- 1Hainan Medical University–The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases & Key Laboratory of Tropical Translational Medicine of Ministry of Education, College of Basic Medical Sciences, Hainan Medical University, Haikou, China
- 2Department of Nutrition and Food Hygiene, School of Public Health, Shenzhen University, Shenzhen, China
- 3Institute of Clinical Medicine, The Second Affiliated Hospital of Hainan Medical University, Haikou, China
- 4The University of Hong Kong, Shenzhen, China
- 5School of Public Health, Xinjiang Medical University, Urumqi, China
- 6Clinical Research Center, The First Affiliated Hospital of Shantou University Medical College, Shantou, China
Background: Tigecycline remains a last-resort antibiotic for treating multidrug-resistant (MDR) Gram-negative pathogens. The emergence of tet(X4)-mediated high-level tigecycline resistance in Escherichia coli has raised global concern, yet its prevalence in healthy human populations remains limited.
Methods: We conducted a community-based surveillance study involving 245 fecal samples from healthy individuals in three urban communities in Shenzhen, China. Tigecycine-resistant strains were isolated using MacConkey agar supplemented with 2 mg/L tigecycline and confirmed by PCR detection of tet(X). Antimicrobial susceptibility testing, whole-genome sequencing (WGS), and phylogenetic analysis were performed.
Results: Tigecycline-resistant E. coli were detected in 1.6% (4/245) of samples. All isolates carried tet(X4) and exhibited an MDR phenotype. WGS revealed that tet(X4) was located on IncY (n=1) and IncFIA8-IncHI1/ST17 plasmids (n=3), which closely resembled previously described plasmids and co-harbored additional resistance genes. The core tet(X4)-carrying region in all four plasmids, associated with ISCR2, was highly similar to that of p47EC—the first tet(X4)-bearing plasmid identified in porcine E. coli in China. Notably, the three IncFIA-IncHI1/ST17 plasmids shared an identical 12,536-bp region structured as IS1–catD–tet(X4)–ISCR2–ΔISCR2–floR–ΔISCR2. Virulence-associated genes involved in adhesion, iron acquisition, biofilm formation, and secretion systems were also identified in four tet(X4)-positive isolates. The four isolates belonged to globally distributed sequence types ST10, ST201, ST877, and ST1308. Phylogenomic analysis demonstrated close genetic relatedness between these community isolates and strains from diverse geographical regions and hosts.
Conclusions: This study reveals silent intestinal colonization by tet(X4)-positive MDR E. coli among healthy urban residents, highlighting the role of community reservoirs in the dissemination of last-resort antibiotic resistance. These findings underscore the urgent need for One Health-oriented antimicrobial resistance surveillance and intervention strategies that extend beyond clinical settings.
Introduction
Tigecycline, a 9-t-butylglycylamido derivative of minocycline, is a glycylcycline antibiotic that inhibits bacterial protein synthesis by binding to the 30S ribosomal subunit (Yaghoubi et al., 2021). This mechanism allows tigecycline to evade traditional tetracycline resistance determinants, making it as a last-resort therapeutic agent for infections caused by multidrug-resistant (MDR) pathogens (Yaghoubi et al., 2021). However, the increasing emergence of tigecycline resistance is compromising its clinical utility.
Of particular concern is the tet(X) family of flavin-dependent monooxygenase genes, which inactivate tigecycline through enzymatic degradation (Aminov, 2021). Among them, the plasmid-mediated tet(X4), first identified in Escherichia coli from swine, confers high-level tigecycline resistance and facilitates horizontal gene transfer across diverse bacterial hosts and ecological inches (He et al., 2019). The widespread detection of tet(X4) in both clinical and agricultural settings, particularly among E. coli strains, underscores its growing public health threat (Li et al., 2023).
Although tet(X4)-positive E. coli has been increasingly reported in food-producing animals—a trend linked to the historical and extensive use of tetracyclines in agriculture—as well as in food products, and human patients (Aminov, 2021; Li et al., 2023), data on its carriage in healthy human populations, especially in community settings, remain limited (Ding et al., 2020, 2024; Dong et al., 2021). The human gut microbiota serves as a significant but underexplored reservoir for antimicrobial resistance genes (ARGs) (Carlet, 2012; Donskey, 2004). Within this niche, horizontal gene transfer can facilitate the dissemination of resistance determinants across environmental, zoonotic, and clinical bacterial populations (McInnes et al., 2020). While antimicrobial misuse and overuse are known drivers of resistance emergence (Allcock et al., 2017; Ferrara et al., 2024), it remains unclear whether AGRs, such as tet(X4), can persist or evolve in community-dwelling individuals without direct antibiotic exposure. This uncertainty is particularly relevant given that tetracyclines are poorly metabolized and can persist in the environment, potentially exerting low-level selective pressure through dietary or environmental exposure, even in the absence of clinical antibiotic use (Allcock et al., 2017).
Given the potential role of asymptomatic carriers in the silent spread of tigecycline resistance, enhanced surveillance of tet(X4) in healthy populations is urgently needed. In this study, we investigated tigecycline-resistant E. coli isolated from fecal samples of 245 asymptomatic adults residing in three urban communities in Shenzhen, China. Our objectives were to assess the prevalence of tigecycline resistance, characterize the genetic and plasmid features of tet(X4)-positive strains, and explore potential epidemiological links to clinical and agricultural sources. These findings offer important insight into the community-level dissemination of tigecycline resistance and highlight the need to broaden antimicrobial resistance (AMR) surveillance beyond clinical settings.
