- 1Laboratory Department, Haikou Maternal and Child Health Hospital, Haikou, Hainan, China
- 2Key Laboratory of Tropical Translational Medicine of Ministry of Education, School of Basic Medicine and Life Sciences, Public Research Center, Hainan Medical University, Haikou, Hainan, China
- 3National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Disease, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 4Department of Biology, McMaster University, Hamilton, ON, Canada
Group B Streptococcus (GBS) is a major cause of pregnancy complication and neonatal morbidity and mortality worldwide, particularly in developing regions. Despite its clinical importance, data on the molecular epidemiology, antibiotic resistance, and virulence factors of GBS in tropical regions are scarce. This study provides the first comprehensive analysis of GBS strains from pregnant women and neonates in Haikou, a tropical city in China, via antibiotic susceptibility testing and whole-genome sequencing. Our results grouped the 138 strains of GBS into seven serotypes and 28 multilocus sequence types (STs). These STs belonged to six clonal complexes (CCs). High antibiotic resistance rates were observed for tetracycline (89.1%) and clindamycin (55.1%) and the commonly detected resistance genes included mreA (100%), ermB (52.9%) and tetM (41.3%). Each strain contained at least one Pili-island (PI) gene and the capsular polysaccharide antigen among the GBS isolates were variably associated with CCs. All strains carried virulence genes cfb and cylE, followed by pavA (99.3%), and lmb (66.7%) etc. Our analyses showed ST862 as a dominant and potentially zoonotic genotype in Haikou, China, with implications for both human and animal health. The high prevalence of tetracycline and clindamycin resistance underscores the need for judicious antibiotic use and the development of region-specific antibiotic treatment guidelines. The discovery of novel STs and broad distributions of several virulence factors provide valuable insights for future vaccine development and targeted interventions in this region.
Introduction
Group B Streptococcus (GBS) is a common type of bacteria frequently referred to as β-hemolytic, Gram-positive coccus that typically appears in pairs or chains. GBS is an opportunistic human pathogen but can colonize the human genital tract and lower digestive tract as part of the normal microflora. Microbiome studies have indicated that imbalances in microbial flora can readily trigger infections at various sites within the body (Dhudasia et al., 2021). Pregnant women, particularly during the perinatal period, represent a special vulnerable population. Hormonal changes during pregnancy often lead to a diversity of physiological shifts, including changes in the vaginal microbiota (Li et al., 2020). For example, elevated estrogen levels during pregnancy cause Lactobacillus to dominate the vaginal microflora, disrupting the vaginal microbial community which can lead to GBS infection of the birth canal. This can lead to adverse pregnancy outcomes, maternal infections, premature birth, miscarriage, stillbirth, and a range of poor maternal and neonatal outcomes such as neonatal sepsis, meningitis, or pneumonia (Okike et al., 2014; Pidwill et al., 2018; Yadeta et al., 2018; HogenEsch et al., 2021; Yuan et al., 2021).
Lancefield defined two types of carbohydrate antigens in the GBS wall: the Group B antigen, which is common to all strains, and the specific capsular polysaccharide antigen (Cieslewicz et al., 2005). Based on serological reactions that target the polysaccharide capsule or multiplex PCR, GBS can be classified into 10 serotypes: Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX. These serotypes exhibit different geographical distributions (Cieslewicz et al., 2005; Slotved et al., 2007; Yao et al., 2013). Previous studies have reported that serotypes Ia, Ib, II, III, and V are commonly involved in invasive GBS infections (López et al., 2018; Huebner et al., 2022; Gharabeigi et al., 2023). Multilocus sequence typing (MLST) has also been used for the classification of GBS strains. Research findings indicate the existence of over 2000 MLST sequence types (STs), and certain serotypes and/or STs are associated with specific disease phenotypes (Luan et al., 2005; Jones et al., 2006; Madrid et al., 2017).
Current clinical treatments for GBS infections primarily rely on antibiotics such as penicillin, ampicillin, vancomycin, levofloxacin, tetracycline, moxifloxacin, linezolid, tigecycline, clindamycin, and quinupristin/dalfopristin. Due to antibiotic misuse and inadequate GBS screening, GBS infections can be hard to treat or prone to being mistreated, leading to high rates of adverse pregnancy outcomes and neonatal mortality (Bianchi-Jassir et al., 2017; Seale et al., 2017; Dong et al., 2020; Gonçalves et al., 2022). To reduce prenatal infections and adverse pregnancy outcomes, there has been increasing emphasis on improving clinical practice, strengthening screening, conducting universal testing for GBS infection among pregnant women, and implementing preventive treatments for potential mother-child infections.
Accurate diagnosis of infectious disease agents and quantitative data on their susceptibilities to antimicrobial agents are crucial for effectively preventing and treating adverse pregnancy outcomes, maternal infections, premature births, miscarriages, stillbirths, and a range of poor maternal and neonatal outcomes caused by GBS. However, much remains unknown about the molecular characteristics, virulence factors, or antibiotic resistance mechanisms of GBS strains from pregnant women and neonates, especially in tropical and developing regions such as Hainan Island in southern China. Here we investigated the molecular characteristics and antibiotic resistance patterns of GBS samples collected from pregnant women and neonates at the Haikou Maternity and Child Health Hospital in Hainan Province, China. For these GBS strains, we obtained data on their antibiotic susceptibilities and whole-genome sequences. This research contributes to the global effort to combat GBS infections and informs the development of targeted interventions, including vaccine developments and region-specific treatment guidelines.
Materials and methods
Sample collection
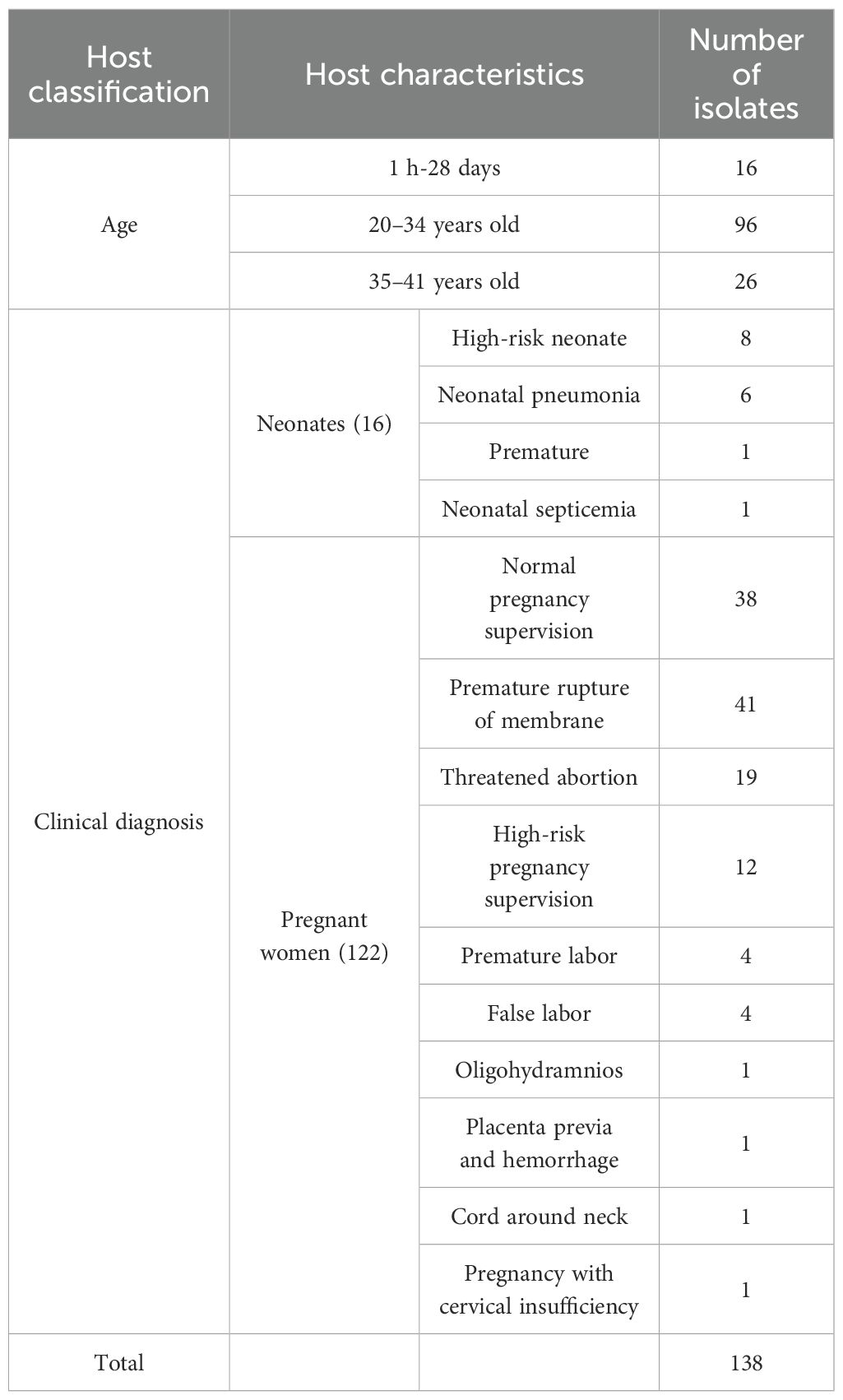
A total of 138 GBS isolates were obtained from 122 pregnant women and 16 neonates at Haikou Maternity and Child Health Hospital in Hainan Province, in tropical China, between 2021 and 2022. The samples from pregnant women included 108 vaginal secretions, 13 cervical secretions, and one amniotic fluid sample. The neonatal samples included 12 sputum samples from the lower respiratory tract, three throat swabs, and one blood sample. Specimen collections were performed in accordance with standard clinical protocols. Each of the 138 samples represented a unique patient visit. The details are provided in Supplementary Table S1.
