conditions in their environment through Anr or Dnr, and regulate the expression of a series of genes to enable the bacteria to colonize and grow in oxygen-limited environments.
4.4.1 Anr
Anr is a well-characterized global transcriptional regulator and a key activator of gene expression under hypoxic conditions. It governs a regulatory network of hypoxia-responsive genes and serves as a hallmark of anaerobic or microaerobic growth (Trunk et al., 2010). Under low oxygen tension, active Anr promotes P. aeruginosa biofilm formation and virulence, playing a crucial role in host colonization (Jackson et al., 2014; Hammond et al., 2015). Deletion of the anr gene results in defective biofilm development and often abolishes anaerobic growth, whereas elevated Anr activity enhances biofilm formation (Jackson et al., 2013). Nevertheless, the precise underlying mechanisms remain incompletely understood. Notably, Anr regulon genes show no significant transcriptional increase under hypoxia, consistent with the findings of Alvarez-Orgeta et al (Alvarez-Ortega and Harwood, 2007b), suggesting Anr regulation involves mechanisms beyond its own regulon transcription.
Anr is indispensable for activating energy metabolism pathways during hypoxia. It strongly induces denitrification and regulates the expression of other transcriptional regulators, including dnr and the narXL two-component system (Jackson et al., 2014). The dnr is essential for activating denitrification during anaerobic growth, controlling a subset of genes involved in nitrate respiration (Ferrara et al., 2021). The NarL response regulator modulates energy metabolism under hypoxic stress by promoting nitrate utilization and repressing less efficient energy-yielding pathways such as pyruvate and arginine fermentation (Schreiber et al., 2007; Benkert et al., 2008). Interestingly, while Anr enhances NarL production, it also independently promotes arginine fermentation by upregulating the arcDABC operon under anaerobic conditions (Filiatrault et al., 2006). Furthermore, Anr activates the ackA-pta operon responsible for pyruvate fermentation in response to low oxygen (Eschbach et al., 2004; Filiatrault et al., 2006).
Anr also upregulates genes encoding high-affinity cytochrome oxidases (hemF and hemN) and CupA fimbriae components (oprG and cupA1-5), facilitating respiratory adaptation during hypoxia and contributing to biofilm development and pathogenicity (Hammond et al., 2015). Additionally, Anr influences quorum sensing by regulating the small regulatory RNA PhrS (Sonnleitner et al., 2011), thereby modulating biofilm adaptability under low oxygen.
A well-studied virulence factor of P. aeruginosa, the hemolytic phospholipase C (PlcH), is tightly regulated by Anr. The catabolism of choline released by PlcH enhances Anr activity (Massimelli et al., 2005; Jackson et al., 2013, 2014). The plcH promoter contains a conserved Anr consensus binding sequence across all P. aeruginosa genomes (Galimand et al., 1991; Trunk et al., 2010). Anr represses plcH expression, maintaining PlcH protein at levels that facilitate effective host-pathogen interactions without compromising biofilm integrity (Jackson et al., 2014). Under oxygen-limited conditions, PlcH production may become disadvantageous, as excessive PlcH protein can compromise the structural integrity of the P. aeruginosa biofilm. Anr likely binds directly to the plcH promoter, which contains a conserved Anr consensus sequence across all P. aeruginosa genomes. Mutations in this conserved sequence result in increased plcH expression under hypoxia. Although Anr shares its consensus binding sequence with the secondary regulator Dnr, their activation mechanisms differ; notably, Dnr does not participate in plcH repression (Jackson et al., 2014). Anr is active when its 4Fe-4S iron-sulfur cluster remains intact, whereas Dnr is activated by the oxidation of its heme cofactor by nitric oxide, which induces a conformational change enabling DNA binding (Rodionov et al., 2005; Trunk et al., 2010).
The small RNA ErsA also plays an important role in anaerobic adaptation. It regulates bacterial-host interactions, including biofilm maturation, motility, and antibiotic resistance (Falcone et al., 2018; Zhang et al., 2017; Sonnleitner et al., 2020). ErsA is transcriptionally induced under oxygen-limited conditions (Ferrara et al., 2015) and transmits low-oxygen signals to the Anr regulon. It positively regulates Anr expression at the post-transcriptional level (Ferrara et al., 2021). Once ErsA surpasses a certain threshold, the RNA-binding protein Hfq acts synergistically with ErsA to activate Anr. This function of Hfq complements its own post-transcriptional regulation of Anr, indicating that ErsA-mediated activation of anr expression depends on Hfq (Ferrara et al., 2021). Hfq’s chaperone activity likely promotes the interaction between ErsA and anr mRNA, enhancing anr mRNA translation, possibly by improving access to its initiation site. This positive regulation by ErsA contributes to the stabilization of anr mRNA, consistent with the observed reduction in anr mRNA levels when ErsA is absent. Additionally, Hfq has been reported to stimulate anr expression via an unknown mechanism (Sonnleitner et al., 2011; Sonnleitner and Bläsi, 2014). Deletion of ErsA leads to reduced virulence of P. aeruginosa both in vitro and in vivo, markedly impaired biofilm formation and maturation (Ferrara et al., 2020), and severely compromised anaerobic growth through denitrification and arginine fermentation. The role of ErsA in biofilm regulation may also involve downregulation of the AlgC enzyme (Ferrara et al., 2015) and the activation of the AmrZ regulon (Falcone et al., 2018). Furthermore, ErsA directly negatively regulates oprD mRNA, affecting the envelope composition of P. aeruginosa (Zhang et al., 2017; Sonnleitner et al., 2020). These findings suggest that P. aeruginosa’s adaptation to the CF lung environment may increase its reliance on ErsA for regulating anaerobic metabolism.
4.4.2 DNR
DNR is a critical transcriptional activator essential for initiating the denitrification process in P. aeruginosa. It was the first protein identified to restore anaerobic respiration and arginine substrate-level phosphorylation growth in anr mutants (Hassett et al., 2009). The expression and activity of Dnr are themselves regulated by Anr, positioning Dnr downstream in the oxygen-sensing regulatory cascade. Under hypoxic conditions, both Anr and Dnr coordinate to activate transcription of genes involved in denitrification and anaerobic respiration. Key operons under their control include narKGHJIm, encoding nitrate reductase; nirSM-CFLGHJEN, encoding nitrite reductase; and ccoN2O2Q2P2, encoding the cbb3–2 cytochrome oxidase complex.