Materials and methods
Bacterial isolation and detection of the tet(X) gene
In October 2022, 245 fecal samples were obtained from healthy individuals aged 16 to 79 years, who had no self-reported symptoms of acute infection (e.g., diarrhea, fever, respiratory or urinary tract infections), no history of hospitalization or surgery in the past three months, and no antibiotic use within the preceding three months, across three residential communities in Shenzhen to investigate the prevalence of tigecycline-resistant Enterobacteriaceae (Supplementary Table S1). Samples were directly inoculated into LB broth and incubated at 37°C for 12–18 h for enrichment. Enriched cultures were then streaked onto LB agar plates containing 2 μg/mL tigecycline and incubated at 37°C for 12–18 h. Presumptive colonies were purified by subculturing, and bacterial species were identified using the VITEK-2 automated microbial identification system (bioMérieux, Lyon, France). The presence of the tet(X) gene was screened by PCR and Sanger sequencing using universal primers tet(X)-F (5′-CCGTTGGACTGACTATGGC-3′) and tet(X)-R (5′- TCAACTTGCGTGTCGGTAA-3′), as previously described (Wang et al., 2019).
Antimicrobial susceptibility testing
Antimicrobial susceptibility profiles were determined using the broth microdilution or the agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Minimum inhibitory concentrations (MICs) were assessed for 14 antibiotics: ampicillin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, tigecycline, chloramphenicol, nalidixic acid, ciprofloxacin, colistin, fosfomycin, and sulfamethoxazole-trimethoprim. MIC breakpoints for streptomycin and tigecycline were interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (https://www.eucast.org/), while those for other agents followed the 33rd edition of the CLSI document M100 (CLSI, 2023). E. coli ATCC 25922 was used as the quality control strain.
Conjugation assay
To assess the horizontal transferability of the tet(X4) gene, conjugation experiments were performed using tet(X)-positive isolates as donor strains and a high-level streptomycin-resistant E. coli strain C600 as the recipient. Briefly, donor and recipient strains were separately cultured in 2 mL LB broth at 37 with shaking (180 rpm) for 4 h, mixed at a 1:4 (v/v) ratio and incubated statically for 24 h. The cultures were then centrifuged at 5,000 × g for 5 min, the supernatant discarded, and the pellet resuspended in sterile PBS. Appropriate dilutions (100 μL) were plated on selective agar containing tigecycline (2 mg/mL) and streptomycin (3000 mg/mL) to select for transconjugants. Colonies were incubated at 37°C for 16–24 h and confirmed as transconjugants by PCR detection of the tet(X) gene as described above. All experiments were performed in triplicate.
Whole-genome sequencing and bioinformatics analysis
The same DNA extraction protocol was applied to both Illumina and Nanopore sequencing to ensure data consistency and comparability. Genomic DNA was extracted from E. coli isolates using the PureLink Genomic DNA Mini Kit (Invitrogen, USA). Short-read sequencing libraries were prepared using the Illumina NovoSeq PE150 platform (2×150 bp paired-end), while long-read sequencing was performed using the Oxford Nanopore MinION platform (Oxford Nanopore Technologies, UK). Hybrid genome assemblies were generated using Unicycler (v 0.5.0) (Wick et al., 2017) and subsequently corrected with Pilon (v 1.24) (Walker et al., 2014). Plasmid replicon types were identified using PlasmidFinder (Carattoli et al., 2014), and antibiotic resistance genes, including chromosomal mutations mediating resistance, were annotated using ResFinder (Bortolaia et al., 2020) and PointFinder (Zankari et al., 2017). Multilocus sequence typing (MLST) were assigned via MLST analysis (Larsen et al., 2012). Virulence factors were identified using ABRicate v0.8 with the VFDB database (updated October 2020). Comparative analysis of tet(X)-carrying plasmids and related plasmids was performed and visualized using BRIG (Alikhan et al., 2011).
A phylogenetic tree based on core genome single nucleotide polymorphism (cgSNP) was constructed using Parsnp v1.5.4 (https://github.com/marbl/parsnp) and visualized with iTOL (https://itol.embl.de/index.shtml). Our tet(X4)-positive E. coli isolate served as the reference genome for phylogenetic tree construction. To identify relevant strains, we retrieved E. coli isolates of the same ST from the NCBI database. cgSNP distances between the isolates sharing the same ST were calculated using Snippy v4.6.0 (https://github.com/tseemann/snippy), and only those with ≤200 SNPs relative to the reference were included in the final phylogenetic analysis.
Nucleotide sequence accession number
The whole-genome sequences of the four tet(X4)-positive isolates have been deposited in GenBank under accession number: PRJNA1288486.
Results
Prevalence of tigecycline-resistant E. coli isolates in healthy individuals
Tigecycline-resistant Enterobacteriaceae were identified in 4 out of 245 fecal samples collected from healthy individuals, yielding a prevalence rate of 1.63%. All isolates were confirmed as E. coli by both the VITEK2 automated identification system. PCR amplification and Sanger sequencing verified the presence of the tet(X4) gene in all four isolates. Notably, three isolates (SZ22HTE1, SZ22HTE2, and SZ22HTE3) were recovered from individuals residing in the same community (Dawang), whereas the fourth isolate (SZ22HTE4) originated from a separate community (Fenghua).
Phenotypic and genotypic characterization of antimicrobial resistance
All four E. coli isolates exhibited resistance to tigecycline, with MICs ranging from 8 to 32 mg/L (Supplementary Table S2). They also displayed resistance to multiple antibiotics, including ampicillin, streptomycin, tetracycline, and chloramphenicol (Table 1). Additionally, three isolates (SZ22HTE1–SZ22HTE3) were resistant to sulfamethoxazole/trimethoprim, whereas SZ22HTE4 remained susceptible. In contrast, all isolates were susceptible to cefotaxime, meropenem, gentamicin, amikacin, nalidixic acid, ciprofloxacin, colistin, and fosfomycin (Supplementary Table S2). According to the standard definition—resistance to at least one agent in three or more antimicrobial classes—all isolates were classified as multidrug-resistant (MDR).
WGS identified resistance genes that largely correlated with phenotypic profiles (Table 1). The presence of tet(X4), previously confirmed by PCR and Sanger sequencing, was further validated by WGS, explaining the observed tigecycline resistance. Additional tetracycline resistance genes, including tet(A), tet(B), and tet(M), were variably present and matched the tetracycline-resistant phenotypes. All isolates carried blaTEM-1B, consistent with ampicillin resistance, and aminoglycoside resistance genes (aadA1, aadA2, aadA22, and strA/strB) aligned with streptomycin resistance. Resistance to chloramphenicol was associated with cmlA1 and/or floR.