Isolation and identification of strains
All suspected GBS strains were transferred to Columbia blood agar plates and GBS chromogenic plates. All GBS isolates were incubated at 35-37 °C and 5% CO2/95% air for 18–24 h. The initial identification was performed via Gram staining, the CAMP test, and colony morphology. For further confirmation, the VITEK 2 Compact System (BioMérieux, Marcy-l’Étoile, France) with a GP colorimetric identification card was utilized to determine the species of GBS. Quality control was maintained via the use of E. casseliflavus ATCC 700327 and E. faecalis ATCC 29212.
Antibiotic susceptibility test
To evaluate antimicrobial susceptibility, an AST GP67 card was employed in conjunction with the BioMérieux VITEK 2 system, following the manufacturer’s guidelines. Quality control was ensured by using E. casseliflavus ATCC 700327 and E. faecalis ATCC 29212 as reference strains. Multi-drug resistance (MDR) was defined as resistance to three or more categories of antimicrobial agents simultaneously.
Whole-genome sequencing
To obtain the whole-genome sequences of the 138 strains, pure cultures of GBS were first cultivated for 18–24 h on Colombian agar with 5% sheep blood. The genomic DNA of each strain was extracted via the Wizard Genomic DNA Purification Kit (Promega, United States). The purified DNA was then sent to the MIGIGENE company for gene library construction and whole-genome sequencing (WGS) on the Illumina HiSeq 2000 platform. The de novo genome was assembled from Illumina data via the SPAdes (v3.13.1) software.
Serotype analysis
Serotype identification via whole-genome sequencing (WGS) was performed following Metcalf’s methodology (Metcalf et al., 2017). For isolates where WGS-based serotype determination was unsuccessful, serotypes were resolved using a multiplex PCR approach (Imperi et al., 2010). Strains that remained serotype-untypable by both WGS and multiplex PCR were subjected to annotation via RAST (Rapid Annotation using Subsystem Technology; https://rast.nmpdr.org/) to screen for the presence of conserved capsular polysaccharide (cps) genes (Cieslewicz et al., 2005).
Multilocus sequence typing
The specific sequence types (STs) and clonal complexes (CCs) were determined based on the gene sequences of the seven housekeeping genes, namely, adhP, pheS, atr, glnA, sdhA, glcK, and tkt (Jones et al., 2003). The STs and CCs were determined or newly identified when the genome sequences were uploaded to the MLST database (https://pubmlst.org/). The MLST minimum spanning tree was constructed via Bionumerics software (Applied Maths, Belgium).
Data analysis
The antibiotic resistance genes and virulence factors were determined based on comparisons with data at the Center for Genomic Epidemiology database (CGE) (http://www.genomicepidemiology.org), the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca), and the Virulence Factor Database (VFDB) (https://www.mgc.ac.cn/VFs/), with a similarity threshold of > 90% and a coverage threshold of 60% compared with the reference sequences in the databases. For serotype and virulence factor gene analyses, we followed the protocols described in a previous study (McGee et al., 2021).
Statistical analysis was performed with GraphPad Prism 9.50 software. Analyses of the prevalences of different antibiotic resistance genes and virulence genes between CCs were conducted via the Chi-square test. Statistical significance was defined as P<0.05.
Results
Strain characteristics
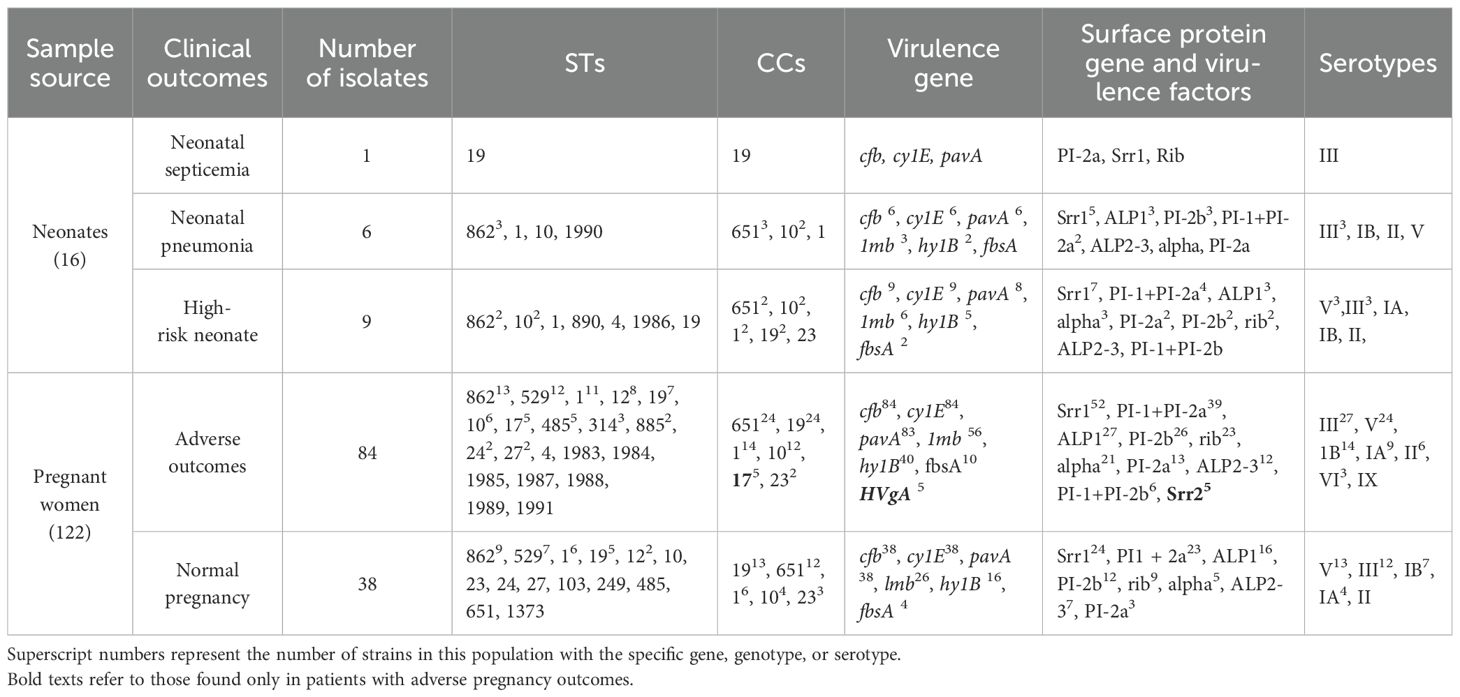
A total of 138 GBS strains were collected from 16 neonates and 122 pregnant women. The neonates’ ages ranged from 1 hour to 28 days, while the pregnant women were categorized into two age groups: 96 were between 20 and 34 years old, and 26 were between 35 and 41 years old. Among the neonates, 8 were identified as high risk, 6 were diagnosed with neonatal pneumonia, 1 was a premature birth, and 1 was diagnosed with neonatal septicemia. Among the pregnant women, 38 were under normal pregnancy supervision, and the remaining 84 experienced adverse pregnancy outcomes. Further details regarding the samples are provided in Table 1 and Supplementary Table S1.