In the presence of nitrate under anaerobic conditions, expression from the narK1 promoter is induced through the combined action of Anr, Dnr, and the nitrate-responsive two-component regulatory system NarXL (Schreiber et al., 2007). The DNA-bending protein integration host factor (IHF) is also crucial for optimal promoter activity. Moreover, Anr and NarXL induce dnr expression, amplifying the regulatory cascade (Schreiber et al., 2007). The cooperative function of NarXL and Dnr, regulated by ANR, is necessary for transcription of the nitrite reductase regulatory gene nirQ under anaerobic conditions (Schreiber et al., 2007; Hassett et al., 2009). Dnr belongs to the Crp-Fnr superfamily of transcriptional regulators and has been reported to activate expression of genes involved in the denitrification pathway, including nir, nor, and nos (Hasegawa et al., 1998; Arai et al., 2003). Transcriptional control of the nar locus occurs via the intergenic region between narXL and narK1. Both nitrate and nitrite induce narK expression, and a basal induction persists even during arginine fermentation (Schreiber et al., 2007). The narK1 promoter activity is modulated by both Anr and Dnr, with Anr being indispensable for its baseline activation, while Dnr enhances promoter activity independently of Anr (Schreiber et al., 2007). Anr indirectly facilitates transcriptional activation of nirS by inducing dnr expression in this regulatory hierarchy. The transcription of NorC also requires Anr and Dnr in the presence of nitrous oxide but is not directly regulated by NarL (Arai et al., 1995).
Although the small RNA ErsA positively regulates Anr post-transcriptional (Ferrara et al., 2021), no direct regulation of Dnr by ErsA has been observed. Nevertheless, dnr transcript levels decrease in the absence of ErsA, likely due to diminished Anr expression. Additionally, under anaerobic conditions, expression of the narXL genes is also upregulated by both Anr and Dnr (Schreiber et al., 2007).
4.5 Two-component regulatory system
Two-component systems (TCSs) are widespread in prokaryotic genomes and constitute a fundamental regulatory network that enables bacteria to adapt, survive, and modulate pathogenicity in response to environmental changes. Functioning as molecular switches, these systems sense external stimuli and regulate gene expression accordingly, facilitating bacterial adaptation to diverse conditions. A canonical TCS consists of a membrane-bound sensor kinase (SK) and a cytoplasmic response regulator (RR), typically encoded adjacently in the genome (Linsky et al., 2020). Upon sensing environmental signals, the sensor domain of the SK undergoes conformational changes that are transmitted through the transmembrane region to its cytoplasmic histidine kinase domain. This triggers autophosphorylation of a conserved C-terminal histidine residue in trans (Mitrophanov and Groisman, 2008; Jacob-Dubuisson et al., 2018). The phosphoryl group is then transferred to a conserved aspartate residue on the RR, activating it. The activated RR modulates the transcription of downstream genes involved in diverse physiological processes, including bacterial virulence, pathogenesis, biofilm formation, cell division, and metabolite production (Tierney and Rather, 2019; Li et al., 2023).
In P. aeruginosa, the NarXL system is a nitrate-responsive two-component regulatory system (Krieger et al., 2002). NarX is the sensor kinase and NarL its response regulator; in the presence of nitrate, NarXL activates and cooperates with Dnr to upregulate nar, nir, and nor genes encoding nitrate, nitrite, and nitric oxide reductases (Hassett et al., 2009). Simultaneously, NarL represses the arginine fermentation pathway by inhibiting the arginine-dependent activation of the arcDABC operon mediated by the transcriptional activator ArgR. Specifically, NarL suppresses the expression of arcA, arcB, and arcC genes without affecting the oxygen tension-dependent activation driven by Anr (Benkert et al., 2008). Under conditions where both nitrate and arginine are present, NarL binding likely interferes with ArgR’s interaction at overlapping DNA binding sites, thereby preventing ArgR-mediated induction of arcDABC transcription (Benkert et al., 2008).
4.6 Other factors
In addition to above factors, studies have identified other specialized genes that promote P. aeruginosa growth and biofilm formation. OmpW is an eight-helix β-barrel outer membrane porin that facilitates the uptake of small hydrophobic molecules (Catel-Ferreira et al., 2016). It has been demonstrated that OmpW participates in bacterial adaptation to various environmental stresses (Brambilla et al., 2014). In P. aeruginosa, OmpW expression is upregulated under hypoxic or anaerobic conditions (McPhee et al., 2009). However, OmpW expression may be downregulated when iron levels are low, as OmpW is implicated in iron uptake (Lin et al., 2008). Concurrently, under hypoxia, the global regulator Lrp is upregulated (Gil-Marqués et al., 2022). The ompW promoter contains an Lrp-binding site, through which Lrp negatively regulates ompW expression (Gil-Marqués et al., 2022).
Another crucial outer membrane protein, OprF, functions as a cytokine and plays a vital role in regulating anaerobic metabolism in P. aeruginosa. It is essential for the optimal survival of anaerobic biofilms (Hassett et al., 2002). The absence of OprF results in severely impaired bacterial growth due to the loss of nitrite reductase activity and defects in anaerobic respiration. Notably, OprF is detectable exclusively in anaerobic biofilms (Beg et al., 2023), and only CF patients with chronic infections possess antibodies against OprF. Bacteria lacking oprF exhibit diminished anaerobic biofilm formation, partly attributable to the lack of NIR activity. Two hypotheses have been proposed regarding OprF’s precise role in anaerobic growth. First, OprF may serve as a porin facilitating nitrate or nitrite transport into the cell and potentially interact directly with NIR to stabilize its enzymatic activity. Second, the absence of OprF may compromise peptidoglycan stability, as OprF has been shown to interact with this essential cell wall component (Rawling et al., 1998). Reduced peptidoglycan integrity renders cells more fragile and susceptible to environmental stress.
5 Clinical significance and future directions
Biofilm formation serves as a crucial protective strategy, enabling pathogenic bacteria to resist various environmental stresses (Ciofu and Tolker-Nielsen, 2019). The hypoxic microenvironment within biofilms facilitates bacterial adaptation through reduced metabolic activity and heightened antibiotic tolerance (Penesyan et al., 2015; Stokes et al., 2019), which contributes to persistent chronic infections. Traditional antibacterial treatments have largely targeted the eradication of pathogens but often overlook the modulation of the infected microenvironment, resulting in issues such as antibiotic resistance and incomplete bacterial clearance (Hou et al., 2021; Zhao et al., 2023b). Recently, strategies aimed at reversing the hypoxic microenvironment have emerged as a promising research focus for combating biofilm-associated infections.
Reversal of hypoxia triggers a cascade of beneficial effects, including the reactivation of suppressed immune responses, promotion of osteogenesis and angiogenesis, induction of cuproptosis-like bacterial death, and stimulation of dendritic cells and macrophages to enhance antibacterial activity via chemotaxis and phagocytosis (Luo et al., 2024). Photodynamic therapy (PDT) is a notable antibacterial approach capable of effectively killing bacteria and preventing multidrug resistance; however, its efficacy is markedly compromised under hypoxic conditions. Enhancing oxygen delivery to treatment sites and alleviating hypoxia significantly improves PDT efficacy, providing a promising avenue for biofilm eradication (Xiu et al., 2020; Bai et al., 2021; Wu et al., 2022).