The plasmid-mediated quinolone resistance gene qnrS1 was identified in all isolates, but no chromosomal mutations were found within the quinolone resistance-determining region, consistent with their susceptibility to fluoroquinolones. Sulfonamide resistance genes (sul2, sul3) and the trimethoprim resistance gene dfrA12 were found only in SZ22HTE1–SZ22HTE3, in agreement with their resistance to sulfamethoxazole/trimethoprim. Additionally, all isolates harbored macrolide-lincosamide resistance genes lnu(G) and/or erm(42).
Virulence gene profiles of tet(X4)-positive E. coli isolates
All four E. coli isolates harbored multiple virulence-associated genes involved in adhesion, iron acquisition, biofilm formation, motility, and secretion systems, which are critical for bacterial colonization and pathogenicity (Table 2). Adhesion genes such as fimH, ehaB, upaG/ehaG, csgG, and cgsF were variably distributed among the isolates, supporting host cell attachment. Iron acquisition genes, including entE, fepD, entC, fepG, fepC, and entB, were also detected in a strain-specific manner, enabling iron scavenging essential for survival in host environments. The toxin gene hlyE/clyA, identified only in SZ22HTE1, may contribute to host cell lysis and tissue damage.
Gene associated with biofilm formation (agn43, cah, and cgsG) were detected in SZ22HTE1 to SZ22HTE3 and may enhance persistence and immune evasion. Motility-related genes (flhA, flhB, and fliA) were present in the same three isolates, likely facilitating bacterial movement and invasion. Secretion system genes (aec27/clpV, aec15, tssM, espX4, espX1, and espX5) were identified in various combinations across all four isolates and may facilitate the delivery of virulence factors into host cells (Table 2). Although the specific virulence gene profiles varied among isolates, each strain possessed multiple functional categories of virulence factors, underscoring their potential to cause clinically relevant infections.
Characterization of the tet(X4)-carrying plasmid in E. coli strains
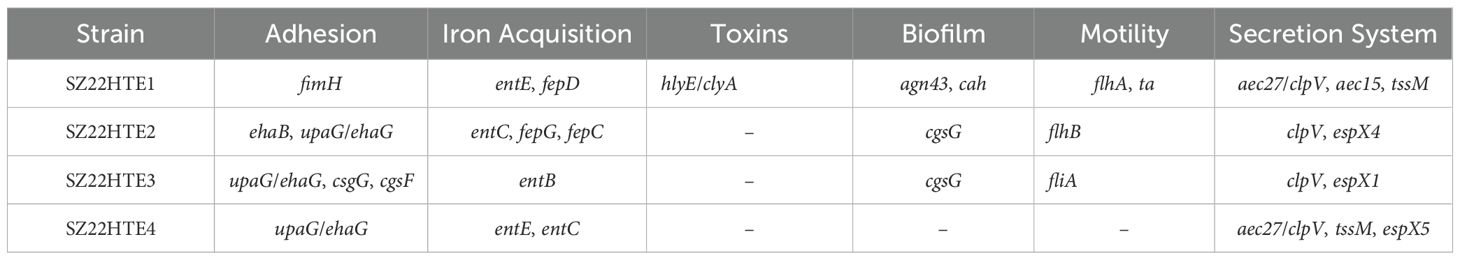
All four isolates carried multiple plasmids with diverse replicon types and antimicrobial resistance genes (Supplementary Table S3). In strain SZ22HTE1, the tet(X4) gene was located on the largest plasmid, pSZ22HT1-1 (IncY, 106,177 bp), which also harbored 11 additional resistance genes blaTEM-1B, aadA1, aadA2, tet(A), tet(M), cmlA1, floR, sul2, sul3, dfrA12, and erm(42). The tet(X4)-carrying IncY plasmid pSZ22HT1–1 exhibited high sequence similarity (>99.9%) to plasmid p803Rt_IncX1 (CP080067) isolated from a human-derived E. coli strain in Shenzhen, China with 94% coverage, and plasmid p13Q15 (ON934549) from an E. coli strain in Guangdong, China with 53% coverage (Figure 1A).
