- 1Hospital of Stomatology, Guangdong Provincial Key Laboratory of Stomatology, Guanghua School of Stomatology, Sun Yat‐sen University, Guangzhou, China
- 2Department of Prosthodontics, School and Hospital of Stomatology, Guangzhou Medical University, Guangzhou, Guangdong, China
- 3School and Hospital of Stomatology, Guangdong Engineering Research Center of Oral Restoration and Reconstruction, Guangzhou Medical University, Guangzhou, Guangdong, China
Dental caries, a prevalent oral disease, has long been attributed primarily to bacteria, but emerging evidence highlights the critical role of fungi in its pathogenesis. Fungal biofilms, predominantly Candida albicans, release extracellular DNA (eDNA) and DNA-carrying extracellular vesicles (EVs). Together with bacterial eDNA, these form the biofilm matrix and can activate the host cGAS-STING signaling pathway. This review systematically elaborates on the molecular architecture and biological functions of the cGAS-STING pathway, comparing mechanistic differences in its activation by viral, bacterial, and fungal DNA. It further explores direct and indirect modes of STING pathway activation by fungal eDNA and EV-carried DNA, along with their immunoregulatory roles. Specifically, it discusses the interactive mechanisms between fungal biofilms and STING activation in root caries onset, emphasizing the dual effects of STING-mediated immune responses—enhancing antifungal immunity while potentially exacerbating tissue damage via excessive inflammation. Finally, this review outlines current knowledge gaps and future research directions, aiming to provide novel insights for precision prevention and treatment of dental caries.
1 Introduction
Dental caries is a highly prevalent chronic oral infectious disease worldwide, with its high incidence and disabling impact posing a serious threat to human oral health and quality of life (Wen et al., 2022; Yirsaw et al., 2024). For a long time, it has been widely accepted that acid production metabolism and biofilm formation by cariogenic bacteria like Streptococcus mutans are the core mechanisms of dental caries. Related antibacterial strategies have mostly focused on bacteria (Bhat et al., 2023; Jiang et al., 2023). However, with the development of microbiome technologies in recent years, growing evidence indicates that fungi represented by Candida albicans play an undeniable role in the occurrence and development of dental caries, especially root caries (Ji et al., 2025; Zhang et al., 2025).
These fungi, together with bacteria, form complex mixed biofilms at carious sites. The extracellular DNA (eDNA) they secrete not only cross-links with bacterial eDNA to form the biofilm matrix skeleton—enhancing community stability and stress resistance—but also acts as a key signaling molecule in host immune regulation (Gallucci, 2024; Geng et al., 2024; Han et al., 2025). Among these, the cGAS-STING signaling pathway serves as a core innate immune hub for cytosolic DNA recognition. The molecular mechanisms by which fungal eDNA and vesicle-carried DNA activate this pathway are critical. They link fungal biofilm colonization to host immune response imbalance (Brown Harding et al., 2024; Yang et al., 2024).
This review systematically clarifies the specific mechanisms by which pathogenic DNA in fungal biofilms—including eDNA and vesicle-carried DNA—activates the STING pathway. It analyzes how these mechanisms regulate immune balance in the caries microenvironment and influence dental hard tissue destruction. It also outlines future research directions. This work lays a theoretical foundation for advancing the understanding of caries etiology and facilitating the development of precise preventive and therapeutic strategies.
2 Molecular architecture and biological functions of the STING signaling pathway
The cGAS-STING signaling pathway acts as a central hub linking cytosolic DNA recognition to innate immune responses (Decout et al., 2021). It plays pivotal roles in defending against pathogenic invasion, regulating anti-tumor immunity, and maintaining autoimmune homeostasis.Cyclic GMP-AMP synthase (cGAS) is a cytosolic DNA sensor, primarily distributed in the cytoplasm and nucleus. Its N-terminal DNA-binding domain (DBD) specifically recognizes double-stranded DNA (dsDNA), including pathogenic DNA, damaged nuclear DNA, and mitochondrial DNA (Gentili et al., 2019). Upon binding to dsDNA, cGAS undergoes conformational changes to activate its C-terminal catalytic domain. This catalyzes the synthesis of the second messenger 2’,3’-cyclic GMP-AMP (2’,3’-cGAMP) from ATP and GTP.As a unique cyclic dinucleotide (CDN), 2’,3’-cGAMP diffuses from its binding site. It then interacts with the transmembrane domain of STING (stimulator of interferon genes), which is localized on the endoplasmic reticulum (ER) (Yu and Liu, 2021; Liu et al., 2022). This interaction induces STING tetramerization and translocation from the ER to the Golgi apparatus and perinuclear vesicles. During translocation, STING recruits the serine/threonine kinase TBK1 (TANK-binding kinase 1). TBK1 phosphorylates the C-terminal tail (CTT) of STING. Activated STING further mediates TBK1-dependent phosphorylation of transcription factors IRF3 (interferon regulatory factor 3) and NF-κB (nuclear factor κB). This drives their nuclear translocation, inducing the expression of type I interferons (IFN-α/β), proinflammatory cytokines (e.g., IL-6, TNF-α), and antiviral proteins (ISGs). This completes the signal cascade amplification (Kwon and Bakhoum, 2020).
Functionally, the cGAS-STING pathway has extensive biological significance. In antiviral immunity, it recognizes viral DNA released by herpesviruses and poxviruses. This induces type I interferons to inhibit viral replication (Decout et al., 2021; Zhang et al., 2024). It also activates innate immune cells such as dendritic cells and macrophages, promoting antigen presentation and initiating adaptive immune responses (Ou et al., 2021; Zhou et al., 2023). In tumor immunoregulation, the pathway recognizes DNA released by necrotic tumor cells. When activated via STING agonists, it enhances the infiltration and activation of cytotoxic T lymphocytes (CTLs). This induces tumor cell apoptosis or autophagy (Samson and Ablasser, 2022). Currently, STING agonists are in clinical trials for melanoma and lung cancer. Their combination with PD-1 inhibitors or chemotherapeutic drugs significantly improves anti-tumor efficacy (Pan et al., 2023; Wang et al., 2024).
However, aberrant activation of the cGAS-STING pathway is linked to various diseases. In autoimmune disorders, self-DNA (e.g., nucleosomal DNA in systemic lupus erythematosus patients) is misrecognized by cGAS. This leads to sustained STING activation and excessive type I interferon production, triggering autoimmune reactions (Zierhut and Funabiki, 2020; Skopelja-Gardner et al., 2022; Liu and Pu, 2023). Patients with Aicardi-Goutières syndrome exhibit abnormal pathway activation due to mutations in cGAS or STING. This results in severe type I interferonopathy (Crow and Manel, 2015). Additionally, mitochondrial DNA leakage into the cytoplasm (e.g., during ischemia-reperfusion injury or neurodegenerative diseases) activates the pathway (Paul et al., 2021; Guo et al., 2024b). This exacerbates inflammatory responses and tissue damage. The pathway also participates in cellular responses to DNA damage. It regulates cell cycle arrest, apoptosis, pyroptosis, and senescence, thereby influencing tissue repair and organismal aging (Bai and Liu, 2019; Schmitz et al., 2023).