Recent advancements include the development of porphyrinic metal-organic framework (MOF)-based metalloantibiotics that catalyze endogenous hydrogen peroxide (H2O2) decomposition to generate oxygen. The resulting oxygen enhances oxygen-dependent sonodynamic therapy (SDT), which disrupts bacterial homeostasis—affecting cell membrane integrity and quorum sensing systems—thereby promoting bacterial killing (Su et al., 2024). Concurrently, sustained oxygen production supports fibroblast survival and migration, stimulates angiogenic growth factors, promotes angiogenesis, and increases secretion of anti-inflammatory cytokines (Su et al., 2024). Nitric oxide (NO) exhibits dual effects depending on its concentration: high levels possess bactericidal activity, whereas low levels induce biofilm dispersion and sensitize bacteria to antibiotics (Cai et al., 2021). In P. aeruginosa models, low-dose NO, which is non-lethal, acts as a signaling molecule triggering biofilm dispersal in ex vivo cystic fibrosis (CF) sputum, reducing bacterial tolerance to tobramycin alone or combined with ceftazidime. This highlights NO’s potential as an adjunct therapy for managing P. aeruginosa biofilm infections in CF patients (Howlin et al., 2017). Consequently, NO-based treatments represent a promising approach against antibiotic-resistant bacteria and biofilm-associated infections. Overall, alleviating biofilm hypoxia holds significant potential for enhancing treatment efficacy and overcoming chronic biofilm-related infections.
6 Conclusions
Biofilm cells exhibit an oxygen gradient that typically reduces their sensitivity to antibiotics, making complete eradication challenging. The adaptation of bacterial biofilms to hypoxic conditions is considered a crucial factor for their prolonged latent persistence in the human body. This hypoxic microenvironment triggers a series of complex bacterial responses. Current studies suggest that under oxygen-limited conditions, P. aeruginosa shifts to anaerobic metabolism, utilizing nitrate for denitrification to support growth. In the absence of nitrate or nitrite, survival is maintained through arginine or pyruvate fermentation pathways. Additionally, P. aeruginosa biofilms produce phenazine compounds to sustain redox balance within the biofilm matrix. Adaptation to oxygen limitation also involves the regulation of virulence gene expression and QS systems. These processes are coordinately controlled by transcriptional regulators such as Anr, Dnr, NarXL, and other specialized genes, collectively promoting bacterial survival and biofilm formation. A deeper understanding of the adaptive mechanisms employed by bacterial biofilms under oxygen-limited conditions can provide new directions for future treatments of biofilm-associated infections, including strategies targeting QS or c-di-GMP pathways, modulation of Anr or Dnr regulators, and nitric oxide-based therapies.
Author contributions
LR: Writing – review & editing, Writing – original draft. YY: Writing – original draft, Writing – review & editing. KF: Writing – review & editing, Data curation, Investigation. XF: Investigation, Data curation, Writing – review & editing. JH: Writing – review & editing, Visualization. BZ: Conceptualization, Writing – review & editing. YL: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Basic Scientific Research Project of Education Department of Liaoning Province (LJKMZ20221190).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alvarez-Ortega, C. and Harwood, C. S. (2007a). Identification of a malate chemoreceptor in pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl. Environ. Microbiol. 73, 7793–7795. doi: 10.1128/AEM.01898-07, PMID: 17933940
Alvarez-Ortega, C. and Harwood, C. S. (2007b). Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165. doi: 10.1111/j.1365-2958.2007.05772.x, PMID: 17581126
Arai, H., Igarashi, Y., and Kodama, T. (1995). Expression of the nir and nor genes for denitrification of pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 371, 73–76. doi: 10.1016/0014-5793(95)00885-d, PMID: 7664887
Arai, H., Mizutani, M., and Igarashi, Y. (2003). Transcriptional regulation of the nos genes for nitrous oxide reductase in pseudomonas aeruginosa. Microbiol. (read. Engl.) 149, 29–36. doi: 10.1099/mic.0.25936-0, PMID: 12576577
Aristotelous, A. C. (2022). Biofilm neutrophils interactions under hypoxia: A mathematical modeling study. Math Biosci. 352, 108893. doi: 10.1016/j.mbs.2022.108893, PMID: 36029807
Bai, Y., Hu, Y., Gao, Y., Wei, X., Li, J., Zhang, Y., et al. (2021). Oxygen self-supplying nanotherapeutic for mitigation of tissue hypoxia and enhanced photodynamic therapy of bacterial keratitis. ACS Appl. Mater Interfaces 13, 33790–33801. doi: 10.1021/acsami.1c04996, PMID: 34254513
Balasubramanian, D., Harper, L., Shopsin, B., and Torres, V. J. (2017). Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 75, ftx005. doi: 10.1093/femspd/ftx005, PMID: 28104617
Baraquet, C. and Harwood, C. S. (2013). Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. U.S.A. 110, 18478–18483. doi: 10.1073/pnas.1318972110, PMID: 24167275
Beg, A. Z., Rashid, F., Talat, A., Haseen, M. A., Raza, N., Akhtar, K., et al. (2023). Functional amyloids in pseudomonas aeruginosa are essential for the proteome modulation that leads to pathoadaptation in pulmonary niches. Microbiol. Spectr. 11, e0307122. doi: 10.1128/spectrum.03071-22, PMID: 36475836
Benkert, B., Quäck, N., Schreiber, K., Jaensch, L., Jahn, D., and Schobert, M. (2008). Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiol. (Reading) 154, 3053–3060. doi: 10.1099/mic.0.2008/018929-0, PMID: 18832311
Borrero-de Acuña, J. M., Molinari, G., Rohde, M., Dammeyer, T., Wissing, J., Jänsch, L., et al. (2015). A periplasmic complex of the nitrite reductase NirS, the chaperone DnaK, and the flagellum protein FliC is essential for flagellum assembly and motility in pseudomonas aeruginosa. J. Bacteriol. 197, 3066–3075. doi: 10.1128/JB.00415-15, PMID: 26170416
Borrero-de Acuña, J. M., Rohde, M., Wissing, J., Jänsch, L., Schobert, M., Molinari, G., et al. (2016). Protein network of the pseudomonas aeruginosa denitrification apparatus. J. Bacteriol. 198, 1401–1413. doi: 10.1128/JB.00055-16, PMID: 26903416