Antimicrobial susceptibility test results for GBS
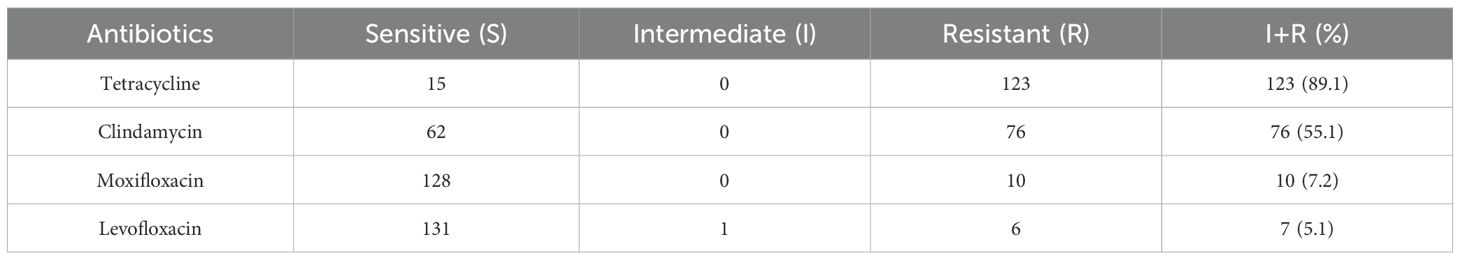
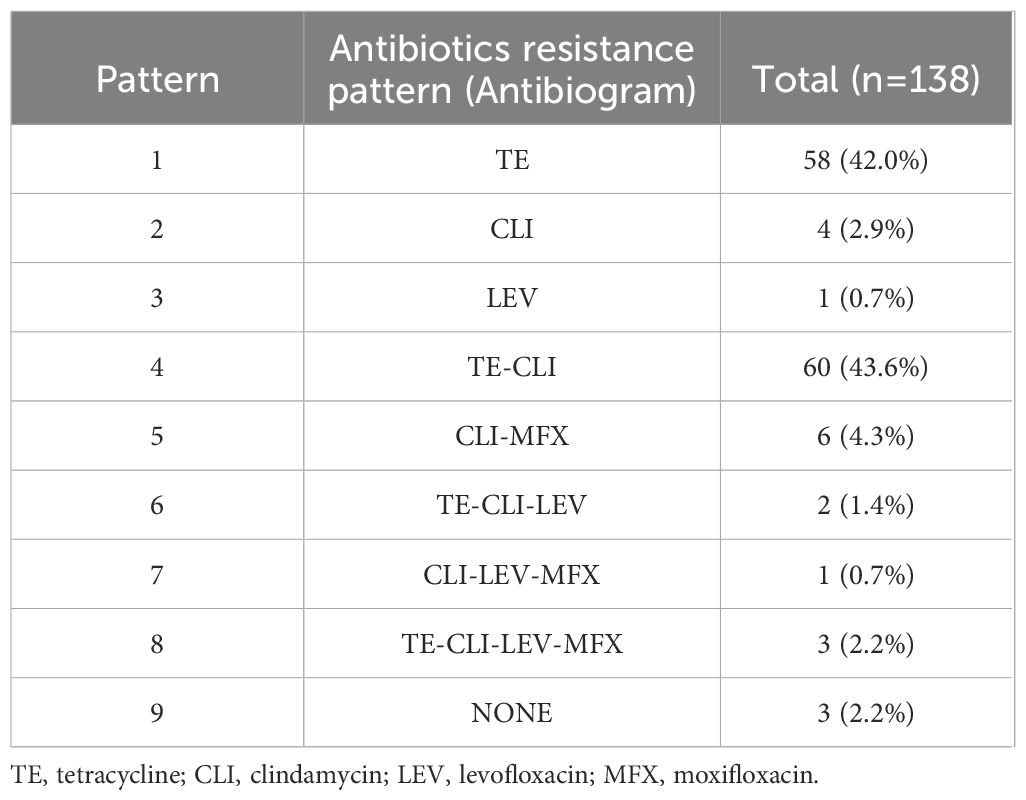
In vitro susceptibility testing of 138 strains of GBS to 10 different antibiotics revealed that all strains were sensitive to the following six drugs: penicillin, ampicillin, vancomycin, linezolid, tigecycline, and quinupristin/dalfopristin. However, varying degrees of resistance/intermediate susceptibility were observed against the remaining four drugs, with the highest resistance rate of 89.1% against tetracycline, followed by clindamycin (55.1%), moxifloxacin (7.2%), and levofloxacin (5.1%) (Table 2). We detected nine resistance patterns among the 138 strains: tetracycline-clindamycin resistance accounted for 43.6% (60/138), followed by tetracycline resistance alone at 42.0% (58/138), and six strains (4.3%) were MDR, as detailed in Table 3.
Distribution of GBS serotypes
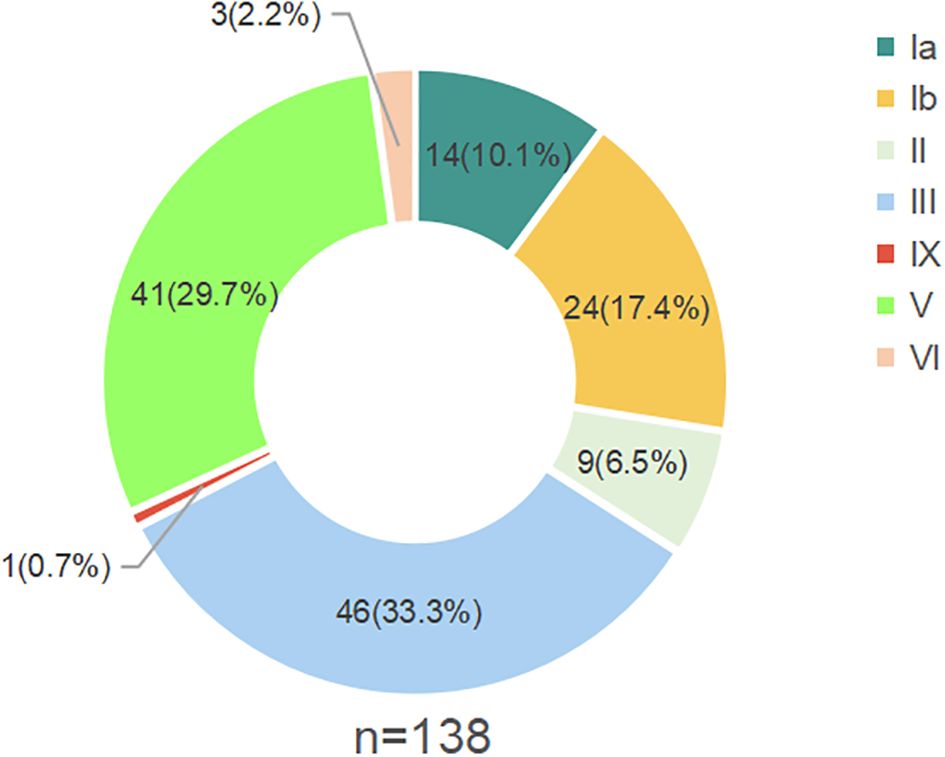
Based on DNA sequences at genes involved in capsular polysaccharide (CPS) synthesis, the 138 GBS strains were classified into seven serotypes: Ia, Ib, II, III, V, IX and VI. The most prevalent serotype was III, representing 33.3% of the strains (46 out of 138), followed by serotype V, which accounted for 29.7% (41 strains). The remaining serotypes included Ib (17.4%), Ia (10.1%), II (6.5%), VI (2.2%), and IX (0.7%) (Figure 1).