3 Mechanistic differences in STING pathway activation by viral and bacterial DNA
Host recognition and clearance of pathogenic microorganisms form a critical defense line of the immune system. Innate immunity acts as the first and fastest barrier against invading microbes. The DNA-activated STING pathway is a key immune mechanism for pathogen recognition. Viruses and bacteria use distinct strategies to deliver DNA into the host cytoplasm, triggering the pathway and inducing immune responses. During viral infection, enveloped viruses (e.g., herpesviruses, poxviruses) release dsDNA into the cytoplasm via membrane fusion or endocytosis. cGAS specifically recognizes the double-stranded structure, length, and conformational features of viral DNA through its N-terminal DBD. Upon binding, it activates the C-terminal catalytic domain to generate 2’,3’-cGAMP.2’,3’-cGAMP then binds to ER-localized STING, inducing its activation and translocation to the Golgi apparatus and perinuclear vesicles. This is followed by TBK1 recruitment, which phosphorylates IRF3 and NF-κB. Ultimately, this induces type I interferons and proinflammatory cytokines (Wu et al., 2022; Patel et al., 2023).Notably, some viruses (e.g., adenoviruses) bypass cGAS. They activate STING via direct binding or interference with nucleic acid metabolism (Lam et al., 2014). Retroviral cDNA intermediates and RNA virus-induced mitochondrial DNA leakage also indirectly activate the pathway. To evade immunity, viruses encode proteins that degrade cGAS or inhibit STING translocation (Lange et al., 2022; Xie and Zhu, 2024).
Bacterial activation of the STING pathway differs mechanistically. Intracellular bacteria (e.g., Listeria monocytogenes) secrete hemolysins to disrupt phagosomal membranes. This releases bacterial DNA rich in unmethylated CpG motifs into the cytoplasm. These motifs enhance DNA-cGAS binding affinity, promoting 2’,3’-cGAMP production (Glomski et al., 2002; Fehér, 2019). Extracellular bacteria (e.g., Escherichia coli) inject DNA into host cells via lysis or type III secretion systems (Cornelis, 2000). Beyond the canonical cGAS-dependent pathway, some bacterial DNA is recognized by endosomally localized TLR9. This synergizes with the STING pathway to activate NF-κB and IRF3, amplifying proinflammatory responses (Temizoz et al., 2022; Danielson et al., 2024). However, bacteria also regulate the STING pathway. For example, Mycobacterium tuberculosis DNA activates STING, but its cell wall components inhibit STING translocation. This attenuates interferon responses to facilitate survival (Marinho et al., 2017; Sun et al., 2020).
In summary, viral and bacterial activation of the STING pathway both start with cGAS recognition of cytosolic DNA. But they differ significantly in DNA sources, cytosolic delivery modes, recognition priorities, downstream effects, and evasion mechanisms. Viruses primarily rely on their genomic DNA or replication intermediates to activate the pathway, inducing type I interferons to inhibit replication (Zevini et al., 2017). Bacteria deliver DNA via phagosomal disruption or secretion systems, synergizing with other pathways to enhance phagocytic killing (Ryan et al., 2023). These mechanisms highlight the complexity of host-pathogen interactions.
A comparative summary of the key mechanisms underlying STING pathway activation by different pathogens is provided in Table 1. Among these pathogens, fungi exhibit unique activation modes of the STING pathway, which are discussed in detail below.
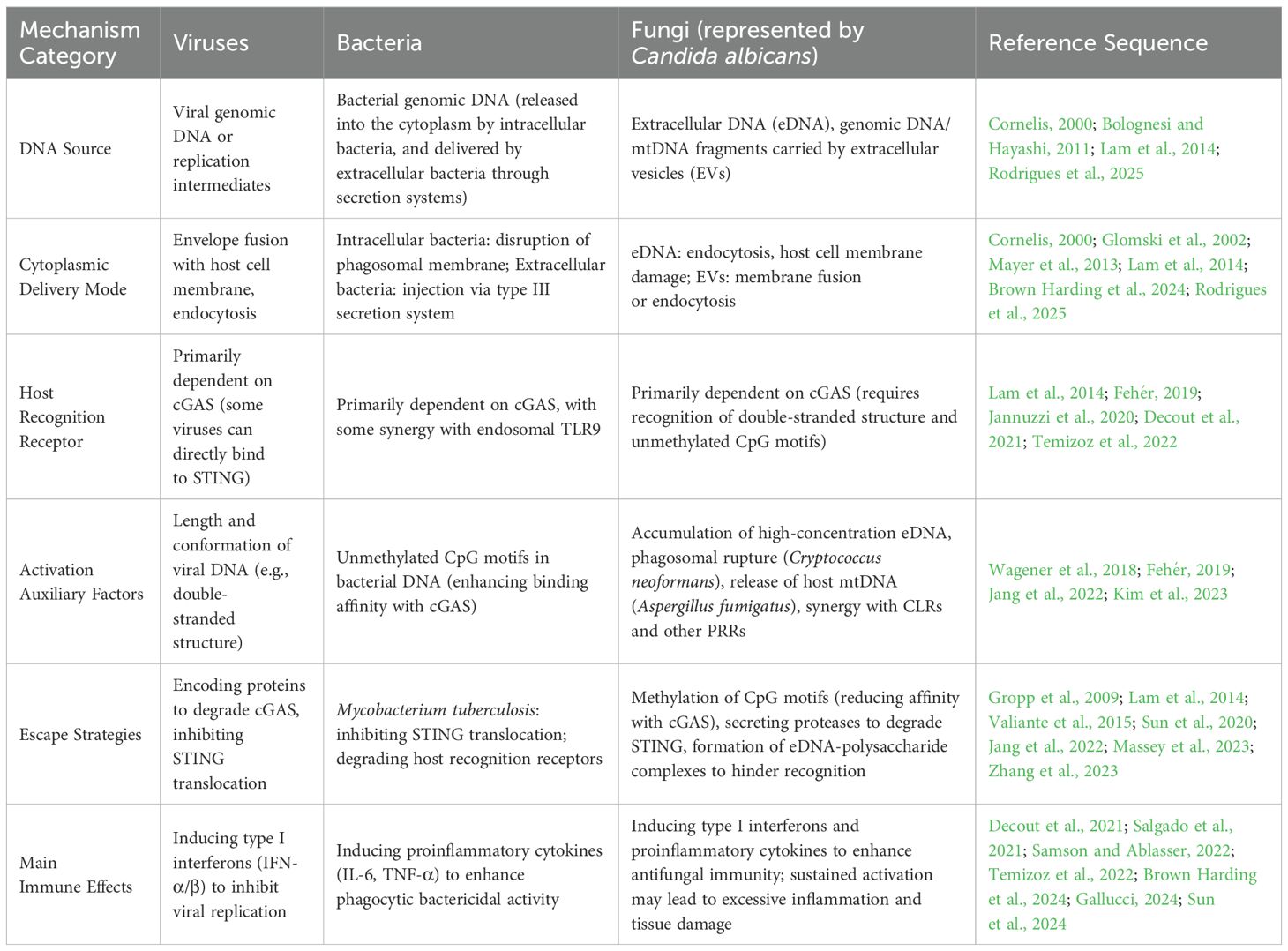
Table 1. Key mechanisms of STING pathway activation by different pathogens (viruses, bacteria, fungi).
4 Activation modes and immunoregulation of host STING pathway in fungal infections
During fungal infections, host cells recognize pathogens and initiate immune responses via multiple signaling pathways. Among these, the STING pathway interacts intricately with other immune signaling networks (Chen et al., 2023). As eukaryotic pathogens, fungi exhibit unique and diverse mechanisms for activating the STING pathway, emerging as a research hotspot in recent years.
In direct activation, invasive fungi (e.g., Candida albicans) release genomic DNA into the host cytoplasm. This occurs via secreted hydrolases (which disrupt cell membranes) or phagosomal rupture (Naglik et al., 2003; Mayer et al., 2013). Host cGAS recognizes the double-stranded structure, unmethylated CpG motifs, or specific conformational features (dsDNA length >40 bp) of fungal DNA. It then catalyzes 2’,3’-cGAMP synthesis. 2’,3’-cGAMP binds to ER-localized STING, inducing conformational changes and translocation to the Golgi apparatus. This is followed by TBK1 recruitment, which phosphorylates IRF3 and NF-κB. Ultimately, this induces the expression of type I interferons (e.g., IFN-β) and proinflammatory cytokines (e.g., IL-6, TNF-α), thereby enhancing antifungal immunity (Yu and Liu, 2021; Yum et al., 2021; Su et al., 2022). However, as eukaryotic DNA, fungal DNA has higher CpG methylation levels. Theoretically, it has lower cGAS binding affinity than bacterial DNA. Thus, it requires higher concentrations or specific conditions (e.g., repeated infections, phagosomal rupture) for effective activation (Jeon et al., 2015; Sarkies, 2022). For example, capsular polysaccharides of Cryptococcus neoformans promote phagosomal rupture. This increases fungal DNA-cGAS interactions to indirectly enhance STING activation (Liu et al., 2023c).