Borriello, G., Werner, E., Roe, F., Kim, A. M., Ehrlich, G. D., and Stewart, P. S. (2004). Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial Agents chemotherapy. 48, 2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004, PMID: 15215123
Brambilla, L., Morán-Barrio, J., and Viale, A. M. (2014). Expression of the Escherichia coli ompW colicin S4 receptor gene is regulated by temperature and modulated by the H-NS and StpA nucleoid-associated proteins. FEMS Microbiol. Lett. 352, 238–244. doi: 10.1111/1574-6968.12385, PMID: 24444297
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x, PMID: 19602144
Cai, Y. M., Zhang, Y. D., and Yang, L. (2021). NO donors and NO delivery methods for controlling biofilms in chronic lung infections. Appl. Microbiol. Biotechnol. 105, 3931–3954. doi: 10.1007/s00253-021-11274-2, PMID: 33937932
Carreau, A., El Hafny-Rahbi, B., Matejuk, A., Grillon, C., and Kieda, C. (2011). Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 15, 1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x, PMID: 21251211
Catel-Ferreira, M., Marti, S., Guillon, L., Jara, L., Coadou, G., Molle, V., et al. (2016). The outer membrane porin OmpW of Acinetobacter baumannii is involved in iron uptake and colistin binding. FEBS Lett. 590, 224–231. doi: 10.1002/1873-3468.12050, PMID: 26823169
Chen, F., Xia, Q., and Ju, L.-K. (2006). Competition between oxygen and nitrate respirations in continuous culture of pseudomonas aeruginosa performing aerobic denitrification. Biotechnol. Bioeng. 93, 1069–1078. doi: 10.1002/bit.20812, PMID: 16435399
Chiba, A., Seki, M., Suzuki, Y., Kinjo, Y., Mizunoe, Y., and Sugimoto, S. (2022). Staphylococcus aureus utilizes environmental RNA as a building material in specific polysaccharide-dependent biofilms. NPJ Biofilms Microbiomes 8, 17. doi: 10.1038/s41522-022-00278-z, PMID: 35379830
Chung, J. C. S., Rzhepishevska, O., Ramstedt, M., and Welch, M. (2013). Type III secretion system expression in oxygen-limited Pseudomonas aeruginosa cultures is stimulated by isocitrate lyase activity. Open Biol. 3, 120131. doi: 10.1098/rsob.120131, PMID: 23363478
Ciemniecki, J. A. and Newman, D. K. (2020). The potential for redox-active metabolites to enhance or unlock anaerobic survival metabolisms in aerobes. J. Bacteriol 202, e00797–e00719. doi: 10.1128/JB.00797-19, PMID: 32071098
Ciofu, O. and Tolker-Nielsen, T. (2019). Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00913, PMID: 31130925
Cowell, B. A., Evans, D. J., and Fleiszig, S. M. J. (2005). Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol. Lett. 250, 71–76. doi: 10.1016/j.femsle.2005.06.044, PMID: 16039071
Cowley, E. S., Kopf, S. H., LaRiviere, A., Ziebis, W., and Newman, D. K. (2015). Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6, e00767. doi: 10.1128/mBio.00767-15, PMID: 26220964
Crabbé, A., Jensen, P.Ø., Bjarnsholt, T., and Coenye, T. (2019). Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 27, 850–863. doi: 10.1016/j.tim.2019.05.003, PMID: 31178124
Crocker, A. W., Harty, C. E., Hammond, J. H., Willger, S. D., Salazar, P., Botelho, N. J., et al. (2019). Pseudomonas aeruginosa ethanol oxidation by AdhA in low-oxygen environments. J. Bacteriol. 201, e00393–e00319. doi: 10.1128/JB.00393-19, PMID: 31527114
Cullen, L. and McClean, S. (2015). Bacterial adaptation during chronic respiratory infections. Pathogens 4, 66–89. doi: 10.3390/pathogens4010066, PMID: 25738646
Cutruzzolà, F. and Frankenberg-Dinkel, N. (2016). Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J. Bacteriol 198, 55–65. doi: 10.1128/JB.00371-15, PMID: 26260455
Das, T., Kutty, S. K., Tavallaie, R., Ibugo, A. I., Panchompoo, J., Sehar, S., et al. (2015). Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci. Rep. 5, 8398. doi: 10.1038/srep08398, PMID: 25669133
Das, T. and Manefield, M. (2012). Pyocyanin promotes extracellular DNA release in pseudomonas aeruginosa. PLoS One 7, e46718. doi: 10.1371/journal.pone.0046718, PMID: 23056420
Debats, I. B. J. G., Booi, D., Deutz, N. E. P., Buurman, W. A., Boeckx, W. D., and van der Hulst, R. R. W. J. (2006). Infected chronic wounds show different local and systemic arginine conversion compared with acute wounds. J. Surg. Res. 134, 205–214. doi: 10.1016/j.jss.2006.03.005, PMID: 16631201
Delic-Attree, I., Toussaint, B., Froger, A., Willison, J. C., and Vignais, P. M. (1996). Isolation of an IHF-deficient mutant of a pseudomonas aeruginosa mucoid isolate and evaluation of the role of IHF in algD gene expression. Microbiol. (read. Engl.) 142, 2785–2793. doi: 10.1099/13500872-142-10-2785, PMID: 8885394
Delic-Attree, I., Toussaint, B., and Vignais, P. M. (1995). Cloning and sequence analyses of the genes coding for the integration host factor (IHF) and HU proteins of pseudomonas aeruginosa. Gene 154, 61–64. doi: 10.1016/0378-1119(94)00875-s, PMID: 7867950
Dietrich, L. E. P., Okegbe, C., Price-Whelan, A., Sakhtah, H., Hunter, R. C., and Newman, D. K. (2013). Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol 195, 1371–1380. doi: 10.1128/JB.02273-12, PMID: 23292774
Dietrich, L. E. P., Teal, T. K., Price-Whelan, A., and Newman, D. K. (2008). Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Sci. (n. Y. N.Y.) 321, 1203–1206. doi: 10.1126/science.1160619, PMID: 18755976
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063, PMID: 12194761
Ehrhardt, K., Becker, A. L., and Grassl, G. A. (2023). Determinants of persistent Salmonella infections. Curr. Opin. Immunol. 82, 102306. doi: 10.1016/j.coi.2023.102306, PMID: 36989589
Eschbach, M., Schreiber, K., Trunk, K., Buer, J., Jahn, D., and Schobert, M. (2004). Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol 186, 4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004, PMID: 15231792
Falcone, M., Ferrara, S., Rossi, E., Johansen, H. K., Molin, S., and Bertoni, G. (2018). The small RNA ErsA of pseudomonas aeruginosa contributes to biofilm development and motility through post-transcriptional modulation of AmrZ. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00238, PMID: 29497413
Fang, F. C. (1997). Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99, 2818–2825. doi: 10.1172/JCI119473, PMID: 9185502