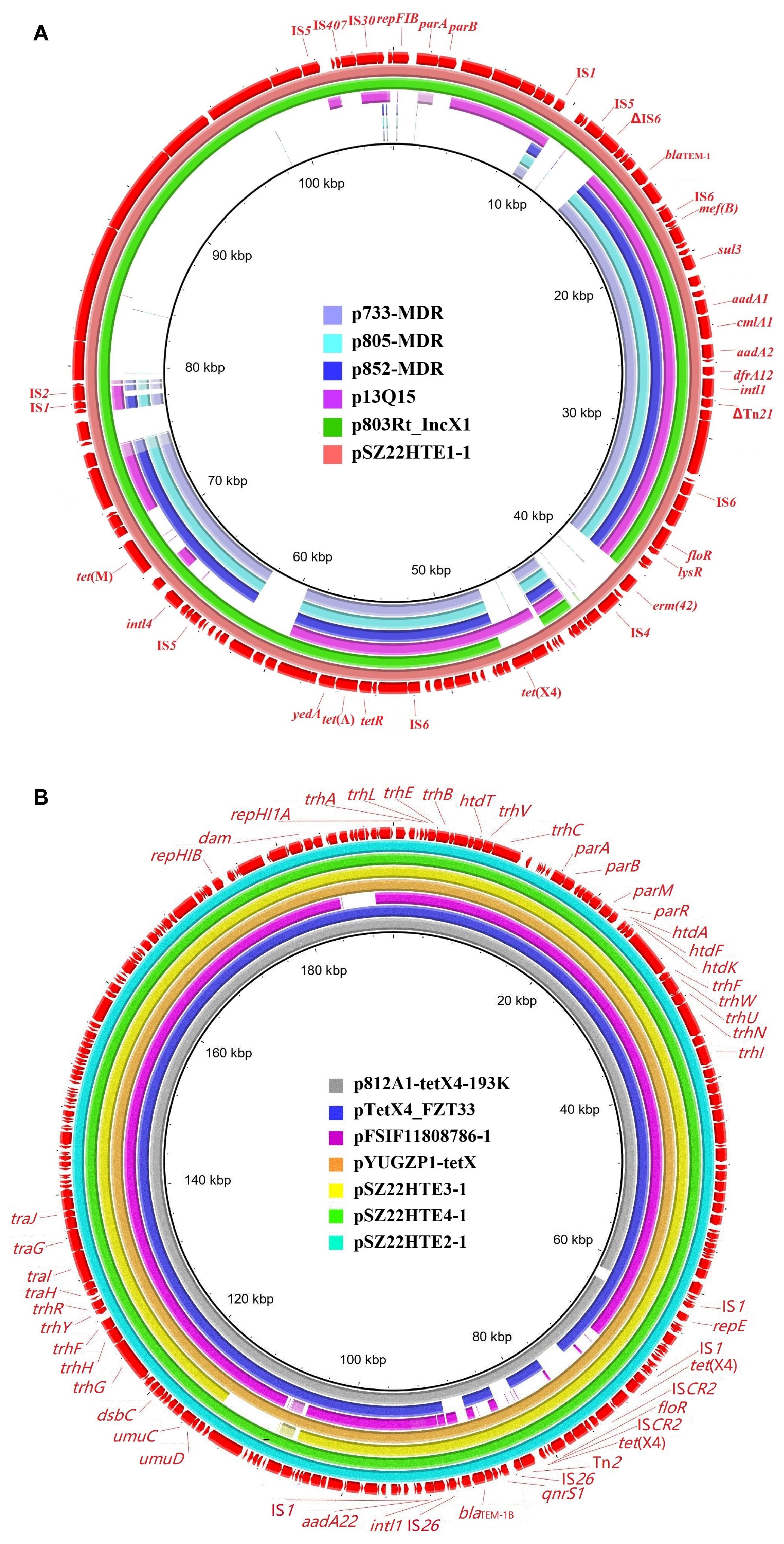
Figure 1. Sequence comparison of tet(X4)-bearing plasmids in this study with other similar plasmids using BRIG. (A) IncY plasmid pSZ22HTE1-1; (B) IncFIA8-IncHI1/ST17 plasmids pSZ22HTE2-1, pSZ22HTE3-1, and pSZ22HTE4-1. The outer circles in red with annotation are the reference plasmids pSZ22HTE1–1 and pSZ22HTE2-1, respectively.
The remaining three isolates (SZ22HTE2, SZ22HTE3, and SZ22HTE4) each carried a tet(X4)-positive hybrid IncFIA8-IncHI1/ST17 plasmid, designated pSZ22HT2-1 (190,711 bp), pSZ22HT3-1 (201,009 bp), and pSZ22HT4-1 (192,057 bp), respectively. These plasmids also carried resistance genes blaTEM-1B, aadA22, qnrS1, and floR. Comparative genomic analysis revealed that the tet(X4)-bearing IncFIA8-IncHI1/ST17 plasmids from SZ22HTE2, SZ22HTE3, and SZ22HTE4 were closely related to multiple plasmids, including tet(X4)-carrying plasmids p812A1-tetX4-193K (CP116047, E. coli, China), pYUGZP1-tetX (pig, E. coli, China) from pig source, and pTetX4_FZT33 (CP132725, E. coli, China) from hospital sewage (Figure 1B).
However, conjugation assays using E. coli C600 as the recipient strain failed to produce transconjugants under the tested conditions, indicating that these tet(X4)-bearing plasmids were either non-conjugative or required specific conditions or helper plasmids for mobilization.
Variation in the genetic environment of tet(X4)
As shown in Figure 2, the tet(X4) gene in the three IncFIA-IncHI1/ST17 plasmids (pSZ22HT2-1, pSZ22HT3-1, and pSZ22HT4-1) was embedded in an identical 12,536-bp region organized as IS1–catD–tet(X4)–ISCR2–ΔISCR2–floR–ΔISCR2. While the core tet(X4)-containing structure closely resembled that of p47EC, the first reported tet(X4)-bearing plasmid isolated from E. coli of porcine origin in China (He et al., 2019), several notable differences were observed. Most prominently, ISCR2 replaced the upstream IS1 element in p47EC. Furthermore, the chloramphenicol resistance gene floR, associated with an incomplete ISCR2, was located downstream of the tet(X4) structure in our plasmids, in contrast to its upstream position in p47EC.