Non-cGAS-dependent STING activation in fungal infections primarily involves indirect activation via induced host mitochondrial DNA (mtDNA) release (Kim et al., 2023). For instance, hyphal invasion by Aspergillus fumigatus causes host mitochondrial damage. Released mtDNA is recognized by cGAS, activating STING. This “self-DNA + pathogen components” dual activation mode amplifies inflammatory responses in chronic infections (e.g., candidemia) (Kim et al., 2023; Peng et al., 2023). Additionally, fungal cell wall components (e.g., β-glucan, mannose) cannot directly activate STING. But they trigger signaling via other pattern recognition receptors (PRRs), synergizing with the STING pathway. Dectin-1, a C-type lectin receptor (CLR), recognizes β-glucan and activates NF-κB via the Syk-Card9 pathway. STING-induced IFN-β upregulates Dectin-1 expression, enhancing phagocytic bactericidal capacity. This “STING-IFN-other PRRs” cascade integrates antifungal immune signals (Wagener et al., 2018; Salgado et al., 2021).
The STING pathway is indispensable for antifungal immunity. On one hand, type I interferons induced by its activation enhance natural killer (NK) cell and T cell activation. They also promote macrophage phagocytosis and bactericidal function (Chen et al., 2023). On the other hand, proinflammatory cytokines recruit neutrophils, forming an inflammatory barrier to restrict fungal diffusion (Robertson et al., 2017). In animal studies, STING-deficient mice show increased susceptibility to Candida albicans and Aspergillus fumigatus infections. They exhibit elevated fungal burden and exacerbated tissue damage, confirming STING’s critical role in host antifungal immunity (Chen et al., 2023). Moreover, the STING pathway interacts with autophagy. STING activation induces autophagy-related genes (e.g., ATG5, ATG7), promoting phagosome-lysosome fusion to accelerate fungal degradation (Liu et al., 2019; Schmid et al., 2024). Autophagy also clears intracellular fungal DNA, avoiding excessive STING activation-induced immunopathological damage. This balances antifungal efficacy and tolerance (Maluquer De Motes, 2022).
Fungi have evolved multiple strategies to evade the STING pathway. At the DNA level, fungi such as Blastomyces dermatitidis methylate CpG motifs in their DNA, reducing cGAS binding efficiency (Yu and Liu, 2021). Cryptococcus neoformans capsular polysaccharides encapsulate DNA, blocking cGAS recognition (Jang et al., 2022). At the signaling level, Candida albicans secretes aspartic proteases (e.g., Sap2) that degrade host STING, inhibiting ER-to-Golgi translocation (Gropp et al., 2009). Mannoproteins in Aspergillus fumigatus cell walls competitively bind 2’,3’-cGAMP with STING, blocking downstream signaling (Valiante et al., 2015). Additionally, fungi such as Talaromyces marneffei inhibit host mtDNA release, reducing cGAS substrates to attenuate STING pathway responses (Zhang et al., 2023).
5 Fungal extracellular DNA: a potential bridge from biofilm matrix to STING pathway activation
Fungal extracellular DNA (eDNA) is a key component of fungal biofilms. It has emerged as a research focus due to its interactions with the host immune system and potential to activate the STING pathway during infections. eDNA refers to DNA actively secreted by fungi or released into the extracellular environment upon cell lysis. It is widely present in biofilm matrices of pathogenic fungi such as Candida albicans and Aspergillus fumigatus (Rajendran et al., 2013; Juszczak et al., 2024). It is generated via two main pathways: active secretion (dependent on specific fungal secretion systems) and passive release (from cell wall/membrane rupture induced by programmed cell death, mechanical damage, or host immune attacks) (Bolognesi and Hayashi, 2011). Fungal eDNA has double-stranded structures and contains unmethylated CpG motifs, providing a structural basis for recognition by host pattern recognition receptors (Panchin et al., 2016).
Fungal eDNA has multiple biological functions. In biofilm construction and protection, eDNA forms a three-dimensional network via physical cross-linking. It connects hyphae, yeast cells, and extracellular polysaccharides to enhance mechanical stability. It also defends against antifungal drug penetration and host immune clearance. For example, Candida albicans eDNA chelates echinocandins, reducing their inhibition of cell wall β-glucan synthase (Rajendran et al., 2013; Sharma and Rajpurohit, 2024). In regulating fungal physiology and virulence, eDNA acts as a signaling molecule to modulate morphological transitions and virulence factor expression. Its specific sequences bind fungal transcription factors, promoting invasion-related gene expression (Campoccia et al., 2021). Additionally, eDNA mediates intercellular communication. It transfers genetic information via horizontal gene transfer, accelerating the spread of drug resistance or virulence genes among fungal populations (Gonçalves and Gonçalves, 2022).
In host interactions, fungal eDNA both activates innate immunity and participates in immune evasion and pathological damage. It is recognized by host surface or intracellular pattern recognition receptors. Endosomally localized TLR9 recognizes unmethylated CpG motifs in eDNA, activating NF-κB to induce proinflammatory cytokines. cGAS, as a cytosolic DNA sensor, also potentially recognizes eDNA (Jannuzzi et al., 2020). Conversely, high eDNA concentrations inhibit immune functions via multiple mechanisms. These include chelating antimicrobial peptides and immune cell surface receptors to impair phagocytosis, or promoting macrophage polarization toward an anti-inflammatory phenotype to reduce antifungal efficiency (Massey et al., 2023). In chronic fungal infections, sustained eDNA stimulation may induce cytokine storms and tissue damage (Conti et al., 2018).
The potential mechanisms of fungal eDNA-activated STING pathway have attracted significant attention, involving direct and indirect activation. In direct activation, eDNA may enter the cytoplasm via endocytosis or host membrane damage. cGAS recognizes its double-stranded structure and unmethylated CpG motifs, catalyzing 2’,3’-cGAMP production. This potentially activates STING, recruits TBK1, phosphorylates IRF3 and NF-κB, and induces type I interferons and proinflammatory cytokines (Liu et al., 2023a; Li et al., 2025). In indirect activation, eDNA may induce host cell damage, promoting mitochondrial DNA (mtDNA) release into the cytoplasm. Or it may synergize with TLR9 and other pattern recognition receptors to enhance STING signaling (Lou and Pickering, 2018; Liu et al., 2023b). Activation efficiency may be influenced by multiple factors. eDNA length, methylation status, and CpG density may affect cGAS binding affinity (Ben Maamar et al., 2023; Dong et al., 2025). eDNA-polysaccharide/protein complexes in biofilms may hinder recognition, while local high concentrations may increase activation probability (LuTheryn et al., 2023). Differences in cGAS-STING pathway sensitivity among host cell types may also impact activation (Li and Bakhoum, 2022).
As a core biofilm component, fungal eDNA’s potential to activate the STING pathway reveals novel interaction modes between fungal infections and host immunity. Targeting the eDNA-STING axis may offer new antifungal strategies, such as developing eDNA-degrading nucleases or STING agonists to enhance immunity. However, eDNA-mediated excessive STING activation may contribute to chronic inflammation and autoimmune diseases, necessitating further research into its regulatory mechanisms in pathological states.