Fang, F. C., Frawley, E. R., Tapscott, T., and Vázquez-Torres, A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20, 133–143. doi: 10.1016/j.chom.2016.07.009, PMID: 27512901
Ferrara, S., Carloni, S., Fulco, R., Falcone, M., Macchi, R., and Bertoni, G. (2015). Post-transcriptional regulation of the virulence-associated enzyme AlgC by the σ22-dependent small RNA ErsA of Pseudomonas aeruginosa. Environ. Microbiol. 17, 199–214. doi: 10.1111/1462-2920.12590, PMID: 25186153
Ferrara, S., Carrubba, R., Santoro, S., and Bertoni, G. (2021). The small RNA ErsA impacts the anaerobic metabolism of Pseudomonas aeruginosa through post-transcriptional modulation of the master regulator anr. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.691608, PMID: 34759894
Ferrara, S., Rossi, A., Ranucci, S., De Fino, I., Bragonzi, A., Cigana, C., et al. (2020). The small RNA ErsA plays a role in the regulatory network of pseudomonas aeruginosa pathogenicity in airway infections. Msphere 5, e00909-20. doi: 10.1128/msphere.00909-20, PMID: 33055260
Ferreira, M. T., Manso, A. S., Gaspar, P., Pinho, M. G., and Neves, A. R. (2013). Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS One 8, e58277. doi: 10.1371/journal.pone.0058277, PMID: 23472168
Filiatrault, M. J., Picardo, K. F., Ngai, H., Passador, L., and Iglewski, B. H. (2006). Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74, 4237–4245. doi: 10.1128/IAI.02014-05, PMID: 16790798
Filiatrault, M. J., Wagner, V. E., Bushnell, D., Haidaris, C. G., Iglewski, B. H., and Passador, L. (2005). Effect of anaerobiosis and nitrate on gene expression in pseudomonas aeruginosa. Infect. Immun. 73, 3764–3772. doi: 10.1128/IAI.73.6.3764-3772.2005, PMID: 15908409
Friedman, L. and Kolter, R. (2004). Genes involved in matrix formation in pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x, PMID: 14731271
Gale, M. J., Maritato, M. S., Chen, Y.-L., Abdulateef, S. S., and Ruiz, J. E. (2015). Pseudomonas aeruginosa causing inflammatory mass of the nasopharynx in an immunocompromised HIV infected patient: a mimic of Malignancy. Idcases 2, 40–43. doi: 10.1016/j.idcr.2015.01.004, PMID: 26793451
Galimand, M., Gamper, M., Zimmermann, A., and Haas, D. (1991). Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in pseudomonas aeruginosa. J. Bacteriol. 173, 1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991, PMID: 1900277
Gardner, A. M., Gessner, C. R., and Gardner, P. R. (2003). Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 278, 10081–10086. doi: 10.1074/jbc.M212462200, PMID: 12529359
Gil-Marqués, M. L., Pachón, J., and Smani, Y. (2022). iTRAQ-Based Quantitative Proteomic Analysis of Acinetobacter baumannii under Hypoxia and Normoxia Reveals the Role of OmpW as a Virulence Factor. Microbiol. Spectr. 10, e0232821. doi: 10.1128/spectrum.02328-21, PMID: 35234505
Glasser, N. R., Kern, S. E., and Newman, D. K. (2014). Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 92, 399–412. doi: 10.1111/mmi.12566, PMID: 24612454
Glasser, N. R., Saunders, S. H., and Newman, D. K. (2017). The colorful world of extracellular electron shuttles. Annu. Rev. Microbiol. 71, 731–751. doi: 10.1146/annurev-micro-090816-093913, PMID: 28731847
Gomila, A., Carratalà, J., Badia, J. M., Camprubí, D., Piriz, M., Shaw, E., et al. (2018). Preoperative oral antibiotic prophylaxis reduces Pseudomonas aeruginosa surgical site infections after elective colorectal surgery: a multicenter prospective cohort study. BMC Infect. Dis. 18, 507. doi: 10.1186/s12879-018-3413-1, PMID: 30290773
Goodman, A. L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R. S., and Lory, S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in pseudomonas aeruginosa. Dev. Cell 7, 745–754. doi: 10.1016/j.devcel.2004.08.020, PMID: 15525535
Goodman, A. L., Merighi, M., Hyodo, M., Ventre, I., Filloux, A., and Lory, S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259. doi: 10.1101/gad.1739009, PMID: 19171785
Grasemann, H. and Ratjen, F. (2012). Nitric oxide and L-arginine deficiency in cystic fibrosis. Curr. Pharm. Des. 18, 726–736. doi: 10.2174/138161212799315911, PMID: 22229575
Green, J., Scott, C., and Guest, J. R. (2001). Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44, 1–34. doi: 10.1016/s0065-2911(01)44010-0, PMID: 11407111
Ha, D.-G., Merritt, J. H., Hampton, T. H., Hodgkinson, J. T., Janecek, M., Spring, D. R., et al. (2011). 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J. Bacteriol 193, 6770–6780. doi: 10.1128/JB.05929-11, PMID: 21965567
Ha, D.-G. and O’Toole, G. A. (2015). c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 3, MB–0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014, PMID: 26104694
Hall-Stoodley, L. and McCoy, K. S. (2022). Biofilm aggregates and the host airway-microbial interface. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.969326, PMID: 36081767
Hammond, J. H., Dolben, E. F., Smith, T. J., Bhuju, S., and Hogan, D. A. (2015). Links between Anr and Quorum sensing in Pseudomonas aeruginosa biofilms. J. Bacteriol 197, 2810–2820. doi: 10.1128/JB.00182-15, PMID: 26078448
Hasegawa, N., Arai, H., and Igarashi, Y. (1998). Activation of a consensus FNR-dependent promoter by DNR of pseudomonas aeruginosa in response to nitrite. FEMS Microbiol. Lett. 166, 213–217. doi: 10.1111/j.1574-6968.1998.tb13892.x, PMID: 9770276
Hassett, D. J., Cuppoletti, J., Trapnell, B., Lymar, S. V., Rowe, J. J., Yoon, S. S., et al. (2002). Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Delivery Rev. 54, 1425–1443. doi: 10.1016/s0169-409x(02)00152-7, PMID: 12458153
Hassett, D. J., Sutton, M. D., Schurr, M. J., Herr, A. B., Caldwell, C. C., and Matu, J. O. (2009). Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 17, 130–138. doi: 10.1016/j.tim.2008.12.003, PMID: 19231190
Høiby, N., Moser, C., and Ciofu, O. (2024). Pseudomonas aeruginosa in the frontline of the greatest challenge of biofilm infection-its tolerance to antibiotics. Microorganisms 12, 2115. doi: 10.3390/microorganisms12112115, PMID: 39597505
Hou, S., Mahadevegowda, S. H., Lu, D., Zhang, K., Chan-Park, M. B., and Duan, H. (2021). Metabolic labeling mediated targeting and thermal killing of gram-positive bacteria by self-reporting Janus magnetic nanoparticles. Small 17, e2006357. doi: 10.1002/smll.202006357, PMID: 33325629