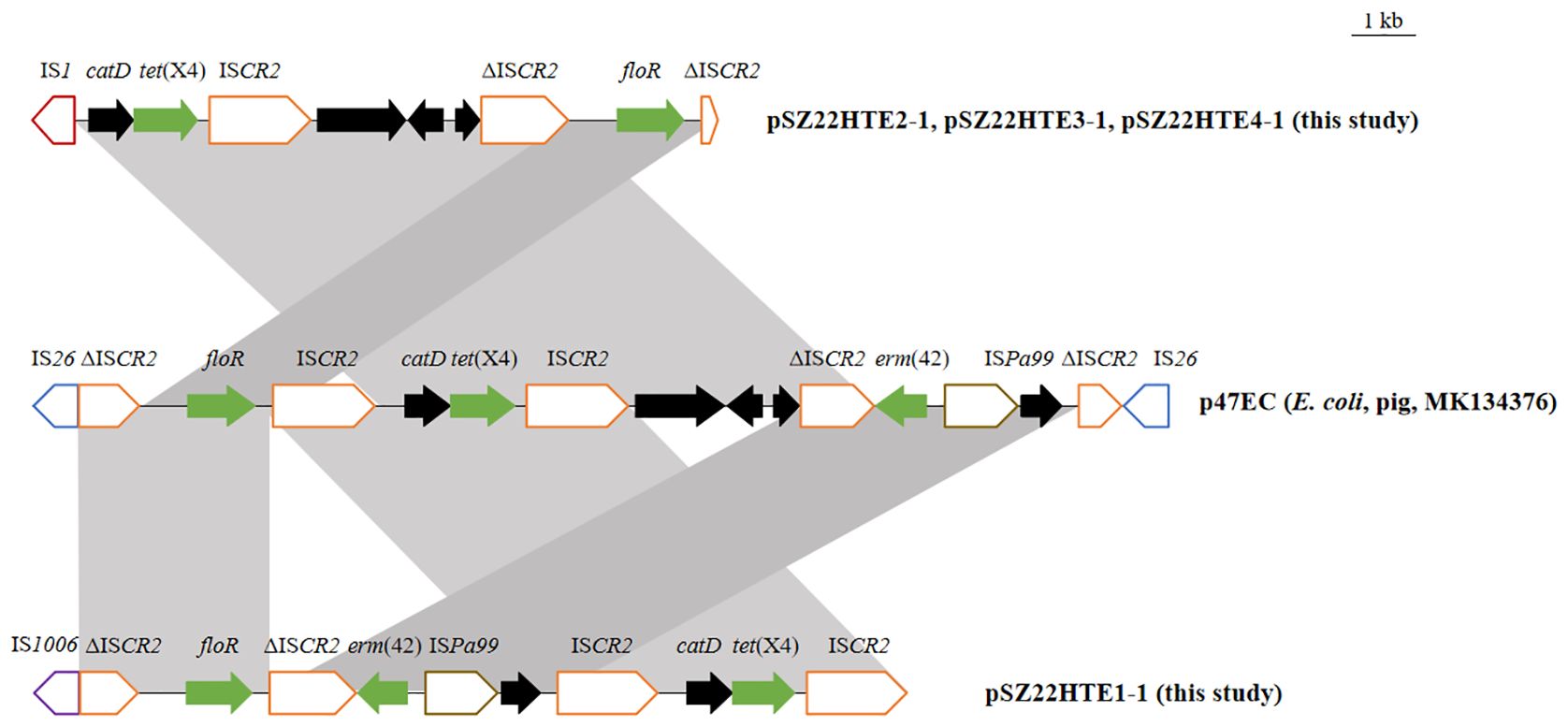
Figure 2. Genetic structures of the tet(X4) gene in this study and comparison with p47EC (MK134376). The extent and directions of genes are indicated by arrows. Antibiotic resistance genes are shown in red. Truncated mobile elements are marked with a “Δ” symbol. Insertion sequences (ISs) are represented as boxes labeled with their names. Regions with >99% identity are shaded in gray.
Plasmid pSZ22HTE1–1 exhibited a different but related arrangement. It retained the conserved ISCR2–catD–tet(X4)–ISCR2 structure found in p47EC, but its upstream region contained an additional resistance module carrying both floR and erm(42). In comparison, p47EC carried the ΔISCR2-erm(42)-ISPa99 segment upstream of the tet(X4) conserved segment, but in the opposite orientation. These structural variations underscore the genetic plasticity of tet(X4)-associated regions and their potential for mobilization and dissemination across diverse plasmid backgrounds driven by mobile genetic elements.
Phylogenomic analysis of tet(X4)-positive E. coli strains
Four tet(X4)-positive E. coli isolates in this study were assigned to four sequence types (STs): ST10, ST201, ST877, and ST1308. To investigate the genetic relatedness between these E. coli isolates and publicly available E. coli strains of the same ST, we conducted a phylogenomic analysis based on cgSNPs. The resulting phylogeny revealed that our community-derived isolates were closely related to E. coli strains from diverse geographical regions and hosts (Figure 3).
Figure 3. Core-genome SNP phylogeny of four tet(X4)-positive E. coli isolates (in red) with the same ST strains with ≤200 SNPs. (A) ST201; (B) ST10; (C) ST1308; (D) ST877. Branch lengths indicate SNP distances; key ARGs are annotated.
Our tet(X4)-positive ST201 isolate (SZ22HTE1) exhibited high genetic similarity to three human clinical ST201 isolates from China (GCA_023516055.1, GCA_019880025.1, and GCA_030325125.1), differing by only 14–25 SNPs and carrying identical or similar antimicrobial resistance genes, suggesting a potential epidemiological link (Figure 3A). SZ22HTE1 also showed relatively limited divergence (105 and 136 SNPs) from E. coli strains of porcine (GCA_037105215.1, Spain), food (GCA_037106035.1, Thailand), and clinical (GCA_028636665.1, China) origin (Figure 3A), further indicating potential inter-host and inter-regional transmission.
Our ST10 tet(X4)-positive isolate, SZ22HTE3, clustered with nine ST10 E. coli isolates from diverse countries and sources. It was most closely related (41 SNPs) to an environmental isolate from South Africa. In contrast, it differed from the remaining eight isolates by 142–199 SNPs, suggesting a certain level of genomic divergence among ST10 isolates (Figure 3B). For ST1308, five isolates, including our community-derived strain SZ22HTE4, formed a tight phylogenetic cluster with minimal genetic SNP differences of only 58 or 63, suggesting recent common ancestry or possible transmission events. Among the four closely related isolates, three were isolated from swine in China (GCA_022538875.1, GCA_022538895.1, GCA_022538855.1), and one had an unreported source (GCA_023272775.1) (Figure 3C). By comparison, the ST877 lineage exhibited greater genetic heterogeneity. Compared with our tet(X4)-positive ST877 isolate (SZ22HTE2), the other 16 ST877 E. coli isolates displayed broader genomic divergence, with SNP differences ranging from 100 to 200, highlighting higher diversity within this ST (Figure 3D).