Multilocus sequence typing of GBS
Based on comparisons with data in the MLST database for GBS at seven gene loci, the 138 GBS strains were identified as belonging to 28 STs, including nine new ones (ST1983–1991). Among these 28 STs, 13 STs were shared by a total of 123 strains, and the remaining 15 STs were each represented by one isolate. The top three most prevalent STs were ST862 (19.6%, 27/138), followed by ST529 (13.8%, 19/138) and ST1 (13.8%, 19/138). Each of the nine new STs was represented by only one strain each. The distributions for all 28 STs are detailed in Figure 2.

Figure 2. MLST results for the 138 GBS strains. The color blocks within circles indicate STs, and the number of circles indicates the proportion of the corresponding STs.
Clonal complexes of the 138 GBS strains
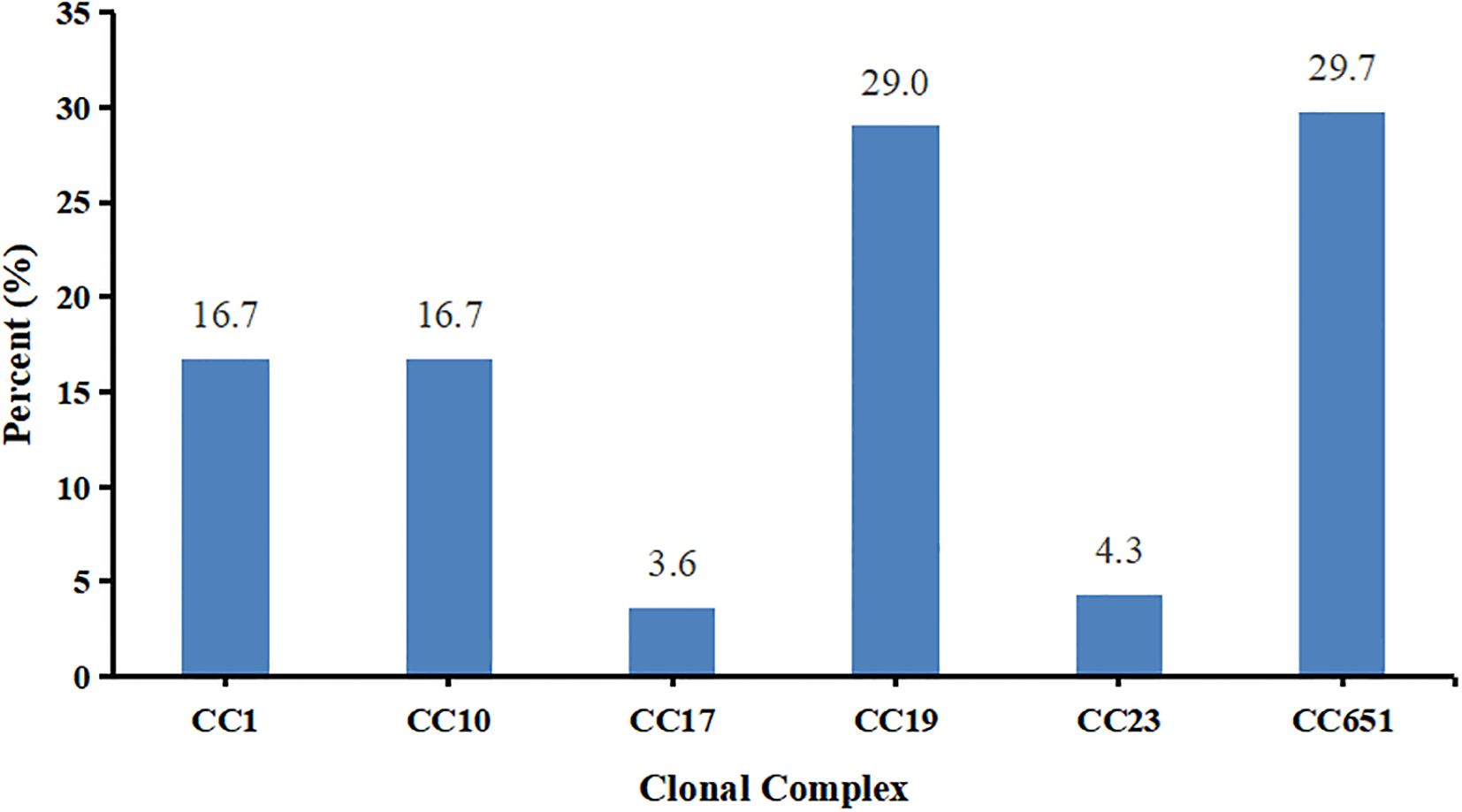
The 28 STs were grouped into 6 clonal complexes (CCs). The relative proportions of these CCs from the most frequent to the least frequent were CC651 with 41 strains (29.7%), CC19 with 40 strains (29.0%), CC1 and CC10 with 23 strains (16.7%), CC23 with 6 strains (4.3%), and CC17 with 5 strains (3.6%) (Figure 3).
The 41 CC651 strains belonged to two serotypes III and Ia, with serotype III being more common than serotype Ia (P<0.001). The 40 CC19 strains belonged to three serotypes II, III, and V, with serotype V being more frequent than serotypes II and III (P<0.001). The 23 CC10 strains belonged to serotypes Ib and II, with serotype Ib being more frequent than serotype II (P<0.05). The 23 CC1 strains belonged to six serotypes, excluding serotype II. The six CC23 strains belonged to serotypes V and Ia and all 5 CC17 strains were classified as serotype III. The detailed data showing the relationships among multilocus sequence type, clonal complex, and serotypes are shown in Figure 4 and Supplementary Table S2.

Figure 4. Distributions and relationships among multilocus sequence type, clonal complex, and serotype in the collection of 138 GBS isolates. Labels within individual circles indicate the STs, the sizes of circles are proportional to the number of isolates, and their colors represent serotypes. The variably shaped colored blocks indicate the clonal complex to which the STs belong. The STs from ST1983 to ST1991 were novel types detected in the study.
Antibiotic resistance genes
A total of 19 antibiotic resistance genes were identified across the 138 strains of GBS. These genes confer resistance to various classes of antibiotics, including macrolides, lincosamides, tetracyclines, chloramphenicols, and aminoglycosides. Specifically, the identified resistance genes included those resistant to (i) macrolide and lincosamide drugs such as ermB, ermA, mreA, mefA, msrD, lunB, lsaC, and lsaE; (ii) tetracycline such as tetM, tetO, tetS, tetL, and tet(O/W/32/O); (iii) chloramphenicol such as cat, catQ, and cat(pC194); and (iv) aminoglycoside such as ant(6)-Ia, aph(3’)-III, and aac(6’)-aph(2’’). Notably, all 138 strains carried the mreA gene, with 52.9% carrying the ermB gene and 41.3% carrying the tetM gene. In addition, all strains in CC19 carried the catQ or the cat(pC194) genes. The resistance gene tet(O/W/32/O) was detected in only one strain. The resistance genes GyrA and ParC were found in only 13.0% and 13.8% of the strains respectively, with these two genes being exclusively (GyrA) or predominantly (ParC) found in CC10 and CC19 strains (Figure 5).

Figure 5. Carriage of antibiotic resistance and virulence genes in 138 strains of GBS from pregnant women and neonates. CS, cervical secretions; SP, sputum; BL, blood; TS, throat swabs; AF, amniotic fluid; VS, vaginal secretions. HN, high-risk neonate; NP, neonatal pneumonia; PN, premature neonate; NS, neonatal septicemia; NPS, normal pregnancy supervision; PROM, premature rupture of membrane; TA, threatened abortion; HPS, high-risk pregnancy supervision; PL, premature labor; FL, false labor; OD, oligohydramnios; PPH, placenta previa and hemorrhage; CAN, cord around neck; PCI, pregnancy with cervical insufficiency.