Howlin, R. P., Cathie, K., Hall-Stoodley, L., Cornelius, V., Duignan, C., Allan, R. N., et al. (2017). Low-dose nitric oxide as targeted anti-biofilm adjunctive therapy to treat chronic Pseudomonas aeruginosa infection in cystic fibrosis. Mol. Ther. 25, 2104–2116. doi: 10.1016/j.ymthe.2017.06.021, PMID: 28750737
Jackson, A. A., Daniels, E. F., Hammond, J. H., Willger, S. D., and Hogan, D. A. (2014). Global regulator Anr represses PlcH phospholipase activity in Pseudomonas aeruginosa when oxygen is limiting. Microbiol. (Reading) 160, 2215–2225. doi: 10.1099/mic.0.081158-0, PMID: 25073853
Jackson, A. A., Gross, M. J., Daniels, E. F., Hampton, T. H., Hammond, J. H., Vallet-Gely, I., et al. (2013). Anr and its activation by PlcH activity in Pseudomonas aeruginosa host colonization and virulence. J. Bacteriol 195, 3093–3104. doi: 10.1128/JB.02169-12, PMID: 23667230
Jacob-Dubuisson, F., Mechaly, A., Betton, J.-M., and Antoine, R. (2018). Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 16, 585–593. doi: 10.1038/s41579-018-0055-7, PMID: 30008469
James, G. A., Ge Zhao, A., Usui, M., Underwood, R. A., Nguyen, H., Beyenal, H., et al. (2016). Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen. 24, 373–383. doi: 10.1111/wrr.12401, PMID: 26748963
Jiménez Otero, F., Newman, D. K., and Tender, L. M. (2023). Pyocyanin-dependent electrochemical inhibition of Pseudomonas aeruginosa biofilms is synergistic with antibiotic treatment. mBio 14, e0070223. doi: 10.1128/mbio.00702-23, PMID: 37314185
Jo, J., Cortez, K. L., Cornell, W. C., Price-Whelan, A., and Dietrich, L. E. (2017). An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6, e30205. doi: 10.7554/eLife.30205, PMID: 29160206
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/c2cs35206k, PMID: 23023210
Kay, E., Humair, B., Dénervaud, V., Riedel, K., Spahr, S., Eberl, L., et al. (2006). Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188, 6026–6033. doi: 10.1128/JB.00409-06, PMID: 16885472
Kempes, C. P., Okegbe, C., Mears-Clarke, Z., Follows, M. J., and Dietrich, L. E. P. (2014). Morphological optimization for access to dual oxidants in biofilms. Proc. Natl. Acad. Sci. U.S.A. 111, 208–213. doi: 10.1073/pnas.1315521110, PMID: 24335705
Kim, B., Park, J.-S., Choi, H.-Y., Yoon, S. S., and Kim, W.-G. (2018). Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP. Sci. Rep. 8, 8617. doi: 10.1038/s41598-018-26974-5, PMID: 29872101
Kolpen, M., Hansen, C. R., Bjarnsholt, T., Moser, C., Christensen, L. D., van Gennip, M., et al. (2010). Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62. doi: 10.1136/thx.2009.114512, PMID: 19846469
Krieger, R., Rompf, A., Schobert, M., and Jahn, D. (2002). The pseudomonas aeruginosa hemA promoter is regulated by anr, dnr, NarL and integration host factor. Mol. Genet. Genom.: MGG 267, 409–417. doi: 10.1007/s00438-002-0672-7, PMID: 12073043
La Rosa, R., Johansen, H. K., and Molin, S. (2018). Convergent metabolic specialization through distinct evolutionary paths in pseudomonas aeruginosa. Mbio 9, e00269–e00218. doi: 10.1128/mBio.00269-18, PMID: 29636437
Lee, J. and Zhang, L. (2015). The hierarchy quorum sensing network in pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x, PMID: 25249263
Li, L., Ma, J., Cheng, P., Li, M., Yu, Z., Song, X., et al. (2023). Roles of two-component regulatory systems in Klebsiella pneumoniae: Regulation of virulence, antibiotic resistance, and stress responses. Microbiol. Res. 272, 127374. doi: 10.1016/j.micres.2023.127374, PMID: 37031567
Lin, M.-H., Shu, J.-C., Huang, H.-Y., and Cheng, Y.-C. (2012). Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS One 7, e34388. doi: 10.1371/journal.pone.0034388, PMID: 22479621
Lin, X., Wu, L., Li, H., Wang, S., and Peng, X. (2008). Downregulation of Tsx and OmpW and upregulation of OmpX are required for iron homeostasis in Escherichia coli. J. Proteome Res. 7, 1235–1243. doi: 10.1021/pr7005928, PMID: 18220334
Line, L., Alhede, M., Kolpen, M., Kühl, M., Ciofu, O., Bjarnsholt, T., et al. (2014). Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00554, PMID: 25386171
Linsky, M., Vitkin, Y., and Segal, G. (2020). A novel legionella genomic island encodes a copper-responsive regulatory system and a single Icm/Dot effector protein transcriptionally activated by copper. mBio 11, e03232–e03219. doi: 10.1128/mBio.03232-19, PMID: 31992628
Luo, Z., Lu, R., Shi, T., Ruan, Z., Wang, W., Guo, Z., et al. (2024). Enhanced bacterial cuproptosis-like death via reversal of hypoxia microenvironment for biofilm infection treatment. Adv. Sci. (Weinh) 11, e2308850. doi: 10.1002/advs.202308850, PMID: 38477452
Lüthi, E., Baur, H., Gamper, M., Brunner, F., Villeval, D., Mercenier, A., et al. (1990). The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene 87, 37–43. doi: 10.1016/0378-1119(90)90493-b, PMID: 2158926
Martin, L. W., Gray, A. R., Brockway, B., and Lamont, I. L. (2023). Pseudomonas aeruginosa is oxygen-deprived during infection in cystic fibrosis lungs, reducing the effectiveness of antibiotics. FEMS Microbiol. Lett. 370, fnad076. doi: 10.1093/femsle/fnad076, PMID: 37516450
Mashruwala, A. A., van de Guchte, A., and Boyd, J. M. (2017). Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 6, e23845. doi: 10.7554/eLife.23845, PMID: 28221135
Massimelli, M. J., Beassoni, P. R., Forrellad, M. A., Barra, J. L., Garrido, M. N., Domenech, C. E., et al. (2005). Identification, cloning, and expression of pseudomonas aeruginosa phosphorylcholine phosphatase gene. Curr. Microbiol. 50, 251–256. doi: 10.1007/s00284-004-4499-9, PMID: 15886911
Mavrodi, D. V., Peever, T. L., Mavrodi, O. V., Parejko, J. A., Raaijmakers, J. M., Lemanceau, P., et al. (2010). Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76, 866–879. doi: 10.1128/AEM.02009-09, PMID: 20008172
McPhee, J. B., Tamber, S., Bains, M., Maier, E., Gellatly, S., Lo, A., et al. (2009). The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol. Lett. 296, 241–247. doi: 10.1111/j.1574-6968.2009.01651.x, PMID: 19486157
Mikkelsen, H., Bond, N. J., Skindersoe, M. E., Givskov, M., Lilley, K. S., and Welch, M. (2009). Biofilms and type III secretion are not mutually exclusive in pseudomonas aeruginosa. Microbiol. (read. Engl.) 155, 687–698. doi: 10.1099/mic.0.025551-0, PMID: 19246740