6 Mechanisms and immunoregulatory roles of fungal extracellular vesicle–carried DNA in STING pathway activation
Fungal extracellular vesicles (EVs) are membrane-bound vesicles (30–1000 nm in diameter) actively secreted by fungal cells. Their carried DNA plays a key role in host-pathogen interactions (Rodrigues et al., 2025). EV DNA primarily includes genomic DNA fragments, mitochondrial DNA, and cDNA derived from non-coding RNA. Most are in double-stranded or circular forms; some contain unmethylated CpG motifs, endowing potential for recognition by host pattern recognition receptors (Guo et al., 2024a; Ghanam et al., 2025). In pathogenic fungi such as Candida albicans and Aspergillus fumigatus, DNA constitutes 10%-15% of total EV content. EV secretion and DNA loading efficiency increase significantly during biofilm formation or environmental stress (Ullah et al., 2023).
Functionally, fungal EV DNA has diverse biological significance. In intercellular communication and genetic information transfer, EVs act as “molecular carriers” to transport DNA to recipient fungal cells. This enables horizontal gene transfer, accelerating the spread of drug resistance or virulence genes (Marcilla and Sánchez-López, 2022; Werner Lass et al., 2024). In regulating fungal physiology and virulence, EV DNA modulates gene expression in recipient fungi. For example, Aspergillus fumigatus EV DNA regulates morphology-related genes to promote hyphal growth and enhance invasiveness (Rizzo et al., 2023). Additionally, EV DNA has dual roles in immune regulation. It acts as a pathogen-associated molecular pattern (PAMP) to activate host immunity. It also mediates immune suppression via associated immunomodulatory components to facilitate fungal evasion (Montanari Borges et al., 2024).
In host interactions, fungal EV DNA has complex immunoregulatory properties. EVs enter host cells via endocytosis or membrane fusion, releasing DNA that is recognized by intracellular pattern recognition receptors. Endosomally localized TLR9 recognizes unmethylated CpG motifs in EV DNA, activating NF-κB to induce proinflammatory cytokines (Higuchi et al., 2024). Conversely, some fungal EVs evade immunity. For example, Cryptococcus neoformans EV DNA binds immunosuppressive miRNAs to downregulate host cell surface MHC-II expression, impairing antigen presentation (Sk Md et al., 2020). Sustained EV DNA stimulation also induces cytokine imbalance, leading to tissue damage (Fan et al., 2025).
The direct activation pathway of STING by fungal EV-carried DNA likely involves EV entry into host cells via membrane fusion or endocytosis, followed by DNA release. Cytosolic cGAS recognizes this DNA, potentially initiating the 2’,3’-cGAMP-STING signaling cascade to induce antiviral immune responses (Brown Harding et al., 2024; Kwaku et al., 2025). Indirect activation may involve EV DNA-induced host mitochondrial damage, releasing mtDNA to activate cGAS. Or it may synergize with EV-carried components (e.g., β-glucan) via TLR pathways to enhance STING signaling (Tao et al., 2024). This process is regulated by multiple factors. EV DNA fragment length, methylation status, and CpG motif density may affect cGAS recognition efficiency. EV source cell types, membrane components, and loaded immunomodulatory molecules may alter DNA delivery efficiency. Host cell type differences influence STING pathway responsiveness, and EV membrane proteins may regulate STING subcellular localization and activation kinetics.
7 Interactive mechanisms between fungal biofilms and STING pathway activation in root caries onset
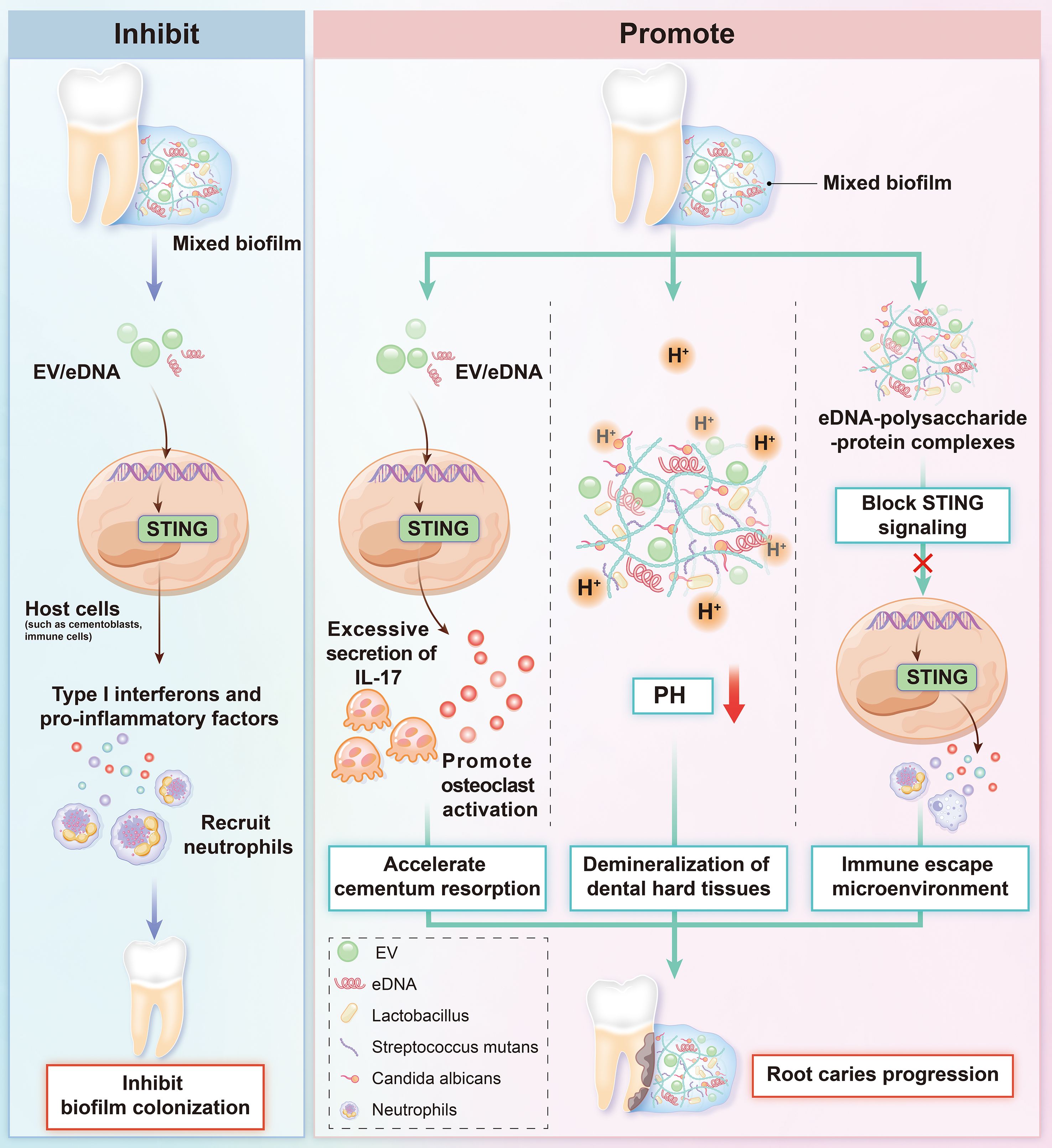
In the pathogenesis of dental caries, particularly root caries, interactions between fungal biofilms and the STING signaling pathway may form a potential pathogenic hub. At carious sites, Candida albicans dominates fungal biofilms and releases extracellular DNA (eDNA) and DNA-carrying extracellular vesicles (EVs). These are recognized by host intracellular cGAS, activating the STING pathway (Tian et al., 2022; Lattar et al., 2024; Han et al., 2025). Candida albicans eDNA, together with eDNA from cariogenic bacteria (e.g., Streptococcus mutans, Lactobacillus), forms a mixed biofilm matrix via physical cross-linking. This creates a dense three-dimensional network that enhances stability and acid tolerance, synergistically producing acids to exacerbate dental hard tissue demineralization (Sampaio et al., 2019; Evans et al., 2025; Han et al., 2025).