Surface protein genes and virulence factors
In this study, virulence factors encompassing various cell surface proteins that facilitate GBS adhesion and invasion were identified. These virulence factors include members of the pilus islands (PI-1, PI-2a, PI-2b), alpha-like protein (Alp) family (Alpha C, Alp1, Alp2/3, Rib), and serine-rich repeat glycoproteins (Srr1 and Srr2). All strains were found to contain at least one Pili-island (PI) gene. The most prevalent combination was PI-1+PI-2a, which was found in 50.0% of the strains (69/138), followed by PI-2b (31.2%, 43/138), PI-2a (13.8%, 19/138), and PI-1+PI-2b (5.1%, 7/138). Among the 138 strains, all CC19 strains were found to carry the PI-1+PI-2a combination. The PI-2a was identified in strains belonging to CC1, CC10, and CC23. The PI-2b was identified in 92.7% of CC651 strains, a rate much higher than that in other clonal complexes (P<0.001). The PI-1+PI-2b was identified in strains belonging to CC1, CC17, and CC651. In contrast, only PI-2b was identified in all CC17 strains, with the exception of one ST17/CC17 strain that carried PI-1+PI-2b, as shown in Figure 5. The Alp1 gene was found in 90.2% of the CC651 strains, which accounted for a significantly greater proportion than those in CC1, CC19, and CC23 (P<0.001). Conversely, the Alp2/3 gene was detected in 73.9% of the CC1 strains, which was much higher than that reported for the other CCs (P<0.001). A total of 64.5% of the isolates in this study contained the Srr1 gene. The strains carrying HvgA or Srr2 were exclusively in CC17, with a 100% carrying rate. The rib gene was found in 75.0% of the CC19 strains, which was a significantly greater percentage than that in the CC17 strains (P<0.001), as illustrated in Figure 5.
The cfb and cylE genes were detected in all 138 strains. The pavA gene was almost universally present (99.3%, 137/138), while the detection rates for the lmb, hylB, and fbsA genes were 66.7%, 45.7%, and 12.3%, respectively.
Distribution and characterization of GBS strains associated with pregnancy outcomes
In six neonatal pneumonia cases, ST862/CC651/serotype III was the predominant genotype, accounting for 50% (3/6) of the cases (Table 4). The only observed case of neonatal septicemia was caused by a strain belonging to ST19/CC19/serotype III. Notably, ST17/CC17/serotype III has been exclusively identified in strains associated with adverse pregnancy outcomes and the only strain harboring the HVgA gene (Table 4). Interestingly, the nine newly identified STs in this study were all associated with invasive diseases and adverse maternal outcomes (Table 4). However, aside from the above, there was no statistically significant difference in the distribution of CCs and serotypes between adverse pregnancy outcomes and normal pregnancies.
Discussion
GBS is a common pathogen that can cause adverse pregnancy outcomes and is a significant cause of neonatal sepsis and meningitis within the first 90 days of life. The integration of GBS screening into pre-pregnancy and prenatal care guidelines by the Obstetrics and Gynecology Branch of the Chinese Medical Association in 2014 underscores the importance of early detection and intervention. Our study provides critical insights into the molecular epidemiology of GBS in Haikou, China, revealing that serotypes Ia, Ib, II, III, and V collectively account for 97.1% of the cases. This distribution aligns with global patterns, highlighting the universality of these serotypes in GBS infections (Teatero et al., 2017; Furfaro et al., 2018). However, the most prevalent GBS serotypes vary among different regions. In our study, serotypes III (33.3%), V (29.7%), and Ib (17.4%) were more prevalent than the global averages of 25%, 18%, and 8%, respectively, while serotypes Ia (10.1%) and II (6.5%) were less prevalent than the global averages of 21% and 11%, respectively (Russell et al., 2017). In Beijing, China, serotype III predominated with a smaller proportion of serotype V (Wang et al., 2015), whereas in Rio de Janeiro, Brazil, serotype Ia was the most common, with serotype III being less prevalent (Costa et al., 2022). India reported a lower prevalence of serotype III (11%) and no serotype V. In Japan, serotypes III and V were less prevalent, whereas VI and VIII were the most prevalent serotypes. These regional differences underscore the necessity for localized surveillance to inform tailored prevention and treatment strategies. Continuous monitoring of serotype shifts is critical for guiding vaccine development and adjusting vaccine compositions to meet evolving prevention and control needs.
Globally, ST17, ST1, ST23, and ST19 are the predominant STs associated with GBS infections in pregnant women (Manning et al., 2008; Shabayek and Spellerberg, 2018). Notably, ST17 and ST19 were more prevalent in strains causing early-onset disease (EOD) and late-onset disease (LOD) respectively (Manning et al., 2009). A total of 28 STs were identified in this study, with ST862 being the most prevalent, followed by ST529 and ST1. Similar to serotype distribution patterns described above, the predominant sequence types vary among geographic areas. For example, previous studies showed that ST19 and ST10 were the most common STs of GBS in pregnant women in China, followed by ST12, ST17 and ST651 (Wang et al., 2023). In our study, ST862 emerged as a dominant sequence type in Haikou, similar to findings from Xiamen and Fuzhou cities in Fujian Province, although it has rarely been reported in other parts of China (Jiang et al., 2016; Wu et al., 2019; Yao et al., 2020; Liang et al., 2023). Importantly, ST862 is also an emerging zoonotic ST that potentially poses a threat to both human health and animal husbandry (Sapugahawatte et al., 2022). These findings suggest that ST862 may possess a high degree of adaptability and transmission capacity in these regions, possibly due to its specific biological characteristics, host preference, or environmental adaptability. Similarly, ST529, another prevalent ST in our study, has been reported as a major sequence type in Linyi, Shandong Province but is rarely observed in other regions (Zhou et al., 2024). Our findings highlight the importance of monitoring spatial and temporal distributions of STs to identify emerging STs, particularly those with zoonotic potential, to mitigate their impact on public health in this and other microbial pathogens (e.g., Dalmieda et al., 2024; Poorrashidi et al., 2024; Meng et al., 2025).
A study by Kimura et al. (2013) revealed that the sensitivity of GBS strains to penicillin is decreasing worldwide, and penicillin is still the first-line choice for the treatment of GBS infection because of its excellent therapeutic effect (Quiroga et al., 2008; Petca et al., 2024). In our study, no penicillin-resistant strains were observed, reinforcing the continued use of penicillin as the primary treatment for GBS infections. However, for patients with severe penicillin allergies, clindamycin is an important alternative. Erythromycin, on the other hand, is less effective because of its poor placental penetration, limiting its clinical use (Elbiss and Osman, 2023). Recent reports indicate that resistance to erythromycin and clindamycin is increasing (Sabroske et al., 2023), emphasizing the need for antibiotic selection based on drug sensitivity testing at regional and local levels. In our study, high resistance rates were observed for tetracycline (89.1%) and clindamycin (55.1%), suggesting that these drugs should not be used without prior susceptibility testing in patients with penicillin allergies in Haikou, China. Additionally, resistance rates for moxifloxacin (7.2%) and levofloxacin (5.1%) in our strains were higher than those reported in Taiwan (Tsai et al., 2022). Variations in antibiotic usage patterns likely explain the geographical differences in resistance prevalence. In summary, while penicillin is still preferred for GBS infections, the high resistance rates to alternative antibiotics, particularly tetracycline and clindamycin, necessitate careful antibiotic selection guided by sensitivity testing to optimize treatment outcomes and reduce resistance.
Tetracycline resistance in streptococci is mediated by ribosome protection genes such as tet(M) and tet(O) or by the efflux pump genes tet(K) and tet(L). Macrolide resistance is due primarily to two mechanisms: ribosomal methylation encoded by the erm gene and active efflux mediated by a membrane-bound protein encoded by the mef gene. The former mechanism often confers high-level resistance to macrolides, lincosamides, and streptogramin B antibiotics. In our study, resistance to tetracycline was attributed to the presence of tet(O), whereas erythromycin resistance was linked to the ermB gene, which encodes erythromycin ribosome methylase. Clindamycin resistance was associated with lnuB, a gene encoding a lincosamide-inactivating nucleotidyl transferase, and kanamycin resistance was linked to aphA3, encoding an aminoglycoside phosphotransferase. The average correlation between resistance phenotypes and resistance genes of GBS was 65.62%, with a 100% correlation for tetracycline resistance. Similar findings have been reported for GBS strains isolated from Brazilian mastitic cows (Silva et al., 2017) and Staphylococcus aureus strains isolated from raw milk (Liu et al., 2017). However, our study revealed that lnuB was detected in only 16 of 76 clindamycin-resistant strains, suggesting that clindamycin resistance may involve other yet unidentified mechanisms.