Mitrophanov, A. Y. and Groisman, E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611. doi: 10.1101/gad.1700308, PMID: 18832064
O’Callaghan, J., Reen, F. J., Adams, C., and O’Gara, F. (2011). Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiol. (Reading) 157, 3417–3428. doi: 10.1099/mic.0.052050-0, PMID: 21873408
Okegbe, C., Fields, B. L., Cole, S. J., Beierschmitt, C., Morgan, C. J., Price-Whelan, A., et al. (2017). Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc. Natl. Acad. Sci. U.S.A. 114, E5236–E5245. doi: 10.1073/pnas.1700264114, PMID: 28607054
Palmer, K. L., Brown, S. A., and Whiteley, M. (2007). Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. bacteriology. 189, 4449–4455. doi: 10.1128/JB.00162-07, PMID: 17400735
Papa, R., Imperlini, E., Trecca, M., Paris, I., Vrenna, G., Artini, M., et al. (2023). Virulence of Pseudomonas aeruginosa in cystic fibrosis: relationships between normoxia and anoxia lifestyle. Antibiotics (Basel) 13, 1. doi: 10.3390/antibiotics13010001, PMID: 38275311
Park, J.-S., Choi, H.-Y., and Kim, W.-G. (2020). The Nitrite Transporter Facilitates Biofilm Formation via Suppression of Nitrite Reductase and Is a New Antibiofilm Target in Pseudomonas aeruginosa. mBio 11, e00878–e00820. doi: 10.1128/mBio.00878-20, PMID: 32636243
Penesyan, A., Gillings, M., and Paulsen, I. T. (2015). Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20, 5286–5298. doi: 10.3390/molecules20045286, PMID: 25812150
Peng, J., Cao, J., Ng, F. M., and Hill, J. (2017). Pseudomonas aeruginosa develops Ciprofloxacin resistance from low to high level with distinctive proteome changes. J. Proteomics 152, 75–87. doi: 10.1016/j.jprot.2016.10.005, PMID: 27771372
Pulukkody, A. C., Yung, Y. P., Donnarumma, F., Murray, K. K., Carlson, R. P., and Hanley, L. (2021). Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer. PLoS One 16, e0250911. doi: 10.1371/journal.pone.0250911, PMID: 34292966
Rada, B., Jendrysik, M. A., Pang, L., Hayes, C. P., Yoo, D.-G., Park, J. J., et al. (2013). Pyocyanin-enhanced neutrophil extracellular trap formation requires the NADPH oxidase. PLoS One 8, e54205. doi: 10.1371/journal.pone.0054205, PMID: 23342104
Rawling, E. G., Brinkman, F. S., and Hancock, R. E. (1998). Roles of the carboxy-terminal half of pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180, 3556–3562. doi: 10.1128/JB.180.14.3556-3562.1998, PMID: 9657997
Richardson, D. J., Berks, B. C., Russell, D. A., Spiro, S., and Taylor, C. J. (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci.: CMLS 58, 165–178. doi: 10.1007/PL00000845, PMID: 11289299
Rodionov, D. A., Dubchak, I. L., Arkin, A. P., Alm, E. J., and Gelfand, M. S. (2005). Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1, e55. doi: 10.1371/journal.pcbi.0010055, PMID: 16261196
Römling, U. and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. doi: 10.1111/joim.12004, PMID: 23025745
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. reviews: MMBR. 77, 1–52. doi: 10.1128/MMBR.00043-12, PMID: 23471616
Rossi, E., La Rosa, R., Bartell, J. A., Marvig, R. L., Haagensen, J. A. J., Sommer, L. M., et al. (2021). Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 19, 331–342. doi: 10.1038/s41579-020-00477-5, PMID: 33214718
Sakuragi, Y. and Kolter, R. (2007). Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol 189, 5383–5386. doi: 10.1128/JB.00137-07, PMID: 17496081
Saunders, S. H., Tse, E. C. M., Yates, M. D., Otero, F. J., Trammell, S. A., Stemp, E. D. A., et al. (2020). Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 182, 919–932.e19. doi: 10.1016/j.cell.2020.07.006, PMID: 32763156
Sawers, R. G. (1991). Identification and molecular characterization of a transcriptional regulator from pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of escherichia coli. Mol. Microbiol. 5, 1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x, PMID: 1787797
Schaible, B., Taylor, C. T., and Schaffer, K. (2012). Hypoxia increases antibiotic resistance in pseudomonas aeruginosa through altering the composition of multidrug efflux pumps. Antimicrob. Agents Chemother. 56, 2114–2118. doi: 10.1128/AAC.05574-11, PMID: 22290986
Schiessl, K. T., Hu, F., Jo, J., Nazia, S. Z., Wang, B., Price-Whelan, A., et al. (2019). Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun. 10, 762. doi: 10.1038/s41467-019-08733-w, PMID: 30770834
Schilcher, K. and Horswill, A. R. (2020). Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84, e00026–e00019. doi: 10.1128/MMBR.00026-19, PMID: 32792334
Schreiber, K., Krieger, R., Benkert, B., Eschbach, M., Arai, H., Schobert, M., et al. (2007). The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol 189, 4310–4314. doi: 10.1128/JB.00240-07, PMID: 17400734
Scribani Rossi, C., Barrientos-Moreno, L., Paone, A., Cutruzzolà, F., Paiardini, A., Espinosa-Urgel, M., et al. (2022). Nutrient sensing and biofilm modulation: the example of L-arginine in pseudomonas. Int. J. Mol. Sci. 23, 4386. doi: 10.3390/ijms23084386, PMID: 35457206
Sharma, V., Noriega, C. E., and Rowe, J. J. (2006). Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72, 695–701. doi: 10.1128/AEM.72.1.695-701.2006, PMID: 16391109
Singh, P. K., Schaefer, A. L., Parsek, M. R., Moninger, T. O., Welsh, M. J., and Greenberg, E. P. (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764. doi: 10.1038/35037627, PMID: 11048725
Sønderholm, M., Kragh, K. N., Koren, K., Jakobsen, T. H., Darch, S. E., Alhede, M., et al. (2017). Pseudomonas aeruginosa aggregate formation in an alginate bead model system exhibits in vivo-like characteristics. Appl. Environ. Microbiol. 83, e00113–e00117. doi: 10.1128/AEM.00113-17, PMID: 28258141
Sonnleitner, E. and Bläsi, U. (2014). Regulation of hfq by the RNA CrcZ in pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 10, e1004440. doi: 10.1371/journal.pgen.1004440, PMID: 24945892
Sonnleitner, E., Gonzalez, N., Sorger-Domenigg, T., Heeb, S., Richter, A. S., Backofen, R., et al. (2011). The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 80, 868–885. doi: 10.1111/j.1365-2958.2011.07620.x, PMID: 21375594