Immunologically, eDNA and EV DNA from mixed biofilms may enter the cytoplasm via endocytosis or membrane damage. This activates the STING pathway, inducing type I interferons and proinflammatory cytokines (Gallucci, 2024). This immune response has dual effects. On one hand, STING activation enhances antifungal immunity to inhibit biofilm colonization. On the other hand, sustained activation may induce excessive secretion of cytokines such as IL-17. This promotes osteoclast activation, accelerates cementum resorption, and drives root caries progression (Mao et al., 2024; Sun et al., 2024). Additionally, eDNA-polysaccharide/protein complexes in the biofilm matrix may block STING signaling, forming an immune-evasive microenvironment (Buzzo et al., 2021). Fungal and cariogenic bacterial metabolic synergies (e.g., carbon source sharing, mutual provision of growth factors) may further amplify inflammatory damage via the STING pathway (Montelongo-Jauregui et al., 2019; Kim et al., 2020). The dynamic interplay between fungal biofilms and the STING pathway offers novel insights into the mechanisms of immune dysregulation and tissue destruction in dental caries (Figure 1).
8 Conclusion and perspective
Fungal DNA released via multiple pathways—including extracellular DNA (eDNA) and extracellular vesicle (EV)-carried DNA—may activate the cGAS-STING signaling pathway to induce host immune responses. This mechanism may play a unique role in dental caries. eDNA and EV DNA from carious fungal biofilms (dominated by Candida albicans) may trigger STING activation. This occurs via direct cGAS stimulation or indirect induction of host mitochondrial DNA release. However, activation efficiency may be influenced by the cariogenic microenvironment (e.g., acidic pH, bacterial metabolites). Additionally, synergies between fungal biofilms and cariogenic bacteria (e.g., enhanced acid production) may amplify inflammatory damage via the STING pathway. Meanwhile, eDNA-polysaccharide complexes in the biofilm matrix may block STING signaling, forming an immune-evasive microenvironment. This complex interplay likely has unique significance in immune dysregulation and dental hard tissue destruction in caries.
Current research has not clarified the precise mechanisms of fungal DNA-activated STING pathway in the caries-specific microenvironment. For example, it remains unclear whether acidic conditions affect cGAS recognition efficiency of fungal DNA, or how the fungal-to-bacterial eDNA ratio in biofilms influences STING activation intensity. Future studies should focus on three areas: (1) developing nucleases targeting eDNA or EV inhibitors in cariogenic fungal biofilms to block excessive STING activation; (2) exploring STING pathway modulators in caries to balance antifungal immunity and inflammatory responses; (3) integrating oral microbiome research to dissect dynamic regulation of the fungal DNA-STING pathway during caries progression, providing a theoretical basis for precision caries prevention and treatment.
Author contributions
YJZ: Investigation, Funding acquisition, Writing – original draft. HJ: Writing – review & editing, Investigation, Conceptualization. YZZ: Writing – review & editing. YL: Writing – review & editing. YN: Supervision, Writing – review & editing, Funding acquisition. PL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 82401116), the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2022A1515110846 and No. 2022A1515111068), the Natural Science Foundation of Guangdong Province (Grant No. 2023A1515012257).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai, J. and Liu, F. (2019). The cGAS-cGAMP-STING pathway: A molecular link between immunity and metabolism. Diabetes 68, 1099–1108. doi: 10.2337/dbi18-0052
Ben Maamar, M., Wang, Y., Nilsson, E. E., Beck, D., Yan, W., and Skinner, M. K. (2023). Transgenerational sperm DMRs escape DNA methylation erasure during embryonic development and epigenetic inheritance. Environ. Epigenet 9, dvad003. doi: 10.1093/eep/dvad003
Bhat, R., Godovikova, V., Flannagan, S. E., Li, Y., Seseogullari-Dirihan, R., González-Cabezas, C., et al. (2023). Targeting cariogenic streptococcus mutans in oral biofilms with charge-switching smart antimicrobial polymers. ACS Biomater Sci. Eng. 9, 318–328. doi: 10.1021/acsbiomaterials.2c01095
Bolognesi, C. and Hayashi, M. (2011). Micronucleus assay in aquatic animals. Mutagenesis 26, 205–213. doi: 10.1093/mutage/geq073
Brown Harding, H., Kwaku, G. N., Reardon, C. M., Khan, N. S., Zamith-Miranda, D., Zarnowski, R., et al. (2024). Candida albicans extracellular vesicles trigger type I IFN signalling via cGAS and STING. Nat. Microbiol. 9, 95–107. doi: 10.1038/s41564-023-01546-0
Buzzo, J. R., Devaraj, A., Gloag, E. S., Jurcisek, J. A., Robledo-Avila, F., Kesler, T., et al. (2021). Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell 184, 5740–5758.e5717. doi: 10.1016/j.cell.2021.10.010
Campoccia, D., Montanaro, L., and Arciola, C. R. (2021). Extracellular DNA (eDNA). A major ubiquitous element of the bacterial biofilm architecture. Int. J. Mol. Sci. 22, 9100. doi: 10.3390/ijms22169100
Chen, T., Feng, Y., Sun, W., Zhao, G., Wu, H., Cheng, X., et al. (2023). The nucleotide receptor STING translocates to the phagosomes to negatively regulate anti-fungal immunity. Immunity 56, 1727–1742.e1726. doi: 10.1016/j.immuni.2023.06.002
Conti, P., Tettamanti, L., Mastrangelo, F., Ronconi, G., Frydas, I., Kritas, S. K., et al. (2018). Impact of fungi on immune responses. Clin. Ther. 40, 885–888. doi: 10.1016/j.clinthera.2018.04.010
Cornelis, G. R. (2000). Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos. Trans. R Soc. Lond B Biol. Sci. 355, 681–693. doi: 10.1098/rstb.2000.0608
Crow, Y. J. and Manel, N. (2015). Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440. doi: 10.1038/nri3850
Danielson, M., Nicolai, C. J., Vo, T. T., Wolf, N. K., and Burke, T. P. (2024). Cytosolic bacterial pathogens activate TLR pathways in tumors that synergistically enhance STING agonist cancer therapies. iScience 27, 111385. doi: 10.1016/j.isci.2024.111385
Decout, A., Katz, J. D., Venkatraman, S., and Ablasser, A. (2021). The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 21, 548–569. doi: 10.1038/s41577-021-00524-z
Dong, L., Hou, Y. R., Xu, N., Gao, X. Q., Sun, Z., Yang, Q. K., et al. (2025). Cyclic GMP-AMP synthase recognizes the physical features of DNA. Acta Pharmacol. Sin. 46, 264–270. doi: 10.1038/s41401-024-01369-7