Consistent with previous studies (Springman et al., 2014; Liu et al., 2023), we observed that PI-1 and PI-2a are the most common PI family genes in GBS strains. In our study, PI-2a was present in 63.8% of the strains, followed by PI-1 (55.1%) and PI-2b (36.2%). Notably, the combination of PI genes was more prevalent than individual PI genes were, a finding supported by previous studies in China (Liu et al., 2023) and Ireland (Meehan et al., 2014). All the isolates in our study carried at least one PI gene, with the PI-1+PI-2a combination being the most common (50.0%). This result aligns with the findings of Khodaei et al (Khodaei et al., 2018). Interestingly, while previous studies suggested that the PI-1+PI-2b combination was exclusive to CC17 isolates, our study revealed that only PI-2b was present in all CC17 strains, except for one ST17/CC17 strain that carried PI-1+PI-2b.
The virulence factor HvgA is an adhesin found in highly virulent strains that are frequently associated with neonatal meningitis. HvgA enhances bacterial adhesion to intestinal epithelial cells, choroidal epithelial cells, and microvascular endothelial cells that constitute the blood-brain barrier (Choi et al., 2022). Similarly, the Rib protein has been implicated in promoting neonatal meningeal infection. These surface-anchored proteins not only facilitate bacterial adhesion but also enable GBS to penetrate the intestinal and blood-brain barriers, allowing migration to the circulatory and central nervous systems (Fischer et al., 2021). In our study, HvgA was predominantly associated with serotype III/CC17 strains, which is consistent with the findings of Bourrel et al (Bourrel et al., 2024), who demonstrated that HvgA enhances the adhesion of CC17 strains, making them more virulent than non-CC17 strains. This highlights the need for enhanced clinical monitoring of CC17 strains to better understand their epidemiology, infection characteristics, and resistance patterns, thereby informing prevention and control strategies.
Another surface adhesin, Srr1, was detected in all CCs except CC17, whereas Srr2 was exclusively found in CC17 strains. This finding is consistent with studies by Lacasse et al. (2022) and Jin et al (Jin et al., 2022), who reported that Srr2 was strongly associated with CC17 strains, particularly in cases of GBS infection in infants. These results suggest that Srr2 may play a role in the virulence of CC17 strains, further emphasizing the importance of monitoring this clonal complex.
The virulence factors identified in this study, including cfb, cylE, pavA, lmb, and hylB, are potential candidates for vaccine development. All the strains carried cfb and cylE, while pavA (99.3%) and lmb (66.7%) were also highly prevalent. In contrast, fbsA was detected in only 12.3% of the strains, making it a less suitable candidate for vaccine epitopes. These findings provide valuable insights for the development of targeted vaccines against GBS.
Overall, our data indicate that multilocus sequence types ST862, ST529, ST1, and ST19 were the predominant GBS genotypes in this region. Additionally, we found nine novel STs (ST1983–1991) never reported before in other geographic regions, all of which were associated with adverse pregnancy outcomes. In this GBS population, the serotype distribution was dominated by III, V, 1b, and 1a and virulence factors such as Srr1, Srr2, ALP1, PI-1, PI-2a, PI-1, and PI-2b were closely associated with maternal infections. Nearly all strains carried virulence genes cfb, cy1E, and pavA. Together, these data provide valuable references for the development of targeted GBS vaccines in this population. A limitation of this study is the absence of longitudinal follow-up with pregnant women and their neonates, which restricts our understanding of maternal colonization by invasive GBS serotypes and subsequent vertical transmission to and potential effects on neonates. Additionally, due to the retrospective nature of this cohort study, we were unable to establish direct causal relationships between specific strains and adverse outcomes. Future studies incorporating longitudinal follow-up and prospective data collection could provide deeper insights into the epidemiology and pathogenesis of GBS infections in this vulnerable population, while mitigating limitations related to causal inference.
Conclusions
This study highlights the emergence of ST862 as a dominant and potentially zoonotic sequence type of GBS in Haikou, China, with significant implications for public health and animal husbandry. The high prevalence of tetracycline and clindamycin resistance underscores the need for judicious antibiotic use and the development of region-specific treatment guidelines. The identification of novel STs and the comprehensive analysis of virulence factors provide valuable insights for future vaccine development and targeted interventions. Continuous resistance monitoring and surveillance are essential to mitigate the impact of GBS infections on maternal and neonatal health, particularly in regions with high rates of antibiotic resistance. These findings emphasize the importance of integrating molecular epidemiology into public health strategies to combat GBS infections effectively.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://nmdc.cn/resource/genomics/project/detail/NMDC10019610.
Ethics statement
The studies involving humans were approved by The Ethics Committees of Haikou Maternal and Child Health Hospital, in Hainan, China. Approval number [2024] 03006. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
WM: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. HW: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. QM: Investigation, Methodology, Writing – review & editing. JZ: Formal analysis, Writing – review & editing. XG: Formal analysis, Writing – review & editing. ZZ: Data curation, Writing – review & editing. JS: Formal analysis, Investigation, Writing – review & editing. XH: Investigation, Writing – review & editing. YW: Investigation, Writing – review & editing. JL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. JX: Project administration, Resources, Supervision, Writing – review & editing. JW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Hainan Province (822RC708, 2019RC227), the National Natural Science Foundation of China (31860035), Hainan Medical University Academic Enhancement Program Project (XSTS2025186) and the Innovative Scientific Research Project at the Province Level of Hainan Province in 2024 (Qhys2024-489).
Acknowledgments
We extend our sincere thanks to the Open Foundation of Key Laboratory of Tropical Translational Medicine of Ministry of Education and the Large Instrument Platform of the Public Research Center at Hainan Medical University for their invaluable support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1655649/full#supplementary-material
References
Bianchi-Jassir, F., Seale, A. C., Kohli-Lynch, M., Lawn, J. E., Baker, C. J., Bartlett, L., et al. (2017). Preterm birth associated with group B Streptococcus maternal colonization worldwide: systematic review and meta - analyses. Clin. Infect. Dis. 65, S133–S142. doi: 10.1093/cid/cix661
Bourrel, A.-S., Picart, A., Fernandez, J.-C., Hays, C., Mignon, V., Saubaméa, B., et al. (2024). Specific interaction between Group B Streptococcus CC17 hypervirulent clone and phagocytes. Infect. Immun. 92, e00062–e00024. doi: 10.1128/iai.00062-24
Choi, Y., Han, H. S., Chong, G. O., Le, T. M., Nguyen, H. D. T., Lee, O. E., et al. (2022). Updates on group B streptococcus infection in the field of obstetrics and gynecology. Microorganisms 10, 2398. doi: 10.3390/microorganisms10122398