Discussion
Since its approval by the U.S. Food and Drug Administration in 2005, tigecycline has served a last-resort antibiotic for the treatment of severe infections caused by MDR bacteria, particularly carbapenem-resistant Enterobacteriaceae (Yaghoubi et al., 2021). However, the growing prevalence of tigecycline resistance has become a significant clinical and public health concern. Among the known resistance mechanisms, the plasmid-mediated tet(X4) gene has gained particular attention due to its ability to confer high-level tigecycline resistance and its rapid dissemination across bacterial species and ecological niches through horizontal gene transfer (Dong et al., 2021; He et al., 2019; Li et al., 2023).
Globally, tet(X4)-carrying E. coli strains have been detected in clinical, animal, food, and environmental settings (Li et al., 2023). However, their presence in healthy human populations, particularly in urban communities, remains insufficiently characterized. Our study identified a tet(X4)-positive E. coli colonization rate of 1.6% (4/245) in fecal samples from healthy individuals in Shenzhen, a densely populated metropolitan area. Although this prevalence is considerably lower than that observed in animal-derived (18.24%) and food-derived (20.6%) E. coli isolates, it exceeds the rates reported in clinical patients (0.07–0.1%) (Bai et al., 2019; He et al., 2019; Li et al., 2021), suggesting that the human gut may serve as a previously underappreciated reservoir for plasmid-mediated tigecycline resistance. These findings highlight the necessity of enhanced AMR surveillance in non-clinical community populations to detect early signs of resistance dissemination.
AMR is an escalating global health crisis, contributing to an estimated 30,000 deaths annually in the EU and nearly 5 million worldwide in 2019 (Cassini et al., 2019; Ho et al., 2025). The human gastrointestinal tract, comprising a dense and diverse microbial community (~10¹4 cells across approximately 1, 000 bacterial species), is a hotspot for the acquisition, persistence, and horizontal transfer of ARGs (Despotovic et al., 2023; Lynch and Pedersen, 2016). The gut microbiome not only reflects the local AMR burden but also influences patient outcomes, particularly in critically ill individuals with gut barrier dysfunction or acute gastrointestinal injury, where microbiome disruption and elevated ARG levels are associated with worse clinical outcomes (Bai et al., 2025). Notably, E. coli, a core member of the intestinal microbiota and a widely used sentinel organism in AMR surveillance, readily acquires ARGs from food, animal, and environmental sources, and can transfer them to pathogenic bacteria such as Shigella and Klebsiella through plasmids or other mobile genetic elements (Thanh Duy et al., 2020). In our study, three isolates (SZ22HTE2, SZ22HTE3, and SZ22HTE4) harbored the epidemic IncFIA8-IncHI1/ST17 plasmid carrying tet(X4), suggesting early-stage horizontal dissemination of tigecycline resistance within the community gut microbiota of an urban community. These plasmids co-harbored additional resistance genes, resulting in MDR phenotypes and enabling co-selection and persistence in diverse hosts (Lu et al., 2018; Yan et al., 2024). Notably, the epidemic IncFIA8-IncHI1/ST17 plasmid identified in this study shows high homology to pRDZ41 (CP139495.1) from Klebsiella pneumoniae, providing direct evidence for its potential to disseminate tet(X4) into this high-risk pathogen.
Importantly, these E. coli isolates possessed a range of virulence-associated genes related to adhesion (e.g., fimH, ehaB, upaG/ehaG), iron acquisition (e.g., entE, fepD), biofilm formation (e.g., agn43, cah), motility (e.g., flhA, fliA), and secretion systems (e.g., aec27/clpV, tssM). The coexistence of virulence factors and resistance determinants in community-derived E. coli is particularly alarming, as it increases the risk of difficult-to-treat infections and facilitates ARG dissemination into high-risk clinical pathogens.
Although the healthy human gut microbiota may provide colonization resistance against invasive AMR bacteria—potentially via mechanisms such as microbiome-mediated nutrient depletion (Isaac et al., 2022; Le Guern et al., 2021)—our findings underscore the vulnerability of even healthy individuals to colonization by plasmid-mediated tigecycline-resistant E. coli strains. This silent carriage may act as a hidden conduit for ARG dissemination between community and healthcare environments, presenting significant challenges for infection prevention and control.
WGS further revealed that our tet(X4)-positive isolates belonged to globally prevalent E. coli STs, e.g., ST10, ST877, and ST1308, frequently associated with MDR phenotypes and detected in both human and animal hosts (Elias et al., 2019; García et al., 2018; Liu et al., 2024). The detection of these epidemic STs in healthy individuals, along with their close genetic relatedness to isolates from clinical, food, animal, and the environmental sources, reinforces a One Health perspective. Phylogenomic analysis demonstrated close genetic relatedness between our community isolates and strains from diverse geographic regions and hosts (e.g., swine, food, environment), reinforcing the role of cross-sectoral transmission in the spread of tet(X4). This underscores the interconnectedness of AMR reservoirs across ecosystems and highlights the urgent need for integrated, cross-sectoral genomic surveillance.
Despite the valuable insights provided, this study has several limitations. Only three urban communities were sampled, and the number of tet(X4)-positive isolates was relatively small, limiting the generalizability of our findings. Additionally, although the plasmid structures were well-characterized, functional studies such as assessing plasmid transfer under different conditions or colonization capacity in vivo were not performed. The observed lack of successful conjugation in vitro highlights the need for further investigation into the mobility mechanisms of tet(X4)-bearing plasmids. Further studies should explore whether these plasmids can successfully conjugate by employing diverse recipient strains, co-introducing helper plasmids, or optimizing mating conditions (e.g., modifying incubation temperature, adjusting donor-to-recipient ratios, or using filter mating assays) under alternative experimental settings. Importantly, future investigations should include larger-scale, longitudinal surveillance, and mechanistic assessments to better understand the dynamics and risks of tet(X4)-positive E. coli colonization in healthy populations.