Evans, D. C. S., Kristensen, M. F., Minero, G. A. S., Palmén, L. G., Knap, I., Tiwari, M. K., et al. (2025). Dental biofilms contain DNase I-resistant Z-DNA and G-quadruplexes but alternative DNase overcomes this resistance. NPJ Biofilms Microbiomes 11, 80. doi: 10.1038/s41522-025-00694-x
Fan, X., Peng, Y., Li, B., Wang, X., Liu, Y., Shen, Y., et al. (2025). Liver-secreted extracellular vesicles promote cirrhosis-associated skeletal muscle injury through mtDNA-cGAS/STING axis. Adv. Sci. (Weinh) 12, e2410439. doi: 10.1002/advs.202410439
Fehér, K. (2019). Single stranded DNA immune modulators with unmethylated cpG motifs: structure and molecular recognition by toll-like receptor 9. Curr. Protein Pept. Sci. 20, 1060–1068. doi: 10.2174/1389203720666190830162149
Gallucci, S. (2024). DNA at the center of mammalian innate immune recognition of bacterial biofilms. Trends Immunol. 45, 103–112. doi: 10.1016/j.it.2023.12.004
Geng, F., Liu, J., Liu, J., Lu, Z., and Pan, Y. (2024). Recent progress in understanding the role of bacterial extracellular DNA: focus on dental biofilm. Crit. Rev. Microbiol. 1-19, 898–916. doi: 10.1080/1040841X.2024.2438117
Gentili, M., Lahaye, X., Nadalin, F., Nader, G. P. F., Puig Lombardi, E., Herve, S., et al. (2019). The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26, 2377–2393.e2313. doi: 10.1016/j.celrep.2019.01.105
Ghanam, J., Lichá, K., Chetty, V. K., Pour, O. A., Reinhardt, D., Tamášová, B., et al. (2025). Unravelling the significance of extracellular vesicle-associated DNA in cancer biology and its potential clinical applications. J. Extracell Vesicles 14, e70047. doi: 10.1002/jev2.70047
Glomski, I. J., Gedde, M. M., Tsang, A. W., Swanson, J. A., and Portnoy, D. A. (2002). The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156, 1029–1038. doi: 10.1083/jcb.200201081
Gonçalves, P. and Gonçalves, C. (2022). Horizontal gene transfer in yeasts. Curr. Opin. Genet. Dev. 76, 101950. doi: 10.1016/j.gde.2022.101950
Gropp, K., Schild, L., Schindler, S., Hube, B., Zipfel, P. F., and Skerka, C. (2009). The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol. Immunol. 47, 465–475. doi: 10.1016/j.molimm.2009.08.019
Guo, S., Wang, X., Shan, D., Xiao, Y., Ju, L., Zhang, Y., et al. (2024a). The detection, biological function, and liquid biopsy application of extracellular vesicle-associated DNA. biomark. Res. 12, 123. doi: 10.1186/s40364-024-00661-2
Guo, X., Yang, L., Wang, J., Wu, Y., Li, Y., Du, L., et al. (2024b). The cytosolic DNA-sensing cGAS-STING pathway in neurodegenerative diseases. CNS Neurosci. Ther. 30, e14671. doi: 10.1111/cns.14671
Han, S. L., Wang, J., Wang, H. S., Yu, P., Wang, L. Y., Ou, Y. L., et al. (2025). Extracellular Z-DNA enhances cariogenicity of biofilm. J. Dent. Res. 104, 774–783. doi: 10.1177/00220345251316822
Higuchi, R., Tanaka, K., Saito, Y., Murakami, D., Nakagawa, T., Nutt, S. L., et al. (2024). Type I interferon promotes the fate of Toll-like receptor 9-stimulated follicular B cells to plasma cell differentiation. PNAS Nexus 3, pgae152. doi: 10.1093/pnasnexus/pgae152
Jang, E. H., Kim, J. S., Yu, S. R., and Bahn, Y. S. (2022). Unraveling capsule biosynthesis and signaling networks in cryptococcus neoformans. Microbiol. Spectr. 10, e0286622. doi: 10.1128/spectrum.02866-22
Jannuzzi, G. P., De Almeida, J. R. F., Paulo, L. N. M., De Almeida, S. R., and Ferreira, K. S. (2020). Intracellular PRRs activation in targeting the immune response against fungal infections. Front. Cell Infect. Microbiol. 10, 591970. doi: 10.3389/fcimb.2020.591970
Jeon, J., Choi, J., Lee, G. W., Park, S. Y., Huh, A., Dean, R. A., et al. (2015). Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae. Sci. Rep. 5, 8567. doi: 10.1038/srep08567
Ji, M., Xiong, K., Fu, D., Chi, Y., Wang, Y., Yao, L., et al. (2025). The landscape of the microbiome at different stages of root caries. Clin. Oral. Investig. 29, 217. doi: 10.1007/s00784-025-06301-9
Jiang, W., Xie, Z., Huang, S., Huang, Q., Chen, L., Gao, X., et al. (2023). Targeting cariogenic pathogens and promoting competitiveness of commensal bacteria with a novel pH-responsive antimicrobial peptide. J. Oral. Microbiol. 15, 2159375. doi: 10.1080/20002297.2022.2159375
Juszczak, M., Zawrotniak, M., and Rapala-Kozik, M. (2024). Complexation of fungal extracellular nucleic acids by host LL-37 peptide shapes neutrophil response to Candida albicans biofilm. Front. Immunol. 15, 1295168. doi: 10.3389/fimmu.2024.1295168
Kim, J., Kim, H. S., and Chung, J. H. (2023). Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway. Exp. Mol. Med. 55, 510–519. doi: 10.1038/s12276-023-00965-7
Kim, H. E., Liu, Y., Dhall, A., Bawazir, M., Koo, H., and Hwang, G. (2020). Synergism of Streptococcus mutans and Candida albicans Reinforces Biofilm Maturation and Acidogenicity in Saliva: An In Vitro Study. Front. Cell Infect. Microbiol. 10, 623980. doi: 10.3389/fcimb.2020.623980
Kwaku, G. N., Jensen, K. N., Simaku, P., Floyd, D. J., Saelens, J. W., Reardon, C. M., et al. (2025). Extracellular vesicles from diverse fungal pathogens induce species-specific and endocytosis-dependent immunomodulation. PloS Pathog. 21, e1012879. doi: 10.1371/journal.ppat.1012879
Kwon, J. and Bakhoum, S. F. (2020). The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 10, 26–39. doi: 10.1158/2159-8290.CD-19-0761
Lam, E., Stein, S., and Falck-Pedersen, E. (2014). Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88, 974–981. doi: 10.1128/JVI.02702-13
Lange, P. T., White, M. C., and Damania, B. (2022). Activation and evasion of innate immunity by gammaherpesviruses. J. Mol. Biol. 434, 167214. doi: 10.1016/j.jmb.2021.167214
Lattar, S. M., Schneider, R. P., Eugenio, V. J., and Padilla, G. (2024). High release of Candida albicans eDNA as protection for the scaffolding of polymicrobial biofilm formed with Staphylococcus aureus and Streptococcus mutans against the enzymatic activity of DNase I. Braz. J. Microbiol. 55, 3921–3932. doi: 10.1007/s42770-024-01550-4
Li, J. and Bakhoum, S. F. (2022). The pleiotropic roles of cGAS-STING signaling in the tumor microenvironment. J. Mol. Cell Biol. 14, mjac019. doi: 10.1093/jmcb/mjac019
Li, L., He, Y., Chen, Y., and Zhou, X. (2025). cGAS-STING pathway’s impact on intestinal barrier. J. Gastroenterol. Hepatol. 40, 1381–1392. doi: 10.1111/jgh.16974
Liu, Y., Chen, X., Zhao, Y., Wang, X. Y., Luo, Y. W., Chen, L., et al. (2023a). Small cytosolic double-stranded DNA represses cyclic GMP-AMP synthase activation and induces autophagy. Cell Rep. 42, 112852. doi: 10.1016/j.celrep.2023.112852
Liu, Y. and Pu, F. (2023). Updated roles of cGAS-STING signaling in autoimmune diseases. Front. Immunol. 14, 1254915. doi: 10.3389/fimmu.2023.1254915