Sonnleitner, E., Pusic, P., Wolfinger, M. T., and Bläsi, U. (2020). Distinctive regulation of carbapenem susceptibility in pseudomonas aeruginosa by hfq. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01001, PMID: 32528439
Stojkova, P., Spidlova, P., and Stulik, J. (2019). Nucleoid-associated protein HU: a lilliputian in gene regulation of bacterial virulence. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00159, PMID: 31134164
Stokes, J. M., Lopatkin, A. J., Lobritz, M. A., and Collins, J. J. (2019). Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259. doi: 10.1016/j.cmet.2019.06.009, PMID: 31279676
Stoodley, P., Toelke, N., Schwermer, C., and de Beer, D. (2024). Bioenergetics of simultaneous oxygen and nitrate respiration and nitric oxide production in a Pseudomonas aeruginosa agar colony biofilm. Biofilm 7, 100181. doi: 10.1016/j.bioflm.2024.100181, PMID: 38425549
Su, Z., Xu, D., Hu, X., Zhu, W., Kong, L., Qian, Z., et al. (2024). Biodegradable oxygen-evolving metalloantibiotics for spatiotemporal sono-metalloimmunotherapy against orthopaedic biofilm infections. Nat. Commun. 15, 8058. doi: 10.1038/s41467-024-52489-x, PMID: 39277594
Taylor, S. J. and Winter, S. E. (2020). Salmonella finds a way: Metabolic versatility of Salmonella enterica serovar Typhimurium in diverse host environments. PLoS Pathog. 16, e1008540. doi: 10.1371/journal.ppat.1008540, PMID: 32525928
Teng, T., Xi, B., Chen, K., Pan, L., Xie, J., and Xu, P. (2018). Comparative transcriptomic and proteomic analyses reveal upregulated expression of virulence and iron transport factors of Aeromonas hydrophila under iron limitation. BMC Microbiol. 18, 52. doi: 10.1186/s12866-018-1178-8, PMID: 29866030
Tierney, A. R. and Rather, P. N. (2019). Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 14, 533–552. doi: 10.2217/fmb-2019-0002, PMID: 31066586
Trunk, K., Benkert, B., Quäck, N., Münch, R., Scheer, M., Garbe, J., et al. (2010). Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12, 1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x, PMID: 20553552
Ueda, A. and Wood, T. K. (2009). Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5, e1000483. doi: 10.1371/journal.ppat.1000483, PMID: 19543378
Vander Wauven, C., Piérard, A., Kley-Raymann, M., and Haas, D. (1984). Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol 160, 928–934. doi: 10.1128/jb.160.3.928-934.1984, PMID: 6438064
Ventre, I., Goodman, A. L., Vallet-Gely, I., Vasseur, P., Soscia, C., Molin, S., et al. (2006). Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 171–176. doi: 10.1073/pnas.0507407103, PMID: 16373506
Vestby, L. K., Grønseth, T., Simm, R., and Nesse, L. L. (2020). Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel) 9, 59. doi: 10.3390/antibiotics9020059, PMID: 32028684
Wagner, V. E. and Iglewski, B. H. (2008). P. aeruginosa biofilms in CF infection. Clin. Rev. Allergy Immunol. 35, 124–134. doi: 10.1007/s12016-008-8079-9, PMID: 18509765
Wang, Z., Liu, X., Duan, Y., and Huang, Y. (2022). Infection microenvironment-related antibacterial nanotherapeutic strategies. Biomaterials 280, 121249. doi: 10.1016/j.biomaterials.2021.121249, PMID: 34801252
Wang, Y. and Newman, D. K. (2008). Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42, 2380–2386. doi: 10.1021/es702290a, PMID: 18504969
Werner, E., Roe, F., Bugnicourt, A., Franklin, M. J., Heydorn, A., Molin, S., et al. (2004). Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004, PMID: 15466566
Williams, H. D., Zlosnik, J. E. A., and Ryall, B. (2007). Oxygen, cyanide and energy generation in the cystic fibrosis pathogen pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71. doi: 10.1016/S0065-2911(06)52001-6, PMID: 17027370
Wu, M., Chen, C., Liu, Z., Tian, J., and Zhang, W. (2022). Regulating the bacterial oxygen microenvironment via a perfluorocarbon-conjugated bacteriochlorin for enhanced photodynamic antibacterial efficacy. Acta Biomater 142, 242–252. doi: 10.1016/j.actbio.2022.02.013, PMID: 35183779
Xiu, W., Gan, S., Wen, Q., Qiu, Q., Dai, S., Dong, H., et al. (2020). Biofilm microenvironment-responsive nanotheranostics for dual-mode imaging and hypoxia-relief-enhanced photodynamic therapy of bacterial infections. Res. (Wash D C) 2020, 9426453. doi: 10.34133/2020/9426453, PMID: 32377640
Xu, K. D., Stewart, P. S., Xia, F., Huang, C. T., and McFeters, G. A. (1998). Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64, 4035–4039. doi: 10.1128/AEM.64.10.4035-4039.1998, PMID: 9758837
Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603. doi: 10.1016/s1534-5807(02)00295-2, PMID: 12408810
Yoon, M. Y., Lee, K.-M., Park, Y., and Yoon, S. S. (2011). Contribution of cell elongation to the biofilm formation of pseudomonas aeruginosa during anaerobic respiration. PLoS One 6, e16105. doi: 10.1371/journal.pone.0016105, PMID: 21267455
Zamudio-Chávez, L., Suesca, E., López, G. D., Carazzone, C., Manrique-Moreno, M., and Leidy, C. (2023). Staphylococcus aureus modulates carotenoid and phospholipid content in response to oxygen-restricted growth conditions, triggering changes in membrane biophysical properties. Int. J. Mol. Sci. 24, 14906. doi: 10.3390/ijms241914906, PMID: 37834354
Zhang, Y., Han, K., Chandler, C. E., Tjaden, B., Ernst, R. K., and Lory, S. (2017). Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 106, 919–937. doi: 10.1111/mmi.13857, PMID: 28976035
Zhao, A., Sun, J., and Liu, Y. (2023a). Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1137947, PMID: 37091673
Keywords: biofilms, oxygen-limited, anaerobic metabolism, transcriptional factors, two-component systems, quorum sensing, Pseudomonas aeruginosa
Citation: Ren L, Yuan Y, Farea K, Feng X, He J, Liu Y and Zheng B (2025) The adaptability of Pseudomonas aeruginosa biofilm in oxygen-limited environments. Front. Cell. Infect. Microbiol. 15:1655335. doi: 10.3389/fcimb.2025.1655335
Received: 27 June 2025; Accepted: 03 September 2025;
Published: 19 September 2025.
Edited by:
Ashwini Chauhan, University of Delhi, IndiaCopyright © 2025 Ren, Yuan, Farea, Feng, He, Liu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Liu, bGl1eWlAY211LmVkdS5jbg==; Bowen Zheng, Ynd6aGVuZ0BjbXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Ling Ren1,2†
Ling Ren1,2† Xu Feng
Xu Feng Yi Liu
Yi Liu