Conclusion
In summary, our findings highlight the emergence of tet(X4)-positive MDR E. coli in the intestinal microbiota of healthy individuals from urban communities. These isolates co-harbored multiple ARGs and virulence determinants, highlighting their potential to cause difficult-to-treat infections and to act as reservoirs for further resistance dissemination. The detection of globally circulating high-risk clones such as ST10 and ST201 in asymptomatic carriers underscores the silent spread of tigecycline resistance in non-clinical settings. These findings call for urgent and coordinated surveillance strategies beyond hospital environments, in alignment with One Health principles, to contain the spread of last-resort antibiotic resistance and safeguard public health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval for human sample collection was obtained from the Shenzhen University Ethics Committee (Reference No. PN-202400026), in compliance with guidelines approved by an independent ethics board comprising non-institutional legal representatives and community stakeholders. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin. Regarding informed consent to participate, this study exclusively utilized anonymized residual pathogen samples obtained from routine clinical diagnostics. These samples were irreversibly de-identified prior to research use, with all personal identifiers (including but not limited to patient names, demographic data, medical records, and clinical history) removed. Only numerical laboratory tracking codes were retained. This research qualified for exemption from informed consent requirements under Clause [Specify Clause Number/Type, e.g., Article 39] of the Ethical Review Measures for Biomedical Research Involving Human Subjects (中国涉及人的生物医学研究伦理审查办法 https://www.gov.cn/zhengce/2016-10/12/content_5713806.htm), as it meets all criteria for exemption: 1. Non-linkability: Samples were completely unlinked from any identifiable individuals; 2.No Additional Risk: The research posed no additional physical, psychological, or social risks to the original providers; The study protocol and this exemption were formally reviewed and approved by the Medical Ethics Committee of Shenzhen University Health Science Center (SUHSC-MEC), with approval number PN-202400026. The committee confirmed that the secondary use of these irreversibly anonymized specimens falls under exempt ethical review categories. We have revised the manuscript’s “Ethics approval and consent to participate” section to clearly state: “Informed consent to participate was not required for this study as it exclusively utilized irreversibly anonymized residual pathogen samples obtained from routine clinical diagnostics. These samples contained no personally identifiable information. This exemption complies with the Ethical Review Measures for Biomedical Research Involving Human Subjects [Reference Number] and was approved by the Medical Ethics Committee of Shenzhen University Health Science Center (PN-202400026).”.
Author contributions
RP: Validation, Data curation, Investigation, Writing – review & editing. PL: Conceptualization, Investigation, Writing – review & editing. WJ: Writing – review & editing, Methodology, Data curation. ZL: Validation, Writing – review & editing, Visualization. LW: Data curation, Writing – review & editing, Resources. YH: Writing – review & editing, Visualization. XZ: Writing – review & editing, Investigation. YG: Writing – review & editing, Validation. YW: Software, Writing – review & editing. JW: Writing – review & editing. JG: Validation, Writing – review & editing, Supervision. FY: Data curation, Conceptualization, Project administration, Writing – review & editing, Supervision. DL: Conceptualization, Writing – original draft, Methodology, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported in part by Guangdong Basic and Applied Basic Research Foundation (2024A1515030195, No. 2023A1515012152 and No. 2022A1515110369), Hainan Provincial International Science and Technology Cooperation Research and Development Project (GHYF2024021), the National Natural Science Foundation of China (82060378), Major Project of Guangzhou National Laboratory (GZNL2023A01001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. The authors verify and take full responsibility for the limited use of generative AI in the preparation of this manuscript. Generative AI tools were used exclusively for grammatical refinement and sentence structure optimization to enhance clarity. All scientific content, data interpretation, conclusions, and writing remain the original work of the authors. No AI was used to generate results, analysis, or critical intellectual contributions.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1667196/full#supplementary-material
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Allcock, S., Young, E. H., Holmes, M., Gurdasani, D., Dougan, G., Sandhu, M. S., et al. (2017). Antimicrobial resistance in human populations: challenges and opportunities. Glob. Health Epidemiol. Genom. 2, e4. doi: 10.1017/gheg.2017.4
Aminov, R. (2021). Acquisition and spread of antimicrobial resistance: A tet(X) case study. Int. J. Mol. Sci. 22, 3905. doi: 10.3390/ijms22083905
Bai, L., Du, P., Du, Y., Sun, H., Zhang, P., Wan, Y., et al. (2019). Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Euro Surveill. 24, 1900340. doi: 10.2807/1560-7917.ES.2019.24.25.1900340
Bai, Y., Hu, Y., Chen, X., Hu, L., Wu, K., Liang, S., et al. (2025). Comparative metagenome-associated analysis of gut microbiota and antibiotic resistance genes in acute gastrointestinal injury patients with the risk of in-hospital mortality. mSystems 10, e0144424. doi: 10.1128/msystems.01444-24
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carlet, J. (2012). The gut is the epicentre of antibiotic resistance. Antimicrob. Resist. Infect. Control 1, 39. doi: 10.1186/2047-2994-1-39
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
CLSI (2023). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 33rd Edn (Wayne, PA: CLSI).