Liu, H., Wang, F., Cao, Y., Dang, Y., and Ge, B. (2022). The multifaceted functions of cGAS. J. Mol. Cell Biol. 14, mjac031. doi: 10.1093/jmcb/mjac031
Liu, Y., Wei, F. Z., Zhan, Y. W., Wang, R., Mo, B. Y., and Lin, S. D. (2023b). TLR9 regulates the autophagy-lysosome pathway to promote dendritic cell maturation and activation by activating the TRAF6-cGAS-STING pathway. Kaohsiung J. Med. Sci. 39, 1200–1212. doi: 10.1002/kjm2.12769
Liu, D., Wu, H., Wang, C., Li, Y., Tian, H., Siraj, S., et al. (2019). STING directly activates autophagy to tune the innate immune response. Cell Death Differ 26, 1735–1749. doi: 10.1038/s41418-018-0251-z
Liu, Y., Zhang, Y., Zhao, X., Lu, W., Zhong, Y., and Fu, Y. V. (2023c). Antifungal peptide SP1 damages polysaccharide capsule of cryptococcus neoformans and enhances phagocytosis of macrophages. Microbiol. Spectr. 11, e0456222. doi: 10.1128/spectrum.04562-22
Lou, H. and Pickering, M. C. (2018). Extracellular DNA and autoimmune diseases. Cell Mol. Immunol. 15, 746–755. doi: 10.1038/cmi.2017.136
LuTheryn, G., Ho, E. M. L., Choi, V., and Carugo, D. (2023). Cationic microbubbles for non-selective binding of cavitation nuclei to bacterial biofilms. Pharmaceutics 15, 1495. doi: 10.3390/pharmaceutics15051495
Maluquer De Motes, C. (2022). Autophagy takes the STING out of DNA sensing. Cell Mol. Immunol. 19, 125–126. doi: 10.1038/s41423-021-00797-3
Mao, H. Q., Zhou, L., Li, J. Q., Wen, Y. H., Chen, Z., and Zhang, L. (2024). STING inhibition alleviates bone resorption in apical periodontitis. Int. Endod. J. 57, 951–965. doi: 10.1111/iej.14057
Marcilla, A. and Sánchez-López, C. M. (2022). Extracellular vesicles as a horizontal gene transfer mechanism in Leishmania. Trends Parasitol. 38, 823–825. doi: 10.1016/j.pt.2022.08.004
Marinho, F. V., Benmerzoug, S., Oliveira, S. C., Ryffel, B., and Quesniaux, V. F. J. (2017). The emerging roles of STING in bacterial infections. Trends Microbiol. 25, 906–918. doi: 10.1016/j.tim.2017.05.008
Massey, J., Zarnowski, R., and Andes, D. (2023). Role of the extracellular matrix in Candida biofilm antifungal resistance. FEMS Microbiol. Rev. 47, fuad059. doi: 10.1093/femsre/fuad059
Mayer, F. L., Wilson, D., and Hube, B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. doi: 10.4161/viru.22913
Montanari Borges, B., Gama De Santana, M., Willian Preite, N., De Lima Kaminski, V., Trentin, G., Almeida, F., et al. (2024). Extracellular vesicles from virulent P. brasiliensis induce TLR4 and dectin-1 expression in innate cells and promote enhanced Th1/Th17 response. Virulence 15, 2329573. doi: 10.3390/ijms22169100
Montelongo-Jauregui, D., Saville, S. P., and Lopez-Ribot, J. L. (2019). Contributions of Candida albicans Dimorphism, Adhesive Interactions, and Extracellular Matrix to the Formation of Dual-Species Biofilms with Streptococcus gordonii. mBio 10, e01179-19. doi: 10.1128/mBio.01179-19
Naglik, J. R., Challacombe, S. J., and Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 400–428. doi: 10.1128/MMBR.67.3.400-428.2003
Ou, L., Zhang, A., Cheng, Y., and Chen, Y. (2021). The cGAS-STING pathway: A promising immunotherapy target. Front. Immunol. 12, 795048. doi: 10.3389/fimmu.2021.795048
Pan, X., Zhang, W., Guo, H., Wang, L., Wu, H., Ding, L., et al. (2023). Strategies involving STING pathway activation for cancer immunotherapy: Mechanism and agonists. Biochem. Pharmacol. 213, 115596. doi: 10.1016/j.bcp.2023.115596
Panchin, A. Y., Makeev, V. J., and Medvedeva, Y. A. (2016). Preservation of methylated CpG dinucleotides in human CpG islands. Biol. Direct 11, 11. doi: 10.1186/s13062-016-0113-x
Patel, D. J., Yu, Y., and Xie, W. (2023). cGAMP-activated cGAS-STING signaling: its bacterial origins and evolutionary adaptation by metazoans. Nat. Struct. Mol. Biol. 30, 245–260. doi: 10.1038/s41594-023-00933-9
Paul, B. D., Snyder, S. H., and Bohr, V. A. (2021). Signaling by cGAS-STING in neurodegeneration, neuroinflammation, and aging. Trends Neurosci. 44, 83–96. doi: 10.1016/j.tins.2020.10.008
Peng, M., Li, X., Zhang, X., and Peng, L. (2023). Inhibition of cGAS aggravated the host inflammatory response to Aspergillus fumigatus. Exp. Lung Res. 49, 86–100. doi: 10.1080/01902148.2023.2211663
Rajendran, R., Williams, C., Lappin, D. F., Millington, O., Martins, M., and Ramage, G. (2013). Extracellular DNA release acts as an antifungal resistance mechanism in mature Aspergillus fumigatus biofilms. Eukaryot Cell 12, 420–429. doi: 10.1128/EC.00287-12
Rizzo, J., Trottier, A., Moyrand, F., Coppée, J. Y., Maufrais, C., Zimbres, A. C. G., et al. (2023). Coregulation of extracellular vesicle production and fluconazole susceptibility in Cryptococcus neoformans. mBio 14, e0087023. doi: 10.1128/mbio.00870-23
Robertson, J. D., Ward, J. R., Avila-Olias, M., Battaglia, G., and Renshaw, S. A. (2017). Targeting neutrophilic inflammation using polymersome-mediated cellular delivery. J. Immunol. 198, 3596–3604. doi: 10.4049/jimmunol.1601901
Rodrigues, M. L., Janbon, G., O’connell, R. J., Chu, T. T., May, R. C., Jin, H., et al. (2025). Characterizing extracellular vesicles of human fungal pathogens. Nat. Microbiol. 10, 825–835. doi: 10.1038/s41564-025-01962-4
Ryan, M. E., Damke, P. P., and Shaffer, C. L. (2023). DNA transport through the dynamic type IV secretion system. Infect. Immun. 91, e0043622. doi: 10.1128/iai.00436-22
Salgado, R. C., Fonseca, D. L. M., Marques, A. H. C., Da Silva Napoleao, S. M., França, T. T., Akashi, K. T., et al. (2021). The network interplay of interferon and Toll-like receptor signaling pathways in the anti-Candida immune response. Sci. Rep. 11, 20281. doi: 10.1038/s41598-021-99838-0
Sampaio, A. A., Souza, S. E., Ricomini-Filho, A. P., Del Bel Cury, A. A., Cavalcanti, Y. W., and Cury, J. A. (2019). Candida albicans Increases Dentine Demineralization Provoked by Streptococcus mutans Biofilm. Caries Res. 53, 322–331. doi: 10.1159/000494033
Samson, N. and Ablasser, A. (2022). The cGAS-STING pathway and cancer. Nat. Cancer 3, 1452–1463. doi: 10.1038/s43018-022-00468-w
Sarkies, P. (2022). Encyclopaedia of eukaryotic DNA methylation: from patterns to mechanisms and functions. Biochem. Soc. Trans. 50, 1179–1190. doi: 10.1042/BST20210725
Schmid, M., Fischer, P., Engl, M., Widder, J., Kerschbaum-Gruber, S., and Slade, D. (2024). The interplay between autophagy and cGAS-STING signaling and its implications for cancer. Front. Immunol. 15, 1356369. doi: 10.3389/fimmu.2024.1356369