Cieslewicz, M. J., Chaffin, D., Glusman, G., Kasper, D., Madan, A., Rodrigues, S., et al. (2005). Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect. Immun. 73, 3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005
Costa, N. S., Rio-Tinto, A., Pinto, I. B. F., Dos Santos Silva Alvim, D. C., de Assis Rocha, A., Oliveira, L. M. A., et al. (2022). Changes in Group B Streptococcus Colonization among Pregnant Women before and after the Onset of the COVID - 19 Pandemic in Brazil. Pathogens 11, 1104. doi: 10.3390/pathogens11101104
Dalmieda, J., Hitchcock, M., and Xu, J. (2024). High diversity within and low but significant genetic differentiation among geographic and temporal populations of the global Streptococcus pneumoniae. Can. J. Microbiol. 70, 226–237. doi: 10.1139/cjm-2023-0155
Dhudasia, M. B., Flannery, D. D., Pfeifer, M. R., and Puopolo, K. M. (2021). Updated guidance: prevention and management of perinatal group B streptococcus infection. Neoreviews 22, e177–e188. doi: 10.1542/neo.22-3-e177
Dong, Y., Basmaci, R., Titomanlio, L., Sun, B., and Mercier, J. C. (2020). Neonatal sepsis: within and beyond China. Chin. Med. J. (Engl). 133, 2219–2228. doi: 10.1097/CM9.0000000000000935
Elbiss, H. and Osman, N. (2023). Placental transport of Erythromycin and its effect on placental inflammatory factors. Pak J. Med. Sci. 39, 75–79. doi: 10.12669/pjms.39.1.6683
Fischer, P., Pawlowski, A., Cao, D., Bell, D., Kitson, G., Darsley, M., et al. (2021). Safety and immunogenicity of a prototype recombinant alpha - like protein subunit vaccine (GBS - NN) against Group B Streptococcus in a randomized placebo - controlled double - blind phase 1 trial in healthy adult women. Vaccine 39, 4489–4499. doi: 10.1016/j.vaccine.2021.06.046
Furfaro, L. L., Chang, B. J., and Payne, M. S. (2018). Perinatal Streptococcus agalactiae epidemiology and surveillance targets. Clin. Microbiol. Rev. 31, e00049–e00018. doi: 10.1128/CMR.00049-18
Gharabeigi, N., Tabatabaee Bafroee, A. S., and Amini, K. (2023). Molecular serotyping and antibiotic resistance profile of group B streptococcus strains isolated from Iranian pregnant women with urinary tract infection. Iran J. Med. Sci. 48, 542–550. doi: 10.30476/ijms.2023.96346.2787
Gonçalves, B. P., Procter, S. R., Paul, P., Chandna, J., Lewin, A., Seedat, F., et al. (2022). Group B streptococcus infection during pregnancy and infancy: estimates of regional and global burden. Lancet Glob Health 10, e807–e819. doi: 10.1016/S2214-109X(22)00093-6
HogenEsch, E., De Mucio, B., Haddad, L. B., Vilajeliu, A., Ropero, A. M., Yildirim, I., et al. (2021). Differences in maternal group B Streptococcus screening rates in Latin American countries. Vaccine 39, B3–B11. doi: 10.1016/j.vaccine.2020.10.082
Huebner, E. M., Gudjónsdóttir, M. J., Dacanay, M. B., Nguyen, S., Brokaw, A., Sharma, K., et al. (2022). Virulence, phenotype, and genotype characteristics of invasive group B Streptococcus isolates obtained from Swedish pregnant women and neonates. Ann. Clin. Microbiol. Antimicrob. 21, 43. doi: 10.1186/s12941-022-00534-2
Imperi, M., Pataracchia, M., Alfarone, G., Baldassarri, L., Orefici, G., and Creti, R. (2010). A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80, 212–214. doi: 10.1016/j.mimet.2009.11.010
Jiang, H., Chen, M., Li, T., Liu, H., Gong, Y., and Li, M. (2016). Molecular characterization of Streptococcus agalactiae causing community - and hospital - acquired infections in Shanghai, China. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01308
Jin, Z., Li, J., Zhou, H., Wang, Z., Yi, L., Liu, N., et al. (2022). Serotype distribution, virulence determinants and antimicrobial susceptibility of Streptococcus agalactiae isolated from young infants. Pathogens 11, 1355. doi: 10.3390/pathogens11111355
Jones, N., Bohnsack, J. F., Takahashi, S., Oliver, K. A., Chan, M. S., Kunst, F., et al. (2003). Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41, 2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003
Jones, N., Oliver, K. A., Barry, J., Harding, R. M., Bisharat, N., Spratt, B. G., et al. (2006). Enhanced invasiveness of bovine - derived neonatal sequence type 17 group B streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42, 915–924. doi: 10.1086/500324
Khodaei, F., Najafi, M., Hasani, A., Kalantar, E., Sharifi, E., Amini, A., et al. (2018). Pilus - encoding islets in S. agalactiae and its association with antibacterial resistance and serotype distribution. Microb. Pathog. 116, 189–194. doi: 10.1016/j.micpath.2018.01.035
Kimura, K., Matsubara, K., Yamamoto, G., Shibayama, K., and Arakawa, Y. (2013). Active screening of group B streptococci with reduced penicillin susceptibility and altered serotype distribution isolated from pregnant women in Kobe, Japan. Jpn J. Infect. Dis. 66, 158–160. doi: 10.7883/yoken.66.158
Lacasse, M., Valentin, A. S., Corvec, S., Bémer, P., Jolivet-Gougeon, A., Plouzeau, C., et al. (2022). Genotypic characterization and biofilm production of group B streptococcus strains isolated from bone and joint infections. Microbiol. Spectr. 10, e02329–e02321. doi: 10.1128/spectrum.02329-21
Li, D., Chi, X. Z., Zhang, L., Chen, R., Cao, J. R., Sun, X. Y., et al. (2020). Vaginal microbiome analysis of healthy women during different periods of gestation. Biosci. Rep. 40, BSR20201766. doi: 10.1042/BSR20201766
Liang, B., Chen, H., Yu, D., Zhao, W., Cai, X., Qiu, H., et al. (2023). Molecular epidemiology of group B streptococcus isolates from pregnant women with premature rupture of membranes in Fuzhou, China. Infect. Drug Resist. 16, 269–278. doi: 10.2147/IDR.S393935
Liu, Z., Jiang, X., Li, J., Ji, W., Zhou, H., Gong, X., et al. (2023). Molecular characteristics and antibiotic resistance mechanisms of clindamycin - resistant Streptococcus agalactiae isolates in China. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1138039
Liu, H., Li, S., Meng, L., Dong, L., Zhao, S., Lan, X., et al. (2017). Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J. Dairy Sci. 100, 8796–8803. doi: 10.3168/jds.2017-13370
López, Y., Parra, E., Cepas, V., Sanfeliú, I., Juncosa, T., Andreu, A., et al. (2018). Serotype, virulence profile, antimicrobial resistance and macrolide - resistance determinants in Streptococcus agalactiae isolates in pregnant women and neonates in Catalonia, Spain. Enferm Infecc Microbiol. Clin. (Engl Ed). 36, 472–477. doi: 10.1016/j.eimc.2017.08.006
Luan, S. L., Granlund, M., Sellin, M., Lagergård, T., Spratt, B. G., and Norgren, M. (2005). Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43, 3727–3733. doi: 10.1128/JCM.43.8.3727-3733.2005
Madrid, L., Seale, A. C., Kohli-Lynch, M., Edmond, K. M., Lawn, J. E., Heath, P. T., et al. (2017). Infant group B streptococcal disease incidence and serotypes worldwide: systematic review and meta - analyses. Clin. Infect. Dis. 65, S160–S172. doi: 10.1093/cid/cix656
Manning, S. D., Lewis, M. A., Springman, A. C., Lehotzky, E., Whittam, T. S., and Davies, H. D. (2008). Genotypic diversity and serotype distribution of group B streptococcus isolated from women before and after delivery. Clin. Infect. Dis. 46, 1829–1837. doi: 10.1086/588296
Manning, S. D., Springman, A. C., Lehotzky, E., Lewis, M. A., Whittam, T. S., and Davies, H. D. (2009). Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47, 1143–1148. doi: 10.1128/JCM.01424-08
McGee, L., Chochua, S., Li, Z., Mathis, S., Rivers, J., Metcalf, B., et al. (2021). Multistate, population - based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive group B streptococci within the United States 2015-2017. Clin. Infect. Dis. 72, 1004–1013. doi: 10.1093/cid/ciaa151