Despotovic, M., de Nies, L., Busi, S. B., and Wilmes, P. (2023). Reservoirs of antimicrobial resistance in the context of One Health. Curr. Opin. Microbiol. 73, 102291. doi: 10.1016/j.mib.2023.102291
Ding, Y., Er, S., Tan, A., Gounot, J.-S., Saw, W.-Y., Tan, L., et al. (2024). Comparison of tet(X4)-containing contigs assembled from metagenomic sequencing data with plasmid sequences of isolates from a cohort of healthy subjects. Microbiol. Spectr. 12, e0396923. doi: 10.1128/spectrum.03969-23
Ding, Y., Saw, W. Y., Tan, L. W. L., Moong, D. K. N., Nagarajan, N., Teo, Y. Y., et al. (2020). Emergence of tigecycline- and eravacycline-resistant Tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75, 3480–3484. doi: 10.1093/jac/dkaa372
Dong, N., Zeng, Y., Cai, C., Sun, C., Lu, J., Liu, C., et al. (2021). Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study. Sci. Total Environ. 818, 151767. doi: 10.1016/j.scitotenv.2021.151767
Donskey, C. J. (2004). The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39, 219–226. doi: 10.1086/422002
Elias, L., Gillis, D. C., Gurrola-Rodriguez, T., Jeon, J. H., Lee, J. H., Kim, T. Y., et al. (2019). The occurrence and characterization of extended-spectrum-beta-lactamase-producing Escherichia coli isolated from clinical diagnostic specimens of equine origin. Animals 10, 28. doi: 10.3390/ani10010028
Ferrara, F., Castagna, T., Pantolini, B., Campanardi, M. C., Roperti, M., Grotto, A., et al. (2024). The challenge of antimicrobial resistance (AMR): current status and future prospects. Naunyn Schmiedebergs Arch. Pharmacol. 397, 9603–9615. doi: 10.1007/s00210-024-03318-x
García, V., García-Meniño, I., Mora, A., Flament-Simon, S. C., Díaz-Jiménez, D., Blanco, J. E., et al. (2018). Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and shiga toxin-producing Escherichia coli in Spain, (2006-2017). Int. J. Antimicrob. Agents 52, 104–108. doi: 10.1016/j.ijantimicag.2018.03.022
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Ho, C. S., Wong, C. T. H., Aung, T. T., Lakshminarayanan, R., Mehta, J. S., Rauz, S., et al. (2025). Antimicrobial resistance: a concise update. Lancet Microbe 6, 100947. doi: 10.1016/j.lanmic.2024.07.010
Isaac, S., Flor-Duro, A., Carruana, G., PuChades-Carrasco, L., Quirant, A., Lopez-Nogueroles, M., et al. (2022). Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus. Nat. Commun. 13, 7718. doi: 10.1038/s41467-022-35380-5
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Le Guern, R., Stabler, S., Gosset, P., Pichavant, M., Grandjean, T., Faure, E., et al. (2021). Colonization resistance against multi-drug-resistant bacteria: a narrative review. J. Hosp Infect. 118, 48–58. doi: 10.1016/j.jhin.2021.09.001
Li, Y., Sun, X., Xiao, X., Wang, Z., and Li, R. (2023). Global distribution and genomic characteristics of tet(X)-positive Escherichia coli among humans, animals, and the environment. Sci. Total Environ. 887, 164148. doi: 10.1016/j.scitotenv.2023.164148
Li, Y., Wang, Q., Peng, K., Liu, Y., Xiao, X., Mohsin, M., et al. (2021). Distribution and genomic characterization of tigecycline-resistant tet(X4)-positive Escherichia coli of swine farm origin. Microb. Genom. 7, 667. doi: 10.1099/mgen.0.000667
Liu, Y. Y., Lu, L., Yue, C., Gao, X., Chen, J., Gao, G., et al. (2024). Emergence of plasmid-mediated high-level tigecycline resistance gene tet(X4) in Enterobacterales from retail aquatic products. Food Res. Int. 178, 113952. doi: 10.1016/j.foodres.2024.113952
Lu, J., Zhang, J., Xu, L., Liu, Y., Li, P., Zhu, T., et al. (2018). Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect. Control 7, 127. doi: 10.1186/s13756-018-0415-0
Lynch, S. V. and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
McInnes, R. S., McCallum, G. E., Lamberte, L. E., and van Schaik, W. (2020). Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43. doi: 10.1016/j.mib.2020.02.002
Thanh Duy, P., Thi Nguyen, T. N., Vu Thuy, D., Chung The, H., Alcock, F., Boinett, C., et al. (2020). Commensal Escherichia coli are a reservoir for the transfer of XDR plasmids into epidemic fluoroquinolone-resistant Shigella sonnei. Nat. Microbiol. 5, 256–264. doi: 10.1038/s41564-019-0645-9
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64, e01326–e01319. doi: 10.1128/AAC.01326-19
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Yaghoubi, S., Zekiy, A. O., Krutova, M., Gholami, M., Kouhsari, E., Sholeh, M., et al. (2021). Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 5, 1–20. doi: 10.1007/s10096-020-04121-1
Yan, Z., Li, Y., Ni, Y., Xia, X., Zhang, Y., Wu, Y., et al. (2024). Plasmid-borne tigecycline resistance gene tet(X4) in Salmonella enterica and Escherichia coli isolates from a pediatric patient with diarrhea. Drug Resist. Updat. 77, 101145. doi: 10.1016/j.drup.2024.101145
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Keywords: community, healthy carriers, tigecycline resistance, E. coli, plasmids
Citation: Peng R, Liang P, Jiang W, Li Z, Wang L, Huang Y, Zhang X, Guo Y, Wang Y, Wang J, Guo J, Yin F and Lin D (2025) Community gut colonization by tet(X4)-positive multidrug-resistant Escherichia coli in healthy individuals from urban residents in Shenzhen, China. Front. Cell. Infect. Microbiol. 15:1667196. doi: 10.3389/fcimb.2025.1667196
Received: 16 July 2025; Accepted: 15 September 2025;
Published: 07 October 2025.
Edited by:
Mattia Pirolo, University of Copenhagen, DenmarkReviewed by:
Marie Louise Guadalupe Attwood, North Bristol NHS Trust, United KingdomRiccardo Polani, Sapienza University of Rome, Italy
Copyright © 2025 Peng, Liang, Jiang, Li, Wang, Huang, Zhang, Guo, Wang, Wang, Guo, Yin and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiubiao Guo, amJndW9Ac3R1LmVkdS5jbg==; Feifei Yin, eWluZmVpZmVpZmZAMTYzLmNvbQ==; Dachuan Lin, bGluZGFjaGF1bkBtdWhuLmVkdS5jbg==
†These authors have contributed equally to this work
 Ruoyan Peng
Ruoyan Peng Pei Liang1†
Pei Liang1† Zhaodong Li
Zhaodong Li Feifei Yin
Feifei Yin Dachuan Lin
Dachuan Lin