Schmitz, C. R. R., Maurmann, R. M., Guma, F., Bauer, M. E., and Barbé-Tuana, F. M. (2023). cGAS-STING pathway as a potential trigger of immunosenescence and inflammaging. Front. Immunol. 14, 1132653. doi: 10.3389/fimmu.2023.1132653
Sharma, D. K. and Rajpurohit, Y. S. (2024). Multitasking functions of bacterial extracellular DNA in biofilms. J. Bacteriol 206, e0000624. doi: 10.1128/jb.00006-24
Sk Md, O. F., Hazra, I., Datta, A., Mondal, S., Moitra, S., Chaudhuri, S., et al. (2020). Regulation of key molecules of immunological synapse by T11TS immunotherapy abrogates Cryptococcus neoformans infection in rats. Mol. Immunol. 122, 207–221. doi: 10.1016/j.molimm.2020.04.021
Skopelja-Gardner, S., An, J., and Elkon, K. B. (2022). Role of the cGAS-STING pathway in systemic and organ-specific diseases. Nat. Rev. Nephrol. 18, 558–572. doi: 10.1038/s41581-022-00589-6
Su, M., Zheng, J., Gan, L., Zhao, Y., Fu, Y., and Chen, Q. (2022). Second messenger 2’3’-cyclic GMP-AMP (2’3’-cGAMP): Synthesis, transmission, and degradation. Biochem. Pharmacol. 198, 114934. doi: 10.1016/j.bcp.2022.114934
Sun, X., Liu, L., Wang, J., Luo, X., Wang, M., Wang, C., et al. (2024). Targeting STING in dendritic cells alleviates psoriatic inflammation by suppressing IL-17A production. Cell Mol. Immunol. 21, 738–751. doi: 10.1038/s41423-024-01160-y
Sun, Y., Zhang, W., Dong, C., and Xiong, S. (2020). Mycobacterium tuberculosis mmsA (Rv0753c) interacts with STING and blunts the type I interferon response. mBio 11, e03254-19. doi: 10.1128/mBio.03254-19
Tao, G., Liao, W., Hou, J., Jiang, X., Deng, X., Chen, G., et al. (2024). Advances in crosstalk among innate immune pathways activated by mitochondrial DNA. Heliyon 10, e24029. doi: 10.1016/j.heliyon.2024.e24029
Temizoz, B., Hioki, K., Kobari, S., Jounai, N., Kusakabe, T., Lee, M. S. J., et al. (2022). Anti-tumor immunity by transcriptional synergy between TLR9 and STING activation. Int. Immunol. 34, 353–364. doi: 10.1093/intimm/dxac012
Tian, X., Liu, C., and Wang, Z. (2022). The induction of inflammation by the cGAS-STING pathway in human dental pulp cells: A laboratory investigation. Int. Endod. J. 55, 54–63. doi: 10.1111/iej.13636
Ullah, A., Huang, Y., Zhao, K., Hua, Y., Ullah, S., Rahman, M. U., et al. (2023). Characteristics and potential clinical applications of the extracellular vesicles of human pathogenic Fungi. BMC Microbiol. 23, 227. doi: 10.1186/s12866-023-02945-3
Valiante, V., Macheleidt, J., Föge, M., and Brakhage, A. A. (2015). The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front. Microbiol. 6, 325. doi: 10.3389/fmicb.2015.00325
Wagener, M., Hoving, J. C., Ndlovu, H., and Marakalala, M. J. (2018). Dectin-1-syk-CARD9 signaling pathway in TB immunity. Front. Immunol. 9, 225. doi: 10.3389/fimmu.2018.00225
Wang, B., Yu, W., Jiang, H., Meng, X., Tang, D., and Liu, D. (2024). Clinical applications of STING agonists in cancer immunotherapy: current progress and future prospects. Front. Immunol. 15, 1485546. doi: 10.3389/fimmu.2024.1485546
Wen, P. Y. F., Chen, M. X., Zhong, Y. J., Dong, Q. Q., and Wong, H. M. (2022). Global burden and inequality of dental caries 1990 to 2019. J. Dent. Res. 101, 392–399. doi: 10.1177/00220345211056247
Werner Lass, S., Smith, B. E., Camphire, S., Eutsey, R. A., Prentice, J. A., Yerneni, S. S., et al. (2024). Pneumococcal extracellular vesicles mediate horizontal gene transfer via the transformation machinery. mSphere 9, e0072724. doi: 10.1128/msphere.00727-24
Wu, Y., Song, K., Hao, W., Li, J., Wang, L., and Li, S. (2022). Nuclear soluble cGAS senses double-stranded DNA virus infection. Commun. Biol. 5, 433. doi: 10.1038/s42003-022-03400-1
Xie, F. and Zhu, Q. (2024). The regulation of cGAS-STING signaling by RNA virus-derived components. Virol. J. 21, 101. doi: 10.1186/s12985-024-02359-1
Yang, H., Yang, S., Guo, Q., Sheng, J., and Mao, Z. (2024). ATP-responsive manganese-based bacterial materials synergistically activate the cGAS-STING pathway for tumor immunotherapy. Adv. Mater 36, e2310189. doi: 10.1002/adma.202310189
Yirsaw, A. N., Bogale, E. K., Tefera, M., Belay, M. A., Alemu, A. T., Bogale, S. K., et al. (2024). Prevalence of dental caries and associated factors among primary school children in Ethiopia: systematic review and meta-analysis. BMC Oral. Health 24, 774. doi: 10.1186/s12903-024-04555-5
Yu, L. and Liu, P. (2021). Cytosolic DNA sensing by cGAS: regulation, function, and human diseases. Signal Transduct Target Ther. 6, 170. doi: 10.1038/s41392-021-00554-y
Yum, S., Li, M., Fang, Y., and Chen, Z. J. (2021). TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. U.S.A. 118, e2100225118. doi: 10.1073/pnas.2100225118
Zevini, A., Olagnier, D., and Hiscott, J. (2017). Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 38, 194–205. doi: 10.1016/j.it.2016.12.004
Zhang, K., Huang, Q., Li, X., Zhao, Z., Hong, C., Sun, Z., et al. (2024). The cGAS-STING pathway in viral infections: a promising link between inflammation, oxidative stress and autophagy. Front. Immunol. 15, 1352479. doi: 10.3389/fimmu.2024.1352479
Zhang, Z., Li, B., Chai, Z., Yang, Z., Zhang, F., Kang, F., et al. (2023). Evolution of the ability to evade host innate immune defense by Talaromyces marneffei. Int. J. Biol. Macromol 253, 127597. doi: 10.1016/j.ijbiomac.2023.127597
Zhang, R. R., Zhang, J. S., Huang, S., Lam, W. Y., Chu, C. H., and Yu, O. Y. (2025). The oral microbiome of root caries: A scoping review. J. Dent. 160, 105899. doi: 10.1016/j.jdent.2025.105899
Zhou, J., Zhuang, Z., Li, J., and Feng, Z. (2023). Significance of the cGAS-STING pathway in health and disease. Int. J. Mol. Sci. 24, 13316. doi: 10.3390/ijms241713316
Keywords: STING pathway, fungal biofilms, extracellular DNA, dental caries, innate immunity
Citation: Zhou Y, Ji H, Zhang Y, Liu Y, Ning Y and Li P (2025) Mechanisms of fungal pathogenic DNA-activated STING pathway in biofilms and its implication in dental caries onset. Front. Cell. Infect. Microbiol. 15:1666965. doi: 10.3389/fcimb.2025.1666965
Received: 16 July 2025; Accepted: 18 August 2025;
Published: 01 September 2025.
Edited by:
Keke Zhang, Wenzhou Medical University, ChinaReviewed by:
Yaling Jiang, VU Amsterdam, NetherlandsHong Chen, Chongqing Medical University, China
Tao Gong, State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases, China
Copyright © 2025 Zhou, Ji, Zhang, Liu, Ning and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Ning, bmluZ3lhbmdAbWFpbC5zeXN1LmVkdS5jbg==; Ping Li, cGluZ2xpQGd6aG11LmVkdS5jbg==
†These authors have contributed equally to this work
 Yujie Zhou
Yujie Zhou Huanzhong Ji1†
Huanzhong Ji1† Yukun Liu
Yukun Liu