Meehan, M., Cunney, R., and Cafferkey, M. (2014). Molecular epidemiology of group B Streptococci in Ireland reveals a diverse population with evidence of capsular switching. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1155–1162. doi: 10.1007/s10096-014-2055-5
Meng, Q., Wang, H., Xiao, W., Mai, W., Liu, Y., Xiao, Y., et al. (2025). Prevalence, drug resistance and genetic diversity of Candida glabrata in the reproductive tract of pregnant women in Hainan and comparison with global multilocus sequence data. Mycology 16, 1–18. doi: 10.1080/21501203.2025.2461725
Metcalf, B. J., Chochua, S., Gertz, R. J., Hawkins, P. A., Ricaldi, J., Li, Z., et al. (2017). Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin. Microbiol. Infect. 23, 574–577. doi: 10.1016/j.cmi.2017.02.021
Okike, I. O., Ribeiro, S., Ramsay, M. E., Heath, P. T., Sharland, M., and Ladhani, S. N. (2014). Trends in bacterial, mycobacterial, and fungal meningitis in England and Wales 2004-11: an observational study. Lancet Infect. Dis. 14, 301–307. doi: 10.1016/S1473-3099(13)70332-3
Petca, A., Șandru, F., Negoiță, S., Dumitrașcu, M. C., Dimcea, D. A., Nedelcu, T., et al. (2024). Antimicrobial resistance profile of group B Streptococci colonization in a sample population of pregnant women from Romania. Microorganisms 12, 414. doi: 10.3390/microorganisms12020414
Pidwill, G. R., Rego, S., Jenkinson, H. F., Lamont, R. J., and Nobbs, A. H. (2018). Coassociation between group B Streptococcus and Candida albicans promotes interactions with vaginal epithelium. Infect. Immun. 86, e00669–e00617. doi: 10.1128/IAI.00669-17
Poorrashidi, M., Hitchcock, M., and Xu, J. (2024). Meta-analyses of the global multilocus genotypes of the human pathogen Campylobacter jejuni. Genome 67, 189–203. doi: 10.1139/gen-2023-0041
Quiroga, M., Pegels, E., Oviedo, P., Pereyra, E., and Vergara, M. (2008). Antibiotic susceptibility patterns and prevalence of group B Streptococcus isolated from pregnant women in Misiones, Argentina. Braz. J. Microbiol. 39, 245–250. doi: 10.1590/S1517-83822008000200009
Russell, N. J., Seale, A. C., O’Driscoll, M., O’Sullivan, C., Bianchi-Jassir, F., Gonzalez-Guarin, J., et al. (2017). Maternal colonization with group B Streptococcus and serotype distribution worldwide: systematic review and meta - analyses. Clin. Infect. Dis. 65, S100–S111. doi: 10.1093/cid/cix658
Sabroske, E. M., Iglesias, M. A. S., Rench, M., Moore, T., Harvey, H., Edwards, M., et al. (2023). Evolving antibiotic resistance in Group B Streptococci causing invasive infant disease: 1970-2021. Pediatr. Res. 93, 2067–2071. doi: 10.1038/s41390-022-02375-3
Sapugahawatte, D. N., Li, C., Dharmaratne, P., Zhu, C., Yeoh, Y. K., Yang, J., et al. (2022). Prevalence and characteristics of Streptococcus agalactiae from freshwater fish and pork in Hong Kong wet markets. Antibiotics (Basel). 11, 397. doi: 10.3390/antibiotics11030397
Seale, A. C., Blencowe, H., Bianchi-Jassir, F., Embleton, N., Bassat, Q., Ordi, J., et al. (2017). Stillbirth with group B streptococcus disease worldwide: systematic review and meta - analyses. Clin. Infect. Dis. 65, S125–S132. doi: 10.1093/cid/cix585
Shabayek, S. and Spellerberg, B. (2018). Group B streptococcal colonization, molecular characteristics, and epidemiology. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00437
Silva, J. R. D., Castro, G. D. A. D. C., Gonalves, M. S., Custódio, D. A., and da Costa, G. M. (2017). In vitro antimicrobial susceptibility and genetic resistance determinants of Streptococcus agalactiae isolated from mastitic cows in Brazilian dairy herds. Semina: Cienc. Agrarias. 38, 2581. doi: 10.5433/1679-0359.2017v38n4SUPLp2581
Slotved, H. C., Kong, F., Lambertsen, L., Sauer, S., and Gilbert, G. L. (2007). Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45, 2929–2936. doi: 10.1128/JCM.00117-07
Springman, A. C., Lacher, D. W., Waymire, E. A., Wengert, S. L., Singh, P., Zadoks, R. N., et al. (2014). Pilus distribution among lineages of group B streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 14, 159. doi: 10.1186/1471-2180-14-159
Teatero, S., Ferrieri, P., Martin, I., Demczuk, W., McGeer, A., and Fittipaldi, N. (2017). Serotype distribution, population structure, and antimicrobial resistance of group B streptococcus strains recovered from colonized pregnant women. J. Clin. Microbiol. 55, 412–422. doi: 10.1128/JCM.01615-16
Tsai, I. A., Su, Y., Wang, Y. H., and Chu, C. (2022). Alterations in Genes rib, scpB and Pilus Island Decrease the Prevalence of Predominant Serotype V, Not III and VI, of Streptococcus agalactiae from 2008 to 2012. Pathogens 11, 1145. doi: 10.3390/pathogens11101145
Wang, P., Tong, J. J., Ma, X. H., Song, F. L., Fan, L., Guo, C. M., et al. (2015). Serotypes, antibiotic susceptibilities, and multi-locus sequence type profiles of Streptococcus agalactiae isolates circulating in Beijing, China. PloS One 10, e0120035. doi: 10.1371/journal.pone.0120035
Wang, J., Zhang, Y., Lin, M., Bao, J., Wang, G., Dong, R., et al. (2023). Maternal colonization with group B Streptococcus and antibiotic resistance in China: systematic review and meta-analyses. Ann. Clin. Microbiol. Antimicrob. 22, 5. doi: 10.1186/s12941-023-00553-7
Wu, B., Su, J., Li, L., Wu, W., Wu, J., Lu, Y., et al. (2019). Phenotypic and genetic differences among group B Streptococcus recovered from neonates and pregnant women in Shenzhen, China: 8 - year study. BMC Microbiol. 19, 185. doi: 10.1186/s12866-019-1551-2
Yadeta, T. A., Worku, A., Egata, G., Seyoum, B., Marami, D., and Berhane, Y. (2018). Vertical transmission of group B Streptococcus and associated factors among pregnant women: a cross - sectional study, Eastern Ethiopia. Infect. Drug Resist. 11, 397–404. doi: 10.2147/IDR.S150029
Yao, Z., Jiayin, W., Xinyi, Z., Ling, C., Mingyuan, H., Simin, M., et al. (2020). Identification of group B streptococcus serotypes and genotypes in late pregnant women and neonates that are associated with neonatal early - onset infection in a south China population. Front. Pediatr. 8. doi: 10.3389/fped.2020.00265
Yao, K., Poulsen, K., Maione, D., Rinaudo, C. D., Baldassarri, L., Telford, J. L., et al. (2013). Capsular gene typing of Streptococcus agalactiae compared to serotyping by latex agglutination. J. Clin. Microbiol. 51, 503–507. doi: 10.1128/JCM.02417-12
Yuan, X. Y., Liu, H. Z., Liu, J. F., Sun, Y., and Song, Y. (2021). Pathogenic mechanism, detection methods and clinical significance of group B Streptococcus. Future Microbiol. 16, 671–685. doi: 10.2217/fmb-2020-0189
Keywords: group B streptococcus, molecular epidemiology, antibiotic resistance, virulence factors, whole-genome sequencing
Citation: Mai W, Wang H, Meng Q, Zhang J, Gong X, Zhuo Z, Sui J, He X, Wang Y, Li J, Xu J and Wu J (2025) Molecular epidemiology and antibiotic resistance of group B Streptococcus in pregnant women and neonates from Haikou, China: implications for vaccine development and antimicrobial stewardship. Front. Cell. Infect. Microbiol. 15:1655649. doi: 10.3389/fcimb.2025.1655649
Received: 28 June 2025; Accepted: 17 August 2025;
Published: 09 September 2025.
Edited by:
Antonella Marangoni, University of Bologna, ItalyReviewed by:
Tina Perme, University Medical Centre Ljubljana, SloveniaKátia Andrea Menezes Torres, Instituto Butantan, Brazil
Copyright © 2025 Mai, Wang, Meng, Zhang, Gong, Zhuo, Sui, He, Wang, Li, Xu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Li, bGlqdWFuQGljZGMuY24=; Jianping Xu, anB4dUBtY21hc3Rlci5jYQ==; Jinyan Wu, d3VqaW55YW5AbXVobi5lZHUuY24=
†These authors have contributed equally to this work
‡ORCID: Juan Li, orcid.org/0000-0001-7951-1783
Jianping Xu, orcid.org/0000-0003-2915-2780
Jinyan Wu, orcid.org/0000-0003-3045-1323
 Wenhui Mai1†
Wenhui Mai1† Huiting Wang
Huiting Wang Juan Li
Juan Li Jianping Xu
Jianping Xu